杨雪琴, 连英丽, 颜庆云

 , 贺志理
, 贺志理

中山大学环境科学与工程学院, 环境微生物组学研究中心, 广东 广州 510006
收稿日期:2017-11-05;修回日期:2017-12-29;网络出版日期:2018-01-18
基金项目:中山大学“****”(38000-18821107);中山大学“****”(38000-18821105)
作者简介:贺志理, 中山大学"****"教授、环境微生物组学研究中心负责人、博士生导师。贺志理教授长期从事交叉学科研究, 以创新生物信息学和微生物生态学理论和方法, 了解微生物驱动的地球化学循环过程, 揭示微生物多样性和生态系统功能的机制, 阐明微生物群落对全球气候变化和环境污染等的响应及其调控机制。已发表专著1部, 220余篇高质量SCI论文在Nature Climate Change、Nature Reviews Microbiology、Nature Communications、PNAS、Nucleic Acids Research、Ecology Letters、ISME Journal、Global Change Biology、mBio等专业期刊, 总引用次数>10000, H指数为56(Google Scholar)。中山大学环境微生物组学研究中心旨在了解微生物在水、土、气环境中的作用, 现有数据科学、环境微生物生态学、系统微生物学和环境应用等核心实验室
*通信作者:颜庆云, Tel:+86-20-31065837;E-mail:yanqy6@mail.sysu.edu.cn
贺志理, Tel: +86-20-31065837;E-mail:hezhili@mail.sysu.edu.cn
摘要:滨海湿地生态系统介于陆地生态系统和海洋生态系统之间,其类型多种多样,环境差异极大,微生物种类丰富。近年来,随着人为氮源的大量输入,造成滨海湿地生态系统富营养化污染问题日趋严重。本文主要总结了滨海湿地生态系统微生物驱动的固氮、硝化、反硝化、厌氧氨氧化、NO3-等主要氮循环过程,并综述了通过功能基因(如nifH、amoA、hzo、nirS、nirK、nrfA)检测微生物群落多样性及其环境影响因素的相关研究,旨在更好理解微生物驱动氮循环过程以去除氮,以期为减轻富营养化和危害性藻类爆发提供科学依据。
关键词: 滨海湿地生态系统 氮循环 固氮 硝化 反硝化 厌氧氨氧化 NO3-还原成铵
Microbially-driven nitrogen cycling in coastal ecosystems
Xueqin Yang, Yingli Lian, Qingyun Yan

 , Zhili He
, Zhili He

Environmental Microbiomics Research Center and School of Environmental Science and Engineering, Sun Yat-sen University, Guangzhou 510006, Guangdong Province, China
Received 5 November 2017; Revised 29 December 2017; Published online 18 January 2018
*Corresponding author: Qingyun Yan, Tel: +86-20-31065837; E-mail: yanqy6@mail.sysu.edu.cn
Zhili He, Tel: +86-20-31065837; hezhili@mail.sysu.edu.cn
Supported by the Hundred Talents Program (38000-18821107) and Thousand Talents Program (38000-18821105) by Sun Yat-sen University
Abstract: Coastal wetlands are ecosystems between terrestrial and marine ecosystems, which are highly diverse and also result in highly diverse microbial communities. In the last several decades, an increasing nitrogen load into coastal wetland ecosystems causes serious environmental problems such as eutrophication, and harmful algal blooms. Here we review the microbial communities involved in nitrogen cycling processes, such as nitrogen fixation, nitrification, denitrification, anaerobic ammonium oxidation, dissimilatory nitrate reduction to ammonium processes, and their key functional genes including nifH, amoA, hzo, nirK, nirS and nrfA as well as related environmental factors to further understand the microbially-driven nitrogen cycling processes for potentially removing nitrogen to mitigate eutrophication and harmful algal blooms.
Key words: coastal ecosystems nitrogen cycling nitrogen fixation nitrification denitrification anaerobic ammonium oxidation dissimilatory nitrate reduction to ammonium (DNRA)
滨海湿地生态系统处于陆地和海洋之间,是指大陆架区域的潮间带和潮下带(水深不超过200 m),包括常被海水淹没的区域和海岸线100 km内的毗邻地。典型的滨海湿地生态系统包括河口、沙滨、红树林、海草等[1]。它们兼具水生和陆生生态系统特征,是一类生物多样性丰富、生产力高、生态服务价值大的生态系统[2-3]。滨海湿地生态系统不仅具有水文防洪功能,同时还能为鱼类和其他野生动物提供栖息场所[4]。滨海湿地生态系统水深较浅,底泥易形成缺氧环境,是发生氧化还原反应的主要场所。随着沿海城市经济的迅速发展,人类活动日渐频繁,滨海湿地的纳污量也随之急剧增加。在过去的30多年的时间里由长江汇入东海的可溶性无机氮增加了3倍多[5]。可溶性无机氮进入海洋后,一方面导致海岸带氮超载水质变差、富营养化[6],出现如波罗的湾、墨西哥湾、切萨皮克湾一样的“死区”[7];另一方面影响生态位及物种多样性[8]。因此了解氮循环过程以及与之相关的微生物作用机理对滨海湿地生态系统具有重要意义。
微生物是生物地球化学循环的主要驱动者,也是地球生态平衡的重要维持者。滨海湿地生态系统具有的典型生态功能与该生境下微生物的群落组成及代谢网络息息相关。近年来新型生物学技术的出现以及不断更新的数据分析方法,使科学家们可以更为深入全面地认识环境微生物群落组成和功能结构,为揭示微生物在生态系统中的重要作用提供了有力的工具。基因芯片和高通量测序是最具代表性的宏基因组学技术,其中基因芯片技术具有方便快捷、精确稳定的特点,可检测包括碳(C)、氮(N)、磷(P)、硫(S)循环、金属还原和抗性、抗生素降解、有机物降解等不同的功能基因[9-13],目前更新到GeoChip5.0,包含167000个寡核苷酸探针,15904个功能基因,可以原位检测微生物群落功能结构和代谢功能;本课题组利用基因芯片研究了滨海湿地生态系统微生物功能菌群[14],南极生态系统的碳、氮循环[15],陆地生态系统[16]、草地生态系统[10]、大豆农田生态系统[11]氮循环基因及微生物群落在CO2升高后的变化,原位添加NO3-后氮循环基因及微生物群落结构和多样性的变化[17]等;高通量测序技术具有全面精准、挖掘新基因和新代谢途径等优点,为未知基因的检测提供了可能。目前关于滨海湿地生态系统微生物功能的分子生态学研究主要是针对C、N、S、P、重金属等物质循环过程,其中氮循环主要包括固氮、硝化、反硝化、厌氧氨氧化等过程(图 1)。
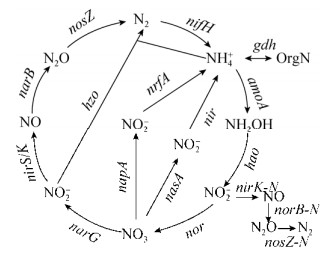 |
| 图 1 氮循环过程及其关键功能基因[10] Figure 1 The nitrogen cycle and involved key functional genes[10]. |
| 图选项 |
本文将主要综述滨海湿地生态系统中微生物作用下氮循环的主要氧化还原过程、微生物群落结构、功能特性以及与之相关的环境因子。
1 微生物固氮 微生物固氮是指将气态N2转化为NH4+的过程,常用功能基因nifH研究固氮过程,因其较为保守,且已知的基因序列非常丰富[18]。根据nifH进行系统发育树分析,可以将固氮微生物分为5个主要的类群(Clusters Ⅰ-Ⅴ),其中滨海湿地生态系统中最常见的固氮菌主要属于Clusters Ⅰ和Clusters Ⅲ[18-19]。Clusters Ⅰ包括蓝细菌/蓝藻(Cyanobacteria)、大部分α-, β-, γ-, δ-变形菌(Proteobacteria)、厚壁菌门(Firmicutes)等多个类群,Clusters Ⅲ主要包括古菌和厌氧细菌,如梭菌(Clostridia)、产甲烷菌(Methanogens)、硫酸盐还原菌(Sulfate-reducing bacteria)等。
****通过测定固氮速率估算微生物固氮在滨海湿地生态系统中的贡献量,发现滨海湿地生态系统中的氮排入和氮损失失衡。如Fitzroy河口的固氮速率为0.99-1.09 μmol N/(m2·h)[20],位置(上游、中部、下游)和季节对其影响不大;Little Lagoon固氮速率相对较高,为1.6-2.6 μmolN/ (m2·h)[21]。Short和Zehr[22]在研究Chesapeake Bay地区时检测到较高的nifH基因多样性,但表达的nifH基因只有两种,且都为蓝细菌型。在2011年Sohm等[23]综述了蓝藻型固氮微生物(如Trichodesmium)固氮作用,并提出异养型固氮微生物的作用还有待研究。Zhou等[24]认为原来的微生物固氮研究局限于水体,基于nifH基因检测沉积物中异养型微生物丰度占总丰度的0.36%-14.95%,并指出沉积物中异养型微生物固氮作用不可忽视。目前已报道的微生物固氮速率仍然较低,有可能是外源性氮排入对滨海湿地生态系统固氮过程的抑制,具体的还有待进一步研究。
研究发现在河口地区,由于沉降作用、流域水文条件和环境选择压力(如盐度、温度、pH等)改变,沿着河头到河嘴再到近海区域的固氮菌皆有所不同[19, 25-26]。Thajudeen等[27]在Cochin河口观察到固氮菌的多样性较高,其中γ-变形菌为优势菌,并指出其固氮速率与K+、Fe2+、总有机碳(TOC)、总氮(TN)、粘土含量为负相关关系。
2 微生物硝化过程 2.1 分步硝化过程与完全硝化过程 硝化过程指氨氧化为NO3-的过程。一个世纪以来,****们普遍认为硝化过程需分步完成,即包括氨氧化为NO2-和NO2-氧化为NO3-两个过程,且分别由两类微生物执行,直到2015年van Kessel等[28]在富集氮循环微生物时发现Candidatus Nitrospira nitrosa和Candidatus Nitrospira nitrificans能执行完整的硝化过程,即完全硝化过程(complete ammonia oxidation,Comammox),同年Daims等[29]在石油勘测井的管壁上富集到中度噬热菌Candidatus Nitrospira inopinata可将NH4+等摩尔地氧化为NO3-,Pinto等[30]在饮用水中发现了类似的Nitrospira,这些发现一方面证实了Comammox理论存在的猜测[28-29],另一方面引起了****对其生态位、在硝化过程中的贡献等氮循环相关问题的广泛关注。近两年有关全程硝化菌的生态生理学研究陆续发表,有研究提到Nitrospira inopinata对NH4+有更强的亲和力,在低浓度NH4+下能缓慢生长[31],适应于贫营养环境,这些研究为滨海湿地系统相关研究提供了重要的理论基础。
下文综述的内容主要为分步硝化过程。分步硝化过程中氨氧化过程是限速步骤[32]。基因amoA编码的氨单加氧酶(amo)是催化第一步氨氧化反应的关键酶。利用amoA可以分析不同环境中氨氧化效率、功能微生物的群落结构及生物多样性等。能执行氨氧化过程的细菌有氨氧化细菌(ammonia oxidizing bacteria,AOB)和氨氧化古菌(ammonia oxidizing archaea,AOA)。人们在很长一段时间里认为只有氨氧化细菌能进行氨氧化,直到2004年科学家在研究海洋基因组学时发现泉古菌具有与细菌相近的amoA基因,随后通过基因转录的方法间接证明了氨氧化古菌也具有氨氧化功能[33-34];通过宏基因组测序的方法,在Cenarchaeum symbiosum的基因组序列中发现与二氧化碳固定相关的基因,同时这些基因也是TCA循环过程的必需基因,推测该菌可能是混合营养型[35],这对碳、氮循环有着重要的意义。
2.2 滨海湿地生态系统中AOA、AOB的丰度及其多样性 滨海湿地生态系统中主要的AOA菌属有Nitrosopumilus、Nitrosophsera、Nitrosocaldus、Nitrosotalea。AOB则主要包括β-变形菌纲的亚硝化单胞菌(Nitrosomonas)与亚硝化螺菌属(Nitrosospira)以及γ-变形菌纲的亚硝化球菌属(Nitrosococcus)[36]。滨海红树林中的AOA的丰度总是高于AOB,就不同红树植物而言,土著红树林的AOA丰度相比于入侵的互花米草型红树林高约100倍,这与其他研究相符[35, 37-39]。相反的研究也有,Wang和Gu[40]发现香港滨海湿地生态系统中AOB的amoA丰度占84%-92%,高于AOA的amoA丰度;Li等[41]和Han等[42]也发现同样的结果。也有研究表明虽然在农耕土壤中AOA的丰度高于AOB[43],但是AOB对氨氧化的贡献更大[44]。Cao等[37]的研究表明红树林AOB多样性高于AOA,以互花米草型红树林和本土红树林AOB生物多样性无明显变化,而互花米草型AOA生物多样性高于本土红树林[45]。这些研究结果表明,滨海湿地生态系统中AOA、AOB的丰度以及其相互关系还需要更多深入的研究来揭示。
2.3 影响AOA和AOB群落的环境因子 Han等[42]利用基因芯片GeoChip4.0检测不同红树林的amoA,发现互花米草、桐花树、秋茄、白骨壤红树林之间的氨氧化功能有显著差异。Wang和Gu[40]研究发现芦苇和莎草对AOB的群落结构有影响,对AOA基本无影响。在淡水中β-AOB丰度显著低于海水的,AOA的丰度基本保持不变[46]。2012年Cao等[47]进一步研究发现从珠江口湿地到东海,β-变形菌纲的优势种由Nitrosomonas变为Nitrosospira,表明盐度和人类活动对AOA和AOB的影响较大。除受盐度和人类活动的影响,滨海湿地生态系统AOA和AOB的相对丰度还受NH4+、pH、SO42-、重金属和有机物等的影响[39]。AOA和NH4+有较高的亲和力,在低NH4+的情况下具有很强的竞争力;AOA还倾向于较低pH [35, 37, 39, 48-49]。综上,AOA比AOB在极端条件或者贫营养环境中更具有生存优势,也使得AOA成为这些环境下主要的氨氧化功能菌。
3 厌氧氨氧化 厌氧氨氧化是指NH4+在厌氧条件下以NO2-为电子受体直接反应生成N2的过程。目前已知厌氧氨氧化过程在水环境中普遍存在,是氮去除的重要过程[50]。2002年Thamdrup和Dalsgaard[51]利用15N稳定性同位素示踪技术首次证实了海洋沉积物中存在厌氧氨氧化过程,并推算出高达67%的N2生成和厌氧氨氧化作用相关,该过程的发现重新定义了氮循环[52]。按16S rRNA基因的系统发育分析,厌氧氨氧化细菌是厌氧化能自养菌[53],浮霉菌门(Planctomycetes) Brocadiales目,5个暂定属分别为:Scalindua、Brocadia、Kuenenia、Jettenia与Anammoxoglobus[50, 52, 54-55]。
3.1 厌氧氨氧化菌的丰度、厌氧氨氧化过程速率 滨海湿地生态系统厌氧氨氧化菌的丰度、厌氧氨氧化过程速率及对氮损失的贡献如表 1所示。厌氧氨氧化菌的丰度与厌氧氨氧化速率及其对氮损失的贡献无明显关系。珠江三角洲的厌氧氨氧化过程对氮损失的贡献率为0.5%-7.0%[56];Hou等[57]利用同位素分析估算长江河口及海岸6.6%-12.9%的氮损失归因为厌氧氨氧化过程;在2015年,同样的基于氮同位素分析发现厌氧氨氧化过程每年可去除滨海湿地生态系统约3.8%-10.7%的氮,结合反硝化作用,去除率达20.7%[58];在Zheng等[59]的研究中厌氧氨氧化对总氮的损失贡献为5%-77%。
表 1. 不同季节各地厌氧氨氧化菌丰度、厌氧氨氧化过程速率及对氮损失的贡献 Table 1. Spatiotemporal variations of anaerobic ammonium oxidation (anammox) bacterial abundance, anammox rates and anammox contribution to N2
| Study sites | Season | Abundance /(copies/g) | Annamox rate/[nmol N/(g·h)] | Contribution /% | References |
| The Yangtze Estuary and adjacent coastal areas | March & July | 3.67×105-8.22×107 | 0.08-6.46 | 5-77 | [59] |
| The coastal wetlands of China | Summer | 2.87 ± 0.80×107 | 0.29±0.06 | 4.5-5.8 | [58] |
| Winter | 2.05 ± 0.60×107 | 0.13±0.03 | 3.8-10.7 | ||
| The Yangtze Estuary | Summer | 4.54×106-1.56×107 | 3.92-6.61 | 6.6-12.9 | [57] |
| Winter | 2.63×106-9.48×106 | 0.94-2.33 | ~6.6-11 | ||
| The Pearl River Delta | Summer | 1.3×106-1.2×107 | 0.04-0.08 | 0.5-0.9 | [56] |
| Winter | 1.4×108-1.0×109 | 0.53-1.38 | 3.8-7.0 | ||
| Assume dry sediment bulk density is 1.88 g/cm3. | |||||
表选项
3.2 厌氧氨氧化菌与其他菌 理论上厌氧氨氧化菌是严格厌氧的,然而在某些有氧的环境中检测到了厌氧氨氧化菌的存在[38, 60]。研究指出可能的原因是氨氧化古菌如Nitrosopumilus maritimus及其他一些细菌如Gammaproteobacteria、Alphaproteobacteria、Bacteroidetes等能为厌氧氨氧化菌创造厌氧微环境,从而满足厌氧氨氧化菌的生长需求[61]。Zhang等[39]在研究高氮的滩涂时发现厌氧氨氧化菌和AOA有相关性,但与AOB无关。有****[60]在研究污水处理中发现AOB是该系统厌氧氨氧化过程NO2-的主要提供者。Wang等[56]分别测定了冬天和夏天的硝化过程和反硝化过程的速率,发现夏天NO2-主要来源于反硝化过程,冬天NO2-主要来源于硝化过程,这一结果也得到了Hou等[58]的支持。反硝化过程提供的NO2-可促进厌氧氨氧化过程[62],众多****研究表明NO2-转化为NO2-的速度要比厌氧氨氧化消耗NO2-的速度快[51, 63-64],这说明NO3-还原为NO2-不是厌氧氨氧化过程的限速过程。
3.3 基于功能基因探究厌氧氨氧化过程 分析厌氧氨氧化常用的功能基因有联氨氧化还原酶基因hzo,厌氧氨氧化细菌特有的亚硝酸还原酶基因nirS,联胺合成酶基因hzsA、hzsB,然而使用不同的功能基因或16S rRNA技术进行研究时可能得出不同的结论[65]。使用16S rRNA在分析厌氧氨氧化菌多样性时有一定的局限性,其一是与目标功能菌不具有直接相关性,其二是已有引物难以覆盖所有厌氧氨氧化菌的多样性[66];有****提出基于功能基因hzo的检测比16S rRNA更详细和高效[67-68];与16S rRNA和hzo相比,nirS表现出一致的系统发育关系,还可量化厌氧氨氧化细菌的丰度、空间分布等,因而被作为新的标记物[67];hzsA是厌氧氨氧化菌系统发育过程中独特的标记物,基于hzsA的检测可涵盖目前已知的所有厌氧氨氧化菌[66]。
3.3.1 厌氧氨氧化菌丰度、群落多样性和活性: 研究表明季节对厌氧氨氧化菌丰度的影响无明显规律。Li等[68]和Bale等[69]基于功能基因hzo、hzsA发现夏天厌氧氨氧化菌丰度高于冬天,而Zheng等[59]发现厌氧氨氧化菌夏季的平均丰度略高于春季,也有****发现季节对无厌氧氨氧化菌丰度无明显影响[57-58]。
厌氧氨氧化菌的多样性受纬度、温度、盐度等因素的影响。Dale等[70]和Fu等[71]在河口和滨海湿地系统中都检测到5个属;Hou等[58]和Li等[41, 72]在黄河长江和珠江滨海湿地以及米埔红树林湿地中检测厌氧氨氧化菌4个属,包括Candidatus Scalindua、Kuenenia、Brocadia、Jettenia,Hou等[57]发现不同取样点的厌氧氨氧化菌的群落组成在不同的季节无显著差异,在纬度上有显著差异,低纬度的多样性高于高纬度,并猜测温度可能是影响厌氧氨氧化菌群落多样性的主要因素。Hu等[73]在研究钱塘江时发现厌氧氨氧化菌有较高的多样性,包括Brocadia、Scalindua、Kueneni和3个新的cluster;当研究区域变成河口时,同样的在长江河口沉积物中检测到Brocadia、Scalindua、Kueneni菌群和2个新cluster[57],在珠江河口中Brocadia、Kueneni为优势菌群[74];范围往海洋区域靠近后,研究发现顺着盐度梯度,低盐度区的河口区的优势菌群为Brocadia,Shen等[65]在淡水区也观察到这样的现象,中盐度区盐度,Scalindua与Kuenenia共存,Scalindua的丰度逐渐高于Brocadia变为优势菌群,至高盐度区时Scalindua几乎占绝对优势[59]。
Jiang等[75]将崇明岛东滩的样品进行室内培养,发现厌氧氨氧化菌的活性和丰度及厌氧氨氧化过程速率随着盐度的上升呈现先增加后减少的趋势。在长江河口和海岸区域厌氧氨氧化菌的速率在0.08到6.46 nmol N/(g·h)之间,其中中盐度区Y7点的厌氧氨氧化速率最高为6.46 nmol N/(g·h),高盐度区厌氧氨氧化速率降低,低盐度区Y4点的厌氧氨氧化速率最低,为0.08 nmol N/(g·h),这说明厌氧氨氧化速率的活性受盐度影响较大[59]。
3.3.2 人为氮源梯度的影响: Han等[74]发现,在珠江河口受人为无机氮污染的区域,珠江水携带大量无机氮经过米埔红树林保护区,后汇入南海浅水区到南海深水区,形成一条无机氮梯度变化带,他们研究这些区域的厌氧氨氧化过程发现米埔的厌氧氨氧化菌多样性最高,接着是南海浅水区、南海深水区,珠江河口的厌氧氨氧化多样性菌最低。在珠江的沉积物中占优势的为Kuenenia、Brocadia,米埔湿地则为Kuenenia和Scalindua。此外,厌氧氨氧化过程还与NH4+、NO3-、NO2-及其之间的比值[68, 76-77]、pH、重金属(如铜[39])、水深[76]、生物扰动[38]等有关。其中水深的影响较为明显,水深超过20 m的区域厌氧氨氧化对氮去除的贡献可达65%以上[76]。Bale等[69]对北海的厌氧氨氧化过程研究发现沉积物特性与该过程的活性丰度有关。红树林对厌氧氨氧化菌也有影响,离红树林越近的区域,hzo丰度越高[68]。
总而言之,厌氧氨氧化的速率较高,但是其对滨海湿地生态系统的氮去除的贡献相对较小,这可能是因为滨海湿地生态系统多为较浅的海洋沿岸和河口,而厌氧氨氧化过程主要发生在沉积物中。
4 反硝化 反硝化是指在厌氧条件下将NO3-还原成N2的过程(NO3-→NO2-→NO→N2O→N2)。其中N2O是近年来研究较多、对臭氧层破坏作用大的温室气体,其单位体积的增温潜能是CO2的300多倍[78]。近年来反硝化型厌氧甲烷氧化过程(denitrifying anaerobic methane oxidation,DAMO)引起了****的广泛关注,该过程以甲烷为电子供体,能达到脱氮、减缓温室效应的目的。
随着生活废水和工业废水大量排入滨海湿地生态系统,大量含氮污染物进入沉积物中[79]。反硝化过程对滨海湿地生态系统中的氮去除起着重要作用,有研究指出滨海湿地生态系统中超过50%的溶解性无机氮是通过反硝化过程去除的[80]。具有反硝化功能的微生物多为兼性厌氧化能异养菌,包括芽孢杆菌属(Bacillus)、假单胞菌属(Pseudomonas)、微球菌(Micrococcus)、产气单胞菌属(Aeromonas)、弧菌属(Vibrio)、气杆菌属(Aerobacter)、产碱杆菌属(Alcalogenes)、短杆菌(Brevibacterium)、黄杆菌属(Flavobactrium)等。
反硝化过程涉及的酶较多。研究反硝化过程常用的功能基因有硝酸还原酶的编码基因narG,亚硝酸还原酶的编码基因nirS与nirK,一氧化氮还原酶的编码基因norB,以及氧化亚氮还原酶的编码基因nosZ[81-82]。但研究发现某些非反硝化菌也具有反硝化功能基因,比如Vibrio succinogenes[83]具有nosZ基因,奇古菌具有nirK的同源基因[84],因此不能单独使用这些功能基因进行功能菌的鉴定。Huang等[85]基于不同的功能基因分析珠江的反硝化过程发现nirS与nirK主要分布于深度0-10 cm处,nosZ分布于深度5-15 cm处,而narG集中分布在15 cm以下;在高NO3-浓度条件下,narG和nirS的丰度较高,而在贫营养环境中则是nosZ具有更高的活性。
4.1 反硝化过程中的关键功能基因 nirS和nirK是反硝化过程中的两个关键功能基因,盐度对nirS和nirK有显著影响[86]。nirK、nirS虽然结构不同但功能一致。目前发现nirS与nirK可在不同环境中可共存,但尚未发现nirS与nirK共存于同一微生物中[86]。在有氧气的环境中nirK占优势[87],推测nirS与nirK型反硝化菌在环境中的生态位有差异[39, 81]。一般情况下nirS型反硝化菌在海洋环境中的丰度更高,而nirK型反硝化菌在河流环境中的丰度更高[80, 88]。莱州湾的nirK型反硝化微生物数量高于nirS型,进一步研究发现nirK型和nirS型反硝化微生物的比值取决于pH、NO2-、NO3-和Cd,当pH低和NOx--N低时nirK/nirS比值高[39]。在芝加哥湾、Fitzroy河,nirS的丰度和多样性比nirK高[20, 80],在Fitzroy河口nirS/nirK为9.9-22.1,而且nirS丰度与反硝化潜能正相关,早些的研究发现在河口沉积物中虽然提取了nirS和nirK,但是只有nirS显著表达[82],这表明在河口地区nirS型反硝化菌相比nirK型更重要。Gao等[88]基于功能基因nirS进一步研究中国滨海湿地生态系统发现nirS型反硝化菌的多样性有空间差异,但无季节差异。
4.2 反硝化速率环境影响因子 盐度、pH、溶解性无机氮(DIN)、溶解性有机碳、温度被认为是影响反硝化过程的重要因素。
有研究发现盐度对NH4+的吸收利用有影响,进而影响反硝化和硝化过程[89]。Neuse河顺着盐度梯度反硝化速率差异较大,从0到275 mmol N/(m2·h),平均为75.8 mmol N/(m2·h),但与盐度关系不显著[90],Magalhaes等[91]和Laverman等[92]的研究结论与此一致。反硝化菌和溶解性无机氮(DIN)有很高的相关性,DIN的变化影响反硝化功能基因的分布、nirS型反硝化菌群落结构[85]。Gao等[88]在中国滨海湿地发现不同纬度nirS型菌的分布有显著性差异,这说明温度影响是nirS型菌分布和组成的主要因素。
季节对反硝化过程的影响见表 2。滨海湿地生态系统土壤中活性溶解有机碳和NO3-的含量较高,反硝化速率较快。以年来看,反硝化的速率为96.2-170.3 g N/(m2·yr)[56]。不同的研究对于反硝化速率的测定数据有所差异,季节对于反硝化速率的影响也有所不同。在很多河口区,反硝化速率呈现春高夏低的季节变化规律[56, 90],这可能是春季时,硝化-反硝化紧密耦合,硝化过程产生的NO3-最终被反硝化还原成N2;夏季时,表层沉积物的低溶氧导致硝化速率降低,尽管反硝化潜力与春季相当,但实际反硝化速率大大降低[93]。也有研究发现冬天的反硝化速率相比于夏天较高[56, 90],9-11月的反硝化速率较高,3-5月较低[20, 94-95],这可能和研究点溶解有机碳的供给有关。
表 2. 不同季节各地反硝化过程速率 Table 2. Spatiotemporal variations of denitrification in different seasons
| Study sites | Season | Denitrification rate/(nmol N/(g·h)) | Refs |
| The Yangtze Estuary and adjacent coastal areas | March & July | 0.02-14.78 | [59] |
| The coastal wetlands of China | Summer | 3.66-7.59 | [58] |
| Winter | 0.87-3.29 | ||
| The Yangtze Estuary | Summer | 41.82-67.13 | [57] |
| Winter | 12.04-20.03 | ||
| The Pearl River Delta | Summer | 16.17-37.04 | [56] |
| Winter | 41.74-65.8 | ||
| Assume dry sediment bulk density is 1.88 g/cm3. | |||
表选项
5 NO3-还原成铵(DNRA) 滨海湿地生态系统中研究集中于NO3-异化还原成NH4+(DNRA)的研究。NO3-还原成铵(dissimilatory nitrate reduction to ammonium,DNRA)是NO3-还原为NH4+的过程,和反硝化、厌氧氨氧化过程一样,DNRA是NO3-还原的重要过程。反硝化和厌氧氨氧化将硝氮转化为N2或N2O,从滨海湿地生态系统中去除氮,而DNRA将硝氮转化为生物可利用的氨氮存留在系统中,供植物、细菌利用,或再被氧化为NO3-。氨氮的产生可提高生态系统初级生产力,NO2-的消除可减少对细胞的毒害作用。该过程严格厌氧,主要发生在沉积物15 cm以下[85]。执行DNRA过程的微生物有以有机物为电子供体的异养型和以H2S、Fe(Ⅱ)或其他无机物为电子供体的化能自养型[96-97]。在滨海湿地生态系统中能执行DNRA过程的微生物包括化能自养型硫细菌如贝扎托菌属(Beggiatoa)和辫硫菌属(Thioploca)[98]、肠原杆菌、Staphylococcus carnosus、Bacillus、Clostridia、Vibrio、Pscudomonas、Azospirillum、Aeromonas[96, 99]等。
DNRA过程分为NO3-还原成NO2-过程和NO2-还原成NH4+过程。执行DNRA过程的微生物种类繁多,常用功能基因nrfA探究该过程[99-100]及研究DNRA微生物多样性。nrfA基因相比于nirB存在于更多的微生物中[99],nrfA基因对低浓度NO3-更敏感[101],nirB适用于NO3-高浓度情况。DNRA过程对Little Lagoon总NH4+通量贡献5%-80%[21]。基于功能基因nrfA的研究发现黄河DNRA速率及nrfA丰度和盐度呈负相关关系[102]。nrfA的分布受NH4+浓度的影响,在碳受限的情况下,DNRA过程被抑制,虽然随着土层往下,nrfA基因丰度减小,但相对丰度增加[85]。功能基因的丰度和相应过程的速率不呈正相关关系。通过nrfA来检测执行DNRA的细菌时,有些细菌(如Shewanella oneidensis MR-1、Thioalkalivibrio nitratireducens)虽可执行DNRA过程,但是其并无nrfA基因[97],因此需要与其他技术相结合。
5.1 DNRA速率 近年来除了被广泛提及的反硝化过程外,许多****提出DRNA对氮去除的贡献不可忽略,甚至起主要作用。nrfA丰度、DNRA过程速率及对硝酸盐还原的贡献如表 3所示。Laverman等[92]用FTRs测定NO3-还原速率为200 nmol N/(cm3·h),其中反硝化为NO3-还原贡献约50%,其余的以DNRA为主,DNRA对氮去除的贡献十分值得重视。以Little Lagoon为例,其DNRA速率平均为54.1 μmol N/(m2·h),远高于反硝化速率(7.7 μmol N/(m2·h))。DNRA对NO3-还原的贡献率为30%-40%,在5月至8月贡献率甚至达到84%,相似的DNRA速率在Florida和Texas河口地区也有观测到[21, 103-104]。研究指出DNRA在电子供体丰富、可利用NO3-浓度低和温度高的环境中占主要优势[105]。在不同的季节下DNRA速率相差较大,Little Lagoon在冬天平均DNRA速率为1 μmol N/(m2·h),而在夏天最高时可达到168 μmol N/(m2·h)[21]。DNRA速率与有机物、可利用NH4+呈显著的正相关关系[102]。盐度对DNRA过程的影响则是矛盾的,一方面有研究表明底泥的盐度增加可能导致NH4+和溶解性有机碳的瞬时释放,增加从底泥到水体NH4+的流出量,提高NO3-还原速率和NO2-产生速率[92];另一方面长江河口DNRA菌的活性和丰度随着盐度梯度变化无明显趋势[102]。
表 3. 不同季节各地nrfA丰度、DNRA过程速率及DNRA对NO3-还原的贡献 Table 3. Spatiotemporal variations of nrfA abundance, DNRA rates and DNRA contribution to total NO2- reduction
| Study sites | Season | DNRA rate/(nmol N/(g·h)) | Abundance/(copies/g) | Contribution/% | Refs |
| The Yangtze Estuary | Summer | 0.71-5.57 | 2.76×108-12.01×108 | 10-30 | [106] |
| Winter | 0.14-2.98 | 1.70×108-9.30×108 | |||
| The Yellow River Estuary | May | 0.10-3.29 | ~3.00×109-2.27×1010 | 8.68-42.07 | [102] |
| The coastal wetlands of China | Summer | 0.48-0.81 | - | - | [58] |
| Winter | 0.15-0.42 | ||||
| Little Lagoon | 2/4/5/8/10/11 | 2.88 | - | 30-40 | [21] |
| Assume dry sediment bulk density is 1.88 g/cm3. | |||||
表选项
5.2 影响DNRA和反硝化的主要因素 同为氮去除过程,DNRA和反硝化常被放在一起比较。研究表明DNRA在电子供体丰富、可利用NO3-浓度低和温度高的环境中占主要优势[105]。在含氧量低或缺氧的环境中影响DNRA和反硝化过程的因素有易降解的有机碳、NO3-可利用性、C/N比、硫化物浓度、土壤性质、pH、温度、微生物世代时间、NO3-/NO2-、盐度[97, 105, 107-108]。在碳受限的情况下,DNRA过程被抑制[85];van den Berg等[109]在氮受限DNRA过程占优势的情况下,提高电子供体富集培养了DNRA菌。在HS-丰富的厌氧环境中,HS-作为电子供体可以干扰硝化过程和限制反硝化过程NO3-的供应,使得DNRA过程在NO3-还原过程中占优势,在Little Lagoon研究中得到了验证,夏天反硝化和DNRA过程速率达到最高时仍相差一个数量级[21]。Decleyre等[110]取潮间带泥滩地沉积物实验表明小范围的土壤异质性会影响DNRA过程速率。一方面HS-浓度与pH、温度正相关[21],通过影响HS-浓度影响DNRA过程和反硝化过程,另一方面温度高、SOD增加为其创造了还原环境[97, 104, 111]。Yoon等[112]通过研究具有反硝化功能和DNRA功能的菌发现NO2-/NO3-比值大小影响Shewanella loihica strain PV-4对氮去除路径的选择,当2-/NO3-比值高时,以DNRA过程为主。盐度的增加会提高DNRA的速率[107, 113-114]。盐度对DNRA过程的影响有两种解释:第一种认为这是由于与DNRA过程竞争的反硝化过程受到盐度的抑制作用,但也有研究表明反硝化对盐度并不敏感,故这一观点还存在争议。另一个解释是随着盐度升高,SO42-含量下降,硫还原细菌的DNRA过程的电子供体(即低价态硫化物)就相应增多了,这既能抑制反硝化过程又能促进DNRA过程。
综上所述,针对氮去除(DNRA、反硝化、厌氧氨氧化)过程,DNRA过程在可利用有机碳多、H2S、低NO3-、温度高、盐度较高的环境下为氮去除的主要方式;可利用的有机碳少的情况下反硝化和厌氧氨氧化过程为氮去除的主要方式,在这种情况下,H2S的存在对厌氧氨氧化过程的影响变小。DNRA在滨海湿地生态系统中比反硝化过程占优势时可能的原因可归纳为:(1) DNRA过程的自由能;(2)相比反硝化菌,NO3-和NO3-铵化细菌的亲和力更高[105]。当DNRA速率高于反硝化速率时,增加的氮排入和氮去除是负反馈关系,被固定的氮仍存于系统中[103-104],可能会加剧水体尤其是海洋富营养化和HABs (harmful algal blooms)。
6 总结和展望 本文综述了滨海湿地生态系统氮循环研究进展,得益于宏基因组学研究的兴起,使得我们对环境微生物的认识不只是局限于有限的可培养的微生物。发展成熟的高通量测序技术和基因芯片技术具有全面、准确、信息深入的特点。本综述中提到的众多研究通过宏基因组学研究解答了传统生物学方法所不能回答的问题,极大地丰富及补充了微生物驱动下氮循环中的理论知识。同时也发现了新的、拟解决以及有待进一步深入探讨的科学问题,概述如下:(1)基于纯培养进行厌氧氨氧化菌的代谢过程及其功能活性特征的研究。厌氧氨氧化菌生长十分缓慢、生长条件要求严格,其纯培养目前仍是尚待解决的难题。(2)优化计算水质模型及检测微生物代谢活性方法。引入多维度、多因子可以更真实全面地分析环境因子与微生物之间的关系,为揭示微生物的代谢规律进而挖掘其功能提供更准确的信息。(3)一定的局限性。需注意引物的选择,针对同一过程,可采用2种以上的功能基因使结果更加真实可信;宏基因组测序可检测环境中所有基因,但常由于测序深度不够,仅能检测到丰度高的基因或种群;单细胞基因组测序有利于氮循环微生物群落结构和功能分析。(4)滨海湿地生态系统中Comammox过程速率及微生物的研究急需开展以重新认识硝化过程。进一步值得研究的还有Comamox微生物和AOA、AOB在滨海湿地生态系统中的生态位以及对硝化作用的贡献。(5)目前滨海湿地生态系统固氮过程的速率相比于其他过程较低,可能是外源性氮排入对滨海湿地生态系统固氮过程的抑制,具体原因有待进一步研究。(6)目前关于环境因子对氮循环过程速率及微生物群落的研究众多,但更深层次的认识仍不够。氮循环与碳、磷、硫循环的偶联机制及生态学意义值得大家进行深入的探索[115]。滨海湿地生态系统是海洋水体最重要的防线之一,国内对滨海湿地生态系统的研究还相对较少,因此深入了解其中的物质循环规律对维系其生态平衡并保护海洋水资源有着重要的参考价值。
References
| [1] | Martínez ML, Intralawan A, Vázquez G, Pérez-Maqueo O, Sutton P, Landgrave R. The coasts of our world:ecological, economic and social importance. Ecological Economics, 2007, 63(2/3): 254-272. |
| [2] | Gabler CA, Osland MJ, Grace JB, Stagg CL, Day RH, Hartley SB, Enwright NM, From AS, McCoy ML, McLeod JL. Macroclimatic change expected to transform coastal wetland ecosystems this century. Nature Climate Change, 2017, 7(2): 142-147. |
| [3] | Costanza R, D'Arge R, de Groot R, Farber S, Grasso M, Hannon B, Limburg K, Naeem S, O'Neill RV, Paruelo J, Raskin RG, Sutton P, van den Belt M. The value of the world's ecosystem services and natural capital. Ecological Economics, 1998, 25(1): 3-15. DOI:10.1016/S0921-8009(98)00020-2 |
| [4] | Keddy PA, Fraser LH, Solomeshch AI, Junk WJ, Campbell DR, Arroyo MTK, Alho CJR. Wet and wonderful:the world's largest wetlands are conservation priorities. Bioscience, 2009, 59(1): 39-51. DOI:10.1525/bio.2009.59.1.8 |
| [5] | Yan WJ, Mayorga E, Li XY, Seitzinger SP, Bouwman AF. Increasing anthropogenic nitrogen inputs and riverine DIN exports from the Changjiang River basin under changing human pressures. Global Biogeochemical Cycles, 2010, 24(24): GB0A06. |
| [6] | Rabalais NN, Turner RE, Díaz RJ, Justi? D. Global change and eutrophication of coastal waters. ICES Journal of Marine Science, 2009, 66(7): 1528-1537. DOI:10.1093/icesjms/fsp047 |
| [7] | Seitzinger S. Nitrogen cycle:out of reach. Nature, 2008, 452(7184): 162-163. DOI:10.1038/452162a |
| [8] | Cai WJ, Hu XP, Huang WJ, Murrell MC, Lehrter JC, Lohrenz SE, Chou WC, Zhai WD, Hollibaugh JT, Wang YC, Zhao PS, Guo XH, Gundersen K, Dai MH, Gong GC. Acidification of subsurface coastal waters enhanced by eutrophication. Nature Geoscience, 2011, 4(11): 766-770. DOI:10.1038/ngeo1297 |
| [9] | He ZL, Gentry TJ, Schadt CW, Wu LY, Liebich J, Chong SC, Huang ZJ, Wu WM, Gu BH, Jardine P, Criddle C, Zhou JZ. GeoChip:a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. The ISME Journal, 2007, 1(1): 67-77. DOI:10.1038/ismej.2007.2 |
| [10] | He ZL, Xu MY, Deng Y, Kang S, Kellogg L, Wu LY, Van Nostrand JD, Hobbie SE, Reich PB, Zhou JZ. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecology Letters, 2010, 13(5): 564-575. DOI:10.1111/j.1461-0248.2010.01453.x |
| [11] | He ZL, Xiong JB, Kent AD, Deng Y, Xue K, Wang GJ, Wu LY, Van Nostrand JD, Zhou JZ. Distinct responses of soil microbial communities to elevated CO2and O3 in a soybean agro-ecosystem. The ISME Journal, 2014, 8(3): 714-726. DOI:10.1038/ismej.2013.177 |
| [12] | Li P, Jiang Z, Wang YH, Deng Y, Van Nostrand JD, Yuan T, Liu H, Wei DZ, Zhou JZ. Analysis of the functional gene structure and metabolic potential of microbial community in high arsenic groundwater. Water Research, 2017, 123: 268-276. DOI:10.1016/j.watres.2017.06.053 |
| [13] | Zhou JZ, He ZL, Yang YF, Deng Y, Tringe SG, Alvarez-Cohen L. High-throughput metagenomic technologies for complex microbial community analysis:open and closed formats. mBio, 2015, 6(1): e02288-14. |
| [14] | Bai SJ, Li JW, He ZL, Van Nostrand JD, Tian Y, Lin GH, Zhou JZ, Zheng TL. GeoChip-based analysis of the functional gene diversity and metabolic potential of soil microbial communities of mangroves. Applied Microbiology and Biotechnology, 2013, 97(15): 7035-7048. DOI:10.1007/s00253-012-4496-z |
| [15] | Yergeau E, Kang S, He ZL, Zhou JZ, Kowalchuk GA. Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. The ISME Journal, 2007, 1(2): 163-179. DOI:10.1038/ismej.2007.24 |
| [16] | Xu MY, He ZL, Deng Y, Wu LY, Van Nostrand JD, Hobbie SE, Reich PB, Zhou JZ. Elevated CO2 influences microbial carbon and nitrogen cycling. BMC Microbiology, 2013, 13: 124. DOI:10.1186/1471-2180-13-124 |
| [17] | Xu MY, Zhang Q, Xia CY, Zhong YM, Sun GP, Guo J, Yuan T, Zhou JZ, He ZL. Elevated nitrate enriches microbial functional genes for potential bioremediation of complexly contaminated sediments. The ISME Journal, 2014, 8(9): 1932-1944. DOI:10.1038/ismej.2014.42 |
| [18] | Zehr JP, Jenkins BD, Short SM, Steward GF. Nitrogenase gene diversity and microbial community structure:a cross-system comparison. Environmental Microbiology, 2003, 5(7): 539-554. DOI:10.1046/j.1462-2920.2003.00451.x |
| [19] | Gaby JC, Buckley DH. A global census of nitrogenase diversity. Environmental Microbiology, 2011, 13(7): 1790-1799. DOI:10.1111/j.1462-2920.2011.02488.x |
| [20] | Abell GCJ, Revill AT, Smith C, Bissett AP, Volkman JK, Robert SS. Archaeal ammonia oxidizers and nirS-type denitrifiers dominate sediment nitrifying and denitrifying populations in a subtropical macrotidal estuary. The ISME Journal, 2010, 4(2): 286-300. DOI:10.1038/ismej.2009.105 |
| [21] | Bernard RJ, Mortazavi B, Kleinhuizen AA. Dissimilatory nitrate reduction to ammonium (DNRA) seasonally dominates NO3- reduction pathways in an anthropogenically impacted sub-tropical coastal lagoon. Biogeochemistry, 2015, 125(1): 47-64. DOI:10.1007/s10533-015-0111-6 |
| [22] | Short SM, Zehr JP. Nitrogenase gene expression in the Chesapeake Bay Estuary. Environmental Microbiology, 2007, 9(6): 1591-1596. DOI:10.1111/emi.2007.9.issue-6 |
| [23] | Sohm JA, Webb EA, Capone DG. Emerging patterns of marine nitrogen fixation. Nature Reviews Microbiology, 2011, 9(7): 499-508. DOI:10.1038/nrmicro2594 |
| [24] | Zhou HX, Dang HY, Klotz MG. Environmental conditions outweigh geographical contiguity in determining the similarity of nifH-harboring microbial communities in sediments of two disconnected marginal seas. Frontiers in Microbiology, 2016: 71111. |
| [25] | Affourtit J, Zehr JP, Paerl HW. Distribution of nitrogen-fixing microorganisms along the Neuse River Estuary, North Carolina. Microbial Ecology, 2001, 41(2): 114-123. |
| [26] | Short SM, Jenkins BD, Zehr JP. Spatial and temporal distribution of two diazotrophic bacteria in the Chesapeake Bay. Applied and Environmental Microbiology, 2004, 70(4): 2186-2192. DOI:10.1128/AEM.70.4.2186-2192.2004 |
| [27] | Thajudeen J, Yousuf J, Veetil VP, Varghese S, Singh A, Abdulla MH. Nitrogen fixing bacterial diversity in a tropical estuarine sediments. World Journal of Microbiology and Biotechnology, 2017, 33(2): 41. DOI:10.1007/s11274-017-2205-x |
| [28] | Van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op Den Camp HJM, Kartal B, Jetten MSM, Lücker S. Complete nitrification by a single microorganism. Nature, 2015, 528(7583): 555-559. DOI:10.1038/nature16459 |
| [29] | Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, Von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. Complete nitrification by Nitrospira bacteria. Nature, 2015, 528(7583): 504-509. DOI:10.1038/nature16461 |
| [30] | Pinto AJ, Marcus DN, Ijaz UZ, Bautista-de Lose Santos QM, Dick GJ, Raskin L. Metagenomic evidence for the presence of comammox Nitrospira-like bacteria in a drinking water system. mSphere, 2015, 1(1): e00054-15. |
| [31] | Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, Daebeler A, Romano S, Albertsen M, Stein LY, Daims H, Wagner M. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature, 2017, 549(7671): 269-272. DOI:10.1038/nature23679 |
| [32] | Pester M, Rattei T, Flechl S, Gr?ngr?ft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environmental Microbiology, 2012, 14(2): 525-539. DOI:10.1111/j.1462-2920.2011.02666.x |
| [33] | Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu DY, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. Environmental genome shotgun sequencing of the Sargasso Sea. Science, 2004, 304(5667): 66-74. DOI:10.1126/science.1093857 |
| [34] | Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environmental Microbiology, 2005, 7(12): 1985-1995. DOI:10.1111/j.1462-2920.2005.00906.x |
| [35] | Prosser JI, Nicol GW. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environmental Microbiology, 2008, 10(11): 2931-2941. DOI:10.1111/emi.2008.10.issue-11 |
| [36] | K?nneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature, 2005, 437(7058): 543-546. DOI:10.1038/nature03911 |
| [37] | Cao HL, Li M, Hong YG, Gu JD. Diversity and abundance of ammonia-oxidizing archaea and bacteria in polluted mangrove sediment. Systematic and Applied Microbiology, 2011, 34(7): 513-523. DOI:10.1016/j.syapm.2010.11.023 |
| [38] | Lipsewers YA, Bale NJ, Hopmans EC, Schouten S, Sinninghe Damsté JS, Villanueva L. Seasonality and depth distribution of the abundance and activity of ammonia oxidizing microorganisms in marine coastal sediments (North Sea). Frontiers in Microbiology, 2014, 5: 472. |
| [39] | Zhang XL, Agogué H, Dupuy C, Gong J. Relative abundance of ammonia oxidizers, denitrifiers, and anammox bacteria in sediments of hyper-nutrified estuarine tidal flats and in relation to environmental conditions. CLEAN-Soil Air Water, 2014, 42(6): 815-823. DOI:10.1002/clen.v42.6 |
| [40] | Wang YF, Gu JD. Higher diversity of ammonia/ammonium-oxidizing prokaryotes in constructed freshwater wetland than natural coastal marine wetland. Applied Microbiology and Biotechnology, 2013, 97(15): 7015-7033. DOI:10.1007/s00253-012-4430-4 |
| [41] | Li M, Cao HL, Hong YG, Gu JD. Spatial distribution and abundances of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in mangrove sediments. Applied Microbiology and Biotechnology, 2011, 89(4): 1243-1254. DOI:10.1007/s00253-010-2929-0 |
| [42] | Han P, Li M, Gu JD. Biases in community structures of ammonia/ammonium-oxidizing microorganisms caused by insufficient DNA extractions from Baijiang soil revealed by comparative analysis of coastal wetland sediment and rice paddy soil. Applied Microbiology and Biotechnology, 2013, 97(19): 8741-8756. DOI:10.1007/s00253-013-5169-2 |
| [43] | Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature, 2006, 442(7104): 806-809. DOI:10.1038/nature04983 |
| [44] | Jia ZJ, Conrad R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environmental Microbiology, 2009, 11(7): 1658-1671. DOI:10.1111/emi.2009.11.issue-7 |
| [45] | Zhang QF, Peng JJ, Chen Q, Li XF, Xu CY, Yin HB, Yu S. Impacts of Spartina alterniflora invasion on abundance and composition of ammonia oxidizers in estuarine sediment. Journal of Soils and Sediments, 2011, 11(6): 1020-1031. DOI:10.1007/s11368-011-0369-9 |
| [46] | Santoro AE, Francis CA, de Sieyes NR, Boehm AB. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environmental Microbiology, 2008, 10(4): 1068-1079. DOI:10.1111/j.1462-2920.2007.01547.x |
| [47] | Cao HL, Hong YG, Li M, Gu JD. Community shift of ammonia-oxidizing bacteria along an anthropogenic pollution gradient from the Pearl River Delta to the South China Sea. Applied Microbiology and Biotechnology, 2012, 94(1): 247-259. DOI:10.1007/s00253-011-3636-1 |
| [48] | Levy-Booth DJ, Prescott CE, Grayston SJ. Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biology and Biochemistry, 2014, 75: 11-25. DOI:10.1016/j.soilbio.2014.03.021 |
| [49] | Prosser JI, Nicol GW. Archaeal and bacterial ammonia-oxidisers in soil:the quest for niche specialisation and differentiation. Trends in Microbiology, 2012, 20(11): 523-531. DOI:10.1016/j.tim.2012.08.001 |
| [50] | Kartal B, Rattray J, van Niftrik LA, van de Vossenberg J, Schmid MC, Webb RI, Schouten S, Fuerst JA, Damsté JS, Jetten MSM, Strous M. Candidatus "Anammoxoglobus propionicus" a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Systematic and Applied Microbiology, 2007, 30(1): 39-49. DOI:10.1016/j.syapm.2006.03.004 |
| [51] | Thamdrup B, Dalsgaard T. Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Applied and Environmental Microbiology, 2002, 68(3): 1312-1318. DOI:10.1128/AEM.68.3.1312-1318.2002 |
| [52] | Kuypers MMM, Sliekers AO, Lavik G, Schmid M, J?rgensen BB, Kuenen JG, Damsté JSS, Strous M, Jetten MSM. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature, 2003, 422(6932): 608-611. DOI:10.1038/nature01472 |
| [53] | Schouten S, Strous M, Kuypers MMM, Rijpstra WIC, Baas M, Schubert CJ, Jetten MSM, Damsté JSS. Stable carbon isotopic Fractionations associated with inorganic carbon fixation by anaerobic ammonium-oxidizing bacteria. Applied and Environmental Microbiology, 2004, 70(6): 3785-3788. DOI:10.1128/AEM.70.6.3785-3788.2004 |
| [54] | Schmid MC, Maas B, Dapena A, van de Pas-Schoonen K, van de Vossenberg J, Kartal B, van Niftrik L, Schmidt I, Cirpus I, Kuenen JG, Wagner M, Damsté JSS, Kuypers M, Revsbech NP, Mendez R, Jetten MSM, Strous M. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Applied and Environmental Microbiology, 2005, 71(4): 1677-1684. DOI:10.1128/AEM.71.4.1677-1684.2005 |
| [55] | Kartal B, Van Niftrik L, Rattray J, Van De Vossenberg JLCM, Schmid MC, Damsté JSS, Jetten MSM, Strous M. Candidatus 'Brocadia fulgida':an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiology Ecology, 2008, 63(1): 46-55. DOI:10.1111/fem.2008.63.issue-1 |
| [56] | Wang SY, Zhu GB, Peng YZ, Jetten MSM, Yin CQ. Anammox bacterial abundance, activity, and contribution in riparian sediments of the Pearl River Estuary. Environmental Science & Technology, 2012, 46(16): 8834-8842. |
| [57] | Hou LJ, Zheng YL, Liu M, Gong J, Zhang XL, Yin GY, You L. Anaerobic ammonium oxidation (anammox) bacterial diversity, abundance, and activity in marsh sediments of the Yangtze Estuary. Journal of Geophysical Research-Biogeosciences, 2013, 118(3): 1237-1246. DOI:10.1002/jgrg.v118.3 |
| [58] | Hou LJ, Zheng YL, Liu M, Li XF, Lin XB, Yin GY, Gao J, Deng FY, Chen F, Jiang XF. Anaerobic ammonium oxidation and its contribution to nitrogen removal in China's coastal wetlands. Scientific Reports, 2015, 5: 15621. DOI:10.1038/srep15621 |
| [59] | Zheng YL, Jiang XF, Hou LJ, Liu M, Lin XB, Gao J, Li XF, Yin GY, Yu CD, Wang R. Shifts in the community structure and activity of anaerobic ammonium oxidation bacteria along an estuarine salinity gradient. Journal of Geophysical Research: Biogeosciences, 2016, 121(6): 1632-1645. DOI:10.1002/2015JG003300 |
| [60] | Wang SY, Peng YZ, Ma B, Wang SY, Zhu GB. Anaerobic ammonium oxidation in traditional municipal wastewater treatment plants with low-strength ammonium loading:widespread but overlooked. Water Research, 2015, 84: 66-75. DOI:10.1016/j.watres.2015.07.005 |
| [61] | Woebken D, Fuchs BM, Kuypers MMM, Amann R. Potential interactions of particle-associated anammox bacteria with bacterial and archaeal partners in the Namibian upwelling system. Applied and Environmental Microbiology, 2007, 73(14): 4648-4657. DOI:10.1128/AEM.02774-06 |
| [62] | Meyer RL, Risgaard-Petersen N, Allen DE. Correlation between anammox activity and microscale distribution of nitrite in a subtropical mangrove sediment. Applied and Environmental Microbiology, 2005, 71(10): 6142-6149. DOI:10.1128/AEM.71.10.6142-6149.2005 |
| [63] | Rysgaard S, Glud RN. Anaerobic N2 production in Arctic sea ice. Limnology and Oceanography, 2004, 49(1): 86-94. DOI:10.4319/lo.2004.49.1.0086 |
| [64] | Trimmer M, Nicholls JC. Production of nitrogen gas via anammox and denitrification in intact sediment cores along a continental shelf to slope transect in the North Atlantic. Limnology and Oceanography, 2009, 54(2): 577-589. DOI:10.4319/lo.2009.54.2.0577 |
| [65] | Shen LD, Cheng HX, Liu X, Li JH, Liu Y. Potential role of anammox in nitrogen removal in a freshwater reservoir, Jiulonghu Reservoir (China). Environmental Science and Pollution Research, 2017, 24(4): 3890-3899. DOI:10.1007/s11356-016-8126-7 |
| [66] | Harhangi HR, Le Roy M, van Alen T, Hu BL, Groen J, Kartal B, Tringe SG, Quan ZX, Jetten MSM, Op Den Camp HJM. Hydrazine synthase, a unique phylomarker with which to study the presence and biodiversity of anammox bacteria. Applied and Environmental Microbiology, 2012, 78(3): 752-758. DOI:10.1128/AEM.07113-11 |
| [67] | Li M, Ford T, Li XY, Gu JD. Cytochrome cd1-containing nitrite reductase encoding gene nirS as a new functional biomarker for detection of anaerobic ammonium oxidizing (anammox) bacteria. Environmental Science & Technology, 2011, 45(8): 3547-3553. |
| [68] | Li M, Hong YG, Cao HL, Gu JD. Mangrove trees affect the community structure and distribution of anammox bacteria at an anthropogenic-polluted mangrove in the Pearl River Delta reflected by 16S rRNA and hydrazine oxidoreductase (HZO) encoding gene analyses. Ecotoxicology, 2011, 20(8): 1780-1790. DOI:10.1007/s10646-011-0711-4 |
| [69] | Bale NJ, Villanueva L, Fan HX, Stal LJ, Hopmans EC, Schouten S, Damsté JSS. Occurrence and activity of anammox bacteria in surface sediments of the southern North Sea. FEMS Microbiology Ecology, 2014, 89(1): 99-110. DOI:10.1111/fem.2014.89.issue-1 |
| [70] | Dale OR, Tobias CR, Song B. Biogeographical distribution of diverse anaerobic ammonium oxidizing (anammox) bacteria in Cape Fear River Estuary. Environmental Microbiology, 2009, 11(5): 1194-1207. DOI:10.1111/emi.2009.11.issue-5 |
| [71] | Fu BB, Liu JW, Yang HM, Hsu TC, He BY, Dai MH, Kao SJ, Zhao MX, Zhang XH. Shift of anammox bacterial community structure along the Pearl Estuary and the impact of environmental factors. Journal of Geophysical Research : Oceans, 2015, 120(4): 2869-2883. DOI:10.1002/2014JC010554 |
| [72] | Li M, Cao HL, Hong YG, Gu JD. Seasonal dynamics of anammox bacteria in estuarial sediment of the Mai Po Nature Reserve revealed by analyzing the 16S rRNA and hydrazine oxidoreductase (hzo) genes. Microbes and Environments, 2011, 26(1): 15-22. DOI:10.1264/jsme2.ME10131 |
| [73] | Hu BL, Shen LD, Zheng P, Hu AH, Chen TT, Cai C, Liu S, Lou LP. Distribution and diversity of anaerobic ammonium-oxidizing bacteria in the sediments of the Qiantang River. Environmental Microbiology Reports, 2012, 4(5): 540-547. DOI:10.1111/emi4.2012.4.issue-5 |
| [74] | Han P, Gu JD. Further analysis of anammox bacterial community structures along an anthropogenic nitrogen-input gradient from the riparian sediments of the Pearl River delta to the deep-ocean sediments of the South China sea. Geomicrobiology Journal, 2015, 32(9): 789-798. DOI:10.1080/01490451.2014.1001502 |
| [75] | Jiang XF, Hou LJ, Zheng YL, Liu M, Yin GY, Gao J, Li XF, Wang R, Yu CD, Lin XB. Salinity-driven shifts in the activity, diversity, and abundance of anammox bacteria of estuarine and coastal wetlands. Physics and Chemistry of the Earth, Parts A/B/C, 2017, 97: 46-53. DOI:10.1016/j.pce.2017.01.012 |
| [76] | Dalsgaard T, Thamdrup B, Canfield DE. Anaerobic ammonium oxidation (anammox) in the marine environment. Research in Microbiology, 2005, 156(4): 457-464. DOI:10.1016/j.resmic.2005.01.011 |
| [77] | Dang HY, Chen RP, Wang L, Guo LZ, Chen PP, Tang ZW, Tian F, Li SZ, Klotz MG. Environmental factors shape sediment anammox bacterial communities in hypernutrified Jiaozhou Bay, China. Applied and Environmental Microbiology, 2010, 76(21): 7036-7047. DOI:10.1128/AEM.01264-10 |
| [78] | Lashof DA, Ahuja DR. Relative contributions of greenhouse gas emissions to global warming. Nature, 1990, 344(6266): 529-531. DOI:10.1038/344529a0 |
| [79] | Nedwell DB, Dong LF, Sage A, Underwood GJC. Variations of the nutrients loads to the mainland U.K. estuaries:correlation with catchment areas, urbanization and coastal eutrophication. Estuarine, Coastal and Shelf Science, 2002, 54(6): 951-970. DOI:10.1006/ecss.2001.0867 |
| [80] | Mosier AC, Francis CA. Denitrifier abundance and activity across the San Francisco Bay estuary. Environmental Microbiology Reports, 2010, 2(5): 667-676. DOI:10.1111/j.1758-2229.2010.00156.x |
| [81] | Zumft WG. Cell biology and molecular basis of denitrification. Microbiology and Molecular Biology Reviews, 1997, 61(4): 533-616. |
| [82] | Nogales B, Timmis KN, Nedwell DB, Osborn AM. Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Applied and Environmental Microbiology, 2002, 68(10): 5017-5025. DOI:10.1128/AEM.68.10.5017-5025.2002 |
| [83] | Yoshinari T. N2O reduction by Vibrio succinogenes. Applied and Environmental Microbiology, 1980, 39(1): 81-84. |
| [84] | Lund MB, Smith JM, Francis CA. Diversity, abundance and expression of nitrite reductase (nirK)-like genes in marine thaumarchaea. The ISME Journal, 2012, 6(10): 1966-1977. DOI:10.1038/ismej.2012.40 |
| [85] | Huang S, Chen C, Yang X, Wu Q, Zhang R. Distribution of typical denitrifying functional genes and diversity of the nirS-encoding bacterial community related to environmental characteristics of river sediments. Biogeosciences, 2011, 8(10): 3041-3051. DOI:10.5194/bg-8-3041-2011 |
| [86] | Jones CM, Hallin S. Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. The ISME Journal, 2010, 4(5): 633-641. DOI:10.1038/ismej.2009.152 |
| [87] | Desnues C, Michotey VD, Wieland A, Zhizang C, Four?ans A, Duran R, Bonin PC. Seasonal and diel distributions of denitrifying and bacterial communities in a hypersaline microbial mat (Camargue, France). Water Research, 2007, 41(15): 3407-3419. DOI:10.1016/j.watres.2007.04.018 |
| [88] | Gao J, Hou LJ, Zheng YL, Liu M, Yin GY, Li XF, Lin XB, Yu CD, Wang R, Jiang XF, Sun XR. nirS-encoding denitrifier community composition, distribution, and abundance along the coastal wetlands of China. Applied Microbiology and Biotechnology, 2016, 100(19): 8573-8582. DOI:10.1007/s00253-016-7659-5 |
| [89] | Rysgaard S, Thastum P, Dalsgaard T, Christensen PB, Sloth NP. Effects of salinity on NH4+ adsorption capacity, nitrification, and denitrification in Danish estuarine sediments. Estuaries, 1999, 22(1): 21-30. DOI:10.2307/1352923 |
| [90] | Fear JM, Thompson SP, Gallo TE, Paerl HW. Denitrification rates measured along a salinity gradient in the eutrophic Neuse River Estuary, North Carolina, USA. Estuaries, 2005, 28(4): 608-619. DOI:10.1007/BF02696071 |
| [91] | Magalh?es CM, Joye SB, Moreira RM, Wiebe WJ, Bordalo AA. Effect of salinity and inorganic nitrogen concentrations on nitrification and denitrification rates in intertidal sediments and rocky biofilms of the Douro River estuary, Portugal. Water Research, 2005, 39(9): 1783-1794. DOI:10.1016/j.watres.2005.03.008 |
| [92] | Laverman AM, Canavan RW, Slomp CP, van Cappellen P. Potential nitrate removal in a coastal freshwater sediment (Haringvliet Lake, The Netherlands) and response to salinization. Water Research, 2007, 41(14): 3061-3068. DOI:10.1016/j.watres.2007.04.002 |
| [93] | Tuominen L, Hein?nen A, Kuparinen J, Nielsen LP. Spatial and temporal variability of denitrification in the sediments of the northern Baltic Proper. Marine Ecology Progress Series, 1998, 172: 13-24. DOI:10.3354/meps172013 |
| [94] | Hietanen S, Kuparinen J. Seasonal and short-term variation in denitrification and anammox at a coastal station on the Gulf of Finland, Baltic Sea. Hydrobiologia, 2008, 596(1): 67-77. DOI:10.1007/s10750-007-9058-5 |
| [95] | Dong LF, Smith CJ, Papaspyrou S, Stott A, Osborn AM, Nedwell DB. Changes in benthic denitrification, nitrate ammonification, and anammox process rates and nitrate and nitrite reductase gene abundances along an estuarine nutrient gradient (the Colne Estuary, United Kingdom). Applied and Environmental Microbiology, 2009, 75(10): 3171-3179. DOI:10.1128/AEM.02511-08 |
| [96] | Burgin AJ, Hamilton SK. Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Frontiers in Ecology and the Environment, 2007, 5(2): 89-96. DOI:10.1890/1540-9295(2007)5[89:HWOTRO]2.0.CO;2 |
| [97] | Giblin AE, Tobias CR, Song B, Weston N, Banta GT, Rivera-Monroy VH. The importance of dissimilatory nitrate reduction to ammonium (DNRA) in the nitrogen cycle of coastal ecosystems. Oceanography, 2013, 26(3): 124-131. DOI:10.5670/oceanog |
| [98] | Sayama M. Presence of nitrate-accumulating sulfur bacteria and their influence on nitrogen cycling in a shallow coastal marine sediment. Applied and Environmental Microbiology, 2001, 67(8): 3481-3487. DOI:10.1128/AEM.67.8.3481-3487.2001 |
| [99] | Mohan SB, Schmid M, Jetten M, Cole J. Detection and widespread distribution of the nrfA gene encoding nitrite reduction to ammonia, a short circuit in the biological nitrogen cycle that competes with denitrification. FEMS Microbiology Ecology, 2004, 49(3): 433-443. DOI:10.1016/j.femsec.2004.04.012 |
| [100] | Darwin A, Hussain H, Griffiths L, Grove J, Sambongi Y, Busby S, Cole J. Regulation and sequence of the structural gene for cytochrome C552 from Escherichia coli:not a hexahaem but a 50 kDa tetrahaem nitrite reductase. Molecular Microbiology, 1993, 9(6): 1255-1265. DOI:10.1111/mmi.1993.9.issue-6 |
| [101] | Wang HN, Gunsalus RP. The nrfA and nirB nitrite reductase operons in Escherichia coli are expressed differently in response to nitrate than to nitrite. Journal of Bacteriology, 2000, 182(20): 5813-5822. DOI:10.1128/JB.182.20.5813-5822.2000 |
| [102] | Bu CN, Wang Y, Ge CH, Ahmad HA, Gao BY, Ni SQ. Dissimilatory nitrate reduction to ammonium in the Yellow River Estuary:rates, abundance, and community diversity. Scientific Reports, 2017, 7(1): 6830. DOI:10.1038/s41598-017-06404-8 |
| [103] | An S, Gardner WS. Dissimilatory nitrate reduction to ammonium (DNRA) as a nitrogen link, versus denitrification as a sink in a shallow estuary (Laguna Madre/Baffin Bay, Texas). Marine Ecology Progress Series, 2002, 237: 41-50. DOI:10.3354/meps237041 |
| [104] | Gardner WS, McCarthy MJ. Nitrogen dynamics at the sediment-water interface in shallow, sub-tropical Florida Bay:why denitrification efficiency may decrease with increased eutrophication. Biogeochemistry, 2009, 95(2/3): 185-198. |
| [105] | Dong LF, Sobey MN, Smith CJ, Rusmana I, Phillips W, Stott A, Osborn AM, Nedwell DB. Dissimilatory reduction of nitrate to ammonium, not denitrification or anammox, dominates benthic nitrate reduction in tropical estuaries. Limnology and Oceanography, 2011, 56(1): 279-291. DOI:10.4319/lo.2011.56.1.0279 |
| [106] | Yin GY, Hou LJ, Liu M, Li XF, Zheng YL, Gao J, Jiang XF, Wang R, Yu CD, Lin XB. DNRA in intertidal sediments of the Yangtze Estuary. Journal of Geophysical Research-Biogeosciences, 2017, 122(8): 1988-1998. DOI:10.1002/jgrg.v122.8 |
| [107] | Gardner WS, McCarthy MJ, An S, Sobolev D, Sell KS, Brock D. Nitrogen fixation and dissimilatory nitrate reduction to ammonium (DNRA) support nitrogen dynamics in Texas estuaries. Limnology and Oceanography, 2006, 51(1): 558-568. |
| [108] | Koop-Jakobsen K, Giblin AE. The effect of increased nitrate loading on nitrate reduction via denitrification and DNRA in salt marsh sediments. Limnology and Oceanography, 2010, 55(2): 789-802. DOI:10.4319/lo.2009.55.2.0789 |
| [109] | van den Berg EM, van Dongen U, Abbas B, van Loosdrecht MCM. Enrichment of DNRA bacteria in a continuous culture. The ISME Journal, 2015, 9(10): 2153-2161. DOI:10.1038/ismej.2015.26 |
| [110] | Decleyre H, Heylen K, van Colen C, Willems A. Dissimilatory nitrogen reduction in intertidal sediments of a temperate estuary:small scale heterogeneity and novel nitrate-to-ammonium reducers. Frontiers in Microbiology, 2015, 6: 1124. |
| [111] | Smyth AR, Thompson SP, Siporin KN, Gardner WS, McCarthy MJ, Piehler MF. Assessing nitrogen dynamics throughout the estuarine landscape. Estuaries and Coasts, 2013, 36(1): 44-55. DOI:10.1007/s12237-012-9554-3 |
| [112] | Yoon S, Sanford RA, L?effler FE. Nitrite control over dissimilatory nitrate/nitrite reduction pathways in shewanella loihica strain PV-4. Applied and Environmental Microbiology, 2015, 81(10): 3510-3517. DOI:10.1128/AEM.00688-15 |
| [113] | Santoro AE. Microbial nitrogen cycling at the saltwater-freshwater interface. Hydrogeology Journal, 2010, 18(1): 187-202. DOI:10.1007/s10040-009-0526-z |
| [114] | Giblin AE, Weston NB, Banta GT, Tucker J, Hopkinson CS. The effects of salinity on nitrogen losses from an oligohaline estuarine sediment. Estuaries and Coasts, 2010, 33(5): 1054-1068. DOI:10.1007/s12237-010-9280-7 |
| [115] | Baker BJ, Lazar CS, Teske AP, Dick GJ. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome, 2015, 3: 14. DOI:10.1186/s40168-015-0077-6 |
