陈柔珂1,2, 吕永新1,2, 张欢欢1,2, 宋庆浩1,2, 徐俊1,2


1. 上海交通大学微生物代谢国家重点实验室, 上海 200240;
2. 上海交通大学生命科学技术学院, 上海 200240
收稿日期:2020-06-12;修回日期:2020-07-24;网络出版日期:2020-08-05
基金项目:国家自然科学基金(41676121,41976085)
*通信作者:徐俊, Tel: +86-21-34207208;E-mail: xujunn@sjtu.edu.cn.
摘要:[目的] 亚氏火球菌(Pyrococcus yayanosii)A1菌株的最适生长温度为98℃,最适生长压力为5.2×104 Pa,是研究此类严格厌氧、超嗜热和兼性嗜压古菌的环境适应性机制的良好材料。本研究基于P.yayanosii A1基因组中Ⅲ-B型CRISPR-Cas系统,旨在建立可应用于此类古菌的基因表达敲降系统(gene knock-down system)。[方法] 人工构建的mini-CRISPR簇,由内源性CRISPR簇Group 1的重复序列(repeats)和待敲降的淀粉酶基因(PYCH_13690)中的原间隔序列(protospacer)组成。将该mini-CRISPR簇插入到具有辛伐他汀抗性的穿梭载体pLMOShhp中,使之转录与目的基因mRNA匹配的crRNA,引导Cmr复合物对其进行切割。[结果] 使用高静水压(HHP)诱导型启动子Phhp诱导mini-CRISPR簇表达时,淀粉酶基因的mRNA数量在5.2×104 Pa静水压时下调到原来的67.05%,在1.0×102 Pa静水压时下调为原来的49.69%;而使用组成型启动子Phmtb诱导mini-CRISPR簇表达时,淀粉酶基因的mRNA数量在5.2×104 Pa静水压时下调为原来的58.48%,在1.0×102 Pa静水压时下调为原来的23.97%。使用鲁戈氏碘液显色法对这两类基因表达敲降突变菌株的培养物中残留的淀粉含量进行分析,结果显示突变菌株的淀粉降解能力明显降低。[结论] 在P.yayanosii A1中建立了基于内源Ⅲ-B型CRISPR-Cas系统的基因敲降系统,可以用于抑制此类超嗜热嗜压古菌体内基因的表达。
关键词:超嗜热古菌嗜压微生物Pyrococcus yayanosiiCRISPR-Cas基因敲降
An endogenous type Ⅲ-B CRISPR-Cas based gene knockdown system in Pyrococcus yayanosii A1
Rouke Chen1,2, Yongxin Lv1,2, Huanhuan Zhang1,2, Qinghao Song1,2, Jun Xu1,2


1. State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University, Shanghai 200240, China;
2. School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China
Received: 12 June 2020; Revised: 24 July 2020; Published online: 5 August 2020
*Corresponding author: Xu Jun, Tel: +86-21-34207208;E-mail: xujunn@sjtu.edu.cn.
Foundation item: Supported by the National Natural Science Foundation of China (41676121, 41976085)
Abstract: [Objective] Pyrococcus yayanosii A1, with its optimum growth condition at 98℃ and 5.2×104 Pa, was a laboratory mutant strain used in the studies of environmental adaptation mechanisms in a group of piezophilic hyperthermophilic archaea in Thermococcaceae. In this study, we established a new genetic tool for P. yayanosii A1, to knock down gene expression based on an endogenous active type Ⅲ-B CRISPR-Cas system. [Methods] The mini-CRISPR cluster was consisted the repeat sequences from endogenous CRISPR arrays Group I and a protospacer from a putative amylase gene (PYCH_13690). We inserted an artificial mini-CRISPR cluster into the shuttle vector pLMOShhp1, with the overexpression of the HMG-CoA (hydroxy methylglutaryl coenzyme A) reductase gene controlled by a constitutive promoter Pgdh, which conferred simvastatin resistance. And the mini-CRISPR cluster transformed crRNA, which guided the Cmr complex cut target mRNA. [Results] The ratio of mRNA cleavage of a high hydrostatic pressure inducible promoter Phhp was 33% at 5.2×104 Pa and 50% at 1.0×102 Pa, while the cleavage of constitutive promoter Phmtb was 42% at 5.2×104 Pa and 76% at 1.0×102 Pa. As a proof, the gene knockdown mutant degraded less starch than its parental strain A1 in vivo. [Conclusion] Thus, we successfully constructed a gene knockdown system based on type Ⅲ-B CRISPR-Cas system in P. yayanosii, which could be used in the genetic studies targeting essential genes in piezophilic hyperthermophiles.
Keywords: hyperthermophilc archaeapiezophilePyrococcus yayanosiiCRISPR-Casgene knock-down
超嗜热微生物的最适生长温度通常高于80 ℃[1]。迄今为止,人们已经从热液、火山口和其他地热环境中,分离培养出超过70个科的超嗜热微生物,其中古菌有20科[2]。严格厌氧的超嗜热古菌热球菌科(Thermococcaceae)是热液环境中的优势微生物,可以适应多种极端环境[3]。热球菌科在高静水压(HHP)和高温(HT)下具有较高的生长速率,对其适应性机制的解析逐渐成为研究的热点[4–6]。
亚氏火球菌(Pyrococcus yayanosii) CH1菌株是从大西洋4100 m深的“Ashadze”热液区域样本中分离获得的热球菌科中第一株专性嗜压古菌,在最适生长温度98 ℃下,其耐受的压力范围为2.0×103–1.2×105 Pa[7]。亚氏火球菌CH1的基因组大小为1.72 Mb,共编码1930个基因[8]。在实验室获得的兼性嗜压菌株亚氏火球菌A1耐受压力的范围扩大为1.0×102–1.2×105 Pa[9]。已经构建成功的亚氏火球菌的遗传操作系统包括:尿嘧啶营养缺陷型菌株A2和As1[10–11],携带有辛伐他汀抗性基因的穿梭质粒(pLMO12103)和整合质粒(pLMO033)[12–13],以及利用高静水压(HHP)诱导启动子Phhp控制的毒素作为新型反向筛选标记等[11]。
对亚氏火球菌在非最适生长压力下的转录组和蛋白质组分析结果显示,非最适压力下的基因差异表达涉及翻译、运动和CRISPR-Cas系统(基因表达量上调),以及能量代谢中的Mbh氢化酶、Mbx硫氢化酶和甲酸代谢等基因(表达显著下调)[14]。由于这些基因多为负责古菌的基础生命过程的必需基因,必须发展基因表达敲降工具才能对其功能进行遗传分析研究。
CRISPR-Cas系统通过crRNA特异性识别并结合来自病毒和质粒的外源DNA或RNA,引导crRNP (CRISPR ribonucleoprotein)复合物对外源核酸序列进行剪切[15]。古菌中主要存在的是Ⅰ型和Ⅲ型CRISPR-Cas系统[16]。其中Ⅰ型仅能剪切DNA,而Ⅲ型可以剪切DNA和RNA[17–19]。目前,古菌中敲降基因表达可通过CRISPRi和剪切目的基因mRNA来实现。CRISPRi通过Cascade蛋白和crRNA的复合体结合在特异性位点阻止转录的起始。如,在Haloferax volcani中一个基于Ⅰ-B型系统的CRISPRi工具抑制编码β半乳糖苷酶的基因bgaHα的转录并降低了操纵子HVO_2526- HVO_2528的转录[20]。而在Sulfolobus solfataricus和S. islandicus中可直接利用该菌内源的Ⅲ-B型CRISPR-Cas系统的Cmr蛋白与mini-CRISPR簇转录的crRNA形成crRNP复合体,结合靶标mRNA并进行剪切[21–25]。
Ⅲ-B型CRISPR-Cas系统有两个亚型,α-亚型系统有6个Cmr蛋白,β-亚型有7个Cmr蛋白[26–28]。在P. furiosus中,Ⅲ-B-α Cmr蛋白复合体呈“海马”结构[27–28]。Ⅲ-B型CRISPR-Cas系统的标志蛋白是Cmr复合物中最大的亚基Cmr2,它具有HD结构域,发挥ssDNase的作用[29–30]。CRISPR核糖核蛋白(crRNPs)由Cmr蛋白复合物和crRNA组成,该复合物与外源RNA结合,然后在外源核苷酸链中进行固定间隔6 nt的剪切[30–32]。在P. furiosus中的crRNA产物有长度为39 nt和45 nt两种长度,crRNA的5′末端具有长度为8 nt的5′-tag序列,即Repeats的3′末端区域上的5′-handle标签[32–33]。与Ⅰ型和Ⅲ-A型CRISPR系统中使用PAM序列识别目的DNA不同,Ⅲ-B型CRISPR-Cas系统不具有PAM序列,仅crRNA匹配目的mRNA即可进行切割,其5′-tag标签可以控制RNA和DNA的剪切,当序列匹配时,仅切割RNA并保护DNA[19, 26]。
基于Ⅲ-B型CRISPR系统独特的切割RNA和DNA的机制,我们在超嗜热古菌亚氏火球菌A1中构建了一个高效率的基因表达敲降系统。通过在穿梭质粒上导入人工构建的mini-CRISPR簇,成功抑制了1个淀粉酶基因的表达。
1 材料和方法 1.1 菌株和质粒 本研究用到的菌株和质粒如表 1所示。
表 1. 本研究中所用菌株和质粒 Table 1. Strains and plasmids used and constructed in this study
| Strains and plasmids | Genotype and features | References |
| P. yayanosii A1 | Li et al.[10] | |
| kdp-1369 | PYCH_13690 gene knockdown mutant generated using pLMOkdp-1369 | This work |
| kdm-1369 | PYCH_13690 gene knockdown mutant generated using pLMOkdm-1369 | This work |
| ?1369- | PYCH_13690 gene deletion mutant of A1 | Song et al.[11] |
| pLMOS776TA | pLMO12102:: HMG-CoA-Pgdh-PhmbB-PF0775-PF0776-Phhp1 | Song et al.[11] |
| pLMOS01 | pLMO12102:: HMG-CoA-Pgdh | Song et al.[11] |
| pLMOShhp | pLMO12102:: HMG-CoA-Pgdh- Phhp | This work |
| pLMOkdp-1369 | pLMOShhp1:: Phhp-mini-CRISPR array | This work |
| pLMOkdm-1369 | pLMOShhp1:: Phmtb-mini-CRSIPR array | This work |
表选项
1.2 合成序列和引物 本文中所用到的合成序列和引物均由生工生物工程(上海)股份有限公司合成,如表 2所示。
表 2. 本研究中所用合成序列和引物 Table 2. Oligonucleotides and primers used and constructed in this study
| Oligonucleotides | Sequences (5′→3′) |
| 1369-hhp | CCTGTCATGAAAGAAGGTGATAAAGGTTCCAATAAGACTCAAAGAGAATTGAAAGCATCAAGT TGATCCGCTCGTCGCTGGATACCCTCTCGGTTCCAATAAGACTCAAAGAGAATTGAAAGGACAC GGGCCCGTCGACTGCAGAGGCCTGCATGCAAGCTTGGCGTAATCATGGTCATAGCTGTTTCC |
| 1369-hmtb | TAGAGTCGACCTGCAGATTTTCGGGTGAGCAAAATGCGCGTTGCGGCAGCTGGACCTGCAGAG CGATATATTTATATAGGGATATAGTAATAGATAATATCACGGAGGTGATGCATGTTCCAATAAGACTCAAAGAGAATTGAAAG CATCAAGTTGATCCGCTCGTCGCTGGATACCCTCTCGGTTCCAATAAGACTCAAAGAGAATTGAAAG GACACGGGCCCGTCGACTGCAGAGGCCTGCATGCAAGCTTGGC GTAATCATGGTCATAGCTGTTTCC |
| pLMOhhp-F | CTTTATCACCTTCTTTCATGACAGG |
| pLMOhhp-R | TCGTAATCATGGTCATAGCTGTTTC |
| CRISPR-hhp-F | CCTGTCATGAAAGAAGGTGATAAAGGTTCCAATAAGACTCAAA |
| CRISPR-hmtb-F | TAGAGTCGACCTGCAGATTTTCGG |
| CRISPR-R | GAAACAGCTATGACCATGATTACGA |
| pLMOS3-F | ATGGAAATAGAGGAGATTATAGAGA |
| pLMOS3-R | TCGTAATCATGGTCATAGCTGTTTC |
| Phmtb-R | CTATGACCATGATTACGAATGCATCACCTCCGTGATATTATCT |
| Phmtb-F | TCTCCTCTATTTCCATGTTCATCCCTCCAAATTAGGTGATT |
| test1-F | CAAAGTTGCGGTCTTCCAC |
| test1-R | AAGGGACCACTCGCTCA |
| test2-F | TTCGTTGTCGATTATAGGGTGA |
| test2-R | TGCGATAGTGTCTTTATGGGAT |
| sim-F | ATGGAAATAGAGGAGATTATAGAGA |
| sim-R | TCATCTCCCAAGCATTTTATGAGCC |
| Cmr1-qF | GGGCACCATCAATCAA |
| Cmr1-qR | ACTCTGCTCCTCCTTGC |
| Cmr2-qF | AACCGCAGTTCTCCCT |
| Cmr2-qR | TTTGTTCTTACCTCGTCATC |
| Cmr3-qF | ATGGCGGTAATGTTCG |
| Cmr3-qR | CTTCCGTCTGCTTTCG |
| Cmr4-qF | GATAGAGCCCACGAACA |
| Cmr4-qR | CCTTTGGCACTCCTTAC |
| Cmr5-qF | GCCTACGCTTACGAGTG |
| Cmr5-qR | TATCTTCCTTGGGTCTTC |
| Cmr6-qF | CGCTGGCGTTCTACTTG |
| Cmr6-qR | TTTGTGGCACCCTATGT |
| 16S-qF | GCCGATTAGGTAGTTGGTGG |
| 16S-qR | CCGTGTCTCAGTGTCCATC |
| Promoterdw-F | CGGAAGCATAAAGTGTAAAG |
| Promoterdw-R | CGACTGGAAAGCGGGC |
| Repeats sequences are underline. | |
表选项
1.3 试剂 聚合酶链式反应(polymerase chain reaction,PCR)中使用3种DNA聚合酶,序列长度大于1000 bp时,使用PrimerSTAR Max DNA聚合酶(TaKaRa)扩增;序列长度小于1000 bp时,使用Pfu DNA聚合酶(Tiangen)扩增;验证重组质粒时,使用Taq PCR Master Mix (Lifefeng)扩增。DNA片段胶回收使用Gel Extraction Kit (Omega)。无缝克隆使用ClonExpress Ⅱ单步克隆试剂盒(Vazyme)。重组质粒在Trans5α Chemically Competent E. coli (Transgene)中复制,使用Plasmid DNA Minikit Ⅰ (Omega)进行提取。总RNA提取中的Trizol、DEPC水和乙醇购自生工生物工程(上海)股份有限公司,三氯甲烷和异戊醇为国产分析纯。DNase Ⅰ购自Thermo Scientific,RNA反转录试剂盒为Revertaid 1st cDNA Synthesis Kit (Thermo Scientific),qPCR中使用染料SYBR Green Master Mix (Thermo Scientific)。鲁戈氏碘液配方为碘5 g,碘化钾10 g,去离子水100 mL。
1.4 生物信息学分析 热球菌科及进化树的外群(Methancoccus voltae A3 and S. islandicus REY15A)的基因组序列来自NCBI数据库(https://www.ncbi.nlm.nih.gov/)。通过BLASTp将基因与COG数据库比对,统计每个物种中的Ⅲ-B型CRISPR-Cas系统的Cmr基因[33]。将这些Cmr基因的串联序列构建进化树。首先使用全局模式MAFFT-linsi将每个Cmr基因的序列(见附表 1),进行比对,然后按照Cmr1至Cmr6的顺序将多序列比对结果串联[34]。随后使用trimAL的atomated1模式对串联序列进行修剪[35],删除gap过多的位置。最后使用IQTREE (TPM3u+F+I+G4模型,1000次快速重复)构建进化树[36]。
附表 1. 具有完整基因组的热球菌科中的Ⅲ-B型CRISPR-Cas系统 Table S1. The putative type Ⅲ-B CRISPR-Cas systems of Thermococcaceae with complete genomes
| Strains | Genes | Functional annotation |
| Pyrococcus furiosus DSM 3638 (NC_003413.1) | WP_081481782.1 | Cmr1 |
| WP_011012264.1 | Cmr6 | |
| WP_011012265.1 | Cmr5 | |
| WP_011012266.1 | Cmr4 | |
| WP_011012268.1 | Cmr3 | |
| WP_011012269.1 | Cmr2 | |
| WP_014835337.1 | Cmr1 | |
| Pyrococcus kukulkanii NCB100 (NZ_CP010835.1) | WP_068320300.1 | Cmr1 |
| WP_068320302.1 | Cmr2 | |
| WP_068320304.1 | Cmr3 | |
| WP_068320310.1 | Cmr4 | |
| WP_068320312.1 | Cmr5 | |
| WP_068320314.1 | Cmr6 | |
| WP_068322519.1 | Cmr2 | |
| Pyrococcus yayanosii CH1 (NC_015680.1) | WP_013905541.1 | Cmr1 |
| WP_013905542.1 | Cmr2 | |
| WP_013905543.1 | Cmr3 | |
| WP_013905545.1 | Cmr4 | |
| WP_013905546.1 | Cmr5 | |
| WP_013905547.1 | Cmr6 | |
| WP_013906170.1 | Cmr2 | |
| Thermococcus barophilus CH5 (NZ_CP013050.1) | WP_056934684.1 | Cmr1 |
| WP_056934685.1 | Cmr2 | |
| WP_056934686.1 | Cmr3 | |
| WP_056934689.1 | Cmr4 | |
| WP_056934690.1 | Cmr5 | |
| WP_056934691.1 | Cmr6 | |
| Thermococcus chitonophagus GC74 (NZ_CP015193.1) | WP_068575899.1 | Cmr2 |
| WP_068576518.1 | Cmr1 | |
| WP_068576520.1 | Cmr2 | |
| WP_088859102.1 | Cmr3 | |
| WP_068576529.1 | Cmr4 | |
| WP_068576531.1 | Cmr5 | |
| WP_068576532.1 | Cmr6 | |
| WP_068577402.1 | Cmr6 | |
| WP_068577403.1 | Cmr5 | |
| WP_157092417.1 | Cmr4 | |
| WP_068577407.1 | Cmr3 | |
| WP_068577409.1 | Cmr2 | |
| WP_157895698.1 | Cmr1 | |
| Thermococcus eurythermalis A501 (NZ_CP008888.1) | WP_050003737.1 | Cmr6 |
| WP_050003738.1 | Cmr5 | |
| WP_050003739.1 | Cmr4 | |
| WP_050003741.1 | Cmr3 | |
| WP_050003742.1 | Cmr2 | |
| WP_050003743.1 | Cmr1 | |
| Thermococcus siculi RG-20 (NZ_CP015103.1) | WP_157727105.1 | Cmr6 |
| WP_088856494.1 | Cmr5 | |
| WP_088856495.1 | Cmr4 | |
| WP_088856496.1 | Cmr3 | |
| WP_157727109.1 | Cmr2 | |
| WP_157727111.1 | Cmr1 | |
| Thermococcus sp. 2319x1 (NZ_CP012200.1) | WP_058947205.1 | Cmr1 |
| WP_058947206.1 | Cmr2 | |
| WP_058947207.1 | Cmr3 | |
| WP_058947209.1 | Cmr4 | |
| WP_058947210.1 | Cmr5 | |
| WP_058947211.1 | Cmr6 | |
| Thermococcus sp. EXT12c (NZ_LT900021.1) | WP_145955410.1 | Cmr6 |
| WP_099211677.1 | Cmr5 | |
| WP_099211679.1 | Cmr4 | |
| WP_099211683.1 | Cmr3 | |
| WP_099211685.1 | Cmr2 | |
| WP_099211687.1 | Cmr1 | |
| Thermococcus sp. IOH1 (NZ_CP040847.1) | WP_139680684.1 | Cmr2 |
| WP_139680987.1 | Cmr1 | |
| WP_139680988.1 | Cmr2 | |
| WP_139680989.1 | Cmr3 | |
| WP_139680991.1 | Cmr4 | |
| WP_139681776.1 | Cmr5 | |
| WP_139680992.1 | Cmr6 | |
| Pyrococcus horikoshii OT3 (NC_000961.1) | WP_048053047.1 | Cmr2 |
| Pyrococcus sp. ST04 (NC_017946.1) | WP_014733238.1 | Cmr2 |
| Thermococcus cleftensis CL1 (NC_018015.1) | WP_014789778.1 | Cmr2 |
| Thermococcus onnurineus NA1 (NC_011529.1) | WP_012571853.1 | Cmr2 |
| Thermococcus piezophilus CDGS (NZ_CP015520.1) | WP_068663804.1 | Cmr2 |
| Thermococcus radiotolerans EJ2 (NZ_CP015106.1) | WP_088866078.1 | Cmr2 |
| Palaeococcus pacificus DY20341 (NZ_CP006019.1) | – | – |
| Pyrococcus abyssi GE5 (NC_001773.1) | – | – |
| Pyrococcus sp. NA2 (NC_015474.1) | – | – |
| Thermococcus barophilus MP (NC_015471.1) | – | – |
| Thermococcus barossii SHCK-94 (NZ_CP015101.1) | – | – |
| Thermococcus celer Vu 13 (NZ_CP014854.1) | – | – |
| Thermococcus gammatolerans EJ3 (NC_012804.1) | – | – |
| Thermococcus gorgonarius W-12 (NZ_CP014855.1) | – | – |
| Thermococcus guaymasensis DSM 11113 (NZ_CP007140.1) | – | – |
| Thermococcus kodakarensis KOD1 (NC_006624.1) | – | – |
| Thermococcus litoralis DSM 5473 (NC_022084.1) | – | – |
| Thermococcus nautili 30-1 (NZ_CP007264.1) | – | – |
| Thermococcus pacificus P-4 (NZ_CP015102.1) | – | – |
| Thermococcus paralvinellae ES1 (NZ_CP006965.1) | – | – |
| Thermococcus peptonophilus OG-1 (NZ_CP014751.1) | – | – |
| Thermococcus profundus DT 5432 (NZ_CP014863.1) | – | – |
| Thermococcus sibiricus MM 739 (NC_012883.1) | – | – |
| Thermococcus sp. 4557 (NC_015865.1) | – | – |
| Thermococcus sp. 5-4 (NZ_CP021848.1) | – | – |
| Thermococcus sp. AM4 (NC_016051.1) | – | – |
| Thermococcus sp. P6 (NZ_CP015104.1) | – | – |
| Thermococcus sp. SY113 (NZ_CP041932.1) | – | – |
| Thermococcus thioreducens OGL-20P (NZ_CP015105.1) | – | – |
| –: the Cmr proteins which is not found. | ||
表选项
1.5 基因表达敲降质粒的构建 该质粒在穿梭质粒骨架上插入不同启动子控制的mini-CRISPR簇元件组成。在目的基因PYCH_13690的非编码链上选取5′-tag序列(或替代序列)下游长度为37 nt的Protospacer,作为特异性间隔序列(spacer)插入重复序列(repeats)中,使得人工构建的mini-CRISPR簇可以转录出与PYCH_13690基因转录出的mRNA配对的crRNA。同时,crRNA上的特定5′-tag序列与PYCH_13690的mRNA结合,不激活crRNP复合体的DNA酶活性,使内源CRISPR系统只剪切mRNA,避免切割DNA。
mini-CRISPR簇中需要人工合成序列(1369-hhp和1369-hmtb)见表 2,其两端具有与质粒重叠的区域。Repeats序列为CRISPR Finder数据库中公告的亚氏火球菌基因组中5′-CTTT CAATTCTCTTTGAGTCTTATTGGAAC-3′[37]。
使用引物pLMOhhp-F/R以质粒pLMOS776TA为模板扩增包括HHP启动子元件和一个HMG-CoA还原酶基因,获得线性化的载体片段pLMOShhp[11];使用引物CRISPR-hhp-F/CRISPR-R以合成的1369-hhp序列为模板扩增包括Phhp启动子重叠区域和mini-CRISPR簇基因,获得片段hhp-array;使用ClonExpress Ⅱ one-step cloning kits (Vazyme)试剂盒将线性化载体片段pLMOShhp与DNA片段hhp-array进行连接,构建敲降质粒pLMOSkdp-1369。
使用引物pLMOS3-F/R以质粒pLMOShhp为模板扩增包含HMG-CoA还原酶基因的线性化载体片段pLMOS01;使用引物Phmtb-F/R以pLMOS776TA为模板扩增包括Phmtb启动子基因及其上游序列,获得片段Phmtb-S1;使用引物CRISPR-hmtb-F/CRISPR-R以合成的1369-hmtb序列为模板扩增包括Phmtb启动子重叠区域和mini-CRSIPR簇基因,获得片段hmtb-array;使用ClonExpress Ⅱ one-step cloning kits (Vazyme)试剂盒将线性化载体pLMOS01、片段Phmtb-S1与片段hmtb-array连接,构建敲降质粒pLMOkdm-1369。
以上2个重组质粒分别转化至E. coli Trans5α中,在含有50 μg/mL的氨苄霉素(Amp)的LB培养基中,37 ℃,培养14 h。通过PCR扩增重组质粒中mini-CRISPR簇,并进行DNA测序分析确保该基因片段的序列无误。
1.6 亚氏火球菌A1菌株培养和转化 亚氏火球菌A1的培养基为TRM培养基(pH 6.8),培养温度为95 ℃,压力为1.0×102 Pa或5.2×104 Pa。将TRM培养基分装在血清瓶中,密封后抽换气3次,使氮气替换氧气,并加入脱氧剂Na2S·9H2O以维持厌氧状态。另外需添加单质硫作为电子受体。固体TRM培养基中使用了多硫化物溶液替代硫和Na2S·9H2O,高温琼脂的浓度为1%。选择培养基中,正筛选标记辛伐他汀(ark pharm)的最终浓度为10 μmol/L。每组均有3个平行样本。使用Plasmid DNA Minikit I试剂盒从E. coli Trans5α提取重组质粒,并使用CaCl2方法将质粒转化至亚氏火球菌A1[10–11],转化后从固体培养基中挑取单克隆,接种在含有10 μmol/L辛伐他汀的培养基中置于1.0×102 Pa,95 ℃培养2 d。使用引物sim-F/R对菌体进行菌落PCR,验证敲降质粒是否转化成功。
1.7 总RNA提取和反转录 亚氏火球菌A1及其敲降菌株在95 ℃、1.0×102 Pa或5.2×104 Pa下培养14 h。在4 ℃下,12000 r/min离心5 min收集菌体,并迅速冻存在–70 ℃。加入Trizol试剂裂解后,使用三氯甲烷、异戊醇和75%乙醇纯化。使用80 μL DEPC水溶解RNA,加入DNase I在37 ℃下反应1 h,降解DNA片段,并用75%乙醇纯化。重新用DEPC水溶解后,测定RNA浓度。取2 μg RNA使用试剂盒RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific)对总RNA进行反转录,PCR程序为:25 ℃ 5 min;42 ℃ 60 min;70 ℃ 5 min。取2 μg RNA加入2× RNA loading buffer用于RNA琼脂糖凝胶电泳(100 V,20 min)检查RNA质量。
1.8 测定mRNA剪切效率 使用ABI 7500/7500 Real-Time PCR System (Thermo Scientific)和SYBR Green Master Mix (Thermo Scientific)进行定量PCR,方法为ΔΔCT。每组qPCR重复3次。PCR条件:95 ℃ 10 min;40循环:95 ℃ 15 s;60 ℃ 1 min;95 ℃ 15 s;60 ℃ 1 min;95 ℃ 30 s;72 ℃ 20 s;72 ℃ 5 min。两对引物设计在基因PYCH_13690上,test 1 (test1-F/R)扩增区域为Protospacer 1剪切区域,test 2 (test2-F/R)扩增cDNA靠近3′末端的对照区域。
1.9 培养物中残留淀粉含量分析 在液体TRM培养基中加入了1‰ (W/V)的可溶性淀粉进行培养。培养12 h后,取1 mL细胞悬浮培养液,加入10 μL的鲁戈氏碘液测定淀粉的残留量。
1.10 测定Cmr蛋白的差异表达量 对Cmr1-6的mRNA进行定量PCR,qPCR方法为ΔΔCT。每组qPCR重复3次。条件及仪器见1.8。引物Cmr1-6-qF/R分别设计在Cmr1-6基因序列上,序列对应编号如下:Cmr1 (PYCH_07990),Cmr2 (PYCH_08000),Cmr3 (PYCH_08010),Cmr4 (PYCH_08030),Cmr5 (PYCH_08040),Cmr6 (PYCH_08050),引物16S-qF/R设计在16S rRNA上,为内参基因。
1.11 测定启动子Phhp和Phmtb下游序列的转录量 对启动子Phhp和Phmtb下游序列进行定量PCR,方法为ΔΔCT。每组qPCR重复3次。条件及仪器见1.8。由于mini-CRISPR簇转录后被Cas6蛋白剪切成crRNA,无法设计qPCR引物,引物Promoterdw-F/R设计在mini-CRISPR簇下游共转录的序列上(长度为200 bp)。引物16S-qF/R设计在16S rRNA上,为内参基因。
2 结果和分析 2.1 热球菌科中的Ⅲ-B型CRISPR-Cas系统 在已公布完整基因组的热球菌科菌株中有3株火球菌(Pyrococcus)和7株热球菌(Thermococcus)存在Ⅲ-B型CRISPR-Cas系统,而在古老球菌(Palaeococcus)中暂未发现。这些火球菌和热球菌基因组中Ⅲ-B型CRISPR-Cas系统均为α亚型,由6个Cmr蛋白(Cmr1-Cmr6)组成,Cmr2 (Cas10)是其中的标志蛋白(图 1-A)。
 |
| 图 1 热球菌科中的Ⅲ-B型CRISPR-Cas编码基因的特征 Figure 1 Characteristics of the gene clusters of Cmr proteins of type Ⅲ-B CRISPR-Cas and the repeats sequences in Thermococcaceae. A: the gene clusters of Cmr proteins in Thermococcaceae; B: the repeats sequences in Thermococcaceae. The 5′-handle is the 8 nucleotides at the end of repeats. |
| 图选项 |
CRISPR基因簇由重复序列(repeats)和间隔序列(spacer)组成,可转录出crRNA。Group 1型repeats长度为30 bp,前导序列具有BRE/TATA box[38]。统计分析热球菌科中所有对应I-A/Ⅲ-B型CRSIRP-Cas系统的repeats序列的偏好性(图 1-B),结果表明Group 1型repeats序列基本保守,其3′末端长度为8 nt的5′-handle序列基本保守,转录得到crRNA上的5′-tag用于保护与之完全匹配的外源DNA。
2.2 构建可敲降淀粉酶编码基因表达的质粒 在亚氏火球菌A1基因组中存在的4条CRISPR簇含有相同的Group 1型repeats序列,可诱导I-A/Ⅲ-B型CRISPR-Cas系统的剪切。邓岭等[26]发现S. islandicus中5′-tag序列可以保护DNA,并得到7个与5′-tag作用相似的6 nt的序列,可以应用于其他的菌株中。我们在PYCH_13690基因的非编码链上找到4处对应的序列,分别位于起始密码子位点(+1)下游的750–756、1530–1535、1616–1621和1889–1894处。我们选取了最靠近起始密码子的“GTAAAG”,在非编码链5′-tag的下游序列选取长度为37 nt的寡聚核苷酸构成protospacer 1片段(图 2-A)。将此片段插入两个Group 1型repeats中成为spacer 1片段,构成mini-CRISPR簇。选取高压诱导型启动子Phhp和组成型启动子Phmtb分别控制mini-CRISPR簇的表达,进一步研究不同启动子对CRISPR-Cas系统剪切效率的影响(图 2-B)。
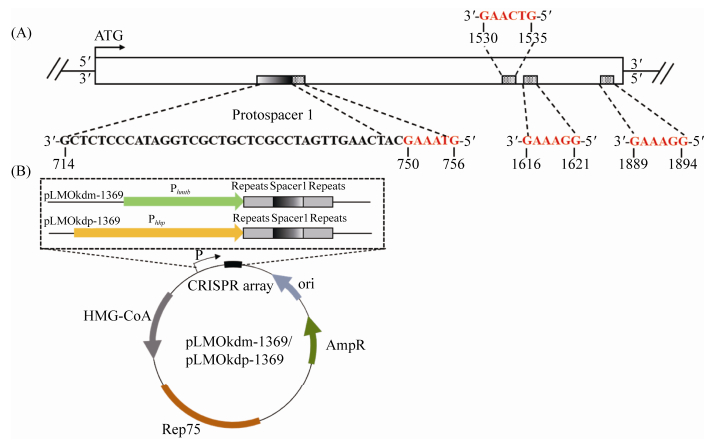 |
| 图 2 mini-CRISPR簇切割位点的设计及敲降质粒的构建 Figure 2 The strategy of cleavage sites and construction of knock down plasmid with artificial mini-CRISPR locus. A: schematic of RNA-tag regions selected for gene silencing of the gene PYCH_13690. The 5′-tag sites were from 750 to 756, 1530 to 1535, 1616 to 1621, and 1889 to 1894 which are red. The protospacer 1 with 37nt from 714 to 750 nt is chosen which is close to the start codon (ATG, +1) of PYCH_13690 in antisense sequence. Then, the spacers 1 was inserted into two repeats. B: the artificial mini-CRISPR locus with an HHP-inducible promoter Phhp and a constitutive promoter Phmtb on the knock down plasmid pLMO-kdp-1369 and pLMO-kdm-1369. |
| 图选项 |
2.3 高压诱导型启动子和组成型启动子控制mini-CRISPR簇进行基因表达敲降的效率差异 CRISPR-Cas系统的剪切位点是crRNA与核酸序列的匹配位点,所以我们在此位点设计了1对qPCR引物test 1,测定未剪切的mRNA。而对照引物test 2设计在剪切位点的下游(图 3-A)。当Ⅲ-B型CRISPR-Cas系统剪切切割位点时,该切割位点上引物无法扩增,而下游对照位点可以正常扩增。设定对照位点test 2为内参,采用相对定量的方法,计算菌株中目标基因mRNA的相对剪切量。将野生型菌株A1中目的引物和对照引物的扩增数量的比例设定为100%,野生型菌株和基因表达敲降菌株之间的比例被定义为剪切效率。
结果表明,目标基因PYCH_13690的mRNA在5.2×104 Pa时降解了32.95%,在1.0×102 Pa下降解了50.31% (图 3-B)。为了提高该CRISPR-Cas系统在嗜热嗜压古菌的最适压力5.2×104 Pa下的基因表达敲除的效率,我们尝试使用了一种来自于P. furiosus的组成型启动子Phmtb替代压力诱导型启动子。结果显示,在不同的生长压力下,组成型启动子控制的CRISPR-Cas系统的敲除效率均更高。以目标基因PYCH_13690为例,其mRNA在常压1.0×102 Pa下的剪切率高达76.03%、在5.2×104 Pa下的剪切率为50.31% (图 3-B)。至此,我们利用超嗜热嗜压亚氏火球菌内源Ⅲ-B型CRISPR-Cas系统,建立了不同抑制效率的基因沉默遗传工具。
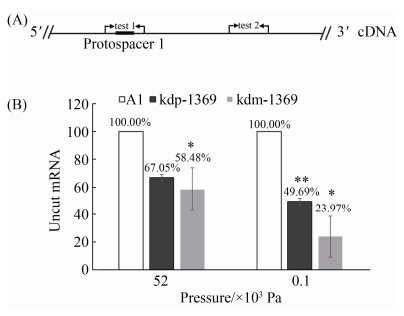 |
| 图 3 qPCR检测淀粉酶编码基因PYCH_13690的mRNA的剪切率 Figure 3 Cleavage analysis of CRISPR-mediated RNA interference of gene PYCH_13690 by real-time qPCR. A: strategy for detecting RNA cleavage at the spacer 1 in mRNA. Primer test 1 set amplified a region containing the predicted RNA cleavage sites while primer test 2 amplified a region positioned upstream the spacer 1 in mRNA. B: results of qPCR showing the ratio of uncut mRNA over total mRNA after reverse transcription of total RNA into cDNA. The strains A1 (white), kdp-1369 (light gray), and kdm-1369 (dark gray) were cultured at different pressures for 14 hours. The results are from three independent experiments, and the error bars represent the standard deviations. *: P < 0.05; **: P < 0.01. The strain kdp-1369 is A1 with plasmid pLMO-kdp-1369 (Figure 2-B, while the strain kdm-1369 is A1 with plasmid pLMO-kdm-1369). |
| 图选项 |
2.4 淀粉酶编码基因表达敲降菌株的表型分析 淀粉酶基因PYCH_13690是淀粉降解途径中表达量最高的基因。在过去的研究中,敲除该基因的突变菌株?1369-在1.0×102 Pa条件下几乎不降解淀粉[11]。
将菌体接种到含有1‰ (W/V)可溶性淀粉的TRM培养基中在95 ℃、1.0×102 Pa下培养16 h,比较野生型菌株A1和淀粉酶基因表达敲降菌株(kdp-1369和kdm-1369)的差异。结果显示,在19 h时野生型菌株A1的培养物中剩余的淀粉含量显著小于敲降菌株,在23 h时野生型菌株A1的培养物中淀粉基本消耗完,而此时淀粉酶基因表达敲降菌株的培养物中淀粉含量开始下降(图 4)。
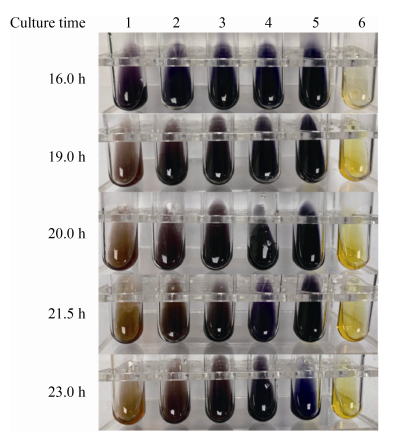 |
| 图 4 淀粉酶编码基因(PYCH_13690)表达敲降菌株的表型分析 Figure 4 Phenotypic analysis of the gene silencing strains of PYCH_13690. Measurement of the residual starch via Lugol's iodine method after at 95 ℃ under 0.1 MP. 1: strain A1 cultured in TRM with 1‰ (W/V) soluble starch; 2: strain kdp1369 cultured in TRM with 1‰ (W/V) soluble starch; 3: strain kdm1369 cultured in TRM with 1‰ (W/V) soluble starch; 4: mutant ?1369- cultured in TRM with 1‰ (W/V) soluble starch; 5: TRM with 1‰ (W/V) soluble starch (negative control); 6: TRM. |
| 图选项 |
3 结论和讨论 Terns研究小组对超嗜热火球菌P. furiosus中的Ⅲ-B型CRISPR-Cas系统的DNA和RNA剪切的机理进行了深入的研究[30–32]。亚氏火球菌中存在有和P. furiosus高度相似的CRISPR-Cas系统,其中Cmr蛋白相似度高于40% (附表 2)。在本研究中,我们通过构建与目的基因匹配的人工mini-CRISPR簇,在超嗜热嗜压亚氏火球菌中实现了基于内源性的Ⅲ-B型CRISPR-Cas对一个淀粉酶基因的mRNA敲降。
附表 2. P. furiosus DSM 3638和P. yayanosii CH1中Cmr蛋白的氨基酸序列相似性 Table S2. The amino acid sequence similarity of Cmr proteins from P. furiosus DSM 3638 and P. yayanosii CH1
| Genes | P. furiosus DSM 3638 | P. yayanosii CH1 | Query cover/% | E value | Identity/% | |||
| Locus | Length | Locus | Length | |||||
| Cmr1 | WP_014835337.1 | 337 | WP_013905541.1 | 359 | 94 | 3e-60 | 38.79 | |
| Cmr2 | WP_011012269.1 | 871 | WP_013905542.1 | 960 | 78 | 2e-98 | 31.14 | |
| Cmr3 | WP_011012268.1 | 322 | WP_013905543.1 | 356 | 98 | 4e-46 | 32.41 | |
| Cmr4 | WP_011012266.1 | 295 | WP_013905545.1 | 301 | 97 | 2e-101 | 52.65 | |
| Cmr5 | WP_011012265.1 | 169 | WP_013905546.1 | 156 | 92 | 7e-32 | 39.38 | |
| Cmr6 | WP_011012264.1 | 340 | WP_013905547.1 | 344 | 72 | 2e-72 | 46.89 | |
表选项
当启动子不同时,上述mRNA敲降系统表现出不同的剪切效率。在最适生长压力(5.2×104 Pa)下,高静水压诱导型启动子Phhp和组成型启动子Phmtb诱导mini-CRISPR簇表达将淀粉酶基因的表达下调的程度与S. solfataricus中41%的剪切率相似[21]。然而,敲降体系在常压下(1.0×102 Pa)的剪切率比最适生长压力(5.2×104 Pa)更高,使用Phhp时基因表达下调了50.31%,使用Phmtb时基因表达下调了76.03%。在不同培养压力下的不同的mRNA剪切率可能与Cmr蛋白的数量和活性有关。有研究表明,CRISPR-Cas系统中核酸酶表达量在非最适生长条件下会提高[14]。我们在1.0×102 Pa和5.2×104 Pa下测定了亚氏火球菌A1菌株中Cmr蛋白的mRNA差异倍数,结果表明在1.0×102 Pa下Cmr蛋白的表达量均提高(图 5)。虽然压力诱导型启动子Phhp控制的mini-CRISPR簇在高压下产生较高的crRNA (附图 1),但在5.2×104 Pa下较低的Cmr蛋白表达水平可能影响了CRISPR-Cas系统的剪切效率。同时,使用组成型启动子Phmtb控制的mini-CRISPR簇在常压和高压下产生的crRNA没有显著差异(附图 1),但Cmr蛋白的表达量在常压下更高(图 5),因此对超嗜热兼性嗜压的亚氏火球菌使用Phmtb诱导的基因表达敲降系统在常压下进行基因沉默遗传操作的效率更高。
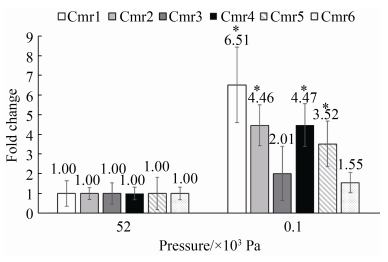 |
| 图 5 亚氏火球菌A1中Cmr1-6蛋白在1.0×102 Pa和5.2×104 Pa下mRNA的差异倍数 Figure 5 The fold changes of Cmr1-6 protein in P. yayanosii A1 under 1.0×102 Pa and 5.2×104 Pa. |
| 图选项 |
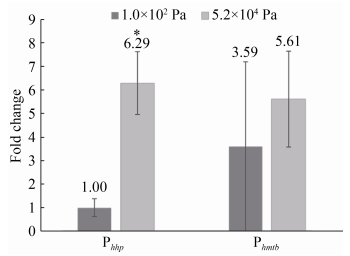 |
| 附图 1 启动子Phhp和Phmtb在1.0×102 Pa和5.2×104 Pa下驱动mini-CRISPR簇转录差异 Figure S1 The fold changes of transcription of mini- CRISPR cluster driven by Promoter Phhp and Phmtb under 1.0×102 Pa and 5.2×104 Pa. |
| 图选项 |
由于最适生长温度和压力的限制,超嗜热嗜压古菌亚氏火球菌A1的遗传操作工具有限。本研究显示基于内源性的Ⅲ-B型CRISPR-Cas系统只需要导入长度较小的mini-CRISPR簇,可以开展必需基因的功能研究。特别是亚氏火球菌中存在有7个5′-tag序列可用来设计剪切位点,可满足构建绝大多数基因表达敲降质粒的需要[26]。此外,可以在一个基因中设计多个剪切位点,进一步提高基因表达的敲降效率;也可以将多个目标基因的序列的spacer连接成CRISPR簇,同时敲降多个基因的表达量。
对已公布基因组的10株超嗜热的热球菌和火球菌中Cmr基因进行系统发育分析,结果显示亚氏火球菌A1基因组中Cmr基因与其他热球菌基因组中的Cmr基因亲缘关系更近(图 6)。推测该系统也容易移植到其他具有Ⅲ-B型CRISPR-Cas系统的菌株中,如T. eurythermalis A501。在其他不具有Ⅲ-B型系统的热球菌科中(如T. kodakarensis及Palaeococcus pacificus),可以使用dCas2的Ⅰ型或dCas9的Ⅱ型CRISPR-Cas系统结合基因启动子区域,抑制基因表达。
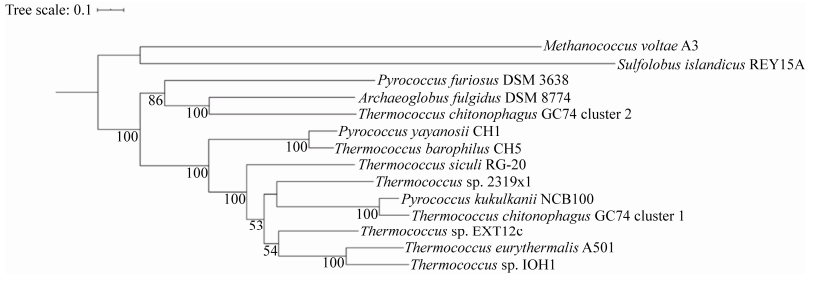 |
| 图 6 具有完整基因组的热球菌中Cmr蛋白的系统发育树 Figure 6 The phylogenetic tree of the Cmr proteins in Thermococcaceae with complete genomes. The maximum likelihood tree is constructed by IQTREE with 'TPM3u+F+I+G4' model, and 1000 ultrafast bootstraps is used for the last construction. The outgroup are Methancoccus voltae A3 and Sulfolobus islandicus REY15A. The access No. of the amino acid sequence of Cmr proteins were listed in Table S1. |
| 图选项 |
References
| [1] | Fang JS, Zhang L, Bazylinski DA. Deep-sea piezosphere and piezophiles: geomicrobiology and biogeochemistry. Trends in Microbiology, 2010, 18(9): 413-422. DOI:10.1016/j.tim.2010.06.006 |
| [2] | Zillig W, Reysenbach AL. Thermococcales//Bergey's manual of systematics of Archaea and Bacteria. Hoboken, NJ: John Wiley & Sons Inc, 2015. |
| [3] | Takai K, Nakamura K. Archaeal diversity and community development in deep-sea hydrothermal vents. Current Opinion in Microbiology, 2011, 14(3): 282-291. DOI:10.1016/j.mib.2011.04.013 |
| [4] | Cario A, Lormières F, Xiang X, Oger P. High hydrostatic pressure increases amino acid requirements in the piezo-hyperthermophilic archaeon Thermococcus barophilus. Research in Microbiology, 2015, 166(9): 710-716. DOI:10.1016/j.resmic.2015.07.004 |
| [5] | Cario A, Jebbar M, Thiel A, Kervarec N, Oger PM. Molecular chaperone accumulation as a function of stress evidences adaptation to high hydrostatic pressure in the piezophilic archaeon Thermococcus barophilus. Scientific Reports, 2016(6): 29483. |
| [6] | Zeng X, Birrien JL, Fouquet Y, Cherkashov G, Jebbar M, Querellou J, Oger P, Cambon-Bonavita MA, Xiao X, Prieur D. Pyrococcus CH1, an obligate piezophilic hyperthermophile: extending the upper pressure-temperature limits for life. The ISME Journal, 2009, 3(7): 873-876. DOI:10.1038/ismej.2009.21 |
| [7] | Birrien JL, Zeng X, Jebbar M, Cambon-Bonavita MA, Quérellou J, Oger P, Bienvenu N, Xiao X, Prieur D. Pyrococcus yayanosii sp. nov., an obligate piezophilic hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent.. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(12): 2827-2881. DOI:10.1099/ijs.0.024653-0 |
| [8] | Xu J, Liu LP, Xu MJ, Philippe O, Wang FP, Mohamed J, Xiao X. Complete genome sequence of the obligate piezophilic hyperthermophilic archaeon Pyrococcus yayanosii CH1. Journal of Bacteriology, 2011, 193(16): 4297-4298. DOI:10.1128/JB.05345-11 |
| [9] | 李学恭. 深海超嗜热嗜压古菌Pyrococcus yayanosii压力适应性机制研究. 中国海洋大学博士学位论文, 2014. |
| [10] | Li XG, Fu L, Li Z, Ma XP, Xiao X, Xu J. Genetic tools for the piezophilic hyperthermophilic archaeon Pyrococcus yayanosii. Extremophiles, 2015, 19(1): 59-67. DOI:10.1007/s00792-014-0705-2 |
| [11] | Song QH, Li Z, Chen RK, Ma XP, Xiao X, Xu J. Induction of a toxin-antitoxin gene cassette under high hydrostatic pressure enables markerless gene disruption in the hyperthermophilic archaeon Pyrococcus yayanosii. Applied and Environmental Microbiology, 2019, 85(4): e02662-18. |
| [12] | Li Z, Li XG, Xiao X, Xu J. An integrative genomic island affects the adaptations of the piezophilic hyperthermophilic archaeon Pyrococcus yayanosii to high temperature and high hydrostatic pressure. Frontiers in Microbiology, 2016(7): 1927. |
| [13] | 李臻. 深海来源嗜压、超嗜热古菌Pyrococcus yayanosii基因组岛PYG1的特征及生理功能研究. 上海交通大学博士学位论文, 2017. |
| [14] | Michoud G, Jebbar M. High hydrostatic pressure adaptive strategies in an obligate piezophile Pyrococcus yayanosii. Scientific Reports, 2016(6): 27289. |
| [15] | Hille F, Richter H, Wong SP, Bratovi? M, Ressel S, Charpentier E. The biology of CRISPR-Cas: backward and forward. Cell, 2018, 172(6): 1239-1259. DOI:10.1016/j.cell.2017.11.032 |
| [16] | Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nature Reviews Microbiology, 2011, 9(6): 467-477. DOI:10.1038/nrmicro2577 |
| [17] | Hochstrasser ML, Taylor DW, Kornfeld JE, Nogales E, Doudna JA. DNA targeting by a minimal CRISPR RNA-guided cascade. Molecular Cell, 2016, 63(5): 840-851. DOI:10.1016/j.molcel.2016.07.027 |
| [18] | Samai P, Pyenson N, Jiang WY, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA cleavage during type Ⅲ CRISPR-cas immunity. Cell, 2015, 161(5): 1164-1174. DOI:10.1016/j.cell.2015.04.027 |
| [19] | Elmore JR, Sheppard NF, Ramia N, Deighan T, Li H, Terns RM, Terns MP. Bipartite recognition of target RNAs activates DNA cleavage by the type Ⅲ-B CRISPR-Cas system. Genes and Development, 2016(30): 447-459. |
| [20] | Stachler AE, Schwarz TS, Schreiber S, Marchfelder A. CRISPRi as an efficient tool for gene repression in archaea. Methods, 2020(172): 76-85. |
| [21] | Zebec Z, Manica A, Zhang J, White MF, Schleper C. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Research, 2014, 42(8): 5280-5288. DOI:10.1093/nar/gku161 |
| [22] | Peng N, Han WY, Li YJ, Liang YX, She QX. Genetic technologies for extremely thermophilic microorganisms of Sulfolobus, the only genetically tractable genus of crenarchaea. Science China Life Sciences, 2017, 60(4): 370-385. DOI:10.1007/s11427-016-0355-8 |
| [23] | Peng WF, Feng MX, Feng X, Liang YX, She QX. An archaeal CRISPR type Ⅲ-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Research, 2015, 43(1): 406-417. DOI:10.1093/nar/gku1302 |
| [24] | Liu T, Pan SF, Li YJ, Peng N, She QX. Type Ⅲ CRISPR-Cas system: introduction and its application for genetic manipulations. Current Issues in Molecular Biology, 2018(26): 1-14. |
| [25] | 彭文舫. Sulfolobus islandicus CRISPR-Cas病毒防御功能的遗传学分析. 华中农业大学博士学位论文, 2015. |
| [26] | Deng L, Garrett RA, Shah SA, Peng X, She QX. A novel interference mechanism by a type Ⅲ-B CRISPR-Cmr module in Sulfolobus. Molecular Microbiology, 2013, 87(5): 1088-1099. DOI:10.1111/mmi.12152 |
| [27] | Osawa T, Inanaga H, Sato C, Numata T. Crystal structure of the CRISPR-CAS RNA silencing cmr complex bound to a target analog. Molecular Cell, 2015, 58(3): 418-430. DOI:10.1016/j.molcel.2015.03.018 |
| [28] | Ramia NF, Spilman M, Tang L, Shao YM, Elmore J, Hale C, Cocozaki A, Bhattacharya N, Terns RM, Terns MP, Li H, Stagg SM. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex. Cell Reports, 2014, 9(5): 1610-1617. DOI:10.1016/j.celrep.2014.11.007 |
| [29] | Shao YM, Cocozaki AI, Ramia NF, Terns RM, Terns MP, Li H. Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA silencing complex. Structure, 2013, 21(3): 376-384. DOI:10.1016/j.str.2013.01.002 |
| [30] | Cocozaki AI, Ramia NF, Shao YM, Hale CR, Terns RM, Terns MP, Li H. Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex. Structure, 2012, 20(3): 545-553. DOI:10.1016/j.str.2012.01.018 |
| [31] | Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA, 2008(14): 2572-2579. |
| [32] | Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell, 2009, 139(5): 945-956. DOI:10.1016/j.cell.2009.07.040 |
| [33] | Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research, 2000, 28(1): 33-36. DOI:10.1093/nar/28.1.33 |
| [34] | Katoh K, Misawa K, Kuma KI, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 2002, 30(14): 3059-3066. DOI:10.1093/nar/gkf436 |
| [35] | Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics, 2009, 25(15): 1972-1973. DOI:10.1093/bioinformatics/btp348 |
| [36] | Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 2015, 32(1): 268-274. DOI:10.1093/molbev/msu300 |
| [37] | Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Research, 2007, 35(S2): W52-W57. |
| [38] | Norais C, Moisan A, Gaspin C, Clouet-d'Orval B. Diversity of CRISPR systems in the euryarchaeal Pyrococcales. RNA Biology, 2013, 10(5): 659-670. DOI:10.4161/rna.23927 |
