Transcription Abundances of CRY1b and CRY2 Genes in Response to Different Light Treatments in Maize
LI Hong-Dan1,2, YAN Lei1,2, SUN Lei1,2, FAN Xiao-Cong1,3, CHEN Shi-Zhan1,3, ZHANG Yan1,3, GUO Lin1, YOU Guang-Xia1, LI Zhuang1,2, YANG Zong-Ju1,2, SU Liang1,*, YANG Jian-Ping1,3,*通讯作者:
第一联系人:
收稿日期:2018-01-25接受日期:2018-06-12网络出版日期:2018-07-02
| 基金资助: |
Received:2018-01-25Accepted:2018-06-12Online:2018-07-02
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (11431KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李红丹, 闫蕾, 孙蕾, 樊晓聪, 陈士瞻, 张燕, 郭林, 游光霞, 李庄, 杨宗举, 苏亮, 杨建平. 玉米隐花色素CRY1b和CRY2基因转录丰度对不同光质处理的响应[J]. 作物学报, 2018, 44(9): 1290-1300. doi:10.3724/SP.J.1006.2018.01290
LI Hong-Dan, YAN Lei, SUN Lei, FAN Xiao-Cong, CHEN Shi-Zhan, ZHANG Yan, GUO Lin, YOU Guang-Xia, LI Zhuang, YANG Zong-Ju, SU Liang, YANG Jian-Ping.
环境中光的辐照度、方向与光质能显著影响植物的株高、株型、产量、品质等[1,2,3,4,5]。光照不仅是植物进行光合作用的能量来源, 更能调控其生长与发育,如光形态建成等。在长期的进化中, 植物逐步形成三大类光受体, 光敏色素(phytochrome), 主要负责感知红光(波长为620~700 nm)和远红光(700~800 nm); 隐花色素(cryptochrome)和向光素(phototropin), 主要感知蓝光(380~500 nm)和UV-A (320~380 nm); UVR8, 能特异性地吸收UV-B (280~ 320 nm)[6,7,8]。其中, 隐花色素是一种分子量为70~80 kD的黄素类蛋白, 有氨基端PHR和羧基端CCE两个明显的功能域, 其氨基端的PHR非共价地结合生色团FAD和叶酸[6]。隐花色素是一种类光解酶的光受体, 参与了动物、植物乃至微生物中的光反应及生物钟的调控[8]。通过研究在不同条件下玉米隐花色素的表达模式, 可为进一步研究其对玉米光形态建成的作用, 进而认知玉米光的反应能力奠定基础。
在拟南芥[9,10]、番茄[2-3,11]、苹果[12]、水稻[13,14]、豌豆[4,15]、蕨类[16]、苔藓[5,17]、大豆[18,19]、甘蓝型油菜[20]等植物中的研究表明, 隐花色素不仅控制下胚轴的伸长、光周期诱导的开花及生物钟的输出, 还参与了向性生长、顶端优势、细胞程序性死亡、种子休眠、根的发育、气孔开张等过程。模式植物拟南芥中存在3种类型的隐花色素, 即CRY1、CRY2和CRY3。在蓝光下, CRY1和CRY2负责调控植物的去黄化与开花期。在蓝光下, CRY1通过抑制COP1/SPA1的E3泛素连接酶活性, 导致正向调控因子HY5的积累, 从而促进光形态建成并抑制下胚轴的伸长, 而cry1突变体则表现为下胚轴伸长且子叶生长缓慢[21,22]。CRY2在蓝光下与互作蛋白CIB结合促进开花, cry2双突变体开花要晚于野生型[23]。CRY1在细胞核和细胞质中来回穿梭[24], CRY2在细胞核中行使功能[25], 而CRY3则主要作为光受体和单链DNA修复酶在线粒体和叶绿体中发挥作用[26]。番茄的隐花色素基因LeCRY1和LeCRY2能促进叶片中花青素苷和叶绿素的积累、促进腋分枝、缩短节间、延迟开花、促进果实中类黄酮和番茄红素的积累[2-3,11]。玉米隐花色素至少存在4个拷贝, 即CRY1a1、CRY1a2、CRY1b和CRY2[27]。玉米ZmCRY1a1基因在白光、红光、蓝光和远红光等条件下的转录丰度存在明显差异, ZmCRY1a2基因对各种光质的响应则比较相似, 暗示着ZmCRY1a1和ZmCRY1a2存在功能上的分工, 可能参与玉米在红光和远红光下的光形态建成的调控[27]。
迄今, 双子叶模式生物拟南芥中隐花色素的研究已较为深入, 而玉米、水稻、马铃薯、小麦等禾本科单子叶作物中隐花色素的研究仅略有涉及, 对于隐花色素是否参与玉米的光形态建成以及是否调控玉米的光温反应也不清楚。通过比较玉米ZmCRY1b和ZmCRY2基因响应不同光质及光周期处理的表达模式, 了解二者在不同条件下的表达差异, 比较二者响应光的能力, 可进一步丰富植物隐花色素基因的研究, 并可为作物的开花期、避荫性、株高、产量等重要农艺性状的改良提供指导意见。因此, 研究玉米ZmCRY1b和ZmCRY2基因在不同光质及光周期处理下的表达模式对于了解玉米对光质及光周期的敏感性、开花机理及玉米的引种和高产栽培都具有重要的理论和实践意义。
1 材料与方法
1.1 试验材料及样品处理
1.1.1 试验材料 以玉米自交系B73的种子为试验材料(B73材料由中国农业科学院作物科学研究所李新海博士赠予)。以上材料适时取样,迅速置于液氮冻存,-80℃保藏备用。1.1.2 组织特异性表达样品的准备 将B73的种子播种于自然条件下一定阶段后, 分别取幼苗的根、茎、叶、叶枕、叶鞘、花丝、花柄、雄花、苞叶和幼穗等部位为材料。
1.1.3 各种持续光质处理 将B73的种子放在28℃, 黑暗(Dk)、蓝光(B, 13 μmol m-2 s-1)、白光(W, 17 μmol m-2 s-1)、红光(R, 22.3 μmol m-2 s-1)和远红光(FR, 0.25 μmol m-2 s-1)等培养箱中, 培养13 d。取幼苗地上部为材料。
1.1.4 黑暗转换其他光质处理 将B73的种子放在28℃、黑暗中生长13 d。一批分别转入白光(W, 17 μmol m-2 s-1)、红光(R, 22.3 μmol m-2 s-11)以及蓝光(B, 13 μmol m-2 s-1)等光质条件下, 并分别于0.25 h、0.5 h、1 h、2 h、4 h、8 h、12 h和24 h时对幼苗地上部分取样。另一批转入远红光(FR, 0.25 μmol m-2 s-1)条件下并于0.16 h、0.25 h、0.5 h、1 h、1.5 h、2 h、3 h、4 h、6 h、8 h、10 h、12 h、24 h和24.08 h时对幼苗地上部取样。
1.1.5 长日照以及短日照处理 将B73种子分别放在28℃, 长日照培养箱(LD, 16 h光照/8 h黑暗)或者短日照培养箱(SD, 8 h光照/16 h黑暗)等条件下生长13 d, 每2 h取一次样, 光照和黑暗互相转换时, 在每个取样节点前5 min对幼苗地上部取样。
1.2 酶、试剂和载体
使用TaKaRa宝生物工程(大连)有限公司生产的PrimeSTAR HS酶和DNase I (RNase-free)酶, Invitrogen (USA)公司生产的TRIzol; Thermo Scientific公司生产的Revert Aid First Strand cDNA Synthesis Kit; TaKaRa宝生物工程(大连)有限公司生产的定量PCR试剂SYBR Premix Ex Taq II, 北京全式金生物技术有限公司生产的pEASY-Blunt-Simple载体。1.3 RNA提取及cDNA合成
用TRIzol法提取各种处理条件下的玉米B73幼苗的总RNA, 用DNase I酶处理并以此为模板, 利用RevertAid First Strand cDNA Synthesis Kit并以Oligo-dT18引物将其反转录成单链cDNA, 保存于-80℃冰箱备用。1.4 ZmCRY1b和ZmCRY2基因的克隆
于NCBI 网站(http://www.ncbi.nlm.nih.gov/)查找ZmCRY1b (ZM02G13620)和ZmCRY2 (ZM09G 09240)序列并设计引物(表1), 以黑暗条件下生长13 d的B73幼苗的cDNA为模板, 经PCR扩增后连接pEASY-Blunt-Simple载体, 经过菌落PCR和双酶切鉴定后, 交北京奥科鼎盛生物科技有限公司测序。PCR程序为94℃ 5 min; 98℃ 10 s, 58℃ 15 s, 72℃ 2 min, 25个循环; 72℃ 10 min。Table 1
表1
表1基因克隆所用引物
Table 1
| 基因名称 Gene name | 基因编号 Accession number | 正向引物序列 Forward primer sequence (5'-3') | 反向引物序列 Reverse primer sequence (5'-3') |
|---|---|---|---|
| ZmCRY1b | ZM02G13620 | CACCGCCTGATGAACTGGA | ATGGATGAGTAGTTCAGTGGACAA |
| ZmCRY2 | ZM09G09240 | TCTGGTTATTGTCATATTGCAGTTCT | TGCCAAGATGTTCCCTTTGAGT |
新窗口打开|下载CSV
1.5 玉米实时荧光定量RT-PCR(qRT-PCR)分析
以不同处理条件下的玉米自交系B73的cDNA第1链为模板, 玉米Tubulin基因为内参基因, 用软件Primer Premier 5.0设计RT-PCR引物(表2)。使用Roche 480荧光定量PCR仪, PCR程序为95℃ 30 s; 95℃ 5 s, 60℃ 20 s, 72℃ 10 s, 50个循环; 60~95℃绘制溶解曲线。采用的定量PCR试剂为SYBR Premix Ex Taq II, 用2-ΔΔCT的方法计算实验结果[28], 经过3次独立的生物学重复后, 计算其标准差。Table 2
表2
表2qRT-PCR所用引物
Table 2
| 基因名称 Gene name | 基因编号 Accession number | 正向引物序列 Forward primer sequence (5'-3') | 反向引物序列 Reverse primer sequence (5'-3') |
|---|---|---|---|
| ZmCRY1b | ZM02G13620 | CACCGCCTGATGAACTGGA | ATGGATGAGTAGTTCAGTGGACAA |
| ZmCRY2 | ZM09G09240 | GAACCACAGGCGAGATGCT | GATCACTACAAACGCACCAGC |
| ZmTubulin | NM_001174192 | ACTTCATGCTTTCGTCCTACGCTCCA | CTGGGAGGCTGGTAGTTGATTC |
新窗口打开|下载CSV
1.6 ZmCRY1b和ZmCRY2蛋白的序列比对、结构域分析与系统发育分析
使用NCBI网站(2 结果与分析
2.1 ZmCRY蛋白和禾本科其他植物CRY蛋白有相同的结构域和较高的同源性
通过RT-PCR方法得到ZmCRY1b和ZmCRY2基因的全长cDNA序列, 其ORF分别含有2067和1965个核苷酸残基, 编码蛋白质的分子式分别为C3464H5295N997O1009S18和C3316H5116N894O990S28, 分别含有688个和654个氨基酸残基, 蛋白质分子量分别为77.6 kD和74.2 kD, 等电点分别为5.89和5.54。利用NCBI网站和SMART网站对拟南芥及小麦、水稻和玉米的CRY蛋白进行结构域分析发现, 四者的CRY1蛋白都包含一个位于N端的PHR (Photolyase Homologous Region)结构域(由1个DNA photolyase结构域以及1个FAD binding 7 结构域组成)和1个位于C端的CCE (Cryptochrome C-terminal Extension)结构域(图1); 而CRY2蛋白则仅包含1个PHR结构域(图2)。玉米CRY1和CRY2蛋白结构域的不同暗示着其功能有异。用NCBI网站获得模式植物拟南芥(Arabidopsis thaliana)、玉米(Zea mays L.)、小麦(Triticum aestivum)、水稻(Oryza sativa L.)、大豆(Giycine max)、番茄(Solanum lycopersicum)等植物的CRY蛋白序列, 用DNAMAN进行系统发育分析发现, ZmCRY1b蛋白与拟南芥、小麦和水稻CRY1蛋白序列的一致性分别为58.6%、72.3%和82.9%; ZmCRY2蛋白与拟南芥、小麦和水稻CRY2蛋白序列的一致性分别为50.2%、73.5%和73.7% (图3)。说明ZmCRY蛋白与禾本科其他植物的一致性更高, 预示它们有更相似的功能。
图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1玉米与拟南芥、小麦和水稻的CRY1蛋白等常见作物的氨基酸序列比对及结构域分析
使用NCBI网站和DNAMAN对其进行序列比对; 使用SMART网站对其进行结构域分析。AtCRY1: 拟南芥CRY1, AAB28724; OsCRY1a: 水稻CRY1a, BAB70686; OsCRY1b: 水稻CRY1b, BAB70688; TaCRY1a: 小麦CRY1a, ABX58028; ZmCRY1b: 玉米CRY1b, ZM02G13620。图中黑色、深灰色、浅灰色及白色分别代表一致性为100%、75%、50%和0。
Fig. 1Multiple sequence alignments and function domains of CRY1 proteins from Zea mays, Arabidopsis thaliana, Oryza sativa, and Triticum aestivum
Multiple sequence alignments at amino acid levels were analyzed by NCBI and DNAMAN, and the function domains were analyzed by SMART. AtCRY1: Arabidopsis thaliana CRY1, AAB28724; OsCRY1a: Oryza sativa CRY1a, BAB70686; OsCRY1b: Oryza sativa CRY1b, BAB70686; TaCRY1a: Triticum aestivum CRY1a, ABX58028; ZmCRY1b: Zea may CRY1b, ZM02G13620. Black, dark grey, light grey, and white in the picture stand for 100%, 75%, 50%, and 0 of consistency, respectively.
图2
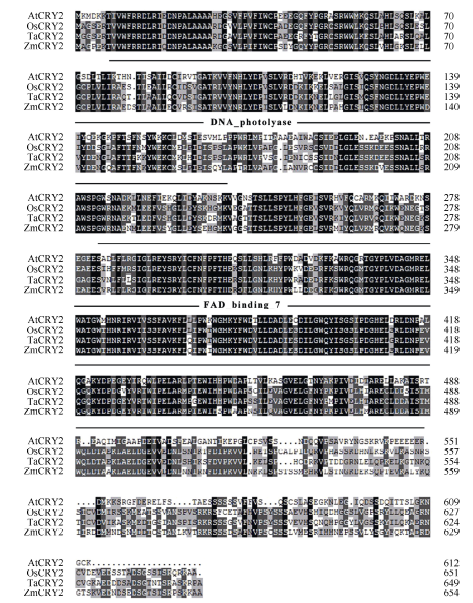 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2玉米与拟南芥、小麦、水稻等常见作物的CRY2蛋白的氨基酸序列比对和结构域分析
使用NCBI网站和DNAMAN对其进行序列比对; 使用SMART网站对其进行结构域分析。AtCRY2: 拟南芥CRY2, AT1G04400.2; OsCRY2: 水稻CRY2, CAC82538.1; TaCRY2: 小麦CRY2, ABX58030.1; ZmCRY2: 玉米CRY2, ZM09G09240。图中黑色、深灰色、浅灰色及白色分别代表一致性为100%、75%、50%和0。
Fig. 2Multiple sequence alignments and function domains of CRY2 proteins from Zea mays, Arabidopsis thaliana, Oryza sativa, and Triticum aestivum
Multiple sequence alignments at amino acid levels were analyzed by NCBI and DNAMAN, and the function domains were analyzed by SMART. AtCRY2: Arabidopsis thaliana CRY2, AT1G04400.2; OsCRY2: Oryza sativa CRY2, CAC82538.1; TaCRY2: Triticum aestivum CRY2, ABX58030.1; ZmCRY2: Zea may CRY2, ZM09G09240. Black, dark grey, light grey, and white in the picture stand for 100%, 75%, 50%, and 0 of consistency, respectively.
2.2 ZmCRY1b和ZmCRY2器官特异性表达分析
基因的表达具有时空特异性, 为研究隐花色素基因在玉米生长和发育过程中的作用, 我们通过qRT-PCR的方法分析了ZmCRY1b和ZmCRY2两个基因在玉米的根、茎、叶、叶枕、叶鞘、花丝、花柄、雄花、苞叶、幼穗等器官中表达丰度的差异。将ZmCRY2基因在根中的表达丰度设为1, 并以此为对照。结果发现ZmCRY1b基因在花丝、幼穗、花柄和雄花和中的表达量较低(为根部ZmCRY2丰度的0.1~0.4倍, P<0.01), 在茎和苞叶中表达丰度与其根中ZmCRY1b的表达丰度相似(1.3~1.9倍), 在叶枕和叶鞘中表达丰度稍高(根中ZmCRY1b表达量的2.5~3.2倍, 差异极显著), 而在叶片中的表达丰度达到最大值, 是根中ZmCRY1b表达量的31.3倍(图4)。ZmCRY2基因在幼穗和苞叶中的表达丰度也比较高(1.1~1.4倍), 且叶片中表达丰度最高(达到根中ZmCRY2的10.4倍, P<0.01), 而在其他器官中的表达丰度仅为根中ZmCRY1b的0.3~0.9倍, 具有显著差异(图4)。说明叶片是ZmCRY1b和ZmCRY2基因在玉米中的主要作用部位, 且ZmCRY1b的表达丰度普遍高于ZmCRY2, 暗示着ZmCRY1b在玉米中起着更重要的作用。
图3
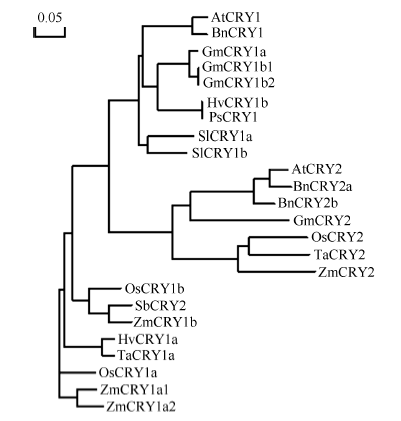 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3玉米与拟南芥、小麦和水稻等常见作物的CRY蛋白在氨基酸水平的系统发育分析
使用NCBI网站获得全长氨基酸系列, 使用DNAMAN对其进行系统发育分析。AtCRY1: 拟南芥, AAB28724; AtCRY2: 拟南芥, AT1G04400.2; BnCRY1: 甘蓝型油菜, CAG28805; BnCRY2a: 甘蓝型油菜, AEA29690.1; BnCRY2b: 甘蓝型油菜油菜, AEA29691.1; GmCRY1a: 大豆, DQ401046; GmCRY1b1: 大豆, AB498929; GmCRY1b2: 大豆, AB498930; GmCRY2: 大豆, XP_006588364.1; OsCRY1a: 水稻, BAB70686; OsCRY1b: 水稻, BAB70688; OsCRY2: 水稻CAC82538.1; HvCRY1a: 大麦, ABB13328; HvCRY1b: 大麦, ABB13331; TaCRY1a: 小麦, ABX58028; TaCRY2: 小麦, ABX58030.1; PsCRY1: 豌豆, AAS79662; SbCRY2: 高粱, AAN37909; SlCRY1a: 番茄, AAD44161; SlCRY1b: 番茄, AAL02092; ZmCRY1a1: 玉米, ZM05G31560; ZmCRY1a2: 玉米, ZM04G17060; ZmCRY1b: 玉米, ZM02G13620; ZmCRY2: 玉米, ZM09G09240。
Fig. 3Phylogenetic analysis of amino acid sequences of ZmCRY proteins and other related CRY proteins
The amino acid sequences were obtained from the NCBI and the neighbor-joining tree was constructed by the DNAMAN. AtCRY1; Arabidopsis thaliana CRY1, AAB28724; AtCRY2: Arabidopsis thaliana CRY2, AT1G04400.2; BnCRY1: Brassica napus CRY1, CAG28805; BnCRY2a: Brassica napus CRY2a, AEA29690.1; BnCRY2b: Brassica napus CRY2b, AEA29691.1; GmCRY1a: Glycine max CRY1a, DQ401046; GmCRY1b1: Glycine max CRY1b1, AB498929; GmCRY1b2: Glycine max CRY1b2, AB498930; GmCRY2: Glycine max CRY2, XP_006588364.1; OsCRY1a: Oryza sativa japonica Group CRY1a, BAB70686; OsCRY1b: Oryza sativa japonica Group CRY1b, BAB70688; OsCRY2: Oryza sativa japonica Group CRY2, CAC82538.1; HvCRY1a: Hordeum vulgare subsp. Vulgare CRY1a, ABB13328; HvCRY1b: Hordeum vulgare subsp. Vulgare CRY1b, ABB13331; TaCRY1a: Triticum aestivum CRY1a, ABX58028; TaCRY2: Triticum aestivum CRY2, ABX58030.1; PsCRY1: Pisum sativum CRY1, AAS79662; SbCRY2: Sorghum bicolor CRY2, AAN37909; SlCRY1a: Solanum lycopersicum CRY1a, AAD44161; SlCRY1b: Solanum lycopersicum CRY1b, AAL02092; ZmCRY1a1: Zea may CRY1a1, ZM05G31560; ZmCRY1a2: Zea may CRY1a2, ZM04G17060; ZmCRY1b: Zea may CRY1b, ZM02G13620; ZmCRY2: Zea may CRY2, ZM09G09240.
图4
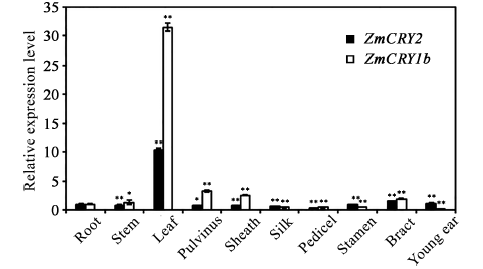 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4ZmCRY1b和ZmCRY2基因在不同器官中的相对表达水平分析
分别取于自然条件下生长一定阶段的B73幼苗的不同器官(根、茎、叶、雄花、叶枕、叶鞘、花丝、幼穗、花柄和苞叶)为材料用于qRT-PCR分析。以根中的ZmCRY1b的转录丰度为对照, 将该ZmCRY1b/Tubulin的比值设为1。柱状图显示了在3次独立的生物学重复下ZmCRY1b/Tubulin比值的平均值, 其中, 误差线代表了标准差。用星号表示差异的显著性, * P < 0.05, ** P < 0.01。
Fig. 4Relative expression analysis of both ZmCRY1b and ZmCRY2 genes in different organs of maize
Different tissues of maize inbred line B73, including root, stem, leaf, stamen, pulvinus, sheath, pedicel, young ear, pistil and bract, were harvested for qRT-PCR assays. The transcription abundance of ZmCRY1b in the root was set as the control, and the ratio of ZmCRY1b/Tubulin in the root was set as 1. Each column shows the mean expression of ZmCRY1b/Tubulin of three biological repeats. Error bars indicate the standard deviation. * P < 0.05, ** P < 0.01.
2.3 ZmCRY1b和ZmCRY2基因转录丰度对不同持续光质处理的响应
为比较ZmCRY1b和ZmCRY2对各种光质响应的差异, 采用qRT-PCR的方法分析了玉米B73幼苗在黑暗(Dk)和持续远红光(FR)、红光(R)、蓝光(B)、白光(W)等条件下ZmCRY1b和ZmCRY2的表达丰度。将ZmCRY2在黑暗条件下的表达量设为1, 并以此为对照。结果发现, ZmCRY1b和ZmCRY2基因对各种持续光质均有较强烈的响应, 尤其是对持续白光的响应最为强烈, 分别为黑暗下ZmCRY2基因表达量的48.0倍和3.7倍(图5, P<0.01)。ZmCRY1b和ZmCRY2基因在持续的远红光下的表达丰度也比较高, 分别为黑暗下ZmCRY2丰度的29.3倍和2.4倍(差异极显著)。在蓝光和红光下, ZmCRY2接近自身黑暗下的丰度, 而ZmCRY1b则为黑暗下ZmCRY2的11.9倍和14.7倍(P<0.01)。结果说明玉米ZmCRY1b和ZmCRY2在远红光、红光、蓝光和白光中均有作用, 而ZmCRY1b的表达丰度普遍高于ZmCRY2, 暗示在各种持续光质下ZmCRY1b更重要。图5
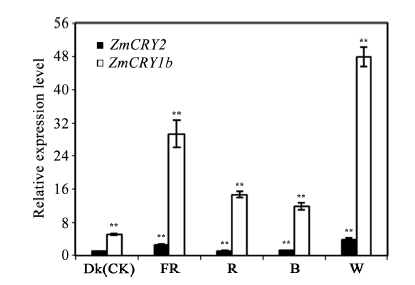 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5ZmCRY1b和ZmCRY2基因对响应不同光质的相对表达水平分析
玉米自交系“B73”的幼苗在黑暗、持续远红光(FR, 0.25 μmol m-2 s-1)、持续红光(R, 22.3 μmol m-2 s-1)、持续蓝光(B, 13 μmol m-2 s-1)或持续白光(W, 17 μmol m-2 s-1)下生长了13 d。以黑暗条件下的ZmCRY2的转录丰度为对照, 并将该ZmCRY2/Tubulin的比值设为1。柱状图代表了在3次独立的生物学重复下ZmCRY2/Tubulin的平均值, 误差线代表了标准差。用星号表示差异的显著性, * P< 0.05, ** P < 0.01。
Fig. 5Relative expression analysis of both ZmCRY1b and ZmCRY2 genes under different continuous light conditions
The seedlings of maize inbred line B73 were grown in continuous far-red light (FR, 0.25 μmol m-2 s-1), red light (R, 22.3 μmol m-2 s-1), blue light (B, 13 μmol m-2 s-1), or white light (W, 170 μmol m-2 s-1) for 13 d. The transcription abundance of ZmCRY2 in the dark was set as the control, and the ratio of ZmCRY2/Tubulin in the dark was set as 1. Each column shows the mean expression of ZmCRY2/Tubulin of three biological repeats. Error bars indicate the standard deviation. * P < 0.05, ** P < 0.01.
2.4 ZmCRY1b和ZmCRY2对黑暗到不同光质转换的响应
将黑暗条件下生长13 d的 B73幼苗依次转入到红光、蓝光和白光下0.25 h、0.5 h、1 h、2 h、4 h、8 h、12 h和24 h以及远红光下0.16 h、0.25 h、0.5 h、1 h、1.5 h、2 h、3 h、4 h、6 h、8 h、10 h、12 h、24 h和24.08 h后取材, 用于进一步研究ZmCRY1b和ZmCRY2的表达丰度响应不同光质的差异。将ZmCRY2基因在黑暗条件下的表达量设为1, 并以此为对照。在由黑暗转换到蓝光0.5 h内, ZmCRY1b的表达丰度持续上升并达到最大值(为黑暗时ZmCRY2的42.9倍)(图6-A); 随后在2 h时下降到自身黑暗条件下的水平并保持平缓; 最终在24 h时其转录丰度又回升至黑暗时ZmCRY2的5.4倍。从黑暗转换至蓝光时, ZmCRY2基因的表达模式与ZmCRY1b比较类似, 只是整体表达丰度为ZmCRY1b的12%~55%, 且在由黑暗转到蓝光12~24 h内, ZmCRY1b的表达丰度略有下降, 而ZmCRY2平缓上升。在由黑暗转换到蓝光0.5 h内, ZmCRY2的表达丰度达到峰值(为自身黑暗时的13倍); 最后在24 h时其丰度又回升到自身黑暗时的2.15倍。结果表明在由黑暗到蓝光光质转换的响应中ZmCRY1b比ZmCRY2更强烈。
ZmCRY1b在由黑暗转换到白光的0.25 h时表达丰度出现最大峰值(为黑暗时ZmCRY2的24.7倍)(图6-B); 之后在1 h时下降至黑暗时ZmCRY2的0.29倍并保持平缓; 在4 h时达到第2个峰值(为黑暗时ZmCRY2的3.4倍); 最终在24 h时其转录丰度上升至黑暗时ZmCRY2的5.3倍。由黑暗转换至白光时, ZmCRY2基因的表达模式与ZmCRY1b比较类似, 但整体表达丰度为ZmCRY1b的20%~55%。在0.25 h时同样出现最大峰值, 为自身黑暗时的5.9倍; 在1 h 时下降至自身黑暗时的0.18倍; 在4 h时达到第2个峰值(为自身黑暗时的0.96倍); 在24 h时上升为自身黑暗时的0.9倍(图6-B)。说明在由黑暗到白光光质转换的响应中ZmCRY1b比ZmCRY2更强烈。
尽管编码的是蓝光受体, 但ZmCRY1b和ZmCRY2基因在持续的远红光和红光光质下也有着相对较高的转录丰度(图5), 于是进一步检测了其对红光和远红光的响应(图6-C和D)。结果发现, 在由黑暗转换至红光时, ZmCRY1b基因的表达丰度维持在较低的水平, 并于1 h和4 h时达到峰值, 分别为黑暗时ZmCRY2的4.8倍和14.2倍(图6-C); 在8 h时下降至2.2倍, 此后开始上升, 并于24 h时达到黑暗时ZmCRY2的58.0倍。在黑暗转换到红光时ZmCRY2表达模式与ZmCRY1b较类似, 但整体表达丰度为ZmCRY1b的12%~55%。在1 h和4 h时达到峰值, 分别为自身黑暗时的1.4倍和2.2倍; 在8 h时下降至自身黑暗时的0.6倍, 并于24 h时达到自身黑暗时的7.4倍。结果说明在由黑暗到红光光质转换的响应中ZmCRY1b也比ZmCRY2更强烈。
在由黑暗转到远红光时, ZmCRY2的丰度整体低于ZmCRY1b, 为其表达丰度的15%~55% (图6-D)。在由黑暗转到远红光0.5 h时, ZmCRY1b的表达丰度下降至黑暗时ZmCRY2的1.8倍; 此后分别于1.5 h、4 h、8 h和24 h时达到峰值(分别为黑暗时ZmCRY2的4.3、8.4、3.0和16.4倍)。ZmCRY2的表达模式与ZmCRY1b整体类似但略有不同: 在0.25 h时下降至自身黑暗时的0.67倍, 此后开始上升并于1 h时达到峰值(自身黑暗时的1.7倍); 此后分别于4 h、8 h和24 h时再次达到峰值(为自身黑暗时的1.3、0.9和1.0倍)。不同之处在于ZmCRY2在0.25~0.50 h内表达丰度上升, 在1.0~1.5 h内下降, 在2~3 h内下降, 这三处与ZmCRY1b正好相反。结果表明在黑暗到远红光转换光质中ZmCRY1b比ZmCRY2更强烈。
图6
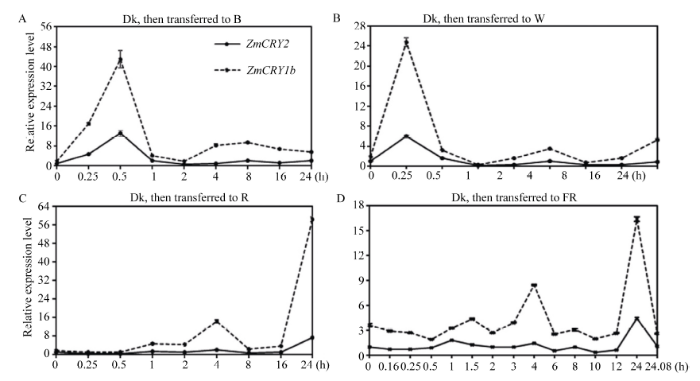 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6ZmCRY1b和ZmCRY2基因由黑暗到不同光质下的相对表达丰度
玉米自交系B73的幼苗在黑暗生长13 d后, 分别转入蓝光(B, 13 μmol m-2 s-1)、白光(W, 17 μmol m-2 s-1)、红光(R, 22.3 μmol m-2 s-1)下0 h、0.25 h、0.5 h、1 h、2 h、4 h、8 h、12 h和24 h以及远红光(FR, 0.25 μmol m-2 s-1)下0 h、0.16 h、0.25 h、0.5 h、1 h、2 h、4 h、8 h、12 h、24 h和24.08 h取材, 用于qRT-PCR分析。把黑暗条件下ZmCRY2/Tubulin的比值设为1, 并以此为对照。折线图代表了在3次独立的生物学重复下ZmCRY2/Tubulin的平均值, 其中误差线代表了标准差。
Fig. 6Relative expression of both ZmCRY1b and ZmCRY2 genes during transitions from darkness to different light conditions
The seedlings of maize inbred line B73 were grown in the dark for 13 d, then transferred to blue light (B, 13 μmol m-2 s-1), white light (W, 17 μmol m-2 s-1), or red light (R, 22.3 μmol m-2 s-1) for 0.25 h, 0.5 h, 1 h, 2 h, 4 h, 8 h, 12 h, or 24 h and far-red light (FR, 0.25 μmol m-2 s-1) for 0.16 h, 0.25 h, 0.5 h, 1 h, 2 h, 4 h, 8 h, 12 h, 24 h, or 24.08 h. The transcription abundance of ZmCRY2 in the dark was set as the control, and the ratio of ZmCRY2/Tubulin in the dark was set as 1. Each line graph shows the mean expression of ZmCRY2/Tubulin of three biological repeats. Error bars indicate the standard deviation.
综合ZmCRY1b和ZmCRY2基因在不同的持续光质以及由黑暗转换到不同光质(白光、蓝光、红光和远红光)下的表达丰度可以发现, 二者均能有效地响应各种不同光质的处理。
2.5 ZmCRY1b和ZmCRY2基因在光周期(长日照和短日照)处理下的表达水平
为了了解玉米对光周期处理的响应能力, 我们采用qRT-PCR分析了ZmCRY1b和ZmCRY2响应长日照和短日照处理的表达模式。将ZmCRY2在黑暗阶段结束时的表达量设为1, 并以此为对照。在长日照条件下, ZmCRY1b和ZmCRY2的表达模式相似, 但整体上ZmCRY2的转录丰度低于ZmCRY1b, 为ZmCRY1b的3%~10%。ZmCRY1b在进入光照阶段2 h、6 h和14 h时出现了3个峰值(分别为黑暗阶段结束时ZmCRY2的78、88和31倍); 在进入黑暗阶段后, 表达丰度迅速上升, 且在18 h时达到峰值(为黑暗阶段结束时ZmCRY2的46倍)。ZmCRY2则在进入光照阶段4 h和12 h时出现了峰值(分别为自身黑暗阶段结束时的5.4倍和4.0倍); 在进入黑暗阶段后, 表达丰度迅速上升, 且在18 h时达到峰值(为自身黑暗阶段结束时的4.5倍)(图7-A)。说明ZmCRY1b和ZmCRY2在由白光转到黑暗时表达丰度均能迅速上升至较高的水平, 但ZmCRY1b能更强烈地响应长日照处理。
在短日照的条件下, ZmCRY1b和ZmCRY2的表达出现了极其相似的模式, 二者的转录水平保持稳定的状态, 但整体上ZmCRY2的转录丰度低于ZmCRY1b, 为其4%~10%。ZmCRY1b和ZmCRY2在光照阶段的峰值均出现在4 h时(此时二者的转录丰度分别为黑暗阶段结束时ZmCRY2的30.2倍和2.7倍)。当转入黑暗时, 二者的转录丰度迅速上升。ZmCRY1b和ZmCRY2分别在转入黑暗阶段12 h和16 h时达到峰值(ZmCRY1b的转录丰度分别为ZmCRY2在黑暗阶段结束时的71.5倍和94.1倍, ZmCRY2则分别为4.4倍和4.3倍)(图7-B)。可见ZmCRY1b和ZmCRY2的表达能响应长日和短日处理, 且ZmCRY1b的响应能力更强。
图7
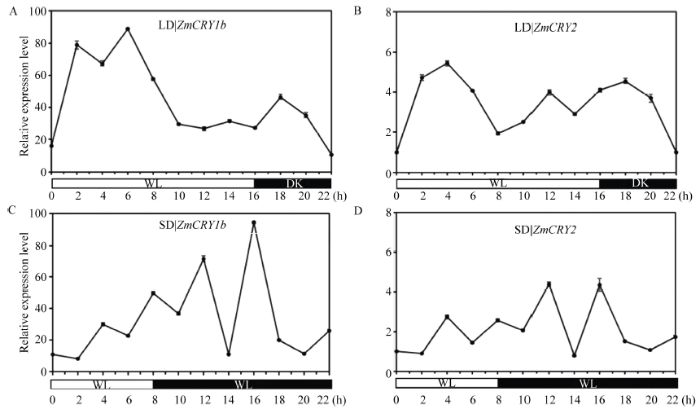 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7ZmCRY1b和ZmCRY2基因在光周期(长日照和短日照)处理的表达水平
“B73”的幼苗分别在长日照条件下(LD, 16 h光照/8 h黑暗)或短日照条件下(SD, 8 h光照/16 h黑暗)生长13 d后, 每2 h取一次样。把ZmCRY2/Tubulin在黑暗结束时期的比值设为1, 并以此为对照。折线图代表了在3次独立的生物学重复下ZmCRY2/Tubulin的平均值, 其中误差线代表了标准差。
Fig. 7Relative expression of both ZmCRY1b and ZmCRY2 genes in response to photoperiod (long-day and short-day) treatment
The seedlings of maize inbred line B73 were grown in long-day condition (LD, 16 h light/8 h dark) or short-day (SD, 8 h light/16 h dark) for 13 d, then sampled every two hours. The transcription abundance of ZmCRY2 in the dark was set as the control, and the ratio of ZmCRY2/Tubulin at the end of the dark period was set as 1. Each line graph shows the mean expression of ZmCRY2/Tubulin of three biological repeats. Error bars indicate the standard deviation.
3 讨论
目前对玉米中隐花色素的研究较少且浅显, 而模式植物拟南芥中的隐花色素研究得较为深入。本研究表明, 玉米与拟南芥及小麦和水稻等禾本科植物的CRY蛋白具有相似的结构域和较高的序列一致性。这暗示着它们可能存在相似的功能, 也为如何深入研究玉米的隐花色素提供了技术思路。作为蓝光受体, ZmCRY1b和ZmCRY2能够强烈地响应各种持续光质、转换光质的处理, 暗示着它们可能参与了白光、蓝光甚至是红光和远红光下的光信号转导途径。此外, 二者还能响应长日照和短日照的处理, 说明其可能参与光周期诱导的开花。拟南芥的CRY1和CRY2在蓝光下可抑制下胚轴的伸长并促进花蕾的形成, 二者分工不同却又功能冗余[29]。在蓝光下, CRY1通过与SPA1结合来抑制COP1/SPA1复合物的活性, 导致HY5的积累以促进光形态建成并抑制下胚轴的伸长, 而CRY2通过结合SPA1以抑制COP1/SPA1复合物的活性, 导致CO蛋白的积累, 或与CIB蛋白结合以促进CO蛋白的积累以及FT基因的转录, 从而加速开花[21,23]。CRY
不仅促进拟南芥的开花, 还能调控植物品种的纬度分布[9]。近期的研究表明, 除拟南芥[9,10]外, 水稻[13,14]、大豆[18,19]、甘蓝型油菜[20]等植物中的CRY均能促进开花。水稻中的OsCRY1介导了蓝光下水稻幼苗的去黄化反应, 而OsCRY2则促进水稻的开花[13,14]。大豆的GmCRY1a和GmCRY2a蛋白与拟南芥的AtCRY1和AtCRY2蛋白序列高度一致, 且均为核蛋白, 但GmCRY1a与AtCRY2的功能类似, 均能促进植物开花[18,19]。本研究显示玉米隐花色素ZmCRY1b和ZmCRY2基因能强烈地响应各种光质及光周期处理, 推测二者也参与了玉米光形态建成和开花期的调控, 但二者对玉米花期的调控作用及其机制尚待验证。
研究表明, 植物中的光敏色素和隐花色素相互作用调控其光形态建成及光周期反应, 进而调控植物的生长和发育过程[10,30-32]。玉米中隐花色素至少存在4个拷贝, 即CRY1a1、CRY1a2、CRY1b和CRY2, 光敏色素则存在6个拷贝, 即PHYA1、PHYA2、PHYB1、PHYB2、PHYC1和PHYC2。它们之间是否存在互作、互作方式及形成的信号网络都值得进一步研究。玉米ZmCRY1b和ZmCRY2基因对玉米开花是否存在调控作用及其与开花的调控基因(GI、FT和CO等)之间的关系也未被阐明。因此, 玉米ZmCRY1b和ZmCRY2基因对玉米光形态建成和光周期的调控作用及机制, 及对玉米的引种和高产的作用也需要进一步研究。
4 结论
玉米CRY1b蛋白与拟南芥、水稻、小麦等CRY1蛋白的结构相似, 均含有PHR和CCE结构域, 而它们的CRY2蛋白则都含有PHR结构域, 且它们的氨基酸序列一致性较高。ZmCRY1b和ZmCRY2基因在叶片中表达丰度最高, 能强烈响应蓝光、白光、红光和远红光持续光质及由黑暗到白光、蓝光、红光、远红光等各种转换光质的处理, 并且对长日照和短日照处理也能很强烈地响应。结果暗示ZmCRY1b和ZmCRY2参与玉米光形态建成及开花调节, 且ZmCRY1b在作物改良上比ZmCRY2更有效, 其在作物改良中的应用价值值得更深入的研究。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
[本文引用: 3]
DOI:10.1104/pp.105.067462URLPMID:16244154 [本文引用: 2]

Cryptochromes are widespread in higher plants but their physiological roles as blue-light photoreceptors have been examined in relatively few species. Screening in a phyA null mutant background has identified several blue-light response mutants in pea (Pisum sativum), including one that carries a substitution of a highly conserved glycine residue in the N-terminal photolyase-homologous domain of the pea CRY1 gene. Analyses of cry1, phyA, and phyB mutants show that all three photoreceptors contribute to seedling photomorphogenesis under high-irradiance blue light, whereas phyA is the main photoreceptor active under low irradiances. Triple phyA phyB cry1 mutants grown under high-irradiance blue light are indistinguishable from dark-grown wild-type plants in length and leaf expansion but show a small residual response to higher-irradiance white light. Monogenic cry1 mutants have little discernable phenotype at the seedling stage, but later in development are more elongated than wild-type plants. In addition, the loss of cry1 moderates the short-internode phenotype of older phyA mutants, suggesting an antagonism between phyA and cry1 under some conditions. Pea cry1 has a small inhibitory effect on flowering under long and short days. However, the phyA cry1 double mutant retains a clear promotion of flowering in response to blue-light photoperiod extensions, indicating a role for one or more additional blue-light photoreceptors in the control of flowering in pea.
[本文引用: 2]
DOI:10.1111/php.12663URLPMID:27861972 [本文引用: 2]

Abstract Blue light affects many aspects of plant growth and development throughout the plant lifecycle. Plant cryptochromes (CRYs) are UV-A/blue light photoreceptors that play pivotal roles in regulating blue light-mediated physiological responses via the regulated expression of more than one thousand genes. Photoactivated CRYs regulate transcription via two distinct mechanisms: indirect promotion of the activity of transcription factors by inactivation of the COP1/SPA E3 ligase complex or direct activation or inactivation of at least two sets of basic helix-loop-helix transcription factor families by physical interaction. Hence, CRYs govern intricate mechanisms that modulate activities of transcription factors to regulate multiple aspects of blue light-responsive photomorphogenesis. Here, we review recent progress in dissecting the pathways of CRY signaling and discuss accumulating evidence that shows how CRYs regulate broad physiological responses to blue light. 2016 The American Society of Photobiology.
[本文引用: 1]
[本文引用: 2]
DOI:10.1073/pnas.1602004113URLPMID:27325772 [本文引用: 3]

Abstract The phenomenon of delayed flowering after the application of nitrogen (N) fertilizer has long been known in agriculture, but the detailed molecular basis for this phenomenon is largely unclear. Here we used a modified method of suppression-subtractive hybridization to identify two key factors involved in N-regulated flowering time control in Arabidopsis thaliana, namely ferredoxin-NADP + -oxidoreductase and the blue-light receptor cryptochrome 1 (CRY1). The expression of both genes is induced by low N levels, and their loss-of-function mutants are insensitive to altered N concentration. Low-N conditions increase both NADPH/NADP + and ATP/AMP ratios, which in turn affect adenosine monophosphate-activated protein kinase (AMPK) activity. Moreover, our results show that the AMPK activity and nuclear localization are rhythmic and inversely correlated with nuclear CRY1 protein abundance. Low-N conditions increase but high-N conditions decrease the expression of several key components of the central oscillator (e.g., CCA1, LHY, and TOC1) and the flowering output genes (e.g., GI and CO). Taken together, our results suggest that N signaling functions as a modulator of nuclear CRY1 protein abundance, as well as the input signal for the central circadian clock to interfere with the normal flowering process.
[本文引用: 3]
[本文引用: 2]
[本文引用: 1]
DOI:10.1111/j.1365-313X.2006.02753.xURLPMID:16805731 [本文引用: 3]

Cryptochromes (CRY) are blue-light photoreceptors that mediate various light responses, such as inhibition of hypocotyl elongation, enhancement of cotyledon expansion, anthocyanin accumulation and stomatal opening in Arabidopsis. The signaling mechanism of Arabidopsis CRY is mediated through direct interaction with COP1, a negative regulator of photomorphogenesis. CRY has now been characterized in tomato, pea, moss and fern, but its function in monocots is largely unknown. Here we report the function and basic signaling mechanism of rice cryptochrome 1 (OsCRY1). Overexpresion of OsCRY1b resulted in a blue light-dependent short hypcotyl phenotype in Arabidopsis, and a short coleoptile, leaf sheath and leaf blade phenotype in rice ( Oryza sativa ). On fusion with -glucuronidase (GUS), the C-terminal domain of either OsCRY1a (OsCCT1a) or OsCRY1b (OsCCT1b) mediated a constitutive photomorphogenic (COP) phenotype in both Arabidopsis and rice, whereas OsCCT1b mutants corresponding to missense mutations in previously described Arabidopsis cry1 alleles failed to confer a COP phenotype. Yeast two-hybrid and subcellular co-localization studies demonstrated that OsCRY1b interacted physically with rice COP1 (OsCOP1). From these results, we conclude that OsCRY1 is implicated in blue-light inhibition of coleoptile and leaf elongation during early seedling development in rice, and that the signaling mechanism of OsCRY1 involves direct interaction with OsCOP1.
DOI:10.1093/pcp/pcj064URLPMID:16760221 [本文引用: 3]

Abstract In order to elucidate the function of cryptochromes (cry) in rice, we have characterized all rice CRY genes, including OsCRY1a, OsCRY1b and OsCRY2. Our studies revealed that OsCRY1 genes were mainly expressed in the green plant tissue, while OsCRY2 gene expression was high in the coleoptile, flower and callus. Light treatment affected neither the expression of any of the OsCRY genes nor the stability of their transcripts. However, it was found that Oscry2 protein was negatively regulated by blue light. Moreover, the level of Oscry2 protein also decreased upon red light treatment, and this red light-dependent degradation was shown to be mediated by phytochrome B. Overexpression of OsCRY1 genes resulted in an increased responsiveness to blue light when measuring coleoptile growth inhibition. Moreover, growth of leaf sheaths and blades was also repressed more in OsCRY1 overexpressers than in wild type (WT) under blue light conditions. These results suggest that Oscry1s are responsible for regulating blue light-mediated de-etiolation in rice. In addition, OsCRY2 antisense transgenic rice flowered later than WT under both long-day and short-day conditions, indicating that Oscry2 is involved in the promotion of flowering time in rice.
DOI:10.1007/s11103-005-0828-zURLPMID:16244915 [本文引用: 1]

The cryptochromes are a family of blue light photoreceptors that play important roles in the control of plant development. We have characterised the cryptochrome gene family in the model legume garden pea ( Pisum sativum L.). Pea contains three expressed cryptochrome genes; a single CRY1 orthologue, and two distinct CRY2 genes that we have termed CRY2a and CRY2b . Genomic southern blots indicate that there are unlikely to be more CRY genes in pea. Each of the three genes encodes a full-length CRY protein that contains all the major domains characteristic of other higher plant cryptochromes. Database searches have identified Medicago truncatula expressed sequence tags (ESTs) corresponding to all three genes, whereas only a single CRY2 is represented in EST collections from the more distantly related legumes soybean and Lotus japonicus . The proteins encoded by the pea and Medicago CRY2b genes are distinguished from other CRY2 proteins by their shorter C-terminus. Expression analyses have identified marked differences in the regulation of the three genes, with CRY2b expression in particular distinguished by high-amplitude diurnal cycling and rapid repression in seedlings transferred from darkness to blue light.
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
[本文引用: 3]
Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation
[本文引用: 2]
DOI:10.1016/j.tplants.2011.09.002URLPMID:3277817 [本文引用: 2]

Cryptochromes (CRY) are blue-light receptors that mediate various light responses in plants. The photoexcited CRY molecules undergo several biophysical and biochemical changes, including electron transfer, phosphorylation and ubiquitination, resulting in conformational changes to propagate light signals. Two modes of CRY signal transduction have recently been discovered: the cryptochrome-interacting basic-helix-loop-helix 1 (CIB)-dependent CRY2 regulation of transcription; and the SUPPRESSOR OF PHYA1/CONSTITUTIVELY PHOTOMORPHOGENIC1 (SPA1/COP1)-dependent cryptochrome regulation of proteolysis. Both CRY signaling pathways rely on blue light-dependent interactions between the CRY photoreceptor and its signaling proteins to modulate gene expression changes in response to blue light, leading to altered developmental programs in plants.
[本文引用: 1]
DOI:10.1016/j.pneurobio.2009.12.005.URL [本文引用: 2]

Among the most devastating diseases of the nervous system that plague today's world with a heavy death toll are those caused by infections, such as cerebral malaria, rabies, toxoplasmosis, bacterial meningitis, arbovirus encephalitis and HIV-associated neurological diseases, to name a few. The burden of these diseases is enhanced because survivors may often be left with neurological sequelae affecting motility, sensory organs and cognitive functions, as well as epilepsy. This in turn seriously impairs socio-economic progress in tropical regions of the world where such diseases are particularly prevalent.
[本文引用: 1]
DOI:10.1105/tpc.107.053017URL [本文引用: 1]

CRY2 is a blue light receptor regulating light inhibition of hypocotyl elongation and photoperiodic flowering in Arabidopsis thaliana. The CRY2 protein is found primarily in the nucleus, and it is known to undergo blue light-dependent phosphorylation and degradation. However, the subcellular location where CRY2 exerts its function or undergoes blue light-dependent phosphorylation and degradation remains unclear. In this study, we analyzed the function and regulation of conditionally nuclear-localized CRY2. Our results show that CRY2 mediates blue light inhibition of hypocotyl elongation and photoperiodic promotion of floral initiation in the nucleus. Consistent with this result and a hypothesis that blue light-dependent phosphorylation is associated with CRY2 function, we demonstrate that CRY2 undergoes blue light-dependent phosphorylation in the nucleus. CRY2 phosphorylation is required for blue light-dependent CRY2 degradation, but only a limited quantity of CRY2 is phosphorylated at any given moment in seedlings exposed to blue light, which explains why continuous blue light illumination is required for CRY2 degradation. Finally, we showed that CRY2 is ubiquitinated in response to blue light and that ubiquitinated CRY2 is degraded by the 26S proteasome in the nucleus. These findings demonstrate that a photoreceptor can complete its posttranslational life cycle (from protein modification, to function, to degradation) inside the nucleus.
[本文引用: 1]
[本文引用: 2]
.
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1111/nph.14886URLPMID:29139123 [本文引用: 1]

Abstract Contents I. II. III. IV. SUMMARY: Plants perceive and respond to light signals by multiple sensory photoreceptors, including phytochromes and cryptochromes, which absorb different wavelengths of light to regulate genome expression and plant development. Photophysiological analyses have long revealed the coordinated actions of different photoreceptors, a phenomenon referred to as the photoreceptor coaction. The mechanistic explanations of photoreceptor coactions are not fully understood. The function of direct protein-protein interaction of phytochromes and cryptochromes and common signaling molecules of these photoreceptors, such as SPA1/COP1 E3 ubiquitin ligase complex and bHLH transcription factors PIFs, would partially explain phytochrome-cryptochrome coactions. In addition, newly discovered proteins that block cryptochrome photodimerization or catalyze cryptochrome phosphorylation may also participate in the phytochrome and cryptochrome coaction. This Tansley insight, which is not intended to make a comprehensive review of the studies of photoreceptor coactions, attempts to highlight those recent findings and their possible roles in the photoreceptor coaction. No claim to original US government works New Phytologist 2017 New Phytologist Trust.
DOI:10.1016/j.molp.2016.02.007URLPMID:26921621 [本文引用: 1]

Light,as both energy source and informational signal,profoundlyinfluences plant growth and development during the whole lifespan from seed germination to flowering.Plant senses lightthrough multiple photoreceptors including the blue/UV-A photo-receptors cryptochromes (CRY,CRY1/2) and phototropins(PHOT,PHOT1/2),the red/far-red photoreceptors phytochromes(PHY,phyA-E),and the UV-B photoreceptor UVR8.In Arabidop-sis,both CRY and PHY are involved in mediating light regulationof hypocotyl elongation,cotyledon/leaf expansion,anthocyaninaccumulation,flowering,stomatal development,and light entrainment of the circadian clock.
