 ,, 王艳娇
,, 王艳娇 ,, 段玉, 马志敏, 宾羽, 周常勇, 宋震
,, 段玉, 马志敏, 宾羽, 周常勇, 宋震 ,西南大学/中国农业科学院柑桔研究所,重庆400712
,西南大学/中国农业科学院柑桔研究所,重庆400712Construction of Genome-Length cDNA of Citrus Vein Enation Virus and Identification of Its Infectivity
XU JianJian ,, WANG YanJiao
,, WANG YanJiao ,, DUAN Yu, MA ZhiMin, BIN Yu, ZHOU ChangYong, SONG Zhen
,, DUAN Yu, MA ZhiMin, BIN Yu, ZHOU ChangYong, SONG Zhen ,Citrus Research Institute, Southwest University/Chinese Academy of Agricultural Sciences, Chongqing 400712
,Citrus Research Institute, Southwest University/Chinese Academy of Agricultural Sciences, Chongqing 400712通讯作者:
责任编辑: 岳梅
收稿日期:2020-06-22接受日期:2020-07-29网络出版日期:2020-09-16
| 基金资助: |
Received:2020-06-22Accepted:2020-07-29Online:2020-09-16
作者简介 About authors
许建建,E-mail:
王艳娇,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2224KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
许建建, 王艳娇, 段玉, 马志敏, 宾羽, 周常勇, 宋震. 柑橘脉突病毒基因组全长cDNA克隆及其侵染性鉴定[J]. 中国农业科学, 2020, 53(18): 3707-3715 doi:10.3864/j.issn.0578-1752.2020.18.007
XU JianJian, WANG YanJiao, DUAN Yu, MA ZhiMin, BIN Yu, ZHOU ChangYong, SONG Zhen.
0 引言
【研究意义】柑橘脉突病毒(citrus vein enation virus,CVEV)是黄矮病毒科(Luteoviridae)豌豆耳突花叶病毒属(Enamovirus)的一种正义单链RNA病毒[1],可引起柑橘脉突病及木瘤病。目前,CVEV在亚洲的日本、印度、土耳其、伊朗,北美的美国,非洲的南非,南美的巴西、秘鲁,欧洲的西班牙,大洋洲的澳大利亚等多个国家均有报道[1,2,3,4,5,6,7],并对南美的秘鲁柑橘产业造成了严重破坏[8]。我国于20世纪90年代在浙江省台州市黄岩区首次检测出该病毒[9],随后在浙江省其他柑橘产区以及四川也检测到该病毒[10,11]。CVEV可以侵染大多数的柑橘品种,除通过嫁接传播外,还可由多种蚜虫如橘蚜(Toxopetera citricida)、棉蚜(Aphis gossypii)、桃蚜(Myzus pcrsicae)等以持久方式进行传播[12,13]。这些蚜虫在中国及世界范围内分布广泛,且种群数量大,因而致使CVEV具有较高的传播流行风险。构建CVEV侵染性克隆将为深入解析其分子特性及致病机理打下基础,对于CVEV的防控具有重要意义。【前人研究进展】1953年,WALLCE等在美国的California酸橙上首次发现柑橘脉突病,并初步证明该病由病毒引起。酸橙和墨西哥莱蒙感病后主要表现为叶片侧脉和支脉产生耳状小突起,叶背对应部位出现凹陷[14]。1959年,在澳大利亚发现一种能引起粗柠檬的根茎部出现木瘤的病害,初步证明该病和脉突病均由CVEV引起[15,16]。CVEV基因组为正义单链RNA,包含5个开放阅读框(open reading frame,ORF)、5′端207 nt和3′端198 nt的两个非翻译区(untranslated region,UTR)。其中,ORF0嵌合于ORF1,编码39 kD多肽(354 aa);ORF1编码100 kD蛋白(902 aa),包含S39肽酶家族特有的丝氨酸蛋白酶结构域和病毒末端结合蛋白;ORF2编码146或148 kD融合蛋白,包含解旋酶和多聚酶结构域包围的甘氨酸-天冬氨酸-天冬氨酸三肽基序;ORF3编码21 kD的外壳蛋白(191 aa),最近被证明是基因沉默抑制子;ORF5编码一个融合蛋白,可能与病毒在植株内的系统移动以及在蚜虫体内的滞留有关[1,17]。CVEV病毒粒子为等径对称多面体,大小约为25或28 nm[7]。CVEV的检测方法有传统指示植物鉴定法、常规反转录多聚酶链式反应RT-PCR法和实时荧光定量RT-PCR法[18,19],但指示植物鉴定法繁琐且易受柑橘衰退病毒(citrus tristeza virus,CTV)干扰[20]。【本研究切入点】CVEV可引起柑橘脉突病,但其柯赫氏法则证明尚未完成。通过构建CVEV基因组全长cDNA克隆,经农杆菌介导接种柑橘,并通过RT-PCR及生物学观察鉴定其侵染性,将为深入了解其病原分子生物学特性、研究其致病机理打下基础。【拟解决的关键问题】构建CVEV的病毒基因组全长RT-PCR扩增体系,获得其侵染性克隆,实现其对柑橘的系统性侵染。1 材料与方法
试验于2017—2020年在西南大学柑桔研究所国家柑桔苗木脱毒中心实验室完成。1.1 供试材料
CVEV毒源植株象山红(Citrus reticulata),柑橘实生苗摩洛哥酸橙(C. aurantium)、邓肯葡萄柚(C. paradisi)、尤力克柠檬(C. limon)、枳柚(C. paradisi × Poncirus trifoliata)、Rusk枳橙(P. trifoliata × C. sinensis)、枣阳小叶枳(P. trifoliata),基因沉默抑制子HC-Pro表达质粒等均由西南大学柑桔研究所提供。pXT1双元表达载体由南京农业大学陶小荣教授惠赠。1.2 酶与试剂
限制性内切酶Stu I、Sma I购自北京NEB公司;LA Taq? Polymerase、In-Fusion HD Cloning Kit、SMARTer? RACE 5′/3′ Kit购自大连TaKaRa公司;SuperScriptTM IV逆转录酶、Trizol试剂购自美国Invitrogen公司;E.Z.N.A.TM Plasmid Mini ProtocolⅠ试剂盒、E.Z.N.A.TM Gel Extraction试剂盒购自Omega公司;大肠杆菌感受态细胞JM 109、农杆菌感受态细胞GV3101购自博迈德生物有限公司。1.3 引物设计
根据NCBI中已报道的CVEV分离株VE-1基因组序列及5′端RACE测序结果,利用Primer 5软件设计扩增CVEV基因组全长cDNA的特异性引物EV25-F/EV5983-R,以及用于RACE的EV532R、EV414R。用于CVEV检测的VE16-F/VE17-R及其他引物见表1。Table 1
表1
表1引物序列
Table 1
| 引物Primer | 序列Sequence (5′→3′) |
|---|---|
| EV532R | CGAGCGGCACAAATTCTAAAGTACCAC |
| EV414R | GACCCCATAGCAAAGCAAGAGCACAT |
| UPM long | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT |
| NUP | AAGCAGTGGTATCAACGCAGAGT |
| SMARTer II A Oligonucleotide | AAGCAGTGGTATCAACGCAGAGTACXXXXX |
| EV25-F | TTTCATTTGGAGAGGAATATAAGCTATAAAAGAAAAGTGCG |
| EV5983-R | ATGCCATGCCGACCCACAAAAGTATGCAAAGAATATG |
| VE16-F | AATTACGGCGTATCTATGGTGAGTCG |
| VE17-R | AATGAGATAGCCCGGTTGTCCAG |
新窗口打开|下载CSV
1.4 CVEV侵染性克隆构建
1.4.1 毒源植株叶片总RNA提取 按照Trizol试剂说明书提取毒源植株的总RNA,琼脂糖凝胶电泳检测RNA完整性。1.4.2 RACE扩增 提取毒源植株总RNA,采用SMARTer? RACE试剂盒对CVEV 5′端序列进行扩增,然后通过常规克隆测序并进行序列分析。第一链cDNA合成:配置预混液RACE Buffer Mix:5× First-Strand Buffer 4.0 μL,SMARTer II A Oligonucleotide 1.0 μL,SMARTScribeTM Reverse Transcriptase(100 U·μL-1)2.0 μL,dNTPs(20 mmol·L-1)1.0 μL,RNase Inhibitor(40 U·μL-1)0.5 μL,DTT(100 mmol·L-1)0.5 μL,总体积9 μL。另取模板RNA 1.75 μL,EV532R 1.0 μL,ddH2O(20 mmol·L-1)1.0 μL,混匀运行程序:72℃ 3 min,迅速置于冰上;加入预混液运行程序:42℃ 60 min;70℃ 10 min。反应所得cDNA备用。
第一轮PCR反应体系:PS Max Premix(2×)12.5 μL,UMP long 1.0 μL,EV532R 1.0 μL,前一步cDNA 2.0 μL,PS Max Premix(2×)12.5 μL,ddH2O 8.5 μL,总体积25 μL;第一轮PCR程序:94℃ 2 min;94℃ 15 s,56℃ 15 s,72℃ 30 s,30个循环;72℃ 5 min。
将上述所得PCR产物稀释50倍,取2.0 μL,再加入EV414R 1.0 μL,NUP 1.0 μL,PS Max Premix(2×)12.5 μL,ddH2O补至总体积25 μL,运行第二轮PCR程序:94℃ 2 min;94℃ 15 s,58℃ 15 s,72℃ 30 s,35个循环;72℃ 5 min。
1.4.3 CVEV基因组全长RT-PCR 以提取的总RNA为模板,使用SuperScriptTM IV逆转录酶合成cDNA,反应体系:在冰上配置以下混合液,50 μmol·L-1 random hexamers 1 μL,10 mmol·L-1 dNTP Mix 1 μL,RNA模板 9 μL,ddH2O补至13 μL;5×SSIV Buffer 4.0 μL,10 mmol·L-1 dTT 1.0 μL,RNase OUTTM Recombinant RNase Inhibitor 1.0 μL,SuperScriptTM IV Reverse Transcriptase(200 U·μL-1)1.0 μL,混匀。运行程序:23℃ 10 min;53℃ 15 min。以产物cDNA为模板,使用引物EV25-F/EV5983-R进行PCR扩增。PCR反应体系为25 μL:ddH2O 4.0 μL,2×GC buffer I 12.5 μL,EV25-F/EV5983-R各1.0 μL,LA Taq酶0.5 μL,dNTP 4.0 μL,模板2.0 μL。反应条件:94℃ 2 min;94℃ 30 s,65℃ 8 min,20个循环;94℃ 30 s,60℃ 30 s,65℃ 8 min,20个循环;68℃ 10 min。
1.4.4 pXT1线性载体获取 使用NEB限制性内切酶Sma I和Stu I酶切pXT1双元表达载体,获得pXT1线性载体,反应体系:双元表达载体pXT1质粒28 μL(1.0 μg),10×Cutsmart Buffer 5.0 μL,限制性内切酶Sma I 1.0 μL,ddH2O 16 μL。反应条件:25℃温育30 min。再加入1 μL Stu I限制性内切酶,37℃温育30 min。电泳验证后利用E.Z.N.A.TM Gel Extraction试剂盒进行产物回收。
1.4.5 CVEV全长cDNA的In-Fusion克隆 使用In-Fusion? HD Cloning Kit将获得的PCR产物与pXT1线性载体连接。反应体系:5×In-Fusion HD Enzyme Premix 2 μL,PCR产物2 μL,线性化pXT1载体2 μL,ddH2O补足至10 μL。反应条件:50℃温育30 min。转化大肠杆菌JM 109,挑选单克隆采用引物VE16-F/VE17-R[1]进行菌液PCR,阳性克隆送华大基因公司测序。
1.5 序列分析
利用DNAMAN 7.0对测序后所得CVEV全长cDNA进行序列分析,利用MAGA X软件构建CVEV进化树(邻接法)。1.6 农杆菌介导真空浸润接种柑橘
提取1.4.5所得阳性克隆质粒,电击法转化农杆菌感受态细胞GV3101,均匀涂布到含有利福平(20 μg·μL-1)和卡那霉素(50 mmol·L-1)抗生素的LB固体培养基上,28℃培养约48 h。挑选阳性单克隆接种含相应抗生素的LB液体培养基(20 mg·L-1 Rif,50 mg·L-1 Kan),200 r/min,28℃振荡培养12—16 h。离心收集菌体,用接种缓冲液悬浮(10 mmol·L-1 MgCl2,10 mmol·L-1 MES,200 μmol·L-1 As),使其OD600为0.8—1.0,同时加入沉默抑制子表达克隆,使样品OD≈1.0,沉默抑制子OD≈0.5,黑暗静置2 h备用。参照文献[21,22]报道的方法进行农杆菌介导真空浸润接种柑橘,即将柑橘实生苗浸入前步骤接种混合液并置于真空干燥仪,抽真空至压力-1.0—-0.8 kg·cm-2,保持5 min后快速释放压力并立即用无菌水冲洗,22℃暗处理24 h。转入正常光周期:22℃ 16 h;20℃ 8 h。一周后开始症状观察记录,并参照王艳娇[23]的方法开展RT-PCR检测。2 结果
2.1 RACE分析
病毒基因组5′端序列对于全长cDNA侵染性克隆的构建具有重要影响,个别碱基突变就可能导致其侵染性降低,甚至失去侵染性。因此,在扩增克隆CVEV基因组之前首先采用RACE技术确定其基因组5′端序列。采集CVEV毒源象山红植株新展开的嫩叶,利用Trizol试剂提取总RNA,对CVEV的5′末端进行RACE(图1-A),获得大小约414 bp的目标条带。扩增产物经切胶回收、连接、转化大肠杆菌JM109,挑取单菌落进行菌液PCR验证(图1-B),获得阳性克隆。随机挑选3个PCR阳性克隆进行测序,并与已报道的CVEV分离株VE-1进行序列比对,在排除RACE试剂的3—5个碱基后,各阳性克隆基因序列及VE-1的序列均为ATATAAGCTATA AAAGAAAAGTGCGTTTACGCTTCCTT···,说明CVEV 5′端前70个碱基序列比较保守,这为利用该保守序列设计引物扩增CVEV全长cDNA,进而构建侵染性克隆打下基础。
图1
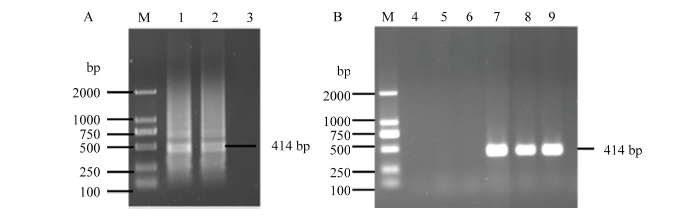 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1CVEV 5′末端RACE及克隆
M:DNA分子标准 DNA marker;1、2:5′末端RACE 5′ RACE ;7—9:5′末端cDNA克隆PCR检测 Detection of cDNA clones of 5′-end sequence using PCR;3—5:阴性对照 Negative control;6:ddH2O
Fig. 15′ rapid amplification of cDNA ends (RACE) and its cloning of CVEV
2.2 CVEV基因组全长RT-PCR
依据5′及3′端保守序列设计直接扩增CVEV基因组全长的引物EV25-F/EV5983-R。以2.1提取的总RNA为模板,进行CVEV的基因组全长RT-PCR。琼脂糖凝胶电泳结果显示(图2),PCR产物片段大小约5 983 bp,符合预期CVEV基因组全长大小,切胶回收备用。图2
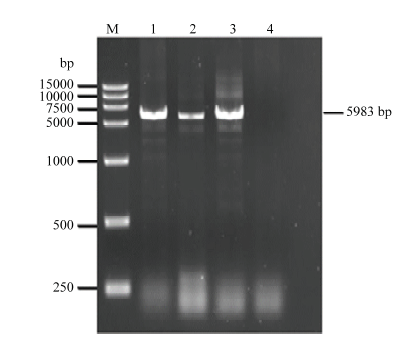 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2CVEV的基因组全长RT-PCR
M:DNA分子标准 DNA marker;1—3:CVEV侵染样品 Samples infected with CVEV;4:阴性对照 Negative control
Fig. 2RT-PCR for full-length genome of CVEV
2.3 CVEV基因组全长cDNA克隆、鉴定及序列分析
使用限制性内切酶Sma I和Stu I酶切pXT1双元表达载体。将2.2获得的RT-PCR产物和pXT1线性载体切胶回收、In-Fusion HD Cloning Kit连接、转化大肠杆菌JM 109。挑取单菌落采用引物VE16-F/VE17-R进行菌液PCR筛选,然后采用引物EV25-F/EV5983-R进行CVEV全长cDNA初步PCR鉴定(图3),共计获得10个CVEV基因组全长cDNA克隆。图3
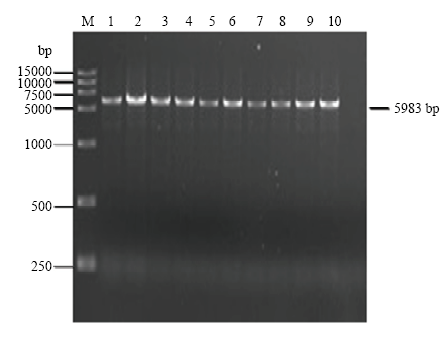 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3CVEV全长cDNA克隆的PCR鉴定
M:DNA分子标准 DNA marker;1—10:CVEV全长cDNA CVEV full-length cDNA
Fig. 3Identification of CVEV full-length cDNA by PCR
随机选取6个阳性克隆,命名为CVEV1901—CVEV1906并进行序列测定。结果表明所获克隆均符合CVEV基因组全长预期,序列之间的一致性为99.35%。其中,CVEV1901全长5 983 nt,由5个ORF、5′端207 nt和3′端198 nt的两个UTR以及在ORF2和ORF3之间的122 nt的基因间隔区组成。进一步分析显示,CVEV-1901与浙江分离株XZG(登录号:MN596377)仅有一个碱基差异,第1 375位碱基G→A,序列一致性99.98%,与四川SM分离株序列一致性99.11%;与西班牙VE-1分离株、美国加州VE701分离株和日本IBK分离株基因组序列一致性分别为98.61%、97.36%和96.89%;与同一属豌豆耳突花叶病毒(pea enation mosaic virus)和紫花苜蓿耳突病毒(alfalfa enamovirus)的序列一致性分别为90.46%和90.26%。
在利用MEGA X构建的系统进化树上,CVEV1901与中国浙江分离株XZG关系最近,并与四川分离株SM一起聚为一簇;与西班牙分离株VE-1、美国加州分离株VE701、日本分离株IBK距离较近;而与同属的豌豆耳突花叶病毒和紫花苜蓿耳突病毒距离相对较远;与黄矮病毒科其他不同属病毒分居在不同的大分支上,这与序列分析结果一致(图4)。
图4
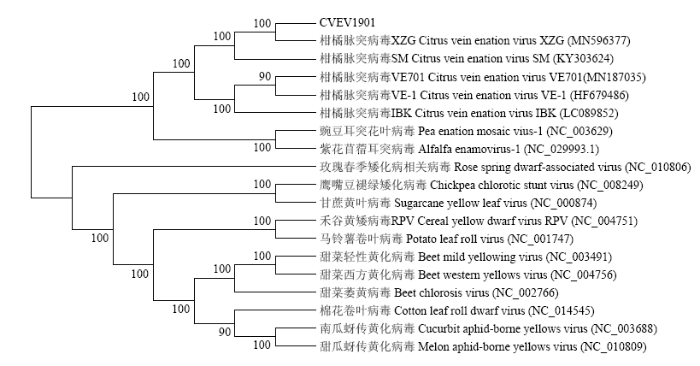 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4CVEV与黄矮病毒科中其他病毒全长基因序列的系统进化树
Fig. 4Polygenetic tree of CVEV and other members of family Luteoviridae
2.4 CVEV基因组全长cDNA克隆的侵染性鉴定
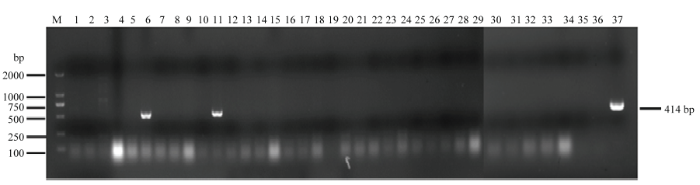
2.4.1 尤力克柠檬接种 将获得的CVEV全长cDNA克隆CVEV1901和CVEV1902分别转化农杆菌GV3101,以pTX1空载体为阴性对照,真空浸润接种尤力克柠檬实生苗。接种30 d后,抽提系统新发叶片RNA进行RT-PCR检测,结果表明两株CVEV1901接种的尤力克植株检测出CVEV特异性条带(图5)。初步说明CVEV侵染性克隆构建成功。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5CVEV全长cDNA克隆接种尤力克柠檬的RT-PCR检测
M:2000 bp分子量marker 2000 bp molecular marker;1—5:pTX1空载接种对照 pTX1 no-load inoculation control;6—19:CVEV1901接种样品 CVEV1901 inoculated samples;20—34:CVEV1902接种样品 CVEV1902 inoculated samples;35:阴性对照Negative control;36:ddH2O;37:阳性对照CVEV XZG Positive control CVEV XZG
Fig. 5RT-PCR detection of C. limon inoculated with full-length cDNA of CVEV
2.4.2 多品种柑橘接种 为进一步确认CVEV1901的侵染性,通过农杆菌介导的真空浸润接种摩洛哥酸橙、邓肯葡萄柚、尤力克柠檬、枳柚、Rusk枳橙、枣阳小叶枳6个柑橘品种,并以pTX1空载体为阴性对照。结果发现部分CVEV1901接种的摩洛哥酸橙出现CVEV侵染的典型症状,叶片侧脉产生耳状小突起,叶背有相应的凹陷,叶片皱缩,部分邓肯葡萄柚和尤力克柠檬叶片出现皱缩现象(图6),而pTX1空载体接种对照则无上述症状表现。CVEV1901接种后120 d的RT-PCR检测结果表明,摩洛哥酸橙、邓肯葡萄柚、尤力克柠檬、枳柚、Rusk枳橙和枣阳小叶枳的CVEV阳性率分别为16/17(94.12%)、12/14(85.71%)、16/21(76.19%)、15/19(78.95%)、13/14(92.86%)和0/18(0),而 pTX1空载体接种对照为0。这进一步表明CVEV1901为具有侵染性的CVEV基因组全长cDNA克隆。
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6CVEV1901接种不同柑橘品种的症状
A、B:CVEV1901接种摩洛哥酸橙的症状Symptom on C. aurantium inoculated with CVEV1901;C:pTX1空载体接种的摩洛哥酸橙C. aurantium inoculated with pTX1 empty vector;D、E:CVEV1901接种邓肯葡萄柚的症状Symptom on C. paradisi inoculated with CVEV1901;F:pTX1空载体接种的邓肯葡萄柚C. paradisi inoculated with pTX1 empty vector;G、H:CVEV1901接种尤力克柠檬的症状Symptom on C. limon inoculated with CVEV1901;I:pTX1空载体接种的尤力克柠檬C. limon inoculated with pTX1 empty vector
Fig. 6Symptoms on different citrus varieties inoculated with CVEV1901
3 讨论
侵染性克隆的构建是深入研究病毒分子生物学特性、致病机理及载体化利用的重要基础[24,25,26,27,28,29]。但相对于侵染草本植物的病毒,目前关于果树病毒侵染性克隆的报道较少,构建难度较大。本研究通过对CVEV病毒全长基因组的一次性RT-PCR扩增,与pXT1载体进行无缝重组克隆,获得了CVEV的侵染性克隆。本研究所获的侵染性克隆CVEV1901与我国已报道的XZG及SM分离株基因组长度一致,结构相同,同源性均高于99%,通过农杆菌介导的真空浸润可以高效侵染多种柑橘品种。这不仅为研究CVEV分子生物学特性如复制、移动、基因功能等提供了单一遗传背景的材料,而且为深入研究CVEV的致病机制打下了坚实基础。在通过突变获取弱毒株系用于交叉保护,或改造为基因沉默载体方面也具有广阔的应用前景。目前,植物RNA病毒侵染性克隆的构建主要还是通过分段扩增、分段克隆来获得[30]。这种方法可能会造成获得的植物病毒全长cDNA为假重组体。本试验采用一步法RT-PCR扩增获得CVEV全长cDNA,则有效避免了这种情况。不过,该方法对RNA模板、反转录酶、DNA聚合酶等有比较严格的要求,尤其是模板质量和反转录酶及聚合酶的扩增和保真能力。多次试验表明,利用Trizol法提取柑橘叶片总RNA可以达到本试验需求,同时为防止RNA降解,提取RNA后立即进行第一链cDNA的合成。不同的反转录酶对于病毒基因组的全长扩增也有不同的影响,通过对不同类型反转录酶多次试验,发现采用SuperScriptTM IV逆转录酶可成功扩增CVEV全长,CVEV1901与毒源植株XZG分离株的全基因组相比仅有一个碱基差异,与中国分离株的同源性高于99%,表明该酶的保真性能也有保证。最后,使用In-Fusion重组连接线性化pXT1和CVEV全长cDNA,精确置于载体的35S后,在启动子与病毒5′端序列之间没有任何非病毒序列,保证了转录出的病毒的精确性,也为所获克隆具备侵染性打下了良好基础。
接种方式对病毒侵染性克隆成功接种具有重要的影响[22]。CVEV1901通过农杆菌介导的真空浸润可高效侵染多个柑橘品种。但在通过农杆菌介导的注射方式对烟草、豌豆、蚕豆、芸豆、昆诺藜等草本植物进行接种时,通过RT-PCR均未检测到CVEV阳性植株。表明CVEV1901可能无法通过注射方式成功侵染上述草本植物,当然部分植物也可能并非CVEV的有效寄主。这一现象在柑橘黄化脉明病毒(citrus yellow vein clearing virus)侵染性克隆接种时也曾出现[22],有待进一步研究。
4 结论
建立了CVEV的基因组全长RT-PCR扩增体系,获得CVEV基因组全长cDNA克隆。通过农杆菌介导的真空浸润接种,RT-PCR检测和生物学症状观察,证明CVEV1901为具有侵染性的CVEV基因组全长cDNA侵染性克隆。致谢:
南京农业大学陶小荣教授惠赠pXT1双元表达载体,在此表示感谢!参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1094/PHYTO-03-13-0068-RURLPMID:23718835 [本文引用: 4]

To identify the causal agent of citrus vein enation disease, we examined by deep sequencing (Solexa-Illumina) the small RNA (sRNA) fraction from infected and healthy Etrog citron plants. Our results showed that virus-derived sRNAs (vsRNAs): (i) represent about 14.21% of the total sRNA population, (ii) are predominantly of 21 and 24 nucleotides with a biased distribution of their 5' nucleotide and with a clear prevalence of those of (+) polarity, and (iii) derive from all the viral genome, although a prominent hotspot is present at a 5'-proximal region. Contigs assembled from vsRNAs showed similarity with luteovirus sequences, particularly with Pea enation mosaic virus, the type member of the genus Enamovirus. The genomic RNA (gRNA) sequence of a new virus, provisionally named Citrus vein enation virus (CVEV), was completed and characterized. The CVEV gRNA was found to be single-stranded, positive-sense, with a size of 5,983 nucleotides and five open reading frames. Phylogenetic comparisons based on amino acid signatures of the RNA polymerase and the coat protein clearly classifies CVEV within the genus Enamovirus. Dot-blot hybridization and reverse transcription-polymerase chain reaction tests were developed to detect CVEV in plants affected by vein enation disease. CVEV detection by these methods has already been adopted for use in the Spanish citrus quarantine, sanitation, and certification programs.
//
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00705-016-3139-6URLPMID:27848014 [本文引用: 1]

The genomic sequences of five Japanese isolates of citrus vein enation virus (CVEV) isolates that induce vein enation were determined and compared with that of the Spanish isolate VE-1. The nucleotide sequences of all Japanese isolates were 5,983 nt in length. The genomic RNA of Japanese isolates had five potential open reading frames (ORF 0, ORF 1, ORF 2, ORF 3, and ORF 5) in the positive-sense strand. The nucleotide sequence identity among the Japanese isolates and Spanish isolate VE-1 ranged from 98.0% to 99.8%. Comparison of the partial amino acid sequences of ten Japanese isolates and three Spanish isolates suggested that four amino acid residues, at positions of 83, 104, and 113 in ORF 2 and position 41 in ORF 5, might be unique to some Japanese isolates.
//
[本文引用: 2]
//
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S2095-3119(14)60903-5URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s42161-018-0163-2URL [本文引用: 1]
DOI:10.16420/j.issn.0513-353x.2016-0150URL [本文引用: 1]

为了快速、灵敏地检测柑橘脉突病毒(Citrus vein enation virus,CVEV),通过设计特异性引物(EVqF4/EVqR4),优化反应条件,建立了CVEV的实时荧光定量RT-PCR检测体系。该方法特异性良好;检测灵敏度比常规RT-PCR高100倍;标准曲线循环阈值与模板浓度呈良好的线性关系,相关系数为0.992,扩增效率101.8%;检测组内和组间变异系数均小于2.85%,重复性较好。利用所建立的实时荧光定量方法对柑橘植株进行检测发现,‘代代酸橙’和‘象山红’植株中CVEV分布不均匀,其中根部病毒滴度最高,分别为162.52和45.32拷贝 · ng-1 RNA,皮及叶部病毒滴度相对较低。
DOI:10.16420/j.issn.0513-353x.2016-0150URL [本文引用: 1]

为了快速、灵敏地检测柑橘脉突病毒(Citrus vein enation virus,CVEV),通过设计特异性引物(EVqF4/EVqR4),优化反应条件,建立了CVEV的实时荧光定量RT-PCR检测体系。该方法特异性良好;检测灵敏度比常规RT-PCR高100倍;标准曲线循环阈值与模板浓度呈良好的线性关系,相关系数为0.992,扩增效率101.8%;检测组内和组间变异系数均小于2.85%,重复性较好。利用所建立的实时荧光定量方法对柑橘植株进行检测发现,‘代代酸橙’和‘象山红’植株中CVEV分布不均匀,其中根部病毒滴度最高,分别为162.52和45.32拷贝 · ng-1 RNA,皮及叶部病毒滴度相对较低。
DOI:10.1094/PDIS.2004.88.12.1328URLPMID:30795193 [本文引用: 1]

Biological indexing for graft-transmissible pathogens of citrus in the presence of additional pathogens was investigated. The probability for symptom expression, the efficacy of the bio-indexing tests, and the number of citrus indicators required for pathogen detection were statistically evaluated. Multiple infections did not preclude symptom expression or reduce the diagnostic efficacy of the primary indexing hosts for Citrus tristeza virus (CTV), Citrus psorosis virus (CPsV), and Citrus tatter leaf virus (Apple stem grooving virus). Symptoms of Citrus vein enation virus (CVEV) and the diagnostic efficacy of Mexican lime were suppressed by the T30 group CTV isolates, but not by other CTV isolates tested. CPsV suppressed symptom expression and diagnostic efficiency of Dweet tangor and sweet orange for concave gum. The application of alternate bioassay hosts for indexing was also investigated. Dweet tangor, sweet orange, and Citrus excelsa are not typically used for bioindexing of CVEV, however, Dweet tangor and C. excelsa detected CVEV in single infections, whereas in sweet orange, CVEV was detected only when CPsV, concave gum, or citrus viroids were present. CTV was readily detected using the alternative indicator C. excelsa, whereas only shock reacting CPsV isolates were effectively indexed by Mexican lime.
[本文引用: 1]
[本文引用: 1]
DOI:10.1128/JVI.00436-12URL [本文引用: 1]

The improvement of the agricultural and wine-making qualities of the grapevine (Vitis vinifera) is hampered by adherence to traditional varieties, the recalcitrance of this plant to genetic modifications, and public resistance to genetically modified organism (GMO) technologies. To address these challenges, we developed an RNA virus-based vector for the introduction of desired traits into grapevine without heritable modifications to the genome. This vector expresses recombinant proteins in the phloem tissue that is involved in sugar transport throughout the plant, from leaves to roots to berries. Furthermore, the vector provides a powerful RNA interference (RNAi) capability of regulating the expression of endogenous genes via virus-induced gene-silencing (VIGS) technology. Additional advantages of this vector include superb genetic capacity and stability, as well as the swiftness of technology implementation. The most significant applications of the viral vector include functional genomics of the grapevine and disease control via RNAi-enabled vaccination against pathogens or invertebrate pests.
DOI:10.1094/PHYTO-02-18-0029-RURLPMID:29726761 [本文引用: 3]

Yellow vein clearing disease (YVCD) causes significant economic losses in lemon and other species of citrus. Usually, citrus yellow vein clearing virus (CYVCV) is considered to be the causal agent of YVCD. However, mixed infection of CYVCV and Indian citrus ringspot virus (ICRSV) or other pathogens is often detected in citrus plants with YVCD. In this study, we re-examined the causal agent of YVCD to fulfill Koch's postulates. First, the full-length genome of CYVCV isolate AY (CYVCV-AY) was amplified by long-distance RT-PCR from a Eureka lemon (Citrus limon) tree with typical YVCD symptoms. The genomic cDNAs were then cloned into a ternary Yeast-Escherichia coli-Agrobacterium tumefaciens shuttle vector, pCY, using transformation-associated recombination (TAR) strategy, and 15 full-length cDNA clones of CYVCV-AY were obtained. Subsequently, four of these clones were selected randomly and inoculated on Jincheng (C. sinensis) seedlings through Agrobacterium-mediated vacuum-infiltration, and it was found that 80 to 100% of inoculated plants were infected with CYVCV by RT-PCR at 20 to 40 days postinoculation (dpi) and by direct tissue blot immunoassay at 60 dpi. The progeny of CYVCV-AY from cDNA clones caused typical symptoms of YVCD such as yellow vein clearing, leaf distortion, and chlorosis, which were the same as that elicited by wild-type virus. Finally, the regeneration of CYVCV-AY genome was confirmed by long-distance RT-PCR in lemon trees inoculated with the infectious cDNA clone. These results proved that CYVCV was the primary causal agent of YVCD. This is the first report on the development of infectious cDNA clones of CYVCV, which lays the foundation for further studies on viral gene functions and virus-host interactions.
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1016/s0042-6822(02)00019-3URLPMID:12620796 [本文引用: 1]

Grapevine virus A (GVA) carries five open reading frames (ORFs). Only the coat protein ORF has been experimentally identified as such; the roles of some of the other ORFs have been deduced by sequence homology to known genes (Minafra et al., 1997). The construction of a full-length, infectious clone of GVA has been previously reported. In an attempt to experimentally define the role of the various genes of GVA, we utilized the infectious clone, inserted mutations in every ORF, and studied the effect on viral replication, gene expression, symptoms and viral movement. Mutations in ORF 1 abolished RNA replication. Mutations in ORF 2 did not affect any of the aforementioned parameters. Mutations in ORFs 3 and 4 restricted viral movement. Mutations in ORF 5 rendered the virus asymptomatic, and partially restricted its movement.
DOI:10.1007/s11262-012-0799-5URL [本文引用: 1]

The complete genome sequence of a Chinese isolate of Apple stem grooving virus (ASGV) was determined to be 6,495 nucleotides long, single-stranded, plus-sense RNA. The viral RNA has two overlapping open reading frames (ORFs): ORF1 and ORF2. Compared with the genome sequences of ASGV isolates available in GenBank, the nucleotide identities ranged from 80.1 to 86.3 %. The amino acid identities of proteins encoded by ORF1 and ORF2 ranged from 79.5 to 86.1 % and 82.0 to 85.9 %, respectively.
[本文引用: 1]
DOI:10.1007/s10658-007-9130-zURL [本文引用: 1]

Papaya ringspot virus (PRSV) HA 5-1, a nitrous acid-induced mild mutant of severe strain HA, widely applied for control of PRSV by cross-protection, was used to study the genetic basis of attenuation. Using infectious clones, a series of recombinants was generated between HA 5-1 and HA and their infectivity was analyzed on the systemic host papaya and the local lesion host Chenopodium quinoa. The recombinants that contained mutations in P1 and HC-Pro genes caused attenuated infection on papaya without conspicuous symptoms, similar to HA 5-1. The recombination and sequence analyses strongly implicated two amino acid changes in the C-terminal region of P1 and two in HC-Pro of HA 5-1 involved in the attenuated infection on papaya. The recombinants that infected C. quinoa plants without local lesions contained the same mutations in the C-terminal region of HC-Pro for attenuated infection on papaya. We conclude that both P1 and HC-Pro bear important pathogenicity determinants for the infection on the systemic host papaya and that the mutations in HC-Pro affecting pathogenicity on papaya are also responsible for the inability to induce hypersensitive reaction on C. quinoa.
DOI:10.1111/pbi.12555URLPMID:26920394 [本文引用: 1]

The long juvenile period of citrus trees (often more than 6 years) has hindered genetic improvement by traditional breeding methods and genetic studies. In this work, we have developed a biotechnology tool to promote transition from the vegetative to the reproductive phase in juvenile citrus plants by expression of the Arabidopsis thaliana or citrus FLOWERING LOCUS T (FT) genes using a Citrus leaf blotch virus-based vector (clbvINpr-AtFT and clbvINpr-CiFT, respectively). Citrus plants of different genotypes graft inoculated with either of these vectors started flowering within 4-6 months, with no alteration of the plant architecture, leaf, flower or fruit morphology in comparison with noninoculated adult plants. The vector did not integrate in or recombine with the plant genome nor was it pollen or vector transmissible, albeit seed transmission at low rate was detected. The clbvINpr-AtFT is very stable, and flowering was observed over a period of at least 5 years. Precocious flowering of juvenile citrus plants after vector infection provides a helpful and safe tool to dramatically speed up genetic studies and breeding programmes.
DOI:10.1016/j.jbiotec.2014.02.010URL [本文引用: 1]

A transient expression vector based on Citrus tristeza virus (CTV) is unusually stable. Because of its stability it is being considered for use in the field to control Huanglongbing (HLB), which is caused by Candidatus Liberibacter asiaticus (CLas) and vectored by Asian citrus psyllid, Diaphorina citri. In the absence of effective control strategies for CLas, emphasis has been on control of D. citri. Coincident cohabitation in phloem tissue by CLas, D. citri and CTV was exploited to develop a novel method to mitigate HLB through RNA interference (RNAi). Since CTV has three RNA silencing suppressors, it was not known if CTV-based vector could induce RNAi in citrus. Yet, expression of sequences targeting citrus phytoene desaturase gene by CTV-RNAi resulted in photo-bleaching phenotype. CTV-RNAi vector, engineered with truncated abnormal wing disc (Awd) gene of D. citri, induced altered Awd expression when silencing triggers ingested by feeding D. citri nymphs. Decreased Awd in nymphs resulted in malformed-wing phenotype in adults and increased adult mortality. This impaired ability of D. citri to fly would potentially limit the successful vectoring of CLas bacteria between citrus trees in the grove. CTV-RNAi vector would be relevant for fast-track screening of candidate sequences for RNAi-mediated pest control. (C) 2014 Published by Elsevier B.V. This is an open access article under the CC BY license (http://creativecommons.orgnicesses/by/3.0/).
DOI:10.3864/j.issn.0578-1752.2018.09.007URL [本文引用: 1]

【目的】构建柑橘叶斑驳病毒(Citrus leaf blotch virus,CLBV)中国分离株的侵染性克隆,为从分子水平解析其致病机理打下基础。【方法】以GeneArt® pYES1L Vector为模板,PYES2117F、PYES2117R为引物扩增含有酵母相关复制起始位点的片段pYES1L-2117,利用限制性内切酶Sac II单酶切双元载体DK1317-2并回收其大片段,利用In-Fusion HD Cloning Kit重组连接双元载体DK1317-2骨架和片段pYES1L-2117,得到可以在酵母-农杆菌-大肠杆菌中复制的三元穿梭载体pCY。以马铃薯X病毒(Potato virus X,PVX)全长cDNA侵染性克隆为模板,pCY-PVX-F、pCY-PVX-R为引物扩增PVX全长,采用限制性内切酶Stu I、Sma I酶切质粒pCY,采用醋酸锂转化法将获得的PVX基因组全长cDNA与线性化的pCY载体共转化酵母菌YPH501,通过同源重组获得PVX基因组全长cDNA克隆。通过农杆菌介导接种本生烟验证所构建克隆的侵染性,从而建立基于酵母同源重组的病毒侵染性克隆快速构建体系。在此基础上,以CLBV中国分离物(CLBV-HBYD)全长cDNA为模板,分段扩增其基因组,得到片段CLBV-1、CLBV-2;利用所建体系对CLBV-1、CLBV-2及pCY载体片段进行同源重组。得到重组质粒pCY-CLBV后,经农杆菌介导接种本生烟和锦橙幼苗,并利用RT-PCR和Northern blot检测其侵染性。【结果】建立了酵母-大肠杆菌-农杆菌的三元穿梭载体PCY,该载体全长10 347 bp,含有酵母、农杆菌、大肠杆菌的复制位点,能在酵母-农杆菌-大肠杆菌稳定复制,可用于在酵母中通过同源重组快速构建病毒基因组全长cDNA克隆,也可用于通过农杆菌介导直接接种植物寄主。利用该体系获得了CLBV中国分离株的基因组全长cDNA克隆16个。随机选取1个克隆进行了序列测定(GenBank登录号:MG572236),其基因组全长8 747 nt,包含3个开放阅读框(open reading frame,ORF),ORF1为5 889 nt、ORF2为1 089 nt、ORF3为1 092 nt。全基因组序列比对显示,CLBV-HBYD与已登录的9个CLBV分离物的核酸序列一致性为79%—98%,其中与柑橘来源的EU857540一致性最高,为98%,与猕猴桃来源的JN983454、JN983455、JN983456和JN900477的一致性均为79%。在利用MEGA6 软件构建的系统发育树上,CLBV-HBYD与柑橘来源的CLBV分离株聚为一簇,而猕猴桃来源的CLBV分离株聚为另一簇。将所获16个CLBV全长cDNA克隆转化农杆菌,以pCY空载体为阴性对照注射接种本生烟。接种20 d后,抽提RNA进行RT-PCR检测,结果表明11个克隆的接种植株检测出CLBV特异性条带;随机选取5个阳性克隆进一步进行了Northern blot杂交,结果显示pCY-CLBV 1、2、3、14、15接种的本生烟可以检测到CLBV特异性条带,而对照样本未检测到任何条带,表明这些克隆均为侵染性克隆。【结论】构建了基于三元穿梭载体pCY的酵母重组克隆体系,利用该体系获得了中国CLBV分离株的适于农杆菌介导接种的基因组全长cDNA侵染性克隆。
DOI:10.3864/j.issn.0578-1752.2018.09.007URL [本文引用: 1]

【目的】构建柑橘叶斑驳病毒(Citrus leaf blotch virus,CLBV)中国分离株的侵染性克隆,为从分子水平解析其致病机理打下基础。【方法】以GeneArt® pYES1L Vector为模板,PYES2117F、PYES2117R为引物扩增含有酵母相关复制起始位点的片段pYES1L-2117,利用限制性内切酶Sac II单酶切双元载体DK1317-2并回收其大片段,利用In-Fusion HD Cloning Kit重组连接双元载体DK1317-2骨架和片段pYES1L-2117,得到可以在酵母-农杆菌-大肠杆菌中复制的三元穿梭载体pCY。以马铃薯X病毒(Potato virus X,PVX)全长cDNA侵染性克隆为模板,pCY-PVX-F、pCY-PVX-R为引物扩增PVX全长,采用限制性内切酶Stu I、Sma I酶切质粒pCY,采用醋酸锂转化法将获得的PVX基因组全长cDNA与线性化的pCY载体共转化酵母菌YPH501,通过同源重组获得PVX基因组全长cDNA克隆。通过农杆菌介导接种本生烟验证所构建克隆的侵染性,从而建立基于酵母同源重组的病毒侵染性克隆快速构建体系。在此基础上,以CLBV中国分离物(CLBV-HBYD)全长cDNA为模板,分段扩增其基因组,得到片段CLBV-1、CLBV-2;利用所建体系对CLBV-1、CLBV-2及pCY载体片段进行同源重组。得到重组质粒pCY-CLBV后,经农杆菌介导接种本生烟和锦橙幼苗,并利用RT-PCR和Northern blot检测其侵染性。【结果】建立了酵母-大肠杆菌-农杆菌的三元穿梭载体PCY,该载体全长10 347 bp,含有酵母、农杆菌、大肠杆菌的复制位点,能在酵母-农杆菌-大肠杆菌稳定复制,可用于在酵母中通过同源重组快速构建病毒基因组全长cDNA克隆,也可用于通过农杆菌介导直接接种植物寄主。利用该体系获得了CLBV中国分离株的基因组全长cDNA克隆16个。随机选取1个克隆进行了序列测定(GenBank登录号:MG572236),其基因组全长8 747 nt,包含3个开放阅读框(open reading frame,ORF),ORF1为5 889 nt、ORF2为1 089 nt、ORF3为1 092 nt。全基因组序列比对显示,CLBV-HBYD与已登录的9个CLBV分离物的核酸序列一致性为79%—98%,其中与柑橘来源的EU857540一致性最高,为98%,与猕猴桃来源的JN983454、JN983455、JN983456和JN900477的一致性均为79%。在利用MEGA6 软件构建的系统发育树上,CLBV-HBYD与柑橘来源的CLBV分离株聚为一簇,而猕猴桃来源的CLBV分离株聚为另一簇。将所获16个CLBV全长cDNA克隆转化农杆菌,以pCY空载体为阴性对照注射接种本生烟。接种20 d后,抽提RNA进行RT-PCR检测,结果表明11个克隆的接种植株检测出CLBV特异性条带;随机选取5个阳性克隆进一步进行了Northern blot杂交,结果显示pCY-CLBV 1、2、3、14、15接种的本生烟可以检测到CLBV特异性条带,而对照样本未检测到任何条带,表明这些克隆均为侵染性克隆。【结论】构建了基于三元穿梭载体pCY的酵母重组克隆体系,利用该体系获得了中国CLBV分离株的适于农杆菌介导接种的基因组全长cDNA侵染性克隆。
