全文HTML
--> --> -->有研究者通过单分子磁镊的方式对DNA双链施加大于8 pN的拉力时, gp5可以持续链置换但是也有持续外切的现象, 而将外力变小, 持续外切的情况就会更加频繁导致gp5难以链置换[11]. 这一结果说明了gp5可以独自链置换, 但是新合成的碱基又会被外切掉. 由于技术局限, 磁镊无法在完全没有力作用下(生理条件下)对gp5的链置换和外切进行测量, 这也留下了疑问: gp5在没有力作用下, 有无一定程度的链置换及外切的现象.
这里我们使用单分子荧光共振能量转移(smFRET)的方法在一个没有力的生理条件下, 并在一个接近单核苷酸尺度的分辨率下对gp5的链置换动力学进行测量. 本工作发现即使没有外力的帮助, gp5也可以链置换, 但是链置换到4 nt左右后就会回退. 通过突变掉外切活性的蛋白, 证明了这种回退是外切导致的. 为了证明退火压力对gp5的影响, 本文使用了纳米张力器的FRET方法, 对DNA双链施加了一个微小的力[12,13], 以此来减弱DNA双链的退火压力. 施加力后外切的长度减小、链置换的长度提高了. 进一步的实验发现gp4和gp5形成了复制体后合成速度会有一定升高, 而其最主要的特征是不再有外切的出现. 这说明了gp4帮助gp5克服了DNA的退火压力, 从而减少了gp5外切的发生.
2.1.聚丙烯酰胺电泳凝胶实验步骤
首先配置12%聚丙烯酰胺电泳凝胶: 48 g尿素, 30 mL 40%丙烯酰胺/甲叉双丙烯酰胺(19∶1)溶液, 10 mL 10 × TBE, H2O(补足100 mL), 10% APS 700 μL, TEMED 66 μL. 配置好后在电泳仪上以400 V的电压预跑30 min. 在这一过程中准备电泳的样品, 分别是样品1: 10 nmol/L DNA; 样品2: 加入40 nmol/L DNA, 40 nmol/L gp5, 100 μmol/L dNTP, 1 mmol/L二硫疏糖醇在37 ℃水浴反应1 min后用100 mmol/L EDTA终止反应; 样品3: 使用exo– gp5, 其余和样品2一样; 样品4: 和样品2一样, 但是反应时间为5 min; 样品5: 和样品3一样, 但是反应时间为5 min. 然后将5个样品加入不同的条带处进行3 h的电泳, 最后用凝胶成像仪分析条带位置.2
2.2.单分子实验玻片的清洗与修饰
首先使用丙酮、甲醇、食人鱼洗液(浓硫酸+双氧水)对玻片进行深度清洁, 接着使用硅烷化试剂进行修饰, 最后使用100∶1的mPEG和biotin-PEG经行修饰. 详情可以见参考文献[14].2
2.3.单分子荧光共振能量实验步骤
首先将上诉的玻片用双面胶粘成具有不同通道的样品池, 接着加入链酶亲和素连接波片的biotin-PEG, 然后加入100 pmol/L DNA, 最后加入1 nmol/L gp5和4 μmol/L dNTP, 收集Cy3和Cy5的光强信息, 并以此计算荧光转移效率. 对于gp5和gp4一起加入的实验过程为, 先体外孵育1 nmol/L gp5, 20 nmol/L gp4和100 μmol/L dTTP形成复合体再加入到样品池中.2
2.4.蛋白和缓冲液
gp5购买于NEB公司, exo– gp5购买于GE health公司. gp4按照文献[15]的方法纯化得到. dNTP购买于Takara公司. DNA购买于上海生工公司, 退火的方法详见文献[16]. 实验时的缓冲体系为: 50 mmol/L NaCl, 20 mmol/L pH为7.9的Tris缓冲液, 10 mmol/L MgCl2, 1 mmol/L DTT.3.1.外切活性对gp5链置换的影响
之前的文献认为持续外切是导致gp5无法链置换的原因[11], 为了直接证明这一点, 本文使用突变了外切活性的exo– gp5作为对照组实验(为了区分, 在后续的结果与讨论部分, 把没有突变的野生型gp5写作exo+ gp5), 并建立了如图1(a)所示的DNA, 其引物链末端标记了Alexa488, 而变性聚丙烯酰胺凝胶(PAGE)被用来分离这些引物链, 最后使用488 nm激光照明就可以特异性地得到引物链的长度, 进而得知DNA聚合酶的链置换情况. 通过对比exo+ gp5和exo– gp5反应产物, 从图1(b)电泳结果的第3道中可以发现在1 min的时间内exo– gp5大部分产物都被全部链置换了, 只有极少数留在原来的位置, 相反的是, 在图1(b)电泳结果的第2道中exo+ gp5只有极少数全部被链置换. 不仅如此, exo+ gp5还外切了一些原有DNA. 通过在图1(b)电泳结果第4道和第5道中展示的延长反应时间后的结果可以发现: 所有exo– gp5都全部合成了, 但是exo+ gp5仍然只有很少的全长延伸产物. 从聚丙烯酰胺凝胶电泳(PAGE)的结果可以得知, exo+ gp5是一个双向的马达, 它既可以链置换也可以外切, 但是由于外切活性的存在导致了其链置换能力很弱, 甚至会把原有的DNA外切的更短. 而如果突变了外切活性, exo– gp5可以独立的打开DNA双链并进行合成. 由于PAGE实验只展示了最后的结果, 无法看到exo+ gp5在链置换与外切中切换的动态过程, 也就无法测量exo+ gp5单次链置换和外切的长度和速度, 所以我们使用了单分子荧光共振能量转移的方法[17,18]来实时观测其转化的方式和原因.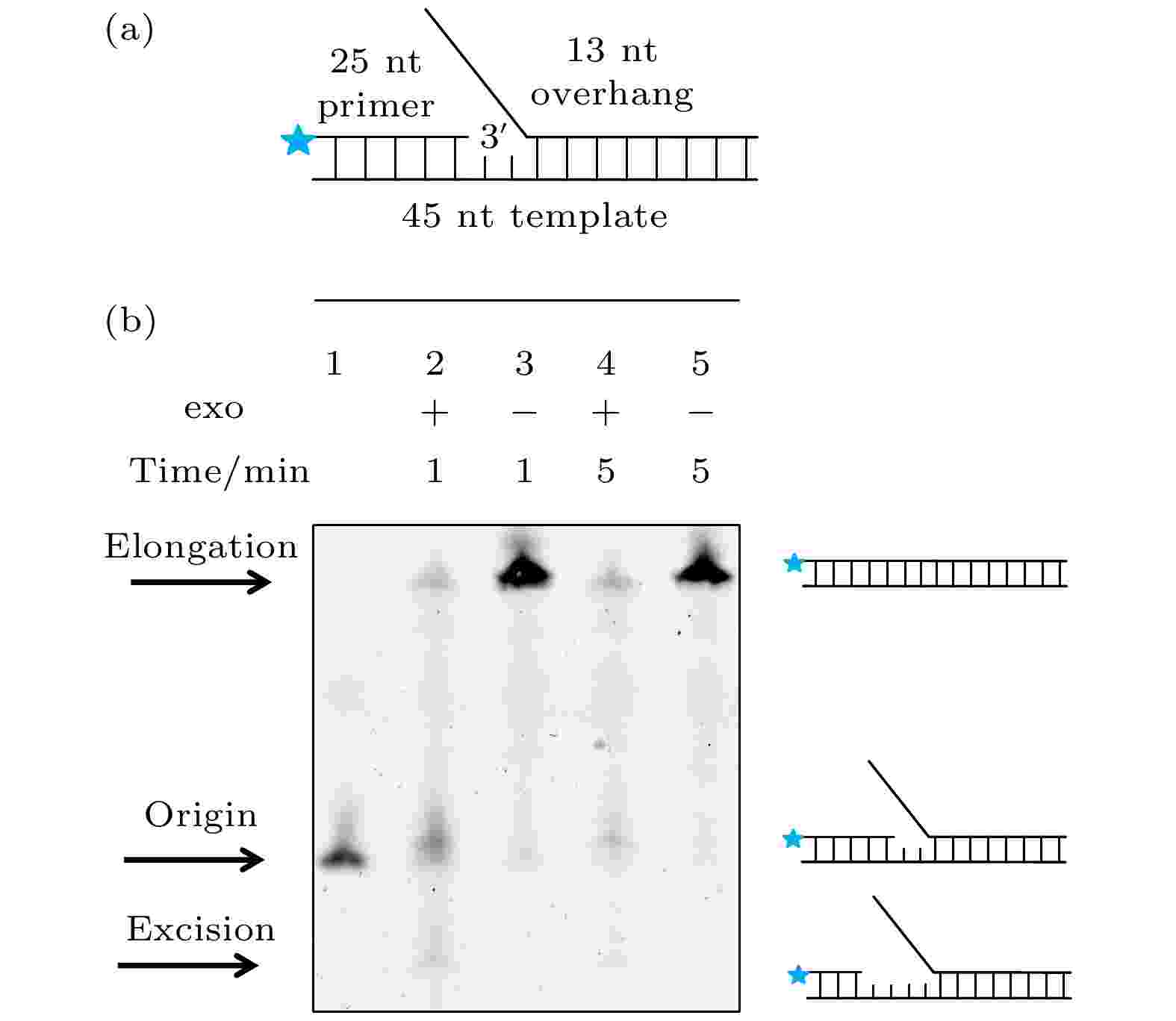 图 1 exo+ gp5和exo– gp5的PAGE实验 (a)电泳实验的DNA, 在引物链5′端标记有Alexa488; (b) 对Alexa488照明得到的结果: 第1个条带为DNA的原始长度, 第2个条带是exo+ gp5合成1 min后的结果, 第3个条带是exo– gp5合成1 min后的结果, 第4个条带是exo+ gp5合成5 min后的结果, 第5个条带是exo– gp5合成5 min后的结果
图 1 exo+ gp5和exo– gp5的PAGE实验 (a)电泳实验的DNA, 在引物链5′端标记有Alexa488; (b) 对Alexa488照明得到的结果: 第1个条带为DNA的原始长度, 第2个条带是exo+ gp5合成1 min后的结果, 第3个条带是exo– gp5合成1 min后的结果, 第4个条带是exo+ gp5合成5 min后的结果, 第5个条带是exo– gp5合成5 min后的结果Figure1. PAGE assays of exo+ gp5 and exo– gp5. (a) Illustration of DNA that used in PAGE assays. Alexa488 is labeled on the 5′ overhand of primer DNA. (b) Results of various condition. The first lane: Original length of the DNA. The second lane: The synthesis-product of exo+ gp5 with 1 minute. The third lane: The synthesis-product of exo– gp5 with 1 minute. The fourth lane: The synthesis-product of exo+ gp5 with 5 minute. The fifth lane: The synthesis-product of exo– gp5 with 5 minute.
2
3.2.gp5外切与链置换的动态过程
构建了如图2(a)所示的Y型DNA, 通过标记在DNA不同单链上的Cy3和Cy5来进行测量DNA聚合酶链置换的长度和速度. 当DNA聚合酶打开DNA双链时, Cy3和Cy5的距离会变远, 进而导致能量转移效率FRET降低, 相反当DNA聚合酶回退时, FRET就会增加. 因为FRET和距离的关系为6次方, 所以这种方法可以达到2—3 ?的精度.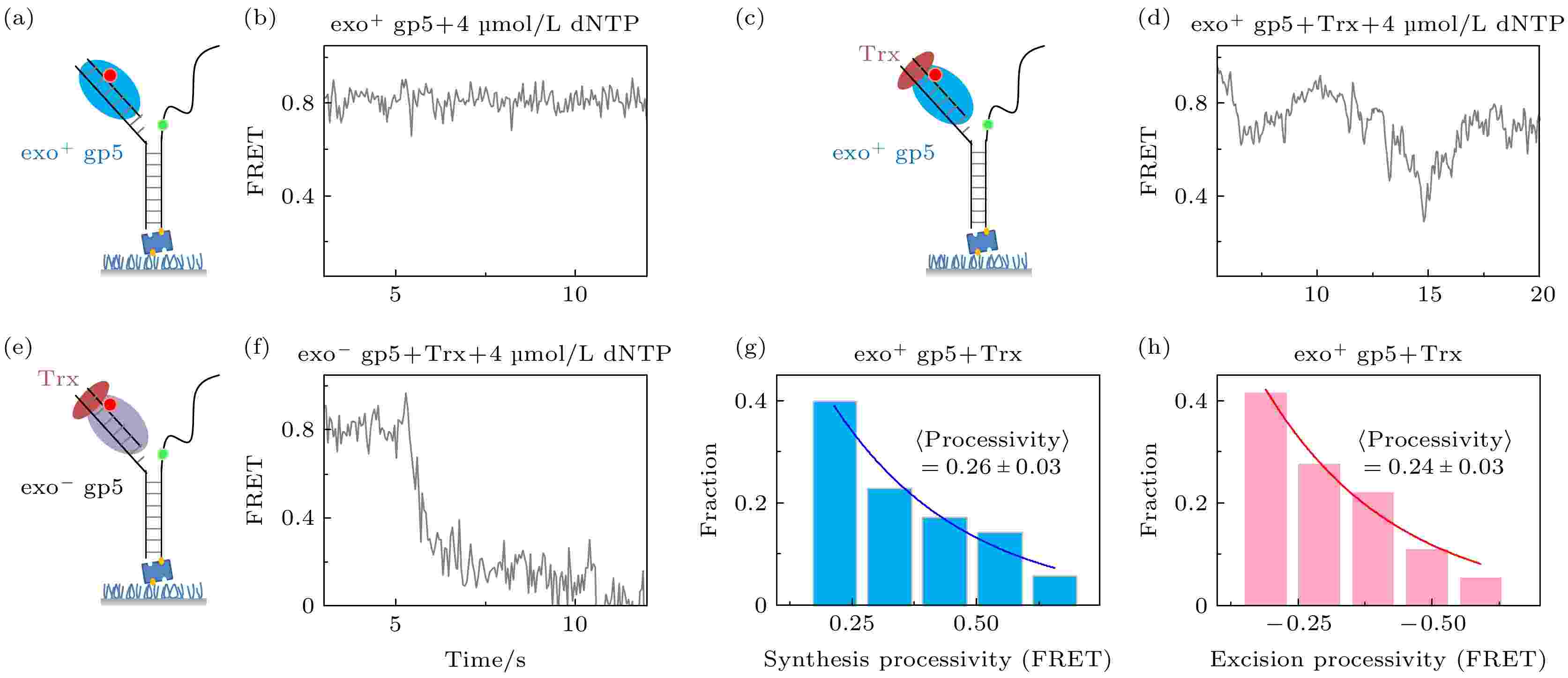 图 2 T7 DNA聚合酶gp5不断重复链置换和外切 (a), (b)没有结合辅助因子Trx时聚合酶无法链置换DNA双链; (c), (d)有辅助因子Trx时聚合酶能够部分链置换DNA, 但是会回退; (e), (f)外切活性突变后的gp5不会再外切; (g) exo+ gp5 + Trx实验中合成长度的统计图, 其分布满足单e指数; (h) exo+ gp5 + Trx实验中外切长度的统计图, 其分布满足单e指数
图 2 T7 DNA聚合酶gp5不断重复链置换和外切 (a), (b)没有结合辅助因子Trx时聚合酶无法链置换DNA双链; (c), (d)有辅助因子Trx时聚合酶能够部分链置换DNA, 但是会回退; (e), (f)外切活性突变后的gp5不会再外切; (g) exo+ gp5 + Trx实验中合成长度的统计图, 其分布满足单e指数; (h) exo+ gp5 + Trx实验中外切长度的统计图, 其分布满足单e指数Figure2. T7 DNA polymerase gp5 repeats in synthesis-excision cycle: (a), (b) exo+ gp5 cannot have displacement synthesis without co-factor Trx; (c), (d) gp5 with Trx repeats in synthesis-excision cycle; (e), (f) exo– gp5 attain full-length displacement synthesis without excision; (g) histogram of synthesis processivity from assay of exo+ gp5 + Trx, the distribution is well fit by an exponential; (h) histogram of excision processivity from assay of exo+ gp5+ Trx, the distribution is well fit by an exponential.
之前报道中发现辅助因子Trx会帮助exo+ gp5结合DNA, 进而延长exo+ gp5在延伸时的合成长度, 使其从数十nt的量级变到数百nt的量级[19]. 图2(a)和图2(b)展示了不加入Trx的对照组实验结果, exo+ gp5几乎无法链置换, 而从图2(c)和图2(d)可以看出加入Trx后exo+ gp5可以打开DNA双链进行链置换, 进而使得FRET下降, 但是FRET下降到一定长度后就会发生回退. 这里我们对曲线中每次下降的长度(synthesis processivity)进行统计, 通过拟合可以发现下降长度的分布是单e指数分布, 其分布的平均长度


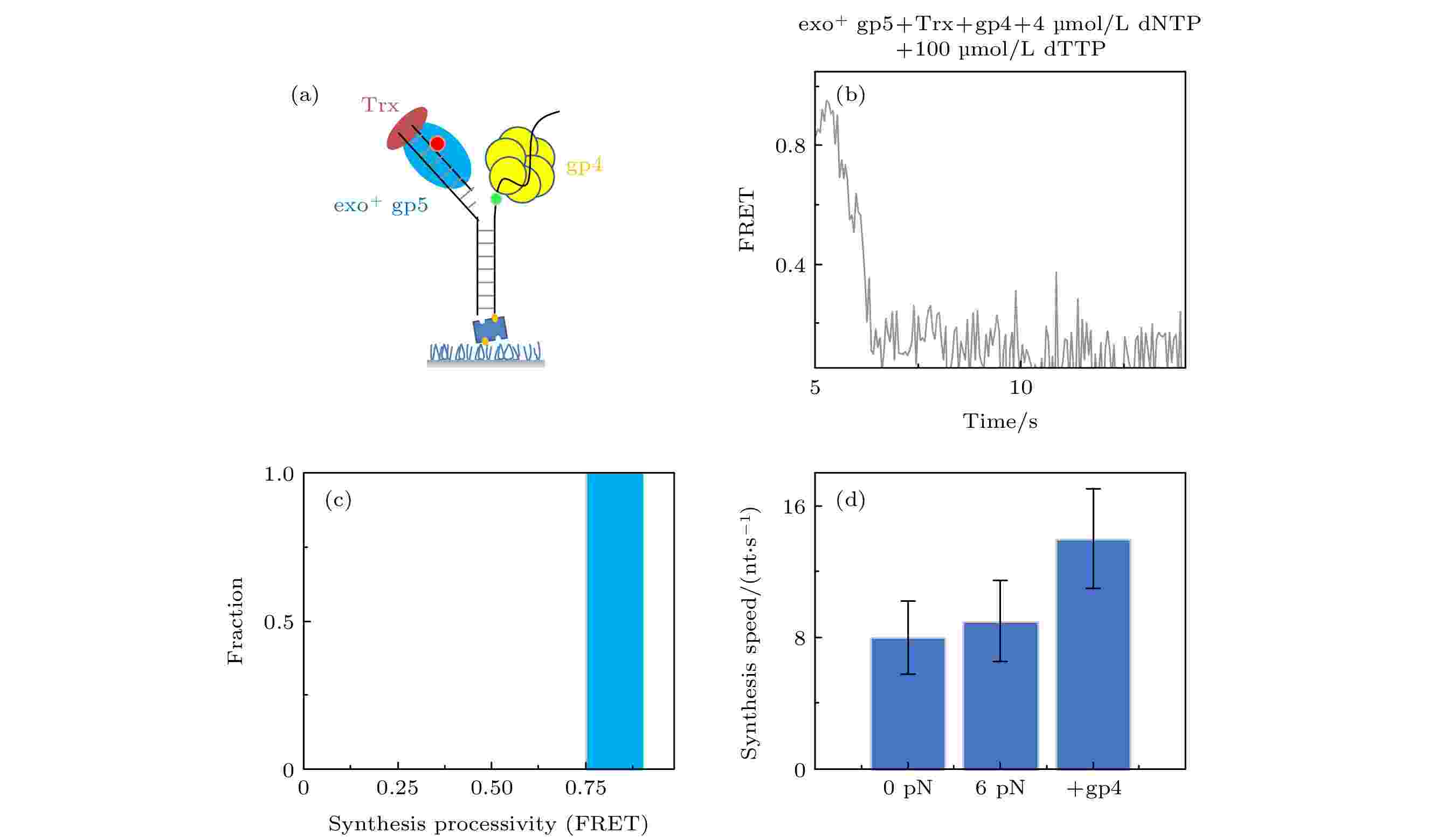 图 4 T7 DNA解旋酶帮助聚合酶克服退火压力 (a) gp4, exo+ gp5和Trx共同链置换的示意图, (b) gp4, exo+ gp5和Trx共同链置换时的典型曲线; (c) exo+ gp5 + Trx + gp4可以完全链置换; (d)不同情况的链置换速度
图 4 T7 DNA解旋酶帮助聚合酶克服退火压力 (a) gp4, exo+ gp5和Trx共同链置换的示意图, (b) gp4, exo+ gp5和Trx共同链置换时的典型曲线; (c) exo+ gp5 + Trx + gp4可以完全链置换; (d)不同情况的链置换速度Figure4. gp4 decrease DNA regression pressure which facilitate gp5 to attain processive strand-displacement synthesis: (a) Illustration of displacement by gp4, exo+ gp5 and Trx; (b) typical trace from assay of gp4, exo+ gp5 and Trx; (c) exo+ gp5 + Trx + gp4 attain processive synthesis; (d) synthesis speed in various condition.
2
3.3.力对野生型gp5链置换长度的影响
通常认为聚合酶的外切功能只会切除错误的核苷酸, 而exo+ gp5只有万分之一的概率插入错误的核苷酸[22], 所以链置换过程中的外切不全是错误导致的. 上面的实验发现外切是要exo+ gp5链置换到一定程度后才会明显出现的, 链置换的过程伴随的是旧双链退火压力. 为了证明是力改变了exo+ gp5的模式, 我们通过图3(a)所示的纳米张力器来研究力对外切的影响. 纳米张力器的原理是通过一段弯曲的双链DNA来对需要链置换的DNA施加张力, 这个张力大概在5—6 pN左右, 而此时根据计算[12] ΔFRET = 0.11为1 nt, 也就对实验精度有进一步的提高. 通过实验发现当张力加到DNA上后, 聚合酶链置换长度和外切长度仍然是单e指数衰减的分布(图3(c)和图3(d)), 经过换算后, 聚合酶的速度稍微有提高到9 nt/s (图3(b)和图4(d)), 而外切的平均长度变为2.9 nt, 比不加力的情况稍微变小, 这意味着外切程度变低了. 另一方面聚合酶的链置换平均长度变为了4.9 nt, 比不加力变长了, 但是这种变长仍然无法完全链置换, 这说明了在6 pN的外力帮助下, gp5仍然无法克服退火压力的影响.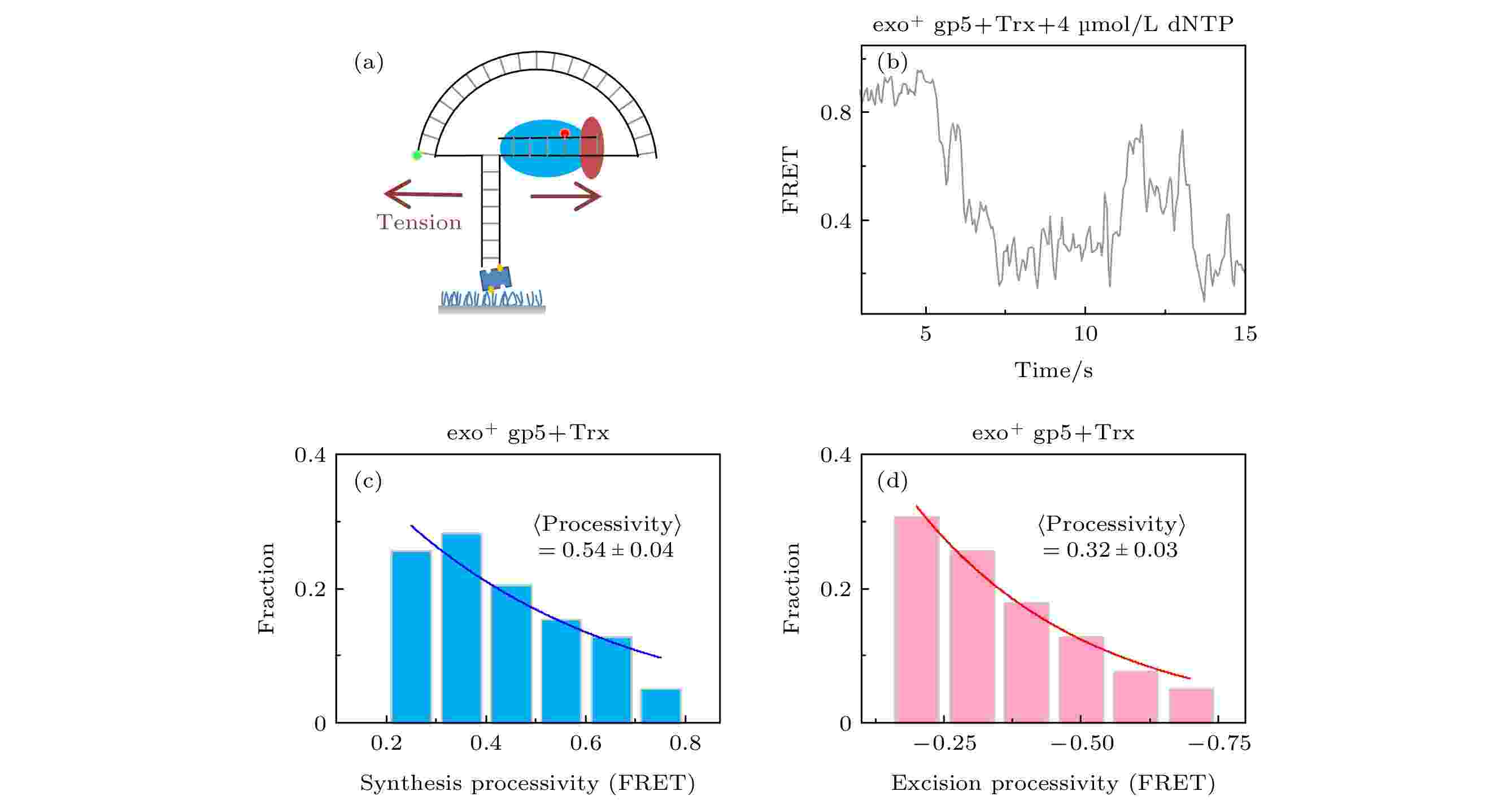 图 3 T7 DNA聚合酶gp5外切的原因是退火压力 (a)纳米张力器示意图; (b) 纳米张力器实验的典型曲线; (c) exo+ gp5 + Trx在受力后合成长度的统计图, 其分布满足单e指数; (d) exo+ gp5 + Trx受力后外切长度的统计图, 其分布满足单e指数
图 3 T7 DNA聚合酶gp5外切的原因是退火压力 (a)纳米张力器示意图; (b) 纳米张力器实验的典型曲线; (c) exo+ gp5 + Trx在受力后合成长度的统计图, 其分布满足单e指数; (d) exo+ gp5 + Trx受力后外切长度的统计图, 其分布满足单e指数Figure3. DNA regression pressure induced exonuclease activity: (a) Illustration of nanotensionior; (b) typical trace from assay of nanotensionior; (c) histogram of synthesis processivity from assay of exo+ gp5 + Trx with tension, the distribution is well fit by an exponential; (d) histogram of excision processivity from assay of exo+ gp5 + Trx with tension, the distribution is well fit by an exponential.
2
3.4.解旋酶帮助聚合酶克服了退火压力解旋酶对聚合酶链置换的影响
我们之前的工作发现单独加入gp4解旋速度是很慢的[16], 而单独加入exo+ gp5就无法完全链置换. 为了研究复制体链置换的情况, exo+ gp5, Trx和gp4以及dTTP被体外孵育好后加入到反应池中(图4(a)), 通过图4(b)所示曲线可发现链置换速度变快了而且也没有外切的出现. 由图4(d)可知, 复制体的速度(14 nt/s)快于聚合酶单独的速度(8 nt/s), 而加入gp4后最大的特点就是聚合酶不再回退(图4(c)). 这说明了聚合酶打开双链后, 由于解旋酶阻挡了退火压力, 聚合酶可以持续链置换而不会外切, 这就使得两者配合在一起, 解旋酶阻挡聚合酶不再外切, 而聚合酶帮助解旋酶打开双链. 这里值得注意的是, 外切对于聚合酶的保真度是有很大帮助的[23,24], 在加入gp4后不再出现外切, 这虽然提高了合成效率但是可能降低了保真度.2
3.5.聚合酶链置换的机制
如图5(a)所示单独的聚合酶面对DNA双链时, 能够打开双链, 但是其合成长度非常的低, 其原因在于退火压力的出现, 这种压力使得聚合酶无法在打开双链后顺利插入核苷酸只能进行外切的反应, 而外切一旦开始就会持续3—4 nt左右导致过度外切的出现, 当外切完后聚合酶又进入了链置换模式. 单独的聚合酶就会一直陷入这种聚合-外切循环. 但是有了解旋酶加入后, 如图5(b)所示解旋酶帮助聚合酶承受了退火压力, 使得聚合酶可以链置换. 另一方面由于聚合酶打开双链的能力很强, 这也就帮助了解旋酶进行解旋. 这种互相配合使得复制体可以快速地进行链置换.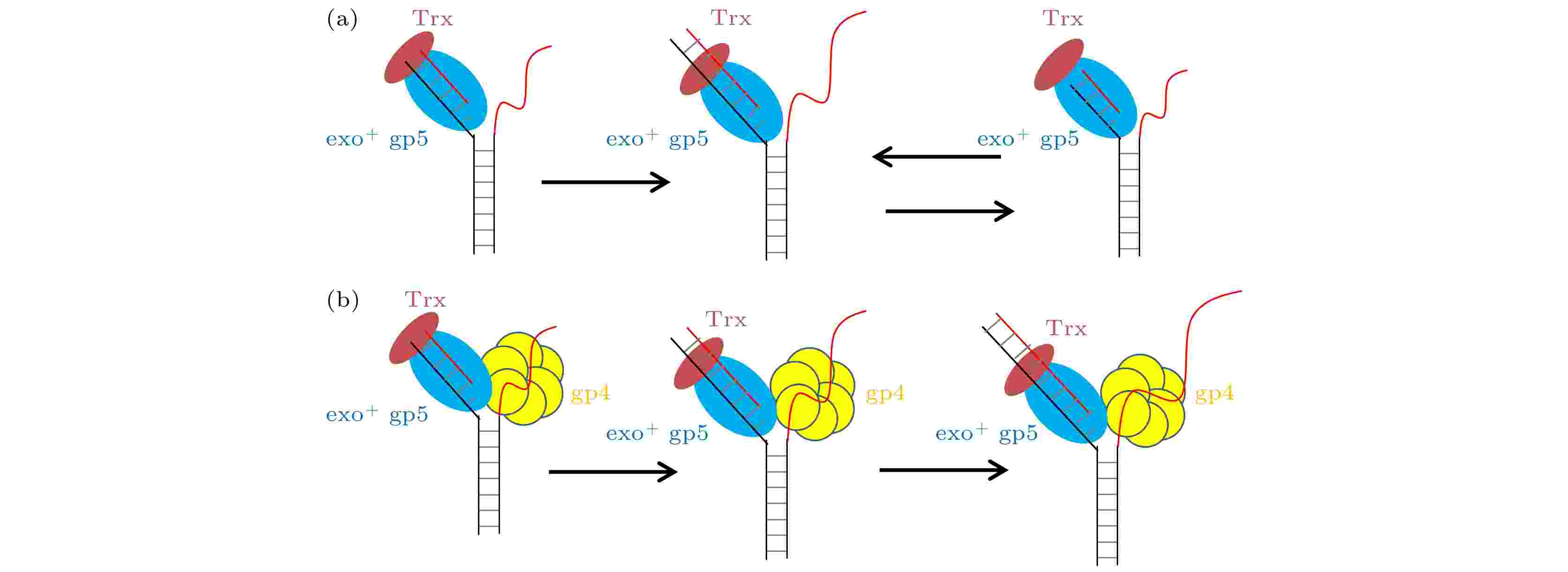 图 5 T7 DNA聚合酶不同情况链置换时的模型 (a) 野生型gp5单独链置换时进入链置换和外切的循环; (b) 野生型gp5在gp4的帮助下, 没有外切的出现可以持续链置换
图 5 T7 DNA聚合酶不同情况链置换时的模型 (a) 野生型gp5单独链置换时进入链置换和外切的循环; (b) 野生型gp5在gp4的帮助下, 没有外切的出现可以持续链置换Figure5. Model for gp5 strand displacement activity in various condition: (a) exo+ gp5 repeats in synthesis-excision cycle; (b) gp4 facilitate exo+ gp5 to attain processive strand-displacement synthesis.
之前的文献证明聚合酶和解旋酶不是一直同步进行的, 而是会有脱耦的出现[25]. 这些情况都需要野生型gp5本身有一定的链置换能力, 在本实验中发现野生型gp5能够独立链置换4 nt左右才容易外切, 也就是说野生型gp5和gp4之间可以存在4 nt的脱耦而不影响复制体的合成. 之前我们的工作也发现了gp4的解旋甚至有换链的情况[26], 这种情况也说明了需要野生型gp5具有一定的独立链置换能力, 否则DNA的复制就会完全停止.
