 ,中国农业科学院果树研究所/农业部园艺作物种质资源利用重点实验室/辽宁省落叶果树矿质营养与肥料高效利用重点实验室,辽宁兴城 125100
,中国农业科学院果树研究所/农业部园艺作物种质资源利用重点实验室/辽宁省落叶果树矿质营养与肥料高效利用重点实验室,辽宁兴城 125100Molecular Cloning and Functional Characterization of VcNAC072 Reveals Its Involvement in Anthocyanin Accumulation in Blueberry
SONG Yang, LIU HongDi, WANG HaiBo, ZHANG HongJun, LIU FengZhi ,Research Institute of Pomology, Chinese Academy of Agricultural Sciences/Key Laboratory of Fruit Germplasm Resources Utilization, Ministry of Agriculture/Laboratory of Mineral Nutrition and Efficient Fertilization for Deciduous Fruits, Liaoning Province, Xingcheng 125100, Liaoning
,Research Institute of Pomology, Chinese Academy of Agricultural Sciences/Key Laboratory of Fruit Germplasm Resources Utilization, Ministry of Agriculture/Laboratory of Mineral Nutrition and Efficient Fertilization for Deciduous Fruits, Liaoning Province, Xingcheng 125100, Liaoning通讯作者:
收稿日期:2018-08-27接受日期:2018-09-28网络出版日期:2019-02-13
| 基金资助: |
Received:2018-08-27Accepted:2018-09-28Online:2019-02-13
| Fund supported: |
作者简介 About authors
宋杨,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2328KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
宋杨, 刘红弟, 王海波, 张红军, 刘凤之. 越橘VcNAC072克隆及其促进花青素积累的功能分析[J]. 中国农业科学, 2019, 52(3): 503-511 doi:10.3864/j.issn.0578-1752.2019.03.010
SONG Yang, LIU HongDi, WANG HaiBo, ZHANG HongJun, LIU FengZhi.
0 引言
【研究意义】越橘亦称蓝莓,作为一种新兴的果树作物之一,近年来其栽培面积和产量稳步增长[1,2]。越橘果实中富含花青素、黄酮醇等次生代谢产物,在提高植物抗逆性和人体免疫力等方面具有一定作用[3,4]。通过分子生物学方法挖掘优异基因,阐释果实花青素积累的调控机理,对越橘新品种选育和品质改良具有重要意义。【前人研究进展】NAC(NAM,ATAF1/2,CUC2)转录因子是一类植物中特有的转录因子,其家族成员最显著的结构特征是其编码蛋白质的N端含有约150个氨基酸的NAC保守结构域。研究表明,NAC转录因子的生物学功能涉及广泛,包括生长发育[5]、信号转导[6]、非生物胁迫[7,8]和次生代谢产物合成[9,10,11]等方面。ODA-YAMAMIZO等[12]发现,拟南芥AtNAC046正调控叶绿素降解和叶片衰老,过量表达AtNAC046使植株表现早衰和叶绿素含量降低。TAKASAKI等[13]也发现,7个拟南芥NAC转录因子均表现受ABA诱导表达并调控叶片衰老。玉米Calcium/calmodulin-dependent protein kinase(CCaMK)可通过磷酸化ZmNAC84的丝氨酸位点,调控植株由ABA介导的抗氧化性[14]。在调控次生代谢物质积累方面,最近研究发现红肉桃PpNAC1可通过激活PpMYB10.1的表达促进果肉中花青素积累[15]。在苹果中也发现MdNAC029可通过激活MdMYB1的转录,促进愈伤组织中花青素合成[16]。研究表明,NAC蛋白可通过结合靶基因的CACG或GATTGGAT(AT)CA位点调控靶基因的表达,从而影响植物的抗性、衰老和次生代谢物质合成[7-8,16]。【本研究切入点】NAC转录因子的研究主要集中在水稻[17]和拟南芥[18]等模式植物。目前,越橘中NAC转录因子的研究尚无报道。【拟解决的关键问题】从‘公爵’越橘中分离并鉴定一个NAC转录因子VcNAC072,通过表达模式分析、转基因分析、酵母单杂交及瞬时表达试验,探讨VcNAC072在调控越橘花青素积累过程中的作用,为揭示NAC转录因子调控花青素积累提供理论基础。1 材料与方法
试验于2016年6月—2018年7月在中国农业科学院果树研究所农业部园艺作物种质资源利用重点实验室和山东农业大学作物生物学国家重点实验室进行。1.1 试验材料
试验所用的植物材料为8年生越橘品种‘公爵’(Vaccinium corymbosum ‘Duke’)、野生型拟南芥(Arabidopsis thaliana)和本氏烟草(Nicotiana benthamiana)。1.2 基因克隆和序列分析
从越橘转录组数据库[19]查找NAC基因,并从中筛选发现VcNAC072高表达且在果实成熟过程中呈持续上调表达。根据筛选到的序列设计引物VcNAC072- F/R扩增CDS(coding domain sequence)序列。以越橘果实的cDNA为模板进行PCR扩增。PCR反应程序为:98℃预变性3 min;98℃变性10 s,57℃退火30 s,72℃延伸2 min,30个循环;72℃延伸10 min。PCR产物用1.2%琼脂糖凝胶电泳并回收目的条带,连接到克隆载体pEAST blunt zero进行测序。所用的引物序列见表1。Table 1
表1
表1本研究中使用的引物
Table 1
| 用途 Use | 引物名称Primer name | 序列(5′-3′)Primer sequence (5′-3′) |
|---|---|---|
| 构建pRI101-VcNAC072表达载体 The construction of pRI101-VcNAC072 expression vector 构建pGADT7-VcNAC072表达载体 The construction of pGADT7-VcNAC072 expression vector 构建pAbAi-AtPAP1pro表达载体 The construction of pAbAi-AtPAP1pro expression vector 构建AtPAP1pro-GUS表达载体 The construction of AtPAP1pro-GUS expression vector 实时荧光定量qRT-PCR Real time fluorescent quantitative PCR | VcNAC072(pRI101)-F VcNAC072(pRI101)-R VcNAC072( pGADT7)-F VcNAC072( pGADT7)-R AtPAP1pro(pAbAi)-F AtPAP1pro(pAbAi)-R AtPAP1pro(GUS)-F AtPAP1pro(GUS)-R VcNAC072-(RT-PCR)-F VcNAC072-(RT-PCR)-R VcGAPDH-(RT-PCR)-F VcGAPDH-(RT-PCR)-R AtUBQ10-(RT-PCR)-F AtUBQ10-(RT-PCR)-R AtPAP1-(RT-PCR)-F AtPAP1-(RT-PCR)-R AtDFR-(RT-PCR)-F AtDFR-(RT-PCR)-R AtANS-(RT-PCR)-F AtANS-(RT-PCR)-R | GGAATTCCATATGATGGGAGTTCAGGAGTCCG CGCGGATCCTCACTGCCGAAACCCGAATCC GGAATTCCATATGATGGGAGTTCAGGAGTCCG CGCGGATCCTCACTGCCGAAACCCGAATCC CGAGCTCCTAATACATAAAATGTGGATATC ACGCGTCGACGGAACAAAGATAGATACGTAA CGGGGTACCCTAATACATAAAATGTGGATATC CATGCCATGGGGAACAAAGATAGATACGTAA GATGAGGTACAATGACATG GAGTTCTGCTGAAACAAC GGTTATCAATGATAGGTTTGGCA CAGTCCTTGCTTGATGGACC CGTTAAGACGTTGACTGGGAAAACT GCTTTCACGTTATCAATGGTGTCA GCTCTGATGAAGTCGATCTTC CTACCTCTTGGCTTTCCTCT GTCGGTCCATTCATCACAAC TGAGCGTTGCATAAGTCGTC TCAAGAAAGCCGGAGAAGAG TTGTCCACTCGCGTTGTTAG |
新窗口打开|下载CSV
利用软件CLC Sequence Viewer 6(
1.3 总RNA提取与实时荧光定量PCR分析
总RNA提取采用TaKaRa公司的植物总RNA提取试剂盒(Code No.9769,Takara,Dalian,China)。以总RNA为模板,反转录合成cDNA。在越橘中,qRT-PCR内参基因为VcGAPDH。在拟南芥中,以AtUBQ10作为内参基因。仪器为Bio-Rad公司的CFX Connect PCR system,试剂为ThermoFisher公司的PowerUpTM SYBR Green Master Mix(Code No.A25742,Thermo Fisher, China)。反应体系:SYBR Mixture 10.0 μL,cDNA 2.0 μL,上、下游引物各0.5 μL,加去离子水至20 μL。PCR反应程序:95℃预变性2 min,95℃变性15 s,58℃退火15 s,72℃延伸1 min,40个循环,每次循环第2步进行荧光采集。最后采用2-ΔΔCT法分析定量数据。所有PCR都设3次重复。实时荧光定量PCR引物见表1。1.4 基因转化拟南芥
构建pRI101-VcNAC072过量表达载体,并将其转化农杆菌GV3101,利用农杆菌侵染花序法转化野生型拟南芥。在含有卡那霉素的MS固体培养基上筛选T1代转基因植株。将抗性苗移栽至基质中并放入光照培养箱中进行培养,收获的T2代种子用于试验。载体构建时使用的引物见表1。1.5 酵母单杂交试验(Y1H)
Y1H具体试验方法参照Clontech说明书,设计引物扩增VcNAC072的CDS序列,重组至pGADT7酵母表达载体,生成pGADT7-VcNAC072。扩增拟南芥中调控花青素合成的MYB转录因子AtPAP1的启动子序列,连接至pAbAi酵母表达载体,生成pAbAi-AtPAP1pro。将pAbAi-AtPAP1pro转化至酵母菌株Y1H Gold中,转化后涂布在氨基酸缺陷培养基SD/-Ura上,并筛选AbA(Aureobasidin A)浓度。再将pGADT7-VcNAC072转化至重组酵母菌株中,转化后涂布在含有AbA的氨基酸缺陷培养基SD/-Leu上进行互作筛选。1.6 烟草瞬时表达试验
具体试验方法参照YIN等[20]和JEFFERSON等[21]。把AtPAP1(GenBank登录号:NM_104541)的启动子序列重组至pCAMBIA1301-GUS载体,生成AtPAP1pro-GUS,转化农杆菌GV3101,再与转化农杆菌的pRI101-VcNAC072共注射烟草叶片。使用荧光分光光度计测定GUS活性。1.7 花青素含量测定
参照PERTUZATTI等[22]方法,利用高效液相色谱-质谱法对花青素总量进行测定。仪器为岛津LP- 10Avp液相色谱仪,SPD-M10Avp二极管阵列检测器。根据色谱峰面积计算花青素总量。1.8 统计学分析
使用SPSS软件进行差异显著性分析。不同字母代表差异显著(P<0.05)。2 结果
2.1 VcNAC072的克隆、保守结构域和系统发生分析
通过RT-PCR技术获得1条大约1 000 bp的条带。对克隆所得片段测序分析,结果显示,VcNAC072的CDS长度为1 032 bp,编码含有343个氨基酸的蛋白质。使用CLC软件分析VcNAC072及其他植物NAC蛋白的保守结构域。结果表明,VcNAC072含有保守的NAC结构域(图1)。将VcNAC072蛋白序列与多个拟南芥NAC蛋白序列进行系统发生分析,以及NCBI序列比对分析发现,VcNAC072与拟南芥ANAC072同源性最高(图2)。因此将该基因命名为VcNAC072(GenBank登录号:MH784502)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1VcNAC072与其他植物NAC蛋白的多序列比较黑框区表示NAC结构域。VcNAC072:越橘 MH784502;VvNAC:葡萄 XP_002284668;AcNAC:中华猕猴桃 PSS06314.1;ANAC072:拟南芥 AT4G27410 The black frame indicated the NAC domain. VcNAC072: Vaccinium corymbosum MH784502; VvNAC: Vitis vinifera XP_002284668; AcNAC: Actinidia chinensis PSS06314.1; Arabidopsis thaliana AT4G27410
Fig. 1Multiple sequence alignment of VcNAC072 and NAC protein from other plants
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2VcNAC072与拟南芥NAC蛋白系统发生分析
Fig. 2Phylogenetic relationships of VcNAC072 of blueberry and NAC protein of Arabidopsis
2.2 VcNAC072的表达
在果实不同发育时期,VcNAC072的相对表达量与花青素含量变化见图3。结果发现,VcNAC072在绿果中表达量最低,随着果实成熟,表达量持续升高,在蓝果中最大。总花青素含量变化趋势与VcNAC072的变化相似,这暗示VcNAC072的持续上调表达可能促进了花青素积累。图3
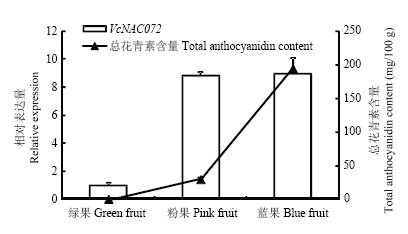 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3果实发育不同阶段VcNAC072表达和花青素含量变化
Fig. 3The change of relative expression of VcNAC072 and anthocyanidin content during fruit development
2.3 异位表达VcNAC072对花青素积累的影响
VcNAC072转基因株系(L1和L2)的种子中明显积累了花青素(图4-A),其花青素含量均显著高于野生型,分别为野生型的38.99倍和32.90倍(图4-B)。qRT-PCR检测花青素合成相关基因的表达,结果发现,过量表达VcNAC072显著促进了AtPAP1及花青素合成基因AtDFR、AtANS的表达(图4-C)。图4
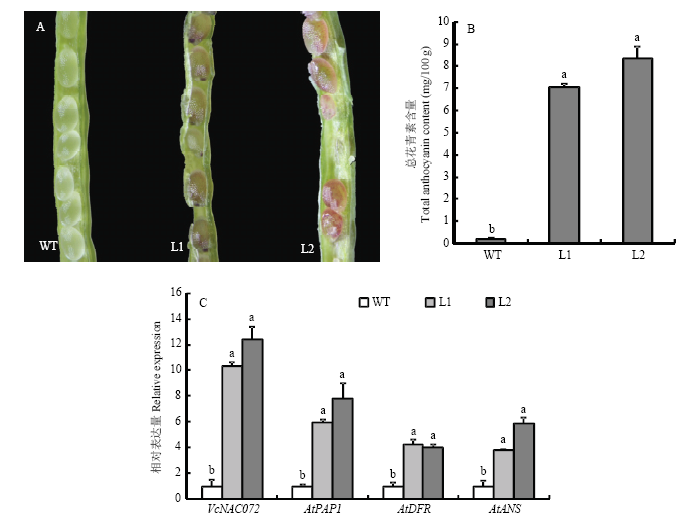 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4VcNAC072在拟南芥中过量表达对花青素积累的影响
A:转基因(L1和L2)和野生型(WT)拟南芥种子中花青素积累情况;B:转基因和野生型拟南芥种子中花青素含量;C:qRT-PCR检测花青素合成相关基因的表达,其在WT中的表达量设为1。不同小写字母表示差异显著(P<0.05)。下同
Fig. 4Effects of VcNAC072 over-expression on anthocyanin accumulation in Arabidopsis
A: Phenotypes of transgenic VcNAC072 and wild-type Arabidopsis on the anthocyanin accumulation; B: Detection of the anthocyanin content; C: qRT-PCR analysis of the expression of anthocyanin biosynthesis genes, the value for WT was set to 1. Different lowercase letters indicate significant differences (P<0.05). The same as below
2.4 VcNAC072对AtPAP1启动子的调控
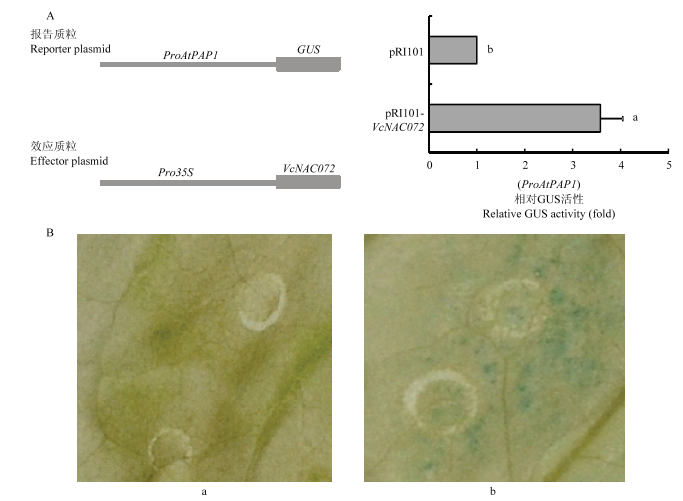
VcNAC072可与AtPAP1的启动子互作(图5)。为进一步研究VcNAC072对AtPAP1的调控方式,构建了pRI101-VcNAC072和AtPAP1pro-GUS载体,在烟草中瞬时表达。结果显示,共转VcNAC072和AtPAP1启动子的烟草叶片中,其GUS相对活性显著高于对照,为对照的3.57倍,说明VcNAC072可激活AtPAP1的表达(图6)。图5
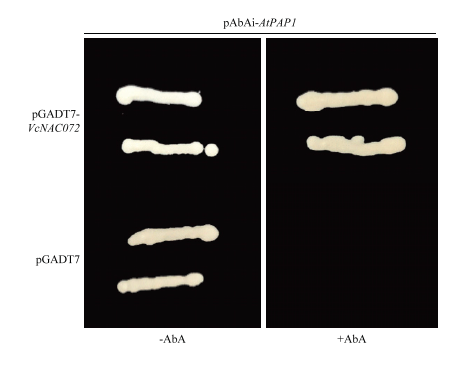 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5酵母单杂交检测VcNAC072与AtPAP1启动子的相互作用
Fig. 5Y1H assay showed the interactions of VcNAC072 with promoter of AtPAP1
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6瞬时表达分析VcNAC072对AtPAP1表达的影响
A:相对GUS活性。B:瞬时表达试验。a:共注射pRI101空载体和AtPAP1pro-GUS;b:共注射pRI101-VcNAC072和AtPAP1pro-GUS
Fig. 6The effect of VcNAC072 on the gene expression of AtPAP1 by transient expression assays
A: Quantitative analysis of relative GUS activity. B: Transient expression assay. a: pRI101 empty vector and AtPAP1pro-GUS. b: pRI101-VcNAC072 and AtPAP1pro-GUS
3 讨论
目前,在拟南芥中已鉴定了117个NAC转录因子[18]。其中,AtNAC072在植物生长发育和胁迫方面发挥重要作用。例如,AtNAC072正调控植物叶绿素降解,促使叶片衰老[23,24]。AtNAC072被发现能够响应病原菌侵染[25]。AtNAC072还可正调控植物的干旱和高盐胁迫响应[26],但NAC072在调控花青素积累过程中的分子机理还不清楚。本研究利用前期转录组测序数据,从越橘中分离出1个编码NAC蛋白的转录因子VcNAC072,该基因在花青素含量最高的蓝果中表达量最高,并通过转基因试验验证了在野生型拟南芥中过量表达VcNAC072促进花青素积累。LI等[27]和SUN等[28]也利用转录测序技术,在越橘中挖掘出多个与果实中花青素积累紧密相关的花青素合成结构基因,如二氢黄酮醇-4-还原酶(Dihydroflavonol-4-Reductase,DFR)和花青素合成酶(Anthocyanin synthase,ANS)等。在拟南芥、苹果和梨等多种植物中,MYB转录因子在调节花青素合成过程中起关键作用。MYB可通过调控DFR、ANS等花青素合成结构基因的表达影响花青素合成[29,30,31]。JAAKOLA等[32]的研究也认为欧洲越橘MADS-box转录因子VmTDR4可直接或间接调控MYB转录因子的表达,进而调控DFR、ANS等基因的转录,从而影响幼嫩果实中种子和胎座中花青素的积累。
在本研究中,对转基因野生型拟南芥进行基因表达分析发现,异位表达VcNAC072可显著促进MYB转录因子AtPAP1及花青素合成结构基因的表达。NAC转录因子可通过与其靶基因上的CACG位点结合,调控靶基因的表达[33,34]。本研究通过Y1H技术验证了VcNAC072能够与包含CACG位点的AtPAP1启动子互作,并通过瞬时表达试验发现VcNAC072可激活AtPAP1的表达。后期将利用凝胶电泳迁移率试验(EMSA),进一步验证VcNAC072对AtPAP1启动子的结合。
4 结论
本研究克隆获得越橘NAC转录因子VcNAC072,该基因转化野生型拟南芥表现出促进花青素积累的表型。Y1H和瞬时表达试验表明VcNAC072可与MYB转录因子AtPAP1的启动子相结合并促进其表达。研究结果为揭示NAC转录因子调控花青素积累提供了参考。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1089/ars.2008.2150URLPMID:2933151 [本文引用: 1]

Endothelial cell neoplasms are the most common soft tissue tumor in infants. Subcutaneous injection of spontaneously transformed murine endothelial (EOMA) cells results in development of hemangioendothelioma (HE). We have previously shown that blueberry extract (BBE) treatment of EOMA cells in vitro prior to injection in vivo can significantly inhibit the incidence and size of developing HE. In this study, we sought to determine whether oral BBE could be effective in managing HE and to investigate the mechanisms through which BBE exerts its effects on endothelial cells. A dose-dependent decrease in HE tumor size was observed in mice receiving daily oral gavage feeds of BBE. Kaplan-Meier survival curve showed significantly enhanced survival for mice with HE tumors given BBE, compared to control. BBE treatment of EOMA cells inhibited both c-Jun N-terminal kinase (JNK) and NF-kappaB signaling pathways that culminate in monocyte chemoattractant protein-1 (MCP-1) expression required for HE development. Antiangiogenic effects of BBE on EOMA cells included decreased proliferation by BrdU assay, decreased sprouting on Matrigel, and decreased transwell migration. Thus, this work provides first evidence demonstrating that BBE can limit tumor formation through antiangiogenic effects and inhibition of JNK and NF-kappaB signaling pathways. Oral administration of BBE represents a potential therapeutic antiangiogenic strategy for treating endothelial cell neoplasms in children.
DOI:10.1111/j.1753-4887.2010.00273.xURLPMID:20384847 [本文引用: 1]

Berries are a good source of polyphenols, especially anthocyanins, micronutrients, and fiber. In epidemiological and clinical studies, these constituents have been associated with improved cardiovascular risk profiles. Human intervention studies using chokeberries, cranberries, blueberries, and strawberries (either fresh, or as juice, or freeze-dried), or purified anthocyanin extracts have demonstrated significant improvements in LDL oxidation, lipid peroxidation, total plasma antioxidant capacity, dyslipidemia, and glucose metabolism. Benefits were seen in healthy subjects and in those with existing metabolic risk factors. Underlying mechanisms for these beneficial effects are believed to include upregulation of endothelial nitric oxide synthase, decreased activities of carbohydrate digestive enzymes, decreased oxidative stress, and inhibition of inflammatory gene expression and foam cell formation. Though limited, these data support the recommendation of berries as an essential fruit group in a heart-healthy diet.
[本文引用: 1]
[本文引用: 1]
DOI:10.3389/fpls.2016.00004URLPMID:26834774 [本文引用: 2]

Abstract The NAC transcription factors play critical roles in regulating stress responses in plants. However, the functions for many of the NAC family members in rice are yet to be identified. In the present study, a novel stress-responsive rice NAC gene, ONAC022, was identified. Expression of ONAC022 was induced by drought, high salinity, and abscisic acid (ABA). The ONAC022 protein was found to bind specifically to a canonical NAC recognition cis-element sequence and showed transactivation activity at its C-terminus in yeast. The ONAC022 protein was localized to nucleus when transiently expressed in Nicotiana benthamiana. Three independent transgenic rice lines with overexpression of ONAC022 were generated and used to explore the function of ONAC022 in drought and salt stress tolerance. Under drought stress condition in greenhouse, soil-grown ONAC022-overexpressing (N22oe) transgenic rice plants showed an increased drought tolerance, leading to higher survival ratios and better growth than wild-type (WT) plants. When grown hydroponically in Hogland solution supplemented with 150 mM NaCl, the N22oe plants displayed an enhanced salt tolerance and accumulated less Na(+) in roots and shoots as compared to WT plants. Under drought stress condition, the N22oe plants exhibited decreased rates of water loss and transpiration, reduced percentage of open stomata and increased contents of proline and soluble sugars. However, the N22oe lines showed increased sensitivity to exogenous ABA at seed germination and seedling growth stages but contained higher level of endogenous ABA. Expression of some ABA biosynthetic genes (OsNCEDs and OsPSY), signaling and regulatory genes (OsPP2C02, OsPP2C49, OsPP2C68, OsbZIP23, OsAP37, OsDREB2a, and OsMYB2), and late stress-responsive genes (OsRAB21, OsLEA3, and OsP5CS1) was upregulated in N22oe plants. Our data demonstrate that ONAC022 functions as a stress-responsive NAC with transcriptional activator activity and plays a positive role in drought and salt stress tolerance through modulating an ABA-mediated pathway.
DOI:10.1007/s10725-015-0124-0URL [本文引用: 2]

<span >Environmental stresses such as drought, salinity, and cold are major factors that significantly limit <span >agricultural productivity. NAC is a plant-specific transcription factor family, which plays essential roles in <span >response to various abiotic stresses. We have identified a <span >novel NAC gene <span >CarNAC5 <span >from chickpea in a previous <span >study, which was induced by drought stress. In the present <span >study, the promoter region of <span >CarNAC5 <span >was isolated. In <span >silico analysis indicated that many basic <span >cis<span >-acting elements, which respond to environmental stresses and plant <span >hormones, were harbored in <span >CarNAC5 <span >promoter region.<span >EMSA and Yeast hybrid assays revealed that CarNAC5 <span >could bind to the core DNA sequence CGT[G/A]. Overexpression of <span >CarNAC5 <span >enhanced drought tolerance in <span >transgenic <span >Ar<span >abidopsis plants, which was simultaneously <span >demonstrated by the enhanced expression of abiotic stressresponsive genes and changes of several physiological <span >indices. Our results indicated that <span >CarNAC5 <span >has potential <span >for utilization in transgenic breeding to improve abiotic <span >stress tolerances in crops.
DOI:10.1111/j.1365-313X.2007.03109.xURLPMID:17565617 [本文引用: 1]

Vascular plants evolved to have xylem that provides physical support for their growing body and serves as a conduit for water and nutrient transport. In a previous study, we used comparative-transcriptome analyses to select a group of genes that were upregulated in xylem of Arabidopsis plants undergoing secondary growth. Subsequent analyses identified a plant-specific NAC-domain transcription factor gene ( ANAC012 ) as a candidate for genetic regulation of xylem formation. Promoter-GUS analyses showed that ANAC012 expression was preferentially localized in the (pro)cambium region of inflorescence stem and root. Using yeast transactivation analyses, we confirmed the function of ANAC012 as a transcriptional activator, and identified an activation domain in the C terminus. Ectopic overexpression of ANAC012 in Arabidopsis ( 35S::ANAC012 plants) dramatically suppressed secondary wall deposition in the xylary fiber and slightly increased cell-wall thickness in the xylem vessels. Cellulose compositions of the cell wall were decreased in the inflorescent stems and roots of 35S::ANAC012 plants, probably resulting from defects in xylary fiber formation. Our data suggest that ANAC012 may act as a negative regulator of secondary wall thickening in xylary fibers.
DOI:10.1105/tpc.106.047399URLPMID:17114348 [本文引用: 1]

Secondary walls in fibers and tracheary elements constitute the most abundant biomass produced by plants. Although a number of genes involved in the biosynthesis of secondary wall components have been characterized, little is known about the molecular mechanisms underlying the coordinated expression of these genes. Here, we demonstrate that the Arabi-dopsis thaliana NAC (for NAM, ATAF1/2, and CUC2) domain transcription factor, SND1 (for secondary wall-associated NAC domain protein), is a key transcriptional switch regulating secondary wall synthesis in fibers. We show that SND1 is expressed specifically in interfascicular fibers and xylary fibers in stems and that dominant repression of SND1 causes a drastic reduction in the secondary wall thickening of fibers. Ectopic overexpression of SND1 results in activation of the expression of secondary wall biosynthetic genes, leading to massive deposition of secondary walls in cells that are normally nonsclerenchymatous. In addition, we have found that SND1 upregulates the expression of several transcription factors that are highly expressed in fibers during secondary wall synthesis. Together, our results reveal that SND1 is a key transcriptional activator involved in secondary wall biosynthesis in fibers.
DOI:10.1111/j.1365-313X.2008.03533.xURLPMID:18445131 [本文引用: 1]

The Arabidopsis thaliana NAC domain transcription factor, VASCULAR-RELATED NAC-DOMAIN7 (VND7), plays a pivotal role in regulating the differentiation of root protoxylem vessels. In order to understand the mechanisms underscoring the function of VND7 in vessel differentiation in more detail, we conducted extensive molecular analyses in yeast ( Saccharomyces cerevisiae ), Arabidopsis, and Nicotiana tabacum L. cv. Bright Yellow 2 (tobacco BY-2) cells. The transcriptional activation activity of VND7 was confirmed in yeast and Arabidopsis, and the C-terminal region was shown to be required for VND7 transcriptional activation. Expression of the C-terminus-truncated VND7 protein under the control of the native VND7 promoter resulted in inhibition of the normal development of metaxylem vessels in roots and vessels in aerial organs, as well as protoxylem vessels in roots. The expression pattern of VND7 overlapped that of VND2 to VND5 in most of the differentiating vessels. Furthermore, a yeast two-hybrid assay revealed the ability of VND7 to form homodimers and heterodimers with other VND proteins via their N-termini, which include the NAC domain. The heterologous expression of VND7 in tobacco BY-2 cells demonstrated that the stability of VND7 could be regulated by proteasome-mediated degradation. Together these data suggest that VND7 regulates the differentiation of all types of vessels in roots and shoots, possibly in cooperation with VND2 to VND5 and other regulatory proteins.
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
DOI:10.1111/tpj.12792URLPMID:25688923 [本文引用: 1]

Summary Anthocyanin pigmentation is an important consumer trait in peach ( Prunus persica ). In this study, the genetic basis of the blood-flesh trait was investigated using the cultivar Dahongpao, which shows high levels of cyanidin-3-glucoside in the mesocarp. Elevation of anthocyanin levels in the flesh was correlated with the expression of an R2R3 MYB transcription factor, PpMYB10.1 . However, PpMYB10.1 did not co-segregate with the blood-flesh trait. The blood-flesh trait was mapped to a 200-kb interval on peach linkage group (LG) 5. Within this interval, a gene encoding a NAC domain transcription factor (TF) was found to be highly up-regulated in blood-fleshed peaches when compared with non-red-fleshed peaches. This NAC TF, designated BLOOD (BL), acts as a heterodimer with PpNAC1 which shows high levels of expression in fruit at late developmental stages. We show that the heterodimer of BL and PpNAC1 can activate the transcription of PpMYB10.1 , resulting in anthocyanin pigmentation in tobacco. Furthermore, silencing the BL gene reduces anthocyanin pigmentation in blood-fleshed peaches. The transactivation activity of the BL-PpNAC1 heterodimer is repressed by a SQUAMOSA promoter-binding protein-like TF, PpSPL1. Low levels of PpMYB10.1 expression in fruit at early developmental stages is probably attributable to lower levels of expression of PpNAC1 plus the presence of high levels of repressors such as PpSPL1. We present a mechanism whereby BL is the key gene for the blood-flesh trait in peach via its activation of PpMYB10.1 in maturing fruit. Partner TFs such as basic helix 搇oop-helix proteins and NAC1 are required, as is the removal of transcriptional repressors.
[本文引用: 2]
[本文引用: 2]
DOI:10.1016/j.gene.2010.06.008URLPMID:20600702 [本文引用: 1]

We investigated 151 non-redundant NAC genes in rice and 117 in Arabidopsis. A complete overview of this gene family in rice is presented, including gene structures, phylogenies, genome localizations, and expression profiles. We also performed a comparative analysis of these genes in rice and Arabidopsis. Conserved amino acid residues and phylogeny construction using the NAC conserved domain sequence suggest that OsNAC gene family was classified broadly into two major groups (A and B) and sixteen subgroups in rice. We presented more specific phylogenetic analysis of OsNAC proteins based on the DNA-binding domain and known gene function, respectively. Loss of introns was observed in the segmental duplication. Homologous, paralogous, and orthologous searches of rice and Arabidopsis revealed that the major functional diversification within the NAC gene family predated the divergence of monocots and dicots. The chromosomal localizations of OsNAC genes indicated nine segmental duplication events involving 18 genes; 32 non-redundant OsNAC genes were involved in tandem duplications. Expression levels of this gene family were checked under various abiotic stresses (cold, drought, submergence, laid-down submergence, osmotic, salinity and hormone) and biotic stresses [infection with rice viruses such as RSV (rice stripe virus) and RTSV (rice tungro spherical virus)]. Biotic stresses are novel work and increase the possibilities for finding the best candidate genes. A preliminary search based on our microarray (22K and 44K) data suggested that more than 45 and 26 non-redundant genes in this family were upregulated in response to abiotic and biotic stresses, respectively. All of the genes were further investigated for their stress responsiveness by RT-PCR analysis. Six genes showed preferential expression under both biotic RSV and RTSV stress. Eleven genes were upregulated by at least three abiotic treatments. Our study provides a very useful reference for cloning and functional analysis of members of this gene family in rice.
DOI:10.1016/j.ygeno.2009.09.003URLPMID:19766710 [本文引用: 2]

Controlled proteolytic activation of membrane-bound transcription factors (MTFs) is recently emerging as a versatile way of rapid transcriptional responses to environmental changes in plants. Here, we report genome-scale identification of putative MTFs in the Arabidopsis and rice genomes. The Arabidopsis and rice genomes have at least 85 and 45 MTFs, respectively, in virtually all major transcription factor families. Of particular interest is the NAC MTFs (designated NTLs): there are at least 18 NTLs in Arabidopsis and 5 NTL members (OsNTLs) in rice. While the full-size OsNTL forms are associated with the membranes, truncated forms lacking the transmembrane domains are detected exclusively in the nucleus. Furthermore, transcript levels of the OsNTL genes were elevated after treatments with abiotic stresses, supporting their roles in plant stress responses. We propose that membrane-mediated transcriptional control is a critical component of gene regulatory network that serves as an adaptive strategy under unfavorable growth conditions.
DOI:10.1016/S2095-3119(16)61461-2URL [本文引用: 1]
[本文引用: 1]
DOI:10.1089/dna.1987.6.583URLPMID:3327686 [本文引用: 1]

We have used the Escherichia coli beta-glucuronidase gene (GUS) as a gene fusion marker for analysis of gene expression in transformed plants. Higher plants tested lack intrinsic beta-glucuronidase activity, thus enhancing the sensitivity with which measurements can be made. We have constructed gene fusions using the cauliflower mosaic virus (CaMV) 35S promoter or the promoter from a gene encoding the small subunit of ribulose bisphosphate carboxylase (rbcS) to direct the expression of beta-glucuronidase in transformed plants. Expression of GUS can be measured accurately using fluorometric assays of very small amounts of transformed plant tissue. Plants expressing GUS are normal, healthy and fertile. GUS is very stable, and tissue extracts continue to show high levels of GUS activity after prolonged storage. Histochemical analysis has been used to demonstrate the localization of gene activity in cells and tissues of transformed plants.
[本文引用: 1]
DOI:10.1111/tpj.13030URLPMID:26407000 [本文引用: 1]

Summary Degreening caused by rapid chlorophyll (Chl) degradation is a characteristic event during green organ senescence or maturation. Pheophorbide a oxygenase gene (PAO) encodes a key enzyme of Chl degradation, yet its transcriptional regulation remains largely unknown. Using yeast one-hybrid screening, coupled with in vitro and in vivo assays, we revealed that Arabidopsis MYC2/3/4 basic helix-loop-helix proteins directly bind to PAO promoter. Overexpression of the MYCs significantly enhanced the transcriptional activity of PAO promoter in Arabidopsis protoplasts, and methyl jasmonate (MeJA) treatment greatly induced PAO expression in wild-type Arabidopsis plants, but the induction was abolished in myc2 myc3 myc4. In addition, MYC2/3/4 proteins could promote the expression of another Chl catabolic enzyme gene, NYC1, as well as a key regulatory gene of Chl degradation, NYE1/SGR1, by directly binding to their promoters. More importantly, the myc2 myc3 myc4 triple mutant showed a severe stay-green phenotype, whereas the lines overexpressing the MYCs showed accelerated leaf yellowing upon MeJA treatment. These results suggest that MYC2/3/4 proteins may mediate jasmonic acid (JA)-induced Chl degradation by directly activating these Chl catabolic genes (CCGs). Three NAC family proteins, ANAC019/055/072, downstream from MYC2/3/4 proteins, could also directly promote the expression of a similar set of CCGs (NYE1/SGR1, NYE2/SGR2 and NYC1) during Chl degradation. In particular, anac019 anac055 anac072 triple mutant displayed a severe stay-green phenotype after MeJA treatment. Finally, we revealed that MYC2 and ANAC019 may interact with each other and synergistically enhance NYE1 expression. Together, our study reveals a hierarchical and coordinated regulatory network of JA-induced Chl degradation.
DOI:10.1007/s00299-016-1991-1URLPMID:27154758 [本文引用: 1]

Key message ANAC072 positively regulates both age- and dark-induced leaf senescence through activating the transcription of NYE1.
DOI:10.1016/j.pmpp.2012.07.002URL [本文引用: 1]

NAC (NAM, ATAF1/2 and CUC2) transcription factors (TFs) play important roles in plant development and environmental stresses, though more recently involvement in biotic stresses has also been observed, but not explored in detail. Based on a differential display library obtained from lipopolysaccharide (LPS)-elicited cells of Arabidopsis thaliana, a differentially expressed sequence tag (EST) was identified, sequenced and found to code for a NAC protein with a N-terminal NAM domain. The corresponding gene, At4g27410.3, encodes a known NAC TF, ANAC072/RD26. A. thaliana callus, cell suspensions and seedlings were used to determine the expression levels of ANAC072 after treatment with Burkholderia cepacia and associated microbe-associated molecular pattern (MAMPs) molecules. ANAC072 showed early responsiveness towards B. cepacia and its LPS and was also responsive to peptidoglycan (PGN) and the flagellin-derived peptide, flg22. The response of cultured cells was characterised by the rapid up-regulation at 10 min post elicitation. Depending on the MAMP, the response was either transient and returned to basal levels, or sustained up to 90 min. The general trend of expression was similar in seedlings compared to that of the undifferentiated cells. Transcript levels of ANAC072 were positively regulated by abscisic acid and hydrogen peroxide, but ethylene, salicylic acid, methyl jasmonate and indole acetic acid had little effect on its expression. Our data indicate that ANAC072 is an early MAMP-responsive TF, part of the altered transcriptome following MAMP perception events.
DOI:10.1105/tpc.104.022699URLPMID:15319476 [本文引用: 1]

Abstract The MYC-like sequence CATGTG plays an important role in the dehydration-inducible expression of the Arabidopsis thaliana EARLY RESPONSIVE TO DEHYDRATION STRESS 1 (ERD1) gene, which encodes a ClpA (ATP binding subunit of the caseinolytic ATP-dependent protease) homologous protein. Using the yeast one-hybrid system, we isolated three cDNA clones encoding proteins that bind to the 63-bp promoter region of erd1, which contains the CATGTG motif. These three cDNA clones encode proteins named ANAC019, ANAC055, and ANAC072, which belong to the NAC transcription factor family. The NAC proteins bound specifically to the CATGTG motif both in vitro and in vivo and activated the transcription of a beta-glucuronidase (GUS) reporter gene driven by the 63-bp region containing the CATGTG motif in Arabidopsis T87 protoplasts. The expression of ANAC019, ANAC055, and ANAC072 was induced by drought, high salinity, and abscisic acid. A histochemical assay using P(NAC)-GUS fusion constructs showed that expression of the GUS reporter gene was localized mainly to the leaves of transgenic Arabidopsis plants. Using the yeast one-hybrid system, we determined the complete NAC recognition sequence, containing CATGT and harboring CACG as the core DNA binding site. Microarray analysis of transgenic plants overexpressing either ANAC019, ANAC055, or ANAC072 revealed that several stress-inducible genes were upregulated in the transgenic plants, and the plants showed significantly increased drought tolerance. However, erd1 was not upregulated in the transgenic plants. Other interacting factors may be necessary for the induction of erd1 in Arabidopsis under stress conditions.
DOI:10.1016/j.gene.2012.09.021URLPMID:22995346 [本文引用: 1]

78 Using RNA-seq, we have generated an extensive map of the blueberry transcriptome. 78 34,464 unique sequences were found to be expressed during the fruit development. 78 1,236 genes related to antioxidants were identified. 78 Comparative analysis of the differentially expressed genes in different tissues.
DOI:10.1186/s12864-015-1842-4URLPMID:4556307 [本文引用: 1]

Cranberries (Vaccinium macrocarpon Ait.), renowned for their excellent health benefits, are an important berry crop. Here, we performed transcriptome sequencing of one cranberry cultivar, from fruits at two different developmental stages, on the Illumina HiSeq 2000 platform. Our main goals were to identify putative genes for major metabolic pathways of bioactive compounds and compare the expression patterns between white fruit (W) and red fruit (R) in cranberry. In this study, two cDNA libraries of W and R were constructed. Approximately 119 million raw sequencing reads were generated and assembled de novo, yielding 57,331 high quality unigenes with an average length of 739 bp. Using BLASTx, 38,460 unigenes were identified as putative homologs of annotated sequences in public protein databases, including NCBI NR, NT, Swiss-Prot, KEGG, COG and GO. Of these, 21,898 unigenes mapped to 128 KEGG pathways, with the metabolic pathways, secondary metabolites, glycerophospholipid metabolism, ether lipid metabolism, starch and sucrose metabolism, purine metabolism, and pyrimidine metabolism being well represented. Among them, many candidate genes were involved in flavonoid biosynthesis, transport and regulation. Furthermore, digital gene expression (DEG) analysis identified 3,257 unigenes that were differentially expressed between the two fruit developmental stages. In addition, 14,473 simple sequence repeats (SSRs) were detected. Our results present comprehensive gene expression information about the cranberry fruit transcriptome that could facilitate our understanding of the molecular mechanisms of fruit development in cranberries. Although it will be necessary to validate the functions carried out by these genes, these results could be used to improve the quality of breeding programs for the cranberry and related species. The online version of this article (doi:10.1186/s12864-015-1842-4) contains supplementary material, which is available to authorized users.
DOI:10.2307/3871236URLPMID:11148285 [本文引用: 1]

Plants produce a wide array of natural products, many of which are likely to be useful bioactive structures. Unfortunately, these complex natural products usually occur at very low abundance and with restricted tissue distribution, thereby hindering their evaluation. Here, we report a novel approach for enhancing the accumulation of natural products based on activation tagging by Agrobacterium-mediated transformation with a T-DNA that carries cauliflower mosaic virus 35S enhancer sequences at its right border. Among 5000 Arabidopsis activation-tagged lines, we found a plant that exhibited intense purple pigmentation in many vegetative organs throughout development. This upregulation of pigmentation reflected a dominant mutation that resulted in massive activation of phenylpropanoid biosynthetic genes and enhanced accumulation of lignin, hydroxycinnamic acid esters, and flavonoids, including various anthocyanins that were responsible for the purple color. These phenotypes, caused by insertion of the viral enhancer sequences adjacent to an MYB transcription factor gene, indicate that activation tagging can overcome the stringent genetic controls regulating the accumulation of specific natural products during plant development. Our findings suggest a functional genomics approach to the biotechnological evaluation of phytochemical biodiversity through the generation of massively enriched tissue sources for drug screening and for isolating underlying regulatory and biosynthetic genes.
DOI:10.1104/pp.106.088104URLPMID:17012405 [本文引用: 1]

Anthocyanins are secondary metabolites found in higher plants that contribute to the colors of flowers and fruits. In apples (Malus domestica Borkh.), several steps of the anthocyanin pathway are coordinately regulated, suggesting control by common transcription factors. A gene encoding an R2R3 MYB transcription factor was isolated from apple (cv Cripps' Pink) and designated MdMYB1. Analysis of the deduced amino acid sequence suggests that this gene encodes an ortholog of anthocyanin regulators in other plants. The expression of MdMYB1 in both Arabidopsis (Arabidopsis thaliana) plants and cultured grape cells induced the ectopic synthesis of anthocyanin. In the grape (Vitis vinifera) cells MdMYB1 stimulated transcription from the promoters of two apple genes encoding anthocyanin biosynthetic enzymes. In ripening apple fruit the transcription of MdMYB1 was correlated with anthocyanin synthesis in red skin sectors of fruit. When dark-grown fruit were exposed to sunlight, MdMYB1 transcript levels increased over several days, correlating with anthocyanin synthesis in the skin. MdMYB1 gene transcripts were more abundant in red skin apple cultivars compared to non-red skin cultivars. Several polymorphisms were identified in the promoter of MdMYB1. A derived cleaved amplified polymorphic sequence marker designed to one of these polymorphisms segregated with the inheritance of skin color in progeny from a cross of an unnamed red skin selection (a sibling of Cripps' Pink) and the non-red skin cultivar Golden Delicious. We conclude that MdMYB1 coordinately regulates genes in the anthocyanin pathway and the expression level of this regulator is the genetic basis for apple skin color.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
