闫雷1,3, 梁斌2, 王爱杰2, 刘双江1, 刘志培1


1. 中国科学院微生物研究所, 微生物资源前期开发国家重点实验室, 北京 100101;
2. 中国科学院生态环境研究中心, 中国科学院环境生物技术重点实验室, 北京 100085;
3. 中国科学院大学, 北京 100049
收稿日期:2019-09-19;修回日期:2019-11-27;网络出版日期:2019-12-04
基金项目:国家自然科学基金(31861133002);中国科学院重点部署项目(KFZD-SW-309)
作者简介:闫雷, 男, 1990年出生于甘肃天水市, 现中国科学院微生物研究所微生物专业博士研究生在读。主要研究方向为抗生素微生物降解的机制、水环境中抗生素及抗性基因的去除等研究。我们课题组常年致力于有机污染物的微生物降解及水土环境修复, 在污染物降解菌剂的研发、应用及降解机制解析方面有富足的经验.
*通信作者:刘志培。Tel:+86-10-64806081;E-mail:liuzhp@im.ac.cn.
摘要:抗生素是一类难降解、低浓度就有高生态毒性效应的化合物,近年来被归为新型环境污染物,其环境残留与去除备受关注。作为广泛使用的抗生素之一,磺胺甲恶唑在水土环境中的残留量不断增加,检出率也越来越高。研究表明,磺胺甲恶唑是少数几种可被微生物降解的抗生素之一,微生物降解法是最具潜力的残留磺胺甲恶唑去除手段。本文总结了磺胺甲恶唑在土壤、沉积物、活性污泥、混合菌群、酶等条件下的降解及已分离的具有降解能力的单菌株对磺胺甲恶唑的降解情况,包括其降解效率、降解条件等,归纳了目前磺胺甲恶唑微生物降解的主要分类,并讨论了影响磺胺甲恶唑降解的两个特有因素。指出从分子生物学及生物信息学角度研究其降解途径,降解菌、降解菌群的人工构建及其在含磺胺甲恶唑污水处理中的应用与效果评价等应为今后磺胺甲恶唑生物降解与应用研究的重点。
关键词:磺胺甲恶唑抗生素微生物降解新型污染物降解机制
Progress in microbial degradation of sulfamethoxazole
Lei Yan1,3, Bin Liang2, Aijie Wang2, Shuangjiang Liu1, Zhipei Liu1


1. State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China;
2. Key Laboratory for Environmental Biotechnology of Chinese Academy of Sciences, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing 100085, China;
3. University of Chinese Academy of Sciences, Beijing 100049, China
Received: 19 September 2019; Revised: 27 November 2019; Published online: 4 December 2019
*Corresponding author: Zhipei Liu, Tel: +86-10-64806081; E-mail: liuzhp@im.ac.cn.
Foundation item: Supported by the NSFC-EC Joint Program (31861133002) and by the Program of Chinese Academy of Sciences (KFZD-SW-309)
Abstract: Antibiotics are regarded as a new group of environmental pollutants in recent years because of they are difficult to degrade and highly ecotoxic even in low concentrations. More and more attentions have been paid to their residues and removals in environmental. With the wide use, sulfamethoxazole residual and the detection frequency in wastewater is increasing. As one of the few microbial degradable antibiotics, sulfamethoxazole degraded by microbes is one of the most promising methods. This article summarizes the degradation efficiency and degradation conditions of sulfamethoxazole by different soil, sediment, activated sludge, mixed flora, enzymes and isolated strains. Meanwhile, degradation pathways reported in the literature are summarized. Two specific factors affecting the degradation of sulfamethoxazole are also discussed. Finally, we look forward to the perspective of molecular biology and bioinformatics in new researches, as well as the construction of degrading bacterial consortium from different pollutants in future applications.
Keywords: sulfamethoxazoleantibacterialmicrobial degradationnew pollutantsdegradation mechanism
随着社会发展和各种需求的不断增加,抗生素在生活中的应用越来越广泛,尤其是在畜禽及水产养殖行业(约84%)[1],导致其在环境中的累积残留引发了越来越多的环保问题,不仅能直接对暴露在环境中的动植物产生毒害作用,更严重的是改变原有的环境菌群结构,筛选出具有抗药性的菌,还能促进抗药基因的产生、传播[2]。抗生素与抗药基因、抗药菌一起构成新的污染体系,互相联系,错综复杂,是环境修复面临的一大难题。
磺胺甲恶唑(Sulfamethoxazole,SMX),又名新诺明,是一种磺胺类的广谱抗生素,纯品为白色结晶粉末,无臭,味微苦,难溶于水、易溶于酸碱,结构式如图 1所示。因其在结构上类似对氨基苯甲酸(PABA),可与PABA竞争作用于细菌体内的二氢叶酸合成酶,阻止细菌二氢叶酸的合成,从而抑制细菌的生长繁殖[3]。主要用于治疗泌尿道感染以及禽霍乱等,对革兰氏阳性菌和革兰氏阴性菌都有良好的抑菌效果,目前广泛应用于人类、畜禽、水产养殖作物等领域。然而由于大量使用,SMX在环境中的残留问题日渐严重,不仅在环境中的检出率非常高,其累积量也越来越多,SMX在水环境中长期存在会对水生生物有严重的危害,对藻类的危险系数 > 7[4-5]。据报道,SMX在中国北方土壤中的残留量达到0.9 mg/kg,粪肥中的残留量高达5.7 mg/kg,而且在使用过粪肥助长的蔬菜中也检测到SMX[6]。在河流及地表水中SMX的含量也处于较高水平,西班牙略夫雷加特河河水中,SMX残留量高达1.49 μg/L[7],甚至研究人员从中国东部经过处理的饮用水中都检测到SMX,而这些饮用水的源头是中国第一大河长江[8]。另外,2017年10月27日,世界卫生组织国际癌症研究机构公布的致癌物清单初步整理,SMX名列3类致癌物清单中。因此,水环境中SMX的残留成为人类及其他动植物健康的一大隐患,而有效去除也逐渐成为一个人们热切关注的焦点。
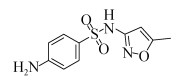 |
| 图 1 磺胺甲恶唑的化学结构 Figure 1 Chemical structure of sulfamethoxazole. |
| 图选项 |
目前,SMX的降解主要集中在物理法(光降解、热降解等)、化学法(芬顿氧化、电化学等)及微生物法。物理或者化学法不仅成本高昂,操作复杂,而且往往只是通过转化、修饰等途径把SMX转变成毒性较小、抑菌效果不明显的化合物,而这些化合物通常是比较稳定的复杂化合物,并不能参与自然界的物质循环,易造成环境中的累积,久而久之会成为自然环境的潜在威胁。相比之下,微生物降解法优势明显,操作简单,价格低廉,环境友好,没有二次污染[9],且已发现能把SMX矿化的菌群[10-11],使得微生物降解法成为非常具有潜力的一种抗生素去除途径。自本世纪初以来,SMX的微生物降解研究开始被广大科研工作者关注,不到20年时间里,已有很多文献报道了SMX在微生物的作用下而降解、代谢或者矿化。
然而,纵观这些文献,大多都是用活性污泥、土壤、沉积物以及混合菌群来降解SMX,在这种复杂的降解过程中,很难了解SMX具体是怎么进行转化、代谢的,更不能掌握在其微生物降解过程中所涉及的酶、关键基因等信息,阻碍了对微生物降解SMX的进一步认识和理解。相比之下,用可培养微生物来降解的研究起步较晚,报道较少。目前的报道主要集中在SMX降解菌筛选、降解产物解析、降解动力学研究以及降解条件优化;而从酶学、分子层面、基因层面去研究其降解机制、代谢途径的报道寥寥无几。这也是为什么SMX降解方法不少,但真正实际运用的却不多的重要原因之一。因此,在这个分子生物学迅速发展的时代,更应注意详细解析SMX微生物降解机制的重要性,以期为实际应用打下坚实的理论基础。
本文详细描述了科研工作者对SMX微生物降解的报道,总结了用土壤、沉积物等自然条件,各种生物反应器中的活性污泥,混合菌及纯菌,还有各种功能酶对SMX的降解效率及条件;归纳了目前已经报道的4种SMX降解方式;探讨了可能影响SMX降解的两个独特因素并指出以后该领域以后科研工作的重点。
1 土壤、沉积物等自然条件下SMX的降解 在自然条件下,残留的SMX也能在微生物、光照、水、热等一系列条件下实现降解(表 1)。但是在不同种类、质地的土壤中有不同的降解速率且容易受环境因素的影响,Yang等发现在壤质土中SMX的降解效果要明显优于粘壤土和壤砂土,初始浓度为10 mg/kg时,20天内降解效率分别为80%、50%、50%以上。而用水-沉积物去除SMX时,其降解效果受到温度及腐殖酸浓度的严重影响[12-13]。另外,在这种条件下SMX的降解通常极其缓慢且不易稳定进行,而在人工干预进行强化后其降解效率大大提高,Chuwen Yang等利用红树林沉积土为降解介质,10 mg/kg的SMX在不同地区的土壤以及不同的季节里的降解效果差异较大,在春季需要10–25 d能降解完全,在秋季30 d的降解效率为40%–60%。加入一定量的蔗糖后,在春季和秋季的完全降解时间分别变为5–15 d和20–30 d[14]。Juying等用未添加过肥料的农业土壤去研究其对SMX的降解发现,不经过人工干预的土壤对4 mg/kg的SMX完全降解需要80 d以上,而在加入5%–10%的生物固体后其降解效率明显提升[15]。
表 1. 菌群及酶对SMX的降解 Table 1. SMX degradation by microbial floras and enzymes
| Microbalfloras/Enryme | Degradation process | Initial concentration | Removal rate or efficiency | Time | Oxygen demand | Degradation products | References |
| Sludge | Sequencing Batch Reactor (SBR) activated sludge | 383 mg/L | 100% | 3 h | Aerobic | – | [16] |
| Photo-Fenton pretreatment followed by a Sequencing Batch Biofilm Reactor | 200 mg/L | nearly 100% | 48 h | 24 h Aerobic/ 24 h Anoxic | mineralized | [17] | |
| Biosolids | 100 μg/L | 100% | 2 d | Anoxic | – | [18] | |
| Activated Sludge Process | 100 μg/L | 17.1% | 48 h | Aerobic | – | [19] | |
| Internal loop photobiodegradation reactor (ILPBR) | 120 mg/L | 100% | 8 h | – | – | [20] | |
| Anaerobic packed and structured-bed reactors | 400 ng/L | (83%±12%)– (85%±10%) | 12 h | Anaerobic | – | [21] | |
| Sulfatereducing bacteria (SRB) sludge | 100 mg/L | 13.2±0.1 mg/(L·d) | Anoxic | – | [22] | ||
| Activated sludge | 10 mg/L | 100% | 14 d | Aerobic | 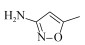 | [23] | |
| Activated sludge cultures | 10 mg/L | 100% | 8 d | Aerobic | – | [24] | |
| Sequencing Batch Reactor sludge | 10–40 mg/L | 59%–100% | 8–41 d | Anaerobic | – | [25] | |
| Sludge | 272 μg/L | 27 μg/(L·d) | 190 h | Aerobic | – | [26] | |
| Bioelectrochemical | 30 mg/L | > 95% | 21 h | – | – | [27] | |
| Sequencing Batch Reactor | 100 μg/L | 86% | 6 h | Aerobic | 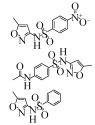 | [28] | |
| 98% | 24 h | ||||||
| Granular and Suspended Activated Sludge Processes | 2 μg/L | 84% in granular, 73% in suspended biomass | 12 h | Anoxic/anaerobic/oxic | – | [29] | |
| Rotating biological contactor (RBC) | 5 to 120 mg/L | 80.3%–89.4% | 60 h | Aerobic | – | [30] | |
| Swine sludge | 2 mg/L | 100% | 2 d | Anaerobic | – | [31] | |
| faster when added lactate, citrate, especially sucrose | |||||||
| Laboratory scale membrane bioreactors | 750 μg/L | 65% in average | 2 d | Aerobic | – | [32] | |
| 50 d | DO=0.5 mg/L | ||||||
| 30 d | DO > 2 mg/L | ||||||
| Microbial fuel cells(MFC) | 0.04/0.08/0.20/0.39/0.79 mmol/L | 0.67/1.37/3.43/7.32/13.36 μm/h | Anaerobic | 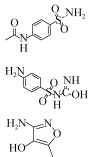 | [33] | ||
| Microbial fuel cell (MFC) reactors | 20 ppm | 100% | 3 d | Anaerobic |  | [34] | |
| 200 ppm | 100% | 96 h | |||||
| Microbial fuel cell | 20 ppm | 85% | 12 h | Anaerobic | 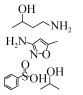 | [10] | |
| Sediments | Mangrove sediments | 10 mg/kg | 100%, | 30d | Anaerobic | [14] | |
| Water–sediment system of a natural river | 20 mg/kg | 90.1% in sediment | – | – | 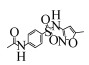 | [12] | |
| 15 mg/L | 86.8% in water | – | |||||
| Soils | Manure-amended soils | 10 mg/kg | > 50%, in clay loam loamy sand soils | 20 d | Faster in anaerobic than aerobic | – | [13] |
| > 80%, in loam soil | 20 d | ||||||
| Soil | 4 mg/kg | (7.6%±0.3%)– (6.8%±0.8%) | 84 d | – | mineralized | [15] | |
| Artificial composite soil treatment system | 5 mg/L | 70% | 10 d | – | – | [35] | |
| Artificial composite soil treatment systems | 15–30 μg/L | < 80% | 1 h | – | – | [36] | |
| Non-sterile soil | 10 mg/kg | 50% | 2 d | Aerobic | – | [37] | |
| 7 d | Anoxic | ||||||
| Laboratory columns/soil column test system | 0.25 mg/L | 60% | 14 d | Aerobic | – | [38] | |
| 0 | Anoxic | ||||||
| Consortium or biomass | Mixed consortium of bacteria | 6 mg/L | (47%±1.2%)– (55%±0.2%) | 300 h | Aerobic | – | [39] |
| Manganese-adapted aerobic biomass | 2 mg/L | > 98% | 2 d | Aerobic | – | [40] | |
| Horizontal-flow anaerobic immobilized biomass reactor | 40 μg/L | 97% | 24 h | Anaerobic | – | [41] | |
| Enzymes | A heme peroxidase, chloroperoxidase (CPO) | 15 μmol/L | 98.64% | 20 min | – | 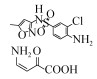 | [42] |
| Anaerobic sludge enzymes | 0.1 ng/μL | > 20% | 72 h | Anaerobic | – | [43] | |
| Others | A municipal sewage treatment plant | 20 mg/L | 100% | 50 d | Aerobic | – | [44] |
| Bioflocculant MFX | 1 mg/L | > 60% | 12 h | Aerobic | – | [45] | |
| Spent mushroom compost (SMC) | 200 mg/kg | 50% | 1.3 d | Aerobic | – | [46] |
表选项
总的来说,这些方法对SMX的去除效率与强度很容易受到土壤理化性质的干扰,其稳定性与持久性都不能保证,如果能针对性地加入SMX降解菌剂或者进行生物强化,其降解效率和强度都会进一步加强。
2 生物反应器中活性污泥对SMX的降解 在传统的污水处理过程中,活性污泥起到不可估量的作用。它对污水中有机污染物的吸附、降解有着重要的意义,可以说活性污泥是污水处理技术的灵魂。因此,在SMX的微生物降解过程中,污泥法研究得最多,配合各种各样的生物反应器(如序批式好氧反应器、生物膜反应器、微生物燃料电池等),在不同的温度、pH、外源碳的添加与否等条件下,从厌氧、兼氧到好氧以及好氧厌氧交替使用情况下,都实现了SMX的高效降解(表 1)。SMX初始浓度从400 ng/L到383 mg/L,大部分都能在2 d内实现80%以上的去除。尤其是Drillia等报道的序批式生物反应器在好氧条件下,能在3 h内将浓度高达383 mg/L的SMX降解完全[16],实现了高浓度SMX的快速、高效去除。
3 已分离菌对SMX的降解 3.1 混合菌对SMX的降解 随着磺胺甲恶唑微生物降解被日渐关注,研究者将目光从复杂环境投向混合菌以及纯菌,可以利用几种不同的菌对SMX进行分步降解,最后彻底矿化或者形成微生物容易利用的小分子物质。Ana C. Reis等发现SMX降解菌Achromobacter denitrificans PR1在单独行使降解功能的时候并不稳定且效率低,而当该菌与Leucobacter sp. GP混合培养时,对SMX的降解能力稳定又高效[47]。Xin等发现SMX能够抑制Pycnoporus sanguineus菌产生的漆酶活性,使得漆酶对SMX降解效率变低,而当把Pycnoporus sanguineus和Alcaligenes faecalis菌混合培养时,SMX降解效率大大提升[48]。我们实验室也曾对降解SMX的富集菌群进行分离培养,从中分离获得了包括SMX-R2A-1~6、SMX-1~3、SMX-LB-1、3A5MI-R2A-1等20多株的细菌,并确定了SMX-R2A-2 (Paenarthrobacter sp.)、SMX-LB-1 (Arthrobacter sp.)菌株对SMX具有降解效果。
3.2 单菌株对SMX的降解 如表 2所示,纯种微生物对SMX的降解主要有细菌、真菌和绿藻,这些微生物可通过生长代谢或共代谢的方式对SMX进行转化或部分降解,而能彻底降解/矿化SMX的菌株少见报道。研究较多的主要是细菌对SMX的降解,包括革兰氏阴性和阳性细菌都对SMX具有转化/降解的能力。从分类学上看,绝大多数SMX降解菌都属于变形菌门和放线菌门,厚壁菌门中只有2株希瓦氏菌具有SMX降解功能[49]。在属水平上,30株不同细菌来自15个属,其中出现频率较高的有假单胞属(Pseudomonas sp.)为6株,细杆菌属(Microbacterium sp.)为4株,红球菌属(Rhodococcus sp.)、无色杆菌属(Achromobacter sp.)各3株,产碱杆菌属(Alcaligenes sp.)、罗尔斯通菌属(Ralstonia sp.)和希瓦氏菌属(Shewanella sp.)各2株。6株真菌除了一株未知分类外,另外5株分别为烟管菌属,侧耳属,密孔菌属,栓菌属和平革菌属。仅有的1株绿藻为四链藻属。从SMX的降解类型来看,产碱杆菌属和红球菌属细菌更倾向于对SMX进行修饰转化,而罗尔斯通菌属、希瓦氏菌属和无色杆菌属细菌更倾向于将SMX降解为小分子化合物,大多数真菌的降解产物并未被鉴定出来。另外,这些纯培养微生物对SMX的降解大多都是在好氧条件下进行的,外加碳源或者维生素会明显提升其降解效率。在真菌界中,白腐真菌(white-rot fungi)在抗生素降解中发挥着重要作用,不仅对SMX有降解效果[50],对四环素及土霉素[51]、氟喹诺酮类药物环丙沙星[52]、正定霉素[53]甚至对农药炔咪菊酯(imiprothrin)、氯氰菊酯(cypermethrin)、卡巴呋喃(carbofuran)都有一定的降解效果[54],而这些降解机制往往都与它们分泌的漆酶有关,意味着这一类菌在环境微生物修复方面有良好的应用前景。
表 2. 纯菌株对SMX的降解 Table 2. SMX degradation by isolated strains
| Classificution | Strains | Initial concentration | Degradation rate, time and oxygen demand | Degradation products | References |
| Actinobacteria | Rhodococcus rhodochrous (ATCC13808) | 30 mg/L | 20%, 35 days, aerobic | 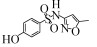 | [55] |
| Rhodococcus equi (ATCC 13557) | 6 mg/L | 29%, 120 h, aerobic | 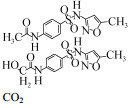 | [56] | |
| Microbacterium sp. JN196543 Rhodococcus sp. JN196542 | 0.5 mmol/L | 52%, 12 h, aerobic | CO2 | [57] | |
| Tsukumurella sp. JN851820 | – | [58] | |||
| Microbacterium sp. HF571532 Microbacterium sp. HF571537 | 10 mg/L | 1.0–1.7 mg/L/d, 10 days, aerobic More faster in R2A- UV and MSM-CN media | – | [59] | |
| Microbacterium sp. strain BR1 | 0.5 mmol/L | 100%, 1 h, aerobic | 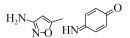 | [60] | |
| Proteobacteria | Gordonia sp. SMX-W2-SCD14 | 5 mg/L | 45.2%/62.2%/51.4%, 288 h, aerobic | 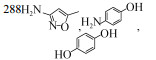 | [61] |
| Pseudomonas psychrophila HA-4 | 100 mg/L | 34.30%, 192 h, aerobic | 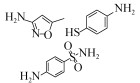 | [62] | |
| Achromobacter denitrificans PR1 | 600 μmoL | 73.6±9.6 μmol SMX/g cell dry weight 100%, 40 h, aerobic | 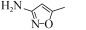 | [63] | |
| Alcaligenes faecalis CGMCC 1.767 | 50 mg/L | > 80%, 16 h, aerobic, 100 mg/L of exogenous VC, VB6 and GSSG enhanced SMX bioremoval |  | [64] | |
| Enterobacter cloacae T2 | 100 mg/kg | 84.14%, 30days, aerobic | 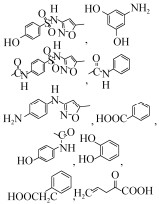 | [49] | |
| Shewanella oneidensis MR-1 Shewanella sp. strain MR-4 | 10 mg/L | 59.88%±1.23% and 63.89%±3.09%, 5 days, aerobic | 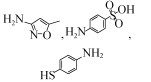 | [60] | |
| Ochrobactrum sp. SMX-PM1-SA1, Labrys sp. SMX-W1-SC11 | 5 mg/L | 45.2%/62.2%, 288 h, aerobic | 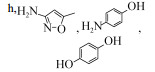 | [65] | |
| Alcaligenes faecalis CGMCC 1.767 | 50 mg/L | 90%, 24 h, aerobic | 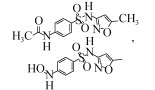 | [66] | |
| Acinetobacter sp. W1 | 5–240 mg/L | 100%, 50 h, aerobic | 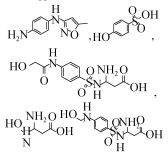 | [57] | |
| Achromobacter sp. JN196540 Achromobacter sp. JN196541 Ralstonia sp. JN196539 Ralstonia sp JN196538 | 0.5 mM | 52%, 12 h, aerobic | CO2 | [58] | |
| Fungus | Brevundimonas sp. HF571531 Pseudomonas sp. HF572913 Pseudomonas sp. HE985241 Pseudomonas sp. HF571533 Pseudomonas sp.HF571535 Pseudomonas sp. HF571536 Variovorax sp. HF571534 | 10 mg/L | 1.0–1.7 mg/(L·d), 10 days, aerobic More faster in R2A-UV and MSM-CN media | – | [67] |
| Trametes versicolor ATCC 42530 and Bjerkandera adusta ATCC 28314 | 25 mg/l | 90–94%, 30 days, anaerobic | – | [50] | |
| white-rot fungus Phanerochaete chrysosporium | 10 mg/L | 74%, 10 days, aerobic | – | [68] | |
| Pleurotus ostreatus | 50 mg/l | 74%, 15days | – | [48] | |
| Pycnoporus sanguineus and Alcaligenes faecalis | 50 mg/L | > 93%, 24 h,aerobic |  | [64] | |
| Alga | Fungal endophyte Z3 | 100 mg/kg | 57.64%, 30days, aerobic | – | [69] |
表选项
4 SMX的降解酶及其酶学研究 由于SMX微生物降解的研究起步较晚,在酶学、基因层面的研究鲜有报道。目前,已知的对SMX具有降解、转化作用的酶有三种:单加氧酶、来源于真菌的漆酶以及过氧化物酶(来源于一株Caldariomyces fumago),另外Gonzalez-Gil等用厌氧污泥中存在的混合酶来处理SMX,也有降解效果[43]。Benjamin Ricken等分离到一株Microbacterium sp.的菌株能以SMX为唯一碳源进行新陈代谢。该细菌中有一个基因簇发挥了降解功能,这个基因簇由3个功能基因组成,其中两个单加氧酶基因(sanA, sadB)和一个NADH+还原酶基因(sadC),通过SadA和SadB酶的作用,在FMNH2辅助下SMX最终生成3-氨基-5-甲基异恶唑(3A5MI)、偏苯三酚和SO2[59, 70-71](图 2),这也是目前为止仅有的从生物化学层面和分子生物学层面都进行验证过的文献报道。此外,该基因簇在放线菌中普遍存在且在该基因簇附近又发现转座子释放酶,这说明这种SMX降解机制可能在环境中普遍存在。漆酶对SMX的转化降解往往发生在真菌中,从效果上看漆酶的活性与SMX的降解效率具有很强的相关性[50, 68],然而其降解机理并不清楚。氯化物过氧化物酶对SMX的降解只有Xiao等[42]研究过,它能对SMX进行氯化修饰,对异恶唑环的打开也有着很好的效果,但是研究人员直接用酶去降解,其运行条件比较严苛,也并不清楚能生产该酶的菌对SMX的降解效果。
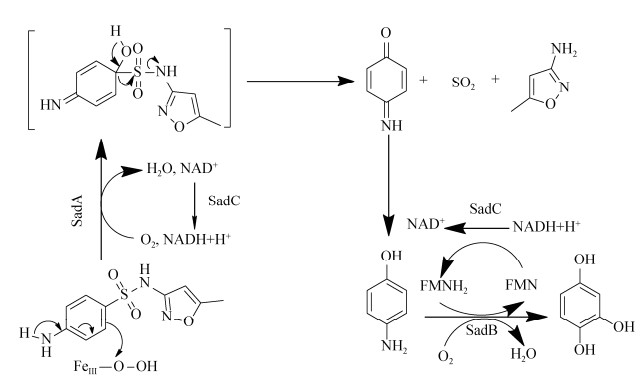 |
| 图 2 已验证的SMX降解途径及需要的酶[59, 70-71] Figure 2 Pathway and enzymes for SMX degradation[59, 70-71]. |
| 图选项 |
相比于已经发现报道的SMX降解菌株资源,其降解功能酶以及基因的深层次研究显得少之又少,尤其是从分子生物学的角度对这些代谢通路研究的缺乏使得我们对细菌降解抗生素的过程缺乏足够的了解和认识。
5 SMX的微生物降解方式 目前已检测到的SMX降解或转化主要有4种方式(如图 3,其中红色标注的是常见的代谢产物),首先是苯环与异恶唑环的断裂,能形成3A5MI、对氨基苯酚、对苯二酚、对巯基苯胺或者对氨基苯磺酸等物质,这是SMX降解最常见的一种形式,其中3A5MI也是最常见的降解产物[55, 61, 71]。第二种转化方式是苯环上氨基的修饰或者去除,乙酰化和羟基化是最常见的修饰方式,粪产碱杆菌对SMX的转化往往都是通过这两种修饰方式进行转化[48, 63, 65]。还有些菌株可以将SMX苯环上的氨基转换为羟基或者直接去掉,或者更复杂的修饰,而部分菌株能在这种转化方式的基础上再进行降解,最终生成分子量较小、容易利用的化合物。但也有很多菌并不能真正意义上降解SMX,而只是简单层次的转化修饰。第三种降解方式是对异恶唑环的作用或者对异恶唑环和苯环同时进行修饰,Shizong等[66]报道了菌株Acinetobacter sp. W1能在修饰SMX苯环氨基的同时将异恶唑环打开,然后再将这两部分断开,这是一种比较少见的降解方式。由于异恶唑环比较稳定、难降解,因此这种方式或许能给SMX的重要降解中间产物3A5MI的进一步降解提供思路。最后一种是苯环和异恶唑环之间连接键的变化,直接脱掉1个硫原子和2个氧原子,这种方式仅有一例报道且是根据降解产物荷质比对产物结构的猜测,并不具备普遍性。
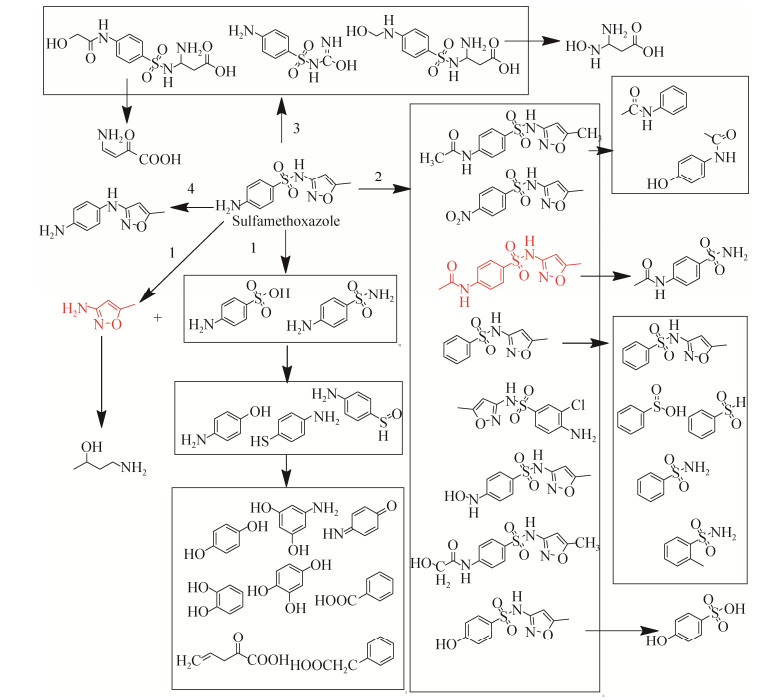 |
| 图 3 已检测到的SMX降解或转化方式 Figure 3 Degradation types of SMX degradation or transformation. |
| 图选项 |
6 影响降解的因素 众所周知,温度、pH、溶氧量、孵育时间、底物初始浓度以及外源碳、氮的添加都会影响细菌的生长繁殖,进而影响降解菌株在降解过程中的效率及强度,这些也都是之前研究比较关注的点。然而抗生素物是一种特殊的污染物,它的降解不光受到这些方面的影响,如SMX的降解与氨氧化过程及抗药性也有着千丝万缕的联系。
6.1 氨氧化过程对SMX降解的影响 很多污水处理厂、畜禽养殖污水处理工艺等在处理含有较高含量SMX污水的过程中,并没有特别添加SMX降解菌剂,但是往往都能取得不错的效果[44]。经研究发现,氨氧化细菌(AOB)和氨氧化古菌(AOA)与PPCPs (pharmaceutical and personal care products)的降解去除有密切关系[72-73]。2016年Kassotaki等发现,氨氧化效率的提升可以促进SMX的去除效率。在具有硝化功能的SBR反应器中,SMX的去除效率能达到98%,而当反应器中加入氨氧化抑制剂(烯丙基硫脲)后,SMX的去除效率直接降为0%[28]。由此可见,氨氧化作用与SMX的降解过程甚至是有机物的降解过程都有着千丝万缕的联系,但是到底这两者之间是如何影响、如何制约的,还需进一步研究。
6.2 耐药性与降解作用之间的关系 抗生素是一类能对微生物生长起到抑制作用的化合物,而在降解过程中这些化合物要对微生物起到促进生长的作用,这是也抗生素降解最难的地方。一般认为,抗生素降解菌是肯定能耐受抗生素的这一命题才合乎情理,即对该抗生素有耐药性才能对进一步降解。然而,有研究发现,4株Klebsiella pneumoniae和1株Escherichia fergusonii能利用氯霉素作为唯一碳源能能源进行生长,但是药敏试验发现这些菌株对氯霉素是敏感的[74],这表明细菌的耐药过程和降解药物过程是两个独立的途径。而又有报道指出,在具有抗药性的菌中,更容易找到能利用该抗生素的表型[75],这意味着抗药性或许对降解过程具有促进作用。那么它们之间到底有没有相关性,或者互相促进、调控的作用,这仍然是一个谜团。
对于SMX,目前已知的抗性基因主要是sul1、sul2和sul3,它们在降解细菌中的存在或者拷贝数的多少和降解菌对SMX的降解效果、效率之间的关系是一个很有意思的方向,需要进一步的研究。
7 总结和展望 随着近年来环境中残留抗生素、抗性基因和抗药菌被称为新型污染体系,用微生物的方法降解抗生素越来越受到大家的关注,尤其是磺胺类药物SMX的微生物降解,已经有很多的科研工作者发现、分离了众多降解菌株。然而,我们应该看到虽然降解菌株资源不断丰富,但是绝大多数科研工作都太过局限于菌株的分离和降解条件的优化,我们对具体的降解途径却知之甚少。迄今为止,只有一篇报道研究了SMX在降解过程中涉及的基因和酶,相比众多且复杂的降解途径来说,这方面的知识显得非常有限。而代谢途径不清楚,则无法详细了解降解过程,更无法将抗生素的微生物降解进一步挖掘和利用。因此,代谢途径的具体阐释及相关基因、酶的探究将会是该研究方向的重要目标。随着生物信息时代的到来,尤其是近年来组学分析、高通量测序等技术飞速发展,已经有很多科研工作者将目光投向了用基因组学的方式去探索、解释微生物对SMX的降解及转化,阅览近年来的文献可以明显看到降解细菌的基因组学分析、转录组学、代谢组学分析不断出现,这将非常准确高效地指导我们对抗生素降解菌资源的进一步开发利用,其中有用的基因、酶等关键信息都将被一一揭示出来,相信在不远的将来,广大科研工作者必将找到能高效、合理降解抗生素的优质方法。
另外,SMX的大部分降解产物都检测到3-氨基-5-甲基异恶唑,后者是一个环境毒性较SMX小的化合物,也具有一定的抗菌效果,化学性质比较稳定,并不容易被微生物降解,受到科研工作者的关注较少,其微生物降解的报道很少见。Lu Wang等发现在微生物燃料电池中,SMX被降解成3A5MI,而3A5MI可进一步降解成4-氨基-2-丁醇[10, 44]。另外,也有报道发现SMX可以先发生异恶唑开环反应再与苯环断裂的降解过程,然后直接生成直连化合物[22, 68]。但这些研究都只是根据质谱鉴定m/z值进行的产物预测,并没有直接证据证实其中发生的降解途径,其中的详细讲解途径和过程并不清楚,这也是往后SMX降解研究过程的一个难点。
磺胺甲恶唑作为一类广谱抗菌药物,其降解过程必然不同于其他传统的有机污染物,尤其是其抗药过程和降解过程这两者之间的联系将是一个非常有意思的研究方向。另外,在找到降解功能基因的基础上,研究其降解途径的调节机制,使其最大化地发挥降解功能,在抗菌药降解菌资源的挖掘和应用上有重要的意义。最后综合工艺使用上的优化参数,将这些抗生素降解菌应用到实际的环境修复当中去,才能使得这些微生物资源得到良好的利用。
本实验室长年致力于有机污染物及难降解污染物的微生物降解,目前已经分离得到了1株能高效降解SMX和1株能高效降解3A5MI的细菌,其中SMX降解菌SMX-R2A-2和3A5MI降解菌3A5MI-R2A-2 (Pimelobacter sp.)分别能在3天内将50 mg/L的底物进行彻底降解,菌SMX-R2A-2的能将SMX降解成3A5MI,而菌3A5MI-R2A-2能将3A5MI彻底矿化。将这两株菌共同培养,SMX能够彻底矿化。通过基因组测序,我们发现菌SMX-R2A-2中存在一个与SadA,SadB,SadC序列相似度高达90%以上的基因簇,其降解机制和途径跟之前科研工作者的报道或许一致。接下来我们将重点对3A5MI的功能酶、基因进行深层次挖掘,以期找到详细的降解途径,并计划进一步分析SMX降解过程和抗药过程之间的联系。结合近年蓬勃发展的微生物3D打印技术,我们还希望能综合多种环境污染物高效降解菌资源,构建能够对复杂污染物、不同类型污染物有降解效果的人工菌群,并投入到实际应用当中。
References
| [1] | Zhang QQ, Zhao JL, Ying GG, Liu YS, Pan CG. Emission estimation and multimedia fate modeling of seven steroids at the river basin scale in China. Environmental Science & Technology, 2014, 48(14): 7982-7992. |
| [2] | Lu X, Jin FG, Xu DY, Wu SY, Yan H, Shi L. The effect of resistance genes horizontal transfer under stimulation of subinhibitory concentrations of antibiotics. Chinese Journal of Antibiotics, 2014, 39(5): 379-384. (in Chinese) 鲁曦, 金发光, 徐冬旸, 吴少云, 闫鹤, 石磊. 3种亚抑菌浓度抗生素对耐药基因水平传播的影响. 中国抗生素杂志, 2014, 39(5): 379-384. |
| [3] | Sk?ld O. Sulfonamide resistance:mechanisms and trends. Drug Resistance Updates, 2000, 3(3): 155-160. DOI:10.1054/drup.2000.0146 |
| [4] | García-Galán M J, Díaz-Cruz M S, Barceló D. Occurrence of sulfonamide residues along the Ebro River basin:removal in wastewater treatment plants and environmental impact assessment. Environment International, 2011, 37(2): 462-473. DOI:10.1016/j.envint.2010.11.011 |
| [5] | Wang JL, Wang SZ. Microbial degradation of sulfamethoxazole in the environment. Applied Microbiology and Biotechnology, 2018, 102(8): 3573-3582. DOI:10.1007/s00253-018-8845-4 |
| [6] | Hua XG, Zhou QX, Luo Y. Occurrence and source analysis of typical veterinary antibiotics in manure, soil, vegetables and groundwater from organic vegetable bases, northern China. Environmental Pollution, 2010, 158(9): 2992-2998. DOI:10.1016/j.envpol.2010.05.023 |
| [7] | Charuaud L, Jarde E, Jaffrezic A, Thomas MF, Le Bot B. Veterinary pharmaceutical residues from natural water to tap water:sales, occurrence and fate. Journal of Hazardous Materials, 2019, 361: 169-186. DOI:10.1016/j.jhazmat.2018.08.075 |
| [8] | Hu YR, Jiang L, Zhang TY, Jin L, Han Q, Zhang D, Lin KF, Cui CZ. Occurrence and removal of sulfonamide antibiotics and antibiotic resistance genes in conventional and advanced drinking water treatment processes. Journal of Hazardous Materials, 2018, 360: 364-372. DOI:10.1016/j.jhazmat.2018.08.012 |
| [9] | Wang JL, Wang SZ. Removal of pharmaceuticals and personal care products (PPCPs) from wastewater:a review. Journal of Environmental Management, 2016, 182: 620-640. DOI:10.1016/j.jenvman.2016.07.049 |
| [10] | Wang L, Liu YL, Ma J, Zhao F. Rapid degradation of sulphamethoxazole and the further transformation of 3-amino-5-methylisoxazole in a microbial fuel cell. Water Research, 2016, 88: 322-328. DOI:10.1016/j.watres.2015.10.030 |
| [11] | Liu QQ, Li M, Liu X, Zhang Q, Liu R, Wang ZL, Shi XT, Quan J, Shen XH, Zhang FW. Removal of sulfamethoxazole and trimethoprim from reclaimed water and the biodegradation mechanism. Frontiers of Environmental Science & Engineering, 2018, 12(6): 6. |
| [12] | Xu BJ, Mao DQ, Luo Y, Xu L. Sulfamethoxazole biodegradation and biotransformation in the water-sediment system of a natural river. Bioresource Technology, 2011, 102(14): 7069-7076. DOI:10.1016/j.biortech.2011.04.086 |
| [13] | Wu Y, Williams M, Smith L, Chen DH, Kookana R. Dissipation of sulfamethoxazole and trimethoprim antibiotics from manure-amended soils. Journal of Environmental Science and Health, Part B, 2012, 47(4): 240-249. DOI:10.1080/03601234.2012.636580 |
| [14] | Yang CW, Tsai LL, Chang BV. Anaerobic degradation of sulfamethoxazole in mangrove sediments. Science of the Total Environment, 2018, 643: 1446-1455. DOI:10.1016/j.scitotenv.2018.06.305 |
| [15] | Li JY, Ye QF, Gan Jay. Influence of organic amendment on fate of acetaminophen and sulfamethoxazole in soil. Environmental Pollution, 2015, 206: 543-550. DOI:10.1016/j.envpol.2015.08.019 |
| [16] | Drillia P, Dokianakis SN, Fountoulakis MS, Kornaros M, Stamatelatou K, Lyberatos G. On the occasional biodegradation of pharmaceuticals in the activated sludge process:the example of the antibiotic sulfamethoxazole. Journal of Hazardous Materials, 2005, 122(3): 259-265. DOI:10.1016/j.jhazmat.2005.03.009 |
| [17] | González O, Esplugas M, Sans C, Torres A, Esplugas S. Performance of a sequencing batch biofilm reactor for the treatment of pre-oxidized sulfamethoxazole solutions. Water Research, 2009, 43(8): 2149-2158. DOI:10.1016/j.watres.2009.02.013 |
| [18] | Wu CX, Spongberg AL, Witter JD. Sorption and biodegradation of selected antibiotics in biosolids. Journal of Environmental Science and Health Part, A, 2009, 44(5): 454-461. DOI:10.1080/10934520902719779 |
| [19] | Li B, Zhang T. Biodegradation and adsorption of antibiotics in the activated sludge process. Environmental Science & Technology, 2010, 44(9): 3468-3473. |
| [20] | Yan N, Xia SQ, Xu LK, Zhu J, Zhang YM, Rittmann BE. Internal loop photobiodegradation reactor (ILPBR) for accelerated degradation of sulfamethoxazole (SMX). Applied Microbiology and Biotechnology, 2012, 94(2): 527-535. DOI:10.1007/s00253-011-3742-0 |
| [21] | Carneiro RB, Sabatini CA, Santos-Neto áJ, Zaiat M. Feasibility of anaerobic packed and structured-bed reactors for sulfamethoxazole and ciprofloxacin removal from domestic sewage. Science of the Total Environment, 2019, 678: 419-429. DOI:10.1016/j.scitotenv.2019.04.437 |
| [22] | Jia YY, Khanal SK, Zhang HQ, Chen GH, Lu H. Sulfamethoxazole degradation in anaerobic sulfate-reducing bacteria sludge system. Water Research, 2017, 119: 12-20. DOI:10.1016/j.watres.2017.04.040 |
| [23] | Müller E, Schüssler W, Horn H, Lemmer H. Aerobic biodegradation of the sulfonamide antibiotic sulfamethoxazole by activated sludge applied as co-substrate and sole carbon and nitrogen source. Chemosphere, 2013, 92(8): 969-978. DOI:10.1016/j.chemosphere.2013.02.070 |
| [24] | Herzog B, Lemmer H, Horn H, Müller E. Screening and monitoring microbial xenobiotics' biodegradation by rapid, inexpensive and easy to perform microplate UV-absorbance measurements. BMC Research Notes, 2014, 7(1): 101. DOI:10.1186/1756-0500-7-101 |
| [25] | Cetecioglu Z, Ince B, Gros M, Rodriguez-Mozaz S, Barceló D, Ince O, Orhon D. Biodegradation and reversible inhibitory impact of sulfamethoxazole on the utilization of volatile fatty acids during anaerobic treatment of pharmaceutical industry wastewater. Science of the Total Environment, 2015, 536: 667-674. DOI:10.1016/j.scitotenv.2015.07.139 |
| [26] | Alvarino T, Nastold P, Suarez S, Omil F, Corvini PFX, Bouju H. Role of biotransformation, sorption and mineralization of 14C-labelled sulfamethoxazole under different redox conditions. Science of the Total Environment, 2016, 542: 706-715. DOI:10.1016/j.scitotenv.2015.10.140 |
| [27] | Zhang S, Yang XL, Li H, Song HL, Wang RC, Dai ZQ. Degradation of sulfamethoxazole in bioelectrochemical system with power supplied by constructed wetland-coupled microbial fuel cells. Bioresource Technology, 2017, 244: 345-352. DOI:10.1016/j.biortech.2017.07.143 |
| [28] | Kassotaki E, Buttiglieri G, Ferrando-Climent L, Rodriguez-Roda I, Pijuan M. Enhanced sulfamethoxazole degradation through ammonia oxidizing bacteria co-metabolism and fate of transformation products. Water Research, 2016, 94: 111-119. DOI:10.1016/j.watres.2016.02.022 |
| [29] | Jafari Kang A, Brown AK, Wong CS, Yuan QY. Removal of antibiotic sulfamethoxazole by anoxic/anaerobic/oxic granular and suspended activated sludge processes. Bioresource Technology, 2018, 251: 151-157. DOI:10.1016/j.biortech.2017.12.021 |
| [30] | Azimi N, Hassani AH, Darzi GN, Borghei SM. Biodegradation of wastewater containing high concentration of sulfamethoxazole by antibiotic adopted biofilm in attached growth bioreactor. Polish Journal of Environmental Studies, 2017, 26(6): 2463-2469. DOI:10.15244/pjoes/67686 |
| [31] | Fan CH, Yang CW, Chang BV. Anaerobic degradation of sulfamethoxazole by mixed cultures from swine and sewage sludge. Environmental Technology, 2019, 40(2): 210-218. DOI:10.1080/09593330.2017.1384510 |
| [32] | Faisal IH, Li XQ, Price WE, Nghiem LD. Removal of carbamazepine and sulfamethoxazole by MBR under anoxic and aerobic conditions. Bioresource Technology, 2011, 102(22): 10386-10390. DOI:10.1016/j.biortech.2011.09.019 |
| [33] | Miran W, Jang J, Nawaz M, Shahzad A, Lee DS. Biodegradation of the sulfonamide antibiotic sulfamethoxazole by sulfamethoxazole acclimatized cultures in microbial fuel cells. Science of the Total Environment, 2018, 627: 1058-1065. DOI:10.1016/j.scitotenv.2018.01.326 |
| [34] | Wang L, Wu YC, Zheng Y, Liu LD, Zhao F. Efficient degradation of sulfamethoxazole and the response of microbial communities in microbial fuel cells. RSC Advances, 2015, 5(69): 56430-56437. DOI:10.1039/C5RA08438E |
| [35] | Liu QQ, Li M, Liu X, Zhang Q, Liu R, Wang ZL, Shi XT, Quan J, Shen XH, Zhang FW. Removal of sulfamethoxazole and trimethoprim from reclaimed water and the biodegradation mechanism. Frontiers of Environmental Science & Engineering, 2018, 12(6): 6. |
| [36] | Liu QQ, Li M, Liu R, Zhang Q, Wu D, Zhu DN, Shen XH, Feng CP, Zhang FW, Liu X. Removal of trimethoprim and sulfamethoxazole in artificial composite soil treatment systems and diversity of microbial communities. Frontiers of Environmental Science & Engineering, 2019, 13(2): 28. |
| [37] | Liu F, Ying GG, Yang JF, Zhou LJ, Tao R, Wang L, Zhang LJ, Peng PA. Dissipation of sulfamethoxazole, trimethoprim and tylosin in a soil under aerobic and anoxic conditions. Environmental Chemistry, 2010, 7(4): 370-376. DOI:10.1071/EN09160 |
| [38] | Baumgarten B, J?ehrig J, Reemtsma T, Jekel M. Long term laboratory column experiments to simulate bank filtration:Factors controlling removal of sulfamethoxazole. Water Research, 2011, 45(1): 211-220. DOI:10.1016/j.watres.2010.08.034 |
| [39] | Larcher S, Yargeau V. The effect of ozone on the biodegradation of 17α-ethinylestradiol and sulfamethoxazole by mixed bacterial cultures. Applied Microbiology and Biotechnology, 2013, 97(5): 2201-2210. DOI:10.1007/s00253-012-4054-8 |
| [40] | Zhang YJ, Zhu H, Szewzyk U, UweGeissen S. Enhanced removal of sulfamethoxazole with manganese-adapted aerobic biomass. International Biodeterioration & Biodegradation, 2017, 116: 171-174. |
| [41] | Chatila S, Amparo MR, Carvalho LS, Penteado ED, Tomita IN, Santos-Neto áJ, Gomes PCFL, Zaiat M. Sulfamethoxazole and ciprofloxacin removal using a horizontal-flow anaerobic immobilized biomass reactor. Environmental Technology, 2016, 37(7): 847-853. DOI:10.1080/09593330.2015.1088072 |
| [42] | Zhang X, Li XH, Jiang YC, Hu MC, Li SN, Zhai QG. Combination of enzymatic degradation by chloroperoxidase with activated sludge treatment to remove sulfamethoxazole:performance, and eco-toxicity assessment. Journal of Chemical Technology and Biotechnology, 2016, 91(11): 2802-2809. DOI:10.1002/jctb.4888 |
| [43] | Gonzalez-Gil L, Krah D, Ghattas AK, Carballa M, Wick A, Helmholz L, Lema JM, Ternes TA. Biotransformation of organic micropollutants by anaerobic sludge enzymes. Water Research, 2019, 152: 202-214. DOI:10.1016/j.watres.2018.12.064 |
| [44] | Pérez S, Eichhorn P, Aga DS. Evaluating the biodegradability of sulfamethazine, sulfamethoxazole, sulfathiazole, and trimethoprim at different stages of sewage treatment. Environmental Toxicology and Chemistry, 2005, 24(6): 1361-1367. DOI:10.1897/04-211R.1 |
| [45] | Xing J, Yang JX, Li A, Ma F, Liu KX, Dan W, Wei W. Removal efficiency and mechanism of sulfamethoxazole in aqueous solution by bioflocculant MFX. Journal of Analytical Methods in Chemistry, 2013, 2013: 568614. |
| [46] | Yang CW, Hsiao WC, Chang BV. Biodegradation of sulfonamide antibiotics in sludge. Chemosphere, 2016, 150: 559-565. DOI:10.1016/j.chemosphere.2016.02.064 |
| [47] | Reis AC, ?van?arová M, Liu Y, Lenz M, Hettich T, Kolvenbach BA, Corvini PFX, Nunes OC. Biodegradation of sulfamethoxazole by a bacterial consortium of Achromobacter denitrificans PR1 and Leucobacter sp. GP. Applied Microbiology and Biotechnology, 2018, 102(23): 10299-10314. DOI:10.1007/s00253-018-9411-9 |
| [48] | Li X, Xu QM, Cheng JS, Yuan YJ. Improving the bioremoval of sulfamethoxazole and alleviating cytotoxicity of its biotransformation by laccase producing system under coculture of Pycnoporus sanguineus and Alcaligenes faecalis. Bioresource Technology, 2016, 220: 333-340. DOI:10.1016/j.biortech.2016.08.088 |
| [49] | Mao F, Liu XH, Wu K, Zhou C, Si YB. Biodegradation of sulfonamides by Shewanella oneidensis MR-1 and Shewanella sp. strain MR-4. Biodegradation, 2018, 29(2): 129-140. |
| [50] | Guo XL, Zhu ZW, Li HL. Biodegradation of sulfamethoxazole by Phanerochaete chrysosporium. Journal of Molecular Liquids, 2014, 198: 169-172. DOI:10.1016/j.molliq.2014.06.017 |
| [51] | Tian QP, Dou X, Huang L, Wang L, Meng D, Zhai LX, Shen Y, You CP, Guan ZB, Liao XR. Characterization of a robust cold-adapted and thermostable laccase from Pycnoporus sp. SYBC-L10 with a strong ability for the degradation of tetracycline and oxytetracycline by laccase-mediated oxidation. Journal of Hazardous Materials, 2020, 382: 121084. DOI:10.1016/j.jhazmat.2019.121084 |
| [52] | Singh SK, Khajuria R, Kaur L. Biodegradation of ciprofloxacin by white rot fungus Pleurotus ostreatus. 3 Biotech, 2017, 7(1): 69. |
| [53] | Kornillowicz-Kowalska T, Ginalska G, Belcarz A, Iglik H. Microbial conversion of daunomycin wastes in unsteril soil inoculated with Bjerkandera adusta R59. Applied Microbiology and Biotechnology, 2006, 70(4): 497-504. |
| [54] | Mir-Tutusaus JA, Masís-Mora M, Corcellas C, Eljarrat E, Barceló D, Sarrà M, Caminal G, Vicent T, Rodríguez-Rodríguez CE. Degradation of selected agrochemicals by the white rot fungus Trametes versicolor. Science of the Total Environment, 2014, 500-501: 235-242. DOI:10.1016/j.scitotenv.2014.08.116 |
| [55] | Gauthier H, Yargeau V, Cooper DG. Biodegradation of pharmaceuticals by Rhodococcus rhodochrous and Aspergillus niger by co-metabolism. Science of the Total Environment, 2010, 408(7): 1701-1706. DOI:10.1016/j.scitotenv.2009.12.012 |
| [56] | Larcher S, Yargeau V. Biodegradation of sulfamethoxazole by individual and mixed bacteria. Applied Microbiology and Biotechnology, 2011, 91(1): 211-218. |
| [57] | Bouju H, Ricken B, Beffa T, Corvini PFX, Kolvenbach B A. Isolation of bacterial strains capable of sulfamethoxazole mineralization from an acclimated membrane bioreactor. Applied and Environmental Microbiology, 2012, 78(1): 277-279. DOI:10.1128/AEM.05888-11 |
| [58] | Herzog B, Lemmer H, Horn H, Müeller E. Characterization of pure cultures isolated from sulfamethoxazole-acclimated activated sludge with respect to taxonomic identification and sulfamethoxazole biodegradation potential. BMC Microbiology, 2013, 13: 276. DOI:10.1186/1471-2180-13-276 |
| [59] | Ricken B, Corvini PFX, Cichocka D, Parisi M, Lenz M, Wyss D, Martínez-Lavanchy PM, Müller JA, Shahgaldian P, Tulli LG, Kohler HPE, Kolvenbach BA. ipso-hydroxylation and subsequent fragmentation:a novel microbial strategy to eliminate sulfonamide antibiotics. Applied and Environmental Microbiology, 2013, 79(18): 5550-5558. DOI:10.1128/AEM.00911-13 |
| [60] | Mulla SI, Hu AY, Sun Q, Li J, Suanon F, Ashfaq M, Yu CP. Biodegradation of sulfamethoxazole in bacteria from three different origins. Journal of Environmental Management, 2018, 206: 93-102. |
| [61] | Jiang BC, Li A, Cui D, Cai R, Ma F, Wang YN. Biodegradation and metabolic pathway of sulfamethoxazole by Pseudomonas psychrophila HA-4, a newly isolated cold-adapted sulfamethoxazole-degrading bacterium. Applied Microbiology and Biotechnology, 2014, 98(10): 4671-4681. DOI:10.1007/s00253-013-5488-3 |
| [62] | Reis PJM, Reis AC, Ricken B, Kolvenbach BA, Manaia CM, Corvini PFX, Nunes OC. Biodegradation of sulfamethoxazole and other sulfonamides by Achromobacter denitrificans PR1. Journal of Hazardous Materials, 2014, 280: 741-749. DOI:10.1016/j.jhazmat.2014.08.039 |
| [63] | Zhang YB, Zhou J, Xu QM, Cheng JS, Luo YL, Yuan YJ. Exogenous cofactors for the improvement of bioremoval and biotransformation of sulfamethoxazole by Alcaligenes faecalis. Science of the Total Environment, 2016, 565: 547-556. DOI:10.1016/j.scitotenv.2016.05.063 |
| [64] | Hu SB, Hu HM, Li WL, Ke YY, Li MH, Zhao YC. Enhanced sulfamethoxazole degradation in soil by immobilized sulfamethoxazole-degrading microbes on bagasse. RSC Advances, 2017, 7(87): 55240-55248. DOI:10.1039/C7RA10150C |
| [65] | Tang MH, Gao N, Zhou J, Zhao Y, Cheng JS, Su WK, Yuan YJ. Improvement of sulfamethoxazole (SMX) elimination and inhibition of formations of hydroxylamine-SMX and N4-acetyl-SMX by membrane bioreactor systems. Biodegradation, 2018, 29(3): 245-258. DOI:10.1007/s10532-018-9826-0 |
| [66] | Wang SZ, Wang JL. Biodegradation and metabolic pathway of sulfamethoxazole by a novel strain Acinetobacter sp.. Applied Microbiology and Biotechnology, 2018, 102(1): 425-432. |
| [67] | Aydin S. Enhanced biodegradation of antibiotic combinations via the sequential treatment of the sludge resulting from pharmaceutical wastewater treatment using white-rot fungi Trametes versicolor and Bjerkandera adusta. Applied Microbiology and Biotechnology, 2016, 100(14): 6491-6499. DOI:10.1007/s00253-016-7473-0 |
| [68] | De Araujo CAV, Maciel GM, Rodrigues EA, Silva LL, Oliveira RF, Brugnari T, Peralta RM, Souza CGM. Simultaneous removal of the antimicrobial activity and toxicity of sulfamethoxazole and trimethoprim by white rot fungi. Water, Air, & Soil Pollution, 2017, 228(9): 341. |
| [69] | Xiong JQ, Kim SJ, Kurade MB, Govindwar S, Abou-Shanab RAI, Kim JR, Roh HS, Khan MA, Jeon BH. Combined effects of sulfamethazine and sulfamethoxazole on a freshwater microalga, Scenedesmus obliquus:toxicity, biodegradation, and metabolic fate. Journal of Hazardous Materials, 2019, 370: 138-146. DOI:10.1016/j.jhazmat.2018.07.049 |
| [70] | Ricken B, Kolvenbach BA, Bergesch C, Benndorf D, Kroll K, Strnad H, Vl?ek C, Adaixo R, Hammes F, Shahgaldia P, Sch?ffer A, Kohle HPE, Corvini PFX. FMNH2-dependent monooxygenases initiate catabolism of sulfonamides in Microbacterium sp. strain BR1 subsisting on sulfonamide antibiotics. Scientific Reports, 2017, 7(1): 15783. |
| [71] | Ricken B, Fellmann O, Kohler HPE, Sch?ffer A, Corvini PFX, Kolvenbach BA. Degradation of sulfonamide antibiotics by Microbacterium sp. strain BR1-elucidating the downstream pathway. New Biotechnology, 2015, 32(6): 710-715. DOI:10.1016/j.nbt.2015.03.005 |
| [72] | Men YJ, Han P, Helbling DE, Jehmlich N, Herbold C, Gulde R, Onnis-Hayden A, Gu AZ, Johnson DR, Wagner M, Fenner K. Biotransformation of two pharmaceuticals by the ammonia-oxidizing archaeon Nitrososphaera gargensis. Environmental Science & Technology, 2016, 50(9): 4682-4692. |
| [73] | Park J, Yamashita N, Wu GX, Tanaka H. Removal of pharmaceuticals and personal care products by ammonia oxidizing bacteria acclimated in a membrane bioreactor:contributions of cometabolism and endogenous respiration. Science of the Total Environment, 2017, 605-606: 18-25. DOI:10.1016/j.scitotenv.2017.06.155 |
| [74] | Zhao X, Tian FW, Wang G, Liu XM, Zhang QX, Zhang H, Chen W. Isolation, identification and characterization of human intestinal bacteria with the ability to utilize chloramphenicol as the sole source of carbon and energy. FEMS Microbiology Ecology, 2012, 82(3): 703-712. DOI:10.1111/j.1574-6941.2012.01440.x |
| [75] | de J Bello González T, Zuidema T, Bor G, Smidt H, Van Passel MWJ. Study of the Aminoglycoside subsistence phenotype of bacteria residing in the gut of humans and zoo animals. Frontiers in Microbiology, 2015, 6: 1550. |
