陈义旺1, 李晓霞1, 周作勇1,2


1.西南大学动物科学学院, 重庆 402460;
2.重庆市兽医科学工程研究中心, 重庆 402460
收稿日期:2017-08-27;修回日期:2017-11-01;网络出版日期:2017-11-29
基金项目:中央高校基本科研业务费专项基金(XDJK2017D078)
*通信作者:周作勇, Tel/Fax:+86-23-46751588;E-mail:zzyxnny@163.com
摘要:胞内寄生病原伪结核棒状杆菌(Corynebacterium pseudotuberculosis,Cp)不仅对全球养殖业造成巨大的经济损失,且可感染人而危害公共卫生安全。组学技术的快速发展促进了Cp致病性的研究。通过基因组测序分析获得72株Cp全基因组序列,初步明确了羊型Cp与马型Cp的结构、进化关系及致病性差异的可能原因;基于蛋白质组学研究,发现了磷脂酶D(PLD)、丝氨酸蛋白酶(CP40)及神经氨酸酶H(NanH)等与Cp生理学、毒性和免疫相关及其他未知功能的蛋白;利用转录组学发现Cp在高渗透压、热休克或酸性条件等恶劣环境中以粘附、应激和氧化还原反应基因表达差异最为典型。本文在介绍Cp基因组测序情况基础上结合作者获得的Cp宣汉株(XH02)基因组序列进行比较基因组学和SNP进化分析,同时对蛋白质组学和转录组学技术在Cp致病机制研究中的应用概况进行综述。
关键词: 伪结核棒状杆菌 基因组学 蛋白质组学 转录组学 致病性 应用
Application of omics technology on Corynebacterium pseudotuberculosis pathogenicity analysis
Yiwang Chen1, Xiaoxia Li1, Zuoyong Zhou1,2


1.College of Animal Science, Southwest University, Chongqing 402460, China;
2.Veterinary Science Engineering Research Center of Chongqing, Chongqing 402460, China
Received 27 August 2017; Revised 1 November 2017; Published online 29 November 2017
*Corresponding author: Zuoyong Zhou, Tel/Fax: +86-23-46751588; E-mail: zzyxnny@163.com
Supported by the Fundamental Research Funds for the Central Universities (XDJK2017D078)
Abstract: Corynebacterium pseudotuberculosis is an interesting bacterium due to its great zoonotic potential and because it causes considerable economic losses worldwide. The rapid development of omics technology has promoted the study of C. pseudotuberculosis pathogenicity. A total of 72 C. pseudotuberculosis genome sequences were obtained by genome sequencing analysis, and the structure, evolution and pathogenicity differences of C. pseudotuberculosis biovar equi or biovar ovis were identified. Based on the proteomics study, proteins such as phospholipase D (PLD), serine protease (CP40) and neuraminidase H (NanH) were found to be related to C. pseudotuberculosis physiology, toxicity and immunity, and other unknown functions. With the transcriptomics, it was found that defense against oxidative stress, adhesion and regulation were most typical in the gene expression difference in harsh environments such as high osmotic pressure, heat shock or acidic conditions. In this paper, we analyzed the C. pseudotuberculosis Xuanhan strain (XH02) we obtained with comparative genomic and SNP evolution, and the application of proteomics and transcriptomics in C. pseudotuberculosis pathogenesis was reviewed.
Key words: Corynebacterium pseudotuberculosis genomics proteomics transcriptomics pathogenicity application
伪结核棒状杆菌(Corynebacterium pseudotuberculosis,Cp)是革兰氏阳性的兼性胞内寄生菌,该菌好氧,多形态,无芽孢,无运动性,具有菌毛[1]。Cp不仅感染山羊、绵羊、马、牛和骆驼等多种动物,同时还可感染人[2]。目前人类感染Cp的病例已超过33例,对公共卫生安全具有潜在的威胁。Cp有两种生物型,羊型(Biovar ovis)(硝酸盐还原实验阴性)和马型(Biovar equi)(硝酸盐还原实验阳性)[3]。其中羊型Cp主要侵染山羊和绵羊的淋巴结和内脏器官,与全球范围内分布的羊干酪样淋巴结炎(caseous lymph adenitis,CLA)相关,该疾病可损害动物的毛皮和胴体,降低产奶量和产肉量,甚至引起死亡,导致巨大的经济损失[4-6]。马型Cp主要感染马和牛等动物,引起牛的溃疡性肉芽肿和乳房炎、马的溃疡性淋巴管炎或鸽热(pigeon fever)[7]及水牛的水肿性皮肤病(oedematous skin disease,OSD)[8]等。Cp引起多种动物持续的慢性感染与其完美的生存机制密切相关,该病原可根据环境灵活调整感染策略,一旦成功感染宿主即能在吞噬细胞中复制并逃避宿主免疫,这种慢性感染可持续发生于动物生命的大部分时间中[9-11]。自2006年对Cp基因组进行测序以来,利用组学技术研究和揭示该病原的分子特点、致病机理及宿主应答机制得到了较大的发展,本文综述了近10年来关于伪结核棒状杆菌的基因组、蛋白质组和转录组的研究概况。
1 伪结核棒杆菌基因组学研究概况 1.1 伪结核棒杆菌基因组测序情况 Dorella等最先尝试进行Cp基因组的测序[12],他们以细菌人工染色体(BAC)载体构建Cp1002基因组文库,获得约18000个克隆,插入片段在25–120 kb。分析发现,Cp基因序列与白喉杆菌(C. diphtheriae)、谷氨酸棒状杆菌(C. glutamicum)、有效棒状杆菌(C. efficiens)和杰氏棒状杆菌(C. jeikeium)之间密切的系统进化关系[12]。由此,2006年由Rede Genoma de Minas Gerais (RGMG-Brazil)启动了针对Cp1002的首个全基因组测序项目,并于2009年完成。
随着测序技术的发展,特别是第二代测序平台如SOLiDTM 550和Ion Torrent PGM的出现,对Cp的测序工作呈上升趋势(图 1-A)。迄今为止,已经有18个国家和地区(图 1-C,表 1)对72株Cp菌株进行了测序(截止时间为2017年6月30日)。其中,包括笔者所在实验室于2016年首次在国内成功获得的伪结核棒状杆菌(XH02株)全基因组序列(NCBI检索号MDWN00000000.1)[13]。从所测Cp全基因组来源国家统计,以美国测序数量最多(16株菌),其次为巴西(15株)、埃及(13株)和墨西哥(6株),而其他国家包括中国、英国、法国和南非等,目前仅1株Cp全基因组测序数据(图 1-C);从基因组测序的Cp宿主来源统计,以感染马的数量最多(28%),其次是绵羊和山羊均占22%,水牛占14% (表 1,图 1-B);根据Cp全基因组序列,发现其GC含量为52.1%–52.2%,总基因组大小为2.28–2.44 Mb。
 |
| 图 1 72株伪结核棒状杆菌的基因组测序完成时间、来源宿主和国家分布 Figure 1 Release date, hosts origin and geographic distribution of 72 C. pseudotuberculosis strains with complete genome (made by author). A: Release date of C. pseudotuberculosis; B: Hosts origin of C. pseudotuberculosis; C: Countries for C. pseudotuberculosis sequencing. |
| 图选项 |
表 1. 2009年到2017年提交到NCBI数据库中的伪结核棒状杆菌的菌株及其结构特征 Table 1. Strains of C. pseudotuberculosis deposited in NCBI between 2009 and 2017 and their structural characteristics
| Strain | Biovar | Animal/Host | Site of isolation | County of isolation | Size/ (Mb) | (G+C)/ % | Genes | Proteins | Accession | Release date |
| C231 | ovis | Sheep | Abscess | Australia | 2.32821 | 52.2 | 2204 | 2091 | CP001829.1 | 2010/8/5 |
| FRC41 | ovis | Human | Inguinal lymph node | Frence | 2.33791 | 52.2 | 2171 | 2110 | CP002097.1 | 2010/7/16 |
| 1002 | ovis | Goat | Abscess | Brazil | 2.33511 | 52.2 | 2203 | 2090 | CP001809.2 | 2010/8/6 |
| I19 | ovis | Bovine | Not specified | Israel | 2.33759 | 52.2 | 2213 | 2095 | CP002251.2 | 2010/10/12 |
| PAT10 | ovis | Sheep | Abscess | Patagonia | 2.33532 | 52.2 | 2200 | 2079 | CP002924.1 | 2011/7/27 |
| 42/02-A | ovis | Sheep | Abscess | Australia | 2.33761 | 52.2 | 2239 | 2142 | CP003062.1 | 2011/10/3 |
| CIP 52.97 | equi | Horse | Not specified | Kenya | 2.32059 | 52.1 | 2194 | 2060 | CP003061.2 | 2011/10/6 |
| 1/06-A | equi | Horse | Abscess | California | 2.27912 | 52.2 | 2127 | 1963 | CP003082.1 | 2011/11/3 |
| 3/99-5 | ovis | Sheep | Abscess | Scotland | 2.33794 | 52.2 | 2239 | 2142 | CP003152.1 | 2012/1/10 |
| 316 | equi | Horse | Subcutaneous abscess | California | 2.31041 | 52.1 | 2234 | 2106 | CP003077.1 | 2012/2/22 |
| P54B96 | ovis | Antelope | Liver, lung, mediastinal lymph node | South Africa | 2.33766 | 52.2 | 2205 | 2084 | CP003385.1 | 2012/3/23 |
| 267 | ovis | Llama | Submandibular abscess | California | 2.33763 | 52.2 | 2249 | 2148 | CP003407.1 | 2012/4/10 |
| 31 | equi | Buffalo | Not specified | Egypt | 2.40296 | 52.1 | 2170 | 2063 | CP003421.3 | 2012/4/18 |
| 258 | equi | Horse | Not specified | Belgium | 2.36982 | 52.1 | 2195 | 2088 | CP003540.2 | 2012/5/22 |
| Cp162 | equi | Camel | Abscess | UK | 2.29346 | 52.2 | 2150 | 2002 | CP003652.2 | 2012/6/11 |
| 48252 | ovis | Human | Not specified | Norwegian | 2.33814 | 52.2 | 2145 | 2011 | CP008922.1 | 2014/7/17 |
| CS_10 | ovis | Goat | Abscess | Norwegian | 2.33814 | 52.2 | 2146 | 2012 | CP008923.1 | 2014/7/17 |
| Ft_2193/67 | ovis | Goat | Abscess | Norwegian | 2.33830 | 52.2 | 2145 | 2020 | CP008924.1 | 2014/7/17 |
| MB20 | equi | Horse | Abscess | California | 2.37090 | 52.1 | 2194 | 1903 | CP016829.1 | 2016/8/11 |
| VD57 | ovis | Goat | Abscess | Brazil | 2.33718 | 52.2 | 2141 | 2015 | CP009927.1 | 2015/1/6 |
| 226 | ovis | Goat | Abscess | California | 2.33782 | 52.2 | 2143 | 1961 | CP010889.1 | 2015/4/16 |
| 12C | ovis | Sheep | Abscess | Brazil | 2.33745 | 52.2 | 2143 | 2012 | CP011474.1 | 2015/6/2 |
| 29156 | ovis | Bovine | Not specified | Israel | 2.33865 | 52.2 | 2147 | 2026 | CP010795.1 | 2015/6/3 |
| 262 | equi | Bostaurus | Not specified | Belgium | 2.32575 | 52.2 | 2121 | 1951 | CP012022.1 | 2015/7/7 |
| E19 | equi | Horse | Not specified | Chile | 2.36796 | 52.1 | 2183 | 2037 | CP012136.1 | 2015/7/24 |
| PO269-5 | ovis | Goat | Abscess | Portugal | 2.33712 | 52.2 | 2145 | 2017 | CP012695.1 | 2015/9/24 |
| 1002B | ovis | Goat | Abscess | Brazil | 2.33511 | 52.2 | 2141 | 2019 | CP012837.1 | 2015/11/5 |
| N1 | ovis | Sheep | Lung | Equatorial Guinea | 2.33785 | 52.2 | 2142 | 2018 | CP013146.1 | 2015/11/23 |
| PA01 | ovis | Sheep | Lymph node | Brazil | 2.33792 | 52.2 | 2141 | 2032 | CP013327.1 | 2015/12/3 |
| MEX25 | ovis | Sheep | Not specified | Mexico | 2.33753 | 52.2 | 2143 | 2016 | CP013697.1 | 2015/12/23 |
| PO222/4-1 | ovis | Goat | Abscess | Portugal | 2.33751 | 52.2 | 2145 | 2026 | CP013698.1 | 2015/12/23 |
| E56 | ovis | Sheep | Not specified | Egypt | 2.33577 | 52.2 | 2141 | 1994 | CP013699.1 | 2015/12/23 |
| MB11 | equi | Horse | Lymph node | California | 2.36342 | 52.1 | 2188 | 1928 | CP013260.1 | 2016/3/3 |
| MB14 | equi | Horse | Lymph node | California | 2.37076 | 52.1 | 2193 | 1967 | CP013261.1 | 2016/3/3 |
| MB30 | equi | Horse | Lymph node | California | 2.36438 | 52.1 | 2188 | 1995 | CP013262.1 | 2016/3/3 |
| MB66 | equi | Horse | Lymph node | California | 2.37220 | 52.1 | 2193 | 1963 | CP013263.1 | 2016/3/3 |
| E55 | ovis | Sheep | Not specified | Egypt | 2.33538 | 52.2 | 2140 | 1994 | CP014341.1 | 2016/5/27 |
| MEX9 | ovis | Goat | Prescapular of a female goat | Mexico | 2.33758 | 52.2 | 2146 | 2025 | CP014543.1 | 2016/5/27 |
| PA02 | equi | Goat | Abscess | Brazil | 2.32843 | 52.2 | 2130 | 2027 | CP015309.1 | 2016/6/16 |
| T1 | ovis | Goat | Granulomatous lesion | Brazil | 2.33720 | 52.2 | 2143 | 2019 | CP015100.1 | 2016/7/8 |
| Cp13 | ovis | Goat | Caseous lymphadenitis | Brazil | 2.34224 | 52.2 | 2149 | 2020 | CP014998.1 | 2016/7/8 |
| MEX29 | ovis | Sheep | Not specified | Mexico | 2.33787 | 52.2 | 2145 | 2032 | CP016826.1 | 2016/11/3 |
| 34 | equi | Bubalus bubalis | Not specified | Egypt | 2.40345 | 52.1 | 2226 | 2076 | CP015192.1 | 2016/11/7 |
| 32 | equi | Bubalus bubalis | Not specified | Egypt | 2.40353 | 52.1 | 2226 | 2071 | CP015183.1 | 2016/11/7 |
| 33 | equi | Bubalus bubalis | Not specified | Egypt | 2.40355 | 52.1 | 2227 | 2068 | CP015184.1 | 2016/11/7 |
| 35 | equi | Bubalus bubalis | Not specified | Egypt | 2.40350 | 52.1 | 2229 | 2073 | CP015185.1 | 2016/11/7 |
| 36 | equi | Bubalus bubalis | Not specified | Egypt | 2.40341 | 52.1 | 2223 | 2065 | CP015186.1 | 2016/11/7 |
| 38 | equi | Bubalus bubalis | Not specified | Egypt | 2.40352 | 52.1 | 2225 | 2065 | CP015187.1 | 2016/11/7 |
| 39 | equi | Bubalus bubalis | Not specified | Egypt | 2.40358 | 52.1 | 2223 | 2069 | CP015188.1 | 2016/11/7 |
| 43 | equi | Bubalus bubalis | Not specified | Egypt | 2.36508 | 52.1 | 2181 | 2033 | CP015189.1 | 2016/11/7 |
| 46 | equi | Bubalus bubalis | Not specified | Egypt | 2.36657 | 52.1 | 2178 | 2030 | CP015190.1 | 2016/11/7 |
| 48 | equi | Bubalus bubalis | Not specified | Egypt | 2.40330 | 52.1 | 2224 | 2069 | CP015191.1 | 2016/11/7 |
| I37 | equi | Cow | Mammary tissue | Israel | 2.37028 | 52.1 | 2172 | 2026 | CP017384.1 | 2016/12/5 |
| MEX30 | equi | Horse | Lymph node, chronical abscesses | Mexico | 2.36814 | 52.1 | 2188 | 2008 | CP017291.1 | 2016/12/27 |
| MEX31 | equi | Horse | Lymph node, chronical abscesses | Mexico | 2.36788 | 52.1 | 2182 | 2058 | CP017292.1 | 2016/12/27 |
| MEX1 | ovis | Goat | Retropharyngeal abscess | Mexico | 2.33709 | 52.2 | 2145 | 2021 | CP017711.1 | 2017/1/30 |
| PA04 | ovis | Sheep | Mandibular lymph node | Brazil | 2.33809 | 52.2 | 2142 | 1987 | CP019587.1 | 2017/2/7 |
| phoP | ovis | Goat | Abscess | Brazil | 2.33930 | 52.2 | 2151 | 2032 | CP019768.1 | 2017/3/2 |
| MIC6 | ovis | Sheep | Abscess | Brazil | 2.33715 | 52.2 | 2143 | 2026 | CP019769.1 | 2017/3/2 |
| SigmaE | ovis | Goat | Abscess | Brazil | 2.33926 | 52.2 | 2146 | 2021 | CP020356.1 | 2017/3/27 |
| PA05 | ovis | Sheep | Not specified | Brazil | 2.43544 | 52.2 | 2247 | 1939 | CP019159.1 | 2017/1/23 |
| PA06 | ovis | Sheep | lymph nodes | Brazil | 2.32043 | 52.2 | 2131 | 1857 | CP019289.1 | 2017/1/23 |
| CCUG 27541 | equi | Horse | Not specified | Norwegian | 2.37942 | 52.1 | 2195 | 2051 | JPJB00000000.1 | 2014/7/15 |
| XH02 | ovis | Goat | Neck subcutaneous abscess | China | 2.35767 | 52.2 | 2157 | 2017 | MDWN00000000.1 | 2016/9/6 |
| PA07 | ovis | Sheep | Caseous secretion | Brazil | 2.32010 | 52.2 | 2128 | 1846 | MQVG00000000.1 | 2017/1/10 |
| MB295 | equi | Horse | Septic arthritis in hock | California | 2.35525 | 52.1 | 2175 | 1843 | MCNY00000000.1 | 2017/1/23 |
| MB336 | equi | Horse | Pectoral abscess | California | 2.36371 | 52.1 | 2195 | 1932 | MCNW00000000.1 | 2017/1/23 |
| MB302 | equi | Horse | Liver abscess | California | 2.36851 | 52.1 | 2189 | 1977 | CP021982.1 | 2017/1/23 |
| MB122 | equi | Horse | Liver abscess | California | 2.37468 | 52.1 | 2144 | 1779 | MCOB01000001.1 | 2017/1/23 |
| MB154 | equi | Horse | Pectoral abscess | California | 2.36317 | 52.1 | 2135 | 1766 | MCOA01000001.1 | 2017/1/23 |
| MB278 | equi | Horse | Renal abscess | California | 2.37002 | 52.1 | 2135 | 1759 | MCNZ01000001.1 | 2017/1/23 |
| MB44 | equi | Horse | Pectoral abscess | California | 2.36223 | 52.1 | 2125 | 1696 | MCOC00000000.1 | 2017/1/23 |
表选项
在已提交的72株Cp全基因组中,羊型有37株,马型35株。感染羊的几乎为羊型菌株(除PA02株为马型)(表 1)。目前尚未检测到羊型和马型菌株在结构特征之间的明显差异,它们在编码区(CDS)、基因和蛋白质的数量上非常相似(表 1)。有****认为Cp不同生物型致病性的差异及感染宿主偏向性可能与其存在特异性表达基因和其他未知过程有关[3]。Oliveira等[14]利用进化上保守基因(rpoB,gapA,fusA和rsmE)及其相应氨基酸序列,分析了18株羊型和马型Cp之间的系统发育关系,认为羊型Cp是由马型缓慢进化而来的。不过,这一进化假说是否成立还需要通过对其他基因和菌株作进一步的分析。虽然感染山羊和绵羊的Cp多为羊型,然而针对绵羊设计的Cp疫苗在山羊中的效果并不是很好。而在不同种生物型之间,针对羊型Cp开发的疫苗对感染了马型Cp的宿主可能不具有同等预防效果,这进一步增加了不同动物育种者对Cp相关疾病治疗的复杂程度[15]。因此,找出羊型与马型菌株之间的差异,开发一种对它们都适用的常规疫苗或诊断方法极为重要。
1.2 比较基因组学 随着大量细菌基因组序列的获得,对相关菌株基因结构之间的比较[16-17]、细菌多样性和进化动力学分析[18-19]成为人们研究的重点。Viana等[8]对引起埃及水牛爆发水肿性皮肤病(OSD)的11株Cp进行比较基因组分析,发现所有感染水牛的分离株中均有16个致病岛/毒力岛(pathogenicity islands,PiCp),其中PiCp12插入了棒状杆菌噬菌体和白喉毒素基因,这可能在Cp适应新宿主水牛中发挥作用。进一步分析显示同一次爆发期间,棒状杆菌噬菌体插入部位的变化。Barauna等[7]对分离自马的7株Cp进行比较基因组分析,表明马型Cp遗传变异性很高。Cp抗性岛(resistance islands)相似性分析确定了导致更严重感染(内脏感染)的菌株之间接近程度(proximity)较高,而抗性岛的高变异性提示Cp基因获取是通过基因水平转移事件实现的。各菌株之间的致病岛大部分保守,Cp致病性相关的基因包括肽酶、重组酶、微球蛋白合成酶,具有抗微生物活性的细菌素等。在引起3种不同类型感染(形成外部脓肿、内部器官脓肿和溃疡性淋巴管炎)的菌株之间没有基因型差异。
笔者对目前18个国家所测的首株Cp基因组进行比较基因组学分析,利用CMG-Biotools构建蛋白质家族比对矩阵(图 2),发现来自苏格兰的Cp3/99-5与来自法国的CpFRC41蛋白质同源性高达98.3%,而来自中国的CpXH02与以上约2/3国家测序的菌株蛋白质同源性达94%以上,其中与以色列Cp I19相似比最高,达97.7%,而与埃及测序的Cp31最低只有82.5%。对这18株Cp全基因组做SNP进化分析(图 3),清楚地看到两种生物型的Cp分界清晰。两种生物型之间的进化关系、确定SNP在基因组上的位置及其导致了蛋白质发生什么变化这是我们今后研究的重点方向。
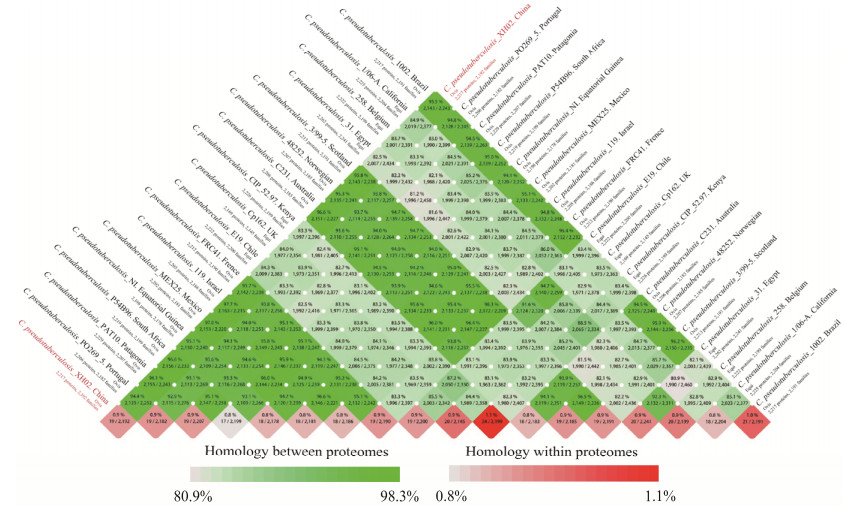 |
| 图 2 18株伪结核棒状杆菌全基因组蛋白质家族比较研究 Figure 2 Genome-wide comparative study of 18 C. pseudotuberculosis strains protein families (made by author). |
| 图选项 |
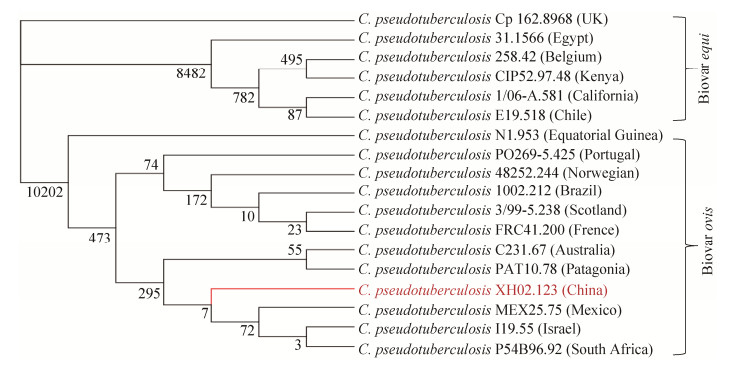 |
| 图 3 18株伪结核棒状杆菌全基因组水平SNP进化分析 Figure 3 Genome-wide SNP analysis of 18 C. pseudotuberculosis strains (made by author). |
| 图选项 |
2 蛋白质组学 2.1 伪结核棒状杆菌的蛋白质组学 蛋白质组学是用来评估细胞、组织或生物体的蛋白质表达谱,研究蛋白质合成、翻译后修饰以及蛋白质之间的相互作用[20]。近年来,蛋白质组学技术被广泛用于研究病原微生物的耐药性、不同生物学特性病原的蛋白表达差异、病原感染与宿主蛋白互作以及病原菌的分泌蛋白质组/胞外蛋白质组等方面,对揭示病原分子结构、致病机制及疫苗抗原筛选等具有重要的指导意义[21-22]。
关于Cp蛋白质研究最初见于Hodgson等[23]的报道,他们为了开发出新的针对羊CLA的免疫诊断方法和疫苗,使用SDS-PAGE和免疫印迹法研究Cp产生的外毒素磷脂酶D(phospholipase D,PLD),报道了PLD基因的克隆、表达和完整的核苷酸序列。然而,随着蛋白质组学研究的深入,发现细菌生长培养基中含有外源蛋白,对所提取的细菌胞外蛋白质产生污染。在这种情况下,培养Cp的限定化学成分培养基(CDM)被开发出来,研究表明使用CDM来鉴定细菌成分具有非常好的应用效果[24]。将Cp在CDM中培养后发现,使用外分泌抗原检测其分泌物中干扰素γ(IFN-γ)的特异性要优于使用全菌粗制抗原,说明使用CDM和分泌蛋白组分进行细菌培养可能是研究Cp免疫原蛋白的有效方法[25]。
蛋白质组学研究的关键是高质量蛋白的提取及蛋白质组学检测方法的合理应用。为了优化提取Cp胞外蛋白质的方法,Paule及其同事[26]制定了一种基于三相分离技术(TPP)的胞外蛋白质提取方案。通过SDS-PAGE和免疫印迹技术分析蛋白提取物,检测出未发现的两种免疫原性分泌蛋白(16 kDa和125 kDa)。然而这些结果仅证明了蛋白质的分子量或蛋白质对受感染动物血清的反应性,而没有质谱(MS)表征。Pacheco等[27]使用2D凝胶电泳来分析菌株的细胞外蛋白,他们利用胰蛋白酶消化,结合MALDI-TOF/TOF分析,并用软件MASCOT?鉴定,获得之前采用TPP-LC/MS方法不能鉴定出的11种新的菌株特异性蛋白,这些新鉴定的蛋白很可能在Cp的生理和毒力方面起重要作用。不过该方法仅鉴定出45种Cp胞外蛋白,而TPP-LC/MS法共检测出93种,在数量上占绝对优势。之后,Silva等[28]对这两种方法进行组合研究,发现这两种技术的组合可以互补,而且将测定的胞外蛋白质数量从93增加到104,针对该结果差异,他们认为可能与分离方法相关,包括用于样品溶解的缓冲液以及每种蛋白质理化性质的不同。
2.2 比较蛋白质组学 比较蛋白质组学是表征细菌蛋白质组很有效的方法。分离自不同宿主(山羊和绵羊)的2个羊型菌株Cp1002和Cp C231是大多数****进行Cp比较蛋白组学研究的对象[29],发现这两种菌株的胞外蛋白质组之间存在定量和定性的变化。共鉴定出93个与Cp的生理学和毒性相关胞外蛋白。包括:Cp的主要毒性因子磷脂酶D(PLD)(与细菌在宿主中的传播有关)、铁载体结合蛋白(FagD)(铁摄取系统的一个组成部分)和丝氨酸蛋白酶(CP40)(对Cp感染起保护作用)等。然而,这些蛋白质仅在Cp C231的胞外蛋白质组中鉴定出来,关于Cp1002菌株是否不会分泌这些蛋白质,以及这些蛋白的缺失是如何影响菌株的致病能力这些方面还尚待研究[27]。
另一种用于分析Cp胞外蛋白质组的方法是血清蛋白质组学分析(SERPA)技术,Seyffert等[30]利用优化后的SERPA技术在Cp1002和Cp C231的培养上清液中共鉴定出13种共同的新型免疫反应性外蛋白(immune-reactive exoproteins)。其中4种,包括神经氨酸酶H (NanH)、细胞表面血红素受体A (HtaA)、海藻糖丙酮转移酶B (CmtB)和海藻糖丙酮转移酶(CmtC),已经在多种细菌病原中鉴定,可作为研发动物疫病防控的广谱疫苗靶标,而另外9种蛋白的功能尚不清楚,且与其他细菌的蛋白质相似度较低,由于这些蛋白可能与CLA诊断不产生交叉反应,因此有希望用于CLA的免疫预防。
最近,Silva等[31]利用Cp1002侵染小鼠,同时对小鼠感染时及恢复后的蛋白组学特征进行比较,发现最初用于实验感染的Cp1002显示低毒力潜力。然而,从感染小鼠脾脏中回收并用于新感染攻击菌株的毒力潜力发生了剧烈变化。在小鼠传代前后的菌株培养物上清液中无标记蛋白质组学分析显示了118个蛋白质差异表达。回收菌株的蛋白质组中包含重要的毒力因子,如CP40蛋白酶和磷脂酶D外毒素等。此外,恢复状态的蛋白质组分析发现了一些涉及解毒过程(detoxification processes)及发病机理等类别的蛋白质,这将有助于我们对该病原体感染过程的毒力因素和不同机制的了解。
3 转录组学 转录组学是一种用于了解生物体内基因结构和RNA调控的有力工具。转录内容、操纵子成员、转录起始位点测定、反义RNA和mRNA丰度都是转录组学研究的重要方面,这些都可以为研究基因功能和调控提供有价值的信息[32-33]。
与结核分枝杆菌相类似,Cp虽不能阻止吞噬体和溶酶体之间的融合,但其可以感染并存在于巨噬细胞中。因为这种细菌在异噬溶酶体中受到不同的胁迫,Pinto及其同事[34]模拟细菌感染宿主环境,将该种细菌在体外条件下暴露于高渗透压(NaCl浓度为2 mol/L)、热休克(50 ℃)或酸性pH值(5.0)等条件,测序发现了474个差异表达基因。在所有模拟条件下改变最典型的生物过程是粘附、应激反应和氧化还原的过程。在这些过程中,涉及生物毒力的基因受到了影响,尚需要进行更详细的研究,以确定它们在细胞中,特别是宿主内部发挥的作用。此外,这些基因的鉴定可能有助于开发更有效的疫苗、诊断试剂盒和CLA治疗方法。此外他们还推测ncRNA与调节有害环境维持自身有关,这还需要进一步研究确证ncRNA在Cp逃避宿主免疫系统中的作用,以及如何帮助其在恶劣环境中存活。
由于细菌等原核生物中含有大量的核糖体RNA基因,因此有效去除核糖体RNA、获得高质量的mRNA是转录组学研究的关键。Castro[35]等以马型Cp为模式菌种,以变性高效液相色谱法为依据,发明了一种可降解78%–92% rRNA的新方法,其效果类似于使用常规降解试剂盒,但减轻了经济负担。另外,由于测序平台产生的数据会显示出质量变化,所以通常使用质量过滤器去除可能含有错误的序列。de Sá等[36]分析了质量过滤值对Cp RNA-Seq数据的影响,认为过滤器参数应根据测序平台产生读数的质量而改变。不过如何定义最佳质量过滤值,无疑是一项复杂的任务。
4 展望 随着新的实验方法、算法和工具的开发以及生物信息学和计算机生物学的进步,对原核生物基因组学、转录组学和蛋白质组学的研究已经得到很大的提高。目前已经完成全基因组测序的72株CP菌株来自于多种宿主,因此有必要利用生物信息学知识进一步深入挖掘这些不同宿主Cp菌株基因组结构特点、进化机制和遗传多样性,促进对Cp基因的重排、缺失、增加和反转等基因组可塑性的理解,进而提供Cp的分子流行病学,种内进化、谱系特有基因以及分离菌株中常见基因等有价值信息[5],有助于开发新的诊断与治疗方法。
此外,基因组学、蛋白质组学和转录组学等组学技术对Cp的研究不是独立的,综合不同的技术成果是阐明Cp的生理、致病力和毒力机制并最终找出有效诊治方法的关键。基因组学研究使我们获得了各种关于Cp的基因组测序信息,从基因组数据中可以推断出Cp的外分泌蛋白质组[29]。然而,关于生物体蛋白是如何相互作用以及它们的生理功能尚未被阐明,这就需要不同领域的研究人员通过相互合作整合资源来完成。与此同时,转录组学也有其自身的局限性,如转录丰度与基因的转录产物含量或者其活性之间没有必然的关联,因此通过与蛋白质组学和代谢组学等其他分子生物学技术以及一些表观的抑菌研究结果相结合可以更好地阐明抗菌物质的抑菌机制,为抗菌物质的进一步应用奠定坚实的理论基础。相信在不久的将来,细菌全转录组学极有可能为有效预防措施的发展提供关键信息,不过研究人员应该先通过整合来自转录组学和蛋白质组学方法得到的有效信息,这样以便于先行一步推动该方面研究的发展。
References
| [1] | Muckle CA, Gyles CL. Characterization of strains of Corynebacterium pseudotuberculosis. Canadian Journal of Comparative Medicine, 1982, 46(2): 206-208. |
| [2] | Peel MM, Palmer GG, Stacpoole AM, Kerr TG. Human lymphadenitis due to Corynebacterium pseudotuberculosis:report of ten cases from Australia and review. Clinical Infectious Diseases, 1997, 24(2): 185-191. DOI:10.1093/clinids/24.2.185 |
| [3] | Dorella FA, Pacheco LGC, Oliveira SC, Miyoshi A, Azevedo V. Corynebacterium pseudotuberculosis:microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Veterinary Research, 2006, 37(2): 201-218. DOI:10.1051/vetres:2005056 |
| [4] | Latif NAA, Abdullah FFJ, Othman AM, Rina A, Chung ELT, Zamri-Saad M, Saharee AA, Haron AW, Lila MAM. Isolation and detection of Corynebacterium pseudotuberculosis in the reproductive organs and associated lymph nodes of non-pregnant does experimentally inoculated through intradermal route in chronic form. Veterinary World, 2015, 8(7): 924-927. DOI:10.14202/vetworld. |
| [5] | Latif NAA, Abba Y, Jesse FFA, Chung ELT, Zamri-Saad M, Saharee AA, Zakaria Z, Haron AW, Mohd-Lila MA. Histopathological assessment of chronic Corynebacterium pseudotuberculosis infection in the reproductive tract and iliac lymph node of Katjang does. Comparative Clinical Pathology, 2017, 26(1): 147-154. DOI:10.1007/s00580-016-2357-3 |
| [6] | Hino C, Crowley D, Chisholm G, Geiger K, Sumida M, Molinder K, Pollock R. Characterizing the murine immune response to Corynebacterium pseudotuberculosis:approaches to developing a C. pseudotuberculosis vaccine (VET1P. 1119). The Journal of Immunology, 2015, 194(S1): 146.7. |
| [7] | Baraúna RA, Ramos RTJ, Veras AAO, Pinheiro KC, Benevides LJ, Viana MVC, Guimar?es LC, Edman JM, Spier SJ, Azevedo V, Silva A. Assessing the genotypic differences between strains of Corynebacterium pseudotuberculosis biovar equi through comparative genomics. PLoS One, 2017, 12(1): e0170676. DOI:10.1371/journal.pone.0170676 |
| [8] | Viana MVC, Figueiredo H, Ramos R, Guimar?es LC, Pereira FL, Dorella FA, Selim SAK, Salaheldean M, Silva A, Wattam AR, Azevedo V. Comparative genomic analysis between Corynebacterium pseudotuberculosis strains isolated from buffalo. PLoS One, 2017, 12(4): e0176347. DOI:10.1371/journal.pone.0176347 |
| [9] | Lima AC, de Moura VA, Pinheiro KD, Paix?o CT, da Costa WL, Folador AR, Guaraldi AL, Ramos RT, Silva A, Marques JM. Draft genome sequence of Corynebacterium pseudotuberculosis strain PA05 isolated from an ovine host in Pará State, Brazil. Genome Announcements, 2017, 5(13): e00082-17. |
| [10] | de Sá PHCG, Veras AAO, Carneiro AR, Barúna RA, Guimar?es LC, Pinheiro KC, Pinto AC, Soares SC, Schneider MPC, Azevedo V, Silva A, Ramos RTJ. Corynebacterium pseudotuberculosis RNA-seq data from abiotic stresses. Data in Brief, 2015, 5: 963-966. DOI:10.1016/j.dib.2015.11.010 |
| [11] | Abdullah FFJ, Latif NAA, Chung ELT, Sarah SA, Zamri-Saad M, Haron AW, Lila MAM, Zakaria Z, Norsidin MJ. Changes in the reproductive hormones of non-pregnant does infected intradermally with Corynebacterium pseudotuberculosis in chronic form. International Journal of Livestock Research, 2015, 5(7): 33-40. DOI:10.5455/ijlr. |
| [12] | Dorella FA, Fachin MS, Billault A, Dias Neto E, Soravito C, Oliveira SC, Meyer R, Miyoshi A, Azevedo V. Construction and partial characterization of a Corynebacterium pseudotuberculosis bacterial artificial chromosome library through genomic survey sequencing. Genetics and Molecular Research, 2006, 5(4): 653-663. |
| [13] | Zhou ZY, Li HX, Zhang MS, Wang ZY, Zhou RQ, Hu SJ, Li XX, Song XY, Zhu Z. Genome sequence of Corynebacterium pseudotuberculosis strain XH02 isolated from a Boer goat in Xuanhan, China. Genome Announcements, 2016, 4(6): e01329-16. |
| [14] | Oliveira A, Teixeira P, Azevedo M, Jamal SB, Tiwari S, Almeida S, Silva A, Barh D, Dorneles EMS, Haas DJ, Heinemann MB, Ghosh P, Lage AP, Figueiredo H, Ferreira RS, Azevedo V. Corynebacterium pseudotuberculosis may be under anagenesis and biovar Equi forms biovar Ovis:a phylogenic inference from sequence and structural analysis. BMC Microbiology, 2016, 16(1): 100. DOI:10.1186/s12866-016-0717-4 |
| [15] | Dorella FA, Pacheco LGC, Seyffert N, Portela RW, Meyer R, Miyoshi A, Azevedo V. Antigens of Corynebacterium pseudotuberculosis and prospects for vaccine development. Expert Review of Vaccines, 2009, 8(2): 205-213. DOI:10.1586/14760584.8.2.205 |
| [16] | Emerson D, Field EK, Chertkov O, Davenport KW, Goodwin L, Munk C, Nolan M, Woyke T. Comparative genomics of freshwater Fe-oxidizing bacteria:implications for physiology, ecology, and systematics. Frontiers in Microbiology, 2013, 4: 254. |
| [17] | Jorda J, Lopez D, Wheatley NM, Yeates TO. Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Science, 2013, 22(2): 179-195. DOI:10.1002/pro.2196 |
| [18] | Lugli GA, Milani C, Turroni F, Duranti S, Ferrario C, Viappiani A, Mancabelli L, Mangifesta M, Taminiau B, Delcenserie V, van Sinderen D, Ventura M. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Applied and Environmental Microbiology, 2014, 80(20): 6383-6394. DOI:10.1128/AEM.02004-14 |
| [19] | Cordero OX, Polz MF. Explaining microbial genomic diversity in light of evolutionary ecology. Nature Reviews Microbiology, 2014, 12(4): 263-273. DOI:10.1038/nrmicro3218 |
| [20] | Gwynne P, Heebner G. Advances in:proteomics. Science, 2005, 308(5722): 707-708, 712, 715. |
| [21] | Bittaye M, Cash P, Forbes K. Proteomic variation and diversity in clinical Streptococcus pneumoniae isolates from invasive and non-invasive sites. PLoS One, 2017, 12(6): e0179075. DOI:10.1371/journal.pone.0179075 |
| [22] | Yimer SA, Birhanu AG, Kalayou S, Riaz T, Zegeye ED, Beyene GT, Holm-Hansen C, Norheim G, Abebe M, Aseffa A, T?njum T. Comparative proteomic analysis of Mycobacterium tuberculosis lineage 7 and lineage 4 strains reveals differentially abundant proteins linked to slow growth and virulence. Frontiers in Microbiology, 2017, 8: 795. DOI:10.3389/fmicb.2017.00795 |
| [23] | Hodgson AL, Bird P, Nisbet IT. Cloning, nucleotide sequence, and expression in Escherichia coli of the phospholipase D gene from Corynebacterium pseudotuberculosis. Journal of Bacteriology, 1990, 172(3): 1256-1261. DOI:10.1128/jb.172.3.1256-1261.1990 |
| [24] | Silva WM, Carvalho RDO, Dorella FA, Folador EL, Souza GHMF, Pimenta AMC, Figueiredo HCP, Le Loir Y, Silva A, Azevedo V. Quantitative proteomic analysis reveals changes in the benchmark Corynebacterium pseudotuberculosis Biovar equi Exoproteome after passage in a murine host. Frontiers in Cellular and Infection Microbiology, 2017, 7: 325. DOI:10.3389/fcimb.2017.00325 |
| [25] | Paule BJ, Azevedo V, Regis LF, Carminati R, Bahia CR, Vale VLC, Moura-Costa LF, Freire SM, Nascimento I, Schaer R, Goes AM, Meyer R. Experimental Corynebacterium pseudotuberculosis primary infection in goats:kinetics of IgG and interferon-γ production, IgG avidity and antigen recognition by Western blotting. Veterinary Immunology and Immunopathology, 2003, 96(3/4): 129-139. |
| [26] | Paule BJA, Meyer R, Moura-Costa LF, Bahia RC, Carminati R, Regis LF, Vale VL, Freire SM, Nascimento I, Schaer R, Azevedo V. Three-phase partitioning as an efficient method for extraction/concentration of immunoreactive excreted-secreted proteins of Corynebacterium pseudotuberculosis. Protein Expression and Purification, 2004, 34(2): 311-316. DOI:10.1016/j.pep.2003.12.003 |
| [27] | Pacheco LGC, Slade SE, Seyffert N, Santos AR, Castro TLP, Silva WM, Santos AV, Santos SG, Farias LM, Carvalho MAR, Pimenta AMC, Meyer R, Silva A, Scrivens JH, Oliveira SC, Miyoshi A, Dowson CG, Azevedo V. A combined approach for comparative exoproteome analysis of Corynebacterium pseudotuberculosis. BMC Microbiology, 2011, 11(1): 12. DOI:10.1186/1471-2180-11-12 |
| [28] | Silva WM, Seyffert N, Santos AV, Castro TLP, Pacheco LGC, Santos AR, Ciprandi A, Dorella FA, Andrade HM, Barh D, Pimenta AMC, Silva A, Miyoshi A, Azevedo V. Identification of 11 new exoproteins in Corynebacterium pseudotuberculosis by comparative analysis of the exoproteome. Microbial Pathogenesis, 2013(61/62): 37-42. |
| [29] | Dorella FA, Gala-Garcia A, Pinto AC, Sarrouh B, Antunes CA, Ribeiro D, Aburjaile FF, Fiaux KK, Guimar?es LC, Seyffert N, El-Aouar RA, Silva R, Hassan SS, Castro TLP, Marques WS, Ramos R, Carneiro A, de Sá P, Miyoshi A, Azevedo V, Silva A. Progression of 'OMICS' methodologies for understanding the pathogenicity of Corynebacterium pseudotuberculosis:the Brazilian experience. Computational and Structural Biotechnology Journal, 2013, 6(7): e201303013. DOI:10.5936/csbj.201303013 |
| [30] | Seyffert N, Silva RF, Jardin J, Silva WM, de Paula Castro TL, Tartaglia NR, de Oliveira Santana KT, Portela RW, Silva A, Miyoshi A, Le Loir Y, Azevedo V. Serological proteome analysis of Corynebacterium pseudotuberculosis isolated from different hosts reveals novel candidates for prophylactics to control caseous lymphadenitis. Veterinary Microbiology, 2014, 174(1/2): 255-260. |
| [31] | Silva WM, Dorella FA, Soares SC, Souza GHMF, Castro TLP, Seyffert N, Figueiredo H, Miyoshi A, Le Loir Y, Silva A, Azevedo V. A shift in the virulence potential of Corynebacterium pseudotuberculosis biovar ovis after passage in a murine host demonstrated through comparative proteomics. BMC Microbiology, 2017, 17(1): 55. DOI:10.1186/s12866-017-0925-6 |
| [32] | Zhang QL, Yuan ML. Progress in insect transcriptomics based on the next-generation sequencing technique. Acta Entomologica Sinica, 2013, 56(12): 1489-1508. |
| [33] | Zhang C, Xu P, Zhu Y. Progress in proteogenomics of prokaryotes. Chinese Journal of Biotechnology, 2014, 30(7): 1026-1035. |
| [34] | Pinto AC, de Sá PHCG, Ramos RTJ, Barbosa S, Barbosa HPM, Ribeiro AC, Silva WM, Rocha FS, Santana MP, de Paula Castro TL, Miyoshi A, Schneider MPC, Silva A, Azevedo V. Differential transcriptional profile of Corynebacterium pseudotuberculosis in response to abiotic stresses. BMC Genomics, 2014, 15(1): 14. DOI:10.1186/1471-2164-15-14 |
| [35] | Castro TLP, Seyffert N, Ramos RTJ, Barbosa S, Carvalho RDO, Pinto AC, Carneiro AR, Silva WM, Pacheco LGC, Downson C, Schneider MPC, Miyoshi A, Azevedo V, Silva A. Ion Torrent-based transcriptional assessment of a Corynebacterium pseudotuberculosis equi strain reveals denaturing high-performance liquid chromatography a promising rRNA depletion method. Microbial Biotechnology, 2013, 6(2): 168-177. DOI:10.1111/1751-7915.12020 |
| [36] | de Sá PHCG, Veras AAO, Carneiro AR, Pinheiro KC, Pinto AC, Soares SC, Schneider MPC, Azevedo V, Silva A, Ramos RTJ. The impact of quality filter for RNA-Seq. Gene, 2015, 563(2): 165-171. DOI:10.1016/j.gene.2015.03.033 |
