 ,*, 王慧
,*, 王慧 ,*华南农业大学 / 国家植物航天育种工程技术研究中心, 广东广州 510642
,*华南农业大学 / 国家植物航天育种工程技术研究中心, 广东广州 510642QTL Mapping for Heading Date in Rice Using High-density Bin Map
DONG Ji-Chi**, YANG Jing**, GUO Tao**, CHEN Li-Kai**, CHEN Zhi-Qiang ,*, WANG Hui
,*, WANG Hui ,*National Engineering Research Centre of Plant Space Breeding / South China Agricultural University, Guangzhou 510642, Guangdong, China
,*National Engineering Research Centre of Plant Space Breeding / South China Agricultural University, Guangzhou 510642, Guangdong, China通讯作者:
第一联系人:
收稿日期:2017-12-14接受日期:2018-03-25网络出版日期:2018-06-12
| 基金资助: |
Received:2017-12-14Accepted:2018-03-25Online:2018-06-12
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (1392KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
董骥驰, 杨靖, 郭涛, 陈立凯, 陈志强, 王慧. 基于高密度Bin图谱的水稻抽穗期QTL定位[J]. 作物学报, 2018, 44(6): 938-946. doi:10.3724/SP.J.1006.2018.00938
DONG Ji-Chi, YANG Jing, GUO Tao, CHEN Li-Kai, CHEN Zhi-Qiang, WANG Hui.
水稻抽穗期决定了品种的种植地区与季节适应性[1], 而且与产量、品质和抗逆性关系密切[2], 相关的研究对指导育种实践、品种改良及品种推广均具有重要意义[3]。自Yano等克隆了第一个水稻抽穗期QTL (Hd1)以来, 目前至少已有734个与水稻抽穗期相关的QTL被报道(http://www.grammene.org/), 用图位克隆的方法至少克隆了14个抽穗期相关的QTL[4,5,6,7,8,9,10,11,12,13,14,15,16,17]。已有研究表明, 水稻有2个成花素基因Hd3a (heading date 3a)和RFT1 (RICE FLOWERING LOCUS T 1), 以及几个抽穗抑制基因(如OsPRR37和OsDof12), 它们形成的调控网络精密控制着抽穗。至少有2个抽穗期调节通路控制着成花素基因表达, 分别是Hd1 (heading date 1)通路和Ehd1 (early heading date 1)通路。大量研究证实, 大多数抽穗期相关基因通过Hd1通路或Ehd1通路来调节成花素基因的表达, OsCO3和DTH2则不同, 它们独立于Hd1和Ehd1通路, 直接调节成花素基因的表达进而促进抽穗[18]。然而, 已定位和克隆的QTL还并不能完全解释水稻抽穗期自然变异的形成, 从根本上理解水稻抽穗期的遗传机理还需更多的深入研究, 从而更好地帮助适宜生育期品种的选育[19,20]。目前的研究大多基于传统分子标记(如AFLP、RFLP、SSR等), 操作起来耗时耗力, 不能实现高通量操作[21]。用传统分子标记构建的遗传图谱密度低、定位区间过大, 导致QTL贡献率估算偏高、基因克隆困难以及难以开发标记应用于聚合育种。此外, 标记密度低的图谱上存在大量Gap, 在QTL定位中不能提供足够的信息, 甚至一些研究最终陷入多态性分子标记缺乏而难以进一步定位的境地[22]。本研究中的Bin标记分布于整个基因组, 构建的Bin图谱标记密度高、位置精确, 达到精细定位要求, 可直接进行分子育种标记的开发和候选基因的筛选。本研究以籼稻玉针香(YZX)和粳稻02428衍生的RIL为作图群体, 采用GBS技术构建包含2711个Bin标记的高密度遗传图谱, 分别于4个环境对水稻抽穗期进行QTL定位, 以期鉴定一些新的、可稳定遗传的QTL位点作为育种资源, 并为水稻抽穗期调控基因克隆及分子标记辅助育种提供更多依据。
1 材料与方法
1.1 试验材料
用籼稻玉针香(YZX)和粳稻02428杂交, 通过单粒传法, 从F2代开始构建玉针香/02428的重组自交系群体。群体包含192个株系, 基因型鉴定为F6世代, 表型调查为F8~F11世代。1.2 田间种植与性状考查
2016年早造(E1环境)、2016年晚造(E2环境)、2017年早造(E3环境)和2017年晚造(E4环境)于华南农业大学试验教学基地(23.17°N, 113.37°E)种植192个株系的RIL群体及两亲本。采用随机区组设计, 田间管理(水、肥、病虫害防治等)按当地大田常规栽培要求实施。E1环境, 于2016年3月5日播种育秧, 4月5日移栽。E2环境, 于2016年7月25日播种育秧, 8月8日移栽。E3环境, 于2017年2月28日播种育秧, 3月31日移栽。E4环境, 于2017年7月22日播种育秧, 8月7日移栽。每个株系6行, 每行6株, 行株距为20 cm × 20 cm, 均单本种植。以单株为单位调查表型, 当单株的第1个稻穗尖露出剑叶叶鞘至少1 cm时, 记为该单株的抽穗日期, 每隔1 d调查一次。剔除异常数据后, 以株系内全部单株从播种到抽穗所经历天数的均值作为该株系的抽穗期表型值。
1.3 DNA提取及高通量测序
采用本实验室自主开发设计的高通量磁珠法制备DNA样品。用Precellys 24研磨仪(4000 × g, 12 s)研磨液氮冷冻过的叶片至粉末, 迅速加入85℃预热的LB提取液(200 mmol L-1 Tris-HCl (pH 7.8), 250 mmol L-1 NaCl, 25 mmol L-1 ethylenediaminetetraacetic acid (EDTA), 0.5% sodium dodecyl sulfate (SDS), 2% polyvinylpyrrolidone (PVP)-40), 确保叶片粉末都浸泡在提取液中, 迅速盖上橡胶盖并混匀, 置于65℃水浴锅中水浴20~25 min后, 置-20℃冰箱放置20 min。将样品转移至96孔深孔板, 每孔加入540 μL异丙醇和2.5 μL磁珠液(100 mg mL-1, 洛阳惠尔纳米科技有限公司), 用自动核酸纯化仪KingFisher Flex进行DNA分离纯化。DNA最终溶于150 μL洗脱液(10 mmol L-1 Tris-HCl, 1 mmol L-1 EDTA (pH 8.3), 100 μg mL-1 RNase A), 于-80℃保存备用。以琼脂糖凝胶电泳检测基因组DNA的质量, Nanodrop ND-1000微量核酸蛋白检测仪检测DNA的浓度与纯度[23]。采用WGS (Whole Genome Sequencing)和GBS (Genotyping-By-Sequencing)技术分别对两亲本及其衍生的RIL群体测序。水稻基因组经电子酶切评估, 选择酶切片段大小适宜, 酶切位点在基因组上分布均匀, 且能有效降低酶切片段中基因组重复序列所占比例的限制性内切酶。基因组加上带有barcode的接头后, 对每个样品进行扩增, 随后混合选择需要的片段构建GBS文库。利用Illumina HiSeq2500测序平台, 进行双末端(Paired-End)测序。测序后的数据分析流程如下: (1)数据质量评估, 去除接头、污染序列及低质量reads; (2)统计酶捕获的Reads数量; (3)参考基因组(http://plants.ensembl.org/Oryza_ sativa/)比对: 利用BWA软件(http://bio-bwa.sourceforge. net/)将测得基因组与参考基因组比对, 统计比对率、覆盖深度、基因组覆盖率等; (4)群体SNP检测; (5)遗传标记开发。基于亲本SNP检测结果, 开发子代标记, 并统计子代测序深度、覆盖度等, 评估测序质量。与亲本玉针香相同的基因型记为“0”, 与亲本02428相同的基因型记为“2”, 部分杂合基因型记为“1”。(6)遗传标记过滤。检查异常碱基, 检查基因型缺失覆盖率并过滤, 检查基因型频率, 过滤偏分离标记。
1.4 遗传重组鉴定和连锁图谱构建
采用ScmapV5软件构建遗传图谱。构建原理是: (1)通过“滑动窗口”法(sliding window approach)寻找交换点; (2)将所有样本中同一段序列上2个交换点之间紧密连锁不发生重组的若干个SNP位点看做一个整体区块, 即“Bin标记”, 以Bin的起点来确定其所在物理位置; (3)根据基因型, 用最大似然函数计算重组率; (4)用Kosambi作图函数将重组率转换为遗传距离; (5)利用两点检验法检验两个标记间是否连锁。(6)通过最短距离法进行聚类分析, 把两两连锁的标记聚为一个类, 得到连锁群; (7)基于三点测验法, 对连锁群内的标记进行排序。1.5 数据的统计分析
在Microsoft Excel 2010中完成表型数据处理和作图。采用BWA软件的默认设置进行基因组比对及相关测序数据统计分析。采用WinQTL Cartographer 2.5软件定位水稻抽穗期QTL, 以LOD=2.5为阈值检测QTL; 应用CIM算法, 扫描区间为1.0 cM, 遗传背景控制选择标准模型(模型6)。QTL定位结果以LOD的峰值作为该QTL的LOD值, 以LOD峰值位置的Bin标记来估计QTL效应, 以LOD值下降1.0的区域作为QTL的置信区间, 遵循McCouch等[24]的原则命名QTL。应用SPSS 19.0软件进行方差分析及计算。应用Origin 8.0进行频次分布图的绘制。2 结果与分析
2.1 遗传连锁图谱构建
亲本玉针香和02428重测序Q20的比例分别为93.72%和94.22%, Q30比例分别为86.44%和87.47%。两亲本测序Reads与Nipponbare (Oryza sativa ssp. japonica)参考基因组的比对率分别为96.29%和97.97%, 平均深度分别达到20.26×和26.21×, 1×以上覆盖度为90.60%和96.36%, 4×以上覆盖度为85.41%和94.53%。两亲本中共发现1 534 036个多态性SNP, 筛选后剩余aa×bb型1 334 454个。利用GBS技术对192个RIL株系进行测序, 测序深度平均值为11.76×, 4×以上覆盖度为7%。分别提取192个子代在上述的1 334 454个亲本多态性标记位点的基因型, 经过分析和筛选最终共获得2711个均匀分布于12条染色体的Bin标记。将Bin标记锚定于连锁群后, 各染色体上标记数量为162~311个, 各染色体的平均遗传距离为195.3 cM, 两标记间遗传距离平均值为0.86 cM, 两标记间物理距离平均值为137.36 kb, 标记整体的分布达到精细作图的要求(图1)。
图1
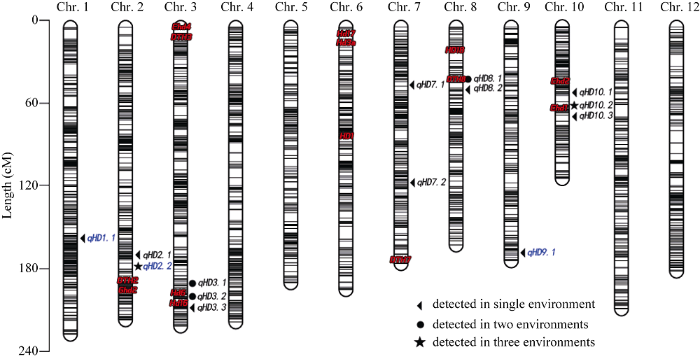 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1遗传标记在染色体上的分布及QTL位置
每条染色体上的黑色线条代表Bin标记所在位置; 红色字体为已克隆QTL; 蓝色字体为新发现的QTL。
Fig. 1Distribution of genetic markers and QTLs on chromosomes
Black lines represent the positions of Bin markers on each linkage group; fonts in red are cloned QTLs; fonts in blue are novel QTLs.
2.2 抽穗期表型特征
方差分析表明两亲本抽穗期天数的均值存在极显著差异, 4个环境下玉针香的抽穗期均显著长于02428 (表1)。Table 1
表1
表102428与玉针香抽穗期天数比较
Table 1
| 环境 Environment | 平均值Average (d) | 变异系数CV (%) | 差异 Difference | |||
|---|---|---|---|---|---|---|
| 02428 | 玉针香 Yuzhenxiang | 02428 | 玉针香 Yuzhenxiang | |||
| E1 | 91.36 | 97.00 | 1.08 | 1.57 | -5.64** | |
| E2 | 64.11 | 69.27 | 3.25 | 3.60 | -5.16** | |
| E3 | 89.20 | 97.03 | 0.84 | 0.76 | -7.83** | |
| E4 | 65.00 | 70.33 | 3.91 | 3.75 | -5.33** | |
新窗口打开|下载CSV
表2表明, 在E1环境中, RIL群体抽穗期的平均值为95.65 d, 抽穗期最短的株系为81.33 d, 抽穗期最长的株系为117.83 d; 在E2环境中, RIL群体抽穗期的平均值为69.15 d, 抽穗期最短的株系为53.13 d, 抽穗期最长的株系为85.4 d; 在E3环境中, RIL群体抽穗期的平均值为95.29 d, 抽穗期最短的株系为81 d, 抽穗期最长的株系为110.3 d; 在E4环境中, RIL群体抽穗期的平均值为70.54 d, 抽穗期最短的株系为61.3 d, 抽穗期最长的株系为87.5 d; 四个环境中RIL群体的抽穗期峰度和偏度绝对值都小于1, 总体呈正态分布(图2), 表现为数量性状遗传模式。
Table 2
表2
表2RIL群体抽穗期性状表现
Table 2
| 环境 Environment | 平均值±标准差 Mean ± SD | 变异范围 Range | 峰度 Kurtosis | 偏度 Skewness |
|---|---|---|---|---|
| E1 | 95.64±6.25 | 81.33-117.83 | 0.44 | 0.60 |
| E2 | 69.15±4.92 | 53.13-85.40 | 0.30 | 0.27 |
| E3 | 95.29±4.99 | 81.00-110.30 | 0.43 | 0.11 |
| E4 | 70.54±4.15 | 61.30-87.50 | 0.48 | 0.46 |
新窗口打开|下载CSV
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2玉针香/02428重组自交系及亲本的抽穗期表型分布特征
Fig. 2Distribution of heading date for YZX/02428 RIL population and parents
2.3 抽穗期QTL定位
在4个环境中共检测到14个抽穗期QTL(表3和图1)。其中, E1环境中检测到9个, 分布于第2、第3、第7、第8和第10染色体, LOD值介于2.9~5.6之间, 贡献率介于4.7%~9.5%之间; E2环境中检测到5个, 分布于第2、第7和第10染色体, LOD值介于3.0~5.0之间, 贡献率介于5.2%~9.5%之间; E3环境中检测到3个, 分布于第1、 第2和第8染色体, LOD值介于2.5~5.9之间, 贡献率介于4.5%~11.2%之间; E4环境中检测到5个, 分布于第3、第9和第10染色体, LOD值介于2.8~9.6之间, 贡献率介于4.6%~17.0%之间。其中, 有2个QTL在3个环境中均检测到。Table 3
表3
表34个环境下检测到的抽穗期QTL
Table 3
| 环境 Environ- ment | QTL | 染色体 Chr. | 位置 Peak position (cM) | LOD | 加性效应1) Additive effect1) | 贡献率 Variation explained (%) | Bin标记区间 Bin marker interval | 置信区间 Confidence interval (Mb) |
|---|---|---|---|---|---|---|---|---|
| E1 | qHD2.2 | 2 | 178.2 | 3.0 | -1.5 | 5.1 | mk487-491 | 28.25-28.65 |
| E1 | qHD3.1 | 3 | 194.2 | 2.9 | -1.4 | 4.7 | mk813-814 | 30.55-30.65 |
| E1 | qHD3.2 | 3 | 205.1 | 3.2 | -1.5 | 5.2 | mk826-831 | 31.85-32.45 |
| E1 | qHD3.3 | 3 | 211.2 | 3.2 | -1.5 | 5.3 | mk836-840 | 33.10-33.55 |
| E1 | qHD7.1 | 7 | 48.8 | 4.2 | 1.7 | 7.0 | mk1618-1620 | 8.65-8.85 |
| E1 | qHD8.1 | 8 | 42.4 | 5.6 | 2.0 | 9.5 | mk1821-1824 | 4.20-4.55 |
| E1 | qHD8.2 | 8 | 49.3 | 4.3 | 1.7 | 7.4 | mk1825-1830 | 4.65-5.15 |
| E1 | qHD10.1 | 10 | 56.7 | 3.5 | -1.6 | 6.7 | mk2244-2247 | 16.35-16.65 |
| E1 | qHD10.2 | 10 | 64.8 | 3.2 | -1.5 | 5.4 | mk2248-2253 | 16.75-17.25 |
| E2 | qHD2.1 | 2 | 173.6 | 3.2 | -1.3 | 5.8 | mk481-488 | 27.60-28.35 |
| E2 | qHD2.2 | 2 | 179.7 | 5.0 | -1.6 | 9.5 | mk489-491 | 28.45-28.65 |
| E2 | qHD7.2 | 7 | 118.4 | 3.0 | -1.2 | 5.2 | mk1723-1727 | 22.05-22.55 |
| E2 | qHD10.2 | 10 | 62.1 | 4.5 | -1.4 | 8.2 | mk2248-2253 | 16.75-17.25 |
| E2 | qHD10.3 | 10 | 67.4 | 4.1 | -1.4 | 7.5 | mk2258-2260 | 17.75-17.95 |
| E3 | qHD1.1 | 1 | 160.1 | 2.5 | -1.3 | 4.4 | mk185-186 | 25.95-26.05 |
| E3 | qHD2.2 | 2 | 175.2 | 5.9 | -1.9 | 11.1 | mk486-491 | 28.15-28.65 |
| E3 | qHD8.1 | 8 | 42.4 | 3.9 | 2.4 | 7.4 | mk1821-1824 | 4.20-4.55 |
| E4 | qHD3.1 | 3 | 197.3 | 3.4 | -1.0 | 5.6 | mk812-818 | 30.45-31.05 |
| E4 | qHD3.2 | 3 | 205.1 | 3.0 | -1.0 | 4.9 | mk826-831 | 31.85-32.45 |
| E4 | qHD9.1 | 9 | 125.4 | 2.8 | -1.0 | 4.6 | mk2088-2090 | 16.55-16.75 |
| E4 | qHD10.2 | 10 | 63.5 | 9.6 | -1.7 | 17.0 | mk2249-2250 | 16.85-16.95 |
| E4 | qHD10.3 | 10 | 70.0 | 7.1 | -1.5 | 13.5 | mk2259-2260 | 17.85-17.95 |
新窗口打开|下载CSV
qHD1.1在第1染色体上, 仅在E3环境中被检测到, 位于160.17 cM, 与mk186连锁。其LOD值为2.5, 置信区间为25.95~26.05 Mb, 表型贡献率为4.5%, 来自于02428的等位基因可缩短生育期1.3 d。
qHD2.1位于第2染色体, 仅在E2环境中被检测到, 在173.61 cM LOD达到峰值3.2, 与mk484紧密连锁, 置信区间为17.60~28.35 Mb, 能解释5.8%的表型变异, 来自于02428的等位基因可缩短抽穗期1.3 d。同一染色体上的qHD2.2在3个环境中均被检测到, 表现出较强的稳定性。在E1环境检测到的位置是178.21 cM, LOD值为3.1, 置信区间为28.25~28.65 Mb, 可解释5.1%表型变异, 来自于02428的等位基因可缩短抽穗期1.5 d; 在E2环境检测到的位置是179.71 cM, LOD值为5.0, 置信区间为28.45~28.65 Mb, 可解释9.5%表型变异, 来自于02428的等位基因可缩短抽穗期1.6 d。在E3环境检测到的位置是175.19 cM, LOD值为5.9, 置信区间为28.15~28.65 Mb, 可解释11.2%的表型变异, 来自于02428的等位基因可缩短抽穗期1.9 d。
第3染色体上共有3个抽穗期QTL, 分别为qHD3.1、qHD3.2和qHD3.3。qHD3.1在E1环境中被检测到, 位于194.21 cM处, LOD值为2.9, 置信区间为30.05~30.65 Mb, 可解释4.7%表型变异, 来自于02428的等位基因可缩短抽穗期1.4 d; 在E4环境中被检测到, 位于197.3 cM处, LOD值为3.4, 置信区间为30.45~31.05 Mb, 可解释5.6%表型变异, 来自于02428的等位基因可缩短抽穗期1.0 d。qHD3.2在E1环境被检测到, 位于205.11 cM处, LOD值为3.2, 置信区间为31.85~32.45 Mb, 可解释5.25%表型变异, 来自于02428的等位基因可缩短抽穗期1.5 d; 在E4环境被检测到, 位于205.11 cM处, LOD值为3.0, 置信区间为31.85~32.45 Mb, 可解释4.85%表型变异, 来自于02428的等位基因可缩短抽穗期1.0 d。qHD3.3仅在E1环境中被检测到, 位于211.21 cM处, LOD值为3.2, 置信区间为33.10~33.55 Mb, 可解释5.3%表型变异, 来自于02428的等位基因可缩短抽穗期1.5 d。
第7染色体上共有2个抽穗期QTL。qHD7.1只在E1环境中被检测到, 位于48.81 cM处, 与mk1681紧密连锁, LOD值为4.2, 置信区间为8.65~8.85 Mb, 可解释7.0%的表型变异, 来自于02428的等位基因可推迟抽穗期1.7 d。qHD7.2只在E2环境中被检测到, 位于118.41 cM处, 与mk1725紧密连锁, LOD值为3.0, 置信区间为22.05~22.55 Mb, 可解释5.2%的表型变异, 来自于02428的等位基因可缩短抽穗期1.2 d。第8染色体上共有2个抽穗期QTL, 即qHD8.1和qHD8.2。在E1环境检测到qHD8.1的位置是42.41 cM, 与mk1823紧密连锁, LOD值为5.6, 置信区间为4.20~4.55 Mb, 可解释9.5%表型变异, 来自于02428的等位基因能推迟抽穗期2 d; 在E3环境检测到qHD8.1的位置是42.44 cM, 与mk1823紧密连锁, LOD值为3.9, 置信区间为4.20~4.55 Mb, 可解释7.5%表型变异, 来自于02428的等位基因能推迟抽穗期2.4 d。qHD8.2仅在E1环境被检测到, 位于56.71 cM处, 与mk1830紧密连锁, LOD值为4.3, 置信区间为4.65~5.15 Mb, 可解释7.4%表型变异, 来自于02428的等位基因能推迟抽穗期1.7 d。
qHD9.1在第9染色体上125.4 cM处, 仅在E4环境中被检测到, 与mk2090连锁。其LOD值为2.8, 置信区间为16.55~16.75 Mb, 表型贡献率为4.6%, 来自于02428的等位基因可缩短生育期1.0 d。
第10染色体56.71 cM处为qHD10.1, 仅在E1环境被检测到, 它与mk2245紧密连锁, LOD值为3.5, 置信区间为16.35~16.65 Mb, 可解释6.7%表型变异, 来自于02428的等位基因可缩短抽穗期1.6 d。同一染色体上的qHD10.2在E1、E2和E4环境被重复检测到, 置信区间为16.75~17.25 Mb, E1环境的LOD值为4.5, 贡献率为8.2%, 来自于02428的等位基因可缩短抽穗期1.4 d; E2环境的LOD值为3.2, 贡献率为5.4%, 来自于02428的等位基因可缩短抽穗期1.5 d; E4环境的LOD值为9.6, 贡献率为17.0%, 来自于02428的等位基因可缩短抽穗期1.7 d。第10染色体67.41 cM处是qHD10.3, 仅在E2环境中被检测到, LOD值为4.2, 可解释7.5%表型变异, 置信区间为17.75~17.95 Mb, 来自于02428的等位基因可缩短抽穗期1.4 d。
2.4 注释基因筛选
在3个环境中可重复检测到qHD2.2, 其置信区间最大时为28.15~28.65 Mb, 在该染色体区间有33个注释基因(The Rice Annotation Project Database), 逐一筛除发现有3个位点很可能影响抽穗期, 分别是LOC_Os02g 46450、LOC_Os02g46710和LOC_Os02g46940。其中, LOC_Os02g46450与拟南芥中PIE1 (PHOTOPERIOD- INDEPENDENT EARLY FLOWERING1)同源, 可影响水稻抽穗期、胚的发育及小花器官数目。LOC_Os02g46710和LOC_Os02g46940的基因产物是可表达的蛋白, 其涉及的生物学进程包括花器官的发育、生殖生长等。DNA双向测序发现, 玉针香和02428的3个候选基因间均有序列差异(图3)。玉针香与02428的LOC_ Os02g46450基因存在23处碱基差异, 共18处为单碱基的转换, 5处为3碱基以内的插入缺失; 其中7处在CDS区变异, 有5处发生错义突变, 分别位于CDS+830 bp (TTT/TCT)、CDS+1008 bp (GAG/GAT)、CDS+150 bp (TTG/TTT)、CDS+2413 bp (CGT/TGT)和CDS+2 418 bp (GAC/GAG)。玉针香与02428的LOC_Os02g46710基因存在23处碱基差异, 其中21处为单碱基的转换, 2处为单碱基插入缺失; 其中2处在CDS区的变异, 有1处为错义突变, 位置是CDS+394 bp (AAT/TAT)。玉针香与02428的LOC_Os02g46940基因存在13处碱基差异, 其中9处为单碱基的转换, 4处为插入缺失; 其中2处在CDS区变异, 有1处为错义突变, 位置是CDS+883 bp (CTT/TTT)。同时, 测序比对发现, 02428中3个候选基因的CDS序列与日本晴参考序列完全相同。
图3
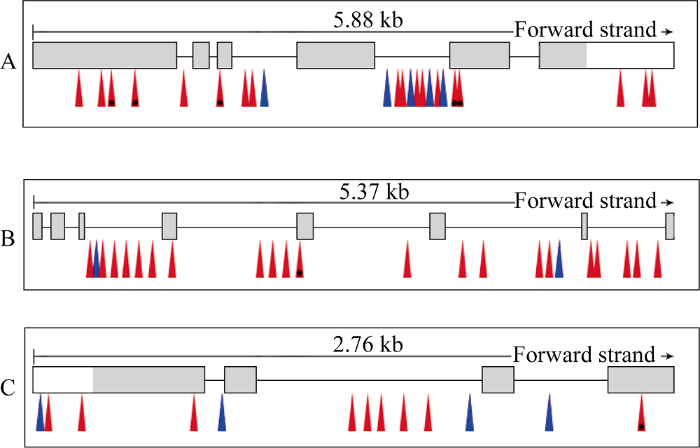 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3玉针香与02428之间候选基因的结构和变异
A: LOC_Os02g46450; B: LOC_Os02g46710; C: LOC_Os02g46940。黑色框: 外显子; 灰色部分: 编码序列; 红色箭头: SNP; 带黑点红色箭头: 错义突变; 蓝色箭头: 插入缺失。
Fig. 3Gene structure and variation of candidate genes between YZX and 02428
A: LOC_Os02g46450; B: LOC_Os02g46710; C: LOC_Os02g46940. Frames with black lines: exon; Grey boxes: protein coding sequence; Red arrow: SNP; Red arrow with a black point: missense mutation; Blue arrow: InDel.
除此之外, 还在其他QTL的置信区间内找到一些与抽穗期相关可能性较大的注释基因, 它们都涉及到花的发育过程(表4)。
Table 4
表4
表4QTL置信区间内注释基因筛选
Table 4
| QTL | 染色体 Chromosome | 置信区间 Confidence interval (Mb) | 抽穗期相关注释(克隆)基因1) Annotated (cloned) gene1) |
|---|---|---|---|
| qHD1.1 | 1 | 25.95-26.05 | LOC_Os01g45760 |
| qHD2.1 | 2 | 27.60-28.35 | OsPIE1 (LOC_Os02g46450) |
| qHD2.2 | 2 | 28.45-28.65 | LOC_Os02g46710; LOC_Os02g46940 |
| qHD3.1 | 3 | 30.45-31.05 | LOC_Os03g53190; LOC_Os03g54160; LOC_Os03g54170 |
| qHD3.2 | 3 | 31.85-32.45 | LOC_Os03g55990 |
| qHD3.3 | 3 | 33.10-33.55 | LOC_Os03g58400; LOC_Os03g58530; LOC_Os03g58530 |
| qHD7.1 | 7 | 8.65-8.85 | |
| qHD7.2 | 7 | 22.05-22.55 | |
| qHD8.1 | 8 | 4.20-4.55 | DTH8 (LOC_Os08g07740) |
| qHD8.2 | 8 | 4.65-5.15 | LOC_Os08g08210; LOC_Os08g08830 |
| qHD9.1 | 9 | 16.55-16.75 | |
| qHD10.1 | 10 | 16.35-16.65 | |
| qHD10.2 | 10 | 16.85-16.95 | Ehd1 (LOC_Os10g32600) |
| qHD10.3 | 10 | 17.85-17.95 |
新窗口打开|下载CSV
3 讨论
3.1 采用Bin图谱定位QTL的优势
目前大多数研究者采用传统分子标记(如SSR、RFLP等)构建遗传图谱[25], 精度约为1~10 Mb, 存在标记数量少且分布不均匀的问题, 影响了QTL定位的精确度, 另外标记密度较小还可能导致某些双交换位点的漏测[26]。Bin图谱与传统的遗传图谱相比, 标记密度高, 精度达100 kb, 且一个Bin内部包含多个不发生重组的SNP, 因而双交换能够被精确检测。同时, Bin图谱是基于测序技术构建的, 能提供准确的物理位置, 可使遗传分析和QTL定位更准确。此外, 测序分型得到的大量SNP标记可快速应用于分子育种。本研究采用GBS技术构建了以SNP为基础的高分辨率Bin图谱, 包含2711个Bin标记, 标记间平均物理距离137.36 kb, 标记整体的分布达到精细作图的密度, 可直接从定位区间筛选候选基因。另外, 定位结果显示, 在LOD值超过阈值的一段区域一般存在多个峰值, 可定位到多个QTL, 如位置相邻的qHD10.1和qHD10.2, 以及qHD3.1、qHD3.2和qHD3.3等, 说明Bin图谱对QTL的检测更精细, 可将位置相邻的QTL有效地分离、解析。3.2 本研究与前人定位QTL的比较
本研究定位的qHD1.1在第1染色体上, 仅在E3环境中被检测到, 其LOD值为2.5, 置信区间为25.95~26.05 Mb, 表型贡献率为4.5%, 在前人的研究中未见报道。qHD2.1位于第2染色体27.95 Mb处, 置信区间为17.60~28.35 Mb, 表型贡献率为5.8%, LOD值为3.2, 在该区间有多篇相关的报道[27,28,29,30,31,32], 其中Thomson等[31]报道的dth2.1表型贡献率为4.4%, LOD为4.06, 在5个环境中检测到1次, 与qHD2.1位置也最近, 它们可能为同一QTL。在第2染色体上, E1环境于28.55 Mb位置检测到1个QTL, 其置信区间为28.25~28.65 Mb; E2环境于28.65 Mb位置检测到1个QTL, 置信区间为28.45~28.65 Mb; E3环境于28.35 Mb位置检测到1个QTL, 置信区间为28.15~28.65 Mb。这3个QTL位置十分相近, 置信区间十分相似, 因此视为同一QTL, 命名为qHD2.2, 它在前人的研究中未见报道, 很可能是与水稻抽穗期相关的新位点。
第3染色体上检测到3个QTL qHD3.1、qHD3.2和qHD3.3。qHD3.1在2个环境中被重复检测到, 在E1环境定位的位置是30.55 Mb, 置信区间分别为30.55~30.65 Mb; E4环境定位的位置是30.95 Mb, 置信区间为30.45~31.05 Mb。qHD3.2也在2个环境中被重复检测到, 定位的位置是32.35 Mb, 置信区间为31.85~32.45 Mb。qHD3.3仅在E1环境被检测到, 位于33.35 Mb, 置信区间为33.10~ 33.55 Mb。这3个QTL所在区域已有相关报道[31,33-34]。
qHD7.1位于第7染色体8.85 Mb处, 置信区间为8.65~8.85 Mb, 已精细定位的HD4包含了该区间[35]。同一染色体上的qHD7.2位于22.35 Mb处, 置信区间为22.05~22.55 Mb, 该区间位于Thomson等[31]定位的dth7.1所在区间RM125-RM336内部, Jiang等[36]定位的qHd-7与qHD7.2的区间也有部分重叠。
qHD8.1位于第8染色体4.45 Mb处, 置信区间为4.20~ 4.55 Mb, 与已克隆的DTH8[39]位置相吻合, 都表现为长日照延长抽穗期。qHD8.2位于第8染色体5.15 Mb处, 置信区间为4.65~5.15 Mb, 在该区段也有多个相关报道[37,38,39,40]。
qHD9.1仅在E4环境被检测到, 位于第9染色体16.75 Mb处, 与mk2090紧密连锁, 置信区间为16.55~16.75 Mb, 未发现相关报道。
3.3 检测到的QTL应用价值及研究价值
本研究在4个环境中共检测到14个影响抽穗期的QTL, 分布于第1、第2、第3、第7、第8、第9和第10染色体上。除qHD2.2、qHD3.1、qHD3.2、qHD8.1和qHD10.2能在2个及以上环境被检测到外, 其他QTL只在单个环境被检测到。其中, qHD8.1和qHD10.2所在的位置分别是克隆基因DTH8和Ehd1。Hori等[20]同时用12个群体进行抽穗期相关的QTL定位, 在其中3个群体中能检测到DTH8, 表型贡献率为7.9%~74.0%; 在其中2个群体中能检测到Ehd1, 表型贡献率为18.2%~18.3%。Cheng等[2]同时用3个群体进行抽穗期相关的QTL定位, 在其中1个群体中能检测到DTH8, 表型贡献率为3.5%; 在3个群体中均能检测到Ehd1, 表型贡献率为3.6%~33.7%。而本研究中检测到DTH8的表型贡献率为7.5%~9.5%; Ehd1的表型贡献率为5.4%~ 17.0%。这些研究表明, 对于不同群体, 由于材料遗传背景影响, QTL贡献率的变化很大; 其次, 图谱的精度对QTL贡献率的估算影响也较大, 传统分子标记构建的图谱精度通常约为4 Mb, 两标记之间经常会存在多个相关基因或QTL簇, 粗定位到的QTL贡献率会较高。因此我们认为, 重点关注QTL在不同环境或不同群体中的重复定位情况对聚合育种的意义更大。
qHD3.1和qHD3.2在定位到的QTL中表型贡献率相对较小, 表达也较不稳定, 只能2次被重复检测到, 因而利用的可能性小。qHD8.1在两年的早造中被重复检测到, 仅在早造环境特异表达, 表现为延长生育期, 这些都与DTH8在长日条件下延长抽穗期的特性相对应, 它对于将早熟品种改良成半晚熟的高产品种具有一定价值。qHD2.2和qHD10.2都能在3个环境中被重复检测到, 且都能够缩短生育期, 说明它们受环境影响较小、表达较稳定, 对光照和温度变化不敏感, 它们的表型贡献率最低时分别为5.1%和5.4%, 表型贡献率最高时分别为11.2%和17.0%, 因此对于QTL聚合改良品种生育期具有重要价值。
此外, qHD2.2的置信区间在3次定位中略有变化, 在E2环境判定的置信区间较其他2个环境中小, LOC_ Os02g46450(PIE1)不在此置信区间内, 但此时qHD2.2对表型的贡献率仍然达到9.5%。因此LOC_Os02g46710和LOC_Os02g46940更有可能是导致抽穗期表型变异的新QTL, 具体情况还需深入研究。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1270/jsbbs.58.367URL [本文引用: 1]
DOI:10.1017/S0016672312000444URLPMID:23298447 [本文引用: 2]

ABSTRACT Summary Two sets of reciprocal introgression lines (ILs) and a population of recombinant inbred lines (RILs) derived from the cross between japonica cultivar Xiushui09 and indica breeding line IR2061-520-6-9 (abbreviated as IR2061) were used to identify QTL for heading date (HD). Phenotyping was conducted in Hainan Island for two winter seasons (2007 and 2009). Nine QTLs were detected in the ILs with Xiushui09 background (XS-ILs), and four of which were repeatedly mapped across 2 years. Five QTLs were identified in the ILs with IR2061 background (IR-ILs), and three of which were commonly detected in 2 years. All commonly detected QTL had the same direction of gene effect. Seven QTL for HD were identified in the RILs in 2009. Only three (25%) QTLs were commonly detected using all the three populations (XS-ILs, IR-ILs and RILs). The number of commonly identified QTLs among populations was related to degree of similarity of their genetic backgrounds, suggesting that the genetic background effect is important for detecting HD QTL. QHd7 and QHd10b stably expressed in different populations and across years thus would be exploited in rice breeding programme. Moreover, lines with both of QHd7 and QHd10b resulted in at least 3 days earlier than lines with only one of them QTL, showing evident pyramiding effect.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1073/pnas.111136798URL [本文引用: 1]
DOI:10.1093/pcp/pcf156URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1104/pp.110.156943URLPMID:20566706 [本文引用: 1]

Abstract The three most important agronomic traits of rice (Oryza sativa), yield, plant height, and flowering time, are controlled by many quantitative trait loci (QTLs). In this study, a newly identified QTL, DTH8 (QTL for days to heading on chromosome 8), was found to regulate these three traits in rice. Map-based cloning reveals that DTH8 encodes a putative HAP3 subunit of the CCAAT-box-binding transcription factor and the complementary experiment increased significantly days to heading, plant height, and number of grains per panicle in CSSL61 (a chromosome segment substitution line that carries the nonfunctional DTH8 allele) with the Asominori functional DTH8 allele under long-day conditions. DTH8 is expressed in most tissues and its protein is localized to the nucleus exclusively. The quantitative real-time PCR assay revealed that DTH8 could down-regulate the transcriptions of Ehd1 (for Early heading date1) and Hd3a (for Heading date3a; a rice ortholog of FLOWERING LOCUS T) under long-day conditions. Ehd1 and Hd3a can also be down-regulated by the photoperiodic flowering genes Ghd7 and Hd1 (a rice ortholog of CONSTANS). Meanwhile, the transcription of DTH8 has been proved to be independent of Ghd7 and Hd1, and the natural mutation of this gene caused weak photoperiod sensitivity and shorter plant height. Taken together, these data indicate that DTH8 probably plays an important role in the signal network of photoperiodic flowering as a novel suppressor as well as in the regulation of plant height and yield potential.
[本文引用: 1]
DOI:10.1093/mp/ssq070URL [本文引用: 1]
DOI:10.1093/pcp/pcs028URLPMID:22399582 [本文引用: 1]

Flowering time of rice depends strongly on photoperiodic responses. We previously identified a quantitative trait locus, Heading date 17 (Hd17), that is associated with a difference in flowering time between Japanese rice (Oryza sativa L.) cultivars. Here, we show that the difference may result from a single nucleotide polymorphism within a putative gene that encodes a homolog of the Arabidopsis EARLY FLOWERING 3 protein, which plays important roles in maintaining circadian rhythms. Our results demonstrate that natural variation in Hd17 may change the transcription level of a flowering repressor, Grain number, plant height and heading date 7 (Ghd7), suggesting that Hd17 is part of rice0964s photoperiodic flowering pathway.
DOI:10.1371/journal.pgen.1003281URLPMID:3578780 [本文引用: 1]

正Land plants have evolved increasingly complex regulatory modes of their flowering time.Rice(Oryza sativa L.) is a short-day plant that flowers more rapidly in short-day but delays under long-day conditions.Previous studies have shown that the CO-FT
[本文引用: 1]
DOI:10.1093/mp/sst088URLPMID:23713079 [本文引用: 1]

Abstract Heading date and photoperiod sensitivity are fundamental traits that determine rice adaptation to a wide range of geographic environments. By quantitative trait locus (QTL) mapping and candidate gene analysis using whole-genome re-sequencing, we found that Oryza sativa Pseudo-Response Regulator37 (OsPRR37; hereafter PRR37) is responsible for the Early heading7-2 (EH7-2)/Heading date2 (Hd2) QTL which was identified from a cross of late-heading rice 'Milyang23 (M23)' and early-heading rice 'H143'. H143 contains a missense mutation of an invariantly conserved amino acid in the CCT (CONSTANS, CO-like, and TOC1) domain of PRR37 protein. In the world rice collection, different types of nonfunctional PRR37 alleles were found in many European and Asian rice cultivars. Notably, the japonica varieties harboring nonfunctional alleles of both Ghd7/Hd4 and PRR37/Hd2 flower extremely early under natural long-day conditions, and are adapted to the northernmost regions of rice cultivation, up to 53 N latitude. Genetic analysis revealed that the effects of PRR37 and Ghd7 alleles on heading date are additive, and PRR37 down-regulates Hd3a expression to suppress flowering under long-day conditions. Our results demonstrate that natural variations in PRR37/Hd2 and Ghd7/Hd4 have contributed to the expansion of rice cultivation to temperate and cooler regions.
[本文引用: 1]
DOI:10.1073/pnas.1213962110URLPMID:23388640 [本文引用: 1]

Flowering time (i.e., heading date in crops) is an important ecological trait that determines growing seasons and regional adaptability of plants to specific natural environments. Rice (Oryza sativa L.) is a short-day plant that originated in the tropics. Increasing evidence suggests that the northward expansion of cultivated rice was accompanied by human selection of the heading date under noninductive long-day (LD) conditions. We report here the molecular cloning and characterization of DTH2 (for Days to heading on chromosome 2), a minor-effect quantitative trait locus that promotes heading under LD conditions. We show that DTH2 encodes a CONSTANS-like protein that promotes heading by inducing the florigen genes Heading date 3a and RICE FLOWERING LOCUS T 1, and it acts independently of the known floral integrators Heading date 1 and Early heading date 1. Moreover, association analysis and transgenic experiments identified two functional nucleotide polymorphisms in DTH2 that correlated with early heading and increased reproductive fitness under natural LD conditions in northern Asia. Our combined population genetics and network analyses suggest that DTH2 likely represents a target of human selection for adaptation to LD conditions during rice domestication and/or improvement, demonstrating an important role of minor-effect quantitative trait loci in crop adaptation and breeding.
[本文引用: 1]
[本文引用: 1]
DOI:10.1186/s12870-015-0501-xURL [本文引用: 2]
DOI:10.1073/pnas.1005931107URLPMID:20498060 [本文引用: 1]

Abstract Bar-coded multiplexed sequencing approaches based on new-generation sequencing technologies provide capacity to sequence a mapping population in a single sequencing run. However, such approaches usually generate low-coverage and error-prone sequences for each line in a population. Thus, it is a significant challenge to genotype individual lines in a population for linkage map construction based on low-coverage sequences without the availability of high-quality genotype data of the parental lines. In this paper, we report a method for constructing ultrahigh-density linkage maps composed of high-quality single-nucleotide polymorphisms (SNPs) based on low-coverage sequences of recombinant inbred lines. First, all potential SNPs were identified to obtain drafts of parental genotypes using a maximum parsimonious inference of recombination, making maximum use of SNP information found in the entire population. Second, high-quality SNPs were identified by filtering out low-quality ones by permutations involving resampling of windows of SNPs followed by Bayesian inference. Third, lines in the mapping population were genotyped using the high-quality SNPs assisted by a hidden Markov model. With 0.05x genome sequence per line, an ultrahigh-density linkage map composed of bins of high-quality SNPs using 238 recombinant inbred lines derived from a cross between two rice varieties was constructed. Using this map, a quantitative trait locus for grain width (GW5) was localized to its presumed genomic region in a bin of 200 kb, confirming the accuracy and quality of the map. This method is generally applicable in genetic map construction with low-coverage sequence data.
[本文引用: 1]
URL [本文引用: 1]
DOI:10.1007/s12284-008-9004-9URL [本文引用: 1]

The Committee on Gene Symbolization, Nomenclature and Linkage (CGSNL) of the Rice Genetics Cooperative has revised the gene nomenclature system for rice ( Oryza ) to take advantage of the completion of the rice genome sequence and the emergence of new methods for detecting, characterizing, and describing genes in the biological community. This paper outlines a set of standard procedures for describing genes based on DNA, RNA, and protein sequence information that have been annotated and mapped on the sequenced genome assemblies, as well as those determined by biochemical characterization and/or phenotype characterization by way of forward genetics. With these revisions, we enhance the potential for structural, functional, and evolutionary comparisons across organisms and seek to harmonize the rice gene nomenclature system with that of other model organisms. Newly identified rice genes can now be registered on-line at http://shigen.lab.nig.ac.jp/rice/oryzabase_submission/gene_nomenclature/ .
DOI:10.1007/978-1-4939-1966-6URL [本文引用: 1]
DOI:10.1111/pbi.12113URL [本文引用: 1]
DOI:10.1007/s001220051517URL [本文引用: 1]

Rice sheath blight, caused by Rhizoctonia solani K眉hn, is one of the three major diseases of rice. The present study was conducted with an F 2 clonal population of Jasmine 85/Lemont. The F 2 population, including 128 clonal families, was inoculated by short toothpicks incubated with a strain, RH-9 of the fungus. Based on field disease evaluations in 2 years and a genetic map with 118 evenly distributed molecular markers, we identified six quantitative trait loci (QTLs) contributing to sheath blight resistance. These QTLs, qSB-2, qSB-3, qSB-7, qSB-9-1, qSB-9-2 and qSB-11, were located on chromosomes 2, 3, 7, 9 and 11, respectively. The respective alleles of qSB-2, qSB-3, qSB-7, and qSB-9-2 from Jasmine 85 could explain 21.2%, 26.5%, 22.2% and 10.1% of the total phenotypic variation, respectively; while the alleles of qSB-9-1 and qSB-11 from Lemont could explain 9.8% and 31.2% of the total phenotypic variation. Of these qSB-2 and qSB-11 could be detected in both years, while remaining loci were detected only in a single year. Furthermore, four QTLs (qHD-2, qHD-3, qHD-5 and qHD-7) controlling heading date and three QTLs (qPH-3, qPH-4 and qPH-11) controlling plant height were also identified. Though rice sheath blight resistance may be influenced by morphological traits, such as heading date and plant height, in the present study most detected resistance loci were not linked to the loci for heading date or plant height.
DOI:10.1007/s001220100539URL [本文引用: 1]
DOI:10.1007/s00122-003-1401-2URLPMID:12961067 [本文引用: 1]

One hundred twenty six doubled-haploid (DH) rice lines were evaluated in nine diverse Asian environments to reveal the genetic basis of genotype × environment interactions (GEI) for plant height (PH) and heading date (HD). A subset of lines was also evaluated in four water-limited environments, where the environmental basis of G × E could be more precisely defined. Responses to the environments were resolved into individual QTL × environment interactions using replicated phenotyping and the mixed linear-model approach. A total of 37 main-effect QTLs and 29 epistatic QTLs were identified. On average, these QTLs were detectable in 56% of the environments. When detected in multiple environments, the main effects of most QTLs were consistent in direction but varied considerably in magnitude across environments. Some QTLs had opposite effects in different environments, particularly in water-limited environments, indicating that they responded to the environments differently. Inconsistent QTL detection across environments was due primarily to non- or weak-expression of the QTL, and in part to significant QTL × environment interaction effects in the opposite direction to QTL main effects, and to pronounced epistasis. QTL × environment interactions were trait- and gene-specific. The greater GEI for HD than for PH in rice were reflected by more environment-specific QTLs, greater frequency and magnitude of QTL × environment interaction effects, and more pronounced epistasis for HD than for PH. Our results demonstrated that QTL × environment interaction is an important property of many QTLs, even for highly heritable traits such as height and maturity. Information about QTL × environment interaction is essential if marker-assisted selection is to be applied to the manipulation of quantitative traits.
DOI:10.1007/BF00223380URLPMID:24166172 [本文引用: 1]

To detect QTLs controlling traits of agronomic importance in rice, two elite homozygous lines 9024 and LH422, which represent the indica and japonica subspecies of rice ( Oryza sativa ), were crossed. Subsequently a modified single-seed-descent procedure was employed to produce 194 recombinant inbred lines (F 8 ). The 194 lines were genotyped at 141 RFLP marker loci and evaluated in a field trial for 13 quantitative traits including grain yield. Transgressive segregants were observed for all traits examined. The number of significant QTLs (LOD 猢 2.0) detected affecting each trait ranged from one to six. The percentage of phenotypic variance explained by each QTL ranged from 5.1% to 73.7%. For those traits for which two or more QTLs were detected, increases in the traits were conditioned by indica alleles at some QTLs Japonica alleles at others. No significant evidence was found for epistasis between markers associated with QTLs and all the other markers. Pleitropic effects of single QTLs on different traits are suggested by the observation of clustering of QTLs. No QTL for traits was found to map to the vicinity of major gene loci governing the same traits qualitatively. Evidence for putative orthologous QTLs across rice, maize, oat, and barley is discussed.
DOI:10.1007/s00122-003-1270-8URLPMID:12736777 [本文引用: 4]

An advanced backcross population between an accession of Oryza rufipogon (IRGC 105491) and the U.S. cultivar Jefferson ( Oryza sativa ssp. japonica ) was developed to identify quantitative trait loci (QTLs) for yield, yield components and morphological traits. The genetic linkage map generated for this population consisted of 153 SSR and RFLP markers with an average interval size of 10.3聽cM. Thirteen traits were examined, nine of which were measured in multiple environments. Seventy-six QTLs above an experiment-wise significance threshold of P 3.6 or a composite interval mapping LOD > 3.9) were identified. For the traits measured in multiple environments, 47% of the QTLs were detected in at least two environments. The O. rufipogon allele was favorable for 53% of the yield and yield component QTLs, including loci for yield, grains per panicle, panicle length, and grain weight. Morphological traits related to the domestication process and/or weedy characteristics, including plant height, shattering, tiller type and awns, were found clustered on chromosomes 1 and 4. Comparisons to previous studies involving wild cultivated crosses revealed O. rufipogon alleles with stable effects in multiple genetic backgrounds and environments, several of which have not been detected in studies between Oryza sativa cultivars, indicating potentially novel alleles from O. rufipogon . Some O. rufipogon -derived QTLs, however, were in similar regions as previously reported QTLs from Oryza sativa cultivars, providing evidence for conservation of these QTLs across the Oryza genus. In addition, several QTLs for grain weight, plant height, and flowering time were localized to putative homeologous regions in maize where QTLs for these traits have been previously reported, supporting the hypothesis of functional conservation of QTLs across the grasses.
DOI:10.1007/s00122-004-1890-7URLPMID:15647921 [本文引用: 1]

To understand the types of gene action controlling seven quantitative traits in rice, QTL mapping was performed to dissect the main effect (M-QTLs) and digenic epistatic (E-QTLs) QTLs responsible for the trait performance of 254 recombinant inbred lines (RILs) of "Lemont/Teqing", and two testcross (TC) F 1 populations derived from these RILs. The correlation analyses reveal a general pattern, i.e. trait heritability in the RILs was negatively correlated to trait heterosis in the TC hybrids. A large number of M-QTLs and E-QTLs affecting seven traits, including heading date (HD), plant height (PH), flag leaf length (FLL), flag leaf width (FLW), panicle length (PL), spikelet number per panicle (SN) and spikelet fertility (SF), were identified and could be classified into two predominant groups, additive QTLs detected primarily in the RILs, and overdominant QTLs identified exclusively in the TC populations. There is little overlap between QTLs identified in the RILs and in the TC populations. This result implied that additive gene action is largely independent from non-additive gene action in the genetic control of quantitative traits of rice. The detected E-QTLs collectively explained a much greater portion of the total phenotypic variation than the M-QTLs, supporting prior findings that epistasis has played an important role in the genetic control of quantitative traits in rice. The implications of these results to the development of inbred and hybrid cultivars were discussed.
DOI:10.1270/jsbbs.51.191URL [本文引用: 1]
DOI:10.1007/s001220050872URL [本文引用: 1]
DOI:10.1270/jsbbs.53.51URL [本文引用: 1]

Fine mapping of Hd4 and Hd5, quantitative trait loci (QTLs) for heading date in rice, was performed by using advanced backcross progeny derived from a cross between a japonica rice variety, Nipponbare, and an indica variety, Kasalath. Hd4 was mapped between restriction fragment length polymorphism (RFLP) markers R46 and C39 in the proximal region of chromosome 7, and Hd5 was mapped between C166 and R902 on the short arm of chromosome 8; both QTLs mapped as single Mendelian factors. We used marker-assisted selection to develop two nearly isogenic lines (NIL), designated NIL(Hd4) and NIL(Hd5), in which small chromosomal segments of Kasalath including Hd4 and Hd5, respectively, each were substituted into the genetic background of Nipponbare. Compared with that of Nipponbare, daysto-heading of NIL(Hd4) and NIL(Hd5) increased under long-day and natural-field conditions, but no differences were observed between those of the two NILs and Nipponbare under short-day conditions. Epistatic interaction was detected between Hd5 and Hdl, a key photoperiod sensitivity QTL, on the basis of an analysis of the F
DOI:10.1016/S1673-8527(07)60006-XURLPMID:17469777 [本文引用: 1]

USSR5, a japonica rice variety from the former Soviet Union, is an extremely early maturing rice variety. To elucidate the genetic basis for its early heading, genetic analysis was carried out by crossing it with a set of major gene nearly isogenic lines (NIL) and QTL-isogenic lines. The early heading of USSR5 was attributed to the presence of photoperiod-insensitive alleles at E 1 and Se-1 gene, the photoperiod-sensitive inhibitor gene i-Se-1 , and the dominant earliness gene Ef-1 . Analysis of a backcrossed population (BC 1 F 1 ) derived from the cross USSR5 脳 N22 indicated that two quantitative trait loci (QTL) for early heading were located on chromosomes 7 and 8, accounting for 27.4% and 11.2% of the phenotypic variance, respectively, with both early alleles originating from USSR5. From an F 2 population of the same cross, early heading QTLs were detected on chromosomes 1, 2, 7, 9, and 10, with individual QTL accounting for between 4.1% and 15.4% of the phenotypic variance. Early heading alleles at four of these five QTLs originated from USSR5. A comparison of chromosomal locations suggests that one of these QTLs may be identical with the known gene Hd4 ( E 1 ). The relationship between the other QTLs and known genes for heading date are not clear. USSR5 is a promising source for propagating earliness for the development of improved early heading rice varieties.
URL [本文引用: 1]

对灿稻窄叶青8号(ZYQ8)和粳稻京系17(JX17)以及由它们构建的加倍单倍体(DH)群体,分别在杭州和海南岛,采用注射器接种法进行纹枯病抗性鉴定,并使用该群体的分子链锁图谱进行数量性状座位(QTL)分析。共检测到4个抗纹枯病的QTL(qSBR-2、qSBR-3、qSBR-7和qSBR-11),分别位于第2、第3、第7和第11染色体。其中qSBR-2、qSBR-3、qSBR-7的抗性基因由抗病亲本ZYQ8贡献,而qSBR-11的抗性基因来自感病亲本JX17。qSBR-2、qSBR-3、qSBR-7在杭州和海南岛都能检测到,而qSBR-11只在杭州检测到。在杭州的实验中,纹枯病病级与秆长和抽穗期呈显著负相关;在控制秆长和抽穗期的QTL中,控制秆长的qCL-3与qSBR-3位于同一染色体区域,其余QTL与抗纹枯病的QTL之间无连锁关系。
URL [本文引用: 1]

对灿稻窄叶青8号(ZYQ8)和粳稻京系17(JX17)以及由它们构建的加倍单倍体(DH)群体,分别在杭州和海南岛,采用注射器接种法进行纹枯病抗性鉴定,并使用该群体的分子链锁图谱进行数量性状座位(QTL)分析。共检测到4个抗纹枯病的QTL(qSBR-2、qSBR-3、qSBR-7和qSBR-11),分别位于第2、第3、第7和第11染色体。其中qSBR-2、qSBR-3、qSBR-7的抗性基因由抗病亲本ZYQ8贡献,而qSBR-11的抗性基因来自感病亲本JX17。qSBR-2、qSBR-3、qSBR-7在杭州和海南岛都能检测到,而qSBR-11只在杭州检测到。在杭州的实验中,纹枯病病级与秆长和抽穗期呈显著负相关;在控制秆长和抽穗期的QTL中,控制秆长的qCL-3与qSBR-3位于同一染色体区域,其余QTL与抗纹枯病的QTL之间无连锁关系。
[本文引用: 1]
DOI:10.1007/BF00223452URLPMID:24162532 [本文引用: 2]

We report here the RFLP mapping of quantitative triat loci (QTLs) that affect some important agronomic traits in cultivated rice. An anther culturederived doubled haploid (DH) population was established from a cross between an indica and a japonica rice variety. On the basis of this population a molecular linkage map comprising 137 markers was constructed that covered the rice genome at intervals of 14.8cM on average. Interval mapping of the linkage map was used to locate QTLs for such important agronomic traits as heading date, plant height, number of spikelets per panicle, number of grains Per panicle, 1000-grain weight and percentage of seed set. Evidence of genotype-byenvironment interaction was found by comparing QTL maps of the same population grown in three diverse environments. A total of 22 QTLs for six agronomic traits were detected that were significant in at least one environment, but only 7 were significant in all three environments, 7 were significant in two environments and 8 could only be detected in a single environment. However, QTL-by-environment interaction was traitdependent. QTLs for spikelets and grains per panicle were common across environments, while traits like heading date and plant height were more sensitive to environment.
DOI:10.1007/BF02951625URL [本文引用: 1]

对水稻籼粳杂交(窄叶青8号×京系17)F_1的花药进行离体培养,建立一个含133个DH系的作图群体,通过构建分子连锁图谱,对水稻上部节间长度、株高和抽穗期的QTL进行区间作图,定位了影响上部节间长度的12个QTL、株高的4个QTL和抽穗期的1个QTL。对这些QTL的遗传效应分析的结果表明,控制抽穗期的1个QTL即Hd8a为主效基因,其余的16个QTL为微效基因。控制上部节间长度单个的QTL对表型的贡献率介于8-18%,其加性效应可使所控制的节间长度增加大约1.6-3.6cm。值得注意的是,一些控制相关性状的、作用方向相同的QTL定位于同一染色体的相同或邻近区段上。这一结果揭示了这些性状相关的遗传基础,在水稻育种中运用这些QTL将有助于对株高进行精细的遗传调控。
DOI:10.1007/BF02951625URL [本文引用: 1]

对水稻籼粳杂交(窄叶青8号×京系17)F_1的花药进行离体培养,建立一个含133个DH系的作图群体,通过构建分子连锁图谱,对水稻上部节间长度、株高和抽穗期的QTL进行区间作图,定位了影响上部节间长度的12个QTL、株高的4个QTL和抽穗期的1个QTL。对这些QTL的遗传效应分析的结果表明,控制抽穗期的1个QTL即Hd8a为主效基因,其余的16个QTL为微效基因。控制上部节间长度单个的QTL对表型的贡献率介于8-18%,其加性效应可使所控制的节间长度增加大约1.6-3.6cm。值得注意的是,一些控制相关性状的、作用方向相同的QTL定位于同一染色体的相同或邻近区段上。这一结果揭示了这些性状相关的遗传基础,在水稻育种中运用这些QTL将有助于对株高进行精细的遗传调控。
