尹华群1,2

 , 刘征华1,2, 刘学端1,2
, 刘征华1,2, 刘学端1,2 1.中南大学资源加工与生物工程学院, 湖南 长沙 410083;
2.教育部生物冶金重点实验室, 湖南 长沙 410083
收稿日期:2017-11-23;修回日期:2018-02-03;网络出版日期:2018-03-02
基金项目:国家自然科学基金(31570113,41573072)
作者简介:尹华群, 博士, 中南大学资源加工与生物工程学院教授, 博士生导师, "湖南省环境微生物组学与应用国际合作基地"负责人, 中国大百科全书编委. Frontiers in microbiology 等国际学术期刊客座主编, 先后主持(参与)973、863、国家支撑计划、国家自然科学基金等项目20多项, 在冶金微生物群落结构与功能分析的基因组学技术、冶金微生物种群的适应机制以及冶金微生物协同促进硫化矿物氧化溶解机理等方面取得了系统性的成果, 相关研究成果在 Applied and Environmental Microbiology 、Environmental Pollution 和 Applied microbiology and Biotechnology 等专业学术期刊上发表SCI论文60多篇, 获湖南省科技进步一等奖1项, 授权发明专利10余项
*通信作者:尹华群, Tel/Fax:+86-731-88830546;E-mail:yinhuaqun_cs@sina.com
摘要:生物冶金是利用微生物铁硫元素代谢活性加速硫化矿物氧化溶解,并对其中有价金属加以提取回收的技术。冶金系统中微生物的代谢多样性及其耦合功能网络,尤其是以铁硫代谢途径为主的功能网络,在硫化矿物加速氧化溶解过程中承担了重要作用,是生物冶金技术理论研究的核心领域。本文归纳了冶金系统中多样化的微生物物种及其铁硫代谢途径,并从微生物代谢耦合角度探讨了微生物代谢多样性与矿物的相互作用。
关键词: 冶金微生物 功能网络 代谢多样性 生物冶金
Diversity of iron and sulfur metabolism in bioleaching microorganisms and their interaction with minerals
Huaqun Yin1,2

 , Zhenghua Liu1,2, Xueduan Liu1,2
, Zhenghua Liu1,2, Xueduan Liu1,2 1.School of Minerals Processing and Bioengineering, Central South University, Changsha 410083, Hunan Province, China;
2.Key Laboratory of Biometallurgy of Ministry of Education, Changsha 410083, Hunan Province, China
Received 23 November 2017; Revised 3 February 2018; Published online 2 March 2018
*Corresponding author: Huaqun Yin, Tel/Fax: +86-731-88830546; E-mail:yinhuaqun_cs@sina.com
Supported by the National Natural Science Foundation of China (31570113, 41573072)
Abstract: Bioleaching is to extract valuable metals from their ores by microbial metabolic activity to take iron/sulfur. Microbial metabolic diversity and coupling function networks in metallurgical systems, especially functional networks dominated by iron and sulfur metabolism, play a major role in driving and accelerating the dissolution process of sulfide ore. Therefore, it is the core research area on bio-metallurgy technology. We summarize here the microbial diversity and iron/sulfur metabolism pathways in the metallurgical system, as well as the interactions between microbial metabolism diversity and minerals from the perspectives of metabolic coupling functional network.
Key words: bioleaching microorganisms functional network metabolic diversity bioleaching
生物冶金技术是利用以矿物为营养基质的微生物,将矿物氧化分解并使有价金属离子进入溶液,然后通过进一步分离、富集、纯化而提取有价金属的高新技术,是低品位、难处理矿产资源清洁高效利用的关键技术之一,也是21世纪矿产资源加工的战略性技术[1-2]。其中,冶金微生物作为生物冶金技术的关键功能角色,促进了整个生物冶金体系的铁、硫元素等物质的循环,并加速了硫化矿物氧化溶解[3-4]。这一研究课题是生物冶金理论研究的核心领域,亦是生物地球化学元素循环的关键科学问题。
生物冶金体系中pH极低,含有浓度极高的硫酸根离子、铁离子及其他重金属离子,具有极低量的有机质[5],这些极端的环境因素让该生境拥有特定的微生物类群及其代谢形式[6-7]。若从冶金微生物类群的铁、氧化代谢多样性及其耦合功能网络的整体角度去研究冶金微生物与矿物的相互作用,能够更好地从总体上把握和调控冶金体系的生态系统功能[8-12]。因此,加强冶金微生物铁、硫氧化代谢多样性及其耦合功能网络与矿物相互作用的研究,有助于生物冶金领域的进一步发展。
1 冶金微生物的多样性 冶金微生物具有极强的耐酸性和重金属抗性,适宜生长温度范围广,能量代谢方式多样,系统发育格局分散[13]。目前为止,在生物浸出系统中已经发现40多种类型的冶金微生物[14]。根据不同的最适生长温度范围,可分为耐寒嗜冷微生物(0–20℃)、嗜温微生物(20–40℃)、中度嗜热微生物(40–60 ℃)和极端嗜热微生物(> 60 ℃)。其中中度嗜热微生物和极端嗜热微生物在生物冶金中应用最为广泛。根据所利用的能源底物,可将它们分为以下几种:(1)氧化Fe2+的铁氧化自养微生物[15];(2)氧化无机硫化合物的硫氧化自养微生物[16];(3)异养或混合营养型的微生物,可以以无机物为电子供体,将Fe3+或SO42–还原[17]。另外,同时也有兼具上述任意多种能量代谢方式的微生物[18]。
根据系统发育信息,冶金微生物主要分布在变形菌门(Proteobacteria)、放线菌门(Actinobacteria)、硝化螺旋菌门(Nitrospira)、厚壁菌门(Firmicutes),以及古菌的热原体属(Thermoplasmales)和硫化叶菌目(Sulfolobales)[19]。其中应用最广泛的物种包括有γ-变形菌纲中的 Acidithiobacillus 属(主要是 At.ferrooxidans 和 At. caldus)[20],硝化螺旋菌门的 Leptospirillum 属[21-25],ɑ-变形菌纲中的 Acidiphilium 属[26-27]和厚壁菌门的 Sulfobacillus 属[28-30]。这些微生物主要是具有铁氧化或硫氧化功能的微生物,且多为可分离的优势菌群,目前已得到大量的基因组学解析,为构建生物冶金人工共培养体系提供了重要的参考信息。
2 冶金微生物的铁硫代谢多样性 生物冶金体系中大部分冶金微生物都是自养微生物,能通过铁或硫的氧化获取维持生命活动所需的能量,并驱动其他碳、氮、磷等元素的循环[8-9, 31]。为了适应这种有机能源底物匮乏的环境,并且最大限度地利用铁硫氧化产生的能量,冶金微生物进化出了多样的与铁、硫元素循环相关的代谢途径。
2.1 铁代谢多样性 在中性环境中,Fe(Ⅱ)暴露在空气中能快速地被氧化成Fe(Ⅲ)。然而,在极端酸性的浸矿体系中(pH < 3),Fe(Ⅱ)即使在有氧条件下也能稳定存在[32]。因此,极端酸性的环境为铁氧化细菌提供了稳定的能源底物,是嗜酸微生物铁代谢途径多样化的基本条件。
2.1.1 铁氧化代谢: 冶金微生物的铁氧化细菌主要包括有 Acidithiobacillusferrooxidans 、At. ferrivorans 、Leptospirillum 属和 Sulfobacillus 属等,而铁氧化古菌主要有 Acidiplasma cupricumulans 、Ferroplasma acidarmanus 、Acidianus 属和 Metallosphaera sedula 等[8, 32]。这些铁氧化细菌和古菌可利用不同的电子传递链捕获Fe(Ⅱ)氧化释放的电子,并供给生命代谢活动所需的能量(表 1)。其中,Acidithiobacillus 属的铁氧化机制研究得较为透彻,目前已证实该菌属参与铁氧化功能代谢的基因有 rusA/B 和 iro 基因[33-34]。rusA/B 基因负责编码铜蓝蛋白A和B,这两种蛋白是细胞周质中负责氧化Fe(Ⅱ)的同工酶。值得注意的是,在 At.ferrivorans GroupⅢ中可检测出 rusA/B 基因,而 At.ferrivorans CF2却没有该基因,但其依然具有铁氧化活性,这表明了该基因至少在 At.ferrivorans 中不是铁氧化代谢途径的核心基因[33]。另一方面,iro 基因负责编码细胞周质中一种具有铁氧化酶活性的高电位铁硫蛋白(high-potentialiron-sulfur protein,HiIPs)[35]。这表明了 Acidithiobacillus 属至少有两种不同的铁氧化代谢途径。相似地,这两种铁氧化途径均是靠细胞外膜上的Cyc2将Fe(Ⅱ)的电子传递到铁氧化酶上,暗示了这两种途径可能来源于同一套铁氧化系统的趋异进化。
表 1. 冶金微生物铁氧化代谢途径多样性 Table 1. Ironmetabolic diversity of bioleaching microorganisms
| Iron metabolic pathways | Gene/Operon | Position | Electron transfer chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| Cyc2 | Outer membrane | Fe(Ⅱ)→Rusticyanin oxidase |  | |||||||||
| Cytochrome 572 (Cyt572) | Outer membrane | Fe(Ⅱ)→Cyt579 | ||||||||||
| Rusticyanin A | rusA | Periplasm | Rusticyanin→Cyc1/CycA1 | |||||||||
| Rusticyanin B | rusB | Periplasm | Rusticyanin→Cyc1/CycA1 | |||||||||
| Iron oxidase | iro | Periplasm | Iron oxidase→Cyc1/CycA1 | |||||||||
| Cytochrome 579 (Cyt579) | Periplasm | Cyt579→Cytochrome c | ||||||||||
| Sulfocyanin | Periplasm | |||||||||||
| aa3 oxidase | Inner membrane | |||||||||||
| Cyc1 | Inner membrane | Cyc1→aa3 oxidase | ||||||||||
| CycA1 | Inner membrane | CycA1→bc1 complex | ||||||||||
| cbb3 oxidase | Inner membrane | |||||||||||
| Cytochrome b | Inner membrane | |||||||||||
| haem-copper terminal | fox cluster | Inner membrane | ||||||||||
| oxidase | ||||||||||||
| bc1 complex | Inner membrane | |||||||||||
| 1. Acidithiobacillusferrooxidans ; 2. At. ferrivorans ; 3. Leptospirillum spp.; 4. Sulfobacillus spp.; 5. Sulfolobus spp.; 6. Acidiplasma spp.; 7. Ferroplasma acidarmanus ; 8. Acidianus spp.; 9. Metallosphaera sedula .White cells: absence ofthe gene/operon; Black cells: presence of the gene/operon. | ||||||||||||
表选项
在 Leptospirillum 属的4个种中,包括 Leptospirillum ferrooxidans (Group Ⅰ)[21]、L. ferriphilum (Group Ⅱ)[22]、L. rubarum (Group Ⅱ)[22]、L. ferrodiazotrophum (Group Ⅲ)和 L. sp. UBA BS (Group Ⅳ)[25],都具有相似的Fe(Ⅱ)氧化代谢途径[23, 25, 36]。与 Acidithiobacillus 属不同的是,Leptospirillum 属是将Fe(Ⅱ)的电子由外膜的Cyt572传递到细胞周质的Cyt579,随后经细胞周质的Cytc传递到内膜的cbb3终端氧化酶,并以氧气为电子受体生成水(“downhill”);或传递到bc1复合体,然后通过QH2转移到NADH脱氢酶中(“uphill”)[35]。
Sulfobacillus 属是一种混合营养型的革兰氏阳性菌,在已分离的 Sulfobacillus 属中均已被证实具有铁氧化能力,但种间的铁氧化代谢途径存在较大差异[37-38]。Nicholas等[37]通过分析比较9株 Sulfobacillus 基因组学发现,与 Leptospirillum 属相似,8株均有完整的downhill铁氧化途径,而 Sulfobacillus sp.AMDSBA1则缺乏关键的编码膜相关c型细胞色素的基因;对于uphill铁氧化途径,只有 Sulfobacillus sp.AMDSBA4具有编码关键复合体bc1的基因,但目前并没验证其具有与铁氧化革兰氏阴性菌相同的功能。此外,我们发现在 S.thermosulfidooxidans ST基因组中,存在编码与铁氧化相关蓝铜蛋白Sulfocyanin的基因,并且其与古菌的铜蓝蛋白编码基因具有较高的相似性,暗示了基因横向转移可能是 Sulfobacillus sp.铁氧化代谢途径趋异进化的重要因素[39]。
在铁氧化古菌 Acidiplasma 属和 Ferroplasma 属中[40],其铁氧化代谢途径与铁氧化细菌 Leptospirillum 属较为相似,但亦有明显的差异。Bulaev等[41]通过基因组分析表明,Acidiplasma sp. MBA-1含有编码与铁氧化相关的铜蓝蛋白。与 Leptospirillum 属相似,Fe(Ⅱ)的电子传递链包括以NADH和氧气为最终电子受体两种。值得注意的是,该菌株对铁离子拥有很高浓度的铁离子耐受性,即使在50 g/L的Fe(Ⅲ)存在下,依然具有很高的铁氧化活性,表明了 Acidiplasma spp.铁氧化系统可能具有其他的特性[42]。加强对该特性的研究,或许可优化出强铁氧化能力的冶金微生物功能类群。在 Ferroplasma spp. JA12的基因组信息中心,Ullrich等[43]发现Fe(Ⅱ)氧化电子传递链与 Leptospirillum spp.相似,但外膜上接收Fe(Ⅱ)电子的蛋白是Cyc2的同源蛋白而非Cyt572。但目前对于细菌和古菌铁氧化代谢途径差异的进化机制仍待深入研究。
2.1.2 铁还原代谢: 铁还原代谢广泛存在于嗜酸的微生物中,铁硫氧化自养菌 At.ferrooxidans 、At. ferrivorans 在厌氧条件下均有铁还原能力,但目前为止依然没有发现与Fe(Ⅲ)还原直接相关的酶。有研究报道Fe(Ⅲ)的还原与 tetH [44]和 arsH 基因[45]的表达量有关,表明Fe(Ⅲ)的最终电子供体可能来源于TetH和ArsH蛋白。另一方面,在 At.ferrooxidans 中,Fe(Ⅲ)也可能自发地参与H2S的厌氧氧化过程[46]。此外,在所有已分离的混合营养型的 Sulfobacillus 属中,即使在没有硫元素存在的条件下,也拥有铁还原能力,表明 Sulfobacillus 属可能具有直接还原铁的能力[47-51],但具体机制仍不明确。而一些异养的细菌和古菌,如 Acidiphilium 属和 Acidiplasma 属等,也都是Fe(Ⅲ)还原的承担者[40, 52-53]。
2.2 硫代谢多样性 相比于铁氧化过程,冶金微生物从S2–/S6+氧化过程中可获得更多的能量。而硫元素具有多的价态以及化合物形态,为了使系统能量输入最大化,冶金微生物的硫代谢途径比铁代谢更为复杂多样[34-35]。
2.2.1 硫氧化代谢: 还原型无机硫化合物(RISCs)的氧化途径分为三种,包括H2S、氧化性谷胱甘肽(GSSH)和S2O32–的氧化[9, 54]。RISCs的氧化涉及到多种微生物不同细胞空间的多种酶、复合体和电子载体(表 2)。在H2S的氧化途径中,细胞周质中的硫化物,经内膜上的硫醌氧化还原酶(SQR)氧化后生成硫[55],硫再经细胞周质硫氧化还原酶(SOR)被氧化生成亚硫酸[56],亚硫酸进一步被亚硫酸氧化酶(SO)或被腺苷磷酸硫酸还原酶(APSR)直接或间接氧化生成硫酸[9, 57]。值得注意的是,编码SOR的 sor 基因存在较高的移动性,例如在 At.Caldus ATCC 51756和 At.thiooxidans A01基因组中含有 sor 基因[58-59],但却在一些同源菌株中,如 At.caldus SM-1[60]和 At.thiooxidans ATCC 19377却没有这个关键的基因[35],同样的现象也发生在混合营养型的 Sulfobacillus spp.种间中[37]。在古菌研究中,最早研究发现的 Acidianus 属和 Sulfolobus 属中部分菌种通过硫氧化还原酶SOR完成第一步硫的氧化还原过程,由亚硫酸受体氧化酶SAOR、SQR、TQO、TetH完成亚硫酸到硫酸的直接氧化过程,由APSR和腺苷酰硫酸磷酸腺苷转移酶(APAT)进行亚硫酸的间接氧化[61-62]。此外,在 Metallosphaera 属菌种的基因组中,均没发现SOR、APSR和APAT及其同源蛋白[63-64],意味着该菌株可能存在其他硫氧化代谢途径。
在氧化性谷胱甘肽(GSSH)的氧化途径中,锚定在内膜上的异二硫化物还原酶(HDR)是参与细胞质中GSSH氧化的关键酶,可将GSSH氧化分解为GSH和SO32-,在大部分嗜酸硫氧化细菌和古菌中具有较高的保守性[65-66]。然而,在混合营养型的 Sulfobacillus spp.中,则存在两种编码异二硫还原酶的 hdr 基因簇:第一种 hdr 基因簇存在于 S. thermosulfidooxidans ST、Sulfobacillus AMDSBA1和 Sulfobacillus AMDSBA5中,而第二种类型的 hdr 基因簇则存在于绝大部分的 Sulfobacillus 属中[37]。与第一种类型相比,第二种类型基因簇在 hdrC 基因上拥有编码黄素蛋白、6个跨膜体和2个富含半胱氨酸结构域的片段[37],这3个基因片段的组合在硫酸盐还原菌 Desulfobacteriumautotrophicum 中也有发现[67],但具体作用仍不明确。GSSH氧化分解产生的SO32–对细胞具有毒害作用,经腺苷硫酸(APS)还原酶催化产生APS,然后被硫酸盐腺苷酰转移酶(SAT)氧化生成硫酸盐[57]。另外,在硫氧化古菌 Metallosphaera 属中,发现了新的能将SO32–氧化成SO42–的亚硫酸盐-受体氧化酶(SAR,表 2)[64, 68]。
表 2. 冶金微生物硫氧化代谢途径多样性 Table 2. Sulfurmetabolic diversity of bioleaching microorganisms
| Sulfur metabolic pathways | Gene/Operon | Position | Electron transfer chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Sox system | SoxXYZAB | Periplasm | S2–/S0/S2O32–/SO32–→SO42– |  | |||||||||
| Tetrathionate hydrolase | tetH | Periplasm | S4O62–→S2O32–+SO42–+S0 | ||||||||||
| Thiosulfate dehydrogenase | tsd | Periplasm | S2O32–→S4O62– | ||||||||||
| Sulfide quinone reductase | sqr | Inner membrane | H2S→S0 | ||||||||||
| Thiosulfate:quinone oxidoreductase | doxDA | Inner membrane | S4O62–→S2O32– | ||||||||||
| Sulfur oxygenase reductase | sor | Cytoplasm | S0→H2S+SO32–+S2O32– | ||||||||||
| Thiosulfate sulfurtransferase | tst | Cytoplasm | S2O32–→SO32–+ S0 | ||||||||||
| Heterodisulfide reductase complex | hdrABC | Cytoplasm | RSSH→RSH+ SO32– | ||||||||||
| Sulfate adenylyltransferase/adenylylsulfate kinase | sat/cysC | Cytoplasm | APS→SO42– | ||||||||||
| sulfite: acceptor oxidoreductase | sar | Periplasm | SO32–→SO42– | ||||||||||
| 1. Acidithiobacilluscaldus ; 2. At. thiobacillus ; 3. At. ferrooxidans ; 4. At.ferrivorans ; 5. Sulfobacillus spp.; 6. Sulfolobus solfataricus ; 7. S. islandicus ; 8. S.acidocaldarius ; 9. Acidianus copahuensis ; 10. Metallosphaera spp..White cells: absence of the gene/operon; Black cells:presence of the gene/operon. | |||||||||||||
表选项
在硫代硫酸盐氧化的相关途径中,硫代硫酸盐被细胞周质中的硫代硫酸盐醌氧化酶(TQO)氧化成连四硫酸盐[69],连四硫酸盐则进一步被连四硫酸盐水解酶(TetH)氧化成硫代硫酸盐、硫酸盐和硫单质[70]。TQO和TetH广泛存在于嗜酸硫氧化细菌中,如 Acidithiobacillus spp.和 Sulfobacillus spp.[70]。此外,具有铁氧化能力的 At. ferrooxidans 和 At. ferrivorans 中还含有硫代硫酸盐脱氢酶(Tsd)[35, 71]。
2.2.2 硫还原代谢: 硫还原微生物在生物冶金体系中扮演着关键的角色,这部分微生物通常以有机物为电子供体,还原高价态的无机硫化合物。冶金体系中硫还原细菌主要包括 Desulfovibrio 属等[72],而古菌主要包括 Acidianus 属[62, 73-74]等。异养的硫还原菌在厌氧条件下,可以利用有机酸和短链醇作为电子供体,在细胞质中将硫酸还原成硫化物[8-9]。在 Desulfovibrio 属中,有机物的电子首先经QmoA/B跨膜复合体将电子传递到ApsA/B还原酶,并将硫酸根还原为亚硫酸根,然后再通过DsrAB/C还原酶以H2为电子供体,进一步将亚硫酸根还原为硫化物,但H2的电子传递路径尚不清楚[72, 75]。在古菌 Acidianus 属基因组中,Urbieta等发现了在其细胞周质中含有Ni/Fe氢化酶,并认为该酶在H2电子转移到硫化合物中发挥了关键作用[62, 74]。另外,铁硫氧化细菌 At.ferrooxidans 在厌氧条件下也具有硫还原的能力,基因组分析表明,At.ferrooxidans 具有编码硫还原酶系统的基因 sreABCD [46],该基因簇可编码细胞内膜上的Sre硫还原酶,并使细胞质中S0还原成H2S[76]。目前,许多硫还原菌的基因组已得到解析,接下来应加强研究硫还原菌与其他菌群的相互作用及其在生态系统的贡献。
3 微生物代谢耦合与矿物的相互作用 硫化矿物的氧化主要分为两种途径,包括硫代硫酸盐途径和多硫化物途径,这两种途径均会产生大量的RISCs中间产物,为硫氧化微生物提供大量的能源物质[77-78]。而硫氧化微生物与其他微生物的代谢耦合,构建起冶金系统中高效的生态功能网络(图 1),可加快矿物的溶解过程[9, 79]。首先,铁氧化微生物氧化亚铁离子生成铁离子,铁离子则作为强氧化剂进攻硫化矿,并被还原成亚铁离子,同时硫化矿物的有价金属溶解和生成RISCs[78, 80];硫氧化微生物进一步将RISCs氧化分解,减少硫膜对硫化矿物溶解的抑制作用,提高硫化矿物的溶解浸出效率[80-81];而铁硫氧化微生物生成的有机物进一步被异养或混合营养型微生物降解,从而降低有机物对铁硫自养微生物的抑制作用[80, 82-84]。选择合适的微生物功能种群,提高生物冶金微生物群落的物种和代谢多样性,是构建高效生态功能网络的关键[30, 80]。
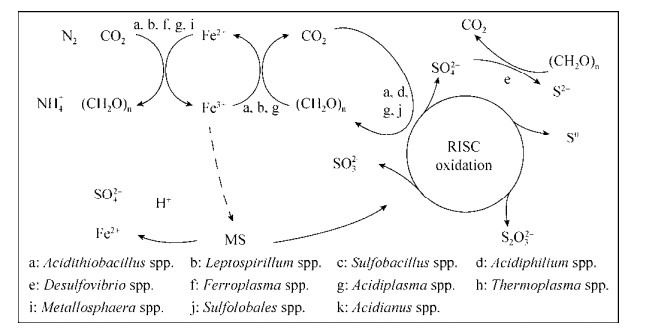 |
| 图 1 生物冶金体系微生物代谢功能网络(MS:金属硫化物;RISC:还原型无机硫化物)[8, 54, 79] Figure 1 Metabolic functionnetwork of microbial community in bioleaching system (MS: metal sulfides; RISC:reduced inorganic sulfur compounds)[8, 54, 79]. |
| 图选项 |
在自然冶金系统的调查中,我们发现江西德兴浸矿堆中的微生物群落比浸出液具有更高的物种多样性,并对黄铁矿有更好的浸出效果[29]。进一步的分析表明,与浸出液微生物群落相比,浸矿堆中除了包含主导微生物 Acidithiobacillus 属和 Leptospirillum 属之外,还具有相对较高丰度的异养微生物,包括 Hydrotalea 属[85]、Frateuria 属[86]、Thermogymnomonas 属[86]和 Thiomonas 属[87],这些菌属可降低有机物对铁硫氧化自养菌的抑制作用。然而,在福建紫金生物冶金系统中,我们却发现了相反的结果,物种多样性较低的浸出堆微生物群落反而有着更高的黄铁矿浸出效率,这可能由于浸出堆系统的细菌群落中拥有更高丰度的铁硫氧化细菌[30]。在德兴浸矿堆与紫金浸矿堆的细菌群落分析比较中,我们发现前者多样性(H=4.4)是后者的(H=3.0)1.5倍,而多样性的这部分差异主要源于上述德兴浸矿堆中异养菌的贡献。在进一步的黄铜矿浸出实验中,我们发现德兴浸矿堆中微生物群落的浸出时间比紫金浸矿堆大约提前了15d,显著提高了浸出效率[29-30]。这表明了生物冶金高效的生态功能网络可能并非只依赖于物种多样性的增加,而是功能微生物种群、代谢途径多样性的有机组合[88-89]。
冶金微生物代谢耦合的功能网络模型,在指导人工构建的高效冶金共培养体系中发挥了重要的作用[9-10]。我们将铁硫氧化细菌 Acidithiobacillusferrooxidans 和异养菌 Acidiphiliumacidophilum 能显著提高整个体系的碳固定速率、铁氧化速率[83]和黄铜矿的浸出速率,并加强了体系对重金属离子的抗性[82]。Li等[90]也发现了铁硫氧化微生物共培养体系比铁氧化微生物具有更高的铀矿浸出速率。此外,我们也开展了铁硫氧化微生物共培养体系中微生物配比对黄铜矿浸出效果的研究,研究结果显示硫氧化微生物比例高的体系中拥有更好的浸出效果[88],这表明了构建高效的共培养体系需要功能微生物特定比例的组合,同时也说明了硫代谢在生物冶金微生物代谢功能网络中的重要性。另一方面,在功能网络的氮循环中,目前只发现了少部分的中温菌能固定大气中的N2,如 At.ferrooxidans 和 Leptospirillum 属[23-24, 35],并未发现极端嗜热的固氮微生物。这暗示了氮源的输入可能会成为生物冶金过程中功能网络运转的重要速限步骤,进而降低硫化矿的浸出速率,添加氮源或引入氮固定功能菌群可能会加速整个群落的物质运转速率[91]。
在生物冶金系统的底泥中,硫还原菌如 Syntrophobacter 属、Desulfosporosinus 属和 Desulfurella 属等,通常占有较高的丰度[92],但由于他们并非直接参与矿物氧化溶解过程,目前对这部分菌属及其在生态功能网络中的作用研究较少。此外,冶金系统中还存在少数的真菌物种[93],这部分真菌亦可通过分泌有机酸参与矿物溶解的过程,如 Aspergillu 和 Penicillium [94]。特别地,也有研究报道了一种具有亚铁氧化酶活性的真菌 Acidomycesacidophilus [95]。这些真菌物种丰富了构建生物冶金微生物功能群和代谢功能网络的选择,但真菌是如何与细菌发生协同作用并形成一个高效的生态功能网络,仍需进一步深入的研究。
4 展望 生物冶金过程中,硫化矿物的生物溶解是一个微生物代谢相互耦合作用的过程,从微生物铁、硫氧化代谢多样性及其耦合功能网络的角度来研究生物冶金的作用机理,还需要强化微生物学、生物信息学和物理化学的结合。未来需要加强以下方面的研究。
(1) 冶金系统中的微生物多样性十分丰富,它们不仅和生物冶金工业应用和矿山环境修复有关,还和极端环境下物种的进化过程与生态过程密切相关。目前,生物冶金系统中仍存在大量未培养且稀有的物种。尽管利用高通量测序、宏基因组、宏转录分析等方法可以避开培养过程,有效分析环境中的微生物组成,预测微生物的代谢途径,但不能通过实验证实未知微生物的生理过程,限制了对于微生物相互耦合、协同作用机制的深入研究。要探索未培养和稀有微生物在生态功能网络中发挥的作用,解析其在生物冶金体系中物种及代谢多样性,需要进一步加强对未培养和稀有物种的鉴定及基因组信息的解析,探寻新的能量代谢途径以完善、构建高效的生态功能网络。
(2) 在生物冶金系统的功能代谢网络中,铁硫循环在物质能量循环中发挥了重要作用。但铁硫循环究竟是怎样与其他碳、氮、磷循环耦合的,其中的关键速率限制环节又是哪个?构建一个模式群落去回答这些科学问题,并探明微生物代谢网络的耦合及其与矿物相互作用的过程,对调控生物冶金微生物群落功能和阐明一些微生物生态学的问题都是极具科学价值的。
References
| [1] | Chen BW, Wen JK, Liu WY. Progress in identification of bioleaching microorganisms. China Mining Magazine, 2007, 16(9): 103-106. (in Chinese) 陈勃伟, 温建康, 刘文彦. 浸矿微生物鉴定研究进展. 中国矿业, 2007, 16(9): 103-106. |
| [2] | Rawlings DE, Johnson DB. The microbiology of biomining:development and optimization of mineral-oxidizing microbial consortia. Microbiology, 2007, 153(2): 315-324. DOI:10.1099/mic.0.2006/001206-0 |
| [3] | Li HX, Wang DZ. Review of investigation on microorganism behaviors in ore bio-leaching. Nonferrous Metals, 2003, 55(2): 58-63. (in Chinese) 李宏煦, 王淀佐. 生物冶金中的微生物及其作用. 有色金属, 2003, 55(2): 58-63. |
| [4] | Watling HR. The bioleaching of sulphide minerals with emphasis on copper sulphides-a review. Hydrometallurgy, 2006, 84(1/2): 81-108. |
| [5] | Akcil A, Koldas S. Acid mine drainage (AMD):causes, treatment and case studies. Journal of Cleaner Production, 2006, 14(12/13): 1139-1145. |
| [6] | Baker BJ, Banfield JF. Microbial communities in acid mine drainage. FEMS Microbiology Ecology, 2003, 44(2): 139-152. DOI:10.1016/S0168-6496(03)00028-X |
| [7] | Kuang JL, Huang LN, Chen LX, Hua ZS, Li SJ, Hu M, Li JT, Shu WS. Contemporary environmental variation determines microbial diversity patterns in acid mine drainage. The ISME Journal, 2013, 7(5): 1038-1050. DOI:10.1038/ismej.2012.139 |
| [8] | Chen LX, Huang LN, Méndez-García C, Kuang JL, Hua ZS, Liu J, Shu WS. Microbial communities, processes and functions in acid mine drainage ecosystems. Current Opinion in Biotechnology, 2016, 38: 150-158. DOI:10.1016/j.copbio.2016.01.013 |
| [9] | Zhang X, Niu JJ, Liang YL, Liu XD, Yin HQ. Metagenome-scale analysis yields insights into the structure and function of microbial communities in a copper bioleaching heap. BMC Genetics, 2016, 17: 21. |
| [10] | Bosse M, Heuwieser A, Heinzel A, Nancucheo I, Dall'Agnol HMB, Lukas A, Tzotzos G, Mayer B. Interaction networks for identifying coupled molecular processes in microbial communities. Biodata Mining, 2015, 8: 21. DOI:10.1186/s13040-015-0054-4 |
| [11] | Méndez-García C, Peláez AI, Mesa V, Sánchez J, Golyshina OV, Ferrer M. Microbial diversity and metabolic networks in acid mine drainage habitats. Frontiers in Microbiology, 2015, 6: 475. |
| [12] | Qi H, Guo X, Liang YL, Hao XD, Ma LY, Yin HQ, Liu XD. Comparative metagenomics reveals microbial community differentiation in a biological heap leaching system. Research in Microbiology, 2015, 166(6): 525-534. DOI:10.1016/j.resmic.2015.06.005 |
| [13] | He ZG, Xiao SM, Xie XH, Hu YH. Microbial diversity in acid mineral bioleaching systems of dongxiang copper mine and Yinshan lead-zinc mine. Extremophiles, 2008, 12(2): 225-234. DOI:10.1007/s00792-007-0130-x |
| [14] | Panda S, Akcil A, Pradhan N, Deveci H. Current scenario of chalcopyrite bioleaching:a review on the recent advances to its heap-leach technology. Bioresource Technology, 2015, 196: 694-706. DOI:10.1016/j.biortech.2015.08.064 |
| [15] | Lang XF, Qiu LN, Ma X, Gong AJ, Wang XN. Bacteria applied in biohydrometallurgy. Metal World, 2009(Z1): 88-91. (in Chinese) 郎序菲, 邱丽娜, 马雪, 弓爱君, 王小宁. 浸矿微生物的研究进展. 金属世界, 2009(Z1): 88-91. DOI:10.3969/j.issn.1000-6826.2009.z1.026 |
| [16] | Dopson M, Johnson DB. Biodiversity, metabolism and applications of acidophilic sulfur-metabolizing microorganisms. Environmental Microbiology, 2012, 14(10): 2620-2631. DOI:10.1111/emi.2012.14.issue-10 |
| [17] | Norlund KL, Southam G, Tyliszczak T, Hu Y, Karunakaran C, Obst M, Hitchcock AP, Warren LA. Microbial architecture of environmental sulfur processes:a novel syntrophic sulfur-metabolizing consortia. Environmental Science & Technology, 2009, 43(23): 8781-8786. |
| [18] | Johnson DB. Biomining-biotechnologies for extracting and recovering metals from ores and waste materials. Current Opinion in Biotechnology, 2014, 30: 24-31. DOI:10.1016/j.copbio.2014.04.008 |
| [19] | Schippers A, Breuker A, Blazejak A, Bosecker K, Kock D, Wright TL. The biogeochemistry and microbiology of sulfidic mine waste and bioleaching dumps and heaps, and novel Fe(Ⅱ)-oxidizing bacteria. Hydrometallurgy, 2010, 104(3/4): 342-350. |
| [20] | Feng SS, Yang HL, Xin Y, Zhang L, Kang WL, Wang W. Isolation of an extremely acidophilic and highly efficient strai. Acidithiobacillus sp. for chalcopyrite bioleaching. Journal of Industrial Microbiology & Biotechnology, 2012, 39(11): 1625-1635. |
| [21] | Hippe H. Leptospirillum gen. (ex Markosyan 1972), nom. rev., includin. Leptospirillum ferrooxidans sp. nov. (ex Markosyan 1972), nom. rev. an. Leptospirillum thermoferrooxidans sp. nov. (Golovachev. et al . 1992). International Journal of Systematic and Evolutionary Microbiology, 2000, 50: 501-503. DOI:10.1099/00207713-50-2-501 |
| [22] | Coram NJ, Rawlings DE. Molecular relationship between two groups of the genu. Leptospirillum and the finding tha. Leptospirillum ferriphilum sp. nov. dominates South African commercial biooxidation tanks that operate at 40℃. Applied and Environmental Microbiology, 2002, 68(2): 838-845. DOI:10.1128/AEM.68.2.838-845.2002 |
| [23] | Goltsman DSA, Denef VJ, Singer SW, VerBerkmoes NC, Lefsrud M, Mueller RS, Dick GJ, Sun CL, Wheeler KE, Zemla A, Baker BJ, Hauser L, Land M, Shah MB, Thelen MP, Hettich RL, Banfield JF. Community genomic and proteomic analyses of chemoautotrophic iron-oxidizing " Leptospirillum rubarum " (Group Ⅱ) and " Leptospirillum ferrodiazotrophum " (Group Ⅲ) bacteria in acid mine drainage biofilms. Applied and Environmental Microbiology, 2009, 75(13): 4599-4615. DOI:10.1128/AEM.02943-08 |
| [24] | Tyson GW, Lo I, Baker BJ, Allen EE, Hugenholtz P, Banfield JF. Genome-directed isolation of the key nitrogen fixe. Leptospirillum ferrodiazotrophum sp. nov. from an acidophilic microbial community. Applied and Environmental Microbiology, 2005, 71(10): 6319-6324. DOI:10.1128/AEM.71.10.6319-6324.2005 |
| [25] | Goltsman DSA, Dasari M, Thomas BC, Shah MB, Verberkmoes NC, Hettich RL, Banfield JF. New Group Ⅰn th. Leptospirillum clade:cultivation-independent community genomics, proteomics, and transcriptomics of the new species " Leptospirillum Group Ⅳ UBA BS". Applied and Environmental Microbiology, 2013, 79(17): 5384-5393. DOI:10.1128/AEM.00202-13 |
| [26] | Harrison AP. Acidiphilium cryptum gen. nov., sp. nov., heterotrophic bacterium from acidic mineral environments. International Journal of Systematic and Evolutionary Microbiology, 1981, 31(3): 327-332. |
| [27] | Hiraishi A, Nagashima KVP, Matsuura K, Shimada K, Takaichi S, Wakao N, Katayama Y. Phylogeny and photosynthetic features o. Thiobacillus acidophilus and related acidophilic bacteria:its transfer to the genu. Acidiphilium a. Acidiphilium acidophilum comb. nov. International Journal of Systematic and Evolutionary Microbiology, 1998, 48(4): 1389-1398. |
| [28] | Cárdenas JP, Valdés J, Quatrini R, Duarte F, Holmes DS. Lessons from the genomes of extremely acidophilic bacteria and archaea with special emphasis on bioleaching microorganisms. Applied Microbiology and Biotechnology, 2010, 88(3): 605-620. DOI:10.1007/s00253-010-2795-9 |
| [29] | Xiao YH, Liu XD, Liang YL, Niu JJ, Zhang X, Ma LY, Hao XD, Gu YB, Yin HQ. Insights into functional genes and taxonomical/phylogenetic diversity of microbial communities in biological heap leaching system and their correlation with functions. Applied Microbiology and Biotechnology, 2016, 100(22): 9745-9756. DOI:10.1007/s00253-016-7819-7 |
| [30] | Xiao YH, Liu XD, Ma LY, Liang YL, Niu JJ, Gu YB, Zhang X, Hao XD, Dong WL, She SY, Yin HQ. Microbial communities from different subsystems in biological heap leaching system play different roles in iron and sulfur metabolisms. Applied Microbiology and Biotechnology, 2016, 100(15): 6871-6880. DOI:10.1007/s00253-016-7537-1 |
| [31] | Denef VJ, Mueller RS, Banfield JF. AMD biofilms:using model communities to study microbial evolution and ecological complexity in nature. The ISME Journal, 2010, 4(5): 599-610. DOI:10.1038/ismej.2009.158 |
| [32] | Bonnefoy V, Holmes DS. Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments. Environmental Microbiology, 2012, 14(7): 1597-1611. DOI:10.1111/j.1462-2920.2011.02626.x |
| [33] | Amouric A, Brochier-Armanet C, Johnson DB, Bonnefoy V, Hallberg KB. Phylogenetic and genetic variation among Fe(Ⅱ)-oxidizing acidithiobacilli supports the view that these comprise multiple species with different ferrous iron oxidation pathways. Microbiology, 2011, 157(1): 111-122. DOI:10.1099/mic.0.044537-0 |
| [34] | Zhang X, Liu XD, Liang YL, Xiao YH, Ma LY, Guo X, Miao B, Liu HW, Peng DL, Huang WK, Yin HQ. Comparative genomics unravels the functional roles of co-occurring acidophilic bacteria in bioleaching heaps. Frontiers in Microbiology, 2017, 8: 790. DOI:10.3389/fmicb.2017.00790 |
| [35] | Zhang X, Liu XD, Liang YL, Fan FL, Zhang XX, Yin HQ. Metabolic diversity and adaptive mechanisms of iron-and/or sulfur-oxidizing autotrophic acidophiles in extremely acidic environments. Environmental Microbiology Reports, 2016, 8(5): 738-751. DOI:10.1111/1758-2229.12435 |
| [36] | Parro V, Moreno-Paz M. Gene function analysis in environmental isolates:th. nif regulon of the strict iron oxidizing bacteriu. Leptospirillum ferrooxidans. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(13): 7883-7888. DOI:10.1073/pnas.1230487100 |
| [37] | Justice NB, Norman A, Brown CT, Singh A, Thomas BC, Banfield JF. Comparison of environmental and isolat. Sulfobacillus genomes reveals diverse carbon, sulfur, nitrogen, and hydrogen metabolisms. BMC Genomics, 2014, 15: 1107. DOI:10.1186/1471-2164-15-1107 |
| [38] | Guo X, Yin HQ, Liang YL, Hu Q, Zhou XS, Xiao YH, Ma LY, Zhang X, Qiu GZ, Liu XD. Comparative genome analysis reveals metabolic versatility and environmental adaptations o. Sulfobacillus thermosulfidooxidans strain ST. PLoS One, 2014, 9(6): e99417. DOI:10.1371/journal.pone.0099417 |
| [39] | Zhang X, Liu XD, Liang YL, Guo X, Xiao YH, Ma LY, Miao B, Liu HW, Peng DL, Huang WK, Zhang YG, Yin HQ. Adaptive evolution of extreme acidophil. Sulfobacillus thermosulfidooxidans potentially driven by horizontal gene transfer and gene loss. Applied and Environmental Microbiology, 2017, 83(7): e03098-16. |
| [40] | Golyshina OV, Yakimov MM, Lünsdorf H, Ferrer M, Nimtz M, Timmis KN, Wray V, Tindall BJ, Golyshin PN. Acidiplasma aeolicum gen. nov., sp. nov., a euryarchaeon of the family Ferroplasmaceae isolated from a hydrothermal pool, and transfer o. Ferroplasma cupricumulans t. Acidiplasma cupricumulans comb. nov. International Journal of Systematic and Evolutionary Microbiology, 2009, 59(11): 2815-2823. DOI:10.1099/ijs.0.009639-0 |
| [41] | Bulaev AG, Kanygina AV, Manolov AI. Genome analysis o. Acidiplasma sp. MBA-1, a polyextremophilic archaeon predominant in the microbial community of a bioleaching reactor. Microbiology, 2017, 86(1): 89-95. DOI:10.1134/S0026261716060059 |
| [42] | Bulaev A. Ferrous iron oxidation in packed-bed reactors at elevated temperatures. Advanced Materials Research, 2015, 1130: 226-229. DOI:10.4028/www.scientific.net/AMR.1130 |
| [43] | Ullrich SR, Poehlein A, Tischler JS, González C, Ossandon FJ, Daniel R, Holmes DS, Schl mann M, Mühling M. Genome analysis of the biotechnologically relevant acidophilic iron oxidising strain JA12 indicates phylogenetic and metabolic diversity within the novel genus " Ferrovum ". PLoS One, 2016, 11(1): e0146832. DOI:10.1371/journal.pone.0146832 |
| [44] | Sugio T, Taha TM, Takeuchi F. Ferrous iron production mediated by tetrathionate hydrolase in tetrathionate-, sulfur-, and iron-grow. Acidithiobacillus ferrooxidans ATCC 23270 Cells. Bioscience, Biotechnology and Biochemistry, 2009, 73(6): 1381-1386. DOI:10.1271/bbb.90036 |
| [45] | Mo HY, Chen Q, Du J, Tang L, Qin F, Miao B, Wu XL, Zeng J. Ferric reductase activity of the ArsH protein fro. Acidithiobacillus ferrooxidans. Journal of Microbiology and Biotechnology, 2011, 21(5): 464-469. DOI:10.4014/jmb |
| [46] | Osorio H, Mangold S, Denis Y, ancucheo I, Esparza M, Johnson DB, Bonnefoy V, Dopson M, Holmes DS. Anaerobic sulfur metabolism coupled to dissimilatory iron reduction in the extremophil. Acidithiobacillus ferrooxidans. Applied and Environmental Microbiology, 2013, 79(7): 2172-2181. DOI:10.1128/AEM.03057-12 |
| [47] | Bogdanova TI, Tsaplina IA, Kondrat'Eva TF, Duda VI, Suzina NE, Melamud VS, Tourova TP, Karavaiko GI. Sulfobacillus thermotolerans sp. nov., a thermotolerant, chemolithotrophic bacterium. International Journal of Systematic and Evolutionary Microbiology, 2006, 56(5): 1039-1042. DOI:10.1099/ijs.0.64106-0 |
| [48] | Melamud VS, Pivovarova TA, Tourova TP, Kolganova TV, Osipov GA, Lysenko AM, Kondrat'Eva TF, Karavaiko GI. Sulfobacillus sibiricus sp. nov., a new moderately thermophilic bacterium. Microbiology, 2003, 72(5): 605-612. DOI:10.1023/A:1026007620113 |
| [49] | Johnson DB, Joulian C, d'Hugues P, Hallberg KB. Sulfobacillus benefaciens sp. nov., an acidophilic facultative anaerobi. Firmicute isolated from mineral bioleaching operations. Extremophiles, 2008, 12(6): 789-798. DOI:10.1007/s00792-008-0184-4 |
| [50] | Bridge TAM, Johnson DB. Reduction of soluble iron and reductive dissolution of ferric iron-containing minerals by moderately thermophilic iron-oxidizing bacteria. Applied and Environmental Microbiology, 1998, 64(6): 2181-2186. |
| [51] | Karavaiko GI, Krasil'Nikova EN, Tsaplina IA, Bogdanova TI, Zakharchuk LM. Growth and carbohydrate metabolism of sulfobacilli. Microbiology, 2001, 70(3): 245-250. DOI:10.1023/A:1010463007138 |
| [52] | Coupland K, Johnson DB. Evidence that the potential for dissimilatory ferric iron reduction is widespread among acidophilic heterotrophic bacteria. FEMS Microbiology Letters, 2008, 279(1): 30-35. DOI:10.1111/fml.2008.279.issue-1 |
| [53] | Küsel K, Dorsch T, Acker G, Stackebrandt E. Microbial reduction of Fe(Ⅲ) in acidic sediments:isolation o. Acidiphilium cryptum JF-5 capable of coupling the reduction of Fe(Ⅲ) to the oxidation of glucose. Applied and Environmental Microbiology, 1999, 65(8): 3633-3640. |
| [54] | Méndez-García C, Mesa V, Sprenger RR, Richter M, Diez MS, Solano J, Bargiela R, Golyshina OV, Manteca , Ramos JL, Gallego JR, Llorente I, Martins dos Santos VA, Jensen ON, Peláez AI, Sánchez J, Ferrer M. Microbial stratification in low pH oxic and suboxic macroscopic growths along an acid mine drainage. The ISME Journal, 2014, 8(6): 1259-1274. DOI:10.1038/ismej.2013.242 |
| [55] | Mangold S, Valdés J, Holmes DS, Dopson M. Sulfur metabolism in the extreme acidophil. Acidithiobacillus caldus. Frontiers in Microbiology, 2011, 2: 17. |
| [56] | Janosch C, Thyssen C, Vera MA, Bonnefoy V, Rohwerder T, Sand W. Sulfur oxygenase reductase in differen. Acidithiobacillus. caldus -like strains. Advanced Materials Research, 2009, 71-73: 239-242. DOI:10.4028/www.scientific.net/AMR.71-73 |
| [57] | Zhou D, Peng TJ, Zhou HB, Liu XD, Gu GH, Chen M, Qiu GZ, Zeng WM. Expression of critical sulfur-and iron-oxidation genes and the community dynamics during bioleaching of chalcopyrite concentrate by moderate thermophiles. Current Microbiology, 2015, 71(1): 62-69. DOI:10.1007/s00284-015-0817-7 |
| [58] | Yin HQ, Zhang X, Li XQ, He ZL, Liang YL, Guo X, Hu Q, Xiao YH, Cong J, Ma LY, Niu JJ, Liu XD. Whole-genome sequencing reveals novel insights into sulfur oxidation in the extremophil. Acidithiobacillus thiooxidans. BMC Microbiology, 2014, 14: 179. DOI:10.1186/1471-2180-14-179 |
| [59] | You XY, Guo X, Zheng HJ, Zhang MJ, Liu LJ, Zhu YQ, Zhu BL, Wang SY, Zhao GP, Poetsch A, Jiang CY, Liu SJ. Unraveling th. Acidithiobacillus caldus complete genome and its central metabolisms for carbon assimilation. Journal of Genetics and Genomics, 2011, 38(6): 243-252. DOI:10.1016/j.jgg.2011.04.006 |
| [60] | Zhang X, Liu XD, He Q, Dong WL, Zhang XX, Fan FL, Peng DL, Huang WK, Yin HQ. Gene turnover contributes to the evolutionary adaptation o. Acidithiobacillus caldus :insights from comparative genomics. Frontiers in Microbiology, 2016, 7: 1960. |
| [61] | Dai X, Wang HN, Zhang ZF, Li K, Zhang XL, Mora-López M, Jiang CY, Liu C, Wang L, Zhu YX, Hernández-Ascencio W, Dong ZY, Huang L. Genome sequencing o. Sulfolobus sp. A20 from costa rica and comparative analyses of the putative pathways of carbon, nitrogen, and sulfur metabolism in variou. Sulfolobus strains. Frontiers in Microbiology, 2016, 7: 1902. |
| [62] | Urbieta MS, Rascovan N, Vázquez MP, Donati E. Genome analysis of the thermoacidophilic archaeo. Acidianus copahuensis focusing on the metabolisms associated to biomining activities. BMC Genomics, 2017, 18: 445. DOI:10.1186/s12864-017-3828-x |
| [63] | Jones DS, Albrecht HL, Dawson KS, Schaperdoth I, Freeman KH, Pi YD, Pearson A, Macalady JL. Community genomic analysis of an extremely acidophilic sulfur-oxidizing biofilm. The ISME Journal, 2012, 6(1): 158-170. DOI:10.1038/ismej.2011.75 |
| [64] | Jiang CY, Liu LJ, Guo X, You XY, Liu SJ, Poetsch A. Resolution of carbon metabolism and sulfur-oxidation pathways o. Metallosphaera cuprina Ar-4 via comparative proteomics. Journal of Proteomics, 2014, 109: 276-289. DOI:10.1016/j.jprot.2014.07.004 |
| [65] | Hedderich R, Klimmek O, Kr ger A, Dirmeier R, Keller M, Stetter KO. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiology Reviews, 1998, 22(5): 353-381. DOI:10.1111/j.1574-6976.1998.tb00376.x |
| [66] | Grein F, Ramos AR, Venceslau SS, Pereira IAC. Unifying concepts in anaerobic respiration:insights from dissimilatory sulfur metabolism. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 2013, 1827(2): 145-160. DOI:10.1016/j.bbabio.2012.09.001 |
| [67] | Strittmatter AW, Liesegang H, Rabus R, Decker I, Amann J, Andres S, Henne A, Fricke WF, Martinez-Arias R, Bartels D, Goesmann A, Krause L, Pühler A, Klenk HP, Richter M, Schüler M, Gl ckner FO, Meyerdierks A, Gottschalk G, Amann R. Genome sequence o. Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environmental Microbiology, 2009, 11(5): 1038-1055. DOI:10.1111/emi.2009.11.issue-5 |
| [68] | Zimmermann P, Laska S, Kletzin A. Two modes of sulfite oxidation in the extremely thermophilic and acidophilic archaeon acidianus ambivalens. Archives of Microbiology, 1999, 172(2): 76-82. DOI:10.1007/s002030050743 |
| [69] | Chi A, Valenzuela L, Beard S, Mackey AJ, Shabanowitz J, Hunt DF, Jerez CA. Periplasmic Proteins of the Extremophil. Acidithiobacillus ferrooxidans :A high throughput proteomics analysis*S. Molecular & Cellular Proteomics, 2007, 6(12): 2239-2251. |
| [70] | Kanao T, Kamimura K, Sugio T. Identification of a gene encoding a tetrathionate hydrolase i. Acidithiobacillus ferrooxidans. Journal of Biotechnology, 2007, 132(1): 16-22. DOI:10.1016/j.jbiotec.2007.08.030 |
| [71] | Kikumot M, Nogami S, Kanao T, Takada J, Kamimura K. Tetrathionate-Forming thiosulfate dehydrogenase from the acidophilic, chemolithoautotrophic bacteriu. Acidithiobacillus. ferrooxidans. Applied and Environmental Microbiology, 2013, 79(1): 113-120. DOI:10.1128/AEM.02251-12 |
| [72] | Zane GM, Yen HCB, Wall JD. Effect of the deletion o. qmoABC and the promoter-distal gene encoding a hypothetical protein on sulfate reduction i. Desulfovibrio vulgaris hildenborough. Applied and Environmental Microbiology, 2010, 76(16): 5500-5509. DOI:10.1128/AEM.00691-10 |
| [73] | Dannenberg S, Kroder M, Dilling W, Cypionka H. Oxidation of H2, organic compounds and inorganic sulfur compounds coupled to reduction of O2 or nitrate by sulfate-reducing bacteria. Archives of Microbiology, 1992, 158(2): 93-99. DOI:10.1007/BF00245211 |
| [74] | Giaveno MA, Urbieta MS, Ulloa JR, Toril EG, Donati ER. Physiologic versatility and growth flexibility as the main characteristics of a novel thermoacidophili. Acidianus strain isolated from Copahue geothermal area in Argentina. Microbial Ecology, 2013, 65(2): 336-346. DOI:10.1007/s00248-012-0129-4 |
| [75] | Pereira IAC, Ramos AR, Grein F, Marques MC, Da Silva SM, Venceslau SS. A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Frontiers in Microbiology, 2011, 2: 69. |
| [76] | Guiral M, Tron P, Aubert C, Gloter A, Iobbi-Nivol C, Giudici-Orticoni MT. A membrane-bound multienzyme, hydrogen-oxidizing, and sulfur-reducing complex from the hyperthermophilic bacteriu. Aquifex aeolicus. Journal of Biological Chemistry, 2005, 280(51): 42004-42015. DOI:10.1074/jbc.M508034200 |
| [77] | Luther Ⅲ GW. Pyrite oxidation and reduction:Molecular orbital theory considerations. Geochimica et Cosmochimica Acta, 1987, 51(12): 3193-3199. DOI:10.1016/0016-7037(87)90127-X |
| [78] | Schippers A, Sand W. Bacterial leaching of metal sulfides proceeds by two indirect mechanisms via thiosulfate or via polysulfides and sulfur. Applied and Environmental Microbiology, 1999, 65(1): 319-321. |
| [79] | Niu JJ, Deng J, Xiao YH, He ZL, Zhang X, van Nostrand JD, Liang YL, Deng Y, Liu XD, Yin HQ. The shift of microbial communities and their roles in sulfur and iron cycling in a copper ore bioleaching system. Scientific Reports, 2016, 6: 34744. DOI:10.1038/srep34744 |
| [80] | Vera M, Schippers A, Sand W. Progress in bioleaching:fundamentals and mechanisms of bacterial metal sulfide oxidation——part A. Applied Microbiology and Biotechnology, 2013, 97(17): 7529-7541. DOI:10.1007/s00253-013-4954-2 |
| [81] | McGuire MM, Edwards KJ, Banfield JF, Hamers RJ. Kinetics, surface chemistry, and structural evolution of microbially mediated sulfide mineral dissolution. Geochimica et Cosmochimica Acta, 2001, 65(8): 1243-1258. DOI:10.1016/S0016-7037(00)00601-3 |
| [82] | Liu HW, Dai YX, Huang W, Yin HQ, Liang YL, Shen L, Liu XD. Influence of acidophilic heterotrophic bacteria on metal resistance and bioleaching b. Acidithiobacillus ferrooxidans . Microbiology, 2012, 39(8): 1069-1078. (in Chinese) 刘宏伟, 戴艳霞, 黄伟, 尹华群, 梁伊丽, 申丽, 刘学端. 嗜酸异养菌对自养菌 Acidithiobacillus ferroxidans 金属离子抗性和生物浸出的影响. 微生物学通报, 2012, 39(8): 1069-1078. |
| [83] | Liu HW, Yin HQ, Dai YX, Dai ZM, Liu Y, Li Q, Jiang HD, Liu XD. The co-culture o. Acidithiobacillus ferrooxidans and Acidiphilium acidophilum enhances the growth, iron oxidation, and CO2 fixation. Archives of Microbiology, 2011, 193(12): 857-866. DOI:10.1007/s00203-011-0723-8 |
| [84] | Fournier D, Lemieux R, Couillard D. Essential interactions betwee. Thiobacillus ferrooxidans and heterotrophic microorganisms during a wastewater sludge bioleaching process. Environmental Pollution, 1998, 101(2): 303-309. DOI:10.1016/S0269-7491(98)00035-9 |
| [85] | Albuquerque L, Rainey FA, Nobre MF, Da Costa MS. Hydrotalea sandarakina sp. nov., isolated from a hot spring runoff, and emended descriptions of the genu. Hydrotalea and the specie. Hydrotalea flava. International Journal of Systematic and Evolutionary Microbiology, 2012, 62(7): 1603-1608. |
| [86] | Joyeux C, Fouchard S, Llopiz P, Neunlist S. Influence of the temperature and the growth phase on the hopanoids and fatty acids content o. Frateuria aurantia (DSMZ 6220). FEMS Microbiology Ecology, 2004, 47(3): 371-379. DOI:10.1016/S0168-6496(03)00302-7 |
| [87] | Slyemi D, Moinier D, Brochier-Armanet C, Bonnefoy V, Johnson DB. Characteristics of a phylogenetically ambiguous, arsenic-oxidizin. Thiomonas sp.,. Thiomonas arsenitoxydans strain 3AsT sp. nov. Archives of Microbiology, 2011, 193(6): 439-449. DOI:10.1007/s00203-011-0684-y |
| [88] | Ma LY, Wang XJ, Feng X, Liang YL, Xiao YH, Hao XD, Yin HQ, Liu HW, Liu XD. Co-culture microorganisms with different initial proportions reveal the mechanism of chalcopyrite bioleaching coupling with microbial community succession. Bioresource Technology, 2017, 223: 121-130. DOI:10.1016/j.biortech.2016.10.056 |
| [89] | Zeng WM, Qiu GZ, Zhou HB, Peng JH, Chen M, Tan SN, Chao WL, Liu XD, Zhang YS. Community structure and dynamics of the free and attached microorganisms during moderately thermophilic bioleaching of chalcopyrite concentrate. Bioresource Technology, 2010, 101(18): 7068-7075. DOI:10.1016/j.biortech.2010.04.003 |
| [90] | Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vazquez-Baeza Y, González A, Morton JT, Mirarab S, Xu ZZ, Jiang LJ, Haroon MF, Kanbar J, Zhu QY, Song SJ, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R. Earth Microbiome Project Consortium. A communal catalogue reveals Earth's multiscale microbial diversity. Nature, 2017, 551(7681): 457-463. |
| [91] | Guo MR, Lin YM, Xu XP, Chen ZL. Bioleaching of iron from kaolin using Fe(Ⅲ)-reducing bacteria with various carbon nitrogen sources. Applied Clay Science, 2010, 48(3): 379-383. DOI:10.1016/j.clay.2010.01.010 |
| [92] | Sánchez-Andrea I, Knittel K, Amann R, Amils R, Sanz JL. Quantification of Tinto River sediment microbial communities:importance of sulfate-reducing bacteria and their role in attenuating acid mine drainage. Applied and Environmental Microbiology, 2012, 78(13): 4638-4645. DOI:10.1128/AEM.00848-12 |
| [93] | Baker BJ, Tyson GW, Goosherst L, Banfield JF. Insights into the diversity of eukaryotes in acid mine drainage biofilm communities. Applied and Environmental Microbiology, 2009, 75(7): 2192-2199. DOI:10.1128/AEM.02500-08 |
| [94] | Valix M, Usai F, Malik R. Fungal bio-leaching of low grade laterite ores. Minerals Engineering, 2001, 14(2): 197-203. DOI:10.1016/S0892-6875(00)00175-8 |
| [95] | Boonen F, Vandamme AM, Etoundi E, Pigneur LM, Housen I. Identification and characterization of a novel multicopper oxidase fro. Acidomyces acidophilus with ferroxidase activity. Biochimie, 2014, 102: 37-46. DOI:10.1016/j.biochi.2014.02.009 |
