 ,**, 姚梦楠
,**, 姚梦楠 ,**, 张晓莉, 曲存民, 卢坤, 李加纳, 梁颖
,**, 张晓莉, 曲存民, 卢坤, 李加纳, 梁颖 ,*西南大学农学与生物科技学院 / 油菜工程研究中心 / 西南大学现代农业科学研究院, 重庆 400715
,*西南大学农学与生物科技学院 / 油菜工程研究中心 / 西南大学现代农业科学研究院, 重庆 400715Prokaryotic expression, subcellular localization and yeast two-hybrid library screening of BnMAPK1 in B. napus
WANG Zhen ,**, YAO Meng-Nan
,**, YAO Meng-Nan ,**, ZHANG Xiao-Li, QU Cun-Min, LU Kun, LI Jia-Na, LIANG Ying
,**, ZHANG Xiao-Li, QU Cun-Min, LU Kun, LI Jia-Na, LIANG Ying ,*College of Agronomy and Biotechnology, Southwest University / Chongqing Engineering Research Center for Rapeseed / Academy of Agricultural Sciences, Southwest University, Chongqing 400715, China
,*College of Agronomy and Biotechnology, Southwest University / Chongqing Engineering Research Center for Rapeseed / Academy of Agricultural Sciences, Southwest University, Chongqing 400715, China通讯作者:
收稿日期:2020-01-26接受日期:2020-04-15网络出版日期:2020-09-12
| 基金资助: |
Received:2020-01-26Accepted:2020-04-15Online:2020-09-12
| Fund supported: |
作者简介 About authors
王珍, E-mail:
姚梦楠, E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (6279KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王珍, 姚梦楠, 张晓莉, 曲存民, 卢坤, 李加纳, 梁颖. 甘蓝型油菜BnMAPK1的原核表达、亚细胞定位及酵母双杂交文库筛选[J]. 作物学报, 2020, 46(9): 1312-1321. doi:10.3724/SP.J.1006.2020.04019
WANG Zhen, YAO Meng-Nan, ZHANG Xiao-Li, QU Cun-Min, LU Kun, LI Jia-Na, LIANG Ying.
油菜是一种重要的油料作物, 占我国的油料供给中较大的比例, 是食用植物油及植物蛋白的重要来源, 在农业生产上占据着举足轻重的地位[1]。甘蓝型油菜(Brassica napus)作为我国广泛种植的栽培种, 气候、环境、病害、虫害及倒伏等问题已经严重制约其产量及品质, 尤其是低温、干旱、渍害、涝害等非生物胁迫及菌核病、霜霉病等, 直接影响国民增收, 制约油料产业的发展[2,3,4]。
MAPKs级联是存在于几乎所有真核生物中的保守信号通路[5], 由下而上的组成分别为MAPKs、MAPKs激酶(MAPKKs)及MAPKKs激酶(MAPKKKs)[6]。MAPKs级联是多种生长信号跨膜传递的交汇点, 植物中的MAPKs级联将外源刺激转入胞内并引发一系列的胞内应答, 参与调控基因表达、生长发育、细胞分裂、分化及凋亡等过程, 尤其在翻译后修饰上具有重要意义[7]。MAPKs是一类Ser/Thr蛋白激酶, 存在保守氨基酸序列TXY[8]。根据氨基酸的类型, 将其分为TEY及TDY两种亚型; 根据序列同源性将其分为A~D族, A~C族属于TEY亚型, 拟南芥中C族MAPKs有4个成员(AtMAPK1/2/7/14), D族属于TDY亚型[9]。植物中JA、ABA、生长素、ET和细胞分裂素的合成与代谢都与MAPKs有关[10]。拟南芥AtMAPK1与AtMAPK2在功能上冗余, 响应盐胁迫、损伤、ROS、JA、ABA, 并参与生长素介导的细胞扩增等[11,12,13,14,15,16]。在蓼科藏边大黄(Rheum australe)中, 低温和JA处理条件下RaMAPK1的转录上调, 而ABA处理后下调[17]。Blanco等[18]发现马铃薯(Solanum tuberosum)块茎中的StMAPK1在JA、ABA及高温处理24 h后表达显著增加, 但其表达水平并不响应SA。Liang等[19]的研究结果显示, 甘蓝型油菜中BnMAPKK9- BnMAPK1-BnWRKY53信号通路参与SA信号转导途径。这些结果表明, 不同物种中的MAPK1基因对相同的刺激的响应各不相同, 且不同的信号通路可能介导不同的应答。本课题组前期研究从甘蓝型油菜中克隆到一个BnMAPK1基因[20], 并发现MeJA、ABA、H2O2、损伤以及核盘菌均能诱导BnMAPK1显著上调[21], BnMAPK1超量表达可有效地提高甘蓝型油菜菌核病抗性[22]和抗旱性[23]。然而, 我们对甘蓝型油菜BnMAPK1参与各类生物学进程及相应信号转导过程的分子机理了解依然有限。
由于MAPKs通过蛋白质的磷酸化将信号级联放大后传递至下游应答分子, 为了进一步找出BnMAPK1的下游靶蛋白, 本试验经过原核表达、亚细胞定位, 通过构建酵母双杂交cDNA文库筛选与其互作的蛋白等深入探索, 证明BnMAPK1与多种生物/非生物胁迫以及生长发育进程均相关。本研究为逐步深入完善甘蓝型油菜MAPKs信号转导网络提供了新的内容, 并为甘蓝型油菜抗性与高产的品种改良奠定理论基础。
1 材料与方法
1.1 试验材料
供试材料甘蓝型油菜黑籽中油821 DH系及本氏烟草(Nicotiana benthamiana), 2019年种植于重庆市油菜工程技术研究中心人工气候光照培养箱, 生长条件为16 h光照(25℃)/8 h黑暗(16℃), 湿度50%。原核表达载体pET28a (+)及亚细胞定位表达载体pEGAD均由本实验室保存, 酵母双杂交载体pGBKT7、pGADT7及酵母菌株Y2Hgold由西南大学梅秀鹏博士馈赠。异丙基硫代半乳糖苷(IPTG)、抗生素、LB/YEB培养基所用试剂购自生工生物工程(上海)股份有限公司, 酵母质粒提取试剂盒(TIANprep Yeast Plasmid DNA Kit)和植物总RNA提取试剂盒(RNAprep pure Plant Kit)购自北京天根生化科技公司, 酵母双杂交文库构建试剂盒(SMART cDNA Library Construction Kit)与酵母培养及转化试剂购自Clontech公司, cDNA均一化试剂盒(Trimmer-Direct cDNA Normalization Kit)购自Evrogen公司。克隆及连接酶、DNA marker及核酸纯化试剂盒购自北京全式金生物公司, Ni-NTA agarose beads购自GE公司, 由上海英潍捷基公司合成引物和测序。1.2 BnMAPK1的生物信息学分析
利用Unipro UGENE开源软件进行序列注释及比对; 利用Primer3web (1.3 BnMAPK1的原核表达
1.3.1 原核表达载体的构建 分别提取甘蓝型油菜中油821 DH系植株苗期(3~4片真叶)根、茎和叶的总RNA, 各取0.5 μg均匀混合进行反转录。以cDNA为模板, 用特异引物P-BnMAPK1-F和P- BnMAPK1-R通过EasyPfu扩增BnMAPK1基因CDS, 加A连接到pGEM-T easy克隆载体, 并转化E. coli DH5α感受态细胞, 对PCR鉴定阳性克隆进行测序。对pGEM-T-BnMAPK1质粒及pET28a (+)载体同时进行Bam H I和Sal I双酶切, 分别回收BnMAPK1片段及pET28a (+)骨架, 采用T4 DNA Ligase过夜连接(16℃), 转化DH5α后PCR验证并测序。将阳性pET28a-BnMAPK1质粒转化至BL21菌株用于原核表达, 所用引物见表1。Table 1
表1
表1所用PCR引物
Table 1
| 引物名称 Primer name | 引物序列 Primer sequences (5°-3°) | 用途 Usage |
|---|---|---|
| P-BnMAPK1 | F: GGATCCATGGCGACACCAGTTGATC R: GTCGACTAGCAGCTCAGATGTTGA | BnMAPK1的原核表达载体构建 Construction of prokaryotic expression vector of BnMAPK1 |
| T7 | F: TAATACGACTCACTATAGGG R: TGCTAGTTATTGCTCAGCGG | 原核表达重组载体的检测 Detection of prokaryotic expression recombinant vectors |
| SC-BnMAPK1 | F: ACGAATTCATGGCGACACCAGTTGATC R: ACGGATCCTAGCAGCTCAGATGTTGA | BnMAPK1的亚细胞定位表达载体构建 Construction of subcellular localization vector of BnMAPK1 |
| P389 | F: ACATGGTCCTGCTGGAGTTC R: ATTGCCAAATGTTTGAACGA | 亚细胞定位重组载体的检测 Detection of subcellular localization recombinant vectors |
| eGFP | F: ATGGTGAGCAAGGGCGAGGAG R: GGACTTGTACAGCTCGTCCATGCC | 亚细胞定位重组载体的检测 Detection of subcellular localization recombinant vectors |
| Y2H-BnMAPK1 | F: CGGAATTCATGGCGACACCAGTTGATC R: CGGGATCCTCATAGCAGCTCAGATGT | BnMAPK1的酵母双杂交载体构建 Construction of Yeast Two-Hybrid vector of BnMAPK1 |
| BD | F: TAATACGACTCACTATAGGG R: TTTTCGTTTTAAAACCTAAGAGTC | Bait载体的检测 Detection of Bait vectors |
| AD | F: TAATACGACTCACTATAGGG R: AGATGGTGCACGATGCACAG | 文库Prey载体的检测 Detection of Prey vectors in library |
新窗口打开|下载CSV
1.3.2 BnMAPK1原核蛋白大小及诱导时间检测
将测序正确的原核表达菌株BL21-pET28a-Bn MAPK1及BL21-pET28a (阴性对照)与BL21 (空白对照)分别按1:1000活化培养后, 再按1:100扩大培养, 摇床37℃, 250转 min-1震荡培养至OD600达到0.6~0.8。加入IPTG至终浓度为1 mmol L-1, 37℃诱导培养, 分别于0、2、4、6和8 h各取500 μL菌液。将收集的所有菌液4℃ 6000×g离心10 min, 弃上清液, 2×SDS Loading buffer重悬后95℃加热10 min, 立即冰浴2 min。4℃ 13,000×g离心1 min, 上清液用于表达产物的SDS-PAGE电泳检测。
1.3.3 BnMAPK1原核蛋白可溶性蛋白纯化 取1 mmol L-1 IPTG, 16℃诱导培养过夜后的400 mL BL21-pET28a-BnMAPK1菌液, 4℃ 6000×g离心10 min后, 菌体用4℃预冷1×PBS (137 mmol L-1 NaCl, 12.7 mmol L-1 KCl, 10 mmol L-1 Na2HPO4, 2 mmol L-1 KH2PO4, pH 5.8)洗涤后再次离心, 用于蛋白纯化。加入20 mL Resuspension buffer A (50 mmol L-1 NaH2PO4, 300 mmol L-1 NaCl, 0.05% Tween-20, pH 8.0), 200 mg L-1 Lysozyme, 1 mmol L-1 PMSF, 20 mg L-1 DNase I及1% Triton-X 100, 4℃孵育30 min。冰浴超声破碎5 min (破碎2 s/间歇5 s, 功率130 W), 处理后于4℃ 13,000×g离心10 min, 并用0.45 μm滤膜过滤杂质, 上清液备用。取1 mLNi-NTA agarose beads加入重力柱, 2 mL Resuspension buffer A洗涤beads两次后, 缓慢加入破碎过滤后的上清液, 4℃孵育30 min。分别用20 mL Resuspension buffer B (Resuspension buffer A with 5 mmol L-1 imidazole)和40 mL Wash buffer (Resuspension buffer A with 20 mmol L-1 imidazole)洗涤beads, 流出速度约为1 mL 20 s-1。最后用500 μL Elution buffer (Resuspension buffer A with 100 mmol L-1 imidazole)洗脱beads, 共洗脱5次, 收集流出蛋白液, 用于纯化产物的SDS-PAGE电泳检测。
1.4 BnMAPK1的亚细胞定位表达分析
用特异引物SC-BnMAPK1-F和SC-BnMAPK1- R, 以pET28a-BnMAPK1测序菌液为模板扩增BnMAPK1 CDS区, 采用Eco R I和Bam H I双酶切法, 构建pEGAD-BnMAPK1的亚细胞定位表达载体, 将测序质粒转化至农杆菌LBA4404菌株, PCR检测后备用, 所用引物见表1。LBA4404-pEGAD- BnMAPK1阳性菌株先以1:1000比例活化培养, 再以1:100比例扩大培养, 摇床28℃, 250转 min-1震荡培养至OD600达到0.8~1.0, 室温6000×g离心5 min收集菌体。参照靳义荣等[24]方法改进, 加入Injection buffer (10 mmol L-1 MgCl2, 10 mmol L-1 MES, 200 μmol L-1 AS)将菌体重悬至OD600为0.4~0.5, 28℃避光静置3~6 h后注射本氏烟草。25℃黑暗培养瞬时表达烟草植株48 h, 将叶片下表皮置共聚焦显微镜下观察并照相。1.5 甘蓝型油菜BnMAPK1酵母双杂交文库筛选
1.5.1 甘蓝型油菜cDNA文库的构建 甘蓝型油菜中油821 DH系植株培养至苗期(3~4片真叶)时, 分别取植株根系、茎(茎尖2~3 cm)和叶片(芯芽叶及幼叶)于液氮中速冻研磨均匀, 各取约100 mg植株组织粉末, 采用RNAprep pure Plant Kit提取各个组织的总RNA后等量混合, 利用SMART cDNA Library Construction Kit反转录合成cDNA后, 使用Trimmer-Direct cDNA Normalization Kit进行均一化处理, 回收的cDNA通过CHROMA SPIN-1000纯化柱去除短片段cDNA。收集的cDNA, 参照SMART cDNA Library Construction Kit操作手册进行酵母文库的构建。1.5.2 BnMAPK1的Bait质粒毒性及自激活活性检测 以pET28a-BnMAPK1测序菌液为模板, 用特异引物Y2H-BnMAPK1-F和Y2H-BnMAPK1-R扩增BnMAPK1 ORF区, 采用Eco R I和Bam H I双酶切法, 构建pGBKT7-BnMAPK1的Bait载体, 所用引物见表1。PCR鉴定后的阳性克隆测序正确后, 参考Matchmaker Gold Yeast Two-Hybrid System操作手册制作酵母感受态细胞。按照小量转化法将pGBKT7- BnMAPK1质粒与pGADT7空白质粒共转化至酵母Y2Hgold感受态中, 分别涂布SD/-Leu/-Trp和SD/-Ade/-His/-Leu/-Trp平板, 30℃倒置培养3~5 d后, 观察菌落生长状况。阳性对照为pGBK-53与pGAD-T, 阴性对照为pGBK-Lam与pGAD-T, 空白对照为Y2Hgold菌株。
1.5.3 文库阳性克隆的筛选、测序及回转验证
参考Matchmaker Gold Yeast Two-Hybrid System操作手册, 按照大量转化/文库筛选法将pGBKT7- BnMAPK1质粒与10 μg文库质粒共转化Y2Hgold感受态细胞中, 涂布于SD/-Leu/-Trp和SD/-Ade/- His/-Leu/-Trp平板上。30℃倒置培养3~5 d后, 从所有SD/-Leu/-Trp平板中随机挑取1000个单克隆转移至SD/-Ade/-His/-Leu/-Trp平板上观察生长情况。然后, 将所有SD/-Ade/-His/-Leu/-Trp平板上的克隆子划线分离纯化, 按照TIANprep Yeast Plasmid DNA Kit说明书, 提取所有SD/-Ade/-His/-Leu/-Trp平板分离纯化后的Y2Hgold-Prey质粒, 并转化E. coli DH5α感受态细胞, 对PCR检测条带> 300 bp的DH5α-Prey进行测序, 并对其进行序列比对及GO聚类等分析。随后, 提取测序的DH5α-Prey质粒, 按照小量转化法, 分别将DH5α-Prey质粒与pGBKT7- BnMAPK1质粒进行点对点回转验证以排除假阳性互作。
2 结果与分析
2.1 BnMAPK1的二级结构与三级结构分析
通过Phyre2、PredictProtein等工具对BnMAPK1及其编码的氨基酸序列进行在线预测分析, GO生物学进程聚类结果显示, BnMAPK1主要参与蛋白质氨基酸磷酸化、生长素介导的信号转导途径、逆境应答、细胞周期及转录等过程, 可信度区间为10%~46% (表2)。BnMAPK1编码369个氨基酸残基, 相对分子质量为42.5 kD, 理论等电点(pI)为6.67。不稳定系数为42.99, 属于不稳定蛋白。平均亲水性系数为-0.248, 推测为亲水性蛋白。由图1-A可以看出, 在氨基酸组成中, 亮氨酸、异亮氨酸和天冬氨酸出现频率最高, 分别占氨基酸总数的12.47%、6.78%和6.23%。带负电荷的氨基酸残基(Asp+Glu)总数为43个, 带正电荷的氨基酸残基(Arg+Lys)为41个。利用Swiss-model在线软件对BnMAPK1蛋白的三级结构进行预测, 得到该蛋白的三维结构(图1-B)。图1
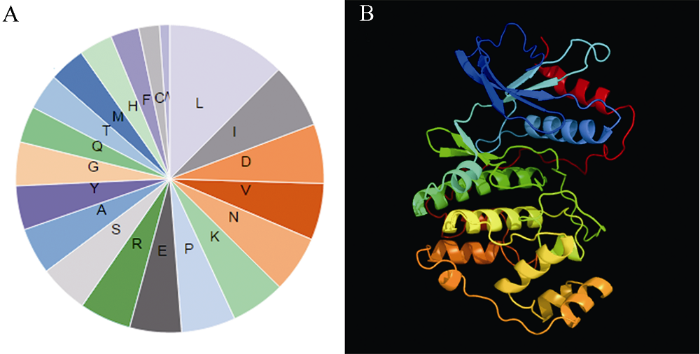 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1BnMAPK1的氨基酸组成分析(A)及三维结构预测(N端到C端) (B)
Fig. 1Amino acid composition analysis (A) and three dimensional structure (rainbow N to C terminus) of BnMAPK1 protein (B)
Table 2
表2
表2BnMAPK1的GO生物学进程聚类分析
Table 2
| 序号 No. | GO编号 GO ID | GO名称 GO term | 可信度 Reliability (%) |
|---|---|---|---|
| 1 | GO:0006468 | 蛋白质氨基酸磷酸化Protein amino acid phosphorylation | 46 |
| 2 | GO:0009734 | 生长素介导的信号途径Auxin mediated signaling pathway | 20 |
| 3 | GO:0006950 | 响应逆境Response to stress | 18 |
| 4 | GO:0007049 | 细胞周期Cell cycle | 10 |
| 5 | GO:0006350 | 转录Transcription | 10 |
新窗口打开|下载CSV
二级结构预测结果如图2所示, BnMAPK1含有15个蛋白-蛋白/蛋白-核酸结合位点(图2-A); BnMAPK1具有37% α螺旋(Alpha helix)、16% β折叠(Beta strand)、14%无规区域(Disordered)及4%跨膜螺旋(TM helix), 且40%以上的氨基酸均为隐藏态(Buried) (图2-B); BnMAPK1无二硫键, PROFbval算法预测的无序蛋白(Disordered protein)区域较另外3种算法明显更多(图2-C)。此外, 对BnMAPK1的核输出信号(Nuclear Export Signal)预测显示, BnMAPK1可能存在NES, 且NES指导BnMAPK1的亚细胞定位及核输出。BnMAPK1的亚细胞定位预测结果显示, BnMAPK1可能主要定位在细胞核内。
图2
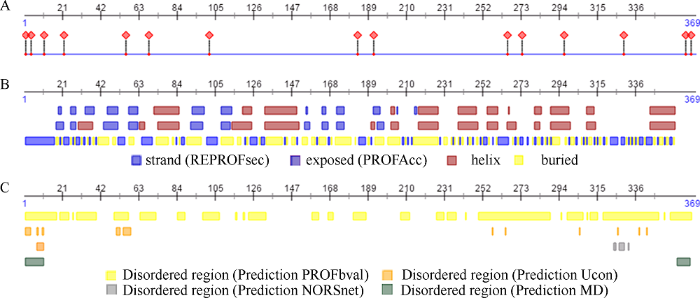 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2BnMAPK1的蛋白结合位点(A)、二级结构及可溶性(B)和蛋白无序性及构象柔性预测(C)
Fig. 2Prediction of protein binding sites (A), secondary structure and solvent accessibility (B), and disorder and flexibility (C) of BnMAPK1
2.2 BnMAPK1的原核表达及可溶性蛋白的纯化
与阴性对照BL21-pET28a与空白对照BL21相比, 重组菌株BL21-pET28a-BnMAPK1在37℃, 1 mmol L-1 IPTG浓度条件下, 诱导2 h已有明显表达, 且表达量随诱导时间逐步递增, 重组蛋白大小约为45 kD, 与预测结果基本一致, 表明pET28a- BnMAPK1融合蛋白在原核细胞中诱导表达成功(图3-A)。通过对温度、IPTG浓度及诱导时间梯度的检测, 发现BL21-pET28a-BnMAPK1在16℃, 1 mmol L-1 IPTG过夜诱导培养条件下可获得可溶性蛋白。将重组菌株BL21-pET28a-BnMAPK1在该条件下扩大培养诱导, 成功纯化出BnMAPK1可溶性蛋白, SDS-PAGE电泳检测纯化结果如图3-B, 纯化产物的Branford染色结果如图3-C。图3
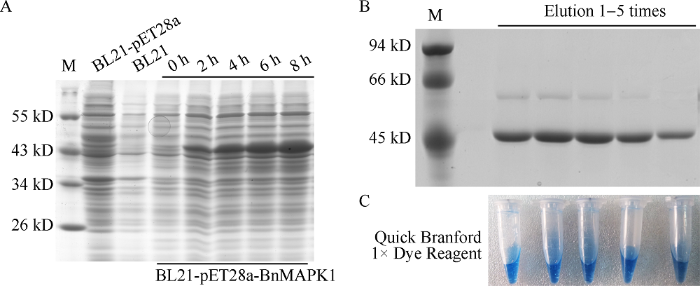 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3BnMAPK1的原核表达(A)、可溶性重组蛋白纯化SDS-PAGE (B)与Branford染色检测(C)
Fig. 3Prokaryotic expression of BnMAPK1 (A), and detection of purified recombinant protein by SDS-PAGE (B), and Branford dye testing (C)
2.3 BnMAPK1的亚细胞定位
通过构建pEGAD-BnMAPK1表达载体, 并通过本氏烟草瞬时表达BnMAPK1-eGFP重组蛋白。共聚焦显微镜观察发现, 只表达空白载体pEGAD的eGFP蛋白时, 烟草表皮细胞中有大量绿色荧光信号, 主要分布在细胞膜、细胞质和细胞核中; 与生物信息预测结果一致的是, BnMAPK1-eGFP融合蛋白在烟草表皮细胞中的荧光信号明显定位在细胞核(图4), 表明BnMAPK1是一种细胞核内的亲水活性蛋白。图4
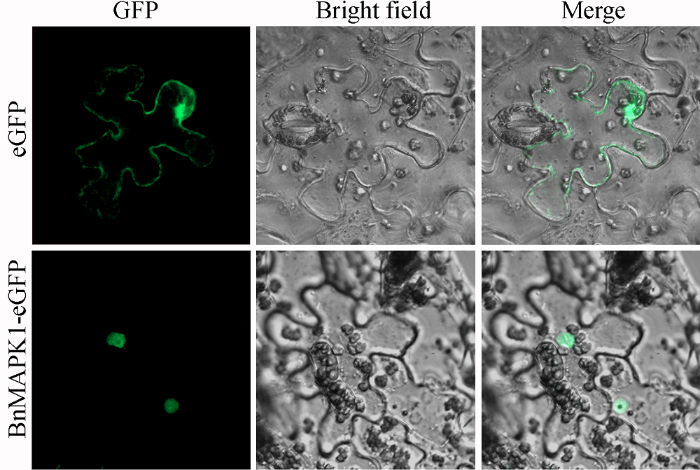 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4BnMAPK1-eGFP融合蛋白的亚细胞定位
Fig. 4Subcellular localization of BnMAPK1-eGFP protein
2.4 甘蓝型油菜cDNA文库的建立
提取甘蓝型油菜苗期各组织的总RNA等量混合后电泳检测RNA质量, 结果28S和18S rRNA条带清晰(图5-A), 表明总RNA完整性好, 无降解, 符合建库要求。对总RNA进一步纯化分离mRNA, 反转录合成cDNA, 如图5-B所示, 得到的基因较完整, 有明显的特异条带。对其进行均一化处理后, 特异高丰度区域削弱明显, 特异条带消失, 效果达到均一化标准(图5-C)。将cDNA过柱纯化以去除短片段, 结果显示, 保留的片段大小为500~3000 bp (图5-D), 有利于文库筛选。cDNA初级文库质量检测结果显示, 初级文库库容 > 1.5×106 cfu, 初级文库的扩增基数 > 11×106 cfu, 提取扩增文库质粒于-80℃保存备用。图5
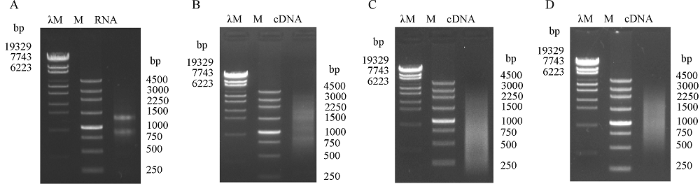 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5总RNA的提取(A)、均一化前(B)后(C)的cDNA及纯化过柱后的cDNA (D)
Fig. 5Analysis of total RNA extraction (A), cDNA synthesis (B), normalization (C), and purification (D)
2.5 Bait-BnMAPK1重组质粒的毒性与自激活检测
采用酵母双杂交核系统检测Bait-BnMAPK1的毒性和自激活活性。通过构建pGBKT7-BnMAPK1重组载体, 将pGBKT7-BnMAPK1和pGADT7空白质粒共同转化Y2Hgold酵母感受态细胞, 分别涂布SD/-Leu-Trp及SD/-Ade-His-Leu-Trp平板。酵母菌落生长结果显示, pGBK-53+pGAD-T (positive)、pGBK- Lam+pGAD-T (negative)和pGBKT7-BnMAPK1+pGA DT7 (Bait-BnMAPK1) 3组均可以在SD/-Leu-Trp平板上正常生长, 表明Bait-BnMAPK1重组质粒没有毒性。Positive在SD/-Ade-His-Leu-Trp平板上正常生长, 而Negative和Bait-BnMAPK1均不能生长, 表明BnMAPK1没有自激活活性(图6)。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6Bait-BnMAPK1的毒性和自激活检测
Fig. 6Toxicity and autoactivation test of Bait-BnMAPK1
2.6 BnMAPK1的酵母双杂交文库筛选及回转验证
以pGBKT7-BnMAPK1为诱饵质粒, 将其与甘蓝型油菜cDNA文库质粒共转化Y2Hgold酵母细胞筛选BnMAPK1的互作蛋白。提取SD/-Ade-His- Leu-Trp平板上分离纯化克隆子的质粒, 转化E. coli DH5α进行PCR检测及质粒扩繁, 除去300 bp以下的克隆子, 将所有pGADT7-Prey与Bait-BnMAPK1进行点对点回转验证及测序, 共获得95个回转阳性互作蛋白(附图1), 验证了阳性克隆的准确性, 同时降低了假阳性的概率。对这些阳性克隆进行序列比对, 基因注释信息见附表1, GO分析结果显示, BnMAPK1可能通过与这些蛋白发生相互作用, 参与信号转导、生长发育、光合作用、次生代谢物的生物合成与代谢、磷酸化与去磷酸化等生物学进程, 对冷害、冻害、盐害、高温、干旱、损伤、病原菌等非生物与生物胁迫环境中的植物体进行保护, 并响应植物激素如ET、JA、SA、ABA等(图7), 表明BnMAPK1在植物的整个生长周期内具有重要的生物学意义。附图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图1BnMAPK1酵母双杂交文库筛选互作蛋白的回转验证
Supplementary Fig. 1Retransformation validation of BnMAPK1-interacting proteins in Y2H screening assays
图7
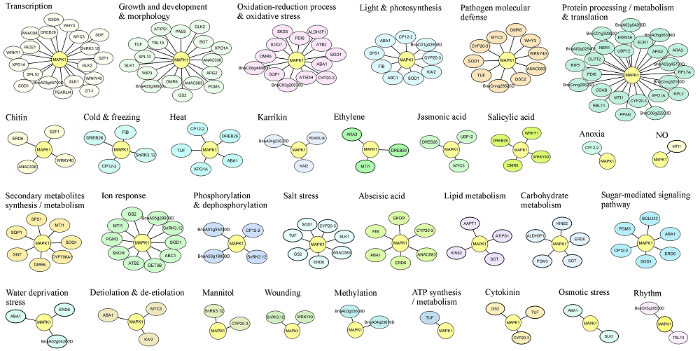 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7BnMAPK1候选阳性互作蛋白的调控网络
Fig. 7Regulatory network of positive BnMAPK1-interacting candidate proteins
3 讨论
在过去的几十年中, 植物MAPKs级联途径在生长周期里如何接收并传递信号已经得到广泛的研究, 其中MAPKs的蛋白磷酸化具有重要生物学意义[8,25-26]。关于植物MAPKs级联的研究, 主要集中在拟南芥、烟草、苜蓿(Medicago sativa)、番茄、欧芹(Petroselinum crispum)、水稻等植物的A族与B族MAPKs (MAPK3/6, MAPK4), 由于它们在植物中的激酶活性较容易检测到, 其研究内容更为透彻[27], 相较而言, C族(MAPK1/2/7/14)的研究还很局限。本研究中, 我们将甘蓝型油菜C族BnMAPK1定位于细胞核, 并在原核系统中成功地表达纯化了BnMAPK1的可溶性活性蛋白, 利用酵母双杂交系统筛选到95个BnMAPAK1相互作用蛋白, 对BnMAPK1可能存在的网络机制进行了解析, 不仅对BnMAPK1在植物抗逆及生长发育分子机制方面有所了解, 还为探索BnMAPK1蛋白的未知生物学功能提供了新线索。植物在不同环境胁迫下, 特异MAPKs通路被激活, 通过调控下游的转录水平或磷酸化相关蛋白, 响应不同的刺激并传递信号以行使生物学功能。MAPKs在被其上游不同的MAPKKs激活时, 参与不同的生物学进程, 表现出不同的激活特性[28]。在哺乳动物和酵母中, MAPKs的激活与其在细胞内定位的动态变化相关, 该信号转导通路中各组分的定位受到严格控制[29]。已有研究表明, NES在蛋白激酶的亚细胞定位及核输出具有重要指导意义[30]。Yoo等[31]发现O3处理拟南芥后, AtMAPK3与AtMAPK6受到激活, 迅速转移至细胞核内。Ahlfors等[32]发现花生中AhMAPK3与AhMAPK6在细胞质和细胞核中均有表达, 但H2O2处理后则仅在细胞核内表达。本研究对BnMAPK1蛋白的结构预测发现其可能含有NES, 且亚细胞定位试验结果显示主要定位在细胞核中。我们推测, BnMAPK1的核移位可能是信号转导过程中非常关键的一步, BnMAPK1蛋白在核内的积累对于胁迫响应是必要的。
在逆境中, 植物进化出一些特殊机制, 通过调控酶活性、转录、翻译及翻译后修饰等, 感知并适应环境。MAPKs级联作为进化保守的信号通路, 将信号转入胞内, 通过活化细胞骨架蛋白、转录因子、磷脂酶、微管相关蛋白, 调控特定的基因簇或翻译后修饰, 参与多种生理、生长、发育及激素等生物学过程[33,34,35,36]。其中, 植物MAPK1响应激素(如ABA、IAA等)与逆境胁迫(如干旱、渗透压、盐害、损伤等)[37]等过程。研究表明, 拟南芥中, LHCB3 (light- harvesting chlorophyll B-binding protein 3)[38]、CURT1B (curvature thylakoid 1B)[39]、SKOR (shaker- like K+ outward rectifying channel)[40]与ERD6 (early response to dehydration 6)[41]等均能够响应ABA且参与调控ABA信号转导途径; ABA1 (ABA deficient 1)基因编码一个玉米黄质环氧酶, 直接控制ABA的生物合成与积累, 进而调控植物对高盐及干旱的耐受性[42,43]。另外, UBP12 (ubiquitin-specific protease 12)和MYC3被报道参与JA介导的信号途径, 前者主要与蛋白泛素化相关[44], 而后者正向调控依赖于JA途径相关基因的转录表达[45]。IAA17 (indole-3-acetic acid inducible 17)被报道调控植物生长素的积累及信号转导, 是叶片衰老的正向调控因子, 可通过与GA信号转导途径交联调控NO介导的盐胁迫途径[46,47]。因此, 本研究推测, BnMAPK1可能通过与上述激素相关蛋白互作, 使得MAPKs级联与多种激素相关的信号转导途径交联, 参与甘蓝型油菜的生长发育及抗逆等过程。
综上, 本研究从甘蓝型油菜cDNA文库中获得多个与BnMAPK1相互作用的靶蛋白。通过分析这些靶蛋白的生物学功能发现, 它们参与植物生长发育及逆境胁迫的响应等过程。初步提出了BnMAPK1参与调控生长周期及提高植物抗逆性的可能分子机制, 对BnMAPK1功能及作用机制的深入研究必将大大推动植物MAPKs级联途径在提高甘蓝型油菜抗逆机制中的研究与应用, 为改良甘蓝型油菜品质, 培养抗逆新品种提供重要理论依据。
4 结论
甘蓝型油菜BnMAPK1编码369个氨基酸, 编码蛋白大小约为45 kD。BnMAPK1主要定位在细胞核内。该蛋白在酵母中不具有毒性及自激活活性。利用酵母双杂交筛选甘蓝型油菜中的BnMAPK1互作蛋白, 获得95个初步验证的阳性克隆。BnM- APK1可能通过与这些蛋白结合或磷酸化, 调控或传递相关信号, 维持植物的正常生长发育并响应逆境等过程。附图和附表 请见网络版: 1) 本刊网站
Supplementary table 1
附表1
附表1BnMAPK1酵母双杂交文库筛选互作蛋白的基因注释
Supplementary table 1
| 编号 No. | 拟南芥基因号 Ath. Gene ID | 油菜基因号 Bna. Gene ID | 出现次数 Frequency | 基因名称 Gene Symbols | 基因注释信息 Description |
|---|---|---|---|---|---|
| 1 | AT3G55980 | BnaC08g26910D | 12 | SZF1 | Salt-inducible zinc finger 1 |
| 2 | AT4G33010 | BnaA01g03860D | 1 | GLDP1 | Glycine decarboxylase P-protein 1 |
| 3 | AT3G10350 | BnaA01g31840D | 3 | GET3B | GUIDED ENTRY OF TAIL-ANCHORED PROTEINS 3B//One of 3 GET paralogs in Arabidopsis. GET3b is a chloroplast localized protein with no obvious role in Tail Anchored (TA) protein insertion. |
| 4 | AT5G54270 | BnaA02g09700D | 1 | LHCB3 | Lhcb3 protein is a component of the main light harvesting chlorophyll a/b-protein complex of Photosystem II (LHC II). |
| 5 | AT4G19420 | BnaC01g11750D | 2 | PAE8 | Pectin acetylesterase 8//Pectin acetylesterase family protein |
| 6 | AT3G05580 | BnaA02g30910D | 1 | TOPP9 | Encodes a Type One Protein Phosphatase that acts as a nucleocytoplasmic negative regulator of tip growth. Mutants affect pollen germination, pollen tube growth, and root hair growth. It acts genetically downstream of ANX1 (AT3G04690) and ANX2 (AT5G28680) |
| 7 | AT3G62410 | BnaA09g39860D | 2 | CP12-2 | CP12-2 encodes a small peptide in the chloroplast stroma. CP12-2 is coordinately regulated by light with the photosynthetic GAPDH and PRK |
| 8 | AT4G04770 | BnaA09g20260D | 2 | ABC1, LAF6, ATNAP1 | Encodes an iron-stimulated ATPase. A member of the NAP subfamily of ABC transporters. Involved in Fe-S cluster assembly. Similar to SufB. Involved in the regulation of iron homeostasis. Able to form homodimers. Interacts with AtNAP7 inside the chloroplast. |
| 9 | AT1G76160 | BnaA07g32400D | 1 | SKS5 | SKU5 similar 5 |
| 10 | AT1G21910 | BnaAnng28960D | 1 | DREB26 | encodes a member of the DREB subfamily A-5 of ERF/AP2 transcription factor family. The protein contains one AP2 domain. |
| 11 | AT4G25520 | BnaA08g30970D | 1 | SLK1 | SEUSS-like 1 |
| 12 | AT5G28050 | BnaA06g29050D | 1 | GSDA | GUANOSINE DEAMINASE, Cytidine/deoxycytidylate deaminase family protein |
| 13 | AT2G17040 | BnaC03g60970D | 1 | ANAC036 | NAC DOMAIN CONTAINING PROTEIN 36, member of the NAC transcription factor family and more specifically, the ONAC022 subfamily |
| 14 | AT2G37130 | BnaC03g20530D | 1 | _ | Peroxidase superfamily protein |
| 15 | AT3G07020 | BnaA05g30520D | 1 | SGT, UGT80A2 | Encodes a 3beta-hydroxy sterol UDP-glucosyltransferase. ugt80a2 mutant plants have reduced steryl glycoside and acyl steryl glycoside levels and reduced seed size. ugt80a2/b1 double mutants have normal levels of celluose and normal cold stress tolerance. |
| 16 | AT3G01280 | BnaA05g37480D | 1 | VDAC1 | Encodes a voltage-dependent anion channel (VDAC: AT3G01280/VDAC1, AT5G67500/VDAC2, AT5G15090/VDAC3, AT5G57490/VDAC4, AT5G15090/VDAC5). VDACs are reported to be porin-type, beta-barrel diffusion pores. They are prominently localized in the outer mitochondrial membrane and are involved in metabolite exchange between the organelle and the cytosol |
| 17 | AT5G67500 | BnaA09g07330D | 1 | VDAC2 | Encodes a voltage-dependent anion channel (VDAC: AT3G01280/VDAC1, AT5G67500/VDAC2, AT5G15090/VDAC3, AT5G57490/VDAC4, AT5G15090/VDAC5). VDACs are reported to be porin-type, beta-barrel diffusion pores. They are prominently localized in the outer mitochondrial membrane and are involved in metabolite exchange between the organelle and the cytosol |
| 18 | AT5G13180 | BnaA10g20110D | 1 | ANAC083, VNI2 | NAC DOMAIN CONTAINING PROTEIN 83, encodes a NAC domain transcription factor that interacts with VND7 and negatively regulates xylem vessel formation |
| 19 | AT5G24150 | BnaC04g28910D | 1 | SQP1,SQE5 | squalene monooxygenase gene homolog |
| 20 | AT5G67030 | BnaA07g12170D | 1 | ABA1, ATZEP, IBS3, LOS6, NPQ2 | Encodes a single copy zeaxanthin epoxidase gene that functions in first step of the biosynthesis of the abiotic stress hormone abscisic acid (ABA) |
| 21 | AT4G39800 | BnaC01g00680D | 1 | ATIPS1, MIPS1 | myo-inositol-1-phosphate synthase |
| 22 | AT1G01140 | BnaA09g51540D | 1 | CIPK9, PKS6, SnRK3.12 | Encodes a CBL-interacting protein kinase with similarity to SOS2 |
| 23 | AT2G01110 | BnaC02g33640D | 1 | APG2,UNE3,PGA2,TATC | Chloroplast Protein Translocation (tatC), mutant is Albino and pale green. Core subunit of the chloroplast Tat translocase. Integral chloroplast thylakoid membrane protein. |
| 24 | AT2G26690 | BnaA07g12830D | 1 | NPF6.2 | Major facilitator superfamily protein |
| 25 | AT5G61770 | BnaA09g05800D | 1 | PPAN, SNAIL1 | A single-copy gene encoding a 346 aa protein with a single Brix domain. Similar to yeast ribosome biogenesis proteins Ssf1/2. |
| 26 | AT4G12060 | BnaC09g23500D | 1 | CLPT2 | Double Clp-N motif protein |
| 27 | AT1G12350 | BnaCnng03720D | 1 | COAB | encodes phosphopantothenoylcysteine synthetase (phosphopantothenoylcysteine ligase) |
| 28 | AT3G58460 | BnaA09g37400D | 1 | RBL11, RBL15 | RHOMBOID-like protein |
| 29 | AT5G43940 | BnaA09g16810D | 1 | ADH2, HOT5 | Encodes a glutathione-dependent formaldehyde dehydrogenase (also known as class III type alcohol dehydrogenase) reduces S-nitrosoglutathione (GSNO), the condensation product of glutathione and NO, that is a naturally occurring NO reservoir and also a reactive nitrogen intermediate. Gene expression is reduced by wounding and induced by salicylic acid. Is required for the acclimation of plants to high temperature and for fertility |
| 30 | AT1G08830 | BnaC08g42970D | 1 | CSD1, SOD1 | Encodes a cytosolic copper/zinc superoxide dismutase CSD1 that can detoxify superoxide radicals. Its expression is affected by miR398-directed mRNA cleavage. Regulated by biotic and abiotic stress. Activation of CSD1 in the cytoplasm involves both a CCS-dependent and -independent pathway. |
| 31 | AT2G42500 | BnaC04g02330D | 1 | PP2A-3 | Encodes one of the isoforms of the catalytic subunit of protein phosphatase 2A: AT1G59830/PP2A-1, AT1G10430/PP2A-2, At2g42500/PP2A-3, At3g58500/PP2A-4 [Plant Molecular Biology (1993) 21:475-485 and (1994) 26:523-528; Note that in more recent publications, there is mixed use of gene names for PP2A-3 and PP2A-4 - some refer to At2g42500 as PP2A-3 and some as PP2A-4]. ACR4 phosphorylates the PROTEIN PHOSPHATASE 2A-3 (PP2A-3) catalytic subunit of the PP2A phosphatase holoenzyme and PP2A dephosphorylates ACR4 |
| 32 | AT2G02740 | BnaC07g21860D | 1 | PTAC11,WHY3 | Encodes a homolog of the potato p24 protein. It shares the conserved KGKAAL domain, a putative DNA-binding domain, with potato p24 |
| 33 | AT3G19850 | BnaC01g32990D | 1 | _ | Phototropic-responsive NPH3 family protein |
| 34 | AT2G04880 | BnaA03g37950D | 1 | ZAP1,WRKY1 | Encodes WRKY1, a member of the WRKY transcription factors in plants involved in disease resistance, abiotic stress, senescence as well as in some developmental processes. WRKY1 is involved in the salicylic acid signaling pathway. The crystal structure of the WRKY1 C-terminal domain revealed a zinc-binding site and identified the DNA-binding residues of WRKY1. |
| 35 | AT3G06080 | BnaA03g57390D | 1 | TBL10 | Encodes a member of the TBL (TRICHOME BIREFRINGENCE-LIKE) gene family containing a plant-specific DUF231 (domain of unknown function) domain. |
| 36 | AT5G23120 | BnaC07g49760D | 2 | HCF136 | encodes a stability and/or assembly factor of photosystem II |
| 37 | AT1G53210 | BnaCnng29300D | 1 | NCL | Encodes a Na+/Ca 2+ exchanger-like protein that participates in the maintenance of Ca 2+ homeostasis |
| 38 | AT5G60210 | BnaA02g06460D | 1 | RIP5 | Encodes RIP5 (ROP interactive partner 5), a putative Rho protein effector, interacting specifically with the active form of ROPs (Rho proteins of plants) |
| 39 | AT3G02850 | BnaCnng06010D | 1 | SKOR | Encodes STELAR K+ OUTWARD RECTIFIER, a member of Shaker family potassium ion (K+) channel. Mediates the delivery of K+ from stelar cells to the xylem in the roots towards the shoot. mRNA accumulation is modulated by abscisic acid. K+ gating activity is modulated by external and internal K+. Involved in response to low potassium. |
| 40 | AT5G54110 | BnaC02g13800D | 1 | MAMI | Encodes a highly polar protein with more than 60% hydrophilic amino acid residues that is associated with the plasma membrane. It has limited secondary structure similarity to VAP-33 from Aplysia, which may be involved in membrane trafficking. |
| 41 | AT5G17020 | BnaC09g40920D | 1 | XPO1A, CRM1, HIT2 | Encodes a member of the exportin protein family (XPO1A) which functions as receptors for nuclear export. Binds to a variety of proteins having leucine rich export signals.Along with XPO1B involved with development of the male and female gametophytes. |
| 42 | AT4G22690 | BnaA08g10430D | 1 | CYP706A1 | Cytochrome P450, family 706, subfamily A, polypeptide 1, member of CYP706A |
| 43 | AT2G35500 | BnaC04g44410D | 1 | SKL2 | Encodes a protein with some sequence similarity to shikimate kinases, but a truncated form of this protein (lacking a putative N-terminal chloroplast transit peptide) does not have shikimate kinase activity in vitro |
| 44 | AT2G38740 | BnaA03g18060D | 1 | _ | Haloacid dehalogenase-like hydrolase (HAD) superfamily protein |
| 45 | AT3G46060 | BnaA06g19870D | 1 | ARA3, RAB8A, RABE1C | small GTP-binding protein (ara-3) |
| 46 | AT1G13560 | BnaC08g40270D | 1 | AAPT1 | Encodes aminoalcoholphosphotransferase AAPT1 |
| 47 | ATCG01310 | BnaC03g31140D | 1 | RPL2 | encodes a chloroplast ribosomal protein L2, a constituent of the large subunit of the ribosomal complex |
| 48 | AT2G16900 | BnaC07g05930D | 1 | PEARLI4 | phospholipase-like protein (PEARLI 4) family protein |
| 49 | AT4G35110 | BnaAnng11620D | 1 | PEARLI 4 | phospholipase-like protein (PEARLI 4) family protein |
| 50 | AT3G07720 | BnaA05g29930D | 1 | _ | Galactose oxidase/kelch repeat superfamily protein |
| 51 | AT1G66720 | BnaA03g03600D | 1 | _ | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
| 52 | AT1G08930 | BnaC08g42900D | 1 | ERD6 | Early response to dehydration 6, encodes a putative sucrose transporter whose gene expression is induced by dehydration and cold |
| 53 | AT5G19510 | BnaA02g04260D | 1 | _ | Translation elongation factor EF1B/ribosomal protein S6 family protein |
| 54 | AT4G16360 | BnaA08g06000D | 1 | KINβ2 | 5'-AMP-activated protein kinase beta-2 subunit protein |
| 55 | AT4G11150 | BnaC03g72900D | 1 | TUF, EMB2448, VHA-E1 | Encodes a vacuolar H+-ATPase subunit E isoform 1 which is required for Golgi organization and vacuole function in embryogenesis |
| 56 | AT5G18370 | BnaC09g39520D | 1 | DSC2 | Dominant supressor of camta3 number 2, Leucine-rich repeat domain (NLR) receptor. Dominant negative alleles suppress catma3 autoimmunity |
| 57 | AT3G13360 | BnaC01g37720D | 1 | WIP3 | WPP domain interacting protein 3, encodes an outer nuclear membrane protein that anchors RanGAP1 to the nuclear envelope. It interacts with SUN proteins and is required for maintaining the elongated nuclear shape of epidermal cells. |
| 58 | AT5G35630 | BnaC08g07930D | 1 | GS2, GLN2, GSL1 | chloroplastic glutamine synthetase |
| 59 | AT1G80750 | BnaC06g19470D | 1 | RPL7A | Ribosomal protein L30/L7 family protein |
| 60 | AT3G50770 | BnaCnng69860D | 1 | CML41 | calmodulin-like 4 |
| 61 | AT4G36250 | BnaC03g61970D | 1 | ALDH3F1 | Encodes a putative aldehyde dehydrogenase 3F1. The gene is not responsive to osmotic stress and is expressed constitutively at a low level in plantlets and root cultures. |
| 62 | AT5G38660 | BnaA07g14540D | 1 | APE1 | Acclimation of photosynthesis to environment, mutant has Altered acclimation responses |
| 63 | AT3G62030 | BnaA09g39610D | 1 | ROC4/CYP20-3 | Nuclear-encoded chloroplast stromal cyclophilin CYP20-3 (also known as ROC4). Protein is tyrosine-phosphorylated and its phosphorylation state is modulated in response to ABA in Arabidopsis thaliana seeds |
| 64 | AT1G60710 | chrC09:10,805,646..10,808,849 | 1 | ATB2 | Encodes ATB2 |
| 65 | AT1G12800 | BnaA06g07940D | 1 | SDP | S1 domain-containing RBP, SDP is a chloroplast localized RNA binding protein that is required for plastid rRNA processing. Plants harboring a mutation in SDP have numerous defects including reduced chlorophyll content, poor growth, yellow leaves and abnormal chloroplasts. |
| 66 | AT4G37470 | BnaA03g53980D | 1 | KAI2 | Karrikin insensitive 2, HTL belonging to the alpha/beta fold hydrolase superfamily. Mutant and over-expression studies indicates its involvement in seedling de-etiolation process. |
| 67 | AT1G04250 | BnaCnng41350D | 1 | IAA17, AXR3 | Transcription regulator acting as repressor of auxin-inducible gene expression. Auxin-inducible AUX/IAA gene. Short-lived nuclear protein with four conserved domains. Domain III has homology to beta alpha alpha dimerization and DNA binding domains. Involved in auxin signaling and is a positive modulator of natural leaf senescence. Auxin induces the degradation of the protein in a dosage-dependent manner in a process mediated by AtRac1. Auxin induced the relocalization of the protein within the nucleus from a diffused nucleoplasmic pattern to a discrete particulated pattern named nuclear protein bodies or NPB in a process also mediated by Rac1. Colocalizes with SCF, CSN and 26S proteasome components. |
| 68 | AT4G04640 | BnaA03g25540D | 1 | ATPC1 | One of two genes (with ATPC2) encoding the gamma subunit of Arabidopsis chloroplast ATP synthase. |
| 69 | AT1G78510 | BnaC06g38940D | 1 | SPS1 | Encodes one of the two paralogous solanesyl diphosphate synthases - SPS1 (At1g78510) and SPS2 (At1g17050) - that assemble the side-chain of plastoquinone-9 in plastids. |
| 70 | AT4G01940 | BnaA03g26440D | 1 | NFU1 | Encodes a protein containing the NFU domain that may be involved in iron-sulfur cluster assembly. Part of a five member gene family, more closely related to NFU2 and 3 than to NFU4 and 5. Targeted to the chloroplast. |
| 71 | AT1G23190 | BnaC08g06210D | 1 | PGM3 | Encodes a cytosolic phosphoglucomutase 3, loss of both PGM2 and PGM3 severely impairs male and female gametophyte development |
| 72 | AT2G05830 | BnaA07g04420D | 1 | MTI1 | Encodes a 5-methylthioribose-1-phosphate isomerase( 5-methylthioribose kinase 1) |
| 73 | AT2G46820 | BnaA05g01060D | 1 | CURT1B, PSI_P | Encodes the P subunit of Photosystem I. About 25% of the TMP14 pool appeared to be phosphorylated, and this ratio is not affected by light. Contains seven phosphorylation sites on threonine residue and chloroplast targeting signal. Located in the proximity of PSI-L, -H and -O subunits. |
| 74 | AT1G80840 | BnaA07g35260D | 1 | WRKY40 | Pathogen-induced transcription factor. Binds W-box sequences in vitro. Forms protein complexes with itself and with WRKY40 and WRKY60. Coexpression with WRKY18 or WRKY60 made plants more susceptible to both P. syringae and B. cinerea. WRKY18, WRKY40, and WRKY60 have partially redundant roles in response to the hemibiotrophic bacterial pathogen Pseudomonas syringae and the necrotrophic fungal pathogen Botrytis cinerea, with WRKY18 playing a more important role than the other two. |
| 75 | AT2G32860 | BnaC04g11860D | 1 | BGLU33 | beta glucosidase 33 |
| 76 | AT1G06650 | BnaC08g44860D | 1 | _ | encodes a protein whose sequence is similar to 2-oxoglutarate-dependent dioxygenase |
| 77 | AT1G56500 | BnaC09g16020D | 1 | SOQ1 | Suppressor of quenching, Encodes a thylakoid membrane protein with thioredoxin-like and beta-propeller domains located in the lumen and a haloacid-dehalogenase domain exposed to the chloroplast stroma. The protein's role may be to prevent formation of a slowly reversible form of antenna quenching, thereby maintaining the efficiency of light harvesting |
| 78 | AT1G13450 | BnaC05g09890D | 1 | GT-1 | Encodes GT-1, a plant transcription factor that binds to one of the cis-acting elements, BoxII, which resides within the upstream promoter region of light-responsive genes. GT-1 was assumed to act as a molecular switch modulated through Ca(2+)-dependent phosphorylation/dephosphorylation in response to light signals. |
| 79 | AT2G44530 | BnaA04g25770D | 1 | PRS5 | Phosphoribosyltransferase family protein |
| 80 | AT3G24150 | BnaC07g07420D | 1 | KIX8 | Kinase-inducible domain interacting 8, hypothetical protein |
| 81 | AT5G19180 | BnaA10g15880D | 1 | ECR1 | E1 C-Terminal related 1, encodes a subunit of a RUB-activating enzyme analogous to the E1 ubiquitin-activating enzyme. ECR1 functions as a heterodimer with AXR1 to activate RUB, a ubiquitin-related protein |
| 82 | AT1G35620 | BnaAnng04610D | 1 | PDI8, PDIL5-2 | Encodes a protein disulfide isomerase-like (PDIL) protein, a member of a multigene family within the thioredoxin (TRX) superfamily. Unlike several other PDI family members, transcript levels for this gene are not up-regulated in response to three different chemical inducers of ER stress (dithiothreitol, beta-mercaptoethanol, and tunicamycin). However, the level of transcripts for this gene is slightly elevated in atbzip60 mutants |
| 83 | AT1G03905 | BnaC08g00400D | 1 | ABCI19, ATNAP4 | ATP-binding cassette I19, non-intrinic ABC protein 4, P-loop containing nucleoside triphosphate hydrolases superfamily protein |
| 84 | AT4G17040 | BnaA01g17990D | 1 | CLPR4 | Encodes the ClpR4 subunit of the chloroplast-localized Clp protease complex. hon mutations disturb plastid protein homeostasis, thereby activating plastid signaling and inducing stress acclimatization. |
| 85 | AT3G22425 | BnaC01g31520D | 1 | HISN5A, IGPD | Encodes imidazoleglycerolphosphate dehydratase |
| 86 | AT3G11770 | BnaC05g41420D | 1 | RICE1 | Encodes a 23kDa protein with 3'-5' exoribonuclease activity. It is expressed ubiquitously and localized to the cytoplasm. |
| 87 | AT5G46760 | BnaC07g19420D | 1 | MYC3 | MYC3 is a JAZ-interacting transcription factor that act together with MYC2 and MYC4 to activate JA-responses |
| 88 | AT5G51970 | BnaCnng32750D | 1 | SDH | Encodes a putative sorbitol dehydrogenase that can be thiolated in vitro |
| 89 | AT4G24690 | BnaC01g16550D | 1 | NBR1 | Encodes NEXT TO BRCA1 GENE 1, a selective autophagy substrate |
| 90 | AT5G44190 | BnaC07g17710D | 1 | GLK2, GPRI2 | Encodes GBF's pro-rich region-interacting factor 2, Golden2-like 2, one of a pair of partially redundant nuclear transcription factors that regulate chloroplast development in a cell-autonomous manner. |
| 91 | AT5G24530 | BnaC02g41200D | 1 | DMR6 | Encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew |
| 92 | AT3G59690 | BnaC08g29930D | 1 | IQD13 | IQ-domain 13 |
| 93 | AT5G06600 | BnaC09g48960D | 1 | UBP12 | Encodes a ubiquitin-specific protease 12 |
| 94 | AT1G27370 | BnaC03g57620D | 1 | SPL10 | Squamosa promoter binding protein-like 10, In conjunction with SPL11 and SPL2, SPL10 redundantly controls proper development of lateral organs in association with shoot maturation in the reproductive phase. SPL10 also controls lamina shape during vegetative development. |
| 95 | AT4G04020 | BnaCnng74600D | 1 | FIB1A, PGL35 | Fibrillin precursor protein. The fibrillin preprotein, but not the mature protein interacts with ABI2. Regulated by abscisic acid response regulators. Involved in abscisic acid-mediated photoprotection. |
新窗口打开|下载CSV
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1038/srep21625URLPMID:26880301 [本文引用: 1]

An optimized plant architecture (PA) is fundamental for high-yield breeding but the genetic control of the important trait is largely unknown in rapeseed. Here plant architecture factors (PAFs) were proposed to consist of main inflorescence length proportion (MILP), branch height proportion (BHP), and branch segment proportion (BSP). Comparison of different genotypes in a DH population grown in diverse environments showed that an optimized PAF performance with MILP and BHP between 0.3-0.4 was important for high yield potential. In total, 163 unique quantitative trait loci (QTLs) for PA- and plant yield (PY)-related traits were mapped onto a high-density genetic map. Furthermore, 190 PA-related candidate genes for 91 unique PA QTLs and 2350 PY epistatic interaction loci-pairs were identified, which explain 2.8-51.8% and 5.2-23.6% of phenotypic variation, respectively. Three gene categories, transcription factor, auxin/IAA, and gibberellin, comprise the largest proportions of candidate genes for PA-related QTLs. The effectiveness of QTL candidate genes prediction was demonstrated by cloning of three candidate genes, Bna.A02.CLV2, Bna.A09.SLY2, and Bna.C07.AHK4. The study thus outlines a gene network for control of PA-related traits and provides novel information for understanding the establishment of ideal PA and for developing effective breeding strategies for yield improvement in rapeseed and other crops.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
华中农业大学博士学位论文,
[本文引用: 1]
PhD Dissertation of Huazhong Agricultural University,
[本文引用: 1]
DOI:10.1128/mcb.20.3.1030-1043.2000URLPMID:10629060 [本文引用: 1]

The c-Jun NH(2)-terminal kinase (JNK) group of mitogen-activated protein kinases (MAPKs) is activated in response to the treatment of cells with inflammatory cytokines and by exposure to environmental stress. JNK activation is mediated by a protein kinase cascade composed of a MAPK kinase and a MAPK kinase kinase. Here we describe the molecular cloning of a putative molecular scaffold protein, JIP3, that binds the protein kinase components of a JNK signaling module and facilitates JNK activation in cultured cells. JIP3 is expressed in the brain and at lower levels in the heart and other tissues. Immunofluorescence analysis demonstrated that JIP3 was present in the cytoplasm and accumulated in the growth cones of developing neurites. JIP3 is a member of a novel class of putative MAPK scaffold proteins that may regulate signal transduction by the JNK pathway.
DOI:10.1105/tpc.112.096156URLPMID:22517321 [本文引用: 1]

Mitogen-activated protein kinases (MAPKs) are evolutionarily conserved proteins that function as key signal transduction components in fungi, plants, and mammals. During interaction between phytopathogenic fungi and plants, fungal MAPKs help to promote mechanical and/or enzymatic penetration of host tissues, while plant MAPKs are required for activation of plant immunity. However, new insights suggest that MAPK cascades in both organisms do not operate independently but that they mutually contribute to a highly interconnected molecular dialogue between the plant and the fungus. As a result, some pathogenesis-related processes controlled by fungal MAPKs lead to the activation of plant signaling, including the recruitment of plant MAPK cascades. Conversely, plant MAPKs promote defense mechanisms that threaten the survival of fungal cells, leading to a stress response mediated in part by fungal MAPK cascades. In this review, we make use of the genomic data available following completion of whole-genome sequencing projects to analyze the structure of MAPK protein families in 24 fungal taxa, including both plant pathogens and mycorrhizal symbionts. Based on conserved patterns of sequence diversification, we also propose the adoption of a unified fungal MAPK nomenclature derived from that established for the model species Saccharomyces cerevisiae. Finally, we summarize current knowledge of the functions of MAPK cascades in phytopathogenic fungi and highlight the central role played by MAPK signaling during the molecular dialogue between plants and invading fungal pathogens.
DOI:10.1016/j.pbi.2009.07.017URLPMID:19716758 [本文引用: 1]

Mitogen-activated protein kinase (MAPK) cascades are signaling modules that transduce extracellular stimuli to a range of cellular responses. Research in yeast and metazoans has shown that MAPK-mediated phosphorylation directly or indirectly regulates the activity of transcription factors. Plant MAPK cascades have been implicated in development and stress responses, but little is known about the specific downstream targets they control. Recent studies have begun to identify direct MAPK transcriptional targets, and provide insights into the mechanisms by which MAPK signaling networks regulate gene expression.
DOI:10.1042/BJ20080625URLPMID:18570633 [本文引用: 2]

Many changes in environmental conditions and hormones are mediated by MAPK (mitogen-activated protein kinase) cascades in all eukaryotes, including plants. Studies of MAPK pathways in genetic model organisms are especially informative in revealing the molecular mechanisms by means of which MAPK cascades are controlled and modulate cellular processes. The present review highlights recent insights into MAPK-based signalling in Arabidopsis thaliana (thale cress), revealing the complexity and future challenges to understanding signal-transduction networks on a global scale.
URLPMID:17433310 [本文引用: 1]
DOI:10.1016/s1360-1385(02)02302-6URLPMID:12119167 [本文引用: 1]

Mitogen-activated protein kinase (MAPK) cascades are universal signal transduction modules in eukaryotes, including yeasts, animals and plants. These protein phosphorylation cascades link extracellular stimuli to a wide range of cellular responses. In plants, MAPK cascades are involved in responses to various biotic and abiotic stresses, hormones, cell division and developmental processes. Completion of the Arabidopsis genome-sequencing project has revealed the existence of 20 MAPKs, 10 MAPK kinases and 60 MAPK kinase kinases. Here, we propose a simplified nomenclature for Arabidopsis MAPKs and MAPK kinases that might also serve as a basis for standard annotation of these gene families in all plants.
DOI:10.1073/pnas.93.2.765URLPMID:8570631 [本文引用: 1]

We describe here the cloning and characterization of a cDNA encoding a protein kinase that has high sequence homology to members of the mitogen-activated protein kinase (MAPK) kinase kinase (MAPKKK or MEKK) family; this cDNA is named cATMEKKI (Arabidopsis thaliana MAP kinase or ERK kinase kinase 1). The catalytic domain of the putative ATMEKK1 protein shows approximately 40% identity with the amino acid sequences of the catalytic domains of MAPKKKs (such as Byr2 from Schizosaccharomyces pombe, Ste11 from Saccharomyces cerevisiae, Bck1 from S. cerevisiae, MEKK from mouse, and NPK1 from tobacco). In yeast cells that overexpress ATMEKK1, the protein kinase replaces Ste11 in responding to mating pheromone. In this study, the expression of three protein kinases was examined by Northern blot analyses: ATMEKK1 (structurally related to MAPKKK), ATMPK3 (structurally related to MAPK), and ATPK19 (structurally related to ribosomal S6 kinase). The mRNA levels of these three protein kinases increased markedly and simultaneously in response to touch, cold, and salinity stress. These results suggest that MAP kinase cascades, which are thought to respond to a variety of extracellular signals, are regulated not only at the posttranslational level but also at the transcriptional level in plants and that MAP kinase cascades in plants may function in transducing signals in the presence of environmental stress.
DOI:10.1126/scisignal.2004651URLPMID:24194583 [本文引用: 1]

During development, differentiation is often initiated by the activation of different receptor tyrosine kinases (RTKs), which results in the tightly regulated activation of cytoplasmic signaling cascades. In the differentiation of neurons and glia in the developing Drosophila eye, we found that the proper intensity of RTK signaling downstream of fibroblast growth factor receptor (FGFR) or epidermal growth factor receptor required two mutually antagonistic feedback loops. We identified a positive feedback loop mediated by the Ras association (RA) domain-containing protein Rau that sustained Ras activity and counteracted the negative feedback loop mediated by Sprouty. Rau has two RA domains that together showed a binding preference for GTP (guanosine 5'-triphosphate)-loaded (active) Ras. Rau homodimerized and was found in large-molecular weight complexes. Deletion of rau in flies decreased the differentiation of retinal wrapping glia and induced a rough eye phenotype, similar to that seen in alterations of Ras signaling. Further, the expression of sprouty was repressed and that of rau was increased by the COUP transcription factor Seven-up in the presence of weak, but not constitutive, activation of FGFR. Together, our findings reveal another regulatory mechanism that controls the intensity of RTK signaling in the developing neural network in the Drosophila eye.
URLPMID:25720833 [本文引用: 1]
DOI:10.1016/j.biotechadv.2013.09.006URLPMID:24091291 [本文引用: 1]

As sessile organisms, plants have developed specific mechanisms that allow them to rapidly perceive and respond to stresses in the environment. Among the evolutionarily conserved pathways, the ABA (abscisic acid) signaling pathway has been identified as a central regulator of abiotic stress response in plants, triggering major changes in gene expression and adaptive physiological responses. ABA induces protein kinases of the SnRK family to mediate a number of its responses. Recently, MAPK (mitogen activated protein kinase) cascades have also been shown to be implicated in ABA signaling. Therefore, besides discussing the role of ABA in abiotic stress signaling, we will also summarize the evidence for a role of MAPKs in the context of abiotic stress and ABA signaling.
DOI:10.1016/j.plantsci.2017.04.006URLPMID:28554467 [本文引用: 1]

Mitogen-Activated Protein Kinase (MAPK) cascades are functional modules widespread among eukaryotic organisms. In plants, these modules are encoded by large multigenic families and are involved in many biological processes ranging from stress responses to cellular differentiation and organ development. Furthermore, MAPK pathways are involved in the perception of environmental and physiological modifications. Interestingly, some MAPKs play a role in several signaling networks and could have an integrative function for the response of plants to their environment. In this review, we describe the classification of MAPKs and highlight some of their biochemical actions. We performed an in silico analysis of MAPK gene expression in response to nutrients supporting their involvement in nutritional signaling. While several MAPKs have been identified as players in sugar, nitrogen, phosphate, iron and potassium-related signaling pathways, their biochemical functions are yet mainly unknown. The integration of these regulatory cascades in the current understanding of nutrient signaling is discussed and potential new avenues for approaches toward plants with higher nutrient use efficiencies are evoked.
DOI:10.1111/tpj.13635URLPMID:28710770 [本文引用: 1]

Mitogen-activated protein kinase (MPK) cascades are conserved mechanisms of signal transduction across eukaryotes. Despite the importance of MPK proteins in signaling events, specific roles for many Arabidopsis MPK proteins remain unknown. Multiple studies have suggested roles for MPK signaling in a variety of auxin-related processes. To identify MPK proteins with roles in auxin response, we screened mpk insertional alleles and identified mpk1-1 as a mutant that displays hypersensitivity in auxin-responsive cell expansion assays. Further, mutants defective in the upstream MAP kinase kinase MKK3 also display hypersensitivity in auxin-responsive cell expansion assays, suggesting that this MPK cascade affects auxin-influenced cell expansion. We found that MPK1 interacts with and phosphorylates ROP BINDING PROTEIN KINASE 1 (RBK1), a protein kinase that interacts with members of the Rho-like GTPases from Plants (ROP) small GTPase family. Similar to mpk1-1 and mkk3-1 mutants, rbk1 insertional mutants display auxin hypersensitivity, consistent with a possible role for RBK1 downstream of MPK1 in influencing auxin-responsive cell expansion. We found that RBK1 directly phosphorylates ROP4 and ROP6, supporting the possibility that RBK1 effects on auxin-responsive cell expansion are mediated through phosphorylation-dependent modulation of ROP activity. Our data suggest a MKK3 * MPK1 * RBK1 phosphorylation cascade that may provide a dynamic module for altering cell expansion.
URLPMID:19688272 [本文引用: 1]
[本文引用: 1]
URLPMID:23758924 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1105/tpc.106.050039URLPMID:17933903 [本文引用: 1]

Although the Arabidopsis thaliana genome contains genes encoding 20 mitogen-activated protein kinases (MAPKs) and 10 MAPK kinases (MAPKKs), most of them are still functionally uncharacterized. In this work, we analyzed the function of the group B MAPK kinase, MKK3. Transgenic ProMKK3:GUS lines showed basal expression in vascular tissues that was strongly induced by Pseudomonas syringae pv tomato strain DC3000 (Pst DC3000) infection but not by abiotic stresses. The growth of virulent Pst DC3000 was increased in mkk3 knockout plants and decreased in MKK3-overexpressing plants. Moreover, MKK3 overexpression lines showed increased expression of several PR genes. By yeast two-hybrid analysis, coimmunoprecipitation, and protein kinase assays, MKK3 was revealed to be an upstream activator of the group C MAPKs MPK1, MPK2, MPK7, and MPK14. Flagellin-derived flg22 peptide strongly activated MPK6 but resulted in poor activation of MPK7. By contrast, MPK6 and MPK7 were both activated by H(2)O(2), but only MPK7 activation was enhanced by MKK3. In agreement with the notion that MKK3 regulates the expression of PR genes, ProPR1:GUS expression was strongly enhanced by coexpression of MKK3-MPK7. Our results reveal that the MKK3 pathway plays a role in pathogen defense and further underscore the importance and complexity of MAPK signaling in plant stress responses.
URLPMID:25301445 [本文引用: 1]
DOI:10.1016/j.pbi.2018.04.012URLPMID:29753266 [本文引用: 1]

Mitogen-activated protein kinase (MAPK) cascades are key signaling modules downstream of receptors/sensors that perceive endogenous and exogenous stimuli such as hormones, peptide ligands, and pathogen-derived patterns/effectors. In this review, we summarize recent advances in the establishment of MAPK cascades as unified signaling modules downstream of receptor-like kinases (RLKs) and receptor-like proteins (RLPs) in plant growth and defense, the identification of components connecting the RLK/RLP receptor complexes to the MAPK cascades, and the interactions between MAPK and hormone signaling pathways. We also propose a set of criteria for defining the physiological substrates of plant MAPKs. With only a limited number of MAPK components, multiple functional pathways often share the same MAPK cascade. As a result, understanding the signaling specificity, which requires detailed information about the spatiotemporal expression of the components involved, their complex formation, and the consequence of substrate phosphorylation, is central to our study of MAPK functions.
[本文引用: 1]
[本文引用: 1]
URLPMID:9927426 [本文引用: 1]
DOI:10.4049/jimmunol.166.7.4416URLPMID:11254696 [本文引用: 1]

The mitogen-activated protein kinase (MAPK) cascade consists of the MAPK (extracellular signal-regulated kinase 2; ERK2) and its activator, MAPK kinase (MAP/ERK kinase; MEK). However, the mechanisms for activation of ERK2 have not been defined yet in cells. Here, we used fluorescent protein-tagged ERK2 and MEK to examine the localization of ERK2 and MEK in living rat basophilic leukemia (RBL-2H3) cells. ERK2 was mainly in the cytoplasm in resting cells but translocated into the nucleus after the ligation of IgE receptors. The import of ERK2 reached the maximum at 6--7 min, and then the imported ERK2 was exported from the nucleus. MEK mainly resided in the cytoplasm, and no significant MEK translocation was detected statically after ligation of IgE receptors. However, analysis of the dynamics of ERK2 and MEK suggested that both of them rapidly shuttle between the cytoplasm and the nucleus and that MEK regulates the nuclear shuttling of ERK2, whereas MEK remains mainly in the cytoplasm. In addition, the data suggested that the sustained calcium increase was required for the optimal translocation of ERK2 into the nucleus in RBL-2H3 cells. These results gave a new insight of the dynamics of ERK2 and MEK in the nuclear shuttling of RBL-2H3 cells after the ligation of IgE receptors.
URLPMID:18273012 [本文引用: 1]
DOI:10.1111/j.1365-313X.2004.02229.xURLPMID:15500467 [本文引用: 1]

Changing environmental conditions, atmospheric pollutants and resistance reactions to pathogens cause production of reactive oxygen species (ROS) in plants. ROS in turn trigger the activation of signaling cascades such as the mitogen-activated protein kinase (MAPK) cascade and accumulation of plant hormones, jasmonic acid, salicylic acid (SA), and ethylene (ET). We have used ozone (O3) to generate ROS in the apoplast of wild-type Col-0 and hormonal signaling mutants of Arabidopsis thaliana and show that this treatment caused a transient activation of 43 and 45 kDa MAPKs. These were identified as AtMPK3 and AtMPK6. We also demonstrate that initial AtMPK3 and AtMPK6 activation in response to O3 was not dependent on ET signaling, but that ET is likely to have secondary effects on AtMPK3 and AtMPK6 function, whereas functional SA signaling was needed for full-level AtMPK3 activation by O3. In addition, we show that AtMPK3, but not AtMPK6, responded to O3 transcriptionally and translationally during O3 exposure. Finally, we show in planta that activated AtMPK3 and AtMPK6 are translocated to the nucleus during the early stages of O3 treatment. The use of O3 to induce apoplastic ROS formation offers a non-invasive in planta system amenable to reverse genetics that can be used for the study of stress-responsive MAPK signaling in plants.
DOI:10.1016/s1369-5266(02)00282-0URLPMID:12183176 [本文引用: 1]

Recent biochemical and genetic studies confirm that hydrogen peroxide is a signalling molecule in plants that mediates responses to abiotic and biotic stresses. Signalling roles for hydrogen peroxide during abscisic-acid-mediated stomatal closure, auxin-regulated root gravitropism and tolerance of oxygen deprivation are now evident. The synthesis and action of hydrogen peroxide appear to be linked to those of nitric oxide. Downstream signalling events that are modulated by hydrogen peroxide include calcium mobilisation, protein phosphorylation and gene expression. Calcium and Rop signalling contribute to the maintenance of hydrogen peroxide homeostasis.
URLPMID:19095804 [本文引用: 1]
DOI:10.1016/j.tplants.2009.12.001URLPMID:20047850 [本文引用: 1]

Although mitogen-activated protein kinase (MAPK) signal transduction cascades are known regulators of various aspects of plant biology, our knowledge of these systems has been largely restricted to a small subset of the MAPKs. However, global analyses are now revealing that many more of these kinases are probably engaged in modulating developmental and fitness adaptation processes in the plant kingdom. In this review, we show how these new findings are beginning to define the overall architecture of plant MAPK signaling, with a particular focus on the interplay between the terminal MPKs and their activators, inactivators and cellular targets.
DOI:10.4161/psb.5.11.13020URLPMID:20980831 [本文引用: 1]

The mitogen-activated protein kinase (MAPK) cascades play diverse roles in intra- and extra-cellular signaling in plants. MAP kinases are the component of kinase modules which transfer information from sensors to responses in eukaryotes including plants. They play a pivotal role in transduction of diverse extracellular stimuli such as biotic and abiotic stresses as well as a range of developmental responses including differentiation, proliferation and death. Several cascades are induced by different biotic and abiotic stress stimuli such as pathogen infections, heavy metal, wounding, high and low temperatures, high salinity, UV radiation, ozone, reactive oxygen species, drought and high or low osmolarity. MAPK signaling has been implicated in biotic stresses and has also been associated with hormonal responses. The cascade is regulated by various mechanisms, including not only transcriptional and translational regulation but through post-transcriptional regulation such as protein-protein interactions. Recent detailed analysis of certain specific MAP kinase pathways have revealed the specificity of the kinases in the cascade, signal transduction patterns, identity of pathway targets and the complexity of the cascade. The latest insights and finding are discussed in this paper in relation to the role of MAPK pathway modules in plant stress signaling.
DOI:10.1016/s1360-1385(01)02103-3URLPMID:11701380 [本文引用: 1]

The Arabidopsis genome encodes approximately 20 different mitogen-activated protein kinases (MAPKs) that are likely to be involved in growth, development and responses to endogenous and environmental cues. Several plant MAPKs are activated by a variety of stress stimuli, including pathogen infection, wounding, temperature, drought, salinity, osmolarity, UV irradiation, ozone and reactive oxygen species. Recent gain-of-function studies show that two tobacco MAPKs induce the expression of defense genes and cause cell death. By contrast, loss-of-function studies of other MAPK pathways revealed negative regulation of disease resistance. This 'push-and-pull' regulation by different MAPK pathways might provide a more precise control of plant defense responses.
DOI:10.1093/jxb/err315URLPMID:22143917 [本文引用: 1]

The light-harvesting chlorophyll a/b binding proteins (LHCB) are perhaps the most abundant membrane proteins in nature. It is reported here that the down-regulation or disruption of any member of the LHCB family, LHCB1, LHCB2, LHCB3, LHCB4, LHCB5, or LHCB6, reduces responsiveness of stomatal movement to ABA, and therefore results in a decrease in plant tolerance to drought stress in Arabidopsis thaliana. By contrast, over-expression of a LHCB member, LHCB6, enhances stomatal sensitivity to ABA. In addition, the reactive oxygen species (ROS) homeostasis and a set of ABA-responsive genes are altered in the lhcb mutants. These data demonstrate that LHCBs play a positive role in guard cell signalling in response to ABA and suggest that they may be involved in ABA signalling partly by modulating ROS homeostasis.
URLPMID:20733066 [本文引用: 1]
DOI:10.1007/s00425-012-1820-xURL [本文引用: 1]

A full-length abscisic acid (ABA) senescence and ripening inducible gene named LcAsr was obtained from litchi. Bioinformatic analysis showed that full-length LcAsr was 1,177 bp and contained an open reading frame (ORF) encoding 153 amino acids, 85- and 146-bp 5' and 3' UTRs, respectively. LcAsr was expressed in all organs, with preferential expression in the flower and low levels in pulp. The expression level of LcAsr in postharvest uncovered fruit reached a maximum at 24 h after harvest. When the litchi fruit was covered with plastic film, the LcAsr expression level remained constant. LcASR protein localized in the nucleus. LcAsr was transformed in Arabidopsis thaliana L. (ecotype Columbia) and four transgenic lines were obtained. One line, 35S::LcAsrD, was selected for drought tolerance analysis and showed higher tolerance to drought than the control. The activities of superoxide dismutase, catalase, ascorbate peroxidase, and glutathione reductase were much higher in the transgenic line than the control under drought conditions. The levels of several ABA/stress-regulated genes were investigated. The transcript level of responsive to ABA (RAB18) remained constant and responsive to dehydration (RD29A) displayed a slight decrease in the Columbia line (Col). However, the transcript levels of LcAsr, RAB18, and RD29A were greatly enhanced in the transgenic 35S::LcAsrD. The transcript levels of KAT1, KAT2, and SKOR were also markedly decreased in the transgenic line. These results suggest an important role of LcAsr as a protective molecule for water deficit and help to understand the molecular mechanism of postharvest litchi fruit dehydration.
DOI:10.1016/s0005-2736(98)00007-8URLPMID:9545564 [本文引用: 1]

Previously, we constructed a cDNA library from Arabidopsis plants that were exposed to dehydration stress for 1 h and obtained the ERD6 clone. Here we report that the ERD6 cDNA consists of 1741 bp and encodes a polypeptide of 496 amino acids having a predicted molecular weight of 54,354. The putative polypeptide of ERD6 is related to those of sugar transporters of bacteria, yeasts, plants and mammals. Hydropathy analysis revealed that ERD6 protein has 12 putative transmembrane domains and a central hydrophilic region. Sequences that are conserved at the ends of the 6th and 12th membrane-spanning domains of sugar transporters are also present in ERD6. These data suggest that ERD6 encodes a sugar transporter. Genomic Southern blots indicate that the ERD6 gene is a member of a multigene family in the Arabidopsis genome. The expression of the ERD6 gene was induced not only by dehydration but also by cold treatment.
DOI:10.1074/jbc.M109275200URLPMID:11779861 [本文引用: 1]

Drought and high salinity induce the expression of many plant genes. To understand the signal transduction mechanisms underlying the activation of these genes, we carried out a genetic screen to isolate Arabidopsis mutants defective in osmotic stress-regulated gene induction. Here we report the isolation, characterization, and cloning of a mutation, los6, which diminished osmotic stress activation of a reporter gene. RNA blot analysis indicates that under osmotic stress the transcript levels for stress-responsive genes such as RD29A, COR15A, KIN1, COR47, RD19, and ADH are lower in los6 plants than in wild type plants. los6 plants were found to have reduced phytohormone abscisic acid (ABA) accumulation and to be allelic to the ABA-deficient mutant, aba1. LOS6/ABA1 encodes a zeaxanthin epoxidase that functions in ABA biosynthesis. Its expression is enhanced by osmotic stress. Furthermore, we found that there exists a positive feedback regulation by ABA on the expression of LOS6/ABA1, which may underscore a quick adaptation strategy for plants under osmotic stress. Similar positive regulation by ABA also exists for other ABA biosynthesis genes AAO3 and LOS5/ABA3 and in certain genetic backgrounds, NCED3. This feedback regulation by ABA is impaired in the ABA-insensitive mutant abi1 but not in abi2. Moreover, the up-regulation of LOS6/ABA1, LOS5/ABA3, AAO3, and NCED3 by osmotic stress is reduced substantially in ABA-deficient mutants. Transgenic plants overexpressing LOS6/ABA1 showed an increased RD29A-LUC expression under osmotic stress. These results suggest that the level of gene induction by osmotic stress is dependent on the dosage of the zeaxanthin epoxidase enzyme.
DOI:10.1105/tpc.000596URLPMID:12045276 [本文引用: 1]
DOI:10.1105/tpc.17.00216URLPMID:28536144 [本文引用: 1]

The transcription factor MYC2 has emerged as a master regulator of jasmonate (JA)-mediated responses as well as crosstalk among different signaling pathways. The instability of MYC2 is in part due to the action of PUB10 E3 ligase, which can polyubiquitinate this protein. Here, we show that polyubiquitinated MYC2 can be deubiquitinated by UBP12 and UBP13 in vitro, suggesting that the two deubiquitinating enzymes can counteract the effect of PUB10 in vivo. Consistent with this view, UBP12 and UBP13 associate with MYC2 in the nucleus. Transgenic Arabidopsis thaliana plants deficient in UBP12 and UBP13 show accelerated decay of MYC2 and are hyposensitive to JA, whereas plants overexpressing UBP12 or UBP13 have prolonged MYC2 half-life and are hypersensitive to JA Our results suggest that there is a genetic link between UBP12, UBP13, and MYC2. Our results identify UBP12 and UBP13 as additional positive regulators of JA responses and suggest that these enzymes likely act by stabilizing MYC2.
DOI:10.1111/nph.13398URLPMID:25817565 [本文引用: 1]

The bHLH transcription factor MYC2, together with its paralogues MYC3 and MYC4, is a master regulator of the response to the jasmonate (JA) hormone in Arabidopsis (Arabidopsis thaliana). In the absence of JA, JASMONATE ZIM (JAZ) proteins interact with the MYC proteins to block their activity. Understanding of the mechanism and specificity of this interaction is key to unravel JA signalling. We generated mutant MYC proteins and assessed their activity and the specificity of their interaction with the 12 Arabidopsis JAZ proteins. We show that the D94N mutation present in the atr2D allele of MYC3 abolishes the interaction between MYC3 and most JAZ proteins. The same effect is observed when the corresponding conserved Asp (D105) was mutated in MYC2. Accordingly, MYC2(D105N) activated target genes in the presence of JAZ proteins, in contrast to wild-type MYC2. JAZ1 and JAZ10 were the only JAZ proteins still showing interaction with the mutant MYC proteins, due to a second MYC interaction domain, besides the classical Jas domain. Our results visualize the divergence among JAZ proteins in their interaction with MYC proteins. Ultimately, the transferability of the Asp-to-Asn amino acid change might facilitate the design of hyperactive transcription factors for plant engineering.
DOI:10.1104/pp.15.00030URLPMID:25818700 [本文引用: 1]

The development of the plant root system is highly plastic, which allows the plant to adapt to various environmental stresses. Salt stress inhibits root elongation by reducing the size of the root meristem. However, the mechanism underlying this process remains unclear. In this study, we explored whether and how auxin and nitric oxide (NO) are involved in salt-mediated inhibition of root meristem growth in Arabidopsis (Arabidopsis thaliana) using physiological, pharmacological, and genetic approaches. We found that salt stress significantly reduced root meristem size by down-regulating the expression of PINFORMED (PIN) genes, thereby reducing auxin levels. In addition, salt stress promoted AUXIN RESISTANT3 (AXR3)/INDOLE-3-ACETIC ACID17 (IAA17) stabilization, which repressed auxin signaling during this process. Furthermore, salt stress stimulated NO accumulation, whereas blocking NO production with the inhibitor N(omega)-nitro-l-arginine-methylester compromised the salt-mediated reduction of root meristem size, PIN down-regulation, and stabilization of AXR3/IAA17, indicating that NO is involved in salt-mediated inhibition of root meristem growth. Taken together, these findings suggest that salt stress inhibits root meristem growth by repressing PIN expression (thereby reducing auxin levels) and stabilizing IAA17 (thereby repressing auxin signaling) via increasing NO levels.
DOI:10.1093/jxb/erw508URLPMID:28158805 [本文引用: 1]

Plants have developed complex mechanisms to respond to salt stress, depending on secondary messenger-mediated stress perception and signal transduction. Nitric oxide (NO) is widely known as a 'jack-of-all-trades' in stress responses. However, NO-mediated crosstalk between plant hormones remains unclear. In this study, we found that salt stabilized both AUXIN/INDOLE-3-ACETIC ACID 17 (Aux/IAA17) and RGA-LIKE3 (RGL3) proteins due to salt-induced NO production. Salt-induced NO overaccumulation and IAA17 overexpression decreased the transcripts of GA3ox genes, resulting in lower bioactive GA4. Further investigation showed that IAA17 directly interacted with RGL3 and increased its protein stability. Consistently, RGL3 stabilized IAA17 protein through inhibiting the interaction of TIR1 and IAA17 by competitively binding to IAA17. Moreover, both IAA17 and RGL3 conferred salt stress resistance. Overexpression of IAA17 and RGL3 partially alleviated the inhibitory effect of NO deficiency on salt resistance, whereas the iaa17 and rgl3 mutants displayed reduced responsiveness to NO-promoted salt resistance. Thus, the associations between IAA17 and gibberellin (GA) synthesis and signal transduction, and between the IAA17-interacting complex and the NO-mediated salt stress response were revealed based on physiological and genetic approaches. We conclude that integration of IAA17 and RGL3 is an essential component of NO-mediated salt stress response.
