张永杰

 , 侯嘉玮, 张姝
, 侯嘉玮, 张姝

山西大学生命科学学院, 山西 太原 030006
收稿日期:2019-05-21;修回日期:2019-08-12;网络出版日期:2019-12-04
基金项目:国家自然科学基金(31872162);山西省回国留学人员科研资助(2017-015)
*通信作者:张永杰, E-mail:zhangyj2008@sxu.edu.cn;
张姝, E-mail:zhangshu@sxu.edu.cn.
摘要:[目的] 明确球孢白僵菌种内线粒体基因组的分化程度。[方法] 从GenBank下载已知的球孢白僵菌6个菌株线粒体基因组序列,详细分析基因组的组成结构,比较外显子区、内含子区和基因间区的碱基变异情况,分析菌株间的系统发育关系。[结果] 球孢白僵菌不同菌株的线粒体基因组大小为28.8-32.3 kb,都有14个常见的核心蛋白编码基因、2个rRNA基因和25个tRNA基因,具有很强的共线性关系。但是,不同菌株含有的线粒体内含子数目存在差异(2-5个/菌株),在cox1、cox2和nad1基因中表现出内含子插入/缺失多态性,这是导致线粒体基因组大小变化的主要因素。对外显子、内含子和基因间区的碱基变异情况进行分析,发现内含子和基因间区相对变异较大,而外显子区相对变异较小。系统发育分析发现,这些球孢白僵菌菌株以很高的支持度聚在一起,具有相同内含子分布规律的菌株也具有较近的聚类关系。[结论] 本研究首次报道球孢白僵菌因内含子数目不同、插入缺失突变和单核苷酸变异等在线粒体基因组上表现出较大程度的遗传分化,为认识真菌种内线粒体基因组分化提供了新的证据。
关键词:球孢白僵菌线粒体基因组遗传分化内含子系统发育
Intraspecific comparison of mitochondrial genomes in the entomopathogenic fungus Beauveria bassiana
Yongjie Zhang

 , Jiawei Hou, Shu Zhang
, Jiawei Hou, Shu Zhang

School of Life Sciences, Shanxi University, Taiyuan 030006, Shanxi Province, China
Received: 21 May 2019; Revised: 12 August 2019; Published online: 4 December 2019
*Corresponding author: Yongjie Zhang, E-mail:zhangyj2008@sxu.edu.cn;
Shu Zhang, E-mail:zhangshu@sxu.edu.cn.
Foundation item: Supported by the National Natural Science Foundation of China (31872162) and by the Research Project Supported by Shanxi Scholarship Council of China (2017-015)
Abstract: [Objective] To understand the intraspecific genetic differentiation of mitochondrial genomes (mitogenomes) in Beauveria bassiana. [Methods] Mitogenomes of six isolates of B. bassiana were downloaded from GenBank. Composition and structure of these mitogenomes, nucleotide variations at exonic, intronic and intergenic regions were compared. Phylogenetic relationship of these isolates was analyzed based on common core protein-coding genes. [Results] The mitogenome size of different isolates of B. bassiana ranged from 28.8 to 32.3 kb. There were 14 common core protein-coding genes, 2 rRNA genes and 25 tRNA genes in all of these mitogenomes, showing a strong collinear relationship. However, the number of introns in different isolates varied (2 to 5 introns per isolate). Intron insertion/deletion polymorphism was found in cox1, cox2 and nad1, as the main factor contributing to variations of mitogenome size. Based on nucleotide variations at exonic, intronic and intergenic regions, the relative variability at intronic and intergenic regions was larger than that at exonic regions. Phylogenetic analysis revealed that these isolates of B. bassiana grouped together with a high support value, and isolates with identical intron distribution tended to cluster closely. [Conclusion] B. bassiana shows genetic differentiation in its mitogenome due to intron number variations, indel and single nucleotide polymorphism. Our data provide new evidence for understanding the differentiation of mitogenomes in fungal species.
Keywords: Beauveria bassianamitochondrial genomegenetic differentiationintronphylogeny
线粒体DNA通常具有多拷贝、基因组较小、单亲遗传、较高的碱基变异速率等特性,常被用作真核生物生态学、群体遗传学等研究的分子标记[1]。真菌属于低等真核生物,在细胞核和线粒体中都含有遗传物质。近年来,随着测序成本的降低,有越来越多真菌的线粒体基因组被测序。真菌线粒体基因组的大小大体上介于动物和植物之间,但其线粒体基因组大小的变化倍数远超过动物和植物[2]。在真菌已知的线粒体基因组中,最小的只有12.1 kb[3],最大的达到235.8 kb[4],相差近20倍。除大小差异外,不同真菌的线粒体基因组还在基因排列顺序、遗传密码、内含子数目和插入位点等方面存在差异[5-6]。在种内水平,真菌线粒体基因组也可能存在较大变化。在开展过种内比较线粒体基因组学分析的真菌中,多数真菌因内含子数目不同引起种内线粒体基因组大小的差异[7-8],而少数真菌未在种内发现内含子数目的变化[9]。
球孢白僵菌是隶属虫草科的一种全球广布的昆虫病原真菌,拥有广泛的寄主范围(可侵染700多种昆虫和10余种蜱螨)[10],易人工大量生产,已在许多国家作为真菌杀虫剂广泛用于害虫生物防治[11]。该菌还能以内生菌的形式存在于多种植物中,可能在植物抵御病虫害中发挥作用[12-13]。中药中的白僵蚕即为球孢白僵菌感染家蚕幼虫而形成的僵虫。将球孢白僵菌开发成真菌杀虫剂时,以及将开发的真菌杀虫剂施用于田间后分析持久性及进行溯源分析时,都需要对菌株的遗传特性有深入的认识,从而有必要开展球孢白僵菌遗传多样性的分析。目前已有大量有关该菌遗传多样性分析的研究工作,主要使用RAPD、ISSR、AFLP等遗传标记。这些研究中的球孢白僵菌种群或大或小(即使用的菌株数量或多或少),但大都表明,球孢白僵菌表现出丰富的遗传多样性,且与地理或寄主的相关性较差[14-16]。
利用RFLP技术对球孢白僵菌线粒体DNA分析的结果显示,该菌在线粒体DNA上也表现出较大的遗传分化[17-18]。截止目前,在GenBank数据库中已有6个球孢白僵菌菌株的线粒体基因组数据,但是,该菌种内线粒体基因组的分化程度尚不清楚。此前,通过对隶属虫草科的蛹虫草和蝉花进行种内比较线粒体基因组分析,发现这两种真菌中都因内含子数目不同引起线粒体基因组的分化[8, 19]。我们推测球孢白僵菌也可能存在内含子插入缺失多态性。为了验证该假设,本文下载了球孢白僵菌已知的6个菌株的线粒体基因组序列,详细分析了基因组的组成结构,比较了外显子区、内含子区和基因间区的碱基变异情况,并分析了这些菌株的系统发育关系,为认识真菌种内线粒体基因组分化提供了新的证据。
1 材料和方法 1.1 菌株信息 目前,GenBank数据库中保存了球孢白僵菌6个菌株的线粒体基因组信息(表 1)。这些菌株来源于多个不同的地点和寄主昆虫,由多家单位提交线粒体基因组序列。
表 1. 球孢白僵菌菌株及其线粒体基因组信息 Table 1. Isolates of Beauveria bassiana and their mitogenome information
| Accession No. | Strain | Host | Locality | Size/bp | Institute | Ref. |
| NC_010652 | Bb13 | Dendrolimus punctatus | Anhui, China | 29961 | Anhui Agricultural University | [20] |
| EU100742 | Bb147 | Ostrinia nubilalis | France | 32263 | University of Athens | [21] |
| ANFO01001745 | D1-5 | Ostrinia furnacalis | Jilin, China | 32315 | Jilin Academy of Agricultural Sciences | N/A |
| KT201148 | K4 | N/A | N/A | 28816 | University of New Zealand | N/A |
| KT201149 | e17 | N/A | N/A | 29944 | University of New Zealand | N/A |
| KU869769 | N/A | N/A | N/A | 29922 | South China Agricultural University | N/A |
| N/A: not available. | ||||||
表选项
1.2 数据分析 为比较6个球孢白僵菌菌株的线粒体基因组,将来自不同菌株的同一线粒体基因的序列整理在一起,使用MAFFT (https://mafft.cbrc.jp/alignment/server/index.html?1801071719)进行序列比对。当发现GenBank数据库中的基因注释结果有误时,使用MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl)重新注释,并辅以必要的人工校对[22]。蛋白编码基因中的内含子采用Zhang & Zhang (2019)推荐的系统命名[23],rRNA中的内含子采用Johansen & Haugen (2001)推荐的系统命名[24]。内含子类型使用RNAweasel (http://megasun.bch.umontreal.ca/cgi-bin/RNAweasel/RNAweaselInterface.pl)分析。使用BRIG[25]对不同菌株的线粒体基因组进行全序列比对和图形化展示。使用DnaSP[26]统计外显子区、内含子区和基因间区的碱基变异情况。将14个蛋白编码基因的核苷酸序列拼接起来,以蛹虫草和蝉花为外群,使用RAxML[27]基于GTR+G模型构建系统发育树。
2 结果和分析 2.1 球孢白僵菌线粒体基因组的组成及大小变化 6个球孢白僵菌菌株的线粒体基因组大小各不相同,从28.8 kb到32.3 kb,最大相差3.5 kb (表 1)。造成不同菌株线粒体基因组大小差异的原因主要是不同菌株含有的内含子数目不同。每个菌株含有2-5个内含子,内含子平均长度1367 bp。在cob、cox1、cox2、nad1和rnl基因中均有内含子插入,每个基因最多存在1个内含子插入位点,不同菌株相同插入位点的内含子基本上属于同一内含子类型(表 2)。6个菌株共展现出3种不同的内含子分布模式(表 2,图 1)。此外,不同菌株间还存在一些短的插入缺失突变,这也是造成线粒体基因组大小差异的原因之一,具体见2.2中的分析。
表 2. 不同球孢白僵菌菌株含有的内含子及其类型 Table 2. Insertional site and type of mitochondrial introns among different isolates of B. bassiana
| Accession No. | cob | cox1 | cox2 | nad1 | rnl | No. introns |
| NC_010652 | P393 (ID) | P228 (IB) | mL2450 (IA) | 3 | ||
| EU100742 | P393 (ID) | P1057 (IB) | P228 (IB) | P636 (I derived, B1) | mL2450 (IA) | 5 |
| ANFO01001745 | P393 (ID) | P1057 (IB) | P228 (IB) | P636 (IB) | mL2450 (IA) | 5 |
| KT201148 | P393 (ID) | mL2450 (IA) | 2 | |||
| KT201149 | P393 (ID) | P228 (IB) | mL2450 (IA) | 3 | ||
| KU869769 | P393 (ID) | P228 (IB) | mL2450 (IA) | 3 | ||
| For protein-coding genes, the number after P represents the intron insertion site (relative to the corresponding genes of Tolypocladium inflatum). For rnl, the number after mL represents the intron insertion site (relative to the corresponding genes of Escherichia coli). Intron types are given in parentheses after insertional sites. | ||||||
表选项
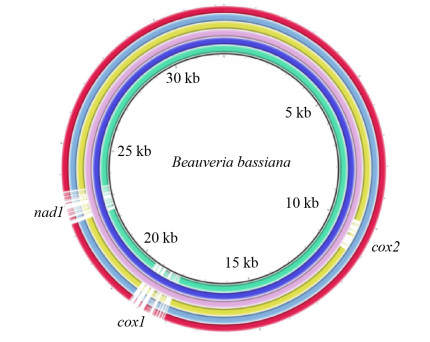 |
| 图 1 不同球孢白僵菌菌株线粒体基因组的比较 Figure 1 Comparison of mitochondrial genomes of different isolates of B. bassiana. Each ring represents a strain (represented by accession number), and they are NC_010652, EU100742, ANFO01001745, KT201148, KT201149 and KU869769 from inside to outside. There are intron presence/absence variations in cox1, cox2 and nad1 among different strains. When drawing the map using BRIG, EU100742 was used as the reference (because it has introns in all 5 intron insertional loci), and all other strains were compared to EU100742 by BLAST. |
| 图选项 |
尽管不同菌株的线粒体基因组大小存在差异,但是线粒体基因数目与排列顺序却非常保守,都含有14个常见的核心蛋白编码基因(排列顺序为nad2、nad3、atp9、cox2、nad4L、nad5、cob、cox1、nad1、nad4、atp8、atp6、cox3、nad6)、2个rRNA基因(rnl和rns)和25个tRNA基因(图 2)。这些tRNA基因主要聚集在rnl基因的上游(trnV、I、S1、W、P)和下游(trnT、E、M1、M2、L1、A、F、K、L2、Q、H、M3),以及rns基因的下游(trnY、D、S2、N)。然而,不同菌株间由于碱基的变化,在trnM1/M2基因间区和cox1/trnR2基因间区存在ORF的有无及其长短方面的差异(图 2)。基于全线粒体基因组序列的比对显示,除3个内含子位点外,这些菌株间具有非常好的共线性关系(图 1)。
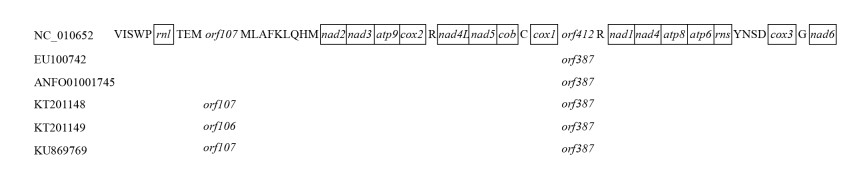 |
| 图 2 不同球孢白僵菌菌株线粒体基因及其排列顺序 Figure 2 Mitochondrial genes and their order in different isolates of B. bassiana. |
| 图选项 |
2.2 球孢白僵菌线粒体基因组的碱基变异 在外显子区,不同菌株间在除nad4L外的其他基因均有碱基变异,在cox1、nad1、nad2、rnl和rns中还存在碱基的插入/缺失突变(表 3)。在这些线粒体基因中,cox1、cox3、nad1、nad2和nad5相比其他基因变异要快一些。在多数蛋白编码基因中,碱基变异引起氨基酸的变化(表 3)。相比外显子区(平均变异频率1.16%),内含子区和基因间区的碱基变异更大(平均变异频率分别为4.51%和5.43%),且多数内含子区和基因间区存在碱基的插入/缺失突变(表 4)。在外显子区,单核苷酸变异(SNP)多于插入/缺失突变,而在内含子区和基因间区正好相反。整体来看,6个菌株线粒体基因组的碱基变异频率是13.06% (4247/32529,包括插入/缺失位点和SNP位点),SNP频率是1.24% (356/28638,不考虑插入/缺失位点)。
表 3. 不同球孢白僵菌菌株在外显子区的碱基和氨基酸变异 Table 3. Variations at exonic regions of each mitochondrial gene among different isolates of B. bassiana
| Gene | Exon | Protein | ||||||||||
| Length/bp | Pi | S | Indel | Total | % | Length/aa | Pi | S | Indel | Total | ||
| atp6 | 792 | 0 | 7 | 0 | 7 | 0.88 | 263 | 0 | 4 | 0 | 4 | |
| atp8 | 147 | 0 | 1 | 0 | 1 | 0.68 | 48 | 0 | 0 | 0 | 0 | |
| atp9 | 225 | 0 | 2 | 0 | 2 | 0.89 | 74 | 0 | 2 | 0 | 2 | |
| cob | 1176 | 0 | 5 | 0 | 5 | 0.43 | 391 | 0 | 3 | 0 | 3 | |
| cox1 | 1590-1608 | 2 | 26 | 24 | 52 | 3.23 | 529-535 | 0 | 8 | 8 | 16 | |
| cox2 | 750 | 0 | 3 | 0 | 3 | 0.40 | 249 | 0 | 2 | 0 | 2 | |
| cox3 | 810 | 2 | 7 | 0 | 9 | 1.11 | 269 | 0 | 6 | 0 | 6 | |
| nad1 | 1104-1107 | 1 | 12 | 3 | 16 | 1.45 | 367-368 | 1 | 6 | 1 | 8 | |
| nad2 | 1698-1704 | 4 | 12 | 6 | 22 | 1.29 | 565-567 | 2 | 7 | 2 | 11 | |
| nad3 | 420 | 0 | 1 | 0 | 1 | 0.24 | 139 | 0 | 0 | 0 | 0 | |
| nad4 | 1449 | 1 | 11 | 0 | 12 | 0.83 | 482 | 0 | 6 | 0 | 6 | |
| nad4L | 270 | 0 | 0 | 0 | 0 | 0.00 | 89 | 0 | 0 | 0 | 0 | |
| nad5 | 1986 | 9 | 22 | 0 | 31 | 1.56 | 661 | 3 | 9 | 0 | 12 | |
| nad6 | 639 | 1 | 2 | 0 | 3 | 0.47 | 212 | 1 | 0 | 0 | 1 | |
| rnl | 3210-3215 | 6 | 8 | 11 | 25 | 0.78 | ||||||
| rns | 1563-1567 | 4 | 8 | 6 | 18 | 1.15 | ||||||
| Combined | 17843-17860 | 30 | 128 | 50 | 208 | 1.16 | 4341-4346 | 7 | 53 | 11 | 71 | |
| Pi: parsimony informative sites; S: singleton sites; indel, insertion/deletion sites; %: the percentage of total variable sites (Pi, S, and indels). | ||||||||||||
表选项
表 4. 不同球孢白僵菌菌株在内含子区和基因间区的碱基变异 Table 4. Variations at intronic and intergenic regions among different isolates of B. bassiana
| Region in comparison | No. strains | Length/bp | Pi | S | Indel | Total | % | |
| Intronic regions | cobP393 | 6 | 1236-1240 | 5 | 14 | 6 | 25 | 2.01 |
| cox1P1057 | 2 | 1265 | 0 | 0 | 0 | 0 | 0.00 | |
| cox2P228 | 5 | 1036-1116 | 0 | 13 | 128 | 141 | 12.57 | |
| nad1P636 | 2 | 1075-1076 | 0 | 5 | 1 | 6 | 0.56 | |
| mL2450 | 6 | 1827-1898 | 3 | 37 | 86 | 126 | 6.60 | |
| Combined | 3113-6565 | 8 | 69 | 221 | 298 | 4.51 | ||
| Intergenic regions | rnl-nad2 | 6 | 1922-1953 | 3 | 26 | 64 | 93 | 4.74 |
| nad3-atp9 | 6 | 260-302 | 0 | 5 | 43 | 48 | 15.89 | |
| atp9-cox2 | 6 | 111-116 | 1 | 7 | 9 | 17 | 14.41 | |
| cox2-nad4L | 6 | 184 | 1 | 0 | 0 | 1 | 0.54 | |
| nad5-cob | 6 | 154 | 1 | 1 | 0 | 2 | 1.30 | |
| cob-cox1 | 6 | 406-412 | 0 | 0 | 8 | 8 | 1.94 | |
| cox1-nad1 | 6 | 1861-1937 | 5 | 16 | 93 | 114 | 5.85 | |
| nad1-nad4 | 6 | 371-386 | 1 | 10 | 22 | 33 | 8.42 | |
| nad4-atp8 | 6 | 71 | 0 | 0 | 0 | 0 | 0.00 | |
| atp8-atp6 | 6 | 106-107 | 1 | 2 | 4 | 7 | 6.42 | |
| atp6-rns | 6 | 429-442 | 2 | 6 | 13 | 21 | 4.75 | |
| rns-cox3 | 6 | 494-496 | 1 | 2 | 3 | 6 | 1.21 | |
| cox3-nad6 | 6 | 169 | 2 | 0 | 0 | 2 | 1.18 | |
| nad6-rnl | 6 | 1248-1287 | 6 | 25 | 54 | 85 | 6.58 | |
| Combined | 7846-7967 | 24 | 100 | 313 | 437 | 5.43 | ||
| The so-called “intergenic regions” in this table include possible tRNA genes and free-standing ORFs (see Figure 2). For all tRNA genes, only 2 and 1 SNP sites are present in trnE and trnM1, respectively, while all other tRNA genes are conserved. | ||||||||
表选项
2.3 不同球孢白僵菌菌株间的系统发育关系 利用14个线粒体蛋白编码基因构建系统发育树,发现球孢白僵菌的6个菌株以100%的支持度聚在一起,但内部各分支除NC_010652和KU869769以100%支持度聚在一起外,其他分支的支持度都不是很高(图 3)。尽管如此,从图中仍可看出,具有相同内含子分布规律的菌株具有较近的亲缘关系。
 |
| 图 3 基于线粒体DNA序列的不同球孢白僵菌菌株间的系统发育关系 Figure 3 Phylogeny of different isolates of B. bassiana based on mitochondrial nucleotide sequences. Nucleotide sequences of 14 protein-coding genes (total length 12889 nt) are used to construct the phylogenetic tree. The dots in the figure represent the presence of introns in corresponding genes (see also Table 2). |
| 图选项 |
3 讨论 真菌线粒体基因组不但在物种间有较大差异,而且在相同物种的不同菌株间也可能存在差异。虫草科目前至少有10种真菌已知线粒体基因组,但只有2种真菌(蛹虫草和蝉花)开展过种内比较线粒体基因组学分析[19]。本研究对球孢白僵菌开展了种内比较线粒体基因组学研究。在这3种真菌中,都发现了内含子插入/缺失多态性现象。由于线粒体内含子通常都比较长(如球孢白僵菌每个内含子平均长度约1.3 kb),因此导致线粒体基因组大小较大的变化。球孢白僵菌不同菌株的线粒体基因组相差可达3.5 kb,而蛹虫草和蝉花不同菌株的线粒体基因组相差最大可达约7 kb[8, 19]。相信随着研究菌株的增多,在这些真菌中可能发现更多线粒体基因组大小变化的信息。
通过对球孢白僵菌线粒体基因组不同区域碱基变异的分析,我们发现内含子区和基因间区的碱基变异频率大于外显子区。该结论与蛹虫草中的报道一致[8]。在球孢白僵菌中,rnl-nad2、nad3-atp9、atp9-cox2、cox1-nad1、nad1-nad4、atp8-atp6、atp6-rns、nad6-rnl这几个基因间区比其他基因间区具有更大的变异频率(表 4)。这些片段有可能作为今后开展球孢白僵菌遗传多样性分析的分子标记。从全线粒体基因组水平看,球孢白僵菌的SNP频率是1.24%,大于蛹虫草中报道的0.7%[8]和蝉花中报道的0.1%[19]。
曾有研究分析不同球孢白僵菌菌株在atp6-rns和nad3-atp9这两个基因间区片段上的变异,发现二者都比核糖体DNA ITS片段的变异大[21, 28]。本研究的6个球孢白僵菌菌株无法从公共数据库中全部找到ITS序列,因此不能进行线粒体DNA与ITS序列变异的比较。有研究通过比较5个球孢白僵菌菌株,发现其在核基因组上的SNP频率(以其中一个指定的菌株作为参照)为2.85-16.8 SNPs/kb[29]。对于本研究分析的6个球孢白僵菌菌株的线粒体基因组,外显子区、内含子区和基因间区的平均SNP频率分别为8.8、11.6和15.4 SNPs/kb (表 3,表 4)。这与报道的球孢白僵菌核基因组SNP频率相当。毫无疑问,若要准确比较核基因组和线粒体基因组的变异频率,使用选定的球孢白僵菌菌株同时进行这种比较才更具说服力。根据已有的报道,真菌线粒体DNA的进化速率可能比核DNA进化快,也可能比核DNA进化慢[2]。
References
| [1] | Galtier N, Nabholz B, Glémin S, Hurst GDD. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Molecular Ecology, 2009, 18(22): 4541-4550. DOI:10.1111/j.1365-294X.2009.04380.x |
| [2] | Sandor S, Zhang YJ, Xu JP. Fungal mitochondrial genomes and genetic polymorphisms. Applied Microbiology and Biotechnology, 2018, 102(22): 9433-9448. DOI:10.1007/s00253-018-9350-5 |
| [3] | James TY, Pelin A, Bonen L, Ahrendt S, Sain D, Corradi N, Stajich JE. Shared signatures of parasitism and phylogenomics unite cryptomycota and microsporidia. Current Biology, 2013, 23(16): 1548-1553. DOI:10.1016/j.cub.2013.06.057 |
| [4] | Losada L, Pakala SB, Fedorova ND, Joardar V, Shabalina SA, Hostetler J, Pakala SM, Zafar N, Thomas E, Rodriguez-Carres M, Dean R, Vilgalys R, Nierman WC, Cubeta MA. Mobile elements and mitochondrial genome expansion in the soil fungus and potato pathogen Rhizoctonia solani AG-3. FEMS Microbiology Letters, 2014, 352(2): 165-173. DOI:10.1111/1574-6968.12387 |
| [5] | Aguileta G, de Vienne DM, Ross ON, Hood ME, Giraud T, Petit E, Gabaldón T. High variability of mitochondrial gene order among fungi. Genome Biology and Evolution, 2014, 6(2): 451-465. DOI:10.1093/gbe/evu028 |
| [6] | Nie Y, Wang L, Cai Y, Tao W, Zhang YJ, Huang B. Mitochondrial genome of the entomophthoroid fungus Conidiobolus heterosporus provides insights into evolution of basal fungi. Applied Microbiology and Biotechnology, 2019, 103(9): 1379-1391. |
| [7] | Xiao SJ, Nguyen DT, Wu BJ, Hao WL. Genetic drift and indel mutation in the evolution of yeast mitochondrial genome size. Genome Biology and Evolution, 2017, 9(11): 3088-3099. DOI:10.1093/gbe/evx232 |
| [8] | Zhang S, Hao AJ, Zhao YX, Zhang XY, Zhang YJ. Comparative mitochondrial genomics toward exploring molecular markers in the medicinal fungus Cordyceps militaris. Scientific Reports, 2017, 7: 40219. DOI:10.1038/srep40219 |
| [9] | Zhang YJ, Yang XQ, Zhang S, Humber RA, Xu J. Genomic analyses reveal low mitochondrial and high nuclear diversity in the cyclosporin-producing fungus Tolypocladium inflatum. Applied Microbiology and Biotechnology, 2017, 101(23/24): 8517-8531. |
| [10] | Imoulan A, Hussain M, Kirk PM, El Meziane A, Yao YJ. Entomopathogenic fungus Beauveria: Host specificity, ecology and significance of morpho-molecular characterization in accurate taxonomic classification. Journal of Asia-Pacific Entomology, 2017, 20(4): 1204-1212. DOI:10.1016/j.aspen.2017.08.015 |
| [11] | de Faria MR, Wraight SP. Mycoinsecticides and Mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biological Control, 2007, 43(3): 237-256. DOI:10.1016/j.biocontrol.2007.08.001 |
| [12] | Vidal S, Jaber LR. Entomopathogenic fungi as endophytes: Plant-endophyte-herbivore interactions and prospects for use in biological control. Current Science, 2015, 109(1): 46-54. |
| [13] | Lefort MC, McKinnon AC, Nelson TL, Glare TR. Natural occurrence of the entomopathogenic fungi Beauveria bassiana as a vertically transmitted endophyte of Pinus radiata and its effect on above and belowground insect pests. New Zealand Plant Protection, 2016, 69: 68-77. DOI:10.30843/nzpp.2016.69.5917 |
| [14] | Serna-Dominguez MG, Andrade-Michel GY, Rosas-Valdez R, Castro-Felix P, Arredondo-Bernal HC, Gallou A. High genetic diversity of the entomopathogenic fungus Beauveria bassiana in Colima, Mexico. Journal of Invertebrate Pathology, 2019, 163: 67-74. DOI:10.1016/j.jip.2019.03.007 |
| [15] | Hu XL, He LM, Chen X, Zhang SL, Luan FG, Li ZZ. Genetic diversity and population genetic structure of Beauveria bassiana in South China. Chinese Journal of Biological Control, 2013, 29(1): 31-41. (in Chinese) 胡晓磊, 何玲敏, 陈雪, 张胜利, 栾丰刚, 李增智. 中国南方球孢白僵菌的遗传多样性和种群遗传结构. 中国生物防治学报, 2013, 29(1): 31-41. DOI:10.3969/j.issn.2095-039X.2013.01.005 |
| [16] | Garrido-Jurado I, Márquez M, Ortiz-Urquiza A, Santiago-álvarez C, Iturriaga EA, Quesada-Moraga E, Monte E, Hermosa R. Genetic analyses place most Spanish isolates of Beauveria bassiana in a molecular group with word-wide distribution. BMC Microbiology, 2011, 11: 84. DOI:10.1186/1471-2180-11-84 |
| [17] | Uribe D, Khachatourians G. Analysis and applications of the molecular typification of the mitochondrial genome of Beauveria bassiana. Revista Colombiana De Entomología, 2007, 33(2): 89-97. |
| [18] | Uribe D, Khachatourians GG. Restriction fragment length polymorphism of mitochondrial genome of the entomopathogenic fungus Beauveria bassiana reveals high intraspecific variation. Mycological Research, 2004, 108(9): 1070-1078. |
| [19] | Fan WW, Zhang S, Zhang YJ. The complete mitochondrial genome of the Chan-hua fungus Isaria cicadae: a tale of intron evolution in Cordycipitaceae. Environmental Microbiology, 2019, 21(2): 864-879. DOI:10.1111/1462-2920.14522 |
| [20] | Xu JZ, Huang B, Qin CS, Li ZZ. Sequence and phylogenetic analysis of Beauveria bassiana with mitochondrial genome. Mycosystema, 2009, 28(5): 718-723. |
| [21] | Ghikas DV, Kouvelis VN, Typas MA. Phylogenetic and biogeographic implications inferred by mitochondrial intergenic region analyses and ITS1-5. 8S-ITS2 of the entomopathogenic fungi Beauveria bassiana and B. brongniartii. BMC Microbiology, 2010, 10: 174. DOI:10.1186/1471-2180-10-174 |
| [22] | Zhang YJ, Zhang S, Zhang GZ, Liu XZ, Wang CS, Xu JP. Comparison of mitochondrial genomes provides insights into intron dynamics and evolution in the caterpillar fungus Cordyceps militaris. Fungal Genetics and Biology, 2015, 77: 95-107. DOI:10.1016/j.fgb.2015.04.009 |
| [23] | Zhang S, Zhang YJ. Proposal of a new nomenclature for introns in protein-coding genes in fungal mitogenomes. IMA Fungus, 2019, 10: 15. DOI:10.1186/s43008-019-0015-5 |
| [24] | Johansen S, Haugen P. A new nomenclature of group Ⅰ introns in ribosomal DNA. RNA, 2001, 7(7): 935-936. DOI:10.1017/S1355838201010500 |
| [25] | Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics, 2011, 12: 402. DOI:10.1186/1471-2164-12-402 |
| [26] | Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 2009, 25(11): 1451-1452. DOI:10.1093/bioinformatics/btp187 |
| [27] | Stamatakis A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 2014, 30(9): 1312-1313. DOI:10.1093/bioinformatics/btu033 |
| [28] | Kouvelis VN, Ghikas DV, Edgington S, Typas MA, Moore D. Molecular characterization of isolates of Beauveria bassiana obtained from overwintering and summer populations of Sunn Pest (Eurygaster integriceps). Letters in Applied Microbiology, 2010, 46(3): 414-420. |
| [29] | Valero-Jiménez CA, Faino L, Veld DSI, Smit S, Zwaan BJ, Van Kan JAL. Comparative genomics of Beauveria bassiana: uncovering signatures of virulence against mosquitoes. BMC Genomics, 2016, 17: 986. DOI:10.1186/s12864-016-3339-1 |
