0 引言
【研究意义】普通野生稻中含有水稻驯化过程中丢失或者削弱的大量优异基因;粒长是水稻首选的育种目标之一。利用以普通野生稻为供体亲本构建的染色体片段置换系进行粒长QTL定位,可以挖掘出野生稻中的优异基因,为水稻育种以及粒长形成的分子机理研究提供新的基因资源。【前人研究进展】水稻是世界上重要的粮食作物之一[1]。中国是稻米消耗大国,65%以上的人口以大米为主食[2]。提高水稻产量对确保中国粮食安全和社会稳定发展有重要意义。水稻产量主要由3个因素构成:单株穗数、每穗粒数和粒重。粒重指标由粒长、粒宽、长宽比和粒厚来综合评价,这四个性状与千粒重成正相关[3,4]。因此,籽粒形状是水稻首选的育种目标之一。与大多数农艺性状一样,水稻粒型是受多基因控制的数量性状[5,6],它不但影响水稻的产量,还影响加工品质与外观品质。对籽粒形状相关数量性状位点(quantitative trait locus,QTL)的鉴定以及新基因的发掘是水稻育种与遗传研究的重要目标。随着分子生物学技术以及现代遗传学的发展,许多研究者通过全面解析粒型相关的遗传机制和挖掘功能基因/QTL位点,定位了越来越多的粒型QTL,并克隆出一些控制粒型性状的重要基因。林鸿宣等[7]利用RFLP图谱对控制籼稻粒型性状QTL进行定位,在特三矮2号/CB1128群体中定位到5个控制粒长QTL;在外引2号/CB1128群体中定位到5个控制粒长QTL,5个控制粒宽QTL。近年来,很多研究通过构建染色体片段置换系进行水稻优异基因的挖掘定位。周丽慧等[8]通过9311/日本晴CSSL群体定位到10个QTL,与单株产量、单株实粒数、千粒质量、每穗颖花数、结实率、单株总颖花数6个性状相关。朱文银等[9]利用染色体片段置换系定位到3个控制水稻粒型的主效QTL,其中有2个粒长相关的QTL,分别为qGL3.1和qGL3.2。qGL3.1被定位在水稻第3染色体RM5551与RM6832之间,遗传距离为14.8 cM;qGL3.2被定位在RM6832与RM3513之间,其遗传距离为5.3 cM。随着第一个控制水稻粒型的基因GS3[10]被克隆,许多控制粒型的基因如GS2[11]、GW2[12]、GL2[13]、GL3.1[14]、GW5[15]、GS5[16]、GL7[17]和GW8[18]等相继被克隆,这些研究为理解调控粒型的机理提供了理论基础,也对作物的遗传改良具有重要意义。【本研究切入点】目前,很多研究都是以栽培稻为研究对象,被挖掘出的QTL大多来源于栽培稻,对于野生稻中优异基因的发掘鲜见报道。野生稻异质性强,构建合适的遗传群体是对其利用的关键。构建一套可以永久性利用的染色体片段群体,不仅建立了一个野生稻基因组学研究的平台,还可以为水稻育种提供新的种质资源。【拟解决的关键问题】本研究以海南普通野生稻为供体亲本,9311为受体亲本,利用中国农业科学院作物科学研究所野生稻实验室已构建的染色体片段置换系群体,通过多年多点表型鉴定试验,选择粒型差异明显的置换系构建次级群体,设计分子标记结合基因型与表型鉴定进行精细定位,发掘与水稻粒长相关的QTL,寻找野生稻中影响粒长的新基因。1 材料与方法
1.1 试验材料
以中国农业科学院作物科学研究所野生稻实验室构建的一套野生稻染色体片段置换系为研究材料。以来自海南的普通野生稻材料为供体亲本,栽培稻品种9311为受体亲本,使用119个SSR标记和62个Indel标记,通过多代回交与分子标记辅助选择,构建出覆盖水稻染色体的200个染色体片段置换系[19]。齐兰等[19]利用构建好的置换系群体定位到了37个粒型相关QTL,其中粒长相关QTL——qGL12定位于RM28621标记附近,且能在4个环境中检测到。选择粒长与受体亲本9311差异显著,且携带qGL12片段的置换系CSSL141与9311回交,构建次级分离群体,分别于2016年南京、2016年三亚、2017年北京和2017年南京进行种植进行粒长QTL——qGL12的精细定位。1.2 田间种植
2016年5月在南京基地种植200株CSSL141/9311 F2群体作为初定位群体,10月单株收获籽粒检测籽粒表型。从初定位结果中挑选出在定位区间内存在基因型差异,而其他区间的基因型为父本或母本的纯合基因型的单株于2016年12月在海南试验基地种植得到400株F3群体,2017年4月收获F3单株,筛选交换单株缩短定位区间,2017年5月从F3中选择目标区间杂合基因型的种子在北京、南京试验基地种植,分别得到2 400株、1 200株F4群体,2017年10月单株收获F4群体籽粒。田间种植规格为,行距16 cm×26 cm,每行种植10株。1.3 表型鉴定
水稻成熟后单株收获种子,自然晒干,籽粒的表型鉴定均用万深SC-G自动考种及千粒重仪(杭州万深检测科技有限公司)进行,测定籽粒的粒长、粒宽以及千粒重。每份材料2次重复,每次测量150—200粒种子。1.4 群体基因型鉴定
所用的SSR引物来源于美国Cornell大学公布的研究成果,定位区间内引物的设计是根据已经测序完成的日本晴和9311的序列信息,使用primer5软件进行引物设计,在Gramene网站进行在线比对,验证其所在染色体位置,确保标记的准确性。利用PCR扩增检验在两亲本间的多态性,选择多态性良好的引物作为精细定位的标记引物。试验自行设计的定位引物序列见表1。Table 1
表1
表1用于精细定位的引物序列
Table 1The primers information of fine mapping
| 标记名称 Marker name | 标记类型 Marker type | 片段大小 Fragment size (bp) | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|---|---|
| DYB1.1 | InDel | 272 | GGAAGGAATCCGCTTTACA | GGGAATATTGCAGGGTTGA |
| DYB6.1 | InDel | 259 | GCCAGAATCTCCACAGATCC | GTTTGGTGGGAAGGTCATCT |
| DYB7.1 | InDel | 283 | GAATCTTTGCAGCTTGGGAA | AGGTGGTTTGGGCATAGTGA |
| DYB9.1 | InDel | 246 | GTTTCAGTTTAGTTTTGGATTGCT | TACTCTCTTATTATCAGCCTA |
新窗口打开
1.5 粒型性状QTL定位
根据SSR分析的结果,与亲本CSSL141一致的带型记为2,与9311带型一致的带型记为0,同时具有两亲本的杂合带型记为1,由于某种原因造成带型缺失或模糊的记为-1。用QTL Ici-Mapping V4.1软件中的完备区间–加性模型(ICIM-ADD)定位方法,扫描步长为1 cM,以LOD值2.5作为阀值,在全基因组范围内进行扫描,判断可能存在的控制粒型性状的QTL,QTL的命名遵循McCouch等的原则。1.6 颖壳细胞扫描电镜观察
选取9311、CSSL141籽粒,F4群体中定位区间基因型分别为野生稻、9311植株的籽粒,将颖壳分离,剪取中间部分的外颖,固定后将颖壳外颖进行喷金镀膜,使用扫描电镜观察颖壳外表面表皮细胞,检测颖壳细胞的长度与宽度。1.7 候选基因预测
根据QTL精细定位结果,利用Rice Genome Browser网站(http://rice.plantbiology.msu.edu/cgi-bin/ gbrowse/rice/#search)查找区间内的所有基因,通过Rice Genome Annotation Project对基因注释进行分析,筛选出与目标性状相关的候选基因。在NCBI数据库(https://www.ncbi.nlm.nih.gov/)中下载候选基因序列,用Primer5设计测序引物对候选基因进行扩增、测序。比较候选基因在9311与CSSL141间的序列差异。2 结果
2.1 亲本CSSL141的选择以及基因型与表型分析
利用染色体片段置换系群体,在多个环境下进行了粒型QTL的定位[19]。在第12染色体RM28621标记附近检测到一个能够在多个环境下稳定表达,与粒长相关的QTL,命名为qGL12(表2)。选择携带qGL12片段且粒型与9311差异显著的置换系CSSL141作为亲本,进行下一步的精细定位。CSSL141染色体基因型如图1所示,携带4个来自野生稻的置换片段,1个片段位于第2染色体,3个片段位于第12染色体,其中第12染色体上的一个置换片段位于RM28621标记附近(图1)。Table 2
表2
表2不同环境下qGL12的检测
Table 2Detection of qGL12 under different environments
| 位点 QTL | 标记 Marker | 染色体 Chr. | 环境 Environment | LOD Threshod | LOD | 贡献率 PVE (%) | 加性效应 Add |
|---|---|---|---|---|---|---|---|
| 2014年南京Nanjing, 2014 | 3.35 | 6.40 | 19.87 | 0.34 | |||
| 2015年北京Beijing, 2015 | 3.14 | 3.24 | 7.14 | 0.17 | |||
| qGL12 | RM28621 | 12 | 2015年南京Nanjing, 2015 | 3.41 | 9.70 | 24.29 | 0.32 |
| 2015年三亚Sanya, 2015 | 3.17 | 5.94 | 14.85 | 0.27 |
新窗口打开
 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图1CSSL141置换片段分布
-->Fig. 1The distribution of substitution segments of CSSL141
-->
根据整套置换系材料在2014—2016年3年不同环境中的调查结果,发现CSSL141和亲本9311在粒型上有明显差异(图2-a和图2-b)。其中,9311在北京、南京、三亚粒长均值分别为9.61、9.77和9.35 mm,CSSL141粒长均值分别为10.10、10.19和9.90 mm;9311千粒重均值分别为31.22、32.60和32.22 g,CSSL141千粒重均值分别为33.83、35.04和35.08 g;粒长、千粒重差异均达到极显著水平(P<0.001)。 9311粒宽均值在北京、南京、三亚分别为2.79、2.79和2.83 mm,CSSL141粒宽均值分别为2.82、2.86和2.93 mm(图2-c)。此外,CSSL141与9311在株高、穗长、有效穗数等农艺性状方面差异不显著。CSSL141中置换片段较少,籽粒表型与9311差异较大而其他农艺性状与9311没有显著差异,而且其置换片段中含有实验室已经定位到的QTL——qGL12,因此选为亲本进行qGL12的精细定位。
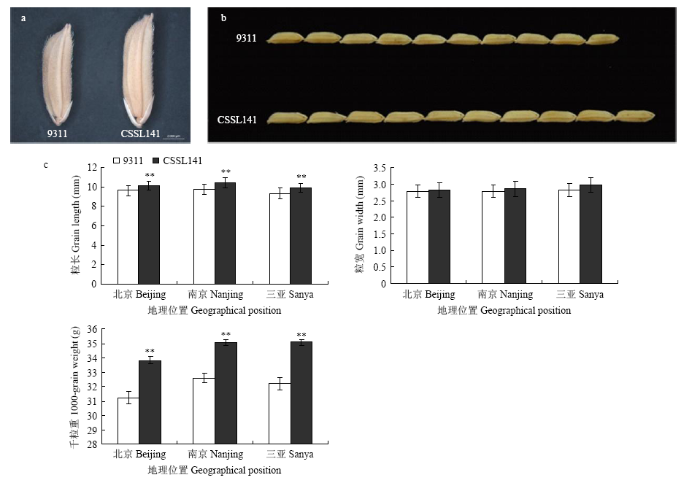 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图29311与CSSL141籽粒性状比较
a:单粒种子;b:粒长比较;c:不同环境下的粒型性状比较。**差异极显著(P<0.001)
-->Fig. 2Comparison of kernel traits between 9311and CSSL141
a: The seeds of 9311 and CSSL141; b: 10 Comparison of grain length; c:Comparison of grain traits under multiple environments. **significantly different at P<0.001
-->
2.2 F2群体定位结果
CSSL141/9311 F2群体种植于南京试验基地。粒长、粒宽以及千粒重3个粒型性状在F2群体的分布均呈连续正态分布(图3)。F2群体粒长介于9.58—10.80 mm,主要集中在9.81—10.27 mm;粒宽介于2.57—2.84 mm,主要集中在2.68—2.72 mm;千粒重介于30.50—36.63 g,主要集中在32.76—35.12 g。说明这三个性状是由多基因调控,属于数量遗传性状,适合QTL分析。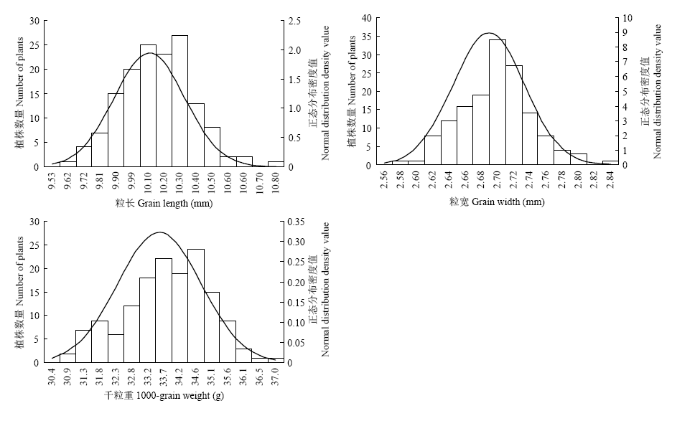 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图3F2群体粒长、粒宽、千粒重频次分布图
-->Fig. 3Frequency distribution diagram of grain length, grain width and 1000 grain weight in CSSL141/9311 F2 population
-->
根据CSSL141染色体基因组中野生稻置换片段的分布,使用置换片段区间或周围的分子标记设引物RM5338、RM5479、RM28621、RM28642、Indel12-12作为初定位标记引物。利用QTL IciMapping分析软件将qGL12定位于第12染色体标记RM5479—RM28621,影响粒长、粒宽以及粒重。对粒长的贡献率最高,为44.61%(表3)。
Table 3
表3
表3CSSL141/9311 F2群体粒型QTL初定位结果
Table 3Mapping of QTLs associated with grain traits using CSSL141/9311 F2 population
| 群体 Population | 性状 Trait | 染色体 Chr. | 标记区间Intervals | 南京 Nanjing | ||||
|---|---|---|---|---|---|---|---|---|
| 左标记 Left marker | 右标记 Right marker | LOD | 贡献率 PVE (%) | 加性 Add | 上位性 EPI | |||
| 粒长Grain length | 12 | RM5479 | RM28621 | 16.78 | 44.61 | 0.20 | 0.0154 | |
| CSSL141 | 粒宽Grain width | 12 | RM5479 | RM28621 | 3.60 | 11.30 | 0.02 | 0.0081 |
| 千粒重Thousand grain weight | 12 | RM5479 | RM28621 | 7.35 | 24.08 | 0.81 | 0.3998 | |
新窗口打开
2.3 qGL12精细定位
利用F2群体将qGL12定位于第12染色体标记RM5479与RM28621之间。F2群体中选择两标记基因型杂合的植株进行种植得到F3。在定位区间RM5479—RM28621两边及中间增加7个多态性分子标记引物,分别为RM28583、RM28586、RM28590、RM28591、RM28597、RM28607和RM28642。对400株海南F3单株进行基因型分析,筛选出18株交换单株。结合交换单株粒长表型与基因型,将qGL12定位于RM5479和RM28586之间,区间长度约为50 kb(图4和图5)。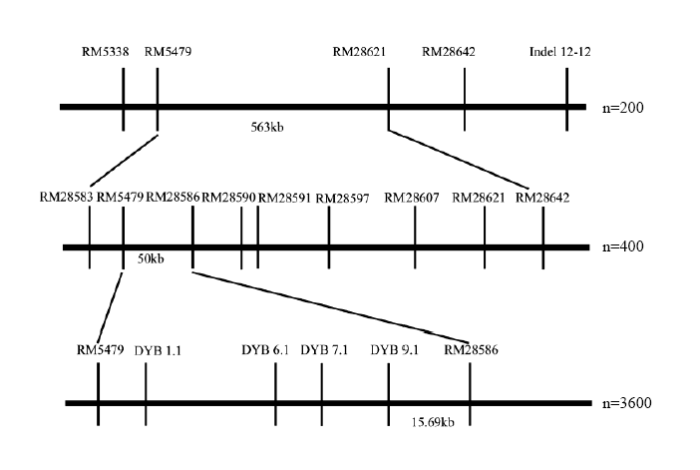 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图4qGL12的精细定位
-->Fig. 4Fine mapping of qGL12
-->
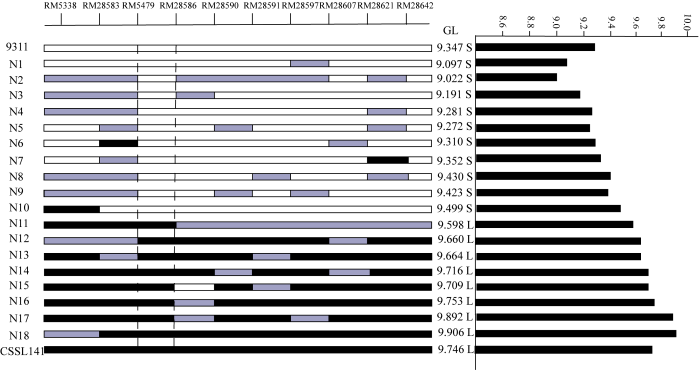 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图5RM5338与RM28642区间内交换单株基因型与表型
S:短粒;L:长粒;白色区域:9311基因型;黑色区域:野生稻基因型;灰色区域:杂合基因型。下同
-->Fig. 5The genotype and phenotype of recombinants in the interval of RM5338-RM28642
S: Short Grain; L: Long grain; White region: The genotype of 9311; Black region: The genotype of wild rice; Grey region: Heterozygous genotype. The same as below
-->
为了进一步精细定位qGL12,将F3群体中目标区间基因型杂合的单株种子分别在北京顺义、南京进行种植得到F4。在NCBI数据库(https://www.ncbi. nlm.nih.gov/)中找到RM5479与RM28586区间片段的序列,利用Primer5软件设计分子标记引物,设计好的引物在两亲本间进行多态性检测,最后得到4个多态性良好的分子标记引物DYB1.1、DYB6.1、DYB7.1和DYB9.1(表1)。对2 400株种植于北京顺义的F4单株以及1 200株种植于南京的F4单株进行定位分析,南京和北京的群体都在RM5479和RM28586之间发现粒长相关的QTL。通过筛选交换单株,得到20株交换单株,结合基因型与表型的结果,根据交换单株信息,将qGL12的左边界定于DYB9.1标记处,右边界定于RM28586,区间长度约15.69 kb(图4和图6)。
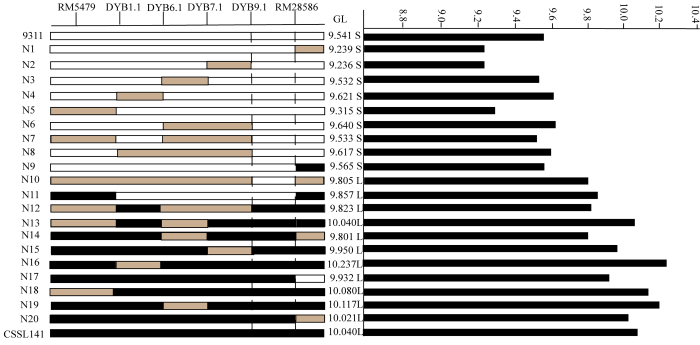 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图6RM5479与RM28586区间内交换单株基因型与表型
-->Fig. 6The genotype and phenotype of recombinants in the interval of RM5479-RM28586
-->
2.4 颖壳细胞学观察
取同一时期、同一环境下9311与CSSL141的籽粒,以及F4群体中2个交换单株的籽粒,2个交换单株目标区间染色体片段分别为野生稻基因型以及9311基因型。扫描电镜观察颖壳的外表皮细胞。结果显示,在400×的视距下,亲本CSSL141的平均细胞长度为40.82 μm(图7-a);9311平均细胞长度为34.48 μm(图7-b);F4交换单株籽粒中标记区间为野生稻基因型的籽粒平均细胞长度为46.51 μm(图7-c);标记区间为9311基因型的交换单株籽粒平均细胞长度为35.71 μm(图7-d)。因此推断qGL12通过调控水稻颖壳细胞大小控制籽粒形状。 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图7颖壳细胞长度比较
a:亲本CSSL141;b:亲本9311;c:标记区间为野生稻基因型的交换单株;d:标记区间为9311基因型的交换单株
-->Fig. 7Comparison of length of lemma cell
a: CSSL141; b: 9311; c: Individual has wild rice genotype in the qGL12 interval; d: Individual has 9311 genotype in the qGL12 interval
-->
2.5 候选基因预测
qGL12最终定位于第12染色体15.69 kb的区间,两侧标记为DYB9.1和RM28586。根据Rice Genome Browser网站(http://rice.plantbiology.msu.edu/cgi-bin/ gbrowse/rice/#search)序列分析,该区间存在3个候选基因,分别为Os12g39650、Os12g39660和Os12g39670。Rice Genome Annotation Project网站基因注释为:Os12g39650编码一种微管蛋白,Os12g39660编码一种质膜钙转运ATP酶,Os12g39670尚未有明确功能(表4)。将9311与CSSL141中3个候选基因的测序结果进行对比,Os12g39650在两亲本编码区中有8个SNP,其中5个导致氨基酸发生改变;第403个碱基中T/A碱基发生替换导致半胱氨酸替换成丝氨酸;第427个碱基中G/T碱基发生替换导致天冬氨酸替换成酪氨酸;第460个碱基中G/A碱基发生替换导致缬氨酸替换成异亮氨酸;第1 450个碱基中G/A碱基发生替换导致丙氨酸替换成苏氨酸;第1 672个碱基中T/C碱基发生替换导致半胱氨酸替换成精氨酸。Os12g39660在两亲本编码区中有17个SNP,其中有2个导致氨基酸发生改变;第433个碱基中T/C碱基发生替换导致半胱氨酸替换成精氨酸;第2 754个碱基中T/A碱基发生替换导致天冬氨酸替换成谷氨酸。Table 4
表4
表4qGL12定位区间内候选基因
Table 4Candidate genes of qGL12
| 基因名称 Locus name | 基因注释 Gene product name |
|---|---|
| LOC_Os12g39650 | 微管蛋白Tubulin, putative, expressed |
| LOC_Os12g39660 | 质膜钙转运ATP酶Calcium-transporting ATPase, plasma membrane-type, putative, expressed |
| LOC_Os12g39670 | 表达蛋白Expressed protein |
新窗口打开
3 讨论
3.1 染色体片段置换系在水稻QTL定位中的应用
传统的QTL定位一般采用的是初级定位群体,例如RIL等,受遗传背景的影响,QTL定位的区间都很大,通常是10 cM以上,难以确定 QTL的作用效应是单个主效QTL的作用还是多个微效QTL的作用。对于精细定位,最关键的环节就是构建合适的定位群体,筛选与目标基因紧密连锁的分子标记。理想的定位群体是除了目标基因所在座位的局部区间外,其他部分的遗传背景是相同的[20]。染色体片段置换系仅在置换片段上发生分离,其余遗传背景与受体亲本完全一致,因此,消除了遗传背景的干扰,尤其是能够检测出遗传效应较小的QTL,是QTL定位的理想工具[21]。本研究中,粒型QTL的定位是建立在染色体片段置换系多年多点田间试验的基础上,利用构建的置换系群体定位到了37个粒型相关QTL,其中粒长相关的QTL qGL12在2年3点多环境试验中在4个环境中均被检测到,说明qGL12是可以在不同环境下稳定遗传的QTL。从CSSL群体中挑选粒型差异与9311较大而且携带qGL12置换片段的CSSL141与9311进行回交,通过F2、F3以及F4群体进行精细定位。最终将qGL12定位于第12染色体15.69 kb的区间,影响粒长、粒宽以及粒重,对粒长的贡献率最高。利用野生稻染色体片段置换系进行精细定位,能够充分消除遗传背景的影响,发掘野生稻中的优异基因,将成为一种主流方法。
3.2 qGL12通过调控颖壳细胞大小影响粒长
稻米的形状和大小受到颖壳形状和大小的严格控制,这表明颖壳的形态影响水稻的外观品质和产量,而颖壳的形态则是由细胞的大小、数目以及排列方式决定的。基因影响水稻粒型的方式可能有两种,一种为影响颖壳细胞的数量。例如GW7[22]通过改变细胞分裂模式来控制籽粒形状,使纵向细胞分裂增加横向细胞分裂减少形成细长的籽粒。一种为影响颖壳细胞的大小。如SRS1[23]通过调控颖壳细胞的长度与宽度影响水稻粒型;SRS5[24]和PGL1[25]主要通过影响颖壳细胞的长度影响籽粒形状。本研究利用扫描电镜对水稻9311、CSSL141以及2个F4交换单株进行水稻颖壳细胞观察,发现9311颖壳细胞的长度与宽度均比CSSL141小,2个交换单株中,目标区间为野生稻基因型的交换单株颖壳细胞的长度与宽度均比目标区间为9311基因型的交换单株大,这表明qGL12通过调控细胞的长度与宽度影响水稻粒型。3.3 qGL12应用前景
qGL12区间内含有3个基因,3个基因均未见报导与水稻粒长有关,测序结果表明2个候选基因在编码区均有非同义突变。下一步的研究将利用转基因等方法对2个候选基因进行功能验证,确定野生稻中控制粒长的基因。置换系CSSL141,或者CSSL141/9311 F4群体中携带qGL12的株系,可以直接作为水稻品种改良的育种材料,DYB9.1与RM28586可以作为分子标记用于辅助选择。本研究将为水稻粒长形成的分子机理研究提供新的基因资源,同时,为水稻粒长的改良提供了新的育种材料。4 结论
qGL12被定位于第12染色体15.69 kb的区间,两侧标记分别为DYB9.1和RM28586。区间内Os12g39650与Os12g39660 2个基因在编码区均有非同义突变,被确定为qGL12的候选基因,并且qGL12通过影响水稻籽粒颖壳细胞的大小影响粒长。The authors have declared that no competing interests exist.
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
| [1] | . 水稻是世界上最重要的粮食作物, 约1 万年前开始被驯化。由于水稻与其祖先野生种存在一定的遗传分化, 水稻的起源和驯化问题长期存在争议。本文综述了水稻起源和驯化方面的研究成果, 特别是近年来基因组学方面的证据, 认为水稻2 个亚种独立起源于野生祖先种内很早就分化的不同类群, 但一些驯化基因——控制重要农艺性状的基因, 可能首先在一个亚种中被驯化, 然后通过基因渐渗, 扩散到另一个亚种中。因此, 水稻驯化的关键是研究驯化基因的起源和扩散方式。随着大规模基因组测序技术的发展和相应数据分析方法的建立, 在全基因组水平对水稻及其祖先进行大规模分析, 已成为揭示水稻起源与驯化之谜的必由之路。 . 水稻是世界上最重要的粮食作物, 约1 万年前开始被驯化。由于水稻与其祖先野生种存在一定的遗传分化, 水稻的起源和驯化问题长期存在争议。本文综述了水稻起源和驯化方面的研究成果, 特别是近年来基因组学方面的证据, 认为水稻2 个亚种独立起源于野生祖先种内很早就分化的不同类群, 但一些驯化基因——控制重要农艺性状的基因, 可能首先在一个亚种中被驯化, 然后通过基因渐渗, 扩散到另一个亚种中。因此, 水稻驯化的关键是研究驯化基因的起源和扩散方式。随着大规模基因组测序技术的发展和相应数据分析方法的建立, 在全基因组水平对水稻及其祖先进行大规模分析, 已成为揭示水稻起源与驯化之谜的必由之路。 |
| [2] | . |
| [3] | . 水稻粒型是决定产量最重要的因素之一。籽粒大小主要是受遗传因子的严格调控。目前,已经克隆了多个控制水稻籽粒大小的( QTL),这些QTL可以作用在独立的遗传途径,与其他已鉴定的粒型基因一起主要参与蛋白酶体降解、激素和G蛋白介导的信号途径,调节细胞增殖和细胞伸长。本文综述了近年来克隆的主要控制水稻籽粒大小的QTL及其调控的分子机制,为理解粒型的分子遗传基础和水稻遗传改良提供理论依据。 . 水稻粒型是决定产量最重要的因素之一。籽粒大小主要是受遗传因子的严格调控。目前,已经克隆了多个控制水稻籽粒大小的( QTL),这些QTL可以作用在独立的遗传途径,与其他已鉴定的粒型基因一起主要参与蛋白酶体降解、激素和G蛋白介导的信号途径,调节细胞增殖和细胞伸长。本文综述了近年来克隆的主要控制水稻籽粒大小的QTL及其调控的分子机制,为理解粒型的分子遗传基础和水稻遗传改良提供理论依据。 |
| [4] | . . |
| [5] | . 用区间作图和混合线性模型的复合区间作图两种方法,对水稻(Oryza sativa L.)珍汕97和明恢63组合的重组自交系群体的谷粒外观性状--粒长、粒宽和粒形进行了数量性状基因(QTL)定位.用区间作图法在LOD≥2.4水平 上(近似于α=0.005),1998年对粒长、粒宽和粒形分别检测到6、2和2个QTLs;1999年对以上3个性状分别检测到3、2和2个QTLs. 其中7个QTLs在两年均检测到.位于第3染色体RG393-C1087区间的QTL效应大,同时影响粒长和粒形,两年贡献率分别为57.5%、 61.4%和26.7%、29.9%.位于第5染色体RG360-C734B区间的QTL效应大,同时影响粒宽和粒形,两年贡献率分别为44.2%、 53.2%和32.1%、36.0%.用混合线性模型的复合区间作图法在P=0.005水平上,对粒长、粒宽和粒形分别检测到8、5和5个QTLs,共解 释各自性状变异的58.81%、44.75%和57.47%.只检测到1个QTL与环境之间存在显著互作. . 用区间作图和混合线性模型的复合区间作图两种方法,对水稻(Oryza sativa L.)珍汕97和明恢63组合的重组自交系群体的谷粒外观性状--粒长、粒宽和粒形进行了数量性状基因(QTL)定位.用区间作图法在LOD≥2.4水平 上(近似于α=0.005),1998年对粒长、粒宽和粒形分别检测到6、2和2个QTLs;1999年对以上3个性状分别检测到3、2和2个QTLs. 其中7个QTLs在两年均检测到.位于第3染色体RG393-C1087区间的QTL效应大,同时影响粒长和粒形,两年贡献率分别为57.5%、 61.4%和26.7%、29.9%.位于第5染色体RG360-C734B区间的QTL效应大,同时影响粒宽和粒形,两年贡献率分别为44.2%、 53.2%和32.1%、36.0%.用混合线性模型的复合区间作图法在P=0.005水平上,对粒长、粒宽和粒形分别检测到8、5和5个QTLs,共解 释各自性状变异的58.81%、44.75%和57.47%.只检测到1个QTL与环境之间存在显著互作. |
| [6] | . 以第1染色体短臂RM1-RM3746和RM151-RM243区间内呈杂合、背景基本纯合的2个水稻剩余杂合体(RHL)衍生两个F6群体,将控制水稻粒长和粒宽的2个粒形QTL(qGL-1和qGW-1)定位于RM3746-RM243区间内。在此基础上,应用SSR标记检测,从其中1个群体中筛选到杂合区间分别为RM151-RM10404、RM10398-RM5359、RM10435-RM259和RM10381-RM243的4个单株,应用SSR标记进一步检测4套F2群体,从每套F2群体中分别筛选到母本珍汕97B和父本密阳46纯合型材料各10株,自交获得4套近等基因系材料并考查其粒长和粒宽。利用交迭重组染色体片段代换系分析法,将控制粒长和粒宽的QTL(qGL-1和qGW-1)界定于437.5kb的RM10390-RM1344区间和392.9kb的RM10376-RM10398区间,增效等位基因均来自母本珍汕97B,表明qGL-1和qGW-1是紧密连锁的不同座位。 . 以第1染色体短臂RM1-RM3746和RM151-RM243区间内呈杂合、背景基本纯合的2个水稻剩余杂合体(RHL)衍生两个F6群体,将控制水稻粒长和粒宽的2个粒形QTL(qGL-1和qGW-1)定位于RM3746-RM243区间内。在此基础上,应用SSR标记检测,从其中1个群体中筛选到杂合区间分别为RM151-RM10404、RM10398-RM5359、RM10435-RM259和RM10381-RM243的4个单株,应用SSR标记进一步检测4套F2群体,从每套F2群体中分别筛选到母本珍汕97B和父本密阳46纯合型材料各10株,自交获得4套近等基因系材料并考查其粒长和粒宽。利用交迭重组染色体片段代换系分析法,将控制粒长和粒宽的QTL(qGL-1和qGW-1)界定于437.5kb的RM10390-RM1344区间和392.9kb的RM10376-RM10398区间,增效等位基因均来自母本珍汕97B,表明qGL-1和qGW-1是紧密连锁的不同座位。 |
| [7] | . 用二个籼型杂交组合的F2群体,构建了二张分别具有89和93个标记位点RFLP图谱。在此基础上,应用方差分析法和区间作图法对控制籼稻粒形性状的QTLS进行定位。 . 用二个籼型杂交组合的F2群体,构建了二张分别具有89和93个标记位点RFLP图谱。在此基础上,应用方差分析法和区间作图法对控制籼稻粒形性状的QTLS进行定位。 |
| [8] | . 为了发掘单株产量及其构成因素相关性状的数量性状基因座(QTL),本研究以9311/日本晴染色体片段置换系群体为材料,调查了单株产量及单株产量构成因素(单株实粒数、单株总颖花数、结实率、每穗实粒数、有效穗数、每穗颖花数、千粒质量等8个性状)。利用Ici Mapping v3.1软件,将分子标记检测结果与田间性状调查值相结合,定位了与单株产量、单株实粒数、千粒质量、每穗颖花数、结实率、单株总颖花数6个性状有关的QTL,未定位到与有效穗数(EPN)和每穗实粒数(GPP)有关的QTL。共定位到的10个相关QTLs,分别分布于第1条、第2条、第5条、第7条、第8条染色体的7个区间,贡献率为7.52%~44.59%,其中4个QTLs的贡献率大于10.00%。单株产量q GY1,单株实粒数q GN1,结实率q SSR1.2、q SSR2和q SSR8,加性效应值为负值,表明9311的等位基因表现为增效作用;单株总颖花数q SN2,结实率q SSR1.1,千粒质量q TGW5,每穗颖花数q SPP5和q SPP7,加性效应值为正值,表明日本晴的等位基因表现为增效作用。10个QTLs位点中,除q SN2、q TGW5、q SPP5与已克隆的LP、q SW5、Os NADH-GOGAT2可能位于同一区域外,其余7个位点均未被克隆或精细定位。 . 为了发掘单株产量及其构成因素相关性状的数量性状基因座(QTL),本研究以9311/日本晴染色体片段置换系群体为材料,调查了单株产量及单株产量构成因素(单株实粒数、单株总颖花数、结实率、每穗实粒数、有效穗数、每穗颖花数、千粒质量等8个性状)。利用Ici Mapping v3.1软件,将分子标记检测结果与田间性状调查值相结合,定位了与单株产量、单株实粒数、千粒质量、每穗颖花数、结实率、单株总颖花数6个性状有关的QTL,未定位到与有效穗数(EPN)和每穗实粒数(GPP)有关的QTL。共定位到的10个相关QTLs,分别分布于第1条、第2条、第5条、第7条、第8条染色体的7个区间,贡献率为7.52%~44.59%,其中4个QTLs的贡献率大于10.00%。单株产量q GY1,单株实粒数q GN1,结实率q SSR1.2、q SSR2和q SSR8,加性效应值为负值,表明9311的等位基因表现为增效作用;单株总颖花数q SN2,结实率q SSR1.1,千粒质量q TGW5,每穗颖花数q SPP5和q SPP7,加性效应值为正值,表明日本晴的等位基因表现为增效作用。10个QTLs位点中,除q SN2、q TGW5、q SPP5与已克隆的LP、q SW5、Os NADH-GOGAT2可能位于同一区域外,其余7个位点均未被克隆或精细定位。 |
| [9] | . . |
| [10] | . The GS3 locus located in the pericentromeric region of rice chromosome 3 has been frequently identified as a major QTL for both grain weight (a yield trait) and grain length (a quality trait) in the literature. Near isogenic lines of GS3 were developed by successive crossing and backcrossing Minghui 63 (large grain) with Chuan 7 (small grain), using Minghui 63 as the recurrent parent. Analysis of a random subpopulation of 201 individuals from the BC 3 F 2 progeny confirmed that the GS3 locus explained 80 90% of the variation for grain weight and length in this population. In addition, this locus was resolved as a minor QTL for grain width and thickness. Using 1,384 individuals with recessive phenotype (large grain) from a total of 5,740 BC 3 F 2 plants and 11 molecular markers based on sequence information, GS3 was mapped to a DNA fragment approximately 7.9 kb in length. A full-length cDNA corresponding to the target region was identified, which provided complete sequence information for the GS3 candidate. This gene consists of five exons and encodes 232 amino acids with a putative PEBP-like domain, a transmembrane region, a putative TNFR/NGFR family cysteine-rich domain and a VWFC module. Comparative sequencing analysis identified a nonsense mutation, shared among all the large-grain varieties tested in comparison with the small grain varieties, in the second exon of the putative GS3 gene. This mutation causes a 178-aa truncation in the C-terminus of the predicted protein, suggesting that GS3 may function as a negative regulator for grain size. Cloning of such a gene provided the opportunity for fully characterizing the regulatory mechanism and related processes during grain development. |
| [11] | . Grain size determines grain weight and affects grain quality.GS2, a dominant QTL, encodes Growth-Regulating Factor 4 (OsGRF4), which acts as a transcriptional regulator. A mutation in this gene at the OsmiR396c target site results in elevated transcript levels ofGS2and accumulation ofGS2leads to enlarged cell size and increased cell number, which in turn results in enhanced grain weight and yield. |
| [12] | . Abstract Grain weight is one of the most important components of grain yield and is controlled by quantitative trait loci (QTLs) derived from natural variations in crops. However, the molecular roles of QTLs in the regulation of grain weight have not been fully elucidated. Here, we report the cloning and characterization of GW2, a new QTL that controls rice grain width and weight. Our data show that GW2 encodes a previously unknown RING-type protein with E3 ubiquitin ligase activity, which is known to function in the degradation by the ubiquitin-proteasome pathway. Loss of GW2 function increased cell numbers, resulting in a larger (wider) spikelet hull, and it accelerated the grain milk filling rate, resulting in enhanced grain width, weight and yield. Our results suggest that GW2 negatively regulates cell division by targeting its substrate(s) to proteasomes for regulated proteolysis. The functional characterization of GW2 provides insight into the mechanism of seed development and is a potential tool for improving grain yield in crops. |
| [13] | . |
| [14] | . Increased crop yields are required to support rapid population growth worldwide. Grain weight is a key component of rice yield, but the underlying molecular mechanisms that control it remain elusive. Here, we report the cloning and characterization of a new quantitative trait locus (QTL) for the control of rice grain length, weight and yield. This locus, GL3.1, encodes a protein phosphatase kelch (PPKL) family - Ser/Thr phosphatase. GL3.1 is a member of the large grain WY3 variety, which is associated with weaker dephosphorylation activity than the small grain FAZ1 variety. GL3.1-WY3 influences protein phosphorylation in the spikelet to accelerate cell division, thereby resulting in longer grains and higher yields. Further studies have shown that GL3.1 directly dephosphorylates its substrate, Cyclin-T1;3, which has only been rarely studied in plants. The downregulation of Cyclin-T1;3 in rice resulted in a shorter grain, which indicates a novel function for Cyclin-T in cell cycle regulation. Our findings suggest a new mechanism for the regulation of grain size and yield that is driven through a novel phosphatase-mediated process that affects the phosphorylation of Cyclin-T1;3 during cell cycle progression, and thus provide new insight into the mechanisms underlying crop seed development. We bred a new variety containing the natural GL3.1 allele that demonstrated increased grain yield, which indicates that GL3.1 is a powerful tool for breeding high-yield crops. |
| [15] | . |
| [16] | . Increasing crop yield is one of the most important goals of plant science research. Grain size is a major determinant of grain yield in cereals and is a target trait for both domestication and artificial breeding(1). We showed that the quantitative trait locus (QTL) GS5 in controls grain size by regulating grain width, filling and weight. GS5 encodes a putative serine carboxypeptidase and functions as a positive regulator of grain size, such that higher expression of GS5 is correlated with larger grain size. Sequencing of the promoter region in 51 accessions from a wide geographic range identified three haplotypes that seem to be associated with grain width. The results suggest that natural variation in GS5 contributes to grain size diversity in and may be useful in improving yield in and, potentially, other crops(2). |
| [17] | . Copy number variants (CNVs) are associated with changes in gene expression levels and contribute to various adaptive traits. Here we show that a CNV at the Grain Length on Chromosome 7 (GL7) locus contributes to grain size diversity in rice (Oryza sativa L.). GL7 encodes a protein homologous to Arabidopsis thaliana LONGIFOLIA proteins, which regulate longitudinal cell elongation. Tandem duplication of a 17.1-kb segment at the GL7 locus leads to upregulation of GL7 and downregulation of its nearby negative regulator, resulting in an increase in grain length and improvement of grain appearance quality. Sequence analysis indicates that allelic variants of GL7 and its negative regulator are associated with grain size diversity and that the CNV at the GL7 locus was selected for and used in breeding. Our work suggests that pyramiding beneficial alleles of GL7 and other yield- and quality-related genes may improve the breeding of elite rice varieties. |
| [18] | . Grain size and shape are important components of grain yield and quality and have been under selection since cereals were first domesticated. Here, we show that a quantitative trait locus GW8 is synonymous with OsSPL16, which encodes a protein that is a positive regulator of cell proliferation. Higher expression of this gene promotes cell division and grain filling, with positive consequences for grain width and yield in rice. Conversely, a loss-of-function mutation in Basmati rice is associated with the formation of a more slender grain and better quality of appearance. The correlation between grain size and allelic variation at the GW8 locus suggests that mutations within the promoter region were likely selected in rice breeding programs. We also show that a marker-assisted strategy targeted at elite alleles of GS3 and OsSPL16 underlying grain size and shape can be effectively used to simultaneously improve grain quality and yield. |
| [19] | . Grain size and weight are important determinants of rice yield. The identification of beneficial genes from wild rice that have been lost or weakened in cultivated rice has become increasingly important for modern breeding strategies. In this study, we constructed a set of chromosome segment substitution lines (CSSLs) of wild rice,Oryza rufipogonwith theindicacultivar 9311 genetic background. Four grain-related traits, i.e., grain length (GL), grain width (GW), length-width ratio (LWR), and thousand grain weight (TGW), were screened across six environments. A total of 37 quantitative trait loci (QTLs) were identified in these environments and mapped to 12 chromosomes. Sixteen QTLs were detected in at least two environments, and two QTL clusters were observed on Chr. 4 and Chr. 8. Based on a comparative analysis with QTLs identified in previous studies, the CSSLs betweenOryza rufipogonaccessions and 9311 had high genetic diversity. Among the sixteen stable QTLs, seven for TGW, LWR, GL, and GW were not previously identified, indicating potentially novel alleles from wild rice. These CSSLs provide powerful tools for functional studies and the cloning of essential genes in rice; furthermore, we identified elite germplasm for rice variety improvement. |
| [20] | . 染色体片段置换系除含有供体基因组中的1个或几个片段外,其余背景与受体亲本完全相同,是用于QTL鉴定、精细定位、图位克隆、互作分析和品种改良等的理想材料.本文概述了作物染色体片段置换系的构建、特点、类型及在作物遗传育种中的应用价值. . 染色体片段置换系除含有供体基因组中的1个或几个片段外,其余背景与受体亲本完全相同,是用于QTL鉴定、精细定位、图位克隆、互作分析和品种改良等的理想材料.本文概述了作物染色体片段置换系的构建、特点、类型及在作物遗传育种中的应用价值. |
| [21] | . |
| [22] | . 正With the support by the National Natural Science Foundation of China,the Ministry of Science and Technology of China and the National Special Project of China,Dr.Fu Xiangdong's laboratory at the Institute of Genetics and Developmental Biology,Chinese Academy of Sciences,reported the OsSPL16—GW7regulatory module simultaneously improves rice yield and grain quality,which was published in Nature Ge- |
| [23] | . Abstract The causal gene of a novel small and round seed mutant 1 (srs1) was identified in rice by map-based cloning and named SMALL AND ROUND SEED 1 (SRS1). The SRS1 gene is identical to the previously identified DENSE AND ERECT PANICLE 2 (DEP2). The SRS1/DEP2 gene encodes a novel protein of 1365 amino acids residues without known functional domains. In the longitudinal direction of the lemma, both cell length and cell number are reduced in srs1-1 compared to the wild type, whereas in the lateral cross section of the lemma, cell length in srs1-1 is greater than that in the wild type, but the cell number in srs1-1 is the same as that in wild type. These results suggest that the small and round seed phenotype of srs1-1 is due to the reduction in both cell length and cell number in the longitudinal direction, and the elongation of the cells in the lateral direction of the lemma. The SRS1 mRNA and proteins are abundant in wild type rice specifically in young organs, namely young leaves, internodes and panicles. Interestingly, the tissues expressing SRS1 are closely related to the tissues that exhibit abnormalities in the srs1 mutants. |
| [24] | . |
| [25] | . Grain size is a major yield component in rice, and partly controlled by the sizes of the lemma and palea. Molecular mechanisms controlling the sizes of these organs largely remain unknown. In this study, we show that an antagonistic pair of basic helix-loop-helix (bHLH) proteins is involved in determining rice grain length by controlling cell length in the lemma/palea. Overexpression of an atypical bHLH, named POSITIVE REGULATOR OF GRAIN LENGTH 1 (PGL1), in lemma/palea increased grain length and weight in transgenic rice. PGL1 is an atypical non-DNA-binding bHLH and assumed to function as an inhibitor of a typical DNA-binding bHLH through heterodimerization. We identified the interaction partner of PGL1 and named it ANTAGONIST OF PGL1 (APG). PGL1 and APG interacted in vivo and localized in the nucleus. As expected, silencing of APG produced the same phenotype as overexpression of PGL1, suggesting antagonistic roles for the two genes. Transcription of two known grain-length-related genes, GS3 and SRS3, was largely unaffected in the PGL1-overexpressing and APG-silenced plants. Observation of the inner epidermal cells of lemma revealed that are caused by increased cell length. PGL1-APG represents a new grain length and weight-controlling pathway in which APG is a negative regulator whose function is inhibited by PGL1. |
