 ,1,2, 华营鹏1,2, 宋海星1,2, 官春云3, 张振华1,2, 周婷
,1,2, 华营鹏1,2, 宋海星1,2, 官春云3, 张振华1,2, 周婷 ,1,2,*
,1,2,*Identification and Bioinformatics Analysis of the PIN Family Gene in Brassica napus
GAO Kun ,1,2, HUA Ying-Peng1,2, SONG Hai-Xing1,2, GUAN Chun-Yun3, ZHANG Zhen-Hua1,2, ZHOU Ting
,1,2, HUA Ying-Peng1,2, SONG Hai-Xing1,2, GUAN Chun-Yun3, ZHANG Zhen-Hua1,2, ZHOU Ting ,1,2,*
,1,2,*通讯作者:
第一联系人:
收稿日期:2018-01-30接受日期:2018-06-12网络出版日期:2018-06-29
| 基金资助: |
Received:2018-01-30Accepted:2018-06-12Online:2018-06-29
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (8218KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
高堃, 华营鹏, 宋海星, 官春云, 张振华, 周婷. 甘蓝型油菜PIN家族基因的鉴定与生物信息学分析[J]. 作物学报, 2018, 44(9): 1334-1346. doi:10.3724/SP.J.1006.2018.01334
GAO Kun, HUA Ying-Peng, SONG Hai-Xing, GUAN Chun-Yun, ZHANG Zhen-Hua, ZHOU Ting.
生长素是具有极性运输特点的植物激素, 主要在植物叶原基、幼叶、根和发育的种子中合成, 通过极性运输到靶细胞调节作物一系列生理反应和生长发育[1]。生长素在细胞间的运输依赖于细胞膜上定位的AUXIN RESISTANT1/LIKE AUX1(AUX/ LAX)和PIN-FORMED(PIN)以及ATP-Binding Cassette subfamily B/P-glycoprotein (PGP/ABCB)生长素运输载体, AUX/LAX属于生长素内向型运输蛋白, 在拟南芥中有AUXIN1(AUX1)、LIKE AUX1 (LAX1)、LAX2和LAX3共4个成员; 而PIN/ PGP/ABCB属于生长素外向型运输蛋白[2]。PIN基因编码的蛋白是最重要的生长素外输载体, 几乎存在于所有单子叶和双子叶植物中[3]。植物PIN1s基因最早在拟南芥中被发现[4], AtPIN1蛋白在细胞膜上呈极性分布, 负责将生长素从胞内运输到胞外[5]。随后发现拟南芥PIN家族蛋白共有8个成员, 命名为PIN1~PIN8[6]。PIN1蛋白在植物发育中具有重要作用, 主要参与胚的发育、叶序和叶脉的形成及维管组织分化, 负责生长素在茎尖分生组织环流及从地上部到根的运输[7]; PIN2蛋白主要参与根的向重力生长, pin2突变体表型为向重力生长缺陷[8]; PIN3s基因主要参与植物的向性生长, 组织化学染色表明其主要在维管组织和黄化苗顶端弯钩表达[9]; PIN4蛋白极性定位在内皮层、皮层和维管组织细胞膜上, 参与生长素向根尖静止中心运输[10]; PIN5蛋白定位在内质网上, 主要负责生长素从胞质到内质网腔的运输[11]; Benková等[12]研究发现, PIN6s基因在整个侧根发育过程中均有表达; PIN7s基因在胚发育早期就有表达, 根尖柱状细胞定位的PIN7蛋白主要参与根的向重力生长[13]; PIN8s基因在花药中特异表达, 调控花粉发育, PIN8蛋白与PIN5蛋白都定位在内质网上, 二者在调控配子体、孢子体发育和植物体内生长素平衡等方面具有拮抗作用[14]; PIN1、PIN2和PIN7蛋白在异源表达系统被证明有向胞外运输生长素的活性, 且该过程不需要其他因子参与[15]。生长素在植物根部有2种运输方式, 一是通过中柱向根尖的向顶运输, 二是根尖生长素经表皮和皮层细胞回运的向基运输[16], PIN1和PIN4蛋白共同调控向顶运输, PIN2和PIN3蛋白参与向基运输[17]。除PIN家族蛋白外, 药物抗性糖蛋白(multidrug-resistance- p-glycoproteins, MDR/PGPs)家族成员MDR1、PGP1、PGP2、PGP4和PGP19也有向胞外输出生长素的功能[18]。
迄今为止, 在玉米、水稻、大豆、小麦等作物上克隆了许多PIN同源基因, 但在甘蓝型油菜中尚未报道。油菜是世界上重要的油料作物, 提供人类所需的主要植物油, 其在保证食用油供给、改善食物结构、促进养殖业和轻纺工业发展等方面具有重要作用[19]。甘蓝型油菜(Brassica napus, AnAnCnCn, ~ 1345 Mb, 2n=4x=38)属于十字花科芸薹属油料作物[20], 是白菜(Brassica rapa, ArAr, ~485 Mb, 2n=2x=20)和甘蓝(Brassica oleracea, CoCo, ~630 Mb, 2n=2x=18)2个二倍体基本种在7500年前通过天然远缘杂交形成的异源四倍体作物, 全基因组包含约十万多个蛋白编码基因[21], 进化、遗传的复杂性在一定程度上影响了各种功能基因在甘蓝型油菜生长发育中的作用。氮是植物生长发育所必需的大量营养元素, 被誉为“生命元素”, 同时, 油菜对氮缺乏极其敏感[22]。已有研究表明, 生长素的极性运输在植物的氮胁迫反应中发挥重要作用[23,24]。目前并不清楚PIN家族基因是如何响应氮素缺乏的。基于上述研究背景, 本研究也开展了甘蓝型油菜PIN家族基因对不同氮素供应水平的转录响应研究。
本研究以拟南芥PIN家族基因为参考序列, 在甘蓝型油菜全基因组数据库中进行了BnPINs家族基因的鉴定, 借助生物信息学方法研究了甘蓝型油菜PIN家族基因的序列信息、分子特征、保守基序特征、染色体定位、系统进化关系、蛋白质结构及低氮胁迫下的表达, 旨在了解甘蓝型油菜PIN家族基因的遗传信息和编码的BnPINs蛋白结构, 为进一步揭示该家族基因的生物学功能奠定研究基础。
1 材料与方法
1.1 试验材料
本研究水培试验所用甘蓝型油菜品种为“湘油15”。1.2 试验方法
1.2.1 甘蓝型油菜PIN家族基因的鉴定 本研究中拟南芥PIN家族蛋白信息参考Paponov[3]分析结果。首先利用AtPINs为原始序列, 使用在线工具Pfam[25] (http://pfam.xfam.org/)和SMART[26] (http:// smart.emblheidelberg.de/)分析AtPINs序列, 分析结果显示AtPINs具有跨膜结构域。其次下载甘蓝型油菜全基因组数据(http://brassicadb.org/brad/), 并利用Bio Edit软件构建本地数据库, 用含有跨膜结构域的AtPINs序列进行本地数据库BLAST检索, 设定检索参数E值<e-6, 手动删除<50个氨基酸和氨基酸同源性<35%的序列, 将鉴定的BnPINs基因编码的氨基酸序列置于Pfam和SMART软件中, 检验候选序列是否具有跨膜结构域。1.2.2 甘蓝型油菜PIN家族基因生物信息学分析
使用ExPASy (http://www.expasy.org/tools)[27]中Prot-Param和Prot Scale工具分析BnPINs蛋白的氨基酸组分和理化性质, 应用TMHMM (http://www. cbs.dtu.dk/services/TMHMM/)[28]预测BnPINs蛋白的跨膜结构。
1.2.3 保守结构域和保守残基的鉴定 使用SMART (http://smart.emblheidelberg.de/)[26]鉴定保守结构域, 并使用Pfam (http://pfam.xfam.org/)注释[25]。使用Vector NTI对保守残基进行多重比对分析。
1.2.4 甘蓝型油菜PIN蛋白保守基序分布信息获取
运用MEME Version 4.9.1 (http://meme.nbcr.net/ meme/cgi-bin/meme.cgi)[29]在线工具分析BnPINs蛋白的保守基序信息, 设置最大基序检索值为10。
1.2.5 甘蓝型油菜PIN家族基因系统进化树构建
运用Clustal X version 2.1[30]对拟南芥、甘蓝、白菜PIN家族蛋白的氨基酸序列和本研究鉴定的BnPINs蛋白氨基酸序列进行多重比对, 使用MEGA6.0[31]分析比对结果并采用邻接法(neighbor- joining, NJ)构建系统进化树, 设定泊松校正法计算进化距离和重复1000次的自展法(bootstrap)检验。
1.2.6 甘蓝型油菜PIN家族基因染色体定位分析
利用Pfam[25] (http://pfam.xfam.org/)对本研究筛选到的BnPINs基因进行结构域判定, 去除冗余基因, 确定位置信息, 利用Circos 0.6[32]构建PIN家族基因染色体分布图谱。
1.2.7 甘蓝型油菜PIN蛋白二级结构分析及三级结构预测 使用2种不同的工具进行二级结构分析: GOR4 (https://npsa-prabi.ibcp.fr/cgibin/page=npsagor4. html/)[33]和PSIPRED (http://bioinf.cs.ucl.ac.uk/ PSIPRED/)[34,35]。使用Phyre2 (http://www.sbg.bio.ic. ac.uk/phyre2/html)[36]预测BnPINs蛋白的三级结构。
1.2.8 低氮胁迫下甘蓝型油菜PIN家族基因时空表达和共表达网络分析
1.2.8.1 营养液培养试验 培养试验在湖南农业大学植物营养与养分高效利用课题组光照培养室进行, 温度设置为22oC, 光照周期为14 h (光照)/10 h (黑暗), 光照强度为300~320 μmol m-2 s-1, 湿度为60%~75%。选取大小一致的油菜种子, 用1%的NaClO灭菌10 min, 将种子表面冲洗干净后, 4oC下用灭菌超纯水(>18.25 MΩ)浸泡24 h, 然后将种子播种到塑料育苗盘固定的纱布上, 育苗盘中加适量超纯水。1周后将长势一致的幼苗移栽至盛有10 L营养液的黑色塑料盆中, 营养液采用Hoagland配方[37]。试验设置2个氮水平: 正常氮(9 mmol L-1 NO3-)和低氮(0.3 mmol L-1 NO3-)。
1.2.8.2 高通量转录组测序 油菜幼苗先在正常氮水平培养10 d, 然后将其移至低氮处理, 地上部(S)和根(R)在0、3和72 h取样, 每个处理准备3个独立的生物学重复。测序平台为Illumina Hiseq 2000[38], 每个样品产生6.0-Gb测序数据, 测序方式为双末端(PE)测序, 读长为150 bp。使用Multiple Experiment Viewer (http://www.tm4.org/mev.html)[39]绘制基因表达谱的热图, 使用错误发现率(FDR)≤0.05和倍数变化(log2)≥1作为鉴定差异表达基因的阈值。
1.2.8.3 基因共表达网络分析 构建基因共表达网络鉴定BnPINs家族基因相互作用的强弱并定位低氮胁迫下连接最相邻的核心基因, 相关值阈值设置为默认参数(http://plantgrn.noble.org/DeGNServer/ Analysis.jsp), 然后用Cytoscapev.3.2.1 (http://www. cytoscape.org/)[40]进行基因共表达网络图谱的绘制。
2 结果与分析
2.1 十字花科作物PIN家族基因拷贝数变异
以AtPINs的氨基酸序列为查询对象, 对NCBI数据库进行BLAST搜索, 在异源四倍体甘蓝型油菜中鉴定了29个BnPINs, 明显多于拟南芥、甘蓝和白菜具有的PIN家族基因数量。然而甘蓝型油菜中PIN1s基因数量远少于甘蓝和白菜中PIN1s的总数, 表明PIN1s在异源多倍体过程中遭受严重的基因损失。29个BnPINs家族基因包括2个BnPIN1s、6个BnPIN2s、4个BnPIN3s、4个BnPIN4s、4个BnPIN5s、2个BnPIN6s、5个BnPIN7s和2个BnPIN8s (图1)。图1
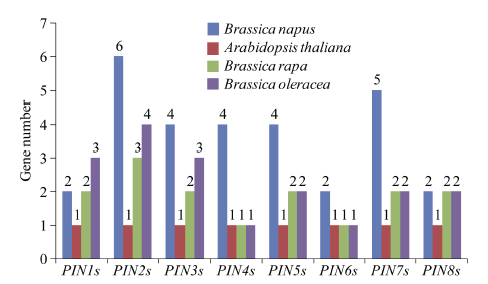 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1甘蓝型油菜、拟南芥、甘蓝、白菜PIN家族基因拷贝数变异
柱状图顶部数值为该物种拷贝的基因数目。
Fig. 1Copy number variation of PIN family genes in Brassica napus, Arabidopsis thaliana, Brassica rapa, and Brassica oleracea
The number at the top of the histogram is the number of genes copied for that species.
2.2 甘蓝型油菜PIN家族基因分子特征
利用AtPINs数据在甘蓝型油菜全基因组数据库中共检索到29条PIN基因序列, 鉴定结果和序列分析表明, 29个BnPINs基因编码的蛋白质平均含有527个氨基酸。氨基酸数目最多的是BnPIN2e蛋白, 长度为643个氨基酸, 预测分子量是69.44 kDa, 理论等电点(pI)为9.24; 氨基酸数目最少的是BnPIN5a蛋白, 长度为348个氨基酸, 预测分子量是38.25 kDa, 理论等电点(pI)为5.96; 29条BnPINs编码的蛋白质等电点变化范围为5.96 (BnPIN5a)~9.38 (BnPIN2f), 大部分编码氨基酸为碱性氨基酸, 仅有BnPIN4d、BnPIN5a、BnPIN5b、BnPIN5c和BnPIN5d蛋白主要由酸性氨基酸组成; BnPIN2e蛋白的外显子/内含子数量最多, 含有13个外显子/12个内含子; 根据亲水性指数介于-0.5~ +0.5为两性蛋白(GRAVY为负值表示亲水性, 正值表示疏水性)的原则[41], 发现仅有BnPIN5a、BnPIN5b、BnPIN5c、BnPIN5d和BnPIN8a、BnPIN8b是疏水性蛋白, 其余均为两性蛋白(表1)。2.3 甘蓝型油菜PIN家族蛋白保守结构域和氨基酸系列比对分析
使用SMART分析BnPINs蛋白保守结构域, 由图2可以看出, 即使BnPIN蛋白保守结构域在同一个类别, 其位置也有差异。29个BnPINs蛋白都具有跨膜结构域, 都在第7~641位的氨基酸残基片段上, BnPIN1a蛋白具有最少的跨膜结构域(5个); BnPIN5d、BnPIN7b、BnPIN7c、BnPIN8b蛋白具有最多的跨膜结构域(均含10个); 绝大部分BnPINs蛋白还有1~2个位于第132~453位氨基酸残基片段的低容量跨膜结构域(除了BnPIN7a蛋白位于第76~88位残基片段上外), 但BnPIN2d、BnPIN3b、BnPIN5a、BnPIN5b、BnPIN5c、BnPIN7c、BnPIN8b蛋白均不含有。通过PIN家族蛋白氨基酸系列比对, 鉴定出BnPINs蛋白高度保守的氨基酸残基, Phe-49、Pro-52、Pro-64、Val-159、Gln-161、Trp-165、Leu-170、Phe-171、Glu-174、Lys-538、Asn-542、Pro-543、Asn-544、Tyr-546、Gly-551、Trp-554、Phe-567、Ser-573、Ser-578、Gly-581、Gly-583、Met-586、Phe-587、Phe-643, 这24个氨基酸残基在所有BnPINs蛋白系列中均高度保守(图3)。这些结果表明, BnPINs蛋白具有参与生长素外向型运输的跨膜结构域。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2甘蓝型油菜PIN家族蛋白结构域
蓝色代表跨膜结构域, 粉色代表低复杂度结构。
Fig. 2Domains architecture of BnPINs
Transmembrane regions are in blue and low complexities are in pink.
图3
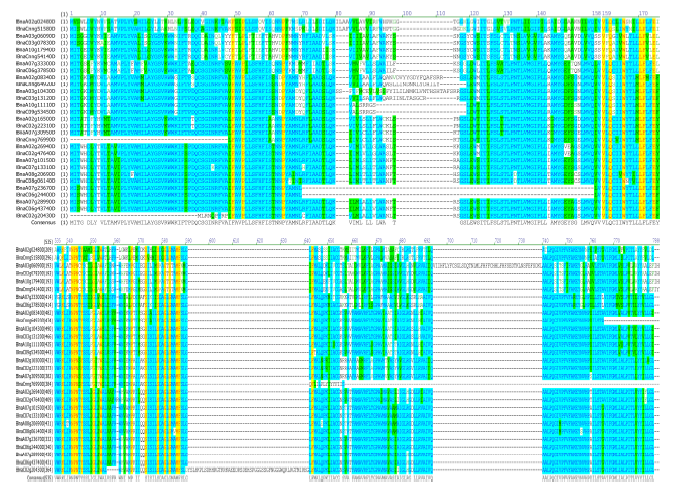 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3甘蓝型油菜PIN家族蛋白氨基酸序列比对和保守结构域分析
黄色区域为高度保守的氨基酸位点。
Fig. 3Amino acid sequence alignment and conserved domain analysis of PIN family in Brassica napus
The yellow area is a highly conserved amino acid site.
2.4 甘蓝型油菜PIN家族基因染色体的位置
染色体定位发现, PIN基因并不是均匀分布在甘蓝型油菜基因组中, 29个BnPINs分布于A/C亚基因组, 13个基因分别位于A亚基因组的A2、A3、A7、A8、A10染色体, 10个基因分别位于C亚基因组的C2、C3、C6、C7、C8、C9染色体。BnPIN8a、BnPIN2f、BnPIN1a、BnPIN4d位于A2染色体, BnPIN5d、BnPIN2e位于A3染色体, BnPIN7e、BnPIN7d、BnPIN4c、BnPIN6b位于A7染色体, BnPIN7c位于A8染色体, BnPIN2d、BnPIN5c位于A10染色体, 而BnPIN3c、BnPIN4b位于C2染色体, BnPIN5b、BnPIN2c位于C3染色体, BnPIN3a、BnPIN3b、BnPIN6a位于C6染色体, BnPIN7b位于C7染色体, BnPIN7a位于C8染色体, BnPIN2b位于C9染色体, 其余6个基因由于基因组拼接及注释不完整具体物理位置尚未确定(图4)。图4
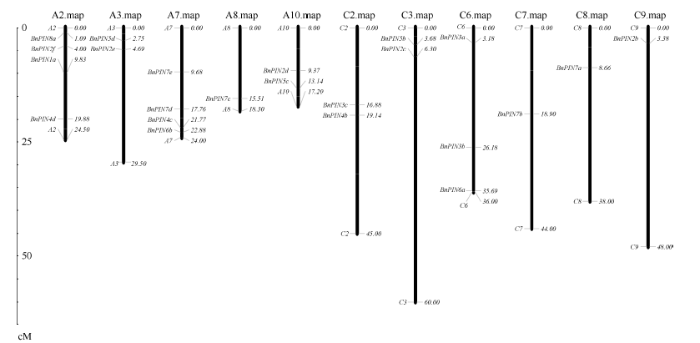 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图423个甘蓝型油菜PIN基因染色体分布
Fig. 4Chromosome mapping of the 23 BnPINs
2.5 甘蓝型油菜PIN家族蛋白保守基序分布
保守基序分析发现, BnPINs蛋白N末端具有保守1 (motif 1)基序。甘蓝型油菜PIN家族蛋白保守基序数量变化具有一定规律, 数量为1~10, 除了BnaC02g20430D、BnaCnng76990D外, 其他序列都含有10个保守基序。BnaCnng54140D、BnaA10g 17940D、BnaC03907830D、BnaA03g06090D、Bna Cnng51580D和BnaA02g02480D缺失8 (Motif 8)基序(图5)。图5
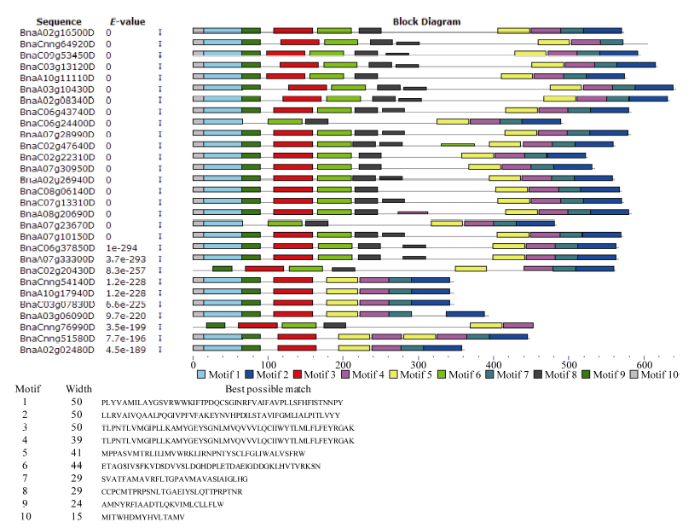 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5甘蓝型油菜PIN家族蛋白保守基序分布
左侧为蛋白系列名称, 右侧不同颜色分别对应不同的保守基序在系列上的位置。
Fig. 5Distribution of conserved motifs in the BnPINs
The different colors on the right correspond to the positions of different conserved motifs on the series, respectively.
2.6 甘蓝型油菜PIN家族基因系统进化分析
利用邻接法构建甘蓝型油菜、拟南芥、甘蓝和白菜PIN家族基因系统进化树。由图6可知: (1)整个进化树可分为两大组, 第一组是PIN1s~PIN4s和PIN7s基因, 第二组为PIN5s、PINs6和PIN8s基因, 该结果与Cazzonelli等[42]将拟南芥PIN蛋白分为PIN1组(AtPIN1~AtPIN4和AtPIN7)和PIN5组(AtPIN5、AtPIN6和AtPIN8)结果一致。(2) BnPINs家族基因与甘蓝和白菜PIN基因进化关系较近, BnPIN2b和BolPIN2a、BnPIN2d和BraPIN2c、BnPIN3a和BolPIN3a、BraPIN7a和BnPIN7c均位于同一个进化分支, BnPIN3b和BraPIN3b进化关系也相近。BnPIN1b、BnPIN4b和BnPIN6b分别与AtPIN1、AtPIN4和AtPIN6基因进化关系密切。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6甘蓝型油菜PIN家族基因系统进化分析
不同颜色代表不同的进化分支, I、II代表PIN家族2个亚组。
Fig. 6Evolutionary relationships of BnPINs taxa
Different clades are in different colors, I and II above clades indicate the two subgroups of PIN family.
2.7 甘蓝型油菜PIN蛋白二级结构分析和三级结构预测
使用GOR4和SPIPRED进行BnPINs蛋白的二级结构分析, GOR4和SPIPRED预测结构类似(图7)。运用Phyre2数据库、分析的保守氨基酸残基和PIN蛋白二级结构对BnPINs蛋白进行三级结构预测(图8)。由BnPIN5b蛋白二级结构和BnPIN2a蛋白三级预测结构可知, α螺旋是BnPINs蛋白的主要结构, BnPIN5b蛋白二级结构特征为, α螺旋>延伸链>无规则卷曲>β转角, 这些结果与PIN蛋白向胞外运输生长素的功能紧密联系。图7
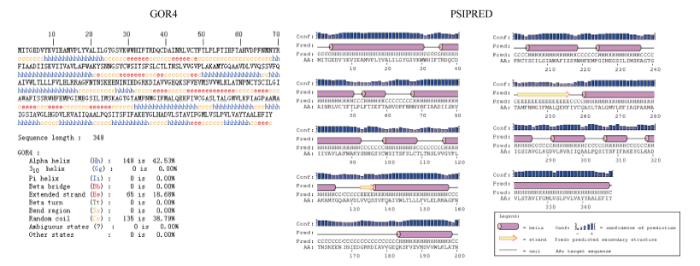 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7甘蓝型油菜BnPIN5b蛋白二级结构分析
GOR4和PSIPRED分别是进行二级结构分析的2种不同的工具。
Fig. 7Secondary structure analysis of BnPIN5b protein
GOR4 and PSIPRED are two tools for the secondary structure analysis.
图8
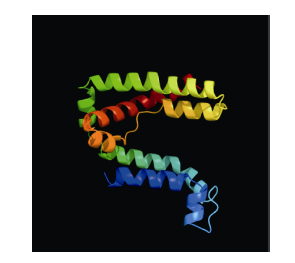 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8甘蓝型油菜BnPIN2a蛋白三级结构预测
三级结构预测该晶体为转运子。
Fig. 8Prediction of three-dimensional structure of BnPIN2a protein
The three-dimensional structure predicts that the crystal is a transporter.
2.8 甘蓝型油菜PIN家族基因在低氮胁迫下时空表达和基因共表达网络分析
由图9可以看出, BnPIN1s、BnPIN2s、BnPIN3s基因主要在甘蓝型油菜根部表达且受长期低氮(72 h)诱导; BnPIN6s和BnPIN8s基因主要在地上部表达, 低氮会抑制BnPIN6s表达。为了进一步确定29个BnPINs中哪一个是核心基因, 又构建了基因共表达网络, BnPIN2a被鉴定为核心基因, 在低氮胁迫下, 29个BnPINs基因在地上部的表达有一定规律(图10-A); 基因表达量几乎都随着低氮胁迫时间延长而先下降后上升, 且在3 h出现最低值, 但在根部未出现这一规律(图10-B)。图9
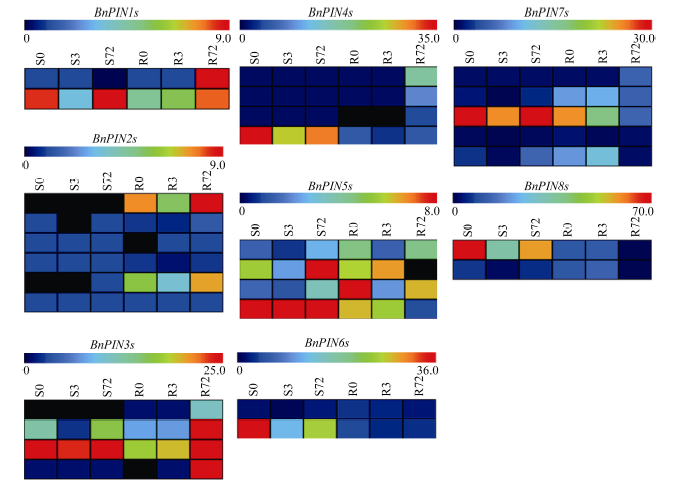 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9低氮胁迫下甘蓝型油菜PIN家族基因转录组分析
S: 地上部; R: 根; 0、3、72 (h)代表缺氮胁迫的时间。图中结果表示3次生物学重复的平均值。
Fig. 9Transcriptome analysis of PIN family genes in Brassica napus under low nitrate concentration conditions
S: shoot; R: root; 0, 3, and 72 (h) represent the time course of nitrogen deficiency. The results represent the average of three biological replicates.
图10
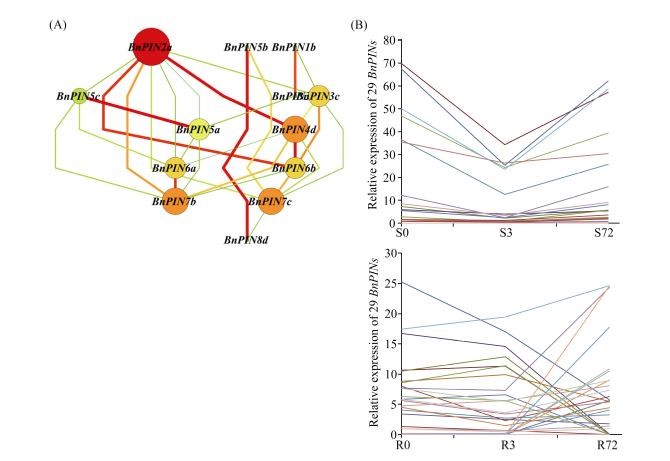 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图10低氮胁迫下甘蓝型油菜PIN家族基因共表达网络分析和表达量分析
A: 共表达网络分析; B: 基因表达量分析; 圆圈节点代表基因, 节点的大小代表节点之间相互作用的能力, 两个节点之间的连线代表基因之间的相互作用; S: 地上部; R: 根; 0、3、72 (h)代表缺氮胁迫的时间。图中结果表示3次生物学重复的平均值。
Fig. 10Co-expression network analysis and expression of PIN family genes in Brassica napus under low nitrate concentration condition
A: co-expression network analysis; B: expression of PIN family genes; cycle nodes represent genes, and the size of the nodes represents the power of the interrelation among the nodes by degree value, edges between two nodes represent interactions between genes; S: shoot; R: root; 0, 3, and 72 (h) represents the time course of nitrogen deficiency. The results represent the average of three biological replicates.
3 讨论
生长素的极性运输与植物生长发育密切相关, 如植物的向光性、向地性、顶端优势、根形成、花发育、胚胎形态建成、维管组织形成等[43]。随着分子生物学和遗传学发展, 在生长素结合蛋白、外输载体和相关基因克隆等领域已取得一系列进展。鉴定了多种拟南芥生长素运输载体, 对其功能和运输机制进行了研究, 而在其他作物中的此类研究十分有限。本研究发现BnPINs家族基因与甘蓝和白菜PIN基因进化关系相近, 但在遗传和进化过程中PIN1s基因遭受严重损失, BnPIN1s基因拷贝数丢失。BnPINs蛋白多为碱性氨基酸组成的稳定蛋白, 定位在细胞质膜上, 具有保守的N末端结构域和5~6次跨膜折叠, 二级结构与AtPIN蛋白相似, 这些结构与BnPINs蛋白向胞外运输生长素的功能紧密相关。因此, 鉴定并获得不同物种PIN家族基因的遗传和进化信息, 了解其编码的PIN蛋白结构, 对于进一步研究PIN家族基因生物学功能和探索生长素外向型运输载体(PIN蛋白)定位机制、影响定位的因素和运输载体之间的相互作用具有重要意义。近年来, PIN蛋白通过调控生长素调控植物逆境胁迫的研究层出不穷, 且在植物响应营养元素缺乏或毒害中也有报道。缺硼时, 拟南芥通过调控PIN1基因的表达调控生长素在根系的分布从而影响根系生长[44]; 磷镁互作时可能通过介导AUX1、PIN2s和PIN3s基因在根中的表达调控根系构型[45]; 超表达水稻PIN2s基因缓解了水稻根系铝毒[46], 延迟了体内磷素由营养器官向生殖器官的转移[47]。
油菜对氮素养分具有较高的营养需求, 但其氮素利用率很低, 缺氮会严重抑制油菜产量和品质的提高[22]。已有研究表明, 缺氮会增加拟南芥主茎中生长素的极性运输水平, 从而增强顶端优势, 造成地上部分枝数减少[23]。氮素缺乏可能通过改变生长素极性运输基因PIN的表达, 从而影响植株的生长表型, 最终影响产量。由此可见, PIN基因与氮素营养的互作在植物生长发育过程中发挥重要作用。本研究利用转录组测序发现, BnPINs基因的表达具有较强组织特异性, BnPIN1s、BnPIN2s、BnPIN3s基因主要在油菜根部表达且受长期低氮(72 h)诱导, BnPIN6s和BnPIN8s基因主要在地上部表达, 低氮会抑制BnPIN6s表达, 反映了PIN家族基因时空表达的特性[48]。上述研究结果为进一步揭示甘蓝型油菜PIN家族基因在低氮胁迫下调控生长素的极性运输作用奠定了研究基础, 为发展氮高效油菜新品种选育提供了新方向。
4 结论
十字花科作物PIN家族基因拷贝数存在差异, 甘蓝型油菜比与它进化关系相近的甘蓝和白菜具有更多PIN同源基因, BnPINs基因分别位于A/C亚基因组的A2、A3、A7、A8、A10、C2、C3、C6、C7、C8、C9染色体。BnPINs蛋白定位在细胞质膜上, 主要是α螺旋结构, 多为两性蛋白, 具有高度保守的氨基酸残基位点、N端保守基序和多个跨膜结构域, 表明BnPINs蛋白可作为细胞质膜上的运输载体参与生长素向胞外运输。BnPINs基因表达具有组织特异性, 低氮胁迫显著改变了BnPINs一些基因的表达丰度, 进而可能影响生长素在作物体内的分配, 最终影响作物生长发育。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
DOI:10.1016/j.tplants.2005.02.009URLPMID:15817418 [本文引用: 2]

It is widely believed that the PIN proteins are crucial for proper cellular coordination. Since the analysis of the Arabidopsis pin-formed mutant in 1991, and the subsequent cloning of AtPIN1, a further seven members of the family have been discovered. Here, we present an overview of this family of auxin efflux facilitators in monocot and dicot plants, summarizing their evolutionary history, expression profiles and, where appropriate, relating them to protein function.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1126/science.282.5397.2226URLPMID:9856939 [本文引用: 1]

Polar auxin transport controls multiple developmental processes in plants, including the formation of vascular tissue. Mutations affecting the PIN-FORMED (PIN1) gene diminish polar auxin transport in Arabidopsis thaliana inflorescence axes. The AtPIN1 gene was found to encode a 67-kilodalton protein with similarity to bacterial and eukaryotic carrier proteins, and the AtPIN1 protein was detected at the basal end of auxin transport-competent cells in vascular tissue. AtPIN1 may act as a transmembrane component of the auxin efflux carrier.
DOI:10.1038/nature03184URLPMID:15635403 [本文引用: 1]

Abstract Local accumulation of the plant growth regulator auxin mediates pattern formation in Arabidopsis roots and influences outgrowth and development of lateral root- and shoot-derived primordia. However, it has remained unclear how auxin can simultaneously regulate patterning and organ outgrowth and how its distribution is stabilized in a primordium-specific manner. Here we show that five PIN genes collectively control auxin distribution to regulate cell division and cell expansion in the primary root. Furthermore, the joint action of these genes has an important role in pattern formation by focusing the auxin maximum and restricting the expression domain of PLETHORA (PLT) genes, major determinants for root stem cell specification. In turn, PLT genes are required for PIN gene transcription to stabilize the auxin maximum at the distal root tip. Our data reveal an interaction network of auxin transport facilitators and root fate determinants that control patterning and growth of the root primordium.
[本文引用: 1]
DOI:10.1016/S0092-8674(02)00656-6URLPMID:11893337 [本文引用: 1]

Abstract In contrast to animals, little is known about pattern formation in plants. Physiological and genetic data suggest the involvement of the phytohormone auxin in this process. Here, we characterize a novel member of the PIN family of putative auxin efflux carriers, Arabidopsis PIN4, that is localized in developing and mature root meristems. Atpin4 mutants are defective in establishment and maintenance of endogenous auxin gradients, fail to canalize externally applied auxin, and display various patterning defects in both embryonic and seedling roots. We propose a role for AtPIN4 in generating a sink for auxin below the quiescent center of the root meristem that is essential for auxin distribution and patterning.
DOI:10.1038/nature08066URLPMID:19506555 [本文引用: 1]

Abstract The plant signalling molecule auxin provides positional information in a variety of developmental processes by means of its differential distribution (gradients) within plant tissues. Thus, cellular auxin levels often determine the developmental output of auxin signalling. Conceptually, transmembrane transport and metabolic processes regulate the steady-state levels of auxin in any given cell. In particular, PIN auxin-efflux-carrier-mediated, directional transport between cells is crucial for generating auxin gradients. Here we show that Arabidopsis thaliana PIN5, an atypical member of the PIN gene family, encodes a functional auxin transporter that is required for auxin-mediated development. PIN5 does not have a direct role in cell-to-cell transport but regulates intracellular auxin homeostasis and metabolism. PIN5 localizes, unlike other characterized plasma membrane PIN proteins, to endoplasmic reticulum (ER), presumably mediating auxin flow from the cytosol to the lumen of the ER. The ER localization of other PIN5-like transporters (including the moss PIN) indicates that the diversification of PIN protein functions in mediating auxin homeostasis at the ER, and cell-to-cell auxin transport at the plasma membrane, represent an ancient event during the evolution of land plants.
DOI:10.1016/S0092-8674(03)00924-3URLPMID:14651850 [本文引用: 1]

Plants, compared to animals, exhibit an amazing adaptability and plasticity in their development. This is largely dependent on the ability of plants to form new organs, such as lateral roots, leaves, and flowers during postembryonic development. Organ primordia develop from founder cell populations into organs by coordinated cell division and differentiation. Here, we show that organ formation in Arabidopsis involves dynamic gradients of the signaling molecule auxin with maxima at the primordia tips. These gradients are mediated by cellular efflux requiring asymmetrically localized PIN proteins, which represent a functionally redundant network for auxin distribution in both aerial and underground organs. PIN1 polar localization undergoes a dynamic rearrangement, which correlates with establishment of auxin gradients and primordium development. Our results suggest that PIN-dependent, local auxin gradients represent a common module for formation of all plant organs, regardless of their mature morphology or developmental origin.
DOI:10.1073/pnas.1013145107URLPMID:21135243 [本文引用: 1]

Auxin is an essential plant-specific regulator of patterning processes that also controls directional growth of roots and shoots. In response to gravity stimulation, the PIN3 auxin transporter polarizes to the bottom side of gravity-sensing root cells, presumably redirecting the auxin flux toward the lower side of the root and triggering gravitropic bending. By combining live-cell imaging techniques with pharmacological and genetic approaches, we demonstrate that PIN3 polarization does not require secretion of de novo synthesized proteins or protein degradation, but instead involves rapid, transient stimulation of PIN endocytosis, presumably via a clathrin-dependent pathway. Moreover, gravity-induced PIN3 polarization requires the activity of the guanine nucleotide exchange factors for ARF GTPases (ARF-GEF) GNOM-dependent polar-targeting pathways and might involve endosome-based PIN3 translocation from one cell side to another. Our data suggest that gravity perception acts at several instances of PIN3 trafficking, ultimately leading to the polarization of PIN3, which presumably aligns auxin fluxes with gravity vector and mediates downstream root gravitropic response.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.1093/aob/mcq202URLPMID:29906712025 [本文引用: 1]

Abstract background and aims: Roots typically respond to localized nitrate by enhancing lateral-root growth. Polar auxin transport has important roles in lateral-root formation and growth; however, it is a matter of debate whether or how auxin plays a role in the localized response of lateral roots to nitrate. METHODS: Treating maize (Zea mays) in a split-root system, auxin levels were quantified directly and polar transport was assayed by the movement of [(3)H]IAA. The effects of exogenous auxin and polar auxin transport inhibitors were also examined. KEY RESULTS: Auxin levels in roots decreased more in the nitrate-fed compartment than in the nitrate-free compartment and nitrate treatment appeared to inhibit shoot-to-root auxin transport. However, exogenous application of IAA only partially reduced the stimulatory effect of localized nitrate, and auxin level in the roots was similarly reduced by local applications of ammonium that did not stimulate lateral-root growth. CONCLUSIONS: It is concluded that local applications of nitrate reduced shoot-to-root auxin transport and decreased auxin concentration in roots to a level more suitable for lateral-root growth. However, alteration of root auxin level alone is not sufficient to stimulate lateral-root growth.
[本文引用: 3]
DOI:10.1093/nar/gku949URLPMID:25300481 [本文引用: 2]

SMART (Simple Modular Architecture Research Tool) is a web resource (http://smart.embl.de/) providing simple identification and extensive annotation of protein domains and the exploration of protein domain architectures. In the current version, SMART contains manually curated models for more than 1200 protein domains, with 200 new models since our last update article. The underlying protein databases were synchronized with UniProt, Ensembl and STRING, bringing the total number of annotated domains and other protein features above 100 million. SMART's 'Genomic' mode, which annotates proteins from completely sequenced genomes was greatly expanded and now includes 2031 species, compared to 1133 in the previous release. SMART analysis results pages have been completely redesigned and include links to several new information sources. A new, vector-based display engine has been developed for protein schematics in SMART, which can also be exported as high-resolution bitmap images for easy inclusion into other documents. Taxonomic tree displays in SMART have been significantly improved, and can be easily navigated using the integrated search engine.
[本文引用: 1]
DOI:10.1056/NEJM199001043220121URL [本文引用: 1]

TMBASE-A database of membrane spanning protein segments HOFMANN K. Biol. Chem. Hoppe-Seyler 374, 166, 1993
URLPMID:7584402 [本文引用: 1]

Abstract The algorithm described in this paper discovers one or more motifs in a collection of DNA or protein sequences by using the technique of expectation maximization to fit a two-component finite mixture model to the set of sequences. Multiple motifs are found by fitting a mixture model to the data, probabilistically erasing the occurrences of the motif thus found, and repeating the process to find successive motifs. The algorithm requires only a set of unaligned sequences and a number specifying the width of the motifs as input. It returns a model of each motif and a threshold which together can be used as a Bayes-optimal classifier for searching for occurrences of the motif in other databases. The algorithm estimates how many times each motif occurs in each sequence in the dataset and outputs an alignment of the occurrences of the motif. The algorithm is capable of discovering several different motifs with differing numbers of occurrences in a single dataset.
[本文引用: 1]
DOI:10.1093/bib/bbn017URLPMID:2562624 [本文引用: 1]

Abstract The Molecular Evolutionary Genetics Analysis (MEGA) software is a desktop application designed for comparative analysis of homologous gene sequences either from multigene families or from different species with a special emphasis on inferring evolutionary relationships and patterns of DNA and protein evolution. In addition to the tools for statistical analysis of data, MEGA provides many convenient facilities for the assembly of sequence data sets from files or web-based repositories, and it includes tools for visual presentation of the results obtained in the form of interactive phylogenetic trees and evolutionary distance matrices. Here we discuss the motivation, design principles and priorities that have shaped the development of MEGA. We also discuss how MEGA might evolve in the future to assist researchers in their growing need to analyze large data set using new computational methods.
[本文引用: 1]
[本文引用: 1]
DOI:10.1006/jmbi.1999.3091URLPMID:10493868 [本文引用: 1]

A two-stage neural network has been used to predict protein secondary structure based on the position specific scoring matrices generated by PSI-BLAST. Despite the simplicity and convenience of the approach used, the results are found to be superior to those produced by other methods, including the popular PHD method according to our own benchmarking results and the results from the recent Critical Assessment of Techniques for Protein Structure Prediction experiment (CASP3), where the method was evaluated by stringent blind testing. Using a new testing set based on a set of 187 unique folds, and three-way cross-validation based on structural similarity criteria rather than sequence similarity criteria used previously (no similar folds were present in both the testing and training sets) the method presented here (PSIPRED) achieved an average Q3 score of between 76.5% to 78.3% depending on the precise definition of observed secondary structure used, which is the highest published score for any method to date. Given the success of the method in CASP3, it is reasonable to be confident that the evaluation presented here gives a fair indication of the performance of the method in general. Copyright 1999 Academic Press.
[本文引用: 1]
DOI:10.1016/j.jmb.2015.10.017URLPMID:26517951 [本文引用: 1]

61We present PhyreStorm, a Web server for fast and comprehensive structural searching.61The PDB is grouped into clusters to vastly reduce the number of alignments required.61PhyreStorm usually takes less than a minute to align a query to the PDB.61PhyreStorm has an average coverage of 93%.
DOI:10.1016/S0140-6736(00)73482-9URL [本文引用: 1]

This is a revised edition of a popular account issued in 1938 [H.A., 10: 28] based on the investigations of the two authors. Since then, experience in the U.S.A. and elsewhere has failed, in the authors' opinion, to support the early exaggerated claims for the value of the technique. Their experience leads to the conclusion that for its successful operation a knowledge of plant physiology is es...
[本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
