Jinqian Ye1, Yongshan Zhao1

 , Yuguang Du2, Jianjun Li2
, Yuguang Du2, Jianjun Li2

1. School of Life Science and Bio-Pharmaceutics, Shenyang Pharmaceutical University, Shenyang 110016, Liaoning Province, China;
2. National Key Laboratory of Biochemical Engineering, National Engineering Research Center for Biotechnology(Beijing), Key Laboratory of Biopharmaceutical Production & Formulation Engineering, PLA, Institute of Process Engineering, Chinese Academy of Sciences, Beijing 100190, China
Received: 20 September 2018; Revised: 28 November 2018; Published online: 28 December 2018
Foundation item: Supported by the National Natural Science Foundation of China (31370799)
Corresponding author: Yongshan Zhao, Tel:+86-24-23986399, Fax:+86-24-23986431, E-mail:zhao09081@163.com; Jianjun Li, Tel/Fax:+86-10-82545039, E-mail:jjli@ipe.ac.cn.
Abstract: [Objective] Isomaltases IMA1 play key roles in full utilization of oligosaccharides containing α-1, 6-O-glucosidic bonds.[Methods] We cloned, overexpressed, purified and characterized four isomaltases IMA1 from four strains of S. cerevisiae including three acidophilic ones.[Results] They showed similar pH and temperature dependence, but different kinetic parameters and thermostability. IMA1-A exhibited the highest binding affinity for α-MG (α-methylglucoside), turnover number, catalytic efficiency, and thermostability. Structure and sequence analysis revealed that even variation in two remote amino acids from the active residues and the substrate binding site could also lead to significantly different kinetic behavior and thermostability of isomaltases IMA1.[Conclusion] Our results will be useful for further investigation into the structure-function relationship of isomaltases IMA1.
Keywords: isomaltase IMA1Saccharomyces cerevisiaekinetic parameterspH-rate profilethermostability
不同酿酒酵母菌株来源异麦芽糖酶IMA1的克隆、表达及表征
叶进前1, 赵勇山1

 , 杜昱光2, 李建军2
, 杜昱光2, 李建军2

1. 沈阳药科大学生命科学与生物制药学院, 辽宁 沈阳 110016;
2. 中国科学院过程工程研究所, 生化工程国家重点实验室, 国家生物技术工程研究中心(北京)全军生物药制造和剂型工程重点实验室, 北京 100190
收稿日期:2018-09-20;修改日期:2018-11-28;网络出版日期:2018-12-28
基金项目:国家自然科学基金(31370799)
通信作者:赵勇山, Tel:+86-24-23986399, Fax:+86-24-23986431, E-mail:zhao09081@163.com; 李建军, Tel/Fax:+86-10-82545039, E-mail:jjli@ipe.ac.cn.
摘要:[目的] 异麦芽糖酶IMA1在充分利用含有α-1,6-O-糖苷键的低聚糖中起着关键作用。[方法] 在本研究中,对来自4株酿酒酵母菌株(包括3株嗜酸性菌株)来源的异麦芽糖酶IMA1进行克隆、表达、纯化和表征。[结果] 研究发现,4种异麦芽糖酶IMA1表现出类似的pH和温度依赖性,但表现出不同的动力学参数和热稳定性。IMA1-A对α-MG(α-甲基葡糖苷)表现出最高的结合亲和力、转换数、催化效率和热稳定性。结构和序列分析表明,2个远离活性位点和底物结合位点的氨基酸的差异对异麦芽糖酶IMA1的动力学参数和热稳定性有重要影响。[结论] 本研究结果对进一步研究异麦芽糖酶IMA1的结构-功能关系奠定了基础。
关键词:异麦芽糖酶IMA1酿酒酵母动力学参数pH值-活性曲线热稳定性
Glucoside hydrolases, which are currently categorized into 156 structural families according to the Carbohydrate-Active enZYmes (CAZy) database (http://www.cazy.org/Glycoside-Hydrolases.html), are key enzymes involved in carbohydrate metabolism[1]. Among them, family 13, includes α-amylase, pullulanase, cyclodextrin glucanotransferase, α-glucosidase, oligo-α-glucosidase, glucodextranase, etc., which specifically act on α-1, 4- and α-1, 6-O-glucosidic linkages[2-3].
Saccharomyces cerevisiae contains two types of α-glucosidases: α-1, 4-glucosidase (E.C. 3.2.1.20, maltase) and oligo-1, 6-glucosidase (E.C. 3.2.1.10, isomaltase)[4]. Maltase preferentially hydrolyzes maltose, but not isomaltose or α-MG (α-methylglucoside), whereas isomaltase prefers to hydrolyze isomaltose and α-MG, but not maltose. Maltose actually acts as a competitive inhibitor for isomaltase[5].
The expression levels of maltase and isomaltase in S. cerevisiae are regulated by the MAL (maltose) and MGL (α-methylglucoside) polymeric genes respectively[6-9]. Maltose utilization in S. cerevisiae is controlled at least by five MAL loci: MAL1–MAL4, and MAL6, whereas utilization of isomaltose and α-MG is controlled by five isomaltase genes (IMA1/YGR287c, IMA2/YOL157c, IMA3/YIL172c, IMA4/YJL221c and IMA5/YJL216c) located in the subtelomeric regions of different chromosomes of S. cerevisiae, with sequence identity from 65% to 99%[8-10]. Deletion studies revealed that IMA1/YGR287c encodes the major isomaltase IMA1[8]. The expression levels of IMA1/YGR287c and IMA5/YJL216c were strongly induced by maltose, isomaltose and α-MG[8]. IMA3 and IMA4 are completely identical and encode the same protein[8, 10]. The IMA2 and IMA3 proteins exhibit only 3 different amino acids, while IMA5 showed 65% sequence identity with other IMAs (IMA1–IMA4)[10]. These four isomaltases demonstrated similarities and differences in the biochemical and enzymological properties[10].
Crystal structures of the major isomaltase IMA1 complexed with maltose and isomaltose have been published respectively, elucidating why isomaltase IMA1 shows the highest activity towards isomaltose and little activity towards longer oligosaccharides[4, 11]. Structure-function relationship of isomaltases IMA1 has recently been explored by Yamamoto et al., who observed that steric hindrance by residues Val216 and Gln279 plays important roles in discriminating the α-1, 4- and α-1, 6-glucosidic linkages[11-12]. A double displacement mechanism was proposed to explain retention of stereochemistry of isomaltase IMA1-catalyzed reaction[11].
When organic acids such as itaconic acid and citric acid were produced through microbial fermentation at pH 2 to 3 using glucose released from starch (through two-step hydrolysis using α-amylase and glucoamylase)[13], residual isomaltose (major) and isomaltotriose (minor) were detected in the final fermentation culture (data not shown). Both isomaltose and isomaltotriose contain α-1, 6-O-glucosidic bonds, which couldn't be completely hydrolyzed into glucose by glucoamylase. Considering that isomaltases generally work under neutral conditions[4, 8, 10-11], acidophilic isomaltases are required to make fully use of the residual oligosaccharides containing α-1, 6-O-glucosidic bonds under acidic fermentation conditions. Acidophilic S. cerevisiae strains are good resource for mining acidophilic isomaltases[14]. Moreover, isomaltases play key roles in full utilization of isomaltose, which is present in grains, potatoes, and other carbohydrate- containing foods. Therefore, isomaltases having different properties such as thermal, acid and alkaline tolerance, especially the major isomaltase IMA1, are preferred for applications.
Considering the fact that IMA1/YGR287c encodes the major isomaltase IMA1, which plays key roles in assimilation of isomaltose and α-MG in S. cerevisiae[8], four IMA1 genes including three from acidophilic S. cerevisiae strains were cloned and overexpressed. Their amino acid sequences were aligned and analyzed. Their kinetic parameters, pH and temperature dependence, and thermal stability were characterized and compared. A few residues which could affect thermostability and kinetic parameters of isomaltase IMA1 were identified.
1 Materials and methods 1.1 Materials Chemicals were from Sigma, Merck or Ameresco. Oligonucleotides were synthesized by Shanghai Sangon Biotech Co. Ltd (China). Taq DNA polymerases and all restriction endonucleases were from Fermentas or TaKaRa Biotechnology. The kits used for molecular cloning were from Omega Bio-tek or TaKaRa Biotechnology. Nickel column was from Novagen. The expression vector pYES2 and S. cerevisiae INVSC1 were from Invitrogen. Antibodies and chemical reagents used for Western blotting were from Tiangen (China).
1.2 Bacterial strains, plasmids and media Three acidophilic strains of S. cerevisiae named ME15, ME16 and ME17, which can grow at pH 2.5, are from our lab. E. coli DH5α was used for routine DNA transformation and plasmid isolation. S. cerevisiae INVSC1 was utilized for IMA1 overexpression. The vector pYES2 was used for subcloning. E. coli strain was routinely grown in Luria-Bertani broth at 37 ℃ or on LB supplemented with 1.5% (W/V) agar. 50 μg/mL Ampicillin was added when required. S. cerevisiae was grown in YPD (1% yeast extract, 2% peptone, 2% D-glucose), and S. cerevisiae transformants harboring IMA1 were selected on SC-U plates or grown in SC-U medium, including 0.67% yeast nitrogen base (without amino acids but with ammonium sulfate), 2% glucose, 0.01% (adenine, arginine, cysteine, leucine, lysine, threonine, tryptophan), 0.005% (aspartic acid, histidine, isoleucine, methionine, phenylalanine, proline, serine, tyrosine, valine) (2% agar added for plates). The IMA1 genes were overexpressed in SC-U induction medium, containing 2% galactose and 1% raffinose.
1.3 DNA manipulations General molecular biology techniques were carried out by standard procedures[15]. Restriction and modification enzymes were used following the recommendations of the manufacturers. DNA fragments were purified from agarose gels using the DNA gel extraction kit. Plasmid DNA was isolated using the plasmid miniprep kit.
The IMA1 genes were amplified by PCR using genomic DNA of different strains of S. cerevisiae as templates. The used primers included the forward one (5′-GCTGGTACCTTCGAATGTCTACTATTTC TGCACATCCAG-3′, the Kpn I restriction site underlined) and the reverse one (5′-ACTCTCG AGCTAGATATGGTGATGATGATGTTCGCTGATA TATATTCTTCCTTC-3′, the Xho I restriction site underlined) respectively. The PCR products were purified from agarose gel, and cloned into the vector pYES2 at the restrictions sites of Kpn I and Xho I. All constructs were confirmed by DNA sequencing.
1.4 Protein over-expression and purification The expression constructs were transformed into competent S. cerevisiae INVSC1. Overexpression of IMA1 was carried out following the standard protocol. A single colony of S. cerevisiae INVSC1 transformant was used to inoculate 15 mL SC-U medium containing 2% glucose and incubated at 225 r/min and 30 ℃ overnight. The cells were harvested, resuspended in SC-U induction medium containing 2% galactose, and incubated at 225 r/min and 30 ℃. At different time points, 5 mL culture was removed and centrifuged, and the pellet was stored at –20 ℃ until use. After the expression conditions were optimized, expression of isomaltase IMA1 was done at large scale.
The cell pellet was suspended in breaking buffer (50 mol/L sodium phosphate, pH 7.4, 1 mmol/L EDTA, 5% glycerol, 1 mmol/L PMSF), and acid-washed glass beads were added. The samples were homogenized using Fastprep FP120 (Bio 101 Savant), and vortexed for 30 s at maximum speed, followed by 5 min on ice. The procedure was repeated for 12 times. The cell lysate was then centrifuged at 13000 g and 4 ℃ for 25 min. Crude samples were analyzed by SDS-PAGE and Western blotting.
All purification procedures were carried out at 4 ℃. Nickel-chelating resin (2 mL) was equilibrated with 5 column volumes of equilibration buffer (buffer A: 75 mmol/L Tris/HCl, pH 8.0, 0.5 mol/L NaCl). The crude supernatant was loaded onto the resin, which was sequentially washed with buffer A containing 20–250 mmol/L imidazole. The enzyme purity was analyzed via SDS-PAGE. The fractions containing IMA1 were pooled, and dialyzed against 0.1 mol/L potassium phosphate buffer, pH 6.8. The protein concentration was determined by the Bradford method using bovine serum albumin as a standard.
1.5 Enzyme assay The enzymatic assays were carried out in triplicate, and performed in a volume of 200 μL of 100 mmol/L B & R (Britton and Robinson) buffer (pH 6.0), containing 12 μg/mL of isomaltase IMA1 and α-MG or isomaltose. After being incubated at 30 ℃ for 1.0, 2.5, 5.0, 7.5, 10.0 min respectively, the reactions were stopped by heating at 80 ℃ for 5 min. The tubes were then centrifuged, and the released glucose was quantified using a biosensor (SBA-40C, Biology Institute of Shandong Academy of Science, China). The Km and Vmax values against α-MG were determined using the Lineweaver-Burk plot. The kcat value was calculated from Vmax on the basis of the molecular weight of isomaltases-68.8 kDa.
1.6 Determination of pH-rate profiles, temperature dependence and thermostability The optimal pH values of isomaltases IMA1 were evaluated in 100 mmol/L B & R buffer at 30 ℃ and pH values between 2.0 to 9.0 using isomaltose (10 mmol/L) as the substrates. The optimal temperature was determined at pH 6.0 (100 mmol/L B & R buffer) with temperatures between 25 ℃ to 60 ℃ using α-MG (0.1 mol/L) as the substrate. Specific activities under different conditions were measured. One unit of enzyme activity (U) is defined as the amount of the enzyme that catalyzes the conversion of 1 μmol substrate per min, and specific activity was defined as units·mg–1 protein.
To determine the thermostability of isomaltases IMA1, they were pre-incubated for different times (5 min to 60 min) at pH 6.0 and 50 ℃ respectively, and the residual activities were measured as above. The half-lives (t1/2) of isomaltases IMA1 at that temperature were calculated.
2 Results 2.1 Gene cloning and sequence analysis of isomaltases IMA1 A pair of primers of IMA1 including the His6 tag at C-terminal were designed based on the sequence of IMA1/YGR287c. Four isomaltases IMA1 genes, including one from S. cerevisiae INVSC1 and three from acidophilic strains of S. cerevisiae ME15, ME16 and ME17, were named as IMA1-A, IMA1-B, IMA1-C and IMA1-D respectively. All of them were amplified by PCR using corresponding genomic DNA of S. cerevisiae as templates and the above-designed primers, then cloned into the pMD18-T simple vector and sequenced respectively. The amino acid sequences of four isomaltases IMA1A-IMA1D were aligned and compared with the sequence of IMA1 (encoded by YGR287c), IMA2, IMA3 and IMA5. IMA1-A showed the identical sequence to IMA1 (data not shown). IMA1-B, IMA1-C, and IMA1-D exhibited high sequence identities to IMA1-A (> 98.6%) (Figure 1). IMA1-A exhibited 93.0%, 92.7% and 65.1% sequence identities to IMA2, IMA3 and IM5 respectively. The key residues, including the catalytic ones (Asp215, Glu277 and Asp352) and those involved in discriminating the α-1, 4- and α-1, 6-glucosidic linkages of substrates (Val216 and Gln279) and isomaltose binding (Tyr158, His280, and loop residues 310–315), were highly conserved (Figure 1). Different amino acids at ten positions were observed among four isomaltases IMA1 (43, Ala or Ser; 54, Ala or Val; 336, Tyr or Phe; 370, Ile or Val; 377, Val or Ile; 380, Ser or Val; 381, Ala or Ser; 387, Tyr or Phe; 388, Val or Ile; 439, Leu or Ile), all of which are located far from the catalytic active sites and the substrate binding pocket (Figure 1, Figure 2).
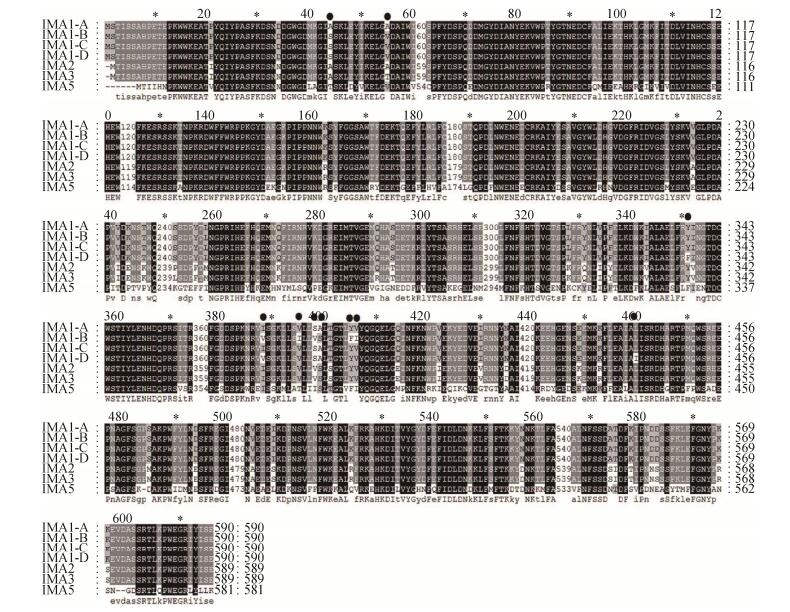 |
| Figure 1 Sequence alignment of isomaltases using ClustalW2. Different amino acids of four isomaltases IMA1 were shown in black dots. |
| 图选项 |
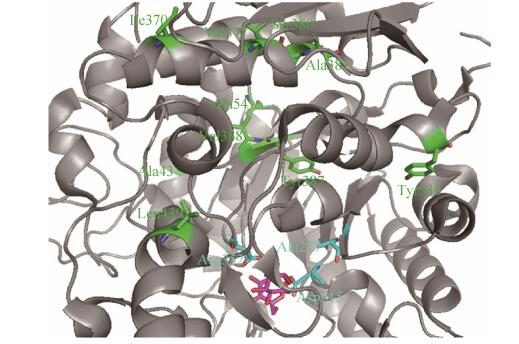 |
| Figure 2 Relative positions of different residues to the active sites and the substrate binding pocket in the crystal structure of isomaltase IMA1 (PDB ID: 3AXH)[11]. Catalytic residues, different ones, and bound isomaltose were colored in cyan, green, and pink respectively. Catalytic site Glu277 was mutated into Ala in the crystal structure[11]. |
| 图选项 |
2.2 Expression and purification of isomaltases IMA1 All four isomaltases IMA1 genes were cloned into the expression vector pYES2 at the restriction sites of Kpn I and Xho I, and were overexpressed in S. cerevisiae INVSC1 respectively. Higher overexpression levels of four IMA1 genes were obtained after 10 h-induction by galactose. All were successfully overexpressed in S. cerevisiae INVSC1. Overexpressed isomaltases IMA1 containing the His6 tag at the C-terminal were purified on nickel column with > 90% purity, and further confirmed by western blotting (Figure 3). Most proteins were present in the soluble form (Figure 3). The protein yield is about 8.0 mg/L.
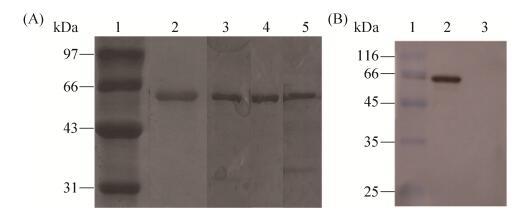 |
| Figure 3 Analysis of isomaltases IMA1 by SDS-PAGE (A) and Western blotting (B). A: lane 1: low protein molecular weight marker; lane 2: c purified IMA1-A; lane 3: purified IMA1-B; lane 4: purified IMA1-C; lane 5: purified IMA1-D. B: lane 1: low protein molecular weight marker; lane 2: crude supernatant for over-expressed IMA1-A; lane 3: crude supernatant for S. cerevisiae INVSC1 carrying the empty pYES2 vector. |
| 图选项 |
2.3 Comparison of kinetic parameters of isomaltases IMA1 All four isomaltases IMA1 could use α-MG and isomaltose as the substrate. The kinetic parameters of four isomaltases IMA1 against α-MG, including Km, kcat and kcat/Km, were determined (Table 1). IMA1-A showed the lowest Km value, the highest kcat value and catalytic efficiency (kcat/Km), whereas IMA1-C had the highest Km value, the lowest kcat value and catalytic efficiency.
Table 1. Comparison of kinetic parameters of four isomaltases IMA1
| Enzymes | Km/(mmol/L) | kcat/s–1 | kcat/Km/(L/(mmol·s)) |
| IMA1-A | 18.9±0.9 | 123.6±2.4 | 6.5±0.1 |
| IMA1-B | 28.7±2.9 | 73.4±2.3 | 2.60±0.08 |
| IMA1-C | 36.3±2.6 | 50.1±1.2 | 1.40±0.03 |
| IMA1-D | 36.1±3.2 | 95.2±3.9 | 2.6±0.1 |
表选项
2.4 Comparison of pH-rate profiles of isomaltases IMA1 The pH dependence of four isomaltases IMA1 was investigated using isomaltose as the substrate in 100 mmol/L B & R (Britton and Robinson) buffer at pH values between 2.0 to 9.0. They demonstrated similar bell-shaped pH-rate profiles, the highest activity was observed at pH 6.0, and activity decreased sharply at pH > 7.0 and pH < 5.0 (Figure 4). IMA1-A showed higher activity than IMA1-B, IMA1-C and IMA1-D at pH values between 4.0 to 8.0 (Figure 4).
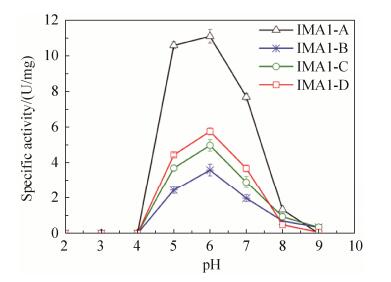 |
| Figure 4 pH-Rate profiles of four isomaltases IMA1. Isomaltose (10 mmol/L) was used as the substrate. Standard errors were obtained from three parallel experiments. |
| 图选项 |
2.5 Comparison of temperature dependence and themostability of isomaltases IMA1 The temperature dependence of four isomaltases IMA1 was investigated using α-MG as the substrate. They exhibited similar temperature dependence, and the highest activity was observed at 45 ℃ (Figure 5). IMA1-A still displayed higher activity than IMA1-B, IMA1-C and IMA1-D at different temperatures (Figure 5). The themostability of four isomaltases IMA1 was evaluated using α-MG as the substrate at pH 6.0. None of them lost 50% of original activities after being incubated at 50 ℃ for 1 h. Therefore, their half-lives (t1/2) at 50 ℃ were determined respectively, which were 35.2±3.3 min for IMA1-A, 18.4±1.9 min for IMA1-B, 8.6±0.9 min for IMA1-C and IMA1-D respectively (Figure 6). IMA1-A showed the highest thermostability, followed by IMA1-B, IMA1-C and IMA1-D.
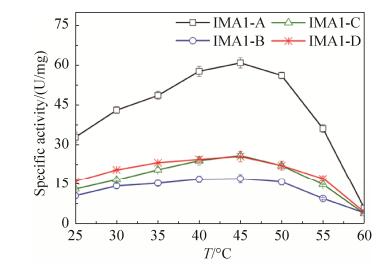 |
| Figure 5 Temperature dependence of four isomaltases IMA1. α-MG (0.1 mol/L) was used as the substrate. Standard errors were obtained from three parallel experiments. |
| 图选项 |
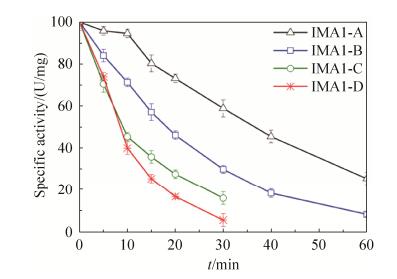 |
| Figure 6 Thermostability of four isomaltases at 50 ℃. α-MG (0.1 mol/L) was used as substrate. The absolute values of 100% relative activity for IMA1-A, IMA1-B, IMA1-C and IMA1-D were 40.5±1.8, 14.4±0.4, 15.7±0.7, and 18.0±0.9 U/mg respectively. Standard errors were obtained from three parallel experiments. |
| 图选项 |
3 Discussion In this study, four isomaltases IMA1 from four strains of S. cerevisiae three from acidophilic ones were cloned, purified, and characterized respectively (Table 1, Figure 4, 5, 6). In comparison with the recently reported Km (17 mmol/L and 27 mmol/L), kcat (103 s–1 and 84 s–1), and kcat/Km (5.7 L/(mmol·s) and 3.1 L/(mmol·s)) values of Ima1p (equivalent to IMA1-A) towards isomaltose and α-MG, IMA1-A exhibited similar kinetic parameters against α-MG (Table 1)[10]. In addition, IMA1-A demonstrated similar pH and temperature dependence and thermal stability to the publisehd ones for Im1p (equivalent to IMA1-A)[10].
It is interesting to see that IMA1-A and IMA1-C displayed completely different kinetic performance and thermostability (Table 1, Figure 6). However, the only difference between them at the protein level lies in positions of 43 (Ala in IMA1-A versus Ser in IMA1-C) and 54 (Ala in IMA1-A versus Val in IMA1-C) (Figure 1, 2). What's more, these two residues are far away from the catalytic sites and the substrate binding pocket according to the crystal structure (Figure 2)[11], and are alanine residues only in IMA1-A (Ser43 and Val54 are completely conserved in the other three isomaltases IMA1, Figure 1). Moreover, residues Ile377, Val380, Ser381, Phe387 and Ile388 are specific for IMA1-B, whereas Phe336 and Ile439 are only present in IMA1-D (Figure 1). These results demonstrated that even a few different amino acids, remote from the active residues and the substrate binding site, can lead to significantly different kinetic behavior and thermostability of isomlatases IMA1. Same situation was noticed when error-prone PCR was used to simultaneously enhance thermostability and catalytic activity of proteins[16-17]. Most of the beneficial residues identified were often far away from the active sites and the substrate binding pocket, which couldn't be readily predicted by rational design[16-17].
Unfortunately, three isomaltases IMA1 from three acidophilic strains of S. cerevisiae didn't demonstrate activity at pH 2.0 (Figure 4). The fact that isomaltases IMA1 were not secreted into the medium and there is a big difference between intracellular pH (around 6.0) and extracellular pH (2.5) can explain the observed pH-rate profiles for them (Figure 4). It has been reported that extracellular glucoamylase and intracellular one from thermoacidophilic Archaea Picrophilus torridus (optimal growth at pH 0.7 and 60 ℃) exhibited completely different optimal pH: 2.0 (extracellular) versus 5.0 (intracellular)[18, 19]. Oshima et al. also observed that glyceraldehyde-3-phosphate dehydrogenase from acidophilic Bacillus acidocaldarius (optimal growth pH 2.5) had the optimum pH 8[20].
In conclusion, four isomaltases IMA1 including three from acidophilic S. cerevisiae strains were cloned, characterized, and compared. They demonstrated similar pH-rate profiles and temperature dependence, but different kinetic parameters and thermostability. IMA1-A performed best in kinetics and thermostability. It was found by structure and sequence analysis that minor difference between isomaltases IMA1 could result in significantly different properties such as kinetics and thermostability. This discovery will be helpful for designing isomaltases IMA1 with novel properties.
References
| [1] | Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enzymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Research, 2009, 37(S1): D233-D238. |
| [2] | Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochemical Journal, 1991, 280(2): 309-316. DOI:10.1042/bj2800309 |
| [3] | Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochemical Journal, 1996, 316(2): 695-696. DOI:10.1042/bj3160695 |
| [4] | Khan NA, Eaton NR. Purification and characterization of maltase andα-methyl glucosidase from yeast. Biochimica et Biophysica Acta, 1967, 146(1): 173-180. DOI:10.1016/0005-2744(67)90084-8 |
| [5] | Yamamoto K, Miyake H, Kusunoki M, Osaki S. Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose. The FEBS Journal, 2010, 277(20): 4205-4214. DOI:10.1111/j.1742-4658.2010.07810.x |
| [6] | Ten Berge AMA. Genes for the fermentation of maltose and α-methylglucoside in Saccharomyces carlsbergensis. Molecular and General Genetics, 1972, 115(1): 80-88. DOI:10.1007/BF00272220 |
| [7] | Khan NA, Haynes RH. Genetic redundancy in yeast: non-identical products in a polymeric gene system. Molecular and General Genetics, 1972, 118(3): 279-285. DOI:10.1007/BF00333464 |
| [8] | Teste MA, Fran?ois JM, Parrou JL. Characterization of a new multigene family encoding isomaltases in the yeast Saccharomyces cerevisiae, the IMA family. Journal of Biological Chemistry, 2010, 285(35): 26815-26824. DOI:10.1074/jbc.M110.145946 |
| [9] | Naumov GI, Naumoff DG. Molecular genetic differentiation of yeastα-glucosidases: Maltase and isomaltase. Microbiology, 2012, 81(3): 276-280. DOI:10.1134/S0026261712030101 |
| [10] | Deng X, Petitjean M, Teste MA, Kooli W, Tranier S, Fran?ois JM, Parrou JL. Similarities and differences in the biochemical and enzymological properties of the four isomaltases from Saccharomyces cerevisiae. FEBS Open Bio, 2014, 4: 200-212. DOI:10.1016/j.fob.2014.02.004 |
| [11] | Yamamoto K, Miyake H, Kusunoki M, Osaki S. Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae. Journal of Bioscience and Bioengineering, 2011, 112(6): 545-550. DOI:10.1016/j.jbiosc.2011.08.016 |
| [12] | Yamamoto K, Nakayama A, Yamamoto Y, Tabata S. Val216 decides the substrate specificity of α-glucosidase in Saccharomyces cerevisiae. European Journal of Biochemistry, 2004, 271(16): 3414-3420. DOI:10.1111/ejb.2004.271.issue-16 |
| [13] | Dwiarti L, Otsuka M, Miura S, Yaguchi M, Okabe M. Itaconic acid production using sago starch hydrolysate by Aspergillus terreus TN484-M1. Bioresource Technology, 2007, 98(17): 3329-3337. DOI:10.1016/j.biortech.2006.03.016 |
| [14] | Sharma A, Kawarabayasi Y, Satyanarayana T. Acidophilic bacteria and archaea: acid stable biocatalysts and their potential applications. Extremophiles, 2012, 16(1): 1-19. DOI:10.1007/s00792-011-0402-3 |
| [15] | Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 1989. |
| [16] | Komeda H, Ishikawa N, Asano Y. Enhancement of the thermostability and catalytic activity of D-stereospecific amino-acid amidase from Ochrobactrum anthropi SV3 by directed evolution. Journal of Molecular catalysis B: Enzymatic, 2003, 21(4/6): 283-290. |
| [17] | Zhao HM, Arnold FH. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Engineering, 1999, 12(1): 47-53. |
| [18] | Serour E, Antranikian G. Novel thermoactive glucoamylases from the thermoacidophilic Archaea Thermoplasma acidophilum, Picrophilus torridus and Picrophilus oshimae. Antonie van Leeuwenhoek, 2002, 81(1/4): 73-83. DOI:10.1023/A:1020525525490 |
| [19] | Schepers B, Thiemann V, Antranikian G. Characterization of a novel glucoamylase from the thermoacidophilic Archaeon Picrophilus torridus heterologously expressed in E. coli. Engineering in Life Sciences, 2006, 6(3): 312-317. |
| [20] | Oshima T, Arakawa H, Baba M. Biochemical studies on an acidophilic, thermophilic bacterium, Bacillus acidocaldarius: isolation of bacteria, intracellular pH, and stabilities of biopolymers. The Journal of Biochemistry, 1977, 81(4): 1107-1113. DOI:10.1093/oxfordjournals.jbchem.a131535 |
