 ,1,2,*, 唐滔1, 李丽丽1, 陈宸1, 陈煜文1, 张亚芳2, 左示敏2
,1,2,*, 唐滔1, 李丽丽1, 陈宸1, 陈煜文1, 张亚芳2, 左示敏2Analysis on the structures of polygalacturonase-inhibiting proteins and the expression profile of its encoding genes in rice
CHEN Xi-Jun ,1,2,*, TANG Tao1, LI Li-Li1, CHEN Chen1, CHEN Yu-Wen1, ZHANG Ya-Fang2, ZUO Shi-Min2
,1,2,*, TANG Tao1, LI Li-Li1, CHEN Chen1, CHEN Yu-Wen1, ZHANG Ya-Fang2, ZUO Shi-Min2通讯作者:
收稿日期:2020-02-16接受日期:2020-07-2网络出版日期:2020-07-17
| 基金资助: |
Received:2020-02-16Accepted:2020-07-2Online:2020-07-17
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (3268KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
陈夕军, 唐滔, 李丽丽, 陈宸, 陈煜文, 张亚芳, 左示敏. 水稻多聚半乳糖醛酸酶抑制蛋白家族OsPGIP结构及基因表达特征
分析 [J]. 作物学报, 2020, 46(12): 1884-1893. doi:10.3724/SP.J.1006.2020.02011
CHEN Xi-Jun, TANG Tao, LI Li-Li, CHEN Chen, CHEN Yu-Wen, ZHANG Ya-Fang, ZUO Shi-Min.
果胶和纤维素是构成植物细胞壁骨架的基本物质, 也是病原菌侵入寄主的首道屏障。在病原菌侵入寄主过程中, PG是其产生的第一个胞壁降解酶[1]。PGIP是植物细胞壁的重要组成部分, 除了可以与胞壁果胶形成复合物加固细胞壁外, 还能特异性结合病菌PG, 减轻PG对寄主细胞的破坏作用, 帮助寄主维护细胞的完整性, 从而阻碍病菌侵入, 降低病害严重度[2,3]。同时, 由于病原菌PG活性被抑制, 其进一步分解果胶裂解产物寡聚半乳糖醛酸苷(oligalacutronides, OG)的能力下降, 从而引起OG在寄主体内富集, 这些具有激发子活性的OG又可激活寄主的防卫反应, 进一步提高寄主抗病性[4,5,6,7]。
要明确某个PGIP对病菌PG活性的抑制作用, 最直接的方法是用纯化的PGIP蛋白进行测定, 但以往研究多为从植物组织中提取PGIP, 这些蛋白可能包含着多个高度相似的PGIP异构体, 所得结果也是这些PGIP共同的作用, 而不是单个个体[8]。因此, 要研究某一个PGIP的作用, 利用外源表达的方法获得纯PGIP蛋白是较好的途径。但至目前为止, 已克隆的170个PGIP基因中仅有少数基因被研究, 因为在许多表达系统中PGIP基因不能表达, 或形成包涵体, 或表达条件要求苛刻, 或表达产物根本没有活性[9,10,11,12]。因此, 在研究这些PGIP之前, 对其相关性质进行生物信息学预测, 为进一步研究提供理论支持非常重要。
在植物体中, PGIP基因多以家族形式存在, 虽然这些基因在遗传上具有高度的相似性, 但其应对不同病原物或其他逆境因子时, 表达水平有显著差异; 即使是相同的基因, 在植物生长的不同发育阶段, 其表达量亦有不同[13,14,15]。如菜豆中有4个PvPGIP, 在其生长发育过程中, 仅PvPGIP2在所有组织中表达, 而PvPGIP3和PvPGIP4只在根部微弱表达, PvPGIP1则在幼叶、下胚轴、根和豆荚中均不表达[16]。在受到病菌侵染和机械损伤等逆境或OG、水杨酸和植物激素等具有诱导作用的因子处理后, 多个PvPGIP基因均有不同的应答, 只有PvPGIP4在所有处理条件下表达量均无明显变化[17,18]。据目前已有报道, 水稻中共存在7个OsPGIP基因[19,20,21]。Lu等[19]研究发现, 在粳稻品种中花11中, 7个OsPGIP基因启动子区顺式调控元件组成多样, 其对各种植物激素处理的反应也各不相同。为了更加明确各OsPGIP基因的表达特征, 本研究选用抗、感纹枯病水稻品种并通过低温、遮光和接种病原菌等逆境处理, 以研究不同条件下各OsPGIP基因的表达水平, 为进一步将这些基因应用于水稻抗病育种提供理论依据。
1 材料与方法
1.1 供试材料
供试菌株: 水稻纹枯病菌YN-7, 由本实验室分离自江苏水稻纹枯病病株。供试水稻: YSBR1 (抗)、徐稻3号(中感)和Lemont (高感)均由扬州大学农学院水稻抗病分子遗传与育种研究组提供。1.2 OsPGIP基因的克隆与测序
水稻基因组DNA提取参照Khanuja等[22]的方法。RNA提取采用赛默飞世尔科技公司(中国)植物RNA提取试剂盒, 反转录cDNA使用天根生化科技(北京)有限公司FastKing RT试剂盒。根据NCBI和参考文献中各基因相关序列, 使用Primer V5.0软件设计本研究所需引物(表1)。所有引物由宝生物工程有限公司(大连TaKaRa公司)合成。分别以水稻基因组DNA和cDNA为底物进行PCR扩增, 扩增产物连接至克隆载体pMD19-T, 阳性克隆子送上海生工生物工程有限公司测序。Table 1
表1
表1扩增各OsPGIP基因所用引物
Table 1
| 基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| OsPGIP1 | TGACTCGCTATTGCATGCG | TGGGAGCTTAATTGCAGGGA |
| OsPGIP2 | ATACACGGCATTGCATGCAC | CTTACACTCGTTCTCCGTAC |
| OsPGIP3 | TAGAAGAGAGGAAGCACGCA | TTGGTGGCCTGAGATAGGT |
| OsPGIP4 | TGTCGTGCACTTGTGTTCAA | GCATTAGCTGGTTGCTTC |
| OsFOR1 | TTCAGGTAGATACAATGGCG | ATGGATGGATGGATGCTC |
| OsPGIP6 | GAGCCGAGACGAGACGA | ATATGTACCCAAGCCCAAA |
| OsPGIP7 | TCCTGCACGGATTTGAGC | TAACAACAGCCAGTCAGCAAT |
新窗口打开|下载CSV
1.3 系统发育树的构建
以“Polygalacturonase inhibiting protein”为关键词从NCBI中搜索相关蛋白序列, 去除非PGIP或未明确释意为PGIP的序列, 将这些序列与本实验中克隆基因的翻译产物进行系统进化分析, 利用MEGA 5.1软件构建系统发育进化树。1.4 OsPGIP结构预测
蛋白质一级结构分析与信号肽预测分析分别使用网站(1.5 OsPGIP性质与功能预测
蛋白质的分子量、等电点、稳定性、脂溶性、疏水性、跨膜结构域、亚细胞定位、N-糖基化位点以及大肠杆菌中表达产物的溶解性等性质与功能分别通过网站(1.6 水稻苗处理与组织取样
不同生育期取样: 选择正常育苗的抗、感纹枯病水稻品种YSBR1、徐稻3号和Lemont不同生育期 (四至五叶期、成株期和穗期)的植株, 取中部叶鞘组织。遮光处理: 取各品种水稻种子经浸种、催芽后, 选取芽长较一致的种子置于平铺在培养皿(150 mm)底部的湿纱布上, 26℃光照培养箱中光暗交替(14 h/10 h)培养14 d后, 遮光黑暗条件下培养至水稻五到六叶期, 取植株中部叶鞘组织, 以正常生长条件下的苗为对照。
低温处理: 将上述培养14 d的水稻苗4℃低温条件下培养4 d, 取植株中部叶鞘组织, 以正常光暗条件下生长苗为对照。
病原菌接种处理: 用灭菌牙签接种的方法[40], 接种纹枯病菌至水稻叶鞘中, 以不接菌作对照, 48 h后取植株中部叶鞘组织。
以上组织取样后立即置于液氮中, -80℃保存, 用于植物总RNA提取。
1.7 qRT-PCR分析
提取各水稻组织样品总RNA, 反转录成cDNA后, 采用CFX96 Real-time检测系统(Bio-Rad)进行检测。20 μL反应体系: AceQqPCR SYBR Green Master Mix 10 μL、Forward Primer 0.4 μL、Reverse Primer 0.4 μL、ROX Reference Dye 1 0.4 μL、模板cDNA 2 μL、ddH2O 6.8 μL。反应过程如下: 95℃ 5 min; 95℃ 15 s, 60℃ 20 s, 40个循环; 95℃ 15 s。以OsACTIN1 (NCBI序列号为X16280)为内参基因, 扩增引物为OsACTIN- F/OsACTIN-R (5′-CAGCATGAAGATCAAGGTGG-3′ / 5′-TTCCTGTGCACAATGGATGG-3′), 扩增各OsPGIP基因的引物序列参见文献[19]。每个样品设3次生物学重复, 每实验重复3次。1.8 数据统计与分析
所有数据采用SPSS 12.05软件进行方差分析, 以Tukey测验进行多重比较, 当P<0.05时, 则表示差异显著。2 结果与分析
2.1 OsPGIP系统进化分析
从水稻cDNA中扩增出7个OsPGIP基因序列并送上海生工生物工程有限公司测序, 经NCBI比对确认扩增产物序列正确。以“Polygalacturonase inhibiting protein”为关键词从NCBI网站中共搜索到蛋白序列333条, 其中明确注明物种来源并释意为PGIP的共258条。将这些蛋白序列与各OsPGIP一起经多序列比对与系统进化分析。结果发现, 大多数相同或相近物种的PGIP相似性较高, 聚集在1到2个分支(群)内, 如十字花科、豆科、禾本科等植物的PGIP (图1-A); 但在这些聚集群中也有例外, 如归为其他类的螺旋狸藻PGIP (NCBI序列号为EPS70789)与水稻的几个PGIP序列高度相似, 而辣椒的PGIP (NCBI序列号为AEX34755)与豆科PGIP归在一起; 同样, 来源于水稻的7个OsPGIP除OsPGIP2、OsPGIP4、OsPGIP6和OsPGIP7序列相似性较高, 被聚在同一分枝, 其他几个OsPGIP分别位于进化树不同的分枝(图1-B)。这些分析结果表明, 尽管同一物种中的PGIP可能是源自1个或少数几个经编码子重排、点突变、小片段插入或缺失的基因编码的产物, 但不同物种中的PGIP也有可能源自同一祖先。图1
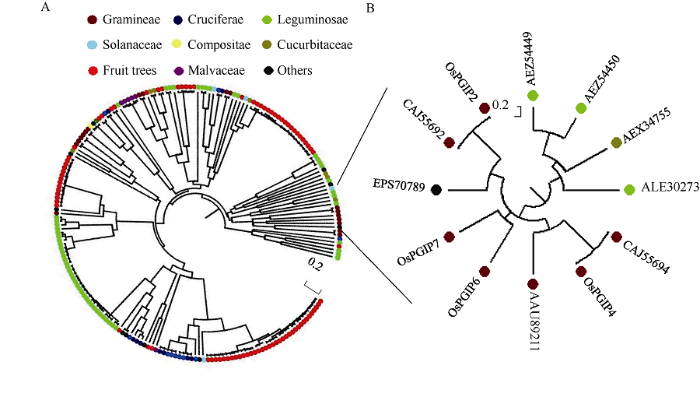 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1OsPGIP和其他物种来源的PGIP系统发育树
图中除OsPGIP为名称缩写外, 其他均为对应PGIP的NCBI序列号。A: 总进化树; B: 子树。
Fig. 1Phylogenetic relationship among OsPGIPs and PGIPs from other plant species
OsPGIPs is the abbreviations of polygalacturonase-inhibiting protein from Oryza sativa, the others are the NCBI accession numbers of the corresponding sequences. A: phylogenetic tree; B: subtree.
2.2 OsPGIP空间结构预测
一级结构分析表明, 7个OsPGIP基因可分别编码含有309~380个氨基酸的OsPGIP蛋白, 每个蛋白均含有一个信号肽, 长度在17~31个氨基酸; 除信号肽外, 各OsPGP还具有N-端、9~11个LRR (Leucine-rich repeat)片段和C-端等典型的PGIP结构域; 在这些LRR片段中, 均存在保守的xxLxLxx序列, L为亮氨酸或脂肪族氨基酸残基, x为任意氨基酸(图2)。OsPGIP的二级结构主要由α-螺旋、β-折叠和随机卷曲组成, 且多以随机卷曲为主; OsPGIP的N-端和C-端分别有8~13个半胱氨酸, 可形成3~5个二硫键, 这些二硫键对稳定OsPGIP的特定空间结构起重要作用(表2)。Table 2
表2
表2OsPGIP二级结构预测结果
Table 2
| 蛋白 Protein | α-螺旋 α-helix | 延伸链 Extended strand | 随机卷曲 Random coil | 半胱氨酸数 Number of cysteine | 二硫键位置 Position of disulfide bond |
|---|---|---|---|---|---|
| OsPGIP1 | 36.25 | 10.68 | 53.07 | 8 | 56-63, 278-298, 300-308 |
| OsPGIP2 | 46.20 | 10.23 | 43.57 | 9 | 34-64, 65-72, 310-323, 331-339 |
| OsPGIP3 | 32.74 | 13.86 | 53.39 | 10 | 17-27, 56-64, 322-328, 330-337 |
| OsPGIP4 | 44.70 | 9.74 | 45.56 | 9 | 33-63, 64-71, 333-339, 341-348 |
| OsFOR1 | 30.12 | 10.24 | 59.64 | 10 | 27-58, 59-66, 312-320 |
| OsPGIP6 | 49.21 | 4.47 | 46.32 | 10 | 64-73, 114-137, 348-370, 372-379 |
| OsPGIP7 | 27.19 | 10.82 | 61.99 | 13 | 16-65, 25-34, 66-73, 322-328, 330-337 |
新窗口打开|下载CSV
图2
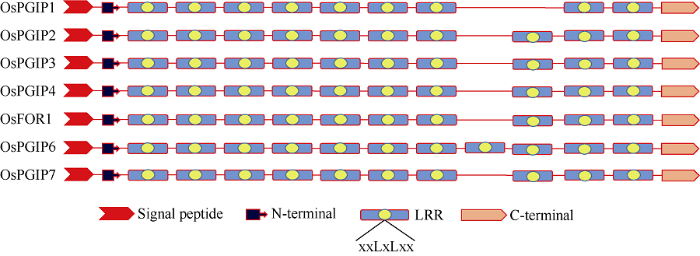 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图27个OsPGIP一级结构
Fig. 2Primary structure of OsPGIPs
利用同源建模的方法预测OsPGIP的三级结构, 结果表明, 利用SWISS-MODEL和CPHmodels预测的所有OsPGIP的三维空间结构均比较相似, 它们由多个随机卷曲-α-螺旋-随机卷曲-β-折叠区组成的线圈状结构按右手螺旋规则形成一个特定的凹面(图3-A)。但不同预测软件得到的同一OsPGIP三维空间结构并不完全相同, 有的甚至差异较大, 特别是SCRATCH的模拟结果与其他两种方法所得结果几乎完全不同。分别以SWISS-MODEL和CPHmodels模拟OsPGIP6的空间结构, 结果显示, 尽管它们参与预测的氨基酸数(34~369、39~354)、氨基酸覆盖率(88.4%、83.2%)及与模板的相似性(30.1%、33.6%)差异均不大, 但其预测结果仍有差异, 以两种方法预测的OsPGIP6空间结构中, 其LRR区分别存在13个和10个β-折叠, 而以SCRATCH模拟的则无典型的PGIP空间结构特征, LRR区亦不存在β-折叠; 同样的方法应用于预测OsPGIP7的空间结构得到的结果相似(图3-B)。以上述3种方式模拟的OsPGIP7结构与水稻纹枯病菌的RsPG2进行蛋白质对接发现, 尽管获得的对接示意图并不完全一致, 但均显示为OsPGIP7的凹面与RsPG2的裂隙区部位进行紧密结合, 说明该部位是两者的互作区域, 这与我们前期研究OsPGIP1、OsPGIP2与RsPG1对接的结果不一致[12], 说明不同PGIP与不同PG的互作方式是多种多样的(图3-C)。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3OsPGIP蛋白质3D结构模拟图
卡通图中红色、黄色和绿色分别表示α-螺旋、β-折叠和随机卷曲。A: 具有典型PGIP空间结构的各OsPGIP3D模型; B: 分别采用SWISS-MODEL、CPHmodels和SCRATCH方法模拟的OsPGIP6和OsPGIP7空间结构模型; C: OsPGIP7与RsPG2的蛋白分子对接, 网眼图为OsPGIP7, 卡通图为RsPG2。
Fig. 33D structure of OsPGIPs
Red, yellow, and green in the cartoon figures mean α-helix, β-sheet, and random coil, respectively. A: 3D structure of OsPGIPs with typical spatial structure of PGIP. B: 3D structures of OsPGIP6 and OsPGIP7 constructed with homologous modeling methods of SWISS-MODEL, CPH model and SCRATCH. C: Protein docking of OsPGIP7 and RsPG2. Mesh and cartoon figures mean OsPGIP7 and RsPG2, respectively.
2.3 OsPGIP性质与功能预测
根据前人报道, 7个OsPGIP的编码基因分别位于3条染色体上, 其中OsPGIP1~OsPGIP4位于水稻5号染色体, OsFOR1和OsPGIP6位于8号染色体, OsPGIP7位于9号染色体[19,20,21]。经预测, 7个OsPGIP的分子量均在35 kD左右(32.75~38.79 kD), 等电点为4.73~8.37。其中, 已报道能抑制病菌PG活性或过表达能提高植物抗性的OsPGIP1、OsPGIP4和OsFOR1[20,23-24]其等电点均近中性或偏碱性。预测的表达产物除OsPGIP2不稳定外, 其他均稳定; 而且所有预测蛋白均为疏水蛋白、脂溶性好、具有跨膜结构、定位于细胞外, 且均存在1到多个N-糖基化位点。另外, 这些蛋白在大肠杆菌中原核表达后均溶解度极低或完全不溶(表3)。Table 3
表3
表3在线软件预测的OsPGIP蛋白性质与功能
Table 3
| 蛋白 Protein | 染色体 位置Location | 分子量 Molecular weight (kD) | 等电点 pI | 不稳定 指数 Instability index | 脂溶性 指数 Aliphatic index | 总平均 亲水性 Grand average of hydropathicity | 跨膜域 Trans- membrane | 亚细胞定位 Subcellular localization | N-糖基化 位点数 Number of N-glycosylation sites | 大肠杆菌中表达的溶解性 Chance of solubility expressed in E. coli (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| OsPGIP1 | Chr. 5 | 32.75 | 6.98 | 31.82 | 98.58 | 0.183 | Yes | Extracellular | 4 | 0 |
| OsPGIP2 | Chr. 5 | 36.97 | 4.73 | 40.16 | 103.33 | 0.080 | Yes | Extracellular | 8 | 0 |
| OsPGIP3 | Chr. 5 | 36.14 | 5.94 | 34.36 | 95.63 | 0.181 | Yes | Extracellular | 5 | 0 |
| OsPGIP4 | Chr. 5 | 37.22 | 8.37 | 33.98 | 101.58 | 0.203 | Yes | Extracellular | 4 | 0 |
| OsFOR1 | Chr. 8 | 35.46 | 7.09 | 36.08 | 99.94 | 0.041 | Yes | Extracellular | 1 | 0 |
| OsPGIP6 | Chr. 8 | 38.79 | 5.86 | 35.69 | 106.42 | 0.286 | Yes | Extracellular | 1 | 0 |
| OsPGIP7 | Chr. 9 | 35.88 | 5.90 | 33.26 | 101.87 | 0.265 | Yes | Extracellular | 3 | 0.7 |
新窗口打开|下载CSV
2.4 OsPGIP基因的表达特征
分别取不同生育期抗、感和高感纹枯病水稻品种YSBR1、徐稻3号和Lemont的叶鞘组织, 提取总RNA, qRT-PCR检测各OsPGIP基因的表达量。数据分析结果表明, 苗期各OsPGIP基因均以在抗性品种YSBR1中表达量最高, 而成株期和穗期则各基因表达规律不明显。但相较于其他基因, OsFOR1和OsPGIP7在成株期水稻叶鞘中表达量较高, 而OsPGIP2、OsPGIP4和OsFOR1则在穗期叶鞘中表达量较高; 以OsPGIP基因的总表达量来比较, 则以高感品种Lemont最高(图4)。此结果与作者前期研究OsPGIP1基因表达特征时的结果亦不一致[25]。图4
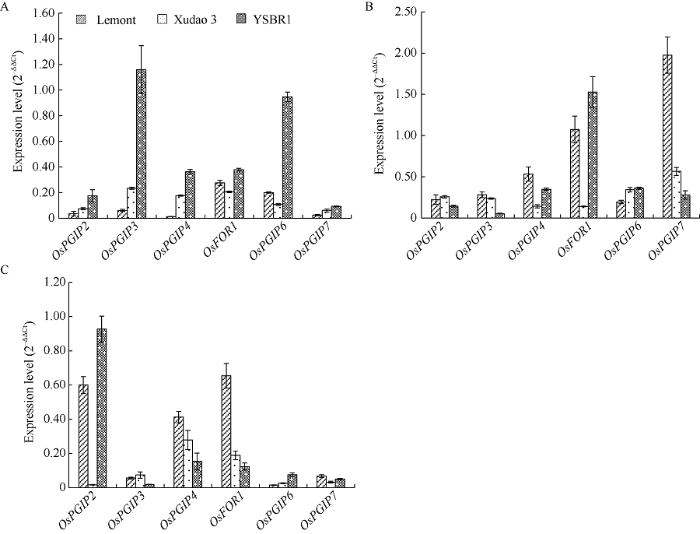 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4不同生育期抗、感纹枯病水稻品种叶鞘中各OsPGIP基因的表达量
A: 苗期; B: 成株期; C: 穗期。
Fig. 4Expression levels of OsPGIP genes in rice cultivars with different resistance to sheath blight at different growth stages
A: seedling stage; B: adult-plant stage; C: spike stage.
分别以接种水稻纹枯病菌、低温和遮光处理等造成生物逆境和非生物逆境, 检测各OsPGIP基因在不同逆境条件下在不同水稻品种植株叶鞘中的表达水平。结果表明, 在低温条件下, 高感品种Lemont中的OsPGIP2、OsPGIP3基因和抗性品种YSBR1中的OsPGIP6基因的表达量均明显下调, 其他各品种中的所有基因均上调表达, 以YSBR1中的OsPGIP4和OsFOR1基因表达量最高(相对表达量分别为8.55和14.53), 且各基因的总体平均上调倍数以抗性品种YSBR1中的为最大; 遮光处理后, 除OsPGIP2基因在Lemont和徐稻3号中表达量无明显变化外(相对表达量分别为0.97和1.10), 其他各OsPGIP基因在各品种中均显著上调表达, 尤以抗性品种YSBR1中的表达量上调倍数最大, 平均为9.09, OsPGIP2的表达量是对照的18.11倍; 接种纹枯病菌后, 不同基因的表达水平亦有上调或下调, 但总体相比对照来说, 有明显上调趋势, 特别是OsFOR1基因和OsPGIP6基因, 最高上调倍数为9.53和6.66。这些结果与作者前期研究OsPGIP1基因表达特征的结果一致, 即在逆境条件下大多数OsPGIP基因可通过提高自身的表达水平来帮助植物增加抗逆性。
图5
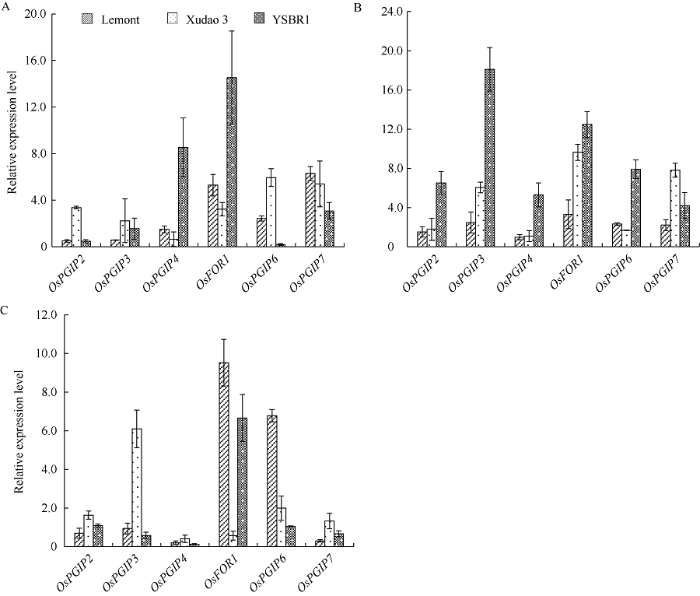 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5不同逆境条件下抗、感纹枯病水稻品种叶鞘中各OsPGIP基因的表达量
A: 低温; B: 遮光; C: 接种。
Fig. 5Expression levels of OsPGIP genes in rice cultivars with different resistance to sheath blight under different stress condition.
A: low temperature; B: dark; C: inoculation.
3 讨论
PGIP作为植物细胞壁的重要组成部分, 其编码基因并不存在大规模的扩张, 只是少数几个或十几个基因以家族的形式存在, 一般为2~16个[26,27,28]。在这些基因中, 除拟南芥2个AtPGIP基因外, 其他均未见有内含子的报道, 但他们可因转座子的插入而导致基因失活[26,28]。水稻中目前共存在7个PGIP基因, 经对DNA和cDNA测序后发现, 7个OsPGIP基因均无内含子, 也不存在转座子插入现象。尽管在这7个OsPGIP中, 有些相似度较高或与其他禾本科植物PGIP在系统发育进化分析时归于同一群组, 但有些OsPGIP的遗传差异较大, 可能来自于不同的祖先基因。Liu等[10]曾对水稻PGIP与其他植物来源的PGIP做过系统发育分析, 认为所有禾本科、豆科、十字花科和林木果树可各自被单独归为一组, 水稻中的7个OsPGIP与小麦、玉米、高粱等的PGIP高度相似。但上述研究仅选用了来自少数几个物种来源的51个PGIP, 如十字花科仅选择了油菜和拟南芥, 豆科也仅选择了大豆和同属不同种的两种菜豆。当将样本量放大, 对NCBI中所有明确释意为PGIP的蛋白序列进行多序列比对与系统进化发育分析时, 我们发现, 尽管相同或相近物种来源的PGIP有归为一类的趋势, 但亦有少数PGIP与其他物种的PGIP反而有着更高的相似性。这些结果说明, 即使是相同或相近物种的PGIP也有可能是来自不同的祖先, 而不同物种中的PGIP亦有来自共同祖先的可能性, 尽管这一比例相对较小。植物细胞壁是高度动态的胞外结构, 在植物生长发育与受到逆境胁迫时, 其可不断被重构, 作为细胞壁的一部分, PGIP被向胞外分泌的能力与其N-端和LRR区有关[29]。而多个LRR片段构成的凹面, 带有大量的负电荷, 负责着与病菌PG的互作[30,31,32]。但不同物种PGIP、甚至同一物种的不同PGIP对不同病原菌或同一病原菌不同PG的抑制能力差异较大, 除因互作方式多样(如竞争性抑制、非竞争性抑制和混合型等) 外, 蛋白质本身氨基酸序列、折叠方式、表面电荷和空间结构等都对互作亦有影响[11,33-35]。只是至目前为止, 由于对PGIP晶体结构的研究还非常少, 关于PGIP-PG互作的晶体结构模型更是还未见报道, 因此对于PGIP功能及其与PG互作的研究多还停留在借助生物信息学分析的结果上。目前进行蛋白结构预测的在线软件很多, 如SWISS- MODEL、CHPmodels、SCRATCH、Modeller、3D-JIGSAW、EsyPred3D、RaptorX、Hhpred等。尽管不同方法预测的结果并不完全相同, 如利用酰胺交换质谱法和小角度X衍射法预测PvPGIP2与FpPG的互作时, 两种方法得到的结果分别是PvPGIP2的凹面与FvPG的N-端外侧的β-折叠区或C-端及活性裂隙区互作, 但这些结果可以通过单氨基酸的定点突变加以验证[31,36-37]。所以, 进行蛋白质空间结构、功能、性质, 以及配体与配基间的对接预测分析等是研究蛋白功能及互作机制的基础, 只是在预测分析时要进行多种方法的比较, 并对预测模型加以修饰和评价。如本研究中分别采用SWISS-MODEL、CHPmodels和SCRATCH在线软件预测了OsPGIP的空间结构, SCRATCH预测的模型显然不符合研究者对植物PGIP空间结构的认知, 因此也不宜用之来进一步研究OsPGIP的结构、性质、功能及与PG的互作等。
PGIP基因除可以组成型表达以外, 还可以受到多种外源因素的诱导表达, 如植物激素、机械损伤、生物和非生物逆境等。菜豆中的4个PvPGIP基因分布在其B2连锁群一个50 kb的区间内, 这些基因在核苷酸水平上的相似性大于80%, 说明其来源于共同的祖先基因, 但它们的表达却差异极大, 如在幼叶、下胚轴、根和荚中PvPGIP1基因均不表达, 而PvPGIP2却在所有器官中表达, PvPGIP3和PvPGIP4则只在根中微量表达[16,18]。受病原菌侵染后, 病菌与寄主的亲和性互作和非亲和性互作则会分别导致PGIP基因的转录产物提早或延迟在寄主特定部位局部积累[16,38-39]。灰霉病菌的侵染可引起拟南芥中两个AtPGIP基因上调表达, 但却是通过不同的信号传导途径[26]。这些结果均表明, 植物中PGIP基因的表达水平受其自身生育期、部位和诸多外源环境的影响。水稻中的7个OsPGIP基因在不同组织器官中的表达水平亦不同; 在赤霉素、激动素和萘乙酸处理后, 籼稻品种明恢63中除OsPGIP2基因在激动素处理后表达量下调外, 其他基因的表达量均略有上调; 而在粳稻品种中花11中, 使用脱落酸、芸薹素内酯、赤霉素、生长素、激动素、茉莉酸和水杨酸处理后, 随着时间的推移, 绝大多数OsPGIP基因的表达量均有上调[19]。除不同类型水稻外, 本研究发现, 对纹枯病具不同抗性的水稻品种在遇到生物与非生物逆境时, 其体内OsPGIP基因的表达亦是有显著差异的。如果能研明这些基因的表达调控及与病原菌PG互作的机制, 将为其在水稻抗病育种中的应用奠定基础。
4 结论
克隆了水稻OsPGIP基因家族除OsPGIP1外的剩余6个基因, 对它们进行了系统进化分析, 预测了其编码产物的一、二、三级结构和生物学功能, 分析了各基因在水稻不同生育期和逆境下的表达特征, 为揭示OsPGIP基因家族不同成员在不同逆境条件下的潜在功能和在水稻抗病育种中的应用奠定了基础。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1111/ppa.1956.5.issue-4URL [本文引用: 1]
DOI:10.1134/s0006297908100015URLPMID:18991551 [本文引用: 1]

It is generally believed that plants
DOI:10.1111/j.1438-8677.2008.00175.xURLPMID:19689781 [本文引用: 1]

Polygalacturonase-inhibiting proteins (PGIPs) are plant cell wall proteins that specifically inhibit the activity of endopolygalacturonases (PGs) produced by fungi during the infection process. The interaction with PGIPs limits the destructive potential of PGs and may trigger plant defence responses through the release of elicitor active oligogalacturonides. In order to pinpoint the residues of PvPGIP2 from Phaseolus vulgaris involved in the interaction with PGs, we used site-directed mutagenesis to mutate the residues D131, D157 and D203, and tested for the inhibitory activity of the mutant proteins expressed in Pichia pastoris against Fusarium phyllophilum and Aspergillus niger PGs. Here, we report that mutation of these residues affects the inhibition capacity of PvPGIP2 against F. phyllophilum PG.
DOI:10.1111/j.1365-313X.2007.03398.xURLPMID:18088306 [本文引用: 1]

In addition to the role of the cell wall as a physical barrier against pathogens, some of its constituents, such as pectin-derived oligogalacturonides (OGA), are essential components for elicitation of defence responses. To investigate how modifications of pectin alter defence responses, we expressed the fruit-specific Fragaria x ananassa pectin methyl esterase FaPE1 in the wild strawberry Fragaria vesca. Pectin from transgenic ripe fruits differed from the wild-type with regard to the degree and pattern of methyl esterification, as well as the average size of pectin polymers. Purified oligogalacturonides from the transgenic fruits showed a reduced degree of esterification compared to oligogalacturonides from wild-type fruits. This reduced esterification is necessary to elicit defence responses in strawberry. The transgenic F. vesca lines had constitutively activated pathogen defence responses, resulting in higher resistance to the necrotropic fungus Botrytis cinerea. Further studies in F. vesca and Nicotiana benthamiana leaves showed that the elicitation capacity of the oligogalacturonides is more specific than previously envisaged.
DOI:10.3389/fpls.2013.00030URLPMID:23440336 [本文引用: 1]

Systemic acquired resistance (SAR) is an inducible defense mechanism in plants that confers enhanced resistance against a variety of pathogens. SAR is activated in the uninfected systemic (distal) organs in response to a prior (primary) infection elsewhere in the plant. SAR is associated with the activation of salicylic acid (SA) signaling and the priming of defense responses for robust activation in response to subsequent infections. The activation of SAR requires communication by the primary infected tissues with the distal organs. The vasculature functions as a conduit for the translocation of factors that facilitate long-distance intra-plant communication. In recent years, several metabolites putatively involved in long-distance signaling have been identified. These include the methyl ester of SA (MeSA), the abietane diterpenoid dehydroabietinal (DA), the dicarboxylic acid azelaic acid (AzA), and a glycerol-3-phosphate (G3P)-dependent factor. Long-distance signaling by some of these metabolites also requires the lipid-transfer protein DIR1 (DEFECTIVE IN INDUCED RESISTANCE 1). The relative contribution of these factors in long-distance signaling is likely influenced by environmental conditions, for example light. In the systemic leaves, the AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (ALD1)-dependent production of the lysine catabolite pipecolic acid (Pip), FLAVIN-DEPENDENT MONOOXYGENASE1 (FMO1) signaling, as well as SA synthesis and downstream signaling are required for the activation of SAR. This review summarizes the involvement and interaction between long-distance SAR signals and details the recently discovered role of Pip in defense amplification and priming that allows plants to acquire immunity at the systemic level. Recent advances in SA signaling and perception are also highlighted.
DOI:10.1186/s12870-016-0959-1URLPMID:28103793 [本文引用: 1]

BACKGROUND: Oligogalacturonides (OGs) are important components of damage-associated molecular pattern (DAMP) signaling and influence growth regulation in plants. Recent studies have focused on the impact of long OGs (degree of polymerization (DP) from 10-15), demonstrating the induction of plant defense signaling resulting in enhanced defenses to necrotrophic pathogens. To clarify the role of trimers (trimeric OGs, DP3) in DAMP signaling and their impact on plant growth regulation, we performed a transcriptomic analysis through the RNA sequencing of Arabidopsis thaliana exposed to trimers. RESULTS: The transcriptomic data from trimer-treated Arabidopsis seedlings indicate a clear activation of genes involved in defense signaling, phytohormone signaling and a down-regulation of genes involved in processes related to growth regulation and development. This is further accompanied with improved defenses against necrotrophic pathogens triggered by the trimer treatment, indicating that short OGs have a clear impact on plant responses, similar to those described for long OGs. CONCLUSIONS: Our results demonstrate that trimers are indeed active elicitors of plant defenses. This is clearly indicated by the up-regulation of genes associated with plant defense signaling, accompanied with improved defenses against necrotrophic pathogens. Moreover, trimers simultaneously trigger a clear down-regulation of genes and gene sets associated with growth and development, leading to stunted seedling growth in Arabidopsis.
DOI:10.1073/pnas.92.10.4095URLPMID:11607534 [本文引用: 1]

Jasmonic acid, synthesized from linolenic acid (the octadecanoid pathway), has been proposed to be part of a signal transduction pathway that mediates the induction of defensive genes in plants in response to oligouronide and polypeptide signals generated by insect and pathogen attacks. We report here that the induction of proteinase inhibitor accumulation in tomato leaves by plant-derived oligogalacturonides and fungal-derived chitosan oligosaccharides is severely reduced by two inhibitors (salicylic acid and diethyldi-thiocarbamic acid) of the octadecanoid pathway, supporting a role for the pathway in signaling by oligosaccharides. Jasmonic acid levels in leaves of tomato plants increased several fold within 2 hr after supplying the polypeptide systemin, oligogalacturonides, or chitosan to the plants through their cut stems, as expected if they utilize the octadecanoid pathway. The time course of jasmonic acid accumulation in tomato leaves in response to wounding was consistent with its proposed role in signaling proteinase inhibitor mRNA and protein synthesis. The cumulative evidence supports a model for the activation of defensive genes in plants in response to insect and pathogen attacks in which various elicitors generated at the attack sites activate the octadecanoid pathway via different recognition events to induce the expression of defensive genes in local and distal tissues of the plants.
DOI:10.1146/annurev.phyto.39.1.313URLPMID:11701868 [本文引用: 1]

Polygalacturonase-inhibiting proteins (PGIPs) are extracellular plant proteins capable of inhibiting fungal endopolygalacturonases (PGs). Plants have evolved different PGIPs with specific recognition abilities against the many PGs produced by fungi. The genes encoding PGIPs are organized into families, and different members of each family may encode proteins with nearly identical characteristics but different specificities and regulation. PGIPs are typically induced by pathogen infection and stress-related signals. The recognition ability of PGIPs resides in their LRR (leucine-rich repeat) structure, where solvent-exposed residues in the beta-strand/beta-turn motifs of the LRRs are determinants of specificity. Manipulation of the primary structure of PGIPs is expected to generate more efficient PGIPs with novel recognition specificities to protect crop plants against pathogens.
DOI:10.1016/S0981-9428(00)01228-6URL [本文引用: 1]
DOI:10.1038/srep39840URLPMID:28079053 [本文引用: 2]

Polygalacturonase-inhibiting protein (PGIP), belonging to a group of plant defence proteins, specifically inhibits endopolygalacturonases secreted by pathogens. Herein, we showed that purified GhPGIP1 is a functional inhibitor of Verticillium dahliae and Fusarium oxysporum f. sp. vasinfectum, the two fungal pathogens causing cotton wilt. Transcription of GhPGIP1 was increased in cotton upon infection, wounding, and treatment with defence hormone and H2O2. Resistance by GhPGIP1 was examined by its virus-induced gene silencing in cotton and overexpression in Arabidopsis. GhPGIP1-silenced cotton was highly susceptible to the infections. GhPGIP1 overexpression in transgenic Arabidopsis conferred resistance to the infection, accompanied by enhanced expression of pathogenesis-related proteins (PRs), isochorismate synthase 1 (ICS1), enhanced disease susceptibility 1 (EDS1), and phytoalexin-deficient 4 (PAD4) genes. Transmission electron microscopy revealed cell wall alteration and cell disintegration in plants inoculated with polygalacturonase (PGs), implying its role in damaging the cell wall. Docking studies showed that GhPGIP1 interacted strongly with C-terminal of V. dahliae PG1 (VdPG1) beyond the active site but weakly interacted with C-terminal of F. oxysporum f. sp. vasinfectum (FovPG1). These findings will contribute towards the understanding of the roles of PGIPs and in screening potential combat proteins with novel recognition specificities against evolving pathogenic factors for countering pathogen invasion.
DOI:10.3389/fpls.2015.00146URLPMID:25852708 [本文引用: 2]

Polygalacturonase inhibiting proteins (PGIPs) are cell wall proteins that inhibit the pectin-depolymerizing activity of polygalacturonases secreted by microbial pathogens and insects. These ubiquitous inhibitors have a leucine-rich repeat structure that is strongly conserved in monocot and dicot plants. Previous reviews have summarized the importance of PGIP in plant defense and the structural basis of PG-PGIP interaction; here we update the current knowledge about PGIPs with the recent findings on the composition and evolution of pgip gene families, with a special emphasis on legume and cereal crops. We also update the information about the inhibition properties of single pgip gene products against microbial PGs and the results, including field tests, showing the capacity of PGIP to protect crop plants against fungal, oomycetes and bacterial pathogens.
DOI:10.1186/s12284-019-0318-6URLPMID:31359264 [本文引用: 2]

BACKGROUND: An economic strategy to control plant disease is to improve plant defense to pathogens by deploying resistance genes. Plant polygalacturonase inhibiting proteins (PGIPs) have a vital role in plant defense against phytopathogenic fungi by inhibiting fungal polygalacturonase (PG) activity. We previously reported that rice PGIP1 (OsPGIP1) inhibits PG activity in Rhizoctonia solani, the causal agent of rice sheath blight (SB), and is involved in regulating resistance to SB. RESULT: Here, we report that OsPGIP2, the protein ortholog of OsPGIP1, does not possess PGIP activity; however, a few amino acid substitutions in a derivative of OsPGIP2, of which we provide support for L233F being the causative mutation, appear to impart OsPGIP2 with PG inhibition capability. Furthermore, the overexpression of mutated OsPGIP2(L233F) in rice significantly increased the resistance of transgenic lines and decreased SB disease rating scores. OsPGIP2(L233F) transgenic lines displayed an increased ability to reduce the tissue degradation caused by R. solani PGs as compared to control plants. Rice plants overexpressing OsPGIP2(L233F) showed no difference in agronomic traits and grain yield as compared to controls, thus demonstrating its potential use in rice breeding programs. CONCLUSIONS: In summary, our results provide a new target gene for breeding SB resistance through genome-editing or natural allele mining.
DOI:10.1007/s10265-012-0515-5URL [本文引用: 1]

Regulation of defense in plants is a complex process mediated by various signaling pathways. Promoter analysis of defense-related genes is useful to understand these signaling pathways involved in regulation. To this end, the regulation of the polygalacturonase-inhibiting protein encoding gene from Vitis vinifera L. (Vvpgip1) was analyzed with regard to expression pattern and induction profile as well as the promoter in terms of putative regulatory elements present, core promoter size and the start of transcription. Expression of Vvpgip1 is tissue-specific and developmentally regulated. Vvpgip1 expression was induced in response to auxin, salicylic acid and sugar treatment, wounding and pathogen infection. The start of transcription was mapped to 17?bp upstream of the ATG and the core promoter was mapped to the 137?bp upstream of the ATG. Fructose- and Botrytis responsiveness were identified in the region between positions –3.1 and –1.5?kb. The analyses showed induction in water when the leaves were submersed and this response and the response to wounding mapped to the region between positions –1.1 and –0.1?kb. In silico analyses revealed putative cis-acting elements in these areas that correspond well to the induction stimuli tested.]]>
DOI:10.1186/s12870-014-0189-3URLPMID:25034494 [本文引用: 1]

BACKGROUND: Polygalacturonase-inhibiting proteins (PGIPs) are leucine-rich repeat (LRR) plant cell wall glycoproteins involved in plant immunity. They are typically encoded by gene families with a small number of gene copies whose evolutionary origin has been poorly investigated. Here we report the complete characterization of the full complement of the pgip family in soybean (Glycine max [L.] Merr.) and the characterization of the genomic region surrounding the pgip family in four legume species. RESULTS: BAC clone and genome sequence analyses showed that the soybean genome contains two pgip loci. Each locus is composed of three clustered genes that are induced following infection with the fungal pathogen Sclerotinia sclerotiorum (Lib.) de Bary, and remnant sequences of pgip genes. The analyzed homeologous soybean genomic regions (about 126 Kb) that include the pgip loci are strongly conserved and this conservation extends also to the genomes of the legume species Phaseolus vulgaris L., Medicago truncatula Gaertn. and Cicer arietinum L., each containing a single pgip locus. Maximum likelihood-based gene trees suggest that the genes within the pgip clusters have independently undergone tandem duplication in each species. CONCLUSIONS: The paleopolyploid soybean genome contains two pgip loci comprised in large and highly conserved duplicated regions, which are also conserved in bean, M. truncatula and C. arietinum. The genomic features of these legume pgip families suggest that the forces driving the evolution of pgip genes follow the birth-and-death model, similar to that proposed for the evolution of resistance (R) genes of NBS-LRR-type.
[本文引用: 1]
URL [本文引用: 3]

The expression analysis of the four polygalacturonase-inhibiting protein (Pgip) genes composing the bean (Phaseolus vulgaris L.) Pgip family showed a pattern of transcriptional variation in young leaves, hypocotyls, roots and pods with Pvpgip1 not expressed, Pvpgip2 expressed in all organs, Pvpgip3 and Pvpgip4 poorly expressed in roots. We compared also transcript accumulation of the four Pvpgip genes during infection of bean plants with the fungal pathogens Botrytis cinerea, Sclerotinia sclerotiorum and Colletotrichum demuthianum. qRT-PCR analyses showed that the transcript level of Pvpgip1, Pvpgip2 and Pvpgip3 increases significantly following fungal infection, whereas Pvpgip4 remains unchanged. The level of induction was different between the three genes, Pvpgip2 exhibiting the strongest transcript accumulation. The induction pattern was similar in the pathosystems bean-S. sclerotiorum, bean-B. cinerea, and in the compatible interaction bean-C. lindemuthianum, with a maximum of transcript accumulation in the late stage of infection. Instead, in the incompatible interaction bean-C. lindemuthianum, Pvpgip1, Pvpgip2 and Pvpgip3 showed an early and transient transcript accumulation, with Pvpgip2 exhibiting an earlier and higher induction. These results extend previous analyses of the whole Pup gip transcript and provide additional evidences of the relevant role of PvPGIP2 in plant defence.
DOI:10.1007/s004250050308URLPMID:9637069 [本文引用: 1]

Polygalacturonase-inhibiting proteins (PGIPs), leucine-rich repeat (LRR) proteins evolutionarily related to several plant resistance genes, bind to and regulate the action of fungal endopolygalacturonases. In Phaseolus vulgaris L., PGIPs are encoded by a gene family comprising at least five members. As a start for a systematic analysis of the regulation of the pgip family, we have analysed the ability of the promoter of the bean gene pgip-1 to direct expression of beta-glucuronidase (GUS) in transfected tobacco protoplasts, microbombarded bean and tobacco leaves, and transgenic tobacco plants. In protoplasts, the pgip-1 gene region from nucleotide (nt) -2004 to nt +27 directed a level of expression that was as high as that directed by the cauliflower mosaic virus (CaMV) 35S promoter and could not be further induced by elicitor treatment; alteration of the region immediately following the TATAA sequence at nt -29 abolished expression. Upon stable integration into tobacco plants of the pgip-1 promoter-GUS construct, as well as of a -394 deletion, expression was detected for both constructs mainly in the stigma and, to a lesser extent, in the anthers and in the conductive vascular tissue. The promoter responded to wounding but not to oligogalacturonides, fungal glucan, salicylic acid, cryptogein, or pathogen infection. This expression pattern does not mirror that of the whole pgip gene family.
DOI:10.1104/pp.104.044644URLPMID:15299124 [本文引用: 2]

Polygalacturonase-inhibiting proteins (PGIPs) are extracellular plant inhibitors of fungal endopolygalacturonases (PGs) that belong to the superfamily of Leu-rich repeat proteins. We have characterized the full complement of pgip genes in the bean (Phaseolus vulgaris) genotype BAT93. This comprises four clustered members that span a 50-kb region and, based on their similarity, form two pairs (Pvpgip1/Pvpgip2 and Pvpgip3/Pvpgip4). Characterization of the encoded products revealed both partial redundancy and subfunctionalization against fungal-derived PGs. Notably, the pair PvPGIP3/PvPGIP4 also inhibited PGs of two mirid bugs (Lygus rugulipennis and Adelphocoris lineolatus). Characterization of Pvpgip genes of Pinto bean showed variations limited to single synonymous substitutions or small deletions. A three-amino acid deletion encompassing a residue previously identified as crucial for recognition of PG of Fusarium moniliforme was responsible for the inability of BAT93 PvPGIP2 to inhibit this enzyme. Consistent with the large variations observed in the promoter sequences, reverse transcription-PCR expression analysis revealed that the different family members differentially respond to elicitors, wounding, and salicylic acid. We conclude that both biochemical and regulatory redundancy and subfunctionalization of pgip genes are important for the adaptation of plants to pathogenic fungi and phytophagous insects.
DOI:10.1007/s00299-012-1239-7URL [本文引用: 5]

All the seven Ospgip genes showed variable expression patterns in Minghui 63 and their expressions were regulated by different phytohormone treatments or fungal infection in Minghui 63 and Zhonghua 11.]]>
DOI:10.1023/B:PLAN.0000006940.89955.f1URL [本文引用: 3]

We have isolated a cDNA clone, OsFOR1, from the immature panicles of rice. The OsFOR1 (Oryza sativa floral organ regulator 1) gene encodes a protein that contains a leucine-rich repeat (LRR) domain. This domain comprises 10 tandem repeats of a canonical 24-amino acid LRR sequence. The structure and the number of LRRs for OsFOR1 are similar to those of polygalacturonase-inhibiting proteins (PGIPs) from various other plant species. Moreover, the OsFOR1 recombinant protein, when fused to maltose-binding protein (MBP), shows PGIP activity against the Aspergillus niger polygalacturonase. OsFOR1 is highly expressed in the calli and immature and mature panicles, while detectable at only low levels in seedling roots and mature stems. Insitu hybridization experiments showed the transcripts of OsFOR1 are present in young spikelet primordia and in almost all of the young floral organs. Transgenic approaches were used to study invivo functioning. Antisense expression of OsFOR1 resulted in an increase in the numbers of floral organs, including the stamen, carpel, palea/lemma, stigma, and lodicule. OsFOR1 transcript was not detected in the frizzy panicle mutant, which is defective in its spikelet formation but normal in inflorescence-meristem initiation and maintenance. Therefore, we suggest that OsFOR1 plays a role in the formation and/or maintenance of floral organ primordia.]]>
DOI:10.1007/s00122-006-0378-zURL [本文引用: 2]

Polygalacturonase-inhibiting proteins (PGIPs) are leucine-rich repeat (LRR) proteins involved in plant defence. A number of PGIPs have been characterized from dicot species, whereas only a few data are available from monocots. Database searches and genome-specific cloning strategies allowed the identification of four rice (Oryza sativa L.) and two wheat (Triticum aestivum L.) Pgip genes. The rice Pgip genes (Ospgip1, Ospgip2, Ospgip3 and Ospgip4) are distributed over a 30kbp region of the short arm of chromosome 5, whereas the wheat Pgip genes, Tapgip1 and Tapgip2, are localized on the short arm of chromosome 7B and 7D, respectively. Deduced amino acid sequences show the typical LRR modular organization and a conserved distribution of the eight cysteines at the N- and C-terminal regions. Sequence comparison suggests that monocot and dicot PGIPs form two separate clusters sharing about 40% identity and shows that this value is close to the extent of variability observed within each cluster. Gene-specific RT-PCR and biochemical analyses demonstrate that both Ospgips and Tapgips are expressed in the whole plant or in a tissue-specific manner, and that OsPGIP1, lacking an entire LRR repeat, is an active inhibitor of fungal polygalacturonases. This last finding can contribute to define the molecular features of PG–PGIP interactions and highlights that the genetic events that can generate variability at the Pgip locus are not only limited to substitutions or small insertions/deletions, as so far reported, but can also involve variation in the number of LRRs.]]>
DOI:10.1023/A:1017213630972URL [本文引用: 1]
DOI:10.1094/PDIS-03-15-0305-REURLPMID:30694142 [本文引用: 1]

Rice sheath blight (SB), caused by necrotrophic pathogen Rhizoctonia solani, is one of the most destructive rice diseases, and no major resistance genes are available. Polygalacturonase-inhibiting proteins (PGIP) are extracellular leucine-rich repeat proteins and play important roles in plant defense against different pathogenic fungi by counteracting secreted fungal polygalacturonases (PG). However, the role of PGIP in conferring resistance to rice SB remains to be thoroughly investigated. Here, we showed that OsPGIP1 is capable of inhibiting PG derived from R. solani. Our real-time reverse-transcription polymerase chain reaction results indicated that resistant rice 'YSBR1' and 'Jasmine 85' express significantly higher levels of OsPGIP1 than susceptible 'Lemont'. Our results also show that OsPGIP1 is most highly expressed at the late tillering stage in the sheath of YSBR1, coinciding with the critical stage of SB development in field. More importantly, the OsPGIP1 level is highly elevated by inoculation with R. solani in resistant cultivars but not in susceptible Lemont. Overexpression of OsPGIP1 significantly increased rice resistance to SB and inhibited tissue degradation caused by R. solani-secreted PG. Furthermore, OsPGIP1 overexpression did not affect rice agronomic traits or yield components. Together, our results not only demonstrate the important role of OsPGIP1 in combatting the rice SB disease but also provide a new avenue to the improvement of rice SB resistance by manipulating an endogenous gene.
DOI:10.1007/s00425-016-2480-zURLPMID:26945855 [本文引用: 1]

MAIN CONCLUSION: OsPGIP4 overexpression enhances resistance to bacterial leaf streak in rice. Polygalacturonase-inhibiting proteins are thought to play important roles in the innate immunity of rice against fungi. Here, we show that the chromosomal location of OsPGIP4 coincides with the major bacterial leaf streak resistance quantitative trait locus qBlsr5a on the short arm of chromosome 5. OsPGIP4 expression was up-regulated upon inoculation with the pathogen Xanthomonas oryzae pv. oryzicola strain RS105. OsPGIP4 overexpression enhanced the resistance of the susceptible rice variety Zhonghua 11 to RS105. In contrast, repressing OsPGIP4 expression resulted in an increase in disease lesions caused by RS105 in Zhonghua 11 and in Acc8558, a qBlsr5a resistance donor. More interestingly, upon inoculation, the activated expression of pathogenesis-related genes was attenuated for those genes involved in the salicylic acid pathway, while the activated expression of jasmonic acid pathway markers was increased in the overexpression lines. Our results not only provide the first report that rice PGIP could enhance resistant against a bacterial pathogen but also indicate that OsPGIP4 is a potential component of the qBlsr5a locus for bacterial leaf streak in rice.
DOI:10.3969/j.issn.10017216.2012.05.018URL [本文引用: 1]

从水稻基因组DNA中扩增出1 184 bp的Ospgip1基因片段。该片段包含930 bp的完整开放阅读框,终止密码子为TAA,无内含子。RTPCR结果表明,Ospgip1基因在水稻抗、感纹枯病品种中均能表达,但不同生育期、不同部位表达量有差异。实时PCR结果显示,抗病品种YSBR1和中感品种Jasmine 85中Ospgip1的表达量要明显高于感病品种Lemont;稻苗黄化处理后,无论是抗病品种还是感病品种的Ospgip1表达量均显著提高;纹枯病菌侵染使得抗病品种Ospgip1表达量大大增加,而对感病品种影响不大。
[本文引用: 1]
DOI:10.1105/tpc.005165URLPMID:12509524 [本文引用: 3]

Polygalacturonase-inhibiting proteins (PGIPs) are plant proteins that counteract fungal polygalacturonases, which are important virulence factors. Like many other plant defense proteins, PGIPs are encoded by gene families, but the roles of individual genes in these families are poorly understood. Here, we show that in Arabidopsis, two tandemly duplicated PGIP genes are upregulated coordinately in response to Botrytis cinerea infection, but through separate signal transduction pathways. AtPGIP2 expression is mediated by jasmonate and requires COI1 and JAR1, whereas AtPGIP1 expression is upregulated strongly by oligogalacturonides but is unaffected by salicylic acid, jasmonate, or ethylene. Both AtPGIP1 and AtPGIP2 encode functional inhibitors of polygalacturonase from Botrytis, and their overexpression in Arabidopsis significantly reduces Botrytis disease symptoms. Therefore, gene duplication followed by the divergence of promoter regions may result in different modes of regulation of similar defensive proteins, thereby enhancing the likelihood of defense gene activation during pathogen infection.
DOI:10.1007/s00425-008-0733-1URL [本文引用: 1]

Most plants encode a limited set of polygalacturonase inhibitor (PGIP) genes that may be involved in aspects of plant development, but more importantly in the inactivation of polygalacturonases (PG) secreted by pathogens. Previously, we characterized two Brassica napus PGIP genes, BnPgip1 and BnPgip2, which were differentially expressed in response to pathogen infection and wounding. Here we report that the B. napus genome encodes a set of at least 16 PGIP genes that are similar to BnPgip1 or BnPgip2. This is the largest Pgip gene family reported to date. Comparison of the BnPGIPs revealed several sites within the xxLxLxx region of leucine rich repeats that form β-sheets along the interacting face of the PGIP that are hypervariable and represent good candidates for generating PGIP diversity. Characterization of the regulatory regions and RT-PCR studies with gene-specific primers revealed that individual genes were differentially responsive to pathogen infection, mechanical wounding and signaling molecules. Many of the BnPgip genes responded to infection by the necrotic pathogen, Sclerotinia sclerotiorum; however, these genes were also induced either by jasmonic acid, wounding and salicylic acid or some combination thereof. The large number of PGIPs and the differential manner in which they are regulated likely ensures that B. napus can respond to attack from a broad spectrum of pathogens and pests.]]>
DOI:10.1007/s00122-008-0719-1URL [本文引用: 2]
DOI:10.3389/fpls.2015.00660URLPMID:26379688 [本文引用: 1]

The plant endomembrane system is massively involved in the synthesis, transport and secretion of cell wall polysaccharides and proteins; however, the molecular mechanisms underlying trafficking toward the apoplast are largely unknown. Besides constitutive, the existence of a regulated secretory pathway has been proposed. A polygalacturonase inhibitor protein (PGIP2), known to move as soluble cargo and reach the cell wall through a mechanism distinguishable from default, was dissected in its main functional domains (A, B, C, D), and C sub-fragments (C1-10), to identify signals essential for its regulated targeting. The secretion patterns of the fluorescent chimeras obtained by fusing different PGIP2 domains to the green fluorescent protein (GFP) were analyzed. PGIP2 N-terminal and leucine-rich repeat domains (B and C, respectively) seem to operate as holding/releasing signals, respectively, during PGIP2 transit through the Golgi. The B domain slows down PGIP2 secretion by transiently interacting with Golgi membranes. Its depletion leads, in fact, to the secretion via default (Sp2-susceptible) of the ACD-GFP chimera faster than PGIP2. Depending on its length (at least the first 5 leucine-rich repeats are required), the C domain modulates B interaction with Golgi membranes allowing the release of chimeras and their extracellular secretion through a Sp2 independent pathway. The addition of the vacuolar sorting determinant Chi to PGIP2 diverts the path of the protein from cell wall to vacuole, suggesting that C domain is a releasing rather than a cell wall sorting signal.
DOI:10.1021/bi0017632URLPMID:11148052 [本文引用: 1]

A detailed analysis of the secondary structure has been carried out on the polygalacturonase-inhibiting protein (PGIP) from Phaseolus vulgaris, a leucine-rich repeat (LRR) protein present in the cell wall of many plants. Far-UV CD and infrared spectroscopies coupled to constrained secondary structure prediction methods indicated the presence of 12 alpha- and 12 beta-segments, thus allowing a schematic representation of three domains of the protein, namely, the central LRR region and the two cysteine-rich flanking domains. Peptides from endoproteinase-degraded PGIP were analyzed by mass spectrometry, and four disulfide bonds were identified. Mass spectrometric analysis in combination with glycosidase treatments revealed two N-linked oligosaccharides located on Asn 64 and Asn 141. The main structure resembled the typical complex plant N-glycan consisting of a core pentasaccharide beta1,2-xylosylated, carrying an alpha1,3-fucose linked to the innermost N-acetylglucosamine and one outer arm N-acetylglucosamine residue. The schematic representation of PGIP structural domains is discussed in the framework of the structure and function of LRR proteins.
DOI:10.1371/journal.pone.0080610URLPMID:24260434 [本文引用: 2]

Polygalacturonases (PGs) are secreted by phytopathogenic fungi to degrade the plant cell wall homogalacturonan during plant infection. To counteract Pgs, plants have evolved polygalacturonase-inhibiting proteins (PGIPs) that slow down fungal infection and defend cell wall integrity. PGIPs favour the accumulation of oligogalacturonides, which are homogalacturonan fragments that act as endogenous elicitors of plant defence responses. We have previously shown that PGIP2 from Phaseolus vulgaris (PvPGIP2) forms a complex with PG from Fusarium phyllophilum (FpPG), hindering the enzyme active site cleft from substrate. Here we analyse by small angle X-ray scattering (SAXS) the interaction between PvPGIP2 and a PG from Colletotrichum lupini (CluPG1). We show a different shape of the PG-PGIP complex, which allows substrate entry and provides a structural explanation for the different inhibition kinetics exhibited by PvPGIP2 towards the two isoenzymes. The analysis of SAXS structures allowed us to investigate the basis of the inability of PG from Fusarium verticilloides (FvPG) to be inhibited by PvPGIP2 or by any other known PGIP. FvPG is 92.5% identical to FpPG, and we show here, by both loss- and gain-of-function mutations, that a single amino acid site acts as a switch for FvPG recognition by PvPGIP2.
URL [本文引用: 1]

Polygalacturonase-inhibiting proteins (PGIPs), extracellular proteins that specifically inhibit fungal endopolygalacturonases (PGs), play a critical role in plant protection by favouring the accumulation of oligogalacturonides (OGs), which are elicitors of plant defence responses. The genes encoding PGIP2 of P. vulgaris and the variant PGIP2.Q224K were subjected to error prone PCR (epPCR) to generate mutated inhibitors with novel and improved recognition capabilities. Using a Pichia pastoris expression library and a high-throughput screening method, two mutated PvPGIP2.Q224K-derived inhibitors active against the PG produced by the phytopathogenic fungus F. phyllophilum (FpPG) were isolated. Both variants were better inhibitors than PGIP2.Q224K and were characterized by the replacement of the lysine in position 224, supporting the view that the absence of this positively charged amino acid at position 224 is a primary requirement for gaining the inhibition capability against FpPG.
DOI:10.1016/j.bbapap.2003.08.012URL [本文引用: 1]
DOI:10.1104/pp.105.067546URLPMID:16244152

Botrytis cinerea is a phytopathogenic fungus that causes gray mold in >1,000 plant species. During infection, it secretes several endopolygalacturonases (PGs) to degrade cell wall pectin, and among them, BcPG1 is constitutively expressed and is an important virulence factor. To counteract the action of PGs, plants express polygalacturonase-inhibiting proteins (PGIPs) that have been shown to inhibit a variety of PGs with different inhibition kinetics, both competitive and noncompetitive. The PG-PGIP interaction promotes the accumulation of oligogalacturonides, fragments of the plant cell wall that are general elicitors of plant defense responses. Here, we characterize the enzymatic activity of BcPG1 and investigate its interaction with PGIP isoform 2 from Phaseolus vulgaris (PvPGIP2) by means of inhibition assays, homology modeling, and molecular docking simulations. Our results indicate a mixed mode of inhibition. This is compatible with a model for the interaction where PvPGIP2 binds the N-terminal portion of BcPG1, partially covering its active site and decreasing the enzyme affinity for the substrate. The structural framework provided by the docking model is confirmed by site-directed mutagenesis of the residues that distinguish PvPGIP2 from the isoform PvPGIP1. The finding that PvPGIP2 inhibits BcPG1 with a mixed-type kinetics further indicates the versatility of PGIPs to evolve different recognition specificities.
DOI:10.1111/j.1438-8677.2008.00175.xURLPMID:19689781 [本文引用: 1]

Polygalacturonase-inhibiting proteins (PGIPs) are plant cell wall proteins that specifically inhibit the activity of endopolygalacturonases (PGs) produced by fungi during the infection process. The interaction with PGIPs limits the destructive potential of PGs and may trigger plant defence responses through the release of elicitor active oligogalacturonides. In order to pinpoint the residues of PvPGIP2 from Phaseolus vulgaris involved in the interaction with PGs, we used site-directed mutagenesis to mutate the residues D131, D157 and D203, and tested for the inhibitory activity of the mutant proteins expressed in Pichia pastoris against Fusarium phyllophilum and Aspergillus niger PGs. Here, we report that mutation of these residues affects the inhibition capacity of PvPGIP2 against F. phyllophilum PG.
DOI:10.1016/j.funbio.2012.04.010URLPMID:22749160 [本文引用: 1]

By using surface plasmon resonance (SPR) technology, the kinetics of the interaction of various fungal endopolygalacturonases (EPGs) (13 EPGs) with Phaseolus vulgaris (bean) PGIP2 was carried out to determine whether or not there is any interaction between polygalacturonases-inhibiting protein (PGIP) and EPG. The effect of polygalacturonic acid (PGA) on these interactions was also evaluated. The results show that all EPGs evaluated bind to PGIP2, except for AnPGb and the strength of the interaction depends on the EPG/PGIP2 pairing. Further, the presence of PGA has a moderate to strong effect on the EPG/PGIP2 interaction and the strength of the effect is dependent on the exact EPG/PGIP2 pairing. The differences in affinity in the absence and presence of PGA, suggest a certain level of cooperativity. These results indicate a three-component complex similar to that observed for the heparin-ATIII-thrombin, the FGF-FGFR-heparin, or the hedgehog-interference hedgehog-heparan complexes. This data points to an architecture in which the inhibitor binds at a location distant from the substrate binding site. Furthermore, we applied differential proteolysis mass spectrometry (DPMS) to study the location of the binding site between EPG and PGIP2. DPMS studies indicate that PGIP2 does not bind AnPGII, AnPGa, and AnPGc directly over the active site but instead binds on the face opposite to the active site, creating an allosteric interaction.
DOI:10.1104/pp.111.181057URL [本文引用: 1]

We report here the low-resolution structure of the complex formed by the endo-polygalacturonase from Fusarium phyllophilum and one of the polygalacturonase-inhibiting protein from Phaseolus vulgaris after chemical cross-linking as determined by small-angle x-ray scattering analysis. The inhibitor engages its concave surface of the leucine-rich repeat domain with the enzyme. Both sides of the enzyme active site cleft interact with the inhibitor, accounting for the competitive mechanism of inhibition observed. The structure is in agreement with previous site-directed mutagenesis data and has been further validated with structure-guided mutations and subsequent assay of the inhibitory activity. The structure of the complex may help the design of inhibitors with improved or new recognition capabilities to be used for crop protection.
DOI:10.1007/s004250050130URL [本文引用: 1]
DOI:10.1006/pmpp.1996.0008URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
