赵欣1,2, 张梁威2, 宋福平2, 张杰2, 李晶1

 , 彭琦2
, 彭琦2

1. 东北农业大学生命科学学院, 黑龙江 哈尔滨 150030;
2. 中国农业科学院植物保护研究所, 植物病虫害生物学国家重点实验室, 北京 100193
收稿日期:2020-05-28;修回日期:2020-10-21;网络出版日期:2020-11-13
基金项目:国家自然科学基金(31772243)
*通信作者:李晶, E-mail: lijing@neau.edu.cn;
彭琦, E-mail: qpeng@ippcaas.cn.
摘要:[目的] rocE基因编码精氨酸降解途径中的精氨酸通透酶,通过分析苏云金芽胞杆菌(Bacillus thuringiensis,Bt) rocE基因的转录活性,明确rocE基因的转录调控机制。[方法] 通过RT-PCR确定rocE基因所在基因簇的转录单元;β-半乳糖苷酶活性测定分析rocE基因启动子(ProcE)的转录活性;采用同源重组技术敲除Bt HD73菌株的rocE基因;通过融合His标签的方法在大肠杆菌中表达纯化RocR蛋白的HTH结构域;通过凝胶阻滞实验明确RocR与rocE基因启动子的结合作用。[结果] 在M9培养基中,精氨酸可诱导ProcE的转录活性;在SSM培养基和精氨酸诱导培养基中,与出发菌株HD73相比,ProcE在sigL (编码Sigma54因子)突变体和rocR突变体中的转录活性显著下降。RocR-HTH蛋白与ProcE有结合作用。rocE基因的缺失对菌体生长和Cry1Ac蛋白产量无显著影响。rocE缺失突变体的芽胞形成率为65.5%,HD73出发菌株为85.7%,显著性分析结果表明差异显著(P < 0.05)。[结论] rocE基因的转录活性受Sigma54的控制,并受RocR正调控。rocE基因的缺失影响菌株的芽胞形成率。
关键词:苏云金芽胞杆菌精氨酸代谢rocE转录调控
Transcription and regulation of rocE in Bacillus thuringiensis
Xin Zhao1,2, Liangwei Zhang2, Fuping Song2, Jie Zhang2, Jing Li1

 , Qi Peng2
, Qi Peng2

1. College of Life Sciences, Northeast Agricultural University, Harbin 150030, Heilongjiang Province, China;
2. State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China
Received: 28 May 2020; Revised: 21 October 2020; Published online: 13 November 2020
*Corresponding author: Jing Li, E-mail: lijing@neau.edu.cn;
Qi Peng, E-mail: qpeng@ippcaas.cn.
Foundation item: Supported by the National Natural Science Foundation of China (31772243)
Abstract: [Objective] rocE encodes arginine permease in arginine degradation pathway. To determine the mechanism of transcriptional regulation of rocE, we analyzed the transcriptional activity of rocE in Bacillus thuringiensis (Bt). [Methods] Transcriptional units of rocE gene cluster were analyzed by RT-PCR. Transcriptional activity of rocE promoter (ProcE) was analyzed by β-galactosidase assay. rocE mutant of Bt HD73 strain was constructed by homologous recombination. The HTH domain of RocR with His fusion protein was purified by HiTrap chelating column. The binding of rocE promoter with RocR-HTH protein was verified by electrophoresis mobility shift assays. [Results] Transcriptional activity of ProcE was induced by arginine in M9 medium. The transcriptional activity of ProcE was sharply decreased in sigL (encodes Sigma 54) and rocR mutants compared with that in HD73 wild-type in Schaeffer's sporulation medium (SSM) and arginine induced medium. ProcE could bind to RocR-HTH protein. Mutation of rocE had no significant differences on growth and Cry1Ac protein production. However, the sporulation efficiency of the rocE mutant was 65.5%, and that of the HD73 strain was 85.7%. The results of significance analysis show that the difference was significant (P < 0.05). [Conclusion] Transcriptional activation of rocE is controlled by Sigma 54, and positive regulated by RocR. rocE gene is related to sporulation efficiency.
Keywords: Bacillus thuringiensisarginine metabolismrocEtranscriptional regulation
精氨酸是许多营养物质代谢的前体物质,其代谢产物谷氨酸更是连接碳代谢与氮代谢的重要物质。在细菌中,精氨酸在通透酶的作用下进入细胞体内;在精氨酸酶的作用下分解成为鸟氨酸;进一步在鸟氨酸转氨酶的作用下生成谷氨酸半醛;再继续酶解最终生成谷氨酸进入细菌的碳循环。精氨酸的分解代谢连接起了氮循环与碳循环[1-3]。精氨酸分解代谢分为两个途径:精氨酸酶途径(arginase pathway)和精氨酸脱亚胺酶途径(arginine deiminase pathway)。其中精氨酸酶代谢途径所产生的尿素能进一步被催化生成氨,作为菌体生长的氮源被利用[4]。精氨酸脱亚胺酶途径则与菌体对恶劣环境的适应相关,能够在菌体受到酸胁迫时提高胞内pH,适应酸性环境[4-5]。以精氨酸作为代谢底物,最终会生成氨,二氧化碳和1分子ATP。在营养缺乏时能够以精氨酸为营养物质为菌体供能[5]。
精氨酸酶途径是枯草芽胞杆菌(Bacillus subtilis,Bs)中唯一的精氨酸代谢途径,其代谢过程中相关的酶主要由roc基因簇编码:rocC和rocE基因编码通透酶(arginine/ornithine permease),使精氨酸由胞外运至胞内;rocF基因编码精氨酸酶(arginase),将精氨酸分解成为鸟氨酸;rocD基因编码鸟氨酸转氨酶(ornithine aminotransferase),将鸟氨酸生成谷氨酸半醛(L-glutamate-5-semialdehyde);谷氨酸半醛与吡咯啉羧酸(Δ1-pyrroline-5-carboxylate)可相互转化;rocA编码吡咯啉羧酸脱氢酶(Δ1-pyrroline-5-carboxylate dehydrogenase),将吡咯啉羧酸生成L-谷氨酸;rocG基因编码谷氨酸脱氢酶(glutamate dehydrogenase),将L-谷氨酸生成α-酮戊二酸,进入三羧酸循环,为菌体生长提供能量。roc基因簇形成3个转录单元:rocABC-rocG-rocDEF。这些基因的转录都依赖于Sigma 54因子,并且受到RocR的正调控。它们的转录会受到精氨酸、鸟氨酸和脯氨酸的诱导,某些基因缺失时会导致菌株在精氨酸或鸟氨酸存在的培养条件中不生长。所以精氨酸代谢途径对枯草芽胞杆菌的生长有着一定的影响[3, 6-9]。
苏云金芽胞杆菌(Bacillus thuringiensis,Bt)是土壤微生物,革兰氏阳性菌,是多种鳞翅目、鞘翅目、双翅目等农业害虫的病原菌[10]。由于其能产生杀虫晶体蛋白的特性而被广泛应用于生物防治领域[11-12]。当Bt菌体进入虫体中肠,分泌的毒素蛋白能够在中肠的碱性条件下被消化成为前毒素,之后在中肠多种酶的作用下溶解,发挥其毒性。在这个过程中Bt能够获取宿主的营养用于自身的生长和繁殖[13]。Sigma 54因子是RNA聚合酶核心酶的亚基,调控许多碳氮源的代谢,它通过与启动子的–12/–24区结合,在增强子结合蛋白(enhance binding protein,EBP)的激活下,起始基因的转录[14]。本实验室前期研究发现,Bt HD73菌株中的Sigma 54因子控制了47个基因的转录[15];8个EBP调控了8个代谢途径,包括:γ-氨基丁酸[16-17]、赖氨酸[18]、肌氨酸[19]、3-羟基丁酮代谢[20]等。其中RocR可能调控精氨酸代谢途径,其调控机制尚不明确,本研究在此基础上,对编码精氨酸通透酶的rocE基因的转录调控进行分析,明确RocR对rocE基因的调控机制。进一步完善Bt中Sigma54和EBPs调控的代谢网络,从代谢调控角度更经济地优化菌株对碳源、氮源的利用,为获得高效的Bt制剂提供科学依据。
1 材料和方法 1.1 材料
1.1.1 菌株和培养条件: 本研究所用菌株和质粒见表 1。大肠杆菌(Escherichia coli,E. coli)在LB培养基中37 ℃培养;Bt的培养分别使用LB培养基、SSM培养基和M9培养基,在30 ℃培养。抗生素的使用终浓度分别为:红霉素5 μg/mL,氨苄青霉素100 μg/mL,卡那霉素100 μg/mL。
表 1. 菌株与质粒 Table 1. Strains and plasmids
| Strains and plasmids | Characterization | Resource |
| Strains | ||
| HD73 | B. thuringiensis subsp. kurstaki carrying the cry1Ac gene | This lab |
| HD(ΔsigL) | B. thuringiensis HD73 sigL gene insertion mutant | [15] |
| HD(ΔrocR) | B. thuringiensis HD73 rocR gene insertion mutant | [15] |
| HD(ProcE) | HD73 strain containing plasmid pHTProcE | This study |
| ΔsigL(ProcE) | HD(ΔsigL) strain containing plasmid pHTProcE | This study |
| ΔrocR(ProcE) | HD(ΔrocR) strain containing plasmid pHTProcE | This study |
| CrocR(ProcE) | ΔrocR(ProcE) strain containing plasmid pHTCrocR | This study |
| HD(ProcEMR) | HD73 strain containing plasmid pHTProcEMR | This study |
| HD(ΔrocE) | B. thuringiensis HD73 rocE gene insertion mutant | This study |
| E. coli TG1 | Δ(lac-proAB) supE thi hsd-5 (F' traD36 proA+ proB+ lacIq lacZΔM15), general purpose cloning host | This lab |
| E. coli ET | F– dam-13: : Tn9 dcm-6 hsdM hsdR recF143 zjj-202: : Tn10 galK2 galT22 ara14 pacY1 xyl-5 leuB6 thi-1, for generation of unmethylated DNA | This lab |
| BL21(DE3) | E. coli B, F–, dcm, ompT hsdS(rB-mB-), gal, λ(DE3) | This lab |
| BL21(pETRocR-HTH) | BL21(DE3) strain containing plasmid pETRocR-HTH | This study |
| BLpET | BL21 strain carrying pET21b | This lab |
| Plasmids | ||
| pHT304-18Z | Promoterless lacZ vector, Eryr, Ampr | This lab |
| pHTProcE | pHT304-18Z carrying promoter upstream from rocE | This study |
| pHTProcEMR | pHT304-18Z carrying promoter upstream from rocE with the mutated sequence of RocR binding site | This study |
| pMAD | ApR, EmR shuttle vector, thermosensitive origin of replication | Institute Pasteur |
| pMADΔrocE | pMAD with rocE insertion fragment | This study |
| pETRocR-HTH | pET21b containing RocR HTH domain, Ampr | This study |
| pHT1618 | ApR, TetR, E. coli-Bt shuttle vector | This lab |
| pHTCrocR | pHT1618 carrying promoter upstream from rocR and rocR gene | This study |
表选项
1.1.2 主要试剂及引物: DNA聚合酶、DNA连接酶和限制性内切酶、PCR产物纯化、DNA回收和质粒提取试剂盒等购自宝生物工程(大连)有限公司和Axygen公司。细菌RNA提取试剂盒购自天根生化科技(北京)有限公司。无缝克隆试剂盒购自中美泰和生物技术(北京)有限公司。镍亲和层析柱填料购自GE公司。poly(dI: dC)购自Sigma公司。引物名称及序列见表 2,参照Bt HD73基因组序列[21]设计引物,引物合成由上海生工生物工程公司完成,序列测定由北京诺赛基因组研究中心有限公司完成。
表 2. 引物序列 Table 2. Primers used in this study
| Primer names | Sequences (5'→3')a |
| RT-1F | CATACGTATACGAGTGGGAGAA |
| RT-1R | TTATTAAAAGATCATCCCTCACTGT |
| RT-2F | CGATGGTTTCCTGATGTACCGT |
| RT-2R | GCAATCATCGTAATTAAAATAGCGG |
| RT-3F | GGACATATGGATGTAGCTGAAGTGA |
| RT-3R | CCCTGTCCTTGCATATGTAAATCA |
| RT-4F | GGAATCAGTGGGTATAGCGAATCAT |
| RT-4R | TCTCCAAGACAGAGCATCGTTAAAT |
| RT-5F | GCAGGGCGGAGATGTGAAG |
| RT-5R | CTGCAACAAATTCTTGCGCC |
| ProcE-F | CCAAGCTTAGATTATCTTCTCCTCATTTC |
| ProcE-R | CGGGATCCTTTATAAATTCCTCCCTCTT |
| rocE-A1 | CGGGAGCTCGAATTCGCGGTTGTGCACACT |
| rocE-A2 | CTCAAATGGTTCGCTGATCCATCATATAAAT |
| rocE-B1 | CCTACGAGGAATTTTTATAATATTGAGTAATTT |
| rocE-B2 | GGCGATATCGGATCCGCGTGCCAGCTTCT |
| E1Kan-1 | ATTTATATGATGGATCAGCGAACCATTTGAG |
| E1Kan-2 | AAATTACTCAATATTATAAAAATTCCTCGTAGG |
| RocR-H1 | CGGGATCCGACTCGCTTTCGCGAACGAAT |
| RocR-H2 | ACGCGTCGACGTGTAAATGCAGTTTTTT |
| rocR-1 | GCTTGCATGCCTGCAGCCTTCTATAACTGTCTTCAC |
| rocR-2 | GGATCCTCTAGAGTCGACTCATATGTGTAAATGCAGTTT |
| a: Restriction enzyme sites are underscored. | |
表选项
1.2 RT-PCR HD73菌株在SSM中培养至T5时期(T0为对数期结束的时期,Tn为T0后的n小时),取菌液低温离心,用TRIzol重悬沉淀,RNA提取方法参照试剂盒说明。以纯化后的RNA为模板合成cDNA,所用引物见表 2。
1.3 rocE基因启动子融合lacZ基因表达载体构建 以Bt HD73基因组为模板,ProcE-F和ProcE-R为引物,PCR扩增rocE (HD73_0562)基因启动子ProcE片段(294 bp),PCR产物经纯化和双酶切(BamH Ⅰ和Hind Ⅲ),连接到穿梭载体pHT304-18Z上,该载体含有lacZ报告基因。连接产物转入E. coli TG1菌株,经鉴定获得重组质粒pHTProcE,再将重组质粒转入E. coli ET 12567菌株中去甲基化,之后通过电击分别转入出发菌株HD73、突变菌株ΔsigL和ΔrocR,转化方法见文献[22],获得菌株HD (ProcE)、ΔsigL (ProcE)和ΔrocR (ProcE)。通过序列合成的方法将ProcE片段上的RocR结合位点进行突变,上述同样方法,PCR获得片段ProcEMR,连接到pHT304-18Z载体,获得重组质粒pHTProcEMR,电击转入HD73出发菌株,获得菌株HD (ProcEMR)。
1.4 rocE基因突变菌株构建及筛选 利用同源重组的原理[23],构建rocE突变体,方法见参考文献[24],以Bt HD73基因组为模板,用引物rocE-A1/rocE-A2扩增rocE基因上游片段(rocE-A,565 bp);用引物rocE-B1/rocE-B2扩增rocE基因下游片段(rocE-B,633 bp)。以含有卡那霉素抗性基因(kan,1473 bp)的Δdxr1突变体[24]为模板,用引物E1Kan-1/E1Kan-2进行PCR扩增。应用无缝克隆试剂盒,将rocE-A、kan和rocE-B三个片段连接至温敏穿梭载体pMAD的BamH I和EcoR I酶切片段上,连接产物转入大肠杆菌TG1菌株,重组质粒pMADΔrocE经鉴定正确后再转入ET去甲基化。之后电击转入HD73菌株,获得具有红霉素和卡那霉素抗性的HD (pMADΔrocE)菌株。突变体筛选方法见参考文献[24],获得的突变菌株命名为HD(ΔrocE)。
1.5 RocR-HTH蛋白表达纯化与凝胶阻滞实验 根据RocR蛋白结构域分析结果,以HD73基因组为模板,以RocR-H1和RocR-H2为引物,PCR扩增RocR-HTH片段(225 bp),PCR产物经纯化、双酶切(BamH I和Sal I),连接到pET21b质粒上,转化E. coli TG1菌株,获得重组质粒pETRocR-HTH。鉴定正确后,转化至E. coli BL21 (DE3)菌株,获得表达菌株BL21(pETRocR-HTH)。RocR-HTH蛋白的表达与纯化见文献[25]。以Bt HD73基因组为模板,用带有羧基荧光素(FAM)标记的引物(表 2),PCR扩增ProcE片段(297 bp)。通过凝胶阻滞实验(EMSA,electrophoresis mobility shift assays),确定ProcE与RocR-HTH蛋白的结合,方法见文献[25]。
1.6 ΔrocR互补菌株构建 以Bt HD73基因组为模板,rocR-1和rocR-2为引物,PCR扩增含有自身启动子片段的rocR基因(1717 bp),应用无缝克隆试剂盒,将纯化后的PCR产物连接到E. coli–Bt穿梭表达载体pHT1618的Pst I和Sal I酶切产物上,转化至E. coli TG1菌株,重组质粒pHTCrocR鉴定正确后转入E. coli ET菌株中去甲基化,之后通过电击转入ΔrocR (ProcE)菌株中,转化方法见文献[22],获得菌株CrocR (ProcE),该菌株中含有表达RocR蛋白的质粒pHTCrocR,同时含有可测定ProcE转录活性的质粒pHTrocE。
1.7 β-半乳糖苷酶活性测定 Bt菌株在SSM培养基中30 ℃、220 r/min振荡培养,从T1开始取样,之后每小时取样1次,取到T8,每次取样2 mL,样品离心后,沉淀用于测定β-半乳糖苷酶活性,方法见参考文献[26]。Bt菌株在LB培养基中30 ℃、220 r/min振荡培养至对数生长期(OD600=2.0),取1%菌液接入M9培养基(含有终浓度为17 mmol/L的葡萄糖和1.15 mmol/L精氨酸),继续振荡培养(30 ℃、220 r/min),2 h后开始取样(记为A1),之后每小时取样1次,共取7个点(A2–A8),每次取样10 mL,离心收集菌体,沉淀用于β-半乳糖苷酶活性测定。每组数据3次独立重复取平均值并计算标准差。
2 结果和分析 2.1 rocE基因簇转录单元确定 对Bt HD73基因组序列进行分析,rocE基因编号为HD73_0562,编码精氨酸通透酶。与Bs 168菌株中的同源基因BSU40330相似性为60%。对rocE基因上下游序列重测序发现,其上游HD73_0560和HD73_0561实际为一个orf,编码未知功能蛋白;rocE下游基因HD73_0563编码乙酰鸟氨酸脱乙酰酶(acetylornitine deacetylase);HD73_0560基因上游是一个反向基因:rocR (HD73_0559)编码依赖于Sigma54的转录调控因子(Sigma54-dependent transcriptional activator)(图 1)。根据HD73菌株基因组序列信息,在rocE基因及其上下游基因内部以及基因之间设计了多对引物,将SSM培养基中T5时期提取并纯化的总RNA反转录成cDNA,再分别扩增1–5个片段(片段1、2、3是基因内部片段;片段4和5是基因间片段)。结果表明(图 1),除了片段5得不到扩增产物外,其余均能得到扩增产物,说明rocE与下游的HD73_0563不共转录,而rocE基因与其上游HD73_0560组成一个转录单元,该转录单元的启动子位于HD73_0560基因的上游
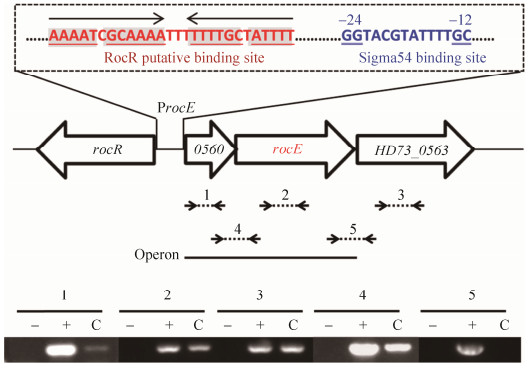 |
| 图 1 Bt HD73中rocE基因簇基因组织及序列分析 Figure 1 Organization and sequence of the rocE cluster in Bt HD73. C: indicated the product amplified with cDNA as template; +: the positive controls (PCR with genomic DNA); –: the negative controls (RT-PCR with RNA). |
| 图选项 |
对rocE基因启动子(ProcE)序列进行分析,通过DBTBS数据库(http://dbtbs.hgc.jp/)中检索ProcE序列上转录调控因子结合位点,发现了Sigma54的结合位点(图 1),具有典型的-12/-24保守序列(BYGGCMYRNNNYYGCW)特征[27],在该序列上游有一段近似回文序列,可能是RocR的结合位点。
2.2 rocE基因转录调控分析 为了研究rocE基因的转录活性,构建了rocE的启动子ProcE融合lacZ基因的表达载体pHTProcE,分别转入Bt HD73菌株、ΔsigL突变体和ΔrocR突变体,获得菌株HD (ProcE)、ΔsigL (ProcE)和ΔrocR (ProcE),通过β-半乳糖苷酶活性测定分析ProcE在不同菌株中的转录情况。结果表明(图 2-A),SSM培养基中,在T1–T8时期(T0为对数生长期结束的时间,Tn为T0之后的n小时),ProcE在HD73菌株中的转录活性基本平稳,在T2时期最高,之后缓慢下降;与出发菌株HD73相比,ProcE在ΔsigL和ΔrocR菌株中的转录活性显著降低,而在ΔrocR互补菌株[CrocR (ProcE)]中,ProcE的活性明显升高。在营养成分贫瘠的M9培养基中对ProcE的诱导转录活性进行分析,β-半乳糖苷酶活性测定结果表明(图 2-B),与无精氨酸的培养基相比,在含有1.15 mmol/L精氨酸的培养基中,HD73出发菌株中的ProcE转录活性明显升高,说明ProcE的转录受精氨酸的诱导;而ProcE在ΔrocR菌株中的诱导转录活性几乎丧失,在ΔrocR互补菌株中恢复了比野生型更高的活性。这些结果说明rocE的转录受精氨酸的诱导,其转录活性受Sigma 54因子控制,并受到RocR的正调控。
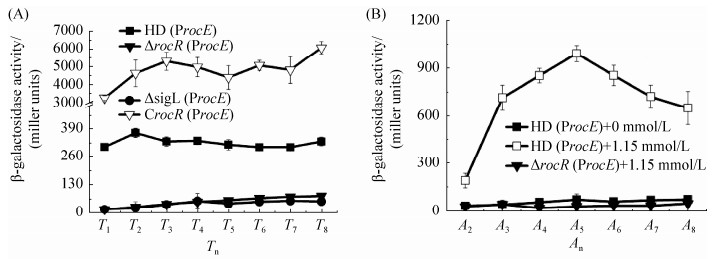 |
| 图 2 ProcE的转录活性 Figure 2 Transcriptional activity of ProcE. The standard deviation reflects the degree of dispersion of the value relative to the mean. A: SSM medium; B: M9 medium. |
| 图选项 |
2.3 RocR对rocE的转录调控作用 通过对RocR氨基酸保守结构域的分析发现,它是一个依赖于Sigma 54的转录调控因子,具有3个典型的保守结构域(图 3-A):与信号识别相关的PAS结构域、与Sigma 54互作的AAA+结构域、与DNA结合的HTH结构域。为了进一步明确RocR对rocE的转录调控作用,表达纯化了RocR-HTH蛋白,通过EMSA实验确定了ProcE与RocR-HTH蛋白的结合。结果表明(图 3-B),凝胶底部为15.7 ng带标记的自由DNA条带,上层为DNA与蛋白结合的条带,随着RocR-HTH蛋白浓度由14 ng增加到112 ng (图 3-B,lane 2–5),底部未与蛋白结合的DNA条带浓度越来越低,上层与蛋白结合的条带浓度逐渐升高,说明ProcE与RocR-HTH蛋白有结合作用;而加入200倍浓度未标记的DNA可以与标记的DNA产生竞争作用(图 3-B,lane 6)。以上结果说明ProcE与RocR-HTH具有特异性的结合作用,表明ProcE的转录受RocR的直接调控。根据rocE启动子序列特征,将预测的RocR结合位点的T碱基替换成C碱基(图 3-C:UAS序列,UAS:upstream activating sequence)。PCR扩增突变片段ProcEMR,将其连接至含有lacZ报告基因的pHT304-18Z载体上,并转入Bt HD73,获得菌株HD (ProcEMR)(图 3-C)。β-半乳糖苷酶活性测定结果表明(图 3-C),SSM培养基中,在T1-T8时期,ProcEMR的转录活性明显低于ProcE的转录活性,说明rocE启动子区域上突变的序列可能是RocR的结合序列。
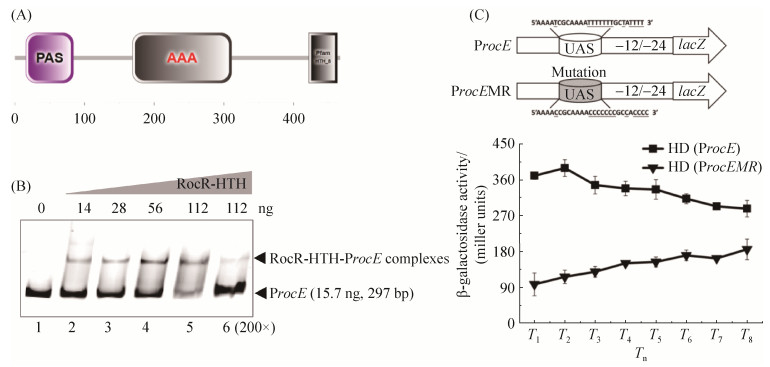 |
| 图 3 RocR对rocE的转录调控 Figure 3 Transcription of rocE is regulated by RocR. A: conserved domain of RocR; B: the binding of RocR-HTH and ProcE; C: transcriptional activity of ProcEMR. The standard deviation reflects the degree of dispersion of the value relative to the mean. |
| 图选项 |
2.4 rocE突变体的表型分析 为了明确rocE基因在Bt中的功能,构建了rocE的突变体,并进行表型分析,比较了出发菌株和突变菌株的生长曲线、芽胞形成率和Cry1Ac蛋白产量,方法参考文献[24]。生长曲线结果表明(图 4-A):无论是在营养丰富的LB培养基,还是在较贫瘠的芽胞形成培养基SSM,rocE基因的缺失对菌体生长均无明显影响。芽胞形成率实验表明(图 4-B),出发菌株HD73的芽胞形成率为85.7(±2.1)%;而ΔrocE突变体的芽胞形成率为65.5(±4.7)%,显著性分析结果显示P-value<0.05,差异显著,说明rocE基因的缺失影响了Bt菌株的芽胞形成率。HD73菌株含有唯一的杀虫晶体蛋白Cry1Ac,为了明确rocE基因的缺失对Cry1Ac蛋白产量的影响,将HD73菌株和ΔrocE突变体在SSM培养基中培养至T24,收集菌体,冻干称重,在总蛋白量相同的条件下,与HD73野生型相比,ΔrocE突变体的Cry1Ac蛋白产量无明显变化(图 4-C),说明rocE基因的缺失对Cry蛋白产量无显著影响。
 |
| 图 4 rocE突变体的表型分析 Figure 4 Phenotype of rocE mutant. A: Growth curve; B: sporulation efficiency; C: Cry1Ac protein production. The standard deviation reflects the degree of dispersion of the value relative to the mean. |
| 图选项 |
3 讨论 本研究发现,在Bt HD73菌株中,编码精氨酸通透酶的rocE基因的转录受精氨酸的诱导、受Sigma 54的控制,并受RocR的正调控。通过EMSA实验,进一步明确了纯化的RocR-HTH结构域可以与rocE的启动子结合,这些结果为细菌精氨酸代谢途径的转录调控机制提供了新的证据。RocR是依赖于Sigma 54的转录激活蛋白,具有3个典型的结构域(图 3-A),结合本研究得到的结果,推测RocR的N端PAS结构域可能与识别精氨酸信号相关;C端的HTH结构域,可能识别rocE启动子上的回文序列并与之相结合。
在Bs中,编码精氨酸分解代谢途径相关酶的基因成簇存在,由roc基因簇中的7个基因组成,形成3个转录单元:rocABC-rocG-rocDEF。这些基因的转录都依赖于Sigma 54因子,并且受到RocR的正调控[3, 6-9]。在Bt HD73基因组中,也存在roc基因簇的同源基因,参与精氨酸分解代谢途径,但这些基因分散排布在Bt HD73基因组上(图 5),只有rocE基因与rocR基因相邻,本研究证实rocE基因的转录受RocR正调控,而其它roc基因的转录是否受到RocR调控,还有待进一步研究。本研究通过rocE突变体的表型分析发现,rocE基因的缺失不影响菌株的生长和Cry1Ac蛋白的产量,但使芽胞形成率略有下降,说明rocE基因参与的精氨酸代谢途径与芽胞形成有关,但其它roc基因的缺失是否影响菌株的表型,还有待进一步研究。
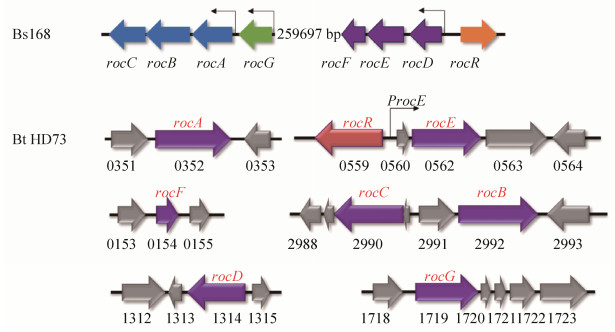 |
| 图 5 Bt和Bs基因组中roc基因的比较 Figure 5 Comparison of roc genes in Bt and Bs genome. |
| 图选项 |
References
| [1] | Belitsky BR, Sonenshein AL. An enhancer element located downstream of the major glutamate dehydrogenase gene of Bacillus subtilis. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(18): 10290-10295. |
| [2] | Xiong LF, Teng JLL, Botelho MG, Lo RC, Lau SKP, Woo PCY. Arginine metabolism in bacterial pathogenesis and cancer therapy. International Journal of Molecular Sciences, 2016, 17(3): 363. DOI:10.3390/ijms1703036310290|1999||| |
| [3] | Gardan R, Rapoport G, Débarbouillé M. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. Journal of Molecular Biology, 1995, 249(5): 843-856. DOI:10.1006/jmbi.1995.0342 |
| [4] | Lu CD. Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains. Applied Microbiology and Biotechnology, 2006, 70(3): 261-272. |
| [5] | Fulde M, Willenborg J, de Greeff A, Benga L, Smith HE, Valentin-Weigand P, Goethe R. ArgR is an essential local transcriptional regulator of the arcABC operon in Streptococcus suis and is crucial for biological fitness in an acidic environment. Microbiology, 2011, 157(2): 572-582. DOI:10.1099/mic.0.043067-0 |
| [6] | Zaprasis A, Hoffmann T, Wünsche G, Flórez LA, Stülke J, Bremer E. Mutational activation of the RocR activator and of a cryptic rocDEF promoter bypass loss of the initial steps of proline biosynthesis in Bacillus subtilis. Environmental Microbiology, 2014, 16(3): 701-717. DOI:10.1111/1462-2920.12193 |
| [7] | Ali NO, Jeusset J, Larquet E, Le Cam E, Belitsky B, Sonenshein AL, Msadek T, Débarbouillé M. Specificity of the interaction of RocR with the rocG-rocA intergenic region in Bacillus subtilis. Microbiology, 2003, 149(3): 739-750. DOI:10.1099/mic.0.26013-0 |
| [8] | Gardan R, Rapoport G, Débarbouillé M. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Molecular Microbiology, 1997, 24(4): 825-837. DOI:10.1046/j.1365-2958.1997.3881754.x |
| [9] | Calogero S, Gardan R, Glaser P, Schweizer J, Rapoport G, Debarbouille M. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. Journal of Bacteriology, 1994, 176(5): 1234-1241. DOI:10.1128/JB.176.5.1234-1241.1994 |
| [10] | Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiology and Molecular Biology Reviews, 1998, 62(3): 775-806. DOI:10.1128/MMBR.62.3.775-806.1998 |
| [11] | Jouzani GS, Valijanian E, Sharafi R. Bacillus thuringiensis: a successful insecticide with new environmental features and tidings. Applied Microbiology and Biotechnology, 2017, 101(7): 2691-2711. DOI:10.1007/s00253-017-8175-y |
| [12] | Heckel DG. How do toxins from Bacillus thuringiensis kill insects? An evolutionary perspective. Archives of Insect Biochemistry and Physiology, 2020, 104(2): e21673. |
| [13] | Raymond B, Johnston PR, Nielsen-LeRoux C, Lereclus D, Crickmore N. Bacillus thuringiensis: an impotent pathogen?. Trends in Microbiology, 2010, 18(5): 189-194. DOI:10.1016/j.tim.2010.02.006 |
| [14] | Danson AE, Jovanovic M, Buck M, Zhang XD. Mechanisms of σ54-dependent transcription initiation and regulation. Journal of Molecular Biology, 2019, 431(20): 3960-3974. DOI:10.1016/j.jmb.2019.04.022 |
| [15] | Peng Q, Wang GN, Liu GM, Zhang J, Song FP. Identification of metabolism pathways directly regulated by sigma54 factor in Bacillus thuringiensis. Frontiers in Microbiology, 2015, 6: 407. |
| [16] | Zhu L, Peng Q, Song FP, Jiang YN, Sun CP, Zhang J, Huang DF. Structure and regulation of the gab gene cluster, involved in the γ-aminobutyric acid shunt, are controlled by a σ54 factor in Bacillus thuringiensis. Journal of Bacteriology, 2010, 192(1): 346-355. DOI:10.1128/JB.01038-09 |
| [17] | Peng Q, Yang M, Wang W, Han LL, Wang GN, Wang PY, Zhang J, Song FP. Activation of gab cluster transcription in Bacillus thuringiensis by γ-aminobutyric acid or succinic semialdehyde is mediated by the sigma 54-dependent transcriptional activator GabR. BMC Microbiology, 2014, 14(1): 306. DOI:10.1186/s12866-014-0306-3 |
| [18] | Zhang Z, Yang M, Peng Q, Wang GN, Zheng QY, Zhang J, Song FP. Transcription of the Lysine-2, 3-aminomutase gene in the kam locus of Bacillus thuringiensis subsp. kurstaki HD73 is controlled by both σ54 and σK factors. Journal of Bacteriology, 2014, 196(16): 2934-2943. DOI:10.1128/JB.01675-14 |
| [19] | Peng Q, Liu CX, Wang B, Yang M, Wu JB, Zhang J, Song FP. Sox transcription in sarcosine utilization is controlled by sigma54 and SoxR in Bacillus thuringiensis HD73. Scientific Reports, 2016, 6(1): 29141. DOI:10.1038/srep29141 |
| [20] | Peng Q, Zhao X, Wen JL, Huang MZ, Zhang J, Song FP. Transcription in the acetoin catabolic pathway is regulated by AcoR and CcpA in Bacillus thuringiensis. Microbiological Research, 2020, 235: 126438. DOI:10.1016/j.micres.2020.126438 |
| [21] | Liu GM, Song L, Shu CL, Wang PS, Deng C, Peng Q, Lereclus D, Wang XM, Huang DF, Zhang J, Song FP. Complete genome sequence of Bacillus thuringiensis subsp. kurstaki strain HD73. Genome Announcements, 2013, 1(2): e00080-13. |
| [22] | Lereclus D, Arantès O, Chaufaux J, Lecadet MM. Transformation and expression of a cloned δ-endotoxin gene in Bacillus thuringiensis. FEMS Microbiology Letters, 1989, 60(2): 211-217. |
| [23] | Arnaud M, Chastanet A, Débarbouillé M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Applied and Environmental Microbiology, 2004, 70(11): 6887-6891. DOI:10.1128/AEM.70.11.6887-6891.2004 |
| [24] | Li N, Wen JL, Song FP, Zhang J, Duan JY, Peng Q. Transcription of dxr1 is controlled by SigH in Bacillus thuringiensis. Acta Microbiologica Sinica, 2020, 60(1): 200-210. (in Chinese) 李娜, 温继龙, 宋福平, 张杰, 段江燕, 彭琦. 苏云金芽胞杆菌dxr1基因的转录受SigH控制. 微生物学报, 2020, 60(1): 200-210. |
| [25] | Cheng HJ, Peng Q, Zhang J, Song FP. Identification of genes regulated by Sigma54 and CcpA in Bacillus thuringiensis. Acta Microbiologica Sinica, 2018, 58(3): 380-390. (in Chinese) 程海舰, 彭琦, 张杰, 宋福平. 苏云金芽胞杆菌Sigma54和CcpA共同调控的基因鉴定. 微生物学报, 2018, 58(3): 380-390. |
| [26] | Lv J, Zhang X, Gao TT, Cui TT, Peng Q, Zhang J, Song FP. Effect of the spoIIID mutation on mother cell lysis in Bacillus thuringiensis. Applied Microbiology and Biotechnology, 2019, 103(10): 4103-4112. DOI:10.1007/s00253-019-09722-1 |
| [27] | Francke C, Groot Kormelink T, Hagemeijer Y, Overmars L, Sluijter V, Moezelaar R, Siezen RJ. Comparative analyses imply that the enigmatic Sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics, 2011, 12(1): 385. DOI:10.1186/1471-2164-12-385 |
