Fei Wang1,2, Xiaodong Yang1,2, Yan Li1,3,4


1. Key Laboratory of Oasis Ecology, Education Ministry, Urumqi 830046, Xinjiang Uygur Autonomous Region, China;
2. College of Resource and Environment Sciences, Xinjiang University, Urumqi 830046, Xinjiang Uygur Autonomous Region, China;
3. Institute of Arid Ecology and Environment, Xinjiang University, Urumqi 830046, Xinjiang Uygur Autonomous Region, China;
4. Ecology Post-doctoral Research Station, Xinjiang University, Urumqi 830046, Xinjiang Uygur Autonomous Region, China
Received: 1 May 2018; Revised: 13 July 2018; Published online: 24 July 2018
Foundation item: Supported by the Chinese Postdoctoral Science Foundation (2016M592866) and by the National Natural Science Foundation of China (U1603241, 41661046)
Corresponding author: Yan Li, Tel:+86-991-8582056; E-mail:liyan1006@ms.xjb.ac.cn.
Abstract: [Objective] Lycium ruthenicum is a halophyte and used to improve saline lands in northwest China. However, little is known about the bacterial community structural dynamics with growth stage.[Methods] We investigated the dynamics of rhizosphere bacterial community structure in three growth stages using Illumina MiSeq high-throughput sequencing.[Results] We obtained a total of 317467 16S rDNA reads, corresponding to 7028 bacterial/archaeal operational taxonomic units. The alpha diversity was higher in the rhizosphere than in bulk soil. The diversity and richness of rhizosphere bacteria were much lower in senescence stage than that in vegetative and flowering/fruiting stages. The relative abundances of Proteobacteria and Acidobacteria gradually decreased, whereas the abundance of Cyanobacteria increased along with growth cycle. The phylum Firmicutes abundance was significantly higher in senescence stage than in other stages. The abundant genera composition also changed with growth stage. Seventeen genera (i.e. Corynebacterium, Acidovorax, Elizabethkingia, Albirhodobacter and Pseudomonas) were abundant at vegetative stage; Sixteen bacterial genera were enriched in flowering/fruiting stage, including Rhodoligotrophos, Geminicoccus, Gracilimonas and Thioprofundum. Four bacterial genera, Exiguobacterium, Citrobacter, Acinetobacter and Pseudomonas, were abundant in senescence stage. In vegetative and flowering/fruiting stages, the rhizosphere bacterial community was of high similarity, and the similarities between rhizosphere communities were higher than that between rhizosphere and bulk soil communities. However, in senescence stage, the rhizosphere bacterial community composition was more different from the communities in previous stages, but turned to be more similar with that of bulk soil.[Conclusion] The rhizosphere bacterial community diversity and composition were changing with growth stage, and great difference was found between senescence stage and previous two stages. Plant growth stage had important effects on structuring the rhizosphere bacterial community of L. ruthenicum.
Keywords: halophyterhizosphere soilbacterial communitygrowth stage dynamics
不同生长季节黑果枸杞的根际细菌群落结构
王飞1,2, 杨晓东1,2, 李岩1,3,4


1. 新疆大学绿洲生态教育部省部共建重点实验室, 新疆 乌鲁木齐 830046;
2. 新疆大学资源与环境科学学院, 新疆 乌鲁木齐 830046;
3. 新疆大学干旱生态环境研究所, 新疆 乌鲁木齐 830046;
4. 新疆大学生态学博士后流动站, 新疆 乌鲁木齐 830046
收稿日期:2018-05-01;修改日期:2018-07-13;网络出版日期:2018-07-24
基金项目:中国博士后项目(2016M592866);国家自然科学基金(U1603241,41661046)
通讯作者:李岩, Tel:+86-991-8582056; E-mail: liyan1006@ms.xjb.ac.cn.
摘要:[目的] 黑果枸杞是一种耐盐植物,是我国西北干旱区盐渍土改良的优良植物物种,其根际土壤细菌群落结构在不同生长时期的变化特征尚不清楚。[方法] 本研究采用Illumina MiSeq高通量测序研究了黑果枸杞3个生长阶段的根际土壤细菌群落结构的动态变化。[结果] 所有样品中共获得317467条序列,对应于7028个细菌/古细菌OTUs。根际土壤细菌群落的α多样性显著高于非根际土壤。衰老期根际细菌的多样性和丰富度明显低于营养生长期和花/果期。变形菌门和酸杆菌门的相对丰度随生长时期的演变而逐渐降低,而蓝细菌门则相反。厚壁菌门的丰度在衰老期明显高于营养生长期和花/果期。优势属的组成也随生长期的演变而改变,营养生长期、花/果期、衰老期的优势属数量分别为17、16、4,且组成也具有差异。相似性分析表明营养生长期和花/果期的根际细菌群落具有很高的相似性,衰老期根际细菌群落组成与生长期和花/果期具有很高差异,然而与非根际土壤的群落结构具有较高的相似性。[结论] 根际土壤细菌群落多样性和组成随生长期的改变而表现出明显的动态变异性,表明黑果枸杞生长时期对根际土壤细菌群落结构具有重要的影响。
关键词:盐生植物根际土壤细菌群落生长阶段动态
Salinization is an important land degradation problem, and high salinity limits plant growth. Due to natural processes such as mineral weathering, dust and precipitation or artificial processes such as irrigation[1], approximately 50% of the world's arable land is estimated to be affected by salinization by 2050[2-3]. In dry regions, salts may accumulate, leading to saline soils and increasing the difficulty for plants to absorb soil moisture. Halophytes are salt-tolerant plants that can grow in saline soil, such as that found in saline semi-deserts, mangrove swamps, marshes, sloughs and seashores. Dominant halophytes play significant role in carbon sequestration, nutrient mineralization, nutrient cycling and improving micro-environment[4].
The rhizosphere represents one of the most diverse habitats on the planet[5]. Rhizosphere microbiomes receive carbon metabolites from the plant through root exudates[6]. In turn, they convert nutrients into more usable forms for assimilation by plants[7] or secreted growth regulators, such as growth-promoting hormones and volatile organic compounds to promote plant growth[8-9]. Some beneficial microbes enhance pathogen resistance, water retention, and the drought and salt resistance ability of plants[10-11].
Rhizosphere microbial community structures are influenced by various factors, among which plant species, soil properties, and growth stage are the most important[12-13]. Plenty of researches have demonstrate that the rhizosphere bacterial community composition is plant-specific, and different plant species tend to shape distinct rhizobacterial community[4, 14-15]. The rhizosphere bacteria associated with halophytes are mostly salt-tolerant or halophilic, which is distinct from non-halophytic plants[16-18]. A large amount of halophilic bacteria have been identified or isolated from halophytic plants, such as species or strains belonging to genera Halomonas, Halobacillus, Brachybacterium, Brevibacterium, Bacillus, Cronobacter, Zhihengliuella, Stenotrophomonas, Alkalimonas, Staphylococcus, Methylibium, Marinococcus, Oceanobacillus, Nesterenkonia and Virgibacillus[16-21].
Considerable studies have evidenced that developmental stage of the plant plays a critical role in deciding the rhizobacterial community structure[22-26]. In different plant growth stages, root physiology, and the quality and quantity of root exudates vary, consequently influencing the rhizosphere soil microenvironment and exerting selective pressure on root-associated microorganisms[27-28].
Lycium ruthenicum is a halophyte that mostly occurs in saline deserts and sands across Europe, Central Asia, the southern part of Russia, and Northwest China (Flora of China). It is capable of migrating and accumulating salt from outside the crown to under the crown or in the rhizosphere soil, which consequently reduces the salt concentration of surrounding soils[29]. They are therefore used as pioneer plants to improve barren hills and saline lands. We have investigated the diversity and structure of rhizobacterial community to gain some insights of the composition of rhizosphere bacterial community and the rhizosphere effect on saline habitat[30]. However, the dynamics of rhizosphere bacterial community with growth stage is not clear. In this study, the rhizosphere bacterial community diversity and structure of L.ruthenicum was investigated over three growth stages (vegetative, flowering/fruiting and senescence) using the Illumina MiSeq sequencing platform. We aim to explore the effect of growth stage on the bacterial community.
1 Materials and methods 1.1 Study areas and sample collection The study area was located at the Ebinur Lake Wetland Nature Reserve at the Western margin of the Gurbantunggut Desert, Xinjiang Uygur Autonomous Region, China. The Reserve has a typical continental climate and is dry and windy, with an annual average precipitation of 105 mm and an evaporation of 1315 mm. The soils are mainly gray desert, gray-brown desert, and sandy soils. The soil in the Reserve is highly salinized and alkalized, and the average electrical conductivity (EC) and pH value of the 0–10 cm soil layer are 5.41 mS/cm and 8.77, respectively, with an average water content of 7.19%[31].
The soil was sampled during three growth stages (vegetative, flowering/fruiting and senescence) of L. ruthenicum. The rhizosphere soil was collected and processed following the protocol of Edwards et al.[32]..Three to four healthy individuals were selected and sampled in each of these stages from the same population. In total, 22 samples, including 11 rhizosphere and 11 bulk soils, were collected. The bulk soils were collected from the 0–40 cm soil layer at least 20–50 cm away from the plant. Roots were discarded, and the remaining soil was processed in the same manner as the rhizosphere soils.
1.2 DNA extraction, amplification, and sequencing Total genomic DNA of each sample was extracted using the E.Z.N.ATM Mag-Bind Soil DNA Kit (OMEGA). Extracted DNA was detected by 1.0% agarose gel and quantified using a Nanodrop 2000 spectrophotometer (Nanodrop Technologies, Wilmington DE). The PCRs were performed with two rounds of amplification. The first round amplified with barcode-fused primers. The 16S rDNA V3–V4 region was amplified with the forward primer 341F [ccctacacgacgctcttccgatctg (barcode) cctacgggnggcwgcag] and reverse primer 805R (gactggagttccttggcacccgagaattccagactachvgggtatctaatcc); Amplification reactions were performed in 30 μL volumes containing 15 μL of 2×Taq Master Mix (Thermo), 1 μL of each primer (10 μmol/L), and 20 ng of template; the procedure began at 94 ℃ for 3 min; followed by 5 cycles of 94 ℃ for 30 s, 45 ℃ for 20 s, and 65 ℃ for 30 s; then 20 cycles of 94 ℃ for 20 s, 55 ℃ for 20 s, and 72 ℃ for 30 s; and a final extension for 10 min at 72 ℃. The second round of amplification was conducted using Illumina bridge PCR-compatible primers, and PCRs were performed using 5 cycles of 95 ℃ for 30 s, 95 ℃ for 15 s, 55 ℃ for 15 s, 72 ℃ for 30 s, and a final extension for 5 min with the same reaction mix as the first round. PCR products were visualized using electrophoresis on 1.5% agarose gels and purified using VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China). The PCR products of all replicates from each stage and each soil type were pooled together as one sample for sequencing, thus each of the six sequencing sample (one rhizosphere and one bulk sample at each of the three growth stage) contained all replicates. Finally, 10 ng of DNA from each sequencing sample was sequenced with the Illumina MiSeq platform by the Sangon Technology Co., Ltd. (Shanghai, China).
1.3 Sequence preprocessing and operational taxonomic unit (OTU) assignment Raw sequence data were first quality controlled as in previous study[33]. Bases with quality scores < 20 were removed from raw reads, paired-end reads were merged into sequences based on overlapping regions with PEAR[34], and the maximum mismatch rate of overlapping areas was constrained to 0.1. Sequences were removed when they contained the ambiguous base Ns or were shorter than 200 bp. Chimeric sequences were identified by UCHIME and discarded. Sequences were assigned to OTUs at a 97% similarity level. Taxonomies of representative OTUs were annotated according to their RDP classifier and BLAST against the Silva and NCBI databases[35]. OTUs with an RDP classification threshold below 0.8 or with identity and coverage lower than 90% were denoted unclassified. All sequencing data in this study were deposited in the NCBI with accession number SAMN06650232-SAMN06650237.
1.4 Statistical analysis Alpha diversity was estimated using the vegan package in R software. Chao1 was used to estimate richness, and Shannon and Simpson indices were used to estimate diversity. Weighted UniFrac distances between samples, based on OTU abundance, were analyzed using the vegan package, and based on which hierarchical clustering maps were used to visualize the relationships and similarities between samples. Pearson's correlation analysis between soil properties and OTUs was performed using the SPSS program.
2 Results 2.1 Soil properties The total organic carbon (TOC), soil organic matter (SOM) and total organic nitrogen (TON) content of rhizosphere soils were higher than those of bulk soils, whereas their EC and pH values were lower than those of bulk soils. The TOC, SOM and TON were significantly different among the three growth stages in both the rhizosphere and bulk soils. TOC, SOM and TON contents decreased from vegetative to flowering/fruiting, and increased in senescence stage (Table 1).
Table 1. Chemical characteristics of rhizosphere and bulk soils associated with L. ruthenicum in the three growth stages
| Soil type | Growth stage | TOC/(g/kg) | SOM/(g/kg) | TON/(g/kg) | pH | EC/(mS/cm) |
| Bulk | Vegetative | 6.46±1.05 | 11.13±1.82 | 0.22±0.17 | 8.78±0.15 | 7.13±1.06 |
| Flowering/fruiting | 5.99±1.28 | 10.32±2.20 | 0.14±0.05 | 8.93±0.61 | 6.37±0.72 | |
| Senescence | 6.71±0.61 | 11.55±1.05 | 0.52±0.23 | 8.90±0.09 | 6.26±1.89 | |
| Rhizosphere | Vegetative | 12.71 | 21.92 | 0.85 | 8.23 | 5.66 |
| Flowering/fruiting | 8.26 | 14.24 | 0.65 | 8.25 | 1.94 | |
| Senescence | 10.25 | 17.68 | 0.76 | 8.09 | 1.96 | |
| Mean and standard deviation values were showed, number of samples (n) of bulk soil was 3–4, while for rhizosphere soil n=1 because rhizosphere soil quantity was too low to test for each individual, thus the rhizosphere soil of all individuals at each stage were mixed together and tested. | ||||||
表选项
2.2 Diversity of microbial community In total, 317467 reads were obtained from samples. After quality control and OTU assignment, 7028 bacteria/archaeal OTUs were obtained. Rarefaction curves (97% identity) in all samples almost approached the plateau (Figure 1), indicating a reasonable representation of bacterial community diversity. The alpha diversity and richness was higher in rhizosphere soils than that in bulk soils. The rhizosphere bacterial/archaeal community diversity peaked in flowering/fruiting stage, and decreased dramatically in senescence stage. For bulk soil, the community diversity in vegetative stage was similar to that in flowering/fruiting stage, and both higher than that in senescence stage (Table 2).
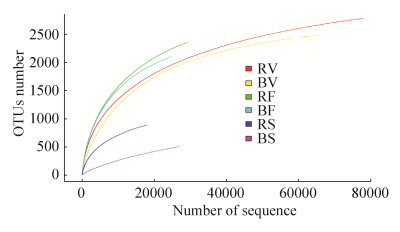 |
| Figure 1 Rarefaction curves for bacterial OTUs clustering at 97 % sequence similarity. |
| 图选项 |
Table 2. Alpha diversity indices and numbers of OTUs in rhizosphere and bulk soil microbial communities in different growth stages.
| Soil type | Sample | OTU number | Shannon index | Simpson index | Chao1 index |
| Bulk | BV | 2662 | 5.03 | 0.03 | 2729.24 |
| BF | 2100 | 5.88 | 0.02 | 2507.88 | |
| BS | 505 | 1.67 | 0.30 | 1006.77 | |
| Rhizosphere | RV | 2778 | 5.98 | 0.01 | 3241.60 |
| RF | 2354 | 6.09 | 0.01 | 2939.34 | |
| RS | 888 | 3.83 | 0.11 | 1134.39 | |
| BV: Bulk soils in vegetative stage; BF: Bulk soils in flowering/fruiting stage; BS: Bulk soils in senescence stage. RV: Rhizosphere soils in vegetative stage; RF: Rhizosphere soils in flowering/fruiting stage; RS: Rhizosphere soils in senescence stage. | |||||
表选项
2.3 Bacterial communities composition Three Archaea phyla (Euryarchaeota, Thaumarchaeota and Crenarchaeota) were detected but accounted for only a very small proportion of the entire microbial community. These three phyla accounted for 0.30% and 0.63% of the total sequences in rhizosphere and bulk soils, respectively (Table 3).
Table 3. Relative abundances of microbial phyla in rhizosphere and bulk soils at three growth stages.
| Phylum | Relative abundance/% | |||||
| RV | RF | RS | BV | BF | BS | |
| Archaea | ||||||
| Thaumarchaeota | 0.01 | 0.01 | 0.02 | 0.09 | 0 | 0.15 |
| Euryarchaeota | 0.07 | 0.01 | 0.18 | 0.21 | 0.06 | 0 |
| Crenarchaeota | 0 | 0 | 0.01 | 0.02 | 0 | 0.10 |
| Bacteria | ||||||
| Proteobacteria | 55.08 | 50.71 | 37.06 | 47.59 | 29.69 | 52.63 |
| Firmicutes | 3.82 | 3.34 | 33.12 | 17.59 | 4.08 | 45.80 |
| Actinobacteria | 8.59 | 17.79 | 5.19 | 15.52 | 29.92 | 0.39 |
| Bacteroidetes | 11.89 | 5.72 | 16.01 | 12.33 | 10.03 | 0.39 |
| Acidobacteria | 7.43 | 5.08 | 0.78 | 1.40 | 1.63 | 0.14 |
| Planctomycetes | 2.00 | 4.70 | 1.04 | 0.20 | 7.83 | 0.05 |
| Chloroflexi | 1.74 | 1.72 | 0.26 | 0.63 | 4.54 | 0.06 |
| Cyanobacteria | 1.30 | 2.73 | 4.46 | 0.16 | 0.02 | 0.03 |
| Deinococcus- Thermus | 0.03 | 0.05 | 0.06 | 2.11 | 0.49 | 0.01 |
| Candidatus Saccharibacteria | 1.80 | 1.77 | 0.30 | 0.13 | 0.38 | 0.03 |
| Verrucomicrobia | 0.69 | 0.62 | 0.27 | 0.78 | 2.05 | 0.04 |
| Parcubacteria | 0.40 | 0.41 | 0.12 | 0.06 | 1.14 | 0.02 |
| Others* | 0.97 | 1.44 | 0.13 | 0. 90 | 0.92 | 0.04 |
| *: phyla with relative abundances less than 0.1% were merged. BV: Bulk soils in vegetative stage; BF: Bulk soils in flowering/ fruiting stage; BS: Bulk soils in senescence stage. RV: Rhizosphere soils in vegetative stage; RF: Rhizosphere soils in flowering/ fruiting stage; RS: Rhizosphere soils in senescence stage. | ||||||
表选项
Forty-four bacterial phyla were detected in all samples. Proteobacteria was the most abundant phylum in rhizosphere soils (Table 3). Alphaproteobacteria and Gammaproteobacteria were the two most abundant classes. The richness of Alpha- and Deltaproteobacteria decreased in senescence stage to a value lower than those of the other two stages, while Gammaproteobacteria was enriched and became the dominant class in senescence stage. Betaproteobacteria was almost disappeared from rhizosphere communities (Figure 2). Bacteroidetes, Actinobacteria, Firmicutes and Planctomycetes were enriched in rhizosphere communities, but their relative abundances varied during the three growth stages. At the genus level, 21 genera were abundant in vegetative stage, they accounted for 48.08% of all OTUs. Gp10, Thioprofundum, Deferrisoma, Haliea and Halomonas were the most abundant genera; 15 genera were abundant in flowering/fruiting, accounting for 37.07% of all OTUs, and Geminicoccus, Pelagibius, Gp10, Thioprofundum and Deferrisoma were the five most abundant genera. In senescence stage, 9 abundant genera were found, accounting for 72.15% of all OTUs, with Planococcus, Halomonas, Pontibacter, Pseudomonas, Salinimicrobium, and Streptophyta being the most abundant (Figure 3).
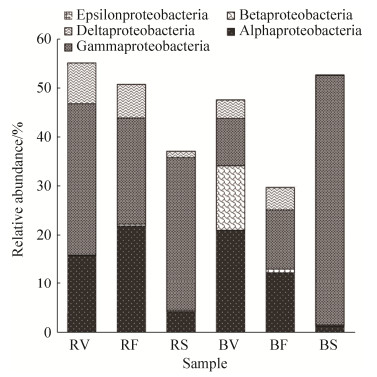 |
| Figure 2 Relative abundance of each class of the phylum Proteobacteria in the soil samples. |
| 图选项 |
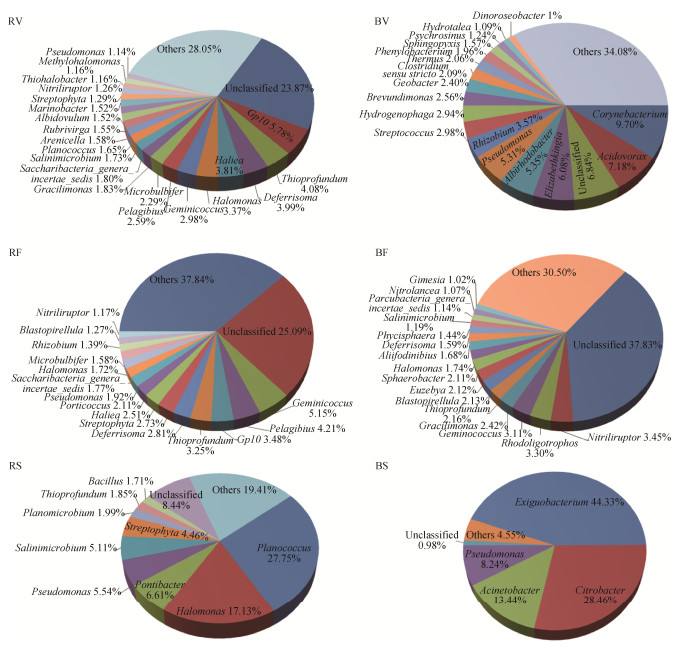 |
| Figure 3 Pie chart of bacterial community composition at genus level in the rhizosphere and bulk soils during the three stages. Only genera with relative abundances ≥1% are presented, and those < 1% are merged into "others". |
| 图选项 |
For bulk soils, Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, Planctomycetes and Cyanobacteria were the most abundant phyla during the growth cycle. Acidobacteria and Chloroflexi were abundant phyla in vegetative and flowering/fruiting stage, but decreased in senescence stage. At genus level, seven genera were abundant in vegetative, for instance, Corynebacterium, Acidovorax, Elizabethkingia, Albirhodobacter and Pseudomonas. Sixteen genera were abundant in flowering/fruiting stage, and Nitriliruptor, Rhodoligotrophos, Geminicoccus, Gracilimonas, Thioprofundum were the most abundant. In senescence stage, four genera, Exiguobacterium, Citrobacter, Acinetobacter and Pseudomonas were abundant.
2.4 Changes of bacterial communities along with growth stages In the rhizosphere communities, relative abundance of phyla Proteobacteria and Acidobacteria gradually decreased, while abundance of Cyanobacteria increased from vegetative to senescence stage. The abundance of Firmicutes was significantly higher in senescence than that in the other stages. The relative abundance of Actinobacteria, Planctomycetes and Gemmatimonadetes were higher in flowering/ fruiting than in the other stages, whereas the Bacteroidetes abundance in rhizosphere soils was lowest in flowering/fruiting stage (Table 3). At the genus level, the bacterial genera Gp10, Thioprofundum, Deferrisoma and Haliea dominated in vegetative stage, while abundances of the Planococcus, Halomonas, Pontibacter, Pseudomonas and Salinimicrobium in senescence stage were significantly higher than those in vegetative and flowering/fruiting. In contrast, the abundances of genera Gp10, Pelagibius, Deferrisoma and Geminicoccus dramatically decreased (Figure 3).
For bulk soil communities, relative abundance of phyla Proteobacteria and Firmicutes decreased from vegetative stage to flowering/fruiting stage, then were enriched and dominated in senescence stage. Conversely, Actinobacteria, Bacteroidetes, Acidobacteria, Planctomycetes and Chloroflexi decreased dramatically in abundance to below than 0.5% (Table 3). The abundant genera number decreased during the growth cycle, twenty-one, eleven and four abundant genera were observed in vegetative, flowering/fruiting and senescence stage, respectively. The composition of abundant genera also changed with growth stage (Figure 3). The abundances of Brevundimonas, Phenylobacterium, and Stenotrophomonas decreased from the vegetative to senescence stage.
2.5 Similarity among samples Hierarchical clustering based on the weighted UniFrac distance metric revealed distinct differences in microbial community structure between senescence stage and the two previous stages. The rhizosphere microbial community in vegetative and flowering/fruiting stage formed clusters that were clearly separated from the bulk soil community. However, in senescence stage, the bacterial communities of rhizosphere and bulk soils clustered together (Figure 4). These results implied that the bacterial communities, both in the rhizosphere and bulk soils, was of high similarity between vegetative and flowering/fruiting stages, but more divergent from the communities in senescence stage.
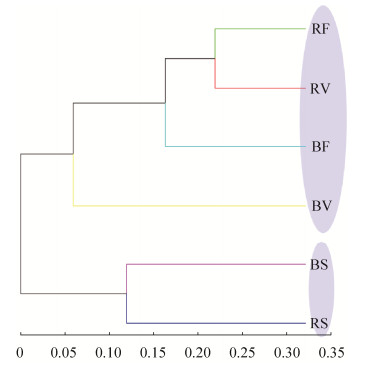 |
| Figure 4 Hierarchical clustering trees based on the weighted UniFrac distance metric. Branch lengths represent distance (indicated by scale bar). |
| 图选项 |
3 Discussion 3.1 Characteristics of the bacterial communities Consistent with result of previous studies on halophytes associated bacterial communities, the enriched or abundant bacteria are classified to salt-tolerant or halophilic genera, such as, Salinimicrobium, Halomonas, Geminicoccus, Pelagibius, Microbulbifer, Planococcus, Rubrivirga, Arenicella, Bacillus and Mesorhizobium. However, the salt-tolerance ability of bacteria species found in rhizosphere of L. ruthenicum are not clear and need further isolation and examination. Their enrichment in rhizosphere indicates that they are adapted to saline environments and their growth are salt dependent[36-43]. Moreover, the high abundance of these bacteria in rhizosphere implies that their reproduction and colonization in rhizosphere soil are driven by nutritional requirements as most of them are positively correlated with soil nutrients, such as TOC and TON (Figure 5). Meanwhile, some of species or strains previously identified from these genera are beneficial to plants, since they can degrade root exudates for root assimilation to help plant growth. For instance, some Planococcus and Microbulbifer members can degrade complex hydrocarbons[44]. Mesorhizobium can fix nitrogen[45].Bacillus members are generally effective for suppressing disease, such as bacterial wilt[46]. We assume that these enriched or abundant bacterial species in rhizosphere may be also beneficial to plant growth therefore they are selected by root and colonize in rhizosphere soils. However, this hypothesis need further determination. In overall, we consider that the enrichment of halophilic bacteria in rhizosphere are deriving from rhizosphere effect and metabolism requirements of bacteria (including salt concentration and nutrients) that are different from glycophytes.
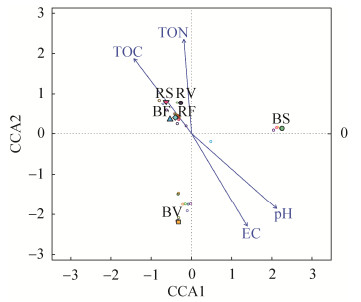 |
| Figure 5 CCA diagram revealing the relationship of bacterial communities and soil properties. |
| 图选项 |
3.2 Rhizosphere effects on soil bacterial communities Many studies have demonstrated the rhizosphere effects on microbial communities which leads to differences of the diversity and community composition in rhizosphere compared to bulk soils[15, 47-48]. In present study, we also find differences of community diversity and composition in rhizosphere and bulk soils, as consistent with our previous study[30]. Bacterial/archaeal diversity is higher in rhizosphere soils than in bulk soils. Most of the abundant phyla differed in abundance between the rhizosphere and bulk soil communities, such as the Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes. Firmicutes was enriched in rhizosphere soil during the growth cycle, mainly attributed by genus Bacillus as reported in plant Aster tripolium[15], whereas the abundance of Acidobacteria was lower in rhizosphere soil than in bulk soil. These results further support that plant species have important influence on composition of soil bacterial community which may be derived by the favorable for the growth and reproduction of some bacteria in the rhizosphere due to the rich of labile organic substrates[28]. The nutrients content in rhizosphere soils are indeed much higher than those in bulk soils associated with L. ruthenicum.
3.3 Growth stage dynamic of bacterial communities Growth stage dynamics are observed in the bacterial diversity and structure both in rhizosphere and bulk soils. In both compartment, the diversity peaks in flower/fruit stage but decrease dramatically in senescence stage. Further, the composition of abundant genus in flower/fruit stage is more diverse than in the senescence stage. The composition in the senescence stage differ apparently from the other two stages. As revealed by hierarchical clustering, the similarity of bacterial structure in vegetative and flowering/fruiting is higher than to that in the senescence stage, both in rhizosphere and bulk soils. Interestingly, we find that in senescence stage, the similarity of rhizosphere community to bulk soil community is higher than to the rhizosphere communities of the former two stages, whereas the similarity of rhizosphere communities is higher than that of rhizosphere and bulk soils in vegetative and flowering/fruiting stages. These results support that the growth stage plays a critical effect on microbial community[24, 27-28].
Some bacterial groups display similar growth stage dynamics in the rhizosphere and bulk soil, for example, the phyla Firmicutes, Actinobacteria, Planctomycetes, and the genera Geminicoccus and Pelagibius. However, most of the microbes, such as the Alpha-, Gamma-, and Deltaproteobacteria, show different temporal dynamics in the rhizosphere and the bulk soil communities. In addition, many of the abundant rhizosphere genera (e.g. genera Streptophyta, Pseudomonas, Halomonas, Planococcus and Salinimicrobium) displayed temporal patterns that differ from their patterns when they are present in bulk soils. These imply that even growth stage play an important role in bacterial community composition, but is not the only dominant factor influencing the microbial structure. Previous studies have revealed that the seasonal shifts and vegetation types affect soil microbial community composition by changing the soil physicochemical properties[49]. In different plant growth stages, root physiology, and the quality and quantity of root exudates vary, consequently influencing the rhizosphere soil microenvironment and exerting selective pressure on root-associated microorganisms[27-28]. We find a negative correlation between nutrients (such as SOM, TOC and TON contents) and community diversity both in rhizosphere and bulk soils. In the flowering/fruit stage, both the rhizosphere and bulk soils have the highest community diversity in spite of the lowest nutrient concentrations, which is also reported in previous research[50]. However, in the senescence stage, the community diversity decrease dramatically even though the SOM, TOC and TON contents are much higher than those in the flowering/fruit stage.
From these results, we assume that other factors may be responsible for changes of community diversity and composition. Soil temperature has been proven to be an important factors influencing soil microbial community structure[51-52]. The average temperature in senescence stage is 3.57 ℃ and 8.64 ℃ lower than that in vegetative stage flowering/fruiting stage according to our successional records of the whole year in the studying regions. We propose that the decrease in temperature might be an important factors causing decline in diversity and change of community composition in the senescence stage. Moreover, biotic factor (such as effects of fungus) might be also a possible deriving force. Previous study show that the enrichment of some pathogenic fungus can cause severe root disease of Panax ginseng and lead to decrease of bacterial diversity and increase of fungal diversity along with growth stages[25]. Their effects are need future research.
4 Conclusion We investigated the growth stage dynamics of bacterial community diversity and structure of a halophytic plant L. ruthencium in three growth stages. The rhizosphere effect causes apparent differences in diversity and composition of rhizosphere community compared to the bulk soils. We observe clear growth stage dynamics of the bacterial community diversity and composition both in the rhizosphere and bulk soils. Specially, the community diversity and structure in the senescence stage differ dramatically from the other two growth stages. These results indicate that growth stage is an important driving forces causing changes in diversity and structure of the bacterial community associated with L. ruthencium. Though we get some meaningful implications regarding the growth stage dynamics of bacterial community, there are some deficiencies in present study. Firstly, there is only one sequencing data for each sample from a mix of multiple individuals in each stage. This omits the difference of plant individuals and the sequencing accuracy tend to be easily influenced by experimental error. Secondly, the potential mechanisms and the determination factors underlying the growth stage dynamics of the bacterial community are not examined and clarified. These questions will be settled in our next research which is ongoing.
Acknowledgements We would like to thank American Journal Experts for providing linguistic assistance during the preparation of this manuscript.
References
| [1] | ILRI. Effectiveness and social/environmental impacts of irrigation projects: a review//Annual Report 1988 of the International Institute for Land Reclamation and Improvement. Wageningen, The Netherlands: ILRI, 1988. |
| [2] | Bartels D, Sunkar R. Drought and salt tolerance in plants. Critical Reviews in Plant Sciences, 2005, 24(1): 23-58. DOI:10.1080/07352680590910410 |
| [3] | Wang WX, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures:towards genetic engineering for stress tolerance. Planta, 2003, 218(1): 1-14. DOI:10.1007/s00425-003-1105-5 |
| [4] | Chaudhary DR, Gautam RK, Yousuf B, Mishra A, Jha B. Nutrients, microbial community structure and functional gene abundance of rhizosphere and bulk soils of halophytes. Applied Soil Ecology, 2015, 91: 16-26. DOI:10.1016/j.apsoil.2015.02.003 |
| [5] | Hinsinger P, Bengough AG, Vetterlein D, Young IM. Rhizosphere:biophysics, biogeochemistry and ecological relevance. Plant and Soil, 2009, 321(1/2): 117-152. |
| [6] | Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and other organisms. Annual Review of Plant Biology, 2006, 57: 233-266. DOI:10.1146/annurev.arplant.57.032905.105159 |
| [7] | Zhang HM, Sun Y, Xie XT, Kim MS, Dowd SE, Paré PW. A soil bacterium regulates plant acquisition of iron via deficiency-inducible mechanisms. Plant Journal, 2009, 58(4): 568-577. DOI:10.1111/tpj.2009.58.issue-4 |
| [8] | Palaniyandi SA, Damodharan K, Yang SH, Suh JW. Streptomyces sp. strain PGPA39 alleviates salt stress and promotes growth of 'Micro Tom' tomato plants. Journal of Applied Microbiology, 2014, 117(3): 766-773. DOI:10.1111/jam.2014.117.issue-3 |
| [9] | Vaishnav A, Kumari S, Jain S, Varma A, Choudhary DK. Putative bacterial volatile-mediated growth in soybean (Glycine max L. Merrill) and expression of induced proteins under salt stress. Journal of Applied Microbiology, 2015, 119(2): 539-551. DOI:10.1111/jam.2015.119.issue-2 |
| [10] | Lee Y, Krishnamoorthy R, Selvakumar G, Kim K, Sa TM. Alleviation of salt stress in maize plant by co-inoculation of arbuscular mycorrhizal fungi and Methylobacterium oryzae CBMB20. Journal of the Korean Society for Applied Biological Chemistry, 2015, 58(4): 533-540. DOI:10.1007/s13765-015-0072-4 |
| [11] | Ngumbi E, Kloepper J. Bacterial-mediated drought tolerance:current and future prospects. Applied Soil Ecology, 2016, 105: 109-125. DOI:10.1016/j.apsoil.2016.04.009 |
| [12] | Tian YQ, Gao LH. Bacterial diversity in the rhizosphere of cucumbers grown in soils covering a wide range of cucumber cropping histories and environmental conditions. Microbial Ecology, 2014, 68(4): 794-806. DOI:10.1007/s00248-014-0461-y |
| [13] | Rodríguez-Blanco A, Sicardi M, Frioni L. Plant genotype and nitrogen fertilization effects on abundance and diversity of diazotrophic bacteria associated with maize (Zea mays L.). Biology and Fertility of Soils, 2015, 51(3): 391-402. DOI:10.1007/s00374-014-0986-8 |
| [14] | Oliveira V, Gomes NCM, Cleary DFR, Almeida A, Silva AMS, Sim?es MMQ, Silva H, Cunha . Halophyte plant colonization as a driver of the composition of bacterial communities in salt marshes chronically exposed to oil hydrocarbons. FEMS Microbiology Ecology, 2014, 90(3): 647-662. DOI:10.1111/fem.2014.90.issue-3 |
| [15] | Szymańska S, Plociniczak T, Piotrowska-Seget Z, Z?och M, Ruppel S, Hrynkiewicz K. Metabolic potential and community structure of endophytic and rhizosphere bacteria associated with the roots of the halophyte Aster tripolium L. Microbiological Research, 2016, 182: 68-79. DOI:10.1016/j.micres.2015.09.007 |
| [16] | Sgroy V, Cassán F, Masciarelli O, del Papa MF, Lagares A, Luna V. Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Applied Microbiology and Biotechnology, 2009, 85(2): 371-381. DOI:10.1007/s00253-009-2116-3 |
| [17] | Siddikee MA, Chauhan PS, Anandham R, Han GH, Sa TM. Isolation, characterization, and use for plant growth promotion under salt stress, of acc deaminase-producing halotolerant bacteria derived from coastal soil. Journal of Microbiology and Biotechnology, 2010, 20(11): 1577-1584. DOI:10.4014/jmb |
| [18] | Ramadoss D, Lakkineni VK, Bose P, Ali S, Annapurna K. Mitigation of salt stress in wheat seedlings by halotolerant bacteria isolated from saline habitats. Springerplus, 2013, 2(1): 6. DOI:10.1186/2193-1801-2-6 |
| [19] | Shi SJ, O'Callaghan M, Jones EE, Richardson AE, Walter C, Stewart A, Condron L. Investigation of organic anions in tree root exudates and rhizosphere microbial communities using in situ and destructive sampling techniques. Plant and Soil, 2012, 359(1/2): 149-163. |
| [20] | Zhou ML, Chen WM, Chen HY, Wei GH. Draft genome sequence of Mesorhizobium alhagi CCNWXJ12-2T, a novel salt-resistant species isolated from the desert of northwestern China. Journal of Bacteriology, 2012, 194(5): 1261-1262. DOI:10.1128/JB.06635-11 |
| [21] | Tang J, Zheng AP, Bromfield ESP, Zhu J, Li SC, Wang SQ, Deng QM, Li P. 16S rRNA gene sequence analysis of halophilic and halotolerant bacteria isolated from a hypersaline pond in Sichuan, China. Annals of Microbiology, 2011, 61(2): 375-381. DOI:10.1007/s13213-010-0137-x |
| [22] | Kim CS, Nam JW, Jo JW, Kim SY, Han JG, Hyun MW, Sung GH, Han SK. Studies on seasonal dynamics of soil-higher fungal communities in Mongolian oak-dominant Gwangneung forest in Korea. Journal of Microbiology, 2016, 54(1): 14-22. |
| [23] | López-Mondéjar R, Vo?í?ková J, Větrovsk T, Baldrian P. The bacterial community inhabiting temperate deciduous forests is vertically stratified and undergoes seasonal dynamics. Soil Biology and Biochemistry, 2015, 87: 43-50. DOI:10.1016/j.soilbio.2015.04.008 |
| [24] | Han LL, Wang JT, Yang SH, Chen WF, Zhang LM, He JZ. Temporal dynamics of fungal communities in soybean rhizosphere. Journal of Soils and Sediments, 2017, 17(2): 491-498. DOI:10.1007/s11368-016-1534-y |
| [25] | Dong LL, Xu J, Zhang LJ, Cheng RY, Wei GF, Su H, Yang J, Qian J, Xu R, Chen SL. Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta Pharmaceutica Sinica B, 2018, 8(2): 272-282. DOI:10.1016/j.apsb.2017.12.011 |
| [26] | Bencherif K, Boutekrabt A, Dalpé Y, Sahraoui ALH. Soil and seasons affect arbuscular mycorrhizal fungi associated with Tamarix rhizosphere in arid and semi-arid steppes. Applied Soil Ecology, 2016, 107: 182-190. DOI:10.1016/j.apsoil.2016.06.003 |
| [27] | Houlden A, Timms-Wilson TM, Day MJ, Bailey MJ. Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiology Ecology, 2008, 65(2): 193-201. DOI:10.1111/fem.2008.65.issue-2 |
| [28] | Li XZ, Rui JP, Mao YJ, Yannarell A, Mackie R. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biology and Biochemistry, 2014, 68: 392-401. DOI:10.1016/j.soilbio.2013.10.017 |
| [29] | Peng F, Huang CH, You QG, Gao TP. Effects of plantation of Lycium ruthenicum on the soil salt distribution in the Minqin Basin. Journal of Desert Research, 2013, 33(5): 1406-1412. (in Chinese) 彭飞, 黄翠华, 尤全刚, 高天鹏. 种植黑果枸杞(Lycium ruthenicum)对盐渍土盐分分布的影响. 中国沙漠, 2013, 33(5): 1406-1412. |
| [30] | Li Y, Yang XD, Qin L, Lv GH, He XM, Zhang XN. The bacterial diversity and community structures in rhizosphere soil of two halophytes, Lycium ruthenicum and Kalidium capsicum. Acta Ecologica Sinica, 2018, 38(9): 3118-3131. (in Chinese) 李岩, 杨晓东, 秦璐, 吕光辉, 何学敏, 张雪妮. 两种盐生植物根际土壤细菌多样性和群落结构. 生态学报, 2018, 38(9): 3118-3131. |
| [31] | Zhang XN, Yang XD, Lyu GH. Diversity patterns and response mechanisms of desert plants to the soil environment along soil water and salinity gradients. Acta Ecologica Sinica, 2016, 36(11): 3206-3215. (in Chinese) 张雪妮, 杨晓东, 吕光辉. 水盐梯度下荒漠植物多样性格局及其与土壤环境的关系. 生态学报, 2016, 36(11): 3206-3215. |
| [32] | Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(8): E911-E920. DOI:10.1073/pnas.1414592112 |
| [33] | Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics, 2011, 27(6): 863-864. DOI:10.1093/bioinformatics/btr026 |
| [34] | Zhang JJ, Kobert K, Flouri T, Stamatakis A. PEAR:a fast and accurate illumina paired-end reAd mergeR. Bioinformatics, 2014, 30(5): 614-620. DOI:10.1093/bioinformatics/btt593 |
| [35] | Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Gl?ckner FO. The SILVA ribosomal RNA gene database project:improved data processing and web-based tools. Nucleic Acids Research, 2013, 41(D1): D590-D596. |
| [36] | Lim JM, Jeon CO, Lee SS, Park DJ, Xu LH, Jiang CL, Kim CJ. Reclassification of Salegentibacter catena Ying et al. 2007 as Salinimicrobium catena gen. nov., comb. nov. and description of Salinimicrobium xinjiangense sp. nov., a halophilic bacterium isolated from Xinjiang province in China. International Journal of Systematic and Evolutionary Microbiology, 2008, 58(Pt 2): 438-442. |
| [37] | Chen YG, Cui XL, Zhang YQ, Li WJ, Wang YX, Kim CJ, Lim JM, Xu LH, Jiang CL. Salinimicrobium terrae sp. nov., isolated from saline soil, and emended description of the genus Salinimicrobium. International Journal of Systematic and Evolutionary Microbiology, 2008, 58(Pt 11): 2501-2504. |
| [38] | Foesel BU, G??ner AS, Drake HL, Schramm A. Geminicoccus roseus gen. nov., sp. nov., an aerobic phototrophic Alphaproteobacterium isolated from a marine aquaculture biofilter. Systematic and Applied Microbiology, 2007, 30(8): 581-586. DOI:10.1016/j.syapm.2007.05.005 |
| [39] | Choi DH, Hwang CY, Cho BC. Pelagibius litoralis gen. nov., sp. nov., a marine bacterium in the family Rhodospirillaceae isolated from coastal seawater. International Journal of Systematic and Evolutionary Microbiology, 2009, 59(Pt 4): 818-823. |
| [40] | Wang YX, Li YP, Liu JH, Xiao W, Lai YH, Li ZY, Ding ZG, Wen ML, Cui XL. Gracilimonas mengyeensis sp. nov., a moderately halophilic bacterium isolated from a salt mine in Yunnan, south-western China. International Journal of Systematic and Evolutionary Microbiology, 2013, 63(Pt 11): 3989-3993. DOI:10.1099/ijs.0.052043-0 |
| [41] | Cho Y, Chung H, Jang GI, Choi DH, Noh JH, Cho BC. Gracilimonas rosea sp. nov., isolated from tropical seawater, and emended description of the genus Gracilimonas. International Journal of Systematic and Evolutionary Microbiology, 2013, 63(Pt 11): 4006-4011. DOI:10.1099/ijs.0.052340-0 |
| [42] | Nedashkovskaya OI, Cleenwerck I, Zhukova NV, Kim SB, de Vos P. Arenicella chitinivorans sp. nov., a gammaproteobacterium isolated from the sea urchin Strongylocentrotus intermedius. International Journal of Systematic and Evolutionary Microbiology, 2013, 63(Pt 11): 4124-4129. DOI:10.1099/ijs.0.051599-0 |
| [43] | Song J, Joung Y, Park S, Cho JC, Kogure K. Rubrivirga profundi sp. nov., isolated from deep-sea water, and emended description of the genus Rubrivirga. International Journal of Systematic and Evolutionary Microbiology, 2016, 66(9): 3253-3257. DOI:10.1099/ijsem.0.001182 |
| [44] | See-Too WS, Chua KO, Lim YL, Chen JW, Convey P, Mohidin TBM, Yin WF, Chan KG. Complete genome sequence of Planococcus donghaensis JH1T, a pectin-degrading bacterium. Journal of Biotechnology, 2017, 252: 11-14. DOI:10.1016/j.jbiotec.2017.05.005 |
| [45] | Ardley JK, Parker MA, de Meyer SE, Trengove RD, O'Hara GW, Reeve WG, Yates RJ, Dilworth MJ, Willems A, Howieson JG. Microvirga lupini sp. nov., Microvirga lotononidis sp. nov. and Microvirga zambiensis sp. nov. are alphaproteobacterial root-nodule bacteria that specifically nodulate and fix nitrogen with geographically and taxonomically separate legume hosts. International Journal of Systematic and Evolutionary Microbiology, 2012, 62(Pt 11): 2579-2588. DOI:10.1099/ijs.0.035097-0 |
| [46] | Okubo A, Matsusaka M, Sugiyama S. Impacts of root symbiotic associations on interspecific variation in sugar exudation rates and rhizosphere microbial communities:a comparison among four plant families. Plant and Soil, 2016, 399(1/2): 345-356. |
| [47] | Li XZ, Rui JP, Xiong JB, Li JB, He ZL, Zhou JZ, Yannarell AC, Mackie RI. Functional potential of soil microbial communities in the maize rhizosphere. PLoS ONE, 2014, 9(11): e112609. DOI:10.1371/journal.pone.0112609 |
| [48] | Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dr?ge J, Pan Y, McHardy AC, Schulze-Lefert P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host & Microbe, 2015, 17(3): 392-403. |
| [49] | Wu ZY, Lin WX, Li JJ, Liu JF, Li BL, Wu LK, Fang CX, Zhang ZX. Effects of seasonal variations on soil microbial community composition of two typical zonal vegetation types in the Wuyi Mountains. Journal of Mountain Science, 2016, 13(6): 1056-1065. DOI:10.1007/s11629-015-3599-2 |
| [50] | Taniuchi Y, Watanabe T, Kakehi S, Sakami T, Kuwata A. Seasonal dynamics of the phytoplankton community in Sendai Bay, northern Japan. Journal of Oceanography, 2017, 73(1): 1-9. DOI:10.1007/s10872-015-0334-0 |
| [51] | Chiriac CM, Szekeres E, Rudi K, Baricz A, Hegedus A, Dragos N, Coman C. Differences in Temperature and water chemistry shape distinct diversity patterns in thermophilic microbial communities. Applied and Environmental Microbiology, 2017, 83(21): e01363-17. |
| [52] | Mateos-Rivera A, Yde JC, Wilson B, Finster KW, Reigstad LJ, ?vre?s L. The effect of temperature change on the microbial diversity and community structure along the chronosequence of the sub-arctic glacier forefield of Styggedalsbreen (Norway). FEMS Microbiology Ecology, 2016, 92(4): fnw038. DOI:10.1093/femsec/fiw038 |
