 ,, 周晓龙, 汪涵, 李向臣, 赵阿勇, 杨松柏
,, 周晓龙, 汪涵, 李向臣, 赵阿勇, 杨松柏 ,浙江农林大学动物科技学院/浙江省畜禽绿色生态健康养殖应用技术研究重点实验室,浙江临安 311300
,浙江农林大学动物科技学院/浙江省畜禽绿色生态健康养殖应用技术研究重点实验室,浙江临安 311300Prediction and Verification of MicroRNAs Targeting Porcine Endoplasmic Reticulum Stress Pathway
ZHU JingJing ,, ZHOU XiaoLong, WANG Han, LI XiangChen, ZHAO AYong, YANG SongBai
,, ZHOU XiaoLong, WANG Han, LI XiangChen, ZHAO AYong, YANG SongBai ,College of Animal Science and Technology, Zhejiang A&F University/Key Laboratory of Applied Technology on Green-Eco-Healthy Animal Husbandry of Zhejiang Province, Lin'an 311300, Zhejiang
,College of Animal Science and Technology, Zhejiang A&F University/Key Laboratory of Applied Technology on Green-Eco-Healthy Animal Husbandry of Zhejiang Province, Lin'an 311300, Zhejiang通讯作者:
责任编辑: 林鉴非
收稿日期:2019-08-30接受日期:2020-03-11网络出版日期:2020-08-01
| 基金资助: |
Received:2019-08-30Accepted:2020-03-11Online:2020-08-01
作者简介 About authors
朱静静,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1384KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
朱静静, 周晓龙, 汪涵, 李向臣, 赵阿勇, 杨松柏. 靶向猪内质网应激通路的microRNAs预测与验证[J]. 中国农业科学, 2020, 53(15): 3169-3179 doi:10.3864/j.issn.0578-1752.2020.15.016
ZHU JingJing, ZHOU XiaoLong, WANG Han, LI XiangChen, ZHAO AYong, YANG SongBai.
0 引言
【研究意义】猪内质网应激过强或持续时间过长会导致一系列疾病的发生,给养猪业造成巨大经济损失。内质网应激是一个复杂的调节过程,因此,从不同角度深入研究猪内质网应激通路至关重要。本研究通过生物信息学和试验方法预测和验证靶向内质网应激通路关键基因的miRNAs,对进一步研究转录后水平调控内质网应激通路具有重要的意义。【前人研究进展】内质网是由细胞内膜构成的网膜系统,主要参与蛋白质合成、转运、折叠、以及脂质和类固醇合成[1,2]。当细胞受到内源性和外源性刺激时,内质网会发生未折叠蛋白或错误折叠蛋白累积,诱导内质网应激(ERS)[3]。在ERS过程中,细胞会产生相应的应答措施,激活未折叠蛋白反应(UPR),减少未折叠蛋白的积累,以恢复内质网的内稳态[4]。内质网启动UPR受到ATF6、IRE1、PERK三条信号传导途径的调控[5]。正常情况下,内质网膜上的ATF6、IRE1、PERK跨膜蛋白与内质网腔内的内稳态感受器葡萄糖调节蛋白78(GRP78)结合,形成稳定的复合物,UPR处于失活状态。当未折叠蛋白或错误折叠蛋白累积时,GRP78与ATF6、IRE1、PERK解离,并与未折叠蛋白或错误折叠蛋白结合,激活三种跨膜蛋白,并启动UPR的ATF6、IRE1、PERK信号通路,从而维持内质网稳态[6,7,8]。同时,激活的IRE1通过剪切XBP1的mRNA内含子,使有转录活性的XBP1s表达,XBP1s能够促进内质网分子伴侣和蛋白折叠相关基因的表达,促进内质网正确折叠能力[9]。研究表明,一定程度ERS有利于增强细胞应对刺激能力,而过强或持续的ERS会导致内质网的内稳态失调,诱导炎症[10]、细胞自噬[11]、细胞凋亡[12]、胰岛素抵抗[13]等一系列反应的发生。在猪养殖过程中,病毒感染[14,15]是引起猪ERS的关键因素。本课题组前期研究结果显示,PRV感染PK15细胞后,PRV能够激活ERS通路[16,17]。miRNA通过抑制靶基因mRNA翻译或诱导靶基因mRNA降解,来调控靶基因的表达。关于miRNA靶向调节ERS通路的机制在人类疾病方面已有研究。miR-30d、miR-181a、miR-199a协同作用能够有效抑制GRP78表达水平和GRP78介导的化学耐药[18]。miR-145、miR-221和miR-494在支气管上皮细胞中调控ATF6[19],在小鼠中miR-702也调控ATF6表达[20]。参与UPR调控的miRNA还包括miR-122和miR-30c-2-3p[21,22]。【本研究切入点】随着高通量miRNA测序技术的飞速发展,大量新的miRNA被陆续发现,靶向不同基因miRNA的预测和验证就显得尤为重要。然而,靶向猪ERS通路的miRNA调节机制尚不清楚。【拟解决的关键问题】研究miRNA对猪ERS通路关键基因的表达调控作用,为后续从转录后水平研究调控ERS的分子机制奠定基础。1 材料与方法
本试验于2019年2—8月在浙江农林大学动物遗传育种与繁殖实验室完成。1.1 试验材料
细胞和病毒株:PK15细胞系和BHK-21细胞系购自中国典型培养物保藏中心(武汉大学);PRV(浙江株)由浙江省农业科学院畜牧兽医研究所张存研究员惠赠。主要试剂:psiCHECK-2质粒、双荧光素酶检测试剂盒均购自美国Promega公司;T4 DNA连接酶、限制性内切酶NotⅠ、XhoⅠ购自美国Thermo公司;miRNA-mimics序列由上海吉玛制药技术有限公司合成;LipofectamineTM 3000购自美国Invitrogen公司;琼脂糖凝胶回收试剂盒、无内毒素质粒中提试剂盒、2×Taq PCR MasterMix、EasyQuick RT MasterMix反转录试剂、BCA试剂盒均购自北京康为世纪生物科技有限公司;荧光定量试剂SYBR? Primix Ex TaqTMⅡ购自宝日医生物技术有限公司;疏水性PVDF膜购自杭州诺扬生物技术有限公司;脱脂奶粉购自美国BD公司;ATF6多克隆抗体购自Abclonal公司;β-Actin鼠源单克隆抗体购自Proteintech公司;HRP标记的羊抗鼠IgG、HRP标记的羊抗兔IgG抗体均购自Abbkine公司。
1.2 试验方法
1.2.1 病毒接种与miRNA测序 PK15细胞接种6 cm培养皿,细胞汇合度80%时接种PRV(TCID50= 10-6.625/0.1 mL)20 μL,同时设空白细胞对照(Control)。感染12 h后收集细胞提取RNA进行测序。miRNA测序试验是由北京博奥晶典生物技术有限公司完成。利用illumina测序平台对空白对照和PRV感染的细胞进行测序,分析其miRNAs表达差异。1.2.2 靶向内质网通路的miRNA预测和分析 利用TargetScan(
1.2.3 双荧光素酶报告基因载体的构建 根据交集miRNA识别并结合靶基因ATF6(Genbank 登录号:XM_021089515.1)、PERK(Genbank 登录号:XM_003124925.4)、IRE1(Genbank 登录号:XM_005668695.3)、GRP78(Genbank 登录号:XM_001927795.7)和XBP1(Genbank 登录号:NM_001142836.1)的3'UTR靶点区域,设计引物(表1)用于扩增包含miRNA靶位点的3′UTR区。以PK15细胞cDNA为模板进行PCR扩增。扩增产物纯化后和psiCHECK-2载体分别用XhoⅠ和NotⅠ于37℃双酶切30min,然后用T4 DNA连接酶于16℃过夜,再将连接产物转化入感受态细胞DH5α中。培养并挑取阳性克隆,送尚亚生物技术(杭州)有限公司测序。鉴定结果正确的阳性克隆,提取质粒,进行酶切验证。
Table 1
表1
表1双荧光素酶报告基因载体构建引物序列
Table 1
| 名称 Names | 引物序列 Primer sequence (5′—3′) | 片段长度 Size (bp) |
|---|---|---|
| ATF6-m142-3′UTR | F:CCGCTCGAGCCGTGGAGTGATTATGTG | 413 |
| R:ATTGCGGCCGCCTTCCTGAAACTTTACCCT | ||
| ATF6-m145-3′UTR | F:CCGCTCGAGCCTGGCTTGAATCTTCCC | 301 |
| R:ATTGCGGCCGCGTTGTGAGTTCGATCCCT | ||
| ATF6-m150-3′UTR | F:CCGCTCGAGTTTTGTCAGTCCTGGGTC | 464 |
| R:ATTGCGGCCGCAAGATACCTTTGTCATTC | ||
| ATF6-m199a-3′UTR | F:CCGCTCGAGTCTTTCTGCCTTCTTGGT | 176 |
| R:ATTGCGGCCGCGGAAGATGTAGGGTAATGTG | ||
| PERK-m145/150-3′UTR | F:CCGCTCGAGCATGGAATAGCCCACCTC | 777 |
| R:ATTGCGGCCGCTTGGGAAAGTACCGACCT | ||
| IRE1-m142/145/199-3′UTR | F:CCGCTCGAGATCGAACCCGAGCCACTG | 1006 |
| R:ATTGCGGCCGCAGACGCCATCATCAATCA | ||
| IRE1-m150-3′UTR | F:CCGCTCGAGAGCCAAGTGCCTTGAGCTG | 256 |
| R:ATTGCGGCCGCACAGGCTGACATCAAACAGGA | ||
| GRP78-m145/199-3′UTR | F:CCGCTCGAGACTGCTCTGCTAGTGTTG | 349 |
| R:ATTGCGGCCGCACCAGTGTAAATAACAAA | ||
| XBP1-m142/145/150/199-3′UTR | F:CCGCTCGAGCTGCTTTCAACCAGCCACT | 596 |
| R:ATTGCGGCCGCCCCTCAGGTAGGCATTCT |
新窗口打开|下载CSV
1.2.4 细胞培养与转染 将BHK-21细胞铺板至24孔板,细胞融合至80%左右,利用LipofectamineTM 3000进行转染,按照说明书操作,将阴性对照(NC)、miR-142-5p mimics、miR-145-5p mimics、miR-150 mimics、miR-199a-5p mimics和双荧光报告载体共转染BHK-21细胞,转染24 h后收集细胞,用于检测细胞荧光素酶活性。
将PK15细胞铺板至6孔板和12孔板,细胞融合至50%左右进行转染,将只加转染试剂空白对照(MOCK)、阴性对照(NC)、miR-142-5p mimics、miR-145-5p mimics、miR-199a-5p mimics转染PK15细胞,共形成5个组合。转染48h后收集细胞,6孔板细胞用于检测ATF6的蛋白水平,12孔板用于检测内质网应激通路关键基因ATF6、PERK、IRE1和XBP1的mRNA表达水平。
1.2.5 双荧光素酶活性检测 按照Dual-Luciferase? Reporter Assay System说明书操作。PBS清洗细胞后,每孔加100 μL 1×Passive Lysis Buffer,放置室温摇床缓慢摇动15 min。吸取5 μL裂解液至96孔黑色酶标板对应孔中,再加入25 μL LAR-II试剂,使用多功能酶标仪测定萤火虫荧光素酶的荧光值。然后每孔加入25 μL 1×Stop &Glo终止液,检测海肾荧光素酶的荧光值。最后以萤火虫荧光作为内参照对结果进行均一化处理。
1.2.6 总RNA提取及cDNA合成 利用Trizol法提取PK15细胞总RNA,使用分光光度计和1.2%琼脂糖凝胶电泳检测RNA的浓度和完整性。以PK15细胞总RNA作为模板进行cDNA的合成,反应体系20 μL:5×EasyQuick RT MasterMix 4 μL,RNA1μg,RNase- free ddH2O补足体系,涡旋震荡混匀,经短暂离心,PCR反应条件:37℃ 15 min;85℃ 5 s,反应结束后-20℃保存。
1.2.7 荧光定量引物设计合成与qRT-PCR反应 根据NCBI已发表的猪ATF6、IRE1、PERK、XBP1、GAPDH序列设计qRT-PCR引物(表2)。以不同处理组的cDNA为模板进行实时荧光定量PCR扩增。扩增结束后分析溶解曲线,采用2-△△Ct法计算ATF6、IRE1、PERK、XBP1的相对表达量。
Table 2
表2
表2qRT-PCR引物序列
Table 2
| 名称 Names | 引物序列 Primer sequence (5′—3′) | 退火温度 Annealing temperature (℃) | 片段长度 Size (bp) |
|---|---|---|---|
| ATF6 | F:CTAAAGCGAGTCCTCTACG | 60 | 241 |
| R:GCCATGCCTGATTTCACA | |||
| XBP1 | F:GGATTCTGACGGTGTTGA | 60 | 161 |
| R:GGAGGCTGGTAAGGAACT | |||
| PERK | F:AGGAGCAAACGGAGCACG | 60 | 187 |
| R:GTCGGTGAGGATGAGGATGG | |||
| IRE1 | F:GGACTGGCGGGAGAACAT | 60 | 272 |
| R:CGTGGTAGTAGGGCTGGAA | |||
| GAPDH | F:GGACTCATGACCACGGTCCAT | 60 | 220 |
| R:TCAGATCCACAACCGACACGT |
新窗口打开|下载CSV
1.2.8 Western blot检测 利用RIPA细胞裂解液提取转染后PK15细胞的总蛋白,BCA试剂盒检测蛋白浓度。每组蛋白上样量相同,经SDS-PAGE电泳分离后,转移到PVDF膜上。将膜置于5%脱脂奶粉中,室温封闭2h。加5%脱脂奶粉稀释ATF6抗体(1:1 000)及β-Actin抗体(1:5 000),4℃孵育过夜,1×TBST充分洗膜3次(10 min/次)后,分别加入HRP标记的二抗(1:10 000),室温孵育1 h,最后扫描分析目的蛋白条带。
1.2.9 数据统计分析 试验数据均用平均值±标准误表示,每个试验3个生物学重复,统计学分析采用T检验,*P<0.05;**P<0.01,具有统计学意义。
2 结果
2.1 PRV感染PK15细胞后miRNA差异表达分析
由笔者所在课题组前期研究结果表明PRV感染能够激活内质网应激,所以为了检测PRV感染引起miRNA表达的变化,用PRV感染PK15细胞,感染后12 h收集细胞提取RNA进行miRNA测序。结果表明,PRV感染PK15细胞后,诱导miRNA差异表达。与PRV未感染的空白对照组相比,PRV感染后共引起35条miRNAs表达变化,其中包括27条miRNAs(miR-132、miR-136-3p、miR-142-3p、miR-142-5p、miR-146b、miR-190b、miR-199a-3p、miR-199a-5p、miR-199b-3p、miR-199b-5p、miR-212、miR-218、miR-218-5p、miR-218b、miR-323、miR-363、miR-369、miR-375、miR-381-3p、miR-382、miR-551a、miR-758、miR-874、miR-9、miR-9-1、miR-9-2、miR-9820-5p)表达量上调,8条miRNAs(miR-10391、miR-2366、miR-27b-5p、miR-30c-1-3p、miR-30c-3p、miR-374b-3p、miR-671-3p、miR-885-3p)表达量下调(图1)。结果表明,PRV感染引起猪内质网应激后,宿主细胞miRNA表达发生变化。图1
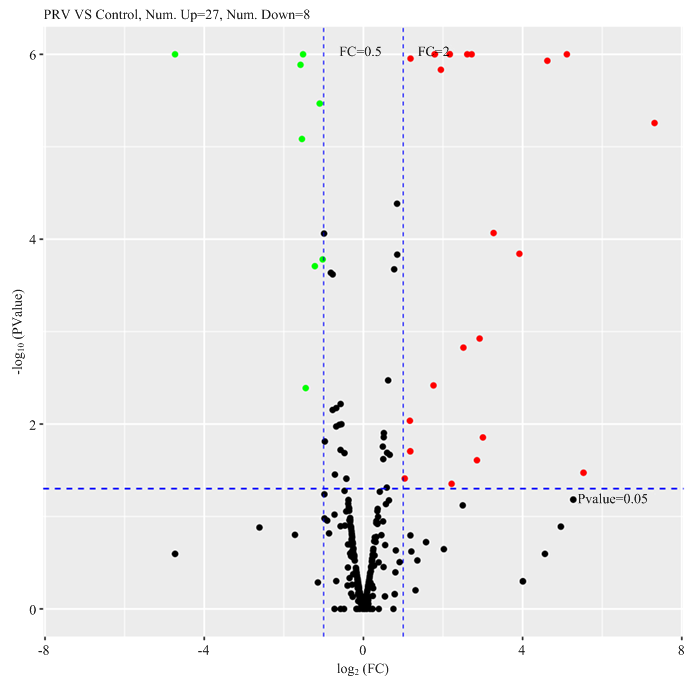 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1PRV感染PK15细胞miRNA测序数据火山图
Fig. 1Volcano plot of miRNA-sequencing data from PRV infected PK15 cells
2.2 靶向内质网通路关键基因的miRNAs预测
利用TargetScan生物信息学软件,预测出与ATF6、IRE1、PERK、GRP78、XBP1具有靶标关系的miRNAs分别有55、14、52、30和21个。用韦恩图取交集,miR-150同时靶向ATF6、IRE1、PERK、GRP78、XBP1五个基因,miR-142-5p同时靶向ATF6、PERK、IRE1、GRP78基因,miR-145-5p和miR-199a-5p同时靶向ATF6、IRE1、GRP78、XBP1基因(图2)。于是,本研究选择miR-142-5p、miR-145-5p、miR-150和miR-199a-5p作为候选miRNAs。图2
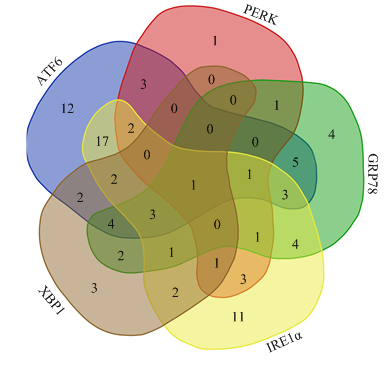 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2与ATF6、IRE1、PERK、GRP78、XBP1具有靶标关系的miRNAs交集预测
Fig. 2The intersection prediction of miRNAs target ATF6, IRE1, PERK, GRP78 and XBP1
2.3 双荧光素酶报告基因载体的构建与鉴定
将ATF6、IRE1、PERK、GRP78、XBP1的3′UTR区插入psiCHECK-2构建双荧光报告载体,再将连接产物转化入大肠杆菌感受态细胞DH5α中,培养并挑取单克隆,送去公司测序。鉴定结果正确的阳性克隆,提取质粒,进行酶切验证(图3)。最终获得重组正确的双荧光报告载体分别命名为psiCHECK-2-ATF6- m142-3′UTR、psiCHECK-2-ATF6-m145-3′UTR、psiCHECK- 2-ATF6-m150-3′UTR、psiCHECK-2-ATF6-m199-3′UTR、psiCHECK-2-IRE1-m150-3′UTR、psiCHECK-2-IRE1- m142/145/199-3′UTR、psiCHECK-2-PERK-m145/150- 3′UTR、psiCHECK-2-XBP1-m142/145/150/199-3′UTR、psiCHECK-2-GRP78-m145/199-3′UTR。图3
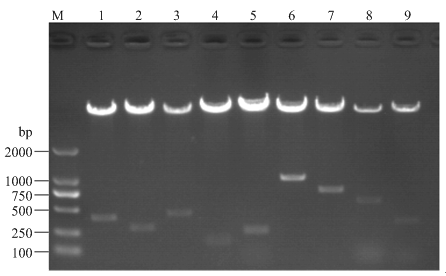 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3双荧光报告基因载体双酶切鉴定
M:DL2000 Marker;1:psiCHECK-2-ATF6-m142-3′UTR,2:psiCHECK- 2-ATF6-m145-3′UTR,3:psiCHECK-2-ATF6-m150-3′UTR,4:psiCHECK- 2-ATF6-m199-3′UTR,5:psiCHECK-2-IRE1-m150-3′UTR,6:psiCHECK- 2-IRE1-m142/145/199-3′UTR,7:psiCHECK-2-PERK-m145/150-3′UTR,8:psiCHECK-2-XBP1-m142/145/150/199-3′UTR,9:psiCHECK-2-GRP78- m145/199-3′UTR
Fig. 3Identification of luciferase reporter gene vector by XhoⅠ and NotⅠ double digestion
2.4 荧光素酶报告基因活性检测
为了进一步确认miR-142-5p、miR-145-5p、miR-150和miR-199a-5p能够靶向作用于内质网通路关键基因,将重组质粒分别与miR-142-5p mimics、miR-145-5p mimics、miR-150 mimics和miR-199a-5p mimics共转染,24 h后检测荧光素酶活性。结果显示,共转染psiCHECK-2-ATF6-m142-3′UTR与miR-142- 5p mimics后,过表达组的荧光素酶活性极显著低于阴性对照组(图4-A)。同时,psiCHECK-2-IRE1-m142/ 145/199-3′UTR分别与miR-142-5p mimics、miR-145-5p mimics、miR-199a-5p mimics共转染,过表达组的荧光素酶活性极显著低于阴性对照组(图4-B),以及psiCHECK-2-XBP1-m142/145/150/199-3′UTR分别与miR-142-5p mimics、miR-199a-5p mimics共转染,过表达组的荧光素酶活性也极显著低于阴性对照组(图4-C)。共转染psiCHECK-2-PERK-m145/150-3′UTR与miR-145-5p mimics,能够显著下调荧光素酶报告基因的表达(图4-D)。结果表明,miR-142-5p与ATF6、IRE1、XBP1的3′UTR靶位点结合,miR-199a-5p与IRE1、XBP1的3′UTR靶位点结合,以及miR-145-5p与IRE1、PERK的3′UTR靶位点结合,从而抑制了荧光信号的产生。图4
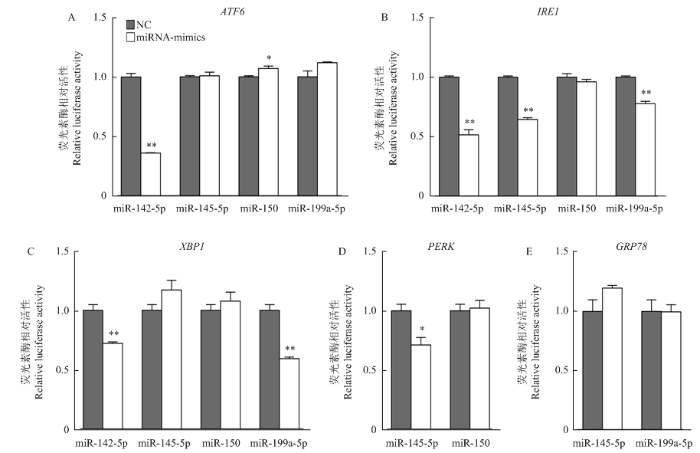 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4miR-142-5p mimics、miR-145-5p mimics、miR-150 mimics和miR-199a-5p mimics对重组载体荧光素酶表达的影响
Fig. 4Influence of miR-142-5p mimics, miR-145-5p mimics, miR-150 mimics and miR-199a-5p mimics on luciferase expression of recombinant vector
2.5 qRT-PCR分析
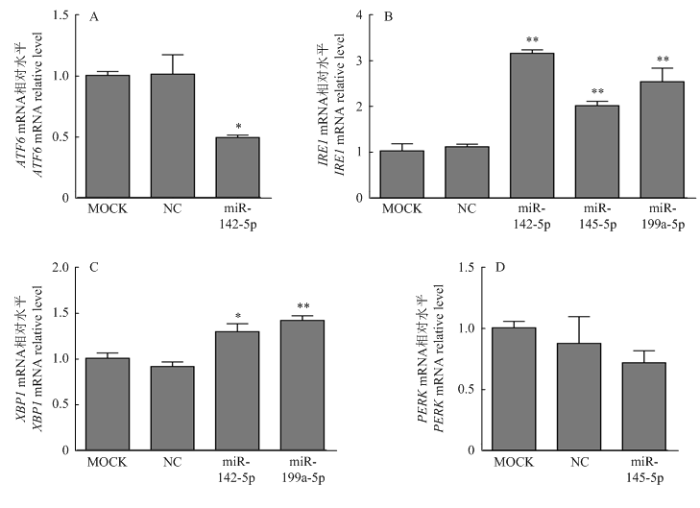
根据荧光素酶报告系统筛选结果,利用qRT-PCR试验来进一步验证。将空白对照(MOCK)、阴性对照(NC)、miR-142-5p mimics、miR-145-5p mimics、miR-199a-5p mimics分别转染PK15细胞,转染48 h后收集细胞,qRT-PCR检测ATF6、IRE1、XBP1和PERK的mRNA表达水平。结果表明,过表达miR-142-5p后,ATF6的表达量显著低于阴性对照组和空白对照组(P<0.05,图5-A)。说明miR-142-5p负调控ATF6的表达。而miR-142-5p、miR-145-5p和miR-199a-5p过表达后,极显著上调IRE1mRNA表达(P<0.01,图5-B)。同样,过表达miR-142-5p和miR-199a-5p后,XBP1的mRNA表达也显著上调(P<0.05,图5-C)。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5过表达miRNAs后ATF6、IRE1、XBP1和PERK mRNA的表达量
Fig. 5The expression of ATF6, IRE1, PERK and XBP1 mRNA after over-expression miRNAs
2.6 过表达miR-142-5p对ATF6蛋白表达的影响
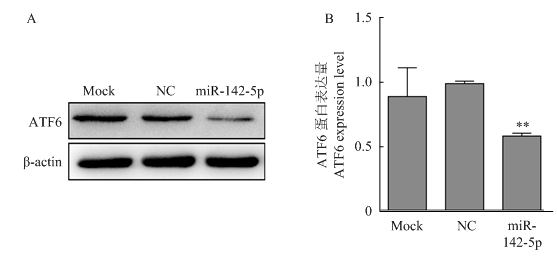
过表达miR-142-5p后,ATF6蛋白的表达情况见图6。结果显示,过表达组中ATF6的表达量显著低于阴性对照组和空白对照组。结果表明,miR-142-5p与ATF6靶位点结合后,抑制ATF6蛋白的表达。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6转染miR-142-5p mimics后PK15细胞中ATF6蛋白表达检测
A.转染miR-142-5p mimics后,PK15细胞中ATF6蛋白Western blotting检测;B.转染miR-142-5p mimics后,PK15细胞中ATF6蛋白相对表达量检测
Fig. 6Western blotting analysis of ATF6 in PK15 cells transfected with miR-142-5p mimics
A. shows the Western blotting analysis of ATF6 protein in PK15 cells transfected with miR-142-5p mimics; B. shows the expression level of ATF6 protein detected by Western blotting after miR-142-5p mimics transfecting into PK15 cells
3 讨论
3.1 miRNA测序分析
miRNA是一类18—25 bp的调节性非编码RNA,主要通过部分碱基互补配对方式将其种子序列与靶基因mRNA的3′UTR结合,从而调控基因表达参与多种重要生理过程。大量研究表明,miRNA在调控内质网应激通路关键基因中发挥重要作用[18,19,20,21,22],但是大部分报道都集中在人和小鼠等哺乳动物的研究上,对猪miRNA的研究尚未报道。因此,开展内质网应激通路相关miRNA的研究,为从转录后水平调控内质网应激通路从而调控PRV感染奠定理论基础。本课题组前期研究结果显示PRV感染激活内质网应激通路。本研究首先利用高通量测序方法发现PRV感染PK15细胞后引起35条miRNAs差异表达,其中包括miR-132、miR-142-5p、miR-199a-5p、miR-374b-3p、miR-382和miR-874。研究表明PRV感染后引起miR-132、miR-199a/b-3p和mir-374b-3p等13条miRNAs在宿主细胞内差异表达,同时通过GO富集分析差异表达miRNAs潜在的宿主靶基因,发现这些靶基因参与生物调节、应激反应、免疫应答和细胞凋亡等信号传导过程[23]。还有研究发现,CSFV感染PK15细胞后引起miR-382和miR-874等69条miRNAs表达变化[24]。推测PRV感染引起猪内质网应激后,宿主miRNA靶向内质网应激通路关键基因,从而参与调控内质网应激过程。
3.2 生物信息学软件预测分析
本研究中,利用生物信息学的方法预测与内质网应激通路关键基因ATF6、IRE1、PERK、GRP78、XBP1具有靶标关系的miRNAs,发现与ATF6、IRE1、PERK、GRP78、XBP1中至少4个基因具有靶标关系的miRNAs,分别是miR-142-5p、miR-145-5p、miR-150、miR-199a-5p。其中miR-142-5p和miR-199a-5p在miRNA测序结果中也显示为差异表达。研究发现ERS传导器IRE1通过下调miR-150介导纤维化,同时IRE1促进XBP1剪接进而激活肌成纤维细胞中内质网扩张[25]。此外,面对各种环境应激,miRNA成为应激后免疫修饰的靶点。miR-142-5p、miR--150在热应激大鼠中差异表达,调控参与大鼠小肠热应激病理及生理反应相关的基因表达[26]。HPV感染细胞后下调miR-145表达,低表达的miR-145使细胞免受环境应激诱导的凋亡[27]。有研究表明,在人肺腺癌细胞中,miR-199a-5p通过调控GRP78转录本使GRP78基因表达水平显著下调[28]。因此推测,miR-142、miR-145、miR-150、miR-199可能在猪内质网应激过程中发挥着调控作用。3.3 候选miRNAs对内质网关键基因表达的影响
根据miRNA预测结果,构建了双荧光素酶报告基因重组载体,并利用双荧光素酶报告系统验证发现miR-142-5p mimics显著抑制psiCHECK-2-ATF6- m142-3′UTR重组载体荧光素酶活性。同时qRT-PCR和Western blot试验再次验证了这一结果,过表达miR-142-5p后,ATF6的mRNA水平和蛋白表达均显著下调。表明miR-142-5p通过降解mRNA的方式靶向猪ATF6。针对靶向人ATF6的miRNA研究结果发现,在UPR过程中,PERK介导的miR-424与ATF6的3′UTR区作用位点结合调节ATF6的表达,并减弱ATF6转录活性[29],miR-145、miR-221和miR-494可以靶向ATF6的3′UTR区进而抑制囊性纤维化中ATF6表达[19]。而本研究中发现miR-145-5p并不靶向猪ATF6,具体机制尚需要进一步研究。本研究中,miR-142-5p、miR-199a-5p均与IRE1、XBP1的3′UTR靶位点结合,从而抑制了荧光素酶活性。但qRT-PCR结果显示,IRE1和XBP1的mRNA表达水平并未下调,而且显著上调,可能是由于miR-142-5p和miR-199a-5p同时激活相关信号通路从而上调IRE1和XBP1表达,如miR-142-5p同时能够激活UPR的ATF6信号传导通路,ATF6可以协同IRE1信号传导通路,促进XBP1的mRNA转录[30]。研究表明,miR-30d、miR-181a和miR-199a-5p三条miRNAs共转录能够明显降低GRP78表达,三条miRNAs的联合作用可能是实现GRP78显著下调的关键[18]。本研究中双荧光素酶报告系统结果显示,单独miR-199a-5p并不靶向猪GRP78 3′UTR。据此推测可能需要多个miRNAs才能有效调控GRP78的表达。
以上结果表明,miR-142-5p可能靶向ATF6达到调控内质网应激反应的作用。
4 结论
本研究通过miRNA测序发现,伪狂犬病毒感染诱导miR-142-5p表达上调,同时,miR-142-5p通过降解mRNA的方式靶向猪ATF6,因此推测miR-142-5p通过靶向ATF6调控伪狂犬病毒增殖。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1194/jlr.R800049-JLR200URL [本文引用: 1]
URLPMID:23637286 [本文引用: 1]
DOI:10.1038/nature17041URLPMID:26791723 [本文引用: 1]

In eukaryotic cells, the endoplasmic reticulum is essential for the folding and trafficking of proteins that enter the secretory pathway. Environmental insults or increased protein synthesis often lead to protein misfolding in the organelle, the accumulation of misfolded or unfolded proteins - known as endoplasmic reticulum stress - and the activation of the adaptive unfolded protein response to restore homeostasis. If protein misfolding is not resolved, cells die. Endoplasmic reticulum stress and activation of the unfolded protein response help to determine cell fate and function. Furthermore, endoplasmic reticulum stress contributes to the aetiology of many human diseases.
URLPMID:32725819 [本文引用: 1]
[本文引用: 1]
DOI:10.1126/science.1209038URLPMID:22116877 [本文引用: 1]

The vast majority of proteins that a cell secretes or displays on its surface first enter the endoplasmic reticulum (ER), where they fold and assemble. Only properly assembled proteins advance from the ER to the cell surface. To ascertain fidelity in protein folding, cells regulate the protein-folding capacity in the ER according to need. The ER responds to the burden of unfolded proteins in its lumen (ER stress) by activating intracellular signal transduction pathways, collectively termed the unfolded protein response (UPR). Together, at least three mechanistically distinct branches of the UPR regulate the expression of numerous genes that maintain homeostasis in the ER or induce apoptosis if ER stress remains unmitigated. Recent advances shed light on mechanistic complexities and on the role of the UPR in numerous diseases.
URLPMID:10854322 [本文引用: 1]
DOI:10.1016/j.molcel.2017.06.017URLPMID:29107536 [本文引用: 1]

The secretory capacity of a cell is constantly challenged by physiological demands and pathological perturbations. To adjust and match the protein-folding capacity of the endoplasmic reticulum (ER) to changing secretory needs, cells employ a dynamic intracellular signaling pathway known as the unfolded protein response (UPR). Homeostatic activation of the UPR enforces adaptive programs that modulate and augment key aspects of the entire secretory pathway, whereas maladaptive UPR outputs trigger apoptosis. Here, we discuss recent advances into how the UPR integrates information about the intensity and duration of ER stress stimuli in order to control cell fate. These findings are timely and significant because they inform an evolving mechanistic understanding of a wide variety of human diseases, including diabetes mellitus, neurodegeneration, and cancer, thus opening up the potential for new therapeutic modalities to treat these diverse diseases.
DOI:10.1128/mcb.23.21.7448-7459.2003URLPMID:14559994 [本文引用: 1]

The mammalian unfolded protein response (UPR) protects the cell against the stress of misfolded proteins in the endoplasmic reticulum (ER). We have investigated here the contribution of the UPR transcription factors XBP-1, ATF6alpha, and ATF6beta to UPR target gene expression. Gene profiling of cell lines lacking these factors yielded several XBP-1-dependent UPR target genes, all of which appear to act in the ER. These included the DnaJ/Hsp40-like genes, p58(IPK), ERdj4, and HEDJ, as well as EDEM, protein disulfide isomerase-P5, and ribosome-associated membrane protein 4 (RAMP4), whereas expression of BiP was only modestly dependent on XBP-1. Surprisingly, given previous reports that enforced expression of ATF6alpha induced a subset of UPR target genes, cells deficient in ATF6alpha, ATF6beta, or both had minimal defects in upregulating UPR target genes by gene profiling analysis, suggesting the presence of compensatory mechanism(s) for ATF6 in the UPR. Since cells lacking both XBP-1 and ATF6alpha had significantly impaired induction of select UPR target genes and ERSE reporter activation, XBP-1 and ATF6alpha may serve partially redundant functions. No UPR target genes that required ATF6beta were identified, nor, in contrast to XBP-1 and ATF6alpha, did the activity of the UPRE or ERSE promoters require ATF6beta, suggesting a minor role for it during the UPR. Collectively, these results suggest that the IRE1/XBP-1 pathway is required for efficient protein folding, maturation, and degradation in the ER and imply the existence of subsets of UPR target genes as defined by their dependence on XBP-1. Further, our observations suggest the existence of additional, as-yet-unknown, key regulators of the UPR.
[本文引用: 1]
[本文引用: 1]
URLPMID:17135238 [本文引用: 1]
URLPMID:27222391 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1631/jzus.B1600208URLPMID:28378569 [本文引用: 1]

Porcine circovirus type 2 (PCV2) has recently been reported to elicit the unfolded protein response (UPR) via activation of the PERK/eIF2alpha (RNA-activated protein kinase-like endoplasmic reticulum (ER) kinase/eukaryotic initiation factor 2alpha) pathway. This study attempted to examine which viral protein might be involved in inducing UPR and whether this cellular event would lead to apoptosis of the cells expressing the viral protein. By transient expression, we found that both replicase (Rep) and capsid (Cap) proteins of PCV2 could induce ER stress as shown by increased phosphorylation of PERK with subsequent activation of the eIF2alpha-ATF4 (activating transcription factor 4)-CHOP (CCAAT/enhancer-binding protein homologous protein) axis. Cap expression, but not Rep, significantly reduced anti-apoptotic B-cell lymphoma-2 (Bcl-2) and increased caspase-3 cleavage, possibly due to increased expression of CHOP. Since knockdown of PERK by RNA interference clearly reduced Cap-induced CHOP expression, caspase-3 cleavage, and apoptotic cell death possibly by partially rescuing Bcl-2 expression, we propose that there is connection between Cap-induced UPR and apoptosis via the PERK/eIF2alpha/ATF4/CHOP/Bcl-2 pathway. This study, together with our earlier studies, provides insight into the mechanisms underlying PCV2 pathogenesis.
DOI:10.3389/fmicb.2017.02129URLPMID:29163417 [本文引用: 1]

Classical swine fever (CSF) is an OIE-listed, highly contagious animal disease caused by classical swine fever virus (CSFV). The endoplasmic reticulum (ER) is an organelle in which the replication of many RNA viruses takes place. During viral infection, a series of events elicited in cells can destroy the ER homeostasis that cause ER stress and induce an unfolded protein response (UPR). In this study, we demonstrate that ER stress was induced during CSFV infection as several UPR-responsive elements such as XBP1(s), GRP78 and CHOP were up-regulated. Specifically, CSFV transiently activated IRE1 pathway at the initial stage of infection but rapidly switched off, likely due to the reduction in cytoplasm Ca(2+) after viral incubation. Additionally, our data show that the ER stress induced by CSFV can promote CSFV production, which the IRE1 pathway play an important role in it. Evidence of ER stress in vivo was also confirmed by the marked elevation of GRP78 in CSFV-infected pig PBMC and tissues. Collectively, these data indicate that the ER stress was induced upon CSFV infection and that the activation of the IRE1 pathway benefits CSFV replication.
DOI:10.1038/srep45922URLPMID:28374783 [本文引用: 1]

Pseudorabies virus (PRV) is one of the most important pathogens of swine, resulting in severe economic losses to the pig industry. To improve our understanding of the host responses to PRV infection, we applied isobaric tags for relative and absolute quantification (iTRAQ) labeling coupled with liquid chromatography-tandem mass spectrometry to quantitatively identify the differentially expressed cellular proteins in PRV-infected PK15 cells. In total, relative quantitative data were identified for 4333 proteins in PRV and mock- infected PK15 cells, among which 466 cellular proteins were differentially expressed, including 234 upregulated proteins and 232 downregulated proteins. Bioinformatics analysis disclosed that most of these differentially expressed proteins were involved in metabolic processes, cellular growth and proliferation, endoplasmic reticulum (ER) stress response, cell adhesion and cytoskeleton. Moreover, expression levels of four representative proteins, beta-catenin, STAT1, GRB2 and PCNA, were further confirmed by western blot analysis. This is the first attempt to analyze the protein profile of PRV-infected PK15 cells using iTRAQ technology, and our findings may provide valuable information to help understand the host response to PRV infection.
URLPMID:31767081 [本文引用: 1]
URLPMID:23085757 [本文引用: 3]
URLPMID:26542291 [本文引用: 3]
DOI:10.1016/j.gene.2013.09.005URLPMID:24035931 [本文引用: 2]

MicroRNAs (miRNAs) are a group of endogenous, small, noncoding RNAs that function as key post-transcriptional regulators. miRNAs are involved in many biological processes including apoptosis. In this study, mouse miR-702 (mmu-miR-702), a mirtron derived from the 13th intron of the Plod3 gene, was identified as a regulator of anti-apoptosis. mmu-miR-702 was down-regulated after treatment with the apoptosis-inducer isoproterenol both in vivo and in vitro. According to over-expression experiments, mmu-miR-702 inhibited apoptosis as well as the expression levels of a subset of apoptosis-related genes including activating transcription factor 6 (ATF6). An interaction between mmu-miR-702 and the ATF6 3'-UTR binding site was confirmed using luciferase reporter and western blot assays. This is the first report of ATF6 interaction with miRNA. Although the possible existence of miR-702 in the human genome is low, our results indicate that mirtrons also participate in the process of apoptosis and may provide a novel study strategy for apoptosis.
URLPMID:21750653 [本文引用: 2]
URLPMID:22431749 [本文引用: 2]
URLPMID:26998839 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:27621275 [本文引用: 1]
URLPMID:21057845 [本文引用: 1]
URLPMID:32742688 [本文引用: 1]
URLPMID:28363780 [本文引用: 1]
DOI:10.1038/srep18304URL [本文引用: 1]
DOI:10.1247/csf.07044URLPMID:18360008 [本文引用: 1]

Eukaryotic cells cope with endoplasmic reticulum (ER) stress by activating the unfolded protein response (UPR), a coordinated system of transcriptional and translational controls, which ensures the integrity of synthesized proteins. Mammalian cells express three UPR transducers in the ER, namely IRE1, PERK and ATF6. The IRE1 pathway, which is conserved from yeast to humans, mediates transcriptional induction of not only ER quality control proteins (molecular chaperones, folding enzymes and components of ER-associated degradation) but also proteins working at various stages of secretion. The PERK pathway, conserved in metazoan cells, is responsible for translational control and also participates in transcriptional control in mammals. ATF6 is an ER-membrane-bound transcription factor activated by ER stress-induced proteolysis which consists of two closely related factors, ATF6alpha and ATF6beta, in mammals. ATF6alpha but not ATF6beta plays an important role in transcriptional control. In this study, we performed a genome-wide search for ATF6alpha-target genes in mice. Only 30 of the 14,729 analyzable genes were identified as specific targets, of which 40% were ER quality control proteins, 20% were ER proteins, while the rest had miscellaneous functions. The negative effects of the absence of PERK on transcriptional induction of ER quality control proteins could be explained by its inhibitory effect on ATF6alpha activation. Further, proteins involved in transport from the ER are not regulated by ATF6alpha, and transport of folded cargo molecules from the ER was not affected by the absence of ATF6alpha. Based on these results, we propose that ATF6 is a transcription factor specialized in the regulation of ER quality control proteins.
