 ,1,2,*, 王亚丽1,2, 陈煜东1,2
,1,2,*, 王亚丽1,2, 陈煜东1,2Transposon-derived Long Noncoding RNA in Plants
Yijun Wang ,1,2,*, Yali Wang1,2, Yudong Chen1,2
,1,2,*, Yali Wang1,2, Yudong Chen1,2通讯作者: *E-mail:wyj@yzu.edu.cn
责任编辑: 朱亚娜
收稿日期:2020-05-26接受日期:2020-09-3网络出版日期:2020-11-01
| 基金资助: |
Corresponding authors: *E-mail:wyj@yzu.edu.cn
Received:2020-05-26Accepted:2020-09-3Online:2020-11-01

摘要
转座子是基因组的重要组分, 影响基因组的结构与稳定。长链非编码RNA (lncRNA)在转录及转录后水平调控多个生物学过程。转座子与lncRNA是物种进化的重要驱动力。含有转座子序列的lncRNA在自然界广泛存在。该文对植物lncRNA的发掘策略和功能研究进行概述, 围绕植物转座子来源lncRNA (TE-lncRNA)的分布和功能展开综述, 并对植物TE-lncRNA的调控机制、表观修饰及育种潜势等进行探讨与展望。
关键词:
Abstract
Transposable element (TE), core component of the genome, influences genome structure and stability. Long noncoding RNA (lncRNA) modulates diverse biological events at transcriptional and post-transcriptional levels. TE and lncRNA are major driving forces of evolution. Emerging evidence has revealed the wide distribution of lncRNA that harbors TE. In this review, we first briefly introduce the methodologies of identification and functional analysis of plant lncRNA. We focus on the distribution and function of transposable element-derived lncRNA (TE-lncRNA). Finally, we discuss the regulatory mechanism, epigenetic modification, and breeding potential of TE-lncRNA in plants.
Keywords:
PDF (770KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
引用本文
王益军, 王亚丽, 陈煜东. 转座子来源的植物长链非编码RNA. 植物学报, 2020, 55(6): 768-776 doi:10.11983/CBB20098
Wang Yijun, Wang Yali, Chen Yudong.

转座子是基因组中可移动的元件。根据不同的转座机制, 转座子可分为逆转座子(Class I)和DNA转座子(Class II) (Feschotte et al., 2002)。逆转座子通过RNA中介物, 在逆转座酶作用下进行转座。逆转座子主要有长末端重复序列(long terminal repeat, LTR)逆转座子和非LTR逆转座子。LTR逆转座子主要包括Copia和Gypsy类。非LTR逆转座子包括长散布重复元件(long interspersed nuclear element, LINE)和短散布重复元件(short interspersed nuclear element, SINE)。DNA转座子是以DNA到DNA的方式在原位置解离, 与新位置整合, 包括Ac/Ds、Spm/dSpm和Mutator转座子等(Dooner and Belachew, 1991; Bunkers et al., 1993; Cong and Li, 2020)。Helitron转座子是一种特殊类型的DNA转座子, 该转座子不具有末端反向重复序列, 也不产生靶点重复序列。Helitron以滚环复制的方式转座(Kapitonov and Jurka, 2001)。
RNA聚合酶II (Pol II)转录产生2种类型的RNA: 编码RNA与非编码RNA (noncoding RNA, ncRNA)。ncRNA也可以由RNA聚合酶I、III、IV、V (Pol I、III、IV、V)转录产生(Ulitsky, 2018)。根据成熟产物的大小, ncRNA分为非编码小RNA (small noncoding RNA, sRNA)和长链非编码RNA (long noncoding RNA, lncRNA)。sRNA长度一般为20-24 nt。不同前体产生不同的sRNA。单链前体产生microRNA (miRNA)和非miRNA。双链和小干扰RNA (small interfering RNA, siRNA)前体产生次级siRNA和异源siRNA等(Axtell, 2013)。lncRNA的长度大于200 nt, 真核生物启动子、增强子和基因间等区域转录可以产生lncRNA。启动子上游区域、增强子、基因间区域及编码蛋白基因转录分别产生启动子上游转录本(promoter upstream transcript, PROMPT)、增强子RNA (enhancer RNA, eRNA)、基因间lncRNA (long intergenic noncoding RNA, lincRNA)和天然反义转录本(natural antisense transcript, NAT) (Wu et al., 2017)。
转座子和lncRNA皆为基因组中重要的调控元件。已有研究表明, 含有转座子序列的lncRNA在自然界广泛存在, 称为转座子来源的lncRNA (transposable element-derived lncRNA, TE-lncRNA) (Wang et al., 2017; Golicz et al., 2018)。在水稻(Oryza sativa)、玉米(Zea mays)和棉花(Gossypium raimondii)等植物中, TE-lncRNA主要来源于逆转座子序列(Wang et al., 2017; Zhao et al., 2018a; Lv et al., 2019)。与来源于编码蛋白基因反义转录的lncRNA相比, 植物TE-lncRNA的功能研究报道较少。拟南芥(Arabidopsis thaliana)和棉花TE-lncRNA的研究表明, TE-lncRNA调控株高和非生物胁迫应答等(Wang et al., 2016, 2017; Zhao et al., 2018a)。重要粮食作物(如水稻、小麦(Triticum aestivum)和玉米)中TE- lncRNA的功能研究报道较少。鉴于TE-lncRNA在植物中广泛分布, 亟待开展TE-lncRNA调控作物株型、产量、品质和抗性等重要性状的机制研究。本文综述了植物TE-lncRNA研究概况, 重点介绍TE-lncRNA在植物中的分布与功能, 并对植物TE-lncRNA的基础研究与应用潜势进行了展望。
1 植物基因组转座子构成与物种进化
转座子是基因组的重要组分, 影响基因组的稳定与进化。以基因组较小的物种拟南芥(~125 Mb)与基因组较大的物种玉米(~2.3 Gb)为例, 介绍植物基因组中转座子分布的特点以及转座子构成在基因组进化中的作用。至少10%的拟南芥基因组序列由转座子构成, 其中逆转座子、DNA转座子以及其它类型转座子的百分比分别约为38%、39%和23% (图1A)。在全部转座子中, LTR逆转座子约占28% (The Arabidopsis Genome Initiative, 2000)。基因组较大的物种玉米, 约85%的基因组为转座子所占据。在全部转座子中, 逆转座子和DNA转座子的百分比分别约为89%和11%。LTR逆转座子占全部转座子的86% (图1B) (Schnable et al., 2009)。通过分析拟南芥和玉米等物种转座子的组成, 发现不同植物中存在特有的转座子类型。转座子的作用方式也存在一定差异。逆转座子, 尤其是LTR逆转座子的“复制-粘贴”式扩增导致一些物种基因组大小增加。图1
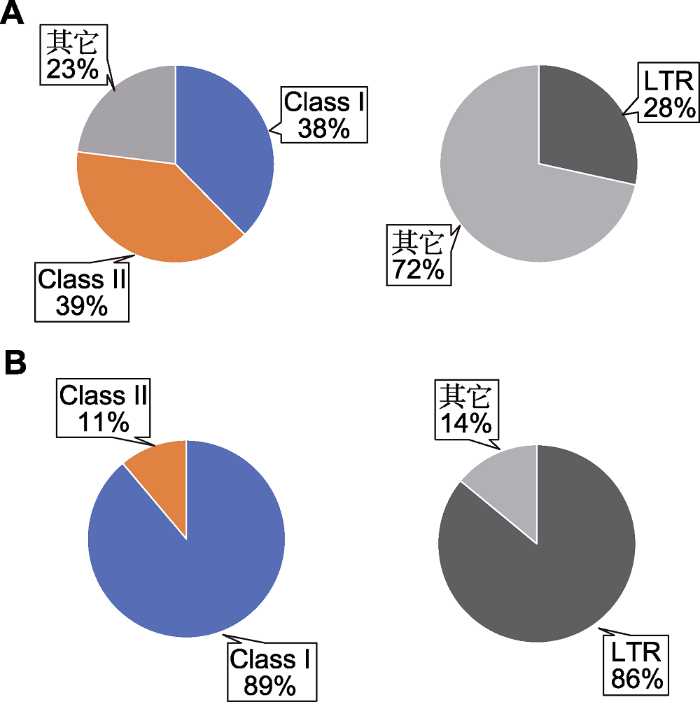 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1拟南芥和玉米基因组中转座子的分布(数据来源: The Arabidopsis Genome Initiative, 2000; Schnable et al., 2009)
(A) 拟南芥基因组中转座子分布(左图)和拟南芥长末端重复序列(LTR)逆转座子占全部转座子的比例(右图); (B) 玉米基因组中转座子分布(左图)和玉米LTR逆转座子占全部转座子的比例(右图)。Class I: 逆转座子; Class II: DNA转座子
Figure 1Transposon composition in the genomes of Arabidopsis and maize (data source: The Arabidopsis Genome Initiative, 2000; Schnable et al., 2009)
(A) Transposon composition in Arabidopsis genome (left) and the proportion of Arabidopsis long terminal repeat (LTR) retrotransposons (right); (B) Transposon composition in maize genome (left) and the proportion of maize LTR retrotransposons (right). Class I: Retrotransposons; Class II: DNA trotransposons
转座子不仅影响基因组大小, 而且影响基因组功能。例如, 通过分析玉米z1C1位点的单倍型, 发现Helitron转座子携带胞苷脱氨酶基因ZmCDA3 (Xu and Messing, 2006)。Copia类LTR逆转座子插入MADS-box转录因子基因J2 (JOINTLESS2), 导致番茄(Solanum lycopersicum)果柄离区消失(Alonge et al., 2020)。可见, 转座子影响基因的结构与功能, 是物种进化的重要驱动力。
2 植物lncRNA发掘
2.1 链特异性转录组测序发掘植物lncRNA
构建链特异性RNA文库并进行转录组测序(strand- specific RNA sequencing, ssRNA-seq), 对lncRNA进行高通量发掘。与普通RNA-seq相比, ssRNA-seq可以区分reads来源于正义链还是反义链, 在转录本定量时可以排除互补链转录本表达的干扰, 使表达定量更精确。此外, ssRNA-seq有助于确定lincRNA的转录方向(Zhao et al., 2015)。采用ssRNA-seq, 已经对拟南芥、水稻和玉米的lncRNA进行了分离。细菌表面翻译延伸因子Tu (translation elongation factor Tu, EF-Tu)被受体识别结合, 激活免疫反应。elf18是EF-Tu的保守肽段(Yang et al., 2020)。用elf18处理拟南芥幼苗, 通过ssRNA- seq发掘elf18诱导的lncRNA, 进而解析植物先天免疫的调控机制(Seo et al., 2017)。通过构建正常生长、干旱、低温和ABA处理等胁迫条件下的拟南芥全细胞、细胞核、细胞质组分的去核糖体、poly(A)和非poly(A)文库, 采用ssRNA-seq共发掘到6 510个拟南芥lncRNA (Zhao et al., 2018b)。选取白叶枯病菌侵染不同时长的水稻叶片, 采用ssRNA-seq检测到567个白叶枯病菌响应的lncRNA (Yu et al., 2020)。对亚洲栽培稻(O. sativa)和普通野生稻(O. rufipogon)发育早期的幼穗进行ssRNA-seq, 发掘到3 363个lncRNA, 包括2 517个lincRNA、200个incRNA (intronic noncoding RNA)和646个NAT-lncRNA。分析亚洲栽培稻和普通野生稻lncRNA序列和表达变异, 有助于揭示稻属驯化进程中lncRNA的选择模式(Zheng et al., 2019)。通过去核糖体文库的ssRNA-seq, 发掘到玉米磷缺乏诱导和赤霉素响应的lncRNA (Du et al., 2018; Wang et al., 2018b)。随着高通量测序技术的发展, 通过ssRNA-seq已经发掘到大量的植物lncRNA。基于发掘的lncRNA进行功能验证和调控机制解析是植物lncRNA研究领域的重要挑战。
2.2 生物信息学分析发掘植物lncRNA
NCBI和EBI等数据库中已有海量的基因表达数据, 包括表达序列标签(expressed sequence tag, EST)、tiling array和转录组数据等。基于数据库中的基因表达数据, 可以通过生物信息学方法发掘lncRNA。采用RepTAS (reproducibility-based tiling array analysis strategy)等方法, 整合拟南芥EST、cDNA、tiling array和基因组数据信息, 发掘到6 480个lincRNA (Liu et al., 2012)。通过分析EST和全长cDNA序列数据库, 发掘到水稻中的lncRNA (Liu et al., 2018)。整合数据库中的全基因组测序、转录组测序、含有全长cDNA的EST数据等信息, 发掘到20 163个玉米lncRNA (Li et al., 2014)。随着多种组学的发展, 有些植物中已有丰富的公共数据可供lncRNA发掘。数据库中的数据质量直接影响到lncRNA发掘的可靠性。lncRNA发掘所用软件种类和阈值设定也会影响lncRNA的发掘结果。此外, 植物lncRNA具有序列保守性较低、丰度低以及组织特异性表达等特点(Huang et al., 2018), 对通过整合不同来源数据进行lncRNA发掘提出了新的挑战。在通过生物学信息学发掘植物lncRNA的过程中, 控制数据质量、权衡阈值设定、采集多维数据, 将有助于获得较为全面且可靠的植物lncRNA信息。
3 植物lncRNA的功能
基于ssRNA-seq和生物信息学分析等方法, 已发掘到大量的植物lncRNA, 但功能及其调控机制得以解析的lncRNA较少(表1)。植物lncRNA调控生长发育、生物/非生物逆境应答和养分平衡等多个生物学过程。Table 1
表1
表1植物长链非编码RNA (lncRNA)功能
Table 1
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | Heo and Sung, 2011 |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | Swiezewski et al., 2009 | |
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | Qin et al., 2017 | |
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | Seo et al., 2017 | |
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | Zhao et al., 2018b | |
| T5120 | NO3-同化 | Liu et al., 2019 | |
| TE-lincRNA11195* | ABA响应 | Wang et al., 2017 | |
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | Yu et al., 2020 |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | Fang et al., 2019 | |
| LAIR (LRK Antisense Intergenic RNA) | 产量 | Wang et al., 2018a | |
| TL (TWISTED LEAF) | 叶片发育 | Liu et al., 2018 | |
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | Du et al., 2018 |
| 棉花 | XLOC_409583* | 苗高 | Zhao et al., 2018a |
| 番茄 | lncRNA-314* | 果实成熟 | Wang et al., 2016 |
| lncRNA16397 | 晚疫病抗性 | Cui et al., 2017 | |
| lncRNA39026 | 晚疫病抗性 | Hou et al., 2020 |
新窗口打开|下载CSV
3.1 植物lncRNA调控生长发育
拟南芥lncRNA功能研究的开创性工作集中于调控FLC (FLOWERING LOCUS C)基因的lncRNA研究。FLC是春化途径关键基因, 调控拟南芥的成花转变。FLC受到多个lncRNA的调控。拟南芥COOLAIR (lncRNA cold induced long antisense intragenic RNA)是FLC的反义转录本, COOLAIR与polycomb复合体互作, 对FLC进行表观抑制(Swiezewski et al., 2009)。与COOLAIR类似, 来自内含子的拟南芥COLDAIR (lncRNA COLD ASSISTED INTRONIC NONCODING RNA)与polycomb复合体互作, 抑制FLC表达, 进而调控春化过程(Heo and Sung, 2011)。拟南芥MAF4 (MADS AFFECTING FLOWERING 4)反义转录产生lncRNA MAS。lncRNA MAS参与春化响应, 为MAF4转录激活所必需(Zhao et al., 2018b)。水稻lncRNA调控叶片发育、开花期和产量等。水稻R2R3-MYB转录因子基因OsMYB60反向转录产生TL (lncRNA TWISTED LEAF), TL调控水稻叶片发育(Liu et al., 2018)。Ef-cd (Early flowering-completely dominant)座位编码lncRNA, 该lncRNA来源于成花激活基因OsSOC1的反义转录。Ef-cd变异可以促进水稻提早开花而不影响产量(Fang et al., 2019; 张硕和吴昌银, 2019)。LRK (leucine-rich repeat receptor kinase)簇反向转录产生LAIR (lncRNA LRK Antisense Intergenic RNA), 过表达LAIR可提高水稻产量(Wang et al., 2018a)。3.2 植物lncRNA参与生物与非生物逆境应答
过表达拟南芥ELENA1 (lncRNA ELF18-INDUCED LONG-NONCODING RNA 1)可增强细菌性疫病菌抗性, ELENA1敲除对细菌性疫病菌敏感, 表明ELENA1正向调控细菌性疫病菌抗性。lncRNA ELENA1与MED19a (Mediator subunit 19a)互作, 通过影响PR1 (PATHOGENESIS-RELATED GENE 1)的表达调控先天免疫反应(Seo et al., 2017)。过表达ALEX1 (lncRNA An Leaf Expressed and Xoo-induced lncRNA 1)可激活茉莉酸途径, 增强水稻白叶枯病的抗性(Yu et al., 2020)。番茄SlGRX22基因反义转录产生lncRNA16397。过表达lncRNA16397诱导SlGRX表达, 进而降低ROS的积累, 减轻细胞膜损伤, 增强对番茄晚疫病的抗性(Cui et al., 2017)。番茄lncRNA39026含有miR168a的拟靶标。过表达lncRNA39026株系其miR168a表达量下降, miR168a靶标基因SlAGO1表达量上升。lncRNA39026可能通过“诱饵”策略抑制miR168a, 诱导病程相关蛋白基因表达, 进而调控番茄晚疫病抗性(Hou et al., 2020)。lncRNA的非生物胁迫响应研究主要集中在拟南芥lncRNA对干旱和盐胁迫应答机制的解析。拟南芥DRIR (DROUGHT INDUCED lncRNA)正向调控干旱和盐胁迫应答(Qin et al., 2017)。拟南芥lncRNA AtR8参与种子萌发过程中的盐胁迫响应(张楠等, 2020)。3.3 植物lncRNA调控养分平衡
植物lncRNA参与调控氮和磷等营养元素的平衡。拟南芥NLP (NIN-like protein)家族成员AtNLP7是氮响应的重要调控因子(Wang et al., 2020)。lncRNA T5120作用于AtNLP7的下游, 调控NO3-同化, 影响氮肥利用率(Liu et al., 2019)。与拟南芥和水稻lncRNA研究相比, 玉米中功能解析的lncRNA较少。miR399/PHO2 (PHOSPHATE2)模块调控磷缺乏响应。磷缺乏诱导的玉米PILNCR1 (lncRNA Pi-deficiency-induced long non-coding RNA 1)抑制miR399介导的PHO2剪切, PILNCR1与miR399协同调控玉米对低磷的适应性(Du et al., 2018)。4 转座子来源的植物lncRNA
4.1 转座子来源植物lncRNA的分布
调控机制已得到解析的植物lncRNA多来源于编码蛋白基因的反义转录(表1)。通过ssRNA-seq和生物信息学分析等方法, 在拟南芥、水稻、玉米、大豆(Glycine max)、棉花和番茄等植物中发掘到一些lncRNA并非来自编码蛋白基因的反义转录本, 而是来源于转座子序列, 这些lncRNA被称为转座子来源的lncRNA (transposable element-derived lncRNA, TE-lncRNA) (图2)。图2
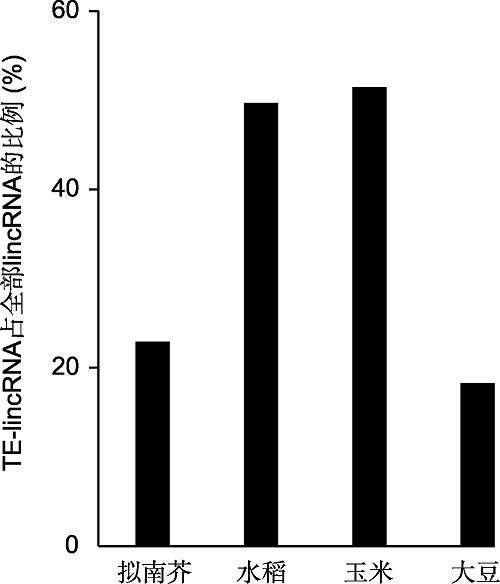 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2植物中转座子来源的基因间长链非编码RNA (TE-lincRNA)占全部lincRNA的比例(数据来源: Wang et al., 2017; Golicz et al., 2018)
Figure 2The proportion of plant transposable element-de- rived long intergenic noncoding RNA (TE-lincRNA) (data source: Wang et al., 2017; Golicz et al., 2018)
通过ssRNA-seq, 在拟南芥、水稻和玉米中分别鉴定到47、611和398个TE-lincRNA, 约占全部lncRNA的23%、50%和51% (Wang et al., 2017)。约18%的大豆lincRNA含有转座子序列, 来源于逆转座子的TE-lincRNA多于来源于DNA转座子的TE-lincRNA (Golicz et al., 2018)。基于PacBio fl-cDNA和转录组等数据, 发掘玉米全基因组的lncRNA。结果表明, 约65%的lncRNA含有转座子序列, 属于TE-lncRNA。玉米TE-lncRNA含有的转座子序列主要来源于LTR逆转座子, LTR逆转座子来源的TE-lncRNA约占全部TE-lncRNA的86% (Lv et al., 2019)。整合ssRNA- seq、NCBI和EBI中的poly(A)转录组数据, 进行番茄lncRNA发掘。结果表明, 85%的番茄lncRNA含有转座子序列, 番茄基因组中存在LTR逆转座子来源的TE-lncRNA (Wang et al., 2016)。在棉花TE-lncRNA中, Gypsy类LTR逆转座子来源的TE-lncRNA占主要部分(Zhao et al., 2018a)。
TE-lncRNA的分布与基因组中转座子类型和数目相关。水稻、玉米、大豆、棉花和番茄中的TE-lncRNA主要来源于逆转座子(Wang et al., 2016, 2017; Golicz et al., 2018; Zhao et al., 2018a; Lv et al., 2019)。在这些物种中, 逆转座子数目的增加导致逆转座子来源的lncRNA在整个TE-lncRNA占优势地位。
4.2 转座子来源植物lncRNA的功能
植物中lncRNA的功能研究集中于编码蛋白基因反义转录产生的lncRNA, 转座子来源lncRNA的功能研究报道较少, 主要集中在拟南芥、番茄和棉花等植物。TE-lncRNA调控株高、果实成熟和非生物胁迫应答等生物学过程(表1)。棉花TE-lncRNA XLOC_409583来源于去甲基化的LINE类逆转座子。与对照相比, 病毒诱导TE-lncRNA XLOC_409583沉默的植株苗高增加, 表明TE- lncRNA XLOC_409583调控棉花株高(Zhao et al., 2018a)。lncRNA-314来源于LTR逆转座子, 在番茄果实中特异表达。lncRNA-314与ABC转运基因共表达。在番茄果实成熟突变体rin和ful1/ful2中, lncRNA-314与ABC转运基因的表达被抑制。LTR逆转座子来源的TE-lncRNA lncRNA-314调控番茄果实成熟(Wang et al., 2016)。高温、低温、盐害、干旱和ABA处理等非生物胁迫诱导TE-lncRNA表达(Wang et al., 2017; Lv et al., 2019)。与野生型相比, 来源于LTR逆转座子的拟南芥TE-lncRNA TE-lincRNA11195突变体根长和幼茎鲜重增加, 对ABA敏感性下降, 表明TE-lncRNA TE-lincRNA11195调控ABA响应。TE-lincRNA11195中转座子的LTR序列缺失导致ABA敏感性下降, 表明转座子的LTR序列为TE-lncRNA TE-lincRNA11195调控非生物胁迫应答所必需(Wang et al., 2017)。
在整个植物lncRNA中, TE-lncRNA是重要组分(图2)。例如, 玉米基因组中, 超过一半的lncRNA属于TE-lncRNA (Wang et al., 2017; Lv et al., 2019)。然而, 功能得以解析的植物TE-lncRNA却较少。开展植物TE-lncRNA功能研究不仅有助于丰富lncRNA调控的理论基础, 而且能进一步揭示lncRNA的起源与进化。
5 结语与展望
5.1 植物转座子与lncRNA的相互作用
转座子是生物体的重要组成部分, 影响基因组的结构与稳定(Kidwell and Lisch, 2000)。lncRNA可以在转录和转录后水平对基因表达进行调控(Wu et al., 2017)。在物种进化过程中, 转座子与lncRNA相互作用。一方面, 转座子影响植物lncRNA的产生。许多植物lncRNA含有转座子序列(图2) (Wang et al., 2016, 2017; Golicz et al., 2018; Zhao et al., 2018a; Lv et al., 2019)。由于非转座子来源的lncRNA比TE-lncRNA受到更强的选择压力, 因此在进化过程中, 植物lncRNA倾向于保留转座子序列(Khanduja et al., 2016; Zhao et al., 2018a)。此外, 植物转座子的表观修饰影响lncRNA的转录活性。例如, 棉花LINE类逆转座子去甲基化, 产生具有转录活性的TE-lncRNA (Zhao et al., 2018a)。TE-lncRNA中的转座子序列形成二级结构(与lncRNA功能相关) (Khanduja et al., 2016)。另一方面, lncRNA影响转座子活性。lncRNA可以作为small RNA前体, small RNA可导致转座子“沉默”(Kidwell and Lisch, 2000; Yoon et al., 2014)。植物转座子以及lncRNA的相互作用不仅增强了各自结构与功能的多样性, 也有助于驱动生物进化和提高适应性。5.2 植物TE-lncRNA调控机制解析
转基因验证是TE-lncRNA功能解析的重要内容。基于CRISPR-Cas的基因编辑技术已成为植物基因功能验证的重要方法(Liu et al., 2020)。植物TE-lncRNA呈现序列保守性低和表达丰度低等特点, 给基因编辑工作带来了挑战。TE-lncRNA的低序列保守性, 可能导致在转基因受体中存在序列变异, 包括插入、缺失和碱基改变等, 从而影响gRNA (guide RNA)设计、编辑事件检测以及转基因结果的可靠性等。TE-lncRNA的低丰度影响编辑效率。低丰度的植物TE-lncRNA要求多靶点敲除并增加转化事件的数量。基于功能验证解析lncRNA的调控机制, 是lncRNA研究的热点与难点。lncRNA多与靶标互作, 调控众多生物学过程。因此, 靶标发掘是lncRNA调控机制解析的关键。已报道调控机制的植物lncRNA多来源于已知编码蛋白基因的反义转录本, lncRNA调控机制的研究多围绕已知编码蛋白基因-已知编码蛋白基因反义转录形成的lncRNA这一模块展开。在该模块中, lncRNA互作靶标为其来源的已知编码蛋白基因(表1)。植物TE-lncRNA来源于转座子, 并非已知编码蛋白基因的转录本。可以通过RNA pull-down联合质谱分析来获得植物TE-lncRNA的靶标(Barnes and Kanhere, 2016)。发掘的植物TE-lncRNA靶标可通过RIP (RNA immunoprecipitation)进一步验证(Gagliardi and Matarazzo, 2016)。基于发掘的靶标, 围绕TE-lncRNA与靶标互作, 解析植物TE-lncRNA的调控机制。
5.3 植物TE-lncRNA表观修饰
转座元件的转座可能会产生突变, 负向的选择压力会限制转座活性, 但转座活性必须保持, 转座子进化就是这两种力量的一个动态平衡(Martin and Garfinkel, 2003)。植物中RNA依赖的DNA甲基化(RNA-directed DNA methylation, RdDM)是转座活性的重要调控方式(Deniz et al., 2019)。TE-lncRNA是来源于转座子的lncRNA, TE-lncRNA中转座子的DNA甲基化程度如何? DNA甲基化与TE-lncRNA活性的相关性如何? 通过比较棉花F1杂交种中全基因组转座子以及lncRNA来源转座子的DNA甲基化水平, 结果表明棉花F1杂交种中, 具有活性的TE-lncRNA主要来自去甲基化的转座子区域, 并且以去甲基化的LINE类逆转座子转录产生为主。此外, 棉花F1中DNA甲基化程度与TE- lncRNA的表达呈负相关(Zhao et al., 2018a)。可见, 转座元件的表观修饰影响植物TE-lncRNA的产生与功能。除了DNA甲基化, siRNA介导的共抑制和组蛋白H3赖氨酸9甲基化(histone H3 lysine-9 methylation, H3K9me)等表观修饰也可以调控植物转座子活性(Lippman et al., 2003)。RNA抑制和组蛋白修饰等表观调控对植物TE-lncRNA的产生、活性以及功能的影响, 尚待进一步研究。5.4 植物TE-lncRNA的应用潜势
TE-lncRNA在植物中广泛存在(图2) (Wang et al., 2016, 2017; Golicz et al., 2018; Zhao et al., 2018a; Lv et al., 2019)。然而, 功能得以解析的植物TE-lncRNA较少。拟南芥、棉花和番茄TE-lncRNA分别调控ABA响应、苗高和果实成熟(表1)。不同植物以及全基因组范围功能性TE-lncRNA的数目有待系统研究。尤其是重要粮食作物, 如水稻、小麦和玉米中TE- lncRNA的功能研究亟待开展。已有研究表明, lncRNA影响作物产量、品质和抗性等重要性状(表1)。因此, 植物lncRNA具有育种应用的潜势。鉴于TE-lncRNA在植物中广泛存在, 开展TE-lncRNA调控株型、产量、品质和抗性等重要性状的机制研究, 发掘具有育种效用的TE-lncRNA, 具有重要的科学意义与应用价值。基于具有应用潜势的TE-lncRNA及其互作靶标, 通过单倍型发掘和基因编辑等策略, 寻找优良变异并进行材料创制, 有望为种质改良与品种选育等育种实践提供新的靶标和路径。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
URLPMID:32553272 [本文引用: 1]
URLPMID:23330790 [本文引用: 1]
[本文引用: 1]
URLPMID:8397136 [本文引用: 1]
DOI:10.16288/j.yczz.19-301URLPMID:32102770 [本文引用: 1]

Transposable elements (TEs) are fragments of DNA sequence, which can mobile from one locus to another within a genome, often replication in the process. Occupying the main component of the genome, TEs can affect the structure and function of gene and/or genome in a variety of ways, and play an important role in the evolution of the host. Based on the transposition intermediate, eukaryotic TEs can be divided into two classes. The Mutator superfamily is found in maize (Zea may L.) in the 1970s. As the member of class II elements, Mutator superfamily transposons are found in all eukaryote genomes and contain many families with clearly distinguishable genetic characteristics. In addition, these TEs transpose at high rates and preferentially insert in gene-rich and low-repetitive genomic regions leading to the rapid generation of massive novel mutations, therefore, they are in great use of both forward and reverse genetics researches. In this review, we summarize the classification, structure characteristic, transposition mechanism, insertion preference and TSD sequence and other autonomous MULEs in maize. Moreover, we discuss the problems faced in TEs' research and research directions in the future, with a view to discuss possible breakthroughs, future development directions and significant impacts with colleagues in the related research field..
URLPMID:27801966 [本文引用: 2]
DOI:10.1038/s41576-019-0106-6URLPMID:30867571 [本文引用: 1]

Maintenance of genome stability requires control over the expression of transposable elements (TEs), whose activity can have substantial deleterious effects on the host. Chemical modification of DNA is a commonly used strategy to achieve this, and it has long been argued that the emergence of 5-methylcytosine (5mC) in many species was driven by the requirement to silence TEs. Potential roles in TE regulation have also been suggested for other DNA modifications, such as N6-methyladenine and oxidation derivatives of 5mC, although the underlying mechanistic relationships are poorly understood. Here, we discuss current evidence implicating DNA modifications and DNA-modifying enzymes in TE regulation across different species.
URLPMID:1661257 [本文引用: 1]
URLPMID:29967097 [本文引用: 3]
URLPMID:31451662 [本文引用: 2]
DOI:10.1038/nrg793URLPMID:11988759 [本文引用: 1]

Transposable elements are the single largest component of the genetic material of most eukaryotes. The recent availability of large quantities of genomic sequence has led to a shift from the genetic characterization of single elements to genome-wide analysis of enormous transposable-element populations. Nowhere is this shift more evident than in plants, in which transposable elements were first discovered and where they are still actively reshaping genomes.
[本文引用: 1]
URLPMID:29284742 [本文引用: 7]
DOI:10.1126/science.1197349URL [本文引用: 2]

Vernalization is an environmentally-induced epigenetic switch in which winter cold triggers epigenetic silencing of floral repressors and thus provides competence to flower in spring. In Arabidopsis, winter cold triggers enrichment of tri-methylated histone H3 Lys(27) at chromatin of the floral repressor, FLOWERING LOCUS C (FLC), and results in epigenetically stable repression of FLC. This epigenetic change is mediated by an evolutionarily conserved repressive complex, polycomb repressive complex 2 (PRC2). Here, we show that a long intronic noncoding RNA [termed COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR)] is required for the vernalization-mediated epigenetic repression of FLC. COLDAIR physically associates with a component of PRC2 and targets PRC2 to FLC. Our results show that COLDAIR is required for establishing stable repressive chromatin at FLC through its interaction with PRC2.
DOI:10.1094/PHYTO-12-19-0445-RURLPMID:31876247 [本文引用: 2]

Our previous study has indicated that a long noncoding RNA (lncRNA), lncRNA39026, can be responsive to Phytophthora infestans infection. However, the function and regulation mechanism of lncRNA39026 during tomato resistance to P. infestans are unknown. In this study, an lncRNA39026 sequence was cloned from tomato Zaofen No. 2, and this sequence contained an endogenous target mimicry for miR168a, which might suppress the expression of miR168a. LncRNA39026 was strongly downregulated at 3 h in the tomato plants infected with P. infestans, and its expression level displayed a negative correlation with the expression level of miR168a and a positive correlation with the expression levels of SlAGO1 genes (target gene of miR168a) upon P. infestans infection. Tomato plants in which lncRNA39026 was overexpressed displayed enhanced resistance to P. infestans, decreased level of miR168a, and increased level of SlAGO1, whereas the resistance was impaired, level of miR168a was increased, and level of SlAGO1 was decreased after lncRNA39026 silencing. In addition, lncRNA39026 could also induce the expression of pathogenesis-related (PR) genes, as shown by increased and decreased expression levels of PR genes in tomato plants with overexpressed and silenced lncRNA39026, respectively. The result demonstrated that lncRNA39026 might function to decoy miR168a and affect the expression of PR genes in tomato plants, increasing resistance to disease.
DOI:10.1111/tpj.14016URLPMID:29975432 [本文引用: 1]

The importance of long non-coding RNAs (lncRNAs) in plant development has been established, but a systematic analysis of lncRNAs expressed during pollen development and fertilization has been elusive. We performed a time series of RNA-seq experiments at five developmental stages during pollen development and three different time points after pollination in Brassica rapa and identified 12 051 putative lncRNAs. A comprehensive view of dynamic lncRNA expression networks underpinning pollen development and fertilization was provided. B. rapa lncRNAs share many common characteristics of lncRNAs: relatively short length, low expression but specific in narrow time windows, and low evolutionary conservation. Gene modules and key lncRNAs regulating reproductive development such as exine formation were uncovered. Forty-seven cis-acting lncRNAs and 451 trans-acting lncRNAs were revealed to be highly coexpressed with their target protein-coding genes. Of particular importance are the discoveries of 14 lncRNAs that were highly coexpressed with 10 function-known pollen-associated coding genes. Fifteen lncRNAs were predicted as endogenous target mimics for 13 miRNAs, and two lncRNAs were proved to be functional target mimics for miR160 after experimental verification and shown to function in pollen development. Our study provides the systematic identification of lncRNAs during pollen development and fertilization in B. rapa and forms the foundation for future genetic, genomic, and evolutionary studies.
DOI:10.1073/pnas.151269298URLPMID:11447285 [本文引用: 1]

All eukaryotic DNA transposons reported so far belong to a single category of elements transposed by the so-called
DOI:10.1016/j.molcel.2016.06.011URLPMID:27392145 [本文引用: 2]

In modern molecular biology, RNA has emerged as a versatile macromolecule capable of mediating an astonishing number of biological functions beyond its role as a transient messenger of genetic information. The recent discovery and functional analyses of new classes of noncoding RNAs (ncRNAs) have revealed their widespread use in many pathways, including several in the nucleus. This Review focuses on the mechanisms by which nuclear ncRNAs directly contribute to the maintenance of genome stability. We discuss how ncRNAs inhibit spurious recombination among repetitive DNA elements, repress mobilization of transposable elements (TEs), template or bridge DNA double-strand breaks (DSBs) during repair, and direct developmentally regulated genome rearrangements in some ciliates. These studies reveal an unexpected repertoire of mechanisms by which ncRNAs contribute to genome stability and even potentially fuel evolution by acting as templates for genome modification.
DOI:10.1016/s0169-5347(99)01817-0URLPMID:10675923 [本文引用: 2]

Several recent reports have challenged the idea that transposable elements (TEs) are mainly 'selfish' or 'junk' DNA with little importance for host evolution. It has been proposed that TEs have the potential to provide host genomes with the ability to enhance their own evolution. They might also be a major source of genetic diversity, allowing response to environmental changes. Because the relationships between TEs and host genomes are highly variable, and because the selfish, junk and beneficial DNA hypotheses are by no means mutually exclusive, a single label for these relationships appears to be inappropriate and potentially misleading.
DOI:10.1186/gb-2014-15-2-r40URLPMID:24576388 [本文引用: 1]

BACKGROUND: Long non-coding RNAs (lncRNAs) are transcripts that are 200 bp or longer, do not encode proteins, and potentially play important roles in eukaryotic gene regulation. However, the number, characteristics and expression inheritance pattern of lncRNAs in maize are still largely unknown. RESULTS: By exploiting available public EST databases, maize whole genome sequence annotation and RNA-seq datasets from 30 different experiments, we identified 20,163 putative lncRNAs. Of these lncRNAs, more than 90% are predicted to be the precursors of small RNAs, while 1,704 are considered to be high-confidence lncRNAs. High confidence lncRNAs have an average transcript length of 463 bp and genes encoding them contain fewer exons than annotated genes. By analyzing the expression pattern of these lncRNAs in 13 distinct tissues and 105 maize recombinant inbred lines, we show that more than 50% of the high confidence lncRNAs are expressed in a tissue-specific manner, a result that is supported by epigenetic marks. Intriguingly, the inheritance of lncRNA expression patterns in 105 recombinant inbred lines reveals apparent transgressive segregation, and maize lncRNAs are less affected by cis- than by trans-genetic factors. CONCLUSIONS: We integrate all available transcriptomic datasets to identify a comprehensive set of maize lncRNAs, provide a unique annotation resource of the maize genome and a genome-wide characterization of maize lncRNAs, and explore the genetic control of their expression using expression quantitative trait locus mapping.
DOI:10.1371/journal.pbio.0000067URLPMID:14691539 [本文引用: 1]

Heritable, but reversible, changes in transposable element activity were first observed in maize by Barbara McClintock in the 1950s. More recently, transposon silencing has been associated with DNA methylation, histone H3 lysine-9 methylation (H3mK9), and RNA interference (RNAi). Using a genetic approach, we have investigated the role of these modifications in the epigenetic regulation and inheritance of six Arabidopsis transposons. Silencing of most of the transposons is relieved in DNA methyltransferase (met1), chromatin remodeling ATPase (ddm1), and histone modification (sil1) mutants. In contrast, only a small subset of the transposons require the H3mK9 methyltransferase KRYPTONITE, the RNAi gene ARGONAUTE1, and the CXG methyltransferase CHROMOMETHYLASE3. In crosses to wild-type plants, epigenetic inheritance of active transposons varied from mutant to mutant, indicating these genes differ in their ability to silence transposons. According to their pattern of transposon regulation, the mutants can be divided into two groups, which suggests that there are distinct, but interacting, complexes or pathways involved in transposon silencing. Furthermore, different transposons tend to be susceptible to different forms of epigenetic regulation.
URLPMID:31264223 [本文引用: 2]
DOI:10.1105/tpc.19.00934URLPMID:32102844 [本文引用: 1]

Maize (Zea mays) is one of the most important crops in the world. However, few agronomically important maize genes have been cloned and used for trait improvement, due to its complex genome and genetic architecture. Here, we integrated multiplexed CRISPR/Cas9-based high-throughput targeted mutagenesis with genetic mapping and genomic approaches to successfully target 743 candidate genes corresponding to traits relevant for agronomy and nutrition. After low-cost barcode-based deep sequencing, 412 edited sequences covering 118 genes were precisely identified from individuals showing clear phenotypic changes. The profiles of the associated gene-editing events were similar to those identified in human cell lines and consequently are predictable using an existing algorithm originally designed for human studies. We observed unexpected but frequent homology-directed repair through endogenous templates that was likely caused by spatial contact between distinct chromosomes. Based on the characterization and interpretation of gene function from several examples, we demonstrate that the integration of forward and reverse genetics via a targeted mutagenesis library promises rapid validation of important agronomic genes for crops with complex genomes. Beyond specific findings, this study also guides further optimization of high-throughput CRISPR experiments in plants.
DOI:10.1105/tpc.112.102855URLPMID:23136377 [本文引用: 1]

Long intergenic noncoding RNAs (lincRNAs) transcribed from intergenic regions of yeast and animal genomes play important roles in key biological processes. Yet, plant lincRNAs remain poorly characterized and how lincRNA biogenesis is regulated is unclear. Using a reproducibility-based bioinformatics strategy to analyze 200 Arabidopsis thaliana transcriptome data sets, we identified 13,230 intergenic transcripts of which 6480 can be classified as lincRNAs. Expression of 2708 lincRNAs was detected by RNA sequencing experiments. Transcriptome profiling by custom microarrays revealed that the majority of these lincRNAs are expressed at a level between those of mRNAs and precursors of miRNAs. A subset of lincRNA genes shows organ-specific expression, whereas others are responsive to biotic and/or abiotic stresses. Further analysis of transcriptome data in 11 mutants uncovered SERRATE, CAP BINDING PROTEIN20 (CBP20), and CBP80 as regulators of lincRNA expression and biogenesis. RT-PCR experiments confirmed these three proteins are also needed for splicing of a small group of intron-containing lincRNAs.
DOI:10.1111/nph.15023URLPMID:29411384 [本文引用: 3]

Natural antisense long noncoding RNAs (lncRNAs) are widespread in many organisms. However, their biological functions remain largely unknown, particularly in plants. We report the identification and characterization of an endogenous lncRNA, TWISTED LEAF (TL), which is transcribed from the opposite strand of the R2R3 MYB transcription factor gene locus, OsMYB60, in rice (Oryza sativa). TL and OsMYB60 were found to be coexpressed in many different tissues, and the expression level of TL was higher than that of OsMYB60. Downregulation of TL by RNA interference (RNAi) and overexpression of OsMYB60 resulted in twisted leaf blades in transgenic rice. The expression level of OsMYB60 was significantly increased in TL-RNAi transgenic plants. This suggests that TL may play a cis-regulatory role on OsMYB60 in leaf morphological development. We also determined that the antisense transcription suppressed the sense gene expression by mediating chromatin modifications. We further discovered that a C2H2 transcription factor, OsZFP7, is an OsMYB60 binding partner and involved in leaf development. Taken together, these findings reveal that the cis-natural antisense lncRNA plays a critical role in maintaining leaf blade flattening in rice. Our study uncovers a regulatory mechanism of lncRNA in plant leaf development.
DOI:10.1186/s12864-019-6245-5URLPMID:31729949 [本文引用: 7]

BACKGROUND: Several studies have mined short-read RNA sequencing datasets to identify long non-coding RNAs (lncRNAs), and others have focused on the function of individual lncRNAs in abiotic stress response. However, our understanding of the complement, function and origin of lncRNAs - and especially transposon derived lncRNAs (TE-lncRNAs) - in response to abiotic stress is still in its infancy. RESULTS: We utilized a dataset of 127 RNA sequencing samples that included total RNA datasets and PacBio fl-cDNA data to discover lncRNAs in maize. Overall, we identified 23,309 candidate lncRNAs from polyA+ and total RNA samples, with a strong discovery bias within total RNA. The majority (65%) of the 23,309 lncRNAs had sequence similarity to transposable elements (TEs). Most had similarity to long-terminal-repeat retrotransposons from the Copia and Gypsy superfamilies, reflecting a high proportion of these elements in the genome. However, DNA transposons were enriched for lncRNAs relative to their genomic representation by ~ 2-fold. By assessing the fraction of lncRNAs that respond to abiotic stresses like heat, cold, salt and drought, we identified 1077 differentially expressed lncRNA transcripts, including 509 TE-lncRNAs. In general, the expression of these lncRNAs was significantly correlated with their nearest gene. By inferring co-expression networks across our large dataset, we found that 39 lncRNAs are as major hubs in co-expression networks that respond to abiotic stress, and 18 appear to be derived from TEs. CONCLUSIONS: Our results show that lncRNAs are enriched in total RNA samples, that most (65%) are derived from TEs, that at least 1077 are differentially expressed during abiotic stress, and that 39 are hubs in co-expression networks, including a small number that are evolutionary conserved. These results suggest that lncRNAs, including TE-lncRNAs, may play key regulatory roles in moderating abiotic responses.
DOI:10.1186/gb-2003-4-4-313URLPMID:12702203 [本文引用: 1]

A report on the Keystone Symposium
DOI:10.1104/pp.17.00574URLPMID:28887353 [本文引用: 2]

Long noncoding RNAs (lncRNAs) affect gene expression through a wide range of mechanisms and are considered as important regulators in many essential biological processes. A large number of lncRNA transcripts have been predicted or identified in plants in recent years. However, the biological functions for most of them are still unknown. In this study, we identified an Arabidopsis (Arabidopsis thaliana) lncRNA, DROUGHT INDUCED lncRNA (DRIR), as a novel positive regulator of the plant response to drought and salt stress. DRIR was expressed at a low level under nonstress conditions but can be significantly activated by drought and salt stress as well as by abscisic acid (ABA) treatment. We identified a T-DNA insertion mutant, drir(D) , which had higher expression of the DRIR gene than the wild-type plants. The drir(D) mutant exhibits increased tolerance to drought and salt stress. Overexpressing DRIR in Arabidopsis also increased tolerance to drought and salt stress of the transgenic plants. The drir(D) mutant and the overexpressing seedlings are more sensitive to ABA than the wild type in stomata closure and seedling growth. Genome-wide transcriptome analysis demonstrated that the expression of a large number of genes was altered in drir(D) and the overexpressing plants. These include genes involved in ABA signaling, water transport, and other stress-relief processes. Our study reveals a mechanism whereby DRIR regulates the plant response to abiotic stress by modulating the expression of a series of genes involved in the stress response.
URLPMID:19965430 [本文引用: 3]
URLPMID:28400491 [本文引用: 3]
DOI:10.1038/nature08618URLPMID:20010688 [本文引用: 2]

Transcription in eukaryotic genomes generates an extensive array of non-protein-coding RNA, the functional significance of which is mostly unknown. We are investigating the link between non-coding RNA and chromatin regulation through analysis of FLC - a regulator of flowering time in Arabidopsis and a target of several chromatin pathways. Here we use an unbiased strategy to characterize non-coding transcripts of FLC and show that sense/antisense transcript levels correlate in a range of mutants and treatments, but change independently in cold-treated plants. Prolonged cold epigenetically silences FLC in a Polycomb-mediated process called vernalization. Our data indicate that upregulation of long non-coding antisense transcripts covering the entire FLC locus may be part of the cold-sensing mechanism. Induction of these antisense transcripts occurs earlier than, and is independent of, other vernalization markers and coincides with a reduction in sense transcription. We show that addition of the FLC antisense promoter sequences to a reporter gene is sufficient to confer cold-induced silencing of the reporter. Our data indicate that cold-induced FLC antisense transcripts have an early role in the epigenetic silencing of FLC, acting to silence FLC transcription transiently. Recruitment of the Polycomb machinery then confers the epigenetic memory. Antisense transcription events originating from 3' ends of genes might be a general mechanism to regulate the corresponding sense transcription in a condition/stage-dependent manner.
DOI:10.1038/35048692URLPMID:11130711 [本文引用: 3]

The flowering plant Arabidopsis thaliana is an important model system for identifying genes and determining their functions. Here we report the analysis of the genomic sequence of Arabidopsis. The sequenced regions cover 115.4 megabases of the 125-megabase genome and extend into centromeric regions. The evolution of Arabidopsis involved a whole-genome duplication, followed by subsequent gene loss and extensive local gene duplications, giving rise to a dynamic genome enriched by lateral gene transfer from a cyanobacterial-like ancestor of the plastid. The genome contains 25,498 genes encoding proteins from 11,000 families, similar to the functional diversity of Drosophila and Caenorhabditis elegans--the other sequenced multicellular eukaryotes. Arabidopsis has many families of new proteins but also lacks several common protein families, indicating that the sets of common proteins have undergone differential expansion and contraction in the three multicellular eukaryotes. This is the first complete genome sequence of a plant and provides the foundations for more comprehensive comparison of conserved processes in all eukaryotes, identifying a wide range of plant-specific gene functions and establishing rapid systematic ways to identify genes for crop improvement.
URLPMID:29749606 [本文引用: 1]
URLPMID:28106309 [本文引用: 13]
DOI:10.1111/nph.13718URLPMID:26494192 [本文引用: 7]

DOI:10.1038/s41467-018-05829-7URLPMID:30158538 [本文引用: 2]

Long noncoding RNAs (lncRNAs) are essential regulators of gene expression in eukaryotes. Despite increasing knowledge on the function of lncRNAs, little is known about their effects on crop yield. Here, we identify a lncRNA transcribed from the antisense strand of neighbouring gene LRK (leucine-rich repeat receptor kinase) cluster named LAIR (LRK Antisense Intergenic RNA). LAIR overexpression increases rice grain yield and upregulates the expression of several LRK genes. Additionally, chromatin immunoprecipitation assay results indicate H3K4me3 and H4K16ac are significantly enriched at the activated LRK1 genomic region. LAIR binds histone modification proteins OsMOF and OsWDR5 in rice cells, which are enriched in LRK1 gene region. Moreover, LAIR is demonstrated to bind 5' and 3' untranslated regions of LRK1 gene. Overall, this study reveals the role of lncRNA LAIR in regulating rice grain yield and lncRNAs may be useful targets for crop breeding.
DOI:10.1186/s12864-020-6769-8URLPMID:32393171 [本文引用: 1]

BACKGROUND: The fluctuation of nitrogen (N) contents profoundly affects the root growth and architecture in maize by altering the expression of thousands of genes. The differentially expressed genes (DEGs) in response to N have been extensively reported. However, information about the effects of N variation on the alternative splicing in genes is limited. RESULTS: To reveal the effects of N on the transcriptome comprehensively, we studied the N-starved roots of B73 in response to nitrate treatment, using a combination of short-read sequencing (RNA-seq) and long-read sequencing (PacBio-sequencing) techniques. Samples were collected before and 30 min after nitrate supply. RNA-seq analysis revealed that the DEGs in response to N treatment were mainly associated with N metabolism and signal transduction. In addition, we developed a workflow that utilizes the RNA-seq data to improve the quality of long reads, increasing the number of high-quality long reads to about 2.5 times. Using this workflow, we identified thousands of novel isoforms; most of them encoded the known functional domains and were supported by the RNA-seq data. Moreover, we found more than 1000 genes that experienced AS events specifically in the N-treated samples, most of them were not differentially expressed after nitrate supply-these genes mainly related to immunity, molecular modification, and transportation. Notably, we found a transcription factor ZmNLP6, a homolog of AtNLP7-a well-known regulator for N-response and root growth-generates several isoforms varied in capacities of activating downstream targets specifically after nitrate supply. We found that one of its isoforms has an increased ability to activate downstream genes. Overlaying DEGs and DAP-seq results revealed that many putative targets of ZmNLP6 are involved in regulating N metabolism, suggesting the involvement of ZmNLP6 in the N-response. CONCLUSIONS: Our study shows that many genes, including the transcription factor ZmNLP6, are involved in modulating early N-responses in maize through the mechanism of AS rather than altering the transcriptional abundance. Thus, AS plays an important role in maize to adapt N fluctuation.
DOI:10.1007/s11103-018-0788-8URLPMID:30341662 [本文引用: 1]

KEY MESSAGE: We report coding and long noncoding RNAs in maize upon phytohormone gibberellin stimulation. Plant hormone gibberellin (GA) orchestrates various facets of biological processes. Dissection the transcriptomic dynamics upon GA stimulation has biological significance. Feature of maize transcriptome in response to GA application remains largely elusive. Herein, two types of plants, one was with normal height, the other was GA-sensitive dwarfism, were selected from advanced backcross population for GA3 treatment with different concentrations. In control and GA3-treated plants, we identified a large number of coding and long noncoding RNAs (lncRNAs) through sequencing eight ribosomal-depleted RNA libraries. Transcripts encoding GA biosynthetic and metabolic enzymes KS, GA20ox, GA3ox, and GA2ox were significantly differentially expressed in GA3-treated samples. A total of 78 protein-coding transcripts were shared between GA3-treated normal height and dwarf plants. Shared transcripts encoding terpene synthase, MYB transcription factor, and receptor-like protein kinase were co-regulated with their corresponding partners. Out of identified lncRNAs, 22 and 34 significantly differentially expressed lncRNAs were responsive to GA application in normal height and dwarf plants, respectively. Shared GA-responsive lncRNAs were found in GA3-treated normal height and dwarf plants. Some lncRNAs corresponded to precursors of known miRNA, such as zma-miR528a and zma-miR528b. Multiple promising targets of significantly differentially expressed lncRNAs were discovered, including Lazy plant1 for auxin- and GA-mediated shoot gravitropism, bZIP transcription factor member for GA-controlled cell elongation. This study will improve our knowledge of GA-triggered transcriptome change and facilitate a comprehensive understanding of regulatory cascade centering on GA.
URLPMID:28629949 [本文引用: 2]
DOI:10.1186/1471-2156-7-52URLPMID:17094807 [本文引用: 1]

BACKGROUND: Genetic maps are based on recombination of orthologous gene sequences between different strains of the same species. Therefore, it was unexpected to find extensive non-collinearity of genes between different inbred strains of maize. Interestingly, disruption of gene collinearity can be caused among others by a rolling circle-type copy and paste mechanism facilitated by Helitrons. However, understanding the role of this type of gene amplification has been hampered by the lack of finding intact gene sequences within Helitrons. RESULTS: By aligning two haplotypes of the z1C1 locus of maize we found a Helitron that contains two genes, one encoding a putative cytidine deaminase and one a hypothetical protein with part of a 40S ribosomal protein. The cytidine deaminase gene, called ZmCDA3, has been copied from the ZmCDA1 gene on maize chromosome 7 about 4.5 million years ago (mya) after maize was formed by whole-genome duplication from two progenitors. Inbred lines contain gene copies of both progenitors, the ZmCDA1 and ZmCDA2 genes. Both genes diverged when the progenitors of maize split and are derived from the same progenitor as the rice OsCDA1 gene. The ZmCDA1 and ZmCDA2 genes are both transcribed in leaf and seed tissue, but transcripts of the paralogous ZmCDA3 gene have not been found yet. Based on their protein structure the maize CDA genes encode a nucleoside deaminase that is found in bacterial systems and is distinct from the mammalian RNA and/or DNA modifying enzymes. CONCLUSION: The conservation of a paralogous gene sequence encoding a cytidine deaminase gene over 4.5 million years suggests that Helitrons could add functional gene sequences to new chromosomal positions and thereby create new haplotypes. However, the function of such paralogous gene copies cannot be essential because they are not present in all maize strains. However, it is interesting to note that maize hybrids can outperform their inbred parents. Therefore, certain haplotypes may function only in combination with other haplotypes or under specialized environmental conditions.
DOI:10.16288/j.yczz.20-015URLPMID:32217513 [本文引用: 1]

In recent years, a great number of plant resistance (R) genes and pathogen avirulence (Avr) genes were identified. Exciting breakthroughs were also made on the structural and functional analysis of R proteins and Avr proteins, and the mechanistic interaction between them. Plants have evolved two layers of the immune system to cope with pathogens in the evolutionary processes, which are pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI). In PTI responses, conserved PAMPs are recognized by plant plasma membrane-localized pattern recognition receptors (PRRs) and disease resistance is activated. Furthermore, the ETI immune signaling is activated by the recognition of pathogen Avr proteins by the host R proteins, which usually results in hypersensitive responses at the infection site. In this review, we summarize the progresses on PTI and ETI, and discuss the genetic mechanism of the interaction between plant R gene and pathogen Avr gene in detail. We also envision the new challenges and propose the new strategies for the future investigations on plant resistance molecular breeding.
DOI:10.1016/j.semcdb.2014.05.015URLPMID:24965208 [本文引用: 1]

In mammals, the vast majority of transcripts expressed are noncoding RNAs, ranging from short RNAs (including microRNAs) to long RNAs spanning up to hundreds of kb. While the actions of microRNAs as destabilizers and repressors of the translation of protein-coding transcripts (mRNAs) have been studied in detail, the influence of microRNAs on long noncoding RNA (lncRNA) function is only now coming into view. Conversely, the influence of lncRNAs upon microRNA function is also rapidly emerging. In some cases, lncRNA stability is reduced through the interaction of specific miRNAs. In other cases, lncRNAs can act as microRNA decoys, with the sequestration of microRNAs favoring expression of repressed target mRNAs. Other lncRNAs derepress gene expression by competing with miRNAs for interaction with shared target mRNAs. Finally, some lncRNAs can produce miRNAs, leading to repression of target mRNAs. These microRNA-lncRNA regulatory paradigms modulate gene expression patterns that drive major cellular processes (such as cell differentiation, proliferation, and cell death) which are central to mammalian physiologic and pathologic processes. We review and summarize the types of microRNA-lncRNA crosstalk identified to-date and discuss their influence on gene expression programs.
DOI:10.1111/pbi.13234URLPMID:31419052 [本文引用: 3]

Plant defence is multilayered and is essential for surviving in a changing environment. The discovery of long noncoding RNAs (lncRNAs) has dramatically extended our understanding of post-transcriptional gene regulation in diverse biological processes. However, the expression profile and function of lncRNAs in disease resistance are still largely unknown, especially in monocots. Here, we performed strand-specific RNA sequencing of rice leaves infected by Xanthomonas oryzae pv. Oryzae (Xoo) in different time courses and systematically identified 567 disease-responsive rice lncRNAs. Target analyses of these lncRNAs showed that jasmonate (JA) pathway was significantly enriched. To reveal the interaction between lncRNAs and JA-related genes, we studied the coexpression of them and found 39 JA-related protein-coding genes to be interplayed with 73 lncRNAs, highlighting the potential modulation of lncRNAs in JA pathway. We subsequently identified an lncRNA, ALEX1, whose expression is highly induced by Xoo infection. A T-DNA insertion line constructed using enhancer trap system showed a higher expression of ALEX1 and exerted a significant resistance to rice bacterial blight. Functional study revealed that JA signalling is activated and the endogenous content of JA and JA-Ile is increased. Overexpressing ALEX1 in rice further confirmed the activation of JA pathway and resistance to bacterial blight. Our findings reveal the expression of pathogen-responsive lncRNAs in rice and provide novel insights into the connection between lncRNAs and JA pathway in the regulation of plant disease resistance.
DOI:10.1186/s12864-015-1876-7URLPMID:26334759 [本文引用: 1]

BACKGROUND: While RNA-sequencing (RNA-seq) is becoming a powerful technology in transcriptome profiling, one significant shortcoming of the first-generation RNA-seq protocol is that it does not retain the strand specificity of origin for each transcript. Without strand information it is difficult and sometimes impossible to accurately quantify gene expression levels for genes with overlapping genomic loci that are transcribed from opposite strands. It has recently become possible to retain the strand information by modifying the RNA-seq protocol, known as strand-specific or stranded RNA-seq. Here, we evaluated the advantages of stranded RNA-seq in transcriptome profiling of whole blood RNA samples compared with non-stranded RNA-seq, and investigated the influence of gene overlaps on gene expression profiling results based on practical RNA-seq datasets and also from a theoretical perspective. RESULTS: Our results demonstrated a substantial impact of stranded RNA-seq on transcriptome profiling and gene expression measurements. As many as 1751 genes in Gencode Release 19 were identified to be differentially expressed when comparing stranded and non-stranded RNA-seq whole blood samples. Antisense and pseudogenes were significantly enriched in differential expression analyses. Because stranded RNA-seq retains strand information of a read, we can resolve read ambiguity in overlapping genes transcribed from opposite strands, which provides a more accurate quantification of gene expression levels compared with traditional non-stranded RNA-seq. In the human genome, it is not uncommon to find genomic loci where both strands encode distinct genes. Among the over 57,800 annotated genes in Gencode release 19, there are an estimated 19 % (about 11,000) of overlapping genes transcribed from the opposite strands. Based on our whole blood mRNA-seq datasets, the fraction of overlapping nucleotide bases on the same and opposite strands were estimated at 2.94 % and 3.1 %, respectively. The corresponding theoretical estimations are 3 % and 3.6 %, well in agreement with our own findings. CONCLUSIONS: Stranded RNA-seq provides a more accurate estimate of transcript expression compared with non-stranded RNA-seq, and is therefore the recommended RNA-seq approach for future mRNA-seq studies.
DOI:10.1186/s13059-018-1574-2URLPMID:30419941 [本文引用: 11]

BACKGROUND: Interspecific hybridization and whole genome duplication are driving forces of genomic and organism diversification. But the effect of interspecific hybridization and whole genome duplication on the non-coding portion of the genome in particular remains largely unknown. In this study, we examine the profile of long non-coding RNAs (lncRNAs), comparing them with that of coding genes in allotetraploid cotton (Gossypium hirsutum), its putative diploid ancestors (G. arboreum; G. raimondii), and an F1 hybrid (G. arboreum x G. raimondii, AD). RESULTS: We find that most lncRNAs (80%) that were allelic expressed in the allotetraploid genome. Moreover, the genome shock of hybridization reprograms the non-coding transcriptome in the F1 hybrid. Interestingly, the activated lncRNAs are predominantly transcribed from demethylated TE regions, especially from long interspersed nuclear elements (LINEs). The DNA methylation dynamics in the interspecies hybridization are predominantly associated with the drastic expression variation of lncRNAs. Similar trends of lncRNA bursting are also observed in the progress of polyploidization. Additionally, we find that a representative novel lncRNA XLOC_409583 activated after polyploidization from a LINE in the A subgenome of allotetraploid cotton was involved in control of cotton seedling height. CONCLUSION: Our results reveal that the processes of hybridization and polyploidization enable the neofunctionalization of lncRNA transcripts, acting as important sources of increased plasticity for plants.
DOI:10.1038/s41467-018-07500-7URLPMID:30498193 [本文引用: 3]

Long non-coding RNAs (lncRNAs) have emerged as important regulators of gene expression and plant development. Here, we identified 6,510 lncRNAs in Arabidopsis under normal or stress conditions. We found that the expression of natural antisense transcripts (NATs) that are transcribed in the opposite direction of protein-coding genes often positively correlates with and is required for the expression of their cognate sense genes. We further characterized MAS, a NAT-lncRNA produced from the MADS AFFECTING FLOWERING4 (MAF4) locus. MAS is induced by cold and indispensable for the activation of MAF4 transcription and suppression of precocious flowering. MAS activates MAF4 by interacting with WDR5a, one core component of the COMPASS-like complexes, and recruiting WDR5a to MAF4 to enhance histone 3 lysine 4 trimethylation (H3K4me3). Our study greatly extends the repertoire of lncRNAs in Arabidopsis and reveals a role for NAT-lncRNAs in regulating gene expression in vernalization response and likely in other biological processes.
DOI:10.1126/sciadv.aax3619URLPMID:32064312 [本文引用: 1]

Genomes carry millions of noncoding variants, and identifying the tiny fraction with functional consequences is a major challenge for genomics. We assessed the role of selection on long noncoding RNAs (lncRNAs) for domestication-related changes in rice grains. Among 3363 lncRNA transcripts identified in early developing panicles, 95% of those with differential expression (329 lncRNAs) between Oryza sativa ssp. japonica and wild rice were significantly down-regulated in the domestication event. Joint genome and transcriptome analyses reveal that directional selection on lncRNAs altered the expression of energy metabolism genes during domestication. Transgenic experiments and population analyses with three focal lncRNAs illustrate that selection on these loci led to increased starch content and grain weight. Together, our findings indicate that genome-wide selection for lncRNA down-regulation was an important mechanism for the emergence of rice domestication traits.
拟南芥AtR8 lncRNA对盐胁迫响应及其对种子萌发的调节作用
1
2020
... 过表达拟南芥ELENA1 (lncRNA ELF18-INDUCED LONG-NONCODING RNA 1)可增强细菌性疫病菌抗性, ELENA1敲除对细菌性疫病菌敏感, 表明ELENA1正向调控细菌性疫病菌抗性.lncRNA ELENA1与MED19a (Mediator subunit 19a)互作, 通过影响PR1 (PATHOGENESIS-RELATED GENE 1)的表达调控先天免疫反应(
长链非编码RNA基因Ef-cd调控水稻早熟与稳产
1
2019
... 拟南芥lncRNA功能研究的开创性工作集中于调控FLC (FLOWERING LOCUS C)基因的lncRNA研究.FLC是春化途径关键基因, 调控拟南芥的成花转变.FLC受到多个lncRNA的调控.拟南芥COOLAIR (lncRNA cold induced long antisense intragenic RNA)是FLC的反义转录本, COOLAIR与polycomb复合体互作, 对FLC进行表观抑制(
Major impacts of widespread structural variation on gene expression and crop improvement in tomato
1
2020
... 转座子不仅影响基因组大小, 而且影响基因组功能.例如, 通过分析玉米z1C1位点的单倍型, 发现Helitron转座子携带胞苷脱氨酶基因ZmCDA3 (
Classification and comparison of small RNAs from plants
1
2013
... RNA聚合酶II (Pol II)转录产生2种类型的RNA: 编码RNA与非编码RNA (noncoding RNA, ncRNA).ncRNA也可以由RNA聚合酶I、III、IV、V (Pol I、III、IV、V)转录产生(
Identification of RNA-protein interactions through in vitro RNA pull-down assays. In: Lanzuolo C, Bodega B, eds. Polycomb Group Proteins
1
2016
... 基于功能验证解析lncRNA的调控机制, 是lncRNA研究的热点与难点.lncRNA多与靶标互作, 调控众多生物学过程.因此, 靶标发掘是lncRNA调控机制解析的关键.已报道调控机制的植物lncRNA多来源于已知编码蛋白基因的反义转录本, lncRNA调控机制的研究多围绕已知编码蛋白基因-已知编码蛋白基因反义转录形成的lncRNA这一模块展开.在该模块中, lncRNA互作靶标为其来源的已知编码蛋白基因(
Maize bronze 1:dSpm insertion mutations that are not fully suppressed by an active Spm
1
1993
... 转座子是基因组中可移动的元件.根据不同的转座机制, 转座子可分为逆转座子(Class I)和DNA转座子(Class II) (
Progress on Mutator superfamily
1
2020
... 转座子是基因组中可移动的元件.根据不同的转座机制, 转座子可分为逆转座子(Class I)和DNA转座子(Class II) (
Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin.
2
2017
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 过表达拟南芥ELENA1 (lncRNA ELF18-INDUCED LONG-NONCODING RNA 1)可增强细菌性疫病菌抗性, ELENA1敲除对细菌性疫病菌敏感, 表明ELENA1正向调控细菌性疫病菌抗性.lncRNA ELENA1与MED19a (Mediator subunit 19a)互作, 通过影响PR1 (PATHOGENESIS-RELATED GENE 1)的表达调控先天免疫反应(
Regulation of transposable elements by DNA modifications
1
2019
... 转座元件的转座可能会产生突变, 负向的选择压力会限制转座活性, 但转座活性必须保持, 转座子进化就是这两种力量的一个动态平衡(
Chromosome breakage by pairs of closely linked transposable elements of the Ac-Ds family in maize
1
1991
... 转座子是基因组中可移动的元件.根据不同的转座机制, 转座子可分为逆转座子(Class I)和DNA转座子(Class II) (
The PILNCR1-miR399 regulatory module is important for low phosphate tolerance in maize
3
2018
... 采用ssRNA-seq, 已经对拟南芥、水稻和玉米的lncRNA进行了分离.细菌表面翻译延伸因子Tu (translation elongation factor Tu, EF-Tu)被受体识别结合, 激活免疫反应.elf18是EF-Tu的保守肽段(
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 植物lncRNA参与调控氮和磷等营养元素的平衡.拟南芥NLP (NIN-like protein)家族成员AtNLP7是氮响应的重要调控因子(
Ef-cd locus shortens rice maturity duration without yield penalty
2
2019
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 拟南芥lncRNA功能研究的开创性工作集中于调控FLC (FLOWERING LOCUS C)基因的lncRNA研究.FLC是春化途径关键基因, 调控拟南芥的成花转变.FLC受到多个lncRNA的调控.拟南芥COOLAIR (lncRNA cold induced long antisense intragenic RNA)是FLC的反义转录本, COOLAIR与polycomb复合体互作, 对FLC进行表观抑制(
Plant transposable elements: where genetics meets genomics
1
2002
... 转座子是基因组中可移动的元件.根据不同的转座机制, 转座子可分为逆转座子(Class I)和DNA转座子(Class II) (
RIP: RNA immunoprecipitation
1
2016
... 基于功能验证解析lncRNA的调控机制, 是lncRNA研究的热点与难点.lncRNA多与靶标互作, 调控众多生物学过程.因此, 靶标发掘是lncRNA调控机制解析的关键.已报道调控机制的植物lncRNA多来源于已知编码蛋白基因的反义转录本, lncRNA调控机制的研究多围绕已知编码蛋白基因-已知编码蛋白基因反义转录形成的lncRNA这一模块展开.在该模块中, lncRNA互作靶标为其来源的已知编码蛋白基因(
The long intergenic noncoding RNA (lincRNA) landscape of the soybean genome
7
2018
... 转座子和lncRNA皆为基因组中重要的调控元件.已有研究表明, 含有转座子序列的lncRNA在自然界广泛存在, 称为转座子来源的lncRNA (transposable element-derived lncRNA, TE-lncRNA) (
... 调控机制已得到解析的植物lncRNA多来源于编码蛋白基因的反义转录(
... ;
... 通过ssRNA-seq, 在拟南芥、水稻和玉米中分别鉴定到47、611和398个TE-lincRNA, 约占全部lncRNA的23%、50%和51% (
... TE-lncRNA的分布与基因组中转座子类型和数目相关.水稻、玉米、大豆、棉花和番茄中的TE-lncRNA主要来源于逆转座子(
... 转座子是生物体的重要组成部分, 影响基因组的结构与稳定(
... TE-lncRNA在植物中广泛存在(
Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA
2
2011
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 拟南芥lncRNA功能研究的开创性工作集中于调控FLC (FLOWERING LOCUS C)基因的lncRNA研究.FLC是春化途径关键基因, 调控拟南芥的成花转变.FLC受到多个lncRNA的调控.拟南芥COOLAIR (lncRNA cold induced long antisense intragenic RNA)是FLC的反义转录本, COOLAIR与polycomb复合体互作, 对FLC进行表观抑制(
LncRNA39026 enhances tomato resistance to Phytophthora infestans by decoying miR168a and inducing PR gene expression
2
2020
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 过表达拟南芥ELENA1 (lncRNA ELF18-INDUCED LONG-NONCODING RNA 1)可增强细菌性疫病菌抗性, ELENA1敲除对细菌性疫病菌敏感, 表明ELENA1正向调控细菌性疫病菌抗性.lncRNA ELENA1与MED19a (Mediator subunit 19a)互作, 通过影响PR1 (PATHOGENESIS-RELATED GENE 1)的表达调控先天免疫反应(
Systematic identification of long non-coding RNAs during pollen development and fertilization inBrassica rapa
1
2018
... 随着多种组学的发展, 有些植物中已有丰富的公共数据可供lncRNA发掘.数据库中的数据质量直接影响到lncRNA发掘的可靠性.lncRNA发掘所用软件种类和阈值设定也会影响lncRNA的发掘结果.此外, 植物lncRNA具有序列保守性较低、丰度低以及组织特异性表达等特点(
Rolling-circle transposons in eukaryotes
1
2001
... 转座子是基因组中可移动的元件.根据不同的转座机制, 转座子可分为逆转座子(Class I)和DNA转座子(Class II) (
Nuclear noncoding RNAs and genome stability
2
2016
... 转座子是生物体的重要组成部分, 影响基因组的结构与稳定(
... ).TE-lncRNA中的转座子序列形成二级结构(与lncRNA功能相关) (
Transposable elements and host genome evolution
2
2000
... 转座子是生物体的重要组成部分, 影响基因组的结构与稳定(
... ).另一方面, lncRNA影响转座子活性.lncRNA可以作为small RNA前体, small RNA可导致转座子“沉默”(
Genome- wide discovery and characterization of maize long non- coding RNAs
1
2014
... NCBI和EBI等数据库中已有海量的基因表达数据, 包括表达序列标签(expressed sequence tag, EST)、tiling array和转录组数据等.基于数据库中的基因表达数据, 可以通过生物信息学方法发掘lncRNA.采用RepTAS (reproducibility-based tiling array analysis strategy)等方法, 整合拟南芥EST、cDNA、tiling array和基因组数据信息, 发掘到6 480个lincRNA (
Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification
1
2003
... 转座元件的转座可能会产生突变, 负向的选择压力会限制转座活性, 但转座活性必须保持, 转座子进化就是这两种力量的一个动态平衡(
The long noncoding RNA T5120 regulates nitrate response and assimilation in Arabidopsis
2
2019
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 植物lncRNA参与调控氮和磷等营养元素的平衡.拟南芥NLP (NIN-like protein)家族成员AtNLP7是氮响应的重要调控因子(
High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize
1
2020
... 转基因验证是TE-lncRNA功能解析的重要内容.基于CRISPR-Cas的基因编辑技术已成为植物基因功能验证的重要方法(
Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis
1
2012
... NCBI和EBI等数据库中已有海量的基因表达数据, 包括表达序列标签(expressed sequence tag, EST)、tiling array和转录组数据等.基于数据库中的基因表达数据, 可以通过生物信息学方法发掘lncRNA.采用RepTAS (reproducibility-based tiling array analysis strategy)等方法, 整合拟南芥EST、cDNA、tiling array和基因组数据信息, 发掘到6 480个lincRNA (
A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice
3
2018
... NCBI和EBI等数据库中已有海量的基因表达数据, 包括表达序列标签(expressed sequence tag, EST)、tiling array和转录组数据等.基于数据库中的基因表达数据, 可以通过生物信息学方法发掘lncRNA.采用RepTAS (reproducibility-based tiling array analysis strategy)等方法, 整合拟南芥EST、cDNA、tiling array和基因组数据信息, 发掘到6 480个lincRNA (
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 拟南芥lncRNA功能研究的开创性工作集中于调控FLC (FLOWERING LOCUS C)基因的lncRNA研究.FLC是春化途径关键基因, 调控拟南芥的成花转变.FLC受到多个lncRNA的调控.拟南芥COOLAIR (lncRNA cold induced long antisense intragenic RNA)是FLC的反义转录本, COOLAIR与polycomb复合体互作, 对FLC进行表观抑制(
Maize transposable elements contribute to long non-coding RNAs that are regulatory hubs for abiotic stress response
7
2019
... 转座子和lncRNA皆为基因组中重要的调控元件.已有研究表明, 含有转座子序列的lncRNA在自然界广泛存在, 称为转座子来源的lncRNA (transposable element-derived lncRNA, TE-lncRNA) (
... 通过ssRNA-seq, 在拟南芥、水稻和玉米中分别鉴定到47、611和398个TE-lincRNA, 约占全部lncRNA的23%、50%和51% (
... TE-lncRNA的分布与基因组中转座子类型和数目相关.水稻、玉米、大豆、棉花和番茄中的TE-lncRNA主要来源于逆转座子(
... 棉花TE-lncRNA XLOC_409583来源于去甲基化的LINE类逆转座子.与对照相比, 病毒诱导TE-lncRNA XLOC_409583沉默的植株苗高增加, 表明TE- lncRNA XLOC_409583调控棉花株高(
... 在整个植物lncRNA中, TE-lncRNA是重要组分(
... 转座子是生物体的重要组成部分, 影响基因组的结构与稳定(
... TE-lncRNA在植物中广泛存在(
Survival strategies for transposons and genomes
1
2003
... 转座元件的转座可能会产生突变, 负向的选择压力会限制转座活性, 但转座活性必须保持, 转座子进化就是这两种力量的一个动态平衡(
A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance
2
2017
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 过表达拟南芥ELENA1 (lncRNA ELF18-INDUCED LONG-NONCODING RNA 1)可增强细菌性疫病菌抗性, ELENA1敲除对细菌性疫病菌敏感, 表明ELENA1正向调控细菌性疫病菌抗性.lncRNA ELENA1与MED19a (Mediator subunit 19a)互作, 通过影响PR1 (PATHOGENESIS-RELATED GENE 1)的表达调控先天免疫反应(
The B73 maize genome: complexity, diversity, and dynamics
3
2009
... 转座子是基因组的重要组分, 影响基因组的稳定与进化.以基因组较小的物种拟南芥(~125 Mb)与基因组较大的物种玉米(~2.3 Gb)为例, 介绍植物基因组中转座子分布的特点以及转座子构成在基因组进化中的作用.至少10%的拟南芥基因组序列由转座子构成, 其中逆转座子、DNA转座子以及其它类型转座子的百分比分别约为38%、39%和23% (
... ;
... (A) 拟南芥基因组中转座子分布(左图)和拟南芥长末端重复序列(LTR)逆转座子占全部转座子的比例(右图); (B) 玉米基因组中转座子分布(左图)和玉米LTR逆转座子占全部转座子的比例(右图).Class I: 逆转座子; Class II: DNA转座子
ELF18-INDUCED LONG-NONCODING RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis
3
2017
... 采用ssRNA-seq, 已经对拟南芥、水稻和玉米的lncRNA进行了分离.细菌表面翻译延伸因子Tu (translation elongation factor Tu, EF-Tu)被受体识别结合, 激活免疫反应.elf18是EF-Tu的保守肽段(
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 过表达拟南芥ELENA1 (lncRNA ELF18-INDUCED LONG-NONCODING RNA 1)可增强细菌性疫病菌抗性, ELENA1敲除对细菌性疫病菌敏感, 表明ELENA1正向调控细菌性疫病菌抗性.lncRNA ELENA1与MED19a (Mediator subunit 19a)互作, 通过影响PR1 (PATHOGENESIS-RELATED GENE 1)的表达调控先天免疫反应(
Cold- induced silencing by long antisense transcripts of an Arabidopsis polycomb target
2
2009
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 拟南芥lncRNA功能研究的开创性工作集中于调控FLC (FLOWERING LOCUS C)基因的lncRNA研究.FLC是春化途径关键基因, 调控拟南芥的成花转变.FLC受到多个lncRNA的调控.拟南芥COOLAIR (lncRNA cold induced long antisense intragenic RNA)是FLC的反义转录本, COOLAIR与polycomb复合体互作, 对FLC进行表观抑制(
Analysis of the genome sequence of the flowering plant Arabidopsis thaliana
3
2000
... 转座子是基因组的重要组分, 影响基因组的稳定与进化.以基因组较小的物种拟南芥(~125 Mb)与基因组较大的物种玉米(~2.3 Gb)为例, 介绍植物基因组中转座子分布的特点以及转座子构成在基因组进化中的作用.至少10%的拟南芥基因组序列由转座子构成, 其中逆转座子、DNA转座子以及其它类型转座子的百分比分别约为38%、39%和23% (
... 拟南芥和玉米基因组中转座子的分布(
... (A) 拟南芥基因组中转座子分布(左图)和拟南芥长末端重复序列(LTR)逆转座子占全部转座子的比例(右图); (B) 玉米基因组中转座子分布(左图)和玉米LTR逆转座子占全部转座子的比例(右图).Class I: 逆转座子; Class II: DNA转座子
Interactions between short and long noncoding RNAs
1
2018
... RNA聚合酶II (Pol II)转录产生2种类型的RNA: 编码RNA与非编码RNA (noncoding RNA, ncRNA).ncRNA也可以由RNA聚合酶I、III、IV、V (Pol I、III、IV、V)转录产生(
Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants
13
2017
... 转座子和lncRNA皆为基因组中重要的调控元件.已有研究表明, 含有转座子序列的lncRNA在自然界广泛存在, 称为转座子来源的lncRNA (transposable element-derived lncRNA, TE-lncRNA) (
... )等植物中, TE-lncRNA主要来源于逆转座子序列(
... ,
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 调控机制已得到解析的植物lncRNA多来源于编码蛋白基因的反义转录(
... The proportion of plant transposable element-de- rived long intergenic noncoding RNA (TE-lincRNA) (data source:
... 通过ssRNA-seq, 在拟南芥、水稻和玉米中分别鉴定到47、611和398个TE-lincRNA, 约占全部lncRNA的23%、50%和51% (
... TE-lncRNA的分布与基因组中转座子类型和数目相关.水稻、玉米、大豆、棉花和番茄中的TE-lncRNA主要来源于逆转座子(
... 棉花TE-lncRNA XLOC_409583来源于去甲基化的LINE类逆转座子.与对照相比, 病毒诱导TE-lncRNA XLOC_409583沉默的植株苗高增加, 表明TE- lncRNA XLOC_409583调控棉花株高(
... 调控非生物胁迫应答所必需(
... 在整个植物lncRNA中, TE-lncRNA是重要组分(
... 转座子是生物体的重要组成部分, 影响基因组的结构与稳定(
... TE-lncRNA在植物中广泛存在(
Expression and diversification analysis reveals transposable elements play important roles in the origin of Lycopersicon-specific lncRNAs in tomato
7
2016
... 转座子和lncRNA皆为基因组中重要的调控元件.已有研究表明, 含有转座子序列的lncRNA在自然界广泛存在, 称为转座子来源的lncRNA (transposable element-derived lncRNA, TE-lncRNA) (
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 通过ssRNA-seq, 在拟南芥、水稻和玉米中分别鉴定到47、611和398个TE-lincRNA, 约占全部lncRNA的23%、50%和51% (
... TE-lncRNA的分布与基因组中转座子类型和数目相关.水稻、玉米、大豆、棉花和番茄中的TE-lncRNA主要来源于逆转座子(
... 棉花TE-lncRNA XLOC_409583来源于去甲基化的LINE类逆转座子.与对照相比, 病毒诱导TE-lncRNA XLOC_409583沉默的植株苗高增加, 表明TE- lncRNA XLOC_409583调控棉花株高(
... 转座子是生物体的重要组成部分, 影响基因组的结构与稳定(
... TE-lncRNA在植物中广泛存在(
aOverexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice
2
2018a
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 拟南芥lncRNA功能研究的开创性工作集中于调控FLC (FLOWERING LOCUS C)基因的lncRNA研究.FLC是春化途径关键基因, 调控拟南芥的成花转变.FLC受到多个lncRNA的调控.拟南芥COOLAIR (lncRNA cold induced long antisense intragenic RNA)是FLC的反义转录本, COOLAIR与polycomb复合体互作, 对FLC进行表观抑制(
High-resolution profile of transcriptomes reveals a role of alternative splicing for modulating response to nitrogen in maize
1
2020
... 植物lncRNA参与调控氮和磷等营养元素的平衡.拟南芥NLP (NIN-like protein)家族成员AtNLP7是氮响应的重要调控因子(
Unveiling gibberellin-responsive coding and long noncoding RNAs in maize
1
2018b
... 采用ssRNA-seq, 已经对拟南芥、水稻和玉米的lncRNA进行了分离.细菌表面翻译延伸因子Tu (translation elongation factor Tu, EF-Tu)被受体识别结合, 激活免疫反应.elf18是EF-Tu的保守肽段(
The diversity of long noncoding RNAs and their generation
2
2017
... RNA聚合酶II (Pol II)转录产生2种类型的RNA: 编码RNA与非编码RNA (noncoding RNA, ncRNA).ncRNA也可以由RNA聚合酶I、III、IV、V (Pol I、III、IV、V)转录产生(
... 转座子是生物体的重要组成部分, 影响基因组的结构与稳定(
Maize haplotype with a helitron- amplified cytidine deaminase gene copy
1
2006
... 转座子不仅影响基因组大小, 而且影响基因组功能.例如, 通过分析玉米z1C1位点的单倍型, 发现Helitron转座子携带胞苷脱氨酶基因ZmCDA3 (
Molecular genetic mechanisms of interaction between host plants and pathogens
1
2020
... 采用ssRNA-seq, 已经对拟南芥、水稻和玉米的lncRNA进行了分离.细菌表面翻译延伸因子Tu (translation elongation factor Tu, EF-Tu)被受体识别结合, 激活免疫反应.elf18是EF-Tu的保守肽段(
Functional interactions among microRNAs and long noncoding RNAs
1
2014
... 转座子是生物体的重要组成部分, 影响基因组的结构与稳定(
Transcriptional landscape of pathogen-responsive lncRNAs in rice unveils the role of ALEX1 in jasmonate pathway and disease resistance
3
2020
... 采用ssRNA-seq, 已经对拟南芥、水稻和玉米的lncRNA进行了分离.细菌表面翻译延伸因子Tu (translation elongation factor Tu, EF-Tu)被受体识别结合, 激活免疫反应.elf18是EF-Tu的保守肽段(
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 过表达拟南芥ELENA1 (lncRNA ELF18-INDUCED LONG-NONCODING RNA 1)可增强细菌性疫病菌抗性, ELENA1敲除对细菌性疫病菌敏感, 表明ELENA1正向调控细菌性疫病菌抗性.lncRNA ELENA1与MED19a (Mediator subunit 19a)互作, 通过影响PR1 (PATHOGENESIS-RELATED GENE 1)的表达调控先天免疫反应(
Comparison of stranded and non-stranded RNA-seq transcriptome profiling and investigation of gene overlap
1
2015
... 构建链特异性RNA文库并进行转录组测序(strand- specific RNA sequencing, ssRNA-seq), 对lncRNA进行高通量发掘.与普通RNA-seq相比, ssRNA-seq可以区分reads来源于正义链还是反义链, 在转录本定量时可以排除互补链转录本表达的干扰, 使表达定量更精确.此外, ssRNA-seq有助于确定lincRNA的转录方向(
LncRNAs in polyploid cotton interspecific hybrids are derived from transposon neofunctionalization
11
2018a
... 转座子和lncRNA皆为基因组中重要的调控元件.已有研究表明, 含有转座子序列的lncRNA在自然界广泛存在, 称为转座子来源的lncRNA (transposable element-derived lncRNA, TE-lncRNA) (
... ;
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 通过ssRNA-seq, 在拟南芥、水稻和玉米中分别鉴定到47、611和398个TE-lincRNA, 约占全部lncRNA的23%、50%和51% (
... TE-lncRNA的分布与基因组中转座子类型和数目相关.水稻、玉米、大豆、棉花和番茄中的TE-lncRNA主要来源于逆转座子(
... 棉花TE-lncRNA XLOC_409583来源于去甲基化的LINE类逆转座子.与对照相比, 病毒诱导TE-lncRNA XLOC_409583沉默的植株苗高增加, 表明TE- lncRNA XLOC_409583调控棉花株高(
... 转座子是生物体的重要组成部分, 影响基因组的结构与稳定(
... ;
... ).此外, 植物转座子的表观修饰影响lncRNA的转录活性.例如, 棉花LINE类逆转座子去甲基化, 产生具有转录活性的TE-lncRNA (
... 转座元件的转座可能会产生突变, 负向的选择压力会限制转座活性, 但转座活性必须保持, 转座子进化就是这两种力量的一个动态平衡(
... TE-lncRNA在植物中广泛存在(
Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA
3
2018b
... 采用ssRNA-seq, 已经对拟南芥、水稻和玉米的lncRNA进行了分离.细菌表面翻译延伸因子Tu (translation elongation factor Tu, EF-Tu)被受体识别结合, 激活免疫反应.elf18是EF-Tu的保守肽段(
... Function of plant long noncoding RNA (lncRNA)
| 物种 | lncRNA ID | lncRNA调控通路 | 参考文献 |
|---|---|---|---|
| 拟南芥 | COLDAIR (COLD ASSISTED INTRONIC NONCODING RNA) | 成花转变 | |
| COOLAIR (cold induced long antisense intragenic RNA) | 成花转变 | ||
| DRIR (DROUGHT INDUCED lncRNA) | 干旱和盐胁迫应答 | ||
| ELENA1 (ELF18-INDUCED LONG-NONCODING RNA1) | 先天免疫反应 | ||
| MAS (lncRNA from MADS AFFECTING FLOWERING4) | 春化响应 | ||
| T5120 | NO3-同化 | ||
| TE-lincRNA11195* | ABA响应 | ||
| 水稻 | ALEX1 (An Leaf Expressed and Xoo-induced lncRNA 1) | 白叶枯病抗性 | |
| Ef-cd (Early flowering-completely dominant) | 开花期和产量 | ||
| LAIR (LRK Antisense Intergenic RNA) | 产量 | ||
| TL (TWISTED LEAF) | 叶片发育 | ||
| 玉米 | PILNCR1 (Pi-deficiency-induced long non-coding RNA 1) | 磷胁迫应答 | |
| 棉花 | XLOC_409583* | 苗高 | |
| 番茄 | lncRNA-314* | 果实成熟 | |
| lncRNA16397 | 晚疫病抗性 | ||
| lncRNA39026 | 晚疫病抗性 |
... 拟南芥lncRNA功能研究的开创性工作集中于调控FLC (FLOWERING LOCUS C)基因的lncRNA研究.FLC是春化途径关键基因, 调控拟南芥的成花转变.FLC受到多个lncRNA的调控.拟南芥COOLAIR (lncRNA cold induced long antisense intragenic RNA)是FLC的反义转录本, COOLAIR与polycomb复合体互作, 对FLC进行表观抑制(
Genome-wide analyses reveal the role of noncoding variation in complex traits during rice domestication
1
2019
... 采用ssRNA-seq, 已经对拟南芥、水稻和玉米的lncRNA进行了分离.细菌表面翻译延伸因子Tu (translation elongation factor Tu, EF-Tu)被受体识别结合, 激活免疫反应.elf18是EF-Tu的保守肽段(
