 ,, 蔡伟聪, 李桂玲, 李健, 刘静雯
,, 蔡伟聪, 李桂玲, 李健, 刘静雯 ,集美大学食品与生物工程学院,厦门 361000
,集美大学食品与生物工程学院,厦门 361000Analysis of microRNA expression profile in Emiliania huxleyi in response to virus infection
Enquan Zhang ,, Weicong Cai, Guiling Li, Jian Li, Jingwen Liu
,, Weicong Cai, Guiling Li, Jian Li, Jingwen Liu ,College of Food and Bioengineering, Jimei University, Xiamen 361021, China
,College of Food and Bioengineering, Jimei University, Xiamen 361021, China通讯作者: 刘静雯,博士,教授,研究方向:海洋微型生物分子生态学。E-mail:ljwsbch@163.com
编委: 赵呈天
收稿日期:2021-04-30修回日期:2021-07-14
| 基金资助: |
Received:2021-04-30Revised:2021-07-14
| Fund supported: |
作者简介 About authors
张恩权,在读硕士研究生,专业方向:海洋微生物组学。E-mail:

摘要
海洋颗石藻病毒-宿主互作是影响海洋碳、硫生物地化循环及全球气候变化的重要环节。作为大的双链DNA病毒,颗石藻病毒进化出一种“病毒细胞代谢”模式,通过重编程宿主代谢途径以满足其代谢需求,但对这一代谢模式的调控机制尚缺乏足够的认识。MicroRNA (miRNA)作为一种基因表达调控的重要因子,能够通过调控代谢过程中的靶基因表达,从而调节相关代谢通路。本研究采用small RNA测序技术分析病毒感染颗石藻差异表达的miRNA及其靶基因功能,鉴定出26条成熟miRNA (包括2条病毒来源的miRNA),均来自23条新的miRNA前体序列,其中5条miRNA显著差异表达,包括4条上调,1条下调。实时荧光定量PCR (quantitative real-time PCR, qRT-PCR)验证结果与miRNA-seq结果基本一致。功能富集分析显示,5个差异表达的miRNA可能参与调节糖代谢、脂代谢和氨基酸等代谢。此外,差异表达miRNA的表达水平与脂质代谢相关靶基因如ACC-1、SPT、ACOX、ACAT、CERS、ACADS等的表达水平呈负相关,说明这些miRNA可能在病毒感染过程中对宿主的脂质代谢发挥重要的调控作用。
关键词:
Abstract
The interactions between Emiliania huxleyi and E. huxleyi virus (EhV) regulate marine carbon and sulfur biogeochemical cycle and play a prominent role in global climate change. As a large DNA virus, EhVs have developed a novel “virocell metabolism” model to meet their higher metabolic needs. However, the regulatory mechanism of this metabolic model is still largely unclear. MicroRNAs (miRNAs) can regulate biological pathways through targeting hub genes in the metabolic processes. Here, we performed high-throughput small RNA sequencing to analyse miRNA expression in EhV99B1 infected E. huxleyi BOF92. A total of 26 miRNAs (including 2 virus-derived miRNAs) were identified, including four up-regulated and one down-regulated miRNAs. These results were further validated through quantitative real-time PCR. Functional enrichment analysis showed that five differentially-expressed miRNAs might be involved in the regulation of carbohydrate metabolism, lipid metabolism and amino acid metabolism. Moreover, the expression levels of differentially-expressed miRNAs were negatively correlated with that of several lipid metabolism-related genes, such as ACC-1, SPT, ACOX, ACAT, CERS and ACADS, indicating that these miRNAs might play an important regulatory role in virus-mediated lipid metabolism.
Keywords:
PDF (2803KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张恩权, 蔡伟聪, 李桂玲, 李健, 刘静雯. 赫氏颗石藻(Emiliania huxleyi)响应病毒感染的microRNA转录组分析. 遗传[J], 2021, 43(11): 1088-1100 doi:10.16288/j.yczz.21-164
Enquan Zhang.
海洋颗石藻(Coccolithophores)是一种全球广泛分布且具有重要生态功能的真核微型浮游植物[1,2,3],其中赫氏颗石藻(Emiliania huxleyi, Eh)具有形成“球石粒”和高产二甲基硫化物(DMSP)的能力,且几乎每年都在大洋(尤其是高纬度海域)中形成大面积赤潮[4],该藻赤潮的迅速大规模消亡被证实是特异性病毒(E. huxleyi virus, EhV)感染和裂解所致[5,6]。因此,EhV-Eh的互作过程是影响海洋碳、硫生物地化循环及全球气候变化的重要环节。颗石藻病毒与宿主间侵染和抵抗的博弈形成了一种新的、稳定的协同进化代谢模式,是研究真核浮游植物宿主-病毒互作的理想模式系统[7]。
脂代谢是当今病毒-宿主互作研究的重要问题。作为大的双链DNA病毒,EhVs进化出一种全新的“病毒细胞代谢(virocell metabolism)”模式,通过重编程宿主代谢途径以满足其更高的代谢需求[8,9]。Evans等[10]2009年首次报道在E. huxleyi赤潮消亡过程中,病毒感染诱导宿主细胞脂肪酸的组成由多不饱和向单不饱和转变,导致被病毒感染的宿主细胞减少多不饱和脂肪酸向食物链中更高级营养水平的传递,从而降低海洋生态系统的总生产力。本课题组前期研究发现,病毒感染早期宿主细胞高饱和三酰基甘油(triacylglycerols, TAGs)合成积累、并形成脂滴聚集在病毒颗粒中促进病毒外壳疏水蛋白的存储,满足病毒复制和组装的需求[11]。最新研究发现,病毒感染颗石藻能够以外泌体形式富集并分泌TAGs,以此加速病毒感染过程[12]。以色列****Carmit团队2016年首次发现,EhV编码的丝氨酸棕榈酰转移酶(viral serine palmitoyltransferase, vSPT:鞘脂从头生物合成途径中的第一个关键限速酶)在感染细胞中能够催化合成病毒特有的新型鞘糖脂(viral-specific glycosphingolipids, vGSLs),该物质被认为是病毒组装的必要条件[9],这表明病毒能够利用编码的辅助代谢基因(virus-encoded auxiliary metabolic genes, vAMGs)重塑宿主鞘脂代谢途径。同时vGSLs还能作为重要的信号分子诱发ROS(H2O2)[13]和NO[14]的产生,进而启动宿主细胞凋亡程序,最终裂解细胞释放病毒粒子[15,16,17]。然而,目前对EhV感染重塑的脂代谢的miRNA调控机制尚缺乏足够的认识。
MicroRNA(miRNA)是一种长度分布在18~26 nt的内源性非编码小RNA。动植物miRNA来自基因组位置略有不同,动物miRNA通常位于内含子区,而植物miRNA更多地来自基因组的基因间区[18]。miRNA通过结合在靶基因的不同位点抑制或激活靶基因的表达水平,作为丰度极高的基因调控因子参与调控多种生物学过程。最近在高等动植物研究中发现,miRNA能够通过调节代谢过程中的靶基因,从而调控相关代谢通路,特别是在脂质合成、脂肪酸氧化及脂蛋白分泌等代谢网络中起重要调节作用[19]。相对于高等动植物而言,由于浮游植物种内和种间的差异性特点,且目前只有少数种类基因组信息得到较完整的诠释,因此对miRNA在海洋浮游植物生物及非生物胁迫中的重要作用了解较少。目前,主要针对氮、磷、硅及铁等营养胁迫条件下miRNA的表达情况进行了初步研究[20,21,22]。有关浮游植物响应病原菌感染相关miRNA的研究国内外尚未报道。
本研究以Eh-EhV模式系统为研究对象,在病毒感染早期(6 h)和晚期(45 h)收集藻细胞样品,采用高通量小RNA测序技术筛选病毒诱导的差异表达miRNA,分析miRNA在病毒重塑宿主脂代谢过程中可能的调控作用,为从表观遗传学角度深入理解EhV-Eh互作的分子机制提供新的理论认识。
1 材料与方法
1.1 赫氏颗石藻的培养及病毒感染
赫氏颗石藻E. huxleyi BOF92及其特异性裂解病毒株系EhV99B1均由挪威卑尔根大学生物系微生物研究所Gunnar Bratbak教授馈赠并保存于本实验室,藻株和病毒株均为纯化株系。颗石藻培养采用70%海水配制的f/2-si加富培养基,培养条件为:温度16℃±2℃,光照强度为60 μmol quanta m-2 s-1,光照周期为14/10 (光/暗)。在2 L指数生长期的藻液中(~106 cells/mL),以1∶50 (EhV∶Eh)体积比加入浓缩病毒裂解液(病毒初始浓度约为107 viruses/mL),另外2 L添加等量高温灭活的病毒作为对照组。基于本课题组前期mRNA转录组和脂质代谢组学分析结果,EhV感染早期(6 h)和中后期(45 h),宿主转录组发生了显著差异表达[23],细胞脂质代谢产物的积累也发生了明显变化[11],因此本研究选择病毒感染的6 h和45 h作为采样时间点。分别于病毒感染6 h和45 h离心收集500 mL藻细胞样品(4℃,7000 r/min,5min),立即置于液氮中速冻,于-80℃保存备用。每个样本设置两个生物学平行,共计8个样本:Con_6 h-1、Con_6 h-2、Exp_6 h-1、Exp_6 h-2、Con_45 h-1、Con_45 h-2、Exp_45 h-1、Exp_45 h-2。1.2 RNA的提取、small RNA文库构建及测序
用mirVana microRNA Isolation Kit (Ambion,美国)试剂盒提取样本总RNA,NanoPhotometer (IMPLEN,德国)检测总RNA的质量和纯度,Agilent 2100 BioAnalyzer系统和RNA 6000 Nano chip (Agilent,美国)分析总RNA的完整性。利用Illumina smallRNA- seq文库构建试剂盒(KAPA Biosystems,美国)制备small RNA测序文库,在Illumina HiSeqTM 2500平台进行测序(华大基因公司,深圳)。参考基因组为近缘颗石藻株系E. huxleyi CCMP1516 (1.3 生物信息学分析
测序获得的原始raw reads过滤筛选后得到clean reads。去除接头序列、长度小于18 bp或者大于30 bp的reads、含有N碱基的reads及低质量的reads。后续所有分析基于clean reads。筛选过程基于Cutadapt 1.7.1[24]和Fastx toolkit 0.0.14[25]软件。Clean reads经过比对Rfam 11.0数据库,过滤掉rRNA、scRNA、snoRNA、snRNA以及tRNA等非编码RNA序列,过滤后的small tags作为预测miRNA的候选序列。使用BLASTN软件将剩余的small tags比对到近缘株系E. huxleyi CCMP1516和EhV86基因组,保留能完全匹配到参考基因组的tags。使用miRDeep2[26]软件对能匹配到参考基因组的tags进行二级结构预测。miRNA的表达水平通过TPM (transcripts per million reads)方法归一化。使用edgeR软件[27]对miRNA进行差异表达分析,差异条件设置为:|log2(foldchange)| >1以及P<0.05。1.4 miRNA保守性分析
使用序列比对工具BLAST,将本实验条件下预测的24条宿主miRNAs序列与miRBase数据库收录的38,589条miRNAs、其他微藻的2650条(数据库尚未收录的miRNAs)以及在E. huxleyi CCMP1516中已鉴定出的18条miRNA[28]进行比对分析。鉴定保守miRNA的条件为:miRNA种子序列(第2到第8个碱基)不允许有错配,且总错配数≤2。由于颗石藻基因组具有明显的种内变异,为了进一步了解E. huxleyi BOF92的miRNAs及其前体的功能和进化情况,分析了miRNA及其前体序列与其他13株测序颗石藻株的保守性[29]。其中,E. huxleyi 92A、Eh2和Van556为深度测序的分离株,其他10株是低覆盖度测序(覆盖度可以达到91%至 95%)。对病毒来源的2条miRNA及其前体序列也与其他13株测序病毒株进行保守性分析。1.5 miRNA靶基因预测及功能富集分析
miRNA的靶基因通过miRanda (v3.3a)[30]软件预测,参数设置为:-sc 140 -en -20 -scale 4 -strict。靶基因的GO (Gene Ontology)和KEGG (Kyoto Encyclopedia of Genes and Genomes)富集通过DAVID[31]在线软件分析,经过超几何分布检验后,P<0.05的term为显著富集。使用R语言程序绘制柱状图及气泡图。1.6 miRNA-脂代谢靶基因互作网络分析
选取差异表达miRNA靶基因中与脂代谢相关的基因进行miRNA-靶基因互作网络分析,包括脂肪酸合成、脂肪酸降解、甘油脂代谢、甘油磷脂代谢以及鞘脂代谢等。使用Cytoscape (v3.3.0)软件绘制互作网络图。1.7 qRT-PCR验证miRNA及其靶基因
对6条差异表达的miRNA进行茎环荧光定量qRT-PCR验证。为了进一步分析病毒诱导的miRNA是否可能参与调节宿主的脂质代谢,从miRNA靶基因中选取了6个与脂代谢相关的关键酶基因,即乙酰辅酶A羧化酶-1 (acetyl-CoA carboxylase-1, ACC-1)、丝氨酸棕榈酰转移酶(serine palmitoyltransferase, SPT)、酰基辅酶A氧化酶(acyl-CoA oxidase, ACOX)、乙酰辅酶A酰基转移酶(acetyl-CoA acyltransferase, ACAT)、鞘氨脂碱N-棕榈转移酶(sphingoid base N-stearoyltransferase, CERS)以及丁酰辅酶A脱氢酶(butyryl-CoA dehydrogenase, ACADS)进行常规荧光定量qRT-PCR检测,并分析它们与miRNA表达水平之间的关系。用Primer 5软件设计引物(表1),其中通用茎环引物序列为:5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACG-3′,均由铂尚生物技术(上海)有限公司合成。提取的RNA经DNA酶消化后反转录为cDNA,反转录试剂盒为FastQuant RT Kit with gDNase试剂盒(TIANGEN,北京)。qRT-PCR反应体系参照Power SYBR® Green PCR Master Mix (Thermo Fisher)荧光定量检测试剂盒说明书。miRNA以U6为内参,靶基因以细胞周期蛋白激酶CDKA为内参[32]。qRT-PCR实验在QuantStudio 7 Flex Real Time PCR system (Thermo Fisher)上进行,每组实验设置3个生物学重复和3个技术重复(所用样本均为测序同批次藻样),并采用2-ΔΔCt法进行相对定量[33]。Table 1
表1
表1miRNA及其靶基因的引物序列
Table 1
| 引物名称 | 序列(5′→3′) | 长度(nt) |
|---|---|---|
| U6-F | CTCGCTTCGGCAGCACA | 17 |
| U6-R | AACGCTTCACGAATTTGCGT | 20 |
| ehx-miR1-5p-F | ATTCCGACCCGTCCCTGA | 18 |
| ehx-miR2-5p-F | GTACTCGCCTGTCGTCCG | 18 |
| ehx-miR3-3p-F | GCGAGGTTGGCGCGGGC | 17 |
| ehv-miR1-3p-F | CTAAGTTCGATCCTCGCC | 18 |
| ehv-miR2-5p-F | CCCTCGTAGCTCAGTGGA | 18 |
| miRNA universal reverse primer | GTGCAGGGTCCGAGGT | 16 |
| CDKA-F | TACGCCGACGAGGACTAC | 18 |
| CDKA-R | GCGATATGCTTGGGCAGG | 18 |
| ACC-1-F | GAACCGGGTGGATGCAGT | 18 |
| ACC-1-R | GAAGGAACATGGCCTTGT | 18 |
| SPT-F | TCTCGGACACGCTCAACC | 18 |
| SPT-R | CCCTCGACGATGATGATG | 18 |
| ACOX-F | GTCCGCTTCACCGTCCAG | 18 |
| ACOX-R | CGACGATCAGTCCAGACA | 18 |
| ACAT-F | GTCAACAAGGTCTGCTCG | 18 |
| ACAT-R | AGGTAGTACGGCACGTTG | 18 |
| CERS-F | GAGGAGCGGCACAAGGA | 17 |
| CERS-R | CCGATGCGGACAAAGTTG | 18 |
| ACADS-F | GAGCACGGGCAACCTCAT | 18 |
| ACADS-R | ATGTCGGCGAGCAGGAAC | 18 |
新窗口打开|下载CSV
1.8 统计学分析
采用SPSS 17.0软件进行统计分析。qRT-PCR的实验数据以平均值±标准差表示,显著性差异采用t检验,P<0.05表示差异显著,P<0.01表示差异极显著。2 结果与分析
2.1 赫氏颗石藻miRNA的基本特征
经Illumina HiSeqTM 2500平台高通量测序后,每个small RNA文库至少获得了两千万个原始reads,其中高质量的reads占比>97%,经过筛选后每个样本中符合条件的clean tags均在70%左右,满足后续分析的需要。原始数据已上传至NCBI的SRA数据库(SRP108676)。通过对small RNA进行过滤、质控、分类以及与参考基因组比对后,共鉴定出26条成熟miRNA (其中包括两条病毒来源的miRNA),23个前体miRNA,且病毒的2条miRNA来自同一个前体miRNA。66.7%的miRNA来自基因间区,29.2%的miRNA来自外显子区,只有1条miRNA来自内含子区。miRNA的长度在18~30 nt之间,峰值为21 nt (图1)。这些miRNA的拷贝数从8到765,680不等,表明在病毒感染条件下miRNA表达水平差异较大。此外,赫氏颗石藻miRNA的首位碱基对C有着较强的偏好性(54.17%)。成熟miRNA的序列、测序丰度以及在基因组的定位见附表1,前体miRNA序列见附表2。
2.2 赫氏颗石藻miRNA的保守性分析
用序列比对工具将预测的26条miRNA序列与miRBase数据库收录的miRNA以及未被录入miRBase数据库的微藻miRNA进行比对分析,结果见附表3。虽然本文中的一些miRNA能够和其他miRNA的部分碱基配对(包括来源于E. huxleyi CCMP1516的18条miRNA),但都不满足鉴定为保守miRNA的条件。赫氏颗石藻BOF92来源的前体miRNA及成熟miRNA与其他13株赫氏颗石藻基因组的比对结果显示(附表4、5),10条miRNA (深灰色标记)与其他所有13株藻株基因组完全匹配,另外14条的前体序列或成熟体序列的匹配较短或无法匹配,表明这10条来源于赫氏颗石藻BOF92的miRNA及其前体比其他miRNAs更为保守。对于病毒的2条miRNA,它们都不能同时比对到其他13株赫氏颗石藻病毒的基因组上,表明EhV99B1编码的miRNA在株系间的变异更为明显,但ehv-miR1-3p比ehv-miR2-5p相对保守(附表6)。2.3 miRNA差异表达分析
对26条miRNA的测序丰度进行差异分析,结果显示共有3条宿主miRNA和2条病毒miRNA在本实验条件下发生了差异表达(表2)。这5条miRNA茎环荧光定量qRT-PCR结果与测序结果基本一致。5条差异miRNA的二级结构如图2所示,其中3条miRNA位于前体二级结构的5′端臂,2条位于3′端臂,且病毒的2条miRNA (ehv-miR1-5p和ehv-miR2- 3p)来自同一个前体二级结构。2.4 差异表达miRNA靶基因的预测及GO富集分析
以同批次样品转录组测序结果中的32,909条宿主mRNA作为靶基因预测的数据库(NCBI登录号为SRP189555)[29],26条miRNA共获得了26,380个靶Table 2
表2
表2差异表达miRNA的测序差异倍数对数值及qRT-PCR的-ΔΔCt结果
Table 2
| 实验组别 | miRNA | log2 (fold change) | -ΔΔCt |
|---|---|---|---|
| Exp_6h vs Con_6h | ehx-miR1-5p | -1.271** | -1.44±0.31* |
| Exp_6h vs Con_6h | ehx-miR2-5p | 1.082* | 2.15±0.68** |
| Exp_6h vs Con_6h | ehx-miR3-3p | 1.545** | 1.88±0.59** |
| Exp_45h vs Con_45h | ehv-miR1-5p | 8.484a, ** | 24.24±1.12b, ** |
| Exp_45h vs Con_45h | ehv-miR2-3p | 11.60a, ** | 30.12±2.08b, ** |
新窗口打开|下载CSV
图2
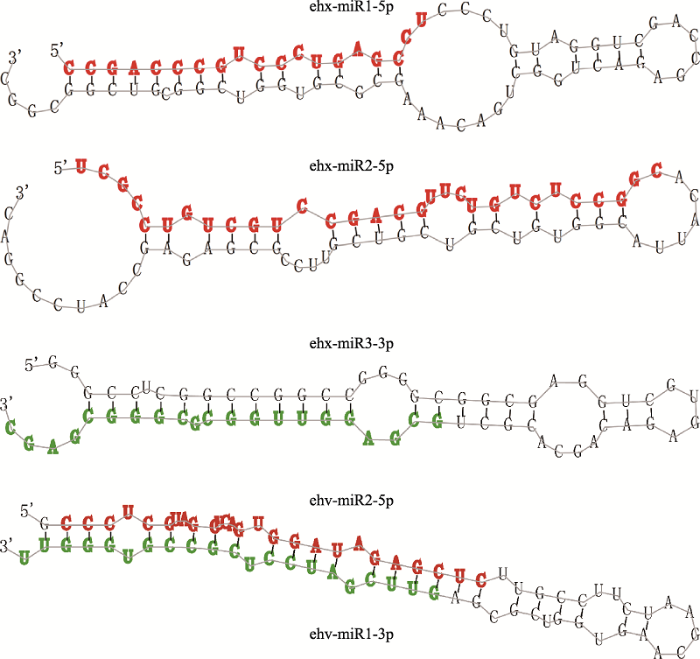 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图25条差异差异表达miRNA的前体二级结构
Fig. 2Precursor structures of five differentially expressed (DE) miRNAs
在每个发夹结构中,红色表示位于5′臂(5p)的成熟miRNA,绿色表示位于3′臂(3p)的成熟miRNAs。
基因。5条差异表达的miRNA 靶向了11,408个靶基因,其中3986个靶基因发生了差异表达(在感染早期和晚期分别为1246和3043个,交集为303个基因)。差异表达的靶基因GO富集结果主要分为3个类别(图3):生物过程(biological process)、细胞组分(cellular component)和分子功能(molecular function)。生物过程中富集到基因数目最多的是代谢过程(metabolic process),还包括刺激应答(response to stimulus)、信号通路(signaling)等;细胞组分中膜(membrane)、膜组分(membrane part)以及细胞(cell)等富集最显著;分子功能中,更多的靶基因与催化活性(catalytic activity)、结合(binding)以及转运活性(transporter activity)相关,此外还包括信号转导活性(signal transducer activity)和抗氧化活性(antioxidant activity)。GO富集结果表明miRNA对颗石藻各种功能基因的调控范围十分广泛。
此外,本研究也对病毒的基因进行了靶向预测分析。结果显示,赫氏颗石藻 BOF92来源的成熟miRNA靶向8个有明确功能的病毒基因(附表7),分别是病毒衣壳蛋白(major capsid protein)、DNA拓扑异构酶(DNA topoisomerase)、核酸内切酶(endonuclease)、核糖核酸酶(ribonuclease)、核糖核苷酸还原酶蛋白(ribonucleoside-diphosphate reductase protein)、DNA依赖的RNA聚合酶II亚基(DNA-dependent RNA polymerase II largest subunit)、DNA指导的RNA聚合酶II亚基(DNA-directed RNA polymerase II subunit)以及丝氨酸蛋白酶(serine protease)。其中,宿主编码的上调ehx-miR2-5p和ehx-miR3-3p靶向病毒衣壳蛋白基因的得分均较高,提示ehx-miR2-5p和ehx-miR3-3p可能在宿主抗病毒防御过程中具有一定的作用。衣壳蛋白的作用是包裹病毒的遗传物质,能够协助病毒感染,且具有免疫原性。而宿主
图3
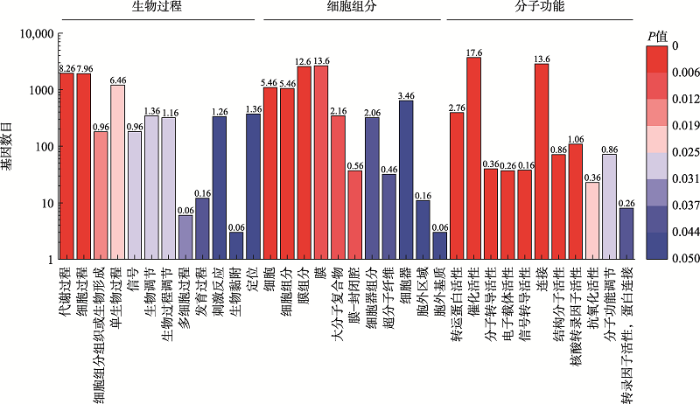 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3miRNA靶基因的GO富集分析
Fig. 3GO enrichment analysis of DE miRNA target genes
横坐标表示不同类别的GO term,纵坐标表示每个GO term中基因的数量,每个柱子上面的数字代表miRNA靶基因占该通路中总基因数的百分比(%);不同的颜色对应不同的显著性P值。
编码的miRNA可能通过靶向病毒衣壳蛋白基因以抑制其翻译,进而影响病毒的装配过程。表明ehx-miR2-5p和ehx-miR3-3p的上调表达可能是宿主的抗病毒策略。由于转录组测序数据中没有检测到病毒的转录本,因此有关宿主miRNA如何影响病毒的生理过程本文将不做进一步分析。
2.5 差异表达miRNA靶基因的KEGG富集分析
差异表达miRNA的靶基因KEGG富集结果见图4。在病毒感染早期,富集最显著的两个通路分别是戊糖葡糖醛酸相互转化(pentose and glucuronate interconversions)和氨酰tRNA合成(aminoacyl-tRNA biosynthesis)此外,还包括脂代谢过程,特别是甘油脂代谢(glycerolipid metabolism)和脂肪酸代谢(fatty acid metabolism)。甘氨酸,丝氨酸和苏氨酸代谢(glycine, serine and threonine metabolism)也富集显著,丝氨酸可作为神经酰胺合成的原料参与鞘脂代谢。在病毒感染晚期,富集最显著的代谢通路包括ABC转运蛋白(ABC transporters)和氨酰tRNA合成(aminoacyl-tRNA biosynthesis),鞘脂代谢(sphingolipid metabolism)、脂肪酸降解(fatty degradation)以及糖酵解/糖异生(glycolysis/gluconeogenesis)等。特别是感染早期和晚期,差异表达miRNA的靶基因均能够显著富集到宿主脂代谢过程,暗示病毒感染重构的脂代谢过程可能存在转录后水平调控。2.6 miRNA与脂代谢靶基因的互作网络
miRNA-脂代谢靶基因的互作网络图能够帮助人们更好地了解miRNA与脂代谢相关基因之间的调控关系。从图5中可以看出,ehx-miR3-3p和ehv- miR1-5p靶基因数量最多,并且这两个miRNA的靶基因在5个脂代谢相关通路中都有分布,表明这两个miRNA可能在脂代谢调控中起到核心作用。同时,部分基因也能被多个miRNA靶向,比如ACSL4 (长链酰基辅酶A合成酶4)能够同时被ehv-miR2-3p和ehx-miR3-3p靶向。此外,病毒miRNA和宿主miRNA也能同时靶向某一代谢途径,表明病毒可能会利用宿主miRNA来共同调节脂代谢过程。图4
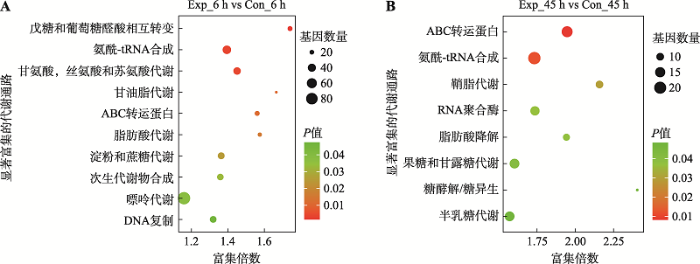 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4病毒感染6 h (A)和45 h (B)差异miRNA靶基因的KEGG富集分析
Fig. 4KEGG enrichment analysis of DE miRNA target genes at 6 (A) and 45 (B) hours post infection
富集倍数为:(某通路中差异基因数目/有注释的差异基因总数)/(某通路中的有注释的基因总数/背景基因中有注释的基因总数);气泡的大小对应该通路中基因数量的多少,颜色表示富集的显著性P值。
图5
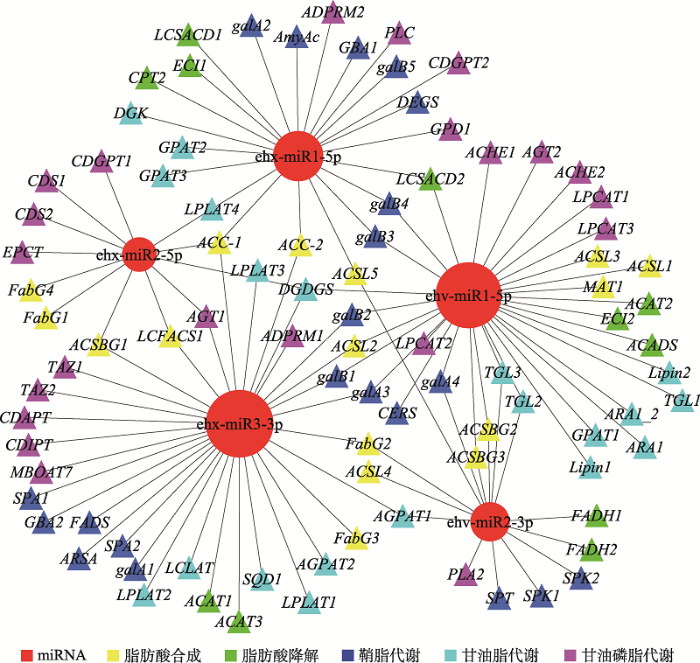 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5miRNA-脂代谢靶基因互作网络图
Fig. 5The correlation network between miRNA and lipid metabolism-related target genes
图中红色圆形表示miRNA,三角形表示脂代谢相关靶基因,不同颜色对应不同脂代谢过程。
2.7 miRNA靶基因qRT-PCR验证
为了进一步了解miRNA与脂代谢相关靶基因的靶向关系,采用qRT-PCR方法验证了miRNA靶基因的表达水平,并分析miRNA与其靶基因表达之间的相关性。选取6个脂代谢关键靶基因(附表8),包括参与脂肪酸合成的ACC-1,参与鞘脂代谢过程的SPT和CERS,以及参与脂肪酸降解过程的ACOX, ACAT和ACADS酶基因。荧光定量结果显示,除了ACC-1基因在病毒感染早期表达水平显著升高外,其他几个脂代谢相关酶基因的表达水平在病毒感染早期(6 h)和晚期(45 h)均不同程度下调(附图1);而且这些酶基因的表达水平与miRNA的qRT-PCR结果均为负相关关系,符合一般的miRNA对靶基因的调控规律,表明这些靶基因的表达水平很可能受到miRNA的调控。3 讨论
miRNA是近年来RNA生物学领域中的重大发现,其通过结合在靶基因的不同位点抑制或激活靶基因的表达水平,作为丰度极高的基因调控因子参与调控多种生物学过程。特别是新近研究发现miRNA与它作用的靶基因是响应生物及非生物环境胁迫的主要调控因子[34]。例如,miRNA作为关键因子在复杂的病毒-宿主互作网络中调控病毒的复制、免疫逃逸及宿主抗病毒等过程,并可能成为病毒感染诊断及治疗的新手段[35]。miRNA的另一个新功能是通过控制代谢过程中的靶基因,从而调控相关代谢通路,特别是在脂质代谢网络中起重要调节作用[19,36]。越来越多的研究表明,病毒感染赫氏颗石藻能够显著重塑宿主脂肪酸代谢、鞘脂代谢及甘油脂等脂代谢过程,且这些相关脂代谢与病毒的侵染、复制、组装及病毒颗粒的释放等过程密切相关[9,10,11]。miRNA是否参与颗石藻病毒感染重塑的脂代谢过程尚不清楚。本研究通过对EhV99B1感染的赫氏颗石藻BOF92细胞样品进行小RNA测序,鉴定出26条新的miRNA,分析其基本特征和保守性,并结合miRNA差异表达谱特征、靶基因功能富集以及qRT-PCR实验等,初步分析了病毒感染诱导的差异表达miRNA在宿主脂代谢过程中可能发挥的作用。3.1 赫氏颗石藻miRNA的基本特征
本研究经测序分析共鉴定出了24条颗石藻miRNA,长度主要集中于18~21 nt,峰值为21 nt,符合一般植物miRNA的特征。E. huxleyi BOF92的miRNA首位碱基对C有着较强的偏好,这一点与营养胁迫条件下E. huxleyi CCMP1516中的miRNA相似[28]。然而,在大部分生物体中,包括其他一些浮游植物,其miRNA的5′端首位碱基通常为U[37,38,39,40,41]。这可能与参与RISC (RNA诱导的沉默复合体)形成过程中的2个重要蛋白即Dicer-like protein (DCL)和 Argonaute (AGO)有关。DCL能够切割初始miRNA进而形成前体miRNA,该蛋白通常包含以下几个特征结构域:DEAD、Hleicase-C、dsRBD及RNA酶Ⅲ结构域[42,43]。E. huxleyi CCMP1516能够编码四种DCL,但是它们都缺少一个或多个特征结构域,有三种包含了DEAD,Helicase-C和DSRM结构域,但不含有RNA酶Ⅲ结构域,该结构与对于RNA的绑定和切割十分重要;另外一种DCL含有DRSM和RNA酶Ⅲ结构域,但不含DEAD和Helicase-C结构域[25],颗石藻DCL结构上的这些差异可能会影响其切割位点的选择,进而影响miRNA首位碱基的偏好性。AGO蛋白位于RISC的中心,其最主要的功能结构域为PAZ和PIWI结构域。E. huxleyi CCMP1516基因组中有2种AGO同源物,虽然它们都含有PAZ和PIWI结构域,但与其他微藻以及部分模式物种的AGO系统发育树的分析结果显示[28],包括颗石藻在内的各种微藻的AGO蛋白都各自聚集成一类,表明这些微藻的AGO蛋白在进化上同源性较低。因此颗石藻可能也存在特殊的RNA沉默机制,使得miRNA的首位碱基不偏好于U。在病毒诱导的E. huxleyi BOF92 miRNA中,66.7%来自基因间区,29.2%来自外显子区,只有1条miRNA来自内含子区(附表1)。这与近缘株系E. huxleyi CCMP1516的miRNA来源分布较为相似,E. huxleyi CCMP1516的miRNA约77%来自基因间区,23%来自内含子区[28]。三角褐指藻(Phaeodactylum tricornutum)和团藻(Volvox carteri)的miRNA也都主要来源于基因间区,只有少部分miRNA比对到了外显子区[38,44],这些微藻的miRNA的来源更类似于陆生高等植物[18]。莱茵衣藻(Chlamydomonas reinhardtii)和长囊水云(Ectocarpus siliculosus)的miRNA来源类似,即大部分位于内含子区,少数位于基因间区[40,39]。综上所述,不同浮游植物miRNA的来源具有多样性,表明miRNA在浮游植物的转录后水平可能具有广泛的调控功能。
图1
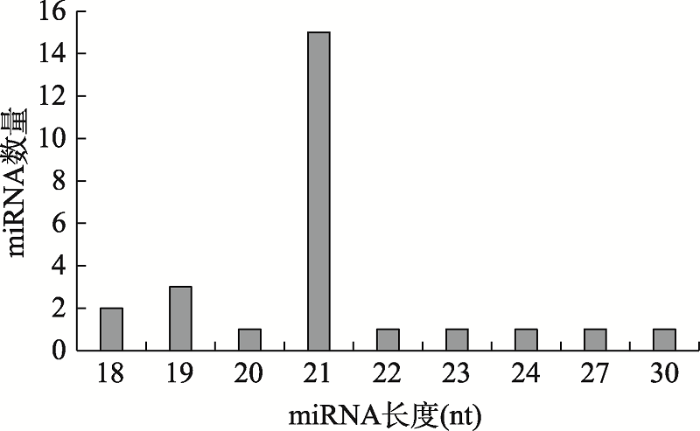 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1miRNA的长度分布
Fig. 1The length distribution of miRNAs
3.2 miRNA保守性特点
针对本研究中鉴定的24条宿主miRNA同源性较低的问题,推测可能有以下几个原因。首先,不同微藻之间的miRNA可能是独立进化的。Zhao等[40]首先对莱茵衣藻的miRNA进行了表征,作者将鉴定出来的莱茵衣藻miRNA与miRBase数据库比对后未发现同源miRNA。随后多位****对不同种的微藻,如假微型海链藻(T. pseudonana)[21],三角褐指藻(P. tricornutum)[44]及赫氏颗石藻CCMP1516[28]等在不同生长条件或环境胁迫条件下的miRNA进行了研究,也均未能在miRBase数据库找到同源的序列。这暗示miRNA可能在浮游植物、动植物中分别有着独立的进化机制。其次,病毒感染诱导产生特异性miRNA的表达。miRNA的表达是动态的,具有时空特异性,不同胁迫条件如营养胁迫、氧化应激、病毒感染等都会影响miRNA的表达,某些特异的miRNA可能表达水平很低,仅在特殊的生理生态条件下表达量才增加[28]。因此,病毒感染可能使赫氏颗石藻BOF92产生了特异性miRNA。另外,赫氏颗石藻miRNA株系间的变异较为明显。根据本文24条宿主miRNA与其他13株赫氏颗石藻基因组的比对结果看,能够全部比对上其他赫氏颗石藻株系的miRNA仅有10条,占miRNA总数的42%左右。赫氏颗石藻作为一种单细胞浮游植物,基因组的种内变异更有利于其适应动态变化的海洋环境以及在与病毒的“arms race”中获得优势。本研究中鉴定的24条宿主miRNA保守性较低可能是这些因素共同作用的结果。此外,本研究也没有找到EhV99B1 miRNA的同源序列。一方面,已鉴定的病毒miRNA非常有限;另一方面,与真核生物相比,病毒基因组有着更高的变异率,这意味着病毒拥有能快速适应宿主和环境条件的进化优势。3.3 miRNA靶向宿主脂代谢途径
miRNA靶基因功能富集结果显示,病毒感染早期(6 h)和晚期(45 h),差异表达的miRNA均能靶向宿主脂肪酸和鞘脂代谢通路(图6)。就脂肪酸代谢而言,感染早期,宿主编码的ehx-miR1-5p下调,其对脂肪酸合成过程的限速酶ACC-1靶基因的抑制作用减弱,与此同时宿主编码的ehx-miR4-3p上调,其对脂肪酸降解过程中的关键酶ACOX和ACAT靶基因的抑制作用增强,从而导致脂肪酸含量的积累(图6A)。感染晚期,则是病毒编码的ehv-miR2-5p 上调,并靶向抑制脂肪酸降解过程的相关酶基因ACADS (图6B)。因此,推测感染过程中,宿主和病毒编码的miRNA协同作用导致宿主细胞中脂肪酸含量的不断积累。脂肪酸在病毒感染过程有着十分重要的作用,一方面大型双链DNA病毒的复制和装配有更高的代谢需求,另一方面脂肪酸可作为甘油脂和鞘脂的合成原料[7]。另外,病毒感染过程中,宿主鞘脂从头合成过程中的两个关键酶基因SPT和CERS的表达分别受到早期宿主上调的ehx-miR3-5p (图6A)和晚期病毒上调的ehv-miR2-5p (图6B)的靶向抑制,因而宿主鞘脂从头合成途径在整个病毒感染过程中被显著抑制,并导致鞘脂类物质含量减少。上述结果可能在一定程度上补充解释了本课题组之前的脂质组学数据,即EhV99B1感染早期(6 h)和晚期(45 h),宿主赫氏颗石藻BOF92细胞中大部分脂肪酸代谢物显著积累,而几乎所有检测到的16种鞘脂类物质的含量均显著降低[11]。众所周知,鞘脂是细胞膜上“脂筏”结构域的关键功能成分,也是病毒包膜结构的主要成分之一,对病毒的侵染、装配及释放过程十分重要,且作为信号分子,在感染晚期能够诱导宿主程序性细胞死亡。对赫氏颗石藻CCMP1516及其特异性裂解病毒EhV86的全基因测序注释结果显示,通过基因水平转移,病毒基因组“截获”了一套鞘脂从头合成途径中的关键酶基因,并通过调节相关基因的表达在一定程度上掌控了宿主的鞘脂代谢过程[45]。在EhV201感染赫氏颗石藻CCMP374过程中发现,随着感染时间的延长(感染0~36 h),宿主基因组编码的hSPT基因表达显著降低,而病毒基因组编码的vSPT在mRNA和蛋白水平的表达均明显上升,从而大量合成并积累病毒性鞘糖脂[9],并最终诱导宿主程序性细胞死亡[15,17]。综上所述,病毒和宿主的miRNA均可能参与宿主脂代谢的重塑过程,其中病毒的miRNA主要在感染晚期通过对脂肪酸降解过程和鞘脂合成过程中的部分基因进行靶向抑制,进而促进宿主脂肪酸的积累和鞘脂水平的降低。图6
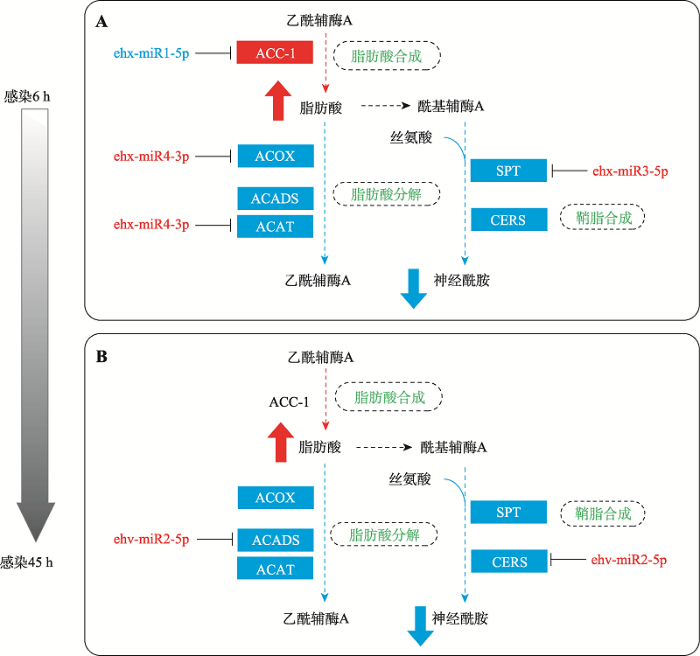 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6miRNA靶向的脂肪酸及鞘脂代谢通路
A和B分别表示病毒感染6 h和45 h差异miRNA对宿主脂肪酸和鞘脂代谢的靶向调控图。红色字体代表miRNA及靶基因高表达,蓝色填充表示miRNA及靶基因低表达;红色箭头代表脂代谢物积累,蓝色箭头表示脂代谢物减少。虚线框中的绿色字体表示不同的代谢通路
Fig. 6Fatty acid and sphingolipid metabolism pathways targeted by miRNAs
(责任编委: 赵呈天)
致谢:
感谢Gunnar Bratbak教授(挪威卑尔根大学生物系)提供的Emiliania huxleyi BOF92藻株和Emiliania huxleyi virus 99B1病毒株。附录:
附加材料详见文章电子版附图1
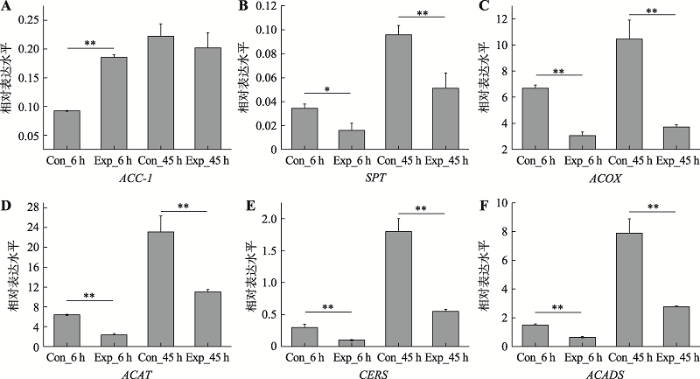 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图16个靶基因的荧光定量qRT-PCR结果
“*”表示 P<0.05;“**”表示P<0.01。
Supplementary Fig. 1The qRT-PCR results of six target genes
Supplementary Table 1
附表1
附表1成熟miRNA的基本信息
Supplementary Table 1
| miRNA ID | miRNA序列 | 长度 (nt) | 表达丰度(FPKM) | 基因组 位置 | |||
|---|---|---|---|---|---|---|---|
| Con_6h | Exp_6h | Con_45h | Exp_45h | ||||
| NW_005195085.1_38173-5p | cgucggcgcaugugguuagag | 21 | 24.23 | 20.52 | 17.56 | 12.32 | Intergenic |
| NW_005195085.1_38180-3p | cuagucguaguaaguguuuga | 21 | 5.51 | 2.53 | 7.48 | 2.66 | Intergenic |
| NW_005196191.1_35170-3p | cagguguauuugugaggccaa | 21 | 2.45 | 1.11 | 3.23 | 2.23 | Intergenic |
| NW_005196603.1_34298-5p (ehx-miR1-5p) | ccgacccgucccugagccu | 19 | 206.33 | 71.57 | 145.54 | 86.69 | intron |
| NW_005196755.1_34052-3p | cgaggagccgccaccagagcu | 21 | 3.42 | 1.57 | 3.56 | 4.59 | Intergenic |
| NW_005196755.1_34028-5p | cgagaugguacugaagagauc | 21 | 240.47 | 205..45 | 266.69 | 116.88 | Intergenic |
| NW_005196755.1_34019-3p | cuuguccgcgucgccucauga | 21 | 1.45 | 2.46 | 1.78 | 2.56 | Intergenic |
| NW_005197715.1_30571-5p | gagguuggcgcgggcgagc | 19 | 2.47 | 4.76 | 7.58 | 3.45 | Exon |
| NW_005198391.1_27186-5p | cgacaccugcuggcuggcgcc | 21 | 3.12 | 1.63 | 1.45 | 1.73 | Exon |
| NW_005198966.1_24265-3p | aacccucgcucggagccgcgc | 21 | 3.32 | 1.86 | 1.73 | 1.43 | Intergenic |
| NW_005198966.1_24265-5p | uugguucugacgauuugu | 18 | 88106.45 | 105030.78 | 101495.53 | 88208.43 | Exon |
| NW_005199363.1_22042-5p | cguucagggcgccgggcacg | 20 | 5.45 | 1.78 | 3.51 | 1.81 | Intergenic |
| NW_005199378.1_21978-3p | gggcuaugcccgcgcgaguuc | 21 | 1.12 | 0.79 | 1.96 | 1.13 | Intergenic |
| NW_005199378.1_21978-5p | guugacguggccgagugguuaag | 23 | 3124.46 | 1430.13 | 1710.79 | 761.41 | Intergenic |
| NW_005199975.1_17848-3p | cagagccacccuccuggacgc | 21 | 2.73 | 1.23 | 1.72 | 0.00 | Intergenic |
| NW_005200282.1_15947-3p | auguugucgccccaguccg | 19 | 24.56 | 11.22 | 17.86 | 19.76 | Exon |
| NW_005200461.1_14543-5p | gccggcuacgccgccgccgggc | 22 | 3.45 | 1.37 | 2.56 | 1.39 | Intergenic |
| NW_005200496.1_14318-5p (ehx-miR2-5p) | ucgccugucguccgacguucugucuccggc | 30 | 25.64 | 52.78 | 14.67 | 7.35 | Exon |
| NW_005200517.1_13952-3p | cggagaagcgucagacucggc | 21 | 17.56 | 18.67 | 13.47 | 13.43 | Intergenic |
| NW_005200535.1_13863-3p | cagucguguguuuggcguccu | 21 | 1.97 | 3.45 | 0.99 | 1.89 | Intergenic |
| NW_005200600.1_13330-3p (ehx-miR3-3p) | gcgagguuggcgcgggcgagc | 21 | 7.36 | 17.41 | 18.22 | 12.56 | Intergenic |
| NW_005200881.1_10661-3p | cgaacuugucggcgcccgcga | 21 | 4.25 | 1.47 | 3.34 | 1.67 | Exon |
| NW_005201234.1_6668-5p | acuccauggccagggccu | 18 | 5.63 | 1.29 | 2.42 | 1.37 | Intergenic |
| NW_005202428.1_1248-3p | gccacggccgcgccgccgcccgcc | 24 | 2.56 | 2.79 | 4.34 | 5.34 | Exon |
| gi|73852470|ref|NC_007346.1|_540-3p (ehv-miR1-3p) | guucgauccucgccguggguu | 21 | 0.01 | 0.01 | 0.01 | 61.35 | Exon |
| gi|73852470|ref|NC_007346.1|_540-5p (ehv-miR2-5p) | cccucguagcucaguggauagagcuc | 26 | 0.01 | 0.01 | 0.01 | 7.64 | Exon |
新窗口打开|下载CSV
Supplementary Table 2
附表2
附表2前体miRNA的完整序列
Supplementary Table 2
| 前体miRNR ID | 长度(nt) | 前体miRNA序列 |
|---|---|---|
| NW_005195085.1_38173 | 74 | cgucggcgcaugugguuagagauacaucuagagauacuucugcauuuuucucuuuccgcacgcaccccgaauuc |
| NW_005195085.1_38180 | 60 | agauguuacgauaacuagcacauuuagaucauucaugugcuagucguaguaaguguuuga |
| NW_005196191.1_35170 | 46 | ggguuggggaaacaucugucaaaugcagguguauuugugaggccaa |
| NW_005196603.1_34298 | 70 | ccgacccgucccugagccucccuguaggucgaccgagacuggcugacaaagcgcguggucggcgucggcg |
| NW_005196755.1_34019 | 52 | aagaggaagggaugugcgagaaagcaaccugcuuguccgcgucgccucauga |
| NW_005196755.1_34028 | 72 | cgagaugguacugaagagauccguuacgcaaaagcaugcgcgacuugcacucuuucgugccacucgcucgcc |
| NW_005196755.1_34052 | 58 | auaacgugugucgggcacucgggcugcuuguacggcucgaggagccgccaccagagcu |
| NW_005197715.1_30571 | 47 | gagguuggcgcgggcgagcuucgacaucuucucgcggcagcggucga |
| NW_005198391.1_27186 | 78 | cgacaccugcuggcuggcgccggccgucgccucugccgcgcgcgcgaggcccacgucggcgagguagccguugccgcg |
| NW_005198966.1_24265 | 53 | uugguucugacgauuuguugcgcgcgcgcaacaacccucgcucggagccgcgc |
| NW_005199363.1_22042 | 52 | cguucagggcgccgggcacgccuuaggggggcgugcgccgugaugcucgggc |
| NW_005199378.1_21978 | 65 | guugacguggccgagugguuaaggcgagggacugcuaaucccuugggcuaugcccgcgcgaguuc |
| NW_005199975.1_17848 | 86 | cuccggcgaggugaggaccucugcacuugugaggaccucugcuuccagcccacaucuuccaggugcagagccacccuccuggacgc |
| NW_005200282.1_15947 | 39 | agcuggggcagcagcgcagcauguugucgccccaguccg |
| NW_005200461.1_14543 | 56 | gccggcuacgccgccgccgggccagcccgccucgccgccucaugcguugccgccgc |
| NW_005200496.1_14318 | 71 | ucgccugucguccgacguucugucuccggcacauuacggugucgucgucguuccgcgagagccauccggac |
| NW_005200517.1_13952 | 57 | gaggucaacauguuucucguagaacacggugacgugcggagaagcgucagacucggc |
| NW_005200535.1_13863 | 70 | gggccgggggggcggcgggcgcggggugcggggugcggugcggugcgugcagucguguguuuggcguccu |
| NW_005200600.1_13330 | 65 | gggccucggccggccggggcggcgaggucgugagacagcacgcugcgagguuggcgcgggcgagc |
| NW_005200881.1_10661 | 55 | accucgcgcggcaggcggcagacgaagauguugccgaacuugucggcgcccgcga |
| NW_005201234.1_6668 | 56 | acuccauggccagggccuaucgucuucaagaggagaagccuaccgccgaggaggac |
| NW_005202428.1_1248 | 53 | cgcagcugcgcgcggccgcggcuaccucggccacggccgcgccgccgcccgcc |
| gi|73852470|ref|NC_007346.1|_540 | 72 | cccucguagcucaguggauagagcucuugccuucuaagcaaguggucgcgaguucgauccucgccguggguu |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表326条成熟miRNA与已鉴定miRNA的比对结果
Supplementary Table 3
| 比对到miRbase数据库中已知miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005196603.1_34298-5p | pal-miR-9987-3p | 93.75 | 16 | 1 | 0 | 2 | 17 | 20 | 5 | 25 | 19.6 |
| NW_005196603.1_34298-5p | hsa-miR-6743-5p | 83.333 | 18 | 3 | 0 | 2 | 19 | 19 | 2 | 80 | 18 |
| NW_005198966.1_24265-3p | hsa-miR-3183 | 83.333 | 18 | 3 | 0 | 4 | 21 | 2 | 19 | 89 | 18 |
| NW_005199363.1_22042-5p | aof-miR160a | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | vca-miR160-5p | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | cpa-miR160d | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | mes-miR160f | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | mes-miR160e | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | cme-miR160d | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | bdi-miR160e-5p | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | tcc-miR160a | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | ahy-miR160-5p | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | rco-miR160c | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | sbi-miR160f | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | vvi-miR160b | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | vvi-miR160a | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | mtr-miR160c | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | ptc-miR160f | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | ptc-miR160e-5p | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199363.1_22042-5p | osa-miR160f-5p | 83.333 | 18 | 3 | 0 | 1 | 18 | 18 | 1 | 84 | 18 |
| NW_005199378.1_21978-5p | cel-miR-8189-5p | 88.889 | 18 | 2 | 0 | 6 | 23 | 21 | 4 | 26 | 19.8 |
| NW_005200461.1_14543-5p | mdo-miR-12392-3p | 88.235 | 17 | 2 | 0 | 6 | 22 | 23 | 7 | 52 | 18.8 |
| NW_005200461.1_14543-5p | bta-miR-11976 | 88.235 | 17 | 2 | 0 | 2 | 18 | 17 | 1 | 52 | 18.8 |
| NW_005200461.1_14543-5p | bta-miR-11975 | 88.235 | 17 | 2 | 0 | 2 | 18 | 17 | 1 | 52 | 18.8 |
| NW_005200461.1_14543-5p | hsa-miR-3960 | 88.235 | 17 | 2 | 0 | 2 | 18 | 17 | 1 | 52 | 18.8 |
| NW_005200461.1_14543-5p | mmu-miR-3960 | 88.235 | 17 | 2 | 0 | 2 | 18 | 17 | 1 | 52 | 18.8 |
| NW_005200461.1_14543-5p | ssc-miR-4332 | 100 | 13 | 0 | 0 | 10 | 22 | 5 | 17 | 69 | 18.4 |
| NW_005200535.1_13863-3p | mle-miR-10a-3p | 80 | 20 | 4 | 0 | 2 | 21 | 3 | 22 | 77 | 18.2 |
| NW_005202428.1_1248-3p | bta-miR-11976 | 85 | 20 | 3 | 0 | 1 | 20 | 20 | 1 | 24 | 20.1 |
| NW_005202428.1_1248-3p | bta-miR-11975 | 85 | 20 | 3 | 0 | 1 | 20 | 20 | 1 | 24 | 20.1 |
| NW_005202428.1_1248-3p | hsa-miR-9899 | 88.235 | 17 | 2 | 0 | 7 | 23 | 6 | 22 | 56 | 18.8 |
| NW_005202428.1_1248-3p | cja-miR-638 | 100 | 13 | 0 | 0 | 12 | 24 | 22 | 10 | 76 | 18.4 |
| NW_005202428.1_1248-3p | mmu-miR-5126 | 77.273 | 22 | 5 | 0 | 2 | 23 | 22 | 1 | 76 | 18.4 |
| NW_005202428.1_1248-3p | mml-miR-638 | 100 | 13 | 0 | 0 | 12 | 24 | 23 | 11 | 76 | 18.4 |
| NW_005202428.1_1248-3p | mdo-miR-7346-3p | 80 | 20 | 4 | 0 | 5 | 24 | 20 | 1 | 87 | 18.2 |
| NW_005195085.1_38173-5p | Vca_miR80 | 85.714 | 14 | 2 | 0 | 6 | 19 | 4 | 17 | 17 | 15.6 |
| NW_005195085.1_38173-5p | Vca_miR116 | 90.909 | 11 | 1 | 0 | 4 | 14 | 9 | 19 | 41 | 14.4 |
| NW_005195085.1_38173-5p | Thalassiosira | 75 | 20 | 4 | 1 | 1 | 19 | 24 | 5 | 47 | 14.2 |
| NW_005195085.1_38173-5p | miR90_0453 | 78.571 | 14 | 3 | 0 | 3 | 16 | 6 | 19 | 63 | 13.7 |
| NW_005195085.1_38173-5p | miR90_0414 | 78.571 | 14 | 3 | 0 | 3 | 16 | 5 | 18 | 63 | 13.7 |
| NW_005195085.1_38173-5p | miR90_0041 | 78.571 | 14 | 3 | 0 | 3 | 16 | 2 | 15 | 63 | 13.7 |
| NW_005195085.1_38173-5p | miR90_0631 | 83.333 | 12 | 2 | 0 | 2 | 13 | 7 | 18 | 73 | 13.5 |
| NW_005195085.1_38173-5p | miR90_0284 | 83.333 | 12 | 2 | 0 | 2 | 13 | 13 | 2 | 73 | 13.5 |
| NW_005195085.1_38173-5p | miR95_0193 | 83.333 | 12 | 2 | 0 | 2 | 13 | 7 | 18 | 73 | 13.5 |
| NW_005195085.1_38173-5p | miR90_0169 | 91.667 | 12 | 0 | 1 | 1 | 12 | 11 | 1 | 85 | 13.3 |
| NW_005195085.1_38173-5p | miR95_0076 | 90 | 10 | 1 | 0 | 1 | 10 | 8 | 17 | 85 | 13.3 |
| NW_005195085.1_38173-5p | CCMP1516-miR8 | 90 | 10 | 1 | 0 | 1 | 10 | 12 | 21 | 85 | 13.3 |
| NW_005195085.1_38173-5p | miR90_0835 | 100 | 8 | 0 | 0 | 1 | 8 | 10 | 17 | 98 | 13.1 |
| NW_005195085.1_38173-5p | miR90_0689 | 100 | 8 | 0 | 0 | 1 | 8 | 12 | 5 | 98 | 13.1 |
| NW_005195085.1_38173-5p | miR90_0682 | 100 | 8 | 0 | 0 | 1 | 8 | 8 | 1 | 98 | 13.1 |
| NW_005195085.1_38173-5p | miR90_0351 | 100 | 8 | 0 | 0 | 1 | 8 | 3 | 10 | 98 | 13.1 |
| NW_005195085.1_38173-5p | miR90_0271 | 100 | 8 | 0 | 0 | 1 | 8 | 2 | 9 | 98 | 13.1 |
| NW_005195085.1_38173-5p | miR90_0220 | 100 | 8 | 0 | 0 | 1 | 8 | 4 | 11 | 98 | 13.1 |
| NW_005195085.1_38180-3p | ccr-miR5 | 85.714 | 14 | 2 | 0 | 5 | 18 | 6 | 19 | 17 | 15.6 |
| NW_005195085.1_38180-3p | miR90_0317 | 84.615 | 13 | 2 | 0 | 5 | 17 | 13 | 25 | 35 | 14.6 |
| NW_005195085.1_38180-3p | miR90_0157 | 78.571 | 14 | 3 | 0 | 4 | 17 | 1 | 14 | 63 | 13.7 |
| NW_005195085.1_38180-3p | ppu-miR21-5p | 83.333 | 12 | 2 | 0 | 1 | 12 | 2 | 13 | 73 | 13.5 |
| NW_005195085.1_38180-3p | miR95_0487 | 68.421 | 19 | 6 | 0 | 2 | 20 | 19 | 1 | 85 | 13.3 |
| NW_005195085.1_38180-3p | Vca_miR107* | 68.421 | 19 | 6 | 0 | 3 | 21 | 19 | 1 | 85 | 13.3 |
| NW_005195085.1_38180-3p | Vca_miR19 | 90 | 10 | 1 | 0 | 2 | 11 | 1 | 10 | 85 | 13.3 |
| NW_005195085.1_38180-3p | SjapMIR84 | 90 | 10 | 1 | 0 | 7 | 16 | 4 | 13 | 85 | 13.3 |
| NW_005195085.1_38180-3p | miR90_0817 | 70.588 | 17 | 5 | 0 | 4 | 20 | 17 | 1 | 98 | 13.1 |
| NW_005195085.1_38180-3p | miR95_0559 | 70.588 | 17 | 5 | 0 | 2 | 18 | 18 | 2 | 98 | 13.1 |
| NW_005196191.1_35170-3p | miR90_0395 | 78.571 | 14 | 3 | 0 | 7 | 20 | 18 | 5 | 63 | 13.7 |
| NW_005196191.1_35170-3p | miR90_0538 | 100 | 8 | 0 | 0 | 3 | 10 | 18 | 11 | 98 | 13.1 |
| NW_005196191.1_35170-3p | miR90_0527 | 70.588 | 17 | 5 | 0 | 2 | 18 | 23 | 7 | 98 | 13.1 |
| NW_005196191.1_35170-3p | miR90_0448 | 70.588 | 17 | 5 | 0 | 2 | 18 | 3 | 19 | 98 | 13.1 |
| NW_005196191.1_35170-3p | miR90_0421 | 100 | 8 | 0 | 0 | 6 | 13 | 1 | 8 | 98 | 13.1 |
| NW_005196191.1_35170-3p | miR95_0114 | 70.588 | 17 | 5 | 0 | 2 | 18 | 20 | 4 | 98 | 13.1 |
| NW_005196603.1_34298-5p | miR90_0660 | 87.5 | 16 | 2 | 0 | 3 | 18 | 4 | 19 | 3.6 | 17.7 |
| NW_005196603.1_34298-5p | miR95_0377 | 100 | 10 | 0 | 0 | 2 | 11 | 14 | 5 | 21 | 15.2 |
| NW_005196603.1_34298-5p | miR95_0344 | 100 | 10 | 0 | 0 | 2 | 11 | 14 | 5 | 21 | 15.2 |
| NW_005196603.1_34298-5p | miR95_0338 | 100 | 10 | 0 | 0 | 2 | 11 | 11 | 2 | 21 | 15.2 |
| NW_005196603.1_34298-5p | miR95_0320 | 100 | 10 | 0 | 0 | 2 | 11 | 10 | 1 | 21 | 15.2 |
| NW_005196603.1_34298-5p | miR90_0701 | 76.471 | 17 | 4 | 0 | 2 | 18 | 17 | 1 | 24 | 15 |
| NW_005196603.1_34298-5p | Vca_miR13* | 76.471 | 17 | 4 | 0 | 2 | 18 | 18 | 2 | 24 | 15 |
| NW_005196603.1_34298-5p | SjapMIR5 | 80 | 15 | 3 | 0 | 5 | 19 | 19 | 5 | 28 | 14.8 |
| NW_005196603.1_34298-5p | miR90_0905 | 90.909 | 11 | 1 | 0 | 7 | 17 | 1 | 11 | 37 | 14.4 |
| NW_005196603.1_34298-5p | ppu-miR35-5p | 100 | 9 | 0 | 0 | 11 | 19 | 16 | 8 | 43 | 14.2 |
| NW_005196603.1_34298-5p | miR90_0203 | 75 | 16 | 4 | 0 | 3 | 18 | 4 | 19 | 50 | 13.9 |
| NW_005196603.1_34298-5p | Vca_miR38 | 77.778 | 18 | 3 | 1 | 1 | 17 | 19 | 2 | 50 | 13.9 |
| NW_005196603.1_34298-5p | miR90_0679 | 78.571 | 14 | 3 | 0 | 6 | 19 | 2 | 15 | 57 | 13.7 |
| NW_005196603.1_34298-5p | miR90_0274 | 78.571 | 14 | 3 | 0 | 1 | 14 | 2 | 15 | 57 | 13.7 |
| NW_005196603.1_34298-5p | miR90_0185 | 78.571 | 14 | 3 | 0 | 6 | 19 | 16 | 3 | 57 | 13.7 |
| NW_005196603.1_34298-5p | SjapMIR6 | 78.571 | 14 | 3 | 0 | 6 | 19 | 2 | 15 | 57 | 13.7 |
| NW_005196603.1_34298-5p | ppu-miR7-5p | 83.333 | 12 | 2 | 0 | 6 | 17 | 18 | 7 | 66 | 13.5 |
| NW_005196603.1_34298-5p | miR95_0036 | 83.333 | 12 | 2 | 0 | 3 | 14 | 7 | 18 | 66 | 13.5 |
| NW_005196603.1_34298-5p | miR90_0147 | 90 | 10 | 1 | 0 | 6 | 15 | 10 | 19 | 77 | 13.3 |
| NW_005196603.1_34298-5p | Vca_miR67 | 90 | 10 | 1 | 0 | 6 | 15 | 10 | 1 | 77 | 13.3 |
| NW_005196603.1_34298-5p | ccr-miR9 | 68.421 | 19 | 6 | 0 | 1 | 19 | 20 | 2 | 77 | 13.3 |
| NW_005196603.1_34298-5p | SjapMIR26_2 | 90 | 10 | 1 | 0 | 7 | 16 | 15 | 6 | 77 | 13.3 |
| NW_005196603.1_34298-5p | SjapMIR26_1 | 90 | 10 | 1 | 0 | 7 | 16 | 15 | 6 | 77 | 13.3 |
| NW_005196603.1_34298-5p | miR90_0579 | 100 | 8 | 0 | 0 | 4 | 11 | 8 | 1 | 89 | 13.1 |
| NW_005196603.1_34298-5p | miR95_0515 | 100 | 8 | 0 | 0 | 10 | 17 | 9 | 2 | 89 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0442 | 82.353 | 17 | 3 | 0 | 4 | 20 | 3 | 19 | 7.1 | 16.9 |
| NW_005196755.1_34052-3p | miR90_0873 | 92.308 | 13 | 1 | 0 | 4 | 16 | 14 | 2 | 9.5 | 16.5 |
| NW_005196755.1_34052-3p | miR90_0824 | 92.308 | 13 | 1 | 0 | 4 | 16 | 20 | 8 | 9.5 | 16.5 |
| NW_005196755.1_34052-3p | miR90_0824 | 90 | 10 | 1 | 0 | 7 | 16 | 14 | 5 | 85 | 13.3 |
| NW_005196755.1_34052-3p | miR90_0764 | 92.308 | 13 | 1 | 0 | 4 | 16 | 19 | 7 | 9.5 | 16.5 |
| NW_005196755.1_34052-3p | miR90_0764 | 90 | 10 | 1 | 0 | 7 | 16 | 13 | 4 | 85 | 13.3 |
| NW_005196755.1_34052-3p | miR90_0529 | 100 | 10 | 0 | 0 | 7 | 16 | 11 | 2 | 23 | 15.2 |
| NW_005196755.1_34052-3p | miR95_0450 | 80 | 15 | 3 | 0 | 6 | 20 | 18 | 4 | 31 | 14.8 |
| NW_005196755.1_34052-3p | Vca_miR44c | 80 | 15 | 3 | 0 | 7 | 21 | 5 | 19 | 31 | 14.8 |
| NW_005196755.1_34052-3p | Vca_miR44b | 80 | 15 | 3 | 0 | 7 | 21 | 5 | 19 | 31 | 14.8 |
| NW_005196755.1_34052-3p | Vca_miR44a | 80 | 15 | 3 | 0 | 7 | 21 | 5 | 19 | 31 | 14.8 |
| NW_005196755.1_34052-3p | miR90_0922 | 84.615 | 13 | 2 | 0 | 1 | 13 | 18 | 6 | 35 | 14.6 |
| NW_005196755.1_34052-3p | miR90_0898 | 84.615 | 13 | 2 | 0 | 7 | 19 | 14 | 2 | 35 | 14.6 |
| NW_005196755.1_34052-3p | miR90_0897 | 84.615 | 13 | 2 | 0 | 3 | 15 | 17 | 5 | 35 | 14.6 |
| NW_005196755.1_34052-3p | miR90_0866 | 84.615 | 13 | 2 | 0 | 1 | 13 | 21 | 9 | 35 | 14.6 |
| NW_005196755.1_34052-3p | miR90_0771 | 84.615 | 13 | 2 | 0 | 3 | 15 | 16 | 4 | 35 | 14.6 |
| NW_005196755.1_34052-3p | miR90_0722 | 84.615 | 13 | 2 | 0 | 9 | 21 | 17 | 5 | 35 | 14.6 |
| NW_005196755.1_34052-3p | miR95_0441 | 84.615 | 13 | 2 | 0 | 8 | 20 | 25 | 13 | 35 | 14.6 |
| NW_005196755.1_34052-3p | miR95_0439 | 84.615 | 13 | 2 | 0 | 3 | 15 | 4 | 16 | 35 | 14.6 |
| NW_005196755.1_34052-3p | miR90_0840 | 90.909 | 11 | 1 | 0 | 1 | 11 | 11 | 1 | 41 | 14.4 |
| NW_005196755.1_34052-3p | miR90_0520 | 90.909 | 11 | 1 | 0 | 5 | 15 | 17 | 7 | 41 | 14.4 |
| NW_005196755.1_34052-3p | miR90_0515 | 100 | 9 | 0 | 0 | 8 | 16 | 10 | 2 | 47 | 14.2 |
| NW_005196755.1_34052-3p | Vca_miR30-5p* | 72.222 | 18 | 5 | 0 | 4 | 21 | 20 | 3 | 47 | 14.2 |
| NW_005196755.1_34052-3p | miR90_0460 | 78.571 | 14 | 3 | 0 | 7 | 20 | 2 | 15 | 63 | 13.7 |
| NW_005196755.1_34052-3p | miR95_0509 | 78.571 | 14 | 3 | 0 | 6 | 19 | 1 | 14 | 63 | 13.7 |
| NW_005196755.1_34052-3p | miR95_0508 | 78.571 | 14 | 3 | 0 | 6 | 19 | 8 | 21 | 63 | 13.7 |
| NW_005196755.1_34052-3p | Vca_miR16 | 78.571 | 14 | 3 | 0 | 8 | 21 | 9 | 22 | 63 | 13.7 |
| NW_005196755.1_34052-3p | miR90_0646 | 83.333 | 12 | 2 | 0 | 4 | 15 | 12 | 1 | 73 | 13.5 |
| NW_005196755.1_34052-3p | Vca_miR98b | 83.333 | 12 | 2 | 0 | 6 | 17 | 17 | 6 | 73 | 13.5 |
| NW_005196755.1_34052-3p | Vca_miR98a | 83.333 | 12 | 2 | 0 | 6 | 17 | 17 | 6 | 73 | 13.5 |
| NW_005196755.1_34052-3p | miR90_0862 | 90 | 10 | 1 | 0 | 1 | 10 | 10 | 1 | 85 | 13.3 |
| NW_005196755.1_34052-3p | miR90_0422 | 90 | 10 | 1 | 0 | 8 | 17 | 8 | 17 | 85 | 13.3 |
| NW_005196755.1_34052-3p | miR95_0300 | 90 | 10 | 1 | 0 | 7 | 16 | 5 | 14 | 85 | 13.3 |
| NW_005196755.1_34052-3p | miR95_0254 | 90 | 10 | 1 | 0 | 7 | 16 | 1 | 10 | 85 | 13.3 |
| NW_005196755.1_34052-3p | miR95_0185 | 90 | 10 | 1 | 0 | 7 | 16 | 9 | 18 | 85 | 13.3 |
| NW_005196755.1_34052-3p | miR95_0164 | 90 | 10 | 1 | 0 | 3 | 12 | 1 | 10 | 85 | 13.3 |
| NW_005196755.1_34052-3p | miR95_0044 | 90 | 10 | 1 | 0 | 7 | 16 | 3 | 12 | 85 | 13.3 |
| NW_005196755.1_34052-3p | ppu-miR7-5p | 100 | 8 | 0 | 0 | 11 | 18 | 8 | 1 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0900 | 100 | 8 | 0 | 0 | 7 | 14 | 3 | 10 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0891 | 100 | 8 | 0 | 0 | 7 | 14 | 3 | 10 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0876 | 100 | 8 | 0 | 0 | 1 | 8 | 12 | 19 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0743 | 100 | 8 | 0 | 0 | 1 | 8 | 11 | 18 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0624 | 100 | 8 | 0 | 0 | 8 | 15 | 9 | 2 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0535 | 100 | 8 | 0 | 0 | 8 | 15 | 8 | 1 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0493 | 100 | 8 | 0 | 0 | 7 | 14 | 8 | 1 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0229 | 70.588 | 17 | 5 | 0 | 4 | 20 | 21 | 5 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0172 | 100 | 8 | 0 | 0 | 7 | 14 | 12 | 5 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0165 | 100 | 8 | 0 | 0 | 7 | 14 | 12 | 5 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0108 | 100 | 8 | 0 | 0 | 7 | 14 | 12 | 5 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR90_0013 | 100 | 8 | 0 | 0 | 7 | 14 | 10 | 17 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0416 | 100 | 8 | 0 | 0 | 2 | 9 | 7 | 14 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0381 | 100 | 8 | 0 | 0 | 8 | 15 | 11 | 18 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0289 | 100 | 8 | 0 | 0 | 7 | 14 | 12 | 5 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0288 | 100 | 8 | 0 | 0 | 7 | 14 | 12 | 5 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0275 | 100 | 8 | 0 | 0 | 8 | 15 | 8 | 1 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0274 | 100 | 8 | 0 | 0 | 8 | 15 | 11 | 18 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0260 | 100 | 8 | 0 | 0 | 8 | 15 | 5 | 12 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0248 | 100 | 8 | 0 | 0 | 7 | 14 | 9 | 16 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0175 | 100 | 8 | 0 | 0 | 8 | 15 | 10 | 17 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0158 | 100 | 8 | 0 | 0 | 7 | 14 | 7 | 14 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0102 | 100 | 8 | 0 | 0 | 7 | 14 | 1 | 8 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0090 | 100 | 8 | 0 | 0 | 7 | 14 | 8 | 1 | 98 | 13.1 |
| NW_005196755.1_34052-3p | miR95_0088 | 100 | 8 | 0 | 0 | 7 | 14 | 8 | 1 | 98 | 13.1 |
| NW_005196755.1_34052-3p | Porphyra | 100 | 8 | 0 | 0 | 6 | 13 | 8 | 1 | 98 | 13.1 |
| NW_005196755.1_34052-3p | SjapMIR103 | 100 | 8 | 0 | 0 | 8 | 15 | 16 | 9 | 98 | 13.1 |
| NW_005196755.1_34028-5p | miR95_0404 | 88.235 | 17 | 0 | 2 | 1 | 17 | 5 | 19 | 35 | 14.6 |
| NW_005196755.1_34028-5p | miR90_0665 | 90.909 | 11 | 1 | 0 | 2 | 12 | 7 | 17 | 41 | 14.4 |
| NW_005196755.1_34028-5p | miR90_0317 | 90.909 | 11 | 1 | 0 | 4 | 14 | 11 | 21 | 41 | 14.4 |
| NW_005196755.1_34028-5p | Vca_miR84 | 90.909 | 11 | 1 | 0 | 4 | 14 | 14 | 4 | 41 | 14.4 |
| NW_005196755.1_34028-5p | Vca_miR58-5p.1 | 75 | 16 | 4 | 0 | 4 | 19 | 8 | 23 | 55 | 13.9 |
| NW_005196755.1_34028-5p | Vca_miR19 | 83.333 | 12 | 2 | 0 | 6 | 17 | 4 | 15 | 73 | 13.5 |
| NW_005196755.1_34028-5p | ppu-miR14-3p | 90 | 10 | 1 | 0 | 12 | 21 | 12 | 21 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0837 | 90 | 10 | 1 | 0 | 2 | 11 | 5 | 14 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0676 | 90 | 10 | 1 | 0 | 10 | 19 | 8 | 17 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0661 | 90 | 10 | 1 | 0 | 2 | 11 | 5 | 14 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0637 | 90 | 10 | 1 | 0 | 2 | 11 | 11 | 2 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0634 | 90 | 10 | 1 | 0 | 2 | 11 | 18 | 9 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0633 | 90 | 10 | 1 | 0 | 2 | 11 | 11 | 2 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0586 | 90 | 10 | 1 | 0 | 2 | 11 | 14 | 5 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0578 | 90 | 10 | 1 | 0 | 2 | 11 | 3 | 12 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0569 | 90 | 10 | 1 | 0 | 4 | 13 | 18 | 9 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0555 | 90 | 10 | 1 | 0 | 2 | 11 | 20 | 11 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0537 | 90 | 10 | 1 | 0 | 2 | 11 | 8 | 17 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0447 | 90 | 10 | 1 | 0 | 2 | 11 | 2 | 11 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0446 | 90 | 10 | 1 | 0 | 2 | 11 | 2 | 11 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0413 | 90 | 10 | 1 | 0 | 2 | 11 | 14 | 5 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0380 | 90 | 10 | 1 | 0 | 2 | 11 | 10 | 19 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0357 | 90 | 10 | 1 | 0 | 2 | 11 | 11 | 2 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0347 | 90 | 10 | 1 | 0 | 2 | 11 | 5 | 14 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0346 | 90 | 10 | 1 | 0 | 2 | 11 | 15 | 6 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0345 | 90 | 10 | 1 | 0 | 2 | 11 | 4 | 13 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0343 | 90 | 10 | 1 | 0 | 2 | 11 | 13 | 4 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0318 | 90 | 10 | 1 | 0 | 2 | 11 | 13 | 4 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0295 | 90 | 10 | 1 | 0 | 2 | 11 | 14 | 5 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0291 | 90 | 10 | 1 | 0 | 2 | 11 | 16 | 7 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0290 | 90 | 10 | 1 | 0 | 2 | 11 | 18 | 9 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0289 | 90 | 10 | 1 | 0 | 2 | 11 | 16 | 7 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0288 | 90 | 10 | 1 | 0 | 2 | 11 | 16 | 7 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0287 | 90 | 10 | 1 | 0 | 2 | 11 | 15 | 6 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0280 | 90 | 10 | 1 | 0 | 2 | 11 | 1 | 10 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0234 | 90 | 10 | 1 | 0 | 2 | 11 | 4 | 13 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0131 | 90 | 10 | 1 | 0 | 2 | 11 | 14 | 5 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0128 | 90 | 10 | 1 | 0 | 2 | 11 | 21 | 12 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0127 | 90 | 10 | 1 | 0 | 2 | 11 | 25 | 16 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0124 | 90 | 10 | 1 | 0 | 2 | 11 | 14 | 5 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0123 | 90 | 10 | 1 | 0 | 2 | 11 | 10 | 1 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0122 | 90 | 10 | 1 | 0 | 2 | 11 | 14 | 5 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0121 | 90 | 10 | 1 | 0 | 2 | 11 | 18 | 9 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0120 | 90 | 10 | 1 | 0 | 2 | 11 | 20 | 11 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0111 | 90 | 10 | 1 | 0 | 2 | 11 | 15 | 6 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0093 | 90 | 10 | 1 | 0 | 2 | 11 | 18 | 9 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0092 | 90 | 10 | 1 | 0 | 2 | 11 | 19 | 10 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0078 | 90 | 10 | 1 | 0 | 2 | 11 | 4 | 13 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0063 | 90 | 10 | 1 | 0 | 2 | 11 | 9 | 18 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0062 | 90 | 10 | 1 | 0 | 2 | 11 | 5 | 14 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0061 | 90 | 10 | 1 | 0 | 2 | 11 | 4 | 13 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0060 | 90 | 10 | 1 | 0 | 2 | 11 | 3 | 12 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0040 | 90 | 10 | 1 | 0 | 2 | 11 | 9 | 18 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR90_0021 | 90 | 10 | 1 | 0 | 2 | 11 | 15 | 6 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0565 | 90 | 10 | 1 | 0 | 2 | 11 | 15 | 6 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0565 | 100 | 8 | 0 | 0 | 13 | 20 | 18 | 11 | 98 | 13.1 |
| NW_005196755.1_34028-5p | miR95_0564 | 90 | 10 | 1 | 0 | 2 | 11 | 20 | 11 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0564 | 100 | 8 | 0 | 0 | 13 | 20 | 23 | 16 | 98 | 13.1 |
| NW_005196755.1_34028-5p | miR95_0443 | 90 | 10 | 1 | 0 | 2 | 11 | 11 | 20 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0305 | 90 | 10 | 1 | 0 | 2 | 11 | 11 | 2 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0305 | 100 | 8 | 0 | 0 | 13 | 20 | 14 | 7 | 98 | 13.1 |
| NW_005196755.1_34028-5p | miR95_0298 | 90 | 10 | 1 | 0 | 2 | 11 | 11 | 20 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0259 | 90 | 10 | 1 | 0 | 2 | 11 | 12 | 3 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0053 | 90 | 10 | 1 | 0 | 2 | 11 | 14 | 5 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0052 | 90 | 10 | 1 | 0 | 2 | 11 | 13 | 4 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0051 | 90 | 10 | 1 | 0 | 2 | 11 | 16 | 7 | 85 | 13.3 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续1
Supplementary Table 3
| NW_005196755.1_34028-5p | miR95_0050 | 90 | 10 | 1 | 0 | 2 | 11 | 12 | 3 | 85 | 13.3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NW_005196755.1_34028-5p | miR95_0049 | 90 | 10 | 1 | 0 | 2 | 11 | 19 | 10 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0026 | 90 | 10 | 1 | 0 | 2 | 11 | 10 | 19 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0010 | 90 | 10 | 1 | 0 | 2 | 11 | 11 | 2 | 85 | 13.3 |
| NW_005196755.1_34028-5p | Vca_miR35-3p* | 91.667 | 12 | 0 | 1 | 10 | 20 | 22 | 11 | 85 | 13.3 |
| NW_005196755.1_34028-5p | Porphyra | 90 | 10 | 1 | 0 | 1 | 10 | 10 | 1 | 85 | 13.3 |
| NW_005196755.1_34028-5p | miR95_0524 | 100 | 8 | 0 | 0 | 4 | 11 | 15 | 8 | 98 | 13.1 |
| NW_005196755.1_34028-5p | miR95_0500 | 100 | 8 | 0 | 0 | 13 | 20 | 12 | 5 | 98 | 13.1 |
| NW_005196755.1_34028-5p | miR95_0480 | 100 | 8 | 0 | 0 | 1 | 8 | 8 | 1 | 98 | 13.1 |
| NW_005196755.1_34028-5p | miR95_0445 | 100 | 8 | 0 | 0 | 13 | 20 | 12 | 5 | 98 | 13.1 |
| NW_005196755.1_34028-5p | SjapMIR20 | 100 | 8 | 0 | 0 | 6 | 13 | 1 | 8 | 98 | 13.1 |
| NW_005196755.1_34019-3p | ppu-miR32-5p | 80 | 20 | 3 | 1 | 2 | 21 | 19 | 1 | 13 | 16.1 |
| NW_005196755.1_34019-3p | miR90_0172 | 76.19 | 21 | 4 | 1 | 1 | 21 | 21 | 2 | 23 | 15.2 |
| NW_005196755.1_34019-3p | miR90_0493 | 76.471 | 17 | 4 | 0 | 1 | 17 | 17 | 1 | 26 | 15 |
| NW_005196755.1_34019-3p | miR95_0289 | 76.471 | 17 | 4 | 0 | 1 | 17 | 21 | 5 | 26 | 15 |
| NW_005196755.1_34019-3p | miR95_0090 | 76.471 | 17 | 4 | 0 | 1 | 17 | 17 | 1 | 26 | 15 |
| NW_005196755.1_34019-3p | miR95_0088 | 76.471 | 17 | 4 | 0 | 1 | 17 | 17 | 1 | 26 | 15 |
| NW_005196755.1_34019-3p | miR95_0468 | 80 | 15 | 3 | 0 | 1 | 15 | 16 | 2 | 31 | 14.8 |
| NW_005196755.1_34019-3p | SjapMIR60 | 90.909 | 11 | 1 | 0 | 5 | 15 | 13 | 3 | 41 | 14.4 |
| NW_005196755.1_34019-3p | miR90_0907 | 75 | 16 | 4 | 0 | 2 | 17 | 2 | 17 | 55 | 13.9 |
| NW_005196755.1_34019-3p | CCMP1516-miR8 | 77.778 | 18 | 3 | 1 | 3 | 20 | 21 | 5 | 55 | 13.9 |
| NW_005196755.1_34019-3p | miR90_0839 | 78.571 | 14 | 3 | 0 | 4 | 17 | 1 | 14 | 63 | 13.7 |
| NW_005196755.1_34019-3p | miR90_0765 | 78.571 | 14 | 3 | 0 | 4 | 17 | 2 | 15 | 63 | 13.7 |
| NW_005196755.1_34019-3p | miR90_0211 | 78.571 | 14 | 3 | 0 | 2 | 15 | 16 | 3 | 63 | 13.7 |
| NW_005196755.1_34019-3p | ppu-miR15-5p | 83.333 | 12 | 2 | 0 | 1 | 12 | 6 | 17 | 73 | 13.5 |
| NW_005196755.1_34019-3p | miR95_0415 | 83.333 | 12 | 2 | 0 | 6 | 17 | 1 | 12 | 73 | 13.5 |
| NW_005196755.1_34019-3p | miR95_0394 | 85.714 | 14 | 1 | 1 | 9 | 21 | 17 | 4 | 73 | 13.5 |
| NW_005196755.1_34019-3p | miR95_0194 | 83.333 | 12 | 2 | 0 | 6 | 17 | 3 | 14 | 73 | 13.5 |
| NW_005196755.1_34019-3p | miR95_0179 | 83.333 | 12 | 2 | 0 | 9 | 20 | 21 | 10 | 73 | 13.5 |
| NW_005196755.1_34019-3p | miR95_0032 | 83.333 | 12 | 2 | 0 | 1 | 12 | 13 | 2 | 73 | 13.5 |
| NW_005196755.1_34019-3p | SjapMIR39 | 83.333 | 12 | 2 | 0 | 7 | 18 | 10 | 21 | 73 | 13.5 |
| NW_005196755.1_34019-3p | miR90_0804 | 90 | 10 | 1 | 0 | 1 | 10 | 15 | 24 | 85 | 13.3 |
| NW_005196755.1_34019-3p | miR90_0691 | 90 | 10 | 1 | 0 | 1 | 10 | 15 | 24 | 85 | 13.3 |
| NW_005196755.1_34019-3p | miR90_0658 | 90 | 10 | 1 | 0 | 12 | 21 | 15 | 6 | 85 | 13.3 |
| NW_005196755.1_34019-3p | miR90_0658 | 100 | 8 | 0 | 0 | 14 | 21 | 4 | 11 | 98 | 13.1 |
| NW_005196755.1_34019-3p | miR90_0457 | 90 | 10 | 1 | 0 | 6 | 15 | 6 | 15 | 85 | 13.3 |
| NW_005196755.1_34019-3p | miR90_0278 | 90 | 10 | 1 | 0 | 7 | 16 | 6 | 15 | 85 | 13.3 |
| NW_005196755.1_34019-3p | miR95_0325 | 90 | 10 | 1 | 0 | 8 | 17 | 17 | 8 | 85 | 13.3 |
| NW_005196755.1_34019-3p | miR95_0269 | 90 | 10 | 1 | 0 | 8 | 17 | 18 | 9 | 85 | 13.3 |
| NW_005196755.1_34019-3p | miR90_0424 | 100 | 8 | 0 | 0 | 14 | 21 | 1 | 8 | 98 | 13.1 |
| NW_005196755.1_34019-3p | miR95_0444 | 100 | 8 | 0 | 0 | 6 | 13 | 11 | 4 | 98 | 13.1 |
| NW_005196755.1_34019-3p | Botryococcus | 70.588 | 17 | 5 | 0 | 1 | 17 | 17 | 1 | 98 | 13.1 |
| NW_005197715.1_30571-5p | miR90_0709 | 91.667 | 12 | 1 | 0 | 8 | 19 | 18 | 7 | 18 | 15.4 |
| NW_005197715.1_30571-5p | miR90_0495 | 100 | 10 | 0 | 0 | 1 | 10 | 18 | 9 | 21 | 15.2 |
| NW_005197715.1_30571-5p | Vca_miR60 | 73.684 | 19 | 5 | 0 | 1 | 19 | 3 | 21 | 21 | 15.2 |
| NW_005197715.1_30571-5p | miR90_0876 | 80 | 15 | 3 | 0 | 4 | 18 | 1 | 15 | 28 | 14.8 |
| NW_005197715.1_30571-5p | Vca_miR82 | 80 | 15 | 3 | 0 | 4 | 18 | 2 | 16 | 28 | 14.8 |
| NW_005197715.1_30571-5p | miR90_0690 | 84.615 | 13 | 2 | 0 | 7 | 19 | 2 | 14 | 32 | 14.6 |
| NW_005197715.1_30571-5p | miR90_0653 | 84.615 | 13 | 2 | 0 | 7 | 19 | 1 | 13 | 32 | 14.6 |
| NW_005197715.1_30571-5p | miR90_0211 | 84.615 | 13 | 2 | 0 | 5 | 17 | 1 | 13 | 32 | 14.6 |
| NW_005197715.1_30571-5p | Vca_miR77 | 84.615 | 13 | 2 | 0 | 7 | 19 | 15 | 3 | 32 | 14.6 |
| NW_005197715.1_30571-5p | miR90_0487 | 90.909 | 11 | 1 | 0 | 9 | 19 | 19 | 9 | 37 | 14.4 |
| NW_005197715.1_30571-5p | ppu-miR9-3p | 100 | 9 | 0 | 0 | 2 | 10 | 3 | 11 | 43 | 14.2 |
| NW_005197715.1_30571-5p | miR90_0862 | 100 | 9 | 0 | 0 | 10 | 18 | 15 | 7 | 43 | 14.2 |
| NW_005197715.1_30571-5p | miR90_0840 | 100 | 9 | 0 | 0 | 10 | 18 | 16 | 8 | 43 | 14.2 |
| NW_005197715.1_30571-5p | miR90_0743 | 100 | 9 | 0 | 0 | 10 | 18 | 6 | 14 | 43 | 14.2 |
| NW_005197715.1_30571-5p | Porphyra | 72.222 | 18 | 5 | 0 | 1 | 18 | 3 | 20 | 43 | 14.2 |
| NW_005197715.1_30571-5p | miR90_0400 | 75 | 16 | 4 | 0 | 1 | 16 | 16 | 1 | 50 | 13.9 |
| NW_005197715.1_30571-5p | miR90_0016 | 75 | 16 | 4 | 0 | 1 | 16 | 17 | 2 | 50 | 13.9 |
| NW_005197715.1_30571-5p | miR95_0258 | 77.778 | 18 | 3 | 1 | 1 | 18 | 19 | 3 | 50 | 13.9 |
| NW_005197715.1_30571-5p | Vca_miR13* | 75 | 16 | 4 | 0 | 4 | 19 | 6 | 21 | 50 | 13.9 |
| NW_005197715.1_30571-5p | miR90_0759 | 78.571 | 14 | 3 | 0 | 4 | 17 | 3 | 16 | 57 | 13.7 |
| NW_005197715.1_30571-5p | SjapMIR60 | 78.571 | 14 | 3 | 0 | 6 | 19 | 2 | 15 | 57 | 13.7 |
| NW_005197715.1_30571-5p | SjapMIR70 | 83.333 | 12 | 2 | 0 | 3 | 14 | 2 | 13 | 66 | 13.5 |
| NW_005197715.1_30571-5p | miR90_0809 | 90 | 10 | 1 | 0 | 7 | 16 | 12 | 3 | 77 | 13.3 |
| NW_005197715.1_30571-5p | miR90_0769 | 90 | 10 | 1 | 0 | 7 | 16 | 11 | 2 | 77 | 13.3 |
| NW_005197715.1_30571-5p | miR90_0754 | 90 | 10 | 1 | 0 | 7 | 16 | 9 | 18 | 77 | 13.3 |
| NW_005197715.1_30571-5p | miR95_0143 | 90 | 10 | 1 | 0 | 1 | 10 | 8 | 17 | 77 | 13.3 |
| NW_005197715.1_30571-5p | SjapMIR102 | 90 | 10 | 1 | 0 | 1 | 10 | 18 | 9 | 77 | 13.3 |
| NW_005197715.1_30571-5p | ppu-miR4-5p | 100 | 8 | 0 | 0 | 12 | 19 | 11 | 18 | 89 | 13.1 |
| NW_005197715.1_30571-5p | miR90_0866 | 100 | 8 | 0 | 0 | 11 | 18 | 25 | 18 | 89 | 13.1 |
| NW_005197715.1_30571-5p | miR90_0706 | 100 | 8 | 0 | 0 | 12 | 19 | 18 | 11 | 89 | 13.1 |
| NW_005197715.1_30571-5p | miR90_0610 | 70.588 | 17 | 5 | 0 | 3 | 19 | 1 | 17 | 89 | 13.1 |
| NW_005197715.1_30571-5p | miR90_0560 | 100 | 8 | 0 | 0 | 1 | 8 | 13 | 6 | 89 | 13.1 |
| NW_005197715.1_30571-5p | miR90_0314 | 100 | 8 | 0 | 0 | 1 | 8 | 14 | 7 | 89 | 13.1 |
| NW_005197715.1_30571-5p | miR90_0217 | 100 | 8 | 0 | 0 | 1 | 8 | 17 | 24 | 89 | 13.1 |
| NW_005197715.1_30571-5p | miR95_0499 | 70.588 | 17 | 5 | 0 | 1 | 17 | 2 | 18 | 89 | 13.1 |
| NW_005197715.1_30571-5p | miR95_0488 | 70.588 | 17 | 5 | 0 | 1 | 17 | 1 | 17 | 89 | 13.1 |
| NW_005197715.1_30571-5p | miR95_0401 | 70.588 | 17 | 5 | 0 | 1 | 17 | 18 | 2 | 89 | 13.1 |
| NW_005198391.1_27186-5p | miR90_0599 | 73.684 | 19 | 5 | 0 | 2 | 20 | 25 | 7 | 23 | 15.2 |
| NW_005198391.1_27186-5p | miR90_0404 | 73.684 | 19 | 5 | 0 | 2 | 20 | 3 | 21 | 23 | 15.2 |
| NW_005198391.1_27186-5p | miR95_0479 | 100 | 10 | 0 | 0 | 12 | 21 | 19 | 10 | 23 | 15.2 |
| NW_005198391.1_27186-5p | miR95_0433 | 100 | 10 | 0 | 0 | 12 | 21 | 18 | 9 | 23 | 15.2 |
| NW_005198391.1_27186-5p | miR95_0337 | 73.684 | 19 | 5 | 0 | 2 | 20 | 22 | 4 | 23 | 15.2 |
| NW_005198391.1_27186-5p | miR95_0199 | 73.684 | 19 | 5 | 0 | 2 | 20 | 6 | 24 | 23 | 15.2 |
| NW_005198391.1_27186-5p | miR95_0197 | 73.684 | 19 | 5 | 0 | 2 | 20 | 7 | 25 | 23 | 15.2 |
| NW_005198391.1_27186-5p | miR95_0082 | 73.684 | 19 | 5 | 0 | 2 | 20 | 19 | 1 | 23 | 15.2 |
| NW_005198391.1_27186-5p | miR95_0023 | 73.684 | 19 | 5 | 0 | 2 | 20 | 25 | 7 | 23 | 15.2 |
| NW_005198391.1_27186-5p | miR95_0019 | 73.684 | 19 | 5 | 0 | 2 | 20 | 19 | 1 | 23 | 15.2 |
| NW_005198391.1_27186-5p | SjapMIR71 | 76.471 | 17 | 4 | 0 | 4 | 20 | 20 | 4 | 26 | 15 |
| NW_005198391.1_27186-5p | Vca_miR112 | 86.667 | 15 | 1 | 1 | 5 | 18 | 16 | 2 | 35 | 14.6 |
| NW_005198391.1_27186-5p | miR90_0778 | 90.909 | 11 | 1 | 0 | 3 | 13 | 2 | 12 | 41 | 14.4 |
| NW_005198391.1_27186-5p | Vca_miR1d | 90.909 | 11 | 1 | 0 | 3 | 13 | 3 | 13 | 41 | 14.4 |
| NW_005198391.1_27186-5p | Vca_miR1c | 90.909 | 11 | 1 | 0 | 3 | 13 | 3 | 13 | 41 | 14.4 |
| NW_005198391.1_27186-5p | Vca_miR1b | 90.909 | 11 | 1 | 0 | 3 | 13 | 3 | 13 | 41 | 14.4 |
| NW_005198391.1_27186-5p | Vca_miR1a | 90.909 | 11 | 1 | 0 | 3 | 13 | 3 | 13 | 41 | 14.4 |
| NW_005198391.1_27186-5p | miR95_0563 | 100 | 9 | 0 | 0 | 12 | 20 | 3 | 11 | 47 | 14.2 |
| NW_005198391.1_27186-5p | miR95_0522 | 100 | 9 | 0 | 0 | 12 | 20 | 1 | 9 | 47 | 14.2 |
| NW_005198391.1_27186-5p | miR95_0481 | 72.222 | 18 | 5 | 0 | 4 | 21 | 1 | 18 | 47 | 14.2 |
| NW_005198391.1_27186-5p | miR95_0335 | 72.222 | 18 | 5 | 0 | 3 | 20 | 1 | 18 | 47 | 14.2 |
| NW_005198391.1_27186-5p | miR90_0616 | 75 | 16 | 4 | 0 | 1 | 16 | 2 | 17 | 55 | 13.9 |
| NW_005198391.1_27186-5p | miR95_0537 | 75 | 16 | 4 | 0 | 2 | 17 | 16 | 1 | 55 | 13.9 |
| NW_005198391.1_27186-5p | miR90_0800 | 78.571 | 14 | 3 | 0 | 2 | 15 | 4 | 17 | 63 | 13.7 |
| NW_005198391.1_27186-5p | miR90_0753 | 81.25 | 16 | 2 | 1 | 4 | 19 | 18 | 4 | 63 | 13.7 |
| NW_005198391.1_27186-5p | miR90_0285 | 78.571 | 14 | 3 | 0 | 2 | 15 | 2 | 15 | 63 | 13.7 |
| NW_005198391.1_27186-5p | miR95_0486 | 78.571 | 14 | 3 | 0 | 8 | 21 | 1 | 14 | 63 | 13.7 |
| NW_005198391.1_27186-5p | miR95_0263 | 78.571 | 14 | 3 | 0 | 1 | 14 | 3 | 16 | 63 | 13.7 |
| NW_005198391.1_27186-5p | miR95_0116 | 78.571 | 14 | 3 | 0 | 1 | 14 | 21 | 8 | 63 | 13.7 |
| NW_005198391.1_27186-5p | miR95_0059 | 78.571 | 14 | 3 | 0 | 1 | 14 | 20 | 7 | 63 | 13.7 |
| NW_005198391.1_27186-5p | Porphyra | 78.571 | 14 | 3 | 0 | 8 | 21 | 1 | 14 | 63 | 13.7 |
| NW_005198391.1_27186-5p | ppu-miR8-5p | 83.333 | 12 | 2 | 0 | 10 | 21 | 9 | 20 | 73 | 13.5 |
| NW_005198391.1_27186-5p | miR90_0913 | 83.333 | 12 | 2 | 0 | 6 | 17 | 2 | 13 | 73 | 13.5 |
| NW_005198391.1_27186-5p | miR90_0730 | 83.333 | 12 | 2 | 0 | 7 | 18 | 14 | 3 | 73 | 13.5 |
| NW_005198391.1_27186-5p | miR90_0680 | 83.333 | 12 | 2 | 0 | 2 | 13 | 7 | 18 | 73 | 13.5 |
| NW_005198391.1_27186-5p | miR90_0400 | 85.714 | 14 | 1 | 1 | 7 | 20 | 18 | 6 | 73 | 13.5 |
| NW_005198391.1_27186-5p | miR90_0258 | 83.333 | 12 | 2 | 0 | 6 | 17 | 16 | 5 | 73 | 13.5 |
| NW_005198391.1_27186-5p | miR95_0247 | 83.333 | 12 | 2 | 0 | 3 | 14 | 1 | 12 | 73 | 13.5 |
| NW_005198391.1_27186-5p | CCMP1516-miR5 | 85.714 | 14 | 1 | 1 | 7 | 19 | 1 | 14 | 73 | 13.5 |
| NW_005198391.1_27186-5p | miR90_0615 | 90 | 10 | 1 | 0 | 7 | 16 | 1 | 10 | 85 | 13.3 |
| NW_005198391.1_27186-5p | miR90_0548 | 91.667 | 12 | 0 | 1 | 5 | 16 | 11 | 1 | 85 | 13.3 |
| NW_005198391.1_27186-5p | Vca_miR98b* | 90 | 10 | 1 | 0 | 10 | 19 | 14 | 5 | 85 | 13.3 |
| NW_005198391.1_27186-5p | Vca_miR98a* | 90 | 10 | 1 | 0 | 10 | 19 | 14 | 5 | 85 | 13.3 |
| NW_005198391.1_27186-5p | SjapMIR39 | 90 | 10 | 1 | 0 | 1 | 10 | 16 | 7 | 85 | 13.3 |
| NW_005198391.1_27186-5p | miR90_0708 | 100 | 8 | 0 | 0 | 10 | 17 | 10 | 17 | 98 | 13.1 |
| NW_005198391.1_27186-5p | miR90_0016 | 100 | 8 | 0 | 0 | 13 | 20 | 14 | 7 | 98 | 13.1 |
| NW_005198391.1_27186-5p | miR95_0485 | 100 | 8 | 0 | 0 | 14 | 21 | 1 | 8 | 98 | 13.1 |
| NW_005198391.1_27186-5p | PC-5p-179621_3 | 70.588 | 17 | 5 | 0 | 2 | 18 | 4 | 20 | 98 | 13.1 |
| NW_005198391.1_27186-5p | SjapMIR88 | 100 | 8 | 0 | 0 | 8 | 15 | 5 | 12 | 98 | 13.1 |
| NW_005198966.1_24265-3p | miR95_0490 | 85.714 | 14 | 2 | 0 | 3 | 16 | 3 | 16 | 17 | 15.6 |
| NW_005198966.1_24265-3p | miR90_0306 | 76.471 | 17 | 4 | 0 | 3 | 19 | 25 | 9 | 26 | 15 |
| NW_005198966.1_24265-3p | miR90_0223 | 84.615 | 13 | 2 | 0 | 7 | 19 | 17 | 5 | 35 | 14.6 |
| NW_005198966.1_24265-3p | miR95_0063 | 84.615 | 13 | 2 | 0 | 4 | 16 | 7 | 19 | 35 | 14.6 |
| NW_005198966.1_24265-3p | miR90_0916 | 90.909 | 11 | 1 | 0 | 4 | 14 | 12 | 2 | 41 | 14.4 |
| NW_005198966.1_24265-3p | miR90_0188 | 70 | 20 | 6 | 0 | 2 | 21 | 22 | 3 | 41 | 14.4 |
| NW_005198966.1_24265-3p | miR90_0640 | 100 | 9 | 0 | 0 | 9 | 17 | 24 | 16 | 47 | 14.2 |
| NW_005198966.1_24265-3p | miR90_0298 | 100 | 9 | 0 | 0 | 9 | 17 | 22 | 14 | 47 | 14.2 |
| NW_005198966.1_24265-3p | miR95_0303 | 72.222 | 18 | 5 | 0 | 1 | 18 | 18 | 1 | 47 | 14.2 |
| NW_005198966.1_24265-3p | miR95_0172 | 72.222 | 18 | 5 | 0 | 1 | 18 | 18 | 1 | 47 | 14.2 |
| NW_005198966.1_24265-3p | miR95_0117 | 75 | 16 | 4 | 0 | 4 | 19 | 2 | 17 | 55 | 13.9 |
| NW_005198966.1_24265-3p | Porphyra | 75 | 16 | 4 | 0 | 5 | 20 | 20 | 5 | 55 | 13.9 |
| NW_005198966.1_24265-3p | miR90_0017 | 78.571 | 14 | 3 | 0 | 5 | 18 | 14 | 1 | 63 | 13.7 |
| NW_005198966.1_24265-3p | Vca_miR77 | 78.571 | 14 | 3 | 0 | 5 | 18 | 4 | 17 | 63 | 13.7 |
| NW_005198966.1_24265-3p | miR95_0292 | 83.333 | 12 | 2 | 0 | 7 | 18 | 12 | 1 | 73 | 13.5 |
| NW_005198966.1_24265-3p | CCMP1516-miR9 | 83.333 | 12 | 2 | 0 | 3 | 14 | 5 | 16 | 73 | 13.5 |
| NW_005198966.1_24265-3p | miR90_0477 | 90 | 10 | 1 | 0 | 9 | 18 | 17 | 8 | 85 | 13.3 |
| NW_005198966.1_24265-3p | miR90_0439 | 68.421 | 19 | 6 | 0 | 3 | 21 | 24 | 6 | 85 | 13.3 |
| NW_005198966.1_24265-3p | miR90_0186 | 71.429 | 21 | 5 | 1 | 1 | 21 | 3 | 22 | 85 | 13.3 |
| NW_005198966.1_24265-3p | miR90_0182 | 90 | 10 | 1 | 0 | 11 | 20 | 2 | 11 | 85 | 13.3 |
| NW_005198966.1_24265-3p | miR95_0165 | 68.421 | 19 | 6 | 0 | 3 | 21 | 19 | 1 | 85 | 13.3 |
| NW_005198966.1_24265-3p | Vca_miR64 | 90 | 10 | 1 | 0 | 12 | 21 | 12 | 21 | 85 | 13.3 |
| NW_005198966.1_24265-3p | SjapMIR10 | 90 | 10 | 1 | 0 | 12 | 21 | 9 | 18 | 85 | 13.3 |
| NW_005198966.1_24265-3p | miR90_0013 | 70.588 | 17 | 5 | 0 | 4 | 20 | 2 | 18 | 98 | 13.1 |
| NW_005198966.1_24265-3p | miR95_0359 | 70.588 | 17 | 5 | 0 | 4 | 20 | 2 | 18 | 98 | 13.1 |
| NW_005198966.1_24265-3p | miR95_0326 | 70.588 | 17 | 5 | 0 | 4 | 20 | 2 | 18 | 98 | 13.1 |
| NW_005198966.1_24265-3p | SjapMIR16 | 70.588 | 17 | 5 | 0 | 3 | 19 | 17 | 1 | 98 | 13.1 |
| NW_005198966.1_24265-5p | SjapMIR63_2 | 84.615 | 13 | 2 | 0 | 2 | 14 | 4 | 16 | 30 | 14.6 |
| NW_005198966.1_24265-5p | SjapMIR63_1 | 84.615 | 13 | 2 | 0 | 2 | 14 | 4 | 16 | 30 | 14.6 |
| NW_005198966.1_24265-5p | miR95_0357 | 90.909 | 11 | 1 | 0 | 1 | 11 | 14 | 4 | 35 | 14.4 |
| NW_005198966.1_24265-5p | miR95_0012 | 90.909 | 11 | 1 | 0 | 8 | 18 | 25 | 15 | 35 | 14.4 |
| NW_005198966.1_24265-5p | Vca_miR53b | 90.909 | 11 | 1 | 0 | 2 | 12 | 1 | 11 | 35 | 14.4 |
| NW_005198966.1_24265-5p | Vca_miR53a | 90.909 | 11 | 1 | 0 | 2 | 12 | 1 | 11 | 35 | 14.4 |
| NW_005198966.1_24265-5p | miR95_0462 | 75 | 16 | 4 | 0 | 1 | 16 | 19 | 4 | 47 | 13.9 |
| NW_005198966.1_24265-5p | miR90_0669 | 78.571 | 14 | 3 | 0 | 2 | 15 | 14 | 1 | 54 | 13.7 |
| NW_005198966.1_24265-5p | miR90_0445 | 78.571 | 14 | 3 | 0 | 3 | 16 | 10 | 23 | 54 | 13.7 |
| NW_005198966.1_24265-5p | miR90_0226 | 78.571 | 14 | 3 | 0 | 3 | 16 | 14 | 1 | 54 | 13.7 |
| NW_005198966.1_24265-5p | miR95_0136 | 78.571 | 14 | 3 | 0 | 3 | 16 | 15 | 2 | 54 | 13.7 |
| NW_005198966.1_24265-5p | Vca_miR94b | 78.571 | 14 | 3 | 0 | 4 | 17 | 2 | 15 | 54 | 13.7 |
| NW_005198966.1_24265-5p | Vca_miR94a | 78.571 | 14 | 3 | 0 | 4 | 17 | 2 | 15 | 54 | 13.7 |
| NW_005198966.1_24265-5p | miR90_0577 | 83.333 | 12 | 2 | 0 | 1 | 12 | 19 | 8 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR90_0360 | 83.333 | 12 | 2 | 0 | 1 | 12 | 7 | 18 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR90_0160 | 83.333 | 12 | 2 | 0 | 1 | 12 | 6 | 17 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR90_0058 | 83.333 | 12 | 2 | 0 | 1 | 12 | 19 | 8 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR90_0057 | 83.333 | 12 | 2 | 0 | 1 | 12 | 13 | 2 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR90_0056 | 83.333 | 12 | 2 | 0 | 1 | 12 | 17 | 6 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0460 | 83.333 | 12 | 2 | 0 | 6 | 17 | 23 | 12 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0424 | 83.333 | 12 | 2 | 0 | 1 | 12 | 1 | 12 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0423 | 83.333 | 12 | 2 | 0 | 1 | 12 | 3 | 14 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0362 | 83.333 | 12 | 2 | 0 | 1 | 12 | 6 | 17 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0331 | 83.333 | 12 | 2 | 0 | 1 | 12 | 25 | 14 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0318 | 83.333 | 12 | 2 | 0 | 1 | 12 | 15 | 4 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0208 | 83.333 | 12 | 2 | 0 | 1 | 12 | 4 | 15 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0207 | 83.333 | 12 | 2 | 0 | 1 | 12 | 2 | 13 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0206 | 83.333 | 12 | 2 | 0 | 1 | 12 | 11 | 22 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0204 | 83.333 | 12 | 2 | 0 | 1 | 12 | 12 | 23 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0201 | 83.333 | 12 | 2 | 0 | 1 | 12 | 9 | 20 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0200 | 83.333 | 12 | 2 | 0 | 1 | 12 | 13 | 24 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0078 | 83.333 | 12 | 2 | 0 | 1 | 12 | 2 | 13 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0077 | 83.333 | 12 | 2 | 0 | 1 | 12 | 6 | 17 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0071 | 83.333 | 12 | 2 | 0 | 1 | 12 | 16 | 5 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0024 | 83.333 | 12 | 2 | 0 | 1 | 12 | 18 | 7 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0022 | 83.333 | 12 | 2 | 0 | 1 | 12 | 12 | 1 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0018 | 83.333 | 12 | 2 | 0 | 1 | 12 | 12 | 1 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR95_0017 | 83.333 | 12 | 2 | 0 | 1 | 12 | 19 | 8 | 63 | 13.5 |
| NW_005198966.1_24265-5p | Vca_miR84 | 83.333 | 12 | 2 | 0 | 2 | 13 | 12 | 1 | 63 | 13.5 |
| NW_005198966.1_24265-5p | miR90_0142 | 90 | 10 | 1 | 0 | 9 | 18 | 20 | 11 | 73 | 13.3 |
| NW_005198966.1_24265-5p | SjapMIR64 | 90 | 10 | 1 | 0 | 2 | 11 | 4 | 13 | 73 | 13.3 |
| NW_005198966.1_24265-5p | miR95_0383 | 100 | 8 | 0 | 0 | 4 | 11 | 2 | 9 | 84 | 13.1 |
| NW_005198966.1_24265-5p | Vca_miR88b | 73.333 | 15 | 4 | 0 | 2 | 16 | 9 | 23 | 97 | 12.9 |
| NW_005198966.1_24265-5p | Vca_miR88a | 73.333 | 15 | 4 | 0 | 2 | 16 | 9 | 23 | 97 | 12.9 |
| NW_005198966.1_24265-5p | Vca_miR68* | 73.333 | 15 | 4 | 0 | 2 | 16 | 4 | 18 | 97 | 12.9 |
| NW_005199363.1_22042-5p | miR90_0108 | 83.333 | 18 | 2 | 1 | 4 | 20 | 2 | 19 | 14 | 15.8 |
| NW_005199363.1_22042-5p | miR90_0013 | 73.684 | 19 | 5 | 0 | 2 | 20 | 21 | 3 | 22 | 15.2 |
| NW_005199363.1_22042-5p | miR90_0172 | 82.353 | 17 | 2 | 1 | 4 | 19 | 2 | 18 | 29 | 14.8 |
| NW_005199363.1_22042-5p | miR90_0165 | 82.353 | 17 | 2 | 1 | 4 | 19 | 2 | 18 | 29 | 14.8 |
| NW_005199363.1_22042-5p | miR95_0448 | 80 | 15 | 3 | 0 | 3 | 17 | 2 | 16 | 29 | 14.8 |
| NW_005199363.1_22042-5p | miR95_0248 | 82.353 | 17 | 2 | 1 | 5 | 20 | 18 | 2 | 29 | 14.8 |
| NW_005199363.1_22042-5p | miR95_0158 | 82.353 | 17 | 2 | 1 | 4 | 19 | 17 | 1 | 29 | 14.8 |
| NW_005199363.1_22042-5p | ppu-miR3-5p | 84.615 | 13 | 2 | 0 | 3 | 15 | 1 | 13 | 34 | 14.6 |
| NW_005199363.1_22042-5p | miR90_0457 | 90.909 | 11 | 1 | 0 | 6 | 16 | 17 | 7 | 39 | 14.4 |
| NW_005199363.1_22042-5p | Vca_miR38 | 90.909 | 11 | 1 | 0 | 2 | 12 | 2 | 12 | 39 | 14.4 |
| NW_005199363.1_22042-5p | miR90_0428 | 100 | 9 | 0 | 0 | 12 | 20 | 24 | 16 | 45 | 14.2 |
| NW_005199363.1_22042-5p | miR95_0522 | 100 | 9 | 0 | 0 | 3 | 11 | 10 | 18 | 45 | 14.2 |
| NW_005199363.1_22042-5p | miR95_0289 | 72.222 | 18 | 5 | 0 | 2 | 19 | 1 | 18 | 45 | 14.2 |
| NW_005199363.1_22042-5p | miR95_0288 | 72.222 | 18 | 5 | 0 | 2 | 19 | 1 | 18 | 45 | 14.2 |
| NW_005199363.1_22042-5p | miR95_0541 | 78.571 | 14 | 3 | 0 | 5 | 18 | 15 | 2 | 60 | 13.7 |
| NW_005199363.1_22042-5p | miR95_0515 | 78.571 | 14 | 3 | 0 | 1 | 14 | 1 | 14 | 60 | 13.7 |
| NW_005199363.1_22042-5p | miR95_0419 | 78.571 | 14 | 3 | 0 | 2 | 15 | 16 | 3 | 60 | 13.7 |
| NW_005199363.1_22042-5p | miR95_0415 | 78.571 | 14 | 3 | 0 | 1 | 14 | 17 | 4 | 60 | 13.7 |
| NW_005199363.1_22042-5p | miR95_0325 | 78.571 | 14 | 3 | 0 | 1 | 14 | 3 | 16 | 60 | 13.7 |
| NW_005199363.1_22042-5p | miR95_0269 | 78.571 | 14 | 3 | 0 | 1 | 14 | 4 | 17 | 60 | 13.7 |
| NW_005199363.1_22042-5p | miR95_0246 | 78.571 | 14 | 3 | 0 | 1 | 14 | 14 | 1 | 60 | 13.7 |
| NW_005199363.1_22042-5p | miR95_0118 | 78.571 | 14 | 3 | 0 | 7 | 20 | 4 | 17 | 60 | 13.7 |
| NW_005199363.1_22042-5p | miR90_0493 | 83.333 | 12 | 2 | 0 | 8 | 19 | 3 | 14 | 70 | 13.5 |
| NW_005199363.1_22042-5p | miR95_0468 | 83.333 | 12 | 2 | 0 | 8 | 19 | 2 | 13 | 70 | 13.5 |
| NW_005199363.1_22042-5p | miR95_0179 | 83.333 | 12 | 2 | 0 | 1 | 12 | 6 | 17 | 70 | 13.5 |
| NW_005199363.1_22042-5p | miR95_0179 | 68.421 | 19 | 6 | 0 | 2 | 20 | 24 | 6 | 81 | 13.3 |
| NW_005199363.1_22042-5p | miR95_0090 | 83.333 | 12 | 2 | 0 | 8 | 19 | 3 | 14 | 70 | 13.5 |
| NW_005199363.1_22042-5p | miR95_0088 | 83.333 | 12 | 2 | 0 | 8 | 19 | 3 | 14 | 70 | 13.5 |
| NW_005199363.1_22042-5p | SjapMIR56 | 83.333 | 12 | 2 | 0 | 1 | 12 | 13 | 2 | 70 | 13.5 |
| NW_005199363.1_22042-5p | miR90_0169 | 90 | 10 | 1 | 0 | 6 | 15 | 9 | 18 | 81 | 13.3 |
| NW_005199363.1_22042-5p | miR95_0188 | 90 | 10 | 1 | 0 | 7 | 16 | 1 | 10 | 81 | 13.3 |
| NW_005199363.1_22042-5p | SjapMIR61 | 90 | 10 | 1 | 0 | 9 | 18 | 12 | 3 | 81 | 13.3 |
| NW_005199363.1_22042-5p | miR90_0876 | 70.588 | 17 | 5 | 0 | 2 | 18 | 1 | 17 | 94 | 13.1 |
| NW_005199363.1_22042-5p | miR90_0726 | 100 | 8 | 0 | 0 | 1 | 8 | 4 | 11 | 94 | 13.1 |
| NW_005199363.1_22042-5p | miR90_0629 | 100 | 8 | 0 | 0 | 1 | 8 | 8 | 15 | 94 | 13.1 |
| NW_005199363.1_22042-5p | miR95_0563 | 100 | 8 | 0 | 0 | 3 | 10 | 12 | 19 | 94 | 13.1 |
| NW_005199363.1_22042-5p | miR95_0388 | 100 | 8 | 0 | 0 | 1 | 8 | 7 | 14 | 94 | 13.1 |
| NW_005199363.1_22042-5p | miR95_0387 | 100 | 8 | 0 | 0 | 1 | 8 | 14 | 21 | 94 | 13.1 |
| NW_005199363.1_22042-5p | miR95_0354 | 100 | 8 | 0 | 0 | 1 | 8 | 3 | 10 | 94 | 13.1 |
| NW_005199363.1_22042-5p | miR95_0133 | 100 | 8 | 0 | 0 | 1 | 8 | 10 | 17 | 94 | 13.1 |
| NW_005199363.1_22042-5p | miR95_0111 | 100 | 8 | 0 | 0 | 1 | 8 | 6 | 13 | 94 | 13.1 |
| NW_005199363.1_22042-5p | miR95_0087 | 100 | 8 | 0 | 0 | 1 | 8 | 15 | 22 | 94 | 13.1 |
| NW_005199363.1_22042-5p | miR95_0083 | 100 | 8 | 0 | 0 | 1 | 8 | 8 | 15 | 94 | 13.1 |
| NW_005199363.1_22042-5p | miR95_0080 | 100 | 8 | 0 | 0 | 1 | 8 | 9 | 16 | 94 | 13.1 |
| NW_005199378.1_21978-3p | ppu-miR5-3p | 73.684 | 19 | 5 | 0 | 3 | 21 | 20 | 2 | 23 | 15.2 |
| NW_005199378.1_21978-3p | Vca_miR22 | 84.615 | 13 | 2 | 0 | 1 | 13 | 13 | 1 | 35 | 14.6 |
| NW_005199378.1_21978-3p | SjapMIR95 | 90.909 | 11 | 1 | 0 | 3 | 13 | 16 | 6 | 41 | 14.4 |
| NW_005199378.1_21978-3p | Vca_miR88b* | 100 | 9 | 0 | 0 | 3 | 11 | 17 | 9 | 47 | 14.2 |
| NW_005199378.1_21978-3p | Vca_miR88a* | 100 | 9 | 0 | 0 | 3 | 11 | 17 | 9 | 47 | 14.2 |
| NW_005199378.1_21978-3p | miR90_0844 | 75 | 16 | 4 | 0 | 1 | 16 | 9 | 24 | 55 | 13.9 |
| NW_005199378.1_21978-3p | miR95_0514 | 75 | 16 | 4 | 0 | 1 | 16 | 9 | 24 | 55 | 13.9 |
| NW_005199378.1_21978-3p | miR95_0390 | 75 | 16 | 4 | 0 | 1 | 16 | 4 | 19 | 55 | 13.9 |
| NW_005199378.1_21978-3p | miR95_0346 | 75 | 16 | 4 | 0 | 1 | 16 | 3 | 18 | 55 | 13.9 |
| NW_005199378.1_21978-3p | miR95_0324 | 75 | 16 | 4 | 0 | 1 | 16 | 5 | 20 | 55 | 13.9 |
| NW_005199378.1_21978-3p | miR90_0701 | 78.571 | 14 | 3 | 0 | 1 | 14 | 1 | 14 | 63 | 13.7 |
| NW_005199378.1_21978-3p | miR95_0369 | 78.571 | 14 | 3 | 0 | 6 | 19 | 19 | 6 | 63 | 13.7 |
| NW_005199378.1_21978-3p | PC-3p-33398_31 | 83.333 | 12 | 2 | 0 | 2 | 13 | 12 | 1 | 73 | 13.5 |
| NW_005199378.1_21978-3p | miR90_0567 | 90 | 10 | 1 | 0 | 6 | 15 | 16 | 7 | 85 | 13.3 |
| NW_005199378.1_21978-3p | miR90_0513 | 90 | 10 | 1 | 0 | 6 | 15 | 11 | 2 | 85 | 13.3 |
| NW_005199378.1_21978-3p | miR90_0012 | 90 | 10 | 1 | 0 | 6 | 15 | 10 | 1 | 85 | 13.3 |
| NW_005199378.1_21978-3p | miR95_0558 | 100 | 8 | 0 | 0 | 1 | 8 | 9 | 16 | 98 | 13.1 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | PC-3p-33398_31 | 92.857 | 14 | 1 | 0 | 6 | 19 | 6 | 19 | 5 | 17.5 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | PC-3p-11444_150 | 92.308 | 13 | 1 | 0 | 7 | 19 | 7 | 19 | 10 | 16.5 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | CCMP1516-miR18 | 80 | 20 | 3 | 1 | 1 | 19 | 2 | 21 | 14 | 16.1 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR90_0571 | 80 | 15 | 3 | 0 | 3 | 17 | 16 | 2 | 33 | 14.8 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | Vca_miR60 | 80 | 15 | 3 | 0 | 1 | 15 | 6 | 20 | 33 | 14.8 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | Vca_miR32* | 80 | 15 | 3 | 0 | 1 | 15 | 7 | 21 | 33 | 14.8 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR90_0599 | 90.909 | 11 | 1 | 0 | 10 | 20 | 11 | 1 | 45 | 14.4 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR90_0378 | 90.909 | 11 | 1 | 0 | 5 | 15 | 17 | 7 | 45 | 14.4 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | ppu-miR15-5p | 75 | 16 | 4 | 0 | 4 | 19 | 17 | 2 | 60 | 13.9 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR90_0701 | 75 | 16 | 4 | 0 | 4 | 19 | 3 | 18 | 60 | 13.9 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR90_0663 | 75 | 16 | 4 | 0 | 5 | 20 | 18 | 3 | 60 | 13.9 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | ppu-miR38-3p | 78.571 | 14 | 3 | 0 | 7 | 20 | 2 | 15 | 69 | 13.7 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR90_0535 | 78.571 | 14 | 3 | 0 | 1 | 14 | 2 | 15 | 69 | 13.7 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR90_0515 | 78.571 | 14 | 3 | 0 | 1 | 14 | 4 | 17 | 69 | 13.7 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR95_0279 | 78.571 | 14 | 3 | 0 | 1 | 14 | 16 | 3 | 69 | 13.7 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR95_0167 | 78.571 | 14 | 3 | 0 | 1 | 14 | 18 | 5 | 69 | 13.7 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR90_0651 | 83.333 | 12 | 2 | 0 | 1 | 12 | 2 | 13 | 80 | 13.5 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | SjapMIR2 | 66.667 | 21 | 7 | 0 | 1 | 21 | 1 | 21 | 80 | 13.5 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR90_0342 | 90 | 10 | 1 | 0 | 6 | 15 | 19 | 10 | 93 | 13.3 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR95_0507 | 90 | 10 | 1 | 0 | 3 | 12 | 15 | 24 | 93 | 13.3 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR95_0451 | 90 | 10 | 1 | 0 | 3 | 12 | 8 | 17 | 93 | 13.3 |
| NW_005199378.1_21978-5p(ehx-miR2-5p) | miR95_0210 | 90 | 10 | 1 | 0 | 3 | 12 | 9 | 18 | 93 | 13.3 |
| NW_005199975.1_17848-3p | miR90_0607 | 88.235 | 17 | 1 | 1 | 5 | 20 | 18 | 2 | 8.2 | 16.7 |
| NW_005199975.1_17848-3p | miR90_0778 | 92.857 | 14 | 0 | 1 | 7 | 19 | 1 | 14 | 20 | 15.4 |
| NW_005199975.1_17848-3p | miR95_0547 | 91.667 | 12 | 1 | 0 | 6 | 17 | 3 | 14 | 20 | 15.4 |
| NW_005199975.1_17848-3p | miR95_0545 | 91.667 | 12 | 1 | 0 | 6 | 17 | 3 | 14 | 20 | 15.4 |
| NW_005199975.1_17848-3p | miR90_0411 | 80 | 15 | 3 | 0 | 1 | 15 | 2 | 16 | 31 | 14.8 |
| NW_005199975.1_17848-3p | miR90_0352 | 86.667 | 15 | 1 | 1 | 3 | 17 | 4 | 17 | 35 | 14.6 |
| NW_005199975.1_17848-3p | miR90_0293 | 86.667 | 15 | 1 | 1 | 6 | 19 | 18 | 4 | 35 | 14.6 |
| NW_005199975.1_17848-3p | miR90_0258 | 84.615 | 13 | 2 | 0 | 6 | 18 | 20 | 8 | 35 | 14.6 |
| NW_005199975.1_17848-3p | miR90_0011 | 86.667 | 15 | 1 | 1 | 6 | 19 | 17 | 3 | 35 | 14.6 |
| NW_005199975.1_17848-3p | miR90_0884 | 92.308 | 13 | 0 | 1 | 7 | 19 | 3 | 14 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0704 | 92.308 | 13 | 0 | 1 | 7 | 19 | 4 | 15 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0583 | 92.308 | 13 | 0 | 1 | 7 | 19 | 10 | 21 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0545 | 92.308 | 13 | 0 | 1 | 7 | 19 | 13 | 24 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0509 | 92.308 | 13 | 0 | 1 | 7 | 19 | 7 | 18 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0431 | 92.308 | 13 | 0 | 1 | 7 | 19 | 7 | 18 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0427 | 92.308 | 13 | 0 | 1 | 7 | 19 | 13 | 2 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0415 | 92.308 | 13 | 0 | 1 | 7 | 19 | 13 | 24 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0292 | 92.308 | 13 | 0 | 1 | 7 | 19 | 9 | 20 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0277 | 92.308 | 13 | 0 | 1 | 7 | 19 | 5 | 16 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0276 | 92.308 | 13 | 0 | 1 | 7 | 19 | 6 | 17 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0139 | 92.308 | 13 | 0 | 1 | 7 | 19 | 16 | 5 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0125 | 90.909 | 11 | 1 | 0 | 1 | 11 | 5 | 15 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0113 | 92.308 | 13 | 0 | 1 | 7 | 19 | 20 | 9 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0081 | 92.308 | 13 | 0 | 1 | 7 | 19 | 5 | 16 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0071 | 92.308 | 13 | 0 | 1 | 7 | 19 | 2 | 13 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0070 | 92.308 | 13 | 0 | 1 | 7 | 19 | 3 | 14 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0069 | 92.308 | 13 | 0 | 1 | 7 | 19 | 3 | 14 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0046 | 92.308 | 13 | 0 | 1 | 7 | 19 | 18 | 7 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0026 | 92.308 | 13 | 0 | 1 | 7 | 19 | 3 | 14 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0022 | 92.308 | 13 | 0 | 1 | 7 | 19 | 17 | 6 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0002 | 92.308 | 13 | 0 | 1 | 7 | 19 | 1 | 12 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0001 | 92.308 | 13 | 0 | 1 | 7 | 19 | 1 | 12 | 41 | 14.4 |
| NW_005199975.1_17848-3p | miR90_0847 | 72.222 | 18 | 5 | 0 | 3 | 20 | 19 | 2 | 47 | 14.2 |
| NW_005199975.1_17848-3p | miR90_0441 | 72.222 | 18 | 5 | 0 | 1 | 18 | 4 | 21 | 47 | 14.2 |
| NW_005199975.1_17848-3p | Vca_miR52 | 100 | 9 | 0 | 0 | 9 | 17 | 18 | 10 | 47 | 14.2 |
| NW_005199975.1_17848-3p | SjapMIR103 | 72.222 | 18 | 5 | 0 | 1 | 18 | 18 | 1 | 47 | 14.2 |
| NW_005199975.1_17848-3p | miR90_0846 | 78.571 | 14 | 3 | 0 | 1 | 14 | 6 | 19 | 63 | 13.7 |
| NW_005199975.1_17848-3p | miR95_0513 | 78.571 | 14 | 3 | 0 | 1 | 14 | 1 | 14 | 63 | 13.7 |
| NW_005199975.1_17848-3p | miR95_0502 | 78.571 | 14 | 3 | 0 | 7 | 20 | 19 | 6 | 63 | 13.7 |
| NW_005199975.1_17848-3p | SjapMIR11 | 78.571 | 14 | 3 | 0 | 4 | 17 | 14 | 1 | 63 | 13.7 |
| NW_005199975.1_17848-3p | miR95_0537 | 83.333 | 12 | 2 | 0 | 3 | 14 | 18 | 7 | 73 | 13.5 |
| NW_005199975.1_17848-3p | miR95_0512 | 83.333 | 12 | 2 | 0 | 3 | 14 | 1 | 12 | 73 | 13.5 |
| NW_005199975.1_17848-3p | miR95_0179 | 83.333 | 12 | 2 | 0 | 10 | 21 | 16 | 5 | 73 | 13.5 |
| NW_005199975.1_17848-3p | miR90_0795 | 90 | 10 | 1 | 0 | 10 | 19 | 2 | 11 | 85 | 13.3 |
| NW_005199975.1_17848-3p | miR90_0698 | 90 | 10 | 1 | 0 | 3 | 12 | 13 | 4 | 85 | 13.3 |
| NW_005199975.1_17848-3p | miR90_0685 | 90 | 10 | 1 | 0 | 3 | 12 | 13 | 4 | 85 | 13.3 |
| NW_005199975.1_17848-3p | miR90_0660 | 90 | 10 | 1 | 0 | 11 | 20 | 17 | 8 | 85 | 13.3 |
| NW_005199975.1_17848-3p | miR90_0268 | 90 | 10 | 1 | 0 | 10 | 19 | 1 | 10 | 85 | 13.3 |
| NW_005199975.1_17848-3p | miR90_0267 | 90 | 10 | 1 | 0 | 10 | 19 | 2 | 11 | 85 | 13.3 |
| NW_005199975.1_17848-3p | miR90_0082 | 90 | 10 | 1 | 0 | 10 | 19 | 1 | 10 | 85 | 13.3 |
| NW_005199975.1_17848-3p | miR95_0543 | 90 | 10 | 1 | 0 | 2 | 11 | 15 | 6 | 85 | 13.3 |
| NW_005199975.1_17848-3p | miR95_0120 | 90 | 10 | 1 | 0 | 9 | 18 | 5 | 14 | 85 | 13.3 |
| NW_005199975.1_17848-3p | miR95_0100 | 90 | 10 | 1 | 0 | 9 | 18 | 8 | 17 | 85 | 13.3 |
| NW_005199975.1_17848-3p | miR90_0316 | 70.588 | 17 | 5 | 0 | 3 | 19 | 2 | 18 | 98 | 13.1 |
| NW_005199975.1_17848-3p | Vca_miR68* | 100 | 8 | 0 | 0 | 1 | 8 | 11 | 4 | 98 | 13.1 |
| NW_005200282.1_15947-3p | Vca_miR32 | 77.778 | 18 | 4 | 0 | 1 | 18 | 19 | 2 | 12 | 16.1 |
| NW_005200282.1_15947-3p | miR95_0534 | 91.667 | 12 | 1 | 0 | 2 | 13 | 4 | 15 | 18 | 15.4 |
| NW_005200282.1_15947-3p | miR95_0532 | 91.667 | 12 | 1 | 0 | 2 | 13 | 3 | 14 | 18 | 15.4 |
| NW_005200282.1_15947-3p | miR95_0538 | 100 | 10 | 0 | 0 | 2 | 11 | 9 | 18 | 21 | 15.2 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续2
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005200282.1_15947-3p | miR95_0479 | 76.471 | 17 | 4 | 0 | 3 | 19 | 18 | 2 | 24 | 15 |
| NW_005200282.1_15947-3p | miR95_0433 | 76.471 | 17 | 4 | 0 | 3 | 19 | 17 | 1 | 24 | 15 |
| NW_005200282.1_15947-3p | miR90_0584 | 80 | 15 | 3 | 0 | 3 | 17 | 15 | 1 | 28 | 14.8 |
| NW_005200282.1_15947-3p | miR95_0485 | 80 | 15 | 3 | 0 | 5 | 19 | 2 | 16 | 28 | 14.8 |
| NW_005200282.1_15947-3p | SjapMIR11 | 90.909 | 11 | 1 | 0 | 9 | 19 | 13 | 3 | 37 | 14.4 |
| NW_005200282.1_15947-3p | Vca_miR1d | 100 | 9 | 0 | 0 | 7 | 15 | 18 | 10 | 43 | 14.2 |
| NW_005200282.1_15947-3p | Vca_miR1c | 100 | 9 | 0 | 0 | 7 | 15 | 18 | 10 | 43 | 14.2 |
| NW_005200282.1_15947-3p | Vca_miR1b | 100 | 9 | 0 | 0 | 7 | 15 | 18 | 10 | 43 | 14.2 |
| NW_005200282.1_15947-3p | Vca_miR1a | 100 | 9 | 0 | 0 | 7 | 15 | 18 | 10 | 43 | 14.2 |
| NW_005200282.1_15947-3p | miR90_0568 | 78.571 | 14 | 3 | 0 | 3 | 16 | 14 | 1 | 57 | 13.7 |
| NW_005200282.1_15947-3p | miR90_0412 | 78.571 | 14 | 3 | 0 | 3 | 16 | 2 | 15 | 57 | 13.7 |
| NW_005200282.1_15947-3p | miR90_0018 | 78.571 | 14 | 3 | 0 | 2 | 15 | 14 | 1 | 57 | 13.7 |
| NW_005200282.1_15947-3p | ppu-miR4-5p | 83.333 | 12 | 2 | 0 | 7 | 18 | 16 | 5 | 66 | 13.5 |
| NW_005200282.1_15947-3p | miR90_0576 | 83.333 | 12 | 2 | 0 | 8 | 19 | 18 | 7 | 66 | 13.5 |
| NW_005200282.1_15947-3p | miR90_0529 | 83.333 | 12 | 2 | 0 | 2 | 13 | 15 | 4 | 66 | 13.5 |
| NW_005200282.1_15947-3p | miR95_0415 | 85.714 | 14 | 1 | 1 | 6 | 19 | 5 | 17 | 66 | 13.5 |
| NW_005200282.1_15947-3p | miR95_0325 | 85.714 | 14 | 1 | 1 | 6 | 19 | 15 | 3 | 66 | 13.5 |
| NW_005200282.1_15947-3p | miR95_0269 | 85.714 | 14 | 1 | 1 | 6 | 19 | 16 | 4 | 66 | 13.5 |
| NW_005200282.1_15947-3p | miR95_0254 | 83.333 | 12 | 2 | 0 | 1 | 12 | 10 | 21 | 66 | 13.5 |
| NW_005200282.1_15947-3p | miR95_0246 | 85.714 | 14 | 1 | 1 | 6 | 19 | 2 | 14 | 66 | 13.5 |
| NW_005200282.1_15947-3p | miR95_0149 | 83.333 | 12 | 2 | 0 | 1 | 12 | 5 | 16 | 66 | 13.5 |
| NW_005200282.1_15947-3p | miR95_0179 | 90 | 10 | 1 | 0 | 4 | 13 | 22 | 13 | 77 | 13.3 |
| NW_005200282.1_15947-3p | Vca_miR88b* | 90 | 10 | 1 | 0 | 9 | 18 | 12 | 3 | 77 | 13.3 |
| NW_005200282.1_15947-3p | Vca_miR88a* | 90 | 10 | 1 | 0 | 9 | 18 | 12 | 3 | 77 | 13.3 |
| NW_005200282.1_15947-3p | Vca_miR45 | 90 | 10 | 1 | 0 | 3 | 12 | 21 | 12 | 77 | 13.3 |
| NW_005200282.1_15947-3p | SjapMIR39 | 90 | 10 | 1 | 0 | 5 | 14 | 12 | 21 | 77 | 13.3 |
| NW_005200282.1_15947-3p | miR90_0445 | 70.588 | 17 | 5 | 0 | 3 | 19 | 18 | 2 | 89 | 13.1 |
| NW_005200282.1_15947-3p | miR90_0402 | 70.588 | 17 | 5 | 0 | 3 | 19 | 25 | 9 | 89 | 13.1 |
| NW_005200282.1_15947-3p | miR90_0239 | 100 | 8 | 0 | 0 | 4 | 11 | 6 | 13 | 89 | 13.1 |
| NW_005200282.1_15947-3p | miR95_0527 | 100 | 8 | 0 | 0 | 2 | 9 | 9 | 16 | 89 | 13.1 |
| NW_005200282.1_15947-3p | miR95_0497 | 100 | 8 | 0 | 0 | 2 | 9 | 10 | 17 | 89 | 13.1 |
| NW_005200282.1_15947-3p | miR95_0484 | 70.588 | 17 | 5 | 0 | 2 | 18 | 18 | 2 | 89 | 13.1 |
| NW_005200282.1_15947-3p | miR95_0251 | 100 | 8 | 0 | 0 | 8 | 15 | 23 | 16 | 89 | 13.1 |
| NW_005200282.1_15947-3p | miR95_0194 | 100 | 8 | 0 | 0 | 6 | 13 | 7 | 14 | 89 | 13.1 |
| NW_005200282.1_15947-3p | miR95_0002 | 100 | 8 | 0 | 0 | 2 | 9 | 11 | 18 | 89 | 13.1 |
| NW_005200461.1_14543-5p | miR90_0642 | 80 | 20 | 4 | 0 | 1 | 20 | 1 | 20 | 3.1 | 18.2 |
| NW_005200461.1_14543-5p | miR90_0642 | 90.909 | 11 | 1 | 0 | 9 | 19 | 3 | 13 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR90_0642 | 90 | 10 | 1 | 0 | 10 | 19 | 1 | 10 | 89 | 13.3 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续3
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005200461.1_14543-5p | miR90_0592 | 80 | 20 | 4 | 0 | 1 | 20 | 1 | 20 | 3.1 | 18.2 |
| NW_005200461.1_14543-5p | miR90_0592 | 90.909 | 11 | 1 | 0 | 9 | 19 | 3 | 13 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR90_0592 | 90 | 10 | 1 | 0 | 10 | 19 | 1 | 10 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0220 | 87.5 | 16 | 2 | 0 | 5 | 20 | 18 | 3 | 4.1 | 17.7 |
| NW_005200461.1_14543-5p | miR90_0657 | 92.857 | 14 | 1 | 0 | 6 | 19 | 1 | 14 | 4.8 | 17.5 |
| NW_005200461.1_14543-5p | miR90_0657 | 77.778 | 18 | 4 | 0 | 3 | 20 | 1 | 18 | 13 | 16.1 |
| NW_005200461.1_14543-5p | miR90_0641 | 92.857 | 14 | 1 | 0 | 6 | 19 | 1 | 14 | 4.8 | 17.5 |
| NW_005200461.1_14543-5p | miR90_0641 | 77.778 | 18 | 4 | 0 | 3 | 20 | 1 | 18 | 13 | 16.1 |
| NW_005200461.1_14543-5p | miR90_0621 | 92.857 | 14 | 1 | 0 | 6 | 19 | 1 | 14 | 4.8 | 17.5 |
| NW_005200461.1_14543-5p | miR90_0621 | 77.778 | 18 | 4 | 0 | 3 | 20 | 1 | 18 | 13 | 16.1 |
| NW_005200461.1_14543-5p | miR90_0727 | 100 | 12 | 0 | 0 | 8 | 19 | 1 | 12 | 5.5 | 17.3 |
| NW_005200461.1_14543-5p | miR90_0727 | 91.667 | 12 | 1 | 0 | 9 | 20 | 5 | 16 | 21 | 15.4 |
| NW_005200461.1_14543-5p | miR90_0580 | 100 | 12 | 0 | 0 | 8 | 19 | 2 | 13 | 5.5 | 17.3 |
| NW_005200461.1_14543-5p | miR90_0580 | 91.667 | 12 | 1 | 0 | 9 | 20 | 6 | 17 | 21 | 15.4 |
| NW_005200461.1_14543-5p | miR90_0580 | 90 | 10 | 1 | 0 | 10 | 19 | 1 | 10 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0434 | 78.947 | 19 | 4 | 0 | 1 | 19 | 19 | 1 | 6.4 | 17.1 |
| NW_005200461.1_14543-5p | miR90_0557 | 82.353 | 17 | 3 | 0 | 6 | 22 | 18 | 2 | 7.4 | 16.9 |
| NW_005200461.1_14543-5p | miR95_0455 | 82.353 | 17 | 3 | 0 | 3 | 19 | 17 | 1 | 7.4 | 16.9 |
| NW_005200461.1_14543-5p | miR90_0466 | 86.667 | 15 | 2 | 0 | 6 | 20 | 4 | 18 | 8.6 | 16.7 |
| NW_005200461.1_14543-5p | miR90_0356 | 86.667 | 15 | 2 | 0 | 6 | 20 | 4 | 18 | 8.6 | 16.7 |
| NW_005200461.1_14543-5p | miR90_0442 | 77.778 | 18 | 4 | 0 | 1 | 18 | 18 | 1 | 13 | 16.1 |
| NW_005200461.1_14543-5p | miR90_0536 | 81.25 | 16 | 3 | 0 | 1 | 16 | 18 | 3 | 15 | 15.8 |
| NW_005200461.1_14543-5p | miR90_0536 | 100 | 9 | 0 | 0 | 12 | 20 | 10 | 2 | 50 | 14.2 |
| NW_005200461.1_14543-5p | miR90_0141 | 81.25 | 16 | 3 | 0 | 1 | 16 | 19 | 4 | 15 | 15.8 |
| NW_005200461.1_14543-5p | miR90_0141 | 100 | 9 | 0 | 0 | 12 | 20 | 11 | 3 | 50 | 14.2 |
| NW_005200461.1_14543-5p | miR95_0004 | 81.25 | 16 | 3 | 0 | 1 | 16 | 19 | 4 | 15 | 15.8 |
| NW_005200461.1_14543-5p | miR95_0004 | 100 | 9 | 0 | 0 | 12 | 20 | 11 | 3 | 50 | 14.2 |
| NW_005200461.1_14543-5p | miR90_0777 | 85.714 | 14 | 2 | 0 | 1 | 14 | 5 | 18 | 18 | 15.6 |
| NW_005200461.1_14543-5p | miR90_0777 | 83.333 | 12 | 2 | 0 | 8 | 19 | 3 | 14 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0271 | 85.714 | 14 | 2 | 0 | 6 | 19 | 18 | 5 | 18 | 15.6 |
| NW_005200461.1_14543-5p | miR90_0271 | 72.222 | 18 | 5 | 0 | 3 | 20 | 18 | 1 | 50 | 14.2 |
| NW_005200461.1_14543-5p | miR90_0271 | 75 | 16 | 4 | 0 | 1 | 16 | 3 | 18 | 57 | 13.9 |
| NW_005200461.1_14543-5p | miR90_0189 | 85.714 | 14 | 2 | 0 | 6 | 19 | 6 | 19 | 18 | 15.6 |
| NW_005200461.1_14543-5p | miR90_0104 | 85.714 | 14 | 2 | 0 | 6 | 19 | 21 | 8 | 18 | 15.6 |
| NW_005200461.1_14543-5p | miR90_0104 | 73.684 | 19 | 5 | 0 | 3 | 21 | 21 | 3 | 24 | 15.2 |
| NW_005200461.1_14543-5p | miR90_0104 | 78.571 | 14 | 3 | 0 | 3 | 16 | 8 | 21 | 66 | 13.7 |
| NW_005200461.1_14543-5p | miR95_0185 | 85.714 | 14 | 2 | 0 | 5 | 18 | 4 | 17 | 18 | 15.6 |
| NW_005200461.1_14543-5p | miR95_0185 | 90.909 | 11 | 1 | 0 | 9 | 19 | 5 | 15 | 43 | 14.4 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续4
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005200461.1_14543-5p | miR95_0076 | 85.714 | 14 | 2 | 0 | 6 | 19 | 2 | 15 | 18 | 15.6 |
| NW_005200461.1_14543-5p | miR95_0076 | 84.615 | 13 | 2 | 0 | 8 | 20 | 1 | 13 | 37 | 14.6 |
| NW_005200461.1_14543-5p | miR95_0076 | 78.571 | 14 | 3 | 0 | 6 | 19 | 18 | 5 | 66 | 13.7 |
| NW_005200461.1_14543-5p | miR90_0689 | 91.667 | 12 | 1 | 0 | 9 | 20 | 5 | 16 | 21 | 15.4 |
| NW_005200461.1_14543-5p | miR90_0689 | 83.333 | 12 | 2 | 0 | 2 | 13 | 4 | 15 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0407 | 91.667 | 12 | 1 | 0 | 9 | 20 | 2 | 13 | 21 | 15.4 |
| NW_005200461.1_14543-5p | miR90_0351 | 91.667 | 12 | 1 | 0 | 8 | 19 | 17 | 6 | 21 | 15.4 |
| NW_005200461.1_14543-5p | miR90_0351 | 83.333 | 12 | 2 | 0 | 9 | 20 | 13 | 2 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0215 | 91.667 | 12 | 1 | 0 | 9 | 20 | 1 | 12 | 21 | 15.4 |
| NW_005200461.1_14543-5p | miR90_0898 | 100 | 10 | 0 | 0 | 9 | 18 | 15 | 6 | 24 | 15.2 |
| NW_005200461.1_14543-5p | miR90_0898 | 90.909 | 11 | 1 | 0 | 9 | 19 | 18 | 8 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR90_0477 | 100 | 10 | 0 | 0 | 11 | 20 | 1 | 10 | 24 | 15.2 |
| NW_005200461.1_14543-5p | miR90_0477 | 90 | 10 | 1 | 0 | 9 | 18 | 2 | 11 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR95_0444 | 100 | 10 | 0 | 0 | 8 | 17 | 17 | 8 | 24 | 15.2 |
| NW_005200461.1_14543-5p | miR95_0444 | 83.333 | 12 | 2 | 0 | 10 | 21 | 18 | 7 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR95_0439 | 100 | 10 | 0 | 0 | 10 | 19 | 8 | 17 | 24 | 15.2 |
| NW_005200461.1_14543-5p | miR95_0439 | 100 | 9 | 0 | 0 | 9 | 17 | 10 | 18 | 50 | 14.2 |
| NW_005200461.1_14543-5p | miR95_0439 | 90 | 10 | 1 | 0 | 10 | 19 | 5 | 14 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR95_0300 | 100 | 10 | 0 | 0 | 9 | 18 | 4 | 13 | 24 | 15.2 |
| NW_005200461.1_14543-5p | miR95_0300 | 90.909 | 11 | 1 | 0 | 9 | 19 | 1 | 11 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR95_0176 | 100 | 10 | 0 | 0 | 10 | 19 | 20 | 11 | 24 | 15.2 |
| NW_005200461.1_14543-5p | miR95_0044 | 100 | 10 | 0 | 0 | 9 | 18 | 2 | 11 | 24 | 15.2 |
| NW_005200461.1_14543-5p | miR95_0044 | 100 | 9 | 0 | 0 | 11 | 19 | 1 | 9 | 50 | 14.2 |
| NW_005200461.1_14543-5p | miR95_0006 | 100 | 10 | 0 | 0 | 10 | 19 | 1 | 10 | 24 | 15.2 |
| NW_005200461.1_14543-5p | Vca_miR44c | 73.684 | 19 | 5 | 0 | 2 | 20 | 21 | 3 | 24 | 15.2 |
| NW_005200461.1_14543-5p | Vca_miR44c | 78.571 | 14 | 3 | 0 | 9 | 22 | 17 | 4 | 66 | 13.7 |
| NW_005200461.1_14543-5p | Vca_miR44b | 73.684 | 19 | 5 | 0 | 2 | 20 | 21 | 3 | 24 | 15.2 |
| NW_005200461.1_14543-5p | Vca_miR44b | 78.571 | 14 | 3 | 0 | 9 | 22 | 17 | 4 | 66 | 13.7 |
| NW_005200461.1_14543-5p | Vca_miR44a | 73.684 | 19 | 5 | 0 | 2 | 20 | 21 | 3 | 24 | 15.2 |
| NW_005200461.1_14543-5p | Vca_miR44a | 78.571 | 14 | 3 | 0 | 9 | 22 | 17 | 4 | 66 | 13.7 |
| NW_005200461.1_14543-5p | miR90_0182 | 76.471 | 17 | 4 | 0 | 1 | 17 | 17 | 1 | 28 | 15 |
| NW_005200461.1_14543-5p | miR90_0208 | 82.353 | 17 | 2 | 1 | 4 | 19 | 1 | 17 | 32 | 14.8 |
| NW_005200461.1_14543-5p | miR90_0208 | 90 | 10 | 1 | 0 | 9 | 18 | 10 | 19 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0809 | 84.615 | 13 | 2 | 0 | 7 | 19 | 1 | 13 | 37 | 14.6 |
| NW_005200461.1_14543-5p | miR90_0809 | 83.333 | 12 | 2 | 0 | 9 | 20 | 6 | 17 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0564 | 84.615 | 13 | 2 | 0 | 6 | 18 | 6 | 18 | 37 | 14.6 |
| NW_005200461.1_14543-5p | miR90_0254 | 84.615 | 13 | 2 | 0 | 7 | 19 | 17 | 5 | 37 | 14.6 |
| NW_005200461.1_14543-5p | miR90_0223 | 84.615 | 13 | 2 | 0 | 6 | 18 | 13 | 1 | 37 | 14.6 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续5
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005200461.1_14543-5p | miR90_0200 | 84.615 | 13 | 2 | 0 | 7 | 19 | 17 | 5 | 37 | 14.6 |
| NW_005200461.1_14543-5p | miR95_0340 | 84.615 | 13 | 2 | 0 | 7 | 19 | 19 | 7 | 37 | 14.6 |
| NW_005200461.1_14543-5p | Vca_miR77 | 84.615 | 13 | 2 | 0 | 8 | 20 | 9 | 21 | 37 | 14.6 |
| NW_005200461.1_14543-5p | miR90_0850 | 90.909 | 11 | 1 | 0 | 11 | 21 | 5 | 15 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR90_0769 | 90.909 | 11 | 1 | 0 | 9 | 19 | 2 | 12 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR90_0769 | 83.333 | 12 | 2 | 0 | 9 | 20 | 5 | 16 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0754 | 90.909 | 11 | 1 | 0 | 9 | 19 | 18 | 8 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR90_0754 | 83.333 | 12 | 2 | 0 | 9 | 20 | 15 | 4 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0644 | 90.909 | 11 | 1 | 0 | 9 | 19 | 8 | 18 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR90_0507 | 90.909 | 11 | 1 | 0 | 10 | 20 | 12 | 2 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR90_0507 | 75 | 16 | 4 | 0 | 1 | 16 | 18 | 3 | 57 | 13.9 |
| NW_005200461.1_14543-5p | miR90_0241 | 90.909 | 11 | 1 | 0 | 10 | 20 | 1 | 11 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR90_0199 | 90.909 | 11 | 1 | 0 | 9 | 19 | 1 | 11 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR90_0199 | 83.333 | 12 | 2 | 0 | 10 | 21 | 8 | 19 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR95_0194 | 90.909 | 11 | 1 | 0 | 4 | 14 | 12 | 2 | 43 | 14.4 |
| NW_005200461.1_14543-5p | miR90_0722 | 100 | 9 | 0 | 0 | 12 | 20 | 17 | 9 | 50 | 14.2 |
| NW_005200461.1_14543-5p | miR90_0294 | 72.222 | 18 | 5 | 0 | 3 | 20 | 18 | 1 | 50 | 14.2 |
| NW_005200461.1_14543-5p | miR95_0254 | 100 | 9 | 0 | 0 | 10 | 18 | 1 | 9 | 50 | 14.2 |
| NW_005200461.1_14543-5p | miR95_0140 | 100 | 9 | 0 | 0 | 11 | 19 | 1 | 9 | 50 | 14.2 |
| NW_005200461.1_14543-5p | miR95_0001 | 72.222 | 18 | 5 | 0 | 1 | 18 | 1 | 18 | 50 | 14.2 |
| NW_005200461.1_14543-5p | miR90_0747 | 75 | 16 | 4 | 0 | 5 | 20 | 1 | 16 | 57 | 13.9 |
| NW_005200461.1_14543-5p | miR90_0169 | 75 | 16 | 4 | 0 | 3 | 18 | 4 | 19 | 57 | 13.9 |
| NW_005200461.1_14543-5p | miR90_0169 | 75 | 16 | 4 | 0 | 1 | 16 | 19 | 4 | 57 | 13.9 |
| NW_005200461.1_14543-5p | miR90_0169 | 83.333 | 12 | 2 | 0 | 9 | 20 | 17 | 6 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0169 | 83.333 | 12 | 2 | 0 | 9 | 20 | 7 | 18 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0169 | 90 | 10 | 1 | 0 | 10 | 19 | 19 | 10 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0157 | 75 | 16 | 4 | 0 | 4 | 19 | 18 | 3 | 57 | 13.9 |
| NW_005200461.1_14543-5p | miR90_0013 | 80 | 20 | 2 | 2 | 1 | 19 | 1 | 19 | 57 | 13.9 |
| NW_005200461.1_14543-5p | miR95_0421 | 75 | 16 | 4 | 0 | 6 | 21 | 3 | 18 | 57 | 13.9 |
| NW_005200461.1_14543-5p | miR95_0326 | 80 | 20 | 2 | 2 | 1 | 19 | 1 | 19 | 57 | 13.9 |
| NW_005200461.1_14543-5p | miR95_0260 | 75 | 16 | 4 | 0 | 4 | 19 | 1 | 16 | 57 | 13.9 |
| NW_005200461.1_14543-5p | miR95_0260 | 68.421 | 19 | 6 | 0 | 1 | 19 | 1 | 19 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0756 | 78.571 | 14 | 3 | 0 | 1 | 14 | 5 | 18 | 66 | 13.7 |
| NW_005200461.1_14543-5p | miR90_0756 | 90 | 10 | 1 | 0 | 9 | 18 | 4 | 13 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0576 | 78.571 | 14 | 3 | 0 | 9 | 22 | 18 | 5 | 66 | 13.7 |
| NW_005200461.1_14543-5p | miR95_0190 | 78.571 | 14 | 3 | 0 | 4 | 17 | 17 | 4 | 66 | 13.7 |
| NW_005200461.1_14543-5p | miR90_0908 | 83.333 | 12 | 2 | 0 | 9 | 20 | 1 | 12 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0887 | 83.333 | 12 | 2 | 0 | 9 | 20 | 2 | 13 | 77 | 13.5 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续6
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005200461.1_14543-5p | miR90_0678 | 83.333 | 12 | 2 | 0 | 2 | 13 | 7 | 18 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0655 | 83.333 | 12 | 2 | 0 | 2 | 13 | 3 | 14 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0520 | 83.333 | 12 | 2 | 0 | 7 | 18 | 18 | 7 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0219 | 83.333 | 12 | 2 | 0 | 10 | 21 | 4 | 15 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR95_0411 | 83.333 | 12 | 2 | 0 | 7 | 18 | 8 | 19 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR95_0373 | 83.333 | 12 | 2 | 0 | 7 | 18 | 7 | 18 | 77 | 13.5 |
| NW_005200461.1_14543-5p | PC-5p-179621_3 | 83.333 | 12 | 2 | 0 | 11 | 22 | 9 | 20 | 77 | 13.5 |
| NW_005200461.1_14543-5p | SjapMIR50 | 83.333 | 12 | 2 | 0 | 4 | 15 | 10 | 21 | 77 | 13.5 |
| NW_005200461.1_14543-5p | miR90_0899 | 90 | 10 | 1 | 0 | 10 | 19 | 9 | 18 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0897 | 90 | 10 | 1 | 0 | 10 | 19 | 10 | 1 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0897 | 90 | 10 | 1 | 0 | 10 | 19 | 13 | 4 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0848 | 90 | 10 | 1 | 0 | 10 | 19 | 4 | 13 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0814 | 90 | 10 | 1 | 0 | 10 | 19 | 8 | 17 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0771 | 90 | 10 | 1 | 0 | 10 | 19 | 12 | 3 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0750 | 90 | 10 | 1 | 0 | 10 | 19 | 8 | 17 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0741 | 90 | 10 | 1 | 0 | 10 | 19 | 3 | 12 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0656 | 90 | 10 | 1 | 0 | 6 | 15 | 17 | 8 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0624 | 68.421 | 19 | 6 | 0 | 1 | 19 | 19 | 1 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0613 | 90 | 10 | 1 | 0 | 6 | 15 | 17 | 8 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0588 | 90 | 10 | 1 | 0 | 6 | 15 | 12 | 3 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0584 | 68.421 | 19 | 6 | 0 | 1 | 19 | 21 | 3 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0336 | 90 | 10 | 1 | 0 | 6 | 15 | 14 | 5 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0331 | 68.421 | 19 | 6 | 0 | 1 | 19 | 1 | 19 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0274 | 90 | 10 | 1 | 0 | 10 | 19 | 6 | 15 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0236 | 90 | 10 | 1 | 0 | 9 | 18 | 17 | 8 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0211 | 90 | 10 | 1 | 0 | 9 | 18 | 12 | 3 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR90_0006 | 90 | 10 | 1 | 0 | 10 | 19 | 1 | 10 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR95_0525 | 68.421 | 19 | 6 | 0 | 1 | 19 | 19 | 1 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR95_0415 | 90 | 10 | 1 | 0 | 4 | 13 | 10 | 1 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR95_0327 | 90 | 10 | 1 | 0 | 9 | 18 | 7 | 16 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR95_0220 | 90 | 10 | 1 | 0 | 10 | 19 | 1 | 10 | 89 | 13.3 |
| NW_005200461.1_14543-5p | miR95_0064 | 90 | 10 | 1 | 0 | 10 | 19 | 8 | 17 | 89 | 13.3 |
| NW_005200496.1_14318-5p | miR90_0521 | 86.667 | 15 | 2 | 0 | 1 | 15 | 1 | 15 | 11 | 16.7 |
| NW_005200496.1_14318-5p | Thalassiosira | 86.667 | 15 | 2 | 0 | 13 | 27 | 2 | 16 | 11 | 16.7 |
| NW_005200496.1_14318-5p | miR90_0808 | 77.778 | 18 | 4 | 0 | 7 | 24 | 2 | 19 | 17 | 16.1 |
| NW_005200496.1_14318-5p | miR90_0808 | 83.333 | 12 | 2 | 0 | 6 | 17 | 12 | 1 | 96 | 13.5 |
| NW_005200496.1_14318-5p | miR90_0496 | 85.714 | 14 | 2 | 0 | 17 | 30 | 2 | 15 | 22 | 15.6 |
| NW_005200496.1_14318-5p | miR90_0379 | 85.714 | 14 | 2 | 0 | 17 | 30 | 2 | 15 | 22 | 15.6 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续7
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005200496.1_14318-5p | miR90_0826 | 91.667 | 12 | 1 | 0 | 7 | 18 | 4 | 15 | 26 | 15.4 |
| NW_005200496.1_14318-5p | miR90_0826 | 83.333 | 12 | 2 | 0 | 6 | 17 | 14 | 3 | 96 | 13.5 |
| NW_005200496.1_14318-5p | ppu-miR36-3p | 73.684 | 19 | 5 | 0 | 1 | 19 | 2 | 20 | 30 | 15.2 |
| NW_005200496.1_14318-5p | SjapMIR6 | 73.684 | 19 | 5 | 0 | 9 | 27 | 3 | 21 | 30 | 15.2 |
| NW_005200496.1_14318-5p | miR95_0193 | 76.471 | 17 | 4 | 0 | 4 | 20 | 4 | 20 | 35 | 15 |
| NW_005200496.1_14318-5p | miR95_0193 | 72.222 | 18 | 5 | 0 | 7 | 24 | 19 | 2 | 62 | 14.2 |
| NW_005200496.1_14318-5p | miR90_0576 | 86.667 | 15 | 1 | 1 | 2 | 16 | 18 | 5 | 46 | 14.6 |
| NW_005200496.1_14318-5p | miR90_0663 | 90.909 | 11 | 1 | 0 | 4 | 14 | 11 | 1 | 54 | 14.4 |
| NW_005200496.1_14318-5p | miR95_0358 | 70 | 20 | 6 | 0 | 7 | 26 | 6 | 25 | 54 | 14.4 |
| NW_005200496.1_14318-5p | SjapMIR23 | 72.222 | 18 | 5 | 0 | 1 | 18 | 4 | 21 | 62 | 14.2 |
| NW_005200496.1_14318-5p | miR90_0566 | 75 | 16 | 4 | 0 | 7 | 22 | 5 | 20 | 72 | 13.9 |
| NW_005200496.1_14318-5p | miR90_0012 | 75 | 16 | 4 | 0 | 7 | 22 | 6 | 21 | 72 | 13.9 |
| NW_005200496.1_14318-5p | SjapMIR17 | 75 | 16 | 4 | 0 | 15 | 30 | 6 | 21 | 72 | 13.9 |
| NW_005200496.1_14318-5p | miR90_0628 | 78.571 | 14 | 3 | 0 | 10 | 23 | 16 | 3 | 83 | 13.7 |
| NW_005200496.1_14318-5p | miR90_0441 | 65.217 | 23 | 8 | 0 | 7 | 29 | 25 | 3 | 83 | 13.7 |
| NW_005200496.1_14318-5p | miR95_0421 | 78.571 | 14 | 3 | 0 | 4 | 17 | 18 | 5 | 83 | 13.7 |
| NW_005200496.1_14318-5p | miR95_0294 | 78.571 | 14 | 3 | 0 | 17 | 30 | 4 | 17 | 83 | 13.7 |
| NW_005200496.1_14318-5p | miR95_0275 | 78.571 | 14 | 3 | 0 | 17 | 30 | 17 | 4 | 83 | 13.7 |
| NW_005200496.1_14318-5p | miR95_0274 | 78.571 | 14 | 3 | 0 | 17 | 30 | 2 | 15 | 83 | 13.7 |
| NW_005200496.1_14318-5p | miR95_0175 | 78.571 | 14 | 3 | 0 | 17 | 30 | 1 | 14 | 83 | 13.7 |
| NW_005200496.1_14318-5p | Vca_miR44c | 78.571 | 14 | 3 | 0 | 17 | 30 | 18 | 5 | 83 | 13.7 |
| NW_005200496.1_14318-5p | Vca_miR44b | 78.571 | 14 | 3 | 0 | 17 | 30 | 18 | 5 | 83 | 13.7 |
| NW_005200496.1_14318-5p | Vca_miR44a | 78.571 | 14 | 3 | 0 | 17 | 30 | 18 | 5 | 83 | 13.7 |
| NW_005200496.1_14318-5p | SjapMIR59 | 78.571 | 14 | 3 | 0 | 17 | 30 | 21 | 8 | 83 | 13.7 |
| NW_005200496.1_14318-5p | miR90_0793 | 83.333 | 12 | 2 | 0 | 2 | 13 | 1 | 12 | 96 | 13.5 |
| NW_005200496.1_14318-5p | miR90_0048 | 83.333 | 12 | 2 | 0 | 7 | 18 | 15 | 4 | 96 | 13.5 |
| NW_005200496.1_14318-5p | miR95_0460 | 66.667 | 21 | 7 | 0 | 10 | 30 | 24 | 4 | 96 | 13.5 |
| NW_005200496.1_14318-5p | miR95_0283 | 83.333 | 12 | 2 | 0 | 7 | 18 | 13 | 2 | 96 | 13.5 |
| NW_005200496.1_14318-5p | PC-5p-179621_3 | 83.333 | 12 | 2 | 0 | 19 | 30 | 1 | 12 | 96 | 13.5 |
| NW_005200496.1_14318-5p | SjapMIR57 | 66.667 | 21 | 7 | 0 | 5 | 25 | 21 | 1 | 96 | 13.5 |
| NW_005200517.1_13952-3p | SjapMIR15_2 | 100 | 9 | 0 | 0 | 3 | 11 | 14 | 6 | 47 | 14.2 |
| NW_005200517.1_13952-3p | SjapMIR15_1 | 100 | 9 | 0 | 0 | 3 | 11 | 14 | 6 | 47 | 14.2 |
| NW_005200517.1_13952-3p | Vca_miR113b | 75 | 16 | 4 | 0 | 1 | 16 | 18 | 3 | 55 | 13.9 |
| NW_005200517.1_13952-3p | Vca_miR113a | 75 | 16 | 4 | 0 | 1 | 16 | 18 | 3 | 55 | 13.9 |
| NW_005200517.1_13952-3p | miR90_0679 | 78.571 | 14 | 3 | 0 | 7 | 20 | 15 | 2 | 63 | 13.7 |
| NW_005200517.1_13952-3p | miR90_0123 | 78.571 | 14 | 3 | 0 | 7 | 20 | 25 | 12 | 63 | 13.7 |
| NW_005200517.1_13952-3p | miR95_0563 | 78.571 | 14 | 3 | 0 | 5 | 18 | 14 | 1 | 63 | 13.7 |
| NW_005200517.1_13952-3p | miR95_0436 | 78.571 | 14 | 3 | 0 | 7 | 20 | 3 | 16 | 63 | 13.7 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续8
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005200517.1_13952-3p | miR95_0407 | 78.571 | 14 | 3 | 0 | 7 | 20 | 3 | 16 | 63 | 13.7 |
| NW_005200517.1_13952-3p | miR95_0379 | 78.571 | 14 | 3 | 0 | 7 | 20 | 2 | 15 | 63 | 13.7 |
| NW_005200517.1_13952-3p | miR95_0370 | 78.571 | 14 | 3 | 0 | 7 | 20 | 3 | 16 | 63 | 13.7 |
| NW_005200517.1_13952-3p | miR95_0314 | 78.571 | 14 | 3 | 0 | 7 | 20 | 2 | 15 | 63 | 13.7 |
| NW_005200517.1_13952-3p | miR95_0094 | 78.571 | 14 | 3 | 0 | 7 | 20 | 23 | 10 | 63 | 13.7 |
| NW_005200517.1_13952-3p | miR95_0091 | 78.571 | 14 | 3 | 0 | 7 | 20 | 23 | 10 | 63 | 13.7 |
| NW_005200517.1_13952-3p | miR95_0007 | 78.571 | 14 | 3 | 0 | 7 | 20 | 2 | 15 | 63 | 13.7 |
| NW_005200517.1_13952-3p | SjapMIR97 | 78.571 | 14 | 3 | 0 | 6 | 19 | 14 | 1 | 63 | 13.7 |
| NW_005200517.1_13952-3p | miR95_0522 | 83.333 | 12 | 2 | 0 | 5 | 16 | 12 | 1 | 73 | 13.5 |
| NW_005200517.1_13952-3p | miR95_0460 | 83.333 | 12 | 2 | 0 | 6 | 17 | 14 | 25 | 73 | 13.5 |
| NW_005200517.1_13952-3p | miR95_0093 | 83.333 | 12 | 2 | 0 | 9 | 20 | 21 | 10 | 73 | 13.5 |
| NW_005200517.1_13952-3p | miR95_0054 | 83.333 | 12 | 2 | 0 | 9 | 20 | 20 | 9 | 73 | 13.5 |
| NW_005200517.1_13952-3p | miR95_0050 | 83.333 | 12 | 2 | 0 | 9 | 20 | 25 | 14 | 73 | 13.5 |
| NW_005200517.1_13952-3p | SjapMIR75 | 90 | 10 | 1 | 0 | 2 | 11 | 1 | 10 | 85 | 13.3 |
| NW_005200517.1_13952-3p | miR90_0591 | 100 | 8 | 0 | 0 | 7 | 14 | 12 | 5 | 98 | 13.1 |
| NW_005200517.1_13952-3p | Vca_miR39 | 100 | 8 | 0 | 0 | 2 | 9 | 13 | 20 | 98 | 13.1 |
| NW_005200535.1_13863-3p | Vca_miR104 | 84.615 | 13 | 2 | 0 | 3 | 15 | 9 | 21 | 35 | 14.6 |
| NW_005200535.1_13863-3p | miR90_0459 | 78.571 | 14 | 3 | 0 | 6 | 19 | 18 | 5 | 63 | 13.7 |
| NW_005200535.1_13863-3p | SjapMIR102 | 78.571 | 14 | 3 | 0 | 8 | 21 | 18 | 5 | 63 | 13.7 |
| NW_005200535.1_13863-3p | miR90_0440 | 83.333 | 12 | 2 | 0 | 7 | 18 | 7 | 18 | 73 | 13.5 |
| NW_005200535.1_13863-3p | miR90_0286 | 83.333 | 12 | 2 | 0 | 7 | 18 | 25 | 14 | 73 | 13.5 |
| NW_005200535.1_13863-3p | miR95_0304 | 83.333 | 12 | 2 | 0 | 3 | 14 | 4 | 15 | 73 | 13.5 |
| NW_005200535.1_13863-3p | miR95_0256 | 83.333 | 12 | 2 | 0 | 7 | 18 | 10 | 21 | 73 | 13.5 |
| NW_005200535.1_13863-3p | Vca_miR58-3p* | 83.333 | 12 | 2 | 0 | 10 | 21 | 15 | 4 | 73 | 13.5 |
| NW_005200535.1_13863-3p | SjapMIR53 | 83.333 | 12 | 2 | 0 | 5 | 16 | 17 | 6 | 73 | 13.5 |
| NW_005200535.1_13863-3p | SjapMIR37_2 | 83.333 | 12 | 2 | 0 | 7 | 18 | 1 | 12 | 73 | 13.5 |
| NW_005200535.1_13863-3p | SjapMIR37_1 | 83.333 | 12 | 2 | 0 | 7 | 18 | 1 | 12 | 73 | 13.5 |
| NW_005200535.1_13863-3p | miR95_0392 | 90 | 10 | 1 | 0 | 1 | 10 | 7 | 16 | 85 | 13.3 |
| NW_005200535.1_13863-3p | Vca_miR111 | 90 | 10 | 1 | 0 | 7 | 16 | 1 | 10 | 85 | 13.3 |
| NW_005200535.1_13863-3p | miR95_0143 | 70.588 | 17 | 5 | 0 | 1 | 17 | 1 | 17 | 98 | 13.1 |
| NW_005200535.1_13863-3p | SjapMIR8 | 100 | 8 | 0 | 0 | 13 | 20 | 7 | 14 | 98 | 13.1 |
| NW_005200600.1_13330-3p | miR90_0709 | 91.667 | 12 | 1 | 0 | 10 | 21 | 18 | 7 | 20 | 15.4 |
| NW_005200600.1_13330-3p | miR90_0495 | 100 | 10 | 0 | 0 | 3 | 12 | 18 | 9 | 23 | 15.2 |
| NW_005200600.1_13330-3p | Vca_miR60 | 73.684 | 19 | 5 | 0 | 3 | 21 | 3 | 21 | 23 | 15.2 |
| NW_005200600.1_13330-3p | miR90_0876 | 80 | 15 | 3 | 0 | 6 | 20 | 1 | 15 | 31 | 14.8 |
| NW_005200600.1_13330-3p | Vca_miR82 | 80 | 15 | 3 | 0 | 6 | 20 | 2 | 16 | 31 | 14.8 |
| NW_005200600.1_13330-3p | miR90_0690 | 84.615 | 13 | 2 | 0 | 9 | 21 | 2 | 14 | 35 | 14.6 |
| NW_005200600.1_13330-3p | miR90_0653 | 84.615 | 13 | 2 | 0 | 9 | 21 | 1 | 13 | 35 | 14.6 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续9
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005200600.1_13330-3p | miR90_0211 | 84.615 | 13 | 2 | 0 | 7 | 19 | 1 | 13 | 35 | 14.6 |
| NW_005200600.1_13330-3p | Vca_miR77 | 84.615 | 13 | 2 | 0 | 9 | 21 | 15 | 3 | 35 | 14.6 |
| NW_005200600.1_13330-3p | miR90_0487 | 90.909 | 11 | 1 | 0 | 11 | 21 | 19 | 9 | 41 | 14.4 |
| NW_005200600.1_13330-3p | ppu-miR9-3p | 100 | 9 | 0 | 0 | 4 | 12 | 3 | 11 | 47 | 14.2 |
| NW_005200600.1_13330-3p | miR90_0862 | 100 | 9 | 0 | 0 | 12 | 20 | 15 | 7 | 47 | 14.2 |
| NW_005200600.1_13330-3p | miR90_0840 | 100 | 9 | 0 | 0 | 12 | 20 | 16 | 8 | 47 | 14.2 |
| NW_005200600.1_13330-3p | miR90_0743 | 100 | 9 | 0 | 0 | 12 | 20 | 6 | 14 | 47 | 14.2 |
| NW_005200600.1_13330-3p | miR95_0499 | 72.222 | 18 | 5 | 0 | 2 | 19 | 1 | 18 | 47 | 14.2 |
| NW_005200600.1_13330-3p | miR95_0258 | 75 | 20 | 4 | 1 | 1 | 20 | 21 | 3 | 47 | 14.2 |
| NW_005200600.1_13330-3p | Porphyra | 72.222 | 18 | 5 | 0 | 3 | 20 | 3 | 20 | 47 | 14.2 |
| NW_005200600.1_13330-3p | miR90_0827 | 75 | 16 | 4 | 0 | 1 | 16 | 3 | 18 | 55 | 13.9 |
| NW_005200600.1_13330-3p | miR90_0559 | 75 | 16 | 4 | 0 | 1 | 16 | 2 | 17 | 55 | 13.9 |
| NW_005200600.1_13330-3p | miR90_0400 | 75 | 16 | 4 | 0 | 3 | 18 | 16 | 1 | 55 | 13.9 |
| NW_005200600.1_13330-3p | miR90_0016 | 75 | 16 | 4 | 0 | 3 | 18 | 17 | 2 | 55 | 13.9 |
| NW_005200600.1_13330-3p | Vca_miR13* | 75 | 16 | 4 | 0 | 6 | 21 | 6 | 21 | 55 | 13.9 |
| NW_005200600.1_13330-3p | miR90_0759 | 78.571 | 14 | 3 | 0 | 6 | 19 | 3 | 16 | 63 | 13.7 |
| NW_005200600.1_13330-3p | miR90_0457 | 78.571 | 14 | 3 | 0 | 2 | 15 | 19 | 6 | 63 | 13.7 |
| NW_005200600.1_13330-3p | miR95_0417 | 78.571 | 14 | 3 | 0 | 1 | 14 | 18 | 5 | 63 | 13.7 |
| NW_005200600.1_13330-3p | SjapMIR60 | 78.571 | 14 | 3 | 0 | 8 | 21 | 2 | 15 | 63 | 13.7 |
| NW_005200600.1_13330-3p | miR90_0777 | 83.333 | 12 | 2 | 0 | 1 | 12 | 18 | 7 | 73 | 13.5 |
| NW_005200600.1_13330-3p | miR90_0363 | 83.333 | 12 | 2 | 0 | 2 | 13 | 6 | 17 | 73 | 13.5 |
| NW_005200600.1_13330-3p | SjapMIR70 | 83.333 | 12 | 2 | 0 | 5 | 16 | 2 | 13 | 73 | 13.5 |
| NW_005200600.1_13330-3p | miR90_0809 | 90 | 10 | 1 | 0 | 9 | 18 | 12 | 3 | 85 | 13.3 |
| NW_005200600.1_13330-3p | miR90_0769 | 90 | 10 | 1 | 0 | 9 | 18 | 11 | 2 | 85 | 13.3 |
| NW_005200600.1_13330-3p | miR90_0754 | 90 | 10 | 1 | 0 | 9 | 18 | 9 | 18 | 85 | 13.3 |
| NW_005200600.1_13330-3p | miR90_0314 | 90 | 10 | 1 | 0 | 1 | 10 | 16 | 7 | 85 | 13.3 |
| NW_005200600.1_13330-3p | miR90_0217 | 90 | 10 | 1 | 0 | 1 | 10 | 15 | 24 | 85 | 13.3 |
| NW_005200600.1_13330-3p | miR95_0549 | 90 | 10 | 1 | 0 | 1 | 10 | 8 | 17 | 85 | 13.3 |
| NW_005200600.1_13330-3p | miR95_0143 | 90 | 10 | 1 | 0 | 3 | 12 | 8 | 17 | 85 | 13.3 |
| NW_005200600.1_13330-3p | SjapMIR102 | 90 | 10 | 1 | 0 | 3 | 12 | 18 | 9 | 85 | 13.3 |
| NW_005200600.1_13330-3p | SjapMIR29 | 68.421 | 19 | 6 | 0 | 1 | 19 | 3 | 21 | 85 | 13.3 |
| NW_005200600.1_13330-3p | ppu-miR4-5p | 100 | 8 | 0 | 0 | 14 | 21 | 11 | 18 | 98 | 13.1 |
| NW_005200600.1_13330-3p | miR90_0866 | 100 | 8 | 0 | 0 | 13 | 20 | 25 | 18 | 98 | 13.1 |
| NW_005200600.1_13330-3p | miR90_0706 | 100 | 8 | 0 | 0 | 14 | 21 | 18 | 11 | 98 | 13.1 |
| NW_005200600.1_13330-3p | miR90_0610 | 70.588 | 17 | 5 | 0 | 5 | 21 | 1 | 17 | 98 | 13.1 |
| NW_005200600.1_13330-3p | miR90_0560 | 100 | 8 | 0 | 0 | 3 | 10 | 13 | 6 | 98 | 13.1 |
| NW_005200600.1_13330-3p | miR95_0488 | 70.588 | 17 | 5 | 0 | 3 | 19 | 1 | 17 | 98 | 13.1 |
| NW_005200600.1_13330-3p | miR95_0401 | 70.588 | 17 | 5 | 0 | 3 | 19 | 18 | 2 | 98 | 13.1 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续10
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005200600.1_13330-3p | miR95_0260 | 70.588 | 17 | 5 | 0 | 1 | 17 | 17 | 1 | 98 | 13.1 |
| NW_005200600.1_13330-3p | SjapMIR98 | 70.588 | 17 | 5 | 0 | 1 | 17 | 2 | 18 | 98 | 13.1 |
| NW_005200881.1_10661-3p | Vca_miR32 | 75 | 20 | 5 | 0 | 2 | 21 | 23 | 4 | 11 | 16.3 |
| NW_005200881.1_10661-3p | miR95_0499 | 85.714 | 14 | 2 | 0 | 8 | 21 | 5 | 18 | 17 | 15.6 |
| NW_005200881.1_10661-3p | miR95_0488 | 85.714 | 14 | 2 | 0 | 8 | 21 | 4 | 17 | 17 | 15.6 |
| NW_005200881.1_10661-3p | miR95_0193 | 76.471 | 17 | 4 | 0 | 2 | 18 | 1 | 17 | 26 | 15 |
| NW_005200881.1_10661-3p | miR95_0534 | 82.353 | 17 | 2 | 1 | 5 | 21 | 2 | 17 | 31 | 14.8 |
| NW_005200881.1_10661-3p | miR95_0532 | 82.353 | 17 | 2 | 1 | 5 | 21 | 1 | 16 | 31 | 14.8 |
| NW_005200881.1_10661-3p | miR90_0351 | 84.615 | 13 | 2 | 0 | 8 | 20 | 4 | 16 | 35 | 14.6 |
| NW_005200881.1_10661-3p | miR90_0271 | 84.615 | 13 | 2 | 0 | 8 | 20 | 3 | 15 | 35 | 14.6 |
| NW_005200881.1_10661-3p | miR90_0457 | 90.909 | 11 | 1 | 0 | 10 | 20 | 9 | 19 | 41 | 14.4 |
| NW_005200881.1_10661-3p | miR95_0386 | 70 | 20 | 6 | 0 | 2 | 21 | 25 | 6 | 41 | 14.4 |
| NW_005200881.1_10661-3p | Vca_miR43d | 90.909 | 11 | 1 | 0 | 4 | 14 | 21 | 11 | 41 | 14.4 |
| NW_005200881.1_10661-3p | Vca_miR43c | 90.909 | 11 | 1 | 0 | 4 | 14 | 21 | 11 | 41 | 14.4 |
| NW_005200881.1_10661-3p | Vca_miR43b | 90.909 | 11 | 1 | 0 | 4 | 14 | 21 | 11 | 41 | 14.4 |
| NW_005200881.1_10661-3p | Vca_miR43a | 90.909 | 11 | 1 | 0 | 4 | 14 | 21 | 11 | 41 | 14.4 |
| NW_005200881.1_10661-3p | miR90_0487 | 100 | 9 | 0 | 0 | 12 | 20 | 11 | 19 | 47 | 14.2 |
| NW_005200881.1_10661-3p | ppu-miR15-5p | 75 | 16 | 4 | 0 | 4 | 19 | 5 | 20 | 55 | 13.9 |
| NW_005200881.1_10661-3p | miR90_0691 | 78.571 | 14 | 3 | 0 | 2 | 15 | 12 | 25 | 63 | 13.7 |
| NW_005200881.1_10661-3p | miR95_0538 | 78.571 | 14 | 3 | 0 | 3 | 16 | 5 | 18 | 63 | 13.7 |
| NW_005200881.1_10661-3p | miR90_0507 | 83.333 | 12 | 2 | 0 | 7 | 18 | 13 | 2 | 73 | 13.5 |
| NW_005200881.1_10661-3p | miR90_0477 | 83.333 | 12 | 2 | 0 | 9 | 20 | 16 | 5 | 73 | 13.5 |
| NW_005200881.1_10661-3p | miR90_0400 | 83.333 | 12 | 2 | 0 | 8 | 19 | 2 | 13 | 73 | 13.5 |
| NW_005200881.1_10661-3p | miR90_0298 | 83.333 | 12 | 2 | 0 | 5 | 16 | 25 | 14 | 73 | 13.5 |
| NW_005200881.1_10661-3p | miR90_0169 | 83.333 | 12 | 2 | 0 | 8 | 19 | 8 | 19 | 73 | 13.5 |
| NW_005200881.1_10661-3p | miR90_0016 | 83.333 | 12 | 2 | 0 | 8 | 19 | 3 | 14 | 73 | 13.5 |
| NW_005200881.1_10661-3p | miR95_0525 | 83.333 | 12 | 2 | 0 | 10 | 21 | 1 | 12 | 73 | 13.5 |
| NW_005200881.1_10661-3p | miR95_0076 | 83.333 | 12 | 2 | 0 | 9 | 20 | 4 | 15 | 73 | 13.5 |
| NW_005200881.1_10661-3p | SjapMIR102 | 83.333 | 12 | 2 | 0 | 2 | 13 | 18 | 7 | 73 | 13.5 |
| NW_005200881.1_10661-3p | miR90_0497 | 90 | 10 | 1 | 0 | 6 | 15 | 2 | 11 | 85 | 13.3 |
| NW_005200881.1_10661-3p | miR90_0315 | 90 | 10 | 1 | 0 | 1 | 10 | 17 | 8 | 85 | 13.3 |
| NW_005200881.1_10661-3p | miR90_0084 | 90 | 10 | 1 | 0 | 1 | 10 | 20 | 11 | 85 | 13.3 |
| NW_005200881.1_10661-3p | miR95_0380 | 90 | 10 | 1 | 0 | 3 | 12 | 11 | 2 | 85 | 13.3 |
| NW_005200881.1_10661-3p | miR95_0118 | 90 | 10 | 1 | 0 | 10 | 19 | 11 | 2 | 85 | 13.3 |
| NW_005200881.1_10661-3p | miR95_0087 | 68.421 | 19 | 6 | 0 | 2 | 20 | 19 | 1 | 85 | 13.3 |
| NW_005200881.1_10661-3p | miR90_0378 | 70.588 | 17 | 5 | 0 | 4 | 20 | 17 | 1 | 98 | 13.1 |
| NW_005200881.1_10661-3p | miR95_0292 | 100 | 8 | 0 | 0 | 9 | 16 | 9 | 2 | 98 | 13.1 |
| NW_005201234.1_6668-5p | miR90_0701 | 80 | 15 | 3 | 0 | 4 | 18 | 4 | 18 | 26 | 14.8 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续11
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005201234.1_6668-5p | miR90_0701 | 90 | 10 | 1 | 0 | 8 | 17 | 11 | 2 | 73 | 13.3 |
| NW_005201234.1_6668-5p | miR90_0701 | 90 | 10 | 1 | 0 | 8 | 17 | 2 | 11 | 73 | 13.3 |
| NW_005201234.1_6668-5p | miR90_0701 | 100 | 8 | 0 | 0 | 4 | 11 | 9 | 2 | 84 | 13.1 |
| NW_005201234.1_6668-5p | miR95_0282 | 84.615 | 13 | 2 | 0 | 3 | 15 | 15 | 3 | 30 | 14.6 |
| NW_005201234.1_6668-5p | miR95_0282 | 78.571 | 14 | 3 | 0 | 2 | 15 | 1 | 14 | 54 | 13.7 |
| NW_005201234.1_6668-5p | miR90_0787 | 90.909 | 11 | 1 | 0 | 6 | 16 | 1 | 11 | 35 | 14.4 |
| NW_005201234.1_6668-5p | miR90_0167 | 90.909 | 11 | 1 | 0 | 5 | 15 | 24 | 14 | 35 | 14.4 |
| NW_005201234.1_6668-5p | miR90_0144 | 90.909 | 11 | 1 | 0 | 5 | 15 | 9 | 19 | 35 | 14.4 |
| NW_005201234.1_6668-5p | miR90_0829 | 100 | 9 | 0 | 0 | 1 | 9 | 7 | 15 | 41 | 14.2 |
| NW_005201234.1_6668-5p | miR90_0829 | 100 | 8 | 0 | 0 | 1 | 8 | 17 | 24 | 84 | 13.1 |
| NW_005201234.1_6668-5p | miR90_0463 | 100 | 9 | 0 | 0 | 1 | 9 | 9 | 1 | 41 | 14.2 |
| NW_005201234.1_6668-5p | miR90_0245 | 100 | 9 | 0 | 0 | 1 | 9 | 9 | 1 | 41 | 14.2 |
| NW_005201234.1_6668-5p | miR90_0245 | 100 | 9 | 0 | 0 | 1 | 9 | 19 | 11 | 41 | 14.2 |
| NW_005201234.1_6668-5p | miR90_0244 | 100 | 9 | 0 | 0 | 1 | 9 | 9 | 1 | 41 | 14.2 |
| NW_005201234.1_6668-5p | miR90_0244 | 100 | 8 | 0 | 0 | 2 | 9 | 18 | 11 | 84 | 13.1 |
| NW_005201234.1_6668-5p | miR90_0193 | 100 | 9 | 0 | 0 | 1 | 9 | 7 | 15 | 41 | 14.2 |
| NW_005201234.1_6668-5p | miR95_0047 | 75 | 16 | 4 | 0 | 3 | 18 | 17 | 2 | 47 | 13.9 |
| NW_005201234.1_6668-5p | miR90_0489 | 78.571 | 14 | 3 | 0 | 4 | 17 | 4 | 17 | 54 | 13.7 |
| NW_005201234.1_6668-5p | miR90_0489 | 78.571 | 14 | 3 | 0 | 4 | 17 | 21 | 8 | 54 | 13.7 |
| NW_005201234.1_6668-5p | miR90_0390 | 78.571 | 14 | 3 | 0 | 4 | 17 | 2 | 15 | 54 | 13.7 |
| NW_005201234.1_6668-5p | miR90_0390 | 78.571 | 14 | 3 | 0 | 4 | 17 | 19 | 6 | 54 | 13.7 |
| NW_005201234.1_6668-5p | miR90_0328 | 78.571 | 14 | 3 | 0 | 4 | 17 | 2 | 15 | 54 | 13.7 |
| NW_005201234.1_6668-5p | Vca_miR90.2 | 78.571 | 14 | 3 | 0 | 3 | 16 | 18 | 5 | 54 | 13.7 |
| NW_005201234.1_6668-5p | miR95_0409 | 83.333 | 12 | 2 | 0 | 7 | 18 | 3 | 14 | 63 | 13.5 |
| NW_005201234.1_6668-5p | Vca_miR82 | 83.333 | 12 | 2 | 0 | 7 | 18 | 4 | 15 | 63 | 13.5 |
| NW_005201234.1_6668-5p | ppu-miR7-5p | 90 | 10 | 1 | 0 | 3 | 12 | 15 | 6 | 73 | 13.3 |
| NW_005201234.1_6668-5p | miR90_0663 | 90 | 10 | 1 | 0 | 9 | 18 | 6 | 15 | 73 | 13.3 |
| NW_005201234.1_6668-5p | miR90_0366 | 90 | 10 | 1 | 0 | 5 | 14 | 10 | 1 | 73 | 13.3 |
| NW_005201234.1_6668-5p | miR95_0482 | 90 | 10 | 1 | 0 | 4 | 13 | 11 | 20 | 73 | 13.3 |
| NW_005201234.1_6668-5p | Vca_miR117 | 90 | 10 | 1 | 0 | 3 | 12 | 16 | 7 | 73 | 13.3 |
| NW_005201234.1_6668-5p | CCMP1516-miR16 | 90 | 10 | 1 | 0 | 9 | 18 | 14 | 5 | 73 | 13.3 |
| NW_005201234.1_6668-5p | SjapMIR58 | 90 | 10 | 1 | 0 | 1 | 10 | 5 | 14 | 73 | 13.3 |
| NW_005201234.1_6668-5p | miR90_0845 | 100 | 8 | 0 | 0 | 9 | 16 | 1 | 8 | 84 | 13.1 |
| NW_005201234.1_6668-5p | miR90_0742 | 70.588 | 17 | 5 | 0 | 2 | 18 | 2 | 18 | 84 | 13.1 |
| NW_005201234.1_6668-5p | miR90_0606 | 100 | 8 | 0 | 0 | 7 | 14 | 8 | 1 | 84 | 13.1 |
| NW_005201234.1_6668-5p | miR90_0251 | 100 | 8 | 0 | 0 | 2 | 9 | 7 | 14 | 84 | 13.1 |
| NW_005201234.1_6668-5p | miR90_0112 | 100 | 8 | 0 | 0 | 2 | 9 | 14 | 21 | 84 | 13.1 |
| NW_005201234.1_6668-5p | miR90_0306 | 73.333 | 15 | 4 | 0 | 4 | 18 | 25 | 11 | 97 | 12.9 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续12
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005201234.1_6668-5p | miR95_0303 | 73.333 | 15 | 4 | 0 | 4 | 18 | 16 | 2 | 97 | 12.9 |
| NW_005201234.1_6668-5p | miR95_0172 | 73.333 | 15 | 4 | 0 | 4 | 18 | 16 | 2 | 97 | 12.9 |
| NW_005202428.1_1248-3p | miR95_0185 | 92.857 | 14 | 1 | 0 | 7 | 20 | 4 | 17 | 5.2 | 17.5 |
| NW_005202428.1_1248-3p | miR95_0185 | 87.5 | 16 | 1 | 1 | 9 | 24 | 3 | 17 | 19 | 15.6 |
| NW_005202428.1_1248-3p | miR90_0642 | 93.333 | 15 | 0 | 1 | 7 | 20 | 1 | 15 | 11 | 16.5 |
| NW_005202428.1_1248-3p | miR90_0642 | 73.684 | 19 | 5 | 0 | 2 | 20 | 19 | 1 | 26 | 15.2 |
| NW_005202428.1_1248-3p | miR90_0642 | 72.222 | 18 | 5 | 0 | 5 | 22 | 3 | 20 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR90_0592 | 93.333 | 15 | 0 | 1 | 7 | 20 | 1 | 15 | 11 | 16.5 |
| NW_005202428.1_1248-3p | miR90_0592 | 73.684 | 19 | 5 | 0 | 2 | 20 | 19 | 1 | 26 | 15.2 |
| NW_005202428.1_1248-3p | miR90_0592 | 72.222 | 18 | 5 | 0 | 5 | 22 | 3 | 20 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR95_0300 | 92.308 | 13 | 1 | 0 | 8 | 20 | 1 | 13 | 11 | 16.5 |
| NW_005202428.1_1248-3p | miR90_0219 | 77.778 | 18 | 4 | 0 | 5 | 22 | 19 | 2 | 15 | 16.1 |
| NW_005202428.1_1248-3p | miR90_0219 | 83.333 | 18 | 2 | 1 | 4 | 20 | 1 | 18 | 17 | 15.8 |
| NW_005202428.1_1248-3p | miR95_0455 | 81.25 | 16 | 3 | 0 | 5 | 20 | 17 | 2 | 17 | 15.8 |
| NW_005202428.1_1248-3p | miR95_0455 | 84.615 | 13 | 2 | 0 | 7 | 19 | 18 | 6 | 40 | 14.6 |
| NW_005202428.1_1248-3p | miR95_0455 | 75 | 16 | 4 | 0 | 5 | 20 | 1 | 16 | 63 | 13.9 |
| NW_005202428.1_1248-3p | miR95_0455 | 75 | 16 | 4 | 0 | 1 | 16 | 3 | 18 | 63 | 13.9 |
| NW_005202428.1_1248-3p | miR95_0001 | 81.25 | 16 | 3 | 0 | 5 | 20 | 3 | 18 | 17 | 15.8 |
| NW_005202428.1_1248-3p | miR90_0576 | 85.714 | 14 | 2 | 0 | 11 | 24 | 18 | 5 | 19 | 15.6 |
| NW_005202428.1_1248-3p | miR90_0557 | 85.714 | 14 | 2 | 0 | 11 | 24 | 15 | 2 | 19 | 15.6 |
| NW_005202428.1_1248-3p | miR90_0274 | 85.714 | 14 | 2 | 0 | 7 | 20 | 1 | 14 | 19 | 15.6 |
| NW_005202428.1_1248-3p | miR90_0274 | 85.714 | 14 | 1 | 1 | 7 | 19 | 6 | 19 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR90_0274 | 68.421 | 19 | 6 | 0 | 1 | 19 | 19 | 1 | 97 | 13.3 |
| NW_005202428.1_1248-3p | Vca_miR89* | 85.714 | 14 | 2 | 0 | 10 | 23 | 20 | 7 | 19 | 15.6 |
| NW_005202428.1_1248-3p | miR90_0636 | 91.667 | 12 | 1 | 0 | 2 | 13 | 8 | 19 | 23 | 15.4 |
| NW_005202428.1_1248-3p | miR90_0636 | 75 | 16 | 4 | 0 | 8 | 23 | 2 | 17 | 63 | 13.9 |
| NW_005202428.1_1248-3p | Vca_miR44c | 91.667 | 12 | 1 | 0 | 12 | 23 | 13 | 2 | 23 | 15.4 |
| NW_005202428.1_1248-3p | Vca_miR44c | 76.471 | 17 | 4 | 0 | 8 | 24 | 20 | 4 | 30 | 15 |
| NW_005202428.1_1248-3p | Vca_miR44b | 91.667 | 12 | 1 | 0 | 12 | 23 | 13 | 2 | 23 | 15.4 |
| NW_005202428.1_1248-3p | Vca_miR44b | 76.471 | 17 | 4 | 0 | 8 | 24 | 20 | 4 | 30 | 15 |
| NW_005202428.1_1248-3p | Vca_miR44a | 91.667 | 12 | 1 | 0 | 12 | 23 | 13 | 2 | 23 | 15.4 |
| NW_005202428.1_1248-3p | Vca_miR44a | 76.471 | 17 | 4 | 0 | 8 | 24 | 20 | 4 | 30 | 15 |
| NW_005202428.1_1248-3p | miR90_0898 | 100 | 10 | 0 | 0 | 11 | 20 | 15 | 6 | 26 | 15.2 |
| NW_005202428.1_1248-3p | miR90_0898 | 85.714 | 14 | 1 | 1 | 11 | 24 | 18 | 6 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR90_0727 | 100 | 10 | 0 | 0 | 11 | 20 | 2 | 11 | 26 | 15.2 |
| NW_005202428.1_1248-3p | miR90_0727 | 78.571 | 14 | 3 | 0 | 2 | 15 | 15 | 2 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR90_0727 | 83.333 | 12 | 2 | 0 | 11 | 22 | 5 | 16 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR90_0657 | 100 | 10 | 0 | 0 | 11 | 20 | 4 | 13 | 26 | 15.2 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续13
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005202428.1_1248-3p | miR90_0657 | 76.471 | 17 | 4 | 0 | 2 | 18 | 17 | 1 | 30 | 15 |
| NW_005202428.1_1248-3p | miR90_0657 | 72.222 | 18 | 5 | 0 | 5 | 22 | 1 | 18 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR90_0657 | 85.714 | 14 | 1 | 1 | 11 | 24 | 1 | 13 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR90_0641 | 100 | 10 | 0 | 0 | 11 | 20 | 4 | 13 | 26 | 15.2 |
| NW_005202428.1_1248-3p | miR90_0641 | 76.471 | 17 | 4 | 0 | 2 | 18 | 17 | 1 | 30 | 15 |
| NW_005202428.1_1248-3p | miR90_0641 | 72.222 | 18 | 5 | 0 | 5 | 22 | 1 | 18 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR90_0641 | 85.714 | 14 | 1 | 1 | 11 | 24 | 1 | 13 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR90_0621 | 100 | 10 | 0 | 0 | 11 | 20 | 4 | 13 | 26 | 15.2 |
| NW_005202428.1_1248-3p | miR90_0621 | 76.471 | 17 | 4 | 0 | 2 | 18 | 17 | 1 | 30 | 15 |
| NW_005202428.1_1248-3p | miR90_0621 | 72.222 | 18 | 5 | 0 | 5 | 22 | 1 | 18 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR90_0621 | 85.714 | 14 | 1 | 1 | 11 | 24 | 1 | 13 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR90_0580 | 100 | 10 | 0 | 0 | 11 | 20 | 3 | 12 | 26 | 15.2 |
| NW_005202428.1_1248-3p | miR90_0580 | 75 | 16 | 4 | 0 | 2 | 17 | 16 | 1 | 63 | 13.9 |
| NW_005202428.1_1248-3p | miR90_0580 | 83.333 | 12 | 2 | 0 | 11 | 22 | 6 | 17 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR95_0044 | 100 | 10 | 0 | 0 | 11 | 20 | 2 | 11 | 26 | 15.2 |
| NW_005202428.1_1248-3p | miR90_0220 | 76.471 | 17 | 4 | 0 | 7 | 23 | 18 | 2 | 30 | 15 |
| NW_005202428.1_1248-3p | miR95_0036 | 76.471 | 17 | 4 | 0 | 2 | 18 | 19 | 3 | 30 | 15 |
| NW_005202428.1_1248-3p | miR90_0701 | 80 | 15 | 3 | 0 | 1 | 15 | 3 | 17 | 35 | 14.8 |
| NW_005202428.1_1248-3p | miR90_0701 | 68.421 | 19 | 6 | 0 | 3 | 21 | 19 | 1 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0451 | 80 | 15 | 3 | 0 | 4 | 18 | 16 | 2 | 35 | 14.8 |
| NW_005202428.1_1248-3p | CCMP1516-miR5 | 80 | 15 | 3 | 0 | 8 | 22 | 15 | 1 | 35 | 14.8 |
| NW_005202428.1_1248-3p | miR90_0908 | 84.615 | 13 | 2 | 0 | 11 | 23 | 1 | 13 | 40 | 14.6 |
| NW_005202428.1_1248-3p | miR90_0887 | 84.615 | 13 | 2 | 0 | 11 | 23 | 2 | 14 | 40 | 14.6 |
| NW_005202428.1_1248-3p | miR90_0756 | 84.615 | 13 | 2 | 0 | 11 | 23 | 4 | 16 | 40 | 14.6 |
| NW_005202428.1_1248-3p | miR90_0747 | 84.615 | 13 | 2 | 0 | 11 | 23 | 5 | 17 | 40 | 14.6 |
| NW_005202428.1_1248-3p | miR90_0706 | 86.667 | 15 | 1 | 1 | 11 | 24 | 3 | 17 | 40 | 14.6 |
| NW_005202428.1_1248-3p | miR90_0696 | 84.615 | 13 | 2 | 0 | 11 | 23 | 17 | 5 | 40 | 14.6 |
| NW_005202428.1_1248-3p | miR90_0203 | 84.615 | 13 | 2 | 0 | 2 | 14 | 16 | 4 | 40 | 14.6 |
| NW_005202428.1_1248-3p | miR90_0182 | 84.615 | 13 | 2 | 0 | 1 | 13 | 13 | 1 | 40 | 14.6 |
| NW_005202428.1_1248-3p | miR95_0327 | 84.615 | 13 | 2 | 0 | 9 | 21 | 5 | 17 | 40 | 14.6 |
| NW_005202428.1_1248-3p | miR95_0327 | 83.333 | 12 | 2 | 0 | 13 | 24 | 6 | 17 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR90_0831 | 90.909 | 11 | 1 | 0 | 14 | 24 | 8 | 18 | 47 | 14.4 |
| NW_005202428.1_1248-3p | miR90_0520 | 90.909 | 11 | 1 | 0 | 10 | 20 | 17 | 7 | 47 | 14.4 |
| NW_005202428.1_1248-3p | miR90_0477 | 90.909 | 11 | 1 | 0 | 13 | 23 | 1 | 11 | 47 | 14.4 |
| NW_005202428.1_1248-3p | miR90_0477 | 90 | 10 | 1 | 0 | 11 | 20 | 2 | 11 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0466 | 90.909 | 11 | 1 | 0 | 12 | 22 | 8 | 18 | 47 | 14.4 |
| NW_005202428.1_1248-3p | miR90_0356 | 90.909 | 11 | 1 | 0 | 12 | 22 | 8 | 18 | 47 | 14.4 |
| NW_005202428.1_1248-3p | miR90_0208 | 90.909 | 11 | 1 | 0 | 1 | 11 | 8 | 18 | 47 | 14.4 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续14
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005202428.1_1248-3p | miR90_0208 | 77.778 | 18 | 3 | 1 | 1 | 17 | 2 | 19 | 63 | 13.9 |
| NW_005202428.1_1248-3p | Vca_miR77 | 90.909 | 11 | 1 | 0 | 13 | 23 | 21 | 11 | 47 | 14.4 |
| NW_005202428.1_1248-3p | miR90_0722 | 100 | 9 | 0 | 0 | 10 | 18 | 18 | 10 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR90_0564 | 100 | 9 | 0 | 0 | 12 | 20 | 10 | 18 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR90_0457 | 72.222 | 18 | 5 | 0 | 1 | 18 | 2 | 19 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR90_0189 | 100 | 9 | 0 | 0 | 12 | 20 | 10 | 18 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR95_0444 | 100 | 9 | 0 | 0 | 11 | 19 | 16 | 8 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR95_0439 | 100 | 9 | 0 | 0 | 12 | 20 | 8 | 16 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR95_0439 | 100 | 9 | 0 | 0 | 11 | 19 | 10 | 18 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR95_0254 | 100 | 9 | 0 | 0 | 12 | 20 | 1 | 9 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR95_0254 | 66.667 | 21 | 7 | 0 | 1 | 21 | 1 | 21 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR95_0176 | 100 | 9 | 0 | 0 | 12 | 20 | 20 | 12 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR95_0118 | 72.222 | 18 | 5 | 0 | 7 | 24 | 18 | 1 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR95_0006 | 100 | 9 | 0 | 0 | 12 | 20 | 1 | 9 | 54 | 14.2 |
| NW_005202428.1_1248-3p | CCMP1516-miR16 | 100 | 9 | 0 | 0 | 1 | 9 | 14 | 6 | 54 | 14.2 |
| NW_005202428.1_1248-3p | SjapMIR71 | 72.222 | 18 | 5 | 0 | 2 | 19 | 21 | 4 | 54 | 14.2 |
| NW_005202428.1_1248-3p | miR90_0644 | 75 | 16 | 4 | 0 | 5 | 20 | 2 | 17 | 63 | 13.9 |
| NW_005202428.1_1248-3p | miR90_0644 | 90 | 10 | 1 | 0 | 2 | 11 | 7 | 16 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0442 | 75 | 16 | 4 | 0 | 5 | 20 | 16 | 1 | 63 | 13.9 |
| NW_005202428.1_1248-3p | miR90_0434 | 75 | 16 | 4 | 0 | 5 | 20 | 17 | 2 | 63 | 13.9 |
| NW_005202428.1_1248-3p | miR90_0434 | 68.421 | 19 | 6 | 0 | 5 | 23 | 1 | 19 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0223 | 77.778 | 18 | 3 | 1 | 4 | 20 | 18 | 1 | 63 | 13.9 |
| NW_005202428.1_1248-3p | miR95_0489 | 75 | 16 | 4 | 0 | 2 | 17 | 6 | 21 | 63 | 13.9 |
| NW_005202428.1_1248-3p | miR95_0415 | 75 | 16 | 4 | 0 | 5 | 20 | 17 | 2 | 63 | 13.9 |
| NW_005202428.1_1248-3p | miR95_0325 | 75 | 16 | 4 | 0 | 5 | 20 | 3 | 18 | 63 | 13.9 |
| NW_005202428.1_1248-3p | miR95_0269 | 75 | 16 | 4 | 0 | 5 | 20 | 4 | 19 | 63 | 13.9 |
| NW_005202428.1_1248-3p | Porphyra | 75 | 16 | 4 | 0 | 9 | 24 | 20 | 5 | 63 | 13.9 |
| NW_005202428.1_1248-3p | Porphyra | 78.571 | 14 | 3 | 0 | 11 | 24 | 15 | 2 | 72 | 13.7 |
| NW_005202428.1_1248-3p | Porphyra | 75 | 16 | 4 | 0 | 6 | 21 | 2 | 17 | 63 | 13.9 |
| NW_005202428.1_1248-3p | Porphyra | 78.571 | 14 | 3 | 0 | 7 | 20 | 15 | 2 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR90_0809 | 78.571 | 14 | 3 | 0 | 5 | 18 | 3 | 16 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR90_0809 | 90 | 10 | 1 | 0 | 11 | 20 | 3 | 12 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0769 | 78.571 | 14 | 3 | 0 | 5 | 18 | 2 | 15 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR90_0769 | 90 | 10 | 1 | 0 | 11 | 20 | 2 | 11 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0754 | 78.571 | 14 | 3 | 0 | 5 | 18 | 18 | 5 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR90_0754 | 90 | 10 | 1 | 0 | 11 | 20 | 18 | 9 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0536 | 78.571 | 14 | 3 | 0 | 5 | 18 | 16 | 3 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR90_0426 | 78.571 | 14 | 3 | 0 | 1 | 14 | 10 | 23 | 72 | 13.7 |
新窗口打开|下载CSV
Supplementary Table 3
附表3
附表3续15
Supplementary Table 3
| 比对其他微藻miRNA的结果 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Query ID (本研究中鉴定的miRNA) | Subject ID (其他已知miRNA) | Identities (%) | Length (nt) | Mismatch | Gap | Query start | Query end | Subject Start | Subject end | E value | Bit score |
| NW_005202428.1_1248-3p | miR90_0141 | 78.571 | 14 | 3 | 0 | 5 | 18 | 17 | 4 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR90_0104 | 78.571 | 14 | 3 | 0 | 5 | 18 | 8 | 21 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR90_0104 | 90 | 10 | 1 | 0 | 11 | 20 | 18 | 9 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR95_0340 | 78.571 | 14 | 3 | 0 | 8 | 21 | 17 | 4 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR95_0292 | 78.571 | 14 | 3 | 0 | 11 | 24 | 1 | 14 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR95_0230 | 78.571 | 14 | 3 | 0 | 6 | 19 | 1 | 14 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR95_0230 | 77.778 | 18 | 2 | 1 | 6 | 21 | 18 | 1 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR95_0004 | 78.571 | 14 | 3 | 0 | 5 | 18 | 17 | 4 | 72 | 13.7 |
| NW_005202428.1_1248-3p | miR90_0794 | 83.333 | 12 | 2 | 0 | 6 | 17 | 15 | 4 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR90_0689 | 83.333 | 12 | 2 | 0 | 11 | 22 | 5 | 16 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR90_0407 | 83.333 | 12 | 2 | 0 | 11 | 22 | 2 | 13 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR90_0215 | 83.333 | 12 | 2 | 0 | 11 | 22 | 1 | 12 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR95_0541 | 83.333 | 12 | 2 | 0 | 4 | 15 | 4 | 15 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR95_0409 | 83.333 | 12 | 2 | 0 | 12 | 23 | 12 | 1 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR95_0194 | 83.333 | 12 | 2 | 0 | 10 | 21 | 2 | 13 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR95_0194 | 90 | 10 | 1 | 0 | 10 | 19 | 11 | 2 | 97 | 13.3 |
| NW_005202428.1_1248-3p | Vca_miR75 | 83.333 | 12 | 2 | 0 | 10 | 21 | 12 | 23 | 84 | 13.5 |
| NW_005202428.1_1248-3p | SjapMIR29 | 83.333 | 12 | 2 | 0 | 3 | 14 | 13 | 2 | 84 | 13.5 |
| NW_005202428.1_1248-3p | miR90_0522 | 90 | 10 | 1 | 0 | 5 | 14 | 6 | 15 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0504 | 90 | 10 | 1 | 0 | 5 | 14 | 8 | 17 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0497 | 90 | 10 | 1 | 0 | 6 | 15 | 15 | 6 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0379 | 90 | 10 | 1 | 0 | 5 | 14 | 12 | 21 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0351 | 90 | 10 | 1 | 0 | 11 | 20 | 16 | 7 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0306 | 90 | 10 | 1 | 0 | 5 | 14 | 7 | 16 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0271 | 90 | 10 | 1 | 0 | 11 | 20 | 15 | 6 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0236 | 90 | 10 | 1 | 0 | 11 | 20 | 17 | 8 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0211 | 90 | 10 | 1 | 0 | 11 | 20 | 12 | 3 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR90_0199 | 90 | 10 | 1 | 0 | 11 | 20 | 1 | 10 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR95_0549 | 90 | 10 | 1 | 0 | 9 | 18 | 10 | 1 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR95_0359 | 90 | 10 | 1 | 0 | 3 | 12 | 9 | 18 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR95_0326 | 90 | 10 | 1 | 0 | 3 | 12 | 9 | 18 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR95_0258 | 68.421 | 19 | 6 | 0 | 3 | 21 | 3 | 21 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR95_0076 | 90 | 10 | 1 | 0 | 11 | 20 | 5 | 14 | 97 | 13.3 |
| NW_005202428.1_1248-3p | miR95_0076 | 90 | 10 | 1 | 0 | 14 | 23 | 5 | 14 | 97 | 13.3 |
| NW_005202428.1_1248-3p | CCMP1516-miR13 | 90 | 10 | 1 | 0 | 1 | 10 | 18 | 9 | 97 | 13.3 |
新窗口打开|下载CSV
Supplementary Table 4
附表4
附表4E. huxleyi BOF92来源的前体miRNA与其他13株颗石藻株基因组比对结果
Supplementary Table 4
| 前体 miRNA ID | 92A (SRX 112489) | EH2 (SRX 112493) | Van556 (SRX 112498) | M217 (SRX 112497) | B39 (SRX 112496) | B11 (SRX 112495) | M219 (SRX 112494) | 12-1 (SRX 112492) | L (SRX 112491) | NZEH (SRX 112490) | 92F (SRX 112499) | 92E (SRX 112488) | 92D (SRX 112487) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NW_005195085.1_38173 | 1.98e-11 | - | - | 2.90e-31 | 1.28e-29 | 1.30e-29 | - | - | 2.41e-25 | - | - | - | - |
| NW_005195085.1_38180 | - | - | - | 1.76e-23 | - | - | - | 9.67e-24 | - | - | - | - | - |
| NW_005196191.1_35170 | - | - | - | 1.07e-15 | - | - | - | - | - | - | - | - | - |
| NW_005196603.1_34298 (ehx-MIR1) | 9.02e-25 | 5.95e-23 | 6.58e-28 | 4.86e-29 | 5.96e-28 | 6.04e-28 | 3.7e-20 | 3.45e-28 | 1.86e-26 | 2.42e-20 | 2.29e-27 | 9.43e-20 | 3.26e-24 |
| NW_005196755.1_34019 | - | - | - | 4.93e-19 | - | 7.93e-17 | - | 2.71e-19 | 5.25e-17 | - | - | 5.68e-17 | 4.24e-18 |
| NW_005196755.1_34028 | 1.16e-28 | 9.96e-21 | 3.08e-21 | 3.76e-30 | 6.00e-23 | 7.87e-22 | 6.14e-23 | 4.50e-22 | 6.74e-21 | 4.02e-23 | 8.29e-22 | 2.03e-21 | 5.45e-22 |
| NW_005196755.1_34052 | 7.02e-21 | - | 2.38e-22 | 2.28e-22 | 2.16e-22 | 1.02e-20 | 2.21e-22 | 1.25e-22 | 1.45e-22 | 6.73e-21 | 1.07e-20 | 2.04e-16 | 1.96e-21 |
| NW_005197715.1_30571 | 1.97e-16 | 2.79e-16 | 1.44e-14 | 2.97e-16 | 2.81e-16 | 2.85e-16 | 2.88e-16 | 1.63e-16 | 1.89e-16 | 1.88e-16 | 3.00e-16 | 2.04e-16 | 1.97e-16 |
| NW_005198391.1_27186 | 3.22e-29 | 3.53e-30 | 8.45e-32 | 8.08e-32 | 7.66e-32 | 7.76e-32 | 7.84e-32 | 7.42e-30 | 3.09e-29 | 3.09e-29 | 1.37e-29 | 2.58e-30 | 3.24e-29 |
| NW_005198966.1_24265 | 9.08e-20 | 1.29e-19 | 1.43e-19 | 1.37e-19 | 1.3e-19 | 1.32e-19 | 1.33e-19 | 7.53e-20 | 8.72e-20 | 8.70e-20 | 1.39e-19 | 9.43e-20 | 9.12e-20 |
| NW_005199363.1_22042 | 3.27e-19 | 4.64e-19 | 1.86e-18 | 4.93e-19 | 4.67e-19 | 4.74e-19 | 4.78e-19 | 2.71e-19 | 3.14e-19 | 3.13e-19 | 6.45e-18 | 3.39e-19 | 3.28e-19 |
| NW_005199378.1_21978 | 1.94e-26 | 2.75e-26 | 3.06e-26 | 2.93e-26 | 2.77e-26 | 2.81e-26 | 2.84e-26 | 1.61e-26 | 1.86e-26 | 1.86e-26 | 2.96e-26 | 2.01e-26 | 9.06e-25 |
| NW_005199975.1_17848 | 2.49e-30 | 1.00e-15 | 3.02e-36 | 6.20e-38 | 5.88e-38 | 5.96e-38 | 2.80e-36 | 1.26e-17 | 1.44e-27 | 1.43e-32 | 1.05e-35 | 1.54e-32 | 1.49e-32 |
| NW_005200282.1_15947 | 5.51e-12 | 3.63e-10 | 8.69e-12 | 8.31e-12 | 7.87e-12 | 7.98e-12 | 8.06e-12 | 4.56e-12 | 5.28e-12 | 5.27e-12 | 8.41e-12 | 5.72e-12 | 5.53e-12 |
| NW_005200461.1_14543 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| NW_005200496.1_14318 (ehx-MIR2) | 4.20e-23 | 1.27e-29 | 6.58e-28 | 1.35e-29 | 2.77e-26 | 2.81e-26 | 6.14e-23 | 4.47e-27 | 1.86e-26 | 4.02e-23 | 1.37e-29 | 4.36e-23 | 3.24e-29 |
| NW_005200517.1_13952 | - | 7.76e-17 | 1.86e-18 | 8.19e-22 | 7.82e-17 | - | 8.00e-17 | 4.53e-17 | 2.44e-15 | 5.24e-17 | 1.79e-18 | - | 1.18e-18 |
| NW_005200535.1_13863 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| NW_005200600.1_13330 (ehx-MIR3) | - | 6.00e-18 | 3.06e-26 | 1.37e-19 | 2.77e-26 | 1.02e-20 | - | 1.61e-26 | - | - | - | 5.68e-17 | - |
| NW_005200881.1_10661 | 7.02e-21 | 9.96e-21 | 1.11e-20 | 1.06e-20 | 1.00e-20 | 1.02e-20 | 1.03e-20 | 5.82e-21 | 6.74e-21 | 6.73e-21 | 1.07e-20 | 7.29e-21 | 7.05e-21 |
| NW_005201234.1_6668 | 1.95e-21 | - | 1.43e-19 | 2.95e-21 | 2.79e-21 | 2.83e-21 | 2.86e-21 | 1.62e-21 | 1.87e-21 | 1.87e-21 | 2.98e-21 | 2.03e-21 | 1.96e-21 |
| NW_005202428.1_1248 | - | - | - | - | 7.87e-12 | - | - | - | - | - | - | - | - |
新窗口打开|下载CSV
Supplementary Table 5
附表5
附表5E. huxleyi BOF92来源的成熟miRNA与其他13株颗石藻基因组的比对结果
Supplementary Table 5
| 成熟 miRNA ID | 92A (SRX 112489) | EH2 (SRX 112493) | Van556 (SRX 112498) | M217 (SRX 112497) | B39 (SRX 112496) | B11 (SRX 112495) | M219 (SRX 112494) | 12-1 (SRX 112492) | L (SRX 112491) | NZEH (SRX 112490) | 92F (SRX 112499) | 92E (SRX 112488) | 92D (SRX 112487) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NW_005195085.1_38173-5p | 71.4% | 81.0% | 76.2% | 100.00% | 100.0% | 100.0% | 81.0% | 71.4% | 100.0% | 71.4% | 71.4% | 66.7% | 66.7% |
| NW_005195085.1_38180-3p | 61.9% | 71.4% | 76.2% | 100.00% | 71.4% | 71.4% | 71.4% | 100.0% | 66.7% | 66.7% | 66.7% | 66.7% | 61.9% |
| NW_005196191.1_35170-3p | 71.4% | 76.2% | 71.4% | 100.00% | 71.4% | 81.0% | 76.2% | 66.7% | 71.4% | 71.4% | 71.4% | 71.4% | 71.4% |
| NW_005196603.1_34298-5p (ehx-miR1-5p) | 89.5% | 89.5% | 94.7% | 100.00% | 94.7% | 89.5% | 94.7% | 89.5% | 89.5% | 89.5% | 89.5% | 89.5% | 89.5% |
| NW_005196755.1_34052-3p | 100.0% | 81.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005196755.1_34028-5p | 95.2% | 71.4% | 66.7% | 100.00% | 100.0% | 66.7% | 76.2% | 76.2% | 66.7% | 66.7% | 71.4% | 66.7% | 66.7% |
| NW_005196755.1_34019-3p | 76.2% | 76.2% | 76.2% | 100.00% | 76.2% | 100.0% | 76.2% | 100.0% | 100.0% | 76.2% | 71.4% | 100.0% | 90.5% |
| NW_005197715.1_30571-5p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005198391.1_27186-5p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005198966.1_24265-3p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005198966.1_24265-5p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005199363.1_22042-5p | 100.0% | 100.0% | 95.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 90.0% | 100.0% | 100.0% |
| NW_005199378.1_21978-3p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005199378.1_21978-5p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005199975.1_17848-3p | 100.0% | 81.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 81.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005200282.1_15947-3p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005200461.1_14543-5p | 100.0% | 81.8% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 86.4% | 100.0% | 100.0% | 86.4% | 100.0% | 86.4% |
| NW_005200496.1_14318-5p (ehx-miR2-5p) | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005200517.1_13952-3p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 90.5% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 81.0% | 100.0% |
| NW_005200535.1_13863-3p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005200600.1_13330-3p (ehx-miR3-3p) | 95.2% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 95.2% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005200881.1_10661-3p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005201234.1_6668-5p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| NW_005202428.1_1248-3p | 100.0% | 100.0% | 100.0% | 100.00% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 83.3% |
新窗口打开|下载CSV
Supplementary Table 6
附表6
附表6EhV99B1来源的成熟miRNA及前体miRNA与其他13株颗石藻病毒基因组的比对结果
Supplementary Table 6
| 前体或成熟 miRNA ID | EhV86 (NC_00 7346) | EhV18 (KF4 81685) | EhV84 (SRX 042587) | EhV88 (SRX 042704) | EhV145 (KF4 81686) | EhV156 (KF4 81687) | EhV164 (KF4 81688) | EhV201 (SRX 042500) | EhV202 (SRX 042425) | EhV203 (SRX 042594) | EhV207 (SRX 042529) | EhV208 (SRX 042474) | EhVPS401 (SRX 042696) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gi|73852470|ref| C_007346.1|_540 | 1.00e-32 | 1.00e-17 | 2.00e-32 | 2.00e-31 | 9.00e-32 | 1.00e-17 | 9.00e-32 | 2.00e-31 | 1.00e-16 | 2.00e-31 | 9.00e-32 | 2.00e-31 | - |
| ehv-miR1-5p | 96.3% | 92.6% | 100.0% | 100.0% | 96.3% | 92.6% | 96.3% | 100.0% | 92.6% | 100.0% | 100.0% | 100.0% | 85.2% |
| ehv-miR2-3p | 100.0% | 47.6% | 100.0% | 100.0% | 100.0% | 47.6% | 100.0% | 100.0% | 52.4% | 100.0% | 100.0% | 100.0% | 61.9% |
新窗口打开|下载CSV
Supplementary Table 7
附表7
附表7宿主或病毒编码的miRNAs及其潜在的病毒靶基因
Supplementary Table 7
| 病毒靶基因ID | NCBI登录号 | 靶基因功能 | miRNA ID | Score | Energy/kCal/Mol |
|---|---|---|---|---|---|
| EhV018 | CAZ69357 | endonuclease | ehx-miR2-5p | 149 | -27.41 |
| EhV022 | CAZ69361 | phosphoglycerate mutase family protein | ehv-miR1-3p | 140 | -23.04 |
| EhV030 | CAZ69369 | DNA polymerase delta catalytic subunit | ehv-miR1-3p | 142 | -26.74 |
| EhV050 | CAZ69386 | serine palmitoyltransferase (vSPT) | ehv-miR1-3p | 146 | -20.96 |
| EhV064 | CAZ69400 | DNA-dependent RNA polymerase II largest subunit | ehx-miR2-5p | 144 | -32.91 |
| EhV085 | CAZ69419 | major capsid protein (mcp) | ehx-miR2-5p | 171 | -37.70 |
| ehx-miR3-3p | 164 | -28.73 | |||
| EhV361 | CAZ69689 | serine protease | ehx-miR2-5p | 140 | -21.53 |
| EhV401 | CAZ69732 | ribonuclease | ehx-miR3-3p | 148 | -29.82 |
| EhV428 | CAZ69758 | ribonucleoside-diphosphate reductase protein | ehx-miR2-5p | 143 | -27.78 |
| EhV434 | CAZ69764 | DNA-directed RNA polymerase II subunit | ehx-miR2-5p | 141 | -25.66 |
| EhV444 | CAZ69774 | DNA topoisomerase | ehx-miR2-5p | 150 | -30.10 |
新窗口打开|下载CSV
Supplementary Table 8
附表8
附表86个靶基因的基本信息
Supplementary Table 8
| 基因名称 | 基因ID | 差异miRNA名称 | miRNA靶基因所属代谢通路 |
|---|---|---|---|
| ACC-1 | 17277436 | ehx-miR1-5p | Fatty acid biosynthesis |
| SPT | 17255778 | ehx-miR2-5p | Sphingolipid metabolism |
| ACOX | 17261797 | ehx-miR3-3p | Fatty acid degradation |
| ACAT | 17283066 | ehx-miR3-3p | Fatty acid degradation |
| CERS | 17255735 | ehv-miR2-5p | Sphingolipid metabolism |
| ACADS | 17256377 | ehv-miR2-5p | Fatty acid degradation |
新窗口打开|下载CSV
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1111/gcb.2018.24.issue-7URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.5194/essd-10-1859-2018URL [本文引用: 1]
DOI:10.3390/v9030061URL [本文引用: 1]
[本文引用: 1]
DOI:10.1105/tpc.114.125641URL [本文引用: 2]
DOI:S0966-842X(16)30069-5PMID:27395772 [本文引用: 1]

Marine viruses are considered to be major ecological, evolutionary, and biogeochemical drivers of the marine environment, responsible for nutrient recycling and determining species composition. Viruses can re-shape their host's metabolic network during infection, generating the virocell-a unique metabolic state that supports their specific requirement. Here we discuss the concept of 'virocell metabolism' and its formation by rewiring of host-encoded metabolic networks, or by introducing virus-encoded auxiliary metabolic genes which provide the virocell with novel metabolic capabilities. The ecological role of marine viruses is commonly assessed by their relative abundance and phylogenetic diversity, lacking the ability to assess the dynamics of active viral infection. The new ability to define a unique metabolic state of the virocell will expand the current virion-centric approaches in order to quantify the impact of marine viruses on microbial food webs.Copyright © 2016. Published by Elsevier Ltd.
DOI:10.1073/pnas.1523168113URL [本文引用: 4]
DOI:10.3354/ame01295URL [本文引用: 2]
DOI:10.1016/j.scitotenv.2019.06.284 [本文引用: 4]

Emiliania huxleyi (Coccolithophore) plays a prominent role in the global carbon cycle and in climate processes. The annual collapse of massive E. huxleyi blooms in the marine environment has been shown to be frequently linked to viral control. These host-virus interactions shape the evolution and dynamics of oceanic microscale ecosystems, yet we still understand little of the molecular mechanism of these virus-mediated processes. Here, we present a detailed characterization of the lipidome of E. huxleyi BOF92 strain, both of uninfected cells and those infected with its specific lytic virus EhV-99B1. Non-targeted lipidomics analysis was performed in order to evaluate the dynamic alterations underlying virus-induced metabolic remodeling. The host lipidome (both lipid content and composition) significantly changed in response to the viral infection. The most statistically significant differential lipids were screened as potential biomarkers for assessing E. huxleyi population sensitivity to EhV infection. Our results reveal that the remodeling of lipid metabolism that underlies the pathogenesis of this infection primarily involved sphingolipid, glycerolipid and fatty acid metabolic pathways. Our study provides insights into how viruses shape their hosts metabolism to support their unique life cycle and a lipid-based chemical arms race during host-virus dynamic interactions in a marine environment. (C) 2019 Published by Elsevier B.V.
DOI:10.1038/s41564-017-0024-3URL [本文引用: 1]
DOI:10.1038/ismej.2015.228PMID:26784355 [本文引用: 1]

The cosmopolitan coccolithophore Emiliania huxleyi is a unicellular eukaryotic alga responsible for vast blooms in the ocean. These blooms have immense impact on large biogeochemical cycles and are terminated by a specific large double-stranded DNA E. huxleyi virus (EhV, Phycodnaviridae). EhV infection is accompanied by induction of hallmarks of programmed cell death and production of reactive oxygen species (ROS). Here we characterized alterations in ROS metabolism and explored its role during infection. Transcriptomic analysis of ROS-related genes predicted an increase in glutathione (GSH) and H2O2 production during infection. In accordance, using biochemical assays and specific fluorescent probes we demonstrated the overproduction of GSH during lytic infection. We also showed that H2O2 production, rather than superoxide, is the predominant ROS during the onset of the lytic phase of infection. Using flow cytometry, confocal microscopy and multispectral imaging flow cytometry, we showed that the profound co-production of H2O2 and GSH occurred in the same subpopulation of cells but at different subcellular localization. Positively stained cells for GSH and H2O2 were highly infected compared with negatively stained cells. Inhibition of ROS production by application of a peroxidase inhibitor or an H2O2 scavenger inhibited host cell death and reduced viral production. We conclude that viral infection induced remodeling of the host antioxidant network that is essential for a successful viral replication cycle. This study provides insight into viral replication strategy and suggests the use of specific cellular markers to identify and quantify the extent of active viral infection during E. huxleyi blooms in the ocean.
DOI:10.1073/pnas.1208895109URL [本文引用: 1]
DOI:10.1073/pnas.0701240104URL [本文引用: 2]
DOI:10.1126/science.1177322URL [本文引用: 1]
DOI:10.1007/s00203-017-1460-4URL [本文引用: 2]
DOI:10.1007/s10142-005-0145-2URL [本文引用: 2]
DOI:10.1097/MOL.0000000000000420URL [本文引用: 2]
DOI:10.1186/1471-2164-15-697URL [本文引用: 1]
DOI:10.1371/journal.pone.0022870URL [本文引用: 2]
DOI:10.1038/ismej.2017.81URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/978-1-62703-302-2_7PMID:23494305 [本文引用: 2]

High-throughput sequencing methods have become invaluable for detection and analysis of small RNAs. The results are millions of sequences that need to be carefully analyzed by computational methods and preferentially verified by different experimental techniques. Here we describe how to use high-throughput sequencing followed by bioinformatics and northern blot to identify one particular class of small RNA, microRNAs.
DOI:10.1093/nar/gkr688PMID:21911355 [本文引用: 1]

microRNAs (miRNAs) are a large class of small non-coding RNAs which post-transcriptionally regulate the expression of a large fraction of all animal genes and are important in a wide range of biological processes. Recent advances in high-throughput sequencing allow miRNA detection at unprecedented sensitivity, but the computational task of accurately identifying the miRNAs in the background of sequenced RNAs remains challenging. For this purpose, we have designed miRDeep2, a substantially improved algorithm which identifies canonical and non-canonical miRNAs such as those derived from transposable elements and informs on high-confidence candidates that are detected in multiple independent samples. Analyzing data from seven animal species representing the major animal clades, miRDeep2 identified miRNAs with an accuracy of 98.6-99.9% and reported hundreds of novel miRNAs. To test the accuracy of miRDeep2, we knocked down the miRNA biogenesis pathway in a human cell line and sequenced small RNAs before and after. The vast majority of the >100 novel miRNAs expressed in this cell line were indeed specifically downregulated, validating most miRDeep2 predictions. Last, a new miRNA expression profiling routine, low time and memory usage and user-friendly interactive graphic output can make miRDeep2 useful to a wide range of researchers.
DOI:10.1093/bioinformatics/btp616URL [本文引用: 1]
DOI:10.1371/journal.pone.0154279URL [本文引用: 6]
DOI:10.1038/nature12221URL [本文引用: 2]
DOI:10.1186/gb-2003-5-1-r1URL [本文引用: 1]
DOI:10.1038/nprot.2008.211PMID:19131956 [本文引用: 1]

DAVID bioinformatics resources consists of an integrated biological knowledgebase and analytic tools aimed at systematically extracting biological meaning from large gene/protein lists. This protocol explains how to use DAVID, a high-throughput and integrated data-mining environment, to analyze gene lists derived from high-throughput genomic experiments. The procedure first requires uploading a gene list containing any number of common gene identifiers followed by analysis using one or more text and pathway-mining tools such as gene functional classification, functional annotation chart or clustering and functional annotation table. By following this protocol, investigators are able to gain an in-depth understanding of the biological themes in lists of genes that are enriched in genome-scale studies.
DOI:10.1111/jam.14958PMID:33275816 [本文引用: 1]

To search for a set of reference genes for reliable gene expression analysis in the globally important marine coccolithophore Emiliania huxleyi-virus model system.Fifteen housekeeping genes (CDKA, CYP15, EFG3, POLAI, RPL30, RPL13, SAMS, COX1, GPB1-2, HSP90, TUA, TUB, UBA1, CAM3 and GAPDH) were evaluated for their stability as potential reference genes for qRT-PCR using ΔCt, geNorm, NormFinder, Bestkeeper and RefFinder software. CDKA, TUA and TUB genes were tested as loading controls for Western blot in the same sample panel. Additionally, target genes associated with cell apoptosis, that is metacaspase genes, were applied to validate the selection of reference genes. The analysis results demonstrated that putative housekeeping genes exhibited significant variations in both mRNA and protein content during virus infection. After a comprehensive analysis with all the algorithms, CDKA and GAPDH were recommended as the most stable reference genes for E huxleyi virus (EhV) infection treatments. For Western blot, significant variation was seen for TUA and TUB, whereas CDKA was stably expressed, consistent with the results of qRT-PCR.CDKA and GAPDH are the best choice for gene and protein expression analysis than the other candidate reference genes under EhV infection conditions.The stable internal control genes identified in this work will help to improve the accuracy and reliability of gene expression analysis and gain insight into complex E. huxleyi-EhV interaction regulatory networks.© 2020 The Society for Applied Microbiology.
PMID:11846609 [本文引用: 1]

The two most commonly used methods to analyze data from real-time, quantitative PCR experiments are absolute quantification and relative quantification. Absolute quantification determines the input copy number, usually by relating the PCR signal to a standard curve. Relative quantification relates the PCR signal of the target transcript in a treatment group to that of another sample such as an untreated control. The 2(-Delta Delta C(T)) method is a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments. The purpose of this report is to present the derivation, assumptions, and applications of the 2(-Delta Delta C(T)) method. In addition, we present the derivation and applications of two variations of the 2(-Delta Delta C(T)) method that may be useful in the analysis of real-time, quantitative PCR data.Copyright 2001 Elsevier Science (USA).
DOI:10.1111/tpj.14369 [本文引用: 1]

MicroRNAs (miRNAs) are major post-transcriptional regulators of gene expression. Their biogenesis relies on the cleavage of longer precursors by a nuclear localized processing machinery. The evolutionary preference of plant miRNAs to silence transcription factors turned these small molecules into key actors during growth and adaptive responses. Furthermore, during their life cycle plants are subject to changes in the environmental conditions surrounding them. In order to face these changes, plants display unique adaptive capacities based on an enormous developmental plasticity, where miRNAs play central roles. Many individual miRNAs have been shown to modulate the plant response to different environmental cues and stresses. In the last few years, increasing evidence has shown that not only individual genes encoding miRNAs but also the miRNA pathway as a whole is subject to regulation in response to external stimulus. In this review, we discuss the current knowledge about the miRNA pathway. We dissect the pathway to analyze the events leading to the generation of these small RNAs and emphasize the regulation of core components of the miRNA biogenesis machinery.
DOI:10.3389/fmicb.2017.00724URL [本文引用: 1]
DOI:10.3389/fmicb.2020.535673URL [本文引用: 1]
DOI:10.1016/S1534-5807(03)00228-4URL [本文引用: 1]
DOI:10.1007/s11427-013-4580-3URL [本文引用: 2]
DOI:10.1093/nar/gkv578PMID:26101255 [本文引用: 2]

There is currently convincing evidence that microRNAs have evolved independently in at least six different eukaryotic lineages: animals, land plants, chlorophyte green algae, demosponges, slime molds and brown algae. MicroRNAs from different lineages are not homologous but some structural features are strongly conserved across the eukaryotic tree allowing the application of stringent criteria to identify novel microRNA loci. A large set of 63 microRNA families was identified in the brown alga Ectocarpus based on mapping of RNA-seq data and nine microRNAs were confirmed by northern blotting. The Ectocarpus microRNAs are highly diverse at the sequence level with few multi-gene families, and do not tend to occur in clusters but exhibit some highly conserved structural features such as the presence of a uracil at the first residue. No homologues of Ectocarpus microRNAs were found in other stramenopile genomes indicating that they emerged late in stramenopile evolution and are perhaps specific to the brown algae. The large number of microRNA loci in Ectocarpus is consistent with the developmental complexity of many brown algal species and supports a proposed link between the emergence and expansion of microRNA regulatory systems and the evolution of complex multicellularity. © The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
DOI:10.1101/gad.1543507URL [本文引用: 3]
PMID:14744438 [本文引用: 1]

MicroRNAs (miRNAs) are endogenous approximately 22 nt RNAs that can play important regulatory roles in animals and plants by targeting mRNAs for cleavage or translational repression. Although they escaped notice until relatively recently, miRNAs comprise one of the more abundant classes of gene regulatory molecules in multicellular organisms and likely influence the output of many protein-coding genes.
DOI:10.1038/nrm3838URL [本文引用: 1]
DOI:10.3724/SP.J.1005.2008.01550URL [本文引用: 1]
DOI:10.3724/SP.J.1005.2008.01550URL [本文引用: 1]
DOI:10.1186/1471-2164-12-337URL [本文引用: 2]
[本文引用: 1]
