 ,, 胡伟, 邢永忠
,, 胡伟, 邢永忠 ,华中农业大学,作物遗传改良国家重点实验室,武汉 430070
,华中农业大学,作物遗传改良国家重点实验室,武汉 430070The history and prospect of rice genetic breeding in China
Bi Wu ,, Wei Hu, Yongzhong Xing
,, Wei Hu, Yongzhong Xing ,National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China
,National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China通讯作者:
编委: 储成才
收稿日期:2018-07-26修回日期:2018-09-4网络出版日期:2018-10-20
| 基金资助: |
Received:2018-07-26Revised:2018-09-4Online:2018-10-20
| Fund supported: |
作者简介 About authors
吴比,博士,研究方向:水稻遗传学E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (564KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
吴比, 胡伟, 邢永忠. 中国水稻遗传育种历程与展望[J]. 遗传, 2018, 40(10): 841-857 doi:10.16288/j.yczz.18-213
Bi Wu, Wei Hu, Yongzhong Xing.
水稻(Oryza sativa L.)是重要的粮食作物,在我国具有悠久的种植和选育历史。《诗经》中就有“丰年多黍多稌”关于水稻丰产的记载,“稌”即为糯稻。水稻品种概念在战国《管子》中就已出现[1]。在水稻种植历史中,无论是常规地方品种、现代品种还是杂交组合,产量一直是重要考量目标之一。特别是20世纪中期以来,我国农业生产力低下,为了解决粮食短缺问题,我国育种家唯产量是举,培育出耐大肥大水的品种,依靠大量使用农药化肥,虽然解决了燃眉之急,但也给我国的生态环境造成了破坏,这种生产模式不具有可持续性。21世纪以来,人们生活水平显著提高,我国的水稻生产渐渐地发生了重大变化,育种目标也从单一高产转向优质高抗高产等复合性状目标,绿色农业指日可待。
1 水稻遗传育种6个重要历程
1.1 水稻第一次“绿色革命”
水稻地方品种基本是高秆类型,耐肥力差,容易倒伏,导致稳产问题。因此,发掘矮秆种质资源,培育矮秆品种变得十分重要。1956年,我国育种家黄耀祥先生以广西农家品种“矮仔占”为材料,选育出“矮仔占4号”,并与高秆品种“广场13”杂交,培育出第一个矮秆籼稻品种“广场矮”[2]。1966年,国际水稻研究所(IRRI)利用我国台湾省地方品种低脚乌尖(Dee-geo-woo-gen)与皮泰(Peta)杂交,育成了半矮化的品种IR8,创造了当时的高产奇迹[3]。我国的矮化育种比国际上整整提前了10年。“广场矮”的培育以及IR8的引进推动了我国水稻育种进入第一次“绿色革命”时代,水稻亩产从50年代的164千克提高到70年代初的238千克,水稻单产提高了将近45%,实现了水稻产量的第一次飞跃[4]。实际上矮秆品种单株产量略微下降[5],但是由于其耐密植和抗倒伏,群体产量大幅度提升;同时矮秆品种的耐肥能力强,氮素吸收利用效率低,大量氮肥的施用在提高产量的同时使土壤和环境受到破坏[6]。1.2 核质互作雄性不育系的培育和水稻三系杂种优势利用
杂种优势是指一个物种的不同品种或者物种间的杂交后代的生物量、发育速度和产量的表型值优于两个亲本的现象[7]。我国稻作科学的奠基人丁颖先生曾用人工办法给水稻“去雄”,但实际效果不佳。1964年,湖南安江农校的袁隆平先生开始杂交水稻育种研究,并提出杂种优势利用的设想[8]。1970年,袁隆平先生和他的助手李必湖等人在海南三亚发现了花粉败育的野生稻,花粉败育是由不育细胞质产生(Wild AbortiveCytoplasmic Male Sterility, CMS- WA),为杂交水稻雄性不育系的选育打开了突破口。1971年,杂交水稻课题被列为全国协作项目,野败材料被分发到国内水稻科研单位[9]。1973年10月,在苏州召开的水稻科研会议上,袁隆平先生发表《利用“野败”选育“三系”进展》的论文,标志中国籼型杂交水稻三系(不育系、保持系和恢复系)配套成功。同时,江西省萍乡市农业科学研究所的颜龙安先生利用“野败”育成不育系珍汕97A。1981年,福建省三明市农业科学研究所的谢华安先生育成具有抗病性强、配合力高、米质优良的恢复系明恢63。明恢63与珍汕97A配制的强优势组合汕优63产量高,且具有抗病、耐低磷钾、耐高低温、米质较好、适应性广等特点,累计推广6287.7万公顷[10,11]。三系杂交稻平均产量比一般普通良种增产20%左右,亩产能达到430公斤[12]。
CMS-WA是细胞质基因和核基因互作导致花粉败育类型,属于孢子体雄性不育。可以通过回交的方法保留细胞质基因组而交换核基因组,达到培育不育系的目的。除CMS-WA外,我国水稻育种学家还创制出不同细胞质来源的核质互作雄性不育系。四川农业大学的周开达先生等用西非品种冈比亚卡与朝阳1号、雅安早等杂交和回交,育成冈型不育系冈12朝阳1号A和冈22雅安早A;同时,周开达先生等从Dissi D52/37//矮脚南特群体中选出不育株,育成D型不育系意大利A。意大利A与珍汕97B中变异株杂交,育成D汕A[13]。湖南省农业科学院利用地理远距离品种间杂交组合,育成印尼水田型不育系[14]。冈型、D型和印尼水田型不育特性以及恢、保关系与野败相似。不同育种单位育成了很多孢子体雄性不育类型的不育系,如四川省农业科学院的K型[15],其胞质不育基因来自于粳稻,恢、保关系与野败相同;安徽省广德县农业科学研究所从江西省引进的矮杆野生稻中发现一株雄性不育株,命名“矮败”,通过核置换育成协青早A,恢、保关系与野败相同[14],K型和矮败型不育系属于孢子体不育。1972年,武汉大学朱英国先生等以红芒野生稻为母本与莲塘早杂交,选育出红莲型细胞质雄性不育系(Honglian-type Cytoplasmic Male Sterility, HL- CMS),为配子体不育类型[11]。包台型(BT)不育系台中65A引入我国,并经湖南省农业科学院转育成BT型黎明A,与C系统恢复系的配组,使得BT型不育系成为我国粳稻杂种优势利用的主要不育系类型[16]。BT型不育系花粉败育属于配子体不育,不育性没有CMS-WA稳定,高温易自交结实,杂交制种种子不纯[17]。滇型不育系是云南省培育出的适应当地高海拔气候环境的粳稻CMS,属于配子体不育类型[18]。HL-CMS的花粉败育特征为圆败型,而BT- CMS和滇型为染败型。
1.3 光温敏雄性核不育系的培育和水稻两系杂种优势利用
1973年,湖北沙湖原种场农技员石明松先生发现水稻农垦58的光敏核不育(photoperiod-sensitive genic male-sterile, PGMS)株,并育成了首个光敏核不育系农垦58S。农垦58S在长日高温条件下表现为雄性不育,作为不育系用于杂交水稻制种;在短日低温条件下可育,用于不育系的繁种。PGMS的敏感阶段为幼穗发育时期,主要是从二次枝梗分化到花粉母细胞形成时期,在长日照条件下绒毡层提前降解,缺乏营养供给导致小孢子败育。温度和光照长度在农垦58S中具有补偿效应,高温可以降低临界日照长度,而低温要提高临界日照长度。利用农垦58S光敏不育的特性,突破三系配套恢保关系束缚,开创了“二系”杂交水稻育种的新阶段[19]。另外一类种质例如安农S-1、衡农S-1和5460S,称为水稻温敏核不育(temperature-sensitive genic male- sterile, TGMS),温度变化可以导致育性的转换,高温不育,低温可育,而光周期长短对育性转换没有影响。TGMS的诱导阶段在花粉母细胞形成和减数分裂时期,小孢子母细胞不能完成减数分裂,败育的花粉都呈现出皱缩的形态[20,21]。两系杂交稻由于冲破了恢保关系的束缚,亲本间的遗传差异变大,两系杂交稻平均产量比三系杂交稻具有较大的提高,代表性的品种如两优培九大面积种植能达到亩产630公斤,比三系对照汕优63增产约10%左右[22]。1.4 籼粳亚种间杂种优势的利用
籼稻和粳稻亚种之间具有更丰富的遗传多样性,杂交组合比籼籼组合具有更强的杂种优势。但是,籼粳杂交种F1不育(或部分可育)限制了籼粳杂种优势的利用。广亲和基因的发现为籼粳亚种间杂种优势利用奠定了理论基础[23]。利用部分粳稻血缘培育的杂交组合例如两优培九、协优9308等表现出很强的杂种优势[24,25]。直到21世纪以来,利用粳稻不育系与籼粳中间型广亲和恢复系配组配制出籼粳亚种间的杂交品种,如“甬优系列”和“春优系列”组合。这些杂交组合在生产上表现出更强的产量优势[26,27]。
1.5 理想株型育种
理想株型是由澳大利亚科学家CM. Donald博士提出,指农作物个体间竞争最小的株型,使每个植株最大限度地获取光照和营养,从而提高群体的收获指数。日本栽培学家松岛省三最早提出高产水稻应该具备“多穗、矮杆、短穗,顶部2、3叶片要短厚直立”的特性。Khush[28]提出少蘖、大穗、茎秆粗壮、株高100~110 cm、叶厚直立、根系发达、晚熟、收获指数高和生产潜力大等特征。我国育种家杨守仁先生等提出高产水稻指标:半矮杆、穗大且直立、分蘖中等,耐肥抗倒、生物量大、谷草比高[29]。袁隆平先生认为超高产杂交水稻在形态上主要特点是上部3片功能叶要长、直、窄、凹、厚,叶面积较大,并且可以两面受光而互不遮蔽,提出库大源足是高产的前提,新株型特征和杂种优势利用相结合是实现超高产水稻育种技术路线[30]。2010年,李家洋先生克隆出理想株型基因IPA1[31]。IPA1植株株型紧凑,茎秆挺直,虽然分蘖能力不强,但成穗率高、穗大、产量潜力大。IPA1的克隆促进了理想株型的育种,培育的理想株型新品种已经表现出巨大的增产潜力。1.6 第二次绿色革命理念及绿色超级稻品种选育
1999年12月14日,西北农林科技大学的李振声先生、华中农业大学的张启发先生和中国农业科学院作物科学研究所贾继增先生在杭州召开的“农作物资源核心种质构建、重要新基因发掘与有效利用”973项目年会中提出了第2次绿色革命的10字口号,“少投入,多产出,保护环境”,并提出为绿色革命准备基因资源。国家973项目“作物高效利用氮磷养分的分子机理”、“主要粮食作物重大病害控制的基础研究”和“害虫爆发成灾的遗传与行为机理”以及农业部948项目“参与全球水稻分子育种计划研究”等,推动了作物营养高效利用和对逆境抗性基因的发掘。2005年和2007年,张启发先生先后两次撰文[32, 33],提出培育绿色超级稻的构想,主要内容包括“少打农药,少施化肥,节水抗旱,优质高产”,将第二次绿色革命的基本理念贯穿始终。经过10年的努力,我国选育出一批绿色超级稻品种,全国累计推广约9000万亩(http://www.hb. xinhuanet.com/2017-10/26/c_1121861611.htm)[34]。
水稻遗传育种的历程就是一个育种理念变迁的过程。我国水稻遗传育种经历了3次大的飞跃(图1),每次飞跃都离不开重要基因资源的发掘和利用。矮秆基因导致“第一次绿色革命”,解决了水稻耐肥和抗倒伏的问题;核质互作不育系和光温敏核不育系的培育,促成了杂种优势的利用。第三次飞跃以理想株型塑造为主要技术路线,以绿色超级稻育种为目标,选育高产优质健康新品种/组合,实现第二次绿色革命。
图1
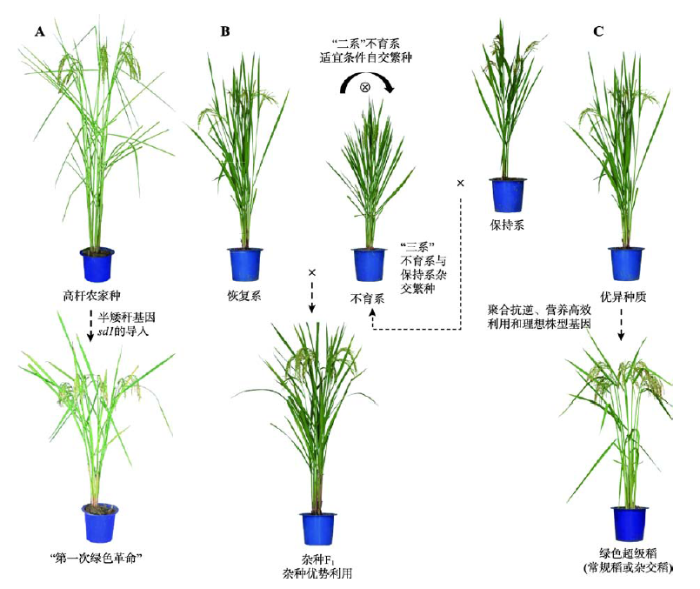 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1水稻遗传育种经历的3次飞跃
A:半矮杆基因sd1利用以及半矮秆水稻品种培育促成“第一次绿色革命”;B:不育系和恢复系配制杂种促进杂种优势的利用;C:抗逆、养分高效利用和理想株型等有利基因的发掘促进绿色超级稻的培育。
Fig. 1Three leaps in rice genetic breeding
2 水稻遗传育种技术的变迁
2.1 常规育种
常规育种包括选择育种、有性杂交育种、物理以及化学诱变育种、离体组织培养育种和细胞杂交育种。常规育种的过程主要是选择合适的亲本,得到分离的群体,利用该育种方法,根据表型从群体后代中选择达到所设定育种目标的个体。这种方法对高产育种效率比较高,但是对稻米品质和非生物逆境的改良效率较低。选择育种是从自然变异中选择优良变异,但是自然变异发生频率低,有价值变异少,育种效率低。诱变育种是通过物理化学处理,增加诱变频率,从大量突变中选择有利突变,由于突变往往是有害的,因此育种效率也低。利用水稻花药培养再生植株,单倍体自然加倍,基因组纯合快,能大量缩短育种历程,但是花药培养严重依赖于基因型,特别是籼稻的花药培养难度较大,因此花药培养育种也受到限制,只有少数单位开展。有性杂交育种是利用不同亲本材料杂交,再通过自交或测交,产生大量的具有丰富表型变异的后代群体,从中选择优良表型的单株。杂交育种充分发挥基因重组的作用,只要亲本间互补性强,杂交育种效率一般比较高,并且很可能育成全新的骨干品种。因此,杂交育种是最主流的水稻育种方法,得到广泛应用。2.2 分子标记辅助选择育种和基因组育种
近30年来水稻功能基因组学的成果为辅助选择育种提供了一系列的功能分子标记。SNP芯片是全基因组选择育种的有效工具,水稻60K SNP芯片的开发和应用为大规模的基因型鉴定提供了便捷的方法[35]。在这些标记辅助选择下,通过回交实现目标性状的定向改良。2017年,缩短作物生长周期的快速育种方法(Speed Breeding)诞生,该方法可以使春小麦(Triticum aestivum)和豌豆(Pisum sativum)等实现一年种植6代,油菜(Brassica napus)一年可以种植4代,加速了育种进程[36]。水稻是短日照植物,结合基因组选择育种和快速育种方法,可充分发挥定向改良的效率。定向改良必须知道哪些基因(等位基因)具有控制有利农艺性状和生物学性状的功能。在育种过程中,水稻材料大多在正常生产条件下种植,抗性性状如抗生物逆境和非生物逆境很难通过田间目测加以选择,而标记辅助选择在苗期就可以进行。因此,利用标记辅助育种和基因组育种定向改良抗逆等性状更有现实意义。随着功能基因的不断挖掘和基因调控网络的建立,全基因组范围的设计育种将有更广阔的天地。
2.3 转基因育种与基因编辑育种
转基因育种是指通过转基因的方法,导入外源的基因,达到性状改良的目标,从而培育出新品种[37]。传统育种只能依靠品种或者种之间的杂交实现重组,选育出具有优良性状的品种。而转基因育种可以实现跨物种的基因交流,对目标性状改良的针对性强,提高育种效率。苏云金芽胞杆菌的Bt毒蛋白基因是目前世界上公认的高效抗虫基因之一,通过转基因的方法将其导入水稻,可以有效提高水稻的抗虫特性[38]。bar基因能使植物特异性获得对除草剂草丁膦的抗性,转bar基因的抗除草剂水稻能获得很好的抗除草剂效果[39]。近10年来,基因编辑技术的突飞猛进,特别是CRISPR/Cas9技术的应用,基因敲除技术已经成为常规技术[40],基因敲入技术也产生了突破[41,42]。因此,定向敲除不良目标基因和定向整合优良目标基因,将大幅提高水稻定向遗传改良效率。并且,该系统获得的植株通过自交重组,容易得到不含转基因的基因编辑品种[43,44]。
以选育矮秆抗病品种为例,3种育种方法各有千秋(图2)。常规育种历时长,但可以培育出全新的矮秆抗病品种;辅助选择快速育种历时大大缩短,定向改良矮秆抗病性状,获得与亲本综合性状类似的矮秆抗病品种;而转基因育种可以在较短的时间(2~3世代)内获得与受体相似的矮秆抗病品种。
图2
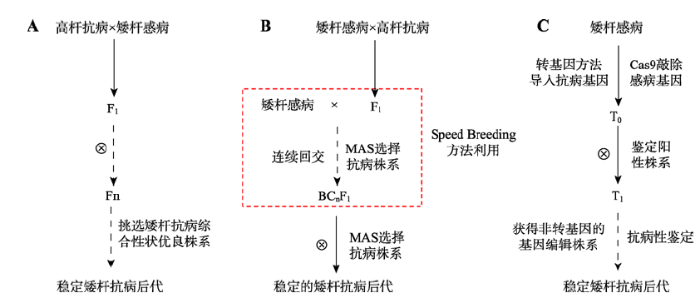 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图23种水稻育种方法流程(以选育矮秆抗病品种为例)
A:常规育种;B:标记辅助快速育种;C:转基因育种。
Fig. 2The flowchart of three types of rice breeding methods (Taking development of a dwarf and disease resistance cultivar as an example)
3 水稻遗传育种分子机制研究进展
水稻基因组测序的完成掀起了水稻基因功能研究的热潮[45,46]。基于自然变异的正向遗传学策略,大量具有重要应用价值的基因相继被克隆,一批功能基因的分子机制得到解析,为水稻的遗传改良提供了重要基因[47]。3.1 雄性不育分子机制解析
近年来,野败型细胞质雄性不育的分子机理得到解析。刘耀光教授课题组报道了一个野生稻线粒体中新近起源的基因WA352,与核编码的线粒体蛋白COX11互作,共同调控水稻CMS-WA[48]。WA352诱导雄性不育可以被两个育性恢复基因Rf3和Rf4恢复,Rf3暂时未被克隆,Rf4编码一个PPR蛋白,可以降低WA352的表达[49]。BT-CMS中orf79与atp6共转录形成B-atp6-orf79,控制包台型CMS。Rf1位点存在两个紧密连锁的恢复基因Rf1A和Rf1B,编码PPR家族蛋白,Rf1A和Rf1B分别参与了对双顺反子mRNA B-atp6-orf79的切割和降解[50]。与BT-CMS不同的是,HL-CMS不育系中orfH79有两种转录产物,一种为atp6-orfH79,另一种为orfH79,并且都能翻译成细胞毒素肽ORFH79[51]。OrFH79蛋白能与电子传递链复合体Ⅲ的亚基P61互作,导致花粉败育[52,53]。CMS中线粒体基因突变改变了线粒体的正常状态,导致不育;而育性恢复基因编码PPR家族蛋白,抑制或者参与对线粒体中CMS基因的mRNA加工,从而导致育性的恢复。我国科学家在光温敏雄性核不育的机理方面也进行了系统的研究。Fan等[54]发现PMS1编码一个长链非编码RNA(long noncoding RNA, lncRNA) PMS1T,在幼穗中表达量较高,是miR2118的作用靶标,切割产生21nt的phasiRNAs。在长日照条件下,农垦58S中miR2118切割位点附近的S2突变,改变了RNA的二级结构,增强了miR2118切割效率,产生更多的phasiRNAs,导致不育。Ding等[55]和Zhou等[56]发现PMS3编码一个lncRNA,分别命名为LDMAR和osa-smR5864w。LDMAR中一个G-C突变导致了RNA二级结构的改变,并且LDMAR的启动子区域甲基化程度升高,导致长日照条件下幼穗中表达量降低,产生农垦58S中的雄性不育[57]。
安农S-1是第一个培育成功的温敏不育系,受一个隐性核不育基因TMS5控制,TMS5编码RNase ZS1,tms5一个SNP的突变导致编码蛋白提前终止。与野生型相比,tms5中3个泛素60S核糖体蛋白UbL401、UbL402和UbL404受高温诱导表达[21],蛋白质活性实验证明,RNase ZS1能特异切割UbL40 mRNA。安农S-1中tms5不能对UbL40 mRNA进行切割降解,导致高温条件下UbL40 mRNA的过度积累,从而影响细胞内泛素平衡,引发花粉母细胞液泡化,最终导致高温条件下花粉败育[21]。
3.2 杂交种F1不育的分子机制
广亲和材料的发掘以及籼粳杂种不育基因的克隆和应用是有效解决籼粳杂种不育的根本出路[58]。Ikehashi和Araki提出广亲和基因的遗传模型,即广亲和的S5n等位基因型、籼稻的S5i等位基因型和粳稻的S5j等位基因型,广亲和的S5n无论是与籼稻还是粳稻杂交均可育。S5位点有3个紧密连锁的基因—ORF3、ORF4和ORF5,其中ORF5和ORF4分别扮演了“杀手(killer)”和“帮凶(partner)”的角色,引起内质网逆境,而ORF3起到了“保镖(protector)”的角色,对内质网逆境有清除作用。典型的籼稻基因型为ORF3+ORF4-ORF5+,典型的粳稻基因型为ORF3-ORF4+ORF5-,而广亲和品种含有ORF5n。ORF4和ORF5为孢子体型作用方式(ORF4和ORF5对所有的配子都能发挥作用),ORF3为配子体型作用方式(只对含有该基因的配子起作用)。籼粳杂种F1雌配子形成过程中,由于籼型配子ORF3+的存在,可以为其正常发育提供保护,粳型配子携带ORF3-不能有效防护ORF4+ORF5+的杀伤,从而导致败育[59,60]。Sa是一个控制杂种雄配子不育基因,由两个紧密连锁的基因SaM和SaF组成,分别编码泛素修饰E3连接酶和F-box蛋白。大多数的籼稻携带SaM+SaF+基因型,粳稻携带SaM-SaF-基因型,SaM-内含子中一个SNP的改变导致剪接模式的改变,从而造成SaM-蛋白翻译提前终止。籼梗杂种F1中存在SaM+的前提下,SaF+能与SaM-互作,导致SaM-基因型花粉败育,称为“两基因/三元件”互作模型。因为SaM+多出一个自我抑制结构域,阻止SaF和SaM+互作,SaF不能与SaM+互作[61]。
杂种雄性不育位点qHMS7也包括两个紧密连锁的基因ORF2和ORF3,分别编码核糖体失活蛋白和含线粒体信号肽的禾本科特异蛋白。滇粳优1号携带有功能的ORF2DORF3D,而南方野生稻(O. meridionalis)只携带一个无功能的ORF2N,完全缺失ORF3。ORF2D是一个毒性蛋白,以孢子体方式发挥作用;ORF3D是一个解毒蛋白,以配子体方式发挥作用。杂合(ORF2DORF3D/ORF2N-)条件下,ORF2D蛋白能杀死不携带ORF3D基因的花粉。因此,只有携带ORF2DORF3D基因型的花粉能遗传到后代,称为自私基因[62]。
日本研究者发现两个杂种不育位点S27和S28存在上位性互作,图位克隆发现S27和S28是重复基因,编码线粒体核糖体蛋白L27。在栽培稻台中65(Taichung 65)中,S27有功能(T+T+)而S28无功能(TSTS),基因型为T+T+|TSTS;在展颖野生稻(O. glumaepatula)中,S27无功能(GSGS)而S28有功能(G+G+),基因型为GSGS|G+G+;后代中基因型为GS|TS的花粉败育[63]。
3.3 穗发育的分子机制
穗长、一次枝梗数、二次枝梗数和着粒密度决定每穗颖花数。Gn1a编码细胞分裂素氧化酶基因OsCKX2,促进细胞分裂素的降解,突变后细胞分裂素得到积累,每穗颖花数增加,从而增产[64]。直立密穗基因DEP1编码G蛋白γ亚基[65]。DEP1蛋白通过调控OsCKX2的表达影响分生组织的活性和细胞的分裂增殖,该基因突变促进细胞分裂,枝梗数增加、每穗粒数增多[66,67]。FZP决定穗分枝向小穗形成的转化,同时还抑制腋分生组织的形成。FZP上游约5 kb处的18 bp串联重复序列抑制其表达,延长了穗分枝历时从而增加每穗颖花数,增产约15%[68]。An-1编码bHLH蛋白,调控细胞分裂,正调控芒长和粒长,负调控每穗颖花数[69]。An-2/ LABA1编码LOG-like蛋白6,催化CK合成的最后一步反应,通过促进细胞分裂增加芒长,同时降低每穗颖花数和分蘖数[70,71]。GAD1也是一个正调控芒长和粒长、负调控每穗颖花数的基因[72]。另外,也发现了正调控每穗颖花数基因,如富含亮氨酸重复受体样激酶基因LRK1,过表达该基因可以增加每穗颖花数[73,74]。Spr3最初由林鸿宣教授课题组定位在4.6 kb区间内,但是这个区间并没有编码基因,通过近等基因系比较发现,SG-64等位基因具有增加每穗颖花数的作用[75];而日本Ishii等[76]将该基因定位在9.3 kb区间,OsLG1位于定位区间下游,通过互补测验验证了9.3 kb的DNA片段具有上调OsLG1的作用,Spr3/OsLG1的克隆证实远端调控对基因功能起到重要的作用。LF1是一个功能获得性突变体,颖花两侧护颖发育成侧生小花,形成簇生小花,从而对每穗颖花数具有重要的调控作用[77]。3.4 粒形发育的分子机制
GS3是粒长的负调控因子,编码G蛋白γ亚基,并且与G蛋白的其他亚基以及OsMADS1共同作用,调控水稻粒型[78,79,80,81]。GW2负调控粒宽,编码一个功能未知的RING-type蛋白,具有E3泛素连接酶活性,可能参与泛素蛋白酶体对蛋白的降解途径[82]。GIF1编码细胞壁蔗糖酶,影响水稻灌浆,表达模式对其功能的发挥具有重要的影响,栽培稻中GIF1自身启动子驱动GIF1的过量表达能增加粒重[83]。BRs是重要的植物激素,其合成和信号转导途径对水稻粒形具有调控作用。GS5编码一个丝氨酸羧肽酶,GS5-1能增强BRs的信号,正调控种子大小[84,85]。GW5/qSW5 (grain width 5/seed width 5)是一个控制粒宽的主效基因,前期研究认为GW5编码一个核定位蛋白,并且1212 bp的缺失增加粒宽[86,87,88];随后,进一步研究发现1212 bp的缺失导致了其5 kb下游处的钙调素结合蛋白(GW5)表达量的降低,增加了粒宽。GW5定位在细胞膜上,直接与GSK2互作并抑制GSK2的激酶活性,激活BRs信号[89]。转录调控因子GRF家族的GL2/GS2/PT2编码OsGRF4,直接与BRs的负调控因子GSK2互作,抑制GSK2的转录激活活性,从而上调受BRs信号诱导的基因表达,激活BRs的响应[90,91,92,93]。qGL3.1/OsPPKL1编码PPKL家族的丝氨酸/苏氨酸磷酸酶OsPPKL1,也可能参与BRs的转导过程[94,95,96]。TGW6编码IAA(indole-3-acetic acid)葡萄糖水解酶,在胚乳发育过程中对保持生长素的稳态起着重要的作用,不仅直接控制胚乳大小,而且间接参与了同化物从源到库的运输及分配[97]。
粒宽QTL GW8编码SPL(squamosa promoter binding protein-like)家族蛋白OsSPL16,通过增加细胞数目增加粒宽和粒重[98]。粒长和粒宽QTL GL7/ GW7编码与拟南芥中LONGIFOLIA蛋白的同源蛋白。GL7位点上17.1 kb片段的串联重复,引起GL7的上调表达,从而增加水稻粒长[99]。GW8中SBP (squamosa promoter-binding protein)结构域直接结合在GW7的启动子,抑制GW7基因的表达,从而粒长变短[100]。粒长粒重基因GLW7编码SPL家族的转录因子OsSPL13,正调控颖壳的细胞大小,具有增加粒长和产量的作用[101]。
3.5 株型的分子机制
自20世纪60年代以来,“绿色革命基因”sd1 (semi-dwarf 1)在水稻矮化育种中得到了广泛的应用,但是直到2002年sd1才被克隆。SD1编码GA20氧化酶蛋白OsGA20ox2,催化GA53~GA44~GA19~ GA20的反应,活性的GA3处理sd1幼苗可以恢复到野生型表型[102,103]。通过对矮杆品种中sd1的比较测序,发现sd1的突变主要有7种等位基因型,分别为IR8的383 bp缺失等位基因型、93-11外显子提前终止等位基因型、矮脚南特中第一个外显子中2 bp缺失等位基因型,以及4种SD1氨基酸改变的等位基因型,它们的广泛利用推动了矮秆育种的进程[104]。Sd1不仅在“第一次绿色革命”中得到选择应用,而且在更早的粳稻驯化过程中就被选择;粳稻中两个功能性的FNPs(SD1-EQ)导致GA20ox2酶活性降低,内源GA含量降低,最终株高降低,而籼稻和野生稻携带了强功能型的SD1(SD1-GR),内源GA含量升高,株高增加[105]。最重要的理想株型基因IPA1(Ideal plant achitecture1)编码OsSPL14,是OsmiRNA156的直接靶标。IPA1的一个点突变阻断了OsmiRNA156介导的OsSPL14调控,使分蘖减少、穗粒数和千粒重增加,同时茎秆变得粗壮,抗倒伏能力增强,产量提高。在营养生长期,OsSPL14控制水稻分蘖;在生殖生长期,OsSPL14高表达促进了穗分支。IPA1可以直接结合在OsTB1的启动子上,负调控水稻分蘖发生,正调控DEP1调节水稻株高和穗长[31,106]。研究还发现,超级稻甬优12的IPA1等位基因上游具有3个3137 bp的串联重复,使启动子区域甲基化程度降低以及染色质结构松散,导致ipa1-2D的表达量上升,分蘖数变少,而穗变大,所以,适度的IPA1表达量能具有最高的产量潜力[107]。
分蘖角是株型的重要性状,普通野生稻具有匍匐生长的特性,分蘖角很大,而栽培稻表现为直立生长,分蘖角较小。PROG1(PROSTRATE GROWTH 1)是一个重要的控制匍匐生长习性的驯化基因,编码Cys2-His2锌指蛋白转录因子。在野生稻到栽培稻的驯化过程中,PROG1基因功能丧失,株型得到改良,每穗粒数增加,产量提高 [108,109]。TAC1是分蘖角主效QTL,编码一个禾本科特有的蛋白,其内含子中SNP的突变导致了tac1表达量的降低,产生了紧凑的株型[110]。
3.6 开花与水稻产量的联动机制
开花期不仅是水稻地域适应性的重要性状,在一定的范围内,开花期与产量正相关。Ghd7编码一个CCT结构蛋白,Ghd8/DTH8编码一个CCAAT盒结合蛋白亚基的OsHAP3,Ghd7.1/DTH7编码一个PRR蛋白,它们都对光照长度敏感,在长日照条件下,通过抑制Ehd1、RFT1和Hd3a延迟抽穗,同时增加株高和每穗颖花数[111,112,113]。抽穗期基因Ghd7、Ghd8、Ghd7.1和Hd1功能的减弱或缺失促成了水稻从低纬度地区向高纬度地区扩散[114]。在长日照条件下,Hd1和Ghd7互作形成Ghd7-Hd1的蛋白复合体,特异性地结合在Ehd1的顺式作用元件上,抑制Ehd1的表达延迟抽穗[115]。在长日照条件下,Hd1也可以直接与Ghd8互作,形成Ghd8-Hd1复合体,抑制Hd3a的转录,从而延迟抽穗。在短日照条件下,Ghd8的转录得不到积累,不能形成Ghd8-Hd1复合体,而Hd1可以激活Hd3a的表达促进抽穗[116]。3.7 水稻重要利用价值的抗逆新基因和营养高效吸收基因发掘
挖掘具有抗生物逆境和非生物逆境的基因并阐明作用机理,对培育高产稳产水稻品种具有重大意义。近20年来,一大批水稻抗病基因被克隆[117],而这些基因具有不同的作用方式,包括作用于病原识别、信号转导、下游防御相关蛋白以及不同信号之间的相互作用[118]。最近几年抗虫基因克隆也取得突破性进展,多个抗褐飞虱基因被克隆,如Bph3[119]、Bph14/Qbp1[120,121]、BPH18[122]和Bph9[123]等,为抗褐飞虱水稻品种的培育提供了优异的基因资源。水稻优良品种应具备抗非生物逆境如高温和冷害等特性。水稻耐寒基因COLD1编码G蛋白信号的调控因子,其与RGA1互作,激活Ca2+通道感受低温,并加速G蛋白GTPase活性。超表达COLD1jap能增强水稻耐寒性[124]。耐冷QTLCTB4a编码富含亮氨酸受体蛋白激酶;上调CTB4a会增强ATP合酶活性,提高ATP含量,在低温条件下提高结实率和产量[125]。耐高温QTL TT1编码26S蛋白酶体α2亚基,参与泛素化蛋白降解途径,非洲栽培稻中的等位基因具有耐高温;高温诱导TT1表达,可以降解有毒蛋白以及维持高温应答过程,从而保护细胞免受损伤[126]。
养份高效利用是绿色水稻品种的重要特性。DEP1/qNGR9编码G蛋白γ亚基,除了控制直立穗的性状外,还影响水稻氮利用效率,DEP1能与Gα (RGA1)和Gβ (RGB1)亚基互作,共同调控氮信号[127]。最近有研究报道GRF4可以结合在控制氮代谢、光合作用、蔗糖代谢、蔗糖转运和细胞分裂等基因的启动子上,上调基因的表达,促进氮素同化、固碳作用以及水稻的生长;而SLR1 (slender rice 1)编码DELLA蛋白,活性GAs可以促进SLR1的降解,SLR1可以与GRF4拮抗,并对GRF4的功能起抑制作用,SLR1积累不仅造成矮化,还降低了氮肥使用效率[6]。因此,可以通过增加GRF4的表达,提高含有半矮杆基因sd1品种的氮利用效率,从而进一步提高产量[6]。PSTOL1/Pup1编码Pup1特异性蛋白激酶,超表达PSTOL1能在磷缺乏条件下显著增加产量。PSTOL1能增强早期根的发育,使植株获得更多的磷和其他营养元素[128]。
3.8 中国在水稻基础研究中的地位和贡献
上述大部分进展是中国科学家完成的。在NCBI中以rice为关键词,提取2003年~2018年7月间发表的所有与水稻相关的论文,选取Nature Index所包含杂志中植物学相关的杂志,共筛选出1286篇论文,其中通讯作者单位为中国的有307篇(图3)。世界每年有60~100篇高水平水稻论文发表,中国发表论文数量在其中所占比例越来越高,从2003年占比12%到2018年7月占比43%。这表明,我国在水稻领域的基础研究突飞猛进,已经处于国际领先水平,中国的重要论文贡献份额近5年稳定在30%左右。图3
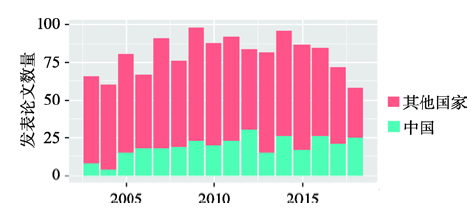 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图32003~2018年发表的与水稻相关的高水平论文
Fig. 3National distribution of high quality papers in rice from 2003 to 2018
4 水稻育种的发展方向
4.1 杂交育种与标记辅助选择技术紧密结合
常规育种、标记辅助选择育种和转基因育种在品种选育中都表现出各自的优势。但是,当前标记辅助选择大多是独立于杂交育种过程,只是对杂交育种等方法选育出的品种的个别缺陷性状进行改良,改良后新品种的基因组结构与原品种变化很小。转基因育种与标记辅助选择育种一样,也只是对现有品种的个别性状加以改良。从育种的效率和效果看,毋庸置疑,水稻杂交育种方法依旧是主流方法。但是,杂交育种一定要在选育过程中与现代育种技术相结合,避免过去仅限于大田的纯表型选择,在杂交育种完成后再开展不良性状的改良。因此,在开展杂交育种之前,一定要先明确育种目标,首先要筛选符合育种目标的材料作为亲本,再明确亲本中调控目标性状的主要基因是否有功能或者功能强弱。这样,在杂交育种的低世代材料中根据农艺性状选择一些优良单株,再对这些单株进行标记辅助选择,从中选择携带无法目测的优良目标基因的单株,进入下一个世代,在杂交育种过程中完成标记辅助选择,使新品种具有育种目标性状,没有明显缺陷,省去辅助选择对某个性状修修补补的必要。4.2 满足消费者需要,培育口感好的品种
随着生活水平的提高,人们对稻米品质有了更高的追求。除了外观品质和营养品质外,口感应该成为品质的重要指标。口感与水稻中直链和支链淀粉的组成比例和蛋白含量密切相关,但是影响口感的遗传基础还不清晰,需要加强遗传学研究。4.3 满足轻简栽培需要,培育适合新的耕作制度的品种
直播和机械化等轻简栽培技术大大降低了劳动力成本,已成为水稻生产的主流技术。然而,轻简栽培出现新的生产问题:直播稻易倒伏,增加机械化收获难度。因此,培育茎秆粗壮、增强抗倒伏特性的品种有利于轻简栽培技术技术推广。另外,当前耕作制度多样化,从两季稻变一季稻,或者一种两收的再生稻。特别是张启发先生提出的长江中下游稻区“双水双绿”稻虾共生的种植/养殖模式,具有较高的经济效益。为满足稻虾连作的需求,应该培育生育期适宜、茎秆粗壮、抗倒伏和抗病虫害的水稻品种。4.4 满足生态环保需要,培育具有特色健康品质的绿色新品种
培育抗病虫害的绿色品种是生产健康稻米的基本条件。水稻中一系列的抗稻瘟病和白叶枯病基因,以及抗飞虱基因得到定位或克隆[129,130]。但是还没有找到有效抗稻曲病和抗螟虫的基因,今后需要在这些方面力争突破。土壤重金属污染特别是镉污染尤其严重,对食品安全和人类健康有重要的影响[131]。2010年,Ueno等[132]克隆出一个控制水稻镉积累的基因OsHMA3,超表达低镉积累的等位基因可以选择性的降低种子中镉的积累,而对其他的微量元素没有影响。选育种子中镉或其他重金属含量低的品种,解决“重金属米”的安全隐患。另外,应该重视培育富含微量元素如富硒水稻品种,重视培育满足特殊人群需要的水稻品种,如适合糖尿病患者食用的抗性淀粉高的稻米等。总之,水稻育种要紧跟耕作制度变化的步伐,培育适合新耕作方法的新品种。一方面要利用基因组育种技术和基因编辑技术,加快水稻功能基因组研究成果向育种应用的转化,另一方面要重视发掘新的重要基因,为设计育种提供元件!培育高产优质的绿色超级稻品种是人心所向,大势所趋。
(责任编委: 储成才)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URL [本文引用: 1]

引言一、考古发掘所见的水稻资源二、历代水稻品种资源的记述和存在的问题1.战国至南北朝的水稻品种2.唐宋时期水稻品种记载存在的问题3.宋元明清地方志中记载的水稻品种三、水稻品种的继承发展与变异1.品种的继承性2.品种的变异性四、文献记述的水稻地方品种特色1.生育期方面 5.肥料反应方面2.株型方面 6.抗虫害兽害方面3.穗型方面 7.抗逆性方面
[本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]

矮秆水稻品种的选育和推广是20世纪水稻育种工作最主要的成就之一.本文综述了水稻矮秆性状的遗传、水稻植株矮化机理以及矮源的育种利用等方面的研究进展,同时分析了水稻矮源利用和矮化育种中存在的问题,展望了其发展前景.
URL [本文引用: 1]

矮秆水稻品种的选育和推广是20世纪水稻育种工作最主要的成就之一.本文综述了水稻矮秆性状的遗传、水稻植株矮化机理以及矮源的育种利用等方面的研究进展,同时分析了水稻矮源利用和矮化育种中存在的问题,展望了其发展前景.
[本文引用: 1]
URLPMID:30111841 [本文引用: 3]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
URL [本文引用: 1]

(一) 农业生产和作物育种的历史表明,凡在育种上有所突破,就会给农业生产带来一次飞跃。如杂交玉米和高粱,矮秆水稻和小麦等的育成和应用,都大幅度地提高了这些作物的产量。 杂交水稻培育成功,属于水稻育种上的一项突破。生产实践证明,在相同条件下,杂交水稻一般比普通良种增
[本文引用: 1]
URL [本文引用: 2]

回顾了籼型“三系”不育系选育的历程。阐述了野败型,矮败型,D型,印尼水田谷型等几个主要类型的籼稻不育的选育与特征特性及其应用状况。对细胞质和细胞核单一,很难提高杂交稻的抗性和品质等问题,提出了今后应培育多种类型的细胞质和细胞核的雄性不育系,并介绍了其方法。
URL [本文引用: 1]

利用粳籼杂交后代出现的不育株作细胞质雄性不育供体,先后育成了K青A、K19A和K17A等籼型不育系。育成的K型杂交水稻新组合已有5个通过四川省品种审定。初步研究发现,K型胞质对提高柱头外露率和开花累积率的作用明显优于WA型胞质;在可恢性及主要数量性状上的胞质效应与WA型和D型胞质差异较小。花药发育的细胞学显示K型不育系花粉败育发生在单核后期,少量花粉发生在二核期,败育花粉以典败为主。K17A和K青A花药的酯酶和过氧化物酶同工酶谱与汕A存在差异。随机扩增多态性NDA(RAPD)分析表明K型胞质与WA型和D型胞质的线粒体基因组存在组织结构差异。
URL [本文引用: 1]

利用粳籼杂交后代出现的不育株作细胞质雄性不育供体,先后育成了K青A、K19A和K17A等籼型不育系。育成的K型杂交水稻新组合已有5个通过四川省品种审定。初步研究发现,K型胞质对提高柱头外露率和开花累积率的作用明显优于WA型胞质;在可恢性及主要数量性状上的胞质效应与WA型和D型胞质差异较小。花药发育的细胞学显示K型不育系花粉败育发生在单核后期,少量花粉发生在二核期,败育花粉以典败为主。K17A和K青A花药的酯酶和过氧化物酶同工酶谱与汕A存在差异。随机扩增多态性NDA(RAPD)分析表明K型胞质与WA型和D型胞质的线粒体基因组存在组织结构差异。
URLMagsci [本文引用: 1]

回顾了“籼粳架桥”育成粳型恢复系C57以来我国北方杂交粳稻的发展历程。通过总结经验,进一步认识到“籼粳架桥”对亚种间杂种优势利用的指导意义,以及广亲和基因单一位点的局限性难以解决籼粳杂种复杂的生理障碍问题。提出了籼粳亚种有利基因集团的构建与配组,偏高秆偏长穗抗倒株型改良以及高产性与抗逆性、适应性相结合的超高产育种思路等问题。文章还叙述了通过“籼粳架桥”育成高配合力的粳型恢复系C418及其配制的系列新组合的推广。北方杂交粳稻进入再发展阶段。
URLMagsci [本文引用: 1]

回顾了“籼粳架桥”育成粳型恢复系C57以来我国北方杂交粳稻的发展历程。通过总结经验,进一步认识到“籼粳架桥”对亚种间杂种优势利用的指导意义,以及广亲和基因单一位点的局限性难以解决籼粳杂种复杂的生理障碍问题。提出了籼粳亚种有利基因集团的构建与配组,偏高秆偏长穗抗倒株型改良以及高产性与抗逆性、适应性相结合的超高产育种思路等问题。文章还叙述了通过“籼粳架桥”育成高配合力的粳型恢复系C418及其配制的系列新组合的推广。北方杂交粳稻进入再发展阶段。
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
URL [本文引用: 1]

To meet the growing food demands in China, a program of super rice breeding was set up by the Chinese Ministry of Agriculture in 1996. The goals of this program is to achieve rice yield targets to 10.5, 12.0, 13.5, and 15.0 t/ha in single season rice growing area through four research phases. A proposed model of combining morphological improvement with the utilization of inter-subspecific(indica/japonica) heterosis, by Prof. Yuan Long Ping in 1997, played a leading contribution for the program of super hybrid rice breeding. Until now, all yield targets of four phases have been achieved. The pioneer super hybrid rice variety, Liangyoupeijiu, met the Phase I yield target in 2000. The Phase II super hybrids were successfully developed in 2004. Y Liangyou No.1(YLY1), the typical Phase II super hybrid rice variety, has a largest annual planting area among all hybrid rice varieties in China since 2010 and has been extended to more than 4 million hectare so far. Y Liangyou No.2(YLY2) met the standard of phase III super rice with a yield record of 926.6 kg/mu(13.9 t/ha) in 2011. The breeding target of Phase IV super hybrid was achieved in 2014. The new variety, Y Liangyou 900(YLY900), created a new yield record of 15.4 t/ha, which doubled the average rice yield in China. Up to now, 125 super rice varieties have been certificated by the Ministry of Agriculture with a total planting area of approximate 80 million hectare in China. However, how to further break the yield ceiling and realize the yield potential of elite varieties in diverse range of target environments remains to be the major challenges for super hybrid rice breeding. Herein, we proposed that utilization of ideotype and inter-subspecific heterosis, assisted with molecular design breeding, is still being the vital way to increase yield ceiling.
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLMagsci [本文引用: 1]

<div><span >以籼粳杂交稻有代表性的品种甬优</span><span >11</span><span >、甬优</span><span >12</span><span >、甬优</span><span >13</span><span >、甬优</span><span >15</span><span >、甬优</span><span >17</span><span >为试材</span><span >, </span><span >通过栽培措施的调控</span><span >, </span><span >形成超高产</span><span >(</span><span >产量</span><span >≥12.0 t hm<sup>-2</sup>)</span><span >和高产</span><span >(10.5 t hm<sup>-2 </sup>≤</span><span >产量<</span><span >12.0 t hm<sup>-2</sup>)</span><span >群体。比较研究其特征表明</span><span >, </span><span >与高产群体比,超高产群体的总颖花量极显著提高</span><span >, </span><span >而结实率和千粒重略降低;平均每穗粒数极显著提高</span><span >, </span><span >并且一、二次枝粳数以及粒数显著或极显著提高;拔节期的干物重略低</span><span >, </span><span >但抽穗期和成熟期的干物重均显著或极显著提高;其有效分蘖临界叶龄期至拔节期的生长平稳</span><span >, </span><span >无效分蘖发生少</span><span >, </span><span >高峰苗低</span><span >, </span><span >拔节期以后茎蘖数下降平缓</span><span >, </span><span >成穗率、叶面积指数、光合势、干物质积累量等均较高;基部第</span><span >1</span><span >、第</span><span >2</span><span >、第</span><span >3</span><span >节间的茎秆粗度和茎壁厚度及基部第</span><span >1</span><span >、第</span><span >2</span><span >、第</span><span >3</span><span >节间的弯曲力矩、抗折力均显著或极显著提高</span><span >, </span><span >且倒伏指数减小。</span></div>
URLMagsci [本文引用: 1]

<div><span >以籼粳杂交稻有代表性的品种甬优</span><span >11</span><span >、甬优</span><span >12</span><span >、甬优</span><span >13</span><span >、甬优</span><span >15</span><span >、甬优</span><span >17</span><span >为试材</span><span >, </span><span >通过栽培措施的调控</span><span >, </span><span >形成超高产</span><span >(</span><span >产量</span><span >≥12.0 t hm<sup>-2</sup>)</span><span >和高产</span><span >(10.5 t hm<sup>-2 </sup>≤</span><span >产量<</span><span >12.0 t hm<sup>-2</sup>)</span><span >群体。比较研究其特征表明</span><span >, </span><span >与高产群体比,超高产群体的总颖花量极显著提高</span><span >, </span><span >而结实率和千粒重略降低;平均每穗粒数极显著提高</span><span >, </span><span >并且一、二次枝粳数以及粒数显著或极显著提高;拔节期的干物重略低</span><span >, </span><span >但抽穗期和成熟期的干物重均显著或极显著提高;其有效分蘖临界叶龄期至拔节期的生长平稳</span><span >, </span><span >无效分蘖发生少</span><span >, </span><span >高峰苗低</span><span >, </span><span >拔节期以后茎蘖数下降平缓</span><span >, </span><span >成穗率、叶面积指数、光合势、干物质积累量等均较高;基部第</span><span >1</span><span >、第</span><span >2</span><span >、第</span><span >3</span><span >节间的茎秆粗度和茎壁厚度及基部第</span><span >1</span><span >、第</span><span >2</span><span >、第</span><span >3</span><span >节间的弯曲力矩、抗折力均显著或极显著提高</span><span >, </span><span >且倒伏指数减小。</span></div>
[本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]

Chinese rice scientists increased rice varietal yields during the 1950s and 1960s by improving the plant type and since the 1970s by exploiting the phenomenon of heterosis in developing F_1 hybrid cultivars. Both approaches seem to have reached a plateau, with yields of 8~9 t/ha. If still higher yields are to be achieved, total biomass yield has to be increased while maintaining a reasonable gram straw ratio. Research efforts should aim at:(1) increasing leaf area; (2) increasing photosynthetic efficiency per unit leaf area and (3) improving fertilizer responsiveness and lodging resistance. This would require combining ideal plant morphology with favorable vigor, Indica—japonica hybridization should meet this objectives. High stomata frequency of indicas could be combined with japonica traits of compact plant type, higher specificleaf weight, higher chlorophyll content per unit leaf area, and higher nitrogen and RUBPC content. All these characteristics are advantageous to close planting and to increasing photosynthetic efficiency of leaves and total biomass yield. Indtca japonica crossing would also result in ideal plant morphology and in increased growth vigor.
URL [本文引用: 1]

杂交水稻超高产育种袁隆平(国家杂交水稻工程技术研究中心410125)1超高产水稻的概念什么叫水稻超高产育种,迄今并没有一个统一的标准和严格的定义,因此各家各派提出的产量指标并不相同。1980年日本制定的水稻超高产育种计划,要求在15a内育成比原有品种...
URLPMID:20495565 [本文引用: 2]

Increasing crop yield is a major challenge for modern agriculture. The development of new plant types, which is known as ideal plant architecture (IPA), has been proposed as a means to enhance rice yield potential over that of existing high-yield varieties. Here, we report the cloning and characterization of a semidominant quantitative trait locus, IPA1 (Ideal Plant Architecture 1), which profoundly changes rice plant architecture and substantially enhances rice grain yield. The IPA1 quantitative trait locus encodes OsSPL14 (SOUAMOSA PROMOTER BINDING PROTEIN-LIKE 14) and is regulated by microRNA (miRNA) OsmiR156 in vivo. We demonstrate that a point mutation in OsSPL14 perturbs OsmiR156-directed regulation of OsSPL14, generating an 'ideal' rice plant with a reduced tiller number, increased lodging resistance and enhanced grain yield. Our study suggests that OsSPL14 may help improve rice grain yield by facilitating the breeding of new elite rice varieties.
URL [本文引用: 1]

在过去的半个世纪中,水稻产量的提高对我国在人口快速增长的形势下成功地解决了人民的吃粮问题做出了巨大的贡献.预计到2030年,我国人口将达到16亿.有分析认为,在未来的30年中,我国的粮食总产必须较现在增产60%,才可满足届时的需求.
URL [本文引用: 1]

在过去的半个世纪中,水稻产量的提高对我国在人口快速增长的形势下成功地解决了人民的吃粮问题做出了巨大的贡献.预计到2030年,我国人口将达到16亿.有分析认为,在未来的30年中,我国的粮食总产必须较现在增产60%,才可满足届时的需求.
URL [本文引用: 1]

From a global viewpoint, a number of challenges need to be met for sustainable rice production: (i) increasingly severe occurrence of insects and diseases and indiscriminate pesticide applications; (ii) high pressure for yield increase and overuse of fertilizers; (iii) water shortage and increasingly frequent occurrence of drought; and (iv) extensive cultivation in marginal lands. A combination of approaches based on the recent advances in genomic research has been formulated to address these challenges, with the long-term goal to develop rice cultivars referred to as Green Super Rice. On the premise of continued yield increase and quality improvement, Green Super Rice should possess resistances to multiple insects and diseases, high nutrient efficiency, and drought resistance, promising to greatly reduce the consumption of pesticides, chemical fertilizers, and water. Large efforts have been focused on identifying germplasms and discovering genes for resistance to diseases and insects, N- and P-use efficiency, drought resistance, grain quality, and yield. The approaches adopted include screening of germplasm collections and mutant libraries, gene discovery and identification, microarray analysis of differentially regulated genes under stressed conditions, and functional test of candidate genes by transgenic analysis. Genes for almost all of the traits have now been isolated in a global perspective and are gradually incorporated into genetic backgrounds of elite cultivars by molecular marker-assisted selection or transformation. It is anticipated that such strategies and efforts would eventually lead to the development of Green Super Rice.
URL [本文引用: 1]

From a global viewpoint, a number of challenges need to be met for sustainable rice production: (i) increasingly severe occurrence of insects and diseases and indiscriminate pesticide applications; (ii) high pressure for yield increase and overuse of fertilizers; (iii) water shortage and increasingly frequent occurrence of drought; and (iv) extensive cultivation in marginal lands. A... [Show full abstract]
URLPMID:24034357 [本文引用: 1]

The advances in genotyping technology provide an opportunity to use genomic tools in crop breeding. As compared to field selections performed in conventional breeding programmes, genomics-based genotype screen can potentially reduce number of breeding cycles and more precisely integrate target genes for particular traits into an ideal genetic background. We developed a whole-genome single nucleotide polymorphism (SNP) array, RICE6K, based on Infinium technology, using representative SNPs selected from more than four million SNPs identified from resequencing data of more than 500 rice landraces. RICE6K contains 5102 SNP and insertion deletion (InDel) markers, about 4500 of which were of high quality in the tested rice lines producing highly repeatable results. Forty-five functional markers that are located inside 28 characterized genes of important traits can be detected using RICE6K. The SNP markers are evenly distributed on the 12 chromosomes of rice with the average density of 12 SNPs per 1 Mb and can provide information for polymorphisms between indica and japonica subspecies as well as varieties within indica and japonica groups. Application tests of RICE6K showed that the array is suitable for rice germplasm fingerprinting, genotyping bulked segregating pools, seed authenticity check and genetic background selection. These results suggest that RICE6K provides an efficient and reliable genotyping tool for rice genomic breeding.
URLPMID:29292376 [本文引用: 1]

The growing human population and a changing environment have raised significant concern for global food security, with the current improvement rate of several important crops inadequate to meet future demand [1]. This slow improvement rate is attributed partly to the long generation times of crop plants. Here we present a method called 'speed breeding', which greatly shortens generation time and accelerates breeding and research programs. Speed breeding can be used to achieve up to 6 generations per year for spring wheat (Triticum aestivum), durum wheat (T. durum), barley (Hordeum vulgare), chickpea (Cicer arietinum), and pea (Pisum sativum) and 4 generations for canola (Brassica napus), instead of 2-3 under normal glasshouse conditions. We demonstrate that speed breeding in fully-enclosed controlled-environment growth chambers can accelerate plant development for research purposes, including phenotyping of adult plant traits, mutant studies, and transformation. The use of supplemental lighting in a glasshouse environment allows rapid generation cycling through single seed descent and potential for adaptation to larger-scale crop improvement programs. Cost-saving through LED supplemental lighting is also outlined. We envisage great potential for integrating speed breeding with other modern crop breeding technologies, including high-throughput genotyping, genome editing, and genomic selection, accelerating the rate of crop improvement.
Magsci [本文引用: 1]

<p>水稻作为世界上超过一半人口的主食, 是最重要的粮食作物之一. 在过去的半个多世纪里, 水稻育种取得了巨大的成功. 水稻的单位产量实现了翻番, 部分地方甚至提高至3倍, 这为保障世界粮食安全作出了巨大的贡献. 但近十几年来水稻的产量停滞不前, 这一方面是由于在育种技术上没有新的突破以及遗传多样性在栽培品种中的逐步变窄, 另一方面也是因为频繁发生的病虫害以及旱灾等自然灾害使得水稻生产损失惨重. 然而, 世界人口的持续增长以及社会经济的快速发展导致对粮食的需求不断增加. 针对这些问题, 我国****提出了培育绿色超级稻的设想, 围绕水稻抗病虫、抗旱、营养高效利用、优质、高产等五大重要性状, 对水稻品种进行全面改良以实现农业的可持续发展. 而转基因技术作为一种新兴的育种手段, 在实现绿色超级稻目标上将发挥重要作用. 水稻的转基因研究始于20世纪80年代末期, 迄今已有大量的转基因水稻研究被报道, 其中大部分的研究内容与绿色超级稻所要实现的目标一致. 本文首先回顾水稻遗传转化技术的发展, 再以抗病虫、抗旱、营养高效利用、优质、高产、外加抗除草剂等几大主要性状为主线对转基因水稻取得的主要研究成果进行逐一阐述, 最后对转基因水稻的发展进行展望.</p>
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:24157548 [本文引用: 1]

Abstract Targeted nucleases are powerful tools for mediating genome alteration with high precision. The RNA-guided Cas9 nuclease from the microbial clustered regularly interspaced short palindromic repeats (CRISPR) adaptive immune system can be used to facilitate efficient genome engineering in eukaryotic cells by simply specifying a 20-nt targeting sequence within its guide RNA. Here we describe a set of tools for Cas9-mediated genome editing via nonhomologous end joining (NHEJ) or homology-directed repair (HDR) in mammalian cells, as well as generation of modified cell lines for downstream functional studies. To minimize off-target cleavage, we further describe a double-nicking strategy using the Cas9 nickase mutant with paired guide RNAs. This protocol provides experimentally derived guidelines for the selection of target sites, evaluation of cleavage efficiency and analysis of off-target activity. Beginning with target design, gene modifications can be achieved within as little as 1-2 weeks, and modified clonal cell lines can be derived within 2-3 weeks.
URLPMID:26028531 [本文引用: 1]

Current hESC knockin strategies typically include a drug-resistance cassette in the donor template for enrichment of correctly targeted clones. Huangfu and colleagues demonstrate a strategy for generating knockin reporter hESC lines without drug selection for both active and silent genes using the CRISPR/Cas9 system. This development may further facilitate the use of hESCs for disease modeling and cell-replacement therapy.
URLPMID:27618611 [本文引用: 1]

Abstract Sequence-specific nucleases have been exploited to create targeted gene knockouts in various plants(1), but replacing a fragment and even obtaining gene insertions at specific loci in plant genomes remain a serious challenge. Here, we report efficient intron-mediated site-specific gene replacement and insertion approaches that generate mutations using the non-homologous end joining (NHEJ) pathway using the clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9) system. Using a pair of single guide RNAs (sgRNAs) targeting adjacent introns and a donor DNA template including the same pair of sgRNA sites, we achieved gene replacements in the rice endogenous gene 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) at a frequency of 2.0%. We also obtained targeted gene insertions at a frequency of 2.2% using a sgRNA targeting one intron and a donor DNA template including the same sgRNA site. Rice plants harbouring the OsEPSPS gene with the intended substitutions were glyphosate-resistant. Furthermore, the site-specific gene replacements and insertions were faithfully transmitted to the next generation. These newly developed approaches can be generally used to replace targeted gene fragments and to insert exogenous DNA sequences into specific genomic sites in rice and other plants.
URLPMID:24906146 [本文引用: 1]

Derived from a microbial defense system, Cas9 can be guided to specific locations within complex genomes by a short RNA. The development, applications, and future directions of the CRISPR-Cas9 system for genome engineering are discussed here.
URL [本文引用: 1]
URLPMID:12447439 [本文引用: 1]

Rice is the principal food for over half of the population of the world. With its genome size of 430 megabase pairs (Mb), the cultivated rice species Oryza sativa is a model plant for genome research
URLPMID:12447438 [本文引用: 1]

Abstract The rice species Oryza sativa is considered to be a model plant because of its small genome size, extensive genetic map, relative ease of transformation and synteny with other cereal crops. Here we report the essentially complete sequence of chromosome 1, the longest chromosome in the rice genome. We summarize characteristics of the chromosome structure and the biological insight gained from the sequence. The analysis of 43.3 megabases (Mb) of non-overlapping sequence reveals 6,756 protein coding genes, of which 3,161 show homology to proteins of Arabidopsis thaliana, another model plant. About 30% (2,073) of the genes have been functionally categorized. Rice chromosome 1 is (G + C)-rich, especially in its coding regions, and is characterized by several gene families that are dispersed or arranged in tandem repeats. Comparison with a draft sequence indicates the importance of a high-quality finished sequence.
URLPMID:29409893 [本文引用: 1]

Abstract Rice (Oryza sativa L.) is a major staple food crop for more than 3.5 billion people worldwide. Understanding the regulatory mechanisms of its complex agronomic traits is critical to global food security. Rice is also a model plant for genomics research of monocotyledons. Thanks to the rapid development of functional genomic technologies, over 2000 genes controlling important agronomic traits have been cloned, and their molecular biological mechanisms have been partially characterized as well. Here, we briefly review the advances in rice functional genomics research during the past ten years, including a summary of the functional genomics platforms, genes and molecular networks that regulate important agronomic traits, and particularly the newly developed tools for gene identification. The achievement in functional genomics research will greatly facilitate the development of green super rice. Finally, future challenges and prospects of rice functional genomics research are proposed.
URLPMID:23502780 [本文引用: 1]

Plant cytoplasmic male sterility (CMS) results from incompatibilities between the organellar and nuclear genomes and prevents self pollination, enabling hybrid crop breeding to increase yields1–6. The Wild Abortive CMS (CMS-WA) has been exploited in the majority of ‘three-line’ hybrid rice production since the 1 1970s, but the molecular basis of this trait remains unknown. Here we report that a new mitochondrial gene, WA352, which originated recently in wild rice, confers CMS-WA because the protein it encodes interacts with the nuclear-encoded mitochondrial protein COX1111. In CMS-WA lines, WA352 accumulates preferentially in the anther tapetum, thereby inhibiting COX11 11 11 function in peroxide metabolism and triggering premature tapetal programmed cell death and consequent pollen abortion. WA352-induced sterility can be suppressed by two restorer-of-fertility (Rf) genes, suggesting the existence of different mechanisms to counteract deleterious cytoplasmic factors. Thus, CMS-related cytoplasmic-nuclear incompatibility is driven by a detrimental interaction between a newly evolved mitochondrial gene and a conserved, essential nuclear gene.
[本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URLPMID:22984070 [本文引用: 1]

Abstract Hybrid sterility is a major form of postzygotic reproductive isolation that restricts gene flow between populations. Cultivated rice (Oryza sativa L.) consists of two subspecies, indica and japonica; inter-subspecific hybrids are usually sterile. We show that a killer-protector system at the S5 locus encoded by three tightly linked genes [Open Reading Frame 3 (ORF3) to ORF5] regulates fertility in indica-japonica hybrids. During female sporogenesis, the action of ORF5+ (killer) and ORF4+ (partner) causes endoplasmic reticulum (ER) stress. ORF3+ (protector) prevents ER stress and produces normal gametes, but ORF3- cannot prevent ER stress, resulting in premature programmed cell death and leads to embryo-sac abortion. Preferential transmission of ORF3+ gametes results in segregation distortion in the progeny. These results add to our understanding of differences between indica and japonica rice and may aid in rice genetic improvement.
[本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]

Selfish genetic elements are pervasive in eukaryote genomes, but their role remains controversial. We show that qHMS7 , a major quantitative genetic locus for hybrid male sterility between wild rice ( Oryza meridionalis ) and Asian cultivated rice ( O. sativa ), contains two tightly linked genes [ Open Reading Frame 2 ( ORF2 ) and ORF3 ]. ORF2 encodes a toxic genetic element that aborts pollen in a sporophytic manner, whereas ORF3 encodes an antidote that protects pollen in a gametophytic manner. Pollens lacking ORF3 are selectively eliminated, leading to segregation distortion in the progeny. Analysis of the genetic sequence suggests that ORF3 arose first, followed by gradual functionalization of ORF2 . Furthermore, this toxin-antidote system may have promoted the differentiation and/or maintained the genome stability of wild and cultivated rice.
URLPMID:20080642 [本文引用: 1]

Abstract Hybrid incompatibility in F(1) hybrids or later generations is often observed as sterility or inviability. This incompatibility acts as postzygotic reproductive isolation, which results in the irreversible divergence of species. Here, we show that the reciprocal loss of duplicated genes encoding mitochondrial ribosomal protein L27 causes hybrid pollen sterility in F(1) hybrids of the cultivated rice Oryza sativa and its wild relative O. glumaepatula. Functional analysis revealed that this gene is essential for the later stage of pollen development, and distribution analysis suggests that the gene duplication occurred before the divergence of the AA genome species. On the basis of these results, we discuss the possible contribution of the "founder effect" in establishing this reproductive barrier.
URLPMID:15976269 [本文引用: 1]

Abstract Most agriculturally important traits are regulated by genes known as quantitative trait loci (QTLs) derived from natural allelic variations. We here show that a QTL that increases grain productivity in rice, Gn1a, is a gene for cytokinin oxidase/dehydrogenase (OsCKX2), an enzyme that degrades the phytohormone cytokinin. Reduced expression of OsCKX2 causes cytokinin accumulation in inflorescence meristems and increases the number of reproductive organs, resulting in enhanced grain yield. QTL pyramiding to combine loci for grain number and plant height in the same genetic background generated lines exhibiting both beneficial traits. These results provide a strategy for tailormade crop improvement.
URLPMID:22748359 [本文引用: 1]

Recently, two important yield quantitative trait loci (QTLs), GS3 and DEP1, have been cloned in rice ( Oryza sativa ). Although their relationship has not been established in the ‘rice literature’, a recent report identified them as heterotrimeric G protein γ subunits. This identification has profound consequences for our current understanding of both QTLs and the plant G protein signaling network and this opinion article discusses how manipulation of G protein signaling may lead to yield improvements in rice and other crop species. Finally, a mechanistic model to explain the seemingly conflicting phenotypes produced by different GS3 and DEP1 alleles is proposed.
URLPMID:25149369 [本文引用: 1]

Abstract Grain size is one of the most important factors determining rice yield. As a quantitative trait, grain size is predominantly and tightly controlled by genetic factors. Several quantitative trait loci (QTLs) for grain size have been molecularly identified and characterized. These QTLs may act in independent genetic pathways and, along with other identified genes for grain size, are mainly involved in the signaling pathways mediated by proteasomal degradation, phytohormones, and G proteins to regulate cell proliferation and cell elongation. Many of these QTLs and genes have been strongly selected for enhanced rice productivity during domestication and breeding. These findings have paved new ways for understanding the molecular basis of grain size and have substantial implications for genetic improvement of crops.
URLPMID:19305410 [本文引用: 1]

Abstract Grain yield is controlled by quantitative trait loci (QTLs) derived from natural variations in many crop plants. Here we report the molecular characterization of a major rice grain yield QTL that acts through the determination of panicle architecture. The dominant allele at the DEP1 locus is a gain-of-function mutation causing truncation of a phosphatidylethanolamine-binding protein-like domain protein. The effect of this allele is to enhance meristematic activity, resulting in a reduced length of the inflorescence internode, an increased number of grains per panicle and a consequent increase in grain yield. This allele is common to many Chinese high-yielding rice varieties and likely represents a relatively recent introduction into the cultivated rice gene pool. We also show that a functionally equivalent allele is present in the temperate cereals and seems to have arisen before the divergence of the wheat and barley lineages.
URLPMID:29085070 [本文引用: 1]

Abstract Transcriptional silencer and copy number variants (CNVs) are associated with gene expression. However, their roles in generating phenotypes have not been well studied. Here we identified a rice quantitative trait locus, SGDP7 (Small Grain and Dense Panicle 7). SGDP7 is identical to FZP (FRIZZY PANICLE), which represses the formation of axillary meristems. The causal mutation of SGDP7 is an 18-bp fragment, named CNV-18bp, which was inserted ~5.3 kb upstream of FZP and resulted in a tandem duplication in the cultivar Chuan 7. The CNV-18bp duplication repressed FZP expression, prolonged the panicle branching period and increased grain yield by more than 15% through substantially increasing the number of spikelets per panicle (SPP) and slightly decreasing the 1,000-grain weight (TGW). The transcription repressor OsBZR1 binds the CGTG motifs in CNV-18bp and thereby represses FZP expression, indicating that CNV-18bp is the upstream silencer of FZP. These findings showed that the silencer CNVs coordinate a trade-off between SPP and TGW by fine-tuning FZP expression, and balancing the trade-off could enhance yield potential.
[本文引用: 1]
URLPMID:26283047 [本文引用: 1]

Abstract A wide range of morphological and physiological traits have changed between cultivated rice Oryza sativa and wild rice Oryza rufipogon under domestication. Here, we report cloning of the An-2 gene, encoding the Lonely Guy Like protein 6 (OsLOGL6), which catalyzes the final step of cytokinin synthesis in O. rufipogon. The near-isogenic line harboring a wild allele of An-2 in the genetic background of the awnless indica Guangluai 4 shows that An-2 promotes awn elongation by enhancing cell division, but decreases grain production by reducing grains per panicle and tillers per plant. We reveal that a genetic variation in the An-2 locus has a large impact on reducing awn length and increasing tiller and grain numbers in domesticated rice. Analysis of gene expression patterns suggests that An-1 regulates the formation of awn primordial, and An-2 promotes awn elongation. Nucleotide diversity of the An-2 locus in cultivated rice was found to be significantly reduced compared with that of wild rice, suggesting that the An-2 locus was subjected to artificial selection. We therefore propose that the selection of genetic variation in An-2 was due to reduced awn length and increased grain yield in cultivated rice. Copyright 2015 The Author. Published by Elsevier Inc. All rights reserved.
URLPMID:26082172 [本文引用: 1]

Common wild rice (Oryza rufipogon), the wild relative of Asian cultivated rice (Oryza sativa), flaunts long, barbed awns, which are necessary for efficient propagation and dissemination of seeds. By contrast, O. sativa cultivars have been selected to be awnless or to harbor short, barbless awns, which facilitate seed processing and storage. The transition from long, barbed awns to short, barbless awns was a crucial event in rice domestication. Here, we show that the presence of long, barbed awns in wild rice is controlled by a major gene on chromosome 4, LONG AND BARBED AWN1 (LABA1), which encodes a cytokinin-activating enzyme. A frame-shift deletion in LABA1 of cultivated rice reduces the cytokinin concentration in awn primordia, disrupting barb formation and awn elongation. Sequencing analysis demonstrated low nucleotide diversity and a selective sweep encompassing an 800-kb region around the derived laba1 allele in cultivated rice. Haplotype analysis revealed that the laba1 allele originated in the japonica subspecies and moved into the indica gene pool via introgression, suggesting that humans selected for this locus in early rice domestication. Identification of LABA1 provides new insights into rice domestication and also sheds light on the molecular mechanism underlying awn development.
[本文引用: 1]
URLPMID:19619185 [本文引用: 1]

Abstract In rice (Oryza sativa L.), the number of panicles, spikelets per panicle and grain weight are important components of grain yield. These characteristics are controlled by quantitative trait loci (QTLs) and are derived from variation inherent in crops. As a result of the complex genetic basis of these traits, only a few genes involved in their control have been cloned and characterized. We have previously map-cloned a gene cluster including eight leucine-rich repeat receptor-like kinase (LRK) genes in Dongxiang wild rice (Oryza rufipogon Griff.), which increased the grain yield by 16%. In the present study, we characterized the LRK1 gene, which was contained in the donor parent (Dongxiang wild rice) genome and absent from the recurrent parent genome (Guichao2, Oryza sativa L. ssp. indica). Our data showed that rice LRK1 is a plasma membrane protein expressed constitutively in leaves, young panicles, roots and culms. The over-expression of rice LRK1 results in increased panicles, spikelets per panicle, weight per grain and enhanced cellular proliferation, leading to a 27.09% increase in total grain yield per plant. The increased number of panicles and spikelets per panicle are associated with increased branch number. Our data suggest that rice LRK1 regulates rice branch number by enhancing cellular proliferation. The functional characterization of rice LRK1 facilitates an understanding of the mechanisms involved in cereal crop yield, and may have utility in improving grain yield in cereal crops.
URL [本文引用: 1]
[本文引用: 1]
URLPMID:23435087 [本文引用: 1]

Abstract Reduction in seed shattering was an important phenotypic change during cereal domestication. Here we show that a simple morphological change in rice panicle shape, controlled by the SPR3 locus, has a large impact on seed-shedding and pollinating behaviors. In the wild genetic background of rice, we found that plants with a cultivated-like type of closed panicle had significantly reduced seed shedding through seed retention. In addition, the long awns in closed panicles disturbed the free exposure of anthers and stigmas on the flowering spikelets, resulting in a significant reduction of the outcrossing rate. We localized the SPR3 locus to a 9.3-kb genomic region, and our complementation tests suggest that this region regulates the liguleless gene (OsLG1). Sequencing analysis identified reduced nucleotide diversity and a selective sweep at the SPR3 locus in cultivated rice. Our results suggest that a closed panicle was a selected trait during rice domestication.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URLPMID:29487318 [本文引用: 1]

Abstract Manipulating grain size is an effective strategy for increasing cereal yields. Here we identify a pathway composed of five subunits of the heterotrimeric G proteins that regulate grain length in rice. The G0205 protein is essential for plant survival and growth. G02± provides a foundation for grain size expansion. Three G0206 proteins, DEP1, GGC2 and GS3, antagonistically regulate grain size. DEP1 and GGC2, individually or in combination, increase grain length when in complex with G0205. GS3, having no effect on grain size by itself, reduces grain length by competitively interacting with G0205. By combining different G-protein variants, we can decrease grain length by up to 35% or increase it by up to 19%, which leads to over 40% decreasing to 28% increasing of grain weight. The wide existence of such a conserved system among angiosperms suggests a possible general predictable approach to manipulating grain/organ sizes.
URLPMID:17417637 [本文引用: 1]

Abstract Grain weight is one of the most important components of grain yield and is controlled by quantitative trait loci (QTLs) derived from natural variations in crops. However, the molecular roles of QTLs in the regulation of grain weight have not been fully elucidated. Here, we report the cloning and characterization of GW2, a new QTL that controls rice grain width and weight. Our data show that GW2 encodes a previously unknown RING-type protein with E3 ubiquitin ligase activity, which is known to function in the degradation by the ubiquitin-proteasome pathway. Loss of GW2 function increased cell numbers, resulting in a larger (wider) spikelet hull, and it accelerated the grain milk filling rate, resulting in enhanced grain width, weight and yield. Our results suggest that GW2 negatively regulates cell division by targeting its substrate(s) to proteasomes for regulated proteolysis. The functional characterization of GW2 provides insight into the mechanism of seed development and is a potential tool for improving grain yield in crops.
URLPMID:18820698 [本文引用: 1]

Abstract Grain-filling, an important trait that contributes greatly to grain weight, is regulated by quantitative trait loci and is associated with crop domestication syndrome. However, the genes and underlying molecular mechanisms controlling crop grain-filling remain elusive. Here we report the isolation and functional analysis of the rice GIF1 (GRAIN INCOMPLETE FILLING 1) gene that encodes a cell-wall invertase required for carbon partitioning during early grain-filling. The cultivated GIF1 gene shows a restricted expression pattern during grain-filling compared to the wild rice allele, probably a result of accumulated mutations in the gene's regulatory sequence through domestication. Fine mapping with introgression lines revealed that the wild rice GIF1 is responsible for grain weight reduction. Ectopic expression of the cultivated GIF1 gene with the 35S or rice Waxy promoter resulted in smaller grains, whereas overexpression of GIF1 driven by its native promoter increased grain production. These findings, together with the domestication signature that we identified by comparing nucleotide diversity of the GIF1 loci between cultivated and wild rice, strongly suggest that GIF1 is a potential domestication gene and that such a domestication-selected gene can be used for further crop improvement.
URLPMID:20 [本文引用: 1]

Increasing crop yield is one of the most important goals of plant science research. Grain size is a major determinant of grain yield in cereals and is a target trait for both domestication and artificial breeding(1). We showed that the quantitative trait locus (QTL) GS5 in controls grain size by regulating grain width, filling and weight. GS5 encodes a putative serine carboxypeptidase and functions as a positive regulator of grain size, such that higher expression of GS5 is correlated with larger grain size. Sequencing of the promoter region in 51 accessions from a wide geographic range identified three haplotypes that seem to be associated with grain width. The results suggest that natural variation in GS5 contributes to grain size diversity in and may be useful in improving yield in and, potentially, other crops(2).
URLPMID:25711711 [本文引用: 1]

Abstract Grain weight is a major determinant of grain yield. GS5 is a positive regulator of grain size such that grain width, filling, and weight are correlated with its expression level. Previous work suggested that polymorphisms of GS5 in the promoter region might be responsible for the variation in grain size. In this study, two single nucleotide polymorphisms (SNPs) between the wide-grain allele GS5-1 and the narrow-grain allele GS5-2 in the upstream region of the gene that were responsible for the differential expression in developing young panicles were identified. These two polymorphs altered the responses of the GS5 alleles to abscisic acid (ABA) treatments, resulting in higher expression of GS5-1 than of GS5-2 in developing young panicles. It was also shown that SNPs in light-responsive elements of the promoter altered the response to light induction, leading to higher expression of GS5-2 than GS5-1 in leaves. Enhanced expression of GS5 competitively inhibits the interaction between OsBAK1-7 and OsMSBP1 by occupying the extracellular leucine-rich repeat (LRR) domain of OsBAK1-7, thus preventing OsBAK1-7 from endocytosis caused by interacting with OsMSBP1, providing an explanation for the positive association between grain size and GS5 expression. These results advanced our understanding of the molecular mechanism by which GS5 controls grain size. The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology.
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URLPMID:28394310 [本文引用: 1]

Abstract Grain size is a major determinant of grain yield in cereal crops. qSW5/GW5, which exerts the greatest effect on rice grain width and weight, was fine-mapped to a 2,263-bp/21-kb genomic region containing a 1,212-bp deletion, respectively. Here, we show that a gene encoding a calmodulin binding protein, located 5 b downstream of the 1,212-bp deletion, corresponds to qSW5/GW5. GW5 is expressed in various rice organs, with highest expression level detected in young panicles. We provide evidence that the 1,212-bp deletion affects grain width most likely through influencing the expression levels of GW5. GW5 protein is localized to the plasma membrane and can physically interact with and repress the kinase activity of rice GSK2 (glycogen synthase kinase 2), a homologue of Arabidopsis BIN2 (BRASSINOSTEROID INSENSITIVE2) kinase, resulting in accumulation of unphosphorylated OsBZR1 (Oryza sativa BRASSINAZOLE RESISTANT1) and DLT (DWARF AND LOW-TILLERING) proteins in the nucleus to mediate brassinosteroid (BR)-responsive gene expression and growth responses (including grain width and weight). Our results suggest that GW5 is a novel positive regulator of BR signalling and a viable target for genetic manipulation to improve grain yield in rice and perhaps in other cereal crops as well.
[本文引用: 1]
[本文引用: 1]
URLPMID:26187814 [本文引用: 1]

Grain size determines grain weight and affects grain quality. Several major quantitative trait loci (QTLs) regulating grain size have been cloned; however, our understanding of the underlying mechanism that regulates the size of rice grains remains fragmentary. Here, we report the cloning and characterization of a dominant QTL, GRAIN SIZE ON CHROMOSOME 2 ( GS2 ), which encodes Growth-Regulating Factor 4 (OsGRF4), a transcriptional regulator. GS2 localizes to the nucleus and may act as a transcription activator. A rare mutation of GS2 affecting the binding site for the regulatory microRNA OsmiR396c causes increased expression of GS2 / OsGRF4 . The increase of GS2 expression leads to larger cells and increased numbers of cells, which thus enhances grain weight and yield. The introduction of this rare allele of GS2 / OsGRF4 into rice cultivars could enhance grain weight and increase grain yield, with possible applications in breeding high-yield rice varieties.
URLPMID:5089622 [本文引用: 1]

Abstract Traits such as grain shape, panicle length and seed shattering, play important roles in grain yield and harvest. In this study, the cloning and functional analysis of PANICLE TRAITS 2 ( PT2 ), a novel gene from the Indica rice Chuandali (CDL), is reported. PT2 is synonymous with Growth-Regulating Factor 4 ( OsGRF4 ), which encodes a growth-regulating factor that positively regulates grain shape and panicle length and negatively regulates seed shattering. Higher expression of OsGRF4 is correlated with larger grain, longer panicle and lower seed shattering. A unique OsGRF4 mutation, which occurs at the OsmiRNA396 target site of OsGRF4 , seems to be associated with high levels of OsGRF4 expression, and results in phenotypic difference. Further research showed that OsGRF4 regulated two cytokinin dehydrogenase precursor genes ( CKX5 and CKX1 ) resulting in increased cytokinin levels, which might affect the panicle traits. High storage capacity and moderate seed shattering of OsGRF4 may be useful in high-yield breeding and mechanized harvesting of rice. Our findings provide additional insight into the molecular basis of panicle growth.
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:26147619 [本文引用: 1]

Copy number variants (CNVs) are associated with changes in gene expression levels and contribute to various adaptive traits. Here we show that a CNV at the Grain Length on Chromosome 7 (GL7) locus contributes to grain size diversity in rice (Oryza sativa L.). GL7 encodes a protein homologous to Arabidopsis thaliana LONGIFOLIA proteins, which regulate longitudinal cell elongation. Tandem duplication of a 17.1-kb segment at the GL7 locus leads to upregulation of GL7 and downregulation of its nearby negative regulator, resulting in an increase in grain length and improvement of grain appearance quality. Sequence analysis indicates that allelic variants of GL7 and its negative regulator are associated with grain size diversity and that the CNV at the GL7 locus was selected for and used in breeding. Our work suggests that pyramiding beneficial alleles of GL7 and other yield- and quality-related genes may improve the breeding of elite rice varieties.
[本文引用: 1]
URLPMID:26950093 [本文引用: 1]

Abstract Although genetic diversity has a cardinal role in domestication, abundant natural allelic variations across the rice genome that cause agronomically important differences between diverse varieties have not been fully explored. Here we implement an approach integrating genome-wide association testing with functional analysis on grain size in a diverse rice population. We report that a major quantitative trait locus, GLW7, encoding the plant-specific transcription factor OsSPL13, positively regulates cell size in the grain hull, resulting in enhanced rice grain length and yield. We determine that a tandem-repeat sequence in the 5' UTR of OsSPL13 alters its expression by affecting transcription and translation and that high expression of OsSPL13 is associated with large grains in tropical japonica rice. Further analysis indicates that the large-grain allele of GLW7 in tropical japonica rice was introgressed from indica varieties under artificial selection. Our study demonstrates that new genes can be effectively identified on the basis of genome-wide association data.
[本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URLPMID:20495564 [本文引用: 1]

Abstract Identification of alleles that improve crop production and lead to higher-yielding varieties are needed for food security. Here we show that the quantitative trait locus WFP (WEALTHY FARMER'S PANICLE) encodes OsSPL14 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 14, also known as IPA1). Higher expression of OsSPL14 in the reproductive stage promotes panicle branching and higher grain yield in rice. OsSPL14 controls shoot branching in the vegetative stage and is affected by microRNA excision. We also demonstrate the feasibility of using the OsSLP14(WFP) allele to increase rice crop yield. Introduction of the high-yielding OsSPL14(WFP) allele into the standard rice variety Nipponbare resulted in increased rice production.
[本文引用: 1]
URLPMID:18820696 [本文引用: 1]

Abstract The closely related wild rice species Oryza rufipogon is considered the progenitor of cultivated rice (Oryza sativa). The transition from the characteristic plant architecture of wild rice to that of cultivated rice was one of the most important events in rice domestication; however, the molecular basis of this key domestication transition has not been elucidated. Here we show that the PROG1 gene controls aspects of wild-rice plant architecture, including tiller angle and number of tillers. The gene encodes a newly identified zinc-finger nuclear transcription factor with transcriptional activity and is mapped on chromosome 7. PROG1 is predominantly expressed in the axillary meristems, the site of tiller bud formation. Rice transformation experiments demonstrate that artificial selection of an amino acid substitution in the PROG1 protein during domestication led to the transition from the plant architecture of wild rice to that of domesticated rice.
URL [本文引用: 1]
URLPMID:17908158 [本文引用: 1]

Summary A critical step during rice ( Oryza sativa ) cultivation is dense planting: a wider tiller angle will increase leaf shade and decrease photosynthesis efficiency, whereas a narrower tiller angle makes for more efficient plant architecture. The molecular basis of tiller angle remains unknown. This research demonstrates that tiller angle is controlled by a major quantitative trait locus, TAC1 ( Tiller Angle Control 1 ). TAC1 was mapped to a 35-kb region on chromosome 9 using a large F 2 population from crosses between an indica rice, IR24, which displays a relatively spread-out plant architecture, and an introgressed line, IL55, derived from japonica rice Asominori, which displays a compact plant architecture with extremely erect tillers. Genetic complementation further identified the TAC1 gene, which harbors three introns in its coding region and a fourth 1.5-kb intron in the 3′-untranslated region. A mutation in the 3′-splicing site of this 1.5-kb intron from ‘AGGA’ to ‘GGGA’ decreases the level of tac1 , resulting in a compact plant architecture with a tiller angle close to zero. Further sequence verification of the mutation in the 3′-splicing site of the 1.5-kb intron revealed that the tac1 mutation ‘GGGA’ was present in 88 compact japonica rice accessions and TAC1 with ‘AGGA’ was present in 21 wild rice accessions and 43 indica rice accessions, all with the spread-out form, indicating that tac1 had been extensively utilized in densely planted rice grown in high-latitude temperate areas and at high altitudes where japonica rice varieties are widely cultivated.
URLPMID:18454147 [本文引用: 1]

Yield potential,plant height and heading date are three classes of traits that determine the productivity of many crop plants. Here we showed that the quantitative trait locus (QTL) Ghd7,isolated from an elite rice hybrid and encoding a CCT domain protein,had major effects on an array of traits in rice,including number of grains per panicle,plant height and heading date. Enhanced expression of Ghd7 under long-day conditions delays heading and increases plant height and panicle size. Natural mutants with reduced function enable rice to be cultivated in temperate and cooler regions. Thus,Ghd7 has played crucial roles for increasing productivity and adaptability of rice globally.
[本文引用: 1]
URLPMID:20566706 [本文引用: 1]

Abstract The three most important agronomic traits of rice (Oryza sativa), yield, plant height, and flowering time, are controlled by many quantitative trait loci (QTLs). In this study, a newly identified QTL, DTH8 (QTL for days to heading on chromosome 8), was found to regulate these three traits in rice. Map-based cloning reveals that DTH8 encodes a putative HAP3 subunit of the CCAAT-box-binding transcription factor and the complementary experiment increased significantly days to heading, plant height, and number of grains per panicle in CSSL61 (a chromosome segment substitution line that carries the nonfunctional DTH8 allele) with the Asominori functional DTH8 allele under long-day conditions. DTH8 is expressed in most tissues and its protein is localized to the nucleus exclusively. The quantitative real-time PCR assay revealed that DTH8 could down-regulate the transcriptions of Ehd1 (for Early heading date1) and Hd3a (for Heading date3a; a rice ortholog of FLOWERING LOCUS T) under long-day conditions. Ehd1 and Hd3a can also be down-regulated by the photoperiodic flowering genes Ghd7 and Hd1 (a rice ortholog of CONSTANS). Meanwhile, the transcription of DTH8 has been proved to be independent of Ghd7 and Hd1, and the natural mutation of this gene caused weak photoperiod sensitivity and shorter plant height. Taken together, these data indicate that DTH8 probably plays an important role in the signal network of photoperiodic flowering as a novel suppressor as well as in the regulation of plant height and yield potential.
[本文引用: 1]
[本文引用: 1]
URLPMID:28549969 [本文引用: 1]

Photoperiodic flowering is one of the most important pathways to govern flowering in rice ( Oryza sativa ), in which Heading date 1 ( Hd1 ), an ortholog of the Arabidopsis CONSTANS gene, encodes a pivotal regulator. Hd1 promotes flowering under short-day conditions but represses flowering under long-day conditions by regulating the expression of Heading date 3a ( Hd3a ), the rice FLOWERING LOCUS T ( FT ) ortholog. However, the molecular mechanism by which Hd1 reverses its activity in response to day length is largely unknown. Here, we demonstrate that the repression of flowering in LD mediated by Hd1 is dependent on the transcription factor DAYS TO HEADING 8 ( DTH8 ). Loss of DTH8 function results in the activation of Hd3a by Hd1 , leading to early flowering. We also show that Hd1 directly interacts with DTH8 and that the formation of the DTH8-Hd1 complex is necessary for the transcriptional repression of Hd3a by Hd1 in LD. We thus detected a switch in function of Hd1 mediated by DTH8 in LD rather than in SD. Further, we reveal that DTH8 associates with the Hd3a promoter to modulate the level of trimethylated H3K27 at the Hd3a genomic locus. H3K27 trimethylation increases at Hd3a in the presence the DTH8-Hd1 complex, while Hd1 attenuates the H3K27me3 level in Hd3a when DTH8 function is lost. Therefore, our findings establish that, in response to day length, DTH8 plays a critical role in mediating Hd1 regulation of Hd3a transcription through the DTH8-Hd1 module to shape epigenetic marks in photoperiodic flowering.
URLPMID:19400646 [本文引用: 1]

Abstract Quantitative disease resistance (QDR) has been observed within many crop plants but is not as well understood as qualitative (monogenic) disease resistance and has not been used as extensively in breeding. Mapping quantitative trait loci (QTLs) is a powerful tool for genetic dissection of QDR. DNA markers tightly linked to quantitative resistance loci (QRLs) controlling QDR can be used for marker-assisted selection (MAS) to incorporate these valuable traits. QDR confers a reduction, rather than lack, of disease and has diverse biological and molecular bases as revealed by cloning of QRLs and identification of the candidate gene(s) underlying QRLs. Increasing our biological knowledge of QDR and QRLs will enhance understanding of how QDR differs from qualitative resistance and provide the necessary information to better deploy these resources in breeding. Application of MAS for QRLs in breeding for QDR to diverse pathogens is illustrated by examples from wheat, barley, common bean, tomato, and pepper. Strategies for optimum deployment of QRLs require research to understand effects of QDR on pathogen populations over time.
URL [本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:27791169 [本文引用: 1]

Abstract Brown planthopper (BPH), Nilaparvata lugens St l, is one of the most devastating insect pests of rice (Oryza sativa L.). Currently, 30 BPH-resistance genes have been genetically defined, most of which are clustered on specific chromosome regions. Here, we describe molecular cloning and characterization of a BPH-resistance gene, BPH9, mapped on the long arm of rice chromosome 12 (12L). BPH9 encodes a rare type of nucleotide-binding and leucine-rich repeat (NLR)-containing protein that localizes to the endomembrane system and causes a cell death phenotype. BPH9 activates salicylic acid- and jasmonic acid-signaling pathways in rice plants and confers both antixenosis and antibiosis to BPH. We further demonstrated that the eight BPH-resistance genes that are clustered on chromosome 12L, including the widely used BPH1, are allelic with each other. To honor the priority in the literature, we thus designated this locus as BPH1/9 These eight genes can be classified into four allelotypes, BPH1/9-1, -2, -7, and -9 These allelotypes confer varying levels of resistance to different biotypes of BPH. The coding region of BPH1/9 shows a high level of diversity in rice germplasm. Homologous fragments of the nucleotide-binding (NB) and leucine-rich repeat (LRR) domains exist, which might have served as a repository for generating allele diversity. Our findings reveal a rice plant strategy for modifying the genetic information to gain the upper hand in the struggle against insect herbivores. Further exploration of natural allelic variation and artificial shuffling within this gene may allow breeding to be tailored to control emerging biotypes of BPH.
URLPMID:25728666 [本文引用: 1]

COLD1 regulates G-protein signaling to confer chilling tolerance in rice, and a SNP in COLD1 underlies the adaptation to cold environment in japonica rice.
URLPMID:28332574 [本文引用: 1]

Abstract Low temperature is a major factor limiting rice productivity and geographical distribution. Improved cold tolerance and expanded cultivation to high-altitude or high-latitude regions would help meet growing rice demand. Here we explored a QTL for cold tolerance and cloned the gene, CTB4a (cold tolerance at booting stage), encoding a conserved leucine-rich repeat receptor-like kinase. We show that different CTB4a alleles confer distinct levels of cold tolerance and selection for variation in the CTB4a promoter region has occurred on the basis of environmental temperature. The newly generated cold-tolerant haplotype Tej-Hap-KMXBG was retained by artificial selection during temperate japonica evolution in cold habitats for low-temperature acclimation. Moreover, CTB4a interacts with AtpB, a beta subunit of ATP synthase. Upregulation of CTB4a correlates with increased ATP synthase activity, ATP content, enhanced seed setting and improved yield under cold stress conditions. These findings suggest strategies to improve cold tolerance in crop plants.
URL [本文引用: 1]
URLPMID:24777451 [本文引用: 1]

The drive toward more sustainable agriculture has raised the profile of crop plant nutrient-use efficiency. Here we show that a major rice nitrogen-use efficiency quantitative trait locus (qNGR9) is synonymous with the previously identified gene DEP1 (DENSE AND ERECT PANICLES 1). The different DEP1 alleles confer different nitrogen responses, and genetic diversity analysis suggests that DEP1 has been subjected to artificial selection during Oryza sativa spp. japonica rice domestication. The plants carrying the dominant dep1-1 allele exhibit nitrogen-insensitive vegetative growth coupled with increased nitrogen uptake and assimilation, resulting in improved harvest index and grain yield at moderate levels of nitrogen fertilization. The DEP1 protein interacts in vivo with both the G02± (RGA1) and G0205 (RGB1) subunits, and reduced RGA1 or enhanced RGB1 activity inhibits nitrogen responses. We conclude that the plant G protein complex regulates nitrogen signaling and modulation of heterotrimeric G protein activity provides a strategy for environmentally sustainable increases in rice grain yield.
URLPMID:22914168 [本文引用: 1]

As an essential macroelement for all living cells, phosphorus is indispensable in agricultural production systems. Natural phosphorus reserves are limited, and it is therefore important to develop phosphorus-efficient crops. A major quantitative trait locus for phosphorus-deficiency tolerance, Pup1, was identified in the traditional aus-type rice variety Kasalath about a decade ago. However, its functional mechanism remained elusive until the locus was sequenced, showing the presence of a Pup1-specific protein kinase gene, which we have named phosphorus-starvation tolerance 1 (PSTOL1). This gene is absent from the rice reference genome and other phosphorus-starvation-intolerant modern varieties. Here we show that overexpression of PSTOL1 in such varieties significantly enhances grain yield in phosphorus-deficient soil. Further analyses show that PSTOL1 acts as an enhancer of early root growth, thereby enabling plants to acquire more phosphorus and other nutrients. The absence of PSTOL1 and other genes--for example, the submergence-tolerance gene SUB1A--from modern rice varieties underlines the importance of conserving and exploring traditional germplasm. Introgression of this quantitative trait locus into locally adapted rice varieties in Asia and Africa is expected to considerably enhance productivity under low phosphorus conditions.
URL [本文引用: 1]

作物抗病机制研究对抗性改良有重要的理论和实践意义。水稻是世界上主要粮食作物之一。目前,越来越多的水稻主效抗病基因和抗病相关基因被克隆。对于这些基因的研究可以提高人们对水稻-病原菌互作的认识,并有利于发掘水稻中新的抗性基因。现主要对近年来在水稻抗病研究中取得的成果进行概述和展望。
URL [本文引用: 1]

作物抗病机制研究对抗性改良有重要的理论和实践意义。水稻是世界上主要粮食作物之一。目前,越来越多的水稻主效抗病基因和抗病相关基因被克隆。对于这些基因的研究可以提高人们对水稻-病原菌互作的认识,并有利于发掘水稻中新的抗性基因。现主要对近年来在水稻抗病研究中取得的成果进行概述和展望。
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:20823253 [本文引用: 1]

Intake of toxic cadmium (Cd) from rice caused Itai-itai disease in the past and it is still a threat for human health. Therefore, control of the accumulation of Cd from soil is an important food-safety issue, but the molecular mechanism for the control is unknown. Herein, we report a gene (OsHMA3) responsible for low Cd accumulation in rice that was isolated from a mapping population derived from a cross between a high and low Cd-accumulating cultivar. The gene encodes a transporter belonging to the P 1B -type ATPase family, but shares low similarity with other members. Heterologous expression in yeast showed that the transporter from the low-Cd cultivar is functional, but the transporter from the high-Cd cultivar had lost its function, probably because of the single amino acid mutation. The transporter is mainly expressed in the tonoplast of root cells at a similar level in both the low and high Cd-accumulating cultivars. Overexpression of the functional gene from the low Cd-accumulating cultivar selectively decreased accumulation of Cd, but not other micronutrients in the grain. Our results indicated that OsHMA3 from the low Cd-accumulating cultivar limits translocation of Cd from the roots to the above-ground tissues by selectively sequestrating Cd into the root vacuoles.
