 ,1,2, 刘泽厚1,2, 万洪深1,2, 魏会廷2,3, 龙海4, 李涛4, 邓光兵4, 李俊
,1,2, 刘泽厚1,2, 万洪深1,2, 魏会廷2,3, 龙海4, 李涛4, 邓光兵4, 李俊 ,1,2, 杨武云
,1,2, 杨武云 ,1,2
,1,2Identification and Pyramiding of QTLs for Traits Associated with Pre-Harvest Sprouting Resistance in Two Wheat Cultivars Chuanmai 42 and Chuannong 16
WANG Qin ,1,2, LIU ZeHou1,2, WAN HongShen1,2, WEI HuiTing2,3, LONG Hai4, LI Tao4, DENG GuangBing4, LI Jun
,1,2, LIU ZeHou1,2, WAN HongShen1,2, WEI HuiTing2,3, LONG Hai4, LI Tao4, DENG GuangBing4, LI Jun ,1,2, YANG WuYun
,1,2, YANG WuYun ,1,2
,1,2通讯作者:
责任编辑: 李莉
收稿日期:2019-10-21接受日期:2020-02-8网络出版日期:2020-09-01
| 基金资助: |
Received:2019-10-21Accepted:2020-02-8Online:2020-09-01
作者简介 About authors
王琴,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2723KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王琴, 刘泽厚, 万洪深, 魏会廷, 龙海, 李涛, 邓光兵, 李俊, 杨武云. 川麦42和川农16抗穗发芽QTL定位及聚合效应分析[J]. 中国农业科学, 2020, 53(17): 3421-3431 doi:10.3864/j.issn.0578-1752.2020.17.001
WANG Qin, LIU ZeHou, WAN HongShen, WEI HuiTing, LONG Hai, LI Tao, DENG GuangBing, LI Jun, YANG WuYun.
0 引言
【研究意义】小麦收获前期如遇连续阴雨或湿热环境,其籽粒在麦穗上萌动、发芽的现象即为穗发芽(pre-harvest sprouting,PHS)[1]。小麦穗发芽在全世界小麦种植区均有发生,全球每年由小麦穗发芽引起的直接经济损失约为10亿美元[2,3];中国约有83%的小麦种植区均发生过严重的穗发芽危害,尤其是长江中下游麦区、西南麦区和东北春麦区频繁发生,黄淮冬麦区和北方冬麦区也时有发生,严重影响中国小麦的安全生产[1,4-6]。四川盆地在小麦成熟期降雨较多,特别是近2年,小麦收获前2—3 d连续阴雨,生产上,穗发芽敏感的品种穗发芽发生严重,引起其内部籽粒蛋白质和储藏物质分解,导致产量和品质严重降低,给种植大户和企业造成巨大的经济损失。因此,挖掘抗穗发芽基因,培育抗穗发芽小麦品种己成为小麦育种的重要目标。【前人研究进展】穗发芽是一个复杂的、由多基因控制的数量性状,遗传基础复杂且易受环境影响[7]。研究表明,种子休眠特性、穗部形态、籽粒性状、颖壳内发芽抑制物质、α-淀粉酶活性以及光照、温度等均不同程度影响穗发芽抗性[8]。目前,与穗发芽抗性相关的QTL已被定位到小麦21条染色体上[9,10,11,12,13,14,15,16],其中,第2、3和4同源群是穗发芽主效抗性基因的富集区域[14-15,17-21]。随着小麦基因组测序和生物信息学的快速发展,7个与穗发芽或休眠相关的基因已被克隆,包括第2同源群上的关键基因TaSdr,第3同源群的关键基因TaVp1、TaMFT、TaMyb10和TaPHS1,第4同源群的关键基因TaPm19和TaMKK3[22,23,24,25,26,27,28,29,30]。【本研究切入点】目前,已定位的小麦穗发芽QTL数量众多,随着候选基因的挖掘,一些与穗发芽抗性相关的功能标记也被开发应用;但不同群体、不同环境下均能有效应用的标记仍较少,且大多标记为与种子休眠特性相关的标记,从而限制了标记检测结果与穗发芽表型的吻合性。因此,穗发芽研究任重道远,挖掘更多穗发芽抗性基因及其优异等位变异,聚合不同效应的抗穗发芽优异等位基因将有助于提高栽培小麦穗发芽抗性。川麦42是四川省农业科学院作物研究所利用CIMMYT人工合成六倍体小麦育成的高产小麦品种,具有大穗大粒、抗条锈病等优异特性,但其对穗发芽和花期低温敏感。为了改良其穗发芽和花期冻害特性,四川省农业科学院小麦种质资源创新团队对收集的资源通过系统评价,发现四川农业大学培育的小麦品种川农16分蘖力强、抗白粉病、对花期低温和穗发芽具有较强抗性,其重要性状与川麦42互补性强。因此,四川省农业科学院小麦种质资源创新团队利用川麦42与川农16杂交,以单粒传的方式构建了重组自交系群体(RILs)。【拟解决的关键问题】本研究通过籽粒发芽和整穗发芽试验,利用中国科学院成都生物研究所构建的川麦42/川农16 RIL群体SNP遗传连锁图谱,挖掘与籽粒发芽和整穗发芽相关的QTL,寻找聚合双亲抗穗发芽QTL且表现抗穗发芽的优异材料。1 材料与方法
1.1 试验材料
小麦品种川麦42、川农16及其构建的127个重组自交系(RIL,F8),以及穗发芽敏感对照川麦45。川麦42和对照川麦45由四川省农业科学院作物研究所提供,川农16由四川农业大学提供;川麦42和川农16为红粒小麦,川麦45为白粒小麦;重组自交系由四川省农业科学院作物研究所利用川麦42与川农16杂交,采用单粒传法构建而成,通过昆明、成都两地穿梭种植稳定成系。川麦42、川农16及其构建的127个重组自交系群体,穗发芽敏感对照川麦45分别种植于广汉和成都。2016年种植于广汉(2016GH),每个小区种植3行,随机区组设计,3次重复,行长1.5 m,行距20 cm,每行15窝,采用免耕、撬窝、稻草覆盖栽培方式;2017和2018年均种植于成都网室(2017CD和2018CD),试验设计和种植方式与广汉相同,播种后采用细土覆盖。田间管理与当地小麦生产一致。
1.2 穗发芽鉴定
籽粒发芽:对2016年和2018年环境下的RIL群体及其亲本进行籽粒发芽测定,于生理成熟期随机取中间一行的5个穗子,室内阴干,7 d后-20℃保存。将保存于-20℃的5个穗子手工脱粒,用0.5%的NaClO消毒冲洗后,取100粒完整种子置于培养皿(培养皿底部置有湿润的滤纸)中;每个材料2次重复,试验第2天开始统计发芽种子数,并将已发芽种子移除,连续计数7 d。计算发芽率(seed germination rate,GR)和发芽指数(seed germination index,GI)。GR=(发芽粒数∕总粒数)×100%;GI=(7×n1+6×n2+5×n3+4×n4+3×n5+2×n6+1×n7)/(7×N)×100%,N表示籽粒总数;n1、n2、…、n7分别表示第1天至第7天的发芽籽粒数。整穗发芽:对2017年和2018年成都点的RIL群体及其亲本进行人工模拟降雨田间整穗发芽测定。采用田间自动喷灌系统人工模拟降雨,于生理成熟期连续喷洒7 d,确保所有试验材料穗子一直保持湿润,待对照穗子籽粒90%以上萌动后,随机收获中间一行的10个穗子,快速烘干后手工脱粒,统计总粒数和发芽粒数,计算整穗发芽率(seed germination rate of in each spike,SGR)。SGR=发芽粒数/总粒数×100%。
1.3 分子标记及遗传图谱的构建
供试材料基因组DNA由中国科学院成都生物研究所采用CTAB方法提取,DNA质检、小麦90K SNP芯片检测工作由北京博奥公司完成。RIL群体SNP数据分型及遗传图谱均由中国科学院成都生物研究所分析完成,遗传连锁图谱采用IciMapping 4.1软件完成。1.4 数据统计及QTL分析
利用SPSS 13.0 软件对表型进行统计分析,通过QTL分析软件IciMapping v3.0(http://www.isbreeding. net/software.html)分别计算各环境下不同性状的广义遗传力。参考前人研究以抗性品种发芽率鉴定筛选抗穗发芽材料[1,5-6,8],结合实际情况,将GI、GR和SGR值低于15%定为高抗穗发芽品种,15%—25%定为抗穗发芽品种。利用QTL分析软件ICIMapping v3.0对RIL群体的GI、GR和SGR进行QTL分析。采用逐步回归法的完备区间作图法(inclusive composite interval mapping,ICIM),基于1 cM的步移速度进行QTL的鉴定分析,最小QTL效应的LOD阈值设为2.5。
2 结果
2.1 穗发芽分析
2016、2017和2018年对亲本川麦42、川农16及其RIL群体进行了穗发芽鉴定,3个指标籽粒发芽指数(GI)、籽粒发芽率(GR)和整穗发芽率(SGR)用于分析其穗发芽抗性(表1)。结果显示,2个环境下,亲本川麦42的3个指标值(GI、GR和SGR)均显著高于亲本川农16,表明川农16穗发芽抗性显著优于川麦42;RIL群体中穗发芽抗性呈连续分布,存在超亲分离现象。不同环境下穗发芽抗性参数的遗传力不同,2016年籽粒发芽率的遗传力最高(0.82)。Table 1
表1
表1RIL群体及其亲本穗发芽性状参数统计
Table 1
| 性状 Trait | 环境 Environment | 亲本Parents | RIL群体 RIL population | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 川麦42 Chuanmai 42 | 川农16 Chuannong 16 | 均值 Mean | 标准差 SD | 最小值 Min | 最大值 Max | 峰度 Kurtosis | 偏度 Skewness | 遗传力Heritability | ||
| GI | 2016GH | 0.74 | 0.11** | 0.57 | 0.10 | 0.06 | 0.98 | -0.02 | -0.27 | 0.57 |
| 2018CD | 0.76 | 0.17** | 0.59 | 0.20 | 0.05 | 0.87 | -0.54 | -0.66 | 0.49 | |
| GR | 2016GH | 0.83 | 0.24** | 0.82 | 0.09 | 0.07 | 1.00 | -0.09 | -0.57 | 0.82 |
| 2018CD | 0.86 | 0.21** | 0.79 | 0.23 | 0.06 | 1.00 | 0.68 | -1.21 | 0.69 | |
| SGR | 2017CD | 0.62 | 0.15** | 0.52 | 0.17 | 0.02 | 0.90 | -0.35 | -0.28 | 0.61 |
| 2018CD | 0.51 | 0.09** | 0.42 | 0.21 | 0.03 | 0.99 | -0.70 | 0.12 | 0.51 | |
新窗口打开|下载CSV
相关性分析表明(表2),所有穗发芽参数间均表现正相关;单环境下,GI、GR和SGR之间表现为显著或极显著正相关,GI与GR相关系数较大,而GI、GR分别与SGR间的相关系数较小;不同环境下籽粒发芽参数相关性不显著。SGR在2个环境下极显著正相关,但相关系数小于0.5。结果表明,小麦穗发芽抗性易受环境影响,但在不同年份间,SGR表现出相对稳定的趋势。
Table 2
表2
表2RIL群体中穗发芽性状间的相关性
Table 2
| 性状Trait | GI-2018CD | GR-2016GH | GR-2018CD | SGR-2017CD | SGR-2018CD |
|---|---|---|---|---|---|
| GI-2016GH | 0.091 | 0.892** | 0.037 | 0.177* | 0.266** |
| GI-2018CD | 0.169 | 0.966** | 0.239** | 0.289** | |
| GR-2016GH | 0.114 | 0.247** | 0.258** | ||
| GR-2018CD | 0.201* | 0.224* | |||
| SGR-2017CD | 0.440** |
新窗口打开|下载CSV
2.2 QTL分析
利用RIL群体在不同环境下穗发芽相关性状的表型数据,结合SNP芯片数据对穗发芽相关QTL进行分析。共检测到11个与穗发芽抗性有关的QTL(表3和图1),贡献率为4.28%—29.03%,主要分布在2B、2D、3A、3D、4A、5A、5B和6B染色体上;除4A和6B染色体上的3个QTL增加穗发芽抗性的等位变异来自川麦42外,其余8个QTL增加穗发芽抗性的等位变异均来自亲本川农16。5B染色体上IWB60217—IWB52109区段检测到的与整穗发芽相关的QTL(QSgr.saas-5B)贡献率最大(29.03%),但仅在单个环境中表达。2017和2018年2个环境下,在2D和3A染色体上均检测到SGR的QTL,不同环境下的QTL贡献率差异明显,最高为10.86%。5A染色体上IWB22536—IWB6853区段检测到与休眠相关的GI和GR的QTL,均能在2个环境下表达,贡献率最高为18.80%。2B和4A染色体上仅检测到在单个环境表达的与休眠相关的微效QTL,其贡献率均较小。Table 3
表3
表3川麦42/川农16重组自交系群体中检测到的穗发芽相关性状QTL
Table 3
| 性状 Trait | QTL | 环境 Environment | 标记区间 Marker interval | LOD | 贡献率 PVE (%) | 加性效应 Additive effecta |
|---|---|---|---|---|---|---|
| GI | QGi.saas-2B | 2016GH | IWB217—IWB12070 | 2.64 | 9.47 | 0.05 |
| QGi.saas-4A | 2016GH | IWB32870—IWB43187 | 2.50 | 4.28 | -0.03 | |
| QGi.saas-5A | 2016GH | IWB22536—IWB43493 | 2.51 | 5.88 | 0.09 | |
| 2018CD | IWB6853—IWB25919 | 3.98 | 14.72 | 0.13 | ||
| GR | QGr.saas-2B | 2016GH | IWB217—IWB12070 | 2.59 | 5.02 | 0.03 |
| QGr.saas-4A | 2016GH | IWB32870—IWB43187 | 2.51 | 4.79 | -0.04 | |
| QGr.saas-5A | 2016GH | IWB22536—IWB43493 | 2.65 | 4.87 | 0.06 | |
| 2018CD | IWB6853—IWB25919 | 5.10 | 18.80 | 0.15 | ||
| QGr.saas-6B | 2016GR | IWB8673—IWB22066 | 11.02 | 14.26 | -0.05 | |
| SGR | QSgr.saas-2D | 2017CD | IWB62848—IWB56618 | 3.29 | 8.06 | 0.05 |
| 2018CD | IWB9680—IWB62848 | 2.54 | 5.73 | 0.05 | ||
| QSgr.saas-3A | 2017CD | IWB10578—IWB30094 | 2.57 | 4.72 | 0.04 | |
| 2018CD | IWB10578—IWB30094 | 3.60 | 10.86 | 0.07 | ||
| QSgr.saas-3D | 2017CD | IWB30686—IWB38826 | 4.69 | 11.96 | 0.06 | |
| QSgr.saas-5B | 2018CD | IWB60217—IWB52109 | 12.36 | 29.03 | 0.16 |
新窗口打开|下载CSV
图1
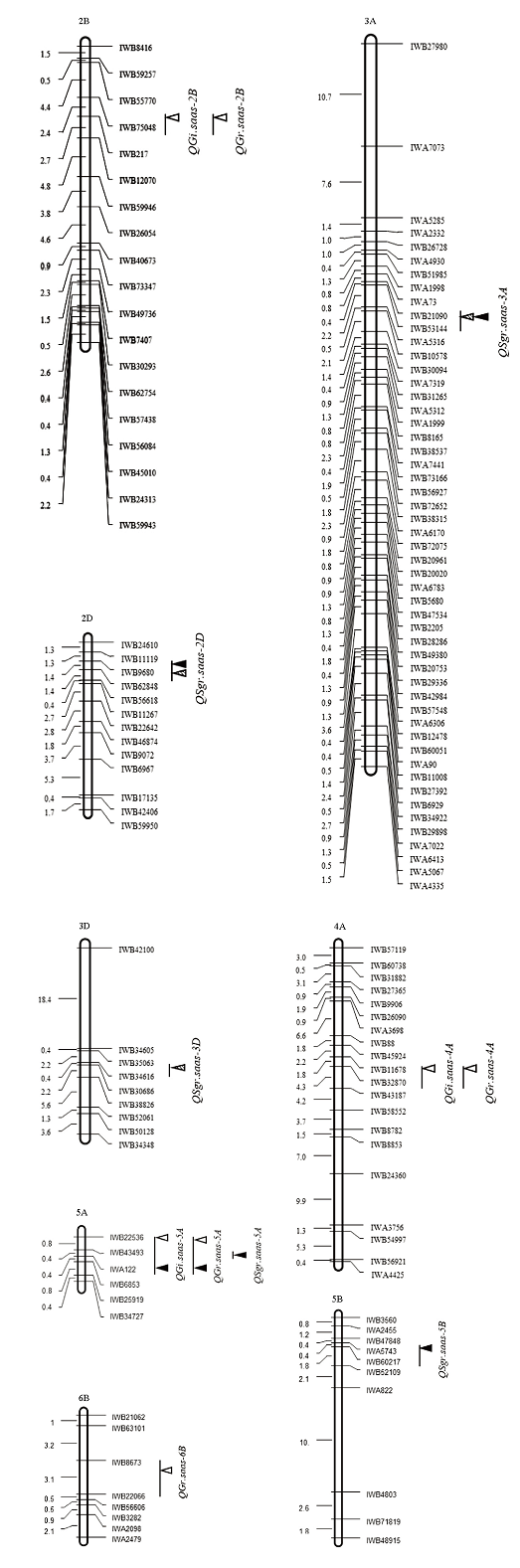 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1穗发芽相关性状QTL在8条染色体上的分布
三角形表示峰值所在位置。无填充的三角形表示2016年的QTL,斜线填充的三角形表示2017年的QTL,黑色填充的三角形表示2018年的QTL
Fig. 1QTLs for PHS related traits on 8 chromosomes
The triangle indicates the position of QTL peak. The unfilled triangles represent QTLs for PHS in 2016, the diagonal filled triangles represent QTLs for PHS in 2017, and the black triangles represent QTLs for PHS in 2018
2.3 抗性QTL聚合效应分析
基因型分析发现,在RIL群体中,普遍存在穗发芽抗性QTL聚合现象,聚合的QTL数量变幅在1—9个,其中,聚合4个抗性QTL的株系数最多(图2)。根据3个环境下穗发芽表型数据,发现RIL群体中GI、GR和SGR值低于15%的株系分别为13、12和14个,GI、GR和SGR值在15%—25%的株系分别为31、32和36个;其中6个株系的GI、GR和SGR值均低于15%,表现为高抗穗发芽(表4)。分析其基因型,发现抗穗发芽的株系携带4—9个穗发芽相关性状的抗性QTL;6个优异株系携带7或8个抗性QTL,编号为12和74的株系携带QTL数最多(8个)。6个高抗株系同时聚合了川麦42在4A染色体上的微效QTL,以及川农16在2D和5B染色体上的主效QTL。图2
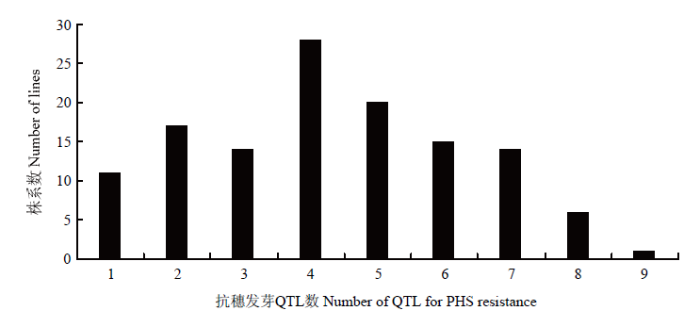 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2川麦42/川农16 RILs中穗发芽抗性QTL聚合频率分布
Fig. 2Frequency distribution of the resistant QTLs pyramiding for PHS in Chuanmai 42/Chuannong 16 RILs
Table 4
表4
表46个高抗穗发芽株系携带的抗性QTL
Table 4
| 株系 Lines | GI (%) | GR (%) | SGR (%) | QTL | |||
|---|---|---|---|---|---|---|---|
| 2016GH | 2018CD | 2016GH | 2018CD | 2017CD | 2018CD | ||
| 12 | 0.14 | 0.12 | 0.12 | 0.15 | 0.07 | 0.05 | QGi.saas-2B, QGr.saas-2B, QSgr.saas-2D, QSgr.saas-3A, QSgr.saas-3D, QGi.saas-4A, QGr.saas-4A, QSgr.saas-5B |
| 74 | 0.13 | 0.10 | 0.13 | 0.15 | 0.02 | 0.03 | QGi.saas-2B, QGr.saas-2B, QSgr.saas-2D, QSgr.saas-3D, QGi.saas-4A, QGr.saas-4A, QSgr.saas-5B, QGr.saas-6B |
| 82 | 0.09 | 0.05 | 0.12 | 0.13 | 0.14 | 0.07 | QGi.saas-2B, QGr.saas-2B, QSgr.saas-2D, QSgr.saas-3A, QGi.saas-4A, QGr.saas-4A, QSgr.saas-5B |
| 104 | 0.07 | 0.12 | 0.10 | 0.14 | 0.13 | 0.11 | QGi.saas-2B, QGr.saas-2B, QSgr.saas-2D, QGi.saas-4A, QGr.saas-4A, QSgr.saas-5B, QGr.saas-6B |
| 112 | 0.06 | 0.09 | 0.13 | 0.11 | 0.14 | 0.15 | QSgr.saas-2D, QSgr.saas-3D, QGi.saas-4A, QGr.saas-4A, QSgr.saas-5B, QGi.saas-5A, QGr.saas-5A |
| 125 | 0.08 | 0.10 | 0.07 | 0.11 | 0.09 | 0.07 | QSgr.saas-2D, QSgr.saas-3A, QSgr.saas-3D, QGi.saas-4A, QGr.saas-4A, QSgr.saas-5B, QGr.saas-6B |
新窗口打开|下载CSV
株系12、74和125在2个环境下SGR值均小于10%,均携带2D、3D和5B染色体上的SGR抗性QTL。高抗穗发芽的株系104已于2012年同时通过国家和四川省审定,定名为川麦104;株系125于2013年通过四川省审定,定名为川麦64。
3 讨论
3.1 人工模拟降雨整穗发芽法效果
小麦穗发芽的鉴定方法有很多,如整穗发芽、籽粒发芽、α-淀粉酶活性鉴定、田间穗发芽鉴定、沉降值测量等,常用的主要是籽粒发芽、整穗发芽法和田间发芽鉴定[1,8]。籽粒发芽法为简化的穗发芽鉴定方法,是生产上大规模鉴定评价的最普遍的方法,但其只能检测种子的休眠特性;整穗发芽法和田间发芽鉴定法既能检测种子的休眠性,又能从穗部形状、颖壳特性、外源抑制物质等方面全面直观地反映品种的抗穗发芽特性,是最符合生产实际的遗传研究方法。本研究同时利用籽粒发芽法和整穗发芽法鉴定评价川麦42/川农16重组自交系群体及其亲本穗发芽发生程度,更能全面地挖掘抗穗发芽QTL,鉴定出更适合大田生产需要的育种材料。本研究同一材料整穗发芽率普遍比籽粒发芽率和发芽指数低,这与前人研究结果一致[31];相关性分析表明,穗发芽抗性易受环境影响,但在不同年份间,SGR表现出相对稳定的趋势。因此,人工模拟降雨整穗发芽法也能较准确地评价穗发芽抗性,通过2种鉴定方法评价筛选抗穗发芽材料,更能优选出适合大田生产实际的资源。3.2 川麦42和川农16抗穗发芽QTL比较
前人利用不同研究群体已将小麦穗发芽和种子休眠相关QTL定位在所有染色体上,其中,第2、3和4同源群上的穗发芽相关基因在不同研究中均被重复定位到[9-16,32-35]。本研究利用中国科学院成都生物研究所构建的SNP遗传连锁图谱,根据穗发芽相关的表型数据,在2B、2D、3A、3D、4A、5A、5B和6B染色体上检测到11个与穗发芽抗性相关的QTL,其中,2D、3A和5A上检测到的主效QTL在2个环境下均能稳定表达。前人利用染色体代换系、DH群体等研究也发现位于2D、3A、4A和5A染色体上与穗发芽相关的主效QTL[7,21,36-41],但由于这些研究所用标记方法与本研究不同,不能直接比较,因此,无法确定本研究在2D、3A、4A和5A染色体上检测到的QTL是否与前人定位的一致。LIN等[42]利用SSR和GBS-SNP技术在5A染色体上发现一个与种子休眠相关的QTL,与SSR标记Xbarc261距离最近;KUMAR等[37]通过籽粒发芽的方法将与籽粒休眠相关的QTL定位在5A染色体上SSR标记Xgwm304—Xcfa2190;GROOS等[36]通过整穗发芽的方法也将穗发芽QTL定位到与KUMAR等[37]发现的5A染色体上同一区域。本研究在5A染色体上检测到的与种子休眠相关的QTL在2个环境下均表达,贡献率为4.87%—18.8%,但是否与前人研究结果位于同一区域还需进一步增加SSR标记扫描验证。ZHANG等[25,26]利用比较基因组学法,在小麦中对水稻休眠基因进行同源克隆,获得休眠基因TaSdr,并开发了TaSdr-A1和TaSdr-B1的分子标记;周勇[43]在3A染色体上发现地方品种中休眠相关QTL与SNP标记AX-111578083连锁,毗邻籽粒颜色相关的转录因子基因TaMyb10。目前,2D、3A和4A染色体上与穗发芽相关基因/QTL多次被报道,其关键基因已经被克隆[3,7,17-19,21,29,38-39,41,44-46],位于第3和第4同源群的TaVp1、TaMFT/TaPHS1、TaMyb10和TaMKK3是目前最有效的小麦穗发芽相关基因[47];本研究在2D、3A和4A染色体上检测到的穗发芽相关主效QTL是否是克隆的基因或邻近已知基因,还需利用有效标记检测及SNP序列比较分析进一步验证。3.3 抗穗发芽QTL聚合及衍生品种川麦104抗性表现
本研究利用双亲染色体重组筛选出一批抗穗发芽材料,聚合了多个穗发芽抗性QTL。其中,株系104于2012年同时通过国家和四川省审定,定名为川麦104;川麦104国家区试平均产量6 130.5 kg·hm-2,比对照川麦42增产8.42%;四川省区试平均产量6 116.1 kg·hm-2,比对照绵麦37增产14.12%。基因型分析发现,川麦104聚合了双亲7个正向穗发芽QTL,具有较强的抗穗发芽能力;李式昭等[48]利用穗发芽抗性等位基因分子标记Vp1B3,对2000年以来四川育成的105份小麦品种进行鉴定,发现川麦104同时含Vp-1Bc抗穗发芽基因。TANG等[49]控制性试验表明,生理成熟期川麦104的籽粒发芽指数在所有参试品种中最低,成熟后延迟1—2周收获,其降落值降低幅度小,仍接近优质小麦标准(300s),显著高于其他多数参试品种。2018年小麦成熟后持续淋雨3 d,30个参试品种平均籽粒发芽率20.8%,最高68.9%,而川麦104籽粒发芽率仅2.3%;2019年成熟期遭遇持续降雨,各地调查川麦104基本未出现发芽,而大多数品种穗发芽严重。由于川麦104产量、品质、抗病及抗穗发芽等优良性状突出,其已成为西南麦区小麦育种的核心亲本,近年来,已育成小麦品种(系)18个。因此,双亲抗穗发芽QTL聚合可以提高穗发芽抗性,本研究鉴定出的抗穗发芽优异株系是小麦穗发芽育种改良的重要基因资源。4 结论
共检测到11个与穗发芽抗性相关的QTL,其中3个来源于川麦42,8个来源于川农16。2D、3A和5A染色体上检测到的穗发芽主效QTL,在2个环境下均能表达,其抗穗发芽等位变异均来源于川农16。在RIL群体中,抗穗发芽的株系均携带4—9个与穗发芽相关的QTL,6个优异株系聚合了7或8个抗性QTL;编号为104和125的优异株系(国家、四川省审定,定名为川麦104和川麦64)聚合了7个正向穗发芽QTL,包括4个来源于川农16的抗性QTL和3个来源于川麦42的QTL。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
北京: 中国农业科学技术出版社,
[本文引用: 4]
Beijing: China Agricultural Science and Technology Press,
[本文引用: 4]
DOI:10.1007/BF00026991URL [本文引用: 1]
DOI:10.1007/s00122-008-0712-8URLPMID:18368385 [本文引用: 2]

The inheritance and genetic linkage analysis for seed dormancy and preharvest sprouting (PHS) resistance were carried out in an F8 recombinant inbred lines (RILs) derived from the cross between
DOI:10.1023/A:1019679924173URL [本文引用: 1]

Most modern white-grained wheat (Triticum aestivum L.) cultivars in China lack sprouting resistance. Some white grain varieties showed lower preharvest sprouting percentage in different years. Tolerance to sprouting in most cultivars comes from the varieties, Wanxian White, Fulingxuxu White, Suiningtuotuo, and Yongchuan White. The α-amylase inhibitor gene under a wheat genetic background inhibited α-amylase activity and delayed sprouting. The falling number of six suspected sources, out of 835Chinese leading varieties since 1950', was independent of late maturityα-amylase.
DOI:10.7606/j.issn.1009-1041.2003.03.107URL [本文引用: 1]

The wheat pre-harvest sprouting is a worldwide natural disaster. The breeders and biochemists have paid more attention to those problems. The conference of pre-harvest sprouting were held for 9 times from 1975 to 2001, and it had brought about great advan
DOI:10.7606/j.issn.1009-1041.2003.03.107URL [本文引用: 1]

The wheat pre-harvest sprouting is a worldwide natural disaster. The breeders and biochemists have paid more attention to those problems. The conference of pre-harvest sprouting were held for 9 times from 1975 to 2001, and it had brought about great advan
URL [本文引用: 2]

Preharvest sprouting resistance in 781 leading wheat varieties since 1950 in Yellow and Huai River Valley Winter Wheat Region,the North Winter Wheat Region,the Southwest Winter Wheat Region,the Yangtze River Winter Wheat Region and the Northeast Spring Wheat Region of China was evaluated by testing the sprouting percentage(SP)and falling number(FN)of mature seeds during 2000–2002 planting seasons. The results indicated that there was highly positive and significant correlation among years. The resistance to preharvest sprouting in the cultivars from the 1980s and the 1990s was lower than those from the 1950s,the 1960s and the 1970s. Cultivars with sprouting resistance from Yellow and Huai River Winter Wheat Region,the Southwest Winter Wheat Region and the Northeast Spring Wheat Region accounted for 1.7%,4.5% and 5.7% of total,respectively. The GP of cultivars in the North Winter Wheat Region and the Yangtze River Winter Wheat Region was above 10%. The sprouting resistance in the Northeast Spring Wheat Region was higher than that in other regions. Meanwhile, 15 cultivars, i.e. Yiyuan 2, Shuwan 24, Shuwan 761, Shan 160, Mengxian 4, Jing 411, Jing 9428, Jian 26, Yanda 1817, Nongda 45, Hengshui 6404, Jinmai 5, Jinmai 8, E’mai 14 and Kehui, in different regions with late maturity α-amylase (LMA) gene were selected based on the analysis of isoelectric focusing(IEF)pattens from those with lower SP and FN (Table 6). Furthermore, the genetic relationship between wheat genotypes prone to LMA was analyzed based on SSR markers. It was showed that some wheat genotypes, which were quite closely related to genotypes with LMA gene, did not carry the LMA gene. These results suggested that wheat genotypes with the LMA gene were distributed widely in Chinese wheat regions but with low frequency (1.9%). So cultivars without LMA gene may be easily selected in wheat breeding programs.
URL [本文引用: 2]

Preharvest sprouting resistance in 781 leading wheat varieties since 1950 in Yellow and Huai River Valley Winter Wheat Region,the North Winter Wheat Region,the Southwest Winter Wheat Region,the Yangtze River Winter Wheat Region and the Northeast Spring Wheat Region of China was evaluated by testing the sprouting percentage(SP)and falling number(FN)of mature seeds during 2000–2002 planting seasons. The results indicated that there was highly positive and significant correlation among years. The resistance to preharvest sprouting in the cultivars from the 1980s and the 1990s was lower than those from the 1950s,the 1960s and the 1970s. Cultivars with sprouting resistance from Yellow and Huai River Winter Wheat Region,the Southwest Winter Wheat Region and the Northeast Spring Wheat Region accounted for 1.7%,4.5% and 5.7% of total,respectively. The GP of cultivars in the North Winter Wheat Region and the Yangtze River Winter Wheat Region was above 10%. The sprouting resistance in the Northeast Spring Wheat Region was higher than that in other regions. Meanwhile, 15 cultivars, i.e. Yiyuan 2, Shuwan 24, Shuwan 761, Shan 160, Mengxian 4, Jing 411, Jing 9428, Jian 26, Yanda 1817, Nongda 45, Hengshui 6404, Jinmai 5, Jinmai 8, E’mai 14 and Kehui, in different regions with late maturity α-amylase (LMA) gene were selected based on the analysis of isoelectric focusing(IEF)pattens from those with lower SP and FN (Table 6). Furthermore, the genetic relationship between wheat genotypes prone to LMA was analyzed based on SSR markers. It was showed that some wheat genotypes, which were quite closely related to genotypes with LMA gene, did not carry the LMA gene. These results suggested that wheat genotypes with the LMA gene were distributed widely in Chinese wheat regions but with low frequency (1.9%). So cultivars without LMA gene may be easily selected in wheat breeding programs.
DOI:10.1534/genetics.107.084939URLPMID:18245824 [本文引用: 3]

Aegilops tauschii, the wild relative of wheat, has stronger seed dormancy, a major component of preharvest sprouting resistance (PHSR), than bread wheat. A diploid Ae. tauschii accession (AUS18836) and a tetraploid (Triticum turgidum L. ssp. durum var. Altar84) wheat were used to construct a synthetic wheat (Syn37). The genetic architecture of PHS was investigated in 271 BC(1)F(7) synthetic backcross lines (SBLs) derived from Syn37/2*Janz (resistant/susceptible). The SBLs were evaluated in three environments over 2 years and PHS was assessed by way of three measures: the germination index (GI), which measures grain dormancy, the whole spike assay (SI), which takes into account all spike morphology, and counted visually sprouted seeds out of 200 (VI). Grain color was measured using both Chroma Meter- and NaOH-based approaches. QTL for PHSR and grain color were mapped and their additive and epistatic effects as well as their interactions with environment were estimated by a mixed linear-model approach. Single-locus analysis following composite interval mapping revealed four QTL for GI, two QTL for SI, and four QTL for VI on chromosomes 3DL and 4AL. The locus QPhs.dpiv-3D.1 on chromosome 3DL was tightly linked to the red grain color (RGC) at a distance of 5 cM. The other locus on chromosome 3D,
[本文引用: 3]
[本文引用: 3]
[本文引用: 2]
DOI:10.1007/s001220051241URL [本文引用: 1]
DOI:10.1007/s10681-012-0705-1URL [本文引用: 1]

Seed dormancy and pre-harvest sprouting are important traits in bread wheat. Bi-parental populations have permitted the identification of several genes/quantitative trait loci controlling these traits, mapping to various bread wheat chromosomes. Here, we report the use of association mapping to uncover the genetic basis of both traits in a panel of 96 diverse winter wheat cultivars to establish the presence of marker-trait associations on many chromosomes. Potential candidate genes were identified by studying the gene content of the chromosome bins into which the major marker trait associations mapped.
DOI:10.1023/A:1019632008244URL [本文引用: 1]

A series of experiments to investigate the genetic basis of pre-harvest sprouting are reported. The results are combined with previously published studies in a composite genetic map for sprout resistance in hexaploid wheat. Different studies, using classical genetics, aneuploids, chromosome substitutions, or QTL mapping, have identified various regions of the A, B, and D genomes affecting dormancy. Comparisons between the available studies lead to the following conclusions: • Different studies often identify different genetic loci, in part reflecting different sampling from the available gene pool. This implies that many loci are involved in determining resistance, and that new loci may be discovered as the number of mapping studies increases.• There are, however, examples where similar map locations are implicated over different crosses. These may reflect the detection of key resistance genes. • Finally, (genotype × environment) interactions are frequently observed. The implications of these observations for crop improvement and research programmes are discussed.
DOI:10.1007/s10681-012-0713-1URL [本文引用: 1]
DOI:10.1007/s10142-004-0105-2URLPMID:14986154 [本文引用: 2]

Quantitative trait loci (QTL) analysis for pre-harvest sprouting tolerance (PHST) in bread wheat was conducted following single-locus and two-locus analyses, using data on a set of 110 recombinant inbred lines (RILs) of the International Triticeae Mapping Initiative population grown in four different environments. Single-locus analysis following composite interval mapping (CIM) resolved a total of five QTLs with one to four QTLs in each of the four individual environments. Four of these five QTLs were also detected following two-locus analysis, which resolved a total of 14 QTLs including 8 main effect QTLs (M-QTLs), 8 epistatic QTLs (E-QTLs) and 5 QTLs involved in QTL x environment (QE) or QTL x QTL x environment (QQE) interactions, some of these QTLs being common. The analysis revealed that a major fraction (76.68%) of the total phenotypic variation explained for PHST is due to M-QTLs (47.95%) and E-QTLs (28.73%), and that only a very small fraction of variation (3.24%) is due to QE and QQE interactions. Thus, more than three-quarters of the genetic variation for PHST is fixable and would contribute directly to gains under selection. Two QTLs that were detected in more than one environment and at LOD scores above the threshold values were located on 3BL and 3DL presumably in the vicinity of the dormancy gene TaVp1. Another QTL was found to be located on 3B, perhaps in close proximity to the R gene for red grain colour. However, these associations of QTLs for PHST with genes for dormancy and grain colour are only suggestive. The results obtained in the present study suggest that PHST is a complex trait controlled by large number of QTLs, some of them interacting among themselves or with the environment. These QTLs can be brought together through marker-aided selection, leading to enhanced PHST.
DOI:10.1007/s10681-009-9927-2URL [本文引用: 2]
DOI:10.1007/s11032-013-0006-yURL [本文引用: 2]

Severe losses attributable to pre-harvest sprouting (PHS) have been reported in Canada in recent years. The genetics of PHS resistance have been more extensively studied in hexaploid wheat and generally not using combinations of elite agronomic parents. The objective of our research was to understand the genetic nature of PHS resistance in an elite durum cross. A doubled haploid (DH) population and checks were phenotyped in replicated trials for grain yield and PHS traits over 3 years in western Canada. The response of intact spikes to sprouting conditions, sampled over two development time points, was measured in a rain simulation chamber. The DH population was genotyped with simple sequence repeat and Diversity Arrays Technology markers. Genotypes were a significant source of variation for grain yield and PHS resistance traits in each tested environment. Transgressive segregant DH genotypes were identified for grain yield and PHS resistance measurements. Low or no correlation was detected between grain yield and PHS, while correlation between PHS resistance measurements was moderate. The heritability of PHS resistance was moderate and higher than grain yield. Significant quantitative trait loci with small effect were detected on chromosomes 1A, 1B, 5B, 7A and 7B. Both parents contributed to the PHS resistance. Promising DH genotypes with high and stable grain yield as well as PHS resistance were identified, suggesting that grain yield and PHS can be improved simultaneously in elite genetic materials, and that these DH genotypes will be useful parental material for durum breeding programs.
DOI:10.1007/BF03195631URL [本文引用: 2]

Based on segregation distortion of simple sequence repeat (SSR) molecular markers, we detected a significant quantitative trait loci (QTL) for pre-harvest sprouting (PHS) tolerance on the short arm of chromosome 2D (2DS) in the extremely susceptible population of F2 progeny generated from the cross of PHS tolerant synthetic hexaploid wheat cultivar ‘RSP’ and PHS susceptible bread wheat cultivar ‘88–1643’. To identify the QTL of PHS tolerance, we constructed two SSR-based genetic maps of 2DS in 2004 and 2005. One putative QTL associated with PHS tolerance, designatedQphs.sau-2D, was identified within the marker intervalsXgwm261-Xgwm484 in 2004 and in the next year, nearly in the same position, between markerswmc112 andXgwm484. Confidence intervals based on the LOD-drop-off method ranged from 9 cM to 15.4 cM and almost completely overlapped with marker intervalXgwm261-Xgwm484. Flanking markers near this QTL could be assigned to the C-2DS1-0.33 chromosome bin, suggesting that the gene(s) controlling PHS tolerance is located in that chromosome region. The phenotypic variation explained by this QTL was about 25.73–27.50%. Genotyping of 48 F6 PHS tolerant plants derived from the cross between PHS tolerant wheat cultivar ‘RSP’ and PHS susceptible bread wheat cultivar ‘MY11’ showed that the allele ofQphs.sau-2D found in the ‘RSP’ genome may prove useful for the improvement of PHS tolerance.
DOI:10.1007/s11032-008-9169-3URL

Pre-harvest sprouting (PHS) is a complex trait controlled by multiple genes with strong interaction between environment and genotype that makes it difficult to select breeding materials by phenotypic assessment. One of the most important genes for pre-harvest sprouting resistance is consistently identified on the long arm of chromosome 4A. The 4AL PHS tolerance gene has therefore been targeted by Australian white-grained wheat breeders. A new robust PCR marker for the PHS QTL on wheat chromosome 4AL based on candidate genes search was developed in this study. The new marker was mapped on 4AL deletion bin 13-0.59-0.66 using 4AL deletion lines derived from Chinese Spring. This marker is located on 4AL between molecular markers Xbarc170 and Xwg622 in the doubled-haploid wheat population Cranbrook×Halberd. It was mapped between molecular markers Xbarc170 and Xgwm269 that have been previously shown to be closely linked to grain dormancy in the doubled haploid wheat population SW95-50213×Cunningham and was co-located with Xgwm269 in population Janz×AUS1408. This marker offers an additional efficient tool for marker-assisted selection of dormancy for white-grained wheat breeding. Comparative analysis indicated that the wheat chromosome 4AL QTL for seed dormancy and PHS resistance is homologous with the barley QTL on chromosome 5HL controlling seed dormancy and PHS resistance. This marker will facilitate identification of the gene associated with the 4A QTL that controls a major component of grain dormancy and PHS resistance.
DOI:10.1007/s11032-007-9135-5URL [本文引用: 1]

Wheat pre-harvest sprouting (PHS) can cause significant reduction in yield and end-use quality of wheat grains in many wheat-growing areas worldwide. To identify a quantitative trait locus (QTL) for PHS resistance in wheat, seed dormancy and sprouting of matured spikes were investigated in a population of 162 recombinant inbred lines (RILs) derived from a cross between the white PHS-resistant Chinese landrace Totoumai A and the white PHS-susceptible cultivar Siyang 936. Following screening of 1,125 SSR primers, 236 were found to be polymorphic between parents, and were used to screen the mapping population. Both seed dormancy and PHS of matured spikes were evaluated by the percentage of germinated kernels under controlled moist conditions. Twelve SSR markers associated with both PHS and seed dormancy were located on the long arm of chromosome 4A. One QTL for both seed dormancy and PHS resistance was detected on chromosome 4AL. Two SSR markers, Xbarc 170 and Xgwm 397, are 9.14cM apart, and flanked the QTL that explained 28.3% of the phenotypic variation for seed dormancy and 30.6% for PHS resistance. This QTL most likely contributed to both long seed dormancy period and enhanced PHS resistance. Therefore, this QTL is most likely responsible for both seed dormancy and PHS resistance. The SSR markers linked to the QTL can be used for marker-assisted selection of PHS-resistant white wheat cultivars.
DOI:10.1007/s00122-014-2345-4URL

Fine mapping by recombinant backcross populations revealed that a preharvest sprouting QTL on 2B contained two QTLs linked in coupling with different effects on the phenotype.
Wheat preharvest sprouting (PHS) occurs when grain germinates on the plant before harvest, resulting in reduced grain quality. Previous mapping of quantitative trait locus (QTL) revealed a major PHS QTL, QPhs.cnl-2B.1, located on chromosome 2B significant in 16 environments that explained from 5 to 31 % of the phenotypic variation. The objective of this project was to fine map the QPhs.cnl-2B.1 interval. Fine mapping was carried out in recombinant backcross populations (BC1F4 and BC1F5) that were developed by backcrossing selected doubled haploids to a recurrent parent and self-pollinating the BC1F4 and BC1F5 generations. In each generation, three markers in the QPhs.cnl-2B.1 interval were used to screen for recombinants. Fine mapping revealed that the QPhs.cnl-2B.1 interval contained two PHS QTLs linked in coupling. The distal PHS QTL, located between Wmc453c and Barc55, contributed 8 % of the phenotypic variation and also co-located with a major seed dormancy QTL determined by germination index. The proximal PHS QTL, between Wmc474 and CNL415-rCDPK, contributed 16 % of the variation. Several candidate genes including Mg-chelatase H subunit family protein, GTP-binding protein and calmodulin/Ca2+-dependent protein kinase were linked to the PHS QTL. Although many recombinant lines were identified, the lack of polymorphism for markers in the QTL interval prevented the localization of the recombination breakpoints and identification of the gene underlying the phenotype.
DOI:10.1071/CP17423URL [本文引用: 3]
[本文引用: 1]
URLPMID:21896881 [本文引用: 1]
URLPMID:23821595 [本文引用: 1]
DOI:10.1007/s00122-014-2262-6URLPMID:24452439 [本文引用: 2]

KEY MESSAGE: After cloning and mapping of wheat TaSdr genes, both the functional markers for TaSdr - B1 and TaVp - 1B were validated, and the distribution of allelic variations at TaSdr - B1 locus in the wheat cultivars from 19 countries was characterized. Seed dormancy is a major factor associated with pre-harvest sprouting (PHS) in common wheat (Triticum aestivum L.). Wheat TaSdr genes, orthologs of OsSdr4 conferring seed dormancy in rice, were cloned by a comparative genomics approach. They were located on homoeologous group 2 chromosomes, and designated as TaSdr-A1, TaSdr-B1 and TaSdr-D1, respectively. Sequence analysis of TaSdr-B1 revealed a SNP at the position -11 upstream of the initiation codon, with bases A and G in cultivars with low and high germination indices (GI), respectively. A cleaved amplified polymorphism sequence marker Sdr2B was developed based on the SNP, and subsequently functional analysis of TaSdr-B1 was conducted by association and linkage mapping. A QTL for GI co-segregating with Sdr2B explained 6.4, 7.8 and 8.7 % of the phenotypic variances in a RIL population derived from Yangxiaomai/Zhongyou 9507 grown in Shijiazhuang, Beijing and the averaged data from those environments, respectively. Two sets of Chinese wheat cultivars were used for association mapping, and results indicated that TaSdr-B1 was significantly associated with GI. Analysis of the allelic distribution at the TaSdr-B1 locus showed that the frequencies of TaSdr-B1a associated with a lower GI were high in cultivars from Japan, Australia, Argentina, and the Middle and Lower Yangtze Valley Winter Wheat Region and Southwest Winter Wheat Region in China. This study provides not only a reliable functional marker for molecular-assisted selection of PHS in wheat breeding programs, but also gives novel information for a comprehensive understanding of seed dormancy.
DOI:10.1007/s00122-016-2793-0URLPMID:27650191 [本文引用: 2]

KEY MESSAGE: We cloned TaSdr - A1 gene, and developed a gene-specific marker for TaSdr - A1 . A QTL for germination index at the TaSdr - A1 locus was identified in the Yangxiaomai/Zhongyou 9507 RIL population. Pre-harvest sprouting (PHS) affects yield and end-use quality in bread wheat (Triticum aestivum L.). In the present study we found an association between the TaSdr-A1 gene and PHS tolerance in bread wheat. TaSdr-A1 on chromosome 2A was cloned using a homologous cloning approach. Sequence analysis of TaSdr-A1 revealed an SNP at position 643, with the G allele being present in genotypes with lower germination index (GI) values and A in those with higher GI. These alleles were designated as TaSdr-A1a and TaSdr-A1b, respectively. A cleaved amplified polymorphism sequence (CAPS) marker Sdr2A based on the SNP was developed, and linkage mapping and QTL analysis were conducted to confirm the association between TaSdr-A1 and seed dormancy. Sdr2A was located in a 2.9 cM interval between SSR markers Xgwm95 and Xgwm372. A QTL for GI at the TaSdr-A1 locus explained 6.6, 7.3, and 8.2 % of the phenotypic variances in a Yangxiaomai/Zhongyou 9507 RIL population grown at Beijing, Shijiazhuang, and the averaged data from the two environments, respectively. Two sets of Chinese wheat cultivars used for validating the TaSdr-A1 polymorphism and the corresponding gene-specific marker Sdr2A showed that TaSdr-A1 was significantly associated with GI. Among 29 accessions with TaSdr-A1a, 24 (82.8 %) were landraces, indicating the importance of Chinese wheat landraces as sources of PHS tolerance. This study identified a novel PHS resistance allele TaSdr-A1a mainly presented in Chinese landraces and it is likely to be the causal gene for QPhs.ccsu-2A.3, providing new information for an understanding of seed dormancy.
DOI:10.1016/S2095-3119(16)61361-8URL [本文引用: 1]
DOI:10.1007/s12041-015-0559-0URLPMID:26440084 [本文引用: 1]

To investigate allelic variation of Myb10-1 genes in Chinese wheat and to examine its association with germination level in wheat, a total of 582 Chinese bread wheat cultivars and 110 Aegilops tauschii accessions were used to identify allelic variations of three Myb10-1 genes. Identification results indicated that there is a novel Tamyb10-B1 allele, designated Tamyb10-B1c, in the five Chinese landraces. The Tamyb10-B1c possibly has a large deletion including Tamyb10-B1 gene. There are three novel Tamyb10-D1 alleles (Aetmyb10-D1c, Aetmyb10-D1d and Aetmyb10-D1e) that were discovered in Aegilops tauschii. Of them, Aetmyb10-D1c allele possessed a 104-bp deletion and this resulted in a frame shift in the open reading frame of the Aetmyb10-D1 gene. AETMYB10-D1d and AETMYB10-D1e proteins possessed three and two different amino acids when compared with TAMYB10-D1b protein, respectively. Association of Tamyb10-1 allelic variation with grain germination level indicated that all five allelic combinations with red grains showed a significantly higher GP (germination percentage) and GI (germination index) values than those of white-grained Tamyb10-A1a/Tamyb10-B1a/Tamyb10-D1a genotype after storing it for one year. Moreover, the Tamyb10-A1b/Tamyb10-B1c/Tamyb10-D1b genotype possesses the significantly highest GP and GI among the six different Tamyb10-1 combinations. This study could provide useful information for wheat breeding programme in terms of grain colour and germination level.
URLPMID:25962727 [本文引用: 2]
URLPMID:26948878 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00122-005-0021-4URLPMID:16133317 [本文引用: 1]

Quantitative trait loci (QTL) analysis was conducted for pre-harvest sprouting tolerance (PHST) in bread wheat for a solitary chromosome 3A, which was shown to be important for this trait in earlier studies. An inter-varietal mapping population in the form of recombinant inbred lines (RILs) developed from a cross between SPR8198 (a PHS tolerant genotype) and HD2329 (a PHS susceptible cultivar) was used for this purpose. The parents and the RIL population were grown in six different environments and the data on PHS were collected in each case. A framework linkage map of chromosome 3A with 13 markers was prepared and used for QTL analysis. A major QTL (QPhs.ccsu-3A.1) was detected on 3AL at a genetic distance of approximately 183 cM from centromere, the length of the map being 279.1 cM. The QTL explained 24.68% to 35.21% variation in individual environments and 78.03% of the variation across the environments (pooled data). The results of the present study are significant on two counts. Firstly, the detected QTL is a major QTL, explaining up to 78.03% of the variation and, secondly, the QTL showed up in all the six environments and also with the pooled data, which is rather rare in QTL analysis. The positive additive effects in the present study suggest that a superior allele of the QTL is available in the superior parent (SPR8198), which can be used for marker-aided selection for the transfer of this QTL allele to obtain PHS-tolerant progeny. It has also been shown that the red-coloured grain of PHS tolerant parent is not associated with the QTL for PHST identified during the present study, suggesting that PHS tolerant white-grained cultivars can be developed.
DOI:10.1007/s10681-014-1266-2URL
DOI:10.1186/s12864-016-3148-6URLPMID:27729004

BACKGROUND: Pre-harvest sprouting (PHS) in wheat can cause substantial reduction in grain yield and end-use quality. Grain color (GC) together with other components affect PHS resistance. Several quantitative trait loci (QTL) have been reported for PHS resistance, and two of them on chromosome 3AS (TaPHS1) and 4A have been cloned. METHODS: To determine genetic architecture of PHS and GC and genetic relationships of the two traits, a genome-wide association study (GWAS) was conducted by evaluating a panel of 185 U.S. elite breeding lines and cultivars for sprouting rates of wheat spikes and GC in both greenhouse and field experiments. The panel was genotyped using the wheat 9K and 90K single nucleotide polymorphism (SNP) arrays. RESULTS: Four QTL for GC on four chromosomes and 12 QTL for PHS resistance on 10 chromosomes were identified in at least two experiments. QTL for PHS resistance showed varied effects under different environments, and those on chromosomes 3AS, 3AL, 3B, 4AL and 7A were the more frequently identified QTL. The common QTL for GC and PHS resistance were identified on the long arms of the chromosome 3A and 3D. CONCLUSIONS: Wheat grain color is regulated by the three known genes on group 3 chromosomes and additional genes from other chromosomes. These grain color genes showed significant effects on PHS resistance in some environments. However, several other QTL that did not affect grain color also played a significant role on PHS resistance. Therefore, it is possible to breed PHS-resistant white wheat by pyramiding these non-color related QTL.
DOI:10.1007/s10681-009-9935-2URL [本文引用: 1]

A framework linkage map comprising 214 molecular marker (SSR, AFLP, SAMPL) loci was prepared using an intervarietal recombinant inbred line (RIL) mapping population of bread wheat. The RIL population that was developed from the cross SPR8198 (red-grained and PHS tolerant genotype)×HD2329 (white-grained and PHS susceptible genotype) following single seed descent segregated for pre-harvest sprouting (PHS). The RIL population and parental genotypes were evaluated in six different environments and the data on PHS were collected. Using the linkage map and PHS data, genome-wide single-locus and two-locus QTL analyses were conducted for PHS tolerance (PHST). Single-locus analysis following composite interval mapping (CIM) detected a total of seven QTL, located on specific arms of five different chromosome (1AS, 2AL, 2DL, 3AL and 3BL). These seven QTL included two major QTL one each on 2AL and 3AL. Two of these seven QTL were also detected following two-locus analysis, which resolved a total of four main-effect QTL (M-QTL), and 12 epistatic QTL (E-QTL), the latter involved in 7 QTL×QTL interactions. Interestingly, none of these M-QTL and E-QTL detected by two-locus analysis was involved in Q×E and Q×Q×E interactions, supporting the results of ANOVA, where genotype×environment interaction were non-significant. The QTL for PHS detected in the present study may be efficiently utilized for marker-aided selection for enhancing PHST in bread wheat.
DOI:10.1007/s001220200004URLPMID:12579426 [本文引用: 2]

In many wheat ( Triticum aestivumL.) growing areas, pre-harvest sprouting (PHS) may cause important damage, and in particular, it has deleterious effects on bread-making quality. The relationship between PHS and grain color is well known and could be due either to the pleiotropic effect of genes controlling red-testa pigmentation ( R) or to linkage between these genes and other genes affecting PHS. In the present work, we have studied a population of 194 recombinant inbred lines from the cross between two cultivars, 'Renan' and 'Recital', in order to detect QTLs for both PHS resistance and grain color. The variety 'Renan' has red kernels and is resistant to PHS, while 'Recital' has white grain and is highly susceptible to PHS. A molecular-marker linkage map of this cross was constructed using SSRs, RFLPs and AFLPs. The population was evaluated over 2 years at Clermont-Ferrand (France). PHS was evaluated on mature spikes under controlled conditions and red-grain color was measured using a chromameter. Over the 2 years, we detected four QTLs for PHS, all of them being co-localized with QTLs for grain color. Three of them were located on the long arm of chromosomes 3 A, 3B and 3D, close to the loci where the genes R and taVp1 were previously mapped. For these three QTLs, the resistance to PHS is due to the allele of the variety 'Renan'. Another co-located QTL for PHS and grain color was detected on the short arm of chromosome 5 A. The resistance for PHS for this QTL is due to the allele of 'Recital'.
DOI:10.1007/s10681-015-1460-xURL [本文引用: 2]
DOI:10.1007/s00122-005-0065-5URLPMID:16133305 [本文引用: 1]

Improved resistance to preharvest sprouting in modern bread wheat (Triticum aestivum. L.) can be achieved via the introgression of grain dormancy and would reduce both the incidence and severity of damage due to unfavourable weather at harvest. The dormancy phenotype is strongly influenced by environmental factors making selection difficult and time consuming and this trait an obvious candidate for marker assisted selection. A highly significant Quantitative Trait Locus (QTL) associated with grain dormancy and located on chromosome 4A was identified in three bread wheat genotypes, two white- and one red-grained, of diverse origin. Flanking SSR markers on either side of the putative dormancy gene were identified and validated in an additional population involving one of the dormant genotypes. Genotypes containing the 4A QTL varied in dormancy phenotype from dormant to intermediate dormant. Based on a comparison between dormant red- and white-grained genotypes, together with a white-grained mutant derived from the red-grained genotype, it is concluded that the 4A QTL is a critical component of dormancy; associated with at least an intermediate dormancy on its own and a dormant phenotype when combined with the R gene in the red-grained genotype and as yet unidentified gene(s) in the white-grained genotypes. These additional genes appeared to be different in AUS1408 and SW95-50213.
[本文引用: 1]
URLPMID:19669633
DOI:10.1007/s11032-010-9448-7URL [本文引用: 2]
URLPMID:25851002 [本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1007/s10681-010-0137-8URL [本文引用: 1]

Pre-harvest sprouting (PHS) in developing wheat (Triticum aestivum L.) spikes is stimulated by cool and wet weather and leads to a decline in grain quality. A low level of harvest-time seed dormancy is a major factor for PHS, which generally is a larger problem in white-grained as compared to red-grained wheat. We have in this study analyzed seed dormancy levels at the 92nd Zadok growth stage of spike development in a doubled-haploid (DH) white wheat population and associated variation for the trait with regions on the wheat genome. The phenotypic data was generated by growing the parent lines Argent (non-dormant) and W98616 (dormant) and 151 lines of the DH population in the field during 2002 and 2003, at two locations each year, followed by assessment of harvest-time seed dormancy by germination tests. A genetic map of 2681cM was constructed for the population upon genotyping 90 DH lines using 361 SSR, 292 AFLP, 252 DArT and 10 EST markers. Single marker analysis of the 90 genotyped lines associated regions on chromosomes 1A, 2B, 3A, 4A, 5B, 6B, and 7A with seed dormancy in at least two out of the four trials. All seven putative quantitative trait loci (QTLs) were contributed by alleles of the dormant parent, W98616. The strongest QTLs positioned on chromosomes 1A, 3A, 4A and 7A were confirmed by interval mapping and markers at these loci have potential use in marker-assisted selection of PHS resistant white-grained wheat.
[本文引用: 1]
DOI:10.1016/j.plantsci.2019.01.004URLPMID:30824050 [本文引用: 1]

Pre-harvest sprouting (PHS) of wheat (Triticum aestivum L.) is an important phenomenon that results in weather dependent reductions in grain yield and quality across the globe. Due to the large annual losses, breeding PHS resistant varieties is of great importance. Many quantitative trait loci have been associated with PHS and a number of specific genes have been proven to impact PHS. TaPHS1, TaMKK3, Tamyb10, and TaVp1 have been shown to have a large impact on PHS susceptibility while many other genes such as TaSdr, TaQSd, and TaDOG1 have been shown to account for smaller, but significant, proportions of variation. These advances in understanding the genetics behind PHS are making molecular selection and loci stacking viable methods for affecting this quantitative trait. The current review article serves to provide a brief synthesis of recent advances regarding PHS, as well as provide unique insight into the genetic mechanisms governing PHS in bread wheat.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
