 ,1, 吕浩1, 郭广雨2, 刘冬旭1, 石岩1, 孙志君1, 张泽鑫1, 张艳娇1, 文莹楠1, 王洁琦1, 刘春燕1, 陈庆山
,1, 吕浩1, 郭广雨2, 刘冬旭1, 石岩1, 孙志君1, 张泽鑫1, 张艳娇1, 文莹楠1, 王洁琦1, 刘春燕1, 陈庆山 ,1, 辛大伟
,1, 辛大伟 ,1, 王锦辉
,1, 王锦辉 ,1
,1Effect of rhcN Gene Mutation on Nodulation Ability of Soybean Rhizobium HH103
LIU HanXi ,1, Lü Hao1, GUO GuangYu2, LIU DongXu1, SHI Yan1, SUN ZhiJun1, ZHANG ZeXin1, ZHANG YanJiao1, WEN YingNan1, WANG JieQi1, LIU ChunYan1, CHEN QingShan
,1, Lü Hao1, GUO GuangYu2, LIU DongXu1, SHI Yan1, SUN ZhiJun1, ZHANG ZeXin1, ZHANG YanJiao1, WEN YingNan1, WANG JieQi1, LIU ChunYan1, CHEN QingShan ,1, XIN DaWei
,1, XIN DaWei ,1, WANG JinHui
,1, WANG JinHui ,1
,1通讯作者:
责任编辑: 李莉
收稿日期:2020-09-3接受日期:2020-10-26网络出版日期:2021-03-16
| 基金资助: |
Received:2020-09-3Accepted:2020-10-26Online:2021-03-16
作者简介 About authors
刘函西,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1648KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
刘函西, 吕浩, 郭广雨, 刘冬旭, 石岩, 孙志君, 张泽鑫, 张艳娇, 文莹楠, 王洁琦, 刘春燕, 陈庆山, 辛大伟, 王锦辉. 大豆根瘤菌HH103 rhcN突变对结瘤能力的影响[J]. 中国农业科学, 2021, 54(6): 1104-1111 doi:10.3864/j.issn.0578-1752.2021.06.003
LIU HanXi, Lü Hao, GUO GuangYu, LIU DongXu, SHI Yan, SUN ZhiJun, ZHANG ZeXin, ZHANG YanJiao, WEN YingNan, WANG JieQi, LIU ChunYan, CHEN QingShan, XIN DaWei, WANG JinHui.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】大豆是世界范围种植广泛的重要经济作物,能够为人类提供丰富的油脂和植物蛋白[1]。随着中国人民生活质量的提高,中国对大豆的需求逐年上升,已成为世界最大的大豆进口国。随着化肥施用量的增加,大豆生产对化肥的依赖越来越大[2,3],导致土壤板结加剧、水体富营养化和重金属富集等环境问题[4,5]。作为高需氮作物,大豆在长期的进化过程中通过与根瘤菌相互作用,在根部发育并形成根瘤。根瘤能够将空气中的氮气高效转化成化合态氮,使大豆适应低氮环境,维持大豆正常的生长发育[6,7]。所以充分发挥大豆根瘤菌共生固氮作用,是减少化肥施用、保障粮食安全、减轻能源压力、减少环境污染和提高土壤肥力、保障农业、环境可持续发展的有效措施[8,9],因此,对大豆-根瘤菌共生体系建立机制及固氮机制的研究具有十分重要的现实意义。【前人研究进展】前人研究发现,大豆-根瘤菌共生体系形成过程中涉及复杂的信号转导和物质交换,根瘤菌Ⅲ型效应因子是由Ⅲ型分泌系统向宿主细胞中分泌的一系列蛋白质,在根瘤菌侵染和根瘤形成过程中能够发挥重要调控作用[10]。根瘤菌Ⅲ型分泌系统(type Ⅲ secretion system,T3SS)是一种类注射器的复杂结构,是由多种蛋白组成[11],其中NopA和NopB是Ⅲ型效应分泌系统中针头的组成部分,NopX是分泌系统的基本构成[12,13,14,15],RhcN和RhcJ也是Ⅲ型分泌系统结构的组成部分之一,是最基本的结构蛋白,它们的缺失会直接造成整个Ⅲ型分泌系统结构的破坏,影响Ⅲ型效应因子的正常分泌[16,17,18]。近年来,对Ⅲ型效应因子研究进展表明,NopC是根瘤菌特异的效应蛋白,NopC位于NopA的上游,由于NopA是T3SS菌毛的结构组成成分,所以NopC也应该是T3SS的组成成分[19]。NopC突变后并不影响其他效应因子的分泌,腺苷酸环化酶测定显示NopC能促进宿主植物结瘤[19]。对于NopP的研究,在根瘤菌HH103中,NopP的表达依赖于黄酮类化合物和转录调节因子TtsI,并影响大豆根和芽中致病相关基因GmPR1的表达[20]。在根瘤菌NGR234中,Ⅲ型效应因子NopP突变后能改变其在非洲山毛豆和扁豆上的结瘤数量[21]。所以根瘤菌Ⅲ型效应因子作用机制及大豆响应Ⅲ型效应因子机理的研究对阐明大豆-根瘤菌共生体系形成机制具有重要作用。【本研究切入点】RhcN是Ⅲ型分泌系统结构的组成部分,能够为根瘤菌Ⅲ型效应因子的分泌提供能量,保证Ⅲ型效应因子的正常分泌[16,17,18]。不同大豆品种具有不同的遗传背景,造成其与根瘤菌的共生固氮具有一定的差异,但根瘤菌Ⅲ型效应因子在该种差异中能发挥的作用并未得到深入研究。【拟解决的关键问题】本研究通过对中华根瘤菌HH103的T3SS结构基因rhcN进行基因突变,得到突变体HH103ΩrhcN,并用PCR和Southern blot鉴定。通过表达分析,确定rhcN能够影响Ⅲ型效应因子的分泌。对HH103ΩrhcN突变体以及野生型菌株HH103进行100份核心种质结瘤试验,通过对两者结瘤结果的比对,进一步明确Ⅲ型效应因子结构基因突变后对共生结瘤的影响。1 材料与方法
1.1 材料
费氏中华根瘤菌HH103(源于西班牙塞维利亚大学Francisco Javier López-Baena实验室),利福平(Rif)抗性;大肠杆菌E.coli DH5α。Williams82和大豆核心种质100份(源于东北农业大学陈庆山实验室保存)。
质粒提取试剂盒(TaKaRa公司,DC201-01),缺氮植物营养液(1 L包含MgSO4 0.5 mol·L-1、Na2MoO4 0.2 mmol·L-1、MnSO4 2 mmol·L-1、H3BO3 4 mmol·L-1、CaCl2 2 mol·L-1、CoSO4 0.2 mmol·L-1、K2SO4 0.5 mol·L-1、CuSO4 4 mmol·L-1、KH2PO4 1 mol·L-1、ZnSO4 1 mmol·L-1和C6H5FeO7 20 mmol·L-1)。
1.2 突变体的构建
1.2.1 重组自杀载体pJQ200SK-rhcN-Km的构建 从NCBI生物信息数据库中,获取HH103基因组中的rhcN序列,提取全基因组序列,进行PCR扩增,片段的引物命名为rhcN-F和rhcN-R。将扩增片段连接pGWC载体,连接体系转化大肠杆菌E.coli DH5α。测序获得pGWC-rhcN。选取rhcN的酶切位点SpeⅠ(5'-ACTAGT-3'),突变位置的上下游各15 bp碱基序列作为上游引物,该引物反向互补序列作为对应的下游引物,以pGWC-rhcN为模板进行PCR扩增,用DpnⅠ消化后转化大肠杆菌E.coli DH5α,测序获得突变结果pGWC-rhcN-DpnⅠ。Km抗性片段PCR扩增以pEASY-Blunt为模板,在扩增引物两端加上KpnⅠ酶切位点序列,引物命名为Km-KpnⅠ-F、Km-KpnⅠ-R。将抗性片段插入,连接体系转化大肠杆菌E.coli DH5α,测序后得到pGWC-rhcN-Km。选取酶切位点SacⅠ和XbaⅠ,用引物SacI-rhcN-XbaⅠ-F(R)扩增pGWC-rhcN-Km,将扩增片段和重组自杀载体分别进行双酶切,将酶切后的片段和重组自杀载体进行连接,经转化DH5α后测序,命名为pJQ200SK-rhcN-Km。1.2.2 三亲杂交 将根瘤菌HH103划线于含Rif的TY固体培养基。将带有Helper质粒的大肠杆菌划线于含Km的LB固体培养基,将带有pJQ200SK-rhcN- Km的大肠杆菌划线于含有Km、Gent的LB固体培养基。过夜培养直至长出单克隆;将3种菌挑单克隆分别于5 mL相应培养基中培养至OD600值为0.6;取3种菌,每个各取1 mL,分别放于1.5 mL离心管中,离心12 000 r/min离心30 s,弃上清,用1 mL无抗TY重悬;取重悬后的HH103菌液200 μL,其余2种菌各100 μL加入到1.5 mL离心管中,离心12 000 r/min离心30 s,弃上清,用20 μL无抗TY重悬;将20 μL的混合菌液滴到无抗TY固体培养基平板上过夜培养;过夜培养后,刮取大斑划线到含Rif/Km的TY固体培养基培养,直至长出单克隆;将单克隆继续划线到含Rif/Km的TY固体培养基进行筛选,重复3次;将经过3次筛选的单克隆划线于含有5%蔗糖的Rif/Km的TY固体培养基进行筛选,重复3次;筛选到的突变体命名为HH103ΩrhcN。
1.3 突变体的验证
1.3.1 突变体的PCR鉴定 利用引物rhcN-F(R)以及rhcN-F和Km-R 2对引物分别扩增野生型根瘤菌HH103及突变菌株HH103ΩrhcN,通过电泳的方法,观察到目的条带,测序验证突变体HH103ΩrhcN。1.3.2 突变体的Southern blot鉴定 提取野生型HH103和突变体HH103ΩrhcN的基因组;标记探针,设计探针引物,扩增突变体基因组,胶回收片段,沸水浴10 min,迅速插入冰中。DIG-High Prime(vial1)与变性的DNA混匀,37℃过夜,65℃水浴终止反应,-20℃保存;消化好的样品琼脂糖凝胶进行电泳;碱变性处理、转膜、预杂交、杂交、洗膜、显影[22],将杂交膜DNA面朝上放在显影夹上,加1 mL CSPD,室温静止避光温浴5 min。37℃避光10 min,获取Southern blot结果验证照片。
1.4 Western blot验证Ⅲ效应因子NopC及NopT的分泌
对野生型根瘤菌HH103和突变体HH103ΩrhcN在含有相应抗生素培养基中进行培养,每种菌培养2组,待OD600=0.8时,一组加入染料木苷,而另一组作为空白对照,28℃,200 r/min培养40 h;4℃,菌液8 000 r/min离心20 min,收集上清液。在上清液中,加入3倍体积的10%三氯乙酸,-20℃静置20 h;将溶液10 000 r/min离心30 min,用事先预冷的80%丙酮对沉淀进行清洗,重复2次,最后将沉淀用适量8 mol·L-1尿素进行稀释;最后利用NopC和NopT的兔源抗体进行Western blot检测[23]。1.5 突变体结瘤能力鉴定及大豆核心种质根瘤表型分析
取Williams82的种子30粒,用氯气灭菌法对大豆种子表面进行灭菌,灭菌时长为12 h。在无菌操作台内吹去种子表面氯气,将种子种入高温灭菌的双层钵中,并在温室中进行培养,培养条件为光16 h/暗/8 h,温度为25℃[24]。待大豆生长到Vc[25]期时,用注射器在大豆根部土层下直接接种2 mL OD600值为0.2的费氏中华根瘤菌HH103菌液和突变体HH103ΩrhcN菌液,保证每个植株接种细菌的个数在109左右,接种30 d后对结瘤数目及根瘤干重等性状进行调查,并利用SPSS软件对不同处理数据进行T-test检验,进行显著性分析。取试验所需100份种质资源的种子各10粒,接种30 d后对结瘤数目及根瘤干重等性状进行调查,并利用SPSS软件对不同处理数据进行T-test检验,进行显著性分析。
2 结果
2.1 突变体HH103ΩrhcN的PCR鉴定
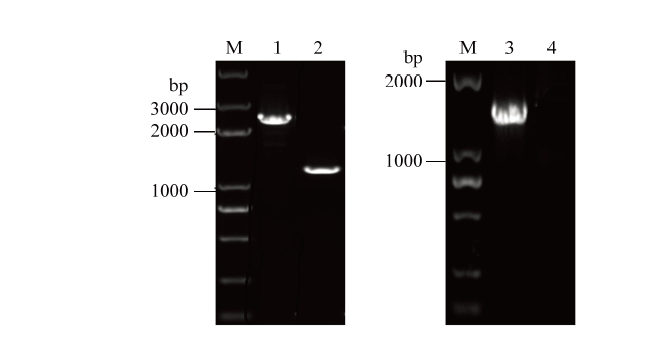
三亲杂交得到的突变体HH103ΩrhcN及野生型菌株HH103,利用引物rhcN-F(R)以及rhcN-F和Km-R扩增HH103ΩrhcN,进行PCR鉴定(图1)。野生型HH103利用引物rhcN-F和rhcN-R扩增的条带大小应为rhcN长度,为1 300 bp,突变型菌株HH103ΩrhcN扩增的条带大小应为rhcN和Km长度,为2 300 bp。野生型HH103利用引物rhcN-F和Km-SpeI-R扩增,由于其不含插入的Km片段结果应无条带,突变型菌株HH103ΩrhcN扩增的条带大小应为rhcN上臂和Km的长度,为1 500 bp(图1)。由结果筛选出正确突变体HH103ΩrhcN,扩增结果与理论相符。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1突变体HH103ΩrhcN的PCR鉴定
Fig. 1PCR analysis of HH103ΩrhcN
M: Trans 2K Plus DNA marker; 1: HH103ΩrhcN(rhcN-F,rhcN-R); 2: HH103(rhcN-F,rhcN-R); 3: HH103ΩrhcN(rhcN-F,Km-SpeⅠ-R); 4: HH103(rhcN-F,Km-SpeⅠ-R)
2.2 突变体HH103ΩrhcN的Southern blot鉴定
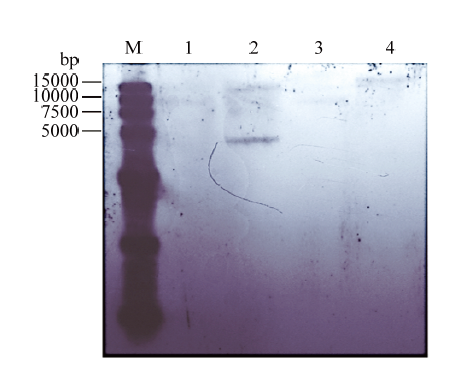
进一步验证突变体HH103ΩrhcN的真实性,采用地高辛标记和检测试剂盒进行突变体Southern blot鉴定。对野生型根瘤菌HH103和HH103ΩrhcN的基因组用2种内切酶酶切。选用内切酶时避免选用目的基因和Km内部的内切酶,以免将突变片段切碎影响验证结果。从杂交结果可以看出每种酶切后突变体的杂交片段比野生型的杂交片段约长1 000 bp(图2),即插入卡那霉素片段的大小,进一步证明突变体HH103ΩrhcN筛选成功。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2突变体HH103ΩrhcN的Southern blot鉴定
1:HH103ΩrhcN突变体基因组HindⅢ酶切结果;2:HH103基因HindⅢ酶切结果;3:HH103ΩrhcN突变体基因组SacⅠ酶切结果;4:HH103基因组SacⅠ酶切结果
Fig. 2Southern blot identification of HH103ΩrhcN
1: The genome of HH103ΩrhcN was digested by HindⅢ; 2: The genome of the wild strain HH103 was digested by HindⅢ; 3: The genome of HH103ΩrhcN was digested by SacⅠ; 4: The genome of the wild strain HH103 was digested by SacⅠ
2.3 rhcN突变抑制Ⅲ型效应因子的分泌
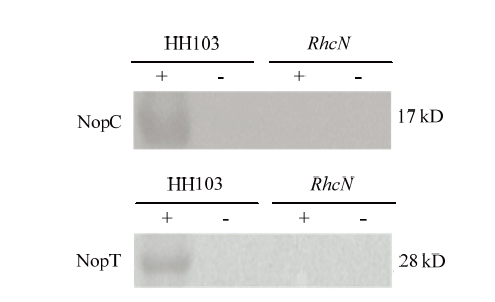
通过对野生型根瘤菌HH103和rhcN突变体中Ⅲ型效应因子NopC和NopT的分泌进行Western blot验证。结果表明,rhcN突变后能够抑制Ⅲ型效应因子NopC和NopT的分泌(图3)。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3Ⅲ型效应因子NopC和NopT的胞外检测
+:存在Genistein的诱导;-:不存在Genistein的诱导
Fig. 3Detection of NopC and NopT in secreted proteins
+: The presence of Genistein; -: The absence of Genistein
2.4 rhcN突变能够抑制根瘤的形成
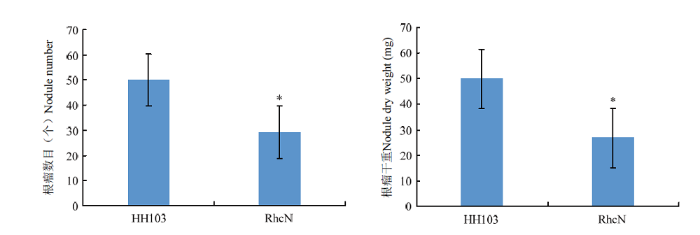
利用Williams82对野生型根瘤菌HH103和根瘤菌HH103ΩrhcN突变体进行结瘤鉴定,大豆植株Vc期时进行根瘤菌接种,接种30 d后对根瘤表型进行统计和分析(图4和图5)。与接种野生型HH103相比,rhcN突变能够引起根瘤数目的显著较少,也能够引起根瘤干重的显著降低。大豆根系中一定数目的根瘤对于大豆的氮源供给是非常必要的,rhcN突变能够引起根瘤菌Ⅲ型效应因子的分泌受到抑制,造成根瘤菌侵染和根瘤发育受到影响,说明根瘤菌Ⅲ型效应因子对于大豆-根瘤菌共生体系的正常建立是必要的。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4HH103和HH103ΩrhcN接种后的根部及根瘤形态
左图为接种HH103时大豆根系和根瘤形态,右图为接种HH103ΩrhcN时大豆根系和根瘤形态
Fig. 4Root morphology and nodules traits after HH103 and HH103ΩrhcN inoculation
The left picture showed the information of soybean roots and nodules inoculated with HH103, and the right picture showed the information of soybean roots and nodules inoculated with HH103Ωrhcn
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5HH103和HH103ΩrhcN接种后根瘤数目及干重
*表示在P<0.05水平差异显著
Fig. 5Nodule number and dry weight of HH103 and HH103ΩrhcN inoculation
*for the difference was significant at P<0.05 level
2.5 HH103ΩrhcN突变体结瘤表型鉴定
利用来自不同积温带的全国大豆种质核心资源的100份品种对野生型根瘤菌HH103、突变型根瘤菌HH103ΩrhcN进行结瘤鉴定,重复10次,对接种后30 d的植株对结瘤数目及根瘤干重等性状进行调查,对表型数据的最大值、最小值、标准差及变异系数结瘤情况进行分析(表1和电子附表1),结果表明,rhcN突变能够引起80份大豆资源根瘤数目减少,13份根瘤数目显著升高,7份不发生显著性变化。说明rhcN在大豆-根瘤菌共生体系形成过程中发挥着重要作用,并且不同的大豆遗传背景具有不同应对反应。Table 1
表1
表1100份种质根瘤性状分析
Table 1
| 参数 Parameter | HH103 | HH103ΩrhcN | ||
|---|---|---|---|---|
| 根瘤数目 NN | 根瘤干重 NDW (mg) | 根瘤数目 NN | 根瘤干重 NDW (mg) | |
| 平均值 Average | 35.52±5.64 | 34.37±6.35 | 17.04±10.56** | 16.32±11.55** |
| 最大值 Maximum | 49.60±4.67 | 50.84±7.49 | 45.20±4.87 | 47.68±6.58 |
| 最小值 Minimum | 11.50±4.84 | 12.98±3.65 | 3.00±0.87 | 0.35±0.26 |
| 标准差 Standard deviation | 5.64 | 6.35 | 10.56 | 11.55 |
| 变异系数 Coefficient of Variation | 54.24 | 61.02 | 101.57 | 111.07 |
新窗口打开|下载CSV
3 讨论
根瘤菌rhcN能够编码RhcN蛋白,是一种ATP转移酶,是根瘤菌Ⅲ型分泌系统重要的结构蛋白,在效应因子的分泌过程中,能够为效应因子提供必不可少的能量,保障效应因子由根瘤菌分泌到宿主细胞中[18]。在其他根瘤菌菌种中,关于RhcN突变的研究较多,根瘤菌NGR234 RhcN突变能够显著抑制豇豆根瘤的产生[21],根瘤菌DOA9 RhcN突变能够引起美洲合萌根瘤的显著减少[26],与本研究根瘤菌HH103 RhcN突变之后对大豆根瘤表型产生影响相似,表明根瘤菌Ⅲ型效应因子在大豆-根瘤菌共生体系建立过程中发挥重要作用。在根瘤菌HH103中,现已报道的Ⅲ型效应因子有10种,分别为NopA、NopB、NopAA(GunA)、NopC、NopD、NopL、NopM、NopP、NopT和NopZ[27],但是HH103不同的效应因子发挥着不同的调控作用。NopAA(GunA)属于一种糖基水解酶,已被证实能够分解细胞壁调控根瘤菌的侵染,其突变情况下抑制根瘤的产生[28]。NopC被证实能够分泌到大豆宿主细胞中,并且在根瘤发育阶段也能发挥调控作用,在根瘤菌侵染过程中,NopC能够诱导大豆病程相关基因GmPR1的表达,并且NopC突变情况下同样抑制根瘤的形成[19]。根瘤菌HH103Ⅲ型效应因子的作用机制并未得到解析,对根瘤菌Ⅲ型效应因子在共生结瘤中的研究,特别是大豆响应根瘤菌Ⅲ型效应因子基因的功能解析能够为研究共生结瘤机制提供重要的理论支持。利用根瘤菌Ⅲ型效应因子及大豆响应Ⅲ型效应因子信号网络对根瘤菌和大豆的相关改造,能够为大豆-根瘤菌共生体系高效结瘤和固氮的研究提供一种新的思路和可能。本研究收集了来自不同地区的100份大豆种质资源,其分别来自东北、黄淮海和华南三大大豆种植区,品种之间具有明显的基因型和生态型差异。利用野生型根瘤菌HH103和根瘤菌HH103ΩrhcN突变体对该100份大豆种质资源进行根瘤表型鉴定,结果表明,rhcN突变能够引起其中80份大豆资源根瘤数目降低,13份根瘤数目显著升高,7份不发生显著性变化,此表型鉴定结果说明RhcN突变情况下能够引起大部分的大豆种质根瘤表型显著降低。而100份种质材料中,13份根瘤数目显著升高,7份不发生显著性变化,表明20份大豆品种的基因型造成了该表型的差异。根瘤菌Ⅲ型效应因子已经被证实能够影响根瘤菌与大豆宿主之间的亲和性,并且利用大豆与根瘤菌相互作用的基因型特异性及共生亲和性能够进行根瘤菌Ⅲ型效应因子相关信号通路研究及实际生产应用[29]。自1966年开始,利用不同大豆材料的基因型差异,越来越多与共生结瘤相关的大豆基因被定位和应用。例如rj1(Rj1)、rj2(Rj2)、rj3(Rj3)、rj4(Rj4)、rj5(Rj5)、rj6(Rj6)、rj7(Rj7)和rj8(Rj8)[30]。其中,rj2(Rj2)和rj4(Rj4)是被研究较多的建立共生密切相关的基因[31,32]。Rj2与根瘤菌Ⅲ型效应因子NopP具有互作关系,并且能够调控宿主大豆对根瘤菌的特异性识别[33]。Rj4能够调控大豆对根瘤菌的亲和敏感性,Rj4可响应Bradyrhizobium elkanii根瘤菌Ⅲ型效应因子影响大豆-根瘤菌共生体系的建立[34]。作者所在实验室在前期的研究中发现,HH103和NopL突变体能够在栽培品种东农594和Charleston引起明显的根瘤表型差异,通过构建以东农594和Charleston为父母本的RIL群体,对大豆响应NopL进行QTL定位和挖掘,鉴定出了大豆能够响应NopL的基因Glyma.13g170500和Glyma.07g099700[35],说明利用大豆基因型差异对Ⅲ型效应因子的研究是可行的。利用本研究筛选的大豆种质材料进行大豆遗传群体的构建可以对大豆共生相关基因,尤其是响应Ⅲ型效应因子的大豆基因进行挖掘,通过进一步的功能解析为探究Ⅲ型效应因子作用机制提供重要支撑,同时利用基因型差异的大豆种质材料可以进行遗传改良,进一步培育高结瘤和高固氮大豆新品种。
4 结论
在共生结瘤过程中,不同基因型大豆在响应根瘤菌Ⅲ型效应因子过程中具有不同的表现。根瘤菌Ⅲ型效应因子在根瘤菌侵染、根瘤发育、固氮过程及根瘤衰老等过程中发挥重要调控作用。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1038/nature10452URLPMID:21993620 [本文引用: 1]

Increasing population and consumption are placing unprecedented demands on agriculture and natural resources. Today, approximately a billion people are chronically malnourished while our agricultural systems are concurrently degrading land, water, biodiversity and climate on a global scale. To meet the world's future food security and sustainability needs, food production must grow substantially while, at the same time, agriculture's environmental footprint must shrink dramatically. Here we analyse solutions to this dilemma, showing that tremendous progress could be made by halting agricultural expansion, closing 'yield gaps' on underperforming lands, increasing cropping efficiency, shifting diets and reducing waste. Together, these strategies could double food production while greatly reducing the environmental impacts of agriculture.
DOI:10.1038/461472aURLPMID:19779433 [本文引用: 1]
DOI:10.1128/mSystems.00516-19URLPMID:32071157 [本文引用: 1]

Rhizobia are soil bacteria able to establish symbiosis with diverse host plants. Specifically, Sinorhizobium fredii is a soil bacterium that forms nitrogen-fixing root nodules in diverse legumes, including soybean. The strain S. fredii CCBAU45436 is a dominant sublineage of S. fredii that nodulates soybeans in alkaline-saline soils in the Huang-Huai-Hai Plain region of China. Here, we present a manually curated metabolic model of the symbiotic form of Sinorhizobium fredii CCBAU45436. A symbiosis reaction was defined to describe the specific soybean-microsymbiont association. The performance and quality of the reconstruction had a 70% score when assessed using a standardized genome-scale metabolic model test suite. The model was used to evaluate in silico single-gene knockouts to determine the genes controlling the nitrogen fixation process. One hundred forty-one of 541 genes (26%) were found to influence the symbiotic process, wherein 121 genes were predicted as essential and 20 others as having a partial effect. Transcriptomic profiles of CCBAU45436 were used to evaluate the nitrogen fixation capacity in cultivated versus in wild soybean inoculated with the microsymbiont. The model quantified the nitrogen fixation activities of the strain in these two hosts and predicted a higher nitrogen fixation capacity in cultivated soybean. Our results are consistent with published data demonstrating larger amounts of ureides and total nitrogen in cultivated soybean than in wild soybean. This work presents the first metabolic network reconstruction of S. fredii as an example of a useful tool for exploring the potential benefits of microsymbionts to sustainable agriculture and the ecosystem.IMPORTANCE Nitrogen is the most limiting macronutrient for plant growth, and rhizobia are important bacteria for agriculture because they can fix atmospheric nitrogen and make it available to legumes through the establishment of a symbiotic relationship with their host plants. In this work, we studied the nitrogen fixation process in the microsymbiont Sinorhizobium fredii at the genome level. A metabolic model was built using genome annotation and literature to reconstruct the symbiotic form of S. fredii Genes controlling the nitrogen fixation process were identified by simulating gene knockouts. Additionally, the nitrogen-fixing capacities of S. fredii CCBAU45436 in symbiosis with cultivated and wild soybeans were evaluated. The predictions suggested an outperformance of S. fredii with cultivated soybean, consistent with published experimental evidence. The reconstruction presented here will help to understand and improve nitrogen fixation capabilities of S. fredii and will be beneficial for agriculture by reducing the reliance on fertilizer applications.
DOI:10.1099/00207713-43-2-305URLPMID:8494742 [本文引用: 1]

The phylogenetic relationships of members of the genera Rhizobium, Agrobacterium, Bradyrhizobium, and Azorhizobium were studied by direct sequencing of their amplified 16S rRNA genes. Comparative analysis of the sequence data confirmed that the genera Bradyrhizobium and Azorhizobium belong to distinct phylogenetic lineages. The genera Rhizobium and Agrobacterium were found to be phylogenetically heterogeneous, and several subgroupings in which Rhizobium and Agrobacterium species were intermixed were evident. The present findings show that the genus and species definitions of these organisms are in need of revision. Different possibilities for this are discussed in the light of sequencing data.
DOI:10.1007/BF00032239URL [本文引用: 1]
[本文引用: 1]
DOI:10.1111/1462-2920.13755URLPMID:28401683 [本文引用: 1]

The type III secretion system (T3SS) is an important genetic determinant that mediates interactions between Gram-negative bacteria and their eukaryotic hosts. Our understanding of the T3SS continues to expand, yet the availability of new bacterial genomes prompts questions about its diversity, distribution and evolution. Through a comprehensive survey of approximately 20 000 bacterial genomes, we identified 174 non-redundant T3SSs from 109 genera and 5 phyla. Many of the bacteria are environmental strains that have not been reported to interact with eukaryotic hosts, while several species groups carry multiple T3SSs. Four ultra-conserved Microsynteny Blocks (MSBs) were defined within the T3SSs, facilitating comprehensive clustering of the T3SSs into 13 major categories, and establishing the largest diversity of T3SSs to date. We subsequently extended our search to identify type III effectors, resulting in 8740 candidate effectors. Lastly, an analysis of the key transcriptional regulators and circuits for the T3SS families revealed that low-level T3SS regulators were more conserved than higher-level regulators. This comprehensive analysis of the T3SSs and their protein effectors provides new insight into the diversity of systems used to facilitate host-bacterial interactions.
[本文引用: 1]
DOI:10.1186/1471-2164-9-55URL [本文引用: 1]
DOI:10.1007/s00284-013-0469-4URLPMID:24121614 [本文引用: 1]

Sinorhizobium fredii USDA257 employs type III secretion system (T3SS) to deliver effector proteins into the host cells through pili. The nopA protein is the major component of USDA257 pili. The promoter region of USDA257 nopA possesses a well conserved tts box. Serial deletion analysis revealed that the tts box is absolutely essential for flavonoid induction of nopA. Deletion of nopA drastically lowered the number of nodules formed by USDA257 on cowpea and soybean cultivar Peking. In contrast to the parental strain, the USDA257 nopA mutant was able to form few nodules on soybean cultivars McCall and Williams 82. Light and transmission electron microscopy examination of these nodules revealed numerous starch grains both in the infected and uninfected cells.
URLPMID:15553251 [本文引用: 1]
DOI:10.1094/MPMI.2003.16.9.743URLPMID:12971597 [本文引用: 1]

The nitrogen-fixing symbiotic bacterium Rhizobium species NGR234 secretes, via a type III secretion system (TTSS), proteins called Nops (nodulation outer proteins). Abolition of TTSS-dependent protein secretion has either no effect or leads to a change in the number of nodules on selected plants. More dramatically, Nops impair nodule development on Crotalaria juncea roots, resulting in the formation of nonfixing pseudonodules. A double mutation of nopX and nopL, which code for two previously identified secreted proteins, leads to a phenotype on Pachyrhizus tuberosus differing from that of a mutant in which the TTSS is not functional. Use of antibodies and a modification of the purification protocol revealed that NGR234 secretes additional proteins in a TTSS-dependent manner. One of them was identified as NopA, a small 7-kDa protein. Single mutations in nopX and nopL were also generated to assess the involvement of each Nop in protein secretion and nodule formation. Mutation of nopX had little effect on NopL and NopA secretion but greatly affected the interaction of NGR234 with many plant hosts tested. NopL was not necessary for the secretion of any Nops but was required for efficient nodulation of some plant species. NopL may thus act as an effector protein whose recognition is dependent upon the hosts' genetic background.
[本文引用: 2]
DOI:10.1094/MPMI.2003.16.7.617URLPMID:12848427 [本文引用: 2]

Several gram-negative plant and animal pathogenic bacteria have evolved a type III secretion system (TTSS) to deliver effector proteins directly into the host cell cytosol. Sinorhizobium fredii USDA257, a symbiont of soybean and many other legumes, secretes proteins called Nops (nodulation outer proteins) into the extracellular environment upon flavonoid induction. Mutation analysis and the nucleotide sequence of a 31.2-kb symbiosis (sym) plasmid DNA region of USDA257 revealed the existence of a TTSS locus in this symbiotic bacterium. This locus includes rhc (rhizobia conserved) genes that encode components of a TTSS and proteins that are secreted into the environment (Nops). The genomic organization of the TTSS locus of USDA257 is remarkably similar to that of another broad-host range symbiont, Rhizobium sp. strain NGR234. Flavonoids that activate the transcription of the nod genes of USDA257 also stimulate the production of novel filamentous appendages known as pili. Electron microscope examination of isolated pili reveals needle-like filaments of 6 to 8 nm in diameter. The production of the pili is dependent on a functional nodD1 and the presence of a nod gene-inducing compound. Mutations in several of the TTSS genes negate the ability of USDA257 to elaborate pili. Western blot analysis using antibodies raised against purified NopX, Nop38, and Nop7 reveals that these proteins were associated with the pili. Mutations in rhcN, rhcJ, rhcC, and ttsI alter the ability of USDA257 to form nodules on Glycine max and Macroptilium atropurpureum.
DOI:10.1046/j.1365-2958.1998.00920.xURLPMID:9680225 [本文引用: 3]

The symbiotic plasmid of Rhizobium sp. NGR234 carries a cluster of genes that encodes components of a bacterial type III secretion system (TTSS). In both animal and plant pathogens, the TTSS is an essential component of pathogenicity. Here, we show that secretion of at least two proteins (y4xL and NolX) is controlled by the TTSS of NGR234 and occurs after the induction with flavonoids. Polar mutations in two TTSS genes, rhcN and the nod-box controlled regulator of transcription y4xl, block the secretion of both proteins and strongly affect the ability of NGR234 to nodulate a variety of tropical legumes including Pachyrhizus tuberosus and Tephrosia vogelii.
DOI:10.1371/journal.pone.0142866URLPMID:26569401 [本文引用: 3]

Sinorhizobium (Ensifer) fredii HH103 is a broad host-range nitrogen-fixing bacterium able to nodulate many legumes, including soybean. In several rhizobia, root nodulation is influenced by proteins secreted through the type 3 secretion system (T3SS). This specialized secretion apparatus is a common virulence mechanism of many plant and animal pathogenic bacteria that delivers proteins, called effectors, directly into the eukaryotic host cells where they interfere with signal transduction pathways and promote infection by suppressing host defenses. In rhizobia, secreted proteins, called nodulation outer proteins (Nops), are involved in host-range determination and symbiotic efficiency. S. fredii HH103 secretes at least eight Nops through the T3SS. Interestingly, there are Rhizobium-specific Nops, such as NopC, which do not have homologues in pathogenic bacteria. In this work we studied the S. fredii HH103 nopC gene and confirmed that its expression was regulated in a flavonoid-, NodD1- and TtsI-dependent manner. Besides, in vivo bioluminescent studies indicated that the S. fredii HH103 T3SS was expressed in young soybean nodules and adenylate cyclase assays confirmed that NopC was delivered directly into soybean root cells by means of the T3SS machinery. Finally, nodulation assays showed that NopC exerted a positive effect on symbiosis with Glycine max cv. Williams 82 and Vigna unguiculata. All these results indicate that NopC can be considered a Rhizobium-specific effector secreted by S. fredii HH103.
DOI:10.1094/MPMI-22-11-1445URLPMID:19810813 [本文引用: 1]

Sinorhizobium fredii HH103 secretes through the type III secretion system at least eight nodulation outer proteins (Nops), including the effector NopP. These proteins are necessary for an effective nodulation of soybean. In this work, we show that expression of the nopP gene depended on flavonoids and on the transcriptional regulators NodD1 and TtsI. Inactivation of nopP led to an increase in the symbiotic capacity of S. fredii HH103 to nodulate Williams soybean. In addition, we studied whether Nops affect the expression of the pathogenesis-related genes GmPR1, GmPR2, and GmPR3 in soybean roots and shoots. In the presence of S. fredii HH103, expression of pathogenesis-related (PR) gene PR1 was induced in soybean roots 4 days after inoculation and it increased 8 days after inoculation. The absence of Nops provoked a higher induction of PR1 in both soybean roots and shoots, suggesting that Nops function early, diminishing plant defense responses during rhizobial infection. However, the inactivation of nopP led to a decrease in PR1 expression. Therefore, the absence of NopP or that of the complete set of Nops seems to have opposite effects on the symbiotic performance and on the elicitation of soybean defense responses.
DOI:10.1111/j.1365-2958.2005.04768.xURLPMID:16102002 [本文引用: 2]

Rhizobium sp. NGR234 nodulates many plants, some of which react to proteins secreted via a type three secretion system (T3SS) in a positive- (Flemingia congesta, Tephrosia vogelii) or negative- (Crotalaria juncea, Pachyrhizus tuberosus) manner. T3SSs are devices that Gram-negative bacteria use to inject effector proteins into the cytoplasm of eukaryotic cells. The only two rhizobial T3SS effector proteins characterized to date are NopL and NopP of NGR234. NopL can be phosphorylated by plant kinases and we show this to be true for NopP as well. Mutation of nopP leads to a dramatic reduction in nodule numbers on F. congesta and T. vogelii. Concomitant mutation of nopL and nopP further diminishes nodulation capacity to levels that, on T. vogelii, are lower than those produced by the T3SS null mutant NGR(Omega)rhcN. We also show that the T3SS of NGR234 secretes at least one additional effector, which remains to be identified. In other words, NGR234 secretes a cocktail of effectors, some of which have positive effects on nodulation of certain plants while others are perceived negatively and block nodulation. NopL and NopP are two components of this mix that extend the ability of NGR234 to nodulate certain legumes.
URL [本文引用: 1]

在综合前人方法的基础上,发展了一种简捷、灵敏、廉价、效果可靠的Southern印迹杂交方案.使用0.4 mol/L NaOH溶液将琼脂糖凝胶上的DNA在变性的同时转移到尼龙膜上,晾干膜后无需固定就可以直接用于杂交.预杂交液和杂交液选用Church缓冲系统,该缓冲系统中仅需10 g/L BSA作为封闭剂即可.整个洗膜过程可简化为一种溶液、2~3次洗涤.碱变性及转移后的尼龙膜,也可以用于地高辛(DIG)等非放射性检测系统,并可耐多轮杂交和洗脱.本方法可适用于包括高等植物基因组在内的多种生物材料的DNA检测.
URL [本文引用: 1]

在综合前人方法的基础上,发展了一种简捷、灵敏、廉价、效果可靠的Southern印迹杂交方案.使用0.4 mol/L NaOH溶液将琼脂糖凝胶上的DNA在变性的同时转移到尼龙膜上,晾干膜后无需固定就可以直接用于杂交.预杂交液和杂交液选用Church缓冲系统,该缓冲系统中仅需10 g/L BSA作为封闭剂即可.整个洗膜过程可简化为一种溶液、2~3次洗涤.碱变性及转移后的尼龙膜,也可以用于地高辛(DIG)等非放射性检测系统,并可耐多轮杂交和洗脱.本方法可适用于包括高等植物基因组在内的多种生物材料的DNA检测.
URL [本文引用: 1]

Western杂交是一种检测蛋白质的免疫化学方法,已在生物科学的许多领域得到广泛应用,但以前的文献介绍的Western杂交程序过于繁琐.本文根据Western杂交的基本原理结合已报道资料进行了一些改进,并应用到猪血清瘦素蛋白的检测.该程序简单易操作,结果清晰,证明改进后的方法完全可行.
URL [本文引用: 1]

Western杂交是一种检测蛋白质的免疫化学方法,已在生物科学的许多领域得到广泛应用,但以前的文献介绍的Western杂交程序过于繁琐.本文根据Western杂交的基本原理结合已报道资料进行了一些改进,并应用到猪血清瘦素蛋白的检测.该程序简单易操作,结果清晰,证明改进后的方法完全可行.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:23468637 [本文引用: 1]
DOI:10.3390/ijms17050755URL [本文引用: 1]
DOI:10.1007/s11104-018-3875-3URL [本文引用: 1]
DOI:10.1146/annurev.mi.46.100192.002151URLPMID:1444262 [本文引用: 1]

An economically important problem in microbial ecology concerns the efficacy of rhizobial inoculants for the formation of nitrogen-fixing root nodules on legume crop plants such as soybean, alfalfa, and clover. Some strains of rhizobia can increase symbiotic nitrogen fixation under controlled conditions. However, attempts to improve nitrogen fixation under agricultural conditions with such strains often fail, usually as a result of the presence of indigenous rhizobia limiting nodulation by the inoculum strains. This problem is referred to as the Rhizobium competition problem, and molecular genetics is being used to address the problem from two perspectives. First, the host specificity of rhizobia is being characterized with the long term goal of developing strains that can nodulate a very strain-specific host-legume genotype. Second, the genetic basis of competitiveness in several strains is being examined. Genetic determinants of nodulation competitiveness have been isolated and mechanisms for their stable integration into the genome of superior nitrogen-fixing strains have been developed. Several phenotypes have been identified as playing an important role in nodulation competitiveness including antibiosis, motility, speed of nodulation, cell-surface characteristics, and nodulation efficiency. Several solutions to this problem are likely to result from these strategies and will be useful for certain legumes in specific locations.
DOI:10.1007/s10681-013-1005-0URL [本文引用: 1]

Soybean research has found that nodule traits, especially nodule biomass, are associated with N-2 fixation ability. Two genotypes, differing in nodule number per plant and individual nodule weight, KS4895 and Jackson, were mated to create 17 F-3- and 80 F-5-derived RILs. The population was mapped with 664 informative markers with an average distance of less than 20 cM between adjacent markers. Nodule traits were evaluated in 3-year field trials. Broad-sense heritability for nodule number (no. plant(-1)), individual nodule dry weight (mg nodule(-1)), individual nodule size (mm nodule(-1)), and total nodule dry weight (g plant(-1)) was 0.41, 0.42, 0.45, and 0.27, respectively. Nodule number was negatively correlated with individual nodule weight and size. Nodule number, individual nodule weight, and size are major components which likely contributed to increased total nodule weight per plant. Composite interval mapping (CIM) identified eight QTLs for nodule number with R-2 values ranging from 0.14 to 0.20. Multiple interval mapping (MIM) identified two QTLs for nodule number, one of which was located close to the QTL identified with CIM. Six QTLs for individual nodule weight were detected with CIM, and one QTL was identified with MIM. For nodule size, CIM identified seven QTLs with R-2 values ranging from 0.14 to 0.27. Five QTLs for total nodule weight were detected with CIM, one of which was located close to a QTL identified with MIM. These results document the first QTL information on nodule traits in soybean from field experiments utilizing a dense, complete linkage map.
[本文引用: 1]
DOI:10.1104/pp.15.01661URLPMID:26582727 [本文引用: 1]

Rj4 is a dominant gene in soybeans (Glycine max) that restricts nodulation by many strains of Bradyrhizobium elkanii. The soybean-B. elkanii symbiosis has a low nitrogen-fixation efficiency, but B. elkanii strains are highly competitive for nodulation; thus, cultivars harboring an Rj4 allele are considered favorable. Cloning the Rj4 gene is the first step in understanding the molecular basis of Rj4-mediated nodulation restriction and facilitates the development of molecular tools for genetic improvement of nitrogen fixation in soybeans. We finely mapped the Rj4 locus within a small genomic region on soybean chromosome 1, and validated one of the candidate genes as Rj4 using both complementation tests and CRISPR/Cas9-based gene knockout experiments. We demonstrated that Rj4 encodes a thaumatin-like protein, for which a corresponding allele is not present in the surveyed rj4 genotypes, including the reference genome Williams 82. Our conclusion disagrees with the previous report that Rj4 is the Glyma.01G165800 gene (previously annotated as Glyma01g37060). Instead, we provide convincing evidence that Rj4 is Glyma.01g165800-D, a duplicated and unique version of Glyma.01g165800, that has evolved the ability to control symbiotic specificity.
URLPMID:30087346 [本文引用: 1]
DOI:10.1128/AEM.01942-15URLPMID:26187957 [本文引用: 1]

Symbioses between leguminous plants and soil bacteria known as rhizobia are of great importance to agricultural production and nitrogen cycling. While these mutualistic symbioses can involve a wide range of rhizobia, some legumes exhibit incompatibility with specific strains, resulting in ineffective nodulation. The formation of nodules in soybean plants (Glycine max) is controlled by several host genes, which are referred to as Rj genes. The soybean cultivar BARC2 carries the Rj4 gene, which restricts nodulation by specific strains, including Bradyrhizobium elkanii USDA61. Here we employed transposon mutagenesis to identify the genetic locus in USDA61 that determines incompatibility with soybean varieties carrying the Rj4 allele. Introduction of the Tn5 transposon into USDA61 resulted in the formation of nitrogen fixation nodules on the roots of soybean cultivar BARC2 (Rj4 Rj4). Sequencing analysis of the sequence flanking the Tn5 insertion revealed that six genes encoding a putative histidine kinase, transcriptional regulator, DNA-binding transcriptional activator, helix-turn-helix-type transcriptional regulator, phage shock protein, and cysteine protease were disrupted. The cysteine protease mutant had a high degree of similarity with the type 3 effector protein XopD of Xanthomonas campestris. Our findings shed light on the diverse and complicated mechanisms that underlie these highly host-specific interactions and indicate the involvement of a type 3 effector in Rj4 nodulation restriction, suggesting that Rj4 incompatibility is partly mediated by effector-triggered immunity.
DOI:10.1007/s11104-018-3745-zURL [本文引用: 1]
