 ,*河南省农业科学院经济作物研究所 / 农业部黄淮海油料作物重点实验室 / 河南省油料作物遗传改良重点实验室, 河南郑州 450002
,*河南省农业科学院经济作物研究所 / 农业部黄淮海油料作物重点实验室 / 河南省油料作物遗传改良重点实验室, 河南郑州 450002Improvement of oleic acid content in peanut (Arachis hypogaea L.) by marker assisted successive backcross and agronomic evaluation of derived lines
HUANG Bing-Yan, QI Fei-Yan, SUN Zi-Qi, MIAO Li-Juan, FANG Yuan-Jin, ZHENG Zheng, SHI Lei, ZHANG Zhong-Xin, LIU Hua, DONG Wen-Zhao, TANG Feng-Shou, ZHANG Xin-You ,*Industrial Crops Research Institute, Henan Academy of Agricultural Sciences / Huanghuaihai Key Laboratory of Oil Crops, Ministry of Agriculture / Henan Provincial Key Laboratory for Oil Crops Improvement, Zhengzhou 450002, Henan, China
,*Industrial Crops Research Institute, Henan Academy of Agricultural Sciences / Huanghuaihai Key Laboratory of Oil Crops, Ministry of Agriculture / Henan Provincial Key Laboratory for Oil Crops Improvement, Zhengzhou 450002, Henan, China通讯作者:
收稿日期:2018-07-10接受日期:2018-12-24网络出版日期:2019-01-04
| 基金资助: |
Received:2018-07-10Accepted:2018-12-24Online:2019-01-04
| Fund supported: |
作者简介 About authors

摘要
关键词:
Abstract
Keywords:
PDF (4870KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
黄冰艳, 齐飞艳, 孙子淇, 苗利娟, 房元瑾, 郑峥, 石磊, 张忠信, 刘华, 董文召, 汤丰收, 张新友. 以分子标记辅助连续回交快速提高花生品种油酸含量及对其后代农艺性状的评价[J]. 作物学报, 2019, 45(4): 546-555. doi:10.3724/SP.J.1006.2019.84096
HUANG Bing-Yan, QI Fei-Yan, SUN Zi-Qi, MIAO Li-Juan, FANG Yuan-Jin, ZHENG Zheng, SHI Lei, ZHANG Zhong-Xin, LIU Hua, DONG Wen-Zhao, TANG Feng-Shou, ZHANG Xin-You.
花生是我国重要的油料和经济作物, 其总产量居我国油料作物之首, 对保障我国食用植物油安全供给具有重要意义。脂肪是花生中含量最大的营养成分, 油酸和亚油酸是两种最主要的脂肪酸, 两者之和约占花生脂肪酸总量的80%, 花生的脂肪酸组成尤其是油酸和亚油酸的比例对花生品质具有重要影响。普通花生的油酸含量在35%~60%, 油酸/亚油酸比值(O/L值)小于1.5, 高油酸花生是指油酸含量超过75%或油酸/亚油酸比值超过10的花生。以往的研究表明, 提高花生的油酸含量能够增强花生及其制品的抗氧化性, 从而提高其货架寿命[1,2]; 食用高油酸花生及其制品, 能够降低人体血液中低密度脂蛋白(LDL)而保持高密度脂蛋白(HDL)水平, 从而减少心血管疾病的发生, 具有一定保健功能[3,4]; 而且随着油酸含量提高, 花生中对人体健康不利的饱和脂肪酸尤其是棕榈酸的含量明显降低, 从而改善了花生脂肪酸组成和营养品质。因此, 将高油酸与其他优良性状有效聚合, 培育高油酸专用型花生品种已经成为当今世界花生育种工作者的重要研究目标之一。
1987年, 美国育种家首次在育种材料中发现了花生高油酸自然突变体F435, 其油酸含量达到80%, 油酸/亚油酸比值超过35 [5]。遗传研究发现, 花生的高油酸性状由2个隐性基因(ol1ol1ol2ol2)控制[6,7]。分子生物学研究表明, 脂肪酸脱氢酶基因(AhFAD2)是控制油酸向亚油酸转化的关键基因, 花生的A亚基因组和B亚基因组分别存在一个AhFAD2A (Ol1Ol1)和一个AhFAD2B (Ol2Ol2)基因, 高油酸性状是这2个基因的功能丧失造成的。F435型高油酸是AhFAD2 A基因448 bp处的G/A突变和AhFAD2B基因442 bp处A插入引起的基因功能丧失, 这一研究结果为开发高油酸花生分子标记提供了可能[8,9,10,11,12]。目前已经报道的花生高油酸分子标记检测方法有Caps [13]、Real-Time PCR[14]、AS-PCR[15]、Taq-man探针法[16]、KASP (Kompetitive Allele-Specific PCR)方法[17]等。
我国高油酸花生育种研究起步于21世纪初, 由于国内缺乏高油酸种质资源, 引自国外的高油酸资源因适应性差等问题无法直接利用, 我国花生育种家以常规育种技术为主, 结合理化诱变等手段, 育成审定了一批高油酸花生品种[18,19]。但常规育种周期长、效率低、聚合多个优良性状困难, 截至目前, 我国审定的高油酸花生品种数量较少、遗传背景狭窄, 不能适应生产上对高油酸花生的多样化需求。
本研究针对常规育种方法对农艺性状的改良效率低、高油酸性状选择准确性差、育成品种遗传背景狭窄等问题, 采用河南省大面积推广的花生当家品种为轮回亲本, 以高油酸品种(系)为高油酸供体, 以分子标记辅助连续回交的育种方法, 结合近红外快速检测技术, 经过本地和海南每年两季节、连续4轮回交和4次自交, 历时5年, 获得以大面积推广品种为遗传背景的高油酸花生BC4F5优良品系, 并进行了纯合稳定品系的农艺性状综合评价, 旨在为快速高效选育高油酸花生新品种提供理论依据和技术支撑。
1 材料与方法
1.1 植物材料
1.1.1 轮回亲本 选用河南省生产上大面积推广的4个不同类型、普通油酸含量、具有不同性状特点的花生品种豫花15、远杂9102、豫花9327、豫花9326作为杂交母本和回交的轮回亲本。豫花15、豫花9327 、豫花9326分别为大果普通型花生品种, 豫花15于2000年通过河南省审定, 2001年通过国家鉴定, 粗脂肪含量高、抗网斑病、耐涝; 豫花9327于2003年通过河南省审定, 2006年通过国家鉴定, 丰产性好; 豫花9326于2005年通过河南省审定, 2006年通过国家鉴定, 粗脂肪含量高。远杂9102为珍珠豆型花生品种, 2000年通过河南省审定, 2001年通过国家鉴定, 适应性广, 粗脂肪含量高, 高抗青枯病[20]。上述品种均为河南省农业科学院选育。1.1.2 高油酸供体亲本 选用开农176、DF12和开选016作为杂交父本和高油酸性状供体亲本。开农176为开封市农林科学研究院育成的高油酸花生新品种, 于2014年通过国家鉴定, 荚果中大。DF12为河南省农业科学院选育的高油酸新品系, 普通型小果。开选016为开封市农林科学研究院选育的高油酸品系, 配合力高, 普通型小果[19]。
1.2 杂交及回交方法
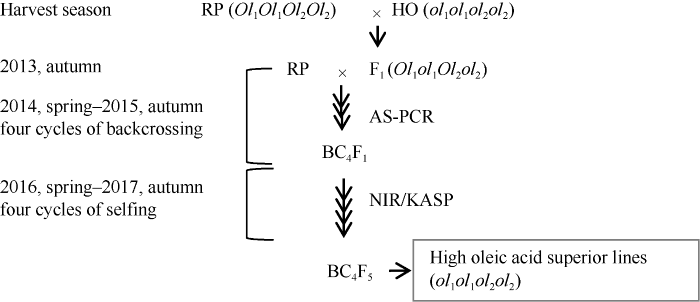
2013年春季分别以普通油酸含量的轮回亲本为母本配置豫花15*开农176、远杂9102*DF12、豫花9327*开农176和豫花9326*开选016四个杂交组合。2013年冬季分别以经过鉴定的F1真杂种与轮回亲本回交获得BC1F1, 2014年至2015年分别以利用AS-PCR-MP方法鉴定出的BC1F1、BC2F1、BC3F1的FAD2位点杂合基因型(Ol1ol1Ol2ol2)连续与轮回亲本回交, 获得BC4F1[21]。2016年至2017年分别根据近红外检测油酸含量和KASP方法FAD2基因型检测进行连续4代的2个基因位点至少各具有1个突变的基因型后代的自交, 获得BC4F5, 从中选出农艺性状优良的高油酸纯合品系扩繁种子参加品种登记多点试验(图1)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1分子标记辅助连续回交高油酸花生育种路线图
RP: 轮回亲本; HO: 高油酸亲本; AS-PCR-MP: 位点特异PCR; NIR: 近红外检测。
Fig. 1Breeding Strategy of marker assisted selection for high oleic acid peanut
RP: recurrent parent; HO: high oleic acid; AS-PCR-MP: allele specific-PCR-mispairing; NIR: near infrared reflectance spectroscopy.
1.3 FAD2基因型鉴定
采取花生未展开幼嫩叶片, 使用DNAsecure Plant Kit (TIANGEN BIOTECH CO., China)提取DNA。1.3.1 杂交和回交F1的FAD2基因型鉴定 采用AS-PCR-MP方法鉴定F1的FAD2基因型。利用4对PCR引物分别扩增FAD2A和FAD2B的野生型和突变型: FAD2A-F(aF19F[21]): 5°-GATTACTGATTATT GACTT-3°, FAD2A-WT-R: 5°-GTTTTGGGACAAAC ACTTCACC-3°, FAD2A-Mu-R: 5°-GTTTTGGGACA AACACTTAGAT-3°; FAD2B-F(bF19F[21]): 5°-CAGA ACCATTAGCTTTG-3°, FAD2B-WT-R: 5°-GACAAA CACTTCGTCGCGTTAG-3°, FAD2B-Ins-R: 5°-GAC AAACACTTCGTCGCGTTAT-3°。
PCR体系为 10 μL, 包含20 ng DNA模板、1 U LA Taq酶(Takara, Japan)、1× PCR buffer、0.4 mmol L-1 dNTPs、正反向引物各3 μmol L-1。PCR程序为94°C 5 min; 94°C 30 s, 45.9°C 30 s, 72°C 45 s, 30或35个循环; 72°C延伸10 min。FAD2A-F/FAD2A- WT-R引物的循环数为30, FAD2A-F/FAD2A-Mu-R、FAD2B-F/FAD2B-WT-R和FAD2B-F/FAD2B-Ins-R引物的循环数为35。PCR产物经琼脂糖凝胶电泳及VersaDoc system (Bio-Rad, USA)紫外检测。
1.3.2 自交后代的FAD2基因型鉴定 采用KASP方法鉴定自交后代BC4F2、BC4F4的FAD2基因型。KASP体系为5 μL, 包括2.5 μL DNA模板(10~20 ng)、2.43 μL 2×KASP Master Mix (LGC, Inc., England)和0.07 μL KASP assay mix。KASPar引物订购自LGC公司, 含有FAM和VIC荧光序列(FAM tail: 5°-GAAGGTGACCAAGTTCATGCT-3°; VIC tail: 5°-GAAGGTCGGAGTCAACGGATT-3°)和3°端FAD2目标片段的SNP。PCR程序为94°C 15 min, 10个循环的降落PCR (94°C 20 s; 然后从61°C开始每个循环降落0.6°C, 60 s), 最后执行26个循环的扩增反应(94°C 20 s; 57°C 60 s)[22]。
1.4 籽仁油酸性状及荚果籽仁外观形态性状测定
选取单株上正常成熟的饱满种子, 利用近红外品质分析仪DA 7200 (Perten, Swiss)测定油酸含量和亚油酸含量, 取3次测定的平均数, 计算O/L。用万深植物种子考种仪SC-G (Wanshen Co., China)采集单株成熟荚果和饱满籽仁的荚果面积、荚果长、荚果宽、荚果长宽比、籽仁面积、籽仁长、籽仁宽、籽仁长宽比8个性状。目测记载荚果果型、荚果网纹和种皮颜色。1.5 农艺性状调查
参照姜慧芳等[23]方法, 从BC4F5的每个株行随机选取10株测量主茎高、侧枝长、总分枝数、结果枝数、单株结果数5个农艺性状。1.6 遗传背景检测
利用经过验证的均匀分布在花生20对染色体上的96个多态性较好的KASP引物进行遗传背景检测。KASP体系同上。1.7 统计分析
利用DPS 14.50数据处理系统统计数据及制作图表完成。2 结果与分析
2.1 杂交及回交F1的基因型鉴定
4个杂交组合收获的杂交F1种子及本地和海南1年2季连续3次回交分别产生的BC1F1、BC2F1和BC3F1种子经AS-PCR-MP鉴定(图2), 选择具有双杂合Ol1ol1Ol2ol2基因型(lean4带型)的真杂种做父本分别与轮回亲本回交直至BC4F1, 鉴定获得Ol1ol1Ol2ol2基因型的BC4F1种子90粒, 全部种植于大田, 收获BC4F2单株。图2
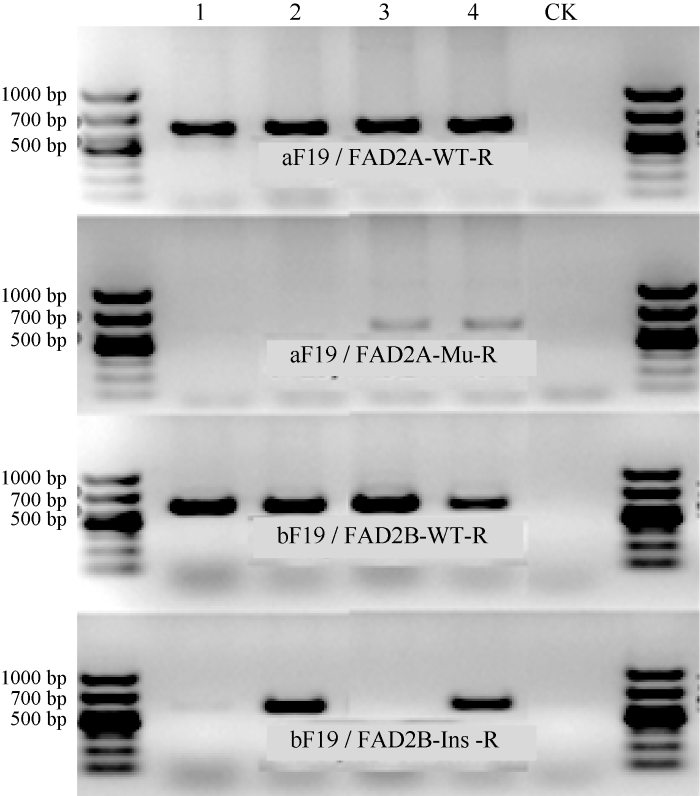 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2AS-PCR-MP扩增的F1的4种基因型带型
Fig. 2F1 genotyping by AS-PCR-MPLane 1: Ol1Ol1Ol2Ol2; lane 2: Ol1Ol1Ol2ol2; lane 3: Ol1ol1Ol2Ol2; lane 4: Ol1ol1Ol2ol2.
2.2 自交后代的基因型及油酸含量筛选
首先进行BC4F2单株FAD2基因的KASP基因型检测(图3)。然后将F2全部单株收获并进行了BC3F2:3籽仁油酸含量的近红外检测分析(表1)。F2的9种基因型后代的油酸含量基本可被分为4类, 最低的为没有隐性基因或者仅有1个ol1隐性基因; 中低油酸含量的为只有1个ol2隐性基因, 或者有2个ol1隐性基因; 中高油酸含量的为至少1对纯合ol2, 或者1个ol2加1对或者1个ol1; 最高油酸含量的为ol1和ol2双纯合的基因型。图3
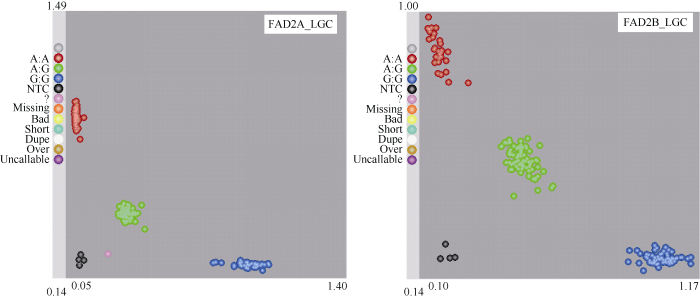 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3KASP检测BC4F2单株FAD2的突变位点SNP
Fig. 3KASP detection of FAD2 SNP in BC4F2 individuals
Table 1
表1
表1远杂9102 BC4F2:3后代油酸含量分布及其F2基因型
Table 1
| F2:3油酸含量 F2:3 oleic acid content | F2基因型 Related F2 genotype | F2:3单株数量 Number of F2:3 plants | F2基因型比例 Genotype ratio |
|---|---|---|---|
| <38% | _ _ _ _*; _ol1 _ _ | 86 | 9.4/16 |
| 38%-48% | _ _ _ol2; ol1ol1 _ _ | 29 | 3.2/16 |
| 48%-68% | ol1ol1 _ol2; _ol1ol2ol2; _ol1 _ol2; __ ol2ol2 | 25 | 2.7/16 |
| >68% | ol1ol1ol2ol2 | 7 | 0.8/16 |
| 合计Total | 147 |
新窗口打开|下载CSV
继续种植种子油酸含量高于60%的单株产生BC4F4, 挑选单株收获, 将种子油酸含量高于70%的21个株行混收扩繁F4:5株系, 从每株系随机取3个混样进行KASP基因型鉴定。其中20个株系可以确定FAD2基因型为ol1ol1ol2ol2高油酸纯合基因型(图4)。
图4
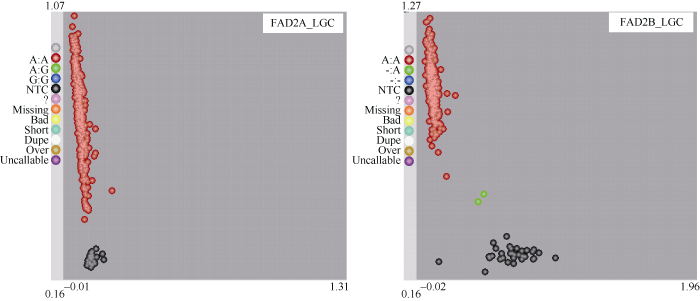 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4KASP检测BC4F4:5 FAD2的突变位点SNP
Fig. 4KASP detection of FAD2 SNPs in BC4F4:5
2.3 BC4F4:5家系的农艺性状表现
2017年共收获64个BC4F4株行的近600个单株。从图5可以看出, 不同轮回亲本后代的株行间及不同性状间变异幅度各不相同, 按照性状统计, 植株形态性状如主茎高、侧枝长、总分枝数等的轮回亲本平均数大都在四分位数(50%的个体平均数)区间内, 只有豫花9327和豫花9326组合的主茎高和侧枝长高于轮回亲本平均数, 豫花15和豫花9327的总分枝数高于轮回亲本平均数。结果习性除豫花9327组合的结果枝数和单株结果数四分位数高于轮回亲本、豫花9326组合的结果枝数四分位数低于轮回亲本以外, 其他杂交组合的轮回亲本平均数也都在四分位数区间内。荚果外观性状除远杂9102组合的荚果面积、荚果长和荚果宽四分位数高于轮回亲本, 远杂9102组合的荚果长宽比四分位数、豫花9327组合的荚果面积、荚果宽和荚果长宽比四分位数低于轮回亲本外, 其他杂交组合的轮回亲本平均数也都在四分位数区间内。图5
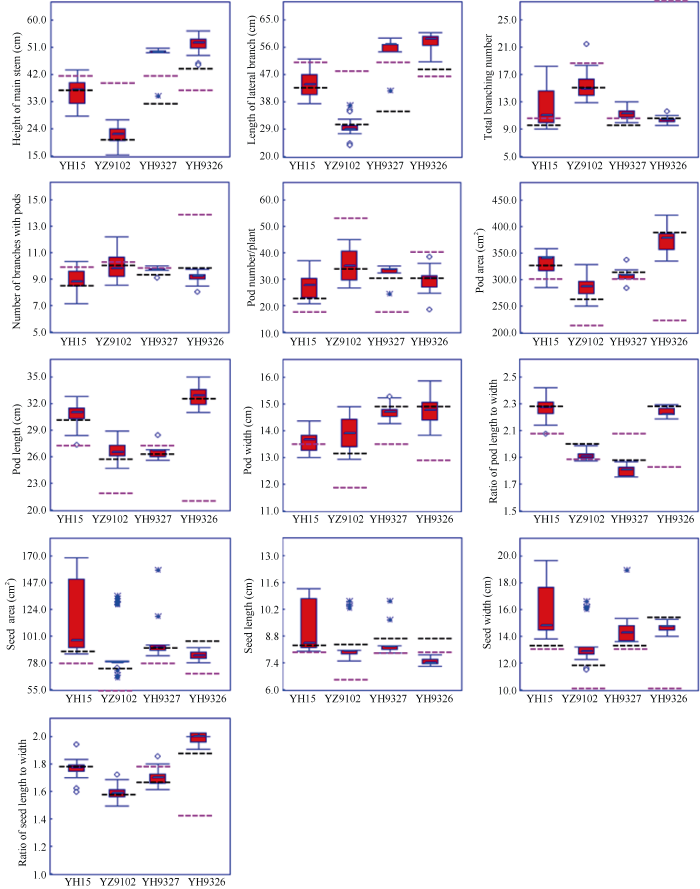 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5不同轮回亲本的BC4F4的性状分布箱线图
蓝色粗实线代表中位数, 黑色虚线代表轮回亲本平均数, 粉色虚线代表非轮回亲本平均数。YH15: 豫花15; YZ9102: 远杂9102; YH9327: 豫花9327; YH9326: 豫花9326。
Fig. 5Box-plot of BC4F4 for different recurrent parents
Heavy blue solid line indicates median, black dotted line indicates mean of recurrent parent, pink dotted line indicates mean of non-recurrent parent. YH15: Yuhua 15; YZ9102: Yuanza 9102; YH9327: Yuhua 9327; YH9326: Yuhua 9326.
籽仁的外观性状在BC4F4株行与轮回亲本平均数之间变异幅度较大, 除豫花15、远杂9102和豫花9327组合的籽仁长宽比、豫花9327组合的籽仁面积、豫花15组合的籽仁宽的轮回亲本平均数在四分位数区间内, 其他杂交组合的轮回亲本平均数都在四分位数区间外。但综合BC4F4各性状分布的中位数以及四分位数分布, 81.67%的性状都更接近于轮回亲本, 表明连续回交对恢复轮回亲本的遗传背景具有明显效果。
2.4 豫花15和远杂9102的BC4高油酸优良株系遗传背景鉴定
于2017年田间分别种植2016年秋季收获的BC4F3和2017年春季海南加代收获的BC4F4株行, 2017年秋季分别收获BC4F4和BC4F5优良单株。96对KASP引物中分别筛选出13对和15对引物在豫花15和开农176之间、远杂9102和DF12 之间具有多态性(附表1)。豫花15和远杂9102的BC4后代的每个株行分别选择1~3个单株的叶片提取DNA, 并利用多态性引物进行遗传背景鉴定。检测结果表明, 2个组合的BC4轮回亲本遗传背景分别为66.67%~92.31%和83.33%~87.50%, BC4F4的杂合率和非轮回亲本遗传背景高于BC4F5 (表2)。结合分子标记对轮回亲本遗传背景的检测, 并综合选择主要农艺性状, 4个轮回选择组合最终决选出24个基因型高纯合单株(表3), 作为对应轮回亲本的高油酸改良系的备选品系, 进一步扩繁参加多点试验。Table 2
表2
表2KASP检测BC4F5株行遗传背景
Table 2
| 遗传背景 Genetic background | YH15 BC4F5 lines | YH15 BC4F4 lines | YZ9102 BC4F5 lines | YZ9102 BC4F4 lines | |||||
|---|---|---|---|---|---|---|---|---|---|
| Z172000 | Z172003 | Z172025 | Z172250 | Z172290 | Z172081 | Z172086 | Z172291 | Z172299 | |
| 轮回亲本RP (%) | 79.49 | 84.62 | 92.31 | 66.67 | 77.78 | 87.50 | 87.50 | 83.33 | 83.33 |
| 杂合率Heterozygous (%) | 7.69 | 7.69 | 7.69 | 22.22 | 0.00 | 6.25 | 6.25 | 16.67 | 16.67 |
| 非轮回亲本NRP (%) | 12.82 | 7.69 | 0.00 | 11.11 | 22.22 | 6.25 | 6.25 | 0.00 | 0.00 |
新窗口打开|下载CSV
Table 3
表3
表3优良单株与亲本荚果及籽仁性状比较
Table 3
| 基因型 Genotype | 优良单株数(个) No. of superior plants | 油酸含量 Oleic acid (%) | 油亚比 O/L | 荚果类型 Pod type | 荚果网纹 Pod reticulation | 种皮颜色 Testa color |
|---|---|---|---|---|---|---|
| 豫花15 Yuhua 15 | 38.71 | 0.96 | 普通型 Virginia | 粗浅 Thick and slight | 粉红色 Tan | |
| 远杂9102 Yuanza 9102 | 34.50 | 0.81 | 珍珠豆型 Spanish | 细深 Thin and prominent | 粉红色 Tan | |
| 豫花9327 Yuhua 9327 | 39.25 | 0.98 | 普通型 Virginia | 粗浅 Thick and slight | 粉红色 Tan | |
| 豫花9326 Yuhua 9326 | 36.01 | 0.82 | 普通型 Virginia | 粗深 Thick and prominent | 粉红色 Tank | |
| 开农176 Kainong 176 | 70.93 | 5.00 | 普通型 Virginia | 粗浅 Thick and slight | 粉红色 Tan | |
| DF12 | 75.09 | 7.52 | 普通型 Virginia | 细浅 Thin and slight | 粉红色 Tan | |
| 开选016 Kaixuan 016 | 70.03 | 4.96 | 普通型 Virginia | 粗浅 Thick and slight | 粉红色 Tan | |
| 高油酸豫花15 HOAP YH 15* | 8 | 70.26-74.21 | 5.14-10.72 | 普通型 Virginia | 粗浅 Thick and slight | 粉红色 Tan |
| 高油酸远杂9102 HOAP YZ 9102* | 8 | 73.37-79.64 | 8.81-42.82 | 珍珠豆型 Spanish | 细深 Thin and prominent | 粉红色 Tan |
| 高油酸豫花9327 HOAP YH 9327* | 2 | 71.90-76.37 | 6.90-11.77 | 普通型 Virginia | 粗浅 Thick and slight | 粉红色 Tan |
| 高油酸豫花9326 HOAP YH 9326* | 6 | 73.63-82.51 | 6.79-34.96 | 普通型 Virginia | 粗深 Thick and prominent | 粉红色 Tan |
新窗口打开|下载CSV
3 讨论
我国花生种质资源中缺乏油酸含量高于75%的材料[24,25,26,27], 引进国外的高油酸资源生育期长, 蔓生或者半蔓, 荚果偏小, 不宜在我国花生生产上直接利用。截至2017年, 我国育成高油酸花生品种43个, 多数为中小果品种, 品种类型较少, 遗传背景不够丰富, 育种手段主要是诱变和常规育种方法[19]。分子标记辅助回交育种技术具有选择效率高、育种成本低的优势。高油酸FAD2分子标记的开发应用经历了Caps方法[13]、real-time PCR[14]、AS-PCR[15]及直接测序[28]等过程, 但均存在或特异性差, 或成本高, 或费时长, 或通量低等不足, FAD2基因SNP的KASP方法为目前最准确的高通量高油酸分子标记鉴定方法, 使FAD2分子标记成为花生分子育种利用最成功的标记之一[16,17]。
分子标记辅助选择已经在我国花生品种的高油酸改良方面开始应用[28,29,30], 本研究将FAD2的KASP引物设计和PCR条件进行了优化, 高油酸FAD2基因的SNP检出率可达到100%, 研究结果为创新高油酸育种方法、拓展高油酸品种遗传背景等方面提供了实践经验和理论依据。创制的高油酸材料分别具有大果、野生种血缘等性状特点和遗传背景。
目前在缺乏遗传背景的全基因组信息的情况下, 本研究仅进行了高油酸分子标记的前景选择, 在连续回交世代对遗传背景没有进行标记辅助的定向选择, 连续回交世代的遗传背景的组成由回交群体的随机重组率决定。经KASP分子标记检测, 本研究中不同回交组合的BC4F5轮回亲本遗传背景为79.49%~92.31%。由于所采用的多态性KASP分子标记较少, 回交后代群体规模也较小, 该数据只是反映了一定的变化趋势, 即总体上来讲, 多基因控制的数量性状的累积效应在回交高世代差异更明显, 但BC4F5比BC4F4的轮回亲本遗传背景比例较高, 一定程度上也反映了自交后代选择过程中人为选择的影响。少数主效基因控制的性状分布更具有随机性, 如籽仁外观性状等, 因此回交后代的表型变异也较大。未来分子育种的发展方向是多基因控制的数量性状更适合通过全基因组选择进行改良, 少数主效基因控制的性状利用目标基因的分子标记辅助选择更直接有效[31]。保持一定的群体大小使遗传重组更加充分也是背景选择或基因组选择的关键因素。
简便无损快速检测籽仁油酸含量的技术的发展也是提高育种效率的主要因素。近红外方法是简便低成本的快速高油酸检测途径, 可在育种高世代进行大量后代的初步筛选[32]。南繁加代结合分子标记辅助连续回交是快速改良适合不同用途和不同生态区需要的花生品种油酸含量的有效途径, 不同的加工用途、生态适应性、抗病性类型的高油酸新品系正在通过鉴定和进一步评价[28-30,33]。本研究还获得了一系列近等基因系, 经进一步分子鉴定后可用于开展相关性状的遗传学研究。
4 结论
利用FAD2分子标记辅助连续回交技术, 结合近红外检测技术和海南加代, 连续回交4代和自交4代, 获得了4个河南省大面积推广花生品种豫花15、远杂9102、豫花9327和豫花9326的高油酸改良候选新品系24个。分析了新品系的13个农艺性状, 所获得的改良系轮回亲本遗传背景为79.49%~92.31%, 证明该技术体系可实现推广品种的高油酸改良。新品系的育成拓宽了现有高油酸品种的遗传背景, 新创制的近等基因系可用于进一步的遗传学研究。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1007/BF02542581URL [本文引用: 1]

A peanut breeding line with high-oleic acid and an isogenic sister line with normal fatty acid composition were obtained. Oil was extracted with dichloromethane and processed in the laboratory by alkali neutralization and bleaching. Fatty acid compositions were determined by gas chromatography and application of theoretical response factors. Oils were extracted and processed in duplicate. The oxidative stability of the oils was measured by the Schall oven test (80C), active oxygen method (AOM) (112C) and by comparison of oxidation rates on thin-layer chromatography-flame ionization detector (TLC-FID) rods (100C). Fatty acid analysis indicated that the high-oleic line had 75.6 and 4.7% oleic and linoleic acids, respectively, compared to 56.1 and 24.2% for the normal line. The induction times for the Schall test were 682 and 47 h for high- and normal-oleic oils ( P <0.01). The AOM induction times were 69 and 7.3 h for high and normal oils, respectively ( P <0.01). The times to reach 50% loss in triglyceride area on TLC-FID were 847 and 247 min for high-oleic compared to normal-oleic oils ( P <0.01). The results indicate that high-oleic peanut oil has much greater autoxidation stability as compared to normal oil.
DOI:10.1016/j.foodchem.2004.02.020URL [本文引用: 1]

The quality of dry roasted peanuts is highly dependent on storage conditions for preventing oxidation of fatty acids; however, changes in polyphenolics affecting antioxidant capacity due to co-oxidative reactions are unknown. The objective of this work was to evaluate oxidative stability of polyphenolics in peanut kernels with naturally varying rates of lipid oxidation. Three peanut varieties containing varying levels of oleic acid (normal, mid, and high) were dry roasted and evaluated for phytochemical changes over four months of storage at 20 and 35 C; analyses included peroxide value, total and individual phenolics and antioxidant capacity. The normal oleic acid peanuts suffered up to 2.6-fold and mid-oleic acid peanuts 2-fold more lipid oxidation than the high-oleic acid peanuts stored at 35 C. Changes in total soluble phenolics were initially similar among cultivars, but antioxidant capacity was found to decrease by 62%, on average, during storage at 35 C, independently of rates of lipid oxidation. Free p-coumaric acid, three esterified derivatives of p-coumaric, and two esterified derivatives of hydroxybenzoic acid were the predominant polyphenolics present and their rates of change were similar among cultivars and independent of storage time or temperature. The high-oleic acid content was essential for prevention of lipid oxidation, but data indicated that co-oxidative reactions, affecting polyphenolic content during storage, were not great enough to significantly alter antioxidant capacity.
DOI:10.1016/j.foodchem.2004.04.011URL [本文引用: 1]

Peanuts are an important food crop with many health benefits of their consumption realized by consumers worldwide. Limited information is available on non-nutrient phytochemical composition of peanuts and their relative antioxidant values, knowledge that may serve to increase overall marketability of the crop. Shelled peanuts from eight cultivars and four experimental genotypes with either high or normal oleic acid contents were evaluated for phytochemical, antioxidant, and sensory properties (roasted only) before and after dry roasting under constant time and temperature conditions. Peanuts were evaluated for color, total and individual phenolics, and antioxidant capacity while a trained sensory panel evaluated the peanuts for roasted and burnt peanut flavor and aroma, sweetness, and bitterness. Overall, no meaningful differences were observed for phytochemical and antioxidant properties between high and normal oleic acid peanuts, but differences were present among cultivars. However, high oleic acid varieties had higher burnt peanut aroma and burnt peanut flavor compared to normal oleic peanuts but were not necessarily independent from roasted peanut aroma and flavor. Numerous polyphenolics were separated and characterized based on spectral similarities to p-hydroxybenzoic acid, tryptophan, and p-coumaric acid in both free and bound (esterified) forms. Peanuts were found to be a good source of antioxidant polyphenolics, such as p-coumaric acid, that may be contributing factors to potential health benefits of their consumption.
DOI:10.1186/1476-511X-8-25URLPMID:19558671 [本文引用: 1]

Background Chronic inflammation is a key player in pathogenesis. The inflammatory cytokine, tumor necrosis factor-alpha is a well known inflammatory protein, and has been a therapeutic target for the treatment of diseases such as Rheumatoid Arthritis and Crohn's Disease. Obesity is a well known risk factor for developing non-insulin dependent diabetes melitus. Adipose tissue has been shown to produce tumor necrosis factor-alpha, which has the ability to reduce insulin secretion and induce insulin resistance. Based on these observations, we sought to investigate the impact of unsaturated fatty acids such as oleic acid in the presence of TNF-?? in terms of insulin production, the molecular mechanisms involved and the in vivo effect of a diet high in oleic acid on a mouse model of type II diabetes, KKAy. Methods The rat pancreatic beta cell line INS-1 was used as a cell biological model since it exhibits glucose dependent insulin secretion. Insulin production assessment was carried out using enzyme linked immunosorbent assay and cAMP quantification with competitive ELISA. Viability of TNF-?? and oleic acid treated cells was evaluated using flow cytometry. PPAR-?? translocation was assessed using a PPRE based ELISA system. In vivo studies were carried out on adult male KKAy mice and glucose levels were measured with a glucometer. Results Oleic acid and peanut oil high in oleic acid were able to enhance insulin production in INS-1. TNF-?? inhibited insulin production but pre-treatment with oleic acid reversed this inhibitory effect. The viability status of INS-1 cells treated with TNF-?? and oleic acid was not affected. Translocation of the peroxisome proliferator- activated receptor transcription factor to the nucleus was elevated in oleic acid treated cells. Finally, type II diabetic mice that were administered a high oleic acid diet derived from peanut oil, had decreased glucose levels compared to animals administered a high fat diet with no oleic acid. Conclusion Oleic acid was found to be effective in reversing the inhibitory effect in insulin production of the inflammatory cytokine TNF-??. This finding is consistent with the reported therapeutic characteristics of other monounsaturated and polyunsaturated fatty acids. Furthermore, a diet high in oleic acid, which can be easily achieved through consumption of peanuts and olive oil, can have a beneficial effect in type II diabetes and ultimately reverse the negative effects of inflammatory cytokines observed in obesity and non insulin dependent diabetes mellitus.
DOI:10.3146/i0095-3679-14-1-3URL [本文引用: 1]

Fatty acid distribution of groundnut (Arachis hypogaea) genotypes (228 in 1984 and 298 in 1985) grown at Gainesville and Marianna (Florida) was determined by gas-liquid chromatography. A wider range in fatty acid composition, especially in oleic and linoleic acids, was found among these genotypes than reported previously for the species. Two closely related experimental lines (435-2--1 and 435-2--2) had 80% oleic and 2% linoleic acid, with iodine values of 74. For the Florida breeding lines, iodine values of the oil ranged from 74 to 107 and the oleic/linoleic ratios from 0.9 to 35.1, compared to 95 and slightly less than 2, respectively, for Florunner. The oleic acid content of different experimental lines ranged from 37% to 80% and the linoleic acid content from 2% to 43%. It is suggested that the magnitude of this variability should permit the development of cultivars with a range of oil composition for various nutritional and industrial purposes. All the oil quality factors studied were highly significantly affected by genotype, and all but 3 of the factors were significantly affected by season.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s004380000243URLPMID:10905348 [本文引用: 1]

A peanut variety with high oleate content has previously been described. When this high oleate variety was used in breeding crosses, the F 2 segregation ratio of high oleate to normal oleate progeny was 3:1 or 15:1 depending on the normal oleate varieties used in the crosses. These data suggested that the high oleate trait is controlled by two recessive genes, and some peanut varieties differ from the high oleate variety by mutations in one gene, while others differ by mutations in two genes. The objective of this study was to understand the molecular nature of the high oleate trait and the various segregation patterns. In the previous paper in this issue, we reported that the level of transcripts expressed by one ( ahFAD2B ) of two homoeologous genes for oleoyl-PC desaturases in cultivated peanut is significantly reduced in high oleate varieties. In this report, we examined gene expression by RT-PCR/restriction digestion in a cross that shows a one-gene segregation pattern for the high oleate trait. Our data showed that the severely reduced level of ahFAD2B transcript correlates absolutely with the high oleate phenotype in this cross, suggesting that the single gene difference is correlated with the ahFAD2B transcript level. When we tested the enzyme activity of the proteins encoded by ahFAD2A and ahFAD2B by expression of the cloned sequences in yeast, only the ahFAD2B gene product showed significant oleoyl-PC desaturase activity. These data, combined with the observation that ahFAD2A shows a change (D150N) in a residue that is absolutely conserved among other desaturases, raised the possibility that the ahFAD2A in these normal and high oleate lines is a mutant allele. In support of this hypothesis, we found that another ahFAD2A allele in a normal oleate peanut line does not have the D150N change. This peanut line displays a two-gene-segregation pattern for the high oleate trait. In conclusion, our results suggest that a mutation in ahFAD2A and a significant reduction in levels of the ahFAD2B transcript together cause the high oleate phenotype in peanut varieties, and that one expressed gene encoding a functional enzyme appears to be sufficient for the normal oleate phenotype.
[本文引用: 1]
DOI:10.2135/cropsci2001.41151xURL [本文引用: 1]

Increasing the ratio of oleic to linoleic acid (O/L) in peanut [groundnut] (Arachis hypogaea) significantly improves the nutritional and quality attributes of the crop. In currently grown cultivars, the O/L ratio ranges from 0.8 to 2.5. Variation in peanut for O/L ratio has been characterized, and the O/L ratio is digenically inherited at two loci designated as Ol1 and Ol2. Previous research has been conducted with Virginia and runner-type peanut; however, there have been no reports regarding the inheritance and allele frequency at these loci in Spanish-type peanut. The objectives of this study were to determine if the inheritance of the high oleate trait in Spanish-type peanut is similar to that previously reported and to determine the allelic composition of Spanish-type peanut at Ol1 and Ol2. Six different Spanish-type peanut cultivars (low oleate) were hybridized with F435-2 - 2 (high oleate). F2 and BC1F1 progenies were evaluated for the O/L ratio. Segregation patterns indicated that high oleic acid content is digenically inherited in Spanish-type peanut, but there seems to be more allelic variation both within and among these cultivars. In addition, variation within the high and low oleate ratio classes indicated that other factors may be involved in determining the precise O/L ratio.
DOI:10.2135/cropsci2001.412522xURL [本文引用: 1]

Commercially important high oleate seed oils, meaning that they are low in polyunsaturated fatty acid content, are resistant to developing rancidity. A peanut cultivar (Arachis hypogaea L.) derived from a naturally occurring peanut has low oleoyl-PC desaturase activity, the key enzyme in the production of linoleate, normally an abundant polyunsaturated fatty acid. Two genes for this enzyme are expressed in peanut seeds, and when they were separately expressed in yeast (Saccharomyces cerevisiae), one produced less linoleate than the other. Although the two cDNA encode similar sequence proteins, they differ by four amino acids. Aspartate at position 150 was asparagine in the low activity copy. An aspartate was present in many membrane desaturases and other related membrane enzymes at an equivalent location suggesting that the mutation at 150 was key to the low activity. In this work, site-specific mutagenesis was used to change the aspartate in the high activity enzyme to asparagine and to change asparagine in the lower activity enzyme to aspartate. These changes, upon expression in yeast, resulted in nearly complete loss of activity of the previously more active desaturase and restored activity to the previously less active desaturase. This decrease in activity of the one gene, a consequence of mutation from aspartate 150 to asparagine, together with reduction in transcript level of the high activity gene in the mutant variety, suggest that these alterations are the molecular basis of the high oleate phenotype in some commercial varieties of peanut.
DOI:10.3835/plantgenome2011.01.0001URL [本文引用: 2]

Despite important strides in marker technologies, the use of marker-assisted selection has stagnated for the improvement of quantitative traits. Biparental mating designs for the detection of loci affecting these traits (quantitative trait loci [QTL]) impede their application, and the statistical methods used are ill-suited to the traits' polygenic nature. Genomic selection (GS) has been proposed to address these deficiencies. Genomic selection predicts the breeding values of lines in a population by analyzing their phenotypes and high-density marker scores. A key to the success of GS is that it incorporates all marker information in the prediction model, thereby avoiding biased marker effect estimates and capturing more of the variation due to small-effect QTL. In simulations, the correlation between true breeding value and the genomic estimated breeding value has reached levels of 0.85 even for polygenic low heritability traits. This level of accuracy is sufficient to consider selecting for agronomic performance using marker information alone. Such selection would substantially accelerate the breeding cycle, enhancing gains per unit time. It would dramatically change the role of phenotyping, which would then serve to update prediction models and no longer to select lines. While research to date shows the exceptional promise of GS, work remains to be done to validate it empirically and to incorporate it into breeding schemes.
DOI:10.1007/s11032-009-9338-zURL [本文引用: 2]

Oleic acid, a monounsaturated, omega-9 fatty acid found in peanut ( Arachis hypogaea L.) oil is an important seed quality trait because it provides increased shelf life, improved flavor, enhanced fatty acid composition, and has a beneficial effect on human health. Hence, a concentrated effort has been put forth on developing peanut cultivars that have high oleic acid (>74%) and a low amount (<10%) of linoleic acid, a polyunsaturated omega-6 fatty acid. A main bottleneck, however, in breeding research is fast selection of the trait(s) of interest. Therefore, in an effort to expedite breeding efforts, a real-time PCR genotyping assay was developed to rapidly identify the wild type and the mutant allele that are responsible for normal or high levels of oleic acid, respectively in peanut seeds. This test utilizes two TaqMan 庐 probes to detect the presence of an indel (insertion/deletion) in FAD2B and can be employed on DNA extracted from either seeds or leaves. The presence of the insertion (mutant allele) in fad2B causes a frameshift downstream in the coding sequence that ultimately alters the mRNA transcript level, and thus, decreases the activity of microsomal oleoyl-PC desaturase enzyme which converts oleic acid (C18:1) to linoleic acid (C18:2). Validation of the real-time assay was carried out by quantitatively evaluating the fatty acid composition by gas chromatography (GC). Overall, this real-time PCR assay facilitates the identification of progeny carrying the high oleic acid alleles, and thus, allows early elimination of undesirable non-high oleic acid lines in segregating populations.
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
DOI:10.1016/j.ejbt.2016.10.010URL [本文引用: 2]

Cultivated peanut (Arachis hypogaea L.) is a major oilseed crop worldwide. Fatty acid composition of peanut oil may affect the flavor and shelf life of the resulting food products. Oleic acid and linoleic acid are the major fatty acids of peanut oil. The conversion from oleic acid to linoleic acid is controlled by the Δ12 fatty acid desaturase (FAD) encoded by AhFAD2A and AhFAD2B, two homoeologous genes from A and B subgenomes, respectively. One nucleotide substitution (G:C02→02A:T) of AhFAD2A and an “A” insertion of AhFAD2B resulted in high-oleic acid phenotype. Detection of AhFAD2 mutation had been achieved by cleaved amplified polymorphic sequence (CAPS), real-time polymerase chain reaction (qRT-PCR) and allele-specific PCR (AS-PCR). However, a low cost, high throughput and high specific method is still required to detect AhFAD2 genotype of large number of seeds. Kompetitive allele specific PCR (KASP) can detect both alleles in a single reaction. The aim of this work is to develop KASP for detection AhFAD2 genotype of large number of breeding materials.ResultsHere, we developed a KASP method to detect the genotypes of progenies between high oleic acid peanut and common peanut. Validation was carried out by CAPS analysis. The results from KASP assay and CAPS analysis were consistent. The genotype of 18 out of 179 BC4F2 seeds was aabb.ConclusionsDue to high accuracy, time saving, high throughput feature and low cost, KASP is more suitable for determining AhFAD2 genotype than other methods.
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
[本文引用: 3]
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
DOI:10.1186/1471-2229-12-14URLPMID:22280551 [本文引用: 1]

Next generation sequencing (NGS) technologies are providing new ways to accelerate fine-mapping and gene isolation in many species. To date, the majority of these efforts have focused on diploid organisms with readily available whole genome sequence information. In this study, as a proof of concept, we tested the use of NGS for SNP discovery in tetraploid wheat lines differing for the previously cloned grain protein content (GPC) geneGPC-B1. Bulked segregant analysis (BSA) was used to define a subset of putative SNPs within the candidate gene region, which were then used to fine-mapGPC-B1. We used Illumina paired end technology to sequence mRNA (RNAseq) from near isogenic lines differing across a ~30-cM interval including theGPC-B1locus. After discriminating for SNPs between the two homoeologous wheat genomes and additional quality filtering, we identified inter-varietal SNPs in wheat unigenes between the parental lines. The relative frequency of these SNPs was examined by RNAseq in two bulked samples made up of homozygous recombinant lines differing for their GPC phenotype. SNPs that were enriched at least 3-fold in the corresponding pool (6.5% of all SNPs) were further evaluated. Marker assays were designed for a subset of the enriched SNPs and mapped using DNA from individuals of each bulk. Thirty nine new SNP markers, corresponding to 67% of the validated SNPs, mapped across a 12.2-cM interval includingGPC-B1. This translated to 1 SNP marker per 0.31 cM defining theGPC-B1gene to within 13-18 genes in syntenic cereal genomes and to a 0.4 cM interval in wheat. This study exemplifies the use of RNAseq for SNP discovery in polyploid species and supports the use of BSA as an effective way to target SNPs to specific genetic intervals to fine-map genes in unsequenced genomes.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
[本文引用: 3]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.1016/j.plantsci.2015.09.009URLPMID:26566828 [本文引用: 1]

Legume crops such as chickpea, pigeonpea and groundnut, mostly grown in marginal environments, are the major source of nutrition and protein to the human population in Asia and Sub-Saharan Africa. These crops, however, have a low productivity, mainly due to their exposure to several biotic and abiotic stresses in the marginal environments. Until 2005, these crops had limited genomics resources and molecular breeding was very challenging. During the last decade (2005-2015), ICRISAT led demand-driven innovations in genome science and translated the massive genome information in breeding. For instance, large-scale genomic resources including draft genome assemblies, comprehensive genetic and physical maps, thousands of SSR markers, millions of SNPs, several high-throughput as well as low cost marker genotyping platforms have been developed in these crops. After mapping several breeding related traits, several success stories of translational genomics have become available in these legumes. These include development of superior lines with enhanced drought tolerance in chickpea, enhanced and pyramided resistance to Fusarium wilt and Ascochyta blight in chickpea, enhanced resistance to leaf rust in groundnut, improved oil quality in groundnut and utilization of markers for assessing purity of hybrids/parental lines in pigeonpea. Some of these stories together with future prospects have been discussed.
DOI:10.1255/jnirs.559URL [本文引用: 1]

The fatty acid composition of ground nuts (Arachis hypogaea L.), commonly known as peanuts, is an important consideration when a new variety is being released. The composition impacts on nutrition and, importantly, shelf-life of peanut products. To select for suitable breeding material, it was necessary to develop a rapid, non-destructive and cost efficient method. Near infrared spectroscopy was chosen as that methodology. Calibrations were developed for two major fatty acid components, oleic and linoleic acids and two minor components, palmitic and stearic acids, as well as total oil content. Partial least squares models indicated a high level of precision with a squared multiple correlation coefficient greater than 0.90 for each constituent. Standard errors of prediction for oleic, linoleic, palmitic, stearic acids and total oil content were 6.4%, 4.5%, 0.8%, 0.9% and 1.3%, respectively. The results demonstrated that suitable calibrations could be developed to predict the oil composition and content of peanuts for a breeding programme.
DOI:10.1016/j.plantsci.2015.08.013URLPMID:26566838 [本文引用: 1]

High oleate peanuts have two marketable benefits, health benefits to consumers and extended shelf life of peanut products. Two mutant alleles present on linkage group a09 ( ahFAD2A) and b09 ( ahFAD2B) control composition of three major fatty acids, oleic, linoleic and palmitic acids which together determine peanut oil quality. In conventional breeding, selection for fatty acid composition is delayed to advanced generations. However by using DNA markers, breeders can reject large number of plants in early generations and therefore can optimize time and resources. Here, two approaches of molecular breeding namely marker-assisted backcrossing (MABC) and marker-assisted selection (MAS) were employed to transfer two FAD2mutant alleles from SunOleic 95R into the genetic background of ICGV 6110, ICGV 6142 and ICGV 6420. In summary, 82 MABC and 387 MAS derived introgression lines (ILs) were developed using DNA markers with elevated oleic acid varying from 62 to 83%. Oleic acid increased by 0.5-1.1 folds, with concomitant reduction of linoleic acid by 0.4-1.0 folds and palmitic acid by 0.1-0.6 folds among ILs compared to recurrent parents. Finally, high oleate ILs, 27 with high oil (53.0-57.9%), and 28 ILs with low oil content (42.4-49.9%) were selected that may be released for cultivation upon further evaluation.
