曾观娣, 徐倩, 刘婉婷, 阳小燕, 孙雪松


暨南大学生命与健康工程研究院, 功能蛋白质研究广东普通高校重点实验室, 广东 广州 510632
收稿日期:2017-05-31;修回日期:2017-06-29;网络出版日期:2017-07-21
基金项目:国家自然科学基金(21571082);广州市科技计划(201607010228)
*通信作者:孙雪松, Tel:+86-20-85226165;Fax:+86-20-85227039;E-mail:tsunxs@jnu.edu.cn
摘要:[目的]比较临床分离的亲缘关系近的多药耐药鲍曼不动杆菌MDR-ZJ06(blaNDM-1-)和ABC3229(blaNDM-1+)的差异蛋白质组,以期发现新德里金属b-内酰胺酶1(New Delhimetallo-β-lactamase-1,NDM-1)对鲍曼不动杆菌生长代谢的影响。[方法]利用2-DE联合MALDI-TOF MS/MS技术鉴定差异表达蛋白,并在GO注析的基础上,对差异蛋白进行通路分析、功能分类和富集分析,并作出蛋白与蛋白相互作用网络。[结果]发现ABC3299相对于MDR-ZJ06有51个差异表达蛋白,其中11个蛋白表达上调,40个蛋白表达下调,并且这些差异蛋白主要涉及降低碳代谢、氨基酸代谢、脂肪酸代谢和细胞壁合成,增加铁离子转运系统形成。[结论]这个结果揭示了NDM-1可能是通过减缓细菌自身的代谢,增加自身铁的摄取使细菌机体系统地抵抗抗生素从而达到耐药。
关键词: 鲍曼不动杆菌 NDM-1 多药耐药 差异蛋白质组学
Comparative proteomics reveals the role of NDM-1 in Acinetobacter baumannii
Guandi Zeng, Qian Xu, Wanting Liu, Xiaoyan Yang, Xuesong Sun


Key Laboratory of Functional Protein Research of Guangdong Higher Education Institutes, Institute of Life and Health Engineering, College of Life Science and Technology, Jinan University, Guangzhou 510632, Guangdong Province, China
Received 31 May 2017; Revised 29 June 2017; Published online 21 July 2017
*Corresponding author: Xuesong Sun, Tel: +86-20-85226165; Fax: +86-20-85227039; E-mail: tsunxs@jnu.edu.cn
Supported by the National Natural Science Foundation of China (21571082) and by the Guangzhou Science Technology and Innovation Plan (201607010228)
Abstract: [Objective]In order to investigate how NDM-1 affect the metabolism of A. baumannii, we analyzed the differentially expressed proteins (DEPs) between clinically isolated multidrug-resistant A. baumannii MDR-ZJ06 (blaNDM-1-) and ABC3229 (blaNDM-1+) which have a close genetic relationship.[Methods]We used 2-DE coupled with MALDI-TOF MS/MS to analyze the differentially expressed proteins (DEPs) between A. baumannii MDR-ZJ06 and ABC3229. Based on GO annotations, we analyze DEPs by KEGG, Function classification, enrichment analysis and protein-protein interaction network.[Results]We found that there were 51 DEPs, including 11 up-regulated proteins and 41 down-regulated proteins in ABC3229. These proteins were predominantly involved in reducing carbohydrate metabolism, amino acid metabolism, fatty acid metabolism, cell wall synthesis, and increasing of iron transport system formed.[Conclusion]These results uncover the effect of NDM-1 on bacteria may be resistant to antibiotics by slowing down the metabolism of bacteria and increasing the uptake of iron, thereby the bacteria will systemic resistance antibiotics.
Key words: Acinetobacter baumannii NDM-1 multidrug resistance comparative proteomics
鲍曼不动杆菌是在医院流行的需氧革兰氏阴性菌[1]。临床很多感染与鲍曼不动杆菌直接相关,如尿路感染、继发性脑膜炎、创伤或烧伤感染、肺炎等[2-3],David首次发现在濒临灭绝的欧洲水貂身上鲍曼不动杆菌感染也会致命[4]。在20世纪70年代,鲍曼不动杆菌对大多数抗生素都敏感。然而,由于抗生素的滥用,鲍曼不动杆菌已经发展到能抵抗几乎所有常用抗生素[5]。在医院的重症监护病房的病人,多发生多药耐药鲍曼不动杆菌感染或致死[6]。
新德里金属β-内酰胺酶1 (New Delhimetallo-β-lactamase-1,NDM-1)是由blaNDM-1基因编码的产物,它能抵抗几乎所有的抗生素,甚至对公认的最后防线的抗生素——碳青霉素也有抵抗作用,因而增大了治疗的难度[7-8]。近年来,除了美国中部和南部以及南极洲外,blaNDM-1通过基因水平在世界快速地传播[7, 9-10]。此外,据报道在印度的饮用水和污水中也分离出了细菌产出的NDM-1[11]。NDM-1被划分为B类金属-β-内酰胺(Class B metallo-β-lactamase,MBL)[12-13],它能通过切割β-内酰胺的酰胺键从而摧毁β-内酰胺类抗生素,除了单环β-内酰胺抗生素外,它能降解所有的β-内酰胺类抗生素[12]。
blaNDM-1和其他抵抗喹诺酮类、磺胺类、大环内酯类和利福平的耐药基因通常位于同一质粒[9],导致非常复杂的耐药机制。为了单一地研究NDM-1对鲍曼不动杆菌生长代谢的影响,我们选择了两个临床分离的多药耐药的鲍曼不动杆菌[14]:没有携带blaNDM-1的MDR-ZJ06和携带blaNDM-1的ABC3229,利用蛋白质组学的方法研究NDM-1的影响。MLST (Mulitlocus sequence typing)分析显示这2个菌株仅有1个位点的差异,表明他们的亲缘关系非常近。联合2-DE (Two-dimensional gel electrophoresis)和MALDI-TOF MS/MS技术分析2个菌株的差异蛋白质组学。基于GO注释,发现被NDM-1改变的生物通路。本研究是第一次通过蛋白质组学的方法去揭示NDM-1对细菌生长代谢的影响。
1 材料和方法 1.1 菌株来源与主要材料 鲍曼不动杆菌MDR-ZJ06和ABC3229是浙江大学俞云松教授课题组提供的,只有ABC3229携带blaNDM-1基因。LB培养基配制所需的Tryptone、Yeast extract购自OXOID公司。尿素、硫脲、CHAPS、SDS购自Sigma公司。IPG buffer和覆盖液购自Amersham Biosciences。DTT、IAA购自Roche公司。咪唑购自GeneBase公司。
1.2 生长条件 MDR-ZJ06和ABC3229都在LB培养基中,37 ℃、220 r/min过夜培养。准备新鲜的肉汤培养基100 mL以1:100的体积比接种,过夜培养。当培养到稳定期早期(OD600=1.6-1.7)时以6000×g、20 min收菌。
1.3 细菌生长曲线的测定 将过夜活化的细菌A. baumannii AB3229、A. baumannii MDR-ZJ06按照1:100接种于100 mL的LB培养基中,37 ℃、240 r/min培养,每隔1-2 h测定1次600 nm处的吸光度,作好记录,用Originpro 8.0绘制生长曲线。
1.4 蛋白提取 用1×PBS缓冲液将收集到的细菌清洗2遍,最后用1-2 mL的溶解缓冲液[7 mol/L尿素,2 mol/L硫脲,4% CHAPS,15 mmol/L Tris-base,蛋白酶抑制剂(Roche,USA),13 mmol/L DTT (Abcam)]将细菌重悬,反复冻融3次,在冰上超声15 min。4 ℃,13200×g离心30 min去除没有破碎的细菌。通过Bradford法测蛋白浓度,并在-80 ℃冰箱中保存,备用。
1.5 双向凝胶电泳(2-DE) 将100 μg的蛋白溶解在水化液(7 mol/L尿素,2 mol/L硫脲,4% CHAPS,20 mmol/L DTT,0.5% IPG pH 4-7,0.002%溴酚蓝)中,使终浓度为250 μL。以1:100 (V/W)加入核酸酶,混匀,在冰上放置1 h,13000 r/min、30 min离心。将处理好的蛋白样品加入清洗干净的胶条槽中,放入pH 4-7 IPG胶条,吸胀30 min。加入1 mL覆盖油,盖槽盖。设置等电聚胶程序并电泳30 V,12 h;500 V,1 h;1000 V,1 h;8000 V,4 h;2000 V,10 h。取出胶条,用电泳缓冲液清洗,放入平衡液A (50 mmol/L Tris-HCl pH 8.8,6 mol/L尿素,30%甘油,2% SDS,0.002%溴酚蓝储液,0.001% DTT)处理胶条15 min,再用平衡液B (50 mmol/L Tris-HCl pH 8.8,6 mol/L尿素,30%甘油,2% SDS,0.002%溴酚蓝储液,0.0025% IAA)平衡15 min。将胶条置于12.5%聚丙烯酰胺凝胶上,并用0.5%低熔点琼脂糖封闭。按每块胶15 mA进行电泳,30 min后再将电流设置为每块胶30 mA并跑到最后。电泳结束后用与质谱兼容的银染溶液对凝胶进行银染[15]。
1.6 图片的获取和2-DE的分析 用Image Scanner (GE)扫描银染的胶,输出tif格式的图片,并用ImageMaster 2D Platinum 6.0对A. baumannii MDR-ZJ06 (blaNDM-1-)和ABC3229 (blaNDM-1+)进行图像分析,参数设置是同一位置两蛋白点差异在1.5倍或以上,差异蛋白质点由软件自动选择合适的蛋白位点后再由手动过滤,确定两蛋白位点确实存在显著性差异的蛋白质位点(大于1.5倍或以上),才能用于质谱分析。
1.7 胶内酶解 手动挖取显著性差异蛋白质位点,并用胶内酶解的方法将胶内的蛋白切为短肽[16]。用质谱级胰蛋白酶(Promega)消化蛋白[17]。消化后,离心取上清到干净的新的离心管中,并在原离心管加入30 μL萃取液(67%乙腈,2.5% TFA),在4 ℃中超声30 min,离心将上清转移到新的离心管中,并用真空泵干燥。
1.8 质谱分析和搜库鉴定 用ABI 4800 plus MALDI TOF/TOF质谱仪(Applied Biosystems, Foster City,CA)分析样品得到肽段信息。将肽段信息通过MASCOT引擎与鲍曼不动杆菌MDR-ZJ06的蛋白数据库进行搜索。肽段的搜库标准是:最多漏掉2个切割位点,可变修饰为半胱氨酸甲酰胺甲基化和甲硫氨酸氧化,无固定修饰。一级质谱的误差为0.01%,二级质谱的误差为0.2 Da。Protein Score C. I. %大于95%,则认为鉴定结果是可信的。肽段鉴定的假阳性率(False discovery rate,FDR)低于1%。
1.9 差异蛋白功能分类、富集分析和相互作用网络构建 基于GO注释,对差异蛋白进行通路分析、功能分类,并编写R语言程序对差异蛋白进行富集分析,最后利用STRING进行构建差异蛋白相互作用的网络[16](http://string-db.org/)。参数设置如下:生物体(Organism):鲍曼不动杆菌19606,可信度阈值(Confidence threshold):0.70,不超10个关联度[18]。Cytoscape网络图代表着分子间的相互作用。
2 结果和分析 2.1 NDM-1对生长无影响 首先,利用紫外分光光度计测定了A. baumannii AB3229和A. baumannii MDR-ZJ06的生长曲线。从图 1看到,两株菌的生长曲线基本重合,无明显差异,表明NDM-1对鲍曼不动杆菌的生长无影响。
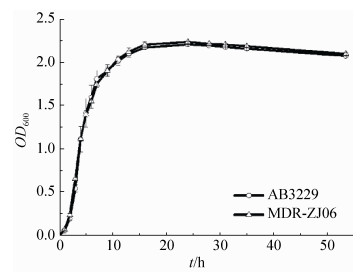 |
| 图 1 A. baumannii AB3229、A. baumannii MDR-ZJ06生长曲线 Figure 1 The growth curves of A. baumannii AB3229 and MDR-ZJ06. |
| 图选项 |
2.2 NDM-1主要下调差异蛋白的表达 为了揭示NDM-1对鲍曼不动杆菌的影响,我们比较分析了临床分离的亲缘关系非常近的鲍曼不动杆菌ABC3229和MDR-ZJ06的蛋白质组学。鲍曼不动杆菌ABC3229包含的blaNDM-1基因产物能抵抗庆大霉素、丁胺卡那霉素、环丙沙星、米诺环素和替加环素[14]。为了弄清楚NDM-1是如何引起细胞代谢的改变,分别提取两株鲍曼不动杆菌的蛋白质,并用双向凝胶电泳对蛋白进行分离。
双向凝胶电泳的结果(图 2)显示这2个菌株的电泳图谱非常相似,进一步说明这两株菌株是高度同源的菌株。蛋白点多集中于pH 5-6。通过Progenesis SameSpots分析,发现相对于鲍曼不动杆菌MDR-ZJ06而言,ABC3229有51个显著性差异的蛋白位点,其中11个上调表达,40个下调表达(表 1),结果表明NDM-1主要降低蛋白的表达,但是铁转运系统的铁蛋白受体(Putative ferric siderophore receptor protein)却上调表达(3.9倍)。
 |
| 图 2 双向电泳图 Figure 2 The 2-DE profiles of whole cell proteins extracted from A. baumannii ABC3229 (A) and A. baumannii MDR-ZJ06 (B). |
| 图选项 |
表 1. 质谱鉴定到的A. baumannii AB3229相对于A. baumannii MDR-ZJ06差异表达蛋白功能分类 Table 1. The detailed information of DEPs identified by MODI-TOF MS/MS
| Spot No.a | Accession No.b | Protein name | Peptidesc | Protein MWd | Protein pIe | F.D.f | Protein score C. I./%h |
| Carbohydrate metabolism | |||||||
| 2 | gb|AEP05957.1 | NAD-dependent aldehyde dehydrogenase | 7 | 76692.2 | 5.94 | -2.2 | 100 |
| 5 | gb|AEP06361.1 | Acetoin: 26-dichlorophenolindophenol oxidoreductase beta subunit | 19 | 41096.1 | 5.90 | -2.4 | 100 |
| 8 | gb|AEP04952.1 | Formyltetrahydrofolate deformylase | 12 | 32673.8 | 5.74 | -1.6 | 100 |
| 10 | gb|AEP05548.1 | Carbonic anhydrase | 12 | 22128.2 | 5.61 | -1.5 | 100 |
| 24 | gb|AEP04987.1 | Bifunctional 4-hydroxy-2-oxoglutarate aldolase/2-dehydro-3-deoxyphosphogluconate aldolase | 8 | 21545.4 | 5.79 | -1.6 | 100 |
| 40 | gb|AEP07193.1 | Putative bifunctional protein (MaeB) | 14 | 82760.9 | 5.52 | -1.5 | 100 |
| 42 | gb|AEP06186.1 | S-adenosyl methionine synthetase | 13 | 41948.2 | 5.44 | -1.5 | 100 |
| 43 | gb|AEP04541.1 | Putative polysaccharide biosynthesis protein | 29 | 40413.1 | 5.46 | -2.0 | 100 |
| 49 | gb|AEP07159.1 | Serine hydroxymethyltransferase | 7 | 44966.7 | 5.44 | +1.9 | 99.463 |
| 51 | gb|AEP07395.1 | Isocitrate dehydrogenase | 9 | 82586.9 | 5.63 | -2.0 | 99.995 |
| Amino acid metabolism | |||||||
| 2 | gb|AEP05957.1 | NAD-dependent aldehyde dehydrogenase | 7 | 76692.2 | 5.94 | +2.2 | 100 |
| 5 | gb|AEP06361.1 | Acetoin:26-dichlorophenolindophenol oxidoreductase beta subunit | 19 | 41096.1 | 5.90 | -2.4 | 100 |
| 3 | gb|AEP08253.1 | Urocanate hydratase | 28 | 61163.7 | 5.64 | -2.1 | 100 |
| 19 | gb|AEP07680.1 | Putative intracellular protease/amidase | 4 | 20995.6 | 5.61 | -1.6 | 99.781 |
| 24 | gb|AEP04987.1 | Bifunctional 4-hydroxy-2-oxoglutarate aldolase/2-dehydro-3-deoxyphosphogluconate aldolase | 8 | 21545.4 | 5.79 | -1.6 | 100 |
| 30 | gb|AEP07715.1 | Gcv-like aminomethyltransferase | 6 | 26768.6 | 5.49 | +2.2 | 100 |
| 33 | gb|AEP04591.1 | Methylmalonate-semialdehydedehydrogenase, oxidoreductase protein | 12 | 55046.3 | 5.40 | -2.3 | 100 |
| 36 | gb|AEP04808.1 | 3-ketoacyl-CoA thiolase | 8 | 41061.9 | 6.09 | +1.8 | 99.993 |
| 42 | gb|AEP06186.1 | S-adenosylmethionine synthetase | 13 | 41948.2 | 5.44 | -1.5 | 100 |
| 48 | gb|AEP05897.1 | Threonine dehydrogenase | 8 | 41812.5 | 5.44 | -1.7 | 100 |
| 49 | gb|AEP07159.1 | Serine hydroxymethyltransferase | 7 | 44966.7 | 5.44 | +1.9 | 99.463 |
| Fatty acid metabolism | |||||||
| 2 | gb|AEP05957.1 | NAD-dependent aldehyde dehydrogenase | 7 | 76692.2 | 5.94 | +2.2 | 100 |
| 7 | gb|AEP07759.1 | Acetyl CoA carboxylase, beta subunit | 18 | 32950.6 | 5.85 | -2.2 | 100 |
| 18 | gb|AEP05106.1 | Acetyl CoA carboxylase alpha subunit | 14 | 29622.0 | 5.60 | -2.3 | 100 |
| 29 | gb|AEP05034.1 | NADH-dependent enoyl-ACP reductase | 17 | 30996.9 | 6.00 | -1.9 | 100 |
| 36 | gb|AEP04808.1 | 3-ketoacyl-CoA thiolase | 8 | 41061.9 | 6.09 | +1.8 | 99.993 |
| 50 | gb|AEP07777.1 | Acyl-CoA dehydrogenase | 18 | 65580.9 | 5.42 | +1.7 | 100 |
| 38 | gb|AEP05798.1 | Alkyl hydroperoxide reductase subunit, FAD/NAD(P)-binding, detoxification of hydroperoxides | 6 | 57261.3 | 4.98 | -1.6 | 99.995 |
| Nucleic acid metabolism | |||||||
| 8 | gb|AEP04952.1 | Formyltetrahydrofolate deformylase | 12 | 32673.8 | 5.74 | -1.6 | 100 |
| 11 | gb|AEP08190.1 | Orotate phosphoribosyltransferase | 10 | 24057.6 | 5.59 | -1.9 | 100 |
| 12 | gb|AEP07873.1 | Xanthine phosphoribosyltransferase | 6 | 20806.2 | 5.83 | -1.9 | 100 |
| 16 | gb|AEP04475.1 | Hypoxanthine phosphoribosyltransferase | 13 | 19576.1 | 4.93 | -1.5 | 100 |
| Translation | |||||||
| 15 | gb|AEP07282.1 | Elongation factor P | 3 | 21205.5 | 4.88 | +2.0 | 99.868 |
| Transcription | |||||||
| 21 | gb|AEP04730.1 | Regulatory protein for nitrogen assimilation by glutamine synthetase, regulates GlnL (NRII) and GlnE | 4 | 12195.6 | 5.41 | +5.9 | 99.966 |
| 25 | gb|AEP05556.1 | Esterase operon transcriptional regulator | 20 | 33751.9 | 5.44 | -1.5 | 100 |
| Iron transport | |||||||
| 1 | gb|AEP04976.1 | Outer membrane receptor for monomeric catechols | 10 | 81197.3 | 5.71 | +2.1 | 100 |
| 28 | gb|AEP08077.1 | Osmolarity response regulator | 23 | 28797.2 | 5.91 | -1.7 | 100 |
| 22 | gb|AEP04581.1 | Putative ferric siderophore receptor protein | 26 | 79483.2 | 5.70 | +3.9 | 100 |
| Cell wall synthesis process | |||||||
| 4 | gb|AEP04540.1 | Putative UDP-N-acetylglucosamine 2-epimerase | 6 | 44276.0 | 5.79 | -2.4 | 99.999 |
| 31 | gb|AEP04542.1 | Putative polysaccharide biosynthesis protein | 17 | 40316.9 | 5.56 | -1.9 | 100 |
| Vitamin synthesis process | |||||||
| 6 | gb|AEP06176.1 | Biotin synthetase | 13 | 37113.5 | 5.46 | -1.9 | 100 |
| 17 | gb|AEP07119.1 | Thiamine biosynthesis protein, thiazole moiety | 6 | 29636.3 | 5.09 | -1.6 | 98.652 |
| Others | |||||||
| 9 | gb|AEP06368.1 | Zn-dependent hydrolase, including glyoxylase | 9 | 26025.7 | 5.20 | -4.0 | 100 |
| 13 | gb|AEP06219.1 | Glucose-inhibited division protein B (methyltransferase) | 8 | 23704.6 | 5.76 | -1.7 | 100 |
| 14 | gb|AEP04726.1 | Conserve hypothetical protein | 6 | 31192.6 | 4.83 | -1.7 | 99.149 |
| 20 | gb|AEP06847.1 | Conserve hypothetical protein | 10 | 37380.5 | 5.49 | -1.6 | 99.996 |
| 23 | gb|AEP04978.1 | ATP-dependent Clp protease proteolytic subunit | 16 | 22525.3 | 5.26 | -1.4 | 100 |
| 26 | gb|AEP06438.1 | Zn-dependent alcohol dehydrogenase, class Ⅲ | 5 | 39225.7 | 5.64 | -1.6 | 99.852 |
| 27 | gb|AEP07535.1 | Nitroreductase | 19 | 22825.7 | 5.13 | -1.6 | 100 |
| 32 | gb|AEP08348.1 | Gentamicin 3′-acetyltransferase (gentamicin acetyltransferase Ⅰ) (aminoglycoside N(3′)-acetyltransferase Ⅰ) (AAC (3)-Ⅰ) | 15 | 19375.9 | 5.78 | -4.6 | 100 |
| 34 | gb|AEP08000.1 | Conserve hypothetical protein | 11 | 18057.1 | 6.06 | -1.4 | 100 |
| 35 | gb|AEP07457.1 | Putative porin protein associated with imipenem resistance | 11 | 26489.1 | 4.80 | -1.9 | 100 |
| 37 | gb|AEP05088.1 | Acyl-CoA synthetase (AMP-forming)/AMP-acid ligase Ⅱ | 10 | 60187.5 | 5.39 | -2.6 | 100 |
| 38 | gb|AEP05798.1 | Alkyl hydroperoxide reductase subunit, FAD/NAD(P)-binding, detoxification of hydroperoxides | 6 | 57261.3 | 4.98 | -1.6 | 99.995 |
| 39 | gb|AEP06215.1 | Parvulin-like peptidyl-prolylisomerase | 18 | 48998.5 | 6.35 | -1.6 | 100 |
| 41 | gb|AEP07733.1 | Outer membrane protein | 15 | 38426.5 | 5.32 | +1.7 | 100 |
| 44 | gb|AEP06279.1 | Scaffold protein | 4 | 13773.0 | 5.38 | -1.8 | 100 |
| 45 | gb|AEP07759.1 | Oxidoreductase | 10 | 39354.9 | 5.83 | -1.8 | 100 |
| 46 | gb|AEP08000.1 | Rossman fold nucleotide-binding protein | 7 | 20885.6 | 5.15 | -1.8 | 100 |
| 47 | gb|AEP06956.1 | Putative kinase | 22 | 49626.5 | 5.60 | +1.7 | 100 |
| a: The spot of 2-DE. b: The number of protein in NCBI. c: The number of peptide. d: The molecular weight of protein. e: The isoelectric point of protein. f: The fold is the ratio of protein expression levels in A. baumannii ABC3229 to A. baumannii MDR-ZJ06. h: The confidence level of peptides matching to protein. | |||||||
表选项
2.3 NDM-1降低了细胞代谢 富集分析(图 3)富集了17个蛋白,其中参与到生物学过程中最多的3个蛋白为锌依赖性乙醇脱氢酶(Zn-dependent alcohol dehydrogenase,class Ⅲ)、丝氨酸羟甲基转移酶(Serine hydroxymethyltransferase,SHMT)、酮脂酰辅酶A硫解酶(3-Ketoacyl-CoA thiolase)。锌依赖性乙醇脱氢酶是细胞内主要短链醇代谢的关键酶[19],利用烟酰胺腺嘌呤二核苷酸(NAD)为辅酶催化伯醇与醛的可逆性反应,可有效消除体内外的甲醛,甚至在酵母中,发现乙醇脱氢酶可将糖类转化为酒精。SHMT除了可催化L-丝氨酸和甘氨酸相互转化[20-21],还可以为很多生物合成反应提供一碳单位,更是氨基酸和核酸转化的节点(连接点)。在辅酶A存在下,酮脂酰辅酶A硫解酶可将β酮酰基辅酶A硫解为乙酰辅酶A和酰基辅酶A。三大营养物质糖、脂肪和蛋白质通过乙酰辅酶A汇聚成一条共同的代谢通路——三羧酸循环和氧化磷酸化,经过这条通路彻底氧化生成二氧化碳和水,释放能量用于ATP合成,用于细胞内的生命活动。乙酰辅酶A是合成脂肪酸、酮体等能源物质的前体物质;酰基辅酶A是脂肪酸合成和分解的活性代谢中间物,水解时生成脂肪酸和辅酶A,从而参与到细胞的脂肪酸代谢。
 |
| 图 3 差异蛋白富集分析 Figure 3 Enrichment analysis of DEPs. Rectangle represents the identified protein, lozenge represents the biological process. |
| 图选项 |
富集分析(图 3、表 2)发现只有2个差异蛋白表达量上调,而这些差异蛋白主要参与了鲍曼不动杆菌的碳代谢、氨基酸代谢、脂肪酸代谢和核酸代谢等生物学过程,这说明NDM-1通过下调表达代谢通路的蛋白使细胞代谢减缓。从而推测耐药菌可能是通过减缓机体的代谢,慢慢调控机体对抗抗生素的能力,从而达到耐药。
表 2. 差异蛋白富集分析富集到的蛋白及参与的通路 Table 2. The biological process of DEPs
| Spot No. | Accession No. | Protein name | Biological process | Count | F.D. |
| 26 | gb|AEP06438.1 | Zn-dependent alcohol dehydrogenase, class Ⅲ | Fatty acid degradation, Biosynthesis of antibiotics, Glycolysis/Glyconeogenesis, Carbon metabolism, Biosynthesis of secondary metabolites, Microbial metabolism in diverse environments, Methane metabolism, Tyrosine metabolism, Degradation of aromatic compounds, Naphthalene degradation, Chloroalkane and chloroalkene degradation | 11 | -1.6 |
| 49 | gb|AEP07159.1 | Serine hydroxymethyltransferase | Biosynthesis of antibiotics, Biosynthesis of secondary metabolites, Carbon metabolism, Biosynthesis of antibiotics, Microbial metabolism in diverse environments, Methane metabolism, Cyanoamino acid metabolism, One carbon pool by folate, Glycine, serine and threonine metabolism, Glyoxylate and dicarboxylate metabolism, | 10 | +1.9 |
| 36 | gb|AEP04808.1 | 3-Ketoacyl-CoA thiolase | Fatty acid biosynthesis, Biosynthesis of secondary metabolites, Biosynthesis of antibiotics, Microbial metabolism in diverse environments, Fatty acid degradation, Benzoate degradation, alpha-Linolenic acid metabolism, Geraniol degradation, Valine, leucine and isoleucine degradation | 9 | +1.8 |
| 18 | gb|AEP05106.1 | Acetyl CoA carboxylase alpha subunit | Fatty acid biosynthesis, Fatty acid metabolism, Propanoate metabolism, Pyruvate metabolism, Biosynthesis of antibiotics, Carbon metabolism, Microbial metabolism in diverse environments, Biosynthesis of secondary metabolites | 8 | -2.3 |
| 7 | gb|AEP07759.1 | Acetyl CoA carboxylase, beta subunit | Fatty acid biosynthesis, Fatty acid metabolism, Propanoate metabolism, Pyruvate metabolism, Biosynthesis of antibiotics, Carbon metabolism, Microbial metabolism in diverse environments | 7 | -2.2 |
| 51 | gb|AEP07395.1 | Isocitrate dehydrogenase | Citrate cycle (TCA cycle), Biosynthesis of secondary metabolites, Carbon metabolism, Biosynthesis of antibiotics, Microbial metabolism in diverse environments, Biosynthesis of amino acids, 2-Oxocarbosylic acid metabolism | 7 | -2.0 |
| 5 | gb|AEP06361.1 | Acetoin:26-dichlorophenolindophenol oxidoreductase beta subunit | Pyruvate metabolism, Biosynthesis of antibiotics, Glycolysis/Glyconeogenesis, Microbial metabolism in diverse environments, Carbon metabolism, Biosynthesis of secondary metabolites, Citrate cycle (TCA cycle) | 6 | -2.4 |
| 33 | gb|AEP04591.1 | Methylmalonate-semialdehydedehydrogenase, oxidoreductase protein | Propanoate metabolism, beta-Alanine metabolism, Inositol phosphate metabolism, Valine, leucine and isoleucine degradation, Pyruvate metabolism | 5 | -2.3 |
| 24 | gb|AEP04987.1 | Bifunctional 4-hydroxy-2-Oxoglutarate aldolase/2-dehydro-3-deoxyphosphogluconate aldolase | Carbon metabolism, Microbial metabolism in diverse environments, Glyoxylate and dicarboxylate metabolism, Pentose phosphate pathway | 4 | -1.6 |
| 40 | gb|AEP07193.1 | Putative bifunctional protein (MaeB) | Pyruvate metabolism, Carbon metabolism, Microbial metabolism in diverse environments | 3 | -1.5 |
| 29 | gb|AEP05034.1 | NADH-dependent enoyl-ACP reductase | Biotin metabolism, Fatty acid biosynthesis, Fatty acid metabolism | 3 | -1.9 |
| 42 | gb|AEP06186.1 | S-adenosyl methionine synthetase | Biosynthesis of secondary metabolites, Biosynthesis of amino acids, Cysteine and methionine metabolism | 3 | -1.5 |
| 8 | gb|AEP04952.1 | Formyltetrahydrofolate deformylase | Glyoxylate and dicarboxylate metabolism, One carbon pool by folate | 2 | -1.6 |
| 12 | gb|AEP07873.1 | Xanthine phosphoribosyltransferase | Purine metabolism, Biosynthesis of secondary metabolites | 2 | -1.9 |
| 2 | gb|AEP05957.1 | NAD-dependent aldehyde dehydrogenase | Microbial metabolism in diverse environments, Phenylalanine metabolism | 2 | -2.2 |
| 16 | gb|AEP04475.1 | Hypoxanthine phosphoribosyltransferase | Purine metabolism, Biosynthesis of secondary metabolites | 2 | -1.5 |
| 6 | gb|AEP06176.1 | Biotin synthetase | Biotin metabolism | 1 | -1.9 |
表选项
2.4 差异蛋白相互作用网络 蛋白相互作用网络(图 4)显示除1个较大的网络外,只有小部分的蛋白形成4个小网络。最大的蛋白相互作用网络的节点蛋白HMPREF0010_00676是丙酮酸脱氢酶复合体的丙酮酸脱氢酶(Pyruvate dehydrogenase complex dihydrolipoamide acetyltransferase),可催化丙酮酸氧化脱羧,将乙酰单位转移到辅酶A形成乙酰辅酶A和降低胞内的NAD+[22-23]。HMPREF0010_00308是乙酰辅酶A羧化酶,生物素羧化酶亚基(Acetyl-CoA carboxylase,biotin carboxylase subunit),催化乙酰辅酶A+ATP+HCO3-→丙二酰辅酶A+ADP+Pi反应的生物素酶,也是降低体内酰辅酶A的途径。此外,共同伴侣蛋白HscA和HscB可促进铁硫簇受体蛋白成熟。结果表明HscA和HscB可促进铁硫簇蛋白成熟。
 |
| 图 4 差异蛋白相互作用网络 Figure 4 The protein-protein network of DEPs. |
| 图选项 |
3 讨论 含有blaNDM-1的鲍曼不动杆菌几乎能抵抗所有的抗生素。然而,blaNDM-1编码产物NDM-1如何影响鲍曼不动杆菌的代谢还是未知的。本研究通过全局分析NDM-1诱导细菌蛋白组学的改变以揭示NDM-1引起细菌耐药的原因。
NDM-1虽然没有影响鲍曼不动杆菌的生长表型(图 1),但是质谱的结果显示NDM-1降低了通路蛋白的表达(表 1),从而使含NDM-1的细菌代谢减缓。这些通路蛋白(差异蛋白)主要参与到细菌碳代谢、氨基酸代谢、脂肪酸代谢、核酸代谢等。
碳代谢是细菌生存最主要的产能途径。有10个差异蛋白参与碳代谢途径,并被blaNDM-1调节。三羧酸循环(Tricarboxylic acid cycle)是生物体最重要的能量代谢途径,并为细菌提供NADH和FADH2。在ABC3229中,三羧酸循环的限制酶——异柠檬酸脱氢酶(Isocitrate dehydrogenase)下调表达意味着在NDM-1存在的情况下,导致三羧酸减弱,从而使细菌产能减弱。
除了三羧酸循环外,脂肪酸代谢是生物体内另一个重要的供能途径。鉴定到6个与脂肪酸代谢相关的蛋白,都是α和β乙酰辅酶A的亚基,参与脂肪酸合成的第一步。在NDM-1存在时,鲍曼不动杆菌中这2个蛋白也下调表达。此外,NDM-1的鲍曼不动杆菌的乙酰辅酶A脱氢酶(Acyl-CoA dehydrogenase)高表达,说明其负责的脂肪酸降解会加快。然而,烷基氢过氧化物还原亚基(Alkyl hydroperoxide reductase subunit)除了参与降解体内的脂肪酸,还在防御宿主的氧化应激方面起着至关重要的作用[24]。结果表明NDM-1降低了脂肪酸的合成,增加脂肪酸的降解,减弱了防御宿主的能力。
脂多糖(Lipopolysaccharide,LPS)是细菌毒力因子,是细菌致病的关键[25]。胞外的多糖聚β-(1-6)-N-乙酰氨基葡萄糖(Polysaccharide poly-beta-(1-6)-N-acetylglucosamine)在细菌细胞壁形成过程中扮演着一个重要的角色。在革兰氏阴性和革兰氏阳性细菌中,UDP-N-乙酰葡糖胺2-差向异构酶(UDP-N-acetylglucosamine 2-epimerase)催化UDP-N-acetylglucosamine (UDP-GlcNAc)和UDP-N-acetylmannosamine (UDP-ManNAc)可逆的差向异构化[26-27]。这2个蛋白的活性形式GlcNAc和ManNAc在这个过程中产生,并参与细胞壁表面多糖的形成[27],这些脂多糖保护细菌对抗宿主的免疫系统[28-29]。在鲍曼不动杆菌ABC3229中UDP-GlcNAc和UDP-GlcNAc下调表达,暗示着细胞壁多聚糖合成减少,从而使细胞壁合成也减缓,最终使含NDM-1的鲍曼不动杆菌的毒力也减弱。这也印证了之前研究的结论,MDR菌株相对于MDS菌株毒力受损[30]。
铁是细菌生存和致病所必需的[31-34]。许多病原菌,比如链球菌分泌铁载体去螯合宿主的铁用于自身的生长。细菌表面特异受体吸收铁[35]。也有研究表明,在缺铁的情况下,相对于敏感株,耐多粘菌素的鲍曼不动杆菌生长严重受限[36],加入铁后耐多粘菌素的鲍曼不动杆菌生长得更好,说明铁是耐药菌的生长必不可少的。Luciana F. Costa发现鼠伤寒沙门氏菌通过摄取铁去帮助扩张宿主发炎的肠道,使宿主致病[31]。本研究发现铁载体(Ferric siderophore)在鲍曼不动杆菌中高表达(3.9倍),这些蛋白将会为鲍曼不动杆菌生长和致病募集更多的铁,将为鲍曼不动杆菌致病和抵抗抗生素作出贡献。
本研究发现含NDM-1会减弱细菌代谢和细胞壁的生物合成,从而减弱细菌的毒力。此外,还通过增加铁的摄入会提高细菌抵抗抗生素的能力,但是NDM-1是如何影响铁的摄取机制还是未知的。未来的研究将使用分子方法去揭示铁的摄取与细菌耐药的关系。本研究将会为NDM-1介导的多药耐药细菌抵抗抗生素提供一个更深的认识。
致谢: 感谢浙江大学俞云松教授课题组提供鲍曼不动杆菌MDR-ZJ06和ABC3229菌株。
References
| [1] | Lin MF, Lan CY. Antimicrobial resistance in Acinetobacter baumannii: from bench to bedside. World Journal of Clinical Cases, 2014, 2(12): 787-814. DOI:10.12998/wjcc.v2.i12.787 |
| [2] | Chen MZ, Hsueh PR, Lee LN, Yu CJ, Yang PC, Luh KT. Severe community-acquired pneumonia due to Acinetobacter baumannii. Chest, 2001, 120(4): 1072-1077. DOI:10.1378/chest.120.4.1072 |
| [3] | Davis KA, Moran KA, McAllister CK, Gray PJ. Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerging Infectious Diseases, 2005, 11(8): 1218-1224. DOI:10.3201/1108.050103 |
| [4] | Cano-Terriza D, Guerra R, Mozos E, Rodríguez-Sánchez B, Borge C, García-Bocanegra I. Fatal Acinetobacter baumannii infection in the critically endangered european mink (Mustela Lutreola). Journal of Zoo and Wildlife Medicine: Official Publication of the American Association of Zoo Veterinarians, 2017, 48(1): 220-223. DOI:10.1638/2016-0082.1 |
| [5] | Kuo LC, Teng LJ, Yu CJ, Ho SW, Hsueh PR. Dissemination of a clone of unusual phenotype of pandrug-resistant Acinetobacter baumannii at a university hospital in Taiwan. Journal of Clinical Microbiology, 2004, 42(4): 1759-1763. DOI:10.1128/JCM.42.4.1759-1763.2004 |
| [6] | Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nature Reviews Microbiology, 2007, 5(12): 939-951. DOI:10.1038/nrmicro1789 |
| [7] | Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. The Lancet Infectious Diseases, 2010, 10(9): 597-602. DOI:10.1016/S1473-3099(10)70143-2 |
| [8] | Poirel L, Lagrutta E, Taylor P, Pham J, Nordmann P. Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrobial Agents and Chemotherapy, 2010, 54(11): 4914-4916. DOI:10.1128/AAC.00878-10 |
| [9] | Nordmann P, Poirel L, Walsh TR, Livermore DM. The emerging NDM carbapenemases. Trends in Microbiology, 2011, 19(12): 588-595. DOI:10.1016/j.tim.2011.09.005 |
| [10] | Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrobial Agents and Chemotherapy, 2009, 53(12): 5046-5054. DOI:10.1128/AAC.00774-09 |
| [11] | Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. The Lancet Infectious Diseases, 2011, 11(5): 355-362. DOI:10.1016/S1473-3099(11)70059-7 |
| [12] | Bebrone C. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochemical Pharmacology, 2007, 74(12): 1686-1701. DOI:10.1016/j.bcp.2007.05.021 |
| [13] | Shimada A, Ishikawa H, Nakagawa N, Kuramitsu S, Masui R. The first crystal structure of an archaeal metallo-β-lactamase superfamily protein; ST1585 from Sulfolobus tokodaii. Proteins, 2010, 78(10): 2399-2402. DOI:10.1002/prot.v78:10 |
| [14] | Chen Y, Zhou ZH, Jiang Y, Yu YS. Emergence of NDM-1-producing Acinetobacter baumannii in China. The Journal of Antimicrobial Chemotherapy, 2011, 66(6): 1255-1259. DOI:10.1093/jac/dkr082 |
| [15] | Sun XS, Ge RG, Cai ZW, Sun HZ, He QY. Iron depletion decreases proliferation and induces apoptosis in a human colonic adenocarcinoma cell line, CaCO3. Journal of Inorganic Biochemistry, 2009, 103(7): 1074-1081. DOI:10.1016/j.jinorgbio.2009.05.004 |
| [16] | Sun XS, Yang XY, Yin XF, Yu GC, Xiao CL, He X, He QY. Proteomic analysis of membrane proteins from streptococcus pneumoniae with multiple separation methods plus high accuracy mass spectrometry. Omics A Journal of Integrative Biology, 2011, 15(10): 683-694. DOI:10.1089/omi.2010.0133 |
| [17] | Sun XS, Jia HL, Xiao CL, Yin XF, Yang XY, Lu J, He X, Li N, Li H, He QY. Bacterial proteome of streptococcus pneumoniae through multidimensional separations coupled with LC-MS/MS. Omics: A Journal of Integrative Biology, 2011, 15(7/8): 477-482. |
| [18] | Yang XY, He K, Du GF, Wu XH, Yu GC, Pan YL, Zhang G, Sun XS, He QY. Integrated translatomics with proteomics to identify novel iron-transporting proteins in Streptococcus pneumoniae. Frontiers in Microbiology, 2016, 7: 78. |
| [19] | Gonzalez-Duarte R, Albalat R. Merging protein, gene and genomic data: the evolution of the MDR-ADH family. Heredity, 2005, 95(3): 184-197. DOI:10.1038/sj.hdy.6800723 |
| [20] | Chang WN, Tsai JN, Chen BH, Huang HS, Fu TF. Serine hydroxymethyltransferase isoforms are differentially inhibited by leucovorin: characterization and comparison of recombinant zebrafish serine hydroxymethyltransferases. Drug Metabolism and Disposition, 2007, 35(11): 2127-2137. DOI:10.1124/dmd.107.016840 |
| [21] | Rao NA, Talwar R, Savithri HS. Molecular organization, catalytic mechanism and function of serine hydroxymethyltransferase-a potential target for cancer chemotherapy. The International Journal of Biochemistry & Cell Biology, 2000, 32(4): 405-416. |
| [22] | Guan Y H, Rawsthorne S, Scofield G, Shaw P, Doonan J. Cloning and characterization of a dihydrolipoamide acetyltransferase (E2) subunit of the pyruvate dehydrogenase complex from Arabidopsis thaliana. Journal of Biological Chemistry, 1995, 270(10): 5412-5417. DOI:10.1074/jbc.270.10.5412 |
| [23] | Wang JJ, Nemeria NS, Chandrasekhar K, Kumaran S, Arjunan P, Reynolds S, Calero G, Brukh R, Kakalis L, Furey W, Jordan F. Structure and function of the catalytic domain of the dihydrolipoyl acetyltransferase component in Escherichia coli pyruvate dehydrogenase complex. The Journal of Biological Chemistry, 2014, 289(22): 15215-15230. DOI:10.1074/jbc.M113.544080 |
| [24] | Chang YY, Cheng TF, Yang XM, Jin LJ, Sun HZ, Li HY. Functional disruption of peroxiredoxin by bismuth antiulcer drugs attenuates Helicobacter pylori survival. Journal of Biological Inorganic Chemistry: A Publication of the Society of Biological Inorganic Chemistry, 2017, 22(5): 673-683. |
| [25] | Luke NR, Sauberan SL, Russo TA, Beanan JM, Olson R, Loehfelm TW, Cox AD, Michael FS, Vinogradov EV, Campagnari AA. Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infection and Immunity, 2010, 78(5): 2017-2023. DOI:10.1128/IAI.00016-10 |
| [26] | Kawamura T, Ishimoto N, Ito E. Enzymatic synthesis of uridine diphosphate N-acetyl-D-mannosaminuronic acid. Journal of Biological Chemistry, 1979, 254(17): 8457-8465. |
| [27] | Kawamura T, Kimura M, Yamamori S, Ito E. Enzymatic formation of uridine diphosphate N-acetyl-D-mannosamine. The Journal of Biological Chemistry, 1978, 253(10): 3595-3601. |
| [28] | Lee CJ, Banks SD, Li JP. Virulence, immunity, and vaccine related to Streptococcus pneumoniae. Critical Reviews in Microbiology, 1991, 18(2): 89-114. DOI:10.3109/10408419109113510 |
| [29] | Morona JK, Morona R, Paton JC. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Molecular Microbiology, 1997, 23(4): 751-763. DOI:10.1046/j.1365-2958.1997.2551624.x |
| [30] | Deptu?a A, Gospodarek E. Reduced expression of virulence factors in multidrug-resistant Pseudomonas aeruginosa strains. Archives of Microbiology, 2010, 192(1): 79-84. DOI:10.1007/s00203-009-0528-1 |
| [31] | Costa LF, Mol JPS, Silva APC, Macêdo AA, Silva TMA, Alves GES, Winter S, Winter MG, Velazquez EM, Byndloss MX, B?umler AJ, Tsolis RM, Paix?o TA, Santos RL. Iron acquisition pathways and colonization of the inflamed intestine by Salmonella enterica serovar Typhimurium. International Journal of Medical Microbiology, 2016, 306(8): 604-610. DOI:10.1016/j.ijmm.2016.10.004 |
| [32] | Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics, 2011, 12: 126. DOI:10.1186/1471-2164-12-126 |
| [33] | Lee CR, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha CJ, Jeong BC, Lee SH. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Frontiers in Cellular and Infection Microbiology, 2017, 7: 55. |
| [34] | Pollack JR, Neilands JB. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochemical and Biophysical Research Communications, 1970, 38(5): 989-992. DOI:10.1016/0006-291X(70)90819-3 |
| [35] | Li H, Li N, Xu Q, Xiao CL, Wang HC, Guo Z, Zhang J, Sun XS, He QY. Lipoprotein FtsB in Streptococcus pyogenes binds ferrichrome in two steps with residues Tyr137 and Trp204 as critical ligands. PLoS One, 2013, 8(6): e65682. DOI:10.1371/journal.pone.0065682 |
| [36] | López-Rojas R, García-Quintanilla M, Labrador-Herrera G, Pachón J, McConnell MJ. Impaired growth under iron-limiting conditions associated with the acquisition of colistin resistance in Acinetobacter baumannii. International Journal of Antimicrobial Agents, 2016, 47(6): 473-477. DOI:10.1016/j.ijantimicag.2016.03.010 |
