Advances in genome-wide association study of chronic obstructive pulmonary disease
Guoqing Qian1,2,3责任编辑: 周钢桥
收稿日期:2020-02-25修回日期:2020-05-29网络出版日期:2020-09-20
| 基金资助: |
Received:2020-02-25Revised:2020-05-29Online:2020-09-20
| Fund supported: |
作者简介 About authors
钱国清,博士,副主任医师,研究方向:呼吸病学。E-mail:

摘要
慢性阻塞性肺疾病(chronic obstructive pulmonary disease, COPD)是一种以不完全可逆的气流受限为主要特征的慢性气道炎症,是一种由遗传因素和环境因素共同作用的复杂疾病,也是世界主要致死疾病之一。近年来,随着全基因组关联研究(genome-wide association study, GWAS)的不断深入,研究者们发现了大量与肺功能或COPD相关的遗传变异或基因位点、药物靶点等。本文综述了2007年以来世界范围内针对肺功能或COPD的GWAS方面的研究工作及其进展综述,分析了可能存在的药物靶点,并探讨了COPD在全基因组关联研究中面临的挑战和困难,为深入研究COPD发病机制提供新思路。
关键词:
Abstract
Chronic obstructive pulmonary disease (COPD) is characterized by irreversible airflow obstruction and chronic airway inflammation, caused by a combination of environmental and genetic factors. It is the third leading cause of death worldwide. In recent years, researchers have applied the genome-wide association study (GWAS) and identified a large number of genetic variants associated with lung functions and potential drug targets for treating COPD. In this review, we summarize the results of GWAS studies and perform a review of the literature since 2007 to highlight the progress of GWAS on COPD. We discuss the challenges, the underlying mechanisms, and the possible drug targets, thereby providing insights on the pathogenesis and potential treatment strategies for COPD.
Keywords:
PDF (685KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
钱国清. 慢性阻塞性肺疾病全基因组关联研究进展. 遗传[J], 2020, 42(9): 832-846 doi:10.16288/j.yczz.19-381
Guoqing Qian.
慢性阻塞性肺疾病(chronic obstructive pulmonary disease, COPD)是一种以气流受限为主要特征的慢性气道炎症。肺功能检查可评估COPD气流受限程度,当吸入支气管扩张剂后,第一秒用力呼气容积(forced expiratory volume in one second, FEV1)和用力肺活量(forced vital capacity, FVC)之间比值<0.7表明存在持续气流受限,排除其他疾病后,可确诊COPD。因此,肺功能是诊断COPD的金标准[1]。据报道我国40岁以上人群COPD患病率高达13.7%,总人数近1亿[2]。2015年,全球预计有320万人死于COPD,其中中国有100多万[3];根据世界卫生组织预测,至2020年COPD将成为全球第3大致死疾病[4]。吸烟、空气污染、职业暴露等多种因素均可导致COPD,其中吸烟患者仅有10%~20%发展成为COPD[5],然而25%的COPD患者终生不吸烟[6],因此表明,COPD是遗传因素与环境因素共同作用的结果。近年,随着全基因组关联研究(genome-wide association study, GWAS)在肺功能和COPD中的应用,发现了大量易感基因(susceptibility gene)或基因座(locus),为进一步阐述COPD的发病机制提供了全新的思路。
GWAS研究是在一定人群中选择病例组和对照组,基于单核苷酸多态性(single nucleotide polymorphism, SNP)作为分子遗传标记,比较全基因组范围内所有SNP位点的等位基因或基因型频率在病例组和对照组间的差异;然后,利用连锁不平衡关系推测可能的疾病或性状的易感基因或区域,从而寻找与疾病发生相关的致病位点。随着人类基因组计划和基于SNP的国际人类单体型图谱(HapMap)构建完成,GWAS得以实现并被广泛应运于糖尿病、精神分裂症和哮喘等疾病[7,8]。经典的GWAS通过建立全基因组高频遗传变异与表型的关联,进行多阶段设计的病例对照研究,并将多个研究结果合并验证。GWAS中P<5.0×10-8的SNP位点才被认为具有全基因组水平阳性。从2005年Science首次发表年龄相关性视网膜黄斑变性的GWAS以来,至今已经发表数千项常见复杂性疾病或性状的研究。2007年来,肺功能和COPD相关GWAS研究层出不穷,特别是COPD遗传流行病学(COPD genetic epidemiology, COPDGene)和英国生物标本库(UK Biobank)等大样本研究,极大地推动了COPD发病机制和寻找有效药物靶点的研究。利用GWAS手段探讨呼吸系统疾病,特别是鉴定COPD的潜在致病基因非常重要:(1)可以更加全面地了解疾病发展和呼吸道正常病理生理功能;(2)有利于根据确定的药物靶标研发新的治疗策略;(3)通过确定一系列风险性和安全性遗传变异,可改善风险评估预防疾病,或者做出更早、更准确的诊断;(4)利用遗传信息将疾病分为不同表型或亚型;(5)利用遗传信息数据,使患者药物治疗获益更多、副作用更少,推动药物遗传学发展[9]。
截至2019年11月21日,通过查询NHGS-GWAS catalog (https://www.ebi.ac.uk/gwas/)网站发现,共有4200余篇文章涉及COPD,有近40项肺功能或COPD的GWAS队列研究,发现大量SNPs与肺功能或COPD相关(图1,图2)。本文将从GWAS与肺功能、GWAS与COPD及COPD亚型的关系进行阐述,并介绍基于GWAS的COPD药物研发,以期为COPD基因和基因组学研究提供参考。
图1
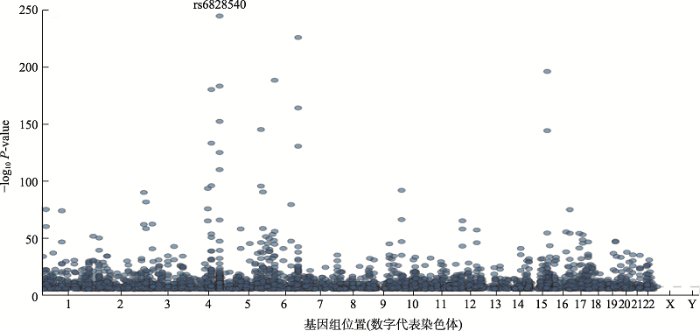 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1与肺功能(Pulmonary function measurement)相关SNPs分布图
以关键词“pulmonary function measurement”使用NHGS-GWAS catalog (
Fig. 1Plots of SNPs associated with pulmonary functions on the human genome (chromosomes)
图2
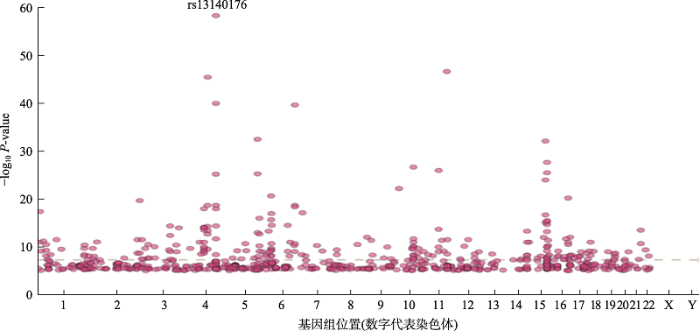 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2与COPD相关的SNPs点状分布图
以关键词“chronic obstructive pulmonary disease”使用NHGS-GWAS catalog (
Fig. 2Plots of SNPs associated with COPD on the human genome (chromosomes)
1 GWAS与肺功能
肺功能检测是诊断COPD的必备条件。据报道,COPD与FEV1和FVC的遗传相关性分别为-0.76和-0.9[10];在欧洲和美国的双胞胎研究中发现,FEV1的遗传力估计达0.77[11]。随着研究的深入,发现肺功能和COPD两者均具有相关性的易感基因位点。Hall等[12]通过文献复习发现28个易感基因位点同时与肺功能和COPD相关(表1)。Wain等[13]分析发现在97个与肺功能相关的致病基因中,95个与COPD有关。这些重叠的遗传变异/致病基因进一步确认了基因座与疾病的相关性,并预计其可能发挥重要的病理生理功能。因此,基于肺功能的GWAS可发现更多致病基因,对于揭示COPD发病机制具有很好的科学意义。Table 1
表1
表1肺功能与COPD重叠致病基因
Table 1
| 基因缩写 | 基因全称 | 位点 |
|---|---|---|
| TRIP1 | Eukaryotic translation initiation factor 3 subunit I | 1p35.2 |
| MFAP2 | Microfibril associated protein 2 | 1p36.13 |
| TNS1 | Mensin 1 | 2q35 |
| PID1 | Phosphotyrosine interaction domain containing 1 | 2q36.3 |
| HDAC4 | Histone deacetylase 4 | 2q37.3 |
| RARB | Retinoic acid receptor beta | 3p24.2 |
| EEFSEC | Eukaryotic elongation factor, selenocysteine-tRNA specific | 3q21.3 |
| FAM13A | Family with sequence similarity 13 member A | 4q22.1 |
| NPNT | Nephronectin | 4q24 |
| GSTCD | Glutathione S-transferase C-terminal domain containing | 4q24 |
| TET2 | Tet methylcytosine dioxygenase 2 | 4q24 |
| HHIP | Hedgehog interacting protein | 4q31.21 |
| HTR4 | 5-hydroxytryptamine receptor 4 | 5q32 |
| ADAM19 | ADAM Metallopeptidase Domain 19 | 5q33.3 |
| NCR3 | Natural cytotoxicity triggering receptor 3 | 6p21.33 |
| AGER | Advanced glycosylation end-product specific receptor | 6p21.32 |
| ARMC2 | Armadillo repeat containing 2 | 6q21 |
| ZKSCAN3 | Zinc finger with KRAB and SCAN domains 3 | 6p22.1 |
| GPR126 | Adhesion G protein-coupled receptor G6 | 6q24.2 |
| ASTN2 | Astrotactin 2 | 9q33.1 |
| CDC123 | Cell division cycle 123 | 10p14-p13 |
| C10orf11 | Leucine rich melanocyte differentiation associated | 10q22.2-q22.3 |
| CCDC38 | Coiled-coil domain containing 38 | 12q23.1 |
| RIN3 | Ras and Rab interactor 3 | 14q32.12 |
| THSD4 | Thrombospondin type 1 domain containing 4 | 15q23 |
| CFDP1 | Craniofacial development protein 1 | 16q23.1 |
| KCNE2 | Potassium voltage-gated channel subfamily E regulatory subunit 2 | 21q22.11 |
| MN1 | MN1 proto-oncogene, transcriptional regulator | 22q12.1 |
新窗口打开|下载CSV
Wilk等[14]于2007年在美国雷明汉心脏研究中心开展了第1项肺功能GWAS研究,该研究涉及10项肺功能测量(表2)和70,987个常染色体SNP,提示GSTO2和IL6R是与肺功能相关的基因。2009年,他和同事又在7691名参与者中研究表型FEV1/FVC,发现在4q31区域上有4个SNP与FEV1/FVC相关,其中SNP rs13147758还与家系的FEV1/FVC显著相关;这些SNPs均与HHIP基因靠近[15]。由此,他们认为4号染色体区域具有影响肺功能的遗传效应,值得进一步研究。
Table 2
表2
表2首项肺功能全基因组关联研究采用的测量指标
Table 2
| 指标 | 指标全称(英文) | 指标全称(中文) |
|---|---|---|
| ppfev1 | Percent predicted FEV1 for latest exam | FEV1占预测值百分比 |
| ppfvc | Percent predicted FVC for latest exam | FVC占预测值百分比 |
| ppratio | Percent predicted FEV1/FVC for latest exam | FEV1/FVC占预测值百分比 |
| ppfef | Percent predicted FEF25-75 for latest exam | FEF25-75占预测值百分比 |
| ppfefrat | Percent predicted FEF25-75/FVC for latest exam | FEF25-75/FVC占预测值百分比 |
| meanfev1 | Mean FEV1 from two exams | FEV1均值 |
| meanfvc | Mean FVC from two exams | FVC均值 |
| meanratio | Mean FEV1/FVC from two exams | FEV1/FVC均值 |
| Fev1slope | Longitudinal slope of FEV1 | FEV1纵向斜率 |
| fefslope | Longitudinal slope of FEV25-75 | FEV25-75纵向斜率 |
新窗口打开|下载CSV
随着meta分析的应用,更多与肺功能相关变异位点被确定。2010年,Hancock等[16]对4个组群
(Atherosclerosis Risk in Communities (ARIC), Cardiovascular Health Study (CHS), Framingham Heart Study (FHS), and Rotterdam Study (RS)) (Cohorts for Heart and Aging Research in Genomic Epidemiology consortium study, CHARGE研究)的20,890名欧洲后裔meta分析,明确8个与FEV1/FVC相关的基因变异位点(HHIP、GPR126、ADAM19、AGER-PPT2、FAM13A、PTCH1、PID1和HTR4)及1个与FEV1相关的基因位点(INTS12-GSTCD-NPNT)。同年,Repapi等[17]对SpiroMeta、CHARGE等研究涉及的74,564名欧洲后裔进行meta分析,明确了5个新的肺功能(FEV1、FEV1/FVC)相关常见变异(common variant),分别为2q35(TNS1)、4q24(GSTCD)、5q33 (HTR4)、6p21(AGER)和15q23(THSD4);并在人类肺组织中验证TNS1、GSTCD、AGER、HTR4和THSD4的mRNA表达。2012年,Hancock等[18]通过meta分析,确定了2个(DNER和HLA-DQB1/ HLA-DQA2)与FEV1/FVC相关、1个(KCNJ2/SOX9)与FEV1相关的新基因座,另外位于DNER、HLA-DQB1、HLA-DQA2和KCNJ2、SOX9的SNPs与FEV1/FVC或FEV1相关。2014年,Loth等[19]发现6个与肺功能FVC相关的SNPs,并定位于EFEMP1、BMP6、MIR-129-2/HSD17B12、PRDM11、WWOX和KCNJ2;同时,在基于多民族动脉粥样硬化研究(multi-ethnic study of atherosclerosis,MESA)的563名中国裔中未发现有与中国裔肺功能相关的SNPs。
随着世界最大队列样本库(UK Biobank)的应用,肺功能GWAS研究获得了更多的成果。2017年,Wain等[13]基于UK BiLEVEL的48,943名参与者和第二阶段基于95,375名来自UK Biobank、SpiroMeta和英国家庭纵向研究(UK household longitudinal study, UKHLS)的参与者,发现与肺功能(FEV1、FVC或FEV1/FVC)相关的遗传变异位点从54个增加至97个,其中新发现43个;经富集分析发现该97个遗传变异位点均与发育、弹性纤维和表观遗传学调节通路相关。因此,随着GWAS研究的深入和生物信息学的应用,将会进一步揭示致病基因的生物学功能。
2019年,Shrine等[20]发表了迄今为止样本量最大、相关位点最多的研究,他们发现有257个位点与肺功能(FEV1、FVC、FEV1/FVC和PEF)相关,其中新发现位点139个,共确认107个参与基因表达、蛋白表达和功能注释的致病基因(causal gene) (表3),8个同时与有害变异(deleterious variant)和表达数量性状基因座(expression quantitative trait locus, eQTL)有关,1个同时与eQTL和蛋白质数量性状基因座(protein quantitative trait locus, pQTL)有关,1个同时与有害变异和pQTL有关,13个仅与有害变异有关,81个与eQTL及3个与pQTL关联。该研究首次利用GWAS发现与最大呼气流量(peak expiratory flow,PEF)关联的致病基因,其中有133个位点与PEF相关(P<10E-5),如SLC26A9已经明确与囊性纤维化相关,同时在UK Biobank中发现与PEF高度相关。将致病基因富集分析,发现有部分致病基因参与纤毛生成(ciliogenesis,包括KIAA0753、CDK2和CEP72)过程,提示纤毛功能的损伤与COPD的发生发展密切相关。因此,肺功能的GWAS研究,不仅局限于肺功能的表型研究,同时可揭示COPD的发病机制和病理生理过程,为后续单基因或多基因的基础提供理论基础。
2 GWAS与COPD
虽然肺功能作为检测COPD气流受限的必备手段,但肺功能相关的遗传变异/致病基因与疾病(COPD)是否相关,仍受到人们的质疑。因此,需要对遗传变异与COPD相关性展开GWAS研究。2009年,Pillai等[32]首次开展COPD的GWAS研究:通过823名COPD患者和810名吸烟者的队列研究,选取前100个SNPs在NETT和NAS研究中验证,发现2个SNPs(rs8034191和rs1051730)位于CHRNA3-CHRNA5-IREB2位点;同时HHIP虽未达到基因组关联统计学意义,但也与FEV1/FVC相关;因此,提示CHRNA3-CHRNA5-IREB2和HHIP均存在COPD的重要风险位点。2010年,Cho等[33]通过2940名COPD和1380名对照组(两组均为既往或正在吸烟者),确认rs7671167和rs1903003位于1个新的致病基因FAM13A(4q22.1)位点。此后陆续有较多COPD的GWAS研究报道,如Van Durme等[34]基于著名的鹿特丹研究,发现了HHIP与COPD风险相关;Brehm等[35]研究表明2个SNPs位于FGF7,SNP的不同可能与FGF7表达有关,肺组织中FGF7表达升高与肺功能恶化有关;Castaldi等[23]研究发现32个SNPs位于/靠近17个致病基因,其中11个为既往4项GWAS研究(NETT/NAS、挪威病例对照研究、ECLIPSE、COPDGene)已报道。2012年,Cho 等[36]基于4项研究(ECLIPSE、NAS和NETT、GenKOLS、COPDGene)发现了一个位于19q13的新基因座,rs7937和rs2604894与严重COPD相关。2012~2017年间,Wilk等[28]、Hanse等l[37]、Cho等[24]、Dijkstra等[26]、Hardin等[38]及其他研究者[31,39~43]又先后发表了多篇GWAS在COPD中的研究,先后发现TMEM26、ANK3、FOXA1、MMP12和TGFB2等易感基因与COPD相关。2017年,Hobbs等[10]发现22个与COPD相关的基因座,其中新发现13个基因座(同时包含9个与肺功能相关),但未发现与哮喘存在基因座重叠。最新研究表明在6p21-22区域、ADAM19、ARMC2、ELAVL2和STAT6存在COPD与哮喘的共享片段,除STAT6外,其余基因座作用方向一致[21]。因此,随着GWAS研究的深入和样本量的增多,COPD相关的致病基因或基因座研究不断取得新进展随后,科学家们又发现了更多的致病基因。Wain等[13]发现的97个致病基因中,51个与COPD定义相关,30个与COPD易感性相关(P<5.26×10-4),但未发现有遗传变异或遗传危险系数与COPD急性发作相关。他们还研究了来自中国慢性病前瞻研究项目协作组(China Kadoorie Biobank cohort, CKB)的7116例COPD (20,919例对照)和5292例急性加重病例(1824 对照组)的71个遗传变异,发现有39个致病基因在欧洲人和中国人样本中作用方向一致,其中7个达到统计学意义(P<0.05) (表4),说明在欧洲人群与中国人群中,某些遗传因素对COPD具有共同的效应,但仍需进一步的实验证明。
2019年,Sakornsakolpat等[21]汇总25项GWAS的35,735例COPD和222,076例对照组,发现82个与肺功能或COPD相关的基因座,其中原有47个、新发现35个;发现156个致病基因位于该82个基因座中,为阐述COPD疾病易感性和临床异质性提供新的视角。35个新基因座中,有9个与Shine等[20]报道相同。包括该82个基因座在内,使用10%的COPD患病率解释了7.0%表型变异(phenotypic variant),与最近的22个基因座所解释的4.7%相比[10],所解释的COPD表型变异增加了48%,可能存在一定的效应高估。通过富集分析发现165个基因集的FDR<5%,其中44%与发育过程相关,如肺发育、肺泡发育和肺形态发生等;还有细胞外基质相关途径,及Wnt、SMAD、MAPK等信号通路[21]。因此,进一步支持早期生活事件在COPD患病风险中的关键作用,COPD患病风险的很大一部分可能在生命的早期即已决定,遗传变异可能会决定初始的肺功能和肺生长模式。
目前已进入后GWAS时代,如何实现GWAS成果转化是当前的研究热点。Zhou等[44]使用shRNA干扰HHIP表达,发现HHIP参与细胞外基质和细胞增殖等过程。COPD易感基因FAM13A在人类气道和肺脏中均有表达,通过使用Fam13a基因敲除(Fam13a-/-)小鼠和在人类肺组织中验证,其可通过抑制β-catenin信号通路促进肺气肿形成[45];另外还在香烟诱导小鼠模型中发现Fam13a可促进脂肪酸- β氧化(fatty acid β-oxidation, FAO)过程[46]。本课题组也先后进一步阐明了GSTCD-INTS12-NPNT[47,48,49]、HTR4[50]和AGER[51]等易感基因的表达和病理生理功能。通过单个基因或多个基因的细胞模型和/或动物模型试验,为揭示COPD潜在发病机制提供了全新的思路。
Table 3
表3
表3107个参与基因表达、蛋白表达和功能注释的致病基因
Table 3
| 基因 | 功能相关基因 | 基因 | 功能相关基因 |
|---|---|---|---|
| DHDDS (3’-UTR) | HMGN2 | NCOR1 | ADORA2B, TTC19 |
| NEXN | - | ASPSCR1 | LRRC45 |
| DENND2D[21] | CEPT1, CHI3L2, DRAM2 | C18orf8 | - |
| C1orf54 | MRPS21, RPRD2, ECM1 | ZFP82 | ZFP14 |
| KRTCAP2 | THBS4 | MFAP2 | - |
| RALGPS2 | ANGPTL1 | LOC101929516 | PABPC4 |
| LMOD1 | SHISA4 | TGFB2[10,22~25] | - |
| ATAD2B | UBXN2A | TRAF3IP1 | ASB1 |
| PKDCC | - | SLMAP | - |
| ITGAV | - | RSRC1 | |
| SPATS2L | - | GSTCD[10,16,17,19,23] | INTS12[16,23] |
| C2orf54 | - | NPNT[5,10,16,23,26,27] | - |
| MIR548G | FILIP1L | AP3B1 | - |
| BCHE | - | SPATA9[22] | RHOBTB3 |
| BTC | - | P4HA2-AS1 | SLC22A5, P4HA2, C1QTNF5 |
| LOC100996325 | CEP72 | CYFIP2 | - |
| RNU6-71P | DST | ADAM19[10,23,28] | - |
| JAZF1 | - | DSP[10] | - |
| MET | - | MIR588 | CENPW |
| IER5L | CRAT, PPP2R4 | GPR126[10,16,27,29] | - |
| DOCK9 | - | C1GALT1 | - |
| CHAC1 | INO80, CHP1, RAD51 | QSOX2 (3’-UTR) | - |
| ATP2A3 | - | DNLZ | SNAPC4, CARD9, INPP5E |
| PITPNM3 | KIAA0753, TXNDC17 | CDC123[22] | NUDT5 |
| TNFSF12-TNFSF13 | TNFSF13, SENP3 | MYPN | - |
| EML3 | EEF1G, ROM1 | SSH2 | EFCAB5 |
| ARHGEF17 | FAM168A | FBXL20 | CRKRS |
| RAB5B | CDK2 | MAPT-AS1 | LRRC37A4, MAPT |
| LRP1[22] | - | TSEN54 | CASKIN2 |
| FGD6 | - | LTBP4[27] | - |
| RPAP1[30] | ITPKA, LTK, TYRO3 | ABHD12 | PYGB |
| AAGAB | SMAD3, IQCH | UQCC1[30] | GDF5 |
| THSD4[10,17,31] | - | SLC2A4RG | LIME1 |
| IL27[10] | SBK1, TUFM, CCDC101[10], SULT1A1, SULT1A2, SH2B1, NPIPL1, CLN3, ATXN2L, EIF3C | SCARF2 | - |
| MMP15 |
新窗口打开|下载CSV
3 GWAS与COPD亚型
虽然肺功能是诊断COPD的金标准,然而依赖肺功能检测指标诊断COPD存在明显缺陷,因为COPD的发病机制存在不同的遗传和环境因素,包含不同的病理生理学机制,因此只有研究基于不同遗传背景、环境因素的病理生理相关的亚型才更具有意义。Table 4
表4
表4欧洲人群与中国人群7个作用方向一致的变异位点
Table 4
| 性状 | 基因 | SNP名称 | P值 |
|---|---|---|---|
| FEV1/FVC | MFAP2* | rs138641402 | 1.00E-04 |
| FEV1/FVC | LOC101929516 (intron) * | rs2070600 | 1.12E-04 |
| FEV1/FVC | CDC7/TGFBR3* | rs2045517 | 7.71E-04 |
| FVC | SPAG17/TBX15* | rs7713065 | 6.39E-03 |
| FEV1/FVC | TGFB2* | rs993925 | 1.49E-02 |
| FEV1/FVC | CHRM3 (intron) * | rs4237643 | 2.62E-02 |
| FEV1 | TNS1* | rs2283847 | 2.82E-02 |
新窗口打开|下载CSV
目前,临床表型分为慢性支气管炎(chronic bronchitis, CB)、肺气肿、频繁急性加重及哮喘-慢阻肺重叠综合症(asthma-COPD overlap syndrome, ACOS)等;亚型包含中性粒细胞、细菌定植、Th2亚型或嗜酸性粒细胞增多亚型等[4,52]。随着基因组学研究的深入,即可实现精准分型,如SERPINA1是
COPD精准治疗需要兼顾亚型和表型,根据分子标志物实现不同亚型的鉴别,实现分层医学(stratified medicine),真正实现个体化和个性化治疗。
4 基于GWAS的COPD药物研发
实现GWAS成果转化可能需要几年甚至十几年的时间,但是基于GWAS靶标设计药物可事半功倍,加速药物研发。目前,共114种临床试验或批准的药物涉及COPD,160个靶标正在开展研究(截至2019年12月15日,https://www.targetvalidation.org/)。比如,由CHRM3编码的毒蕈碱型乙酰胆碱受体M3是明确的药物靶标,针对该药物靶标已有许多批准的药物,包括用于治疗哮喘和慢性阻塞性肺疾病的药物[13]。编码5-羟色胺受体的HTR4在早期的肺功能GWAS中就已被发现。INPP5E与FVC(和FEV1)相关,编码肌醇多磷酸5磷酸酶E,另一成分磷酸肌醇3激酶(PI3K)δ是正在开发中的COPD和哮喘的药物靶标[13]。肺组织中FAM13A的表达减少和COPD风险降低有关,因此,抑制FAM13A表达可能具有保护性作用。Hedgedog信号通路在肺脏早期发育中发挥重要的作用,其中PTCH1、TGFB2和HHIP作为该信号通路分子,它们是肺功能相关联的潜在致病基因[61]。另外,据报道新发现有7个与COPD和肺功能相关的可治疗靶标,包括ABHD6、CDKL2、GSTO2、KCNC4、PDHB、SLK和TRPM7[21];目前许多靶标仅依赖于细胞模型的初步探索,但其在细胞内或体内的相互作用和效果仍需进一步通过开展临床试验评估。随着后GWAS时代转化研究的深入,必将发现更多的药物靶标及开发新型高效药物提供基础。
5 结语与展望
尽管在COPD中成功地开展了全基因组关联研究,但仍存在如下挑战:(1) GWAS通过病例对照寻找差异SNPs,但大部分SNPs位于非编码区,仅有一部分基因变异与疾病表型相关。(2)通过GWAS识别的遗传变异仅占遗传力的一小部分,已知肺功能信号分别占FEV1、FVC和FEV1 / FVC的遗传力的9.6%、6.4%和14.3%[13]。因此,GWAS仅识别常见变异,而忽略了罕见变异、基因拷贝数量变异等其他类型变异。(3)当前商业化的SNPs芯片多基于高加索人群设计,亟需设计基于亚洲人群遗传多态性的芯片。(4)基于肺功能诊断的COPD,受多种因素的影响,如对样本量、资料收集的完整性、研究对象的选择及诊断方法等。(5)在COPD研究中,大多数研究均聚焦于肺通气功能测量,而非COPD本身,使得在疾病中的应用和转化面临困难。(6)无法完全匹配病例组与对照组参与者,而且COPD患者可能存在多种并发症,如心血管疾病、2型糖尿病等其他疾病。在欧美等国家,开展了大规模的COPD基因和基因组学研究,但目前国内COPD基因组学研究严重落后,尚无一项特定的针对COPD开展大规模GWAS研究[62]。因此,应该抓住机遇,加强国际合作,建立以中国汉族人群为基础的慢性阻塞性肺疾病全基因组关联研究,利用先进的分子生物学技术,阐明中国人COPD的发病机制、寻找有效治疗靶点、发现新型生物标志物和改善COPD和亚型的预测。推进个性化预防和治疗COPD,降低我国COPD患病率和死亡率,全面提高全民健康水平。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URL [本文引用: 1]
DOI:10.1016/S0140-6736(18)30841-9URLPMID:29650248 [本文引用: 1]

BACKGROUND: Although exposure to cigarette smoking and air pollution is common, the current prevalence of chronic obstructive pulmonary disease (COPD) is unknown in the Chinese adult population. We conducted the China Pulmonary Health (CPH) study to assess the prevalence and risk factors of COPD in China. METHODS: The CPH study is a cross-sectional study in a nationally representative sample of adults aged 20 years or older from ten provinces, autonomous regions, and municipalities in mainland China. All participants underwent a post-bronchodilator pulmonary function test. COPD was diagnosed according to 2017 Global Initiative for Chronic Obstructive Lung Disease (GOLD) criteria. FINDINGS: Between June, 2012, and May, 2015, 57 779 individuals were invited to participate, of whom 50 991 (21 446 men and 29 545 women) had reliable post-bronchodilator results and were included in the final analysis. The overall prevalence of spirometry-defined COPD was 8.6% (95% CI 7.5-9.9), accounting for 99.9 (95% CI 76.3-135.7) million people with COPD in China. Prevalence was higher in men (11.9%, 95% CI 10.2-13.8) than in women (5.4%, 4.6-6.2; p<0.0001 for sex difference) and in people aged 40 years or older (13.7%, 12.1-15.5) than in those aged 20-39 years (2.1%, 1.4-3.2; p<0.0001 for age difference). Only 12.0% (95% CI 8.1-17.4) of people with COPD reported a previous pulmonary function test. Risk factors for COPD included smoking exposure of 20 pack-years or more (odds ratio [OR] 1.95, 95% CI 1.53-2.47), exposure to annual mean particulate matter with a diameter less than 2.5 mum of 50-74 mug/m(3) (1.85, 1.23-2.77) or 75 mug/m(3) or higher (2.00, 1.36-2.92), underweight (body-mass index <18.5 kg/m(2); 1.43, 1.03-1.97), sometimes childhood chronic cough (1.48, 1.14-1.93) or frequent cough (2.57, 2.01-3.29), and parental history of respiratory diseases (1.40, 1.23-1.60). A lower risk of COPD was associated with middle or high school education (OR 0.76, 95% CI 0.64-0.90) and college or higher education (0.47, 0.33-0.66). INTERPRETATION: Spirometry-defined COPD is highly prevalent in the Chinese adult population. Cigarette smoking, ambient air pollution, underweight, childhood chronic cough, parental history of respiratory diseases, and low education are major risk factors for COPD. Prevention and early detection of COPD using spirometry should be a public health priority in China to reduce COPD-related morbidity and mortality. FUNDING: Ministry of Health and Ministry of Science and Technology of China.
DOI:10.1016/S2213-2600(17)30293-XURLPMID:28822787 [本文引用: 1]

BACKGROUND: Chronic obstructive pulmonary disease (COPD) and asthma are common diseases with a heterogeneous distribution worldwide. Here, we present methods and disease and risk estimates for COPD and asthma from the Global Burden of Diseases, Injuries, and Risk Factors (GBD) 2015 study. The GBD study provides annual updates on estimates of deaths, prevalence, and disability-adjusted life years (DALYs), a summary measure of fatal and non-fatal disease outcomes, for over 300 diseases and injuries, for 188 countries from 1990 to the most recent year. METHODS: We estimated numbers of deaths due to COPD and asthma using the GBD Cause of Death Ensemble modelling (CODEm) tool. First, we analysed data from vital registration and verbal autopsy for the aggregate category of all chronic respiratory diseases. Subsequently, models were run for asthma and COPD relying on covariates to predict rates in countries that have incomplete or no vital registration data. Disease estimates for COPD and asthma were based on systematic reviews of published papers, unpublished reports, surveys, and health service encounter data from the USA. We used the Global Initiative of Chronic Obstructive Lung Disease spirometry-based definition as the reference for COPD and a reported diagnosis of asthma with current wheeze as the definition of asthma. We used a Bayesian meta-regression tool, DisMod-MR 2.1, to derive estimates of prevalence and incidence. We estimated population-attributable fractions for risk factors for COPD and asthma from exposure data, relative risks, and a theoretical minimum exposure level. Results were stratified by Socio-demographic Index (SDI), a composite measure of income per capita, mean years of education over the age of 15 years, and total fertility rate. FINDINGS: In 2015, 3.2 million people (95% uncertainty interval [UI] 3.1 million to 3.3 million) died from COPD worldwide, an increase of 11.6% (95% UI 5.3 to 19.8) compared with 1990. There was a decrease in age-standardised death rate of 41.9% (37.7 to 45.1) but this was counteracted by population growth and ageing of the global population. From 1990 to 2015, the prevalence of COPD increased by 44.2% (41.7 to 46.6), whereas age-standardised prevalence decreased by 14.7% (13.5 to 15.9). In 2015, 0.40 million people (0.36 million to 0.44 million) died from asthma, a decrease of 26.7% (-7.2 to 43.7) from 1990, and the age-standardised death rate decreased by 58.8% (39.0 to 69.0). The prevalence of asthma increased by 12.6% (9.0 to 16.4), whereas the age-standardised prevalence decreased by 17.7% (15.1 to 19.9). Age-standardised DALY rates due to COPD increased until the middle range of the SDI before reducing sharply. Age-standardised DALY rates due to asthma in both sexes decreased monotonically with rising SDI. The relation between with SDI and DALY rates due to asthma was attributed to variation in years of life lost (YLLs), whereas DALY rates due to COPD varied similarly for YLLs and years lived with disability across the SDI continuum. Smoking and ambient particulate matter were the main risk factors for COPD followed by household air pollution, occupational particulates, ozone, and secondhand smoke. Together, these risks explained 73.3% (95% UI 65.8 to 80.1) of DALYs due to COPD. Smoking and occupational asthmagens were the only risks quantified for asthma in GBD, accounting for 16.5% (14.6 to 18.7) of DALYs due to asthma. INTERPRETATION: Asthma was the most prevalent chronic respiratory disease worldwide in 2015, with twice the number of cases of COPD. Deaths from COPD were eight times more common than deaths from asthma. In 2015, COPD caused 2.6% of global DALYs and asthma 1.1% of global DALYs. Although there are laudable international collaborative efforts to make surveys of asthma and COPD more comparable, no consensus exists on case definitions and how to measure disease severity for population health measurements like GBD. Comparisons between countries and over time are important, as much of the chronic respiratory burden is either preventable or treatable with affordable interventions. FUNDING: Bill & Melinda Gates Foundation.
DOI:10.1016/bs.adgen.2015.12.002URLPMID:26915270 [本文引用: 2]

Chronic respiratory diseases are a major cause of worldwide mortality and morbidity. Although hereditary severe deficiency of alpha1 antitrypsin (A1AD) has been established to cause emphysema, A1AD accounts for only approximately 1% of Chronic Obstructive Pulmonary Disease (COPD) cases. Genome-wide association studies (GWAS) have been successful at detecting multiple loci harboring variants predicting the variation in lung function measures and risk of COPD. However, GWAS are incapable of distinguishing causal from noncausal variants. Several approaches can be used for functional translation of genetic findings. These approaches have the scope to identify underlying alleles and pathways that are important in lung function and COPD. Computational methods aim at effective functional variant prediction by combining experimentally generated regulatory information with associated region of the human genome. Classically, GWAS association follow-up concentrated on manipulation of a single gene. However association data has identified genetic variants in >50 loci predicting disease risk or lung function. Therefore there is a clear precedent for experiments that interrogate multiple candidate genes in parallel, which is now possible with genome editing technology. Gene expression profiling can be used for effective discovery of biological pathways underpinning gene function. This information may be used for informed decisions about cellular assays post genetic manipulation. Investigating respiratory phenotypes in human lung tissue and specific gene knockout mice is a valuable in vivo approach that can complement in vitro work. Herein, we review state-of-the-art in silico, in vivo, and in vitro approaches that may be used to accelerate functional translation of genetic findings.
DOI:10.1016/S2213-2600(15)00283-0URLPMID:26423011 [本文引用: 2]

BACKGROUND: Understanding the genetic basis of airflow obstruction and smoking behaviour is key to determining the pathophysiology of chronic obstructive pulmonary disease (COPD). We used UK Biobank data to study the genetic causes of smoking behaviour and lung health. METHODS: We sampled individuals of European ancestry from UK Biobank, from the middle and extremes of the forced expiratory volume in 1 s (FEV1) distribution among heavy smokers (mean 35 pack-years) and never smokers. We developed a custom array for UK Biobank to provide optimum genome-wide coverage of common and low-frequency variants, dense coverage of genomic regions already implicated in lung health and disease, and to assay rare coding variants relevant to the UK population. We investigated whether there were shared genetic causes between different phenotypes defined by extremes of FEV1. We also looked for novel variants associated with extremes of FEV1 and smoking behaviour and assessed regions of the genome that had already shown evidence for a role in lung health and disease. We set genome-wide significance at p<5 x 10(-8). FINDINGS: UK Biobank participants were recruited from March 15, 2006, to July 7, 2010. Sample selection for the UK BiLEVE study started on Nov 22, 2012, and was completed on Dec 20, 2012. We selected 50,008 unique samples: 10,002 individuals with low FEV1, 10,000 with average FEV1, and 5002 with high FEV1 from each of the heavy smoker and never smoker groups. We noted a substantial sharing of genetic causes of low FEV1 between heavy smokers and never smokers (p=2.29 x 10(-16)) and between individuals with and without doctor-diagnosed asthma (p=6.06 x 10(-11)). We discovered six novel genome-wide significant signals of association with extremes of FEV1, including signals at four novel loci (KANSL1, TSEN54, TET2, and RBM19/TBX5) and independent signals at two previously reported loci (NPNT and HLA-DQB1/HLA-DQA2). These variants also showed association with COPD, including in individuals with no history of smoking. The number of copies of a 150 kb region containing the 5' end of KANSL1, a gene that is important for epigenetic gene regulation, was associated with extremes of FEV1. We also discovered five new genome-wide significant signals for smoking behaviour, including a variant in NCAM1 (chromosome 11) and a variant on chromosome 2 (between TEX41 and PABPC1P2) that has a trans effect on expression of NCAM1 in brain tissue. INTERPRETATION: By sampling from the extremes of the lung function distribution in UK Biobank, we identified novel genetic causes of lung function and smoking behaviour. These results provide new insight into the specific mechanisms underlying airflow obstruction, COPD, and tobacco addiction, and show substantial shared genetic architecture underlying airflow obstruction across individuals, irrespective of smoking behaviour and other airway disease. FUNDING: Medical Research Council.
DOI:10.1016/S0140-6736(09)61303-9URLPMID:19716966 [本文引用: 1]

Chronic obstructive pulmonary disease (COPD) is a leading cause of morbidity and mortality worldwide. Tobacco smoking is established as a major risk factor, but emerging evidence suggests that other risk factors are important, especially in developing countries. An estimated 25-45% of patients with COPD have never smoked; the burden of non-smoking COPD is therefore much higher than previously believed. About 3 billion people, half the worldwide population, are exposed to smoke from biomass fuel compared with 1.01 billion people who smoke tobacco, which suggests that exposure to biomass smoke might be the biggest risk factor for COPD globally. We review the evidence for the association of COPD with biomass fuel, occupational exposure to dusts and gases, history of pulmonary tuberculosis, chronic asthma, respiratory-tract infections during childhood, outdoor air pollution, and poor socioeconomic status.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/ng.3752URLPMID:28166215 [本文引用: 12]

Chronic obstructive pulmonary disease (COPD) is a leading cause of mortality worldwide. We performed a genetic association study in 15,256 cases and 47,936 controls, with replication of select top results (P < 5 x 10(-6)) in 9,498 cases and 9,748 controls. In the combined meta-analysis, we identified 22 loci associated at genome-wide significance, including 13 new associations with COPD. Nine of these 13 loci have been associated with lung function in general population samples, while 4 (EEFSEC, DSP, MTCL1, and SFTPD) are new. We noted two loci shared with pulmonary fibrosis (FAM13A and DSP) but that had opposite risk alleles for COPD. None of our loci overlapped with genome-wide associations for asthma, although one locus has been implicated in joint susceptibility to asthma and obesity. We also identified genetic correlation between COPD and asthma. Our findings highlight new loci associated with COPD, demonstrate the importance of specific loci associated with lung function to COPD, and identify potential regions of genetic overlap between COPD and other respiratory diseases.
DOI:10.1164/arrd.1982.125.4.409URLPMID:7200340 [本文引用: 1]

As part of the NHLBI Twin Study, pulmonary function tests were successfully administered to 127 monozygotic and 141 dizygotic white male twin pairs 42 to 56 yr of age. Values for forced vital capacity (FVC) and forced expiratory volume in one second (FEV1) were obtained using a standardized protocol for spirometry. Initial twin analyses showed significant genetic variance (p less than 0.001) for both FVC and FEV1, whether or not adjustments were made for individual differences in age and body size. After adjustment, heritability estimates were 0.91 and 0.77 for FVC and FEV1, respectively. Further analyses indicated that the observed heritability of FVC resulted from the effects of pack-years of smoking as well as from genetic factors related to body size. These findings suggest that there were no other significant genetic determinants of FVC. In contrast, heritability of FEV1 could not be explained by constitutional factors, such as height and weight, or by cigarette smoking or propensity for cardiopulmonary disease symptoms. Additional analyses were done based on frequency of twin contact, which served as an indirect measure of environmental similarity between cotwins. Results suggested that there was a shared environmental variation in FEV1, as well as genetic variation, that could not be attributed to subpopulation differences in measured characteristics. The findings of this study are consistent with theories of genetic influences on alveolar and airway development and argue in favor of early as well as adult environmental influences on pulmonary function.
DOI:10.1111/resp.13436URLPMID:30421854 [本文引用: 1]

Chronic respiratory diseases are a major cause of morbidity and mortality. Asthma and chronic obstructive pulmonary disease (COPD) combined affect over 500 million people worldwide. While environmental factors are important in disease progression, asthma and COPD have long been known to be heritable with genetic components playing an important role in the risk of developing disease. Identification of genetic variation contributing to disease progression is important for a number of reasons including identification of risk alleles, understanding underlying disease mechanisms and development of novel therapies. Genome-wide association studies (GWAS) have been successful in identifying many loci associated with lung function, COPD and asthma. In recent years, meta-analyses and improved imputation have facilitated the growth of GWAS in terms of numbers of subjects and the number of single nucleotide polymorphisms (SNP) that can be interrogated. As a consequence, there has been a significant increase in the number of signals associated with asthma, COPD and lung function. SNP that have shown association with lung function reassuringly show a significant overlap with SNP associated with COPD giving a glimpse at pathways that may be involved in COPD mechanisms including genes in, for example, developmental pathways. In asthma, association signals are often in or near genes involved in both adaptive and innate immune response pathways, epithelial cell homeostasis and airway structural changes. The challenges now are translating these genetic signals into a new understanding of lung biology, understanding how variants impact health and disease and how they may provide opportunities for therapeutic intervention.
DOI:10.1038/ng.3787URLPMID:28166213 [本文引用: 6]

Chronic obstructive pulmonary disease (COPD) is characterized by reduced lung function and is the third leading cause of death globally. Through genome-wide association discovery in 48,943 individuals, selected from extremes of the lung function distribution in UK Biobank, and follow-up in 95,375 individuals, we increased the yield of independent signals for lung function from 54 to 97. A genetic risk score was associated with COPD susceptibility (odds ratio per 1 s.d. of the risk score ( approximately 6 alleles) (95% confidence interval) = 1.24 (1.20-1.27), P = 5.05 x 10(-49)), and we observed a 3.7-fold difference in COPD risk between individuals in the highest and lowest genetic risk score deciles in UK Biobank. The 97 signals show enrichment in genes for development, elastic fibers and epigenetic regulation pathways. We highlight targets for drugs and compounds in development for COPD and asthma (genes in the inositol phosphate metabolism pathway and CHRM3) and describe targets for potential drug repositioning from other clinical indications.
DOI:10.1186/1471-2350-8-S1-S8URL [本文引用: 2]
DOI:10.1371/journal.pgen.1000429URLPMID:19300500 [本文引用: 1]

The ratio of forced expiratory volume in one second to forced vital capacity (FEV(1)/FVC) is a measure used to diagnose airflow obstruction and is highly heritable. We performed a genome-wide association study in 7,691 Framingham Heart Study participants to identify single-nucleotide polymorphisms (SNPs) associated with the FEV(1)/FVC ratio, analyzed as a percent of the predicted value. Identified SNPs were examined in an independent set of 835 Family Heart Study participants enriched for airflow obstruction. Four SNPs in tight linkage disequilibrium on chromosome 4q31 were associated with the percent predicted FEV(1)/FVC ratio with p-values of genome-wide significance in the Framingham sample (best p-value = 3.6e-09). One of the four chromosome 4q31 SNPs (rs13147758; p-value 2.3e-08 in Framingham) was genotyped in the Family Heart Study and produced evidence of association with the same phenotype, percent predicted FEV(1)/FVC (p-value = 2.0e-04). The effect estimates for association in the Framingham and Family Heart studies were in the same direction, with the minor allele (G) associated with higher FEV(1)/FVC ratio levels. Results from the Family Heart Study demonstrated that the association extended to FEV(1) and dichotomous airflow obstruction phenotypes, particularly among smokers. The SNP rs13147758 was associated with the percent predicted FEV(1)/FVC ratio in independent samples from the Framingham and Family Heart Studies producing a combined p-value of 8.3e-11, and this region of chromosome 4 around 145.68 megabases was associated with COPD in three additional populations reported in the accompanying manuscript. The associated SNPs do not lie within a gene transcript but are near the hedgehog-interacting protein (HHIP) gene and several expressed sequence tags cloned from fetal lung. Though it is unclear what gene or regulatory effect explains the association, the region warrants further investigation.
DOI:10.1038/ng.500URLPMID:20010835 [本文引用: 5]

Spirometric measures of lung function are heritable traits that reflect respiratory health and predict morbidity and mortality. We meta-analyzed genome-wide association studies for two clinically important lung-function measures: forced expiratory volume in the first second (FEV(1)) and its ratio to forced vital capacity (FEV(1)/FVC), an indicator of airflow obstruction. This meta-analysis included 20,890 participants of European ancestry from four CHARGE Consortium studies: Atherosclerosis Risk in Communities, Cardiovascular Health Study, Framingham Heart Study and Rotterdam Study. We identified eight loci associated with FEV(1)/FVC (HHIP, GPR126, ADAM19, AGER-PPT2, FAM13A, PTCH1, PID1 and HTR4) and one locus associated with FEV(1) (INTS12-GSTCD-NPNT) at or near genome-wide significance (P < 5 x 10(-8)) in the CHARGE Consortium dataset. Our findings may offer insights into pulmonary function and pathogenesis of chronic lung disease.
DOI:10.1038/ng.501URLPMID:20010834 [本文引用: 3]

Pulmonary function measures are heritable traits that predict morbidity and mortality and define chronic obstructive pulmonary disease (COPD). We tested genome-wide association with forced expiratory volume in 1 s (FEV(1)) and the ratio of FEV(1) to forced vital capacity (FVC) in the SpiroMeta consortium (n = 20,288 individuals of European ancestry). We conducted a meta-analysis of top signals with data from direct genotyping (n < or = 32,184 additional individuals) and in silico summary association data from the CHARGE Consortium (n = 21,209) and the Health 2000 survey (n < or = 883). We confirmed the reported locus at 4q31 and identified associations with FEV(1) or FEV(1)/FVC and common variants at five additional loci: 2q35 in TNS1 (P = 1.11 x 10(-12)), 4q24 in GSTCD (2.18 x 10(-23)), 5q33 in HTR4 (P = 4.29 x 10(-9)), 6p21 in AGER (P = 3.07 x 10(-15)) and 15q23 in THSD4 (P = 7.24 x 10(-15)). mRNA analyses showed expression of TNS1, GSTCD, AGER, HTR4 and THSD4 in human lung tissue. These associations offer mechanistic insight into pulmonary function regulation and indicate potential targets for interventions to alleviate respiratory disease.
DOI:10.1371/journal.pgen.1003098URLPMID:23284291 [本文引用: 1]

Genome-wide association studies have identified numerous genetic loci for spirometic measures of pulmonary function, forced expiratory volume in one second (FEV(1)), and its ratio to forced vital capacity (FEV(1)/FVC). Given that cigarette smoking adversely affects pulmonary function, we conducted genome-wide joint meta-analyses (JMA) of single nucleotide polymorphism (SNP) and SNP-by-smoking (ever-smoking or pack-years) associations on FEV(1) and FEV(1)/FVC across 19 studies (total N = 50,047). We identified three novel loci not previously associated with pulmonary function. SNPs in or near DNER (smallest P(JMA = )5.00x10(-11)), HLA-DQB1 and HLA-DQA2 (smallest P(JMA = )4.35x10(-9)), and KCNJ2 and SOX9 (smallest P(JMA = )1.28x10(-8)) were associated with FEV(1)/FVC or FEV(1) in meta-analysis models including SNP main effects, smoking main effects, and SNP-by-smoking (ever-smoking or pack-years) interaction. The HLA region has been widely implicated for autoimmune and lung phenotypes, unlike the other novel loci, which have not been widely implicated. We evaluated DNER, KCNJ2, and SOX9 and found them to be expressed in human lung tissue. DNER and SOX9 further showed evidence of differential expression in human airway epithelium in smokers compared to non-smokers. Our findings demonstrated that joint testing of SNP and SNP-by-environment interaction identified novel loci associated with complex traits that are missed when considering only the genetic main effects.
DOI:10.1038/ng.3011URLPMID:24929828 [本文引用: 2]

Forced vital capacity (FVC), a spirometric measure of pulmonary function, reflects lung volume and is used to diagnose and monitor lung diseases. We performed genome-wide association study meta-analysis of FVC in 52,253 individuals from 26 studies and followed up the top associations in 32,917 additional individuals of European ancestry. We found six new regions associated at genome-wide significance (P < 5 x 10(-8)) with FVC in or near EFEMP1, BMP6, MIR129-2-HSD17B12, PRDM11, WWOX and KCNJ2. Two loci previously associated with spirometric measures (GSTCD and PTCH1) were related to FVC. Newly implicated regions were followed up in samples from African-American, Korean, Chinese and Hispanic individuals. We detected transcripts for all six newly implicated genes in human lung tissue. The new loci may inform mechanisms involved in lung development and the pathogenesis of restrictive lung disease.
DOI:10.1038/s41588-018-0321-7URLPMID:30804560 [本文引用: 2]

Reduced lung function predicts mortality and is key to the diagnosis of chronic obstructive pulmonary disease (COPD). In a genome-wide association study in 400,102 individuals of European ancestry, we define 279 lung function signals, 139 of which are new. In combination, these variants strongly predict COPD in independent populations. Furthermore, the combined effect of these variants showed generalizability across smokers and never smokers, and across ancestral groups. We highlight biological pathways, known and potential drug targets for COPD and, in phenome-wide association studies, autoimmune-related and other pleiotropic effects of lung function-associated variants. This new genetic evidence has potential to improve future preventive and therapeutic strategies for COPD.
DOI:10.1038/s41588-018-0342-2URLPMID:30804561 [本文引用: 5]

Chronic obstructive pulmonary disease (COPD) is the leading cause of respiratory mortality worldwide. Genetic risk loci provide new insights into disease pathogenesis. We performed a genome-wide association study in 35,735 cases and 222,076 controls from the UK Biobank and additional studies from the International COPD Genetics Consortium. We identified 82 loci associated with P < 5 x 10(-8); 47 of these were previously described in association with either COPD or population-based measures of lung function. Of the remaining 35 new loci, 13 were associated with lung function in 79,055 individuals from the SpiroMeta consortium. Using gene expression and regulation data, we identified functional enrichment of COPD risk loci in lung tissue, smooth muscle, and several lung cell types. We found 14 COPD loci shared with either asthma or pulmonary fibrosis. COPD genetic risk loci clustered into groups based on associations with quantitative imaging features and comorbidities. Our analyses provide further support for the genetic susceptibility and heterogeneity of COPD.
DOI:10.1164/rccm.201102-0192OCURLPMID:21965014 [本文引用: 4]

RATIONALE: Genomic loci are associated with FEV1 or the ratio of FEV1 to FVC in population samples, but their association with chronic obstructive pulmonary disease (COPD) has not yet been proven, nor have their combined effects on lung function and COPD been studied. OBJECTIVES: To test association with COPD of variants at five loci (TNS1, GSTCD, HTR4, AGER, and THSD4) and to evaluate joint effects on lung function and COPD of these single-nucleotide polymorphisms (SNPs), and variants at the previously reported locus near HHIP. METHODS: By sampling from 12 population-based studies (n = 31,422), we obtained genotype data on 3,284 COPD case subjects and 17,538 control subjects for sentinel SNPs in TNS1, GSTCD, HTR4, AGER, and THSD4. In 24,648 individuals (including 2,890 COPD case subjects and 13,862 control subjects), we additionally obtained genotypes for rs12504628 near HHIP. Each allele associated with lung function decline at these six SNPs contributed to a risk score. We studied the association of the risk score to lung function and COPD. MEASUREMENTS AND MAIN RESULTS: Association with COPD was significant for three loci (TNS1, GSTCD, and HTR4) and the previously reported HHIP locus, and suggestive and directionally consistent for AGER and TSHD4. Compared with the baseline group (7 risk alleles), carrying 10-12 risk alleles was associated with a reduction in FEV1 (beta = -72.21 ml, P = 3.90 x 10(-4)) and FEV1/FVC (beta = -1.53%, P = 6.35 x 10(-6)), and with COPD (odds ratio = 1.63, P = 1.46 x 10(-5)). CONCLUSIONS: Variants in TNS1, GSTCD, and HTR4 are associated with COPD. Our highest risk score category was associated with a 1.6-fold higher COPD risk than the population average score.
DOI:10.1165/rcmb.2011-0055OCURLPMID:21659657 [本文引用: 5]

Two recent metaanalyses of genome-wide association studies conducted by the CHARGE and SpiroMeta consortia identified novel loci yielding evidence of association at or near genome-wide significance (GWS) with FEV(1) and FEV(1)/FVC. We hypothesized that a subset of these markers would also be associated with chronic obstructive pulmonary disease (COPD) susceptibility. Thirty-two single-nucleotide polymorphisms (SNPs) in or near 17 genes in 11 previously identified GWS spirometric genomic regions were tested for association with COPD status in four COPD case-control study samples (NETT/NAS, the Norway case-control study, ECLIPSE, and the first 1,000 subjects in COPDGene; total sample size, 3,456 cases and 1,906 controls). In addition to testing the 32 spirometric GWS SNPs, we tested a dense panel of imputed HapMap2 SNP markers from the 17 genes located near the 32 GWS SNPs and in a set of 21 well studied COPD candidate genes. Of the previously identified GWS spirometric genomic regions, three loci harbored SNPs associated with COPD susceptibility at a 5% false discovery rate: the 4q24 locus including FLJ20184/INTS12/GSTCD/NPNT, the 6p21 locus including AGER and PPT2, and the 5q33 locus including ADAM19. In conclusion, markers previously associated at or near GWS with spirometric measures were tested for association with COPD status in data from four COPD case-control studies, and three loci showed evidence of association with COPD susceptibility at a 5% false discovery rate.
DOI:10.1016/S2213-2600(14)70002-5URLPMID:24621683 [本文引用: 1]

BACKGROUND: The genetic risk factors for susceptibility to chronic obstructive pulmonary disease (COPD) are still largely unknown. Additional genetic variants are likely to be identified by genome-wide association studies in larger cohorts or specific subgroups. We sought to identify risk loci for moderate to severe and severe COPD with data from several cohort studies. METHODS: We combined genome-wide association analysis data from participants in the COPDGene study (non-Hispanic white and African-American ethnic origin) and the ECLIPSE, NETT/NAS, and Norway GenKOLS studies (self-described white ethnic origin). We did analyses comparing control individuals with individuals with moderate to severe COPD and with a subset of individuals with severe COPD. Single nucleotide polymorphisms yielding a p value of less than 5 x 10(-7) in the meta-analysis at loci not previously described were genotyped in individuals from the family-based ICGN study. We combined results in a joint meta-analysis (threshold for significance p<5 x 10(-8)). FINDINGS: Analysis of 6633 individuals with moderate to severe COPD and 5704 control individuals confirmed association at three known loci: CHRNA3 (p=6.38 x 10(-14)), FAM13A (p=1.12 x 10(-14)), and HHIP (p=1.57 x 10(-12)). We also showed significant evidence of association at a novel locus near RIN3 (p=5.25 x 10(-9)). In the overall meta-analysis (ie, including data from 2859 ICGN participants), the association with RIN3 remained significant (p=5.4 x 10(-9)). 3497 individuals were included in our analysis of severe COPD. The effect estimates for the loci near HHIP and CHRNA3 were significantly stronger in severe disease than in moderate to severe disease (p<0.01). We also identified associations at two additional loci: MMP12 (overall joint meta-analysis p=2.6 x 10(-9)) and TGFB2 (overall joint meta-analysis p=8.3 x 10(-9)). INTERPRETATION: We have confirmed associations with COPD at three known loci and identified three new genome-wide significant associations. Genetic variants other than in alpha-1 antitrypsin increase the risk of COPD. FUNDING: US National Heart, Lung, and Blood Institute; the Alpha-1 Foundation; the COPD Foundation through contributions from AstraZeneca, Boehringer Ingelheim, Novartis, and Sepracor; GlaxoSmithKline; Centers for Medicare and Medicaid Services; Agency for Healthcare Research and Quality; and US Department of Veterans Affairs.
DOI:10.1186/s12863-015-0299-4URLPMID:26634245 [本文引用: 1]

BACKGROUND: Pulmonary function decline is a major contributor to morbidity and mortality among smokers. Post bronchodilator FEV1 and FEV1/FVC ratio are considered the standard assessment of airflow obstruction. We performed a genome-wide association study (GWAS) in 9919 current and former smokers in the COPDGene study (6659 non-Hispanic Whites [NHW] and 3260 African Americans [AA]) to identify associations with spirometric measures (post-bronchodilator FEV1 and FEV1/FVC). We also conducted meta-analysis of FEV1 and FEV1/FVC GWAS in the COPDGene, ECLIPSE, and GenKOLS cohorts (total n = 13,532). RESULTS: Among NHW in the COPDGene cohort, both measures of pulmonary function were significantly associated with SNPs at the 15q25 locus [containing CHRNA3/5, AGPHD1, IREB2, CHRNB4] (lowest p-value = 2.17 x 10(-11)), and FEV1/FVC was associated with a genomic region on chromosome 4 [upstream of HHIP] (lowest p-value = 5.94 x 10(-10)); both regions have been previously associated with COPD. For the meta-analysis, in addition to confirming associations to the regions near CHRNA3/5 and HHIP, genome-wide significant associations were identified for FEV1 on chromosome 1 [TGFB2] (p-value = 8.99 x 10(-9)), 9 [DBH] (p-value = 9.69 x 10(-9)) and 19 [CYP2A6/7] (p-value = 3.49 x 10(-8)) and for FEV1/FVC on chromosome 1 [TGFB2] (p-value = 8.99 x 10(-9)), 4 [FAM13A] (p-value = 3.88 x 10(-12)), 11 [MMP3/12] (p-value = 3.29 x 10(-10)) and 14 [RIN3] (p-value = 5.64 x 10(-9)). CONCLUSIONS: In a large genome-wide association study of lung function in smokers, we found genome-wide significant associations at several previously described loci with lung function or COPD. We additionally identified a novel genome-wide significant locus with FEV1 on chromosome 9 [DBH] in a meta-analysis of three study populations.
DOI:10.1183/09031936.00093314URLPMID:25234806 [本文引用: 2]

Smoking is a notorious risk factor for chronic mucus hypersecretion (CMH). CMH frequently occurs in chronic obstructive pulmonary disease (COPD). The question arises whether the same single-nucleotide polymorphisms (SNPs) are related to CMH in smokers with and without COPD. We performed two genome-wide association studies of CMH under an additive genetic model in male heavy smokers (>/=20 pack-years) with COPD (n=849, 39.9% CMH) and without COPD (n=1348, 25.4% CMH), followed by replication and meta-analysis in comparable populations, and assessment of the functional relevance of significantly associated SNPs. Genome-wide association analysis of CMH in COPD and non-COPD subjects yielded no genome-wide significance after replication. In COPD, our top SNP (rs10461985, p=5.43x10(-5)) was located in the GDNF-AS1 gene that is functionally associated with the GDNF gene. Expression of GDNF in bronchial biopsies of COPD patients was significantly associated with CMH (p=0.007). In non-COPD subjects, four SNPs had a p-value <10(-5) in the meta-analysis, including a SNP (rs4863687) in the MAML3 gene, the T-allele showing modest association with CMH (p=7.57x10(-6), OR 1.48) and with significantly increased MAML3 expression in lung tissue (p=2.59x10(-12)). Our data suggest the potential for differential genetic backgrounds of CMH in individuals with and without COPD.
DOI:10.1038/ncomms9658URLPMID:26635082 [本文引用: 3]

Lung function measures are used in the diagnosis of chronic obstructive pulmonary disease. In 38,199 European ancestry individuals, we studied genome-wide association of forced expiratory volume in 1 s (FEV1), forced vital capacity (FVC) and FEV1/FVC with 1000 Genomes Project (phase 1)-imputed genotypes and followed up top associations in 54,550 Europeans. We identify 14 novel loci (P<5 x 10(-8)) in or near ENSA, RNU5F-1, KCNS3, AK097794, ASTN2, LHX3, CCDC91, TBX3, TRIP11, RIN3, TEKT5, LTBP4, MN1 and AP1S2, and two novel signals at known loci NPNT and GPR126, providing a basis for new understanding of the genetic determinants of these traits and pulmonary diseases in which they are altered.
DOI:10.1164/rccm.201202-0366OCURLPMID:22837378 [本文引用: 2]

RATIONALE: Genome-wide association studies (GWAS) have identified loci influencing lung function, but fewer genes influencing chronic obstructive pulmonary disease (COPD) are known. OBJECTIVES: Perform meta-analyses of GWAS for airflow obstruction, a key pathophysiologic characteristic of COPD assessed by spirometry, in population-based cohorts examining all participants, ever smokers, never smokers, asthma-free participants, and more severe cases. METHODS: Fifteen cohorts were studied for discovery (3,368 affected; 29,507 unaffected), and a population-based family study and a meta-analysis of case-control studies were used for replication and regional follow-up (3,837 cases; 4,479 control subjects). Airflow obstruction was defined as FEV(1) and its ratio to FVC (FEV(1)/FVC) both less than their respective lower limits of normal as determined by published reference equations. MEASUREMENTS AND MAIN RESULTS: The discovery meta-analyses identified one region on chromosome 15q25.1 meeting genome-wide significance in ever smokers that includes AGPHD1, IREB2, and CHRNA5/CHRNA3 genes. The region was also modestly associated among never smokers. Gene expression studies confirmed the presence of CHRNA5/3 in lung, airway smooth muscle, and bronchial epithelial cells. A single-nucleotide polymorphism in HTR4, a gene previously related to FEV(1)/FVC, achieved genome-wide statistical significance in combined meta-analysis. Top single-nucleotide polymorphisms in ADAM19, RARB, PPAP2B, and ADAMTS19 were nominally replicated in the COPD meta-analysis. CONCLUSIONS: These results suggest an important role for the CHRNA5/3 region as a genetic risk factor for airflow obstruction that may be independent of smoking and implicate the HTR4 gene in the etiology of airflow obstruction.
DOI:10.1164/rccm.201707-1493OCURLPMID:29394082 [本文引用: 1]

RATIONALE: Lung function and chronic obstructive pulmonary disease (COPD) are heritable traits. Genome-wide association studies (GWAS) have identified numerous pulmonary function and COPD loci, primarily in cohorts of European ancestry. OBJECTIVES: Perform a GWAS of COPD phenotypes in Hispanic/Latino populations to identify loci not previously detected in European populations. METHODS: GWAS of lung function and COPD in Hispanic/Latino participants from a population-based cohort. We performed replication studies of novel loci in independent studies. MEASUREMENTS AND MAIN RESULTS: Among 11,822 Hispanic/Latino participants, we identified eight novel signals; three replicated in independent populations of European Ancestry. A novel locus for FEV1 in ZSWIM7 (rs4791658; P = 4.99 x 10(-9)) replicated. A rare variant (minor allele frequency = 0.002) in HAL (rs145174011) was associated with FEV1/FVC (P = 9.59 x 10(-9)) in a region previously identified for COPD-related phenotypes; it remained significant in conditional analyses but did not replicate. Admixture mapping identified a novel region, with a variant in AGMO (rs41331850), associated with Amerindian ancestry and FEV1, which replicated. A novel locus for FEV1 identified among ever smokers (rs291231; P = 1.92 x 10(-8)) approached statistical significance for replication in admixed populations of African ancestry, and a novel SNP for COPD in PDZD2 (rs7709630; P = 1.56 x 10(-8)) regionally replicated. In addition, loci previously identified for lung function in European samples were associated in Hispanic/Latino participants in the Hispanic Community Health Study/Study of Latinos at the genome-wide significance level. CONCLUSIONS: We identified novel signals for lung function and COPD in a Hispanic/Latino cohort. Including admixed populations when performing genetic studies may identify variants contributing to genetic etiologies of COPD.
DOI:10.12688/wellcomeopenres.12583.3URLPMID:30175238 [本文引用: 2]

Background: Over 90 regions of the genome have been associated with lung function to date, many of which have also been implicated in chronic obstructive pulmonary disease. Methods: We carried out meta-analyses of exome array data and three lung function measures: forced expiratory volume in one second (FEV 1), forced vital capacity (FVC) and the ratio of FEV 1 to FVC (FEV 1/FVC). These analyses by the SpiroMeta and CHARGE consortia included 60,749 individuals of European ancestry from 23 studies, and 7,721 individuals of African Ancestry from 5 studies in the discovery stage, with follow-up in up to 111,556 independent individuals. Results: We identified significant (P<2.8x10 (-7)) associations with six SNPs: a nonsynonymous variant in RPAP1, which is predicted to be damaging, three intronic SNPs ( SEC24C, CASC17 and UQCC1) and two intergenic SNPs near to LY86 and FGF10. Expression quantitative trait loci analyses found evidence for regulation of gene expression at three signals and implicated several genes, including TYRO3 and PLAU. Conclusions: Further interrogation of these loci could provide greater understanding of the determinants of lung function and pulmonary disease.
DOI:10.1186/s12920-014-0072-yURL [本文引用: 2]
DOI:10.1371/journal.pgen.1000421URLPMID:19300482 [本文引用: 1]

There is considerable variability in the susceptibility of smokers to develop chronic obstructive pulmonary disease (COPD). The only known genetic risk factor is severe deficiency of alpha(1)-antitrypsin, which is present in 1-2% of individuals with COPD. We conducted a genome-wide association study (GWAS) in a homogenous case-control cohort from Bergen, Norway (823 COPD cases and 810 smoking controls) and evaluated the top 100 single nucleotide polymorphisms (SNPs) in the family-based International COPD Genetics Network (ICGN; 1891 Caucasian individuals from 606 pedigrees) study. The polymorphisms that showed replication were further evaluated in 389 subjects from the US National Emphysema Treatment Trial (NETT) and 472 controls from the Normative Aging Study (NAS) and then in a fourth cohort of 949 individuals from 127 extended pedigrees from the Boston Early-Onset COPD population. Logistic regression models with adjustments of covariates were used to analyze the case-control populations. Family-based association analyses were conducted for a diagnosis of COPD and lung function in the family populations. Two SNPs at the alpha-nicotinic acetylcholine receptor (CHRNA 3/5) locus were identified in the genome-wide association study. They showed unambiguous replication in the ICGN family-based analysis and in the NETT case-control analysis with combined p-values of 1.48 x 10(-10), (rs8034191) and 5.74 x 10(-10) (rs1051730). Furthermore, these SNPs were significantly associated with lung function in both the ICGN and Boston Early-Onset COPD populations. The C allele of the rs8034191 SNP was estimated to have a population attributable risk for COPD of 12.2%. The association of hedgehog interacting protein (HHIP) locus on chromosome 4 was also consistently replicated, but did not reach genome-wide significance levels. Genome-wide significant association of the HHIP locus with lung function was identified in the Framingham Heart study (Wilk et al., companion article in this issue of PLoS Genetics; doi:10.1371/journal.pgen.1000429). The CHRNA 3/5 and the HHIP loci make a significant contribution to the risk of COPD. CHRNA3/5 is the same locus that has been implicated in the risk of lung cancer.
DOI:10.1038/ng.535URLPMID:20173748 [本文引用: 1]

We performed a genome-wide association study for chronic obstructive pulmonary disease (COPD) in three population cohorts, including 2,940 cases and 1,380 controls who were current or former smokers with normal lung function. We identified a new susceptibility locus at 4q22.1 in FAM13A and replicated this association in one case-control group (n = 1,006) and two family-based cohorts (n = 3,808) (rs7671167, combined P = 1.2 x 10(-11), combined odds ratio in case-control studies 0.76, 95% confidence interval 0.69-0.83).
DOI:10.1183/09031936.00129509URLPMID:19996190 [本文引用: 1]

The Hedgehog signalling pathway plays an important role in lung morphogenesis and cellular responses to lung injury. A genome-wide association study has demonstrated that two single nucleotide polymorphisms (SNPs) near the Hedgehog-interacting protein (Hip) gene, SNP identifiers rs1828591 and rs13118928, are associated with risk of chronic obstructive pulmonary disease (COPD). The aim of the present study was to validate the observed association between genetic variation near the Hip gene and COPD, and to investigate whether risk estimates were modified by smoking behaviour. The association between the Hip gene SNPs and COPD was investigated in the Rotterdam Study by logistic regression analyses, adjusted for several covariates. In addition, an association meta-analysis was performed that included data from the genome-wide association study on COPD. Both SNPs were significantly associated with risk of COPD (OR 0.80; 95% CI 0.72-0.91). Homozygosity for the minor G allele resulted in a decreased risk of COPD of approximately 40% (95% CI 0.47-0.78). There was a significant interaction with the number of pack-years of smoking (p = 0.004). The meta-analysis yielded an odds ratio for COPD of 0.80 per additional G allele (p = 3.4 x 10(-9)). Genetic variation near the Hip gene was significantly associated with risk of COPD, depending on the number of pack-years of smoking.
DOI:10.1136/thoraxjnl-2011-200017URL [本文引用: 1]

Rationale Traditional genome-wide association studies (GWASs) of large cohorts of subjects with chronic obstructive pulmonary disease (COPD) have successfully identified novel candidate genes, but several other plausible loci do not meet strict criteria for genome-wide significance after correction for multiple testing.
Objectives The authors hypothesise that by applying unbiased weights derived from unique populations we can identify additional COPD susceptibility loci.
Methods The authors performed a homozygosity haplotype analysis on a group of subjects with and without COPD to identify regions of conserved homozygosity haplotype (RCHHs). Weights were constructed based on the frequency of these RCHHs in case versus controls, and used to adjust the p values from a large collaborative GWAS of COPD.
Results The authors identified 2318 RCHHs, of which 576 were significantly (p<0.05) over-represented in cases. After applying the weights constructed from these regions to a collaborative GWAS of COPD, the authors identified two single nucleotide polymorphisms (SNPs) in a novel gene (fibroblast growth factor-7 (FGF7)) that gained genome-wide significance by the false discovery rate method. In a follow-up analysis, both SNPs (rs12591300 and rs4480740) were significantly associated with COPD in an independent population (combined p values of 7.9E-7 and 2.8E-6, respectively). In another independent population, increased lung tissue FGF7 expression was associated with worse measures of lung function.
Conclusion Weights constructed from a homozygosity haplotype analysis of an isolated population successfully identify novel genetic associations from a GWAS on a separate population. This method can be used to identify promising candidate genes that fail to meet strict correction for multiple testing.
DOI:10.1093/hmg/ddr524URL [本文引用: 1]

The genetic risk factors for chronic obstructive pulmonary disease (COPD) are still largely unknown. To date, genome-wide association studies (GWASs) of limited size have identified several novel risk loci for COPD at CHRNA3/CHRNA5/IREB2, HHIP and FAM13A; additional loci may be identified through larger studies. We performed a GWAS using a total of 3499 cases and 1922 control subjects from four cohorts: the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE); the Normative Aging Study (NAS) and National Emphysema Treatment Trial (NETT); Bergen, Norway (GenKOLS); and the COPDGene study. Genotyping was performed on Illumina platforms with additional markers imputed using 1000 Genomes data; results were summarized using fixed-effect meta-analysis. We identified a new genomewide significant locus on chromosome 19q13 (rs7937, OR = 0.74, P = 2.9 x 10(-9)). Genotyping this single nucleotide polymorphism (SNP) and another nearby SNP in linkage disequilibrium (rs2604894) in 2859 subjects from the family-based International COPD Genetics Network study (ICGN) demonstrated supportive evidence for association for COPD (P = 0.28 and 0.11 for rs7937 and rs2604894), pre-bronchodilator FEV1 (P = 0.08 and 0.04) and severe (GOLD 3&4) COPD (P = 0.09 and 0.017). This region includes RAB4B, EGLN2, MIA and CYP2A6, and has previously been identified in association with cigarette smoking behavior.
DOI:10.1165/rcmb.2014-0198OCURLPMID:25514360 [本文引用: 1]

Increased airway responsiveness is linked to lung function decline and mortality in subjects with chronic obstructive pulmonary disease (COPD); however, the genetic contribution to airway responsiveness remains largely unknown. A genome-wide association study (GWAS) was performed using the Illumina (San Diego, CA) Human660W-Quad BeadChip on European Americans with COPD from the Lung Health Study. Linear regression models with correlated meta-analyses, including data from baseline (n = 2,814) and Year 5 (n = 2,657), were used to test for common genetic variants associated with airway responsiveness. Genotypic imputation was performed using reference 1000 Genomes Project data. Expression quantitative trait loci (eQTL) analyses in lung tissues were assessed for the top 10 markers identified, and immunohistochemistry assays assessed protein staining for SGCD and MYH15. Four genes were identified within the top 10 associations with airway responsiveness. Markers on chromosome 9p21.2 flanked by LINGO2 met a predetermined threshold of genome-wide significance (P < 9.57 x 10(-8)). Markers on chromosomes 3q13.1 (flanked by MYH15), 5q33 (SGCD), and 6q21 (PDSS2) yielded suggestive evidence of association (9.57 x 10(-8) < P
DOI:10.1183/09031936.00216013URL [本文引用: 1]

Individuals with chronic obstructive pulmonary disease (COPD) and asthma are an important but poorly characterised group. The genetic determinants of COPD and asthma overlap have not been studied. The aim of this study was to identify clinical features and genetic risk factors for COPD and asthma overlap.
Subjects were current or former smoking non-Hispanic whites or African-Americans with COPD. Overlap subjects reported a history of physician-diagnosed asthma before the age of 40 years. We compared clinical and radiographic features between COPD and overlap subjects. We performed genome-wide association studies (GWAS) in the non-Hispanic whites and African-American populations, and combined these results in a meta-analysis. More females and African-Americans reported a history of asthma. Overlap subjects had more severe and more frequent respiratory exacerbations, less emphysema and greater airway wall thickness compared to subjects with COPD alone.
The non-Hispanic white GWAS identified single nucleotide polymorphisms in the genes CSMD1 (rs11779254, p=1.57 x 10(-6)) and SOX5 (rs59569785, p=1.61 x 10-6) and the meta-analysis identified single nucleotide polymorphisms in the gene GPR65 (rs6574978, p=1.18 x 10(-7)) associated with COPD and asthma overlap.
Overlap subjects have more exacerbations, less emphysema and more airway disease for any degree of lung function impairment compared to COPD alone. We identified novel genetic variants associated with this syndrome. COPD and asthma overlap is an important syndrome and may require distinct clinical management.
DOI:10.1186/s12931-014-0113-2URL [本文引用: 2]
DOI:10.1513/AnnalsATS.201408-380OCURLPMID:25584925

RATIONALE: Genome-wide association studies (GWAS) of chronic obstructive pulmonary disease (COPD) have identified disease-susceptibility loci, mostly in subjects of European descent. OBJECTIVES: We hypothesized that by studying Hispanic populations we would be able to identify unique loci that contribute to COPD pathogenesis in Hispanics but remain undetected in GWAS of non-Hispanic populations. METHODS: We conducted a metaanalysis of two GWAS of COPD in independent cohorts of Hispanics in Costa Rica and the United States (Multi-Ethnic Study of Atherosclerosis [MESA]). We performed a replication study of the top single-nucleotide polymorphisms in an independent Hispanic cohort in New Mexico (the Lovelace Smokers Cohort). We also attempted to replicate prior findings from genome-wide studies in non-Hispanic populations in Hispanic cohorts. MEASUREMENTS AND MAIN RESULTS: We found no genome-wide significant association with COPD in our metaanalysis of Costa Rica and MESA. After combining the top results from this metaanalysis with those from our replication study in the Lovelace Smokers Cohort, we identified two single-nucleotide polymorphisms approaching genome-wide significance for an association with COPD. The first (rs858249, combined P value = 6.1 x 10(-8)) is near the genes KLHL7 and NUPL2 on chromosome 7. The second (rs286499, combined P value = 8.4 x 10(-8)) is located in an intron of DLG2. The two most significant single-nucleotide polymorphisms in FAM13A from a previous genome-wide study in non-Hispanics were associated with COPD in Hispanics. CONCLUSIONS: We have identified two novel loci (in or near the genes KLHL7/NUPL2 and DLG2) that may play a role in COPD pathogenesis in Hispanic populations.
DOI:10.1165/rcmb.2014-0198OCURLPMID:25514360

Increased airway responsiveness is linked to lung function decline and mortality in subjects with chronic obstructive pulmonary disease (COPD); however, the genetic contribution to airway responsiveness remains largely unknown. A genome-wide association study (GWAS) was performed using the Illumina (San Diego, CA) Human660W-Quad BeadChip on European Americans with COPD from the Lung Health Study. Linear regression models with correlated meta-analyses, including data from baseline (n = 2,814) and Year 5 (n = 2,657), were used to test for common genetic variants associated with airway responsiveness. Genotypic imputation was performed using reference 1000 Genomes Project data. Expression quantitative trait loci (eQTL) analyses in lung tissues were assessed for the top 10 markers identified, and immunohistochemistry assays assessed protein staining for SGCD and MYH15. Four genes were identified within the top 10 associations with airway responsiveness. Markers on chromosome 9p21.2 flanked by LINGO2 met a predetermined threshold of genome-wide significance (P < 9.57 x 10(-8)). Markers on chromosomes 3q13.1 (flanked by MYH15), 5q33 (SGCD), and 6q21 (PDSS2) yielded suggestive evidence of association (9.57 x 10(-8) < P
DOI:10.1165/rcmb.2014-0135OCURLPMID:24825563

Hypoxemia is a major complication of chronic obstructive pulmonary disease (COPD) that correlates with disease prognosis. Identifying genetic variants associated with oxygenation may provide clues for deciphering the heterogeneity in prognosis among patients with COPD. However, previous genetic studies have been restricted to investigating COPD candidate genes for association with hypoxemia. To report results from the first genome-wide association study (GWAS) of resting oxygen saturation (as measured by pulse oximetry [Spo2]) in subjects with COPD, we performed a GWAS of Spo2 in two large, well characterized COPD populations: COPDGene, including both the non-Hispanic white (NHW) and African American (AA) groups, and Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE). We identified several suggestive loci (P < 1 x 10(-5)) associated with Spo2 in COPDGene in the NHW (n = 2810) and ECLIPSE (n = 1758) groups, and two loci on chromosomes 14 and 15 in the AA group (n = 820) from COPDGene achieving a level of genome-wide significance (P < 5 x 10(-8)). The chromosome 14 single-nucleotide polymorphism, rs6576132, located in an intergenic region, was nominally replicated (P < 0.05) in the NHW group from COPDGene. The chromosome 15 single-nucleotide polymorphisms were rare in subjects of European ancestry, so the results could not be replicated. The chromosome 15 region contains several genes, including TICRR and KIF7, and is proximal to RHCG (Rh family C glyocoprotein gene). We have identified two loci associated with resting oxygen saturation in AA subjects with COPD, and several suggestive regions in subjects of European descent with COPD. Our study highlights the importance of investigating the genetics of complex traits in different racial groups.
[本文引用: 1]
DOI:10.1016/j.ygeno.2013.02.010URL [本文引用: 1]

Hedgehog interacting protein (HHIP) was implicated in chronic obstructive pulmonary disease (COPD) by genome-wide association studies (GWAS). However, it remains unclear how HHIP contributes to COPD pathogenesis. To identify genes regulated by HHIP, we performed gene expression microarray analysis in a human bronchial epithelial cell line (Beas-2B) stably infected with HHIP shRNAs. HHIP silencing led to differential expression of 296 genes; enrichment for variants nominally associated with COPD was found. Eighteen of the differentially expressed genes were validated by real-time PCR in Beas-2B cells. Seven of 11 validated genes tested in human COPD and control lung tissues demonstrated significant gene expression differences. Functional annotation indicated enrichment for extracellular matrix and cell growth genes. Network modeling demonstrated that the extracellular matrix and cell proliferation genes influenced by HHIP tended to be interconnected. Thus, we identified potential HHIP targets in human bronchial epithelial cells that may contribute to COPD pathogenesis. (c) 2013 Elsevier Inc.
DOI:10.1164/rccm.201505-0999OCURLPMID:26862784 [本文引用: 1]

RATIONALE: A genetic locus within the FAM13A gene has been consistently associated with chronic obstructive pulmonary disease (COPD) in genome-wide association studies. However, the mechanisms by which FAM13A contributes to COPD susceptibility are unknown. OBJECTIVES: To determine the biologic function of FAM13A in human COPD and murine COPD models and discover the molecular mechanism by which FAM13A influences COPD susceptibility. METHODS: Fam13a null mice (Fam13a(-/-)) were generated and exposed to cigarette smoke. The lung inflammatory response and airspace size were assessed in Fam13a(-/-) and Fam13a(+/+) littermate control mice. Cellular localization of FAM13A protein and mRNA levels of FAM13A in COPD lungs were assessed using immunofluorescence, Western blotting, and reverse transcriptase-polymerase chain reaction, respectively. Immunoprecipitation followed by mass spectrometry identified cellular proteins that interact with FAM13A to reveal insights on FAM13A's function. MEASUREMENTS AND MAIN RESULTS: In murine and human lungs, FAM13A is expressed in airway and alveolar type II epithelial cells and macrophages. Fam13a null mice (Fam13a(-/-)) were resistant to chronic cigarette smoke-induced emphysema compared with Fam13a(+/+) mice. In vitro, FAM13A interacts with protein phosphatase 2A and recruits protein phosphatase 2A with glycogen synthase kinase 3beta and beta-catenin, inducing beta-catenin degradation. Fam13a(-/-) mice were also resistant to elastase-induced emphysema, and this resistance was reversed by coadministration of a beta-catenin inhibitor, suggesting that FAM13A could increase the susceptibility of mice to emphysema development by inhibiting beta-catenin signaling. Moreover, human COPD lungs had decreased protein levels of beta-catenin and increased protein levels of FAM13A. CONCLUSIONS: We show that FAM13A may influence COPD susceptibility by promoting beta-catenin degradation.
DOI:10.1165/rcmb.2016-0282OCURLPMID:28199134 [本文引用: 1]

Bioenergetics homeostasis is important for cells to sustain normal functions and defend against injury. The genetic controls of bioenergetics homeostasis, especially lipid metabolism, remain poorly understood in chronic obstructive pulmonary disease (COPD), the third leading cause of death in the world. Additionally, the biological function of most of the susceptibility genes identified from genome-wide association studies (GWASs) in COPD remains unclear. Here, we aimed to address (1) how fatty acid oxidation (FAO), specifically beta-oxidation, a key lipid metabolism pathway that provides energy to cells, contributes to cigarette smoke (CS)-induced COPD; and (2) whether-and if so, how-FAM13A (family with sequence similarity 13 member A), a well-replicated COPD GWAS gene, modulates the FAO pathway. We demonstrated that CS induced expression of carnitine palmitoyltransferase 1A (CPT1A), a key mitochondrial enzyme for the FAO pathway, thereby enhancing FAO. Pharmacological inhibition of FAO by etomoxir blunted CS-induced reactive oxygen species accumulation and cell death in lung epithelial cells. FAM13A promoted FAO, possibly by interacting with and activating sirutin 1, and increasing expression of CPT1A. Furthermore, CS-induced cell death was reduced in lungs from Fam13a(-/-) mice. Our results suggest that FAM13A, the COPD GWAS gene, shapes the cellular metabolic response to CS exposure by promoting the FAO pathway, which may contribute to COPD development.
DOI:10.1371/journal.pone.0074630URLPMID:24058608 [本文引用: 1]

Genome-Wide Association Study (GWAS) meta-analyses have identified a strong association signal for lung function, which maps to a region on 4q24 containing two oppositely transcribed genes: glutathione S-transferase, C-terminal domain containing (GSTCD) and integrator complex subunit 12 (INTS12). Both genes were found to be expressed in a range of human airway cell types. The promoter regions and transcription start sites were determined in mRNA from human lung and a novel splice variant was identified for each gene. We obtained the following evidence for GSTCD and INTS12 co-regulation and expression: (i) correlated mRNA expression was observed both via Q-PCR and in a lung expression quantitative trait loci (eQTL) study, (ii) induction of both GSTCD and INTS12 mRNA expression in human airway smooth muscle cells was seen in response to TGFbeta1, (iii) a lung eQTL study revealed that both GSTCD and INTS12 mRNA levels positively correlate with percent predicted FEV1, and (iv) FEV1 GWAS associated SNPs in 4q24 were found to act as an eQTL for INTS12 in a number of tissues. In fixed sections of human lung tissue, GSTCD protein expression was ubiquitous, whereas INTS12 expression was predominantly in epithelial cells and pneumocytes. During human fetal lung development, GSTCD protein expression was observed to be highest at the earlier pseudoglandular stage (10-12 weeks) compared with the later canalicular stage (17-19 weeks), whereas INTS12 expression levels did not alter throughout these stages. Knowledge of the transcriptional and translational regulation and expression of GSTCD and INTS12 provides important insights into the potential role of these genes in determining lung function. Future work is warranted to fully define the functions of INTS12 and GSTCD.
URLPMID:31646334 [本文引用: 1]

Nephronectin (NPNT) is a novel extracellular matrix protein and a new ligand of integrin alpha8beta1. Recent studies showed that NPNT is highly expressed in kidney, lung, thyroid, etc, and it may play an important role in many pathological conditions. NPNT is involved in the process of kidney development and acute kidney injury, regulates proliferation and differentiation of osteoblast, and induces the vasculogenesis in vitro. NPNT may play a key role in pathological osteoporosis and therefore be a new therapeutic target of bone diseases. NPNT gene variants are not only associated with lung function, but also potentially implicated in chronic airway diseases development. Moreover, NPNT is also an important factor that mediates pathology of cardiac, epidermis, breast, liver and teeth diseases. In this paper, we reviewed some research progresses on the structure, distribution, physiological and pathophysiological functions of NPNT.
[本文引用: 1]
DOI:10.1186/1465-9921-14-77URL [本文引用: 1]
DOI:10.1371/journal.pone.0164041URLPMID:27755550 [本文引用: 1]

INTRODUCTION: Genome-Wide Association Studies have identified associations between lung function measures and Chronic Obstructive Pulmonary Disease (COPD) and chromosome region 6p21 containing the gene for the Advanced Glycation End Product Receptor (AGER, encoding RAGE). We aimed to (i) characterise RAGE expression in the lung, (ii) identify AGER transcripts, (iii) ascertain if SNP rs2070600 (Gly82Ser C/T) is associated with lung function and serum sRAGE levels and (iv) identify whether the Gly82Ser variant is functionally important in altering sRAGE levels in an airway epithelial cell model. METHODS: Immunohistochemistry was used to identify RAGE protein expression in 26 human tissues and qPCR was used to quantify AGER mRNA in lung cells. Gene expression array data was used to identify AGER expression during lung development in 38 fetal lung samples. RNA-Seq was used to identify AGER transcripts in lung cells. sRAGE levels were assessed in cells and patient serum by ELISA. BEAS2B-R1 cells were transfected to overexpress RAGE protein with either the Gly82 or Ser82 variant and sRAGE levels identified. RESULTS: Immunohistochemical assessment of 6 adult lung samples identified high RAGE expression in the alveoli of healthy adults and individuals with COPD. AGER/RAGE expression increased across developmental stages in human fetal lung at both the mRNA (38 samples) and protein levels (20 samples). Extensive AGER splicing was identified. The rs2070600T (Ser82) allele is associated with higher FEV1, FEV1/FVC and lower serum sRAGE levels in UK smokers. Using an airway epithelium model overexpressing the Gly82 or Ser82 variants we found that HMGB1 activation of the RAGE-Ser82 receptor results in lower sRAGE production. CONCLUSIONS: This study provides new information regarding the expression profile and potential role of RAGE in the human lung and shows a functional role of the Gly82Ser variant. These findings advance our understanding of the potential mechanisms underlying COPD particularly for carriers of this AGER polymorphism.
DOI:10.1164/rccm.200912-1843CCURLPMID:20522794 [本文引用: 1]

Significant heterogeneity of clinical presentation and disease progression exists within chronic obstructive pulmonary disease (COPD). Although FEV(1) inadequately describes this heterogeneity, a clear alternative has not emerged. The goal of phenotyping is to identify patient groups with unique prognostic or therapeutic characteristics, but significant variation and confusion surrounds use of the term
DOI:10.1371/journal.pgen.1003585URLPMID:23990791 [本文引用: 1]

Several infrequent genetic polymorphisms in the SERPINA1 gene are known to substantially reduce concentration of alpha1-antitrypsin (AAT) in the blood. Since low AAT serum levels fail to protect pulmonary tissue from enzymatic degradation, these polymorphisms also increase the risk for early onset chronic obstructive pulmonary disease (COPD). The role of more common SERPINA1 single nucleotide polymorphisms (SNPs) in respiratory health remains poorly understood. We present here an agnostic investigation of genetic determinants of circulating AAT levels in a general population sample by performing a genome-wide association study (GWAS) in 1392 individuals of the SAPALDIA cohort. Five common SNPs, defined by showing minor allele frequencies (MAFs) >5%, reached genome-wide significance, all located in the SERPINA gene cluster at 14q32.13. The top-ranking genotyped SNP rs4905179 was associated with an estimated effect of beta = -0.068 g/L per minor allele (P = 1.20*10(-12)). But denser SERPINA1 locus genotyping in 5569 participants with subsequent stepwise conditional analysis, as well as exon-sequencing in a subsample (N = 410), suggested that AAT serum level is causally determined at this locus by rare (MAF<1%) and low-frequent (MAF 1-5%) variants only, in particular by the well-documented protein inhibitor S and Z (PI S, PI Z) variants. Replication of the association of rs4905179 with AAT serum levels in the Copenhagen City Heart Study (N = 8273) was successful (P<0.0001), as was the replication of its synthetic nature (the effect disappeared after adjusting for PI S and Z, P = 0.57). Extending the analysis to lung function revealed a more complex situation. Only in individuals with severely compromised pulmonary health (N = 397), associations of common SNPs at this locus with lung function were driven by rarer PI S or Z variants. Overall, our meta-analysis of lung function in ever-smokers does not support a functional role of common SNPs in the SERPINA gene cluster in the general population.
DOI:10.1164/rccm.201004-0541OCURLPMID:20709820 [本文引用: 1]

RATIONALE: chronic obstructive pulmonary disease (COPD), characterized by airflow limitation, is a disorder with high phenotypic and genetic heterogeneity. Pulmonary emphysema is a major but variable component of COPD; familial data suggest that different components of COPD, such as emphysema, may be influenced by specific genetic factors. OBJECTIVES: to identify genetic determinants of emphysema assessed through high-resolution chest computed tomography in individuals with COPD. METHODS: we performed a genome-wide association study (GWAS) of emphysema determined from chest computed tomography scans with a total of 2,380 individuals with COPD in three independent cohorts of white individuals from (1) a cohort from Bergen, Norway, (2) the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) Study, and (3) the National Emphysema Treatment Trial (NETT). We tested single-nucleotide polymorphism associations with the presence or absence of emphysema determined by radiologist assessment in two of the three cohorts and a quantitative emphysema trait (percentage of lung voxels less than -950 Hounsfield units) in all three cohorts. MEASUREMENTS AND MAIN RESULTS: we identified association of a single-nucleotide polymorphism in BICD1 with the presence or absence of emphysema (P = 5.2 x 10(-7) with at least mild emphysema vs. control subjects; P = 4.8 x 10(-8) with moderate and more severe emphysema vs. control subjects). CONCLUSIONS: our study suggests that genetic variants in BICD1 are associated with qualitative emphysema in COPD. Variants in BICD1 are associated with length of telomeres, which suggests that a mechanism linked to accelerated aging may be involved in the pathogenesis of emphysema. Clinical trial registered with www.clinicaltrials.gov (NCT00292552).
DOI:10.1164/rccm.201306-1061OCURLPMID:24383474 [本文引用: 1]

RATIONALE: Pulmonary emphysema overlaps partially with spirometrically defined chronic obstructive pulmonary disease and is heritable, with moderately high familial clustering. OBJECTIVES: To complete a genome-wide association study (GWAS) for the percentage of emphysema-like lung on computed tomography in the Multi-Ethnic Study of Atherosclerosis (MESA) Lung/SNP Health Association Resource (SHARe) Study, a large, population-based cohort in the United States. METHODS: We determined percent emphysema and upper-lower lobe ratio in emphysema defined by lung regions less than -950 HU on cardiac scans. Genetic analyses were reported combined across four race/ethnic groups: non-Hispanic white (n = 2,587), African American (n = 2,510), Hispanic (n = 2,113), and Chinese (n = 704) and stratified by race and ethnicity. MEASUREMENTS AND MAIN RESULTS: Among 7,914 participants, we identified regions at genome-wide significance for percent emphysema in or near SNRPF (rs7957346; P = 2.2 x 10(-8)) and PPT2 (rs10947233; P = 3.2 x 10(-8)), both of which replicated in an additional 6,023 individuals of European ancestry. Both single-nucleotide polymorphisms were previously implicated as genes influencing lung function, and analyses including lung function revealed independent associations for percent emphysema. Among Hispanics, we identified a genetic locus for upper-lower lobe ratio near the alpha-mannosidase-related gene MAN2B1 (rs10411619; P = 1.1 x 10(-9); minor allele frequency [MAF], 4.4%). Among Chinese, we identified single-nucleotide polymorphisms associated with upper-lower lobe ratio near DHX15 (rs7698250; P = 1.8 x 10(-10); MAF, 2.7%) and MGAT5B (rs7221059; P = 2.7 x 10(-8); MAF, 2.6%), which acts on alpha-linked mannose. Among African Americans, a locus near a third alpha-mannosidase-related gene, MAN1C1 (rs12130495; P = 9.9 x 10(-6); MAF, 13.3%) was associated with percent emphysema. CONCLUSIONS: Our results suggest that some genes previously identified as influencing lung function are independently associated with emphysema rather than lung function, and that genes related to alpha-mannosidase may influence risk of emphysema.
DOI:10.1164/rccm.201501-0148OCURLPMID:26030696 [本文引用: 2]

RATIONALE: Chronic obstructive pulmonary disease (COPD) is defined by the presence of airflow limitation on spirometry, yet subjects with COPD can have marked differences in computed tomography imaging. These differences may be driven by genetic factors. We hypothesized that a genome-wide association study (GWAS) of quantitative imaging would identify loci not previously identified in analyses of COPD or spirometry. In addition, we sought to determine whether previously described genome-wide significant COPD and spirometric loci were associated with emphysema or airway phenotypes. OBJECTIVES: To identify genetic determinants of quantitative imaging phenotypes. METHODS: We performed a GWAS on two quantitative emphysema and two quantitative airway imaging phenotypes in the COPDGene (non-Hispanic white and African American), ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints), NETT (National Emphysema Treatment Trial), and GenKOLS (Genetics of COPD, Norway) studies and on percentage gas trapping in COPDGene. We also examined specific loci reported as genome-wide significant for spirometric phenotypes related to airflow limitation or COPD. MEASUREMENTS AND MAIN RESULTS: The total sample size across all cohorts was 12,031, of whom 9,338 were from COPDGene. We identified five loci associated with emphysema-related phenotypes, one with airway-related phenotypes, and two with gas trapping. These loci included previously reported associations, including the HHIP, 15q25, and AGER loci, as well as novel associations near SERPINA10 and DLC1. All previously reported COPD and a significant number of spirometric GWAS loci were at least nominally (P < 0.05) associated with either emphysema or airway phenotypes. CONCLUSIONS: Genome-wide analysis may identify novel risk factors for quantitative imaging characteristics in COPD and also identify imaging features associated with previously identified lung function loci.
DOI:10.1371/journal.pone.0091621URLPMID:24714607 [本文引用: 1]

BACKGROUND: Chronic mucus hypersecretion (CMH) is associated with an increased frequency of respiratory infections, excess lung function decline, and increased hospitalisation and mortality rates in the general population. It is associated with smoking, but it is unknown why only a minority of smokers develops CMH. A plausible explanation for this phenomenon is a predisposing genetic constitution. Therefore, we performed a genome wide association (GWA) study of CMH in Caucasian populations. METHODS: GWA analysis was performed in the NELSON-study using the Illumina 610 array, followed by replication and meta-analysis in 11 additional cohorts. In total 2,704 subjects with, and 7,624 subjects without CMH were included, all current or former heavy smokers (>/=20 pack-years). Additional studies were performed to test the functional relevance of the most significant single nucleotide polymorphism (SNP). RESULTS: A strong association with CMH, consistent across all cohorts, was observed with rs6577641 (p = 4.25x10(-6), OR = 1.17), located in intron 9 of the special AT-rich sequence-binding protein 1 locus (SATB1) on chromosome 3. The risk allele (G) was associated with higher mRNA expression of SATB1 (4.3x10(-9)) in lung tissue. Presence of CMH was associated with increased SATB1 mRNA expression in bronchial biopsies from COPD patients. SATB1 expression was induced during differentiation of primary human bronchial epithelial cells in culture. CONCLUSIONS: Our findings, that SNP rs6577641 is associated with CMH in multiple cohorts and is a cis-eQTL for SATB1, together with our additional observation that SATB1 expression increases during epithelial differentiation provide suggestive evidence that SATB1 is a gene that affects CMH.
DOI:10.1002/jcsm.12171URLPMID:28044437 [本文引用: 1]

BACKGROUND: There have been a number of candidate gene association studies of cancer cachexia-related traits, but no genome-wide association study (GWAS) has been published to date. Cachexia presents in patients with a number of complex traits, including both cancer and COPD. The objective of the current investigation was to search for a shared genetic aetiology for change in body mass index (DeltaBMI) among cancer and COPD by using GWAS data in the Framingham Heart Study. METHODS: A linear mixed effects model accounting for age, sex, and change in smoking status was used to calculate DeltaBMI in participants over 40 years of age with three consecutive BMI time points (n = 4162). Four GWAS of DeltaBMI using generalized estimating equations were performed among 1085 participants with a cancer diagnosis, 204 with gastrointestinal (GI) cancer, 112 with lung cancer, and 237 with COPD to test for association with 418 365 single-nucleotide polymorphisms (SNPs). RESULTS: Two SNPs reached a level of genome-wide significance (P < 5 x 10(-8) ) with DeltaBMI: (i) rs41526344 within the CNTN4 gene, among COPD cases (beta = 0.13, P = 4.3 x 10(-8) ); and (ii) rs4751240 in the gene Dedicator of Cytokinesis 1 (DOCK1) among GI cancer cases (beta = 0.10, P = 1.9 x 10(-8) ). The DOCK1 SNP association replicated in the DeltaBMI GWAS among COPD cases (betameta-analyis = 0.10, Pmeta-analyis = 9.3 x 10(-10) ). The DOCK1 gene codes for the dedicator of cytokinesis 1 protein, which has a role in myoblast fusion. CONCLUSIONS: In sum, one statistically significant common variant in the DOCK1 gene was associated with DeltaBMI in GI cancer and COPD cases providing support for at least partially shared aetiology of DeltaBMI in complex diseases.
DOI:10.1165/rcmb.2014-0210OCURLPMID:25101718 [本文引用: 1]

Pulmonary hypertension is associated with advanced chronic obstructive pulmonary disease (COPD), although pulmonary vascular changes occur early in the course of the disease. Pulmonary artery (PA) enlargement (PAE) measured by computed tomography correlates with pulmonary hypertension and COPD exacerbation frequency. Genome-wide association studies of PAE in subjects with COPD have not been reported. To investigate whether genetic variants are associated with PAE within subjects with COPD, we investigated data from current and former smokers from the COPDGene Study and the Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints study. The ratio of the diameter of the PA to the diameter of the aorta (A) was measured using computed tomography. PAE was defined as PA/A greater than 1. A genome-wide association study for COPD with PAE was performed using subjects with COPD without PAE (PA/A
[本文引用: 1]
[本文引用: 1]
DOI:10.1136/jmedgenet-2014-102525URL [本文引用: 1]

Variants in FAM13A have been found in genome-wide association studies (GWAS) to associate with lung function in the general population as well as in several common chronic lung diseases (CLD) such as chronic obstructive pulmonary disease (COPD), asthma, as well as in idiopathic interstitial pneumonias (IIP). The gene was cloned in 2004, yet the encoded protein has not been characterised and its function is unknown. The redundancy of its genetic contribution in CLD suggests a major function of this gene both in lung physiology and CLD. This review provides a brief summary of the current knowledge of FAM13A, and demonstrates the necessity to resolve its biological function besides its well accepted genetic contribution. Further interpretations of FAM13A variants may help in the understanding of CLD mechanisms and reveal opportunity for intervention.
[本文引用: 1]
