 ,, 李梦玮, 王颖, 陈权, 徐寒梅
,, 李梦玮, 王颖, 陈权, 徐寒梅 ,中国药科大学江苏省合成多肽药物发现与评价工程中心,南京 211198
,中国药科大学江苏省合成多肽药物发现与评价工程中心,南京 211198Progress on sORF-encoded micropeptides
Xiangying Chen ,, Mengwei Li, Ying Wang, Quan Chen, Hanmei Xu
,, Mengwei Li, Ying Wang, Quan Chen, Hanmei Xu ,Engineering Research Center of Synthetic Peptide Drug Discovery and Evaluation of Jiangsu Province, China Pharmaceutical University, Nanjing 211198, China
,Engineering Research Center of Synthetic Peptide Drug Discovery and Evaluation of Jiangsu Province, China Pharmaceutical University, Nanjing 211198, China通讯作者: 徐寒梅,博士,教授,研究方向:多肽类药物研究与开发。E-mail:13913925346@126.com
编委: 薛宇
收稿日期:2021-05-8修回日期:2021-07-6网络出版日期:2021-08-20
| 基金资助: |
Received:2021-05-8Revised:2021-07-6Online:2021-08-20
| Fund supported: |
作者简介 About authors
陈相颖,在读硕士研究生,专业方向:微生物与生化药学。E-mail:

摘要
已有的研究表明,生命体中存在着大量的非编码RNA (non-coding RNA, ncRNA),先前被错误注释为ncRNA的分子序列中实际上包含小的开放阅读框(short open reading frame, sORF),部分sORF可转录并翻译成进化保守的微肽(micropeptide),这些sORF由于序列较短和研究技术手段的限制而被忽略。迄今为止,已在生命体中发现一些sORF编码的功能各异的微肽,它们对生命活动的调控起着重要作用。本文对近年来发现的功能性微肽进行综述,介绍了本课题组发现新型微肽MIAC (micropeptide inhibiting actin cytoskeleton)的过程,同时总结了研究潜在微肽的相关技术,以期为研究人员利用相关技术发现新微肽提供借鉴和参考。
关键词:
Abstract
Existing research has shown that there are a large amount of non-coding RNAs (ncRNAs) in organisms. Short open reading frames (sORFs) abundantly exist in molecular sequences inaccurately annotated as ncRNAs. Several sORFs can be transcribed and translated into evolutionarily conserved micropeptides, which were ignored in previous studies due to short sequence lengths and the limitations of research techniques. To date, sORF-encoded micropeptides with various functions have been found to play important roles in regulating vital biological activities. This article reviews the functional micropeptides which have been found in recent years, introduces the new micropeptide designated as MIAC that we have discovered and describes the related technologies for mining potential micropeptides, thereby providing insights and references for new micropeptide discovery for researchers.
Keywords:
PDF (737KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
陈相颖, 李梦玮, 王颖, 陈权, 徐寒梅. 小开放阅读框编码微肽的研究进展. 遗传[J], 2021, 43(8): 737-746 doi:10.16288/j.yczz.21-167
Xiangying Chen.
随着科学技术的发展,人们对于生物复杂性有了更进一步的认识。中心法则指出,遗传信息通常经历由脱氧核苷酸转录到核糖核苷酸再翻译为蛋白质的过程[1]。人类基因组计划证明,人类基因组的3/4能够被转录,但只有约1.5%的基因具有编码蛋白的能力[2]。这就引发了人们对基因组中剩余的大量非蛋白编码基因的思考,这些非蛋白编码基因是否包含更多的遗传信息。DNA元件百科全书(encyclopedia of DNA elements, ENCODE)的数据表明, 80%的基因具有特定的生物学功能,而大部分基因处于非蛋白质编码区域,这部分基因转录产生大量的ncRNA[3]。随着高通量测序技术的发展,越来越多的实验证明ncRNA分子序列上含有小的开放阅读框序列(short open reading frame, sORF),可编码小于100个氨基酸的微小蛋白,被人们称为微肽(micropeptide),加工修饰后的微肽可通过与其他蛋白相互作用而发挥其生理或病生理的作用[4]。研究表明,包括果蝇(Drosophila melanogaster)、小鼠(Mus musculus)、人(Homo spaiens)在内的许多动物基因组中包含数百万个sORF,其中一些具有关键的生理或病生理功能,如钙离子稳态、代谢、成肌细胞融合和肌肉发育、胚胎发育、物质降解、癌症等[5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24]。本文主要介绍了近年来发现的功能性微肽、本课题组发现新型微肽MIAC (micropeptide inhibiting actin cytoskeleton)的过程以及研究潜在微肽的技术,期望为进行相关微肽研究的科研人员提供新思路。
1 sORF和微肽简介
开放阅读框(open reading frame, ORF)最初被定义为起始密码子与终止密码子间的潜在翻译序列[25]。可翻译的ORF通常是指mRNA上的编码序列(coding sequences, CDS),该序列翻译产生具有生物学功能的蛋白质[26]。由于ORF编码蛋白质的可能性随着其长度的增加而增加,查找ORF的算法大多都以300个密码子或100个氨基酸为阈值作为最短的检测长度[27]。sORF在序列长度上区别于ORF,理论上sORF的大小可以从最低限制的2个密码子到100个密码子,sORF由于其极短的长度在最初被认为是非编码的[28]。最近研究发现,真核基因组中存在数百万个sORF序列,并且有些sORF序列可以定位到转录本,这部分sORF具有编码并翻译产生蛋白的能力[28,29]。因此,微肽被定义为长度小于100个氨基酸的蛋白质。根据果蝇和哺乳动物中sORF的位置、大小、保守性和翻译方式等特性,sORF可分为五类(图1):基因间ORF (intergenic ORF)、上游ORF (upstream ORF, uORF)、长非编码ORF (long non-coding ORF, lncORF)、短编码序列(short coding sequence, short CDS)和短同工型ORF (short isoform ORF)。其中,基因间ORF占sORF的96%,但其并不会进行转录和翻译。uORF位于5′端非翻译区(5′ untranslated region, 5′ UTR),具有较低效率的翻译功能并能调节转录本中下游的ORF。lncORF存在于lncRNA (long non-coding RNA)中,与uORF相似具有较低的翻译效率。最近发现,几种lncRNA可编码和翻译为具有生物学功能的微肽,并且在进化中高度保守。短CDS是在单顺反子转录本中发现的,具有与ORF类似的翻译效率,在果蝇和哺乳动物中存在着数百种短CDS。短同工型ORF是sORF中占比最少的一类,由mRNA的选择性剪接产生[4]。
图1
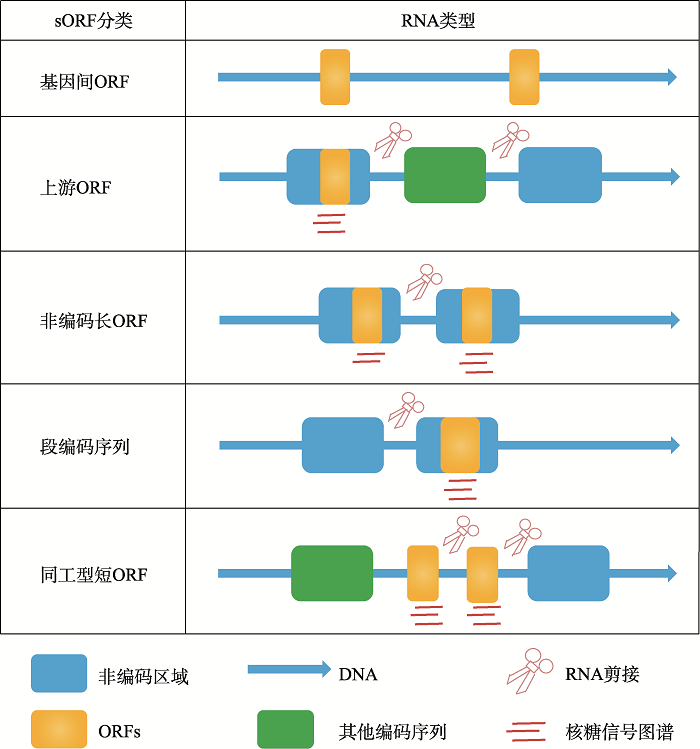 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1sORF的分类
Fig. 1sORF classification
关于微肽的翻译机制,目前有如下几种可能的解释:根据核糖体的扫描模型,mRNA的5′端帽子结构与核糖体40S小亚基结合以复合物形式向3′端扫描,若遇起始密码子,核糖体40S小亚基便与60S大亚基形成80S核糖体,从而介导5′UTR中sORF的翻译,遇到终止密码子时翻译结束,大小亚基解离;而40S小亚基则继续向前扫描,当遇到ORF的起始密码子时重新结合核糖体60S大亚基。第二种可能的机制是只有部分40S小亚基结合在5' UTR中sORF的起始密码子处,另一部分继续向前扫描至ORF的起始密码子,这种机制被称为核糖体的泄漏扫描[30]。但上述两种机制只适用于uORF产生的微肽,关于其他类型微肽的翻译机制还有待研究,还有一种猜想是RNA编辑促成了微肽的翻译,即在转录后将A-C/G/A-G修改为A-U-G[31]。未来可以通过基因敲除RNA编辑的关键酶来研究RNA编辑在RNA水平上产生sORF起始密码子的作用。
2 功能性微肽的发现
基于生物信息学及高通量测序技术对微肽的深入研究,越来越多的微肽被证明在生命活动的许多过程中起着重要的调节作用,包括钙离子稳态、代谢、成肌细胞融合和肌肉发育、胚胎发育、物质降解、癌症等(图2)[5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24]。下面将对近年来功能性微肽的发现进行介绍,并将其总结在表1中。图2
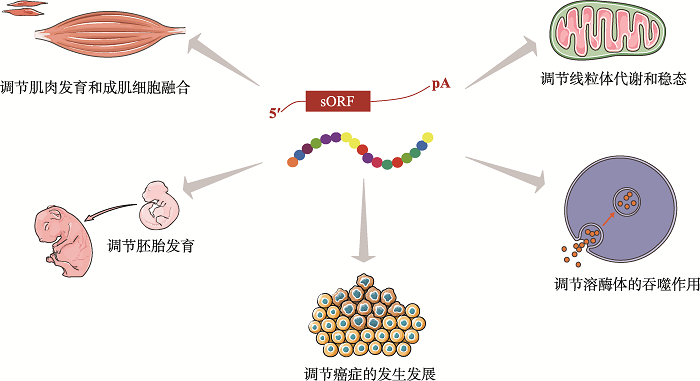 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2微肽的生理与病生理功能
Fig. 2Physiological and Pathophysiological functions of micropeptides
Table 1
表1
表1功能性微肽的发现
Table 1
| 基因 | 微肽 | 长度(氨基酸) | 作用 | 啊啊啊啊 |
|---|---|---|---|---|
| 鼠AK009351、人LINC00948 | MLN | 46 | 抑制SERCA,调节钙离子转运 | [6] |
| 1110017F19Rik/SMIM6 | ELN | 56 | 抑制SERCA,调节钙离子转运 | [5] |
| 1810037I17Rik | ALN | 65 | 抑制SERCA,调节钙离子转运 | [5] |
| 鼠NONMMUG026737、人LOC100507537 | DWORF | 34 | 激活SERCA,调节钙离子转运 | [8] |
| pncr003:2L | Scl | 28/29 | 调节钙离子转运,影响肌肉收缩 | [7] |
| 鼠1500011K16Rik、人LINC00116 | MOXI | 56 | 增强脂肪酸β-氧化作用 | [10] |
| LINC00116 | Mtln | 56 | 增强呼吸效率 | [11] |
| 12S rRNA | MOTS-c | 16 | 调节胰岛素敏感性 | [9] |
| LOC101929726 | Minion | 84 | 促进成肌细胞融合和肌肉发育 | [12] |
| LOC101929726 | Myomixer | 84 | 促进成肌细胞融合和肌肉发育 | [13] |
| LOC100506013 | Toddler | 54 | 激活APJ/Apelin受体促进胚胎发育 | [14] |
| polished rice(pri) | Pri | 11/32 | 促进胚胎发育中的表皮形成 | [15] |
| Tarsal-less(tal) | Tal | 11 | 控制基因表达和组织折叠 | [16] |
| LINC00961 | SPAR | 90 | 抑制mTORC1和肌肉再生 | [17] |
| hemotion | Hemotion | 88 | 促进吞噬细胞吞噬作用 | [18] |
| PIGBOS | PIGBOS | 54 | 调节内质网应激反应 | [19] |
| SMIM22 | CASIMO1 | 83 | 促进乳腺癌 | [20] |
| HOXB-AS3 | HOXB-AS3 | 53 | 抑制结肠癌 | [21] |
| LINC00998 | SMIM30 | 59 | 促进肝癌 | [22] |
| LINC00278 | YY1BM | 21 | 抑制食管鳞状细胞癌 | [23] |
| AC025154.2 | MIAC | 51 | 抑制头颈鳞状细胞癌 | [24] |
| LINC01420 | NoBody | 68 | 促进无义介导的mRNA衰变 | [38] |
| MIR155HG | miPEP155(P155) | 17 | 调节抗原呈递细胞的抗原转运和呈递 | [45] |
新窗口打开|下载CSV
2.1 钙离子稳态相关微肽
Ca2+是肌肉收缩的主要调节因子,控制着肌肉的生长、代谢和病理重塑[32]。美国德克萨斯大学西南医学中心Eric N. Olson实验室的研究结果显示,微肽对于Ca2+稳态的调节起重要作用[5,6,8]。肌调素(myoregulin, MLN)是由骨骼肌特异性lncRNA编码的46个氨基酸的微肽,它可直接与肌浆网Ca2+-ATP酶(sarcoplasmic reticulum Ca2+-ATPase, SERCA)相互作用以降低SERCA对Ca2+的亲和力,从而减少Ca2+摄入肌浆网和肌细胞的收缩性[6]。因此,将MLN鉴定为骨骼肌中的SERCA抑制性微肽。相反,DWORF (dwarf open reading frame)可解除SERCA抑制性微肽的作用而增强肌浆网摄取Ca2+的能力,它是由心肌特异性lncRNA编码的34个氨基酸的微肽[8]。随后,Anderson等[5]在非肌肉细胞中发现两种SERCA抑制性微肽ELN (endoregulin)和ALN (another-regulin),这两种微肽具有与MLN相似的结构和功能,表明Ca2+相关微肽在不同的细胞类型中保守,Ca2+稳态的调节对于许多细胞功能具有重要意义。另外,Magny等[7]在果蝇的pncr003:2L基因中也发现编码SERCA抑制性微肽的序列,该微肽可影响果蝇心肌中Ca2+的运输。跨物种的氨基酸相关序列分析表明,Ca2+相关微肽的结构和功能在果蝇到脊椎动物中具有高度保守性,与其在SERCA中调节Ca2+摄取的生物学功能相关[5,7]。
2.2 线粒体代谢相关微肽
线粒体作为一种功能性细胞器,在新陈代谢及能量供应方面起着重要的作用,大量的研究表明线粒体DNA中也存在sORFs[9,33]。Makarewich等[10]在线粒体内膜中发现由lncRNA编码的微肽MOXI (micropeptide regulator of β-oxidation),MOXI与催化长链脂肪酸氧化的线粒体三功能蛋白(mitochondrial trifunctional protein, MTP)结合,可增强脂肪酸的β氧化作用。Stein等[11]在骨骼肌和心脏中发现由lncRNA LINC00116编码的线粒体跨膜蛋白Mtln (mitoregulin),Mtln作为粘性分子可通过增强线粒体蛋白复合物的装配和稳定性从而提高线粒体的呼吸效率。此外,在线粒体中还发现由12S rRNA编码的16个氨基酸的微肽MOTS-c (mitochondrial open reading frame of the 12S rRNA-c),它可抑制叶酸循环及嘌呤核苷酸的从头合成途径而导致AMPK (AMP-activated protein kinase)活化,从而调节胰岛素的敏感性[9]。这些结果表明,线粒体可通过微肽在细胞和机体水平上主动调控代谢稳态。2.3 成肌细胞融合和肌肉发育相关微肽
骨骼肌的形成需要单核成肌细胞融合形成多核细胞肌管以产生收缩性肌纤维,Myomaker是成肌细胞融合所需的肌肉特异性蛋白[34,35]。最近研究发现,有多种肌肉特异性的微肽在哺乳动物成肌细胞融合的过程中也起着关键作用[12,13]。Zhang等[12]发现一种sORF编码的新微肽Minion (microprotein inducer of fusion),Minion与Myomaker共表达可诱导细胞融合和细胞骨架的快速重排。Myomixer是长为84个氨基酸的肌肉特异性微肽,可促进成肌细胞融合,Myomixer与Myomaker结合还可诱导成纤维细胞间的融合及成纤维细胞和成肌细胞的融合[13]。因此,sORF编码的微肽对于肌肉发育过程中的肌纤维形成具有重要调控作用。2.4 胚胎发育相关微肽
Toddler是在斑马鱼(Danio rerio)中发现的由lncRNA LOC100506013编码的长为58个氨基酸的微肽,研究发现它作为APJ/Apelin受体信号转导的激活剂,可促进原肠胚的形成[14]。先前的研究表明APJ/Apelin受体信号转导在心血管发育和生理调节等多种生物过程中发挥着重要作用[36]。Pauli等[14]发现Toddler功能缺失的斑马鱼没有正常的心脏和血液循环,这些研究表明Toddler在早期胚胎发育过程中是不可或缺的。另外,在果蝇中还发现与胚胎发育相关的微肽[15,16]。Kondo等[15]在果蝇的上皮组织中发现lncRNA polished rice(pri)实际上被转录成多顺反子mRNA,可编码长为11或32个氨基酸的微肽(Pri)。Pri通过调节F-actin在上皮形态的发生中起重要作用,而Pri功能的丧失可完全消除果蝇的表皮结构。Galindo等[16]在果蝇中发现基因tarsal-less(tal)对果蝇的胚胎发育和形态发生至关重要,tal可翻译为短至11个氨基酸的微肽,控制着果蝇的基因表达和组织折叠。这些结果表明,极短的sORF具有翻译功能并在发育过程中具有重要调控作用。
2.5 物质降解相关微肽
近年来,在生物技术的驱动下,与物质降解作用相关的微肽也不断被发现,它们在废物和毒素的降解方面发挥着重要的作用[17,18,19]。SPAR (small regulatory polypeptide of amino acid response)是由lncRNA LINC00961编码的长为90个氨基酸的保守性微肽,定位于晚期内体及溶酶体。SPAR与溶酶体表面v-ATPase复合物的四个亚基相互作用,负性调节mTORC1的活化而抑制肌肉再生[17]。此外,Pueyo等[18]在果蝇中发现一个组织特异性的sORF基因hemotion,其编码果蝇巨噬细胞中长为88个氨基酸的跨膜微肽(Hemotin)。实验研究表明,Hemotin peptide结合并抑制衔接蛋白14-3-3ζ,进而促进磷脂酰肌醇的磷酸化而调节吞噬作用中的内体成熟。并且,研究人员在脊椎动物中还发现Hemotin的功能同源物Stannin,表明这种吞噬作用的新型调节因子具有物种间保守性[18]。
未折叠蛋白反应(unfolded protein response, UPR)是真核细胞内质网(endoplasmic reticulum, ER)中的一个基本过程,在ER中只有正确组装和折叠的蛋白质才能分泌到胞外或展示在细胞表面,而不折叠的蛋白将被内质网相关蛋白所降解[37]。位于线粒体外膜的微肽PIGBOS与ER蛋白CLCC1结合从而调节内质网中的UPR,而PIGBOS的缺失会导致UPR升高和细胞死亡[19]。由此可见,微肽对细胞器间的通讯、体内的平衡以及细胞的存亡至关重要。
2.6 癌症相关微肽
微肽在癌症的发生发展中也具有重要的调控作用[20,21,22,23,24]。CASIMO1 (cancer-associated small integral membrane open reading frame 1)是第一个被发现具有致癌作用的功能性微肽,它与胆固醇合成的关键酶角鲨烯环氧化酶(squalene epoxidase, SQLE)相互作用从而调节癌细胞的代谢稳态,敲低CASIMO1可导致乳腺癌细胞的增殖减少[20]。在结肠癌(colon cancer, CRC)方面,Huang等[21]发现由lncRNA HOXB-AS3编码的长为53个氨基酸的保守微肽(HOXB-AS3)的缺失是CRC代谢中的关键致癌因素,HOXB-AS3能抑制结肠癌的生长。Pang等[22]还发现由lncRNA LINC00998编码的59个氨基酸的微肽SMIM30可通过调节细胞增殖和迁移促进肝癌的发生发展。Wu等[23]通过对281对男性食管鳞状细胞癌(esophageal squamous cell carcinoma, ESCC)组织样本中lncRNA的差异表达分析发现与癌旁组织相比,LINC00278在ESCC组织中显著下调,进一步研究发现LINC00278编码微肽YY1BM (Yin Yang 1 (YY1)-binding micropeptide),YY1BM可与雄激素受体(androgen receptor, AR)结合并下调eEF2K的表达从而导致癌细胞凋亡,由此可见,YY1BM可作为一种潜在的抗癌微肽。另外,Li等[24]证明MIAC能抑制头颈鳞状细胞癌(head and neck squamous cell carcinoma, HNSCC)的生长和转移。此外,还有一些微肽与癌症并不直接相关,例如NoBody (non-annotated P-body dissociating polypeptide)是由LINC01420编码的68个氨基酸的微肽,它与mRNA脱帽蛋白相互作用,促进无义介导的mRNA衰变(nonsense mediated decay, NMD),癌细胞可能利用此过程降解抑制肿瘤的mRNA[38]。总而言之,这些新发现的转录本丰富了肿瘤调控分子,并为癌症的临床诊断和治疗提供新的潜在靶标。3 微肽MIAC的发现
本课题组研究发现,lncRNA AC025154.2可能编码长为51个氨基酸的内源性微肽[24]。在验证这段序列的翻译编码能力时,我们通过体外翻译实验和体内细胞构建实验加以证明,结果表明lncRNA AC025154.2能够编码一种新型微肽,我们将其命名为MIAC。在微肽MIAC的功能研究中,我们构建了MIAC稳定过表达和敲除的CAL27细胞系,实验发现MIAC通过负调控癌细胞的增殖和转移而抑制HNSCC的发生发展。接下来,我们对MIAC抑制HNSCC相关机制做了进一步的研究。通过质谱鉴定与MIAC相互作用的蛋白,并结合50个HNSCC临床样本和50个正常样本中蛋白的表达情况,最后聚焦于其中的这三种蛋白:水通道蛋白2 (aquaporin 2, AQP2)、ITGB4 (integrin beta 4)和SEPT2 (septin 2)。进一步的机制探究表明MIAC直接与AQP2相互作用,通过调控SEPT2/ITGB4抑制骨架蛋白重排,最终抑制HNSCC的生长和转移。由此可见,MIAC在HNSCC中具有调控作用,为开发治疗HNSCC的药物提供新的研究方向,而AQP2作为MIAC的作用靶点对于研究HNSCC的药物同样存在重要意义。
为进一步探究MIAC的临床和治疗意义,我们分析TCGA数据库中500个HNSCC临床样本和44个正常样本中MIAC的相对表达情况,发现MIAC在HNSCC中呈下调趋势,并且MIAC表达水平的降低与HNSCC患者的整体生存率差呈正相关。我们进一步分析94对HNSCC临床样本中MIAC的相对表达量,分析结果也与数据库中的情况一致,相比于在正常样本中,MIAC在HNSCC样本中的表达量降低。因此,MIAC是由lncRNA AC025154.2编码的一种新型内源性微肽,对于HNSCC的发生发展起着重要的调控作用。
创新型药物作为自主研发和具有自主知识产权的药物,对于我国建设创新型国家的进一步发展具有重要意义。MIAC作为HNSCC的调控分子,在创新型HNSCC药物的开发中具有重大的研究意义:MIAC可作为潜在诊断标志物来制备诊断HNSCC的试剂盒,为HNSCC的诊断和预防提供新的途径;而MIAC作为调控HNSCC的小分子多肽,也可通过偶联化学药物的方式来提高治疗HNSCC药物的靶向性和稳定性。此外,MIAC在其他肿瘤和疾病中的意义还有待探究。
4 研究潜在微肽的相关技术
当前的研究表明,在动物基因组中约1.2%的sORF可被转录,其中只有约1/3能被翻译[4]。这些占比很小的功能性sORF理论上也可产生成千上万个未被表征的微肽,即使这些微肽中只有小部分具有生物活性,仍意味着可能存在数百甚至数千种有生物学功能的微肽。因此,当前面临的挑战是如何识别具有生物活性的sORF及其微肽。下面将总结介绍研究潜在微肽的相关技术,这些技术可用于鉴定可能编码微肽的sORF。4.1 生物信息学分析
生物信息学(bioinformatics)是利用生物数据来开发算法和软件的交叉学科,目前运用生物信息分析技术,基于保守序列可从非编码区域预测具有编码蛋白能力的sORFs,生物信息分析技术还依据sORFs序列中的密码子含量和编码特征以区分sORFs编码区与非编码区[39]。我们可以利用生物信息数据库挖掘相关数据,如ATCG、UCSC等,而常用于预测sORFs的分析软件有CPAT、ORFfinder、PhyloCSF、uPEPperoni[40,41,42,43,44]等。Niu等[45]运用ORFfinder在人源MIR155HG基因中预测到一条54个碱基的sORF,后续实验证明该sORF可编码长为17个氨基酸的功能性微肽miPEP155。miPEP155可调节抗原呈递细胞(antigen-presenting cells, APC)中的抗原转运和呈递,可作为自身免疫性疾病的候选药物[45]。4.2 核糖体图谱分析
核糖体图谱分析(ribosome profiling)可用来识别具有翻译潜力的sORF,该技术的原理是翻译核糖体可保护长为20~30个核苷酸的mRNA片段免受核酸酶的消化[46]。然而,Wilson等[47]的研究表明某些sORF虽然与核糖体结合但并不进行翻译。于是,在Ribo-Seq的基础上改良而开发了多聚核糖体分析(Poly-Ribo-Seq),使用这种技术可以分离由多个核糖体结合并被主动翻译的mRNA,由此可将不进行翻译的单核糖体-mRNA复合物区分开[48]。此外,Guttman等[49]还开发了核糖体释放分数(ribosome release score, RRS)作为翻译的度量指标,相比于终止密码子下游的非编码区,编码区与核糖体具有更高的相关性,由此可区分编码转录本和非编码转录本。Chen等[26]利用核糖体图谱分析发现了3455个非经典CDS,其中的96%是编码小于100个氨基酸的微肽。4.3 质谱和蛋白质组学
最近基于质谱(mass spectrometry, MS)的蛋白质组学(proteomics)也用于发现和验证内源表达的微肽。该技术的基本原理是通过测量气态的离子化肽或蛋白质的质荷比来研究蛋白质的表达和相互作用,因此MS通过检测从sORF翻译的微肽,从而直接验证转录产物编码蛋白质的潜力[50]。基于MS的蛋白质组学在研究和鉴定新型微肽方面已取得实质性的进展,Chen等[26]通过基于MS的HLA-I肽组学,发现240个微肽可被HLA-I提呈,表明这些肽会进入HLA-I呈递途径并可能拥有免疫原性。但MS在技术上仍然存在一定限制,样品制备过程中的消化酶决定微肽片段化的方式,片段过小不能产生足够的检测信号,片段过大则无法用于MS分析,小片段的微肽在样品制备过程中还存在丢失的可能[39]。因此,需要进一步结合核糖体图谱分析等其他分析方法以确定新型微肽的存在。4.4 蛋白质基因组学
蛋白质基因组学(proteogenomics)是在基于蛋白质组学分析的基础上结合基因组学和转录组学的分析方法,通过追溯基因组和转录本中的蛋白质/微肽的预测序列,来鉴定基因的翻译和表达情况[51]。在蛋白质基因组学研究中,Slavoff等[31]从人白血病细胞系K562细胞中发现了86个未报道过的微肽。4.5 其他相关技术
为证实sORF是否具有编码蛋白产生微肽的能力,可以使用以下几种方法来进行验证。在理想的状态下,可以设计目的微肽的抗体并通过免疫组化或蛋白质印记来验证其特异性[52]。例如,Li等[24]通过制备MIAC的单克隆抗体以检测MIAC的内源表达。对于不能产生抗体的目的微肽而言,也可采用CRISPR/Cas9基因编辑技术。该技术通过同源定向修复将FLAG/MYC或其他标签添加到预测的sORF,从而产生融合蛋白,再通过检测融合蛋白以验证目的微肽的存在[52]。为确定CASIMO1转录本中的sORF是否翻译为微肽,Polycarpou-Schwarz等[20]在CASIMO1编码序列的C端插入了一个Flag标签,并通过anti-Flag抗体检测到了CASIMO1-Flag的表达。此外,还可通过体外翻译来评估sORF编码蛋白的能力,通过多方面的验证以确定sORF是否具有编码能力。5 结语与展望
大规模基因组测序的迅速发展促进人们对基因组的深入研究,揭示sORF序列的复杂性。微肽的发现使人们认识到这些重要小肽的生物学作用,它们在生命活动及疾病的发展进程中起着重要调控作用。微肽可以以配体或信号分子的形式发挥作用,也可与其他蛋白质相结合,通过遮蔽受体蛋白的关键位点或影响受体蛋白的活性从而发挥调控作用,如前所述的HOXB-AS3 peptide[21]通过竞争性结合hnRNP A1中RGG基序的精氨酸残基,阻断精氨酸残基与丙酮酸激酶M(pyruvate kinase M, PKM)的结合,从而抑制结肠癌细胞的葡萄糖代谢进程。SMIM30[22]与非受体酪氨酸激酶SRC/YES1结合,驱动其膜锚定和磷酸化,激活下游丝裂原活化蛋白激酶(mitogen-activated protein kinase, MAPK)信号通路,通过调节细胞增殖和迁移促进肝癌的发生发展。然而,对于功能性微肽及其作用机制的探索仍处于起步状态,虽然已经存在许多挖掘未知微肽的生物技术,但由于微肽本身分子量小、表达丰度低等特点,这些生物技术的应用仍然存在局限性,生命体中仍有大量的微肽等待被发现。相信在未来的研究中,能克服检测障碍,进一步拓展和优化挖掘微肽的技术与方法。另一方面,还需要进行大量的工作以阐明微肽的生物学作用,并对其作用机制开展进一步的研究,以便应用于正常生理功能的探索及疾病的临床诊疗。
(责任编委: 薛宇)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1038/227561a0URL [本文引用: 1]
DOI:10.1038/nature11233URL [本文引用: 1]
DOI:10.1038/nature11247URL [本文引用: 1]
DOI:10.1038/nrm.2017.58URL [本文引用: 3]
DOI:10.1126/scisignal.aaj1460URL [本文引用: 5]
DOI:S0092-8674(15)00010-0PMID:25640239 [本文引用: 4]

Functional micropeptides can be concealed within RNAs that appear to be noncoding. We discovered a conserved micropeptide, which we named myoregulin (MLN), encoded by a skeletal muscle-specific RNA annotated as a putative long noncoding RNA. MLN shares structural and functional similarity with phospholamban (PLN) and sarcolipin (SLN), which inhibit SERCA, the membrane pump that controls muscle relaxation by regulating Ca(2+) uptake into the sarcoplasmic reticulum (SR). MLN interacts directly with SERCA and impedes Ca(2+) uptake into the SR. In contrast to PLN and SLN, which are expressed in cardiac and slow skeletal muscle in mice, MLN is robustly expressed in all skeletal muscle. Genetic deletion of MLN in mice enhances Ca(2+) handling in skeletal muscle and improves exercise performance. These findings identify MLN as an important regulator of skeletal muscle physiology and highlight the possibility that additional micropeptides are encoded in the many RNAs currently annotated as noncoding. Copyright © 2015 Elsevier Inc. All rights reserved.
DOI:10.1126/science.1238802URL [本文引用: 4]
DOI:10.1126/science.aad4076URL [本文引用: 4]
DOI:10.1016/j.cmet.2015.02.009URL [本文引用: 4]
DOI:S2211-1247(18)30822-2PMID:29949755 [本文引用: 3]

Micropeptide regulator of β-oxidation (MOXI) is a conserved muscle-enriched protein encoded by an RNA transcript misannotated as non-coding. MOXI localizes to the inner mitochondrial membrane where it associates with the mitochondrial trifunctional protein, an enzyme complex that plays a critical role in fatty acid β-oxidation. Isolated heart and skeletal muscle mitochondria from MOXI knockout mice exhibit a diminished ability to metabolize fatty acids, while transgenic MOXI overexpression leads to enhanced β-oxidation. Additionally, hearts from MOXI knockout mice preferentially oxidize carbohydrates over fatty acids in an isolated perfused heart system compared to wild-type (WT) animals. MOXI knockout mice also exhibit a profound reduction in exercise capacity, highlighting the role of MOXI in metabolic control. The functional characterization of MOXI provides insight into the regulation of mitochondrial metabolism and energy homeostasis and underscores the regulatory potential of additional micropeptides that have yet to be identified.Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
DOI:10.1016/j.celrep.2018.06.002URL [本文引用: 3]
DOI:10.1038/ncomms15664PMID:28569745 [本文引用: 4]

Although recent evidence has pointed to the existence of small open reading frame (smORF)-encoded microproteins in mammals, their function remains to be determined. Skeletal muscle development requires fusion of mononuclear progenitors to form multinucleated myotubes, a critical but poorly understood process. Here we report the identification of Minion (microprotein inducer of fusion), a smORF encoding an essential skeletal muscle specific microprotein. Myogenic progenitors lacking Minion differentiate normally but fail to form syncytial myotubes, and Minion-deficient mice die perinatally and demonstrate a marked reduction in fused muscle fibres. The fusogenic activity of Minion is conserved in the human orthologue, and co-expression of Minion and the transmembrane protein Myomaker is sufficient to induce cellular fusion accompanied by rapid cytoskeletal rearrangement, even in non-muscle cells. These findings establish Minion as a novel microprotein required for muscle development, and define a two-component programme for the induction of mammalian cell fusion. Moreover, these data also significantly expand the known functions of smORF-encoded microproteins.
DOI:10.1126/science.aam9361URL [本文引用: 4]
DOI:10.1126/science.1248636URL [本文引用: 4]
DOI:10.1038/ncb1595URL [本文引用: 4]
DOI:10.1371/journal.pbio.0050106URL [本文引用: 4]
DOI:10.1038/nature21034URL [本文引用: 4]
DOI:10.1371/journal.pbio.1002395URL [本文引用: 5]
DOI:10.1038/s41467-019-12816-zURL [本文引用: 4]
DOI:10.1038/s41388-018-0281-5PMID:29765154 [本文引用: 5]

Breast cancer is a leading cause of cancer-related death in women. Small open reading frame (sORF)-encoded proteins or microproteins constitute a new class of molecules often transcribed from presumed long non-coding RNA transcripts (lncRNAs). The translation of some of these sORFs has been confirmed, but their cellular function and importance remains largely unknown. Here, we report the identification and characterization of a novel microprotein of 10?kDa, which we named Cancer-Associated Small Integral Membrane Open reading frame 1 (CASIMO1). CASIMO1 RNA is overexpressed predominantly in hormone receptor-positive breast tumors. Its knockdown leads to decreased proliferation in multiple breast cancer cell lines. Its loss disturbs the organization of the actin cytoskeleton, leads to inhibition of cell motility, and causes a G/G cell cycle arrest. The proliferation phenotype upon overexpression is observed only with CASIMO1 protein expression, but not with a non-translatable mutant attributing the effects to the sORF-derived protein rather than a lncRNA function. CASIMO1 microprotein interacts with squalene epoxidase (SQLE), a key enzyme in cholesterol synthesis and a known oncogene in breast cancer. Overexpression of CASIMO1 leads to SQLE protein accumulation without affecting its RNA levels and increased lipid droplet clustering, while knockdown of CASIMO1 decreased SQLE protein abundance and ERK phosphorylation downstream of SQLE. Importantly, SQLE knockdown mimicked the CASIMO1 knockdown phenotype and in turn SQLE overexpression fully rescued the effect of CASIMO1 knockdown. These findings establish CASIMO1 as the first functional microprotein that plays a role in carcinogenesis and is implicated in the cell lipid homeostasis.
DOI:10.1016/j.molcel.2017.09.015URL [本文引用: 5]
DOI:10.1016/j.jhep.2020.05.028URL [本文引用: 5]
DOI:10.1158/0008-5472.CAN-19-3440URL [本文引用: 4]
DOI:10.1021/jacs.0c00706URL [本文引用: 6]
DOI:10.1016/j.tig.2017.12.009URL [本文引用: 1]
DOI:10.1126/science.aay0262PMID:32139545 [本文引用: 3]

Ribosome profiling has revealed pervasive but largely uncharacterized translation outside of canonical coding sequences (CDSs). In this work, we exploit a systematic CRISPR-based screening strategy to identify hundreds of noncanonical CDSs that are essential for cellular growth and whose disruption elicits specific, robust transcriptomic and phenotypic changes in human cells. Functional characterization of the encoded microproteins reveals distinct cellular localizations, specific protein binding partners, and hundreds of microproteins that are presented by the human leukocyte antigen system. We find multiple microproteins encoded in upstream open reading frames, which form stable complexes with the main, canonical protein encoded on the same messenger RNA, thereby revealing the use of functional bicistronic operons in mammals. Together, our results point to a family of functional human microproteins that play critical and diverse cellular roles.Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
DOI:10.1038/s41586-018-0794-7URL [本文引用: 1]
DOI:10.1093/nar/gkz734URL [本文引用: 2]
DOI:10.1186/gb-2011-12-11-r118URL [本文引用: 1]
DOI:10.1038/nrg3520PMID:24514441 [本文引用: 1]

Short open reading frames (sORFs) are a common feature of all genomes, but their coding potential has mostly been disregarded, partly because of the difficulty in determining whether these sequences are translated. Recent innovations in computing, proteomics and high-throughput analyses of translation start sites have begun to address this challenge and have identified hundreds of putative coding sORFs. The translation of some of these has been confirmed, although the contribution of their peptide products to cellular functions remains largely unknown. This Review examines this hitherto overlooked component of the proteome and considers potential roles for sORF-encoded peptides.
DOI:10.1038/nchembio.1120PMID:23160002 [本文引用: 2]

The complete extent to which the human genome is translated into polypeptides is of fundamental importance. We report a peptidomic strategy to detect short open reading frame (sORF)-encoded polypeptides (SEPs) in human cells. We identify 90 SEPs, 86 of which are previously uncharacterized, which is the largest number of human SEPs ever reported. SEP abundances range from 10-1,000 molecules per cell, identical to abundances of known proteins. SEPs arise from sORFs in noncoding RNAs as well as multicistronic mRNAs, and many SEPs initiate with non-AUG start codons, indicating that noncanonical translation may be more widespread in mammals than previously thought. In addition, coding sORFs are present in a small fraction (8 out of 1,866) of long intergenic noncoding RNAs. Together, these results provide strong evidence that the human proteome is more complex than previously appreciated.
DOI:10.1152/ajpcell.00285.2015URL [本文引用: 1]
DOI:10.3389/fgene.2020.00095URL [本文引用: 1]
DOI:10.1101/cshperspect.a029298URL [本文引用: 1]
DOI:10.1073/pnas.1600101113URL [本文引用: 1]
DOI:10.1124/pr.119.017533PMID:31492821 [本文引用: 1]

The predicted protein encoded by the APJ gene discovered in 1993 was originally classified as a class A G protein-coupled orphan receptor but was subsequently paired with a novel peptide ligand, apelin-36 in 1998. Substantial research identified a family of shorter peptides activating the apelin receptor, including apelin-17, apelin-13, and [Pyr]apelin-13, with the latter peptide predominating in human plasma and cardiovascular system. A range of pharmacological tools have been developed, including radiolabeled ligands, analogs with improved plasma stability, peptides, and small molecules including biased agonists and antagonists, leading to the recommendation that the APJ gene be renamed APLNR and encode the apelin receptor protein. Recently, a second endogenous ligand has been identified and called Elabela/Toddler, a 54-amino acid peptide originally identified in the genomes of fish and humans but misclassified as noncoding. This precursor is also able to be cleaved to shorter sequences (32, 21, and 11 amino acids), and all are able to activate the apelin receptor and are blocked by apelin receptor antagonists. This review summarizes the pharmacology of these ligands and the apelin receptor, highlights the emerging physiologic and pathophysiological roles in a number of diseases, and recommends that Elabela/Toddler is a second endogenous peptide ligand of the apelin receptor protein.Copyright © 2019 by The Author(s).
DOI:10.1126/science.1209038PMID:22116877 [本文引用: 1]

The vast majority of proteins that a cell secretes or displays on its surface first enter the endoplasmic reticulum (ER), where they fold and assemble. Only properly assembled proteins advance from the ER to the cell surface. To ascertain fidelity in protein folding, cells regulate the protein-folding capacity in the ER according to need. The ER responds to the burden of unfolded proteins in its lumen (ER stress) by activating intracellular signal transduction pathways, collectively termed the unfolded protein response (UPR). Together, at least three mechanistically distinct branches of the UPR regulate the expression of numerous genes that maintain homeostasis in the ER or induce apoptosis if ER stress remains unmitigated. Recent advances shed light on mechanistic complexities and on the role of the UPR in numerous diseases.
DOI:10.1038/nchembio.2249PMID:27918561 [本文引用: 1]

Proteomic detection of non-annotated microproteins indicates the translation of hundreds of small open reading frames (smORFs) in human cells, but whether these microproteins are functional or not is unknown. Here, we report the discovery and characterization of a 7-kDa human microprotein we named non-annotated P-body dissociating polypeptide (NoBody). NoBody interacts with mRNA decapping proteins, which remove the 5' cap from mRNAs to promote 5'-to-3' decay. Decapping proteins participate in mRNA turnover and nonsense-mediated decay (NMD). NoBody localizes to mRNA-decay-associated RNA-protein granules called P-bodies. Modulation of NoBody levels reveals that its abundance is anticorrelated with cellular P-body numbers and alters the steady-state levels of a cellular NMD substrate. These results implicate NoBody as a novel component of the mRNA decapping complex and demonstrate potential functionality of a newly discovered microprotein.
DOI:S0962-8924(17)30064-8PMID:28528987 [本文引用: 2]

Advances in computational biology and large-scale transcriptome analyses have revealed that a much larger portion of the genome is transcribed than was previously recognized, resulting in the production of a diverse population of RNA molecules with both protein-coding and noncoding potential. Emerging evidence indicates that several RNA molecules have been mis-annotated as noncoding and in fact harbor short open reading frames (sORFs) that encode functional peptides and that have evaded detection until now due to their small size. sORF-encoded peptides (SEPs), or micropeptides, have been shown to have important roles in fundamental biological processes and in the maintenance of cellular homeostasis. These small proteins can act independently, for example as ligands or signaling molecules, or they can exert their biological functions by engaging with and modulating larger regulatory proteins. Given their small size, micropeptides may be uniquely suited to fine-tune complex biological systems.Copyright © 2017 Elsevier Ltd. All rights reserved.
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/nar/gkt006URL [本文引用: 1]
DOI:10.1093/bioinformatics/btp688URL [本文引用: 1]
DOI:10.1093/bioinformatics/btr209URL [本文引用: 1]
DOI:10.1186/1471-2105-15-36PMID:24484385 [本文引用: 1]

Background: Several small open reading frames located within the 5' untranslated regions of mRNAs have recently been shown to be translated. In humans, about 50% of mRNAs contain at least one upstream open reading frame representing a large resource of coding potential. We propose that some upstream open reading frames encode peptides that are functional and contribute to proteome complexity in humans and other organisms. We use the term uPEPs to describe peptides encoded by upstream open reading frames. Results: We have developed an online tool, termed uPEPperoni, to facilitate the identification of putative bioactive peptides. uPEPperoni detects conserved upstream open reading frames in eukaryotic transcripts by comparing query nucleotide sequences against mRNA sequences within the NCBI RefSeq database. The algorithm first locates the main coding sequence and then searches for open reading frames 5' to the main start codon which are subsequently analysed for conservation. uPEPperoni also determines the substitution frequency for both the upstream open reading frames and the main coding sequence. In addition, the uPEPperoni tool produces sequence identity heatmaps which allow rapid visual inspection of conserved regions in paired mRNAs. Conclusions: uPEPperoni features user-nominated settings including, nucleotide match/mismatch, gap penalties, Ka/Ks ratios and output mode. The heatmap output shows levels of identity between any two sequences and provides easy recognition of conserved regions. Furthermore, this web tool allows comparison of evolutionary pressures acting on the upstream open reading frame against other regions of the mRNA. Additionally, the heatmap web applet can also be used to visualise the degree of conservation in any pair of sequences. uPEPperoni is freely available on an interactive web server at http://upep-scmb. biosci. uq. edu. au.
[本文引用: 2]
DOI:10.1016/j.cell.2016.02.066URL [本文引用: 1]
DOI:10.1093/gbe/evr099URL [本文引用: 1]
DOI:10.7554/eLife.03528URL [本文引用: 1]
DOI:10.1016/j.cell.2013.06.009URL [本文引用: 1]
DOI:10.1038/nature19949URL [本文引用: 1]
DOI:10.1002/mas.21483PMID:26670565 [本文引用: 1]

Proteogenomics is a research area that combines areas as proteomics and genomics in a multi-omics setup using both mass spectrometry and high-throughput sequencing technologies. Currently, the main goals of the field are to aid genome annotation or to unravel the proteome complexity. Mass spectrometry based identifications of matching or homologues peptides can further refine gene models. Also, the identification of novel proteoforms is also made possible based on detection of novel translation initiation sites (cognate or near-cognate), novel transcript isoforms, sequence variation or novel (small) open reading frames in intergenic or un-translated genic regions by analyzing high-throughput sequencing data from RNAseq or ribosome profiling experiments. Other proteogenomics studies using a combination of proteomics and genomics techniques focus on antibody sequencing, the identification of immunogenic peptides or venom peptides. Over the years, a growing amount of bioinformatics tools and databases became available to help streamlining these cross-omics studies. Some of these solutions only help in specific steps of the proteogenomics studies, e.g. building custom sequence databases (based on next generation sequencing output) for mass spectrometry fragmentation spectrum matching. Over the last few years a handful integrative tools also became available that can execute complete proteogenomics analyses. Some of these are presented as stand-alone solutions, whereas others are implemented in a web-based framework such as Galaxy. In this review we aimed at sketching a comprehensive overview of all the bioinformatics solutions that are available for this growing research area. © 2015 Wiley Periodicals, Inc. Mass Spec Rev 36:584-599, 2017.© 2015 Wiley Periodicals, Inc.
DOI:10.1016/j.bbagrm.2015.07.017PMID:26265145 [本文引用: 2]

Long noncoding RNAs (lncRNAs) are a diverse class of RNAs with increasingly appreciated functions in vertebrates, yet much of their biology remains poorly understood. In particular, it is unclear to what extent the current catalog of over 10,000 annotated lncRNAs is indeed devoid of genes coding for proteins. Here we review the available computational and experimental schemes for distinguishing between coding and noncoding transcripts and assess the conclusions from their recent genome-wide applications. We conclude that the model most consistent with the available data is that a large number of mammalian lncRNAs undergo translation, but only a very small minority of such translation events results in stable and functional peptides. The outcomes of the majority of the translation events and their potential biological purposes remain an intriguing topic for future investigation. This article is part of a Special Issue entitled: Clues to long noncoding RNA taxonomy1, edited by Dr. Tetsuro Hirose and Dr. Shinichi Nakagawa. Copyright © 2015 Elsevier B.V. All rights reserved.
DOI:10.1080/15476286.2017.1391443PMID:29028415

Cancer is characterized by multiple genetic and epigenetic alterations, including a higher prevalence of mutations of oncogenes and/or tumor suppressors. Mounting evidences have shown that noncoding RNAs (ncRNAs) are involved in the epigenetic regulation of cancer genes and their associated pathways. The clustered regularly interspaced short palindromic repeats (CRISPR)-associated nuclease 9 (CRISPR/Cas9) system, a revolutionary genome-editing technology, has shed light on ncRNA-based cancer therapy. Here, we briefly introduce the classifications and mechanisms of CRISPR/Cas9 system. Importantly, we mainly focused on the applications of CRISPR/Cas9 system as a molecular tool for ncRNA (microRNA, long noncoding RNA and circular RNA, etc.) editing in human cancers, and the novel techniques that are based on CRISPR/Cas9 system. Additionally, the off-target effects and the corresponding solutions as well as the challenges toward CRISPR/Cas9 were also evaluated and discussed. Long- and short-ncRNAs have been employed as targets in precision oncology, and CRISPR/Cas9-mediated ncRNA editing may provide an excellent way to cure cancer.
