 ,*西南大学水稻研究所 / 西南大学农业科学研究院, 重庆 400715
,*西南大学水稻研究所 / 西南大学农业科学研究院, 重庆 400715Physiological and biochemical analysis and gene mapping of a novel short radicle and albino mutant sra1 in rice
ZHANG Li-Sha, MI Sheng-Nan, WANG Ling, WEI Gang, ZHENG Yao-Jie, ZHOU Kai, SHANG Li-Na, ZHU Mei-Dan, WANG Nan ,*Rice Research Institute, Southwest University / Academy of Agricultural Sciences, Southwest University, Chongqing 400715, China
,*Rice Research Institute, Southwest University / Academy of Agricultural Sciences, Southwest University, Chongqing 400715, China通讯作者:
收稿日期:2018-07-29接受日期:2018-12-24网络出版日期:2019-01-07
| 基金资助: |
Received:2018-07-29Accepted:2018-12-24Online:2019-01-07
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (8923KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张莉莎, 米胜南, 王玲, 委刚, 郑尧杰, 周恺, 尚丽娜, 朱美丹, 王楠. 水稻短根白化突变体sra1生理生化分析及基因定位[J]. 作物学报, 2019, 45(4): 556-567. doi:10.3724/SP.J.1006.2019.82041
ZHANG Li-Sha, MI Sheng-Nan, WANG Ling, WEI Gang, ZHENG Yao-Jie, ZHOU Kai, SHANG Li-Na, ZHU Mei-Dan, WANG Nan.
据统计, 水稻90%~95%碳水化合物由叶片光合作用产生, 叶绿体除了参与光合作用外, 还参与一些次生代谢物的合成[1]。通常情况下, 叶色突变体叶绿素含量降低导致光合作用效率下降进而影响作物品质, 但有报道称茶叶由于叶绿素含量降低表现出特殊品质反而优化了茶叶的种质资源[2]。大多数叶色突变体在苗期就表现出肉眼可区分的性状且突变类型多样, 因此可以利用各种叶色突变体研究叶绿体发育分化机理、叶绿素生物合成和降解途径、光合作用机制以及叶绿体结构和功能基因组学[3,4]。
高等植物叶绿体由不含片层结构的前质体, 经组织分化、光诱导及叶绿体基因与核基因互作摄取必要蛋白和叶绿素最终发育而成。叶绿体基因分为3类: 第一类主要与光系统I (PSI)和光系统II (PSII)相关, 包括编码Rubisco大亚基(rbcL)、Rubisco小亚基(rbcS)和ATP酶的基因; 第二类主要包括DNA聚合酶基因ssb、RNA聚合酶亚基基因、rRNA基因rps; 第三类包括NADH脱氢酶各亚基基因、转运蛋白基因、固氮酶基因等。调控叶绿体基因表达至少需要两类酶参与即核基因编码的RNA聚合酶(NEP)和叶绿体基因编码的RNA聚合酶(PEP)[5]。NEP主要负责管家基因如rRNA、tRNA的转录; PEP具有多个亚基, 分别由叶绿体基因rpoA、rpoB、rpoC1和rpoC2编码, 主要负责与光合作用相关基因的转录[6]。叶绿体基因表达受阻导致植株叶色变异, 利用T-DNA插入得拟南芥突变体pdm1, 该基因突变造成rpoA转录本缺失, 导致pdm1叶片出现白化表型[7]。但是叶绿体自身编码的蛋白较少, 大多数叶绿体蛋白由核基因编码。水稻CFM3蛋白参与叶绿体基因trnG-UCC、rps16和rpl16转录本的剪切, T-DNA插入得cfm3, 体内trnG-UCC、rps16和rpl16转录本不能被剪切, cfm3幼苗期致死且叶片始终表现白化[8]; 水稻AL1蛋白以剂量形式介导叶绿体发育, 轻微下调AL1表达幼苗能够存活但叶片表现黄化, 而al1叶片白化[9]; T-DNA插入水稻AL2第9个外显子获得al2, 叶片白化, 叶绿体被破坏[10]; 水稻核基因ASL2编码一个多肽, 该多肽是叶绿体核糖体大亚基的一个组分, asl2幼苗白化死亡, 几乎检测不到其叶绿素a、叶绿素b以及类胡萝卜素含量[11]。
叶绿素的生物合成始于谷氨酰-tRNA合成酶催化谷氨酸与tRNA的结合, 从L-谷氨酞-tRNA合成叶绿素a需14步酶促反应, 合成叶绿素b需16步酶促反应[12,13]。整个生物过程非常复杂, 由20多个基因编码15种酶共同参与(附表1)[14,15], 包含谷氨酰-tRNA还原酶(HEMA)、镁离子螯合酶(CHL)和叶绿素a加氧酶1 (CAO1) 3个关键酶[16]。拟南芥编码谷氨酰-tRNA还原酶AtHEMA基因突变后, 植株出现白化表型[17]。镁离子螯合酶由I亚基、D亚基及H亚基组成, 该酶催化镁离子插入原卟啉Ⅸ生成镁原卟啉Ⅸ, 水稻ChlH基因编码镁离子螯合酶H亚基, 该基因突变出现白化致死表型[18], 水稻黄绿叶突变体chl1、chl9分别由编码镁离子螯合酶D亚基和I亚基的基因突变所致[19]。水稻叶绿素a加氧酶1 (CAO1)催化叶绿素a转化为叶绿素b, T-DNA或Tos17插入获得的cao1突变体, 其叶色变淡、株高变矮[20]。此外, 叶绿素生物合成途径中其他基因突变也会产生相应的叶色突变体。植物血红素对叶绿素含量具有反馈抑制作用, 过量的血红素能抑制谷氨酰-tRNA还原酶的活性从而影响叶绿素的合成, 引起叶色变异[21]。类胡萝卜素是一类多异戊间二烯化合物, 主要由胡萝卜素和叶黄素组成的, 其生物合成受损可造成光氧化损伤和脱落酸ABA缺陷, ABA含量降低会出现穗发芽、叶片白化等表型[22]。
Table 1
表1
表1野生型(WT)与突变体sra1光合特性
Table 1
| 材料 Material | 净光合速率 Net photosynthetic rate (μmol CO2 m-2 s-1) | 气孔导度 Stomatal conductance (mol H2O m-2 s-1) | 蒸腾速率 Transpiration rate (mol H2O m-2 s-1) | 胞间CO2浓度 Intercellular CO2 concentration (μmol CO2 L-1) |
|---|---|---|---|---|
| WT | 11.77±0.1091 | 0.30±0.0019 | 3.29±0.0090 | 298.44±2.9901 |
| sra1 | -3.61±0.0680** | 0.18±0.0008** | 1.28±0.0070** | 424.52±5.7801** |
新窗口打开|下载CSV
白化突变体的形成机制不尽相同, 但最终都会造成叶绿素代谢中某个或多个酶失活, 参与合成叶绿素、类胡萝卜素等色素中的酶活性发生变化是目前发现较多的一类引起白化苗产生的原因[23]。我们利用化学诱变剂EMS处理西农1B, 获得一个苗期短根白化突变体sra1, 通过分析sra1光合色素含量、酶活性等生理参数以及遗传分析和基因定位等实验, 为候选基因的确定及进一步功能分析奠定了基础。
Supplementary table 1
附表1
附表1编码被子植物叶绿素合成途径关键酶的基因
Supplementary table 1
| 编号 Code | 酶 Enzyme | 基因 Gene | 水稻 Rice |
|---|---|---|---|
| 1 | 谷氨酰-tRNA合成酶 Glutamyl-tRNAsynthetase | GltX | AK099931 |
| 2 | 谷氨酰-tRNA还原酶 Glutamyl-tRNAreductase | HEMA | AK099393 |
| 3 | 谷氨酸-1-半醛转氨酶 Glutamate-1-semialdehyde aminotransferase | GSA | AK064826 |
| 4 | 5-氨基乙酰丙酸脱水酶 5-aminolevulinate dehydratase | HEMB | AK101836 |
| 5 | 羟甲基后胆色素原合酶 Hydroxymethylbilane synthase | HEMC | AK060914 |
| 6 | 尿卟啉原III合酶 Uroporphyrinogen III synthase | HEMD | AK107127 |
| 7 | 尿卟啉原III脱羧酶 Uroporphyrinogen III decarboxylase | HEME | AK070859 |
| 8 | 粪卟啉原III氧化酶 Copro-porphyrinogen III oxidase | HEMF | AK070391 |
| 9 | 原卟啉原氧化酶 Protoporphyrinogen IX oxidase | HEMG | AK108365 |
| 10 | 镁螯合酶D亚基 Mg chelatase D subunit | CHLD | AK072463 |
| 镁螯合酶H亚基 Mg chelatase H subunit | CHLH | AK067323 | |
| 镁螯合酶I亚基 Mg chelatase I subunit | CHLI | AK060389 | |
| 11 | 镁原卟啉IX甲基转移酶 Mg-protoporphyrin IX methyltransferase | CHLM | AK059151 |
| 12 | 镁原卟啉 IX单甲酯环化酶 Mg-protoporphyrinogen IX monomethylester cyclase | CHL27 | AK069333 |
| 13 | 二乙烯还原酶 3,8-divinyl reductase protochlorophyll a-8-vinyl reductase | DVR | AK103940 |
| 14 | 原叶绿素酸酯氧化还原酶 Protochlorophyllide oxidoreductase | POR | AK068143 |
| 15 | 叶绿素合酶 Chlorophyll synthase | CHLG | AK068855 |
| 16 | 叶绿素a加氧酶1 Chlorophyllide a oxygenase | CAO1 | AF284781/AK063367 |
新窗口打开|下载CSV
1 材料与方法
1.1 试验材料
EMS处理籼稻西农1B, 通过多代自交分离筛选出稳定的短根白化突变体sra1。由于sra1三叶期致死, 从自交分离群体的正常株收种用于保存突变体, 并于次年分单株种植。田间材料按常规管理, 均种植于重庆北碚歇马基地。1.2 表型观察及根系长度测定
从浸种出芽开始到突变体第三叶期死亡结束, 田间观察野生型及突变体的叶色变化, 并在播种后第3、10、20 d随机选取50株野生型单株和突变体单株测量根系长度, 取平均值。1.3 叶绿体形态与结构观察
取三叶期长势一致的野生型和突变体叶片, 将相同位置的叶片切成2 mm × 2 mm的小片, 根据洪建等人提出的方法包埋、切片及染色并观察叶绿体的形态与结构[24]。1.4 光合色素含量的测定
取20株长势相对一致的野生型和突变体, 分别称取0.2 g叶片剪碎, 加入20 mL丙酮∶无水乙醇( 1∶1, v/v)混合液, 黑暗封口处理48 h, 期间摇动使其充分混匀。使用分光光度计测定波长在470 nm、645 nm、663 nm的光吸收值, 参考Wellburn方法计算光合色素含量[25], 对每个样品重复3次, 取平均值。1.5 光合特定参数和叶绿素荧光参数的测定
取长势良好的野生型和突变植株, 测定叶片面积, 利用LI-6400型便携式光合测定仪测定光合特性参数。材料经暗适应20 min后弱光照射测定初始荧光Fo, 用一定强度的饱和脉冲光照射0.8 s测定暗适应最大荧光Fm; 材料光适应30 min后打开光稳态荧光, 用一定强度的饱和脉冲光照射0.8 s测定光适应下最大荧光Fm′, 关闭饱和脉冲光打开远红光测定光适应最小荧光Fo′[26], 对每个样品重复测定3次, 取平均值。1.6 酶活性测定
剪取等量的野生型和突变体叶片, 参照南京建成生物工程研究所研发的试剂盒分别采取比色法、羟胺法、可见光法测定抗氧化过氧化物酶、总超氧化物歧化酶和过氧化氢酶活性, 采取TBA法测定氧化降解产物丙二醛(MDA)含量。对每个样品重复测定3次, 取平均值。1.7 SRA1遗传分析和基因定位
单株种植用于保存突变体而收获的自交分离群体的种子, 随机选取10个正常单株收种, 次年种植于北碚基地, 田间统计后代发生突变分离的株系中正常株与突变株的总数, 进行卡方验证完成遗传分析。单株种植用于保存突变体而收获的自交分离群体的种子, 随机选取10个正常单株与籼稻西大1S (不育系)杂交得F1代种子, 次年单株种植F1种子获得各单株的F2代种子, 播种并筛选出短根白化表型分离的群体作为连锁分析群体。剪取F2等量的正常单株和突变单株的叶片, 采用CTAB法提取DNA分别构建正常基因池和突变基因池, 同时采用CTAB法提取亲本和F2代分离群体的DNA。用均匀分布于水稻12条染色体上的350对SSR引物和100对InDel引物, 以亲本西大1S和突变亲本的DNA为模板进行PCR扩增。通过10%非变性聚丙烯酰胺凝胶电泳和快速银染法筛选出两个亲本间多态性分子标记, 再利用该标记分析正常基因池与突变基因池之间的多态性, 对可能与目标基因连锁的分子标记进行F2隐性单株验证。在确定该分子标记连锁后, 再在连锁标记附近寻找多态性分子标记用于基因初步定位, 待连锁区间确定后, 进一步开发新的多态性分子标记, 用于基因精细定位。用于基因定位的部分多态性引物(附表2)由上海英骏技术公司合成。
Supplementary table 2
附表2
附表2用于基因定位的部分多态性引物序列
Supplementary table 2
| 标记 Marker | 正向引物序列 Forward primer sequence (5′-3′) | 反向引物序列 Reverse primer sequence (5′-3′) |
|---|---|---|
| Z-12 | ACGCCGTCTCCTTGGTAATC | CTTACATTGGAAAGCGGAGG |
| Z-20 | TTGTAGTGCGGGCAGTTCTC | GTACTGTTTGCCTCTGCTTGC |
| Z-42 | CCTTAATCTTGTCGCACAAC | GCTTCTCTCCATCAACCATAT |
| Z-78 | ACGCAGGAGAAGCTAGATAGG | AACCATCTTTTAGTGTCGGGT |
| Z-83 | TACCCTTGGAGCACATATAGTTAA | TTGGTCACCAACATATCTTGAA |
| Z-112 | GGCTCTGCTAGCGAAATACTAG | GCTCACCATACTCACCAATTACT |
| ZMD5-18 | TCAGGTCTTACGACGGTATGG | GGACACACTAGAATCTACGCACG |
| ZMD5-44 | GAAGCTATTAGCCGGGATCG | GCCAAGGCAAAGCTCTCTT |
| ZTQ43 | TGCAGAGACAAGGAAGCGG | TCCTGATCGTTGAGCAGGC |
| RM8208 | TGTAAATGCCTGAGTGCCTACCC | AGCTAAACCGCTAGGGCCTTCC |
| RM3400 | GGGTGCACCTTTGTATCTGTGC | TGACAGAGGTAAAGCAGCAGTAGTCG |
| RM15177 | TCCTGTGTTGGACGGAGTATGC | GCCTCAGAGGTTAGAAGACAGACAGC |
| RM405 | TATGCTTTCTGTCAGCTTCC | CTGCTGTGAAAGAGTTGACG |
| RM18053 | GAGACCAGAGGGAGACAAAGAGAGG | CTTAGGTCTCCCGACAGTCACG |
新窗口打开|下载CSV
1.8 候选基因的预测
根据精细定位的结果, 利用基因预测网站(1.9 RNA的提取与cDNA的合成
用天根RNA prep pure Plant试剂盒提取野生型和突变体同一时期叶片的RNA, 用Invitrogen公司RNA反转录试剂盒反转录合成cDNA。1.10 定量RT-PCR表达分析
以Actin为内参, 使用SYBR Green染料实时荧光定量PCR分析叶绿体发育(叶绿体早期发育部分核基因FtsZ、OsPOLP、OsRpoTp、Rpob、V2、V3)、叶绿素合成(光合色素合成部分基因HEMA1、HEMD、CHLD、CHLM、CAO1、PSY1、PSY2)、光合作用(光合作用部分基因RbcL、RbcS、Cab1R、Cab2R、PsaA、PsbA、PetA、PetB) 3个途径中几个基因的表达量(附表3)。用于定量分析的引物序列(附表4)由上海英骏技术公司合成。25 μL反应体系含cDNA模板2 μL、10 μmol L-1上下游引物各2 μL、SYBR Green荧光染料12.5 μL、RNase-out H2O 8.5 μL。每个样品重复3次, 在Bio-Rad荧光定量PCR仪上进行PCR, 利用CFX-Manager软件收集和处理数据, 采用2-ΔΔCt的方法[27]计算目的基因的相对表达量。Supplementary table 3
附表3
附表3定量PCR的相关基因描述
Supplementary table 3
| 基因 Gene | 基因产物 Product | RAP位点 RAP_locus |
|---|---|---|
| OsPOLP | PolI-like DNA polymerase, plastidal DNA polymerase 1 | Os08g0175300 |
| FtsZ | Plastid division protein FtsZ1 | Os04g0665400 |
| OsRPOTP | RNA polymerase | Os06g0652000 |
| RpoB | RNA polymerase beta subunit | CAA33986 |
| V2 | Virscent 2 | Os03g0320900 |
| V3 | Virscent 3 | Os06g0168600 |
| HEMA1 | Glutamyl-tRNA reductase 1 | Os10g0502400 |
| HEMD | Uroporphyrinogen-III synthase | Os03g0186100 |
| CHLD | Magnesium chelatase subunit | Os03g0811100 |
| CHLM | Mg-protoporphyrin IX methyltransferase | Os06g0132400 |
| CAO1 | Chlorophyll a oxygenase 1 | Os10g0567400 |
| PSY1 | Phytoene Synthase 1 | Os06g0729000 |
| PSY2 | Phytoene synthase 2 | Os12g0626400 |
| carb1R | Chlorophyll A/B binding protein 1R | Os09g0346500 |
| carb2R | Chlorophyll A/B binding protein 2R | Os01g0600900 |
| petA | petA, cytochrome f | CAA33961 |
| petB | petB, cytochrome B6 | CAA33977 |
| psaA | psaA, PSI P700 apoprotein A1 | CAA33996 |
| psbA | psbA, PSII 32 kDa protein | CAA34007 |
| RbcL | RbcL, Ribulose bisphosphate carboxylase large chain | CAA34004 |
| RbcS | Rubisco small subunit | Os12g0274700 |
新窗口打开|下载CSV
Supplementary table 4
附表4
附表4用于定量的引物序列
Supplementary table 4
| 基因 Gene | 正向引物 Forward sequence (5′-3′) | 反向引物 Reverse sequence (5′-3′) |
|---|---|---|
| Actin | GACCCAGATCATGTTTGAGACCT | CAGTGTGGCTGACACCATCAC |
| OsPOLP | ACCGGTGCTTTCAGGCTTG | GCTGACTGATAATCACACG |
| FtsZ | AAAGGACATAACCTTGCAAG | AGTTTTCCTATTGAACCGTG |
| OsRPOTP | TCCTCATGTCGAGCAAGGAT | GAAAGAATGTCTGGACTTTG |
| RpoB | TATGGTCTAATTCCGAGCGGT | TATGGTCTAATTCCGAGCGG |
| V2 | GAGGAGTTCCTCACGATGAT | AGCATCAATGATAGACTCC |
| V3 | GTTAGATGCTTCACTACACAG | GTACCATTGCCAACATGGCAAC |
| HEMA1 | CGCTATTTCTGATGCTATGGGT | TCTTGGGTGATGATTGTTTGG |
| HEMD | AGGGATGGAAGGCTGCTGGA | CAGTGGTCCTTGGAAGCTCTG |
| CHLD | GCTTGCAGAAAGCTACACAAGC | AGGCCGTGAGCTAAAGGAGA |
| CHLM | CCATCCATTGGTCTCCTTATGACA | GTAGCCTACTTACCATCAATGAGTC |
| CAO1 | GACACCTTCATCTGGGCTTCAA | CGAGAGACATCCGGTAGAGC |
| PSY1 | GTGACCGAGCTCAGCCAG | TTAATCGGAAGGCAGATGCT |
| PSY2 | GGGCGTTGCGCATCTAGA | CAAGAAATCGTGCGAAATCA |
| carb1R | AGATGGGTTTAGTGCGACGAG | TTTGGGATCGAGGGAGTATTT |
| carb2R | TGTTCTCCATGTTCGGCTTCT | GCTACGGTCCCCACTTCACT |
| petA | TGAATGTGGGTGCTGTTCTTATTT | TCGGGCGGCGCTAAT |
| petB | TCCTCGGTTCAATACATAATGACCG | CGTGCAGGATCATCATTAGAACCA |
| psaA | GCGAGCAAATAAAACACCTTTC | GTACCAGCTTAACGTGGGGAG |
| psbA | CCCTCATTAGCAGATTCGTTTT | ATGATTGTATTCCAGGCAGAGC |
| RbcL | CTTGGCAGCATTCCGAGTAA | ACAACGGGCTCGATGTGATA |
| RbcS | TCCGCTGAGTTTTGGCTATTT | GGACTTGAGCCCTGGAAGG |
新窗口打开|下载CSV
2 结果与分析
2.1 突变体和野生型表观性状比较
与野生型相比, 突变体从出芽(图1-A~C)到第三叶期(图1-D~E)叶片始终为白色, 且胚根较同时期野生型胚根明显变短: 发芽后第3、第10、第20天突变体胚根长度分别下降了39.53%、37.75%和42.78% (图2-H); 不定根数目与长度、胚根中部侧根的平均长度和密度较野生型无明显差异(图2-I~J)。由于白化严重, 突变体三叶期后出现完全死亡现象, 暂时将该突变体命名为短根白化突变体sra1 (short radicle and albino 1)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1野生型(WT)和sra1表型特征
A~C: 萌发3 d野生型和sra1植株; D: 萌发10 d野生型和sra1植株; E: 萌发20 d野生型和sra1植株; 标尺: ABC中标尺为1 cm, DE中标尺为2 cm。
Fig. 1Plant morphology of the wide type (WT) and the sra1 mutant
A-C: 3-day seeding of the wild type and the sra1; D: 10-day seeding of the wild type and the sra1; E: 20-day seeding of the wild type and the sra1; Scale bars: 1 cm in A, B, and C, 2 cm in D and E.
图2
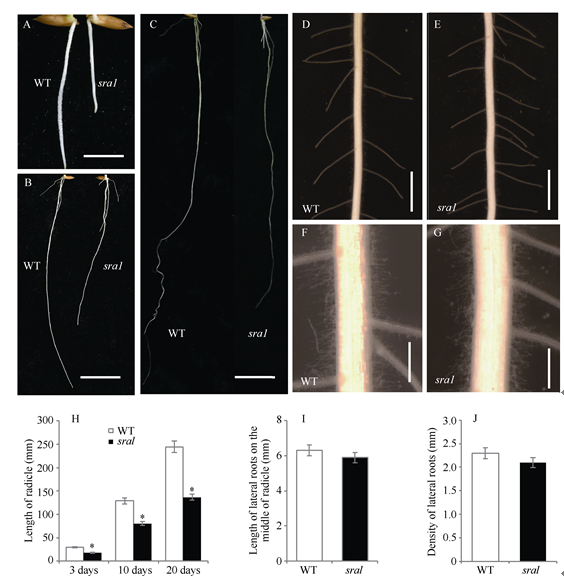 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2野生型(WT)和sra1突变体根系长度变化
A: 萌发3 d野生型和sra1植株根系; B: 萌发10 d野生型和sra1植株根系; C: 萌发20 d野生型和sra1植株根系; D~G: 萌发10 d野生型和sra1胚根中部; 标尺: 标尺在ABC中为1 cm, DE中为0.5 cm, FG中为0.1 cm。H: 野生型和sra1植株胚根长度; I: 野生型和sra1植株胚根中部侧根长度; J: 野生型和sra1植株侧根密度。*显著差异(P < 0.005)。
Fig. 2Length of roots of the wild type (WT) and the sra1 mutant
A: 3-day seeding roots of the wild type and the sra1 mutant; B: 10-day seeding roots of the wild type and the sra1 mutant; C: 20-day seeding roots of the wild type and the sra1 mutant; D-G: the middle of 10-day seeding roots of the wild type and the sra1 mutant; Scale bars: 1 cm in A, B, and C, 0.5 cm in D and E, 0.1 cm in F and G. H: length of radicle of the wild type and the sra1 mutant; I: length of lateral roots on the middle of radicle of the wild type and the sra1 mutant; J: density of lateral roots of the wild type and the sra1 mutant. *Significantly different at P < 0.05 by t-test.
2.2 sra1叶绿体形态和结构分析
利用透射电子显微镜对野生型和sra1三叶期相同位置的叶片观察发现(图3), 野生型叶肉细胞液泡体积一定, 叶绿体数量丰富且膜系统发育完整, 其基粒类囊体清晰且排列较为整齐; 而sra1植株叶肉细胞数目增多, 细胞体积变小, 液泡体积明显增大, 叶绿体明显变少或没有, 内部的基粒类囊体膜松散, 类囊体发育不完整, 只能观察到类似于类囊体的间质类囊体。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3野生型(WT)与sra1叶绿体透射电镜观察
1: 叶绿体; 2: 液泡; 3: 基粒; 4: 类囊体; 5: 间质类囊体。A~C: 野生型植株叶绿体透射电镜分析; D~F: sra1植株叶绿体透射电镜分析; 标尺: A, D中标尺为2 μm; B, E中标尺为500 nm; C, F中标尺为200 nm。
Fig. 3Chloroplast transmission electron microscope observation in the wild type (WT) and the sra1 mutant
1: chloroplast; 2: cell vacuole; 3: grana lamella; 4: thylakoid; 5: stroma thylakoids-like. A-C: transmission electron microscopy observation of chloroplasts of wild type plants; D-F: transmission electron microscopy observation of chloroplasts of sra1 plants; Scale: 2 μm in A and D, 500 nm in B and E, 200 nm in C and F.
2.3 sra1光合色素含量分析
图4表明相比野生型, sra1叶绿素a、叶绿素b和类胡萝卜素含量接近于零, 这可能与其叶肉细胞内叶绿体急剧减少, 类囊体膜遭到破坏有关。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4野生型(WT)和sra1突变体光合色素含量
Fig. 4Photosynthetic pigments contents of the wild type and the sra1 mutant
2.4 sra1光合特性参数和叶绿素荧光参数分析
sra1净光合速率为负值, 气孔导度和蒸腾速率较野生型分别下降40.00%和61.09%, 胞间CO2浓度极显著升高(表1); 与野生型相比, 在暗适应和光适应条件下几乎检测不到sra1的初始荧光和最大荧光(图5), 这与sra1叶绿素a、叶绿素b和类胡萝卜素的含量接近于零的结果相一致。图5
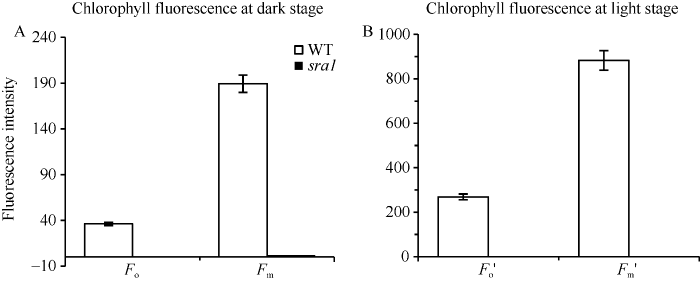 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5野生型(WT)与突变体sra1叶绿素荧光参数
A: 暗适应条件下野生型和sra1最大荧光量Fm和最小荧光量Fo; B: 光适应条件下野生型和sra1最大荧光量Fm′和最小荧光量Fo′。
Fig. 5Chlorophyll fluorescence of the wild type and the sra1 mutant
A: maximum and minimum fluorescence amounts of wild type and sra1 under dark adaptation; B: maximum and minimum fluorescence of wild type and sra1 under light adaptation conditions.
2.5 sra1和野生型酶活性及MDA含量比较
SOD是机体内天然存在的超氧自由基清除因子, 把有害的超氧自由基转化为过氧化氢。机体内的过氧化氢酶(CAT)和过氧化物酶(POD)会立即将其分解为完全无害的水。这3种酶组成一个完整的防氧化链条在机体中发挥着重要作用。MDA是膜脂过氧化最重要的产物之一, 它的产生能加剧膜的损伤, 通过MDA了解膜脂过氧化程度以间接测定膜受损程度。图6所示, 与野生型相比, sra1叶片中SOD和POD活性分别升高397.89%和157.67%, CAT活性下降55.70%, MDA含量升高223.91%。说明sra1叶肉细胞的叶绿体膜和类囊体膜受到极大程度的损伤。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6野生型(WT)和sra1中SOD、POD、CAT活性和MDA含量** 在0.001水平上显著差异。**Significantly different at P < 0.001.
Fig. 6Activities of SOD, POD, CAT, and MDA content in sra1 and wild type
2.6 SRA1遗传分析
随机选取的10个正常单株收种, 田间统计后代发生突变分离的株系中正常株与突变株的分离, 发现3706株中, 正常单株2818株、短根白化单株888株, 经卡平方测验, 符合隐性基因3∶1的分离比例(χ2 = 2.03 < χ20.05 = 3.84), 表明sra1的短根白化性状受1对隐性核基因控制。2.7 SRA1基因定位
选取在亲本西大1S和突变亲本间出现多态性的引物扩增正常基因池和突变基因池, 第3染色体SSR标记RM15177、RM3400和RM8208表现多态性。随机选取32株F2代短根白化苗验证连锁, 并扩大群体至208株缩小定位区间, 统计RM15177、RM3400和RM8208的重组子个数分别为13、12和6个, 遗传距离分别为4.2、2.9和1.5 cM, 其中RM15177的重组子包含RM3400的重组子, 且RM3400和RM8208的重组子不同。因此初步把SRA1定位在SSR标记RM3400和RM8208之间。初步定位区间内设计的60对InDel引物和30对SSR引物, 有6对在亲本间表现出多态性, 利用这6对多态性引物精细定位888株F2分离群体。最终将SRA1定位于第3染色体长臂InDel标记Z-20和Z-42之间, 遗传距离分别为0.05 cM和0.11 cM, 物理距离约为657 kb, InDel标记Z-78和Z-83与SRA1基因发生共分离(图7)。图7
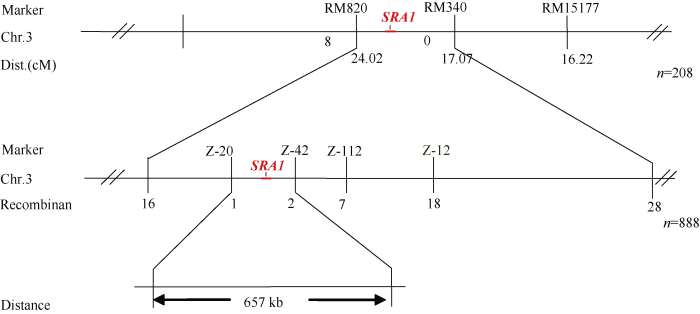 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7SRA1基因在第3染色体上的基因定位图谱
图中的粗黑线代表染色体, 线上方标记的是基因定位使用的SSR标记, 线下方数值代表两标记间的遗传距离, n为F2定位群体总株数。
Fig. 7Gene mapping of SRA1 on rice chromosome 3
The thick black line in the figure represents the chromosome, the SSR markers used for gene mapping are marked above the line, the value below the line represents the genetic distance between the two markers, and n is the total number of F2 localized populations.
2.8 候选基因的预测
利用基因预测网站(http://rice.plantbiology.msu. edu/cgi-bin/gbrowse/rice/#search?tdsourcetag=spcqqaiomsg)对基因预测显示, 目标区域共有101个基因, 包含62个有功能的候选基因。其中大多数属于反转座子基因和转座子基因, 还有一些基因表达酶蛋白或结构域蛋白如肌醇-1-单磷酸酶、谷胱甘肽S-转移酶、锌指结构域、RNA聚合酶结构域、蛋白激酶结构域。只有极少数基因如细胞色素P450、叶绿素a/b结合蛋白与光合作用相关。2.9 相关基因的表达分析
SRA1基因突变可能影响到叶绿体发育、叶绿素合成、光合作用途径中某些基因的表达, sra1叶绿体早期发育部分基因如核基因FtsZ (编码一个类似于维管蛋白结构的原核细胞骨架GTP酶)、OsPOLP (编码一个与原核DNA酶同源的DNA聚合酶)、OsRpoTp (编码NEP)、Rpoβ (RNA聚合酶β亚基)、V2 (鸟苷酸激酶2)、V3 (鸟苷酸激酶3)表达上调(图8-A); 光合色素合成的部分基因如HEMA1 (谷氨酰-tRNA还原酶1)、HEMD (尿卟啉原-III合成酶)、CHLD (镁螯合酶D亚基)、CHLM (镁螯合酶M亚基)、CAO1 (叶绿素a加氧酶1)、PSY1 (八氢番茄红素合成酶1)、PSY2 (八氢番茄红素合成酶2)表达下调(图8-B); 光合作用的部分基因如RbcL (核酮糖二磷酸羧化酶大亚基)、RbcS (核酮糖二磷酸羧化酶小亚基)、Cab1R和Cab2R (PSII光捕获叶绿素a/b结合蛋白1R和2R)、PsaA (光系统I叶绿素脱辅基蛋白)、PsbA (光系统II醌结合蛋白)、PetA (细胞色素f亚基)、PetB (细胞色素b6亚基)在sra1中几乎检测不到(图8-C)。光合作用的几个基因在sra1中几乎不表达, 很有可能是由于sra1光合色素合成基因表达下调影响了光合色素的合成, 在光合色素缺乏的情况下无法转录光合作用相关的基因。叶绿体早期发育部分基因FtsZ、OsPOLP、OsRpoTp、Rpoβ、V2、V3在sra1上调表达, 说明sra1叶绿体早期发育是正常的(透射电子显微镜观察到sra1有极少数量的叶绿体也证实了这一点), 但由于取材时叶绿体已经处于完全形成期, sra1中观察不到完整的叶绿体。我们推测SRA1基因突变可能不影响叶绿体早期发育相关基因, 但可能影响其他叶绿体发育基因的表达, 造成sra1突变体叶绿体发育不完整, 无法进行光合作用, 田间存活1个月后耗尽体内营养最终凋亡。
图8
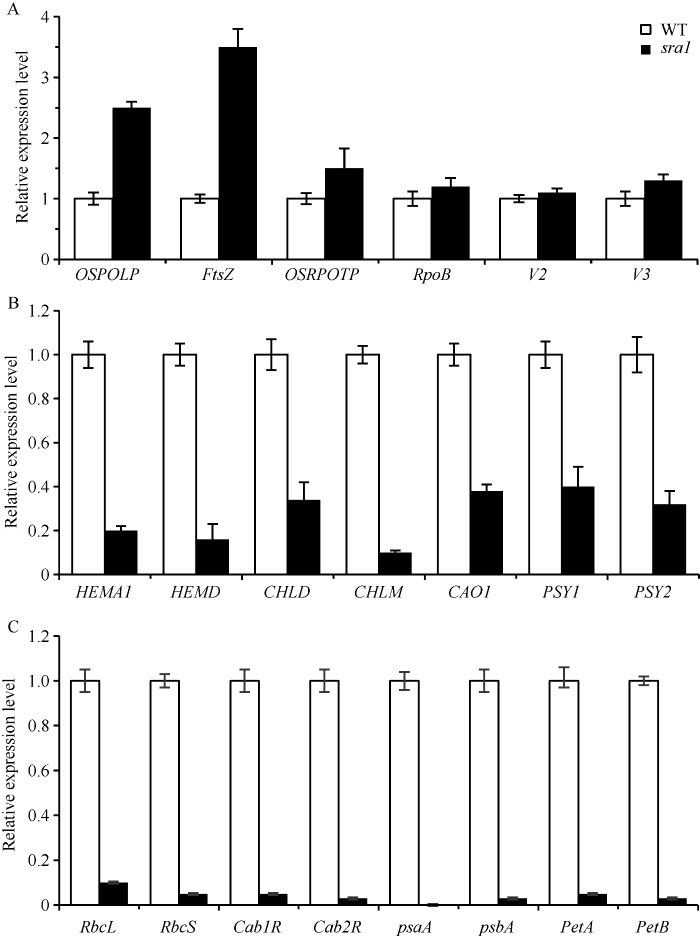 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8野生型(WT)和sra1突变体相关基因的表达
A: 野生型和sra1植株叶绿体早期发育部分基因OsPOLP、FtsZ、OsRpoTp、Rpob、V2、V3表达量; B: 野生型和sra1植株光合色素合成部分基因HEMA1、HEMD、CHLD、CHLM、CAO1、PSY1、PSY2 表达量; C: 野生型和sra1植株光合作用部分基因RbcL、RbcS、Cab1R、Cab2R、PsaA、PsbA、PetA、PetB表达量。
Fig. 8Expression levels of genes associated in the wild type (WT) and sra1 mutant
A: expression levels of OsPOLP, FtsZ, OsRpoTp, Rpob, V2, V3 in the wild type and sra1; B: expression levels of HEMA1, HEMD, CHLD, CHLM, CAO1, PSY1, PSY2 in the wild type and sra1; C: expression levels of RbcL, RbcS, Cab1R, Cab2R, PsaA, PsbA, PetA, PetB in the wild type and sra1.
3 讨论
近年来, 叶色突变体具有一定研究价值而备受关注, 国内外己报道80多个水稻苗期叶色突变, 在12条染色体上均有分布, 但是仅有少数被具体定位和克隆出来[28]。白化突变体alb23叶绿体体积变小,具有双层膜结构但基质中没有片层结构, 只有淀粉粒、脂肪粒以及一些类似于前质体的结构, 该基因定位到第2染色体分子标记R2M501和R2M502之间约280 kb范围内[29]。白化致死突变体abl4叶片从发芽至三叶期一直表现为白化, 三叶期后逐渐死亡,透射电镜观察abl4叶绿体结构遭到破坏, 导致abl4无法进行光合作用而死亡, 图位克隆最后将ABL4定位于第4染色体201 kb范围内[30]。白化致死突变体asl1叶片幼苗期表现为白化, 四叶期后死亡, 结合SSR和InDel分子标记将ASL1基因定位在第1染色体长臂分子标记P4和P6之间, 最后证实该基因编码质体核糖体小亚基S20蛋白[31]。
我们通过EMS处理西农1B得到sra1, 叶片从出芽到第三叶期始终表现为白化且胚根长度明显变短, 这在已报道过的水稻白化苗中是少有的现象, 并且在已报道的短根表型中也很少出现白化表型。水稻突变体cyt-inv1在正常培养条件下表现短根、开花延迟和部分不育, 切片分析发现cyt-inv1根部细胞纵向变短, 伸长区细胞皱缩。突变体cyt-inv1蔗糖含量增加而己糖含量降低, 外施葡萄糖能恢复其根部的生长[32]。SRT5编码中性/碱性转化酶催化蔗糖水解成葡萄糖和果糖, 是水稻根发育中提供能量的重要酶类, srt5短根表型受一个隐性基因控制, 一个核苷酸替换导致其编码的蛋白从Gly变成Arg[33]。
我们推测sra1叶绿体早期发育正常, 后期由于SRA1突变影响到光合色素的合成, 植株无法进行有效的光合作用待体内营养物质耗尽后死亡。sra1胚根明显变短可能是根部营养向叶部运输使得根部营养缺乏, 胚根无法正常生长。前人报道的白化致死突变体最终均只与叶绿体缺陷有关, 并未出现短根表型, 而我们获得sra1突变体胚根明显变短, 因此推测sra1可能是一个新型的短根白化致死突变体, SRA1基因可能同时参与水稻叶绿体及主根发育, 但是关于突变体sra1真正形成机制还需要进一步分析和研究。
4 结论
sra1 (short radicle and albino 1)自出芽到第三叶期叶片始终表现为白色且胚根变短, 叶绿体极少或没有, 叶绿体膜和类囊体膜受到极大程度的损伤, 叶绿素a、叶绿素b和类胡萝卜素含量几乎为零, 无法进行有效的光合作用。该短根白化性状由单个隐性核基因控制, SRA1基因位于水稻第3染色体长臂InDel标记Z-20和Z-42之间, 物理距离约657 kb, 推测sra1是一个新型的短根白化致死突变体, SRA1基因可能同时参与水稻叶绿体及主根发育。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/S0168-9525(02)00003-3URLPMID:12493248 [本文引用: 1]

Chloroplast research takes significant advantage of genomics and genome sequencing, and a new picture is emerging of how the chloroplast functions and communicates with other cellular compartments. In terms of evolution, it is now known that only a fraction of the many proteins of cyanobacterial origin were rerouted to higher plant plastids. Reverse genetics and novel mutant screens are providing a growing catalogue of chloroplast protein–function relationships, and the characterization of plastid-to-nucleus signalling mutants reveals cell–organelle interactions. Recent advances in transcriptomics and proteomics of the chloroplast make this organelle one of the best understood of all plant cell compartments.
[本文引用: 1]
[本文引用: 1]
DOI:10.1104/pp.113.217604URLPMID:20202020202020202020202020202020202020 [本文引用: 1]

The plastidic caseinolytic protease (Clp) of higher plants is an evolutionarily conserved protein degradation apparatus composed of a proteolytic core complex (the P and R rings) and a set of accessory proteins (ClpT, ClpC, and ClpS). The role and molecular composition of Clps in higher plants has just begun to be unraveled, mostly from studies with the model dicotyledonous plant Arabidopsis (Arabidopsis thaliana). In this work, we isolated a virescent yellow leaf (vyl) mutant in rice (Oryza sativa), which produces chlorotic leaves throughout the entire growth period. The young chlorotic leaves turn green in later developmental stages, accompanied by alterations in chlorophyll accumulation, chloroplast ultrastructure, and the expression of chloroplast development-and photosynthesis-related genes. Positional cloning revealed that the VYL gene encodes a protein homologous to the Arabidopsis ClpP6 subunit and that it is targeted to the chloroplast. VYL expression is constitutive in most tissues examined but most abundant in leaf sections containing chloroplasts in early stages of development. The mutation in vyl causes premature termination of the predicted gene product and loss of the conserved catalytic triad (serine-histidine-aspartate) and the polypeptide-binding site of VYL. Using a tandem affinity purification approach and mass spectrometry analysis, we identified OsClpP4 as a VYL-associated protein in vivo. In addition, yeast two-hybrid assays demonstrated that VYL directly interacts with OsClpP3 and OsClpP4. Furthermore, we found that OsClpP3 directly interacts with OsClpT, that OsClpP4 directly interacts with OsClpP5 and OsClpT, and that both OsClpP4 and OsClpT can homodimerize. Together, our data provide new insights into the function, assembly, and regulation of Clps in higher plants.
[本文引用: 1]
[本文引用: 1]
DOI:10.1111/j.1365-313X.2005.02362.xURLPMID:15807777 [本文引用: 1]

Eubacterial-type multi-subunit plastid RNA polymerase (PEP) is responsible for the principal transcription activity in chloroplasts. PEP is composed of plastid-encoded core subunits and one of multiple nuclear-encoded sigma factors that confer promoter specificity on PEP. Thus, the replacement of sigma factors associated with PEP has been assumed to be a major mechanism for the switching of transcription patterns during chloroplast development. The null mutant ( sig6-1 ) of plastid sigma factor gene AtSIG6 exhibited a cotyledon-specific pale green phenotype. Light-dependent chloroplast development was significantly delayed in the sig6-1 mutant. Genetic complementation of the mutant phenotype by the AtSIG6 cDNA demonstrated that AtSIG6 plays a key role in light-dependent chloroplast development. Northern and array-based global analyses for plastid transcripts revealed that the transcript levels of most PEP-dependent genes were greatly reduced in the sig6-1 mutant, but that the accumulation of nuclear-encoded RNA polymerase (NEP)-dependent transcripts generally increased. As the PEP α subunit and PEP-dependent trnV accumulated at normal levels in the sig6-1 mutant, the AtSIG6 knockout mutant probably retained functional PEP, and the transcriptional defects are likely to have been directly caused by AtSIG6 deficiency. Most of the AtSIG6-dependent genes are preceded by σ 70 -type promoters comprised of conserved 6135/6110 elements. Thus, AtSIG6 may act as a major general sigma factor in chloroplasts during early plant development. On the other hand, the mutant phenotype was restored in older seedlings. Arabidopsis probably contains another late general sigma factor, the promoter specificity of which widely overlaps with that of AtSIG6.
[本文引用: 1]
[本文引用: 1]
DOI:10.1105/tpc.107.055160URLPMID:18065687 [本文引用: 1]

The CRM domain is a recently recognized RNA binding domain found in three group II intron splicing factors in chloroplasts, in a bacterial protein that associates with ribosome precursors, and in a family of uncharacterized proteins in plants. To elucidate the functional repertoire of proteins with CRM domains, we studied CFM2 (for CRM Family Member 2), which harbors four CRM domains. RNA coimmunoprecipitation assays showed that CFM2 in maize (Zea mays) chloroplasts is associated with the group I intron in pre-trnL-UAA and group II introns in the ndhA and ycf3 pre-mRNAs. T-DNA insertions in the Arabidopsis thaliana ortholog condition a defective-seed phenotype (strong allele) or chlorophyll-deficient seedlings with impaired splicing of the trnL group I intron and the ndhA, ycf3-int1, and clpP-int2 group II introns (weak alleles). CFM2 and two previously described CRM proteins are bound simultaneously to the ndhA and ycf3-int1 introns and act in a non-redundant fashion to promote their splicing. With these findings, CRM domain proteins are implicated in the activities of three classes of catalytic RNA: group I introns, group II introns, and 23S rRNA.
.
DOI:10.1104/pp.16.00325URLPMID:27208287 [本文引用: 1]

Abstract Chloroplast, the photosynthetic organelle in plants, plays a crucial role in plant development and growth through manipulating the capacity of photosynthesis. However, the regulatory mechanism of chloroplast development still remains elusive. Here, we characterized a mutant with defective in chloroplasts in rice, termed albino leaf 1 (al1), which exhibits distinct albino phenotype in leaves, eventually leading the al1 seedling to lethal. Electronic microscopy observation demonstrated that the numbers of thylakoids was reduced and the structure of thylakoids was disrupted in the al1 mutant during rice development, which eventually led to the breakdown of chloroplast. Molecular cloning revealed that AL1 encodes the sole Octotricopeptide Repeat Protein (RAP) in rice. Genetic complementation of the Arabidopsis rap mutants indicated that AL1 protein is a functional RAP. Further analysis illustrated that three transcript variants were present in the AL1 gene and the altered splices were occurred at the 3'UTR of AL1 transcript. In addition, our results also indicate that disruption of AL1 gene results in the altered expression of chloroplast associated genes. Consistently, proteomic analysis demonstrated that the abundances of photosynthesis associated proteins are significantly altered, as well as a group of metabolism associated proteins. More specifically, we found that the loss of AL1 resulted in the altered abundances of ribosomal proteins, suggesting that RAP likely also regulates the homeostasis of ribosomal proteins in rice in addition to the rRNA. Taken together, we conclude that the AL1, particularly the AL1a and c isoform, plays an essential role in chloroplast development in rice. 2016 American Society of Plant Biologists. All rights reserved.
DOI:10.1093/jxb/erw296URLPMID:27543605 [本文引用: 1]

TheAlbino Leaf 2(AL2) gene is involved in the splicing of chloroplast group I and II introns and is responsible for chloroplast development in rice. Chloroplasts play an essential role in plant growth and development through manipulating photosynthesis and the production of hormones and metabolites. Although many genes or regulators involved in chloroplast biogenesis and development have been isolated and characterized, identification of novel components is still lacking. We isolated a rice (Oryza sativa) mutant, termedalbino leaf 2(al2), using genetic screening. Phenotypic analysis revealed that theal2mutation caused obvious albino leaves at the early developmental stage, eventually leading toal2seedling death. Electron microscopy investigations indicated that the chloroplast structure was disrupted in theal2mutants at an early developmental stage and subsequently resulted in the breakdown of the entire chloroplast. Molecular cloning illustrated thatAL2encodes a chloroplast group IIA intron splicing facilitator (CRS1) in rice, which was confirmed by a genetic complementation experiment. Moreover, our results demonstrated thatAL2was constitutively expressed in various tissues, including green and non-green tissues. Interestingly, we found that the expression levels of a subset of chloroplast genes that contain group IIA and IIB introns were significantly reduced in theal2mutant compared to that in the wild type, suggesting thatAL2is a functional CRS1 in rice. Differing from the orthologous CRS1 in maize and Arabidopsis that only regulates splicing of the chloroplast group II intron, our results demonstrated that theAL2gene is also likely to be involved in the splicing of the chloroplast group I intron. They also showed that disruption ofAL2results in the altered expression of chloroplast-associated genes, including chlorophyll biosynthetic genes, plastid-encoded polymerases and nuclear-encoded chloroplast genes. Taken together, these findings shed new light on the function of nuclear-encoded chloroplast group I and II intron splicing factors in rice.
DOI:10.1111/plb.12271URLPMID:25280352 [本文引用: 1]

Abstract The plastid ribosome proteins (PRPs) play important roles in plastid protein biosynthesis, chloroplast differentiation and early chloroplast development. However, the specialised functions of individual protein components of the chloroplast ribosome in rice ( Oryza sativa ) remain unresolved. In this paper, we identified a novel rice PRP mutant named asl2 ( Albino seedling lethality 2 ) exhibiting an albino, seedling death phenotype. In asl2 mutants, the alteration of leaf colour was associated with chlorophyll (Chl) content and abnormal chloroplast development. Through map-based cloning and complementation, the mutated ASL2 gene was isolated and found to encode the chloroplast 50S ribosome protein L21 (RPL21c), a component of the chloroplast ribosome large subunit, which was localised in chloroplasts. ASL2 was expressed at a higher level in the plumule and leaves, implying its tissue-specific expression. Additionally, the expression of ASL2 was regulated by light. The transcript levels of the majority of genes for Chl biosynthesis, photosynthesis and chloroplast development were strongly affected in asl2 mutants. Collectively, the absence of functional ASL2 caused chloroplast developmental defects and seedling death. This report establishes the important role of RPL21c in chloroplast development in rice.
[本文引用: 1]
DOI:10.1007/PL00022068URLPMID:17370354 [本文引用: 1]

Abstract The importance of chlorophyll (Chl) to the process of photosynthesis is obvious, and there is clear evidence that the regulation of Chl biosynthesis has a significant role in the regulation of assembly of the photosynthetic apparatus. The understanding of Chl biosynthesis has rapidly advanced in recent years. The identification of genetic loci associated with each of the biochemical steps has been accompanied by a greater appreciation of the role of Chl biosynthesis intermediates in intracellular signaling. The purpose of this review is to provide a source of information for all the steps in Chl and bacteriochlorophyll a biosynthesis, with an emphasis on steps that are believed to be key regulation points.
DOI:10.1093/pcp/pcp138URLPMID:19808806 [本文引用: 1]

For systematic and genome-wide analyses of rice gene functions, we took advantage of the full-length cDNA overexpresser (FOX) gene-hunting system and generated >12000 independent FOX-rice lines from >25000 rice calli treated with the rice-FOXAgrobacteriumlibrary. We found two FOX-rice lines generating green calli on a callus-inducing medium containing 2,4-D, on which wild-type rice calli became ivory yellow. In both lines,OsGLK1cDNA encoding a GARP transcription factor was ectopically overexpressed. Using rice expression-microarray and northern blot analyses, we found that a large number of nucleus-encoded genes involved in chloroplast functions were highly expressed and transcripts of plastid-encoded genes,psaA,psbAandrbcL, increased in theOsGLK1-FOX calli. Transmission electron microscopy showed the existence of differentiated chloroplasts with grana stacks inOsGLK1-FOX calli cells. However, in darkness,OsGLK1-FOX calli did not show a green color or develop grana stacks. Furthermore, we found developed chloroplasts in vascular bundle and bundle sheath cells of coleoptiles and leaves fromOsGLK1-FOX seedlings. TheOsGLK1-FOX calli exhibited high photosynthetic activity and were able to grow on sucrose-depleted media, indicating that developed chloroplasts inOsGLK1-FOX rice calli are functional and active. We also observed that the endogenousOsGLK1mRNA level increased synchronously with the greening of wild-type calli after transfer to plantlet regeneration medium. These results strongly suggest that OsGLK1 regulates chloroplast development under the control of light and phytohormones, and that it is a key regulator of chloroplast development.
DOI:10.2307/3870056URLPMID:20417532 [本文引用: 1]

Abstract Chlorophyll captures and redirects light-energy and is thus essential for photosynthetic organisms. The demand for chlorophyll differs throughout the day and night and in response to changing light conditions. Moreover, the chlorophyll biosynthesis pathway is up to certain points shared between the different tetrapyrroles; chlorophyll, heme, siroheme and phytochromobilin, for which the cell has different requirements at different time points. Combined with the phototoxic properties of tetrapyrroles which, if not properly protected, can lead to formation of reactive oxygen species (ROS), the need for a strict regulation of the chlorophyll biosynthetic pathway is obvious. Here we describe the current knowledge on regulation of chlorophyll biosynthesis in plants by the chloroplast redox state with emphasis on the Mg-chelatase situated at the branch point between the heme and the chlorophyll pathway. We discuss the proposed role of the Mg-chelatase as a key regulator of the tetrapyrrole pathway by its effect on enzymes both up- and downstream in the pathway and we specifically describe how redox state might regulate the Mg-branch. Finally, we propose that a recently identified NADPH-dependent thioredoxin reductase (NTRC) could be involved in redox regulation or protection of chlorophyll biosynthetic enzymes and describe the possible modes of action by this enzyme. Copyright 2010 Elsevier Ltd. All rights reserved.
[本文引用: 1]
DOI:10.1104/pp.122.1.49URLPMID:10631248 [本文引用: 1]

5-Aminolevulinic acid (ALA) is a precursor in the biosynthesis of tetrapyrroles including chlorophylls and heme. The formation of ALA involves two enzymatic steps which take place in the chloroplast in plants. The first enzyme, glutamyl-tRNA reductase, and the second enzyme, glutamate-1-semialdehyde-2,1-aminomutase, are encoded by the nuclear HEMA and GSA genes, respectively. To assess the significance of the HEMA gene for chlorophyll and heme synthesis, transgenic Arabidopsis plants that expressed antisense HEMA1 mRNA from the constitutive cauliflower mosaic virus 35S promoter were generated. These plants exhibited varying degrees of chlorophyll deficiency, ranging from patchy yellow to total yellow. Analysis indicated that these plants had decreased levels of chlorophyll, non-covalently bound hemes, and ALA; their levels were proportional to the level of glutamyl-tRNA reductase expression and were inversely related to the levels of antisense HEMA transcripts. Plants that lacked chlorophyll failed to survive under normal growth conditions, indicating that HEMA gene expression is essential for growth.
DOI:10.1093/pcp/pcg064URLPMID:12773632 [本文引用: 1]

We have previously generated a large pool of T-DNA insertional lines in rice. In this study, we screened those T-DNA pools for rice mutants that had defective chlorophylls. Among the 1,995 lines examined in the T2 generation, 189 showed a chlorophyll-deficient phenotype that segregated as a single recessive locus. Among the mutants, 10 lines were -glucuronidase (GUS)-positive in the leaves. Line 9-07117 has a T-DNA insertion into the gene that is highly homologous to XANTHA-F in barley and CHLH in Arabidopsis. This OsCHLH gene encodes the largest subunit of the rice Mg-chelatase, a key enzyme in the chlorophyll branch of the tetrapyrrole biosynthetic pathway. In the T2 and T3 generations, the chlorina mutant phenotypes are co-segregated with the T-DNA. We have identified two additional chlorina mutants that have a Tos17 insertion in the OsCHLH gene. Those phenotypes were co-segregated with Tos17 in the progeny. GUS assays and RNA blot analysis showed that expression of the OsCHLH gene is light inducible, while TEM analysis revealed that the thylakoid membrane of the mutant chloroplasts is underdeveloped. The chlorophyll content was very low in the OschlH mutants. This is the first report that T-DNA insertional mutagenesis can be used for functional analysis of rice genes.
[本文引用: 1]
DOI:10.1093/jxb/erv529URLPMID:4762379 [本文引用: 1]

Abstract Chlorophyll (Chl) b is a ubiquitous accessory pigment in land plants, green algae, and prochlorophytes. This pigment is synthesized from Chl a by chlorophyllide a oxygenase and plays a key role in adaptation to various environments. This study characterizes a rice mutant, pale green leaf (pgl), and isolates the gene PGL by using a map-based cloning approach. PGL, encoding chlorophyllide a oxygenase 1, is mainly expressed in the chlorenchyma and activated in the light-dependent Chl synthesis process. Compared with wild-type plants, pgl exhibits a lower Chl content with a reduced and disorderly thylakoid ultrastructure, which decreases the photosynthesis rate and results in reduced grain yield and quality. In addition, pgl exhibits premature senescence in both natural and dark-induced conditions and more severe Chl degradation and reactive oxygen species accumulation than does the wild-type. Moreover, pgl is sensitive to heat stress. The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology.
DOI:10.1016/S1875-2780(09)60022-5URL [本文引用: 1]

To understand the mechanism of grain yield in response to heat stress in rice (Oryza sativa L.), the development of anther and pollen, yield components, and some physiological parameters under the heat-stress during meiosis were investigated in a pot experiment. Two indica rice varieties differing in heat tolerance, Shuanggui 1 (heat-sensitive) and Huanghuazhan (heat-tolerant), were treated with heat stress (average temperature during the day above 35C) and natural temperature (average temperature during the day below 33C, control). The heat stress significantly reduced anther dehiscence and pollen fertility rate in Shuanggui 1, whereas, its effects were much smaller in Huanghuazhan. Number of spikelet per panicle, seed-setting rate, 1000-grain weight, and grain yield significantly decreased in both varieties, but the yield reduction in Shuanggui 1 was greater than that in Huanghuazhan. Moreover, significant narrowness of grain width and significant enlargement of grain length/width ratio were observed in Shuanggui 1. However, such effect was much smaller in Huanghuazhan. In physiological parameters that are associated with heat stress, the oxidation activity in roots and the RNA content in young panicles reduced significantly and the malondialdehyde (MDA) content in leaves and the ethylene evolution rate in young panicles increased significantly. These physiological characteristics were more sensitive to heat stress in Shuanggui 1 than in Huanghuazhan. The results indicate that the heat tolerant in rice is subject to high activity of roots, strong antioxidative defense system, high RNA content, little ethylene synthesis, and low MDA content in plants during meiosis.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S0176-1617(11)81192-2URL [本文引用: 1]

Specific absorption (α) coefficients for individual carotenoids and chlorophylls a and b, as well as the E1%1cm values for combined carotenoids, have been (re)estimated using 6 solvents (80 % acetone, chloroform, diethyl ether, dimethyl formamide, dimethyl sulphoxide, and methanol) using 2 different types of spectrophotometer (0.1—0.5 nm and 1—4 nm band pass resolution). From these values, 2 sets of equations to calculate concentrations of chlorophyll a (Ca), chlorophyll b (Cb) and total carotenoids (Cx+c) in γg mL-1 for the different instrument types were freshly derived or confirmed from earlier publications. These were then tested with 3 different types of spectrophotometers (the two variable types plus 2 nm fixed resolution diode array) using equal aliquots of a mixed extract in the 6 different solvents. These showed that the concentrations and ratios derived by the 2 sets of equations were comparable when used with their own type of spectrophotometer but less so if the inappropriate equations were used. Measurements taken with the diode array spectrophotometer, however, did not give accurate concentrations or ratios of chlorophylls and carotenoids
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S1673-8527(08)60098-3URLPMID:19232310 [本文引用: 1]

Green-revertible albino is a novel type of chlorophyll deficiency in rice ( Oryza sativa L.), which is helpful for further research in chlorophyll synthesis and chloroplast development to illuminate their molecular mechanism. In the previous study, we had reported a single recessive gene, gra(t), controlling this trait on the long arm of chromosome 2. In this paper, we mapped the gra(t) gene using 1,936 recessive individuals with albino phenotype in the F 2 population derived from the cross between themo-photoperiod-sensitive genic male-sterile (T/PGMS) line Pei'ai 64S and the spontaneous mutant Qiufeng M. Eventually, it was located to a confined region of 42.4 kb flanked by two microsatellite markers RM2-97 and RM13553. Based on the annotation results of RiceGAAS system, 11 open reading frames (ORFs) were predicted in this region. Among them, ORF6 was the most possible gene related to chloroplast development, which encoded the chloroplast protein synthesis elongation factor Tu in rice. Therefore, we designated it as the candidate gene of gra(t). Sequence analysis indicated that only one base substitution C to T occurred in the coding region, which caused a missense mutation (Thr to Ile) in gra(t) mutant. These results are very valuable for further study on gra(t) gene.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1534/g3.113.007856URLPMID:3789801 [本文引用: 1]

Plastid ribosomal proteins (PRPs) are essential for ribosome biogenesis, plastid protein biosynthesis, chloroplast differentiation, and early chloroplast development. This study identifies the first rice PRP mutant, asl1 (albino seedling lethality1), which exhibits an albino lethal phenotype at the seedling stage. This albino phenotype was associated with altered chlorophyll (Chl) content and chloroplast development. Map-based cloning revealed that ASL1 encodes PRP S20 (PRPS20), which localizes to the chloroplast. ASL1 showed tissue-specific expression, as it was highly expressed in plumule and young seedlings but expressed at much lower levels in other tissues. In addition, ASL1 expression was regulated by light. The transcript levels of nuclear genes for Chl biosynthesis and chloroplast development were strongly affected in asl1 mutants; transcripts of some plastid genes for photosynthesis were undetectable. Our findings indicate that nuclear-encoded PRPS20 plays an important role in chloroplast development in rice.
DOI:10.1007/s00425-008-0718-0URLPMID:18317796 [本文引用: 1]

A short root mutant was isolated from an EMS-generated rice mutant library. Under normal growth conditions, the mutant exhibited short root, delayed flowering, and partial sterility. Some sections of the roots revealed that the cell length along the longitudinal axis was reduced and the cell shape in the root elongation zone shrank. Genetic analysis indicated that the short root phenotype was controlled by a recessive gene. Map-based cloning revealed that a nucleotide substitution causing an amino acid change from Gly to Arg occurred in the predicted rice gene (Os02g0550600). It coded an alkaline/neutral invertase and was homologous to Arabidopsis gene AtCyt-inv1. This gene was designated as OsCyt-inv1. The results of carbohydrate analysis showed an accumulation of sucrose and reduction of hexose in the Oscyt-inv1 mutant. Exogenously supplying glucose could rescue the root growth defects of the Oscyt-inv1 mutant. These results indicated that OsCyt-inv1 played important roles in root cell development and reproductivity in rice.
[本文引用: 1]
