 ,1,*
,1,*GhP4H2 encoding a prolyl-4-hydroxylase is involved in regulating cotton fiber development
GAO Lu1,2, XU Wen-Liang ,1,*
,1,*通讯作者:
收稿日期:2020-09-22接受日期:2020-12-1网络出版日期:2020-12-29
| 基金资助: |
Received:2020-09-22Accepted:2020-12-1Online:2020-12-29
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1467KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
高璐, 许文亮. 脯氨酸羟化酶GhP4H2在棉花纤维发育中的功能研究[J]. 作物学报, 2020, 47(7): 1239-1247. doi:10.3724/SP.J.1006.2021.04217
GAO Lu, XU Wen-Liang.
棉花作为世界上最大的纤维作物和纺织工业原料, 品质是其经济价值的重要考量因素。棉纤维是由棉花胚珠外珠被表皮层的单细胞发育而成, 是自然界最长的单细胞之一。细胞壁作为棉纤维的重要组分, 其合成过程直接决定着棉纤维的品质参数[1], 它是一个复杂的动态结构, 由90%高分子量的多聚物和少量糖蛋白组成[2]。虽然糖蛋白组分占比很少, 却是细胞壁形态构成和功能发挥的重要因子。富含羟脯氨酸糖蛋白(hydroxyproline (Hyp)-rich glycoproteins, HRGP)代表细胞壁糖蛋白的一个大家族, 根据其核心蛋白的糖基化程度, 可分为高度糖基化的阿拉伯半乳聚糖蛋白(arabinogalactan proteins, AGP), 中度糖基化的伸展蛋白(extensins, EXT)以及轻度糖基化的富含脯氨酸蛋白(proline-rich proteins, PRPs) 3类[3,4,5]。PRP广泛存在于不同作物中且功能不等, 如参与玉米胚的发育[6,7]、响应大豆干旱和盐胁迫应答[8]等。我们前期从棉花cDNA文库中分离了3个PRP家族基因, 命名为GhPRP1、GhPRP2和GhPRP5, 发现GhPRP1在下胚轴和根中高量表达, GhPRP2在棉花开花后10 d (days post anthesis, DPA)胚珠中优势表达[9]。在拟南芥中表达GhPRP5基因增强了转基因拟南芥对盐和ABA的敏感性[10]。GhPRP5-RNAi转基因棉纤维的长绒和短绒均增长, 酵母双杂交表明, GhPRP5可形成同源和异源二聚体发挥功能, 推测GhPRP5通过复杂的调控网络影响细胞壁的合成从而影响棉纤维的发育[11]。AGP可能是自然界中翻译后修饰程度最高的蛋白, 其合成过程包括N末端信号肽序列的切割、由脯氨酸羟化酶(prolyl 4-hydroxylases, P4Hs)催化的脯氨酸残基的羟基化、GPI锚的修饰和羟脯氨酸残基上的阿拉伯半乳聚糖糖基化。它是一类O-糖基化蛋白, O-糖基化部位主要发生在含有丝氨酸(Ser)和羟脯氨酸(Hyp)的肽链中[12], 广泛参与细胞膨胀、细胞分化、生殖发育、体细胞胚胎发育、木质部分化、非生物胁迫应答、激素信号应答等过程[5]。AGP功能的发挥很大程度上依赖于占到整个分子量90%以上的糖侧链[13], 拟南芥mur1突变体中AGP分子的糖侧链缺失岩藻糖, 使得根长明显变短[14]。在N. alata中, TTS蛋白参与花粉管的伸长, 但去糖基化的TTS蛋白丧失该功能[15]。近年来, 已经筛选出大量编码AGP合成的基因, 根据核心蛋白中氨基酸的组成, AGP可分为典型性和非典型性两大类, 成束蛋白样阿拉伯半乳聚糖蛋白(Fasciclin-like arabinogalactan proteins, FLAs)作为典型性AGP的代表, 广泛存在于拟南芥[16]、水稻[17]、小麦[18]、毛果杨[19]、大麻[20]、陆地棉[21]、海岛棉[22]等多种植物中。我们之前从棉花中分离了19个编码FLA的基因, 其中的3个(GhFLA1/2/4)在10 DPA纤维中优势表达。我们进一步的研究发现, 过表达GhFLA1可以促进纤维伸长, 而抑制GhFLA1表达减缓了纤维起始和伸长。免疫组学分析显示, GhFLA1影响AGP组分尤其是AG多糖侧链的合成[21]。随后, 我们又分离了糖基转移酶基因GT31家族成员GhGalT1, 发现其参与AGP (主要是FLA) II型AG糖链β-1,3-半乳糖苷链的合成, 通过调控GhFLA的表达, 导致转基因株系棉纤维细胞壁的形态结构发生明显变化[23]。综上, AGP多糖侧链在棉纤维发育过程中发挥重要的作用, 但对于催化脯氨酸羟基化的脯氨酸羟化酶在棉纤维发育过程中的作用还不清楚。
P4H是普遍存在于动物和植物中的一种2-氧化戊二酸双加氧酶, 主要定位于内膜系统, 其功能的发挥需要Fe2+、α-酮戊二酸、O2等的参与[24]。动物等高级生物中P4H分为两大类, 影响胶原蛋白合成的C-P4H和缺氧诱导HIF-P4H。C-P4H是胶原蛋白合成的关键因子, 这类P4H以α2β2四聚体结构发挥功能, β-基序形成二硫键异构酶, α-基序则形成催化亚基, 催化底物氨基酸序列-X-Pro-Gly-中的Pro羟基化[25,26]。目前植物中关于P4H的报道还较少, 与动物中C-P4H不同的是其只含有α催化亚基, 催化底物中位于Ala、Gln、Hyp、Pro、Ser、Thr和Val之后的Pro羟基化, 但位于其他氨基酸之后的第1个Pro不能被羟基化。P4H催化的底物EXT中的Pro为连续的2~4个单元, 并且始终毗邻Ser(Ser-(Pro)2-4), 羟基化后形成Ser-(Hyp)2-4。当AGP作为催化底物时, AP/PA/SP/TP重复序列是其羟基化的敏感位点。而PRPs的PPVX [KT]、KKPCPP和PPV序列以及其他富含Pro的嵌合蛋白XPnY基序是其催化的靶序列。光谱测定和生物信息学方法预测到了拟南芥中AtPRP4 (富含32.5% Pro)、At1g09750、At1g31580 (含27%~28% Pro)、At3g08030、At2g10940 (含6%~8% Pro)催化底物序列Pro的定位, 证明了Pro羟基化序列的多样性和特异性[27]。莱茵衣藻中共有10个P4H基因, 但只有Cr-P4H1的功能得到证实, 缺陷型Cr-P4H1导致细胞壁的合成受阻[28]。
棉花是世界上最重要的经济作物之一, 目前关于棉花中GhP4H的报道还尚属空白。P4H功能的发挥离不开保守残基与2-氧戊二酸盐(2-oxoglutarate)和Fe2+的结合。3,4-二羟基苯甲酸乙酯(ethyl-3,4-dihydroxybenzoate, EDHB)和α,α-联吡啶(α,α-Dipyridyl, DP)是P4H酶的2种抑制剂, EDHB可以特异结合2-oxoglutarate, DP则通过螯合Fe2+抑制P4H酶活[29]。我们前期用不同浓度梯度的EDHB和DP处理棉花离体胚珠后, 棉纤维伸长受到严重抑制, 且抑制程度与抑制剂浓度呈正相关, 说明GhP4H参与棉纤维生长发育[12]。随后, 我们从棉花基因组中识别了30个可能编码P4H的基因, 许多P4H家族基因在纤维发育的不同阶段高量表达, 进一步表明这些基因可能在纤维发育过程中起作用。为研究P4H在棉纤维发育过程中的功能, 本研究克隆了一个在棉纤维伸长期高量表达的P4H家族基因GhP4H2, 通过农杆菌介导法转化棉花, 获得GhP4H2转基因棉花植株, 旨在了解P4H在棉纤维发育过程中的功能, 为棉纤维发育的分子机制提供新的视野。
1 材料与方法
1.1 植物材料
本研究选用陆地棉(Gossypium hirsutum L.) Corker 312品种为棉花遗传转化受体。1/2 MS培养基培养棉花无菌苗5~6 d, 将棉花下胚轴切成10 mm左右的小段, 并用含有GhP4H2过量表达载体或RNAi载体的农杆菌LBA4404悬浮液浸染15 min, 然后将外植体转移到共培养培养基上(不含任何抗生素)在28℃ (16 h光照/8 h黑暗)下共培养2 d, 后续详细继代培养步骤参照Qin等[23]的方法。获得的转基因幼苗转移至温室土壤中用于目的基因的鉴定和遗传分析。1.2 棉花基因组DNA提取
取棉花幼苗2 g左右幼叶, 利用CTAB法提取基因组DNA[23]。1.3 载体构建
设计带有Xba I和Sal I酶切位点的GhP4H2 OE引物(表1), 利用Primer STAR高保真酶扩增GhP4H2开放阅读框序列, 连接到克隆载体pSK上, 测序验证没有碱基突变后构建到表达载体pBI121上获得pBI121-GhP4H2过表达载体。两端酶切位点为Bam H I和Xba I的L1引物(表1)扩增正义链, 酶切位点为Sac I和Not I的L2引物(表1)扩增反义链, 正义链和反义链中间有一个GhTUA基因内含子(Intron), 两端酶切位点分别为Xba I和Not I。将GhP4H2正义链-Intron-反义链序列构建到pBI121质粒中, 获得pBI121-GhP4H2 RNAi载体[23]。
1.4 RNA提取和实时定量RT-PCR
利用TianGen RNA试剂盒提取棉花RNA。逆转录获得cDNA, 将各个样品的cDNA稀释到浓度相当后对基因进行实时定量RT-PCR分析[23]。棉花泛素基因(Ubiquitin1, GhUBI1)作为内参, 本研究所用基因RT-PCR引物见表1。Table 1
表1
表1本研究所用引物
Table 1
| 引物 Primer | 正向序列 Forward sequence (5'-3') | 反向序列 Reverse sequence (5'-3') |
|---|---|---|
| Gh_D12G1409RT | TAGAACCCAAGCTGGTCAGG | TCGATTTAGACCCTGTCGATG |
| Gh_D01G1917RT | AGATCTCATGCGCCGTCA | AGGCAAAGAAAGAAACAAGTGTG |
| Gh_D07G2484RT | TTTGCCTTTTTCTGTGAGCA | GCTGGTTGGGATGAACACTT |
| Gh_D10G0598RT | CTATGAGTGTGATTGGCGTACAGGTAA | AATCCGTGGTCACGTTATATTTGG |
| Gh_D08G2475RT | TAACCCACCAAGTGGGAAAC | TGAGCTTAAGGGTGGTGCAT |
| Gh_A07G0433RT | TGAAGAAGACGAAGAACCATCACCA | AGAGCCAGAGCTTTCAACTGATGT |
| Gh_D05G0131RT | TTAAGCCACCCATTGACCTC | TCCTGAAAGAACCGGACAAC |
| Gh_D12G2178RT | TCTTTTCCCTGGTGAGTGCT | AACGGCAGCTTCAAGATCAG |
| GhGalT1RT | TCCCTCCTCACCTTCGCCATTG | CCTTGACGATCAGAAGGCATCCAC |
| GhGalT4RT | GATGATACTTTGAAAATCGTTGCT | ATTTTCTTCAAAGTTTCACCGCT |
| GhGalT6RT | TCAGGAGTATGTACCCCAGCTTGC | TGCAAATGAAGTCACATGTCGAC |
| GhGalT7RT | CCTCACCAGATCAACAGCCCTCT | CCGTTGAAGGCCCTGATGATC |
| GhP4H2RT | ATCGGAATGTGCAAAGAAGG | AGCCAGGAAGTTCTGCAGTT |
| GhUBI1RT | CTGAATCTTCGCTTTCACGTTATC | GGGATGCAAATCCTTCGTGAAAAC |
| GhP4H2 RNAiL1 | CTTGGATCCGTGCAAAGAAGGGAATTG | GGGTCTAGAAATAGCCAGGAAGTTCTG |
| GhP4H2RNAiL2 | GGGGAGCTCGTGCAAAGAAGGGAATTG | CTTGCGGCCGCAATAGCCAGGAAGTTCT |
| GhP4H2OE | GGGTCTAGAATGGCTATTGAGAGGATTT | CTTGTCGACTCAACATACTTTGCAGCTT |
新窗口打开|下载CSV
1.5 棉纤维AGP的分离、纯化及定量分析
按照Qin等[23]的方法, 取10 g冷冻干燥的10 DPA纤维, 加入液氮研磨成粉末后, 加入10 mL缓冲液(50 mmol L-1 Tris-Cl, 10 mmol L-1 pH 8.0 EDTA, 0.1% β-巯基乙醇, 1% (w/v) Triton X-100), 4℃孵育3 h。14,000×g离心10 min, 取上清加入10 mL乙醇, 4℃孵育过夜。离心弃上清, 5 mL 50 mmol L-1 Tris-Cl重悬沉淀, 冷冻干燥过夜。350 μL 1% NaCl重悬样品, 加入350 μL的β-Yariv, 混匀, 4℃过夜。14,000×g离心1 h, 1% NaCl洗涤沉淀3次, 甲醇洗涤沉淀2次去除β-Yariv, 静置干燥, 加入适量二甲基亚砜至沉淀完全溶解, 加入微量亚硫酸钠, 再加入适量超纯水至溶液呈亮黄色, 溶液过PD-10柱脱盐, 所得即为AGP溶液。AGP浓度测定按Qin等[23]的方法, 配制浓度为1%琼脂糖, 0.02% 叠氮钠(NaN3), 0.15 mol L-1 NaCl以及10 μg mL-1 β-Yariv的溶液20 mL, 加热至沸腾, 待琼脂糖完全融化后, 将混合液倒入培养皿中, 冷却。用直径为2 mm的玻璃吸管对冷却的凝胶打孔。取1 μL AGP点样, 均匀点入每个孔中, 并选择浓度为0.1~0.6 μg μL-1的阿拉伯树胶作为标样对照。将平板置于黑暗潮湿的环境中过夜。测量每个扩散色圈的直径, 并计算色圈面积, 根据标样绘制标准曲线并计算出各个待测样品的浓度。
1.6 Immuno Dot blot分析
根据Qin等[23]的方法进行Dot blot分析野生型与转基因棉花株系中AGP的差异。分别利用JIM8和JIM13作为一抗, 碱性磷酸酶标记的山羊抗大鼠IgG(H+L)作为二抗进行免疫组织化学反应。1.7 转录组分析
收取野生型和GhP4H2过表达株系5 DPA纤维, 利用Spectrum plant total RNA Kit试剂盒提取总RNA, 纯化后检测RNA的完整性。将完整性好的RNA进行RAN-seq (北京诺禾致源基因公司)。GO (Gene ontology)预测差异基因功能并分类, 以P值为依据, 筛选出差异表达基因进行RT-PCR验证。2 结果与分析
2.1 GhP4H2蛋白的系统进化分析和基因表达谱
利用Clustalx、MEGA7.0 (为研究GhP4H2基因在棉花中的表达模式, 提取棉花的根、下胚轴、子叶、真叶、花瓣、花药、15 d胚珠及不同发育时期纤维的RNA, 逆转录后进行实时定量RT-PCR分析。GhP4H2基因在子叶、真叶、花药中高量表达, 在纤维发育的不同时期(0~20 DPA)均有较高水平的表达, 且在5 DPA和9 DPA纤维中保持较高的表达量(图1-B), 暗示GhP4H2可能参与棉纤维伸长过程。
图1
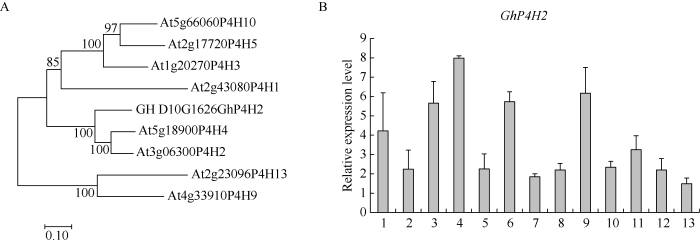 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1GhP4H2的系统进化分析和GhP4H2基因表达谱
A: 棉花GhP4H2与拟南芥GhP4H蛋白系统进化关系; B: GhP4H2基因在棉花各组织的表达谱。1: 根; 2: 下胚轴; 3: 子叶; 4: 真叶; 5: 花瓣; 6: 花药; 7: 15 DPA胚珠; 8: 0 DPA纤维(含胚珠); 9: 3 DPA纤维(含胚珠); 10: 5 DPA纤维; 11: 9 DPA纤维; 12: 15 DPA纤维; 13: 20 DPA纤维。DPA: 开花后天数。误差线代表标准误差。
Fig. 1Phylogenetic analysis of GhP4H2 and expression profile of GhP4H2
A: phylogenetic relationship of GhP4H2 and Arabidopsis P4Hs; B: expression analysis of GhP4H2 in different cotton tissues. 1: root; 2: hypocotyl; 3: cotyledon; 4: leaves; 5: petals; 6: anthers; 7: 15 DPA ovule; 8: 0 DPA fiber (with ovule); 9: 3 DPA fiber (with ovule); 10: 5 DPA fiber; 11: 9 DPA fiber; 12: 15 DPA fiber; 13: 20 DPA fiber. DPA: days post anthesis. Error bar represents the standard deviation.
2.2 转基因棉花株系中GhP4H2的表达分析
为研究GhP4H2在棉纤维生长发育过程中的功能, 本研究分别构建了由35S启动子驱动的过表达(overexpression, OE)和RNA interference (RNAi)载体, 通过农杆菌介导法转化棉花下胚轴, 获得转基因棉花植株。取不同转基因株系5 DPA和10 DPA纤维进行表达量鉴定表明, GhP4H2基因在RNAi (Ri)株系L16和L28中表达量显著低于WT, 在过表达株系L10和L38中表达量显著增加(图2)。我们后续研究选择RiL16、RiL28、OEL10、OEL38这4个转基因株系进行表型分析。图2
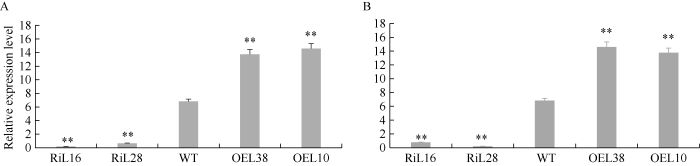 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2GhP4H2在野生型和转基因棉花株系中的表达分析
A: GhP4H2在不同株系5 DPA纤维中表达分析; B: GhP4H2在不同株系10 DPA纤维中表达分析。RiL16、RiL28表示2个独立的GhP4H2 RNAi株系; WT表示野生型; OEL10、OEL38表示2个独立的GhP4H2过表达株系。**表示在0.01水平差异显著。
Fig. 2Expression analysis of GhP4H2 in wild type and transgenic cotton lines
A and B: quantitative RT-PCR analysis of GhP4H2 expression in 5 DPA (A) and 10 DPA (B) fibers from independent transgenic cotton lines and wild type. RiL16 and RiL28 represent two independent GhP4H2-RNAi lines; WT represents the wild type; OEL10 and OEL38 represent two independent GhP4H2 overexpression lines. ** means significant difference at the 0.01 probability level.
2.3 GhP4H2转基因棉花成熟纤维表型分析
分析T1~T3代成熟纤维表型变化发现, 与野生型相比, GhP4H2过表达株系的成熟棉纤维长度显著变短, 而RNAi株系没有明显变化。脱绒后比较不同株系种子形态大小, 未发现明显差异(图3), 表明GhP4H2主要影响纤维发育而对胚珠影响较小。图3
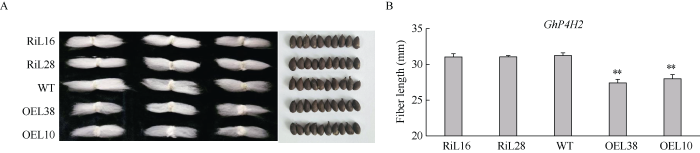 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3GhP4H2转基因与野生型棉花成熟纤维表型分析
A: GhP4H2转基因棉花与WT成熟种子和纤维表型比较; B: GhP4H2转基因棉花与WT成熟纤维长度比较。**表示在0.01水平差异显著。株系名称缩写同
Fig. 3Comparison of mature fibers from transgenic cotton lines and wild type
A and B: measurement and statistical analysis of mature fiber length and seed size of the transgenic cotton plants and wild type. ** means significant difference at the 0.01 probability level. Abbreviations of lines name are the same as those given in
2.4 GhP4H2转基因棉纤维AGP含量分析
考虑到AGP基因数目占了棉纤维HRGP基因的绝大多数[23], 我们推测AGP可能是P4H的首选底物。为研究GhP4H2对AGP合成的影响, 本研究提取了10 DPA纤维AGP进行定量检测。以0.1~0.6 μg μL-1浓度梯度的阿拉伯树胶作对照, 绘制折线图并计算各转基因株系中AGP样品浓度(图4)。RiL28中AGP含量明显低于WT, RiL16中AGP含量有轻微减少; 而过表达株系OEL38中含量显著增加, OEL10中AGP含量有轻微增加, 但未见显著差异。图4
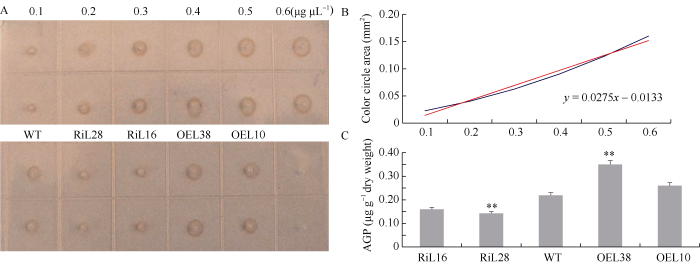 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4GhP4H2转基因与野生型棉花AGP定量分析
A: 不同样品AGP特异反应色圈。B: 阿拉伯树胶标准曲线; 横坐标代表AGP浓度, 纵坐标代表对应的色圈面积。C: 不同样品AGP含量。0.1~0.6 (μg μL-1): 阿拉伯树胶浓度。**表示在0.01水平差异显著。株系名称缩写同
Fig. 4AGP content in fibers of GhP4H2 transgenic cotton lines and wild type
A: halos of different samples from transgenic lines and wild type. B: standard curve; Abscissa: the AGPs concentration; Ordinate: the area of the corresponding halos. C: the AGPs content of different samples from transgenic lines and wild type. 0.1-0.6 (μg μL-1): gum arabic concentration. ** means significant difference at the 0.01 probability level. Abbreviations of lines name are the same as those given in
2.5 GhP4H2影响AGPs多糖侧链的合成
为研究GhP4H2对AGP糖侧链合成的影响, 本研究用不同的抗体进行了Dot blot分析。其中, JIM8、JIM13能够与AGP糖侧链抗原决定簇特异性结合, JIM8特异结合阿拉伯半乳糖侧链, JIM13特异结合β-GlcA-(1,3)-α-Gal-(1,2)-Rha侧链。由图5可知, RNAi株系杂交信号与WT相比减弱, 并且JIM8杂交信号弱于JIM13。过表达株系则呈现相反趋势。说明转基因株系中GhP4H2影响了AGP糖侧链的组成。图5
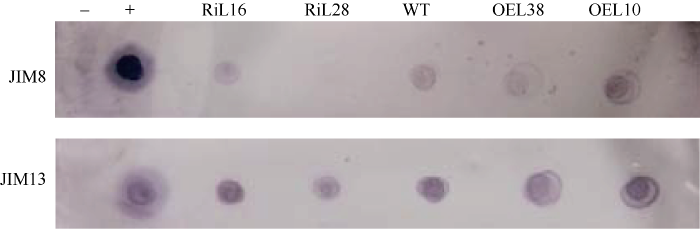 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5AGP糖链抗原决定簇在GhP4H2转基因与野生型棉花纤维中的分布与丰度
-: PBS阴性对照。+: 1 mol L-1 arabic gum阳性对照。株系名称缩写同
Fig. 5Distribution and abundance of AGP epitopes in GhP4H2 transgenic cotton and wild type
-: PBS negative control. +: 1 mol L-1 arabic gum positive control. Abbreviations of lines name are the same as those given in
2.6 GhP4H2影响棉纤维细胞壁相关基因的表达
为研究GhP4H2在棉纤维发育过程中的调控通路, 取野生型和过表达株系OEL38的5 DPA纤维进行转录组测序分析。本研究主要关注细胞壁相关基因。与WT相比, OEL38株系202个上调差异表达基因中与细胞壁合成相关的有15个(图6-A), 其中编码HRGP的基因有8个, 占到细胞壁基因的53.3%, Gh_D01G1917、Gh_D12G1409属于AGP类, Gh_D07G2484、Gh_D10G0598属于EXT类, Gh_D08G2475、Gh_A07G0433、Gh_D05G0131、Gh_D12G2178属于PRP类。因此推测这些编码HRGP的基因很可能是GhP4H2作用的靶基因。选择这8个基因进行RT-PCR表达验证(图6-B)发现, 这8个基因在OEL38中表达量均显著升高, 说明GhP4H2可能通过催化这些底物来调控棉纤维发育。图6
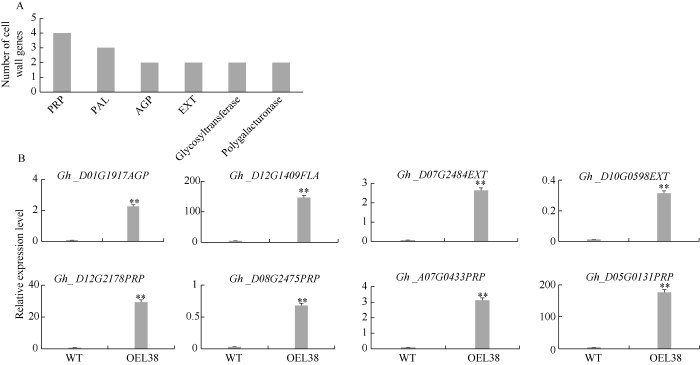 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6细胞壁相关差异表达基因分析
A: 细胞壁相关基因的主要类别; B: 糖蛋白相关基因的表达验证。PRP: 富含脯氨酸蛋白; AGP: 阿拉伯半乳聚糖蛋白; EXT: 伸展蛋白; PAL: 苯丙氨酸解氨酶。**表示在0.01水平差异显著。株系名称缩写同
Fig. 6Analysis of differentially expressed genes related cell wall
A: major classes of the upregulated cell wall genes in transgenic fibers; B: expression analysis of genes encoding glycoproteins. PRP: proline-rich protein; AGP: arabinogalactan protein; EXT: extensin; PAL: phenylalanine ammonia-lyase. ** means significant difference at the 0.01 probability level. Abbreviations of lines name are the same as those given in
2.7 GhP4H2影响糖基转移酶基因(GhGalTs)的表达
我们之前的研究显示, GhGalTs参与棉纤维发育过程[23]。GhGalTs位于GhP4H下游, 为研究GhP4H2对GhGalT表达的影响, 本研究选择GhGalT家族部分基因进行表达分析(图7)。选择的7个基因中有4个GhGalT基因(GhGalT1、GhGalT4、GhGalT6和GhGalT7)在GhP4H2过表达株系中的表达量发生变化, 表明GhP4H2可能影响了AGP糖基化过程, 这可能是AGP糖侧链发生改变的原因。图7
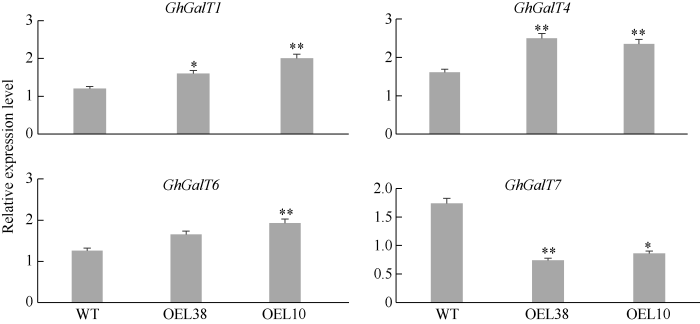 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7部分GhGalT基因表达分析
*、**分别表示在0.05和0.01水平差异显著。株系名称缩写同
Fig. 7Expression analysis of several GhGalT genes
* and ** mean significant differences at the 0.05 and 0.01 probability levels, respectively. Abbreviations of lines name are the same as those given in
3 讨论
先前有研究发现, AtP4H1参与拟南芥低氧应答, 过量表达AtP4H1致使拟南芥发生根毛增长, 毛状体缺失, 种子变小等变化, 同时导致与缺氧应答相关基因表达量升高。转录组数据显示, AtP4H1还参与光合作用、JA信号转导、植物激素调控等过程[30]。另外, 有研究表明, AtP4H5分别通过与AtP4H2和AtP4H13形成异源二聚体发挥Pro羟基化功能, 并且优先羟基化EXT, 三者共同调控拟南芥根毛的伸长[31]。敲除SlP4H1会促进番茄根的伸长和叶子的发育, 免疫组学分析显示, 与JIM8特异性结合的AGP糖链组分缺失[32]。拟南芥Atp4h2、Atp4h5和Atp4h13突变体中, 根毛相对于野生型都变短, DP和EDHB处理也会抑制根毛的伸长[31]。前期我们分别用DP和EDHB处理棉花离体胚发现, 棉纤维的生长发育受到了明显的抑制, 推测GhP4Hs参与调控棉纤维的生长发育[12]。本研究克隆了GhP4H2基因获得过表达和RNAi不同株系转基因棉花, 对T1~T3代成熟棉铃进行表型分析发现, GhP4H2-RNAi株系长绒与WT相比并没有明显变化, 这可能是由于基因功能冗余的原因。而GhP4H2 OE株系的成熟纤维明显短于WT。Dot blot分析显示, JIM8和JIM13的杂交信号均增强。我们前期证实GhFLA1促进棉纤维的生长发育, 免疫组学分析显示, GhFLA1转基因株系AGP侧链糖组分发生不同程度的变化, 其中过表达株系中JIM13杂交信号减弱, 说明GhFLA1过表达株系中与JIM13特异结合的AGP表位抗原决定簇减少[21]。随后我们发现过量表达GhGalT1的棉纤维伸长受到抑制, 经免疫组学分析, JIM8和JIM13的杂交信号均增强, 表明过量表达GhGalT1会增加与JIM8和JIM13特异结合的AGP表位抗原决定簇丰度[23]。本研究与GhGalT1过表达棉纤维取得的结果一致, 我们推测GhP4H2过表达转基因棉纤维变短的原因与GhGalT1类似。而在RNAi株系中AGP变化的程度可能不足以明显影响纤维发育。前期我们的结果显示, GhGalT1正调控FLA基因的表达[23], 本研究依据转录组及RT-PCR结果, 证实筛选到的8个表达AGP、EXT、PRP的差异基因受GhP4H2调控, 可能是其下游靶基因, 后续又发现GhP4H2影响下游GalTs的表达, 暗示GhP4H2不仅仅影响HRGP羟基化, 对参与糖基化的糖基转移酶基因的表达也有影响, 而GhP4H2转基因株系中AGP的变化很可能正是由于GhP4H2与GhGalTs协同调控引起。但过表达株系成熟纤维表型差异是否与AGP核心蛋白变化有关?AGP可能是GhP4H2优先催化的底物, EXT或PRP也是其作用底物吗?这些问题都有待深入研究。4 结论
本研究克隆了一个脯氨酸羟化酶基因GhP4H2, 该基因通过影响HRGP基因和糖基转移酶基因的表达来影响AGP的合成。过表达株系成熟纤维变短与AGP含量增加和糖侧链组分改变相关, 而RNAi株系成熟纤维长度未发生明显变化, 可能是由于基因功能冗余导致。推测过表达基因株系出现的表型差异是由AGP糖侧链组分变化所致, GhP4H2影响棉花纤维发育的分子机制还需进一步研究。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.3389/fpls.2012.00104URLPMID:22661979 [本文引用: 1]

Cotton fibers are single-celled extensions of the seed epidermis. They can be isolated in pure form as they undergo staged differentiation including primary cell wall synthesis during elongation and nearly pure cellulose synthesis during secondary wall thickening. This combination of features supports clear interpretation of data about cell walls and cellulose synthesis in the context of high throughput modern experimental technologies. Prior contributions of cotton fiber to building fundamental knowledge about cell walls will be summarized and the dynamic changes in cell wall polymers throughout cotton fiber differentiation will be described. Recent successes in using stable cotton transformation to alter cotton fiber cell wall properties as well as cotton fiber quality will be discussed. Futurec prospects to perform experiments more rapidly through altering cotton fiberwall properties via virus-induced gene silencing will be evaluated.
URLPMID:11842138 [本文引用: 1]
DOI:10.1104/pp.110.156000URLPMID:20388666 [本文引用: 1]
DOI:10.1105/tpc.5.1.9URLPMID:8439747 [本文引用: 1]
DOI:10.1104/pp.110.156554URLPMID:20395450 [本文引用: 2]

Hydroxyproline-rich glycoproteins (HRGPs) are a superfamily of plant cell wall proteins that function in diverse aspects of plant growth and development. This superfamily consists of three members: hyperglycosylated arabinogalactan proteins (AGPs), moderately glycosylated extensins (EXTs), and lightly glycosylated proline-rich proteins (PRPs). Hybrid and chimeric versions of HRGP molecules also exist. In order to
DOI:10.1105/tpc.4.4.413URLPMID:1498600 [本文引用: 1]

A gene from maize that encodes a hybrid proline-rich protein (HyPRP) formed by two well-defined domains, proline-rich and hydrophobic, respectively, has been characterized at the level of its structure and expression. The proline-rich domain is composed of elements PPYV and PPTPRPS, similar to those found in PRP proteins from soybean. The hydrophobic domain is rich in cysteine and is similar to seed proteins, mainly to a soybean hydrophobic seed protein. In maize, HyPRP is encoded by a single gene, and its mRNA accumulates in immature maize zygotic embryos, with a maximum accumulation between 12 and 18 days after pollination. The HyPRP mRNA can also be detected in ovary prior to pollination. In situ hybridization experiments on embryo sections show an expression of the gene in scutellum and in nonvascular cells from the embryo axis. Functional hypotheses related to HyPRP are discussed.
DOI:10.1104/pp.116.2.485URLPMID:9490753 [本文引用: 1]

The pattern of expression of two genes coding for proteins rich in proline, HyPRP (hybrid proline-rich protein) and HRGP (hydroxyproline-rich glycoprotein), has been studied in maize (Zea mays) embryos by RNA analysis and in situ hybridization. mRNA accumulation is high during the first 20 d after pollination, and disappears in the maturation stages of embryogenesis. The two genes are also expressed during the development of the pistillate spikelet and during the first stages of embryo development in adjacent but different tissues. HyPRP mRNA accumulates mainly in the scutellum and HRGP mRNA mainly in the embryo axis and the suspensor. The two genes appear to be under the control of different regulatory pathways during embryogenesis. We show that HyPRP is repressed by abscisic acid and stress treatments, with the exception of cold treatment. In contrast, HRGP is affected positively by specific stress treatments.
URLPMID:12582622 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s11103-013-0066-8URLPMID:23625445 [本文引用: 1]

Proline-rich proteins contribute to cell wall structure of specific cell types and are involved in plant growth and development. In this study, a fiber-specific gene, GhPRP5, encoding a proline-rich protein was functionally characterized in cotton. GhPRP5 promoter directed GUS expression only in trichomes of both transgenic Arabidopsis and tobacco plants. The transgenic Arabidopsis plants with overexpressing GhPRP5 displayed reduced cell growth, resulting in smaller cell size and consequently plant dwarfs, in comparison with wild type plants. In contrast, knock-down of GhPRP5 expression by RNA interference in cotton enhanced fiber development. The fiber length of transgenic cotton plants was longer than that of wild type. In addition, some genes involved in fiber elongation and wall biosynthesis of cotton were up-regulated or down-regulated in the transgenic cotton plants owing to suppression of GhPRP5. Collectively, these data suggested that GhPRP5 protein as a negative regulator participates in modulating fiber development of cotton.
[本文引用: 3]
[本文引用: 3]
DOI:10.1073/pnas.96.26.14736URLPMID:10611282 [本文引用: 1]

Design of hydroxyproline (Hyp)-rich glycoproteins (HRGPs) offers an approach for the structural and functional analysis of these wall components, which are broadly implicated in plant growth and development. HRGPs consist of multiple small repetitive
DOI:10.1046/j.1365-313x.2002.01406.xURLPMID:12366804 [本文引用: 1]

The Arabidopsis thaliana mutant mur1 is affected in the biosynthesis of l-fucose and has less than 2% of the normal amounts of this sugar in the cell walls of its aerial parts. Although in roots the reduction of l-fucose is only 40%, this causes a decrease of about 50% in root cell elongation. Since arabinogalactan-proteins (AGPs) are known to play a role in plant cell expansion we studied the composition of mur1 root AGPs. Arabidopsis root AGPs were shown to contain l-fucose, which was reduced in level in mur1 AGPs. In wild-type plants, an l-fucose containing epitope is present in AGPs in the cell wall of differentiating root cells. Addition of eel lectin, which specifically recognizes this epitope, and not fucose in other wall polymers, can phenocopy mur1 roots. Several lines of evidence are presented to support the contention that l-fucose containing root AGPs are required for the full elongation of root cells.
DOI:10.1046/j.1365-313x.2000.00731.xURLPMID:10792832 [本文引用: 1]

Upon germination on the stigma, pollen tubes elongate in the stylar transmitting tract, aided by female factors, with speed and directionality not mimicked in in vitro pollen tube growth cultures. We have shown that a stylar transmitting tissue arabinogalactan protein (AGP) from Nicotiana tabacum (tobacco), TTS protein, stimulates pollen tube growth in vivo and in vitro and attracts pollen tubes grown in a semi-in vivo culture system. It has been reported that the self-incompatible Nicotiana alata produced a stylar glycoprotein, GaRSGP, which had a backbone polypeptide that shared 97% identity with those of TTS proteins but some of its properties were different from those described for TTS proteins. We report here the characterization of a family of stylar transmitting tissue glycoproteins from N. alata that is virtually identical to tobacco TTS proteins and which we refer to as NaTTS proteins. Like their tobacco counterparts, NaTTS proteins are recognized by the traditional AGP-diagnostic reagent beta-glucosyl Yariv reagent, and they are also recognized by JIM13, a monoclonal antibody against AGP. NaTTS proteins also stimulate pollen tube elongation in vitro and attract pollen tubes in a semi-in vivo pollen tube culture system. Biochemical and immunological characterization of NaTTS proteins revealed that they have extraordinary variability in the extent of sugar modifications of their polypeptide backbones. The extent of sugar modifications on NaTTS proteins significantly affects their biochemical properties, influences how they interact with the transmitting tissue extracellular matrix, and affects their solubility from this matrix. Our results suggest that the strategy used to purify GaRSGP only recovered a less glycosylated, more tightly extracellular matrix-bound sub-population of the entire spectrum of N. alata TTS proteins.
DOI:10.1104/pp.003459URLPMID:12177459 [本文引用: 1]

Arabinogalactan proteins (AGPs) are extracellular hydroxyproline-rich proteoglycans implicated in plant growth and development. The protein backbones of AGPs are rich in proline/hydroxyproline, serine, alanine, and threonine. Most family members have less than 40% similarity; therefore, finding family members using Basic Local Alignment Search Tool searches is difficult. As part of our systematic analysis of AGP function in Arabidopsis, we wanted to make sure that we had identified most of the members of the gene family. We used the biased amino acid composition of AGPs to identify AGPs and arabinogalactan (AG) peptides in the Arabidopsis genome. Different criteria were used to identify the fasciclin-like AGPs. In total, we have identified 13 classical AGPs, 10 AG-peptides, three basic AGPs that include a short lysine-rich region, and 21 fasciclin-like AGPs. To streamline the analysis of genomic resources to assist in the planning of targeted experimental approaches, we have adopted a flow chart to maximize the information that can be obtained about each gene. One of the key steps is the reformatting of the Arabidopsis Functional Genomics Consortium microarray data. This customized software program makes it possible to view the ratio data for all Arabidopsis Functional Genomics Consortium experiments and as many genes as desired in a single spreadsheet. The results for reciprocal experiments are grouped to simplify analysis and candidate AGPs involved in development or biotic and abiotic stress responses are readily identified. The microarray data support the suggestion that different AGPs have different functions.
DOI:10.1093/jxb/erq104URLPMID:20423940 [本文引用: 1]

Arabinogalactan proteins (AGPs) comprise a family of hydroxyproline-rich glycoproteins that are implicated in plant growth and development. In this study, 69 AGPs are identified from the rice genome, including 13 classical AGPs, 15 arabinogalactan (AG) peptides, three non-classical AGPs, three early nodulin-like AGPs (eNod-like AGPs), eight non-specific lipid transfer protein-like AGPs (nsLTP-like AGPs), and 27 fasciclin-like AGPs (FLAs). The results from expressed sequence tags, microarrays, and massively parallel signature sequencing tags are used to analyse the expression of AGP-encoding genes, which is confirmed by real-time PCR. The results reveal that several rice AGP-encoding genes are predominantly expressed in anthers and display differential expression patterns in response to abscisic acid, gibberellic acid, and abiotic stresses. Based on the results obtained from this analysis, an attempt has been made to link the protein structures and expression patterns of rice AGP-encoding genes to their functions. Taken together, the genome-wide identification and expression analysis of the rice AGP gene family might facilitate further functional studies of rice AGPs.
DOI:10.1007/s00438-006-0159-zURLPMID:16944204 [本文引用: 1]

Putative plant adhesion molecules include arabinogalactan-proteins having fasciclin-like domains. In animal, fasciclin proteins participate in cell adhesion and communication. However, the molecular basis of interactions in plants is still unknown and none of these domains have been characterized in cereals. This work reports the characterization of 34 wheat (Triticum aestivum) and 24 rice (Oryza sativa) Fasciclin-Like Arabinogalactan-proteins (FLAs). Bioinformatics analyses show that cereal FLAs share structural characteristics with known Arabidopsis FLAs including arabinogalactan-protein and fasciclin conserved domains. At least 70% of the wheat and rice FLAs are predicted to be glycosylphosphatidylinositol-anchored to the plasma membranes. Expression analyses determined from the relative abundance of ESTs in the publicly available wheat EST databases and from RNA gel blots indicate that most of these genes are weakly expressed and found mainly in seeds and roots. Furthermore, most wheat genes were down regulated by abiotic stresses except for TaFLA9 and 12 where cold treatment induces their expression in roots. Plant fasciclin-like domains were predicted to have 3-D homology with FAS1 domain of the fasciclin I insect neural cell adhesion molecule with an estimated precision above 70%. The structural analysis shows that negatively charged amino acids are concentrated along the beta1-alpha3-alpha4-beta2 edges, while the positively charged amino acids are concentrated on the back side of the folds. This highly charged surface distribution could provide a way of mediating protein-protein interactions via electrostatic forces similar to many other adhesion molecules. The identification of wheat FLAs will facilitate studying their function in plant growth and development and their role in stress response.
DOI:10.1186/s12870-016-0912-3URLPMID:27769192 [本文引用: 1]

BACKGROUND: Hydroxyproline-rich glycoproteins (HRGPs) constitute a plant cell wall protein superfamily that functions in diverse aspects of growth and development. This superfamily contains three members: the highly glycosylated arabinogalactan-proteins (AGPs), the moderately glycosylated extensins (EXTs), and the lightly glycosylated proline-rich proteins (PRPs). Chimeric and hybrid HRGPs, however, also exist. A bioinformatics approach is employed here to identify and classify AGPs, EXTs, PRPs, chimeric HRGPs, and hybrid HRGPs from the proteins predicted by the completed genome sequence of poplar (Populus trichocarpa). This bioinformatics approach is based on searching for biased amino acid compositions and for particular protein motifs associated with known HRGPs with a newly revised and improved BIO OHIO 2.0 program. Proteins detected by the program are subsequently analyzed to identify the following: 1) repeating amino acid sequences, 2) signal peptide sequences, 3) glycosylphosphatidylinositol lipid anchor addition sequences, and 4) similar HRGPs using the Basic Local Alignment Search Tool (BLAST). RESULTS: The program was used to identify and classify 271 HRGPs from poplar including 162 AGPs, 60 EXTs, and 49 PRPs, which are each divided into various classes. This is in contrast to a previous analysis of the Arabidopsis proteome which identified 162 HRGPs consisting of 85 AGPs, 59 EXTs, and 18 PRPs. Poplar was observed to have fewer classical EXTs, to have more fasciclin-like AGPs, plastocyanin AGPs and AG peptides, and to contain a novel class of PRPs referred to as the proline-rich peptides. CONCLUSIONS: The newly revised and improved BIO OHIO 2.0 bioinformatics program was used to identify and classify the inventory of HRGPs in poplar in order to facilitate and guide basic and applied research on plant cell walls. The newly identified poplar HRGPs can now be examined to determine their respective structural and functional roles, including their possible applications in the areas plant biofuel and natural products for medicinal or industrial uses. Additionally, other plants whose genomes are sequenced can now be examined in a similar way using this bioinformatics program which will provide insight to the evolution of the HRGP family in the plant kingdom.
URLPMID:28931375 [本文引用: 1]
DOI:10.1104/pp.112.203760URLPMID:23349362 [本文引用: 3]

Arabinogalactan proteins (AGPs) are involved in many aspects of plant development. In this study, biochemical and genetic approaches demonstrated that AGPs are abundant in developing fibers and may be involved in fiber initiation and elongation. To further investigate the role of AGPs during fiber development, a fasciclin-like arabinogalactan protein gene (GhFLA1) was identified in cotton (Gossypium hirsutum). Overexpression of GhFLA1 in cotton promoted fiber elongation, leading to an increase in fiber length. In contrast, suppression of GhFLA1 expression in cotton slowed down fiber initiation and elongation. As a result, the mature fibers of the transgenic plants were significantly shorter than those of the wild type. In addition, expression levels of GhFLAs and the genes related to primary cell wall biosynthesis were remarkably enhanced in the GhFLA1 overexpression transgenic fibers, whereas the transcripts of these genes were dramatically reduced in the fibers of GhFLA1 RNA interference plants. An immunostaining assay indicated that both AGP composition and primary cell wall composition were changed in the transgenic fibers. The levels of glucose, arabinose, and galactose were also altered in the primary cell wall of the transgenic fibers compared with those of the wild type. Together, our results suggested that GhFLA1 may function in fiber initiation and elongation by affecting AGP composition and the integrity of the primary cell wall matrix.
DOI:10.1371/journal.pone.0070185URLPMID:23875019 [本文引用: 1]

Fasciclin-like arabinogalactan (FLA) protein is a cell-wall-associated protein playing crucial roles in regulating plant growth and development, and it was characterized in different plants including Upland cotton (Gossypium hirsutum L.). In cDNA-AFLP analysis of 25 DPA (days post anthesis) fiber mRNA, two FLA gene-related transcripts exhibit differential expression between Sea Island cotton (G. barbadense L.) and Upland cotton. Based on the transcript-derived fragment, RACE-PCR and realtime PCR technique, GbFLA5 full-length cDNA was isolated and its expression profiles were characterized in both cotton plant tissues and secondary cell wall (SCW) fibers in this study. The 1154 bp GbFLA5 cDNA contains an ORF of 720 bp, encoding GbFLA5 protein of 239 amino acids residues in length with an estimated molecular mass of 25.41 kDa and isoelectric point of 8.63. The deduced GbFLA5 protein contains an N-terminal signal sequence, two AGP-like domains, a single fasciclin-like domain, and a GPI anchor signal sequence. Phylogenetic analysis shows that GbFLA5 protein is homologous to some known SCW-specific expressed FLAs of plant developing xylem, tension wood and cotton fibers. In the SCW deposition stage from 15 to 45 DPA detected, FLA5 maintains a significantly higher expression level in Sea Island cotton fibers than in Upland cotton fibers. The increasing FLA5 transcript abundance coincided with the SCW deposition process and the expression intensity differences coincided with their fiber strength differences between Sea Island cotton and Upland cotton. These expression profile features of GbFLA5 in cotton fibers revealed its tissue-specific and SCW developmental stage-specific expression characters. Further analysis suggested that GbFLA5 is a crucial SCW-specific protein which may contribute to fiber strength by affecting cellulose synthesis and microfibril deposition orientation.
DOI:10.1111/tpj.13434URLPMID:27888523 [本文引用: 12]

Arabinogalactan proteins (AGPs) are highly glycosylated proteins that play pivotal roles in diverse developmental processes in plants. Type-II AG glycans, mostly O-linked to the hydroxyproline residues of the protein backbone, account for up to 95% w/w of the AGP, but their functions are still largely unclear. Cotton fibers are extremely elongated single-cell trichomes on the seed epidermis; however, little is known of the molecular basis governing the regulation of fiber cell development. Here, we characterized the role of a CAZy glycosyltransferase 31 (GT31) family member, GhGalT1, in cotton fiber development. The fiber length of the transgenic cotton overexpressing GhGalT1 was shorter than that of the wild type, whereas in the GhGalT1-silenced lines there was a notable increase in fiber length compared with wild type. The carbohydrate moieties of AGPs were altered in fibers of GhGalT1 transgenic cotton. The galactose: arabinose ratio of AG glycans was higher in GhGalT1 overexpression fibers, but was lower in GhGalT1-silenced lines, compared with that in the wild type. Overexpression of GhGalT1 upregulates transcript levels of a broad range of cell wall-related genes, especially the fasciclin-like AGP (FLA) backbone genes. An enzyme activity assay demonstrated that GhGalT1 is a beta-1,3-galactosyltransferase (beta-1,3-GalT) involved in biosynthesis of the beta-1,3-galactan backbone of the type-II AG glycans of AGPs. We also show that GhGalT1 can form homo- and heterodimers with other cotton GT31 family members to facilitate AG glycan assembly of AGPs. Thus, our data demonstrate that GhGalT1 influences cotton fiber development via controlling the glycosylation of AGPs, especially FLAs.
DOI:10.1016/j.molp.2018.03.003URLPMID:29530817 [本文引用: 1]

Extensins (EXTs) are highly repetitive plant O-glycoproteins that require several post-translational modifications (PTMs) to become functional in plant cell walls. First, they are hydroxylated on contiguous proline residues; then they are O-glycosylated on hydroxyproline and serine. After secretion into the apoplast, O-glycosylated EXTs form a tridimensional network organized by inter- and intra-Tyr linkages. Recent studies have made significant progress in the identification of the enzymatic machinery required to process EXTs, which includes prolyl 4-hydroxylases, glycosyltransferases, papain-type cysteine endopeptidases, and peroxidases. EXTs are abundant in plant tissues and are particularly important in rapidly expanding root hairs and pollen tubes, which grow in a polar manner. Small changes in EXT PTMs affect fast-growing cells, although the molecular mechanisms underlying this regulation are unknown. In this review, we highlight recent advances in our understanding of EXT modifications throughout the secretory pathway, EXT assembly in cell walls, and possible sensing mechanisms involving the Catharanthus roseus cell surface sensor receptor-like kinases located at the interface between the apoplast and the cytoplasmic side of the plasma membrane.
DOI:10.1016/s0945-053x(98)90009-9URLPMID:9524356 [本文引用: 1]

Prolyl 4-hydroxylases (EC 1.14,11.2) catalyze the formation of 4-hydroxyproline in collagens and other proteins with collagen-like sequences. The vertebrate type I and type II enzymes are [alpha (I)]2 beta 2 and [alpha (II)]2 beta 2 tetramers, respectively, whereas the enzyme from the nematode Caenorhabditis elegans is an alpha beta dimer. The type I enzyme is the major form in most but not all vertebrate tissues. The catalytic properties of the various enzyme forms are highly similar, but there are distinct, although small, differences in K(m) values for various peptide substrates between the enzyme forms and major differences in Ki values for the competitive inhibitor, poly(L-proline). Prolyl 4-hydroxylase requires Fe2+, 2-oxoglutarate, O2 and ascorbate. Kinetic studies and theoretical considerations have led to elucidation of the reaction mechanism, and recent extensive site-directed mutagenesis studies have identified five critical residues at the cosubstrate binding sites. A number of compounds have been characterized that inhibit it competitively with respect to some of the cosubstrates, and three groups of suicide inactivators have also been identified. The beta subunit in all forms of prolyl 4-hydroxylase is identical to protein disulfide isomerase (PDI), a multifunctional polypeptide that also serves as a subunit in the microsomal triglyceride transfer protein, as a chaperone-like polypeptide that probably assists folding of a number of newly synthesized proteins, and in several other functions. The main role of the PDI polypeptide as a protein subunit is probably related to its chaperone function. Recent expression studies of recombinant human prolyl 4-hydroxylase subunits in a yeast have indicated that the formation of a stable enzyme tetramer in vivo requires coexpression of collagen polypeptide chains.
DOI:10.1002/9780470123188.ch9URLPMID:9559057 [本文引用: 1]

Prolyl 4-hydroxylases catalyze the formation of 4-hydroxyproline in collagens and other proteins with an appropriate collagen-like stretch of amino acid residues. The enzyme requires Fe(II), 2-oxoglutarate, molecular oxygen, and ascorbate. This review concentrates on recent progress toward understanding the detailed mechanism of 4-hydroxylase action, including: (a) occurrence and function of the enzyme in animals; (b) general molecular properties; (c) intracellular sites of hydroxylation; (d) peptide substrates and mechanistic roles of the cosubstrates; (e) insights into the development of antifibrotic drugs; (f) studies of the enzyme's subunits and their catalytic function; and (g) mutations that lead to Ehlers-Danlos Syndrome. An account of the regulation of collagen hydroxylase activities is also provided.
DOI:10.3389/fpls.2017.01802URLPMID:29089960 [本文引用: 1]

Cell wall proteins (CWPs) play critical and dynamic roles in plant cell walls by contributing to developmental processes and response to environmental cues. Since the CWPs go through the secretion pathway, most of them undergo post-translational modifications (PTMs) which can modify their biological activity. Glycosylation is one of the major PTMs of CWPs and refers to N-glycosylation, O-glycosylation and glypiation. Each of these PTMs occurs in different amino acid contexts which are not all well defined. This article deals with the hydroxylation of Pro residues which is a prerequisite for O-glycosylation of CWPs on hydroxyproline (Hyp) residues. The location of Hyp residues is well described in several structural CWPs, but yet rarely described in other CWPs. In this article, it is studied in detail in five Arabidopsis thaliana proteins using mass spectrometry data: one of them (At4g38770, AtPRP4) is a structural CWP containing 32.5% of Pro residues arranged in typical motifs, the others are either rich (27-28%, At1g31580 and At2g10940) or poor (6-8%, At1g09750 and At3g08030) in Pro residues. The known rules of Pro hydroxylation allowed a good prediction of Hyp location in AtPRP4. However, they could not be applied to the other proteins whatever their Pro content. In addition, variability of the Pro hydroxylation patterns was observed within some amino acid motifs in all the proteins and new patterns of Pro hydroxylation are described. Altogether, this work shows that Hyp residues are present in more protein families than initially described, and that Pro hydroxylation patterns could be different in each of them. This work paves the way for completing the existing Pro hydroxylation code.
DOI:10.1105/tpc.106.042739URLPMID:17220203 [本文引用: 1]

Prolyl 4-hydroxylases (P4Hs) catalyze formation of 4-hydroxyproline (4Hyp), which is found in many plant glycoproteins. We cloned and characterized Cr-P4H-1, one of 10 P4H-like Chlamydomonas reinhardtii polypeptides. Recombinant Cr-P4H-1 is a soluble 29-kD monomer that effectively hydroxylated in vitro both poly(l-Pro) and synthetic peptides representing Pro-rich motifs found in the Chlamydomonas cell wall Hyp-rich glycoprotein (HRGP) GP1. Similar Pro-rich repeats that are likely to be Cr-P4H-1 substrates are also present in the cell wall HRGP GP2 and probably GP3. Suppression of the gene encoding Cr-P4H-1 by RNA interference led to a defective cell wall consisting of a loose network of fibrils resembling the inner and outer W1 and W7 layers of the wild-type wall, while the layers forming the dense central triplet were absent. The lack of Cr-P4H-1 most probably affected 4Hyp content of the major HRPGs of the central triplet, GP1, GP2, and GP3. The reduced 4Hyp levels in these HRGPs can also be expected to affect their glycosylation and, thus, the interactive properties and stabilities of their fibrous shafts. Interestingly, our RNA interference data indicate that the nine other Chlamydomonas P4H-like polypeptides could not fully compensate for the lack of Cr-P4H-1 activity and are therefore likely to have different substrate specificities and functions.
DOI:10.1074/jbc.M706554200URLPMID:17940281 [本文引用: 1]

Prolyl 4-hydroxylases (P4Hs) are 2-oxoglutarate dioxygenases that catalyze the hydroxylation of peptidyl prolines. They play an important role in collagen synthesis, oxygen homeostasis, and plant cell wall formation. We describe four structures of a P4H from the green alga Chlamydomonas reinhardtii, two of the apoenzyme at 1.93 and 2.90 A resolution, one complexed with the competitive inhibitor Zn2+, and one with Zn2+ and pyridine 2,4-dicarboxylate (which is an analogue of 2-oxoglutarate) at 1.85 A resolution. The structures reveal the double-stranded beta-helix core fold (jellyroll motif), typical for 2-oxoglutarate dioxygenases. The catalytic site is at the center of an extended shallow groove lined by two flexible loops. Mutagenesis studies together with the crystallographic data indicate that this groove participates in the binding of the proline-rich peptide-substrates. It is discussed that the algal P4H and the catalytic domain of collagen P4Hs have notable structural similarities, suggesting that these enzymes form a separate structural subgroup of P4Hs different from the hypoxia-inducible factor P4Hs. Key structural differences between these two subgroups are described. These studies provide first insight into the structure-function relationships of the collagen P4Hs, which unlike the hypoxia-inducible factor P4Hs use proline-rich peptides as their substrates.
DOI:10.1007/s10142-009-0118-yURLPMID:19277739 [本文引用: 1]

Proline hydroxylation is an important phenomenon of a living cell. Prolyl-4-hydroxylases (P4H) responsible for this process have been characterized from animals, and one of its forms, HIF-P4H, is regarded as an oxygen sensor. In plants, P4H has been partially characterized from few species, and one of the Arabidopsis P4H (AtP4H1) has been shown to hydroxylate proline-rich peptides in vitro. In order to study its function in planta, we have overexpressed AtP4H1 in Arabidopsis. The AtP4H1oexp plants showed hypoxia-in-normoxia phenotype with strict requirement for carbon source for its growth, increased root hair, absence of trichome, and reduction in seed size. Genome-wide expression analyses suggest that expression of several genes related to hypoxia as well as plant growth and development are upregulated in AtP4H1oexp lines. Based on our studies on AtP4H1oexp lines, we speculate a direct role of AtP4H1 in hypoxia stress and in different stages of plant growth and development.
DOI:10.1016/j.molp.2014.11.017URLPMID:25655826 [本文引用: 2]

Root hairs are single cells that develop by tip growth, a process shared with pollen tubes, axons, and fungal hyphae. However, structural plant cell walls impose constraints to accomplish tip growth. In addition to polysaccharides, plant cell walls are composed of hydroxyproline-rich glycoproteins (HRGPs), which include several groups of O-glycoproteins, including extensins (EXTs). Proline hydroxylation, an early post-translational modification (PTM) of HRGPs catalyzed by prolyl 4-hydroxylases (P4Hs), defines their subsequent O-glycosylation sites. In this work, our genetic analyses prove that P4H5, and to a lesser extent P4H2 and P4H13, are pivotal for root hair tip growth. Second, we demonstrate that P4H5 has in vitro preferred specificity for EXT substrates rather than for other HRGPs. Third, by P4H promoter and protein swapping approaches, we show that P4H2 and P4H13 have interchangeable functions but cannot replace P4H5. These three P4Hs are shown to be targeted to the secretory pathway, where P4H5 forms dimers with P4H2 and P4H13. Finally, we explore the impact of deficient proline hydroxylation on the cell wall architecture. Taken together, our results support a model in which correct peptidyl-proline hydroxylation on EXTs, and possibly in other HRGPs, is required for proper cell wall self-assembly and hence root hair elongation in Arabidopsis thaliana.
DOI:10.1007/s11103-014-0197-6URLPMID:24803411 [本文引用: 1]

Proline hydroxylation is a major posttranslational modification of hydroxyproline-rich glycoproteins (HRGPs) that is catalyzed by prolyl 4-hydroxylases (P4Hs). HRGPs such as arabinogalactan proteins (AGPs) and extensios play significant roles on cell wall structure and function and their implication in cell division and expansion has been reported. We used tobacco rattle virus (TRV)-based virus induced gene silencing to investigate the role of three tomato P4Hs, out of ten present in the tomato genome, in growth and development. Eight-days old tomato seedlings were infected with the appropriate TRV vectors and plants were allowed to grow under standard conditions for 6 weeks. Lower P4H mRNA levels were associated with lower hydroxyproline content in root and shoot tissues indicating successful gene silencing. P4H-silenced plants had longer roots and shoots and larger leaves. The increased leaf area can be attributed to increased cell division as indicated by the higher leaf epidermal cell number in SlP4H1- and SlP4H9-silenced plants. In contrast, SlP4H7-silenced plants had larger leaves due to enhanced cell expansion. Western blot analysis revealed that silencing of SlP4H7 and SlP4H9 was associated with reduced levels of JIM8-bound AGP and JIM11-bound extensin epitopes, while silencing of SlP4H1 reduced only the levels of AGP proteins. Collectively these results show that P4Hs have significant and distinct roles in cell division and expansion of tomato leaves.
