张艳敏, 吴耿, 蒋宏忱


中国地质大学(武汉), 生物地质与环境地质国家重点实验室, 湖北 武汉 430074
收稿日期:2017-11-28;修回日期:2018-01-24;网络出版日期:2018-02-05
基金项目:科技基础性工作专项(2015FY110100);国家自然科学基金(41422208,41502318,41521001);科技部国际合作重点项目(2013DFA31980);中央高校基本研科业务费专项
作者简介:蒋宏忱, 博士, 中国地质大学(武汉)生物地质与环境地质国家重点实验室教授, 2007年毕业于美国迈阿密大学, 博士。2012年入选教育部"新世纪优秀人才支持计划", 2014年获国家优秀青年基金。现任中国微生物学会地质微生物专业委员会委员、Frontiers in Microbiology副主编、《盐湖研究》和《微生物学报》编委。致力于盐湖和热泉等极端环境的微生物生态及其相关的生源元素循环过程研究。先后主持了国家基金委重大研究计划重点项目、国家优秀青年基金项目等重要课题, 已在Environmental Microbiology、Applied and Environmental Microbology等专业期刊发表科研论文80余篇。2015年获中国地质学会青年地质科技奖-银锤奖, 2017年获云南省自然科学二等奖
*通信作者:蒋宏忱, Tel:+86-27-67883452;Fax:+86-27-67883451;E-mail:jiangh@cug.edu.cn
摘要:热泉微生物是驱动热泉氮(N)循环的主导力量,开展热泉生态系统中驱动氮循环微生物种群构成及其与环境响应的研究,对于探索热泉中氮的生物地球化学循环、生命进化、生物修复等方面都具有重要的理论和应用价值。本文综合阐述了热泉生态系统驱动氮循环的功能微生物(如固氮菌、氨氧化菌、厌氧氨氧化菌、反硝化菌、异化硝酸盐还原菌)在系统发育学上的分布、功能基因的相对丰度、活性及其与环境因子(如温度、pH)的相关性等方面的研究现状和亟待解决的问题。并展望了热泉生境中驱动氮循环微生物未来的研究方向。
关键词: 热泉微生物 氮(N)循环 功能基因 环境因子
Research progress in microorganisms involved in nitrogen cycles in hot springs
Yanmin Zhang, Geng Wu, Hongchen Jiang


State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, Wuhan 430074, Hubei Province, China
Received 28 November 2017; Revised 24 January 2018; Published online 5 February 2018
*Corresponding author: Hongchen Jiang, Tel: +86-27-67883452; Fax: +86-27-67883451; E-mail: jiangh@cug.edu.cn
Supported by the National Natural Science Foundation of China (41422208, 41502318, 41521001), by the Key Project of International Cooperation of China Ministry of Science and Technology (2013DFA31980) and by the Fundamental Research Funds for the Central Universities, China University of Geosciences (Wuhan)
Abstract: Microorganisms are the dominant players driving the nitrogen cycles in hot springs. Thus, exporing microbial community composition of nitrogen-related microorganisms and their ecological response to environmental factors are of important theoretical and practical value for the study of biogeochemical cycling of nitrogen, life evolution and bioremediation. This review focuses on the distribution of functional microorganisms involved in the nitrogen cyclings and how their functional gene abundance and activity responded to environmental factors (e.g., temperature, pH). So far, microbial functional groups that have been investigated in hot springs included nitrogen-fixation, aerobic (anaerobic) ammonia oxidation and denitrification. Finally, prospect was given on the microbial studies on nitrogen cycles in hot springs.
Key words: microorganisms in hot springs nitrogen cycles functional genes environmental variables
热泉是现存与地球早期环境最为相似的生态系统,也是研究极端环境的模式生态系统之一,具有群落组成简单(主要为细菌和古菌、病毒)、自我维持、生物地球化学过程活跃等特点[1]。在热泉生境中,氮元素是控制初级生产力的关键元素之一。微生物在各种形态氮的转化过程中发挥着重要作用,是驱动氮循环的强大引擎[2](图 1)。因此,对热泉中驱动氮循环微生物的研究既有助于揭示极端环境微生物驱动氮循环功能及其在地热环境改变过程中的重要作用,又有助于获得地球早期生命起源、进化的启示作用。
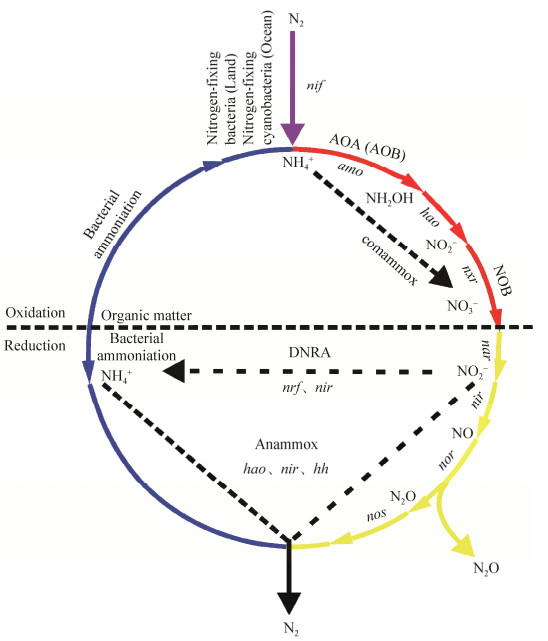 |
| 图 1 微生物驱动的氮循环 Figure 1 Nitrogen cycling driven by microorganisms in hot springs[2]. nif, nitrogenase; amo, ammonia monooxygenase; hao, hydroxyamine oxidoreductase; nxr, nitrate oxidoreductase; nar, nitrate reductase; nir and nrf, nitrite reductase; hh, hydrazine hydrolase; nor, nitric oxide reductase; nos, nitrous oxide reductase. The purple line represented the process of nitrogen fixation, the red line represented the process of nitrification, the yellow line represented the process of denitrification, the blue line represented the process of ammonification, the black line represented the process of anaerobic ammonification, and the dotted line represented the process of dissimilation nitrate reduction. |
| 图选项 |
微生物驱动的氮循环主要包括氮固定作用、硝化作用、反硝化作用、异化硝酸盐还原成铵作用和厌氧氨氧化作用[2](图 1)。相对于其他生境(如土壤、海洋、湖泊),热泉生境中有关氮循环微生物的研究相对较少,本综述将针对热泉中氮循环过程、驱动氮循环的微生物群落组成及其影响因素等方面的研究现状进行阐述,并对其存在的问题和未来的研究方向提出展望。
1 固氮作用 氮固定作用是将N2转化为生物可利用的铵态氮(NH4+)的过程,由固氮微生物执行。固氮微生物在细菌域和古菌域中广泛分布[3]。微生物固氮作用由高度保守的固氮酶nif基因(包括nifH、nifD、nifK)催化完成,其中nifH (固氮酶铁蛋白编码基因)已被广泛用于检测环境中固氮微生物的多样性[4]。据nifH基因构建的系统发育树,可将固氮微生物分为5个类群(Clusters Ⅰ-Ⅴ)。Cluster Ⅰ中大多分布于Proteobacteria和Cyanobacteria门,少量分布于Firmicutes (Paenibacillus)和Actinobacteria (Frankia)门;Cluster Ⅱ中主要包括具有固氮功能的产甲烷古菌;Cluster Ⅲ中主要包括厌氧的细菌或古菌,如产甲烷菌、产醋酸菌、硫酸盐还原细菌及梭菌属等;Cluster Ⅳ和Ⅴ中主要是含有和nifH基因同源的非固氮菌[5]。
热泉中的固氮微生物主要分布于Cluster Ⅰ中的Cyanobacteria、α-和β-proteobacteria门。其中,Cyanobacteria是热泉中分布最广和研究最多的固氮菌。至今,只有少数高温固氮微生物被成功分离,包括从智利热泉中分离出的Mastigocladus sp. strain CHP1(最适生长温度45-50 ℃)[6]及从澳大利亚热泉中分离出的Ewamiania thermalis (生长温度48.5-62.7 ℃)[7],二者均属于Cyanobacteria。目前,从深海热液中分离的类似于Methanocaldococcu sjannaschii的古菌菌株FS406-22C是所有固氮菌中生长温度最高的,可在92 ℃的高温下发挥固氮功能[8]。
热泉中固氮菌的分布受温度和pH的影响。例如,在低温热泉(40-60 ℃)中的优势种群为丝状的异形细胞Mastigocladus属;高温热泉中(>60 ℃)的优势种群为单细胞蓝藻Synechococcus属。后来Alcamán等(2015)证实Mastigocladus属的固氮微生物具有更广泛的温度分布范围(38-69 ℃)[9]。
前人研究证明nifH在热泉环境中广泛存在(温度16.0-89.0 ℃,pH 1.9-9.8),环境因子(如温度、pH)和地理距离等均会影响nifH基因的相对丰度和表达[10-11]。如图 2所示,nifH的相对丰度随温度的升高呈现先升高后降低的趋势,并在温度为50-60 ℃时达到峰值。在pH>5时,nifH基因相对丰度随pH升高呈现升高的趋势,而在pH<5时,只有少数样点中检测到nifH的存在(图 2)。Loiacono等(2012)研究发现温度为57.2-80.2 ℃的中性热泉中均可检测到nifH基因的存在,并且随着温度的降低nifH基因的表达也有所降低。然而,当温度高于72.7 ℃时,并未观察到nifH的表达[4]。由此可见nifH的存在并不完全意味着环境中固氮功能的存在。此外,还有****研究发现微生物固氮作用(如nifH基因丰度和表达)除受温度、pH等环境因子影响外,还遵循一定的昼夜周期[12-13]。因此关于热泉环境微生物固氮作用的影响因素和固氮机制尚有诸多未知领域需要探索。
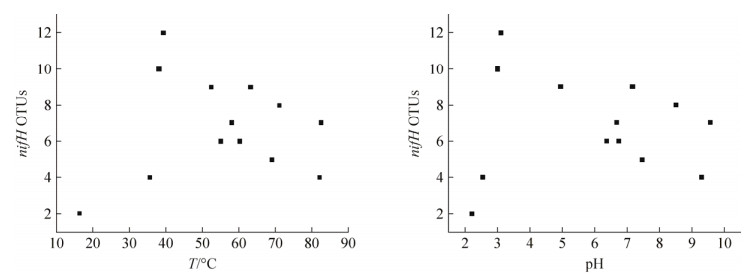 |
| 图 2 热泉nifH基因相对丰度随温度、pH的变化 Figure 2 Relative abundance of nifH gene in response to temperature and pH of hot springs[3]. |
| 图选项 |
2 硝化作用 硝化作用是指氨氧化为硝酸盐的过程(图 1)。前人普遍认为硝化作用需要氨氧化菌和亚硝酸盐氧化菌分工完成。氨氧化作用是硝化作用的第一步,也是限速步骤。氨氧化作用的功能基因amoA编码的氨单加氧酶(ammonia monooxygenase)是催化氨氧化的关键酶,amoA已被广泛用来检测环境样品中氨氧化微生物的群落组成。可执行氨氧化功能的微生物包括氨氧化细菌(ammonia oxidizing bacteria,AOB)和氨氧化古菌(ammonia oxidizing archaea,AOA),属于好氧化能自养或兼性自养型微生物[14]。在很长一段时间内,人们认为AOB是环境中氨氧化功能的主要执行者,直到Treusch等(2005)[15]提出广泛分布于陆地和海洋环境的泉古菌门(Crenarchaeota)微生物中有类似amoA基因的表达,后来的研究证实,在土壤、海洋等环境中,AOA比AOB发挥着更重要的氨氧化作用[16-17]。直到2015年,****们发现Nitrospira (sublineage Ⅱ)属的细菌可单独将氨完全氧化成NO3-,这类微生物被称做“comammox”[18-20]。最新研究发现,在寡营养的环境中,comammox对NH4+的亲和力要高于AOA,比AOA更有竞争力,暗示了comammox是寡营养、低NH4+环境中氨氧化功能的主要执行者[21]。前人研究结果证实AOA、AOB和comammox在不同环境因子的氨氧化功能具有差异性,他们之间是否存在密切的竞争、协作机制还有待进一步的探讨。由于comammox的amoA含有两个单系的基因群(clade A和clade B),与AOA和AOB的amoA基因具有明显差异[19-20],因此,无法通过amoA基因的扩增来检测comammox的存在。目前,已有关于检测环境中comammox群落组成的引物发表[18],这将有助于检测不同环境中comammox的存在和深入了解全球氮循环机制。
2.1 AOB和AOA 热泉中的AOB主要分布在Nitrospira属(Nitrospirae门),生长温度大约为55 ℃[22];AOA主要分布在Crenarchaeota和Thaumarchaeota门[15, 22-24]。迄今为止,热泉中可培养的氨氧化菌只有AOA[25]。可培养AOA仅限自养型的“Candidatus Nitrososphaera gargensis”(温度46.0 ℃)和“Candidatus Nitrosocaldus yellowstonii”(温度60.0-76.0℃),二者均属于Crenarchaeota门,类似于这两类的AOA也是热泉中丰度最高的种群[23, 26-27]。
前人研究发现AOA在热泉环境中广泛存在,且amoA的相对丰度会受到温度、pH和地理距离的影响[28-30](图 3)。如图 3所示,在温度为41.0-94.0℃和在pH为6.0-9.5,amoA的相对丰度随温度或pH的递增均呈现先升高后降低的趋势,而在pH<5时,只有少量amoA被检测到[29-30](图 3)。
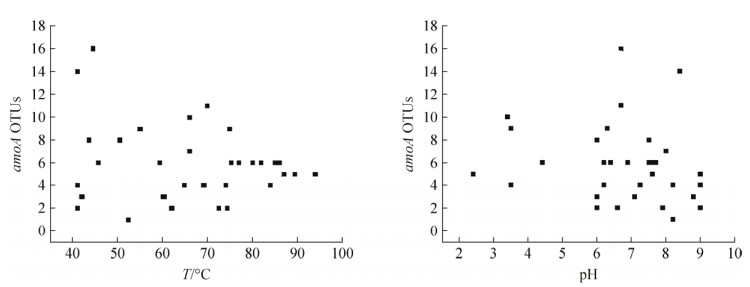 |
| 图 3 热泉amoA基因相对丰度随温度和pH的变化 Figure 3 Relative abundance of amoA gene in response to temperature and pH of hot springs[28, 31]. |
| 图选项 |
****们在关注amoA的存在及相对丰度的同时,也初步探讨了氨氧化功能及其与环境因子(如温度)的响应关系。如Jiang等(2010)[31]研究发现amoA可在94 ℃的高温下发生转录[32]。Li等(2015)[31]发现在50-70 ℃,AOA的氨氧化活性与温度正相关[17-52 pmol NO2-/(cell·d)]。Dodsworth等(2011)[27]发现80.0 ℃中性热泉沉积物中氨氧化速率为5.5-8.6 nmol N/(g·h),同时该研究发现Candidatus Nitrosocaldus yellowstonii的相对丰度最高,为(3.5-6.4)×108基因拷贝/g。
2.2 亚硝酸盐氧化菌(nitrite oxidizing bacteria,NOB) 硝化作用的第二步是将NO2-氧化生成NO3-,由好氧、化能自养型的NOB来完成。催化NO2-氧化的关键酶是NXR(亚硝酸盐氧化还原酶),该酶具有胞浆型(cytoplasmic type)和胞质型(periplasmic type)两个不同的发育谱系,前者主要分布于Nitrobacter、Nitrococcus和Nitrolancetus属,后者主要分布于Nitrospira和Nitrospina属[33]。NXR功能基因由NxrA、NxrB和NxrC组成。目前,可通过nxrB基因的扩增来检测土壤中NOB的存在[33]。热泉中关于NOB的研究非常少,这与NOB难以分离培养有关[33-34]。
常温环境中的NOB主要分布在Nitrospira和Proteobacteria门(α-、β-、δ-Proteobacteria),热泉中的NOB主要分布在Nitrospira门中的Nitrospira属[22, 35-36]。目前,热泉中分离出的NOB纯菌株只有Nitrospira calida(生长温度48 ℃,pH 8.6)[37]。Edwards等(2012)[36]通过培养的方法从美国大盆地、中国和亚美尼亚热泉中富集到了NOB,并证明Nitrospira属NOB的温度上限为60-65 ℃,NO2-氧化速率为7.53-23.00 fmoles/(cell·h),且Nitrospira的分布具有明显的生物地理学特征。
值得指出的是,可单独执行完整硝化过程的comammox均分布于常温的Nitrospira属[18],且前人研究认为comammox在寡营养的环境中更具有竞争力[21],由此推断,在热泉高温、寡营养的环境中很可能有comammox的存在。然而热泉中Nitrospira属的NOB是否具有comammox的特性有待进一步的研究[22, 38]。
3 厌氧氨氧化 厌氧氨氧化是指厌氧条件下以NO2-作为氧化剂,将NH4+氧化成N2的过程,这一过程由厌氧氨氧化菌(Anammox)催化完成。Anammox主要分布于海洋和淡水生态系统,是全球氮循环及废水氮污染物去除的主要驱动者[39-41]。目前,可用厌氧氨氧化第一步的功能基因nirS (Anammox特有的亚硝酸还原酶基因)来检测环境中Anammox的存在[41-43]。
Anammox生长缓慢且难培养,至今无纯培养物,只能以富集产物的形式生长于序批式反应器(SBR)或生物膜反应器(MBR)中[44]。Anammox在海洋和淡水生态系统中主要分布在浮霉状菌目(Planctomycetales)的5个属Candidatus Brocadia, Candidatus Kuenenia,Candidatus Scalindua,Candidatus Jettenia和Candidatus Anammoxoglobus[45]。热泉中至今未有可培养Anammox的报道。
Jaeschke等(2009)[46]通过16S rRNA和Anammox特有的脂质核心结构——阶梯烷(Ladderane core lipids)相结合的分析方法,检测到美国加州和内华达州热泉中Anammox的存在,分布于Planctomycetales目(类似于Candidatus Brocadia fulgida, Candidatus Brocadia anammoxidans和Candidatus Kuenenia stuttgartiensis)。该研究还发现热泉中Anammox存在的温度上限为52.1 ℃,高温热泉中(96.6和89.1 ℃)未检测到阶梯烷的存在[46]。Rattray等(2010)[47]通过实验室模拟温度对阶梯烷结构变化的影响,发现随着温度的升高,C20的含量随之升高,而C18的含量却随之降低,并由此推测不同温度下分布着不同Anammox类群。
4 反硝化 反硝化是指在厌氧条件下将硝酸盐还原成氮气的过程(NO3-→NO2-→NO→N2O→N2),是氮循环的最后阶段,也是负责将氮气返回到大气中的主要生物学过程。
反硝化过程涉及的功能基因有narG (硝酸还原酶的编码基因)、nirS与nirK (亚硝酸还原酶的编码基因)、norB (一氧化氮还原酶的编码基因)以及nosZ(氧化亚氮还原酶的编码基因)[48-49]。分子生物学中,常用nirS与nirK检测环境中反硝化菌的存在[48, 50-51]。目前,尚未在同一微生物中同时发现nirS和nirK的存在,由此推测二者在环境中代表着不同的反硝化种群类型[52]。也有****指出nirS存在于厌氧环境中,而nirK倾向于生存在好氧环境中[53]。
高温条件下具有反硝化功能的微生物主要分布在古菌域的Crenarchaeota和Euryarchaeota门,以及细菌域的Aquificae、Firmicutes和Thermus门。分离自深海热液的纯菌株Pyrobaculum aerophilum (Crenarchaeota)[54]、Ferroglobus placidus (Euryarchaeota)[55]和Pyrolobus fumarii (Crenarchaeota)[56],分别可在80、95和113℃的温度下将NO3-还原[25]。细菌域中的Thermales目(属Thermus门)是研究最为透彻的反硝化菌,一些纯培养物如Thermus thermophilus可以把NO3-彻底还原为N2[57]。但是在Thermales目中,大部分的兼性厌氧菌,如Meiothermus、Oceanithermus和Vulcanithermu属的微生物只能把NO3-还原为NO2-[57]。
热泉中关于反硝化菌活性的研究较少,目前只有少量****对美国大盆地80 ℃热泉中反硝化速率进行检测,反硝化速率分别为(67.8±2.6)μmol N2O-N/(m2·d)和15.8-51.0 N/(g·h)[27, 57],有关其他热泉及不同环境因子下的反硝化速率还缺乏相应的研究。
5 异化硝酸盐还原 异化硝酸盐还原成铵(Dissimilatory nitrate reduction to ammonium,DNRA)是微生物在厌氧条件下降解有机质的同时将硝酸盐或亚硝酸盐异化还原成铵的过程。DNRA是硝酸盐还原的重要过程,与反硝化、厌氧氨氧化过程相比,该过程没有氮的损失,而是将硝态氮转化为生物可吸收、利用的氨态氮。常温环境下,可用功能基因nrfA (编码亚硝酸盐还原酶NrfA)来检测环境中异化硝酸盐还原菌的存在[58-60]。热泉中还未有扩增nrfA基因引物的报道。
Edwards等(2012)[61]发现在美国大盆地热泉中,当温度>84.4 ℃时,DNRA过程会受到抑制。但从深海热液中分离出的Pyrolobus fumarii也可在113 ℃时将NO3-还原为NH4+[56]。高温条件下的异化硝酸盐还原菌主要有从深海热液分离出的Thermovibrio ammonificans[62]、Caldithrix abysii[63-64]及热泉中分离出的Ammonifex degensii[55]。热泉中关于DNRA过程速率的研究也仅限于Dodsworth等(2011)[27]对美国大盆地热泉的研究,该研究检测到80 ℃时DNRA的速率为315.0 N/(g·h),而对于其他热泉中DNRA的活性和温度上限我们还无从知晓。
6 总结和展望 综合热泉微生物中参与氮循环微生物的研究发现,固氮微生物、氨氧化菌、厌氧氨氧化菌、反硝化菌和异化硝酸盐还原菌等在热泉环境中均有存在,这些微生物是驱动热泉环境氮循环的主导力量。目前可以通过功能基因检测到部分氮循环微生物功能群(如固氮菌、氨氧化菌等)的相对丰度和初步探讨其对环境因子(如温度、pH、水化学条件)和空间因子(如地理距离)的响应机制。然而对热泉驱动氮循环微生物的研究尚存在诸多问题亟待解决,如:(1)纯培养物有限(如AOA、NOB);(2)分类地位尚不明确(如AOA);(3)大多参与氮循环的微生物功能群尚未发现成熟的引物(特别是AOB、NOB、Anammox、反硝化菌和异化硝酸盐还原菌等)来检测其在环境中的多样性;(4)热泉中驱动氮循环微生物在不同环境因子下的活性(表 1)和各功能群间的协作竞争机制尚不清楚等。热泉生态系统与地球早期生命起源时的环境及其相似,对热泉微生物的研究是窥探地球早期生命的一个窗口,但是我们如何将现代地质过程中这些微生物在氮循环中的重要作用和各地质历史时期相结合,如何评估热泉微生物对古今生态环境的影响,以及如何推衍地球早期生命的生存演化过程,均需进一步的探索和突破。
表 1. 热泉中微生物驱动氮循环功能活性的温度上限 Table 1. Upper temperature limits of the microbial activity on driving nitrogen cycles
| Microbial functional communities | Upper limits of field measurement/℃ | Upper limits of pure isolates(or enrichment)/℃ |
| Nitrogen-fixation | 72.7[4] | 62.7[7] |
| Ammonia oxidation | 94.0[31] | 76.0[23, 26-27] |
| Nitrite oxidation | - | 65.0[36] |
| Anaerobic ammonia oxidation | 52.1[46] | - |
| Nitrite reduction | 80.0[27, 57] | 80.0[57] |
| Dissimilatory nitrate reduction to ammonium | 84.4[61] | 80.0[27] |
| “-”, no such data in published literatures. | ||
表选项
References
| [1] | Amend JP, Shock EL. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiology Reviews, 2001, 25(2): 175-243. DOI:10.1111/j.1574-6976.2001.tb00576.x |
| [2] | Gruber N, Galloway JN. An Earth-system perspective of the global nitrogen cycle. Nature, 2008, 451(7176): 293-296. DOI:10.1038/nature06592 |
| [3] | Hamilton TL, Lange RK, Boyd ES, Peters JW. Biological nitrogen fixation in acidic high-temperature geothermal springs in Yellowstone National Park, Wyoming. Environmental Microbiology, 2011, 13(8): 2204-2215. DOI:10.1111/j.1462-2920.2011.02475.x |
| [4] | Loiacono ST, Meyer-Dombard DAR, Havig JR, Poret-Peterson AT, Hartnett HE, Shock EL. Evidence for high-temperature in situ nifH transcription in an alkaline hot spring of Lower Geyser Basin, Yellowstone National Park. Environmental Microbiology, 2012, 14(5): 1272-1283. DOI:10.1111/emi.2012.14.issue-5 |
| [5] | Gaby JC, Buckley DH. A global census of nitrogenase diversity. Environmental Microbiology, 2011, 13(7): 1790-1799. DOI:10.1111/j.1462-2920.2011.02488.x |
| [6] | Alcamán ME, Alcorta J, Bergman B, Vásquez M, Polz M, Díez B. Physiological and gene expression responses to nitrogen regimes and temperatures in Mastigocladus sp. strain CHP1, a predominant thermotolerant cyanobacterium of hot springs. Systematic and Applied Microbiology, 2017, 40(2): 102-113. DOI:10.1016/j.syapm.2016.11.007 |
| [7] | McGregor GB, Sendall BC. Ewamiania thermalis gen. et sp. nov. (Cyanobacteria, Scytonemataceae), a new cyanobacterium from Talaroo thermal springs, north-eastern Australia. Australian Systematic Botany, 2017, 30(1): 38-47. DOI:10.1071/SB16039 |
| [8] | Mehta MP, Baross JA. Nitrogen fixation at 92 C by a hydrothermal vent archaeon. Science, 2006, 314(5806): 1783-1786. DOI:10.1126/science.1134772 |
| [9] | Alcamán ME, Fernandez C, Delgado A, Bergman B, Díez B. The cyanobacterium Mastigocladus fulfills the nitrogen demand of a terrestrial hot spring microbial mat. The ISME Journal, 2015, 9(10): 2290-2303. DOI:10.1038/ismej.2015.63 |
| [10] | Hall JR, Mitchell KR, Jackson-Weaver O, Kooser AS, Cron BR, Crossey LJ, Takacs-Vesbach CD. Molecular characterization of the diversity and distribution of a thermal spring microbial community by using rRNA and metabolic genes. Applied and Environmental Microbiology, 2008, 74(15): 4910-4922. DOI:10.1128/AEM.00233-08 |
| [11] | Hamilton TL, Boyd ES, Peters JW. Environmental constraints underpin the distribution and phylogenetic Diversity of nifH in the Yellowstone geothermal complex. Microbial Ecology, 2011, 61(4): 860-870. DOI:10.1007/s00248-011-9824-9 |
| [12] | Steunou AS, Bhaya D, Bateson MM, Melendrez MC, Ward DM, Brecht E, Peters JW, Kühl M, Grossman AR. In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(7): 2398-2403. DOI:10.1073/pnas.0507513103 |
| [13] | Steunou AS, Jensen SI, Brecht E, Becraft ED, Bateson MM, Kilian O, Bhaya D, Ward DM, Peters JW, Grossman AR, Kühl M. Regulation of nif gene expression and the energetics of N2 fixation over the diel cycle in a hot spring microbial mat. The ISME Journal, 2008, 2(4): 364-378. DOI:10.1038/ismej.2007.117 |
| [14] | Prosser JI, Nicol GW. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environmental Microbiology, 2008, 10(11): 2931-2941. DOI:10.1111/emi.2008.10.issue-11 |
| [15] | Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environmental Microbiology, 2005, 7(12): 1985-1995. DOI:10.1111/j.1462-2920.2005.00906.x |
| [16] | Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature, 2006, 442(7104): 806-809. DOI:10.1038/nature04983 |
| [17] | Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environmental Microbiology, 2007, 9(5): 1162-1175. DOI:10.1111/emi.2007.9.issue-5 |
| [18] | Pjevac P, Schauberger C, Poghosyan L, Herbold CW, van Kessel MAHJ, Daebeler A, Steinberger M, Jetten MSM, Lücker S, Wagner M, Daims H. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. Frontiers in Microbiology, 2017, 8: 1508. DOI:10.3389/fmicb.2017.01508 |
| [19] | Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. Complete nitrification by Nitrospira bacteria. Nature, 2015, 528(7583): 504-509. DOI:10.1038/nature16461 |
| [20] | van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, Jetten MSM, Lücker S. Complete nitrification by a single microorganism. Nature, 2015, 528(7583): 555-559. DOI:10.1038/nature16459 |
| [21] | Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, Daebeler A, Romano S, Albertsen M, Stein LY, Daims H, Wagner M. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature, 2017, 549(7671): 269-272. DOI:10.1038/nature23679 |
| [22] | Lebedeva EV, Alawi M, Fiencke C, Namsaraev B, Bock E, Spieck E. Moderately thermophilic nitrifying bacteria from a hot spring of the Baikal rift zone. FEMS Microbiology Ecology, 2005, 54(2): 297-306. DOI:10.1016/j.femsec.2005.04.010 |
| [23] | De La Torre JR, Walker CB, Ingalls AE, K?nneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environmental Microbiology, 2008, 10(3): 810-818. DOI:10.1111/emi.2008.10.issue-3 |
| [24] | Gerbl FW, Weidler GW, Wanek W, Erhardt A, Stan-Lotter H. Thaumarchaeal ammonium oxidation and evidence for a nitrogen cycle in a subsurface radioactive thermal spring in the Austrian Central Alps. Frontiers in Microbiology, 2014, 5: 225. |
| [25] | Dodsworth JA, Hungate B, De La Torre JR, Jiang HC, Hedlund BP. Measuring nitrification, denitrification, and related biomarkers in terrestrial geothermal ecosystems. Methods in Enzymology, 2011, 486: 171-203. DOI:10.1016/B978-0-12-381294-0.00008-0 |
| [26] | Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(6): 2134-2139. DOI:10.1073/pnas.0708857105 |
| [27] | Dodsworth JA, Hungate BA, Hedlund BP. Ammonia oxidation, denitrification and dissimilatory nitrate reduction to ammonium in two US Great Basin hot springs with abundant ammonia-oxidizing archaea. Environmental Microbiology, 2011, 13(8): 2371-2386. DOI:10.1111/j.1462-2920.2011.02508.x |
| [28] | Zhang CL, Ye Q, Huang ZY, Li WJ, Chen JQ, Song ZQ, Zhao WD, Bagwell C, Inskeep WP, Ross C, Gao L, Wiegel J, Romanek CS, Shock EL, Hedlund BP. Global occurrence of archaeal amoA genes in terrestrial hot springs. Applied and Environmental Microbiology, 2008, 74(20): 6417-6426. DOI:10.1128/AEM.00843-08 |
| [29] | Reigstad LJ, Richter A, Daims H, Urich T, Schwark L, Schleper C. Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiology Ecology, 2008, 64(2): 167-174. DOI:10.1111/fem.2008.64.issue-2 |
| [30] | Jiang HC, Huang QY, Dong HL, Wang P, Wang FP, Li WJ, Zhang CL. RNA-based investigation of ammonia-oxidizing archaea in hot springs of Yunnan Province, China. Applied and Environmental Microbiology, 2010, 76(13): 4538-4541. DOI:10.1128/AEM.00143-10 |
| [31] | Li HZ, Yang QH, Li J, Gao H, Li P, Zhou HY. The impact of temperature on microbial diversity and AOA activity in the Tengchong Geothermal Field, China. Scientific Reports, 2015, 5: 17056. DOI:10.1038/srep17056 |
| [32] | Jiang HC, Huang LQ, Deng Y, Wang S, Zhou Y, Liu L, Dong HL. Latitudinal distribution of ammonia-oxidizing bacteria and archaea in the agricultural soils of Eastern China. Applied and Environmental Microbiology, 2014, 80(18): 5593-5602. DOI:10.1128/AEM.01617-14 |
| [33] | Pester M, Maixner F, Berry D, Rattei T, Koch H, Lücker S, Nowka B, Richter A, Spieck E, Lebedeva E, Loy A, Wagner M, Daims H. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environmental Microbiology, 2014, 16(10): 3055-3071. DOI:10.1111/emi.2014.16.issue-10 |
| [34] | Edwards TA, Calica NA, Huang DA, Manoharan N, Hou WG, Huang LQ, Panosyan H, Dong HL, Hedlund BP. Cultivation and characterization of thermophilic Nitrospira species from geothermal springs in the US Great Basin, China, and Armenia. FEMS Microbiology Ecology, 2013, 85(2): 283-292. DOI:10.1111/femsec.2013.85.issue-2 |
| [35] | Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Sinninghe Damsté JS, Spieck E, Le Paslier D, Daims H. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(30): 13479-13484. DOI:10.1073/pnas.1003860107 |
| [36] | Edwards TA. Nitrite oxidation in geothermal springs: evidence of an upper temperature limit for thermophilic nitrite-oxidizing bacteria of 60-65℃. Master Dissertation of University of Nevada, 2012. |
| [37] | Lebedeva EV, Off S, Zumbr?gel S, Kruse M, Shagzhina A, Lücker S, Maixner F, Lipski A, Daims H, Spieck E. Isolation and characterization of a moderately thermophilic nitrite-oxidizing bacterium from a geothermal spring. FEMS Microbiology Ecology, 2011, 75(2): 195-204. DOI:10.1111/fem.2010.75.issue-2 |
| [38] | Weidler GW, Dornmayr-Pfaffenhuemer M, Gerbl FW, Heinen W, Stan-Lotter H. Communities of Archaea and Bacteria in a subsurface radioactive thermal spring in the Austrian Central Alps, and evidence of ammonia-oxidizing Crenarchaeota. Applied and Environmental Microbiology, 2007, 73(1): 259-270. DOI:10.1128/AEM.01570-06 |
| [39] | Burgin AJ, Hamilton SK. Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Frontiers in Ecology and the Environment, 2007, 5(2): 89-96. DOI:10.1890/1540-9295(2007)5[89:HWOTRO]2.0.CO;2 |
| [40] | Dalsgaard T, Thamdrup B, Canfield DE. Anaerobic ammonium oxidation (anammox) in the marine environment. Research in Microbiology, 2005, 156(4): 457-464. DOI:10.1016/j.resmic.2005.01.011 |
| [41] | Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Médigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJM, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, Van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes HW, Weissenbach J, Jetten MSM, Wagner M, Le Paslier D. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature, 2006, 440(7085): 790-794. DOI:10.1038/nature04647 |
| [42] | Li M, Ford T, Li XY, Gu JD. Cytochrome cd1-containing nitrite reductase encoding gene nirS as a new functional biomarker for detection of anaerobic ammonium oxidizing (anammox) bacteria. Environmental Science & Technology, 2011, 45(8): 3547-3553. |
| [43] | Francis CA, Beman JM, Kuypers MMM. New processes and players in the nitrogen cycle:the microbial ecology of anaerobic and archaeal ammonia oxidation. The ISME Journal, 2007, 1(1): 19-27. DOI:10.1038/ismej.2007.8 |
| [44] | Kartal B, Geerts W, Jetten MSM. Cultivation, detection, and ecophysiology of anaerobic ammonium-oxidizing bacteria. Methods in Enzymology, 2011, 486: 89-108. DOI:10.1016/B978-0-12-381294-0.00004-3 |
| [45] | Jetten MSM, Op den Camp HJM, Kuenen JG, Strous M. Taxonomic description of the family Anammoxaceae. Bergery's Manual of Systematic Bacteriology. 2 Ed. New-York: Springer, 2005. |
| [46] | Jaeschke A, Op den Camp HJM, Harhangi H, Klimiuk A, Hopmans EC, Jetten MSM, Schouten S, Sinninghe Damsté JS. 16S rRNA gene and lipid biomarker evidence for anaerobic ammonium-oxidizing bacteria (anammox) in California and Nevada hot springs. FEMS Microbiology Ecology, 2009, 67(3): 343-350. DOI:10.1111/fem.2009.67.issue-3 |
| [47] | Rattray JE, van de Vossenberg J, Jaeschke A, Hopmans EC, Wakeham SG, Lavik G, Kuypers MMM, Strous M, Jetten MSM, Schouten S, Sinninghe Damsté JS. Impact of temperature on ladderane lipid distribution in anammox bacteria. Applied and Environmental Microbiology, 2010, 76(5): 1596-1603. DOI:10.1128/AEM.01796-09 |
| [48] | Zumft WG. Cell biology and molecular basis of denitrification. Microbiology and Molecular Biology Reviews, 1997, 61(4): 533-616. |
| [49] | Nogales B, Timmis KN, Nedwell DB, Osborn AM. Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Applied and Environmental Microbiology, 2002, 68(10): 5017-5025. DOI:10.1128/AEM.68.10.5017-5025.2002 |
| [50] | Priemé A, Braker G, Tiedje JM. Diversity of nitrite reductase (nirK and nirS) gene fragments in forested upland and wetland soils. Applied and Environmental Microbiology, 2002, 68(4): 1893-1900. DOI:10.1128/AEM.68.4.1893-1900.2002 |
| [51] | Heylen K, Gevers D, Vanparys B, Wittebolle L, Geets J, Boon N, de Vos P. The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environmental Microbiology, 2006, 8(11): 2012-2021. DOI:10.1111/emi.2006.8.issue-11 |
| [52] | Jones CM, Hallin S. Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. The ISME Journal, 2010, 4(5): 633-641. DOI:10.1038/ismej.2009.152 |
| [53] | Desnues C, Michotey VD, Wieland A, Cui ZZ, Four?ans A, Duran R, Bonin PC. Seasonal and diel distributions of denitrifying and bacterial communities in a hypersaline microbial mat (Camargue, France). Water Research, 2007, 41(15): 3407-3419. DOI:10.1016/j.watres.2007.04.018 |
| [54] | V?lkl P, Huber R, Drobner E, Rachel R, Burggraf S, Trincone A, Stetter KO. Pyrobaculum aerophilum sp. nov., a novel nitrate-reducing hyperthermophilic archaeum. Applied and Environmental Microbiology, 1993, 59(9): 2918-2926. |
| [55] | Hafenbradl D, Keller M, Dirmeier R, Rachel R, Rossnagel P, Burggraf S, Huber H, Stetter KO. Ferroglobus placidus gen. nov., sp. nov., a novel hyperthermophilic archaeum that oxidizes Fe2+ at neutral pH under anoxic conditions. Archives of Microbiology, 1996, 166(5): 308-314. DOI:10.1007/s002030050388 |
| [56] | Bl?chl E, Rachel R, Burggraf S, Hafenbradl D, Jannasch HW, Stetter KO. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113℃.. Extremophiles, 1997, 1(1): 14-21. DOI:10.1007/s007920050010 |
| [57] | Hedlund BP, McDonald AI, Lam J, Dodsworth JA, Brown JR, Hungate BA. Potential role of Thermus thermophilus and T. oshimai in high rates of nitrous oxide (N2O) production in 80℃ hot springs in the US Great Basin. Geobiology, 2011, 9(6): 471-480. DOI:10.1111/j.1472-4669.2011.00295.x |
| [58] | Mohan SB, Schmid M, Jetten M, Cole J. Detection and widespread distribution of the nrfA gene encoding nitrite reduction to ammonia, a short circuit in the biological nitrogen cycle that competes with denitrification. FEMS Microbiology Ecology, 2004, 49(3): 433-443. DOI:10.1016/j.femsec.2004.04.012 |
| [59] | Dong LF, Smith CJ, Papaspyrou S, Stott A, Osborn AM, Nedwell DB. Changes in benthic denitrification, nitrate ammonification, and anammox process rates and nitrate and nitrite reductase gene abundances along an estuarine nutrient gradient (the Colne Estuary, United Kingdom). Applied and Environmental Microbiology, 2009, 75(10): 3171-3179. DOI:10.1128/AEM.02511-08 |
| [60] | Welsh A, Chee-Sanford JC, Connor LM, L?ffler FE, Sanford RA. Refined NrfA phylogeny improves PCR-based nrfA gene detection. Applied and Environmental Microbiology, 2014, 80(7): 2110-2119. DOI:10.1128/AEM.03443-13 |
| [61] | Everroad RC, Otaki H, Matsuura K, Haruta S. Diversification of bacterial community composition along a temperature gradient at a thermal spring. Microbes and Environments, 2012, 27(4): 374-381. DOI:10.1264/jsme2.ME11350 |
| [62] | Vetriani C, Speck MD, Ellor SV, Lutz RA, Starovoytov V. Thermovibrio ammonificans sp. nov., a thermophilic, chemolithotrophic, nitrate-ammonifying bacterium from deep-sea hydrothermal vents. International Journal of Systematic and Evolutionary Microbiology, 2004, 57(Pt 1): 175-181. |
| [63] | Miroshnichenko ML, Kostrikina NA, Chernyh NA, Pimenov NV, Tourova TP, Antipov AN, Spring S, Stackebrandt E, Bonch-Osmolovskaya EA. Caldithrix abyssi gen. nov., sp. nov., a nitrate-reducing, thermophilic, anaerobic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent, represents a novel bacterial lineage. International Journal of Systematic and Evolutionary Microbiology, 2003, 53(Pt 1): 323-329. |
| [64] | Huber R, Rossnagel P, Woese CR, Rachel R, Langworthy TA, Stetter KO. Formation of ammonium from nitrate during chemolithoautotrophic growth of the extremely thermophilic bacterium Ammonifex degensii gen. nov. sp. nov. Systematic and Applied Microbiology, 1996, 19(1): 40-49. DOI:10.1016/S0723-2020(96)80007-5 |
