0 引言
【研究意义】MHC(major histocompatibility complex)是所有生物主要组织相容性复合体的统称,常见类型有3类,即Class-Ⅰ、Class-Ⅱ和Class-Ⅲ类基因。MHC不仅控制着同种移植排斥反应,更重要的是与生物机体免疫应答、免疫调节及某些病理状态的产生均密切相关。不同种类哺乳动物的MHC基因序列结构、名称存在差异[1,2,3,4]。双峰驼因其应对恶劣环境挑战的能力而闻名。关于MHC的研究,双峰驼是一种具有实际意义的生物模型[5,6]。定位并注释双峰驼中的MHC基因,对双峰驼MHC的进一步研究,以及对双峰驼机体免疫机制的研究有重要的科学意义。【前人研究进展】对于MHC的研究工作最早可以追溯到1936年,国外****在研究小鼠肿瘤细胞排斥反应中首次发现了MHC基因;1951年,研究者发现了小鼠的MHC-H2系统;1958年,DAUSSET首次发现了人类的白细胞抗原;1961年SCHIERMAN和NORDSKOG确定了家禽中的MHC为一种红细胞抗原,并将其命名为B[2,7];到了20世纪70代,人们发现在遗传上与H2同源的系统同样存在于其它动物中[8,9,10];近年来已确定几乎所有的脊椎动物都存在MHC基因,且各国****不断丰富着不同种属动物MHC基因的研究。2012年吉日木图教授等首次完成双峰驼全基因组序列图谱绘制和解析。2016年捷克科学家通过FISH技术将单峰驼MHC基因定位到了20号染色体短臂上(20q12)[11]。【本研究切入点】目前双峰驼参考基因组的组装及注释水平不完善,在双峰驼基因组中MHC基因并没有得到组装和注释[12]。传统的基因定位方法普遍采用的是杂交,侧交和自交,分别求出基因间的交换率和相对距离,然后在染色体上确定基因间的排列顺序,这些方法操作繁琐、复杂,成本也很昂贵。【拟解决的关键问题】本研究通过利用目前已知组装水平最好的人类MHC基因和与双峰驼亲缘关系较近的牛MHC基因为种子序列,采用比较基因组学方法来探究物种间基因的同源性关系,准确定位并注释到双峰驼中的MHC基因,为双峰驼MHC基因的进一步研究奠定理论基础。1 材料与方法
试验于2017年6—9月在内蒙古农业大学的内蒙古自治区基础兽医学重点实验室(农业部动物疾病临床诊疗技术重点实验室)进行。1.1 序列来源
双峰驼MHC基因序列从家养阿拉善双峰驼的参考基因组中获得,参考基因组Camelus bactrianus (assembly Ca_bactrianus_MBC_1.0)在NCBI上下载得到,人类HLA、牛BoLA基因序列都是从ensemble网站下载得到。相关信息如下(表1)。Table 1
表1
表1双峰驼、人类、牛基因序列及数据来源
Table 1Gene sequences and data sources of Bactrian camel, human, bovine
| 名称 Name | 来源 Source | 网址 URL |
|---|---|---|
| 家养双峰驼参考基因组 Reference genome of domestic Bactrian camels | NCBI | ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/767/855/GCF_000767855.1_Ca_bactrianus_MBC_1.0/GCF_000767855.1_Ca_bactrianus_MBC_1.0_genomic.fna.gz |
| 人类HLA编码序列 Human HLA coding sequence | ensemble | ftp://ftp.ensembl.org/pub/release-90/fasta/homo_sapiens/cdna/ |
| 人类HLA全基因组序列 Human HLA whole genome sequence | ensemble | ftp://ftp.ensembl.org/pub/release-90/fasta/homo_sapiens/dna/ |
| 人类HLA注释文件 Human HLA annotation file | ensemble | ftp://ftp.ensembl.org/pub/release-90/gtf/homo_sapiens |
| 牛BoLA编码序列 Cattle BoLA coding sequence | ensemble | ftp://ftp.ensembl.org/pub/release-90/fasta/bos_taurus/cdna/ |
| 牛BoLA全基因组序列 Cattle BoLA whole genome sequence | ensemble | ftp://ftp.ensembl.org/pub/release-90/fasta/bos_taurus/dna/ |
| 牛BoLA注释文件 Cattle BoLA annotation file | ensemble | ftp://ftp.ensembl.org/pub/release-90/gtf/bos_taurus |
新窗口打开
1.2 方法
1.2.1 利用blast v2.3.0软件将分别从表1所示网址提取的HLA、BoLA基因编码序列,分别与双峰驼转录本进行blastn序列比对,识别出相似度较高的scaffolds;再通过分析HLA、BoLA比对在scaffolds的位置顺序,从而对多条scaffold进行拼接。1.2.2 分别提取HLA、BoLA基因组序列与双峰驼已拼接的scaffolds进行基因组共线性分析,利用lastz v.1.04.00软件建立起已拼接的scaffolds与HLA、BoLA基因组序列的线性关系,以判断筛选出的scaffolds是否准确。
1.2.3 通过分析MHC在两物种间的线性关系,利用BLAT v.35软件提取出MHC在双峰驼基因组的基因序列,并对这些基因序列进行基因注释。
1.2.4 利用MEGA 7.0.26软件对得到的双峰驼MHC基因绘制系统进化树。
2 结果
2.1 blastn序列对比及拼接
分别以人类HLA、牛BoLA基因编码序列与双峰驼转录本进行blastn序列对比,结果识别出了相似度较高的3条scaffold,即NW_011511766.1(全长4.1M)、NW_011515227.1(全长1.2M)和NW_011514613.1(全长15K),进一步通过分析HLA、BoLA比对在scaffolds的位置顺序,发现CBLAⅠ类和Ⅲ类基因分布在NW_011515227.1上,而CBLAⅡ类基因分布在NW_011511766.1和NW_011514613.1上对上述3条scaffolds进行拼接,得到双峰驼MHC的Pseudo chromosome,其结构示意图如图1。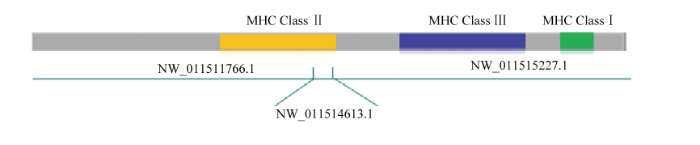 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图1双峰驼MHC-Pseudo chromosome结构示意图
-->Fig. 1Schematic diagram of structure for Bactrian camel MHC-Pseudo chromosome
-->
2.2 共线性分析
2.2.1 人类HLA与双峰驼Pseudo chromosome共线性分析 提取人类MHC基因(HLA)的基因组DNA序列,与双峰驼基因组进行共线性分析。在基因组共线性分析中一个共线性基因对应一个点,而密集的共线性基因可以连接成片段,并以线条的形式呈现在图中。绘制双峰驼MHC基因与人类HLA基因共线性关系图(图2)。可以看出,有很多双峰驼MHC基因片段和人类HLA基因片段是共线性关系,即存在较长的线性片段,有力地支持了本研究所定位到的双峰驼MHC基因的准确性,从而说明双峰驼的MHC基因家族主要分布在笔者拼接的scaffolds上。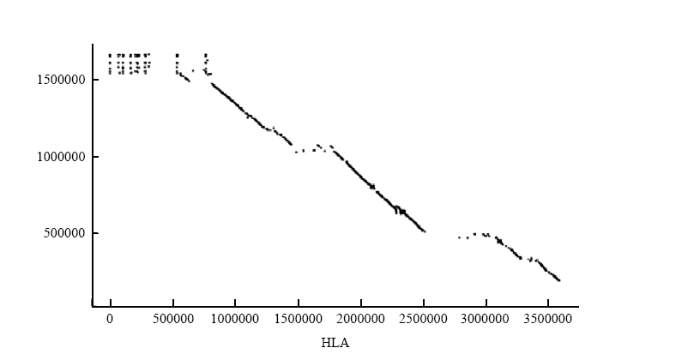 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图2人类HLA基因与双峰驼MHC基因的共线性关系比较
横轴为人HLA的基因序列,纵轴为双峰驼MHC的基因序列
-->Fig. 2Comparison of collinearity between human HLA gene and Bactrian camel MHC gene
The horizontal axis is the gene sequence of human HLA, and the vertical axis is the gene sequence of Bactrian camel MHC
-->
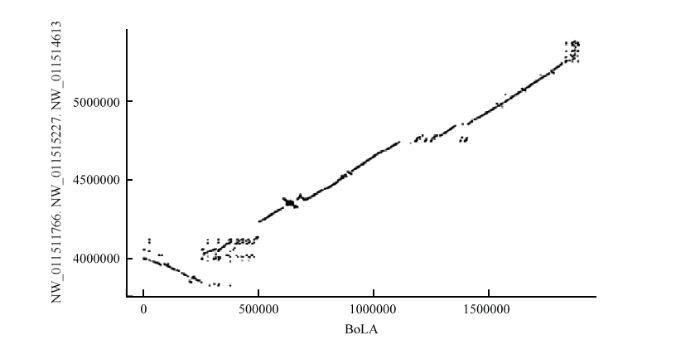
2.2.2 牛BoLA与双峰驼pseudo chromosome 共线性分析 同理,提取与双峰驼亲缘关系最近的牛的MHC基因(BoLA)[13]的基因组DNA序列,与双峰驼基因组进行共线性分析,结果显示牛BoLA基因与双峰驼的NW_011511766.1(全长4.1M)、NW_011515227.1(全长1.2M)和NW_011514613.1(全长15K)相似度最高,且Ⅰ类和Ⅲ类基因分布在NW_011515227.1上,而Ⅱ类基因分布在NW_011511766.1和NW_011514613.1上,进一步分析得知Ⅱ类基因主要分布在NW_011511766.1 的3.5—4.1M的位置。在基因组共线性分析中一个共线性基因对应一个点,密集的共线性基因可以连接成片段,并以线条的形式呈现在图中。绘制双峰驼MHC基因与牛BoLA基因共线性关系图(图3)。可以清楚的表明,有很多双峰驼MHC基因片段和牛BoLA基因片段是共线性关系,即存在较长的线性片段,有力地支持了本研究所定位到的双峰驼MHC基因的准确性,更加说明双峰驼的MHC基因家族主要分布在拼接的scaffolds上,该结果与结果1的一致。但在局部双峰驼的片段呈现更多的碎片,即当纵坐标低于4 000 000时牛BoLA与双峰驼pseudo chromosome 共线性区域出现倒置现象。
 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图3牛BoLA基因与双峰驼MHC基因的共线性关系比较
横轴为牛BoLA的基因序列,纵轴为双峰驼MHC的基因序列
-->Fig. 3Comparison of collinearity between bovine BoLA gene and Bactrian camel MHC gene
The horizontal axis is the gene sequence of bovine BoLA, and the vertical axis is the gene sequence of Bactrian camel MHC
-->
2.3 BLAT分析
将存在共线性区域的基因组序列提取出来,与比对到双峰驼上的CBLA基因的编码序列运用BLAT v.35进行分析,从而把双峰驼CBLA编码定位在基因组上。比对分析结果显示,在双峰驼基因组中共识别出24个与牛BoLA基因高度相似的基因,即在本次试验中发现双峰驼的CBLA包含24个基因,其中Ⅰ类基因1个,Ⅱ类10个, Ⅲ类基因13个(表2)。并由此可绘制人/牛/双峰驼 MHC基因个数统计表(表3)。Table 2
表2
表2BLAT分析找到的CBLA基因
Table 2The CBLA gene found by BLAT analysis
| 基因区Class | Ⅰ | Ⅱ | Ⅲ |
|---|---|---|---|
| 基因编码 Genetic code | XM 010961543.1 | XM 010949122.1 XM 010958395.1 XM 010949120.1 XM 010949126.1 XM 010949114.1 XM 010949113.1 XM 010949105.1 XM 010949127.1 XM 010949125.1 XM 010949153.1 | XM 010961490.1 XM 010961476.1 XM 010961495.1 XM 010961533.1 XM 010961451.1 XM 010961470.1 XM 010961594.1 XM 010961447.1 XM 010961538.1 XM 010961482.1 XM 010961484.1 XM 010961536.1 XM 010961596.1 |
新窗口打开
Table 3
表3
表3人/牛/双峰驼 MHC基因个数统计表
Table 3Statistics table of MHC gene number in Human / bovine / Bactrian camels
| 基因区Class | 人类 MHC基因HLA | 牛MHC基因BoLA | 双峰驼MHC基因CBLA |
|---|---|---|---|
| Ⅰ | 36/106 | 1 | 1 |
| Ⅱ | 33/59 | 11 | 10 |
| Ⅲ | 59/59 | 13 | 13 |
新窗口打开
2.4 基因注释
对照从ensemble上获取的人类HLA、牛BoLA已知MHC基因注释信息对找到的这24个基因进行注释,可绘制双峰驼CBLA基因注释信息表(表4)。Table 4
表4
表4双峰驼CBLA基因注释信息
Table 4CBLA gene annotation information of Bactrian camel
| 基因编码 Gene id | 长度 Length | 外显子 Exon | 位置 Scaffold | 起始点 Start | 终止点 End | 注释信息 Description |
|---|---|---|---|---|---|---|
| XM 010949105.1 | 2866 | 5 | NW 011511766.1 | 3837314 | 3842070 | PREDICTED:CBLA-DOA |
| XM 010949113.1 | 1189 | 5 | NW 011511766.1 | 3874933 | 3878475 | PREDICTED:CBLA-DMA |
| XM 010949114.1 | 1391 | 6 | NW 011511766.1 | 3885523 | 3891578 | PREDICTED:CBLA-DMB |
| XM 010949120.1 | 1381 | 6 | NW 011511766.1 | 3965547 | 3972059 | PREDICTED:CBLA-DOB |
| XM 010949122.1 | 813 | 5 | NW 011511766.1 | 4106164 | 4114129 | PREDICTED:CBLA-DRB2 |
| XM 010949125.1 | 1177 | 5 | NW 011511766.1 | 4047230 | 4051352 | PREDICTED:CBLA-DQA |
| XM 010949126.1 | 1033 | 4 | NW 011511766.1 | 4031979 | 4039091 | PREDICTED:CBLA-DQB |
| XM 010949127.1 | 1307 | 5 | NW 011511766.1 | 4120230 | 4124972 | PREDICTED:CBLA-DRA |
| XM 010949153.1 | 759 | 4 | NW 011511766.1 | 3992073 | 3995513 | PREDICTED:CBLA-DYA |
| XM 010958395.1 | 1315 | 7 | NW 011514613.1 | 5057 | 14519 | PREDICTED:CBLA-DRB3 |
| XM 010961447.1 | 1407 | 9 | NW 011515227.1 | 42577 | 46241 | PREDICTED:PBX2 |
| XM 010961451.1 | 1602 | 8 | NW 011515227.1 | 67307 | 73614 | PREDICTED:PPT2 |
| XM 010961470.1 | 1452 | 8 | NW 011515227.1 | 175625 | 182554 | PREDICTED:STK19 |
| XM 010961476.1 | 2773 | 18 | NW 011515227.1 | 207277 | 218549 | PREDICTED:C2 |
| XM 010961482.1 | 2376 | 1 | NW 011515227.1 | 303003 | 305378 | PREDICTED:HSPA1A |
| XM 010961484.1 | 2500 | 2 | NW 011515227.1 | 305769 | 309435 | PREDICTED:HSPA1L |
| XM 010961490.1 | 2657 | 25 | NW 011515227.1 | 340559 | 355003 | PREDICTED:MSH5 |
| XM 010961495.1 | 1158 | 6 | NW 011515227.1 | 357933 | 363242 | PREDICTED:CLIC1 |
| XM 010961533.1 | 668 | 6 | NW 011515227.1 | 462534 | 464279 | PREDICTED:AIF1 |
| XM 010961536.1 | 688 | 4 | NW 011515227.1 | 488003 | 489391 | PREDICTED:LTA |
| XM 010961538.1 | 1318 | 5 | NW 011515227.1 | 499275 | 507275 | PREDICTED:NFKBIL1 |
| XM 010961543.1 | 1513 | 9 | NW 011515227.1 | 1073777 | 1105682 | PREDICTED:CBLA-A |
| XM 010961594.1 | 6598 | 30 | NW 011515227.1 | 17926 | 38510 | PREDICTED:NOTCH4 |
| XM 010961596.1 | 12304 | 45 | NW 011515227.1 | 105330 | 155862 | PREDICTED:TNXB |
新窗口打开
2.5 双峰驼MHC基因绘制系统进化树
对得到的双峰驼MHC基因绘制系统进化树,结果显示注释的CBLA Class-Ⅰ类和Class-Ⅱ类基因在同一分支(图4),说明CBLA Class-Ⅰ类和Class-Ⅱ类基因进化关系最近。 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图4双峰驼CBLA基因系统进化树
蓝色线表示CBLA-Ⅰ类基因,红色线表示CBLA-Ⅱ基因,黑色线表示CBLA-Ⅲ类基因
-->Fig. 4Phylogenetic tree of Bactrian camel CBLA gene
The blue line indicates the CBLA class- I gene, the red line indicates the CBLA-II gene, and the black line indicates the CBLA class-Ⅲ gene
-->
3 讨论
近年来出现了很多MHC基因的定位方法,常用的方法有体细胞杂交法、克隆嵌板法、原位杂交和荧光原位杂交法(FISH)、连锁分析法。如Martin Plasil在2016年通过荧光原位杂交法将单峰驼的12个MHC基因定位到了20号染色体短臂(20q12)上的具体位置,并绘制了物理图谱。本研究通过利用目前已知组装水平最好的人类HLA基因和与双峰驼亲缘关系较近的牛BoLA基因[14]作为种子序列来定位并注释双峰驼的MHC基因。通过与两个参考物种的比较基因组学分析,最终在双峰驼基因组序列中找到了24个MHC基因(其中Ⅰ类基因1个,Ⅱ类10个, Ⅲ类基因13个),并绘制出了双峰驼MHC的Pseudo chromosome。相比于以前的MHC基因的定位方法,该方法操作简便、检测迅速、成本低廉,有望推广应用于其他物种MHC基因的定位和注释。共线性描述了由同一祖先型分化而来的不同物种间基因的类型及相对顺序的保守性,是物种间的一种关系,体现了基因的同源性以及基因的排列顺序。本研究通过使用lastz软件分别对人类HLA、牛BoLA基因与拼接出来的双峰驼CBLA基因序列进行共线性分析发现,其绝大部分基因具有很高的共线性,说明这3种动物MHC基因组同源性很高[15]。对比图2、3,可以看到牛BoLA基因序列与拼接出来的双峰驼CBLA基因序列具有更好的共线性,即牛与双峰驼的的MHC基因同源性更高。说明了相比于人类,牛与双峰驼亲缘关系更近。这一结果与WU等[16]所得结论一致,从而证明本试验所用方法的结果是准确的。再对3个物种进行BLAT分析,发现其MHC基因组上的等位基因种类、结构和位置也极其相似,因此可以认定MHC基因为直向同源基因的一种[17,18]。对于像MHC基因这种功能基因的研究,通过选取其他近缘生物基因组为参照进行比较基因组学研究,利用计算机的强大处理信息能力鉴别存在于各物种的直向同源基因,并选择具有重要生物学功能或价值的候选标记基因进行试验研究,可以迅速实现动物基因组比较信息的物种间转移,发掘更多新的功能基因和定位信息,从而加快对其他物种基因组学研究[19]。
目前,人类已知的HLA-Ⅰ类基因共有106个,其中36个编码基因,其余为假基因;HLA-Ⅱ类基因59个,其中编码基因33个,其余为假基因;HLA-Ⅲ类基因59个,均为编码基因[20,21]。与人类HLA基因数量相比,双峰驼中所注释到的MHC基因数量相差较大[22,23],推测原因可能有以下几点:1)双峰驼参考基因组的组装水平较差;2)双峰驼MHC基因具有低多样性特点[24,25];3)人类与双峰驼亲缘关系较远,基因在物种进化过程中发生改变[26,27,28]。而与亲缘关系较近的牛BoLA相比,双峰驼MHC基因数量基本一致(表3)。并且,从表3中能够明显观察到牛和双峰驼的MHCⅠ类基因的个数显著少于人类MHCⅠ类基因的个数[29,30,31]。因此,比较研究双峰驼与人类MHC基因在进化过程中发生的差异将是一个重要的研究课题,这对于进一步揭示双峰驼MHC的作用具有重要意义,也将是今后重点研究的新方向,同时,也将对牛和双峰驼的基因组研究起推动作用。
在对牛BoLA与双峰驼pseudo chromosome 进行共线性分析后发现当纵坐标低于4 000 000时牛BoLA与双峰驼pseudo chromosome 共线性区域出现倒置现象,说明双峰驼CBLA部分基因出现倒置。这与2016年PLASIL通过FISH技术绘制的物理图谱结果相似[11]。因此推测该部分基因出现倒置的原因可能是在物种进化过程中发生倒置改变,这种倒置改变增加了MHC基因的多态性。关于这一现象的发生机制以及意义仍需进一步的研究。
4 结论
本研究运用比较基因组学方法,利用目前已知组装水平最好的人类HLA基因和与双峰驼亲缘关系较近的牛BoLA基因作为种子序列,成功构建了一种新型的用于定位并注释双峰驼的MHC基因的方法。使用该方法找到了存在于双峰驼基因组中的24个MHC基因,其中Ⅰ类基因有1个,Ⅱ类基因有10个,Ⅲ类基因有13个。分别分布在NW_011511766.1(全长4.1M)、NW_011515227.1(全长1.2M)和NW_011514613.1(全长15K)3条scaffolds上,且Ⅰ类和Ⅲ类基因分布在NW_011515227.1上,而Ⅱ类基因分布在NW_011511766.1和NW_011514613.1上,进一步分析得知Ⅱ类基因主要分布在NW_011511766.1 的3.5—4.1M的位置,可绘制双峰驼MHC的Pseudo chromosome长约1.8M,并对双峰驼这24个MHC基因进行了信息注释,为双峰驼MHC的进一步研究奠定了理论基础。The authors have declared that no competing interests exist.
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
| [1] | . . |
| [2] | . 主要组织相容性复合体(Major Histocompatibility Complex,MHC)是脊椎动物体内与免疫功能密切相关的一类庞大的基因家族,在调节免疫系统识别自身和异物之间起着重要的作用。最近对其在动物体内遗传进化等方面进行了广泛的研究,本文从鱼类、两栖类、鸟类和哺乳类几个方面分别介绍MHC基因的研究进展。本文从MHC基因的结构、检测方法、功能等方面进行了相关的阐述。 . 主要组织相容性复合体(Major Histocompatibility Complex,MHC)是脊椎动物体内与免疫功能密切相关的一类庞大的基因家族,在调节免疫系统识别自身和异物之间起着重要的作用。最近对其在动物体内遗传进化等方面进行了广泛的研究,本文从鱼类、两栖类、鸟类和哺乳类几个方面分别介绍MHC基因的研究进展。本文从MHC基因的结构、检测方法、功能等方面进行了相关的阐述。 |
| [3] | . |
| [4] | . The MHC, the most polymorphic and gene dense region in the vertebrate genome, contains many loci essential to immunity. In mammals, this region spans approximately 4 Mb. Studies of avian species have found the MHC to be greatly reduced in size and gene content with an overall locus organization differing from that of mammals. The chicken MHC has been mapped to two distinct regions (MHC-B and -Y) of a single chromosome. MHC-B haplotypes possess tightly linked genes encoding the classical MHC molecules and few other disease resistance genes. Furthermore, chicken haplotypes possess a dominantly expressed class I and class II B locus that have a significant effect on the progression or regression of pathogenic disease. In this study, we present the MHC-B region of the turkey (Meleagris gallopavo) as a similarly constricted locus, with 34 genes identified within a 0.2-Mb region in near-perfect synteny with that of the chicken MHC-B. Notable differences between the two species are three BG and class II B loci in the turkey compared with one BG and two class II B loci in the chicken MHC-B. The relative size and high level of similarity of the turkey MHC in relation to that of the chicken suggest that similar associations with disease susceptibility and resistance may also be found in turkey. |
| [5] | . |
| [6] | . Bactrian camels serve as an important means of transportation in the cold desert regions of China and Mongolia. Here we present a 2.0165Gb draft genome sequence from both a wild and a domestic bactrian camel. We estimate the camel genome to be 2.3865Gb, containing 20,821 protein-coding genes. Our phylogenomics analysis reveals that camels shared common ancestors with other even-toed ungulates about 55-60 million years ago. Rapidly evolving genes in the camel lineage are significantly enriched in metabolic pathways, and these changes may underlie the insulin resistance typically observed in these animals. We estimate the genome-wide heterozygosity rates in both wild and domestic camels to be 1.0 × 10(-3). However, genomic regions with significantly lower heterozygosity are found in the domestic camel, and olfactory receptors are enriched in these regions. Our comparative genomics analyses may also shed light on the genetic basis of the camel's remarkable salt tolerance and unusual immune system. |
| [7] | . |
| [8] | . Major histocompatibility complex (MHC) is widely found in vertebrate. Because of its high polymorphism, MHC genes have become the focus of attentionhop in relation to genetics and breeding and disease-resistance etc. This article make a retrospective conclusion on the major histocompatibility complex genes polymorphism and the relationship between this gene and disease, and other aspects on the domestic and foreign researchers’ papers in recent decades.Some of these contents as a reference for the next correlation experiments. . Major histocompatibility complex (MHC) is widely found in vertebrate. Because of its high polymorphism, MHC genes have become the focus of attentionhop in relation to genetics and breeding and disease-resistance etc. This article make a retrospective conclusion on the major histocompatibility complex genes polymorphism and the relationship between this gene and disease, and other aspects on the domestic and foreign researchers’ papers in recent decades.Some of these contents as a reference for the next correlation experiments. |
| [9] | . Comments on the major histocompatibility complex in genetic field. Development of immunological means to protect mice against the growth of transplanted syngeneic leukemias; Test of segregans for the presence of antigen II; Outcome of the linkage test. |
| [10] | . Abstract In the past few years the DNA sequence database for molecules of the MHC (major histocompatibility complex) has expanded greatly, yielding a more complete picture of the long-term rates and patterns of evolution of the MHC in vertebrates. Sharing of MHC allelic lineages between long-diverged species (trans-species evolution) has been detected virtually wherever it is sought, but new analyses of linked neutral regions and the complexities of sequence convergence and microrecombination in the peptide binding region challenge traditional phylogenetic analyses. Methods for estimating the intensity of selection on MHC genes suggest that viability is important, but recent studies in natural populations of mammals give inconsistent results concerning mate choice. The complex and interacting roles of microrecombination, parasite-mediated selection and mating preferences for maintaining the extraordinary levels of MHC polymorphism observed are still difficult to evaluate. |
| [11] | . The Major Histocompatibility Complex (MHC) is a genomic region containing genes with crucial roles in immune responses. MHC class I and class II genes encode antigen-presenting molecules expressed on the cell surface. To counteract the high variability of pathogens, the MHC evolved into a region of considerable heterogeneity in its organization, number and extent of polymorphism. Studies of MHCs in different model species contribute to our understanding of mechanisms of immunity, diseases and their evolution. Camels are economically important domestic animals and interesting biomodels. Three species of Old World camels have been recognized: the dromedary (Camelus dromedarius), Bactrian camel (Camelus bactrianus) and the wild camel (Camelus ferus). Despite their importance, little is known about the MHC genomic region, its organization and diversity in camels. The objectives of this study were to identify, map and characterize the MHC region of Old World camelids, with special attention to genetic variation at selected class MHC II loci. Physical mapping located the MHC region to the chromosome 20 in Camelus dromedarius. Cytogenetic and comparative analyses of whole genome sequences showed that the order of the three major sub-regions is entromere - Class II Class III Class I .DRA, DRB, DQAandDQBexon 2 sequences encoding the antigen binding site of the corresponding class II antigen presenting molecules showed high degree of sequence similarity and extensive allele sharing across the three species. Unexpectedly low extent of polymorphism with low numbers of alleles and haplotypes was observed in all species, despite different geographic origins of the camels analyzed. TheDRAlocus was found to be polymorphic, with three alleles shared by all three species.DRAandDQAsequences retrieved from ancient DNA samples ofCamelus dromedariussuggested that additional polymorphism might exist. This study provided evidence that camels possess an MHC comparable to other mammalian species in terms of its genomic localization, organization and sequence similarity. We described ancient variation at theDRAlocus, monomorphic in most species. The extent of molecular diversity of MHC class II genes seems to be substantially lower in Old World camels than in other mammalian species. The online version of this article (doi:10.1186/s12864-016-2500-1) contains supplementary material, which is available to authorized users. |
| [12] | . Abstract Cytogenetic chromosome maps offer molecular tools for genome analysis and clinical cytogenetics and are of particular importance for species with difficult karyotypes, such as camelids (2n = 74). Building on the available human-camel zoo-fluorescence in situ hybridization (FISH) data, we developed the first cytogenetic map for the alpaca (Lama pacos, LPA) genome by isolating and identifying 151 alpaca bacterial artificial chromosome (BAC) clones corresponding to 44 specific genes. The genes were mapped by FISH to 31 alpaca autosomes and the sex chromosomes; 11 chromosomes had 2 markers, which were ordered by dual-color FISH. The STS gene mapped to Xpter/Ypter, demarcating the pseudoautosomal region, whereas no markers were assigned to chromosomes 14, 21, 22, 28, and 36. The chromosome-specific markers were applied in clinical cytogenetics to identify LPA20, the major histocompatibility complex (MHC)-carrying chromosome, as a part of an autosomal translocation in a sterile male llama (Lama glama, LGL; 2n = 73,XY). FISH with LPAX BACs and LPA36 paints, as well as comparative genomic hybridization, were also used to investigate the origin of the minute chromosome, an abnormally small LPA36 in infertile female alpacas. This collection of cytogenetically mapped markers represents a new tool for camelid clinical cytogenetics and has applications for the improvement of the alpaca genome map and sequence assembly. The American Genetic Association. 2012. All rights reserved. For permissions, please email: journals.permissions@oup.com. |
| [13] | . |
| [14] | . |
| [15] | . 由于硬骨鱼类代表了一半的脊椎动物,是具有MHCⅠ基因的低等脊椎动物,其MHCI基因结构与高等脊椎动物相比,存在较大差异,因而已经成为比较基因组学研究的焦点。对硬骨鱼类MHCⅠ基因的研究始于1990年,Hashimoto等通过比较人、鼠、鸡的MHCⅠ类α3结构域和Ⅱ类β结构域氨基酸序列,设计高度简并性引物,扩增得到鲤鱼部分MHCⅠ基因序列。这一研究成果揭开了鱼类MHC研究的序幕。 . 由于硬骨鱼类代表了一半的脊椎动物,是具有MHCⅠ基因的低等脊椎动物,其MHCI基因结构与高等脊椎动物相比,存在较大差异,因而已经成为比较基因组学研究的焦点。对硬骨鱼类MHCⅠ基因的研究始于1990年,Hashimoto等通过比较人、鼠、鸡的MHCⅠ类α3结构域和Ⅱ类β结构域氨基酸序列,设计高度简并性引物,扩增得到鲤鱼部分MHCⅠ基因序列。这一研究成果揭开了鱼类MHC研究的序幕。 |
| [16] | . Abstract Bactrian camel (Camelus bactrianus), dromedary (Camelus dromedarius) and alpaca (Vicugna pacos) are economically important livestock. Although the Bactrian camel and dromedary are large, typically arid-desert-adapted mammals, alpacas are adapted to plateaus. Here we present high-quality genome sequences of these three species. Our analysis reveals the demographic history of these species since the Tortonian Stage of the Miocene and uncovers a striking correlation between large fluctuations in population size and geological time boundaries. Comparative genomic analysis reveals complex features related to desert adaptations, including fat and water metabolism, stress responses to heat, aridity, intense ultraviolet radiation and choking dust. Transcriptomic analysis of Bactrian camels further reveals unique osmoregulation, osmoprotection and compensatory mechanisms for water reservation underpinned by high blood glucose levels. We hypothesize that these physiological mechanisms represent kidney evolutionary adaptations to the desert environment. This study advances our understanding of camelid evolution and the adaptation of camels to arid-desert environments. |
| [17] | . |
| [18] | . pAbstract/p pBackground/p pThe major histocompatibility complex (MHC) is a group of genes with a variety of roles in the innate and adaptive immune responses. MHC genes form a genetically linked cluster in eutherian mammals, an organization that is thought to confer functional and evolutionary advantages to the immune system. The tammar wallaby it(Macropus eugenii/it), an Australian marsupial, provides a unique model for understanding MHC gene evolution, as many of its antigen presenting genes are not linked to the MHC, but are scattered around the genome./p pResults/p pHere we describe the core tammar wallaby MHC region on chromosome 2q by ordering and sequencing 33 BAC clones, covering over 4.5 MB and containing 129 genes. When compared to the MHC region of the South American opossum, eutherian mammals and non-mammals, the wallaby MHC has a novel gene organization. The wallaby has undergone an expansion of MHC class II genes, which are separated into two clusters by the class III genes. The antigen processing genes have undergone duplication, resulting in two copies of TAP1 and three copies of TAP2. Notably, Kangaroo Endogenous Retroviral Elements are present within the region and may have contributed to the genomic instability./p pConclusions/p pThe wallaby MHC has been extensively remodeled since the American and Australian marsupials last shared a common ancestor. The instability is characterized by the movement of antigen presenting genes away from the core MHC, most likely via the presence and activity of retroviral elements. We propose that the movement of class II genes away from the ancestral class II region has allowed this gene family to expand and diversify in the wallaby. The duplication of TAP genes in the wallaby MHC makes this species a unique model organism for studying the relationship between MHC gene organization and function./p |
| [19] | . . |
| [20] | . |
| [21] | . The genomic organisation of the Major Histocompatibility Complex (MHC) varies greatly between different vertebrates. In mammals, the classical MHC consists of a large number of linked genes (e.g. greater than 200 in humans) with predominantly immune function. In some birds, it consists of only a small number of linked MHC core genes (e.g. smaller than 20 in chickens) forming a minimal essential MHC and, in fish, the MHC consists of a so far unknown number of genes including non-linked MHC core genes. Here we report a survey of MHC genes and their paralogues in the zebrafish genome. Using sequence similarity searches against the zebrafish draft genome assembly (Zv4, September 2004), 149 putative MHC gene loci and their paralogues have been identified. Of these, 41 map to chromosome 19 while the remaining loci are spread across essentially all chromosomes. Despite the fragmentation, a set of MHC core genes involved in peptide transport, loading and presentation are still found in a single linkage group. The results extend the linkage information of MHC core genes on zebrafish chromosome 19 and show the distribution of the remaining MHC genes and their paralogues to be genome-wide. Although based on a draft genome assembly, this survey demonstrates an essentially fragmented MHC in zebrafish. |
| [22] | . |
| [23] | . |
| [24] | .[D]. |
| [25] | . <FONT face=Verdana>作者应用比较基因组学和生物信息学方法比较了绵羊、牛、鹿和狗MHC-DQA1基因部分序列,并对该基因的遗传多样性及分子系统发育进行了分析。结果表明,38个基因序列检测到107个多态位点,共生成38个单倍型。物种间及物种内MHCDQA1存在较丰富的遗传多样性。<BR> </FONT> . <FONT face=Verdana>作者应用比较基因组学和生物信息学方法比较了绵羊、牛、鹿和狗MHC-DQA1基因部分序列,并对该基因的遗传多样性及分子系统发育进行了分析。结果表明,38个基因序列检测到107个多态位点,共生成38个单倍型。物种间及物种内MHCDQA1存在较丰富的遗传多样性。<BR> </FONT> |
| [26] | . |
| [27] | . The northern pig-tailed macaque(Macaca leonina) has been identified as an independent species from the pig-tailed macaque group.The species is a promising animal model in HIV/AIDS pathogenesis and vaccine studies due to susceptibility to HIV-1.However,the major histocompatibility complex(MHC) genetics in northern pig-tailed macaques remains poorly understood.We have previously studied the MHC class Ⅰ genes in northern pig-tailed macaques and identified 39 novel alleles.Here,we describe the MHC class Ⅱ alleles in all six classical loci(DPA,DPB,DQA,DQB,DRA,and DRB) from northern pig-tailed macaques using a sequence-based typing method for the first time.A total of 60 MHCII alleles were identified of which 27 were shared by other macaque species.Additionally,northern pig-tailed macaques expressed a single DRA and multiple DRB genes similar to the expression in humans and other macaque species.Polymorphism and positive selection were detected,and phylogenetic analysis suggested the presence of a common ancestor in human and northern pig-tailed macaque MHC class Ⅱ allelic lineages at the DQA,DQB and DRB loci.The characterization of full-length MHC class Ⅱ alleles in this study significantly improves understanding of the immunogenetics of northern pig-tailed macaques and provides the groundwork for future animal model studies. |
| [28] | . Twenty new world camelidae primer pairs were selected for cross-species amplification of loci in the old world camelids, the dromedary (Camelus dromedarius) and the bacterian (camelus bactrianus). A panel of DNA samples from 34 unrelated dromedaries (Kenya) and from 34 unrelated bactrians (32 domestic bactrians and two wild bactrians, China) were amplified. DNA samples from two llamas (Lama glama) and two alpacas (Lama pacos) were used as positive controls. PCR conditions; amplification and polymorphism, and microsatellite sequences were assessed and results presented. |
| [29] | . Both archaeological data and the presence of few mitochondrial DNA lineages suggest that most widespread domestic mammals (cattle, sheep, goats, pigs and dogs) derive from only a handful of domestication events. However, each of these species shows a high level of diversity at the nuclear genes of the major histocompatibility complex (MHC). Through simulations incorporating various degrees of population subdivision, growth rate and selection, we demonstrate that the numerous MHC DRB alleles that are present in modern domestic mammals implies that substantial backcrossing with wild ancestors, either accidental or intentional, has been important in shaping the genetic diversity of our domesticates. These results support the view that, contrary to common assumption, domestic and wild lineages might not have been clearly separated throughout their history. |
| [30] | . |
| [31] | . The single‐humped dromedary (Camelus dromedarius) is the most numerous and widespread of domestic camel species and is a significant source of meat, milk, wool, transportation and sport for millions of people. Dromedaries are particularly well adapted to hot, desert conditions and harbour a variety of biological and physiological characteristics with evolutionary, economic and medical importance. To understand the genetic basis of these traits, an extensive resource of genomic variation is required. In this study, we assembled at 65× coverage, a 2.0602Gb draft genome of a female dromedary whose ancestry can be traced to an isolated population from the Canary Islands. We annotated 2102167 protein‐coding genes and estimated ~33.7% of the genome to be repetitive. A comparison with the recently published draft genome of an Arabian dromedary resulted in 1.9102Gb of aligned sequence with a divergence of 0.095%. An evaluation of our genome with the reference revealed that our assembly contains more error‐free bases (91.2%) and fewer scaffolding errors. We identified ~1.4 million single‐nucleotide polymorphisms with a mean density of 0.7102×0210613per base. An analysis of demographic history indicated that changes in effective population size corresponded with recent glacial epochs. Ourde novoassembly provides a useful resource of genomic variation for future studies of the camel's adaptations to arid environments and economically important traits. Furthermore, these results suggest that draft genome assemblies constructed with only two differently sized sequencing libraries can be comparable to those sequenced using additional library sizes, highlighting that additional resources might be better placed in technologies alternative to short‐read sequencing to physically anchor scaffolds to genome maps. |
