 ,1, 冯晋川2, 李贵林1, 王讯1, 李明洲1, 刘海峰
,1, 冯晋川2, 李贵林1, 王讯1, 李明洲1, 刘海峰 ,1
,1The effect of lnc-RAP3 on 3T3-L1 preadipocyte differentiation in mouse
Huan Li ,1, Jinchuan Feng2, Guilin Li1, Xun Wang1, Mingzhou Li1, Haifeng Liu
,1, Jinchuan Feng2, Guilin Li1, Xun Wang1, Mingzhou Li1, Haifeng Liu ,1
,1通讯作者:
编委: 宋旭
收稿日期:2018-03-1修回日期:2018-05-7网络出版日期:2018-09-20
| 基金资助: |
Editorial board:
Received:2018-03-1Revised:2018-05-7Online:2018-09-20
| Fund supported: |
作者简介 About authors
作者简介:李欢,硕士研究生,专业方向:分子遗传与动物育种E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (533KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李欢, 冯晋川, 李贵林, 王讯, 李明洲, 刘海峰. Lnc-RAP3对小鼠3T3-L1前脂肪细胞分化的影响[J]. 遗传, 2018, 40(9): 758-766 doi:10.16288/j.yczz.18-053
Huan Li, Jinchuan Feng, Guilin Li, Xun Wang, Mingzhou Li, Haifeng Liu.
随着人们生活水平的不断提高,肥胖及相关代谢性疾病的发生已经成为危害人类健康的重要因素之一。研究表明,脂肪细胞内甘油三酯含量聚集过多可能会引发肥胖,从而增加胰岛素抵抗、2型糖尿病、心血管疾病、高血压、癌症等许多疾病的患病风险[1,2,3,4]。脂肪细胞的形成和成熟需要大量基因的时序表达,经过不同的发育阶段,涉及多种生物学进程。同时,脂肪组织通过自分泌、旁分泌及内分泌方式分泌大量的脂肪因子影响整个机体的一系列生物学活动。
长链非编码RNA (long non-coding RNA, lncRNA)是在真核生物中发现的一类长度大于200nt、没有长开放阅读框架但往往具有mRNA结构特征(甲基鸟苷帽子和多聚腺苷酸尾)的RNA[5],广泛参与生物 体的各种生理及病理过程,如细胞周期调控[6]、肿瘤发生及转移[7]、神经系统发育[8]等。研究发现,lncRNA可以通过多种方式在表观遗传、转录及转录后水平参与基因的表达调控。如lncRNA TARID可定位于肿瘤抑制因子TCF21启动子区,并招募GADD45A结合到该区域,促使DNA去甲基化,从而激活靶基因的表达[9];Tsai等[10]研究发现,lncRNA CRG通过募集RNA聚合酶Ⅱ到CASK上游启动子区域,从而调节CASK基因的表达;在转录后水平调控研究中,Beltran等[11]发现Zeb2的反义lncRNA能够与其mRNA 5°端内含子剪切位点所在区域形成互补双链,阻止该内含子剪切,从而保证Zeb2的正常表达。此外,也有部分研究表明lncRNA密切参与脂肪细胞分化过程的调节[12,13,14,15]。
Lnc-RAP3位于小鼠(Mus musculus)17号染色体,但到目前为止,关于lnc-RAP3生物学功能的研究鲜有报道,仅有Sun等[16]通过siRNA敲降实验对lnc-RAP3在小鼠皮下前脂肪细胞内脂滴集聚情况进行了研究。本课题组在前期实验中对诱导前后的3T3-L1前脂肪细胞进行转录组测序,发现lnc-RAP3在3T3-L1细胞诱导分化后呈现高丰度表达。为了进一步探究该lncRNA在小鼠3T3-L1前脂肪细胞成脂分化过程中的作用,本研究采用过表达和敲降lnc-RAP3的方式首次揭示了其对3T3-L1前脂肪细胞内脂滴聚集、成脂分化相关基因表达等的影响,为进一步研究lnc-RAP3在小鼠脂肪细胞分化中的功能机制提供理论依据。
1 材料和方法
1.1 细胞培养
小鼠3T3-L1前脂肪细胞(购自中国科学院上海生命科学研究院细胞库)采用含10%胎牛血清(Fetal Bovine Serum, FBS) (Gibco,美国)和双抗(青霉素100 U/mL、链霉素0.1 mg/mL) (北京索莱宝科技有限公司)的DMEM高糖培养基(Gibco,美国)培养,培养条件为37℃、5% CO2及饱和湿度的无菌培养箱(Thermo,美国)。1.2 重组pcDNA3.1-RAP3真核表达载体的构建
根据GenBank中lnc-RAP3序列(登录号NR_ 045146.1)设计引物,并在上下游引物5°端分别引入Hind Ⅲ和Kpn Ⅰ (TaKaRa,日本)限制性酶切位点(表1),通过PCR扩增lnc-RAP3目的片段并进行胶回收,用于TA克隆。将获得的lnc-RAP3目的片段与pMD19-T载体相连,连接产物转化入DH5α感受态细胞并涂板,挑取阳性单克隆进行PCR鉴定、测序(成都擎科梓熙生物技术有限公司)。将测序正确的对应菌体放大培养后用快速质粒小提试剂盒(北京天根生化科技有限公司)提取质粒pMD19-T-RAP3。用限制性内切酶Hind Ⅲ和Kpn Ⅰ将pMD19-T-RAP3与真核表达载体pcDNA3.1分别进行酶切,酶切产物经凝胶电泳检测后回收目的片段,用T4连接酶过夜连接后进行转化涂板并挑选阳性克隆进行测序,将测序正确的菌体放大培养,用无内毒素质粒大提试剂盒提取重组质粒pcDNA3.1-RAP3。Table 1
表1
表1 本文所用的引物信息
Table 1
| 基因 | GenBank登录号 | 引物序列(5°→3°) | 引物用途 |
|---|---|---|---|
| lnc-RAP3 | NR_045146.1 | F: CCCAAGCTTTCCACCCTAGGAGGGTAAGG | 基因克隆 |
| R: CGGGGTACCGGGGGAACCTTTATTTCAGGGA | |||
| lnc-RAP3 | NR_045146.1 | F: ACACGAACACACGCATACAA | qRT-PCR |
| R: TCATAACGACAGTGGTGCAG | |||
| PPARγ | NM_011146.3 | F: AAGAGCTGACCCAATGGTTG | qRT-PCR |
| R: ACCCTTGCATCCTTCACAAG | |||
| C/EBPα | NM_001287523.1 | F: GGTTTTGCTCTGATTCTTGCC | qRT-PCR |
| R: CGAAAAAACCCAAACATCCC | |||
| Glut4 | AB008453.1 | F: CAGATCGGCTCTGACGATGG | qRT-PCR |
| R: TCAATCACCTTCTGTGGGGC | |||
| FAS | NM_007988.3 | F: GGAGGTGGTGATAGCCGGTAT | qRT-PCR |
| R: TGGGTAATCCATAGAGCCCAG | |||
| LPL | NM_008509.2 | F: AGGACCCCTGAAGACACAGCT | qRT-PCR |
| R: TGTACAGGGCGGCCACAAGT | |||
| ACTB | NM_007393.5 | F: CAGCCTTCCTTCTTGGGTAT | qRT-PCR |
| R: TGGCATAGAGGTCTTTACGG |
新窗口打开|下载CSV
1.3 细胞转染与分化
选用生长状态良好的3T3-L1细胞接种于12孔细胞培养板(Corning,美国),当细胞汇合度达70%时,将RAP3-Smart Silencer(产品编号lnc3160612112429,以下统一用si-RAP3表示,广州锐博生物科技有限公司)、si-NC(产品编号siN05815122147,广州锐博生物科技有限公司)以及pcDNA3.1(湖南优宝生物)和构建的pcDNA3.1-RAP3用LipofectamineTM 2000转染试剂(Thermo,美国)转染到3T3-L1细胞中,转染方法按说明书操作。转染2 d后(此时记为诱导分化第0 d),将培养液换成含5 μg/mL胰岛素、0.5 mmol/L 3-异丁基-1-甲基黄嘌呤(3-isobutyl- 1-methylxanthine, IBMX)和1 μmol/L地塞米松(Sigma,美国)的DMEM高糖培养基进行诱导分化,诱导分化2 d后将培养液换成含5 μg/mL胰岛素的培养基继续培养至分化第4 d。每一处理组均设置3个重复。1.4 油红O染色
将转染后的各组细胞于诱导分化的第4 d取出,经油红O染色后置于倒置显微镜下观察、拍照;用异丙醇萃取脂滴中的油红O,用酶标仪(Promega,美国)测定OD510 nm值。1.5 实时荧光定量PCR
当3T3-L1细胞汇合度达70%时进行转染与诱导分化,方法同上。对不同处理的3T3-L1细胞分别于诱导分化第0 d、2 d、4 d采用Trizol试剂提取细胞总RNA(重复3次,并等量混合构建RNA pool),用PrimeScriptTM反转录试剂盒(TaKaRa,日本)进行cDNA合成,采用SYBR Green法定量检测各组中lnc-RAP3及成脂分化标志基因PPARγ、C/EBPα、Glut4、FAS、LPL的表达水平,以ACTB (β-actin)为内参,相关基因引物信息见表1。1.6 统计分析
采用2-ΔΔCT法计算基因的相对表达量,对si-NC与si-RAP3、pcDNA3.1与pcDNA3.1-RAP3两两处理间的差异显著性分析采用t检验,对同一基因相同处理不同时间点间的多重比较采用DUNCAN法,结果以均值±SD表示,由GraphPad Prism 6.0统计软件完成并作图。2 结果与分析
2.1 重组pcDNA3.1-RAP3真核表达载体构建结果
Lnc-RAP3基因PCR扩增产物经1%琼脂糖凝胶电泳检测,条带在700~900 bp之间,与预期产物(833 bp)相符(图1A)。对TA克隆所筛选的阳性克隆进行测序,测序结果与理论预期完全一致,进一步说明PCR扩增条带为目的条带。对测序正确的菌体放大培养后提取质粒pMD19-T-RAP3,经电泳检测出现两个条带(图1B)。其中,较亮的条带为超螺旋结构,较暗的一条为疏松结构,均为正确质粒条带。将质粒pMD19-T-RAP3与pcDNA3.1分别用限制性内切酶Hind Ⅲ和Kpn Ⅰ进行双酶切,经电泳检测片段大小与预期相符(图1C)。将酶切后的lnc-RAP3片段与同样经过酶切处理的pcDNA3.1载体纯化回收,并用T4连接酶连接转化入DH5α感受态细胞,挑取阳性单克隆放大培养并测序,用无内毒素质粒大提试剂盒提取重组质粒pcDNA3.1-RAP3。测序结果与NCBI数据库中lnc-RAP3基因序列比对相符,说明重组pcDNA3.1-RAP3真核表达载体构建成功。图1
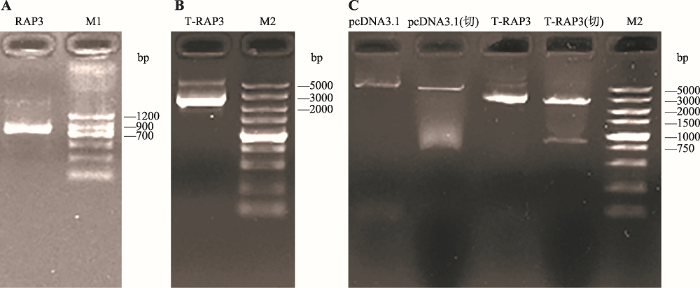 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1重组pcDNA3.1-RAP3真核表达载体的构建
A:lnc-RAP3基因PCR产物电泳结果;B:质粒pMD19-T-RAP3电泳结果;C:双酶切鉴定结果。M1/M2:DL1200/5000 DNA marker;T-RAP3:pMD19-T-RAP3;限制性内切酶为Hind Ⅲ和Kpn Ⅰ。
Fig. 1Construction of recombinant eukaryotic expression vector pcDNA3.1-RAP3
2.2 细胞转染效率测定
采用si-RAP3转染细胞对lnc-RAP3进行敲降处理,与转染si-NC组相比,lnc-RAP3的相对表达量在诱导分化第0 d、2 d和4 d分别下降了52.5%、50.3%和18.4%,其中第0 d和2 d细胞内lnc-RAP3的表达量在两处理间存在极显著差异(P<0.01),而第4 d存在显著差异(P<0.05) (图2A)。通过向细胞内转染pcDNA3.1-RAP3过表达lnc-RAP3,与转染pcDNA3.1的对照组相比,lnc-RAP3的相对表达量在诱导分化第0 d、2 d和4 d分别是对照组的5.4、2.6和2.1倍,在3个不同时间点两处理间细胞内lnc-RAP3的表达量均存在极显著差异(P<0.01) (图2B)。此外,每种处理在诱导分化第2 d时细胞内lnc-RAP3的相对表达量均出现极显著上升,说明在3T3-L1前脂肪细胞诱导分化过程中lnc-RAP3的表达量会随之而增加。图2
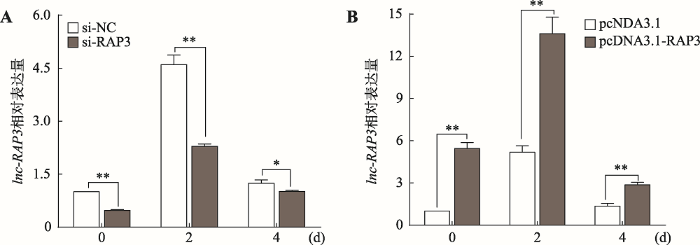 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2转染细胞中lnc-RAP3的表达量
A:抑制lnc-RAP3的表达量;B:过表达lnc-RAP3的表达量。*表示P<0.05,差异显著;**表示P<0.01,差异极显著。
Fig. 2The expression level of lnc-RAP3 in transfected cells
2.3 油红O染色结果
对4种不同处理的细胞培养至诱导分化第4 d时进行油红O染色。结果显示,si-NC组与pcDNA3.1组的脂滴大小和数目比较相似,且它们在510 nm处的吸光度值分别为0.263±0.033和0.268±0.029,差异不显著;与si-NC组相比,抑制lnc-RAP3表达的si-RAP3组细胞内脂滴大小与数目显著增加,吸光度值也显著升高(OD510 nm=0.323) (P<0.05) (图3A);相反,与空载pcDNA3.1组相比,过表达lnc-RAP3的pcDNA3.1-RAP3组细胞内脂滴大小与数目显著减少,吸光度值也显著下降(OD510 nm=0.213) (P<0.05) (图3B),说明lnc-RAP3参与3T3-L1前脂肪细胞的分化成熟,并抑制细胞内脂滴的集聚。图3
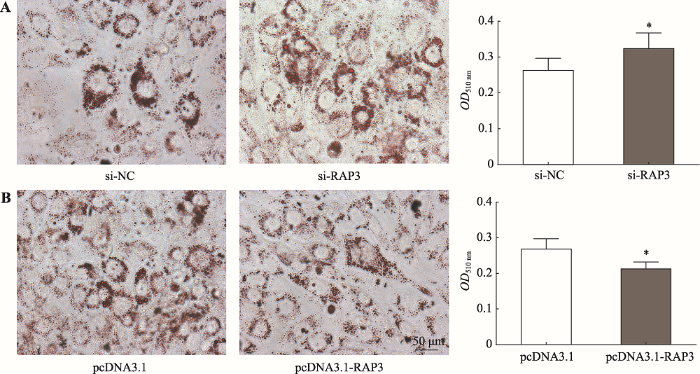 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3油红O染色及吸光度测定结果
A:抑制lnc-RAP3后经油红O染色(×400)及定量结果;B:过表达lnc-RAP3后经油红O染色(×400)及定量结果。*表示P<0.05,差异显著。
Fig. 3Results of Oil Red O staining and absorbance measure
2.4 脂肪分化相关标志基因的表达变化
脂肪分化相关基因C/EBPα、PPARγ、FAS、LPL和Glut4的相对表达量均以si-NC组诱导分化第0 d的对应表达量为对照(设为1),并对si-NC与si-RAP3、pcDNA3.1与pcDNA3.1-RAP3两两处理间各基因相对表达量进行差异显著性分析(图4),以及对每一基因每一处理在诱导分化第0 d、2 d、4 d等3个不同时间点间的相对表达量进行多重比较(表2)。从整体趋势看PPARγ、C/EBPα、FAS、LPL和Glut4等5个与成脂分化相关的基因在诱导分化第2 d的相对表达量均较诱导分化前(第0 d)有显著或极显著的增加,但过表达lnc-RAP3后FAS出现极显著下降(P<0.01),PPARγ出现显著下降(P<0.05)。在抑制lnc-RAP3表达的si-RAP3组中各基因的相对表达量均较si-NC组有一定程度升高(C/EBPα和FAS第4 d除外),在诱导分化第0 d时两组各基因相对表达量间均存在显著或极显著差异,但到诱导分化第2 d时C/EBPα和FAS两基因已无显著性差异,到诱导分化第4 d时除LPL还存在显著差异外,其他4个基因在两组间的相对表达量均无显著性差异。在过表达lnc- RAP3的pcDNA3.1-RAP3组中,除PPARγ基因的相对表达量在诱导分化第0 d略有升高但差异不显著外,其他各基因的相对表达量在3个不同时间点均较pcDNA3.1对照组显著或极显著降低,在诱导分化第2 d时PPARγ、FAS、LPL、Glut4和C/EBPα等基因在过表达lnc-RAP3的pcDNA3.1-RAP3组中的相对表达量较pcDNA3.1对照组分别降低了79.8%、71%、69.5%、39.2%和31.2%。这些结果说明lnc-RAP3能通过抑制成脂分化相关标志基因的表达而抑制3T3-L1前脂肪细胞的成脂分化。图4
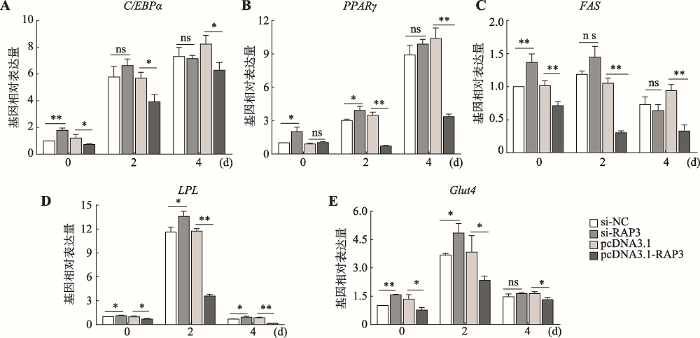 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4转染细胞中脂肪分化相关基因的表达
A~E:敲降/过表达lnc-RAP3后C/EBPα、PPARγ、FAS、LPL、Glut4的表达水平。*表示P<0.05,差异显著;**表示P<0.01,差异极显著;ns表示P>0.05,差异不显著。
Fig. 4The expression of adipose differentiation genes in transfected cells
Table 2
表2
表2 成脂分化相关标志基因mRNA相对表达量的均值及多重比较
Table 2
| 基因 | 分化时间 | si-NC | si-RAP3 | pcDNA3.1 | pcDNA3.1-RAP3 |
|---|---|---|---|---|---|
| PPARγ | 0 d | 1.00±0.00C | 2.01±0.39C | 0.90±0.07C | 1.02±0.08Ba |
| 2 d | 3.01±0.13B | 3.93±0.35B | 3.48±0.28B | 0.70±0.07Bb | |
| 4 d | 8.90±0.86A | 9.88±0.44A | 10.39±0.93A | 3.35±0.25A | |
| C/EBPα | 0 d | 1.00±0.00B | 1.79±0.16B | 1.21±0.25C | 0.73±0.04C |
| 2 d | 5.78±0.78Ab | 6.63±0.49Aa | 5.69±0.44B | 3.92±0.54B | |
| 4 d | 7.32±0.67Aa | 7.15±0.25Aa | 8.24±0.66A | 6.29±0.57A | |
| FAS | 0 d | 1.00±0.00Ab | 1.37±0.13Aa | 1.02±0.07Aa | 0.71±0.06A |
| 2 d | 1.19±0.05Aa | 1.45±0.16Aa | 1.05±0.08Aa | 0.30±0.03Ba | |
| 4 d | 0.73±0.12B | 0.64±0.09B | 0.94±0.09Aa | 0.33±0.09Ba | |
| LPL | 0 d | 1.00±0.00Ba | 1.12±0.06Ba | 0.97±0.11Ba | 0.69±0.08B |
| 2 d | 11.62±0.60A | 13.59±0.64A | 11.74±0.31A | 3.58±0.21A | |
| 4 d | 0.69±0.03Ba | 0.94±0.10Ba | 0.85±0.07Ba | 0.15±0.00C | |
| Glut4 | 0 d | 1.00±0.00C | 1.57±0.03Ba | 1.34±0.24Ba | 0.77±0.13C |
| 2 d | 3.68±0.08A | 4.85±0.50A | 3.84±0.87A | 2.34±0.22A | |
| 4 d | 1.47±0.15B | 1.64±0.03Ba | 1.63±0.10Ba | 1.31±0.10B |
新窗口打开|下载CSV
每组基因同列数据后所标字母相同表示差异不显著(P>0.05),字母不同表示差异显著(P<0.05,小写字母)或极显著(P<0.01,大写字母)。
3 讨 论
由于人们生活方式及饮食结构的改变,肥胖现象越来越普遍,与肥胖相关的疾病发病率也急剧升高。研究表明,肥胖及相关代谢疾病的发生与脂肪细胞的分化及调控失常密切相关,深入研究脂肪细胞分化过程的作用机制,对控制和治疗肥胖及相关代谢疾病具有重要意义。PPARγ和C/EBPα作为最主要的两个核心转录因子,在脂肪分化调控网络中起着不可或缺的协同作用[17]。FAS作为内源性脂肪酸合成的关键酶,在脂肪细胞分化过程中,其主要作用是促进胞内脂滴的形成和甘油三酯的积累,其表达调控主要受到PPARγ的激活[18, 19]。LPL作为脂肪沉积过程中的关键酶,在脂肪细胞分化过程中主要表达于终末分化早期,随细胞内甘油三酯含量的增加而逐渐上升,其表达也受PPARγ的调控[20]。此外,Glut4在脂肪和肌肉组织中具有较高表达,其表达主要受C/EBPα的调控[21]。研究表明,lncRNA可以靶向作用于脂肪细胞分化过程中相关通路的分子组件而影响下游转录因子的表达,或是直接作用于C/EBPs和PPARs等关键转录因子调控下游基因的表达,从而调节脂肪细胞分化。如linc-RAP1,也称Firre,能与核基质因子核糖核蛋白U (heterogeneous nuclear ribonucleoprotein U, hnRNP U)相互作用调节脂肪形成关键基因的表达[22];lncRNA SRA对脂肪生成也具有调控作用,它能与PPARγ结合,从而增加其转录活性,促进3T3-L1前脂肪细胞的分化[23, 24]。Xiao等[25]通过功能缺失实验发现敲降lncRNA ADINR导致人间充质干细胞内脂滴形成显著减少,成脂分化相关标志基因C/EBPα、PPARγ、FABP4和LPL的表达量也显著下降,进一步研究发现lncRNA ADINR可通过招募染色体修饰复合物MLL3/4调节C/EBPα基因启动子区域的H3K4me3修饰,从而转录激活C/EBPα正向调节脂肪形成。
本研究为探讨lnc-RAP3对3T3-L1前脂肪细胞分化的影响,通过比较诱导分化前后细胞内lnc- RAP3的相对表达量发现,诱导分化第2 d细胞内lnc-RAP3的表达量较诱导分化前显著增加,这与Sun等[16]的研究结果相一致。采用干扰细胞内lnc-RAP3的表达和构建lnc-RAP3的真核过表达载体并转染3T3-L1前脂肪细胞的方法,比较不同处理组间细胞内脂滴集聚情况、成脂分化相关标志基因PPARγ、C/EBPα、FAS、LPL和Glut4等的相对表达量差异发现lnc-RAP3可抑制3T3-L1前脂肪细胞的成脂分化,这与Sun等[16]所报道结果相反,究其原因,可能存在以下几方面的影响:(1)本研究所用细胞为3T3-L1前脂肪细胞系,细胞单一,而Sun等所用细胞为小鼠皮下脂肪来源的原代分离培养细胞,所分离细胞里可能混杂部分脂肪细胞和其他类型细胞如成纤维细胞等,可能对实验结果产生一定影响;(2)本研究采用对lnc-RAP3敲降和过表达相结合的方式均发现lnc-RAP3可抑制前脂肪细胞的成脂分化(均持续转染48 h),而Sun等只进行了敲降试验,且转染siRNA只持续了24 h,并且本研究中发现Sun等所用3条siRNA对lnc-RAP3的抑制效率均低于50%。同时,本研究所用si-RAP3在分化第0 d时(转染2 d后)其抑制效率为52.5%,分化第2 d时(转染4 d后)抑制效率为50.3%,到分化第4 d时(转染6 d后)抑制效率出现急剧下降,仅有18.4%,这可能与siRNA作用的时效性有关;(3) 3T3-L1前脂肪细胞在含胰岛素、3-异丁基-1-甲基黄嘌呤和地塞米松的诱导分化培养液的诱导刺激下,PPARγ、C/EBPα、FAS、LPL和Glut4等成脂分化相关标志基因的表达量本应上升[26,27,28,29],但本研究中过表达lnc-RAP3后脂肪酸合成酶基因FAS的表达量较诱导分化前极显著下降了57.7%,PPARγ显著下降了31.4%,进一步说明lnc-RAP3可通过抑制FAS和PPARγ等成脂分化相关基因的表达而抑制3T3-L1前脂肪细胞的成脂分化。此外,本研究结果显示lnc-RAP3的表达量在诱导分化后先升高后降低,猜测lnc-RAP3可能属于阶段性表达基因,即随着3T3-L1前脂肪细胞诱导分化,细胞会发生一系列改变与适应。而lnc-RAP3可能扮演着拮抗者的角色,在细胞内刚开始成脂分化时,为了维系细胞稳态而出现高表达,当细胞的成脂分化已不可逆时,其表达量又自然回落,但lnc-RAP3在脂肪生成中的具体作用机制还有待进一步研究。
本研究结果表明lnc-RAP3可通过抑制FAS和PPARγ等成脂分化相关基因的表达而抑制3T3-L1前脂肪细胞的成脂分化,为进一步探讨lncRNA对脂肪细胞分化的分子机制提供了理论依据。
(责任编委: 宋旭)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:21717813 [本文引用: 1]

Abstract The metabolic syndrome is a clinically important but often misunderstood topic. A concept in evolution, controversy surrounds the metabolic syndrome. The medical literature documents disagreement in the criteria for diagnosis, a shift in emphasis as to the roles of obesity and insulin resistance as underlying causes, and questions about the fundamental clinical utility of the syndrome. The practical strengths and limitations of the metabolic syndrome in clinical practice are reviewed.
URLPMID:2360167 [本文引用: 1]

We performed a meta-analysis of studies of the association between excess body weight and risk of gallbladder cancer identified from MEDLINE and EMBASE databases from 1966 to February 2007 and the references of retrieved articles. A random-effects model was used to combine results from eight cohort studies and three case-control studies, with a total of 3288 cases. Compared with individuals of 'normal weight', the summary relative risk of gallbladder cancer for those who were overweight or obese was 1.15 (95% CI, 1.01-1.30) and 1.66 (95% CI, 1.47-1.88) respectively. The association with obesity was stronger for women (relative risk, 1.88; 95% CI, 1.66-2.13) than for men (relative risk, 1.35; 95% CI, 1.09-1.68). There was no statistically significant heterogeneity among the results of individual studies. This meta-analysis confirms the association between excess body weight and risk of gallbladder cancer.
URL [本文引用: 1]
URLPMID:24290031 [本文引用: 1]

Large-scale mapping of transcriptomes has revealed significant levels of transcriptional activity within both unannotated and annotated regions of the genome. Interestingly, many of the novel transcripts demonstrate tissue-specific expression and some level of sequence conservation across species, but most have low protein-coding potential. Here, we describe progress in identifying and characterizing long noncoding RNAs (lncRNAs) and review how these transcripts interact with other biological molecules to regulate diverse cellular processes. We also preview emerging techniques that will help advance the discovery and characterization of novel transcripts. Finally, we discuss the role of lncRNAs in disease and therapeutics.
URL [本文引用: 1]

肿瘤转移是影响肿瘤患者预后的重要因素,但其作用机制复杂,涉及多个生物学过程。长链非编码RNA(Lnc RNA)作为人类基因组重要的调控分子,可以通过多种作用机制在肿瘤的转移过程中发挥重要的调控功能。并且Lnc RNA的异常表达与肿瘤患者临床因素密切相关,有望成为新型肿瘤标志物在临床治疗中发挥重要的作用。文章主要综述Lnc RNA在肿瘤转移中的分子作用机制及相关临床应用。
URL [本文引用: 1]

肿瘤转移是影响肿瘤患者预后的重要因素,但其作用机制复杂,涉及多个生物学过程。长链非编码RNA(Lnc RNA)作为人类基因组重要的调控分子,可以通过多种作用机制在肿瘤的转移过程中发挥重要的调控功能。并且Lnc RNA的异常表达与肿瘤患者临床因素密切相关,有望成为新型肿瘤标志物在临床治疗中发挥重要的作用。文章主要综述Lnc RNA在肿瘤转移中的分子作用机制及相关临床应用。
URLPMID:27338628 [本文引用: 1]

Background: Long-non-coding RNAs (lncRNAs), RNA molecules longer than 200 nucleotides, have been involved in several biological processes and in a growing number of diseases, controlling gene transcription, pre-mRNA processing, the transport of mature mRNAs to specific cellular compartments, the regulation of mRNA stability, protein translation and turnover. The fundamental role of lncRNAs in central nervous system (CNS) is becoming increasingly evident. LncRNAs are abundantly expressed in mammalian CNS in a specific spatio-temporal manner allowing a quick response to environmental/molecular changes. <br/> Methods: This article reviews the biology and mechanisms of action of lncRNAs underlying their potential role in CNS and in some neurodegenerative diseases. <br/> Results: an increasing number of studies report on lncRNAs involvement in different molecular mechanisms of gene expression modulation in CNS, from neural stem cell differentiation mainly by chromatin remodeling, to control of neuronal activities. More recently, lncRNAs have been implicated in neurodegenerative diseases, including Alzheimer090005s Disease, where the role of BACE1-AS lncRNA has been widely defined. BACE1-AS levels are up-regulated in AD brains where BACE1-AS acts by stabilizing BACE1 mRNA thereby increasing BACE1 protein content and A020542 formation. In Frontotemporal dementia and Amyotrophic lateral sclerosis the lncRNAs NEAT1_2 and MALAT1 co-localize at nuclear paraspeckles with TDP-43 and FUS proteins and their binding to TDP-43 is markedly increased in affected brains. In Parkinson090005s Disease the lncRNA UCHL1-AS1 acts by directly promoting translation of UCHL1 protein leading to perturbation of the ubiquitin-proteasome system. Different lncRNAs, such as HTT-AS, BDNF-AS and HAR1, were found to be dysregulated in their expression also in Huntington090005s Disease. In Fragile X syndrome (FXS) and Fragile X tremor/ataxia syndrome (FXTAS) patients, the presence of CGG repeats expansion alters the expression of the lncRNAs FMR1-AS1 and FMR6. Interestingly, they are expressed in peripheral blood leukocytes, suggesting these lncRNAs may represent biomarkers for FXS/FXTAS early detection and therapy. Finally, the identification of the antisense RNAs SCAANT1-AS and ATXN8OS in spinocerebellar ataxia 7 and 8, respectively, suggests that very different mechanisms of action driven by lncRNAs may trigger neurodegeneration in these disorders. <br/> Conclusion: The emerging role of lncRNAs in neurodegenerative diseases suggests that their dysregulation could trigger neuronal death via still unexplored RNA-based regulatory mechanisms which deserve further investigation. The evaluation of their diagnostic significance and therapeutic potential could also address the setting up of novel treatments in diseases where no cure is available to date.
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:26459763 [本文引用: 1]

More than 40% of the U.S. population are clinically obese and suffer from metabolic syndrome with an increased risk of postmenopausal estrogen receptor-positive breast cancer. Adipocytes are the primary component of adipose tissue and are formed through adipogenesis from precursor mesenchymal stem cells. While the major molecular pathways of adipogenesis are understood, little is known about the noncoding RNA signaling networks involved in adipogenesis. Using adipocyte-derived stem cells (ADSCs) isolated from wild-type and microRNA 140 (miR-140) knockout mice, we identify a novel miR-140/long noncoding RNA (lncRNA) NEAT1 signaling network necessary for adipogenesis. miR-140 knockout ADSCs have dramatically decreased adipogenic capabilities associated with downregulation of NEAT1 expression. We identified a miR-140 binding site in NEAT1 and found that mature miR-140 in the nucleus can physically interact with NEAT1, leading to increased NEAT1 expression. We demonstrated that reexpression of NEAT1 in miR-140 knockout ADSCs is sufficient to restore their ability to undergo differentiation. Our results reveal an exciting new noncoding RNA signaling network that regulates adipogenesis and that is a potential new target in the prevention or treatment of obesity.
URL [本文引用: 3]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
URLPMID:24463464 [本文引用: 1]

Abstract RNA, including long noncoding RNA (lncRNA), is known to be an abundant and important structural component of the nuclear matrix. However, the molecular identities, functional roles and localization dynamics of lncRNAs that influence nuclear architecture remain poorly understood. Here, we describe one lncRNA, Firre, that interacts with the nuclear-matrix factor hnRNPU through a 156-bp repeating sequence and localizes across an ~5-Mb domain on the X chromosome. We further observed Firre localization across five distinct trans-chromosomal loci, which reside in spatial proximity to the Firre genomic locus on the X chromosome. Both genetic deletion of the Firre locus and knockdown of hnRNPU resulted in loss of colocalization of these trans-chromosomal interacting loci. Thus, our data suggest a model in which lncRNAs such as Firre can interface with and modulate nuclear architecture across chromosomes.
URLPMID:10199399 [本文引用: 1]

Nuclear receptors play critical roles in the regulation of eukaryotic gene expression. We report the isolation and functional characterization of a novel transcriptional coactivator, termed steroid receptor RNA activator (SRA). SRA is selective for steroid hormone receptors and mediates transactivation via their amino-terminal activation function. We provide functional and mechanistic evidence that SRA acts as an RNA transcript; transfected SRA, unlike other steroid receptor coregulators, functions in the presence of cycloheximide, and SRA mutants containing multiple translational stop signals retain their ability to activate steroid receptor-dependent gene expression. Biochemical fractionation shows that SRA exists in distinct ribonucleoprotein complexes, one of which contains the nuclear receptor coactivator steroid receptor coactivator 1. We suggest that SRA may act to confer functional specificity upon multiprotein complexes recruited by liganded receptors during transcriptional activation.
URLPMID:21152033 [本文引用: 1]

Peroxisome proliferator-activated receptor-γ (PPARγ) is a master transcriptional regulator of adipogenesis. Hence, the identification of PPARγ coactivators should help reveal mechanisms controlling gene expression in adipose tissue development and physiology. We show that the non-coding RNA, Steroid receptor RNA Activator (SRA), associates with PPARγ and coactivates PPARγ-dependent reporter gene expression. Overexpression of SRA in ST2 mesenchymal precursor cells promotes their differentiation into adipocytes. Conversely, knockdown of endogenous SRA inhibits 3T3-L1 preadipocyte differentiation. Microarray analysis reveals hundreds of SRA-responsive genes in adipocytes, including genes involved in the cell cycle, and insulin and TNFα signaling pathways. Some functions of SRA may involve mechanisms other than coactivation of PPARγ. SRA in adipocytes increases both glucose uptake and phosphorylation of Akt and FOXO1 in response to insulin. SRA promotes S-phase entry during mitotic clonal expansion, decreases expression of the cyclin-dependent kinase inhibitors p21Cip1 and p27Kip1, and increases phosphorylation of Cdk1/Cdc2. SRA also inhibits the expression of adipocyte-related inflammatory genes and TNFα-induced phosphorylation of c-Jun NH(2)-terminal kinase. In conclusion, SRA enhances adipogenesis and adipocyte function through multiple pathways.
URLPMID:4649137 [本文引用: 1]

Chen and colleagues show that the lncRNAADINRregulates the development of hMSCs along the adipogenic pathway through transcriptional regulation ofC/EBPαexpression by exerting its effect onC/EBPα’s promoter.
URL [本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
