2.
Histone ubiquitylation and its roles in DNA damage response
Qingyi Zhang1, Yingzi Zhang2, Kai Shen1, Shuyu Zhang2, Jianping Cao21. 2.
编委: 朱卫国
收稿日期:2018-07-10修回日期:2018-09-4网络出版日期:2019-01-20
| 基金资助: |
Received:2018-07-10Revised:2018-09-4Online:2019-01-20
| Fund supported: |
作者简介 About authors
张卿义,本科在读,专业方向:临床医学E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (886KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张卿义, 张樱子, 沈凯, 张舒羽, 曹建平. 组蛋白泛素化修饰及其在DNA损伤应答中的作用[J]. 遗传, 2019, 41(1): 29-40 doi:10.16288/j.yczz.18-112
Qingyi Zhang, Yingzi Zhang, Kai Shen, Shuyu Zhang, Jianping Cao.
DNA是真核生物遗传信息的载体,其遗传保守性是维持物种相对稳定的基础。然而,各种内源性或外源性因素造成的DNA损伤如不能及时修复,将导致细胞凋亡甚至癌变。因此,生物体在进化过程中形成了复杂的DNA损伤应答(DNA damage response, DDR)机制,以应对各种DNA损伤压力。组蛋白是真核生物染色质中主要的蛋白成分,包括H1、H2A、H2B、H3和H4五种类型。约146 bp的DNA通过左手螺旋的方式环绕由2分子H2A-H2B二聚体和1分子H3-H4四聚体组成的核心颗粒1.75圈,形成核小体辅助DNA折叠[1,2]。
组蛋白N端含有大量精氨酸、赖氨酸残基,是主要的翻译后修饰位点。组蛋白在各修饰酶、去修饰酶共同编译下形成组蛋白密码(histone code),构成了DDR中精密的信号网络[3]。近年来,组蛋白泛素化修饰在DDR中的作用愈发受到关注。组蛋白泛素化修饰可以通过调节核小体结构、激活细胞周期检查点、影响修复因子的招募与装配等途径参与DDR。此外,组蛋白修饰之间还存在交互作用,一位点修饰能促进或抑制其他位点的修饰[4,5]。组蛋白泛素化修饰同样可以通过这种串扰(crosstalk)作用调节其他类型翻译后修饰作用于DDR。本文介绍了组蛋白泛素化修饰的各主要位点和相关的E3连接酶、去泛素化酶与效应分子,以及这些修饰作用共同编译形成的信号网络在DDR中的作用。
1 泛素与泛素化修饰
泛素(ubiquitin, ub)是由76个氨基酸组成的多肽,广泛存在于真核生物体内。泛素分子高度保守,在动物、植物和酵母菌中其一级结构仅有1~3个氨基酸残基不同,三级结构基本相同[6]。泛素分子在激活酶(E1)、结合酶(E2)和连接酶(E3)的作用下连接于底物,形成单泛素化修饰。泛素分子还可依次连接于前一泛素分子的赖氨酸或甲硫氨酸残基形成泛素链,称多聚泛素化修饰。连接位点有第6、11、27、29、33、48、63位赖氨酸残基和第1位甲硫氨酸残基(K6/11/27/29/33/48/63-linked and M1-linked) 8种类型。泛素化修饰是泛素-蛋白酶体途径(ubiquitin- proteasome pathway, UPP)的重要步骤,介导了细胞内短寿蛋白和错误折叠蛋白通过26S蛋白酶体降解。此外,泛素化修饰还能调节蛋白在细胞内的定位与活性,广泛参与DNA复制转录、损伤应答、炎症反应、免疫应答、细胞周期与凋亡、囊泡运输等诸多生理过程[7,8,9,10,11]。不同的效应往往与泛素链不同的拓扑结构有关,例如K11/48连接的泛素链主要参与蛋白质降解,K63连接的泛素链主要参与DNA损伤应答与信号转导,而M1连接的泛素链则主要参与免疫应答与炎症反应[12]。
2 H2A多位点泛素化修饰参与DDR
H2A泛素化修饰最早于1975年发现,首个修饰位点定位于K119 (第119位赖氨酸,同下),后又陆续在K13/15和K127/129等位点发现泛素化修饰[13,14]。H2A各位点泛素化修饰介导多种生物学效应,对修复进程进行调控,在DNA双链断裂(double-strand breaks, DSBs)和紫外线造成的DNA损伤(UV-induced DNA damage)应答中起重要作用。2.1 沉默损伤周围基因
哺乳动物细胞核内约5%~15%的H2A处于单泛素化修饰状态,其中以H2AK119单泛素化修饰为主。H2AK119单泛素化修饰参与抑制RNA Pol Ⅱ延伸、多梳蛋白家族(polycomb group proteins, PcG)基因沉默、X染色体失活和抑制趋化因子基因表达等诸多生理过程[15,16,17]。多梳抑制复合体1 (polycomb repressive complex 1, PRC1)是PcG家族成员,其亚基环指蛋白2 (ring finger protein 2, RING2)是单泛素化修饰H2AK119的E3连接酶,通过第98位精氨酸残基嵌入H2A-H2B二聚体间缝隙定位催化反应。由于缺乏活性精氨酸/赖氨酸残基,RING2还需与PRC1另一亚基BMI-1结合形成异二聚体才能充分发挥其催化活性。异二聚体形成有助于稳定E2~ub结构,促进泛素分子传递,BMI-1缺失将严重影响RING2活性[18]。H2AK119单泛素化修饰可与PcG家族另一成员PRC2催化的H3K27三甲基化修饰相互串扰,即PRC1的CBX亚基识别H3K27三甲基化修饰定位,单泛素化修饰H2AK119;PRC2的JARID2亚基识别H2AK119单泛素化修饰定位,三甲基化修饰H3K27[19]。这种交叉招募可极大地提高损伤位点附近H2AK119单泛素化修饰水平。
近年来,已有关于RING2/BMI-1催化的H2AK119单泛素化修饰参与DSBs修复的研究报道,如在损伤早期调节γH2AX生成、影响修复因子招募以及辅助DNA定位于核仁周围等,但具体机制尚未明确[20,21]。目前,H2AK119 单泛素化修饰在DSBs修复中较为明确的作用是实现损伤位点周围数千碱基对范围内基因沉默,抑制损伤区域的复制、转录行为,减少错误产物的生成,为DNA修复创造条件[22,23]。然而,Chandler等[24]在AsiSI限制酶诱导的DSBs周围并未发现PRC1聚集,对上述理论提出挑战。此外,还有证据表明除参与DSBs修复外,细胞核内储备的K119单泛素化修饰的H2A还可在分子伴侣CAF-1的辅助下定位于UV损伤周围,并通过共济失调毛细血管扩张症Rad3相关蛋白激酶(ATM and Rad3 related kinase, ATR)依赖的途径参与核苷酸切除修复(nucleotide excision repair, NER)后染色质重塑[25,26]。
2.2 调节DNA断端剪切
同源重组修复(homologous recombination, HR)和非同源断端连接(non-homologous end joining, NHEJ)是细胞修复DSBs的两种重要方式。BRCA1和53BP1分别是HR和NHEJ中重要的效应分子,二者相互竞争,又彼此协同。BRCA1通过易化DNA断端剪切,促进HR;相反,53BP1则在DNA断端两侧限制DNA剪切长度,防止过度剪切造成的DNA单链复性和染色体重排,易化NHEJ。BRCA1和53BP1在不同位点被招募并级联不同的后续效应,共同决定两种修复方式间的平衡[27](图1A)。图1
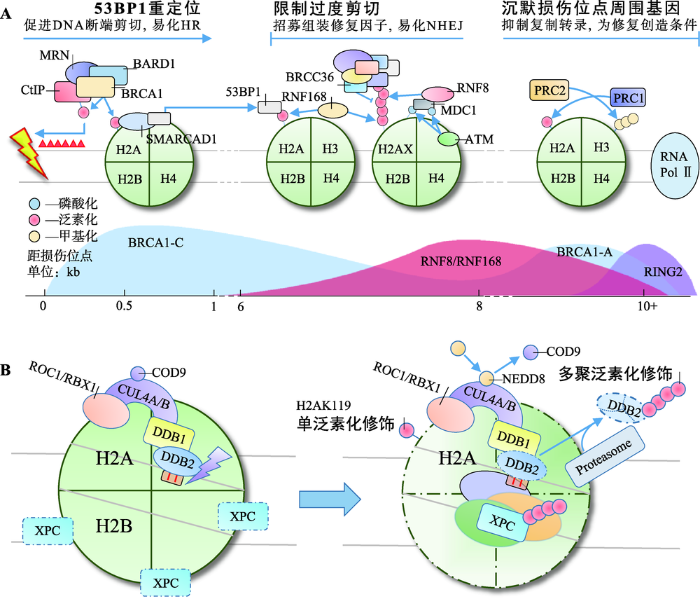 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1H2A各位点泛素化修饰在HR/NHEJ与NER中的作用
A:H2A多位点泛素化修饰共同参与DSBs修复;B:H2AK119单泛素化修饰参与NER。
Fig. 1Model of site-specific H2A ubiquitylation in the HR/NHEJ and NER
环指蛋白8 (ring finger protein 8, RNF8)和环指蛋白168 (ring finger protein 168, RNF168)催化的H2A/H2AXK13/15泛素化修饰,是BRCA1和53BP1招募过程中的重要信号。RNF8和RNF168的聚集,依赖于ATM和DNA损伤检测点介质1 (mediator of DNA damage checkpoint 1, MDC1)的作用。ATM检测到DSBs后,磷酸化修饰H2AX和MDC1。磷酸化MDC1的BRCT结构域识别γH2AX定位于损伤位点,作为脚手架招募RNF8[28,29]。早期认为由RNF8直接多聚泛素化修饰H2A/H2AXK13/15招募修复因子[30],或首先由RNF8单泛素化修饰H2A/H2AXK13/ 15招募RNF168,再由RNF168延伸K63连接的泛素链招募修复因子[31]。然而实验中发现RNF8在体内对核小体中H2A/H2AX缺乏亲和力,却具有延伸K63连接的泛素链的能力。近年来的研究对这一过程逐渐有了清晰的认识:RNF8被招募至损伤位点后首先多聚泛素化修饰H1(K63连接的泛素链)招募RNF168,由后者单泛素化修饰H2A/H2AXK13/15,最后再由RNF8延伸K63连接的泛素链,招募修复因子[32,33]。
BRCA1是H2A/H2AXK13/15多聚泛素化修饰招募的修复因子之一,实际上BRCA1与RAP80、Abraxas、MERIT40、BRCC36、BRCC45和BARD1形成BRCA-A复合体共同被招募[34]。该复合体以Abraxas为核心组装,RAP80负责识别泛素链定位[35]。BRCA1-A复合体能限制DNA断端剪切,防止HR过度激活[34]。去泛素化酶BRCC36清除K63连接的泛素链,被认为在该过程中发挥主要作用[36]。53BP1是另一个被招募的修复因子,与BRCA1不同,53BP1通过识别H2AK15单泛素化修饰定位[37,38]。53BP1可在损伤位点与剪切酶竞争,调整DNA断端剪切长度。同时,53BP1还可作为脚手架,促进修复因子组装[39]。BRCA1-A复合体和53BP1的协同作用有效避免了DNA断端过度剪切,使细胞倾向于通过NHEJ途径修复DSBs。
K127/129单泛素化修饰是新近发现的第3个参与DSBs修复的H2A泛素化修饰类型,由BRCA1-C复合体催化。BRCA1-C复合体包含BRCA1/BARD1二聚体、DNA内切酶CtIP和MRN复合体3种成分,BRCA1是主要的E3活性单位[40]。与RING2/BMI-1 二聚体相似,BRCA1需与BARD1结合才能充分发挥其催化活性[41]。BRCA1/BARD1在异染色质蛋白HP1的辅助下定位至损伤中心区域催化H2AK127/ 129单泛素化修饰,后者可被SMARCAD1的CUE结构域识别[42]。SMARCAD1依赖其ATP酶活性将53BP1重新定位于损伤外围,易化CtIP在MRN复合体辅助下剪切DNA断端,促进以高保真的HR修复DSBs[41,43~45]。值得一提的是,BRCA1还可形成BRCA1-B/D复合体,分别通过调节细胞周期和促进链侵入的方式参与DDR[34]。
2.3 辅助起始NER
除RING2/BMI-1外,H2AK119单泛素化修饰还可由DDB1-CUL4DDB2复合体催化,辅助起始 NER[46]。NER是应对UV造成的DNA损伤的主要方式,除修复环丁烷嘧啶二聚体(cyclobutane pyrimidine dimers, CPDs)和6-4光产物[(6-4) photoproducts, 6-4 PPs]外,NER还可广泛地识别多种损伤,与其特殊的识别机制有关。XPC是全基因组NER中主要的识别因子,可识别DNA发生修饰(损伤)且Waston-Crick碱基配对破坏时形成的不稳定结构,而非损伤本身[47]。CPDs本身不足以引起DNA螺旋结构的不稳定,因此还需DDB1-CUL4DDB2复合体的辅助才能激活NER。该复合体由E3连接酶CUL4和紫外线损伤DNA结合蛋白(UV-damaged DNA-binding protein, UV-DDB)组成,其中UV-DDB包括DDB1与DDB2两种类型,DDB1位于CUL4的N端,是连接CUL4与DDB2的桥梁。DDB2通过WD40结构域识别CPDs后招募DDB1-CUL4至损伤位点[48]。正常情况下,CUL4的活性受COP9信号体抑制,激活则依赖NEDD8类泛素化修饰[49]。正常细胞在UV损伤后H2AK119单泛素化修饰水平迅速下降,DDB1-CUL4DD2复合体在H2AK119单泛素化修饰水平的恢复中起重要作用。H2AK119单泛素化修饰能有效促进H2A/H2A-H2B从核小体解离,加剧DNA螺旋结构的不稳定性,促进XPC识别[50]。同时,DDB2、XPC均是DDB1- CUL4DDB2泛素化修饰的底物,DDB2泛素化修饰后可被分子伴侣VCP/p97识别,介导DDB2通过UPP降解,解除位阻效应,促进后续NER进程[51,52]。XPC泛素化修饰可增强其与DNA的亲和力,促进其与损伤DNA结合[52,53](图1B)。
目前认为,DDB1-CUL4DDB2单泛素化修饰H2AK119是发生在NER早期的事件,辅助XPC对损伤DNA的识别,激活NER。RING2/BMI-1单泛素化修饰H2AK119则更多的以剪切后事件的形式发生于NER后期,通过CAF-1和ATR依赖的途径参与染色质重塑[25,26]。
3 H2B单泛素化修饰串扰其他修饰
RNF20-RNF40催化的H2BK120单泛素化修饰是参与哺乳动物DDR的主要类型[54]。在正常细胞中,H2BK120单泛素化修饰还参与了基因转录的起始、延伸和转录后mRNA的剪切,并能选择性地促进或抑制基因的表达[55,56,57]。转录相关的H2BK120单泛素化修饰高背景为研究H2B泛素化修饰在DDR中的作用提高了难度,直至2011年Moyal等[58]才证实DNA损伤可提高局部H2BK120单泛素化修饰水平,确认了H2BK120单泛素化修饰同样参与DDR。H2BK120单泛素化修饰参与DDR,与组蛋白翻译后修饰间的串扰作用密切相关。以串扰H3K79甲基化修饰为例,H2BK120单泛素化修饰能促进H3K79甲基化修饰,特别是H3K79二甲基化修饰,对53BP1等修复因子的招募具有重要意义[59,60]。关于H2BK120单泛素化修饰是如何串扰H3K79甲基化修饰的,Zhou等[61]提出了“占位诱导”学说,即H2BK120单泛素化修饰在空间上封闭核小体表面无功能位点,促进类端粒沉默干扰体1 (disruptor of telomeric silencing 1-like, Dot1L)在效应位点聚集并甲基化修饰H3K79。同时,H2BK120单泛素化修饰通过串扰作用还能改变染色质高度压缩的结构,例如串扰H3K4甲基化修饰可协同染色质重塑因子SNF2h调节核小体结构[62,63];串扰H4K16乙酰化修饰可开放约30 nm长度的染色质纤维[64,65];串扰H3K56乙酰化修饰同样可以促进促转录因子复合体FACT调节染色质结构,为修复因子装配提供条件[66,67]。
相似的作用同样存在于酵母菌中,由E3连接酶Bre1催化的H2BK123单泛素化修饰同样可以提高Dot1甲基化修饰H3K79的效率,促进53BP1、Ku80和XRCC4的招募[68,69]。同时,H3K79甲基化修饰还可作为修复因子的停靠位点参与NER[70]。H2BK123单泛素化修饰还参与了复制过程中DNA损伤耐受机制的调控,可能同样与调节核小体结构,促进复制叉恢复与缺损区段DNA的填补有关[71,72]。
另一方面,H2BK120单泛素化修饰在DDR中的作用还体现在激活细胞周期检查点中。Kari等[66]敲除RNF40并采用新制霉菌素处理细胞后发现,与对照组相比,G2/M:G1从5.02下降至2.29,S期占比从2.93%升至5.66%,提出H2BK120单泛素化修饰对细胞周期检查点的激活和维持具有重要作用。但Moyal等[58]在实验中沉默RNF20后并未发现细胞周期检查点激活异常,认为RNF20-RNF40并非通过激活细胞周期检查点的方式参与DDR。上述差异可能与分别沉默RNF40和RNF20有关,其具体作用还有待进一步研究。
4 H3、H4泛素化修饰协同参与DDR
正常生理状态下细胞内仅有约0.3%的H3和0.1%的H4处于泛素化修饰状态。UV损伤后,H3、H4泛素化修饰水平迅速升高,并于1~2 h内达到峰值[73]。Wang等[73]通过层析与质谱分析,提纯并确认了泛素化修饰H3和H4的E3连接酶复合体——CUL4- DDB-ROC1 (即DDB1-CUL4DDB2复合体)。UV损伤后,泛素化修饰的H3在胞浆和核浆中比例分别从5%升至19%,12%升至40%;相应地,在核颗粒中的比例从83%跌至41%,表明泛素化修饰可促进H3从核小体解离,易化XPC识别,激活NER[73]。除上述作用外,CUL4-DDB-ROC1复合体还可促进NER后H3K56乙酰化修饰水平恢复,促进核小体组装[74];以及在修复前辅助H3.3在损伤部位沉积,为修复后转录恢复打下基础[75]。
通常认为,组蛋白修饰位点位于伸出核小体外的肽链N端,但近年来研究发现组蛋白核心区域也是翻译后修饰的热点部位[76]。H4K91处于H2A-H2B二聚体与H3-H4四聚体连接的核心区域,可由B细胞淋巴瘤和BAL相关蛋白(B-lymphoma and BAL- associated protein, BBAP)单泛素化修饰。H4K91单泛素化修饰可串扰H4K20甲基化修饰影响53BP1的招募[77]。实验表明H4K91单泛素化修饰可以提高赖氨酸甲基转移酶PR-Set7/Set8的聚集效率,促进H4K20二甲基化修饰。BBAP敲除后,53BP1在损伤部位的聚集显著降低。
此外,BBAP还可通过多聚泛素化修饰底物招募修复因子,并且这是一种独立于RNF8/RNF168轴介导的H2A/H2AXK13/15多聚泛素化修饰的招募模式[63]。该过程中,多聚ADP-核糖聚合酶1 [poly (ADP-ribose) polymerase 1, PARP1]首先识别DNA损伤,在损伤部位多聚ADP-核糖基化修饰底物,后者可被BBAP在BAL1的辅助下识别并定位,再由BBAP催化产生泛素链,招募BRCA1等修复因子[78] (图2B)。分析认为,BBAP催化的多聚泛素化修饰发生于DNA损伤早期,较RNF8/RNF168催化的H2A/H2AXK13/15多聚泛素化修饰更为简单快捷。
图2
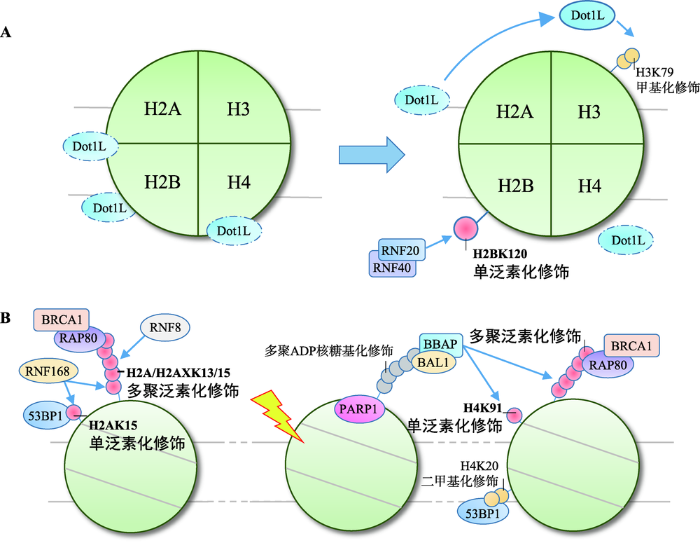 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2H2B和H4泛素化修饰在DNA损伤应答中的作用
A: H2BK120单泛素化修饰串扰H3K79甲基化修饰;B: H4泛素化修饰招募修复因子。
Fig. 2Roles of H2B and H4 ubiquitylation in the DDR
5 组蛋白去泛素化修饰
组蛋白密码编译过程中,修饰与去修饰总是对应存在的。无一例外地,DDR过程中泛素化修饰也必然伴随着去泛素化修饰。上调去泛素化酶(deubiquitinating enzymes, DUBs)能抑制修复因子的招募,延缓DDR进程;下调DUBs则引起自发性染色体断裂,更加强调了泛素化修饰与去泛素化修饰间动态平衡在维持基因组稳定性中的重要性[79]。泛素特异性蛋白酶(ubiquitin-specific proteases, USPs)是DUBs家族中成员最多的一类。USPs的3个结构域空间构象类似于“手指-手掌-拇指”(fingers-palm-thumb),其中palm和thumb构成活性中心,fingers负责定位[80]。H2AK119可由USP16去泛素化修饰,USP16敲除将抑制修复完成后转录的重启[22,81]。BAP1是另一个作用于H2AK119的DUB,其C端与核小体结合,可在ASXL1的辅助下完成去泛素化修饰[82]。在最新的研究中,Jullien等[83]还发现USP21亦能解除H2AK119单泛素化修饰产生的基因抵抗作用。USP3、USP51均是参与H2A/H2AXK13/15去泛素修饰的DUBs,区别在于USP3过表达能降低RNF168在损伤位点的招募[84];而USP51依赖于RNF168定位,过表达仅影响RNF168下游修复因子如53BP1、BRCA1的招募,不影响上游分子ATM、MDC1、RNF168的聚集[85]。USP3、USP51产生不同效应或与拮抗不同的E3连接酶有关:USP3可能直接拮抗RNF8,抑制H1多聚泛素化修饰而影响RNF168的招募及后续修饰,USP51则可能与RNF168拮抗,影响H2A/H2AXK13/ 15单泛素化修饰。USP48是新近发现的DUB,去泛素化修饰H2AK127/129,通过限制MARCAD1对53BP1的重定位限制DNA断端的剪切,对HR起负性调控作用[86]。值得注意的是,USP48的激活还需H2A上其他位点泛素化修饰的辅助,可能与改变USP48构象形成活性中心有关[86]。
转录辅助复合体SAGA是参与DDR过程中H2BK120 (H2BK123)去泛素化修饰的DUB。在哺乳动物中SAGA亚基USP22起主要催化作用,而在酵母菌中以Ubp8为主[87]。实验表明敲除USP22将严重影响细胞通过HR或NHEJ修复DSBs,表明H2BK120去泛素化修饰在DDR中同样起重要作用[88]。
6 结语与展望
DDR是一个复杂的过程,涵盖了损伤位点的识别、细胞周期检查点激活、DNA修复和染色质重塑等诸多环节,组蛋白翻译后修饰在该过程中扮演重要角色。本文总结了组蛋白泛素化修饰/去泛素化修饰的各位点和相关组分,以及这些修饰作用共同编译形成的信号网络在DDR中的作用(表1)。Table 1
表1
表1 组蛋白泛素化/去泛素化修饰在DNA损伤应答中的作用
Table 1
| 修饰位点 | 催化酶 | 作用 | 参考文献 |
|---|---|---|---|
| H2A | |||
| K119 | RING2/BMI-1 | 串扰H3K27三甲基化修饰、沉默损伤位点周围基因、染色质重塑 | [19, 22, 23, 25, 26] |
| DDB1-CUL4DDB2 | 松散染色质结构,加剧DNA不稳定性,促进XPC识别,辅助起始NER | [46, 48~50] | |
| USP16、USP21、 BAP1/ASXL1 | 去泛素化修饰H2AK119 | [22, 81~83] | |
| K13/15 | RNF8 | 延伸K-63连接的泛素链,招募BRCA1-A复合体,限制DNA断端剪切,促进NHEJ | [32~35] |
| RNF168 | 单泛素化修饰H2A/H2AXK13/15,H2AK15单泛素化修饰招募53BP1,限制DNA断端剪切 | [32, 33, 37, 38] | |
| USP3、USP51 BRCC36 | 去泛素化修饰H2A/H2AXK13/15 | [36, 84, 85] | |
| 修饰位点 | 催化酶 | 作用 | 参考文献 |
| K127/129 | BRCA1/BARD1 | 重定位53BP1,易化DNA断端剪切,促进HR | [40, 42~45] |
| USP48 | 去泛素化修饰H2AK127/129 | [86] | |
| H2B | |||
| K120 | RNF20-RNF40 Bre1(酵母菌) | 激活细胞周期检查点、串扰H3K4甲基化修饰、H4K16乙酰化修饰、H3K56乙酰化修饰和H3K79甲基化修饰影响染色质结构与修复因子招募 | [58~72] |
| USP22 Ubp8(酵母菌) | 去泛素化修饰H2BK120(H2BK123酵母菌) | [87, 88] | |
| H3 | |||
| ? | CUL4-DDB-ROC1 | 松散染色质结构,加剧DNA不稳定性,促进XPC识别,辅助起始NER | [73] |
| H4 | |||
| ? | CUL4-DDB-ROC1 | 松散染色质结构,加剧DNA不稳定性,促进XPC识别,辅助起始NER | [73] |
| K91 | BBAP/BAL1 | BBAP催化H4K91单泛素化修饰,并提高PR-Set7/Set8聚集效率,促进H4K20甲基化修饰,间接招募53BP1;催化形成泛素链,直接招募BRCA1 | [77, 78] |
新窗口打开|下载CSV
近年来,人们对于组蛋白泛素化修饰在DDR中的作用有了深入的了解。这得益于研究手段的进步与高度特异性抗体的制备,使人们能够排除高背景的干扰,直接观察局部DNA损伤后细胞的应答情况[58]。方法的创新也同样至关重要,在组蛋白上人为连接泛素分子为探究位阻效应在组蛋白翻译后修饰间的串扰作用提供了新的思路[61,68]。然而,现阶段的研究还存在一定的问题:某些泛素化修饰的位点、类型尚未确定[77];某些泛素化修饰的具体作用仍不够清晰[40];泛素化与去泛素化修饰间关系混乱[79];与其他翻译后修饰间串扰的具体作用及机制不明等。此外,从现有的研究来看,仍有其他泛素化修饰位点尚未发现[86]。对于上述问题的深入研究,必将为全面系统地阐述组蛋白密码在DDR中的作用奠定坚实的基础。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:1831843 [本文引用: 1]

Histone proteins play essential structural and functional roles in the transition between active and inactive chromatin states. Although histones have a high degree of conservation due to constraints to maintain the overall structure of the nucleosomal octameric core, variants have evolved to assume diverse roles in gene regulation and epigenetic silencing. Histone variants, post-translational modifications and interactions with chromatin remodeling complexes influence DNA replication, transcription, repair and recombination. The authors review recent findings on the structure of chromatin that confirm previous interparticle interactions observed in crystal structures.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:19013272 [本文引用: 1]

Histone modifications play a complex role in the regulation of transcription. Recent studies ( Duncan et al., 2008; Lee et al., 2007; Li et al., 2008) reveal that regulation of histone modifications can be functionally linked to reinforce the activation or repression of gene expression.
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
URLPMID:16064136 [本文引用: 1]

Protein ubiquitylation is a recognized signal for protein degradation. However, it is increasingly realized that ubiquitin conjugation to proteins can be used for many other purposes. Furthermore, there are many ubiquitin-like proteins that control the activities of proteins. The central structural element of these post-translational modifications is the ubiquitin superfold. A common ancestor based on this superfold has evolved to give various proteins that are involved in diverse activities in the cell.
URLPMID:12860974 [本文引用: 1]

Receptor signaling at the plasma membrane often releases calcium from intracellular stores. For example, inositol triphosphate (IP3) produced by receptor-coupled phospholipase C activates an intracellular store calcium channel, the IP3R. Conversely, stores can induce extracellular calcium to enter the cell through plasma membrane channels, too. How this "reverse" coupling works was unclear, but store IP(3)Rs were proposed to bind and regulate plasma membrane TRIP cation channels. Here, we demonstrate that the adaptor protein, termed Homer, facilitates a physical association between TRPC1 and the IP3R that is required for the TRP channel to respond to signals. The TRPC1-Homer-IP3R complex is dynamic and its disassembly parallels TRPC1 channel activation. Homer's action depends on its ability to crosslink and is blocked by the dominant-negative immediate early gene form, H1a. Since H1a is transcriptionally regulated by cellular activity, this mechanism can affect both short and long-term regulation of TRPC1 function.
URL [本文引用: 1]
URLPMID:1165239 [本文引用: 1]

In earlier studies, the nucleolar levels of protein A24 were found to be markedly decreased in the nucleolar hypertrophy induced by thioacetamide or during liver regeneration (Ballal, N.R., Goldknopf, I.L., Goldberg, D.A., and Busch, H. (1974) Life Sci. 14, 1835-1845; Ballal, N.R., Kang, Y.-J., Olson, M.O.-J., and Busch, H.J. Biol. Chem. 250, 5921-5925). To determine the role of protein A24, methods were developed for its isolation in highly purified form. Milligram quantities of highly purified protein A24 were isolated from the 0.4 N H2SO4-soluble proteins of calf thymus chromatin by exclusion chromatography on Sephadex G-100, followed by preparative polyacrylamide gel electrophoresis. Protein A24 was highly purified as shown by its migration as a single spot on two-dimensional polyacrylamide gel electrophoresis, its single NH2-terminal amino acid, methionine, and the production of approximately 50 peptides by tryptic digestion. Like histones 2A, 2B, 3, and 4. A24 was extractable from chromatin with 0.4 N H2SO4 or 3 M NaCl/7 M urea, but unlike most non-histone proteins or histone 1, protein A24 was not extracted with 0.35 M NaCl, 0.5 M HClO4, or 0.6 M NaCl. Protein A24 was present in only 1.9% of the total amount of histones 2A, 2B, 3 and 4; its molecular weight is 27,000.
URLPMID:265581 [本文引用: 1]

Chromosomal protein A24 has a unique structure inasmuch as it contains histone 2A and a nonhistone polypeptide the sequence of which has been partially determined. Comparative analysis of the ninhydrin-insensitive amino-terminal tryptic peptides of protein A24 and histone 2A and a quantitative analysis of their carboxyl-terminal amino acid indicated that protein A24 has two amino termini and one carboxyl terminus. The amino acid sequence analysis of tryptic peptide 17 f protein A24: [Note: See the image of page 864 for this formatted text]$ \matrix\format\ l \\ \quad \quad \quad \quad \ \ \overset \text{O}\to{||}\,\overset \text{H}\to{|}\quad \quad \ \ \overset \text{O}\to{||} \\ \text{H}_{2}\text{N-CH}_{2}\text{-C-N-CH}_{2}\text{-C-NH} \\ \quad \quad (\text{Gly})\quad \quad \, (\text{Gly})\ \ (\underset|\to{\overset|\to{\text{C}}}\text{H}_{2})_{4} \\ \quad \quad \quad \quad \quad \quad \quad \ \ \text{H}_{2}\text{N-CH-}\text{C}^{^{\text{O}}}\text{-Thr-Glu-Ser-His-His-Lys} \\ \quad \quad \quad \quad \quad \quad \quad \quad \quad \ (\text{Lys}119) \endmatrix $showed it contains tryptic peptide 17 of histone 2A, Lys-Thr-Glu-Ser-His-His-Lys. Lysine 119, the amino terminus of this peptide, which is derived from the histone 2A portion of protein A24, is linked by an isopeptide bond to the carboxyl group of a glycine residue. Accordingly, the branched structure of protein A24 proposed is:$ \matrix\format\ l \\ \quad \quad \quad \quad \,\overset \text{H}\to{|}\quad \quad \ \ \overset \text{O}\to{||}\,\overset \text{H}\to{|}\quad \quad \ \ \overset \text{O}\to{||} \\ \text{Met - - - N-CH}_{2}\text{-C-N-CH}_{2}\text{-C-NH} \\ \quad \quad \quad \quad \quad (\text{Gly})\quad \quad (\text{Gly})\ \ (\underset|\to{\overset|\to{\text{C}}}\text{H}_{2})_{4} \\ N\text{-Acetylserine - - - - - -}\overset \text{H}\to{\text{N}}\text{-CH-}\text{C}^{^{\text{O}}}\text{- - - Lys} \\ \text{Histone}\ 2\text{A}\colon 1\quad \quad \quad \quad \quad \ \ \text{Lys}\ 199\quad \quad \quad \ 129 \endmatrix $
URLPMID:15386022 [本文引用: 1]

Abstract Covalent modification of histones is important in regulating chromatin dynamics and transcription. One example of such modification is ubiquitination, which mainly occurs on histones H2A and H2B. Although recent studies have uncovered the enzymes involved in histone H2B ubiquitination and a 'cross-talk' between H2B ubiquitination and histone methylation, the responsible enzymes and the functions of H2A ubiquitination are unknown. Here we report the purification and functional characterization of an E3 ubiquitin ligase complex that is specific for histone H2A. The complex, termed hPRC1L (human Polycomb repressive complex 1-like), is composed of several Polycomb-group proteins including Ring1, Ring2, Bmi1 and HPH2. hPRC1L monoubiquitinates nucleosomal histone H2A at lysine 119. Reducing the expression of Ring2 results in a dramatic decrease in the level of ubiquitinated H2A in HeLa cells. Chromatin immunoprecipitation analysis demonstrated colocalization of dRing with ubiquitinated H2A at the PRE and promoter regions of the Drosophila Ubx gene in wing imaginal discs. Removal of dRing in SL2 tissue culture cells by RNA interference resulted in loss of H2A ubiquitination concomitant with derepression of Ubx. Thus, our studies identify the H2A ubiquitin ligase, and link H2A ubiquitination to Polycomb silencing.
URL [本文引用: 1]
URLPMID:18206970 [本文引用: 1]

Solving the biological roles of covalent histone modifications, including monoubiquitination of histone H2A, and the molecular mechanisms by which these modifications regulate specific transcriptional programs remains a central question for all eukaryotes. Here we report that the N-CoR/HDAC1/3 complex specifically recruits a specific histone H2A ubiquitin ligase, 2A-HUB/hRUL138, to a subset of regulated gene promoters. 2A-HUB catalyzes monoubiquitination of H2A at lysine 119, functioning as a combinatoric component of the repression machinery required for specific gene regulation programs. Thus, 2A-HUB mediates a selective repression of a specific set of chemokine genes in macrophages, critically modulating migratory responses to TLR activation. H2A monoubiquitination acts to prevent FACT recruitment at the transcriptional promoter region, blocking RNA polymerase II release at the early stage of elongation. We suggest that distinct H2A ubiquitinases, each recruited based on interactions with different corepressor complexes, contribute to distinct transcriptional repression programs.
URLPMID:3133356 [本文引用: 1]

Abstract DNA damage activates signaling pathways that lead to modification of local chromatin and recruitment of DNA repair proteins. Multiple DNA repair proteins having ubiquitin ligase activity are recruited to sites of DNA damage, where they ubiquitinate histones and other substrates. This DNA damage-induced histone ubiquitination is thought to play a critical role in mediating the DNA damage response. We now report that the polycomb protein BMI1 is rapidly recruited to sites of DNA damage, where it persists for more than 8 h. The sustained localization of BMI1 to damage sites is dependent on intact ATM and ATR and requires H2AX phosphorylation and recruitment of RNF8. BMI1 is required for DNA damage-induced ubiquitination of histone H2A at lysine 119. Loss of BMI1 leads to impaired repair of DNA double-strand breaks by homologous recombination and the accumulation of cells in G(2)/M. These data support a crucial role for BMI1 in the cellular response to DNA damage.
URLPMID:24856970 [本文引用: 1]

Formation of repressive polycomb domains depends on histone ubiquitination catalyzed by variant PRC1 complexes at unmethylated CpG islands followed by PRC2 recruitment and methylation, rather than exclusively through initial association of PRC2.
URLPMID:2953429 [本文引用: 1]

Polycomb group (PcG) proteins are major determinants of cell identity, stem cell pluripotency, and epigenetic gene silencing during development. The polycomb repressive complex 1, which contains BMI1, RING1, and RING2, functions as an E3-ubuiquitin ligase. We found that BMI1 and RING2 are recruited to sites of DNA double-strand breaks (DSBs) where they contribute to the ubiquitylation of -H2AX. In the absence of BMI1, several proteins dependent on ubiquitin signaling, including 53BP1, BRCA1, and RAP80, are impaired in recruitment to DSBs. Loss of BMI1 sensitizes cells to ionizing radiation to the same extent as loss of RNF8. The simultaneous depletion of both proteins revealed an additive increase in radiation sensitivity. These data uncover an unexpected link between the polycomb and the DNA damage response pathways, and suggest a novel function for BMI1 in maintaining genomic stability.
URLPMID:5458174 [本文引用: 1]

One of the major cellular DNA repair pathways is nucleotide excision repair (NER). It is the primary pathway for repair of various DNA lesions caused by exposure to ultraviolet (UV) light, such as cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts. Although lesion-containing DNA associates with the nuclear matrix after UV irradiation it is still not understood how nuclear organization affects NER. Analyzing unscheduled DNA synthesis (UDS) indicates that NER preferentially occurs in specific nuclear areas, viz the nucleolus. Upon inducing localized damage, we observe migration of damaged DNA towards the nucleolus. Employing a LacR-based tethering system we demonstrate that H2A-ubiquitylation via the UV-RING1B complex localizes chromatin close to the nucleolus. We further show that the H2A-ubiquitin binding protein ZRF1 resides in the nucleolus, and that it anchors ubiquitylated chromatin along with XPC. Our data thus provide insight into the sub-nuclear organization of NER and reveal a novel role for histone H2A-ubiquitylation.
URLPMID:20550933 [本文引用: 2]

78 Transcriptional silencing occurs on chromatin in cis to DNA double-strand breaks 78 Silencing extends multiple kilobases from the site of damage 78 Silencing is dependent on the ATM kinase 78 Reversal of silencing requires histone H2A deubiquitylation
URLPMID:4157577 [本文引用: 1]

Cells silence ongoing transcription in response to a neighboring DNA double-strand break. Kakarougkas et02al. show that the PBAF chromatin remodeling complex is important for this event and that failure to silence transcription leads to a delay in repair.
URLPMID:25057768 [本文引用: 1]

Abstract A growing body of evidence suggests that Polycomb group (PcG) proteins, key regulators of lineage specific gene expression, also participate in the repair of DNA double-strand breaks (DSBs) but evidence for direct recruitment of PcG proteins at specific breaks remains limited. Here we explore the association of Polycomb repressive complex 1 (PRC1) components with DSBs generated by inducible expression of the AsiSI restriction enzyme in normal human fibroblasts. Based on immunofluorescent staining, the co-localization of PRC1 proteins with components of the DNA damage response (DDR) in these primary cells is unconvincing. Moreover, using chromatin immunoprecipitation and deep sequencing (ChIP-seq), which detects PRC1 proteins at common sites throughout the genome, we did not find evidence for recruitment of PRC1 components to AsiSI-induced DSBs. In contrast, the S2056 phosphorylated form of DNA-PKcs and other DDR proteins were detected at a subset of AsiSI sites that are predominantly at the 5' ends of transcriptionally active genes. Our data question the idea that PcG protein recruitment provides a link between DSB repairs and transcriptional repression.
URLPMID:16702407 [本文引用: 2]

Chromatin changes within the context of DNA repair remain largely obscure. Here we show that DNA damage induces monoubiquitylation of histone H2A in the vicinity of DNA lesions. Ultraviolet (UV)-induced monoubiquitylation of H2A is dependent on functional nucleotide excision repair and occurs after incision of the damaged strand. The ubiquitin ligase Ring2 is required for the DNA damage-induced H2A ubiquitylation. UV-induced ubiquitylation of H2A is dependent on the DNA damage signaling kinase ATR (ATM- and Rad3-related) but not the related kinase ATM (ataxia telangiectasia-mutated). Although the response coincides with phosphorylation of variant histone H2AX, H2AX was not required for H2A ubiquitylation. Together our data show that monoubiquitylation of H2A forms part of the cellular response to UV damage and suggest a role of this modification in DNA repair-induced chromatin remodeling.
URLPMID:2718537 [本文引用: 2]

Restoration of functionally intact chromatin structure following DNA damage processing is crucial for maintaining genetic and epigenetic information in human cells. Here, we show the UV-induced uH2A foci formation in cells lacking XPC, DDB2, CSA or CSB, but not in cells lacking XPA, XPG or XPF indicating that uH2A incorporation relied on successful damage repair occurring through either GGR or TCR sub-pathway. In contrast, XPA, XPG or XPF were not required for formation of H2AX foci in asynchronous cells. Notably, the H2A ubiquitin ligase Ring1B, a component of Polycomb repressor complex 1, did not localize at DNA damage sites. However, histone chaperone CAF-1 showed distinct localization to the damage sites. Knockdown of CAF-1 p60 abolished CAF-1 as well as uH2A foci formation. CAF-1 p150 was found to associate with NER factors TFIIH, RPA p70 and PCNA in chromatin. These data demonstrate that successful NER of genomic lesions and prompt CAF-1-mediated chromatin restoration link uH2A incorporation at the sites of damage repair within chromatin.
URLPMID:28624371 [本文引用: 1]

DNA double strand breaks need to be repaired in an organized fashion to preserve genomic integrity. In the organization of faithful repair, histone ubiquitination plays a crucial role. Recent findings suggest an integrated model for DNA repair regulation through site-specific histone ubiquitination and crosstalk to other posttranslational modifications. Here we discuss how site-specific histone ubiquitination is achieved on a molecular level and how different multi-protein complexes work together to integrate different histone ubiquitination states. We propose a model where site-specific H2A ubiquitination organizes the spatio-temporal recruitment of DNA repair factors which will ultimately contribute to DNA repair pathway choice between homologous recombination and non-homologous end joining.
URLPMID:16377563 [本文引用: 1]

Cell. 2005 Dec 29;123(7):1213-26. Research Support, Non-U.S. Gov't
URLPMID:18001825 [本文引用: 1]

DNA-damage signaling utilizes a multitude of posttranslational modifiers as molecular switches to regulate cell-cycle checkpoints, DNA repair, cellular senescence, and apoptosis. Here we show that RNF8, a FHA/RING domain-containing protein, plays a critical role in the early DNA-damage response. We have solved the X-ray crystal structure of the FHA domain structure at 1.35 . We have shown that RNF8 facilitates the accumulation of checkpoint mediator proteins BRCA1 and 53BP1 to the damaged chromatin, on one hand through the phospho-dependent FHA domain-mediated binding of RNF8 to MDC1, on the other hand via its role in ubiquitylating H2AX and possibly other substrates at damage sites. Moreover, RNF8-depleted cells displayed a defective G2/M checkpoint and increased IR sensitivity. Together, our study implicates RNF8 as a novel DNA-damage-responsive protein that integrates protein phosphorylation and ubiquitylation signaling and plays a critical role in the cellular response to genotoxic stress.
URLPMID:18001824 [本文引用: 1]

Accumulation of repair proteins on damaged chromosomes is required to restore genomic integrity. However, the mechanisms of protein retention at the most destructive chromosomal lesions, the DNA double-strand breaks (DSBs), are poorly understood. We show that RNF8, a RING-finger ubiquitin ligase, rapidly assembles at DSBs via interaction of its FHA domain with the phosphorylated adaptor protein MDC1. This is accompanied by an increase in DSB-associated ubiquitylations and followed by accumulation of 53BP1 and BRCA1 repair proteins. Knockdown of RNF8 or disruption of its FHA or RING domains impaired DSB-associated ubiquitylation and inhibited retention of 53BP1 and BRCA1 at the DSB sites. In addition, we show that RNF8 can ubiquitylate histone H2A and H2AX, and that its depletion sensitizes cells to ionizing radiation. These data suggest that MDC1-mediated and RNF8-executed histone ubiquitylation protects genome integrity by licensing the DSB-flanking chromatin to concentrate repair factors near the DNA lesions.
URLPMID:19203579 [本文引用: 1]

DNA double-strand breaks (DSBs) not only interrupt the genetic information, but also disrupt the chromatin structure, and both impairments require repair mechanisms to ensure genome integrity. We showed previously that RNF8-mediated chromatin ubiquitylation protects genome integrity by promoting the accumulation of repair factors at DSBs. Here, we provide evidence that, while RNF8 is necessary to trigger the DSB-associated ubiquitylations, it is not sufficient to sustain conjugated ubiquitin in this compartment. We identified RNF168 as a novel chromatin-associated ubiquitin ligase with an ability to bind ubiquitin. We show that RNF168 interacts with ubiquitylated H2A, assembles at DSBs in an RNF8-dependent manner, and, by targeting H2A and H2AX, amplifies local concentration of lysine 63-linked ubiquitin conjugates to the threshold required for retention of 53BP1 and BRCA1. Thus, RNF168 defines a new pathway involving sequential ubiquitylations on damaged chromosomes and uncovers a functional cooperation between E3 ligases in genome maintenance.
[本文引用: 1]
URLPMID:26503038 [本文引用: 1]

DNA double-strand breaks (DSBs) are highly cytotoxic DNA lesions that trigger non-proteolytic ubiquitylation of adjacent chromatin areas to generate binding sites for DNA repair factors. This depends on the sequential actions of the E3 ubiquitin ligases RNF8 and RNF168 (refs 1-6), and UBC13 (also known as UBE2N), an E2 ubiquitin-conjugating enzyme that specifically generates K63-linked ubiquitin chains. Whereas RNF168 is known to catalyse ubiquitylation of H2A-type histones, leading to the recruitment of repair factors such as 53BP1 (refs 8-10), the critical substrates of RNF8 and K63-linked ubiquitylation remain elusive. Here we elucidate how RNF8 and UBC13 promote recruitment of RNF168 and downstream factors to DSB sites in human cells. We establish that UBC13-dependent K63-linked ubiquitylation at DSB sites is predominantly mediated by RNF8 but not RNF168, and that H1-type linker histones, but not core histones, represent major chromatin-associated targets of this modification. The RNF168 module (UDM1) recognizing RNF8-generated ubiquitylations is a high-affinity reader of K63-ubiquitylated H1, mechanistically explaining the essential roles of RNF8 and UBC13 in recruiting RNF168 to DSBs. Consistently, reduced expression or chromatin association of linker histones impair accumulation of K63-linked ubiquitin conjugates and repair factors at DSB-flanking chromatin. These results identify histone H1 as a key target of RNF8-UBC13 in DSB signalling and expand the concept of the histone code by showing that posttranslational modifications of linker histones can serve as important marks for recognition by factors involved in genome stability maintenance, and possibly beyond.
URLPMID:25400280 [本文引用: 3]

BRCA1 is a major breast and ovarian cancer susceptibility gene, with mutations in this gene predisposing women to a very high risk of developing breast and ovarian tumours. BRCA1 primarily functions to maintain genomic stability via critical roles in DNA repair, cell cycle checkpoint control, transcriptional regulation, apoptosis and mRNA splicing. As a result, BRCA1 mutations often result in defective DNA repair, genomic instability and sensitivity to DNA damaging agents. BRCA1 carries out these different functions through its ability to interact, and form complexes with, a vast array of proteins involved in multiple cellular processes, all of which are considered to contribute to its function as a tumour suppressor. This review discusses and highlights recent research into the functions of BRCA1-related protein complexes and their roles in maintaining genomic stability and tumour suppression.
URLPMID:17525341 [本文引用: 1]

Mutations affecting the BRCT domains of the breast cancer--associated tumor suppressor BRCA1 disrupt the recruitment of this protein to DNA double--strand breaks (DSBs). The molecular structures at DSBs recognized by BRCA1 are presently unknown. We report the interaction of the BRCA1 BRCT domain with RAP80, a ubiquitin-binding protein. RAP80 targets a complex containing the BRCA1-BARD1 (BRCA1-associated ring domain protein 1) E3 ligase and the deubiquitinating enzyme (DUB) BRCC36 to MDC1-γH2AX--dependent lysine68- and$\text{lysine}^{63}$-linked ubiquitin polymers at DSBs. These events are required for cell cycle checkpoint and repair responses to ionizing radiation, implicating ubiquitin chain recognition and turnover in the BRCA1-mediated repair of DSBs.
URL [本文引用: 1]
URLPMID:23760478 [本文引用: 1]

53BP1 (also called TP53BP1) is a chromatin-associated factor that promotes immunoglobulin class switching and DNA double-strand-break (DSB) repair by non-homologous end joining. To accomplish its function in DNA repair, 53BP1 accumulates at DSB sites downstream of the RNF168 ubiquitin ligase. How ubiquitin recruits 53BP1 to break sites remains unknown as its relocalization involves recognition of histone H4 Lys 20 (H4K20) methylation by its Tudor domain. Here we elucidate how vertebrate 53BP1 is recruited to the chromatin that flanks DSB sites. We show that 53BP1 recognizes mononucleosomes containing dimethylated H4K20 (H4K20me2) and H2A ubiquitinated on Lys 15 (H2AK15ub), the latter being a product of RNF168 action on chromatin. 53BP1 binds to nucleosomes minimally as a dimer using its previously characterized methyl-lysine-binding Tudor domain and a carboxy-terminal extension, termed the ubiquitination-dependent recruitment (UDR) motif, which interacts with the epitope formed by H2AK15ub and its surrounding residues on the H2A tail. 53BP1 is therefore a bivalent histone modification reader that recognizes a histone `code' produced by DSB signalling.
URLPMID:27462807 [本文引用: 1]

Abstract DNA double-strand breaks (DSBs) elicit a histone modification cascade that controls DNA repair. This pathway involves the sequential ubiquitination of histones H1 and H2A by the E3 ubiquitin ligases RNF8 and RNF168, respectively. RNF168 ubiquitinates H2A on lysine 13 and lysine 15 (refs 7, 8) (yielding H2AK13ub and H2AK15ub, respectively), an event that triggers the recruitment of 53BP1 (also known as TP53BP1) to chromatin flanking DSBs. 53BP1 binds specifically to H2AK15ub-containing nucleosomes through a peptide segment termed the ubiquitination-dependent recruitment motif (UDR), which requires the simultaneous engagement of histone H4 lysine 20 dimethylation (H4K20me2) by its tandem Tudor domain. How 53BP1 interacts with these two histone marks in the nucleosomal context, how it recognizes ubiquitin, and how it discriminates between H2AK13ub and H2AK15ub is unknown. Here we present the electron cryomicroscopy (cryo-EM) structure of a dimerized human 53BP1 fragment bound to a H4K20me2-containing and H2AK15ub-containing nucleosome core particle (NCP-ubme) at 4.5 resolution. The structure reveals that H4K20me2 and H2AK15ub recognition involves intimate contacts with multiple nucleosomal elements including the acidic patch. Ubiquitin recognition by 53BP1 is unusual and involves the sandwiching of the UDR segment between ubiquitin and the NCP surface. The selectivity for H2AK15ub is imparted by two arginine fingers in the H2A amino-terminal tail, which straddle the nucleosomal DNA and serve to position ubiquitin over the NCP-bound UDR segment. The structure of the complex between NCP-ubme and 53BP1 reveals the basis of 53BP1 recruitment to DSB sites and illuminates how combinations of histone marks and nucleosomal elements cooperate to produce highly specific chromatin responses, such as those elicited following chromosome breaks.
URLPMID:27348077 [本文引用: 1]

Repair of DNA double-strand breaks (DSBs) in mammals is coordinated by the ubiquitin-dependent accumulation of 53BP1 at DSB-flanking chromatin. Owing to its ability to limit DNA-end processing, 53BP1 is thought to promote nonhomologous end-joining (NHEJ) and to suppress homology-directed repair (HDR). Here, we show that silencing 53BP1 or exhausting its capacity to bind damaged chromatin changes limited DSB resection to hyper-resection and results in a switch from error-free gene conversion by RAD51 to mutagenic single-strand annealing by RAD52. Thus, rather than suppressing HDR, 53BP1 fosters its fidelity. These findings illuminate causes and consequences of synthetic viability acquired through 53BP1 silencing in cells lacking the BRCA1 tumor suppressor. We show that such cells survive DSB assaults at the cost of increasing reliance on RAD52-mediated HDR, which may fuel genome instability. However, our findings suggest that when challenged by DSBs, BRCA1- and 53BP1-deficient cells may become hypersensitive to, and be eliminated by, RAD52 inhibition.
URLPMID:25131202 [本文引用: 2]

The ubiquitin ligase (E3) activity of BRCA1 is its only known biochemical activity in02vitro. Kalb et02al. report that the heterodimeric complex formed by BRCA1/BARD1 ubiquitylates histone H2A in nucleosomes specifically at its C-terminal tail. Moreover, in02vitro and in02vivo assays identify H2A lysines127 and 129 as the target lysines. These results help to explain the localization and activity of BRCA1/BARD1 on chromatin in cells.
URLPMID:5403137 [本文引用: 2]

The protein product of the breast and ovarian cancer gene, BRCA1, is part of an obligate heterodimer with BARD1. Together these RING bearing proteins act as an E3 ubiquitin ligase. Several functions have been attributed to BRCA1 that contribute to genome integrity but which of these, if any, require this enzymatic function was unclear. Here we review recent studies clarifying the role of BRCA1 E3 ubiquitin ligase in DNA repair. Perhaps the most surprising finding is the narrow range of BRCA1 functions this activity relates to. Remarkably ligase activity promotes chromatin remodelling and 53BP1 positioning through the remodeller SMARCAD1, but the activity is dispensable for the cellular survival in response to cisplatin or replication stressing agents. Implications for therapy response and tumor susceptibility are discussed.
URLPMID:27239795 [本文引用: 1]

The opposing activities of 53BP1 and BRCA1 influence pathway choice in DNA double-strand-break repair. How BRCA1 counteracts the inhibitory effect of 53BP1 on DNA resection and homologous recombination is unknown. Here we identify the site of BRCA1-BARD1 required for priming ubiquitin transfer from E2 biquitin and demonstrate that BRCA1-BARD1's ubiquitin ligase activity is required for repositioning 53BP1 on damaged chromatin. We confirm H2A ubiquitination by BRCA1-BARD1 and show that an H2A-ubiquitin fusion protein promotes DNA resection and repair in BARD1-deficient cells. BRCA1-BARD1's function in homologous recombination requires the chromatin remodeler SMARCAD1. SMARCAD1 binding to H2A-ubiquitin and optimal localization to sites of damage and activity in DNA repair requires its ubiquitin-binding CUE domains. SMARCAD1 is required for 53BP1 repositioning, and the need for SMARCAD1 in olaparib or camptothecin resistance is alleviated by 53BP1 loss. Thus, BRCA1-BARD1 ligase activity and subsequent SMARCAD1-dependent chromatin remodeling are critical regulators of DNA repair.
URLPMID:21346409 [本文引用: 1]

Polycomb group proteins, which have well-established roles in gene regulation, were recently found to accumulate on chromatin surrounding DNA damage and to contribute up to 40 percent of the radiation resistance of cell lines. The oncogenic polycomb protein, BMI-1, was additionally shown to be essential for the increased radiation resistance observed in stem cells and cancer stem cells relative to their more differentiated counterparts. BMI-1, is a very early DNA damage response protein that accumulates through a H2AX/RNF8-independent, but poly(ADP-ribosyl)ation-dependent mechanism at DNA double-strand breaks. BMI-1 acts together with RING2 and other components of the PRC1 histone H2A E3 ubiquitin ligase to ubiquitylate histones H2A and H2AX in response to DNA damage. BMI-1 dependent ubiquitin modifications are at the base of an ubiquitin pathway that enhances radioresistance through the accumulation of RAP80, 53BP1, and BRCA1. Members of the PRC2 histone H3 lysine 27 methyltransferase complex are also recruited to sites of DSBs but it remains to be determined whether the histone methyltransferase and histone E3 ubiquitin ligase polycomb complexes function in concert or independently during DNA repair. Understanding the contribution of polycomb group proteins to the DNA damage response may lead to novel therapeutic strategies that increase the response of human cancers to therapies that work through DNA damage, while simultaneously sensitizing the cancer stem cell population that would otherwise lead to relapse.
URLPMID:25310973

CtIP is a key factor regulating DNA-end resection, an early step in DNA repair that controls the choice of repair pathway. CtIP interacts with BRCA1, but the exact role of BRCA1 in DNA-end resection is unclear. Cruz-García et02al. develop a high-resolution method to measure the extent of DNA resection following DNA breaks. They find that resection occurs in the absence of a BRCA1-CtIP interaction, but the rate of resection is slower, suggesting that BRCA1 modulates its speed.
URLPMID:4042650 [本文引用: 1]

Abstract Homologous recombination (HR) is initiated by DNA end resection, a process in which stretches of single-strand DNA (ssDNA) are generated and used for homology search. Factors implicated in resection include nucleases MRE11, EXO1, and DNA2, which process DNA ends into 3' ssDNA overhangs; helicases such as BLM, which unwind DNA; and other proteins such as BRCA1 and CtIP whose functions remain unclear. CDK-mediated phosphorylation of CtIP on T847 is required to promote resection, whereas CDK-dependent phosphorylation of CtIP-S327 is required for interaction with BRCA1. Here, we provide evidence that CtIP functions independently of BRCA1 in promoting DSB end resection. First, using mouse models expressing S327A or T847A mutant CtIP as a sole species, and B cells deficient in CtIP, we show that loss of the CtIP-BRCA1 interaction does not detectably affect resection, maintenance of genomic stability or viability, whereas T847 is essential for these functions. Second, although loss of 53BP1 rescues the embryonic lethality and HR defects in BRCA1-deficient mice, it does not restore viability or genome integrity in CtIP(-/-) mice. Third, the increased resection afforded by loss of 53BP1 and the rescue of BRCA1-deficiency depend on CtIP but not EXO1. Finally, the sensitivity of BRCA1-deficient cells to poly ADP ribose polymerase (PARP) inhibition is partially rescued by the phospho-mimicking mutant CtIP (CtIP-T847E). Thus, in contrast to BRCA1, CtIP has indispensable roles in promoting resection and embryonic development.
URLPMID:26344709 [本文引用: 1]

61Comprehensive overview of the shared and distinct functions of CUL4A and CUL4B61Table of all known CRL4 substrates with supporting evidence and references61Up-to-date information on CRL4s' roles in human diseases and mammalian development
URLPMID:9192622 [本文引用: 1]

Mammalian nucleotide excision repair (NER) eliminates carcinogen-DNA adducts by double endonucleolytic cleavage and subsequent release of 24-32 nucleotide-long single-stranded fragments. Here we manipulated the deoxyribose-phosphate backbone of DNA to analyze the mechanism by which damaged strands are discriminated as substrates for dual incision. We found that human NER is completely inactive on DNA duplexes containing single C4′-modified backbone residues. However, the same C4′ backbone variants, which by themselves do not perturb complementary hydrogen bonds, induced strong NER reactions when incorporated into short segments of mispaired bases. No oligonucleotide excision was detected when DNA contained abnormal base pairs without concomitant changes in deoxyribose-phosphate composition. Thus, neither C4′ backbone lesions nor improper base pairing stimulated human NER, but the combination of these two substrate alterations constituted an extremely potent signal for double DNA incision. In summary, we used C4′-modified backbone residues as molecular tools to dissect DNA damage recognition by human NER into separate components and identified a bipartite discrimination mechanism that requires changes in DNA chemistry with concurrent disruption of Watson-Crick base pairing.
URLPMID:3320950 [本文引用: 1]

Background:The compaction of DNA into nucleosomes interferes with DNA repair. Results:Monoubiquitination of core histone H2A destabilizes nucleosomes containing UV-damaged DNA. Conclusion:Destabilized nucleosomes enable the release of the DNA damage-binding complex DDB1-CUL4BDDB2, which assists in histone ubiquitination. Significance:This mechanism explains how the ubiquitination of histone H2A, in addition to chromatin remodeling, promotes repair and facilitates genome stability. How the nucleotide excision repair (NER) machinery gains access to damaged chromatinized DNA templates and how the chromatin structure is modified to promote efficient repair of the non-transcribed genome remain poorly understood. The UV-damaged DNA-binding protein complex (UV-DDB, consisting of DDB1 and DDB2, the latter of which is mutated in xeroderma pigmentosum group E patients, is a substrate-recruiting module of the cullin 4B-based E3 ligase complex, DDB1-CUL4BDDB2. We previously reported that the deficiency of UV-DDB E3 ligases in ubiquitinating histone H2A at UV-damaged DNA sites in the xeroderma pigmentosum group E cells contributes to the faulty NER in these skin cancer-prone patients. Here, we reveal the mechanism by which monoubiquitination of specific H2A lysine residues alters nucleosomal dynamics and subsequently initiates NER. We show that DDB1-CUL4BDDB2E3 ligase specifically binds to mononucleosomes assembled with human recombinant histone octamers and nucleosome-positioning DNA containing cyclobutane pyrimidine dimers or 6-4 photoproducts photolesions. We demonstrate functionally that ubiquitination of H2A Lys-119/Lys-120 is necessary for destabilization of nucleosomes and concomitant release of DDB1-CUL4BDDB2from photolesion-containing DNA. Nucleosomes in which these lysines are replaced with arginines are resistant to such structural changes, and arginine mutants prevent the eviction of H2A and dissociation of polyubiquitinated DDB2 from UV-damaged nucleosomes. The partial eviction of H3 from the nucleosomes is dependent on ubiquitinated H2A Lys-119/Lys-120. Our results provide mechanistic insight into how post-translational modification of H2A at the site of a photolesion initiates the repair process and directly affects the stability of the human genome.
URLPMID:2858918 [本文引用: 1]

Eukaryotic cells repair ultraviolet light (UV)- and chemical carcinogen-induced DNA strand-distorting damage through the nucleotide excision repair (NER) pathway. Concurrent activation of the DNA damage checkpoints is also required to arrest the cell cycle and allow time for NER action. Recent studies uncovered critical roles for ubiquitin-mediated post-translational modifications in controlling both NER and checkpoint functions. In this review, we will discuss recent progress in delineating the roles of cullin-RING E3 ubiquitin ligases in orchestrating the cellular DNA damage response through ubiquitination of NER factors, histones, and checkpoint effectors.
URLPMID:16473935 [本文引用: 1]

Xeroderma pigmentosum (XP) is a heritable human disorder characterized by defects in nucleotide excision repair (NER) and the development of skin cancer. Cells from XP group E (XP-E) patients have a defect in the UV-damaged DNA-binding protein complex (UV-DDB), involved in the damage recognition step of NER. UVDDB comprises two subunits, products of the DDB1 and DDB2 genes, respectively. Mutations in the DDB2 gene account for the underlying defect in XP-E. The UV-DDB complex is a component of the newly identified cullin 4A-based ubiquitin E3 ligase, $DDB1-CUL4A^{DDB2}$. The E3 ubiquitin ligases recognize specific substrates and mediate their ubiquitination to regulate protein activity or target proteins for degradation by the proteasomal pathway. In this study, we have addressed the role of the UV-DDB-based E3 in NER and sought a physiological substrate. We demonstrate that monoubiquitinated histone H2A in native chromatin coimmunoprecipitates with the endogenous $DDB1-CUL4A^{DDB2}$ complex in response to UV irradiation. Further, mutations in DDB2 alter the formation and binding activity of the $DDB1-CUL4A^{DDB2}$ ligase, accompanied by impaired monoubiquitination of H2A after UV treatment of XP-E cells, compared with repair-proficient cells. This finding indicates that DDB2, as the substrate receptor of the DDB1-CUL4A-based ligase, specifically targets histone H2A for monoubiquitination in a photolesion-binding-dependent manner. Given that the loss of monoubiquitinated histone H2A at the sites of UV-damaged DNA is associated with decreased global genome repair in XP-E cells, this study suggests that histone modification, mediated by the XPE factor, facilitates the initiation of NER.
URLPMID:4007632 [本文引用: 1]

DNA damage recognition subunits such as DDB2 and XPC protect the human skin from ultraviolet (UV) light-induced genome instability and cancer, as demonstrated by the devastating inherited syndrome xeroderma pigmentosum. Here we show that the beneficial DNA repair response triggered by these two genome caretakers critically depends on a dynamic spatiotemporal regulation of their homeostasis. The prolonged retention of DDB2 and XPC in chromatin, because of a failure to readily remove both recognition subunits by the ubiquitin-dependent p97/VCP/Cdc48 segregase complex, leads to impaired DNA excision repair of UV lesions. Surprisingly, the ensuing chromosomal aberrations in p97-deficient cells are alleviated by a concomitant downregulation of DDB2 or XPC. Also, genome instability resulting from an excess of DDB2 persisting in UV-irradiated cells is prevented by concurrent p97 overexpression. Our findings demonstrate that DNA damage sensors and repair initiators acquire unexpected genotoxic properties if not controlled by timely extraction from chromatin.
[本文引用: 2]
URLPMID:15882621 [本文引用: 1]

The xeroderma pigmentosum group C (XPC) protein complex plays a key role in recognizing DNA damage throughout the genome for mammalian nucleotide excision repair (NER). Ultraviolet light (UV)-damaged DNA binding protein (UV-DDB) is another complex that appears to be involved in the recognition of NER-inducing damage, although the precise role it plays and its relationship to XPC remain to be elucidated. Here we show that XPC undergoes reversible ubiquitylation upon UV irradiation of cells and that this depends on the presence of functional UV-DDB activity. XPC and UV-DDB were demonstrated to interact physically, and both are polyubiquitylated by the recombinant UV-DDB-ubiquitin ligase complex. The polyubiquitylation altered the DNA binding properties of XPC and UV-DDB and appeared to be required for cell-free NER of UV-induced (6-4) photoproducts specifically when UV-DDB was bound to the lesion. Our results strongly suggest that ubiquitylation plays a critical role in the transfer of the UV-induced lesion from UV-DDB to XPC.
URLPMID:26422137 [本文引用: 1]

DNA in human cells is constantly assaulted by endogenous and exogenous DNA damaging agents. It is vital for the cell to respond rapidly and precisely to DNA damage to maintain genome integrity and reduce the risk of mutagenesis. Sophisticated reactions occur in chromatin surrounding the damaged site leading to the activation of DNA damage response (DDR), including transcription reprogramming, cell cycle checkpoint, and DNA repair. Histone proteins around the DNA damage play essential roles in DDR, through extensive post-translational modifications (PTMs) by a variety of modifying enzymes. One PTM on histones, mono-ubiquitylation, has emerged as a key player in cellular response to DNA damage. In this review, we will (1) briefly summarize the history of histone H2A and H2B ubiquitylation (H2Aub and H2Bub, respectively), (2) discuss their roles in transcription, and (3) their functions in DDR.
URLPMID:21827756 [本文引用: 1]

The DNA damage response (DDR) is emerging as a vast signaling network that temporarily modulates numerous aspects of cellular metabolism in the face of DNA lesions, especially critical ones such as the double strand break (DSB). The DDR involves extensive dynamics of protein post-translational modifications, most notably phosphorylation and ubiquitylation. The DSB response is mobilized primarily by the ATM protein kinase, which phosphorylates a plethora of key players in its various branches. It is based on a core of proteins dedicated to the damage response, and a cadre of proteins borrowed temporarily from other cellular processes to help meet the challenge. A recently identified novel component of the DDR pathway histone H2B monoubiquitylation exemplifies this principle. In mammalian cells, H2B monoubiquitylation is driven primarily by an E3 ubiquitin ligase composed of the two RING finger proteins RNF20 and RNF40. Generation of monoubiquitylated histone H2B (H2Bub) has been known to be coupled to gene transcription, presumably modulating chromatin decondensation at transcribed regions. New evidence indicates that the regulatory function of H2Bub on gene expression can selectively enhance or suppress the expression of distinct subsets of genes through a mechanism involving the hPAF1 complex and the TFIIS protein. This delicate regulatory process specifically affects genes that control cell growth and genome stability, and places RNF20 and RNF40 in the realm of tumor suppressor proteins. In parallel, it was found that following DSB induction, the H2B monoubiquitylation module is recruited to damage sites where it induces local H2Bub, which in turn is required for timely recruitment of DSB repair protein and, subsequently, timely DSB repair. This pathway represents a crossroads of the DDR and chromatin organization, and is a typical example of how the DDR calls to action functional modules that in unprovoked cells regulate other processes.
URLPMID:24476359 [本文引用: 1]

Commitment to splicing occurs co-transcriptionally, but a major unanswered question is the extent to which various modifications of chromatin, the template for transcription in vivo, contribute to the regulation of splicing.ResultsHere, we perform genome-wide analyses showing that inhibition of specific marks 090009 H2B ubiquitylation, H3K4 methylation and H3K36 methylation 090009 perturbs splicing in budding yeast, with each modification exerting gene-specific effects. Furthermore, semi-quantitative mass spectrometry on purified nuclear mRNPs and chromatin immunoprecipitation analysis on intron-containing genes indicated that H2B ubiquitylation, but not Set1-, Set2- or Dot1-dependent H3 methylation, stimulates recruitment of the early splicing factors, namely U1 and U2 snRNPs, onto nascent RNAs.ConclusionsThese results suggest that histone modifications impact splicing of distinct subsets of genes using distinct pathways.
URLPMID:18374642 [本文引用: 1]

Recently, many of the enzymes responsible for the addition and removal of ubiquitin from the histones H2A and H2B have been identified and characterized. From these studies, it has become clear that H2A and H2B ubiquitination play critical roles in regulating many processes within the nucleus, including transcription initiation and elongation, silencing, and DNA repair. In this review, we present the enzymes involved in H2A and H2B ubiquitination and discuss new evidence that links histone ubiquitination to other chromatin modifications, which has provided a model for the role of H2B ubiquitination, in particular, in transcription initiation and elongation.
URLPMID:3397146 [本文引用: 3]

The cellular response to DNA double-strand breaks (DSBs) is mobilized by the protein kinase ATM, which phosphorylates key players in the DNA damage response (DDR) network. A major question is how ATM controls DSB repair. Optimal repair requires chromatin relaxation at damaged sites. Chromatin reorganization is coupled to dynamic alterations in histone posttranslational modifications. Here, we show that in human cells, DSBs induce monoubiquitylation of histone H2B, a modification that is associated in undamaged cells with transcription elongation. We find that this process relies on recruitment to DSB sites and ATM-dependent phosphorylation of the responsible E3 ubiquitin ligase: the RNF20-RNF40 heterodimer. H2B monoubiquitylation is required for timely recruitment of players in the two major DSB repair pathways—nonhomologous end-joining and homologous recombination repair—and optimal repair via both pathways. Our data and previous data suggest a two-stage model for chromatin decondensation that facilitates DSB repair.Graphical AbstractView high quality image (332K)
URL [本文引用: 1]
URLPMID:22373577 [本文引用: 1]

The methyltransferase DOT1L methylates histone H3 at K79 to facilitate specific biological events. H3K79 dimethylation (H3K79-2Me) by DOT1L influences the DNA damage response by promoting 53BP1 recruitment to DNA damage sites; however, it is unclear if this methylation is required as 53BP1 interacts with dimethylated H4 (H4K20-2Me) with a much higher affinity. We demonstrate that H3K79-2Me, while negligible during S-phase, is required for ionizing radiation (IR)-induced 53BP1 foci formation during G1/G2-phases when H4K20-2Me levels are low. Further, we describe an essential role for HLA-B-associated transcript 3 (Bat3) in regulating this process in U2OS cells. Bat3 co-localizes with DOT1L at histone H3, and Bat3 knockdown results in decreased DOT1L090009H3 interaction and H3K79-2Me, leading to a reduction in IR-induced 53BP1 foci formation, defects in DNA repair and increased sensitivity to IR. We demonstrate that a conserved Bat3 ubiquitin-like motif and a conserved DOT1L ubiquitin-interacting motif promote DOT1L090009Bat3 interaction to facilitate efficient H3K79-2Me and IR-induced 53BP1 foci formation during G1/G2-phases. Taken together, our findings identify a novel role for Bat3 in regulating DOT1L function, which plays a critical role in DNA damage response.
URL [本文引用: 2]
URLPMID:4584319 [本文引用: 1]

Rapid progress in the study on the association of histone modifications with chromatin remodeling factors has broadened our understanding of chromatin dynamics in DNA transactions. In DNA double-strand break (DSB) repair, the well-known mark of histones is the phosphorylation of the H2A variant, H2AX, which has been used as a surrogate marker of DSBs. The ubiquitylation of histone H2B by RNF20 E3 ligase was recently found to be a DNA damage-induced histone modification. This modification is required for DSB repair and regulated by a distinctive pathway from that of histone H2AX phosphorylation. Moreover, the connection between H2B ubiquitylation and the chromatin remodeling activity of SNF2H has been elucidated. In this review, we summarize the current knowledge of RNF20-mediated processes and the molecular link to H2AX-mediated processes during DSB repair.
URLPMID:19410543 [本文引用: 2]

H2B ubiquitylation has been implicated in active transcription but is not well understood in mammalian cells. Beyond earlier identification of hBRE1 as the E3 ligase for H2B ubiquitylation in human cells, we now show (1) that hRAD6 serves as the cognate E2-conjugating enzyme; (2) that hRAD6, through direct interaction with hPAF-bound hBRE1, is recruited to transcribed genes and ubiquitylates chromatinized H2B at lysine 120; (3) that hPAF-mediated transcription is required for efficient H2B ubiquitylation as a result of hPAF-dependent recruitment of hBRE1-hRAD6 to the Pol II transcription machinery; (4) that H2B ubiquitylation per se does not affect the level of hPAF-, SII-, and p300-dependent transcription and likely functions downstream; and (5) that H2B ubiquitylation directly stimulates hSET1-dependent H3K4 di- and trimethylation. These studies establish the natural H2B ubiquitylation factors in human cells and also detail the mechanistic basis for H2B ubiquitylation and function during transcription.
URLPMID:21196936 [本文引用: 1]

Regulation of chromatin structure involves histone post-translational modifications which can modulate intrinsic properties of the chromatin fiber to change the chromatin state. We used chemically defined nucleosome arrays to demonstrate that H2B ubiquitylation (uH2B), a modification associated with transcription, interferes with chromatin compaction and leads to an open and biochemically accessible fiber conformation. Importantly, these effects were specific for ubiquitin, as compaction of chromatin modified with a similar ubiquitin-sized protein, Hub1, was only weakly affected. Applying a fluorescence based method we found that uH2B acts through a mechanism distinct from H4 tail acetylation (acH4), a modification known to disrupt chromatin folding. Finally, incorporation of both uH2B and acH4 in nucleosomes resulted in synergistic inhibition of higher order chromatin structure formation, possibly a result of their distinct mode of action.
URLPMID:18653199 [本文引用: 1]

The mechanism by which chromatin is decondensed to permit access to DNA is largely unknown. Here, using a model nucleosome array reconstituted from recombinant histone octamers, we have defined the relative contribution of the individual histone octamer N-terminal tails as well as the effect of a targeted histone tail acetylation on the compaction state of the 30 nm chromatin fiber. This study goes beyond previous studies as it is based on a nucleosome array that is very long (61 nucleosomes) and contains a stoichiometric concentration of bound linker histone, which is essential for the formation of the 30 nm chromatin fiber. We find that compaction is regulated in two steps: Introduction of H4 acetylated to 30% on K16 inhibits compaction to a greater degree than deletion of the H4 N-terminal tail. Further decompaction is achieved by removal of the linker histone.
URLPMID:22031019 [本文引用: 2]

Many anticancer therapies function largely by inducing DNA double-strand breaks (DSBs) or altering the ability of cancer cells to repair them. Proper and timely DNA repair requires dynamic changes in chromatin assembly and disassembly characterized by histone H3 lysine 56 acetylation (H3K56ac) and phosphorylation of the variant histone H2AX ( H2AX). Similarly, histone H2B monoubiquitination (H2Bub1) functions in DNA repair, but its role in controlling dynamic changes in chromatin structure following DSBs and the histone chaperone complexes involved remain unknown. Therefore, we investigated the role of the H2B ubiquitin ligase RNF40 in the DSB response. We show that RNF40 depletion results in sustained H2AX phosphorylation and a decrease in rapid cell cycle checkpoint activation. Furthermore, RNF40 knockdown resulted in decreased H3K56ac and decreased recruitment of the facilitates chromatin transcription (FACT) complex to chromatin following DSB. Knockdown of the FACT component suppressor of Ty homolog-16 (SUPT16H) phenocopied the effects of RNF40 knockdown on both H2AX and H3K56ac following DSB induction. Consistently, both RNF40 and SUPT16H were required for proper DNA end resection and timely DNA repair, suggesting that H2Bub1 and FACT cooperate to increase chromatin dynamics, which facilitates proper checkpoint activation and timely DNA repair. These results provide important mechanistic insights into the tumor suppressor function of H2Bub1 and provide a rational basis for pursuing H2Bub1-based therapies in conjunction with traditional chemo- and radiotherapy.
URLPMID:3142300 [本文引用: 1]

Abstract Q308K allele is synthetically lethal with an allele of histone H4 that prevents the diacetylation of newly synthesized molecules. We have analyzed the genetic interactions between the Q308K allele of and mutations in all of the sites of acetylation that have been identified on newly synthesized histones. Genetic interactions were observed between and sites of acetylation on the NH-terminal tails of H3 and H4. For histone H3, lysine residues 14 and 23 were particularly important when activity is compromised. Surprisingly, synthetic defects observed when the Q308K allele was combined with mutations of H4 lysines 5 and 12, were not phenocopied by deletion of , which encodes the enzyme that is thought to generate this pattern of acetylation on H4. Genetic interactions were also observed between and sites of acetylation found in the core domain of newly synthesized histones H3 and H4. These include synthetic lethality with an allele of H4 lysine 91 that mimics constitutive acetylation. While the mutations that alter H4 lysines 5, 12 and 91 do not affect binding to Pob3p, mutation of histone H3 lysine 56 decreases the association of histones with Pob3p. These results support the model that the yFACT complex plays a central role in chromatin assembly pathways regulated by acetylation of newly synthesized histones.
URLPMID:25141862 [本文引用: 2]

Abstract Histone H2B ubiquitination is a dynamic modification that promotes methylation of histone H3K79 and H3K4. This crosstalk is important for the DNA damage response and has been implicated in cancer. Here, we show that in engineered yeast strains, ubiquitins tethered to every nucleosome promote H3K79 and H3K4 methylation from a proximal as well as a more distal site, but only if in a correct orientation. This plasticity indicates that the exact location of the attachment site, the native ubiquitin-lysine linkage and ubiquitination cycles are not critical for trans-histone crosstalk in vivo . The flexibility in crosstalk also indicates that other ubiquitination events may promote H3 methylation. Synopsis
URLPMID:19667127 [本文引用: 1]

Histone H2B monoubiquitination by Rad6/Bre1 is required for the trimethylation of both histone H3K4 and H3K79 by COMPASS and Dot1 methyltransferases, respectively. The dependency of methylation at H3K4 and H3K79 on the monoubiquitination of H2BK123 was recently challenged, and extragenic mutations in the strain background used for previous studies or epitope-tagged proteins were suggested to be the sources of this discrepancy. In this study, we show that H3K4 and H3K79 methylation is solely dependent on H2B monoubiquitination regardless of any additional alteration to the H2B sequence or genome. Furthermore, we report that Y131, one of the yeast histone H2A/H2B shuffle strains widely used for the last decade in the field of chromatin and transcription biology, carries a wild-type copy of each of theHTA2andHTB2genes under theGAL1/10promoter on chromosome II. Therefore, we generated the entire histone H2A and H2B alanine-scanning mutant strains in another background, which does not express wild-type histones.
URLPMID:21460225 [本文引用: 1]

Global genomic repair (GGR) and transcription coupled repair (TCR) are two pathways of nucleotide excision repair (NER) that differ in the damage recognition step. How NER factors, especially GGR factors, access DNA damage in the chromatin of eukaryotic cells has been poorly understood. Dot1, a histone methyltransferase required for methylation of histone H3 lysine 79 (H3K79), has been shown to confer yeast cells with resistance to DNA-damaging agents and play a role in activation of DNA damage checkpoints. Here, we show that Dot1 and H3K79 methylation are required for GGR in both nucleosomal core regions and internucleosomal linker DNA, but play no role in TCR. H3K79 trimethylation contributes to but is not absolutely required for GGR, and lower levels of H3K79 methylation (mono- and dimethylation) also promote GGR. Our results also indicate that the roles of Dot1 and H3K79 methylation in GGR are not achieved by either activating DNA damage checkpoints or regulating the expression of the GGR-specific factor Rad16. Rather, the methylated H3K79 may serve as a docking site for the GGR machinery on the chromatin. Our studies identified a novel GGR-specific NER factor and unveiled the critical link between a covalent histone modification and GGR.
URLPMID:28246327 [本文引用: 1]

DNA lesion bypass is mediated by DNA damage tolerance (DDT) pathways and homologous recombination (HR). The DDT pathways, which involve translesion synthesis and template switching (TS), are activated by the ubiquitylation (ub) of PCNA through components of the RAD6-RAD18 pathway, whereas the HR pathway is independent of RAD18. However, it is unclear how these processes are coordinated within the context of chromatin. Here we show that Bre1, an ubiquitin ligase specific for histone H2B, is recruited to chromatin in a manner coupled to replication of damaged DNA. In the absence of Bre1 or H2Bub, cells exhibit accumulation of unrepaired DNA lesions. Consequently, the damaged forks become unstable and resistant to repair. We provide physical, genetic, and cytological evidence that H2Bub contributes toward both Rad18-dependent TS and replication fork repair by HR. Using an inducible system of DNA damage bypass, we further show that H2Bub is required for the regulation of DDT after genome duplication. We propose that Bre1-H2Bub facilitates fork recovery and gap-filling repair by controlling chromatin dynamics in response to replicative DNA damage.
URLPMID:5100568 [本文引用: 1]

Abstract Histone modifications play an important role in regulating access to DNA for transcription, DNA repair and DNA replication. A central player in these events is the mono-ubiquitylation of histone H2B (H2Bub1), which has been shown to regulate nucleosome dynamics. Previously, it was shown that H2Bub1 was important for nucleosome assembly onto nascent DNA at active replication forks. In the absence of H2Bub1, incomplete chromatin structures resulted in several replication defects. Here, we report new evidence, which shows that loss of H2Bub1 contributes to genomic instability in yeast. Specifically, we demonstrate that H2Bub1-deficient yeast accumulate mutations at a high frequency under conditions of replicative stress. This phenotype is due to an aberrant DNA Damage Tolerance (DDT) response upon fork stalling. We show that H2Bub1 normally functions to promote error-free translesion synthesis (TLS) mediated by DNA polymerase eta (Pol02·). Without H2Bub1, DNA polymerase zeta (Pol0209) is responsible for a highly mutagenic alternative mechanism. While H2Bub1 does not appear to regulate other DDT pathways, error-free DDT mechanisms are employed by H2Bub1-deficient cells as another means for survival. However, in these instances, the anti-recombinase, Srs2, is essential to prevent the accumulation of toxic HR intermediates that arise in an unconstrained chromatin environment. 0008 The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
URL [本文引用: 3]
URL [本文引用: 1]

Acetylated histone H3 lysine 56 (H3K56Ac) diminishes in response to DNA damage but is restored following DNA repair. Here, we report that CRL4DDB2ubiquitin ligase preferentially regulates post-repair chromatin restoration of H3K56Ac through recruitment of histone chaperon CAF-1. We show that H3K56Ac accumulates at DNA damage sites. The restoration of H3K56Ac but not H3K27Ac, H3K18Ac and H3K14Ac depends on CAF-1 function, whereas all these acetylations are mediated by CBP/p300. The CRL4DDB2components, DDB1, DDB2 and CUL4A, are also required for maintaining the H3K56Ac and H3K9Ac level in chromatin, and for restoring H3K56Ac following induction of DNA photolesions and strand breaks. Depletion of CUL4A decreases the recruitment of CAF-1 p60 and p150 to ultraviolet radiation- and phleomycin-induced DNA damage. Neddylation inhibition renders CRL4DDB2inactive, decreases H3K56Ac level, diminishes CAF-1 recruitment and prevents H3K56Ac restoration. Mutation in the PIP box of DDB2 compromises its capability to elevate the H3K56Ac level but does not affect XPC ubiquitination. These results demonstrated a function of CRL4DDB2in differential regulation of histone acetylation in response to DNA damage, suggesting a novel role of CRL4DDB2in repair-driven chromatin assembly.
URLPMID:24074863 [本文引用: 1]

Transient targeting of HIRA to damaged chromatin regions deposits newly synthesized H3.3 histones, “bookmarking” the chromatin substrate for reactivation of transcription once repair is complete.
URLPMID:23739170 [本文引用: 1]

Histones have two structurally and functionally distinct domains: globular domains forming the nucleosomal core around which DNA is wrapped and unstructured tails protruding from the nucleosomal core. Whereas post-translational modifications (PTMs) in histone tails are well studied, much less is currently known about histone-core PTMs. Many core PTMs map to residues located on the lateral surface of the histone octamer, close to the DNA, and they have the potential to alter intranucleosomal histone-DNA interactions. Here we discuss recent advances in understanding the function of lateral-surface PTMs. Whereas modifications in the histone tails might have limited structural impact on the nucleosome itself and function as signals to recruit specific binding proteins, PTMs in the lateral surface can have a direct structural effect on nucleosome and chromatin dynamics, even in the absence of specific binding proteins, which adds a twist to the debate on the functionality and causality of PTMs.
URLPMID:19818714 [本文引用: 2]

Although the BBAP E3 ligase and its binding partner BAL are overexpressed in chemotherapy-resistant lymphomas, the role of these proteins in DNA damage responses remains undefined. Because BAL proteins modulate promoter-coupled transcription and contain structural motifs associated with chromatin remodeling and DNA repair, we reasoned that the BBAP E3 ligase might target nucleosomal proteins. Herein, we demonstrate that BBAP selectively monoubiquitylates histone H4 lysine 91 and protects cells exposed to DNA-damaging agents. Disruption of BBAP-mediated monoubiquitylation of histone H4K91 is associated with the loss of chromatin-associated H4K20 methylase, mono- and dimethyl H4K20, and a delay in the kinetics of 53BP1 foci formation at sites of DNA damage. Because 53BP1 localizes to DNA damage sites, in part, via an interaction with dimethyl H4K20, these data directly implicate BBAP in the monoubiquitylation and additional posttranslational modification of histone H4 and an associated DNA damage response.
URLPMID:23230272 [本文引用: 1]

The BAL1 macrodomain-containing protein and its partner E3 ligase, BBAP, are overexpressed in chemotherapy-resistant lymphomas. BBAP selectively ubiquitylates histone H4 and indirectly promotes early 53BP1 recruitment to DNA damage sites. However, neither BBAP nor BAL1 has been directly associated with a DNA damage response (DDR), and the function of BAL1 remains undefined. Herein, we describe a direct link between rapid and short-lived poly(ADP-ribose) (PAR) polymerase 1 (PARP1) activation and PARylation at DNA damage sites, PAR-dependent recruitment of the BAL1 macrodomain-containing protein and its partner E3 ligase, local BBAP-mediated ubiquitylation, and subsequent recruitment of the checkpoint mediators 53BP1 and BRCA1. The PARP1-dependent localization of BAL1-BBAP functionally limits both early and delayed DNA damage and enhances cellular viability independent of ATM, MDC1, and RNF8. These data establish that BAL1 and BBAP are bona fide members of a DNA damage response pathway and are directly associated with PARP1 activation, BRCA1 recruitment, and double-strand break repair.
URLPMID:25113974 [本文引用: 2]

Abstract Histone ubiquitination at DNA breaks is required for activation of the DNA damage response (DDR) and DNA repair. How the dynamic removal of this modification by deubiquitinating enzymes (DUBs) impacts genome maintenance in vivo is largely unknown. To address this question, we generated mice deficient for Ub-specific protease 3 (USP3; Usp30200/0200), a histone H2A DUB which negatively regulates ubiquitin-dependent DDR signaling. Notably, USP3 deletion increased the levels of histone ubiquitination in adult tissues, reduced the hematopoietic stem cell (HSC) reserves over time, and shortened animal life span. Mechanistically, our data show that USP3 is important in HSC homeostasis, preserving HSC self-renewal, and repopulation potential in vivo and proliferation in vitro. A defective DDR and unresolved spontaneous DNA damage contribute to cell cycle restriction of Usp30200/0200 HSCs. Beyond the hematopoietic system, Usp30200/0200 animals spontaneously developed tumors, and primary Usp30200/0200 cells failed to preserve chromosomal integrity. These findings broadly support the regulation of chromatin ubiquitination as a key pathway in preserving tissue function through modulation of the response to genotoxic stress. 0008 2014 Lancini et al.
URLPMID:12507430 [本文引用: 1]

The ubiquitin-specific processing protease (UBP) family of deubiquitinating enzymes plays an essential role in numerous cellular processes. HAUSP, a representative UBP, specifically deubiquitinates and hence stabilizes the tumor suppressor protein p53. Here, we report the crystal structures of the 40 kDa catalytic core domain of HAUSP in isolation and in complex with ubiquitin aldehyde. These studies reveal that the UBP deubiquitinating enzymes exhibit a conserved three-domain architecture, comprising Fingers, Palm, and Thumb. The leaving ubiquitin moiety is specifically coordinated by the Fingers, with its C terminus placed in the active site between the Palm and the Thumb. Binding by ubiquitin aldehyde induces a drastic conformational change in the active site that realigns the catalytic triad residues for catalysis.
URLPMID:17914355 [本文引用: 1]

Post-translational histone modifications have important regulatory roles in chromatin structure and function. One example of such modifications is histone ubiquitination, which occurs predominately on histone H2A and H2B. Although the recent identification of the ubiquitin ligase for histone H2A has revealed important roles for H2A ubiquitination in Hox gene silencing as well as in X-chromosome inactivation, the enzyme(s) involved in H2A deubiquitination and the function of H2A deubiquitination are not known. Here we report the identification and functional characterization of the major deubiquitinase for histone H2A, Ubp-M (also called USP16). Ubp-M prefers nucleosomal substrates in vitro, and specifically deubiquitinates histone H2A but not H2B in vitro and in vivo. Notably, knockdown of Ubp-M in HeLa cells results in slow cell growth rates owing to defects in the mitotic phase of the cell cycle. Further studies reveal that H2A deubiquitination by Ubp-M is a prerequisite for subsequent phosphorylation of Ser 10 of H3 and chromosome segregation when cells enter mitosis. Furthermore, we demonstrate that Ubp-M regulates Hox gene expression through H2A deubiquitination and that blocking the function of Ubp-M results in defective posterior development in Xenopus laevis. This study identifies the major deubiquitinase for histone H2A and demonstrates that H2A deubiquitination is critically involved in cell cycle progression and gene expression.
URLPMID:26739236 [本文引用: 1]

The deubiquitinating enzyme BAP1 is an important tumor suppressor that has drawn attention in the clinic since its loss leads to a variety of cancers. BAP1 is activated by ASXL1 to deubiquitinate mono-ubiquitinated H2A at K119 in Polycomb gene repression, but the mechanism of this reaction remains poorly defined. Here we show that the BAP1 C-terminal extension is important for H2A deubiquitination by auto-recruiting BAP1 to nucleosomes in a process that does not require the nucleosome acidic patch. This initial encounter-like complex is unproductive and needs to be activated by the DEUBAD domains of ASXL1, ASXL2 or ASXL3 to increase BAP1's affinity for ubiquitin on H2A, to drive the deubiquitination reaction. The reaction is specific for Polycomb modifications of H2A as the complex cannot deubiquitinate the DNA damage-dependent ubiquitination at H2A K13/15. Our results contribute to the molecular understanding of this important tumor suppressor. The tumor suppressor BAP1 is activated by ASXL1 to deubiquitinate mono-ubiquitinated H2A at K119 in Polycomb gene repression. Here, the authors show how BAP1's C-terminal extension auto-recruits it to nucleosomes, where the DEUBAD domain of ASXL1 increases BAP1's affinity for ubiquitin to drive deubiquitination.
URLPMID:28257702 [本文引用: 1]

Understanding the mechanism of resistance of genes to reactivation will help improve the success of nuclear reprogramming. Using mouse embryonic fibroblast nuclei with normal or reduced DNA methylation in combination with chromatin modifiers able to erase H3K9me3, H3K27me3, and H2AK119ub1 from transplanted nuclei, we reveal the basis for resistance of genes to transcriptional reprogramming by oocyte factors. A majority of genes is affected by more than one type of treatment, suggesting that resistance can require repression through multiple epigenetic mechanisms. We classify resistant genes according to their sensitivity to 11 chromatin modifier combinations, revealing the existence of synergistic as well as adverse effects of chromatin modifiers on removal of resistance. We further demonstrate that the chromatin modifier USP21 reduces resistance through its H2AK119 deubiquitylation activity. Finally, we provide evidence that H2A ubiquitylation also contributes to resistance to transcriptional reprogramming in mouse nuclear transfer embryos. Graphical Abstract 61Identification of genes resistant to direct transcriptional reprogramming61Determination of resistant gene sensitivity to 11 chromatin modifier combinations61USP21 removes resistance through its H2AK119 deubiquitylation activity61USP21 improves the reprogramming of gene expression in two-cell-stage mouse embryos Identification of genes resistant to direct transcriptional reprogramming Determination of resistant gene sensitivity to 11 chromatin modifier combinations USP21 removes resistance through its H2AK119 deubiquitylation activity USP21 improves the reprogramming of gene expression in two-cell-stage mouse embryos Using nuclear transfer toXenopusoocytes, Jullien et02al. identify genes in MEF nuclei whose epigenetic configuration directly resists transcriptional reprogramming by oocyte factors. They show that loss of resistance can be achieved by a combinatorial action of chromatin modifiers, which alter H2A ubiquitylation, H3K9, H3K27, and DNA methylation levels.
URLPMID:3925719 [本文引用: 1]

Histone ubiquitination plays a vital role in DNA damage response (DDR), which is important for maintaining genomic integrity in eukaryotic cells. In DDR, ubiquitination of histone H2A and 0206H2AX by the concerted action of ubiquitin (Ub) ligases, RNF168 and RNF8, generates a cascade of ubiquitination signaling. However, little is known about deubiquitinating enzymes (DUBs) that may catalyze the removal of Ub from these histones. This study demonstrated that USP3, an apparent DUB for mono-ubiquitinated H2A, is indeed the enzyme for deubiquitinating Ub conjugates of 0206H2AX and H2A from lysine sites, where the ubiquitination is initiated by RNF168. Here, we showed that ectopic expression of USP3 led to the deubiquitination of both H2A and 0206H2AX in response to UV-induced DNA damage. Moreover, ectopic USP3 expression abrogated FK2 antibody-reactive Ub-conjugate foci, which co-localize with damage-induced 0206H2AX foci. In addition, USP3 overexpression impaired the accumulation of downstream repair factors BRCA1 and 53BP1 at the damage sites in response to both UV and 0206-irradiation. We further identified that the USP3 removes Ub at lysine 13 and 15 of H2A and 0206H2AX, as well as lysine 118 and 119 of H2AX in response to DNA damage. Taken together, the results suggested that USP3 is a negative regulator of ubiquitination signaling, counteracting RNF168- and RNF8-mediated ubiquitination.
URLPMID:27083998 [本文引用: 1]

Dynamic regulation of RNF168-mediated ubiquitylation of histone H2A Lys13,15 (H2AK13,15ub) at DNA double-strand breaks (DSBs) is crucial for preventing aberrant DNA repair and maintaining genome stability. However, it remains unclear which deubiquitylating enzyme (DUB) removes H2AK13,15ub. Here we show that USP51, a previously uncharacterized DUB, deubiquitylates H2AK13,15ub and regulates DNA damage response. USP51 depletion results in increased spontaneous DNA damage foci and elevated levels of H2AK15ub and impairs DNA damage response. USP51 overexpression suppresses the formation of ionizing radiation-induced 53BP1 and BRCA1 but not RNF168 foci, suggesting that USP51 functions downstream from RNF168 in DNA damage response. In vitro, USP51 binds to H2A-H2B directly and deubiquitylates H2AK13,15ub. In cells, USP51 is recruited to chromatin after DNA damage and regulates the dynamic assembly/disassembly of 53BP1 and BRCA1 foci. These results show that USP51 is the DUB for H2AK13,15ub and regulates DNA damage response.
URL [本文引用: 3]

BRCA1-BARD1-catalyzed ubiquitination of histone H2A is an important regulator of the DNA damage response, priming chromatin for repair by homologous recombination. However, no specific deubiquitinating enzymes (DUBs) are known to antagonize this function. Here we identify ubiquitin specific protease-48 (USP48) as a H2A DUB, specific for the C-terminal BRCA1 ubiquitination site. Detailed biochemical analysis shows that an auxiliary ubiquitin, an additional ubiquitin that itself does not get cleaved, modulates USP48 activity, which has possible implications for its regulation in vivo. In cells we reveal that USP48 antagonizes BRCA1 E3 ligase function and in BRCA1-proficient cells loss of USP48 results in positioning 53BP1 further from the break site and in extended resection lengths. USP48 repression confers a survival benefit to cells treated with camptothecin and its activity acts to restrain gene conversion and mutagenic single-strand annealing. We propose that USP48 promotes genome stability by antagonizing BRCA1 E3 ligase function. BRCA1 ligase activity is tightly regulated to maintain genome stability and confer DNA double strand repair. Here the authors identify USP48 as a H2A deubiquitinating enzyme that acts as a BRCA1 E3 ligase antagonist and characterize its role during DNA repair.
URLPMID:26912860 [本文引用: 1]

Monoubiquitinated histone H2B plays multiple roles in transcription activation. H2B is deubiquitinated by the Spt-Ada-Gcn5 acetyltransferase (SAGA) coactivator, which contains a four-protein subcomplex known as the deubiquitinating (DUB) module. The crystal structure of the Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to a ubiquitinated nucleosome reveals that the DUB module primarily contacts H2A/H2B, with an arginine cluster on the Sgf11 zinc finger domain docking on the conserved H2A/H2B acidic patch. The Ubp8 catalytic domain mediates additional contacts with H2B, as well as with the conjugated ubiquitin. We find that the DUB module deubiquitinates H2B both in the context of the nucleosome and in H2A/H2B dimers complexed with the histone chaperone, FACT, suggesting that SAGA could target H2B at multiple stages of nucleosome disassembly and reassembly during transcription. Authors: Michael T. Morgan, Mahmood Haj-Yahya, Alison E. Ringel, Prasanthi Bandi, Ashraf Brik, Cynthia Wolberger
URLPMID:27160905 [本文引用: 1]

Activation-induced deaminase induces class switch recombination by inducing DNA breaks at the immunoglobulin locus. Ramachandran et02al. showed that the SAGA deubiquitination module is critical for this process. They showed that this complex is required for optimal γH2AX formation and functions upstream of various double-stranded DNA repair pathways.
