任浩, 王绿菁, 戴楚涵, 吕镇梅


浙江大学生命科学学院, 生命系统稳态与保护教育部重点实验室, 浙江杭州 310058
收稿日期:2019-10-29;修回日期:2019-11-29;网络出版日期:2019-12-06
基金项目:国家自然科学基金(41630637,41721001)
作者简介:任浩, 男, 1991年12月出生, 2017年9月入浙江大学生命科学学院攻读博士学位, 主要研究方向为呋喃类难降解有机污染物微生物降解机制探究.
*通信作者:吕镇梅, Tel:+86-571-88206279;E-mail:lzhenmei@zju.edu.cn.
摘要:含氧杂环化合物(Oxygen heterocycles)是污染环境中常见的一类难降解有机污染物,具有毒性和致癌特性,其所引发的环境问题受到人们广泛关注。本文综述了典型含氧单杂环化合物四氢呋喃、1,4-二氧六环以及含氧稠杂环化合物二苯并呋喃、二苯并对二噁英的生物降解研究进展,主要包括降解菌降解性能、降解途径和降解相关基因。此外,本文还对二噁英类衍生物的研究现状进行了讨论,并展望了含氧杂环及二噁英类衍生物生物降解的未来研究方向。
关键词:含氧杂环化合物降解菌降解途径降解基因二噁英
Research advances in biodegradation of oxygen heterocycles and their derivatives
Hao Ren, Lüjing Wang, Chuhan Dai, Zhenmei Lü


Key Laboratory of Biosystems Homeostasis and Protection, Ministry of Education, College of Life Sciences, Zhejiang University, Hangzhou 310058, Zhejiang Province, China
Received: 29 October 2019; Revised: 29 November 2019; Published online: 6 December 2019
*Corresponding author: Zhenmei Lü, Tel: +86-571-88206279; E-mail: lzhenmei@zju.edu.cn.
Foundation item: Supported by the National Natural Science Foundation of China (41630637, 41721001)
Abstract: Oxygen heterocycles are a kind of refractory organic pollutants commonly found in polluted environments. These compounds are of immense concern from point view of the environment because they are known for their toxic and carcinogenic properties. Here, we review the research progress in the biodegradation of typical oxygen-containing monoheterocyclic compounds tetrahydrofuran, 1, 4-dioxane and oxygen-containing fused heterocyclic compounds dibenzofuran and dibenzo-p-dioxin. We address the degradation performance of bacteria, the degradation pathways and degradation-related genes. In addition, we discuss the research status of dioxins, and propose further research directions of biodegradation of oxygen heterocycles and their dioxin derivatives.
Keywords: oxygen heterocyclesdegradation bacteriadegradation pathwaysdegradation genesdioxins
杂环化合物(Heterocyclic compounds)是一类具有环状结构,成环原子除碳原子外,还有其他元素的有机化合物。分子中含有的环状骨架叫作杂环,参与杂环组成的非碳原子统称为杂原子,最常见的杂原子有氧(O)、氮(N)和硫(S)[1-2]。杂环化合物的种类繁多,占已知有机化合物的一半以上[3],按杂原子的不同常分为含氧杂环化合物、含氮杂环化合物和含硫杂环化合物三大类,按含环数目可分为单杂环和稠杂环两大类。最常见单杂环有五元环和六元环,而稠杂环由苯环与单杂环或者由两个以上的单杂环稠合而成。杂环化合物因其独特的结构和特性在有机化合物合成中占重要地位,常作为合成中间体、保护基、手性助剂和有机催化剂[3-5]。杂环化合物应用十分广泛,如在医药领域常用于合成抗肿瘤、抗疟疾和抗糖尿病类药物[6-8];在农药领域可作为除草剂和杀虫剂[9];在材料领域因其光致变色及生物和化学发光性质常用于染料、荧光传感器和分析试剂[3, 10]。杂环化合物及其衍生物也是一类重要的有毒、难降解有机物,大多数具有致癌、致畸和致突变性[11],它们所引起的环境污染和健康问题也日益受到关注。
含氧杂环化合物常见如呋喃、四氢呋喃、1, 4-二氧六环、苯并呋喃、1, 4-苯并二噁烷,二苯并呋喃和二苯并对二噁英等(图 1)。其中稠杂环化合物如二苯并呋喃和二苯并对二噁英苯环上的氢原子都可被1–4个氯原子取代,并且由于取代位置和数量的不同可产生多种异构体,形成二噁英类杂环衍生物[12]。二噁英类化合物(Dioxins)是一类持久性有机污染物,一般包括多氯代二苯并呋喃(Polychlorinated dibenzofuran, PCDFs)和多氯代二苯并二噁英(Polychlorinated dibenzo-p-dioxin, PCDDs)两类物质。它们分别具有二苯并呋喃和二苯并对二噁英类似的骨架结构,在大气、土壤以及水体中通常以微小颗粒的形态存在,是目前已知化合物中毒性最强的污染物[13-15]。
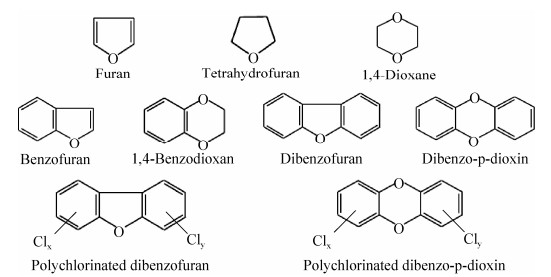 |
| 图 1 环境污染物中典型的含氧杂环化合物及其衍生物 Figure 1 Examples of oxygen heterocycles and their derivatives that are commonly found in environmental pollutants. |
| 图选项 |
1 含氧单杂环化合物 1.1 典型含氧单杂环化合物结构及特性 四氢呋喃和1, 4-二氧六环是含氧单杂环化合物的典型代表,其生物降解研究受到广泛关注。四氢呋喃(Tetrahydrofuran,THF),别名1, 4-环氧丁烷,为饱和四碳环醚,是结构最简单的呋喃类化合物。THF是一种重要的有机合成原料和性能优良的溶剂[16],广泛应用于医药和化工领域,常作为三氯乙烯和三氯乙烷的稳定剂以及高分子聚合物如聚氯乙烯的反应介质。THF具有肝脏毒性与致癌活性,多项小鼠体外和体内毒理学实验表明THF可通过诱导细胞增殖、DNA损伤和引起神经系统功能障碍等多种方式诱发肿瘤形成[17-19]。
1, 4-二氧六环(1, 4-Dioxane,14D),别名1, 4-二氧己环、二噁烷,为无色透明的杂环有机化合物,含两个对称醚键,具高水溶性,能迅速从土壤迁移到地表和地下水。14D常作为三氯乙烷的稳定剂,也常存在于化妆品、洗涤剂的表面活性剂中,最终释放到生活和工业废水中造成环境污染[20-22]。国际癌症研究机构将THF和14D归为B2类致癌物,大量研究表明长期暴露于14D会造成肾衰竭、肝损伤甚至诱发癌症[21, 23]。世界卫生组织建议将饮用水中14D含量的安全标准定为50 μg/L[24]。我国国家质量监督检验检疫总局于2009年对化妆品中14D含量限量要求低于100 mg/kg。近年来,14D用量显著增加,欧盟成员国每年的14D产量至少达到1000 t[25]。2015年,Adamson等[26]研究发现加利福尼亚州2000多处地下水中有194处含14D污染,其中95%的含14D样品同时含有含氯污染物如三氯乙烯,且样品中14D污染浓度远超美国EPA制定的14D浓度应低于3 μg/L的饮用水水质安全标准[27]。2017年,对美国公共饮用水供应中心收集的数据分析发现4864处供水系统中有21%的水体样本含14D,其中6.9%的样品超过了健康参考浓度,位列28种污染物的第二位[28]。THF和14D因具有高水溶性而难以阻滞或吸附到固相,利用常规膜处理技术很难有效去除此类污染物[29],因此寻求高效的生物降解技术日益迫切。
1.2 THF和14D降解菌 THF和14D均属于难降解有机污染物,目前共分离纯化获得18株以THF为唯一碳源和能源生长的降解菌和12株以14D为唯一碳源和能源生长的降解菌(表 1)。THF和14D降解菌主要来源于革兰氏阳性菌Rhodococcus sp.、Pseudonocardia sp.和Mycobacterium sp.,其中R.ruber 219[30]、P. dioxanivorans CB1190[31]和M. dioxanotrophicus PH-06[32]可降解THF和14D两种底物。另外,革兰氏阴性菌Afipia sp. D1[33]、Flavobacterium sp.[34]和真菌Cordyceps sinensis[35]也具有降解THF和14D的能力。自1991年Bernhardt等[36]发现第一株THF和14D降解菌R. ruber 219以来,研究者一直致力于从环境中分离纯化具有强耐受能力的高效降解菌。2009年,Yao等[37]从活性污泥中分离纯化获得Rhodococcus sp. YYL,其最大THF耐受浓度达200 mmol/L且能在32 h内将6 mmol/L THF完全降解。2010年,Chen等[38]从制药废水中分离纯化获得Pseudomonas oleovorans DT4,该菌株最大THF耐受浓度达100 mmol/L,最大降解速率达203.9 mg THF/(h g dry weight),明显高于其他已报道THF降解菌。此外,混合菌群也具有良好的THF降解能力。2019年,Huang等[39]通过富集驯化培养获得了一个稳定的THF降解混合菌群H-1,其最大THF耐受浓度和最大THF降解率分别达200 mmol/L、208.522 mg THF/(h g dry weight)。14D与THF结构类似,多数THF降解菌同时具有单独或共代谢降解14D的能力。1994年,Parales等[40]利用14D作为唯一碳源连续富集培养,最终分离获得一株能以14D、THF、1, 3-二氧六环、四氢吡喃和2-甲基-1, 3-二氧戊环为唯一碳源生长的好氧放线菌P. dioxanivorans CB1190,其14D最大降解速率达0.33 mg dioxane/(min mg protein)。相较于已知代表性14D降解菌P. dioxanivorans CB1190[31]、P. benzenivorans B5[41]和Afipia sp. D1[33],Xanthobacter flavus DT8[42]具有更强的降解性能,其最大生长速率(0.15 h–1)和细胞产率[0.35 mg protein/(mg dioxane)]优于其他菌株,且能完全降解1200 mg/L 14D。
表 1. 目前已报道的四氢呋喃和1, 4-二氧六环降解菌 Table 1. Tetrahydrofuran and 1, 4-dioxane degradation microorganisms reported so far
| Classified | Species | Strains | Degradation substrate (sole carbon) | Tolerated concentration | Degradation ability | Cometabolic substrates | References |
| G+ | Rhodococcus sp. | R. ruber M2 | THF | ND | ND | Daye et al., 2003 | |
| R. ruber 219 | THF, 14D | Above 10 mmol/L THF | 28.5 mg THF/ (h g dry weight) | Bernhardt et al., 1991 | |||
| Rhodococcus sp. YYL | THF | 200 mmol/L THF | 137.60 mg THF/ (h g dry weight) | Yao et al., 2009 | |||
| Rhodococcus sp. DTB | THF | ND | ND | Horn et al., 2005 | |||
| R. aetherivorans M8 | THF | 35 mmol/L THF | 5 mmol/L THF within 5 h | Tajima et al., 2012 | |||
| R. aetherivorans JCM14343 | 14D | 1100 mg/L 14D | 0.0073 mg dioxane/ (h mg protein) | THF/14D 1, 4- Butanediol/THF | Inoue et al., 2018 | ||
| R. ruber T1 | THF | Above 400 mg/L | ND | THF/14D | Sei et al., 2013 | ||
| R. jostii RHA1 | 1-Butanol/14D Propane/14D | Hand et al., 2015 | |||||
| R. ruber T5 | THF | Above 300 mg/L | ND | THF/14D | Sei et al., 2013 | ||
| R. ruber ENV425 | Propane/14D | David et al., 2015 | |||||
| G+ | Pseudonocardia sp. | P. benzenivorans B5 | 14D | ND | 0.1 mg dioxane/ (h mg protein) | Mahendra et al., 2006 | |
| Pseudonocardia sp. M1 | THF | ND | ND | Daye et al., 2003 | |||
| P. dioxanivorans JCM13855 | 14D | ND | 20 mg/L dioxane within 4 d | THF/14D | Inoue et al., 2016 | ||
| P. acaciae JCM 16707 | THF/14D | Inoue et al., 2016 | |||||
| P. asaccharolytica JCM 10410 | THF/14D | Inoue et al., 2016 | |||||
| P. dioxanivorans CB1190 | 14D, THF | ND | 0.33 mg dioxane/ (min mg protein) | Parales et al., 1994 | |||
| Pseudonocardia sp. ENV478 | THF | ND | ND | THF/14D | Masuda et al., 2012 | ||
| P. antarctica DVS 5a1 | 14D | ND | ND | Li et al., 2010 | |||
| Pseudonocardia sp. K1 | THF | Above 30 mmol/L THF | 20 mmol/L THF within 72 h | THF/14D | Kohlweyer et al., 2000 Mahendra et al., 2006 | ||
| G+ | Mycobacterium sp. | M. dioxanotrophicus PH-06 | THF, 14D | Above 900 mg/L 14D Above 500 mg/L THF | 900 mg/L 14D with 15 d | Kim et al., 2009 | |
| M. vaccae JOB5 | Propane/THF Propane/14D 1-Butanol/14D | Lan et al., 2013 Hand et al., 2015 | |||||
| G- | Acinetobacter sp. | A. baumannii DD1 | THF, 14D | 1000 mg/L | 100 mg/L 1, 4- dioxane within 42 h | Huang et al., 2014 | |
| G- | Afipia sp. | Afipia sp. D1 | THF, 14D | Above 100 mg/L 14D | 0.263 mg dioxane/ (h mg protein) | Sei et al., 2013 | |
| G- | Pseudomonas sp. | P. oleovorans DT4 | THF | 100 mmol/L THF | 203.9 mg THF/ (h g dry weight) | Chen et al., 2010 | |
| P. mendocina KR1 | Toluene/14D | Mahendra et al., 2013 | |||||
| G- | Azoarcus sp. | Azoarcus sp. DD4 | Propane/14D | Deng et al., 2018 | |||
| G- | Flavobacterium sp. | Flavobacterium sp. | THF, 14D | ND | ND | THF/14D | Sun et al., 2010 |
| G- | Xanthobacter sp. | X. flavus DT8 | 14D | 1200 mg/L 14D | 100 mg/L 14D within 50 h | THF/14D | Chen et al., 2016 |
| Fungus | Cordyceps sp. | C. sinensis | 14D, THF | ND | 0.034 mol/L 14D within 3 d | Nakamiya et al., 2005 | |
| Fungus | Graphium sp. | Graphium sp. (ATCC 58400) | THF | ND | 20 mmol/L THF within 9 d | Propane/14D THF/14D | Skinner et al., 2009 |
| ND: Not determined | |||||||
表选项
共代谢现象广泛存在于有机污染物降解过程,目前已报道有超过16株菌能共代谢降解14D,主要分布于Rhodococcus sp.、Pseudonocardia sp.、Mycobacterium sp.、Pseudomonas sp.、Flavobacterium sp.和Xanthobacter sp.。共代谢一般需要基础生长底物的诱导,常用的诱导底物包括THF、甲烷、丙烷、甲苯、1, 4-丁二醇和乙醇[38, 43-45]。低浓度诱导底物能促进14D的降解,而当诱导底物浓度高于14D浓度时,诱导底物能够竞争性抑制14D降解[46-47]。另外,在共代谢过程中,一些含氯污染物如三氯乙烷会竞争性抑制14D降解或产生有害中间产物二氯乙烯影响14D降解过程[48-49]。
目前研究仍存在一些不足之处:(1)已报道THF和14D降解菌主要为革兰氏阳性菌,当面对较高浓度THF和14D时,目前的降解菌株普遍出现延滞期延长或者无法完全矿化底物的现象,严重影响污染物降解效率,革兰氏阴性及真菌降解菌株资源有待进一步挖掘。(2)面对实际复杂的复合污染环境,单一降解菌难以发挥理想效果,利用微生物共代谢开发复合菌群的相关研究有待加强。(3)未培养微生物在污染物降解中发挥着不可忽视的作用,目前对未培养微生物的降解性能研究缺乏。
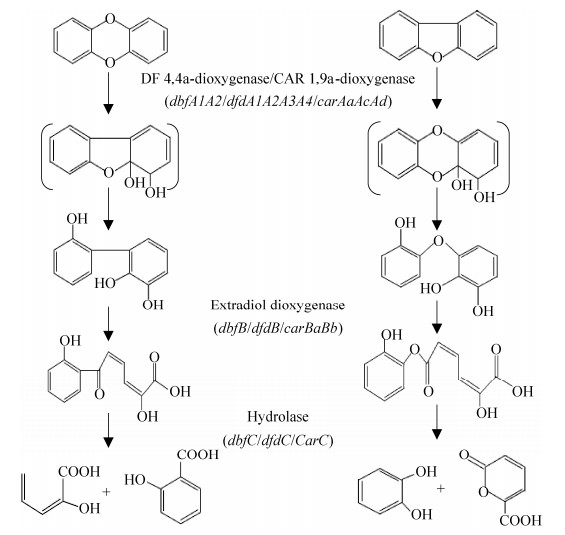
1.3 THF和14D降解途径 目前推测微生物主要通过氧化途径(Oxidation pathway)降解THF(图 2)。1991年,Bernhardt等[36]首次在R.ruber 219中提出THF降解氧化途径,THF经氧原子邻位碳原子羟基化生成2-羟基四氢呋喃,再经氧化反应生成γ-丁内酯,然后开环生成4-羟基丁酸,4-羟基丁酸进一步氧化为丁二酸进入三羧酸循环,进而彻底矿化。另外,2-羟基四氢呋喃也可首先生成异构体4-羟基丁醛,然后再经氧化生成4-羟基丁酸[50]。THF降解主要在胞内进行,产生的关键代谢产物含量低且转化快,难以进行检测和鉴定。研究者分别在细菌Rhodococcus sp. DTB[51]和真菌Graphium sp. ATCC58400[50]降解THF的过程中发现THF降解过程中的下游关键代谢产物γ-丁内酯。2012年,Tajima等[52]利用静息细胞反应在R. aetherivorans M8降解THF的过程中检测到了4-羟基丁酸。目前研究的不足之处:至今未明确检测到THF代谢过程中关键的代谢产物2-羟基四氢呋喃,更未有利用GC-MS进行鉴定的相关报道。
 |
| 图 2 目前已报道THF和14D降解途径[50, 58] Figure 2 Tetrahydrofuran and 1, 4-dioxane degradation pathways reported so far[50, 58]. The structures shown in brackets have not been characterized yet. |
| 图选项 |
14D降解过程中的关键中间代谢产物已陆续报道,其主要代谢途径如图 2所示。2005年,Nakamiya等[35]以重氢标记的14D-d8为底物在真菌C. sinensis共代谢14D过程中检测到代谢产物乙烯乙二醇、乙醇酸和草酸。2006年,Vainberg等[53]发现Pseudonocardia sp. ENV478菌株在降解14D过程中积累中间产物2-羟基乙氧基乙酸(2-HEAA)。2007年,Mahendra等[54]利用三重四极杆质谱、傅里叶变换离子回旋共振质谱和14C标记技术发现单加氧酶表达菌P. dioxanivorans CB1190降解14D时产生的中间产物主要为HEAA、乙烯乙二醇、乙醇酸和草酸。2009年,Kim等[32]利用同位素标记法并结合GC-MS分析在Mycobacterium sp. PH-06降解14D过程中检测到1, 4-二氧六环-2-酮和乙烯乙二醇。2012年,Grostern等[55]将P. dioxanivorans CB1190菌株的乙醛酸聚醛酶基因于R. jostii RHA1中异源表达,从细胞提取液中检测到酶活性,证实了乙醛酸途径在14D最终降解过程中的重要作用。2014年,Huang等[56]从A. baumannii DD1降解14D过程中检测到了1, 4-二噁烯,但未检测到HEAA,检测结果表明2-羟基14D也可还原成1, 4-二噁烯。2018年,Deng等[44]在Azoarcus sp. DD4中发现HEAA在酸性条件下会形成衍生物1, 4-二氧六环-2-酮,而中性条件下未检测到此衍生物。目前有关14D代谢途径的研究比较成熟,主要代谢途径已经明确。
1.4 THF和14D降解基因 可溶性二铁单加氧酶(Soluble di-iron monooxygenases,SDIMOs)是参与细菌起始氧化烃类化合物的一类多组分蛋白酶,具有广泛的底物范围[57],根据其亚基数目、亚基排列方式、底物专一性和序列相似性划分为六类[58]。编码单加氧酶的基因簇在THF/14D降解菌中广泛存在[59],常见的单加氧酶有THF单加氧酶(Pseudonocardia sp. K1)、甲苯单加氧酶(Burkholderia cepacia G4、Ralstonia picketii、P. mendocina KR1)、甲烷单加氧酶(Methylococcus capsulatus Bath)、丙烷单加氧酶(M. vaccae JOB5)等。目前已报道的THF和14D降解酶主要来源于革兰氏阳性细菌,催化THF和14D第一步降解反应的单加氧酶为THF单加氧酶和丙烷单加氧酶,它们在THF和14D降解过程参与催化第一步在C原子上加-OH的反应。2003年,Thiemer等[60]在降解菌Pseudonocardia sp. K1中首次成功克隆出THF降解相关基因thmADBC、sad及aldh,分别编码THF单加氧酶、琥珀酸半醛脱氢酶和4-羟基丁醛脱氢酶。THF单加氧酶体外蛋白纯化进行酶活检测十分困难,目前只有将thmD导入大肠杆菌中异源表达并成功获得纯化蛋白的报道[61]。2012年,Masuda等[62]在Pseudonocardia sp. ENV478中首次证实了thm基因簇参与THF和14D的降解,通过敲除thmB和反义RNA技术降低thmB转录水平,thmB功能缺失导致Pseudonocardia sp. ENV478无法降解THF和14D。另外,全基因组测序分析发现P. dioxanivorans CB1190和R. ruber YYL中均存在thm基因簇,且与P.tetrahydrofuranoxydans K1中的thmADBC高度相似[63-64]。2013年,Sales等[58]将P. dioxanivorans CB1190和P.tetrahydrofuranoxydans K1中的thmADBC基因簇导入到Rhodococcus sp. RHA1中成功实现了异源表达,表达株具备降解THF和14D能力。2019年,Qi等[65]利用宏基因组测序技术分析THF降解混菌样品,最终拼接获得一条全长9387 bp的THF降解基因簇thmX,该基因簇与已报道thm基因簇的氨基酸相似性达79%–93%。
除已广泛报道的THF单加氧酶外,Mathieu等在M. dioxanotrophicus PH-06中发现一个新的丙烷单加氧酶基因簇prmABCD,转录组和RT-PCR分析均证实prmABCD能在底物14D和THF诱导下高表达[66]。2018年,He等[67]通过DNA测序分析发现在降解14D微生物菌群中存在其他的group 5和group 6可溶性二铁单加氧酶基因富集,这可能暗示着存在更多种类的单加氧酶催化14D起始降解。另外,降解菌基因组中常同时含有编码多种单加氧酶的基因簇,如P. dioxanivorans CB1190基因组中同时含8个编码不同单加氧酶的基因簇[55, 63]。降解菌中单加氧酶种类的多样性可能与其能共代谢降解多种有机污染物密切相关。不同于细菌,真菌Graphium sp. ATCC58400中催化THF第一步羟基化反应的关键酶是细胞色素P450,该酶由基因CYP52L1编码,系统发育分析表明其属于CYP52细胞色素P450家族[68]。
目前研究的不足之处:(1)暂无thmADBC/ prmABCD单加氧酶体外酶活检测及酶动力学分析的报道。(2)目前已报道催化THF/14D第一步反应的单加氧酶基因全部来源于革兰氏阳性菌,革兰氏阴性菌中是否存在有别于革兰氏阳性菌的降解基因簇有待进一步挖掘。(3)单加氧酶种类多样,细菌基因组中常含有不止一类的单加氧酶基因簇,降解菌基因组中单加氧酶基因簇种类的多样性与其降解底物的广谱性密切相关,此方面研究有待加强。(4)除催化第一步反应的单加氧酶外,代谢途径中参与其他中间代谢产物转化的酶和基因报道较少,仍待进一步挖掘。
2 含氧稠杂环化合物 2.1 二苯并呋喃和二苯并对二噁英降解菌 二苯并呋喃(Dibenzofuran,DF)和二苯并对二噁英(Dibenzo-p-dioxin,DD)是二噁英的非氯代类似物,尽管它们比氯代形式毒性更低,但作为研究二噁英类含氧杂环污染物生物降解的模式化合物,其生物降解机制研究具有重要意义。自1991年Botanik等首次从环境中分离获得了一株能以DF和DD为唯一碳源生长的降解菌Sphingomonas sp. RW1以来[69],研究者陆续从DF或DD污染环境中筛选获得多株降解菌。目前已报道有超过20株DF或DD降解菌,主要集中在革兰氏阳性菌Rhodococcus sp. (SAO101、HA01、p52)[70-72]和Paenibacillus sp. (YK5、4B1)[73-74]及革兰氏阴性菌Terrabacter sp. (DBF63、DPO360、YK3)[75-77]、Pseudomonas sp. (CA10、FA-HZ1)[78-79]和Comamonas sp. (KD7、JB)[80-81](表 2)。2013年,Peng等[72]筛选获得一株DF降解菌Rhodococcus sp. p52,该菌株能在48 h内完全降解500 mg/L DF,且能在96 h内去除70% 100 mg/L 2-氯二苯并呋喃,此外该菌株还可代谢DD、2, 8-二氯二苯并呋喃、二苯并噻吩和联苯等芳香族化合物。2019年,Ali等[79]分离得到一株DF降解菌P. aeruginosa FA-HZ1,其在最适生长条件30 ℃、pH 5.0下能利用0.1 mmol/L DF。2019年,Thanh等[74]从越南二噁英污染土壤中分离得到一株嗜热DF降解菌Paenibacillus naphthalenovorans 4B1,其最适生长温度45 ℃,代时2.7 h,能在48 h内降解89% 1000 mg/L DF,降解基因的高水平表达可能是其生长降解快速的原因。
表 2. 目前已报道的二苯并呋喃、二苯并对二噁英和二噁英降解菌 Table 2. Dibenzofuran, dibenzo-p-dioxin and dioxins degradation microorganisms reported so far
| Classified | Species | Strains | Substrates | Initial dioxygenation | Enzymes | Genes | References |
| G– | Sphingomonas sp. | S. wittichii RW1 | DD, DF | Angular | DF 4, 4a-dioxygenase | dxnA1A2, dbfB | Chai et al., 2016 |
| G– | Terrabacter sp. | Terrabacter sp. DBF63 | DF, 2-CDF, 2-CDD, 2, 8-diCDF, 2, 3-diCDF | Angular | DF 4, 4a-dioxygenase, Extradiol dioxygenase | dbfA1A2, dbfBC, dfdA1A2A3A4, dfdBC | Kasuga et al., 2013 |
| Terrabacter sp. DPO360 | DF | Angular | Extradiol dioxygenase | BphC1C2 | Schmid et al., 1997 | ||
| Terrabacter sp. YK3 | DF | Angular | dfdA1A2A3A4 | Iida et al., 2002 | |||
| G– | Pseudomonas sp. | Pseudomonas sp. CA10 | DF, DD | Angular | Car 1, 9a-dioxygenase | carAaAcAd | Sato, et al., 1997 |
| P. aeruginosa FA-HZ1 | DF | Angular, lateral | New HZ6359 dioxygenase | Ali et al., 2019 | |||
| G– | Comamonas sp. | Comamonas sp. KD7 | DF | Angular,lateral | Wang et., 2004 | ||
| Comamonas sp. JB | DF, DD | Ji et al., 2017 | |||||
| G– | Dehalococcoides sp. | Dehalococcoides sp. CBDB1 | PeCDD | Michael et al., 2003 | |||
| G+ | Microbacterium sp. | Microbacterium sp. YK18 | |||||
| G+ | Paenibacillus sp. | Paenibacillus sp. strain YK5 | DF | Aromatic hydrocarbon dioxygenase (AhDOa), Extradiol dioxygenase, Hydrolase | dbfA1A2, dbfB, dbfC | Iida et al., 2006 | |
| P. naphthalenovorans 4B1 | DF | dbfA1A2BC | Thanh et al., 2019 | ||||
| G+ | Rhodococcus sp. | Rhodococcus sp. YK2 | DF | THB-dioxygenase | dfdB | Iida et al., 2002 | |
| R. opacus SAO101 | DD, DF, 1-CDD, 2, 3-diCDD, 2, 7-diCDD, 2, 8-diCDD, 1, 2, 3-triCDD, 2, 3, 7-triCDD | Angular, lateral | Nar dioxygenase | NarAaAb | Kimura et al., 2006 | ||
| Rhodococcus sp. HA01 | DF, DD, 3-CDF | Angular | DF 4, 4a-dioxygenase | dfdA1A2A3A4 dbfA1A2 | Aly et al., 2008 | ||
| Rhodococcus sp. p52 | DF, DD, 2-CDF, 2, 8-diDCDF | Angular | DF 4, 4a-dioxygenase | dbfA, dfdA | Peng et al., 2013 | ||
| G+ | Nocardioides sp. | Nocardioides sp. DF412 | DF | DF 4, 4a-dioxygenase, THB-dioxygenase, Extradiol dioxygenase | dfdA1A2A3A4, dfdBC | Miyauchi et al., 2008 | |
| G+ | Geobacillus sp. | Geobacillus sp. UZO 3 | 2, 3, 7, 8-TCDD | Dioxin reductive etherase | dreE | Suzuki et al., 2016 Suzuki et al., 2019 | |
| Fungus | Trichosporon sp. | T. mucoides SBUG801 | DF | Hammer et al., 1998 | |||
| DF 4, 4a-dioxygenase: Dibenzofuran 4, 4a-dioxygenase; Car 1, 9a-dioxygenase: Carbazole 1, 9a-dioxygenase; THB dioxygenase: 2, 2′, 3-Trihydroxybiphenyl dioxygenase; PeCDD: 1, 2, 3, 7, 8-Pentachlorodibenzo-p-dioxin. | |||||||
表选项
虽然已报道降解菌种类繁多,但它们对DF或DD的耐受能力和降解效率普遍较低,寻求更加高效耐受的降解菌一直是研究者关注的热点。且目前报道的降解菌主要是DF降解菌,DD降解菌较少,可能与DD毒性高于DF有关。2016年,Chai等[82]分别利用DF、DD和琥珀酸作为唯一碳源对菌株Sphingomonas wittichii RW1进行了转录组分析,结果表明DD引起300多个基因差异表达,上调基因数目最多,涉及分解代谢、底物识别、转运和解毒多个途径。相较于细菌,目前已报道的真菌中只有酵母菌Trichosporon mucoides SBUG 801能利用DF作为唯一碳源生长[83],真菌降解DF和DD的研究较少,有待进一步深入探究。
2.2 DF和DD降解途径 已报道微生物对DF和DD的降解途径以好氧降解为主,参与二者降解的主要反应类似。微生物降解DF的关键步骤是起始的二羟基化反应,由双加氧酶催化完成,可分为有角度双加氧和侧向双加氧两种类型。侧向双加氧是在侧向双加氧酶催化作用下,DF与氧气的两个氧原子发生结合,结合位点常在碳原子的1, 2或2, 3位。1998年,Hammer等[83]利用LC-MS从DF降解菌Trichosporon mucoides SBUG 801中检测到了6种主要的代谢产物,其中4种为不同的单羟基BF,另外2种为单羟基BF进一步氧化形成的2, 3-二羟基二苯并呋喃和它的开环产物2-(1-羧基亚甲基)-2, 3二氢苯并呋喃亚烷基乙醇酸。2017年,Ji等[81]发现一株Comamonas sp. JB能够在萘为诱导底物下降解DF和二苯并噻吩,其萘双加氧酶在DF的1, 2位碳原子上双加氧生成1, 2-二羟基二苯并呋喃,随后降解为2-羟基-4-(3′-氧代-3′H-苯并呋喃)丁-2-烯酸,最后降解为邻苯二酚。
有角度双加氧是在有角度双加氧酶催化作用下,将两个氧原子催化结合到DF或DD分子上,其常与4, 4a位的碳原子相结合,生成中间产物4, 4a-二羟基-二苯并呋喃,该物质不稳定会立即转化为2, 2′, 3-三羟基联苯(2, 2′, 3-Trihydroxybiphenyl,THB)。有角度双加氧方式在目前已报道DF和DD降解菌中普遍存在,是降解DF和DD的主要途径[84] (图 3)。1992年,Wittich等[69]证实Sphingomonas sp. RW1降解DD时产生中间产物三羟基二苯醚和邻苯二酚。1997年,Kasuga等[75]证实在Terrabacter sp. DBF63中有角度双加氧酶首先氧化DF形成THB,然后THB分别在外二醇双加氧酶和水解酶作用下降解为水杨酸,并最终转化为邻苯二酚和龙胆酸。DD同样首先在双加氧酶作用下形成三羟基二苯醚,然后与DF降解反应过程类似,最终转化为邻苯二酚进入三羧酸循环。在菌株R. opacus SAO101[85]、P. aeruginosa FA-HZ1[79]和Comamonas sp. KD7[80]中,存在有角度双加氧和侧向双加氧两种方式。
 |
| 图 3 二苯并呋喃和二苯并对二噁英有角度双加氧降解途径[84] Figure 3 The angular dioxygenation degradation pathways of dibenzofuran and dibenzo-p-dioxin[84]. The structures shown in brackets have not been characterized yet. |
| 图选项 |
目前研究不足之处:(1)研究主要集中在有角度双加氧降解途径,而侧向双加氧降解途径中仍有部分中间代谢产物未鉴定,多步参与催化反应的酶仍未明确。(2)部分降解菌中同时存在两种不同的降解方式是否受底物调控?是否暗示其具有更强的抵抗污染物胁迫能力?这些问题仍需进一步探究。
2.3 DF和DD降解基因 目前已对多种属好氧细菌参与DF和DD降解的分子机制广泛研究。1993年,Bunz等[86]从Sphingomonas sp.中首次纯化获得了一个能催化DF形成THB的三组分酶系统4, 4a-双加氧酶,该酶由负责电子传递的黄素蛋白还原酶RedA2、一个铁氧还原蛋白Fdx1和一个异多聚体加氧酶dxnA1A2组成,每个催化反应消耗1 mmol/L NADH和O2并将DF双加氧成1 mmol/L THB。编码双加氧酶的基因dxnA1A2和其他编码电子传递蛋白的基因不在一个操纵子上,而是分布在整个染色体上[87]。dbfA1A2和dfdA1A2A3A4是近年来研究最为广泛的能够编码DF和DD有角度双加氧酶的基因。Kasuga等在DF降解菌Terrabacter sp. DBF63中先后发现编码二苯并呋喃4, 4a-双加氧酶的基因簇dbfA1A2和dfdA1A2A3A4,其中dfdA1A2A3A4与Terrabacter sp. YK3和Rhodococcus sp. HA01中dfdA氨基酸序列高度相似[88-89]。2008年,Aly等[71]将Rhodococcus sp. HA01中两个双加氧酶基因簇dbfA和dfdA分别进行了异源表达,两种表达株产生的双加氧酶都能催化DF和DD双加氧,但是对氯代二苯并呋喃表现出互补的底物特异性,即dfdA表达株能转化3-氯二苯并呋喃,不能转化2-氯二苯并呋喃;而dbfA表达株能转化2-氯二苯并呋喃,不能转化3-氯二苯并呋喃。DF降解菌Rhodococcus sp. p52的双加氧酶基因dfdA和dbfA分别分布在两个可接合转移的质粒pDF01和pDF02上[90],Ren等[91]在添加p52菌株的序批式反应器中进行了活性污泥的基因强化,具备降解能力的转化结合子有效提升了反应器中污染物降解效率。另外,咔唑-1, 9a双加氧酶也能以有角度的方式催化DF和DD的起始羟基化,在菌株Pseudomonas sp. CA10中该酶由基因簇carAaAcAd编码[92]。在侧向双加氧途径中,由narAaAb编码环羟基化双加氧酶起始降解,narAaAb基因于Rhodococcus sp.中表达能降解DF并转化为中间产物二氢二醇[85]。
目前已经在多株DF和DD降解菌中克隆获得编码外二醇双加氧酶和水解酶的基因,其中最具代表性的是dbfBC和dfdBC[75, 89]。此外,Schmid等[76]从Terrabacter sp. DPO360菌株中扩增获得的编码外二醇双加氧酶基因BphC1和BphC2,也参与中间产物THB的间位裂解反应。DF的细菌降解同样受转录调控因子的调控,Iida等在DF降解菌Rhodococcus sp. YK2和Terrabacter sp. YK3中发现了一个含GAF结构域的转录调控因子dfdR,dfdR和DF能够激活dfdA1启动子PdfdA1,且dfdR缺失将导致dfdA1无法受底物诱导转录而无法降解DF[93]。此外,随着组学技术的发展,研究者陆续发现一些新的降解基因。2019年,Ali等[79]通过组学分析在P. aeruginosa FA-HZ1中发现了一个编码HZ6359脱卤素酶的基因并在大肠杆菌中进行基因表达,表达株具备降解2, 8-二溴二苯并呋喃的能力。目前有角度双加氧途径中相关降解基因的研究已经比较清楚,侧向双加氧途径中降解相关的基因仍有待挖掘。
3 二噁英类化合物 3.1 二噁英危害及污染现状 二噁英是对人体健康有很大威胁的环境污染物,长久暴露于二噁英污染环境会引起皮肤损伤、肝脏损伤、内分泌系统异常和神经系统功能障碍[94-95]。它具有多种同分异构体,其中多氯代二苯并噁英有75种异构体,而多氯代二苯并呋喃有135种异构体。二噁英中氯原子的取代位置和数量不同,其毒性也不同,目前研究主要集中在2、3、7、8位碳原子被氯代的污染物,其中最具代表性的是剧毒物质2, 3, 7, 8-四氯代二苯并二噁英(2, 3, 7, 8-Tetrachlorodibenzo-p-dioxin, TCDD)。二噁英类化合物的污染主要由人为造成,是人类工业生产过程中产生的副产物,如冶金工业、造纸工业、化工工业等[96-97]。另外,垃圾焚化、柴油燃烧、煤炭燃烧等的不完全燃烧均会产生二噁英[98-99]。二噁英类污染物在世界各地广泛存在,严重威胁人类健康。2014年,Urban等[100]分别对美国城市、郊区和农村地区土壤中二噁英类污染物的含量及毒性进行了评估,表明城市土壤中二噁英毒性当量更高且变化更大达7–186 ng/kg,明显高于美国环保局建议的土壤修复目标水平。2013年,Pan等[101]从中国15个城市商业固体废物焚化厂采集飞灰样品,对17种多氯二苯并二噁英和多氯二苯并呋喃浓度进行了分析,研究表明二噁英浓度达2.8–190 ng/g,其中六氯同系物是主要化合物。尽管2012年欧洲食品安全局对26个欧洲国家全面检测数据评估显示在2002–2004年到2008–2010年间,人们膳食中二噁英和多氯联苯的摄入量下降了16.6%–79.3%,但是由于二噁英在环境中潜伏期长,在人体内半衰期长达7–11年,其造成的危害不容忽视[102]。
3.2 二噁英降解菌及降解机制 利用微生物降解二噁英类污染物一直是国内外研究者关注的热点。研究表明微生物对多氯联苯、多氯二苯并对二噁英和多氯呋喃的脱氯作用广泛存在于沉积物、下水道、垃圾填埋场和地下水中[103],微生物在实际污染物处理中具有广泛的应用前景。二噁英的生物降解主要包括脱氯作用、氧化作用、开环作用和酶催化降解等。相较于DF和DD,二噁英类化合物因苯环上氯原子的取代而对微生物产生不同程度的毒性,目前已报道降解二噁英的菌株较少且降解活性随氯原子的增加而减少。2002年,Habe等[104]研究发现Terrabacter sp. DBF63能够降解土样污染物中2-氯二苯并呋喃(2-CDF)、2-氯二苯并二噁英(2-CDD)、2, 8-二氯二苯并呋喃(2, 8-DCDF)和2, 3-二氯二苯并呋喃(2, 3-DCDF),菌株培养7 d后能分别降解90%的1 ppm 2, 8-DCDF和40%的1 ppm 2, 3-DCDD。另外,对菌株Sphingomonas sp. RW1降解特性分析表明其具有广泛的底物谱,能降解单氯二苯并呋喃和单氯二苯并二噁英(2-CDF、3-CDF、4-CDF、1-CDD、2-CDD)、二氯二苯并呋喃和二氯二苯并二噁英(2, 3-DCDF、2, 8-DCDF、3, 7-DCDF、2, 3-DCDD,2, 7-DCDD)以及三氯二苯并二噁英(1, 2, 4-TrCDD),但不能降解三氯二苯并呋喃(2, 4, 8-TrCDF)[105]。目前已知的具有最高脱氯能力的菌株为Dehalococcoides sp. CBDB1,其能够对1, 2, 3, 7, 8-五氯二苯并二噁英(PeCDD)进行还原脱氯[106]。
二噁英生物降解过程中的主要催化反应与DF和DD类似,研究发现咔唑1, 9-双加氧酶和DF 4, 4a-双加氧酶均能催化部分二噁英降解,如将2-CDF和2, 8-DCDF转化为5-氯水杨酸,将2-CDD和2, 3-DCDD分别转化为4-氯邻苯二酚和4, 5-二氯邻苯二酚[78]。有关二噁英代谢中间产物检测的报道较少,Suzuki等[107]利用GC-MS检测了嗜热菌Geobacillus sp. UZO3降解2, 3, 7, 8-四氯二苯并二噁英(2, 3, 7, 8-TCDD)的中间产物,发现该菌能将2, 3, 7, 8-TCDD先转化为3′, 4′, 4, 5-四氯-2-羟基二苯醚(TCDE),再进一步转化成二氯苯酚。另外,研究报道了一个新的编码2, 3, 7, 8-TCDD降解酶的基因dreE,氨基酸序列分析表明其含有一个CoA结合位点,和枯草芽孢杆菌核黄素合成操纵子上一个未知功能的基因高度相似。将dreE在大肠杆菌中进行表达,表达蛋白可还原性切割2, 3, 7, 8- TCDD的二芳基醚键并产生TCDE[108]。二噁英类污染物种类繁多,目前已报道具有降解能力的降解菌株较少,且降解能力及降解底物的范围有限。另外,降解途径仍不明确,与关键脱氯反应相关的基因和酶有待进一步挖掘。
4 问题和展望 含氧杂环化合物种类繁多,不同含氧杂环化合物生物降解效果受杂环结构、杂原子数目、取代基位置等多种因素影响。目前研究者以典型代表性难降解污染物THF、14D、DF和DD为主要研究对象,对其降解途径及降解机制开展了深入探究,但是含氧杂环化合物尤其是二噁英类化合物的微生物降解仍存在以下问题:(1)目前已报道降解菌对毒性较强的二噁英类化合物的降解效率普遍较低,降解底物种类有限,已筛选到的降解菌主要为细菌,真菌资源有待进一步挖掘。另外由于环境中微生物菌群结构的复杂性和难培养性,面对未培养微生物急需开发新的降解菌筛选方法。(2) 14D降解菌普遍存在对多种底物的共代谢,可能存在更多种类的单加氧酶催化起始降解过程。DD和DF降解菌在有角度双加氧途径中可选择利用不同降解基因以应对底物的差异,更多不同的降解基因可能参与不同途径的降解过程。二噁英类化合物降解过程中的中间产物及降解基因仍未明确。以上研究均表明传统降解途径及降解机制的研究仍需深入。(3)微生物常通过与周围的生物和非生物因素组成代谢网络的方式共同实现含氧杂环化合物的降解转化,目前研究侧重于单一降解菌降解特性及降解机制研究,对降解菌群中菌株间复杂的代谢网络研究较少,人工构建微生物降解菌群处理环境污染物的研究也有待增强。
随着微生物多组学技术的发展、多学科方法手段的交叉融合,一些新的研究方向逐渐引起研究者的关注。(1)利用基因强化技术提高二噁英降解效率。基因强化技术可通过强化降解基因在土著菌群中的水平迁移和传播,促进土著降解菌群的进化,进而提高难降解污染物的生物去除效果。(2)开发靶向降解基因的基因探针,评估环境样品中降解菌丰度及污染物降解活性,为筛选分离高效降解菌提供指导。(3)环境样品中有丰富的未培养微生物资源,其在污染物降解中发挥着不可忽视的作用。利用宏转录组技术分析复杂环境样品应对不同污染物的转录水平响应,并结合宏基因组测序分析获得未培养微生物中可能的降解基因,最后利用传统的分子生物学方法进行基因功能验证,以上研究思路使得从未培养微生物基因组中挖掘新的污染物降解基因成为可能。(4)微生物在实际污染物降解过程中常与周围其他微生物组成复杂的代谢网络,探究各种环境胁迫下降解菌株与其他菌株互作关系,解析降解菌群菌株间互作代谢网络,最终人工构建降解混合菌群用于实际环境污染物修复也是未来研究热点。
References
| [1] | Eicher T, Hauptmann S, Speicher A. The structure of heterocyclic compounds//The Chemistry of Heterocycles: Structure, Reactions, Syntheses, and Applications. 3rd ed. Weinheim, Germany: Wiley-VCH, 2012: 1-4. |
| [2] | Dua R, Shrivastava S, Sonwane SK, Srivastava SK. Pharmacological significance of synthetic heterocycles scaffold:A review. Advances in Biological Research, 2011, 5(3): 120-144. |
| [3] | Eftekhari-Sis B, Zirak M, Akbari A. Arylglyoxals in synthesis of heterocyclic compounds. Chemical Reviews, 2013, 113(5): 2958-3043. DOI:10.1021/cr300176g |
| [4] | Gomtsyan A. Heterocycles in drugs and drug discovery. Chemistry of Heterocyclic Compounds, 2012, 48(1): 7-10. |
| [5] | Manjal SK, Pathania S, Bhatia R, Kaur R, Kumar K, Rawal RK. Diversified synthetic strategies for pyrroloindoles:An overview. Journal of Heterocyclic Chemistry, 2019, 56(9): 2318-2332. DOI:10.1002/jhet.3661 |
| [6] | Martins P, Jesus J, Santos S, Raposo LR, Roma-Rodrigues C, Baptista PV, Fernandes AR. Heterocyclic anticancer compounds:recent advances and the paradigm shift towards the use of nanomedicine's tool box. Molecules, 2015, 20(9): 16852-16891. DOI:10.3390/molecules200916852 |
| [7] | Kalaria PN, Karad SC, Raval DK. A review on diverse heterocyclic compounds as the privileged scaffolds in antimalarial drug discovery. European Journal of Medicinal Chemistry, 2018, 158: 917-936. |
| [8] | Dhameja M, Gupta P. Synthetic heterocyclic candidates as promising α-glucosidase inhibitors:An overview. European Journal of Medicinal Chemistry, 2019, 176: 343-377. DOI:10.1016/j.ejmech.2019.04.025 |
| [9] | Lamberth C. Heterocyclic chemistry in crop protection. Pest Management Science, 2013, 69(10): 1106-1114. DOI:10.1002/ps.3615 |
| [10] | Anderson RG, Nickless G. Heterocyclic azo dyestuffs in analytical chemistry. A review. Analyst, 1967, 92(1093): 207-238. DOI:10.1039/an9679200207 |
| [11] | Eisentraeger A, Brinkmann C, Hollert H, Sagner A, Tiehm A, Neuwoehner J. Heterocyclic compounds:toxic effects using algae, daphnids, and the Salmonella/microsome test taking methodical quantitative aspects into account. Environmental Toxicology and Chemistry, 2008, 27(7): 1590-1596. DOI:10.1897/07-201.1 |
| [12] | Zheng GJ, Leung AOW, Jiao LP, Wong MH. Polychlorinated dibenzo-p-dioxins and dibenzofurans pollution in China:sources, environmental levels and potential human health impacts. Environment International, 2008, 34(7): 1050-1061. DOI:10.1016/j.envint.2008.02.011 |
| [13] | EF SA. Update of the monitoring of levels of dioxins and PCBs in food and feed. European Food Safety Authority Journal, 2012, 10(7): 2832. |
| [14] | Kakimoto H, Oka H, Harada Y, Ushijima S, Toriba A, Kizu R, Hayakawa K. Comparison of compositions of polychlorinated dibenzo-p-dioxins (PCDDs) and dibenzofurans (PCDFs) in air and soil samples collected in Ishikawa. Journal of Health Science, 2004, 50(1): 58-65. DOI:10.1248/jhs.50.58 |
| [15] | Morales L, Dachs J, González-Gaya G, Hernán G, ábalos M, Abad E. Background concentrations of polychlorinated dibenzo-p-dioxins, dibenzofurans, and biphenyls in the global oceanic atmosphere. Environmental Science & Technology, 2014, 48(17): 10198-10207. |
| [16] | Liu D, Liu C, Li H, Lei AW. Direct functionalization of tetrahydrofuran and 1, 4-dioxane:nickel-catalyzed oxidative C(sp3)-H arylation. Angewandte Chemie International Edition, 2013, 52(16): 4453-4456. DOI:10.1002/anie.201300459 |
| [17] | Gamer AO, Jaeckh R, Leibold E, Kaufmann W, Gembardt C, Bahnemann R, van Ravenzwaay B. Investigations on cell proliferation and enzyme induction in male rat kidney and female mouse liver caused by tetrahydrofuran. Toxicological Sciences, 2002, 70(1): 140-149. DOI:10.1093/toxsci/70.1.140 |
| [18] | Hermida SAS, Possari EPM, Souza DB, de Arruda Campos IP, Gomes OF, Di Mascio P, Medeiros MHG, Loureiro APM. 2'-Deoxyguanosine, 2'-deoxycytidine, and 2'-deoxyadenosine adducts resulting from the reaction of tetrahydrofuran with DNA bases. Chemical Research in Toxicology, 2006, 19(7): 927-936. DOI:10.1021/tx060033d |
| [19] | Malley LA, Christoph GR, Stadler JC, Hansen JF, Biesemeier JA, Jasti SL. Acute and subchronic neurotoxicological evaluation of tetrahydrofuran by inhalation in rats. Drug and Chemical Toxicology, 2001, 24(3): 201-219. DOI:10.1081/DCT-100103720 |
| [20] | Tahara M, Obama T, Ikarashi Y. Development of analytical method for determination of 1, 4-dioxane in cleansing products. International Journal of Cosmetic Science, 2013, 35(6): 575-580. DOI:10.1111/ics.12079 |
| [21] | Xu XY, Liu SM, Smith K, Wang YJ, Hu HY. Light-driven breakdown of 1, 4-dioxane for potable reuse:A review. Chemical Engineering Journal, 2019, 373: 508-518. DOI:10.1016/j.cej.2019.05.063 |
| [22] | Guo WQ, Brodowsky H. Determination of the trace 1, 4-dioxane. Microchemical Journal, 2000, 64(2): 173-179. DOI:10.1016/S0026-265X(99)00026-0 |
| [23] | Nannelli A, De Rubertis A, Longo V, Gervasi PG. Effects of dioxane on cytochrome P450 enzymes in liver, kidney, lung and nasal mucosa of rat. Archives of Toxicology, 2005, 79(2): 74-82. DOI:10.1007/s00204-004-0590-z |
| [24] | Mohr TKG. Environmental Investigation and Remediation: 1, 4-Dioxane and Other Solvent Stabilizers. Boca Raton: CRC Press, 2010: 46. |
| [25] | Stepien DK, Diehl P, Helm J, Thoms A, Püttmann W. Fate of 1, 4-dioxane in the aquatic environment:from sewage to drinking water. Water Research, 2014, 48: 406-419. DOI:10.1016/j.watres.2013.09.057 |
| [26] | Adamson DT, Mahendra S, Walker KL Jr, Rauch SR, Sengupta S, Newell CJ. A multisite survey to identify the scale of the 1, 4-dioxane problem at contaminated groundwater sites. Environmental Science & Technology Letters, 2014, 1(5): 254-258. |
| [27] | Chiang SYD, Anderson RH, Wilken M, Walecka-Hutchison C. Practical perspectives of 1, 4-dioxane investigation and remediation. Remediation, 2016, 27(1): 7-27. |
| [28] | Adamson DT, Pi?a EA, Cartwright AE, Rauch SR, Anderson RH, Mohr T, Connor JA. 1, 4-Dioxane drinking water occurrence data from the third unregulated contaminant monitoring rule. Science of the Total Environment, 2017, 596-597: 236-245. DOI:10.1016/j.scitotenv.2017.04.085 |
| [29] | Cashman MA, Kirschenbaum L, Holowachuk J, Boving TB. Identification of hydroxyl and sulfate free radicals involved in the reaction of 1, 4-dioxane with peroxone activated persulfate oxidant. Journal of Hazardous Materials, 2019, 380: 120875. DOI:10.1016/j.jhazmat.2019.120875 |
| [30] | Bock C, Kroppenstedt RM, Diekmann H. Degradation and bioconversion of aliphatic and aromatic hydrocarbons by Rhodococcus ruber 219. Applied Microbiology and Biotechnology, 1996, 45(3): 408-410. |
| [31] | Mahendra S, Alvarez-Cohen L. Pseudonocardia dioxanivorans sp. nov., a novel actinomycete that grows on 1, 4-dioxane. International Journal of Systematic and Evolutionary Microbiology, 2005, 55(2): 593-598. DOI:10.1099/ijs.0.63085-0 |
| [32] | Kim YM, Jeon JR, Murugesan K, Kim EJ, Chang YS. Biodegradation of 1, 4-dioxane and transformation of related cyclic compounds by a newly isolated Mycobacterium sp. PH-06. Biodegradation, 2009, 20(4): 511-519. DOI:10.1007/s10532-008-9240-0 |
| [33] | Sei K, Miyagaki K, Kakinoki T, Fukugasako K, Inoue D, Ike M. Isolation and characterization of bacterial strains that have high ability to degrade 1, 4-dioxane as a sole carbon and energy source. Biodegradation, 2013, 24(5): 665-674. DOI:10.1007/s10532-012-9614-1 |
| [34] | Sun BZ, Ko K, Ramsay JA. Biodegradation of 1, 4-dioxane by a Flavobacterium. Biodegradation, 2011, 22(3): 651-659. DOI:10.1007/s10532-010-9438-9 |
| [35] | Nakamiya K, Hashimoto S, Ito H, Edmonds JS, Morita M. Degradation of 1, 4-dioxane and cyclic ethers by an isolated fungus. Applied and Environmental Microbiology, 2005, 71(3): 1254-1258. DOI:10.1128/AEM.71.3.1254-1258.2005 |
| [36] | Bernhardt D, Diekmann H. Degradation of dioxane, tetrahydrofuran and other cyclic ethers by an environmental Rhodococcus strain. Applied Microbiology and Biotechnology, 1991, 36(1): 120-123. |
| [37] | Yao YL, Lv ZM, Min H, Lv ZH, Jiao HP. Isolation, identification and characterization of a novel Rhodococcus sp. strain in biodegradation of tetrahydrofuran and its medium optimization using sequential statistics-based experimental designs. Bioresource Technology, 2009, 100(11): 2762-2769. DOI:10.1016/j.biortech.2009.01.006 |
| [38] | Chen MJ, Zhou YY, Chen DZ, Jin XJ. A newly isolated strain capable of effectively degrading tetrahydrofuran and its performance in a continuous flow system. Bioresource Technology, 2010, 101(16): 6461-6467. DOI:10.1016/j.biortech.2010.03.064 |
| [39] | Huang H, Yu HX, Qi MB, Liu ZB, Wang HX, Lu ZM. Enrichment and characterization of a highly efficient tetrahydrofuran-degrading bacterial culture. Biodegradation, 2019, 30(5): 467-479. |
| [40] | Parales RE, Adamus JE, White N, May HD. Degradation of 1, 4-dioxane by an actinomycete in pure culture. Applied and Environmental Microbiology, 1994, 60(12): 4527-4530. DOI:10.1128/AEM.60.12.4527-4530.1994 |
| [41] | Mahendra S, Alvarez-Cohen L. Kinetics of 1, 4-dioxane biodegradation by monooxygenase-expressing bacteria. Environmental Science & Technology, 2006, 40(17): 5435-5442. |
| [42] | Chen ZD, Jin XJ, Chen J, Ye JX, Jiang NX, Chen JM. Intermediates and substrate interaction of 1, 4-dioxane degradation by the effective metabolizer Xanthobacter flavus DT8. International Biodeterioration & Biodegradation, 2016, 106: 133-140. |
| [43] | Inoue D, Tsunoda T, Yamamoto N, Ike M, Sei K. 1, 4-Dioxane degradation characteristics of Rhodococcus aetherivorans JCM 14343. Biodegradation, 2018, 29(3): 301-310. DOI:10.1007/s10532-018-9832-2 |
| [44] | Deng DY, Li F, Wu C, Li MY. Synchronic biotransformation of 1, 4-dioxane and 1, 1-dichloroethylene by a Gram-negative propanotroph Azoarcus sp. DD4. Environmental Science & Technology Letters, 2018, 5(8): 526-532. |
| [45] | Hand S, Wang BX, Chu HK. Biodegradation of 1, 4-dioxane:effects of enzyme inducers and trichloroethylene. Science of the Total Environment, 2015, 520: 154-159. DOI:10.1016/j.scitotenv.2015.03.031 |
| [46] | Guan XY, Liu F, Wang J, Li CX, Zheng XX. Mechanism of 1, 4-dioxane microbial degradation revealed by 16S rRNA and metatranscriptomic analyses. Water Science & Technology, 2018, 77(1): 123-133. |
| [47] | Li MY, Liu YY, He Y, Mathieu J, Hatton J, DiGuiseppi W, Alvarez PJJ. Hindrance of 1, 4-dioxane biodegradation in microcosms biostimulated with inducing or non-inducing auxiliary substrates. Water Research, 2017, 112: 217-225. DOI:10.1016/j.watres.2017.01.047 |
| [48] | Mahendra S, Grostern A, Alvarez-Cohen L. The impact of chlorinated solvent co-contaminants on the biodegradation kinetics of 1, 4-dioxane. Chemosphere, 2013, 91(1): 88-92. DOI:10.1016/j.chemosphere.2012.10.104 |
| [49] | Zhang S, Gedalanga PB, Mahendra S. Biodegradation kinetics of 1, 4-dioxane in chlorinated solvent mixtures. Environmental Science & Technology, 2016, 50(17): 9599-9607. |
| [50] | Skinner K, Cuiffetti L, Hyman M. Metabolism and cometabolism of cyclic ethers by a filamentous fungus, a Graphium sp.. Applied and Environmental Microbiology, 2009, 75(17): 5514-5522. DOI:10.1128/AEM.00078-09 |
| [51] | Moreno-Horn M, Garbe LA, Tressl R, G?risch H. Transient accumulation of γ-butyrolactone during degradation of bis (4-chloro-n-butyl) ether by diethylether-grown Rhodococcus sp. strain DTB. Applied Microbial and Cell Physiology, 2005, 69(3): 335-340. |
| [52] | Tajima T, Hayashida N, Matsumura R, Omura A, Nakashimada Y, Kato J. Isolation and characterization of tetrahydrofuran-degrading Rhodococcus aetherivorans strain M8. Process Biochemistry, 2012, 47(11): 1665-1669. DOI:10.1016/j.procbio.2011.08.009 |
| [53] | Vainberg S, McClay K, Masuda H, Root D, Condee C, Zylstra GJ, Steffan RJ. Biodegradation of ether pollutants by Pseudonocardia sp. strain ENV478. Applied and Environmental Microbiology, 2006, 72(8): 5218-5224. DOI:10.1128/AEM.00160-06 |
| [54] | Mahendra S, Petzold CJ, Baidoo EE, Keasling JD, Alvarez-Cohen L. Identification of the intermediates of in vivo oxidation of 1, 4-dioxane by monooxygenase-containing bacteria. Environmental Science & Technology, 2007, 41(21): 7330-7336. |
| [55] | Grostern A, Sales CM, Zhuang WQ, Erbilgin O, Alvarez-Cohen L. Glyoxylate metabolism is a key feature of the metabolic degradation of 1, 4-dioxane by Pseudonocardia dioxanivorans strain CB1190. Applied and Environmental Microbiology, 2012, 78(9): 3298-3308. DOI:10.1128/AEM.00067-12 |
| [56] | Huang HL, Shen DS, Li N, Shan D, Shentu JL, Zhou YY. Biodegradation of 1, 4-dioxane by a novel strain and its biodegradation pathway. Water, Air, & Soil Pollution, 2014, 225(9): 2135. |
| [57] | Coleman NV, Bui NB, Holmes AJ. Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environmental Microbiology, 2006, 8(7): 1228-1239. DOI:10.1111/j.1462-2920.2006.01015.x |
| [58] | Sales CM, Grostern A, Parales JV, Parales RE, Alvarez-Cohen L. Oxidation of the cyclic ethers 1, 4-dioxane and tetrahydrofuran by a monooxygenase in two Pseudonocardia species. Applied and Environmental Microbiology, 2013, 79(24): 7702-7708. DOI:10.1128/AEM.02418-13 |
| [59] | Li MY, Mathieu J, Yang Y, Fiorenza S, Deng Y, He ZL, Zhou JZ, Alvarez PJJ. Widespread distribution of soluble di-iron monooxygenase (SDIMO) genes in arctic groundwater impacted by 1, 4-dioxane. Environmental Science & Technology, 2013, 47(17): 9950-9958. |
| [60] | Thiemer B, Andreesen JR, Schr?der T. Cloning and characterization of a gene cluster involved in tetrahydrofuran degradation in Pseudonocardia sp. strain K1. Archives of Microbiology, 2003, 179(4): 266-277. DOI:10.1007/s00203-003-0526-7 |
| [61] | Oppenheimer M, Pierce BS, Crawford JA, Ray K, Helm RF, Sobrado P. Recombinant expression, purification, and characterization of ThmD, the oxidoreductase component of tetrahydrofuran monooxygenase. Archives of Biochemistry and Biophysics, 2010, 496(2): 123-131. DOI:10.1016/j.abb.2010.02.006 |
| [62] | Masuda H, McClay K, Steffan RJ, Zylstra GJ. Biodegradation of tetrahydrofuran and 1, 4-dioxane by soluble di-iron monooxygenase in Pseudonocardia sp. strain ENV478. Journal of Molecular Microbiology and Biotechnology, 2012, 22(5): 312-316. DOI:10.1159/000343817 |
| [63] | Sales CM, Mahendra S, Grostern A, Parales RE, Goodwin LA, Woyke T, Nolan M, Lapidus A, Chertkov O, Ovchinnikova G, Sczyrba A, Alvarez-Cohen L. Genome sequence of the 1, 4-dioxane-degrading Pseudonocardia dioxanivorans strain CB1190. Journal of Bacteriology, 2011, 193(17): 4549-4550. DOI:10.1128/JB.00415-11 |
| [64] | He ZX, Zhang K, Wang HX, Lv ZM. Trehalose promotes Rhodococcus sp. strain YYL colonization in activated sludge under tetrahydrofuran (THF) stress. Frontiers in Microbiology, 2015, 6: 438. |
| [65] | Qi MB, Huang H, Zhang Y, Wang HX, Li HB, Lu ZM. Novel tetrahydrofuran (THF) degradation-associated genes and cooperation patterns of a THF-degrading microbial community as revealed by metagenomic. Chemosphere, 2019, 231: 173-183. DOI:10.1016/j.chemosphere.2019.05.137 |
| [66] | He Y, Mathieu J, Yang Y, Yu PF, da Silva MLB, Alvarez PJJ. 1, 4-Dioxane biodegradation by Mycobacterium dioxanotrophicus PH-06 is associated with a group-6 soluble di-iron monooxygenase. Environmental Science & Technology Letters, 2017, 4(11): 494-499. |
| [67] | He Y, Mathieu J, da Siva MLB, Li MY, Alvarez PJJ. 1, 4-Dioxane-degrading consortia can be enriched from uncontaminated soils:prevalence of Mycobacterium and soluble di-iron monooxygenase genes. Microbial Biotechnology, 2018, 11(1): 189-198. |
| [68] | Trippe KM, Wolpert TJ, Hyman MR, Ciuffetti LM. RNAi silencing of a cytochrome P450 monoxygenase disrupts the ability of a filamentous fungus, Graphium sp., to grow on short-chain gaseous alkanes and ethers. Biodegradation, 2014, 25(1): 137-151. DOI:10.1007/s10532-013-9646-1 |
| [69] | Wittich RM, Wilkes H, Sinnwell V, Francke W, Fortnagel P. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Applied and Environmental Microbiology, 1992, 58(3): 1005-1010. DOI:10.1128/AEM.58.3.1005-1010.1992 |
| [70] | Kimura N, Urushigawa Y. Metabolism of dibenzo-p-dioxin and chlorinated dibenzo-p-dioxin by a gram-positive bacterium, Rhodococcus opacus SAO101. Journal of Bioscience and Bioengineering, 2001, 92(2): 138-143. DOI:10.1016/S1389-1723(01)80214-0 |
| [71] | Aly HAH, Huu NB, Wray V, Junca H, Pieper DH. Two angular dioxygenases contribute to the metabolic versatility of dibenzofuran-degrading Rhodococcus sp. strain HA01. Applied and Environmental Microbiology, 2008, 74(12): 3812-3822. DOI:10.1128/AEM.00226-08 |
| [72] | Peng P, Yang HY, Jia RB, Li L. Biodegradation of dioxin by a newly isolated Rhodococcus sp. with the involvement of self-transmissible plasmids. Applied Microbiology and Biotechnology, 2013, 97(12): 5585-5595. DOI:10.1007/s00253-012-4363-y |
| [73] | Iida T, Nakamura K, Izumi A, Mukouzaka Y, Kudo T. Isolation and characterization of a gene cluster for dibenzofuran degradation in a new dibenzofuran-utilizing bacterium, Paenibacillus sp. strain YK5. Archives of Microbiology, 2006, 184(5): 305-315. DOI:10.1007/s00203-005-0045-9 |
| [74] | Thanh LTH, Thi TVN, Shintani M, Moriuchi R, Dohra H, Loc NH, Kimbara K. Isolation and characterization of a moderate thermophilic Paenibacillus naphthalenovorans strain 4B1 capable of degrading dibenzofuran from dioxin-contaminated soil in Vietnam. Journal of Bioscience and Bioengineering, 2019, 128(5): 571-577. |
| [75] | Kasuga K, Nojiri H, Yamane H, Kodama T, Omori T. Cloning and characterization of the genes involved in the degradation of dibenzofuran by Terrabacter sp. strain DBF63. Journal of Fermentation and Bioengineering, 1997, 84(5): 387-399. DOI:10.1016/S0922-338X(97)81997-6 |
| [76] | Schmid A, Rothe B, Altenbuchner J, Ludwig W, Engesser KH. Characterization of three distinct extradiol dioxygenases involved in mineralization of dibenzofuran by Terrabacter sp. strain DPO360. Journal of Bacteriology, 1997, 179(1): 53-62. DOI:10.1128/JB.179.1.53-62.1997 |
| [77] | Iida T, Mukouzaka Y, Nakamura K, Kudo T. Plasmid-borne genes code for an angular dioxygenase involved in dibenzofuran degradation by Terrabacter sp. strain YK3. Applied and Environmental Microbiology, 2002, 68(8): 3716-3723. DOI:10.1128/AEM.68.8.3716-3723.2002 |
| [78] | Habe H, Chung JS, Lee JH, Kasuga K, Yoshida T, Nojiri H, Omori T. Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by two types of bacteria having angular dioxygenases with different features. Applied and Environmental Microbiology, 2001, 67(8): 3610-3617. DOI:10.1128/AEM.67.8.3610-3617.2001 |
| [79] | Ali F, Hu HY, Wang WW, Zhou ZK, Shah SB, Xu P, Tang HZ. Characterization of a dibenzofuran-degrading strain of Pseudomonas aeruginosa, FA-HZ1. Environmental Pollution, 2019, 250: 262-273. DOI:10.1016/j.envpol.2019.04.026 |
| [80] | Wang YX, Yamazoe A, Suzuli S, Liu CT, Aono T, Oyaizu H. Isolation and characterization of dibenzofuran-degrading Comamonas sp. strains isolated from white clover roots. Current Microbiology, 2004, 49(4): 288-294. DOI:10.1007/s00284-004-4348-x |
| [81] | Ji XJ, Xu J, Ning SX, Li N, Tan L, Shi SN. Cometabolic degradation of dibenzofuran and dibenzothiophene by a naphthalene-degrading Comamonas sp. JB. Current Microbiology, 2017, 74(12): 1411-1416. DOI:10.1007/s00284-017-1334-7 |
| [82] | Chai BL, Tsoi TV, Iwai S, Liu C, Fish JA, Gu C, Johenson TA, Zylstra G, Teppen BJ, Li H, Hashasham SA, Boyd SA, Cole JR, Tiedje JM. Sphingomonas wittichii strain RW1 genome-wide gene expression shifts in response to dioxins and clay. PLoS One, 2016, 11(6): e0157008. DOI:10.1371/journal.pone.0157008 |
| [83] | Hammer E, Krowas D, Sch?fer A, Specht M, Fpancke W, Schauer F. Isolation and characterization of a dibenzofuran-degrading yeast:identification of oxidation and ring cleavage products. Applied and Environmental Microbiology, 1998, 64(6): 2215-2219. DOI:10.1128/AEM.64.6.2215-2219.1998 |
| [84] | Nojiri H, Omori T. Molecular bases of aerobic bacterial degradation of dioxins:involvement of angular dioxygenation. Bioscience, Biotechnology, and Biochemistry, 2002, 66(10): 2001-2016. DOI:10.1271/bbb.66.2001 |
| [85] | Kimura N, Kitagawa W, Mori T, Nakashima N, Tamura T, Kamagata Y. Genetic and biochemical characterization of the dioxygenase involved in lateral dioxygenation of dibenzofuran from Rhodococcus opacus strain SAO101. Applied Microbiology and Biotechnology, 2006, 73(2): 474-484. DOI:10.1007/s00253-006-0481-8 |
| [86] | Bünz PV, Cook AM. Dibenzofuran 4, 4a-dioxygenase from Sphingomonas sp. strain RW1:angular dioxygenation by a three-component enzyme system. Journal of Bacteriology, 1993, 175(20): 6467-6475. DOI:10.1128/JB.175.20.6467-6475.1993 |
| [87] | Armengaud J, Happe B, Timmis KN. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1:catabolic genes dispersed on the genome. Journal of Bacteriology, 1998, 180(15): 3954-3966. DOI:10.1128/JB.180.15.3954-3966.1998 |
| [88] | Kasuga K, Habe H, Chung JS, Yoshida T, Nojiri H, Yamane H, Omori T. Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochemical and Biophysical Research Communications, 2001, 283(1): 195-204. DOI:10.1006/bbrc.2001.4763 |
| [89] | Kasuga K, Nitta A, Kobayashi M, Habe H, Nojiri H, Yamane H, Omori T, Kojima I. Cloning of dfdA genes from Terrabacter sp. strain DBF63 encoding dibenzofuran 4, 4a-dioxygenase and heterologous expression in Streptomyces lividans. Applied Microbiology and Biotechnology, 2013, 97(10): 4485-4498. DOI:10.1007/s00253-012-4565-3 |
| [90] | Sun J, Qiu YL, Ding PF, Peng P, Yang HY, Li L. Conjugative transfer of dioxin-catabolic megaplasmids and bioaugmentation prospects of a Rhodococcus sp.. Environmental Science & Technology, 2017, 51: 6298-6307. |
| [91] | Ren CY, Wang YY, Tian LL, Chen M, Sun J, Li L. Genetic bioaugmentation of activated sludge with dioxin-catabolic plasmids harbored by Rhodococcus sp. strain p52. Environmental Science & Technology, 2018, 52(9): 5339-5348. |
| [92] | Sato SI, Nam JW, Kasuga K, Nojiri H, Yamane H, Omori T. Identification and characterization of genes encoding carbazole 1, 9a-dioxygenase in Pseudomonas sp. strain CA10. Journal of Bacteriology, 1997, 179(15): 4850-4858. DOI:10.1128/JB.179.15.4850-4858.1997 |
| [93] | Iida T, Waka T, Nakamura K, Mukouzaka Y, Kudo T. The GAF-like-domain-containing transcriptional regulator dfdR is a sensor protein for dibenzofuran and several hydrophobic aromatic compounds. Journal of Bacteriology, 2009, 191(1): 123-134. DOI:10.1128/JB.01112-08 |
| [94] | Nadal M, Marauès M, Mari M, Domingo JL. Climate change and environmental concentrations of POPs:A review. Environmental Research, 2015, 143: 177-185. DOI:10.1016/j.envres.2015.10.012 |
| [95] | Wang B, Wang PL, Xie LH, Lin RB, Lv J, Li JR, Chen BL. A stable zirconium based metal-organic framework for specific recognition of representative polychlorinated dibenzo-p-dioxin molecules. Nature Communications, 2019, 10: 3861. DOI:10.1038/s41467-019-11912-4 |
| [96] | Amirova Z, Weber R. Massive PCDD/F contamination at the Khimprom organochlorine plant in Ufa-A review and recommendations for future management. Environmental Science and Pollution Research, 2015, 22(19): 14416-14430. DOI:10.1007/s11356-015-5048-8 |
| [97] | Iyer R, Aggarwal J, Iken B. A review of the Texas, USA San Jacinto Superfund site and the deposition of polychlorinated dibenzo-p-dioxins and dibenzofurans in the San Jacinto River and Houston Ship Channel. Environmental Science and Pollution Research, 2016, 23(23): 23321-23338. DOI:10.1007/s11356-016-7501-8 |
| [98] | Awasthi AK, Zeng XL, Li JH. Environmental pollution of electronic waste recycling in India:A critical review. Environmental Pollution, 2016, 211: 259-270. DOI:10.1016/j.envpol.2015.11.027 |
| [99] | Gao XB, Ji BJ, Yan DH, Huang DF, Zhu XM. A full-scale study on thermal degradation of polychlorinated dibenzo-p-dioxins and dibenzofurans in municipal solid waste incinerator fly ash and its secondary air pollution control in China. Waste Management & Research, 2017, 35(4): 437-443. |
| [100] | Urban JD, Wikoff DS, Bunch ATG, Harris MA, Haws LC. A review of background dioxin concentrations in urban/suburban and rural soils across the United States:Implications for site assessments and the establishment of soil cleanup levels. Science of the Total Environment, 2014, 466-467: 586-597. DOI:10.1016/j.scitotenv.2013.07.065 |
| [101] | Pan Y, Yang LB, Zhou JZ, Liu JY, Qian GR, Ohtsuka N, Motegi M, Oh K, Hosono S. Characteristics of dioxins content in fly ash from municipal solid waste incinerators in China. Chemosphere, 2013, 92(7): 765-771. DOI:10.1016/j.chemosphere.2013.04.003 |
| [102] | Malisch R, Kotz A. Dioxins and PCBs in feed and food-review from European perspective. Science of the Total Environment, 2014, 491-492: 2-10. DOI:10.1016/j.scitotenv.2014.03.022 |
| [103] | Rodenburg LA, Krumins V, Curran JC. Microbial dechlorination of polychlorinated biphenyls, dibenzo-p-dioxins, and-furans at the Portland harbor superfund site, Oregon, USA. Environmental Science & Technology, 2015, 49(12): 7227-7235. |
| [104] | Habe H, Ide K, Yotsumoto M, Tsuji H, Yoshida T, Nojiri H, Omori T. Degradation characteristics of a dibenzofuran- degrader Terrabacter sp. strain DBF63 toward chlorinated dioxins in soil. Chemosphere, 2002, 48(2): 201-207. DOI:10.1016/S0045-6535(02)00064-4 |
| [105] | Wilkes H, Wittich R, Timmis KN, Fortnagel P, Francke W. Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by Sphingomonas sp. strain RW1. Applied and Environmental Microbiology, 1996, 62(2): 367-371. DOI:10.1128/AEM.62.2.367-371.1996 |
| [106] | Bunge M, Adrian L, Kraus A, Opel M, Lorenz WG, Andressen JR, G?risch H, Lechner U. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature, 2003, 421(6921): 357-360. DOI:10.1038/nature01237 |
| [107] | Suzuki Y, Nakamura M, Otsuka Y, Suzuki N, Ohyama K, Kawakami T, Umeka Y, Maninang JS, Sato KI, Hishiyama S, Inoue K, Kameyama T, Takahashi A, Katayama Y. 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) degradation by the thermophilic Geobacillus sp. UZO3. Journal of Environmental Biotechnology, 2016, 15(2): 105-108. |
| [108] | Suzuki Y, Nakamura M, Otsuka Y, Suzuki N, Ohyama K, Kawakami T, Sato-Izawa K, Navarro RR, Hishiyama S, Inoue K, Kameyama T, Takahashi A, Katayama Y. Cloning and sequencing of the gene encoding the enzyme for the reductive cleavage of diaryl ether bonds of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin in Geobacillus thermodenitrificans UZO3. Journal of Bioscience and Bioengineering, 2018, 126(4): 488-496. DOI:10.1016/j.jbiosc.2018.04.013 |
