 ,1,2, 杨启台2,3, 刘章雄2, 郭勇2, 李英慧2, 关荣霞2, 邱丽娟2,*
,1,2, 杨启台2,3, 刘章雄2, 郭勇2, 李英慧2, 关荣霞2, 邱丽娟2,*Genotyping of SCN, SMV Resistance, Salinity Tolerance and Screening of Pyramiding Favorable Alleles in Introduced Soybean Accessions
YE Jun-Hua ,1,2, YANG Qi-Tai2,3, LIU Zhang-Xiong2, GUO Yong2, LI Ying-Hui2, GUAN Rong-Xia2, QIU Li-Juan2,*
,1,2, YANG Qi-Tai2,3, LIU Zhang-Xiong2, GUO Yong2, LI Ying-Hui2, GUAN Rong-Xia2, QIU Li-Juan2,*第一联系人:
收稿日期:2018-03-4接受日期:2018-06-12网络出版日期:2018-07-02
| 基金资助: |
Received:2018-03-4Accepted:2018-06-12Online:2018-07-02
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (894KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
叶俊华, 杨启台, 刘章雄, 郭勇, 李英慧, 关荣霞, 邱丽娟. 大豆引进种质抗胞囊线虫病、抗花叶病毒病和耐盐基因型鉴定及优异等位基因聚合种质筛选[J]. 作物学报, 2018, 44(9): 1263-1273. doi:10.3724/SP.J.1006.2018.01263
YE Jun-Hua, YANG Qi-Tai, LIU Zhang-Xiong, GUO Yong, LI Ying-Hui, GUAN Rong-Xia, QIU Li-Juan.
引进种质在我国大豆品种改良中发挥了重要作用, 通过对1923?1995年间育成和推广的651份大豆品种统计, 有224份品种可追溯到46个国外种质[1]。其中包括绥农14、合丰25等优良品种。绥农14拥有国外品种十胜长叶、Amsoy的血缘, 具有良好的遗传基础和农艺特征, 曾获得国家科技进步二等奖; 合丰25具有早熟、高产稳产、广适、抗逆性强等特点, 曾连续13年保持全国大豆品种播种面积第一的记录。分子标记遗传多样性分析结果表明, 美国和日本的品种与中国品种间存在明显遗传差异, 是重要的优异基因来源, 有利于改善我国大豆育种遗传基础狭窄的问题[2,3,4,5]。系统比较分析发现, 引进种质株型好、抗倒伏、抗病性较好, 对提高大豆产量、增强抗病、抗逆性、改善品质具有重要意义[6]。因此, 对引进种质的研究将促进其有效利用[7]。
大豆种质的鉴定评价是合理利用的前提。传统的种质鉴定是表型鉴定, 其鉴定不仅耗时费力, 鉴定结果准确性易受到环境条件的影响。如大豆胞囊线虫(soybean cyst nematode, SCN)鉴定至少在30 d以上[8]且鉴定数量有限。由于抗病、抗逆性鉴定条件及人力物力的限制, 难以对庞大数目的种质进行系统鉴定。因此, 自“八五”以来, 我国对引进国外大豆种质抗SCN、抗SMV (soybean mosaic virus)和耐盐性的鉴定数目不到总数的10%。
大豆基因组测序的完成促进了重要性状基因定位和克隆, 促进了基于抗病、耐逆性状的分子标记鉴定。抗大豆胞囊线虫方面, 已克隆SCN抗性的主效基因rhg1和Rhg4, 发现了微效基因GmSNAP11[9]。Kadam等[10]利用KASPar (Kompetitive Allele-Specific PCR)技术明确了95份大豆种质的rhg1、Rhg4及其他QTL基因型。史学晖等[11]对105份种质鉴定发现, Rhg4-389鉴定的抗性等位变异对抗病种质的选择效率为97.1%。在抗大豆花叶病毒方面, Zheng等[12]定位了东北最强株系的抗性基因, 开发并验证了与SMV紧密连锁的SCAR标记——SCN11。利用SCN11对中品95-5117的12份系谱材料进行检测, 抗性条带的选择效率约为63.6%[13]。在耐盐方面, Guan等[14]图位克隆了大豆耐盐基因GmSALT3, 从53份大豆种质中鉴定出9种单倍型, 包括2种耐盐单倍型和7种盐敏感单倍型, 通过对172份微核心种质及12份美国大豆种质进行鉴定, 单倍型H1对耐盐性的选择效率为91.9%[14]。
为了加快大豆优异种质鉴定效率, 本研究开发了大豆抗胞囊线虫病3个基因(rhg1、Rhg4、SCN3-11)和耐盐基因(GmSALT3)的KASP标记, 并利用与大豆花叶病毒病抗性相关的SCAR标记(SCN11), 对1489份引进种质进行了分析, 鉴定出携带优异等位基因且已具有抗性的种质, 同时, 筛选出少量携带优异等位基因的种质, 为表型高效鉴定提供了依据, 发掘出具有抗性但不携带优异等位基因的种质, 是新抗性(等位)基因挖掘的重要材料。本研究结果为大豆新基因发掘和新品种培育提供了材料和技术支撑。
1 材料与方法
1.1 试验材料
本研究利用引进大豆种质共1489份, 分别来自亚洲、欧洲和美洲。其中以来自美洲(包括美国、加拿大和巴西3个国家)的种质数目最多, 达935份, 来自欧洲(包括俄罗斯、瑞典、德国等13个国家)的有371份, 来自亚洲(包括日本、泰国、韩国等6个国家或地区)的种质共91份, 解放前留下的大豆种质共27份, 另有65份无来源记载。1.2 大豆基因组DNA的提取与质量检测
将参试种质每份播种5粒, 收集新鲜叶片。在TECAN液体自动化工作站平台进行大豆基因组DNA提取, 以96孔板为单位, 用改良的CTAB法[15]从大豆叶片中提取基因组DNA。提取后的DNA, 每96孔板抽检12个样品, 进行琼脂糖电泳检测, 并取2 μL DNA在BioTeK Synergy HIM上测定OD260、OD230值以及DNA浓度质检。1.3 标记的开发与鉴定
大豆SCN抗性及耐盐相关的5个SNP位点采用KASP标记进行检测。根据选取的5个SNP位点及其侧翼序列(表1), 用Primer软件设计3°末端PCR扩增引物, Tm值在55~65°C之间。每个SNP位点设计两条SNP特异性引物和一条通用引物[16]。标记的验证与检测在DouglasArray Tape平台上进行, 在SOELLEX高通量PCR水浴中完成PCR反应。PCR反应体系5 μL, 包括2 μL模板DNA (浓度约20 ng L-1)、2.5 μL 2×Master Mix (LGC Genomics, Hoddeston, UK)、0.07 μL Primer Mix、0.43 μL ddH2O。反应程序为94°C热激15 min; 94°C变性20 s, 65°C退火和延伸60 s, 10个循环, 每循环降低0.8°C; 94°C变性20 s, 57°C退火和延伸60 s, 26个循环。使用ARAYA荧光阅读仪读取荧光信号, 由Krake软件根据读取结果按照分型明确、NTC (无样品阴性对照)无特异性扩增的原则进行样品SNP分型。Table 1
表1
表1抗SCN、抗SMV和耐盐相关标记的类型及引物序列
Table 1
| 性状 Trait | 标记 Marker | 标记类型 Type | 基因 Gene | 引物序列信息 Primer sequence (5°-3°) | 等位基因 Allele | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 大豆胞囊线虫抗性 SCN resistance | ||||||
| Rhg4-389 | KASP | Glyma08g11490 | Rhg4-COM: CTACACCGCCGTCCTCAAC | [17] | ||
| Rhg4-FAM: GAGGTGGCCGCCGGAGC | G/G | |||||
| Rhg4-HEX: GAGGTGGCCGCCGGAGG | C/C | |||||
| rhg1 | KASP | Glyma18g02590 | rhg1-COM: CTGGCATCTGCCAACTCTGTAAAGA | [18] | ||
| rhg1-FAM: TTCTAATGCATTGGTTATAGCAACAACG | C/C | |||||
| rhg1-HEX: TTCTAATGCATTGGTTATAGCAACAACC | G/G | |||||
| SCN3-11 | KASP | Glyma11g35820 | SCN3-11-COM: CAGAAGTCGATTGAGATTTACGAAGAGATA | [9] | ||
| SCN3-11-FAM: ATTATTGTTGAGGGATTGGCGAGC | C/C | |||||
| SCN3-11-HEX: AAATTATTGTTGAGGGATTGGCGAGT | T/T | |||||
| 大豆耐盐性 Soybean salinity tolerance | ||||||
| SALT3 | KASP | Glyma03g32900 | GmSALT3-COM: GATCATTGGATGTAATTGGGTGGAGAA | [14] | ||
| GmSALT3-FAM: GATACCAGCAAATATTAAATGTGTGTTTTT | T/T | |||||
| GmSALT3-HEX: GATACCAGCAAATATTAAATGTGTGTTTTA | -/- | |||||
| Ncl-5 | KASP | Glyma03g32900 | Ncl-5-COM: AGGTACTTACCCTTATGAAGAAAACA | [14] | ||
| Ncl-5-FAM: AGAACTCGTATTTTATTTTGGTTGAC | G/G | |||||
| Ncl-5-HEX: AGAACTCGTATTTTATTTTGGTTGAT | A/A | |||||
| 大豆花叶病毒抗性 SMV resistance | ||||||
| SCN11 | SCAR | — | SCN11-F: TTCACGTGGCCCTCCTATC | — | [12] | |
| SCN11-R: CGCCGCAAACTCACAGGAC | — | |||||
新窗口打开|下载CSV
与大豆花叶病毒抗性相关的位点用SCAR标记[12]进行检测。以基因组DNA为模板, 反应体系10 μL, 包括50 ng基因组DNA 2 μL、10×PCR缓冲液1 μL、2 mmol L-1的dNTPs 1 μL、2 μmol L-1上下游引物各0.2 μL、Taq聚合酶0.2 μL (全式金生物技术有限公司)、ddH2O 5.4 μL。PCR在ABI (Applied Biosystems, 美国)公司的PCR扩增热循环仪上进行, 反应程序为95°C预变性5 min; 95°C变性30 s, 59°C退火30 s, 72°C延伸70 s, 34个循环; 最后72°C延伸8 min, 于4°C保存。采用浓度为1.5%的琼脂糖凝胶电泳, 130V电压下电泳25 min, 经EB染色后在紫外灯下观察结果, 记录结果。
1.4 数据分析
使用Microsoft Excel 2007进行等位基因分布频率计算。2 结果与分析
2.1 5个位点的优异等位基因频率分析
利用抗大豆胞囊线虫病基因(rhg1、Rhg4、SCN3-11)和耐盐基因GmSALT3的KASP标记对引进种质的分型结果显示, 两种纯合基因型和杂合基因型呈明显的簇状分离(图1-A~E), 证明了开发KASP标记的有效性。通过PCR扩增检测结果可以看出, 大豆花叶病毒病抗性相关标记SCN将参试种质分为扩增片段980 bp的抗病优异等位基因和扩增片段1070 bp的感病等位基因(图1-F)。标记SALT3和Ncl-5源自同一个位点(Glyma03g32900), 共同决定了耐盐单倍型H1, 记作SALT3(H1)。用6个分子标记对1489份引进大豆种质鉴定, 除去分型结果不明确的种质, 对有效鉴定的种质分析结果显示, 功能标记rhg1、Rhg4-389、SCN3-11、SALT3优异等位基因的频率较低(分别为0.022、0.025、0.049和0.170), 而连锁标记SCN11的优异等位基因频率较高(0.645) (表2)。图1
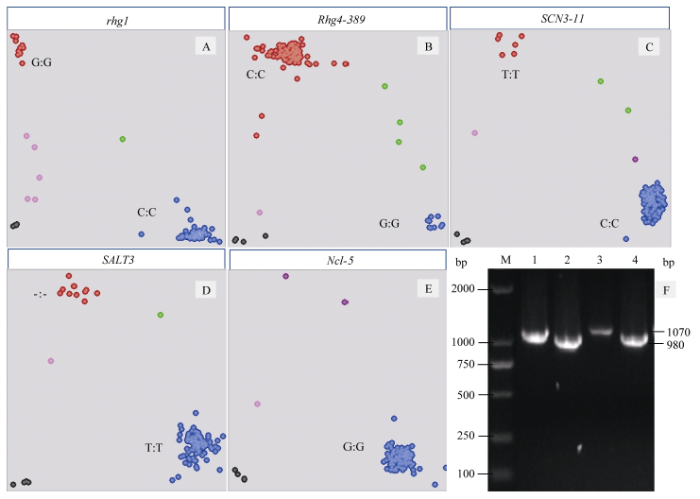 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1KASP和SCAR标记对部分种质基因型检测结果
A~E: 每个圆点各对应一份检测种质。红色或蓝色表示纯合基因型; 绿色表示杂合基因型; 粉色表示检测无信号或信号较弱; 紫色表示有信号但无明确分型; 黑色代表NTC, 即无模板对照。F: M是DL2000; 1、3为感SMV基因型; 2、4为抗SMV基因型。
Fig. 1Partial germplasms genotyping results with KASP and SCAR marker
A-E: each dot corresponds to an accession tested. Red or blue dots represent homozygous genotypes; green dots represent heterozygotes; pink dots represent no signal detected or weak signals; purple dots represent signals which cannot be classified; black dots represent NTC, no template control. F: M is DL2000; 1, 3 represent sensitivity to soybean mosaic virus; 2, 4 represent resistance to soybean mosaic virus.
2.2 5个位点抗性种质的来源分布规律
各位点的抗性种质来源分布显示, SCN主效位点rhg1、Rhg4的抗性种质主要来自美洲, 分别为24份和30份; 少量来自亚洲, 分别为4份和2份。微效基因SCN3-11的抗性种质多数(37份)来自美洲, 但部分种质(16份)来自欧洲。耐盐单倍型SALT3(H1)的大豆种质中来自美洲的最多(53.19%), 其次是欧洲(31.06%), 亚洲最少(4.68%)。SCAR标记SCN11鉴定出的抗SMV种质960份, 占总数的64.47%, 且美洲(700份)多于欧洲(147份)和亚洲(57份) (图2)。Table 2
表2
表26个抗性标记对引进种质的分型结果
Table 2
| 性状 Trait | 标记名称 Marker | 种质有效鉴定数目 Accessions effectively identified No. | 等位基因 Allele | 对应表型1) Corresponding phenotype 1) | 种质Accessions | |
|---|---|---|---|---|---|---|
| 数量 Size (No.) | 比例 Ratio (%) | |||||
| 大豆胞囊线虫抗性 SCN resistance | rhg1 | 1383 | G/G | R | 31 | 2.24 |
| C/C | S | 1352 | 97.76 | |||
| Rhg4-389 | 1480 | G/G | R | 37 | 2.50 | |
| C/C | S | 1441 | 97.37 | |||
| SCN3-11 | 1356 | T/T | R | 66 | 4.87 | |
| C/C | S | 1290 | 95.13 | |||
| 大豆耐盐性 Soybean salinity tolerance | SALT3 | 1380 | -/- | — | 236 | 17.10 |
| T/T | S | 1127 | 81.67 | |||
| Ncl-5 | 1479 | G/G | — | 1384 | 93.58 | |
| A/A | S | 85 | 5.75 | |||
| SALT3, Ncl-5 | 1380 | SALT3(H1) | T | 235 | 17.03 | |
| 大豆花叶病毒抗性 SMV resistance | SCN11 | 1489 | — | R | 960 | 64.47 |
| — | S | 529 | 35.53 | |||
新窗口打开|下载CSV
图2
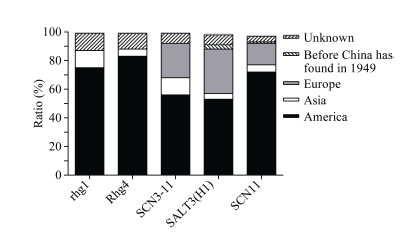 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图25个位点抗性等位基因或单倍型分布及来源
Fig. 2Distribution and origins of resistant alleles or haplotype in five loci
2.3 基于优异等位基因的优异种质筛选
按表型特性进行分析, 筛选出仅具有单一性状抗性(单抗)的种质共907份, 包括抗SCN种质21份、抗SMV种质796份、耐盐种质90份。筛选出具有2种抗性(双抗)种质160份, 兼抗SCN和SMV种质32份; 抗SCN且耐盐的种质13份; 抗SMV且耐盐的种质115份。筛选出同时具有3种抗性(三抗)种质17份。按基因型进行分析, 鉴定出携带优异等位基因的种质1084份, 包括具有单位点优异等位基因种质901份, 以SCN11标记筛选出的种质数目最多; 携带2个位点优异等位基因种质147份, 抗SCN双位点叠加种质最少, 而耐盐和抗SMV的基因聚合种质数量最多(表3); 携带3个或3个以上优异等位基因的种质36份, 其中有聚合3个位点的种质19份、聚合4个位点的种质9份、聚合5个位点的种质8份(表4)。
Table 3
表3
表35个位点对引进种质的筛选
Table 3
| 抗性特性 Resistance | 优异基因型 Favorable allele | 携带优异等位基因的种质 Germplasm with favorable alleles | |||
|---|---|---|---|---|---|
| 数目 Size (No.) | 比例 Ratio (%) | 代表性种质 Representative germplasm | |||
| 单位点 Single locus | 901 | 60.51 | |||
| SCN | SCN3-11 | 15 | 1.01 | 中特1号 Zhongte 1 | |
| SMV | SCN11 | 796 | 53.46 | Provar | |
| Salt | SALT3(H1) | 90 | 6.04 | Harosoy 71 | |
| 双位点 Two loci | 147 | 9.87 | |||
| SCN | rhg1/Rhg4 | 1 | 0.07 | M044 | |
| SCN | Rhg4/SCN3-11 | 2 | 0.13 | PR 149-3 | |
| SCN, SMV | rhg1/SCN11 | 2 | 0.13 | PI576145 | |
| SCN, SMV | Rhg4/SCN11 | 5 | 0.34 | Mack | |
| SCN, SMV | SCN3-11/SCN11 | 12 | 0.81 | CA50 | |
| SCN, Salt | SCN3-11/SALT3(H1) | 8 | 0.54 | PI590579 | |
| SCN, Salt | Rhg4/SALT3(H1) | 2 | 0.13 | D85-10412 | |
| SMV, Salt | SALT3(H1)/SCN11 | 115 | 7.72 | Lee 68 | |
| 3个位点 Three loci | 19 | 1.28 | |||
| SCN | rhg1/Rhg4/SCN3-11 | 3 | 0.20 | Centennial | |
| SCN, SMV | rhg1/Rhg4/SCN11 | 2 | 0.13 | Delsoy4900 | |
| SCN, SMV | rhg1/SCN3-11/SCN11 | 4 | 0.27 | AGS175 | |
| SCN, SMV | Rhg4/SCN3-11/SCN11 | 1 | 0.07 | 9234 | |
| SCN, Salt | rhg1/Rhg4/SALT3(H1) | 1 | 0.07 | T221 | |
| SCN, Salt | Rhg4/SCN3-11/SALT3(H1) | 1 | 0.07 | LINE272H选 LINE272H Xuan | |
| SCN, SMV, Salt | rhg1/SALT3(H1)/SCN11 | 2 | 0.13 | H6255RR | |
| SCN, SMV, Salt | Rhg4/SALT3(H1)/SCN11 | 2 | 0.13 | Lamar | |
| SCN, SMV, Salt | SCN3-11/SALT3(H1)/SCN11 | 3 | 0.20 | P951341RR | |
| 4个位点 Four loci | 9 | 0.60 | |||
| SCN, SMV | rhg1/Rhg4/SCN3-11/SCN11 | 6 | 0.40 | Newton | |
| SCN, Salt | rhg1/Rhg4/SCN3-11/SALT3(H1) | 1 | 0.07 | PI90763 | |
| SCN, SMV, Salt | rhg1/SCN3-11/SALT3(H1)/SCN11 | 1 | 0.07 | S01-9364 | |
| SCN, SMV, Salt | Rhg4/SCN3-11/SALT3(H1)/SCN11 | 1 | 0.07 | S-10-1 | |
| 5个位点 Five loci | 8 | 0.54 | |||
| SCN, SMV, Salt | rhg1/Rhg4/SCN3-11/SALT3(H1)/SCN11 | 8 | 0.54 | Pin-din-guan | |
新窗口打开|下载CSV
Table 4
Table 4Thirty-six accessions with three or more resistance alleles
| 携带抗性位点数 Number of resistant loci | 统一编号 Accession No. | 名称 Name | 优异等位基因 Favorable alleles | 抗性表型 Resistance | ||||
|---|---|---|---|---|---|---|---|---|
| 3个位点 Three loci | WDD00828 | Centennial | rhg1, Rhg4, SCN3-11 | SCN* | ||||
| WDD01994 | M87-1569 | rhg1, Rhg4, SCN3-11 | SCN* | |||||
| WDD00691 | AGS272+ | rhg1, Rhg4, SCN3-11 | SCN* | |||||
| WDD03200 | LINE272H Xuan# | Rhg4, SCN3-11, SALT3(H1) | SCN, Salt | |||||
| WDD00289 | T221 | rhg1, Rhg4, SALT3(H1) | SCN*, Salt | |||||
| WDD00858 | N80-2317# | rhg1, SCN3-11, SCN11 | SCN, SMV | |||||
| WDD00859 | N80-50232# | rhg1, SCN3-11, SCN11 | SCN, SMV | |||||
| WDD00666 | AGS65+ | rhg1, SCN3-11, SCN11 | SCN, SMV | |||||
| 携带抗性位点数 Number of resistant loci | 统一编号 Accession No. | 名称 Name | 优异等位基因 Favorable alleles | 抗性表型 Resistance | ||||
| WDD00683 | AGS175+ | rhg1, SCN3-11, SCN11 | SCN, SMV | |||||
| WDD02019 | 9234 | Rhg4, SCN3-11, SCN11 | SCN*, SMV | |||||
| WDD01607 | Delsoy4900 | rhg1, Rhg4, SCN11 | SCN*, SMV | |||||
| WDD03086 | G04-Ben229lR-M | rhg1, Rhg4, SCN11 | SCN, SMV | |||||
| WDD02211 | H6255 | rhg1, SALT3(H1), SCN11 | SCN, Salt, SMV | |||||
| WDD03205 | M017-1# | rhg1, SALT3(H1), SCN11 | SCN, Salt, SMV | |||||
| WDD00916 | SRE-D-14A# | Rhg4, SALT3(H1), SCN11 | SCN, Salt, SMV | |||||
| WDD01640 | Lamar | Rhg4, SALT3(H1), SCN11 | SCN*, Salt, SMV | |||||
| WDD00334 | Clark-G | SCN3-11, SALT3(H1), SCN11 | SCN, Salt, SMV | |||||
| WDD02252 | P951341 | SCN3-11, SALT3(H1), SCN11 | SCN, Salt, SMV | |||||
| WDD00335 | Clark-S | SCN3-11, SALT3(H1), SCN11 | SCN, Salt, SMV | |||||
| 4个位点 Four loci | WDD02102 | PI90763 | rhg1, Rhg4, SCN3-11, Salt3(H1) | SCN*, Salt | ||||
| WDD00926 | TGX814-26D# | rhg1, Rhg4, SCN3-11, SCN11 | SCN, SMV | |||||
| WDD01583 | Newton | rhg1, Rhg4, SCN3-11, SCN11 | SCN*, SMV | |||||
| WDD00595 | Custer | rhg1, Rhg4, SCN3-11, SCN11 | SCN*, SMV | |||||
| WDD00602 | Franklin | rhg1, Rhg4, SCN3-11, SCN11 | SCN*, SMV | |||||
| WDD02015 | TBD | rhg1, Rhg4, SCN3-11, SCN11 | SCN, SMV | |||||
| WDD01538 | rhg1, Rhg4, SCN3-11, SCN11 | SCN, SMV | ||||||
| WDD03083 | S01-9364 | rhg1, SCN3-11, SALT3(H1), SCN11 | SCN*, Salt*, SMV | |||||
| WDD00774 | S-10-1 | Rhg4, SCN3-11, SALT3(H1), SCN11 | SCN, Salt, SMV | |||||
| 5个位点 Five loci | WDD03084 | S01-9391 | rhg1, Rhg4, SCN3-11, SALT3(H1), SCN11 | SCN*, Salt, SMV | ||||
| WDD01632 | Bryan | rhg1, Rhg4, SCN3-11, SALT3(H1), SCN11 | SCN*, Salt, SMV | |||||
| WDD01614 | Rhodes | rhg1, Rhg4, SCN3-11, SALT3(H1), SCN11 | SCN*, Salt, SMV | |||||
| WDD00721 | Forrest | rhg1, Rhg4, SCN3-11, SALT3(H1), SCN11 | SCN*, Salt, SMV | |||||
| WDD01971 | D83-3349 | rhg1, Rhg4, SCN3-11, SALT3(H1), SCN11 | SCN*, Salt, SMV | |||||
| WDD00661 | A-66 Jia+ | rhg1, Rhg4, SCN3-11, SALT3(H1), SCN11 | SCN, Salt, SMV | |||||
| WDD02989 | Pin-din-guan | rhg1, Rhg4, SCN3-11, SALT3(H1), SCN11 | SCN*, Salt*, SMV | |||||
| WDD03094 | S99-2281 | rhg1, Rhg4, SCN3-11, SALT3(H1), SCN11 | SCN*, Salt, SMV | |||||
* represents the resistance or tolerance of the accession has been reported.
新窗口打开|下载CSV
3 讨论
3.1 基于分子标记的基因型检测为提高种质鉴定和利用效率创造了条件
20世纪40年代以来, 我国多次引入国外大豆种质[5], 目前引进大豆种质已超过3000余份, 并保存在国家种质库[19]。20世纪80年代, 我国开始对大豆种质的农艺性状、抗病性及抗逆性进行评价, 而美国已从种质中筛选出了抗病、抗逆及抗虫种质[6]。由于基于表型鉴定筛选种质受到人力、财力、物力的影响, 鉴定结果受环境影响很大, 鉴定种质的数量非常有限。迄今为止, 仅对不超过500份引入种质的抗病性、抗逆性等性状进行了不完全鉴定。目前基于大豆重要性状相关的分子标记对种质进行基因型鉴定的报道较少。例如,利用大豆灰斑病抗性基因连锁的3个SSR标记, 对45份地方种质进行检测, 鉴定效率分别为72.7%、81.8%、83.3%[20];用大豆白粉病基因连锁标记Sat_366和Sat_393检测2个杂交分离F2群体, 鉴定效率分别为92.7%和60.3%[21]。用大豆黄种皮基因紧密连锁的标记ls1-22对355分析大豆核心种质等材料,鉴定选择效率为76.23%[21]。利用色素合成基因的4个功能标记分析272份种质, 对茸毛色的检测效率在80%以上[22]。利用与生育期相关的SSR标记Satt431、Satt215和Satt557对47份黄淮育成品种进行鉴定, 对熟期组MGIII和MGV的选择效率分别为83.33%和90.00%[23]。本研究选用的功能标记Rhg4-389及GmSALT3单倍型Salt3(H1)的鉴定效率在90%以上, 连锁标记SCN11的鉴定效率在60%以上, 揭示了在分子水平上直接对目的性状进行选择的可能性。通过高通量基因型鉴定明确了1489份引入种质的3个性状基因型, 鉴定种质数量占引进种质总数近50%, 选择出具有优异等位基因的种质, 其中已证明的有耐盐种质20份, 抗SCN种质21份, 抗SMV3号小种的种质12份, 可直接利用[25]。携带3个或3个以上抗性等位基因的优异种质36份, 虽然有33份表型尚需进一步验证, 但提高了筛选优异种质的目标性。本研究除了抗大豆花叶病毒SCAR标记SCN11外, 抗SCN和耐盐基因开发了可进行高通量检测的KASP标记, 降低了检测成本, 为上万份种质资源的高效发掘与利用提供了新思路。
3.2 KASP技术在大豆基因型鉴定中的应用
KASP技术是由LGC公司(http://www.lgcgroup. com/)设计和创制, 与芯片技术同属于高通量的SNP测序技术。与下一代测序(NGS)技术相比, 高通量SNP检测具有快速高效、敏感度高、使用方便、结果可靠、价格低廉等优势。芯片技术适用于对100到1 000 000个以上的SNP进行检测, 对少量SNP进行检测时, KASP技术具有经济高效的优势[26]。与基于芯片的Illumina GoldenGate相比, KASP技术分型的错误率(0.7%~1.6%)低于芯片GoldeGate平台(2.0%~2.4%), 且用于分子标记辅助回交选择时, 使用KASP技术分型将比使用其他高通量平台节省7.9%~46.1%的费用[27]。KASP技术以价格低廉、高效灵活性[28], 已在水稻[29]、玉米[30,31]、小麦[32,33,34,35,36]、蚕豆[37]等作物的研究中应用。在大豆研究中,Patil等[38]开发了基于种子组成性状相关等位基因KASP标记, 并应用于明确突变来源和基因定位。Patil等[39] 还对耐盐基因Glyma03g32900多个与结构变异相关的SNP进行KASP检测, 验证了这些SNP与表型的高度相关。Pham等[40]定位来自PI594891和PI594774中抗灰斑病的2个候选基因, 开发KASP标记用于检测群体基因型与表型间的关联。本研究利用抗胞囊线虫病基因和耐盐基因开发的5个KASP标记, 系统检测了1489份引进种质基因型,占引进种质的46.2%,开创了我国大豆种质重要性状基因型快速鉴定的新局面。3.3 优异等位基因种质的发掘为遗传育种提供了材料支撑
利用目的基因或与目的基因紧密连锁的分子标记可聚合多个优异等位基因。Maroof等[41]利用SSR标记对SMV感病大豆Essex的近等基因系中含有Rsv位点进行检测, 发现Rsv1Rsv3、Rsv1Rsv4 和Rsv1Rsv3Rsv4可通过两基因和三基因等位基因聚合可产生抗性, 但Rsv3Rsv4则表现出晚期易感。Wang等[42]用SMV抗性基因RSC、RSC8、RSC14Q连锁的10个SSR标记对杂交后代群体检测, 并对标记聚合株系进行SMV抗性评价, F7代选出抗21个SMV菌株的纯合株系5个。分子标记辅助选择不仅应用于聚合同一性状的多个基因, 还可以聚合多个性状的多个基因。Kumar等[43]通过分子辅助回交育种将抗细菌性枯萎病基因Xa21和抗稻瘿蚊的基因Gm4、Gm8聚合到水稻恢复系RPHR-1005; Hur等[44]利用标记鉴定了BC4F6群体, 筛选出聚合水稻细菌枯萎病基因Xa3和Xa4及与耐冷QTL的株系ABL132-36。姚姝等[45]通过对杂交分离后代进行分子标记辅助选择, 创制出聚合水稻抗稻瘟病基因Pi-ta、Pi-b和低直链淀粉含量基因Wx-mq的新品系“南粳0051”。分子标记辅助选择在多基因聚合育种的成功应用, 提高了育种效率, 但在种质资源基因型鉴定方面鲜见报道。本研究检测的5个位点涉及抗SCN、抗SMV和耐盐3个性状, 筛选出携带抗SCN的3个位点优异等位基因(Peking型)种质19份, 其中15份具有抗SCN特性[46,47,48,49,50], 包括Centennial、Franklin、Forrest等; 用耐盐基因的2个标记筛选出235份种质, 其中20份已证明有耐盐性[51], 包括Altona、Mansoy、Baekun Kong、Lee68等; 用抗SMV标记筛选出960份大豆种质, 其中12份已证明表现为抗SMV3号株系[52,53], 包括L88-8440、L82-951、L84-2112、Columbia和L93-3327。除此之外, 仍有81份种质具有耐盐性, 但经标记检测不属于GmSALT3的耐盐单倍型H1, 推断少数种质[14]可能为耐盐单倍型H2, 还可能存在影响耐盐性的新位点; 有5份引进种质经SCN11检测为SMV感病型, 却表现为抗SMV3号株系, 包括L88-8431[54]、L78-379[55]、L83-4744 (内部资源)、L83-4483 (内部资源)和新八达2号[56], 一个原因可能是标记与基因间发生了交换, 这也是功能标记比连锁标记检测效率高的原因, 另一个原因可能是存在控制SMV抗性的新位点。有6份抗SCN种质(Bedford、Yale、Fayette、PI 209332、Cartter、Bell)在SCN三个位点鉴定结果显示均为感病基因型, 与它们的实际抗性表型不符[57,58,59,60,61,62], 其原因是这些种质为PI88788型, 在rhg1的另一处发生变异所致[63]。与现有资源相比, 新增可能抗SCN的种质4份, 可能具有耐盐特性的种质215份, 可能抗SMV的种质948份。携带3个或3个以上抗性等位基因的36份种质中, 其中16份种质的一种或两种特性已被报道, 且可能存在新特性; 17份种质尚未有3种特性的鉴定报道, 可能存在抗SCN、SMV或耐盐性。
本研究基于主要性状已知基因(rhg1、Rhg4、SCN3-11、GmSALT3)开发的功能标记鉴定出的优异种质数目较少, 分别占鉴定种质总量的2.24%、2.50%、4.87%和17.03%; 而基于主要性状紧密连锁的标记(SCN11)鉴定出的种质数目较多, 占鉴定种质总量的64.47%。这是由于基因的连锁标记的遗传效应值依赖于该标记与基因的连锁紧密程度[64], 而功能标记的遗传效应具有可靠性和普适性[65], 能够准确地检测目的基因, 因此, 基于功能基因筛选的优异种质效率高于与基因连锁的标记。然而, 无论是用功能基因标记还是连锁标记, 一旦选择出优异种质, 其分子标记的选择效率都会提高, 可应用于分子标记辅助选择育种。
4 结论
抗病耐逆性是与大豆产量、品质相关的重要性状, 而通过分子标记进行基因型鉴定是进行优异种质筛选的有效手段。本研究鉴定出携带至少1种优异等位基因的种质1084份, 44份已由前人证明相应的抗性; 携带3个或3个以上优异等位基因的种质有36份, 其中52.78%种质的一种或两种优异特性已被报道。在不携带抗性优异等位变异的种质中, 93份具有耐盐性或SMV3号株系抗性报道, 这些种质可能存在新的抗性(等位)基因。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
DOI:10.1007/PL00002917URL [本文引用: 1]

Advanced-cycle pedigree breeding has caused maize ( Zea mays L.) inbreds to become more-elite but more-narrow genetically. Our objectives were to evaluate the genetic distance among current and historical maize inbreds, and to estimate how much genetic diversity has been lost among current inbreds. We selected eight maize inbreds (B14, B37, B73, B84, Mo17, C103, Oh43 and H99) that largely represented the genetic background of current elite inbreds in the U.S. seed industry. A total of 32 other inbreds represented historical inbreds that were once important in maize breeding. Cluster analysis of the inbreds, using data for 83 SSR marker loci, agreed well with pedigree information. Inbreds from Iowa Stiff Stalk Synthetic (BSSS), Reid Yellow Dent, and Lancaster clustered into separate groups with only few exceptions. The average number of alleles per locus was 4.9 among all 40 inbreds and 3.2 among the eight current inbreds. The reduction in the number of alleles per locus was not solely due to sample size. The average genetic distance ( D ij ) was 0.65 among the eight current inbreds, 0.67 among the 32 historical inbreds, and 0.67 among all 40 inbreds. These differences were statistically insignificant. We conclude that genetic diversity among current inbreds has been reduced at the gene level but not at the population level. Hybrid breeding in maize maintained, rather than decreased, genetic diversity, at least during the initial subdivision of inbreds into BSSS and non-BSSS heterotic groups. We speculate, however, that exploiting other germplasm sources is necessary for sustaining long-term breeding progress in maize.
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1111/tpj.12695URLPMID:25292417 [本文引用: 3]

Summary The identification of genes that improve the salt tolerance of crops is essential for the effective utilization of saline soils for agriculture. Here, we use fine mapping in a soybean ( Glycine max (L.) Merr.) population derived from the commercial cultivars Tiefeng 8 and 85–140 to identify GmSALT3 (salt tolerance-associated gene on chromosome 3), a dominant gene associated with limiting the accumulation of sodium ions (Na+) in shoots and a substantial enhancement in salt tolerance in soybean. GmSALT3 encodes a protein from the cation/H+ exchanger family that we localized to the endoplasmic reticulum and which is preferentially expressed in the salt-tolerant parent Tiefeng 8 within root cells associated with phloem and xylem. We identified in the salt-sensitive parent, 85–140, a 3.78-kb copia retrotransposon insertion in exon 3 of Gmsalt3 that truncates the transcript. By sequencing 31 soybean landraces and 22 wild soybean ( Glycine soja ) a total of nine haplotypes including two salt-tolerant haplotypes and seven salt-sensitive haplotypes were identified. By analysing the distribution of haplotypes among 172 Chinese soybean landraces and 57 wild soybean we found that haplotype 1 (H1, found in Tiefeng 8) was strongly associated with salt tolerance and is likely to be the ancestral allele. Alleles H2–H6, H8 and H9, which do not confer salinity tolerance, were acquired more recently. H1, unlike other alleles, has a wide geographical range including saline areas, which indicates it is maintained when required but its potent stress tolerance can be lost during natural selection and domestication. GmSALT3 is a gene associated with salt tolerance with great potential for soybean improvement.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
.
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s11032-015-0368-4URL [本文引用: 1]

Soybean is a primary source of vegetable oil, accounting for 53% of the total vegetable oil consumption in the USA in 2013. Soybean oil with high oleic acid and low linolenic acid content is desired, because it not only improves the oxidative stability of the oil, but also reduces the amount of undesirable trans-fat production by the hydrogenation processing. The mutant FAD2-1A and FAD2-1B alleles contribute to an elevated oleic acid level, while mutations in FAD3A and FAD3C lead to a reduction in linolenic acid levels of soybean seed oil. Although SNP (Single Nucleotide Polymorphism) SimpleProbe assays have been developed for these genes, breeder friendly and high-throughput marker systems are needed for large scale selection in soybean breeding programs. TaqMan or Kompetitive Allele Specific PCR (KASP) assays were successfully developed to detect and discriminate mutant alleles of FAD2-1A (17D and PI 603452 sources) and FAD2-1B (PI 283327 source) for the high oleic acid trait, as well as those of FAD3A (CX1512-44 and C1640 sources) and FAD3C (CX1512-44 source) for low linolenic acid content soybean. All of the assays have been proven to be robust in distinguishing different genotypes in a high-throughput setting. The accuracy of the assays was validated using multiple populations as well as a panel of diverse soybean lines and a significant correlation was observed between the SNP alleles and their corresponding fatty acid phenotypes. These marker assays appear to be very reliable for detecting mutant and wild type alleles, and thus will assist forward and backcrossing breeding of soybean lines with desired oil profiles in an accurate and efficient manner.
,
URL [本文引用: 1]
[本文引用: 1]
DOI:10.1186/s12870-017-1197-xURL [本文引用: 1]

KASP (KBioscience Competitive Allele Specific PCR) and Amplifluor (Amplification with fluorescence) SNP markers are two prominent technologies based upon a shared identical Allele-specific PCR platform. Amplifluor-like SNP and KASP analysis was carried out using published and own design of Universal probes (UPs) and Gene-specific primers (GSPs). Advantages of the Amplifluor-like system over KASP include the significantly lower costs and much greater flexibility in the adjustment and development of ‘self-designed’ dual fluorescently-labelled UPs and regular GSPs. The presented results include optimisation of ‘tail’ length in UPs and GSPs, protocol adjustment, and the use of various fluorophores in different qPCR instruments. The compatibility of the KASP Master-mix in both original and Amplifluor-like systems has been demonstrated in the presented results, proving their similar principles. Results of SNP scoring with rare alleles in addition to more frequently occurring alleles are shown. The Amplifluor-like system produces SNP genotyping results with a level of sensitivity and accuracy comparable to KASP but at a significantly cheaper cost and with much greater flexibility for UPs with self-designed GSPs. The online version of this article (10.1186/s12870-017-1197-x) contains supplementary material, which is available to authorized users.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1155/2017/6572969URLPMID:28630621 [本文引用: 1]

Seed composition is one of the most important determinants of the economic values in soybean. The quality and quantity of different seed components, such as oil, protein, and carbohydrates, are crucial ingredients in food, feed, and numerous industrial products. Soybean researchers have successfully developed and utilized a diverse set of molecular markers for seed trait improvement in soybean breeding programs. It is imperative to design and develop molecular assays that are accurate, robust, high-throughput, cost-effective, and available on a common genotyping platform. In the present study, we developed and validated KASP (Kompetitive allele-specific polymerase chain reaction) genotyping assays based on previously known functional mutant alleles for the seed composition traits, including fatty acids, oligosaccharides, trypsin inhibitor, and lipoxygenase. These assays were validated on mutant sources as well as mapping populations and precisely distinguish the homozygotes and heterozygotes of the mutant genes. With the obvious advantages, newly developed KASP assays in this study can substitute the genotyping assays that were previously developed for marker-assisted selection (MAS). The functional gene-based assay resource developed using common genotyping platform will be helpful to accelerate efforts to improve soybean seed composition traits.
[本文引用: 1]
[本文引用: 1]
DOI:10.2135/cropsci2007.08.0479URL [本文引用: 1]

Soybean mosaic virus (SMV) causes a disease of soybean [Glycine max (L.) Men.] that is prevalent throughout the United States. The disease can be effectively managed through the deployment of single-dominant resistance genes known as Rsv genes that confer resistance to different strains of SMV. Pyramiding respective Rsv genes from different loci (Rsv1, Rsv3, and Rsv4) through marker-assisted selection (MAS) is an ideal method for creating durable and wide spectrum resistance to all strains of SMV. In this study, simple sequence repeat markers were used to create isogenic lines of the susceptible cultivar Essex containing one, two, or three Rsv loci for observing background and epistatic effects of Rsv1, Rsv3, and Rsv4 on inoculation with six strains of SMV. Results indicate that an Essex background or modifier genes from the donor source had effects on reactions of Rsv3 and Rsv4 genes, causing the isogenic lines to be more susceptible than the Rsv donor parents. Two-gene and three-gene isolines of Rsv1Rsv3, Rsv1Rsv4 and Rsv1Rsv3Rsv4, acted in a complementary manner, conferring resistance against all strains of SMV, whereas isolines of Rsv3Rsv4 displayed a late susceptible reaction to selected SMV strains. We demonstrate with MAS and three near-isogenic lines, each containing a different SMV-resistance gene, that pyramided lines can be generated in a straightforward manner into two- or three-gene-containing lines with high levels of resistance to SMV.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.2135/cropsci1995.0011183X003500050064xURL [本文引用: 1]

Two hundred soil samples from the A(p) horizon of a reed canarygrass field overlaying several different but related soils in northern Minnesota were analyzed for plant-parasitic nematodes and 22 edaphic factors. Pratylenchus penetrans was the predominant nematode taxon. Others were Aglenchus agricola, Tylenchorhynchus spp., Heterodera trifolii, Paratylenchus spp., Tylenchus maius, and... [Show full abstract]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/jhered/92.1.51URLPMID:11336229 [本文引用: 1]

The gene symbol Rsv2 was previously assigned to the gene in the soybean [Glycine max (L.) Merr.] line OX670 for resistance to soybean mosaic virus (SMV). The Rsv2 gene was reported to be derived from the Raiden soybean (PI 360844) and to be independent of Rsv1. Accumulated data from our genetic experiments were in disagreement with this conclusion. In this study, Raiden and L88-8431, a Williams BC5 isoline with SMV resistance derived from Raiden, were crossed with two SMV-susceptible cultivars to investigate the mode of inheritance of SMV resistance in Raiden. They were also crossed with five resistant cultivars to examine the allelomorphic relationships of the Raiden gene with other reported genes at the Rsv1 locus. F1 plants, F2 populations, and F2-derived F3 (F2:3) lines were tested with SMV strains G1 or G7 in the greenhouse or in the field. The individual plant reactions were classified as resistant (R, symptomless), necrotic (N, systemic necrosis), or susceptible (S, mosaic). The F2 populations from R x S crosses segregated in a ratio of 3 (R + N):1 S and the F2:3 lines from Lee 68 (S) x Raiden (R) exhibited a segregation pattern of 1 (all R):2 segregating:1 (all S). The F2 populations and F2:3 progenies from all R x R crosses did not show any segregation for susceptibility. These results demonstrate that the resistance to SMV in Raiden and L88-8431 is controlled by a single dominant gene and the gene is allelic to Rsv1. The heterozygous plants from R x S and R x N crosses exhibited systemic necrosis when inoculated with SMV G7, indicating a partial dominance nature of the resistance gene. Raiden and L88-8431 are both resistant to SMV G1-G4 and G7, but necrotic to G5, G6, and G7A. Since the resistance gene in Raiden is clearly an allele at the Rsv1 locus and it exhibits a unique reaction to the SMV strain groups, assignment of a new gene symbol, Rsv1-r, to replace Rsv2 would seem appropriate. Further research is ongoing to investigate the possible existence of
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1534/g3.116.035964URL [本文引用: 1]

The soybean cyst nematode (SCN)Heterodera glycinesis a major threat to soybean production, made more challenging by the current limitations of natural resistance for managing this pathogen. The use of resistant host cultivars is effective, but, over time, results in the generation of virulent nematode populations able to robustly parasitize the resistant host. In order to understand how virulence develops in SCN, we utilized a single backcross BC1F2strategy to mate a highly virulent inbred population (TN20), capable of reproducing on all current sources of resistance, with an avirulent one (PA3), unable to reproduce on any of the resistant soybean lines. The offspring were then investigated to determine how virulence is inherited on the main sources of SCN resistance, derived from soybean lines Peking, PI 88788, PI 90763, and the broad spectrum resistance source PI 437654. Significantly, our results suggest virulence on PI 437654 is a multigenic recessive trait that allows the nematode to reproduce on all current sources of resistance. In addition, we examined how virulence on different sources of resistance interact by placing virulent SCN populations under secondary selection, and identified a strong counter-selection between virulence on PI 88788- and PI 90763-derived resistances, while no such counter-selection existed between virulence on Peking and PI 88788 resistance sources. Our results suggest that the genes responsible for virulence on PI 88788 and PI 90763 may be different alleles at a common locus. If so, rotation of cultivars with resistance from these two sources may be an effective management protocol.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[J].
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
