 ,苏州大学,剑桥-苏大基因组资源中心,苏州 215123
,苏州大学,剑桥-苏大基因组资源中心,苏州 215123MDMPR: Mouse Developmental and Metabolic Phenotype Repository
Wenjing Zhu, Zhiwei Liu ,Cambridge-Suda Genomic Resource Center, Soochow University, Suzhou 215123, China
,Cambridge-Suda Genomic Resource Center, Soochow University, Suzhou 215123, China通讯作者: 刘志玮,博士,研究方向:生物化学与分子生物学。E-mail:liuzhiwei@cam-su.org
编委: 陈帅
收稿日期:2021-01-5修回日期:2021-03-4网络出版日期:2021-04-16
| 基金资助: |
Received:2021-01-5Revised:2021-03-4Online:2021-04-16
| Fund supported: |
作者简介 About authors
朱文静,硕士,实验技术员,研究方向:生物化学与分子生物学。E-mail:

摘要
小鼠发育代谢表型库(Mouse Developmental and Metabolic Phenotype Repository, MDMPR)是一个致力于小鼠资源和表型数据实时共享的开放性平台,它依托于科技部重点研发计划“发育编程及其代谢调节”专项项目“建立小鼠发育代谢表型库”。该项目预计在5年内完成500个发育代谢相关小鼠敲除模型的建立,并对其表型数据进行标准化的解析、建立表型数据库。MDMPR作为一个资源及数据集成的库,由多个子系统作为支撑,包括ES细胞数据库、项目管理系统、繁育管理系统、精子库管理系统、表型分析系统,信息化管理深入到项目中每个环节,从基因突变ES细胞制备、基因突变小鼠制备、小鼠繁育,精子冻存到最终的表型分析、数据处理及展示,保证了MDMPR产生数据的真实性及实时性。MDMPR除了不断地推进项目进行,增加自身产生的数据外,也在积极的整合其他的资源及数据,如人特异性基因敲除ES细胞库、蛋白相互作用数据库(STRING)、核心转录调节环路(dbCoRc)和Enhancer-Indel数据库,今后还将进一步整合,帮助发育代谢及其他领域的研究人员能够一站式的获取所需资源和数据、加快研究进程,最终服务于全人类的医疗事业。
关键词:
Abstract
Mouse Developmental and Metabolic Phenotype Repository (MDMPR) is an open access, real-time database which dedicates to share mouse resources and phenotype data. MDMPR is supported by the National Key Research and Development Project “Establishment of Mouse Developmental and Metabolic Phenotype Repository” within the Key Project of “Developmental Programming and Its Metabolic Regulation” from the Ministry of Science and Technology of the People’s Republic of China’s program. In the next 5 years, MDMPR will create 500 mutant mouse models related to development and metabolism, perform standard phenotyping analysis, and establish a phenotype database. MDMPR is a combination of resources and data repository, has several sub-systems, including the ES cell database, the project management system, the breeding management system, the sperm bank management system and the phenotyping database. These systems digitalize all data and ensure their authenticity in real-time. Besides the gradual increase of data during the project, MDMPR will also integrate other resources, such as human KO ES cell database, STRING database, database of Core Transcriptional Regulatory Circuitries and Enhancer-Indel database. MDMPR is anticipated to contribute to various areas of developmental and metabolic research to investigators through more convenient accesses to the resources and data in one-stop manner, thereby accelerating the research processes and ultimately serving the medical causes of human health.
Keywords:
PDF (1001KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
朱文静, 刘志玮. MDMPR:小鼠发育代谢表型库. 遗传[J], 2021, 43(4): 375-384 doi:10.16288/j.yczz.21-005
Wenjing Zhu.
作为哺乳动物,小鼠(Mus musculus)和人(Homo sapiens)的基因组同源性高达90%以上,在组织结构和功能、生理学稳态、生殖、行为和疾病发生机制等方面也高度相似。更重要的是,临床疾病大多有其遗传基础,基因突变小鼠模型可以真实模拟人类基因组变异导致的疾病,是研究病理机制和发现潜在靶标的最佳模式动物。人类和小鼠基因组计划完成后,美国、欧盟、加拿大、日本等批准了一系列重大工程项目,系统化建立基因突变小鼠模型,这些项目包括美国国立卫生院的小鼠基因敲除计划(Knockout Mouse Project, KOMP)、欧盟第七框架的欧洲小鼠条件性敲除计划(The European Conditional Mouse Mutagenesis Program,EUCOMM),和加拿大的北美小鼠条件性敲除计划(The North American Conditional Mouse Mutagenesis Project, NorCOMM)等,中国也有支撑计划(如The Chinese Conditional Mouse Mutagenesis, ChiCOMM)[1,2,3,4,5,6]。随着小鼠敲除模型的不断积累,国际上成立了敲除小鼠联盟(The International Knockout Mouse Consortium, IKMC)[7])。在2011年更是将单纯制备基因修饰小鼠模型的IKMC联盟,发展为系统性无偏见功能性解析小鼠表型的国际小鼠表型分析联盟(International Mouse Phenotyping Consortium, IMPC)[8,9,10,11])。迄今为止,全球14个国家的25个研究机构参与了这个国际大科学计划,Nature杂志将该计划和国际核聚变计划、国际加速器计划并列为三大国际大科学计划,并指出系统分析基因功能对疾病解析及新药开发将起到核心推动作用[12]。该计划也是G7国家认可的生命科学领域最大的合作项目,受到领域内研究机构的空前重视。为实施该计划,美国国立卫生院、欧盟、英国医学研究理事会、加拿大卫生部、日本理化研究院及Wellcome Trust基金会都投入了大量经费,目前由现任NIH院长Francis Collins博士担任IMPC领导者。
在我国,疾病模型研究和资源平台建设在过去几年也取得了长足的发展,依托国家遗传工程小鼠资源库等小鼠基因组改造技术平台,科技部于2006年启动了国家科技支撑计划重点项目“人类重大疾病小鼠模型的建立与应用”,使得我国与世界同步开展了基因敲除计划(ChiCOMM),主要承担方为南京大学模式动物研究所(Model Animal Research Center, Nanjing University, MARC),也借此发展成为国内最大的遗传突变小鼠资源库。ChiCOMM形成了与国际对接的小鼠表型分析技术体系及相关技术标准,显著推动了国内敲除小鼠模型的共享及开发。随后在2014年,由苏州大学和英国剑桥大学Sanger研究所共同成立了剑桥-苏大基因组资源中心(Cambridge- Suda Genomic Resource Center, CAM-SU GRC),一个隶属于苏州大学的校级研究机构。剑桥-苏大基因组资源中心依托Sanger研究所提供的资源优势,建立亚太地区的干细胞资源中心,同时建立小鼠标准化基因组功能解析平台,从此成为一个提供小鼠突变胚胎干细胞、疾病模型以及基因功能解析技术的生物技术平台。CAM-SU GRC同时成为继英国、德国、美国后第4个小鼠突变干细胞资源中心,也是亚洲唯一的代表,负责亚太地区48个国家的资源分发工作。在多年的运行期间,剑桥-苏大基因组资源中心已经向国内外上百个课题组提供了细胞及小鼠资源,极大地促进了研究的进展。在国际上,剑桥-苏大基因组资源中心也与南京大学模式动物研究所一起在国际小鼠表型分析联盟指导委员会成员中占得两个席位,这不仅是我国对人类科学事业应尽的责任和义务,同时也是我国社会经济发展的客观需要。
紧跟国际生命科学研究的发展,科技部于2018年启动了国家重点研发计划“发育编程及其代谢调节”重点专项,提出需建立小鼠突变品系的发育与代谢特征的系统分析,对现有和新创建的小鼠突变品系的发育与代谢进行系统性分析,发现重要基因的新功能;建立和实时更新突变品系的表型数据,形成有效的数据共享网络系统。在此背景下,由苏州大学剑桥-苏大基因组资源中心牵头,南京大学模式动物研究所作为主要参与方,联合复旦大学、中国科学院遗传与发育生物学研究所、东华大学、军事医学科学院等单位申报并获批了“建立小鼠发育代谢表型库”项目,本项目的核心就是联合并利用国内优势资源和技术,在5年内建立不少于500个具有自主知识产权的小鼠品系及其发育、代谢表型的数据库,同时重点开展小鼠表型数据与临床特征之间的关联研究,促进人类疾病遗传基础研究领域的变革型发展。小鼠发育代谢表型库(Mouse Developmental and Metabolic Phenotype Repository, MDMPR)作为项目最为主要的成果展示,也于2020年底正式上线(小鼠发育代谢表型库访问地址:
1 国内外同类资源库现状
国外的同类数据库起步较早,目前数据量较大,应用比较广泛的数据库有Mouse Genome Informatics (MGI,国内的此类数据库起步较晚,而且大部分尚处在资源展示的阶段,如国家遗传工程小鼠资源库(
2 MDMPR项目基因选择
小鼠中大约有20,000个基因,而在有限的项目经费支持下,我们只能完成约500个基因的小鼠制备以及功能解析,这样对于目标基因的挑选提出了很大的挑战性,怎么样能从有限的数量中找到尽可能多的有改变的基因。因此,在项目启动初期,我们也做了很多工作,首先利用国际公认权威的人类基因组元件、远端转录调控数据库进行比对整合获得人类4D调控组数据库,进一步与人类疾病相关基因组突变数据库(Motif Break、Mutation Targeted Pair)及对应转录组数据进行关联分析,得到存在潜在逻辑调控关系的人类疾病关联基因数据库,而后再根据GWAS对基因进行深层筛选和分类得到1000多个人类疾病相关的发育代谢相关基因。通过与小鼠基因组进行比对,结合国际表型分析联盟的研究进展并与小鼠基因组进行比对从中深层筛选得到514个小鼠同源基因。另外也从国内的发育、代谢领域专家中征集了部分基因,共同构成MDMPR候选基因。项目进展至今,早期获得的一些基因突变小鼠的初步表型数据也证实了我们筛选基因的方式是有效的,已经发现了数个控制小鼠存活的关键基因,并且是国际上尚未有相关报道的,相信随着项目的深入进行,更多有意义的表型改变会出现。3 MDMPR技术路线
3.1 标准化基因遗传工程小鼠敲除策略
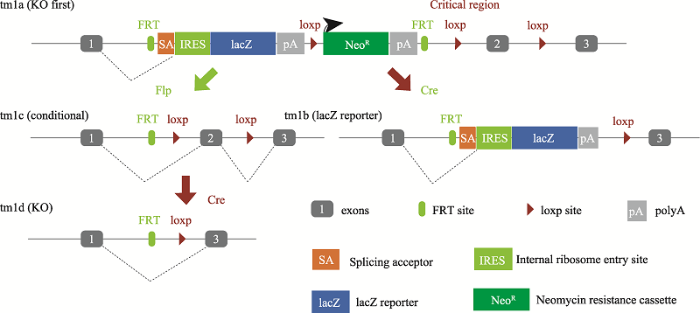
MDMPR同时采用了两种不同的技术手段去建立基因遗传工程小鼠,其中剑桥-苏大基因组资源中心主要基于现有的突变小鼠胚胎干细胞资源(图1),南京大学模式动物研究所基于CRISPR/Cas9敲除技术(图2),分别建立基因敲除小鼠模型。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1基于突变小鼠胚胎干细胞的敲除策略
“Knockout First” allele (tm1a)是在目的基因内含子中插入一个SA-IRES-reporter片段(含有报告基因lacZ和一个启动子驱动的Neo),lacZ的转录在SA的作用下受目的基因启动子控制,并在polyA处终止,从而达到终止目的基因转录、破坏基因表达的目的。Flp的表达可以删除SA-IRES-reporter片段,将tm1a转变成条件敲除基因(tm1c),从而恢复基因功能。在tm1a的基础上,Cre的表达可以删除Neo和critical region从而变成lacZ标记的allele (tm1b)同时敲除基因;在tm1c的基础上,Cre的表达可以删除tm1c allele上的critical region而变成移码突变的tm1d,从而使该基因不表达。
Fig. 1Knockout first targeting strategy based on mutant mouse ES cells
小鼠的遗传背景和微生物级别是影响实验结果的重要因素:不同遗传背景的小鼠对同一个基因的改变可能呈现不同量甚至是不同质的反应;同样,小鼠的微生物级别反映小鼠的生理健康状态,其差异也会造成实验结果的波动。对小鼠遗传背景和微生物进行质量控制可以保证使用最少的实验资源完成既定实验目标,最大限度实现实验效益。这部分工作主要包括:(1)小鼠模型制备过程中遗传质控;(2)小鼠繁育过程中的遗传质控;(3)小鼠繁育过程中的微生物质控。
在项目进行期间,依据现有实验动物遗传质量控制和微生物学等级的相应国标[13,14],各单位根据实际情况制定规范,定期对小鼠的遗传背景(C57BL/6)和微生物级别(无特定病原体动物)进行质控,保障了实验动物的可靠性与实验数据的可重复性。截止2020年11月,检测结果见表1,CAM-SU GRC及MARC的微生物质控标准参见附表1及附表2。
图2
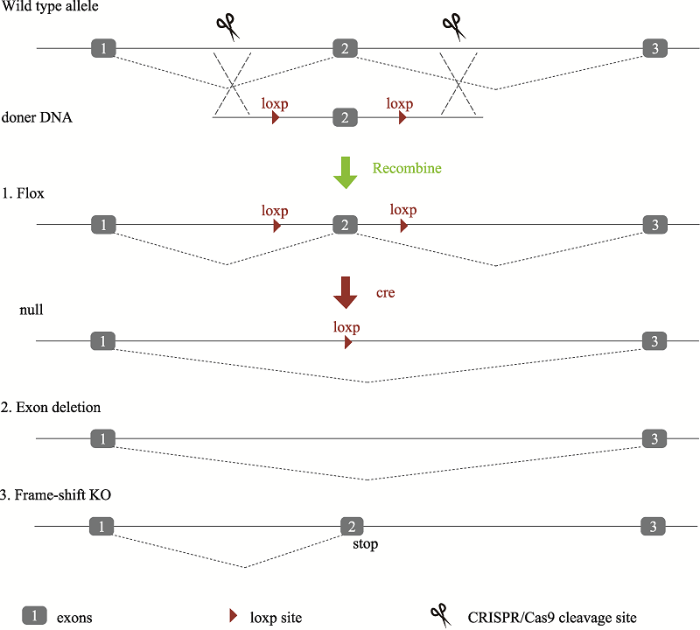 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2基于CRISPR/Cas9的敲除策略
基于CRISPR/Cas9的敲除策略,针对目标外显子两侧设计一对sgRNA,通过Cas9介导的同源重组方式将供体DNA(目标外显子外侧包含一对loxp序列及外侧的同源臂)形成Flox基因,在Cre作用下能够切除目标外显子,造成基因敲除。该策略可能存在两种副产物,其一是同源重组未能发生,造成目标外显子缺失;另可能造成目标外显子部分缺失,也可能引起frame-shift而造成基因敲除。
Fig. 2CRISPR/Cas9 Knockout strategy
3.2 标准化的发育、代谢表型分析流程
以“国际小鼠表型分析联盟”的生理病理表型分析技术流程为基础,本项目发展了重点针对代谢和发育功能的专有分析流程(图3)。在交配获得进入流程的实验小鼠过程中,利用纯和的雌性或雄性与杂合鼠进行交配,对其生殖能力进行评估;另外存活检测作为一个重要的检查点,决定该基因敲除小鼠品系是否会进入胚胎分析流程以及是用杂合还是纯和敲除小鼠进行成体流程分析。Table 1
表1
表1遗传背景和微生物级别质控情况汇总
Table 1
| 质控 | 单位 | 检测方 | 检测频次 | 总检测次数 | 检测结果 |
|---|---|---|---|---|---|
| 遗传背景质控 | CAM-SU GRC | 苏州西山生物有限公司 | 半年 | 2 | 合格 |
| MARC | 江苏集萃药康生物科技有限公司 | 半年 | 2 | 合格 | |
| 微生物质控 | CAM-SU GRC | 苏州西山生物有限公司 | 隔月 | 8 | 合格 |
| MARC | 江苏集萃药康生物科技有限公司 | 季度 | 4 | 合格 |
新窗口打开|下载CSV
为了保证小鼠在表型分析流程中产生数据的一致性以及可重复性,所有的表型分析操作基于SOP (Standard Operating Procedure)来进行,采集的具体参数,包括实验设置相关参数(metadata)以及具体实验参数(parameter)的参数种类、数据类型、数值选项也通过SOP进行规范,从制度上保障了产生数据的可靠性(图4)。目前已经建立了111个SOP、492个参数,随着项目进行,SOP及参数数量还将进一步增加,MDMPR即是建立在这些基础参数之上。
图3
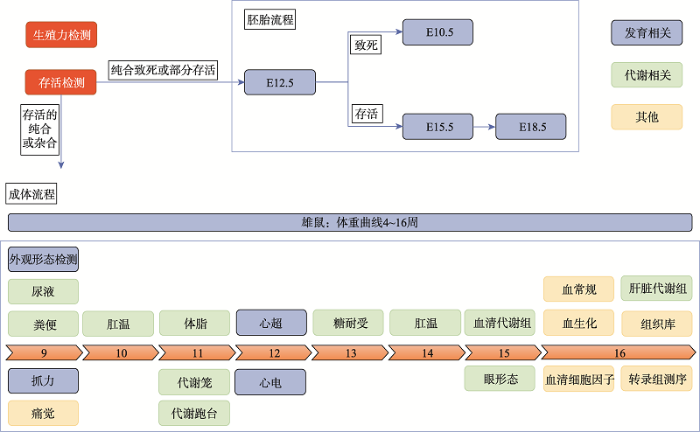 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3标准化的发育、代谢表型分析流程
Fig. 3Standardized phenotyping pipeline of development and metabolism
4 MDMPR系统建设
4.1 MDMPR系统架构
建立的细胞、小鼠资源及表型数据通过数据库进行存储,进而通过网页前端将信息通过合理的方式开放给公众进行查询及使用。尽管数据库的目的在于存储细胞、小鼠及表型的信息,但构建时并不仅仅局限于此,还包含了小鼠制备、繁育、品系保种等过程中相关的实验记录及数据。小鼠发育代谢表型库是一个非常庞大的信息化系统,主要包括ES细胞数据库、项目管理系统、繁育管理系统、精子库管理系统及表型分析系统,各系统间信息互相交互,共同形成MDMPR中的对外展示信息。同时,为了更加方便用户使用,我们在进行多个数据库的整合工作,目前已经将蛋白相互作用数据库(STRING)、核心专利调节环路(a database of Core transcriptional Regulatory Circuitries, dbCoRC),Enhancer-Indel数据库进行了整合,使用户能够获取更多的关注基因的咨询,将MDMPR发展成为一站式的小鼠信息资源数据库(图5)。图4
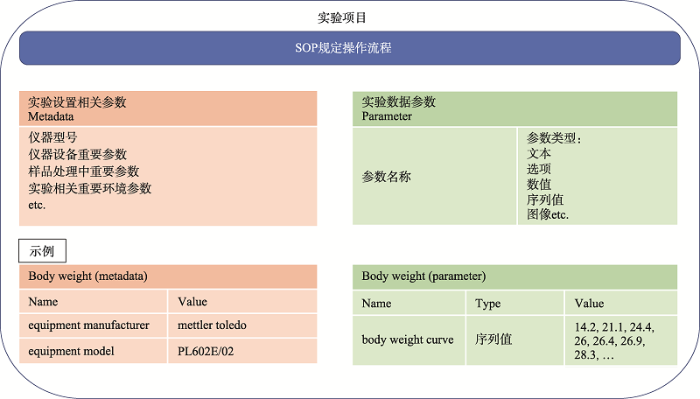 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4实验参数结构
Fig. 4Structure of experimental parameter
4.2 MDMPR数据流
数据起始源头来自于ES细胞数据库,项目启动,开始复苏后,克隆信息进入项目管理系统,对复苏状态,QC结果进行记录,相关克隆信息同时返回ES细胞数据库;QC通过的克隆会进行注射,经项目管理系统记录注射数据及嵌合鼠结果,产生的雄性嵌合鼠自动在繁育管理系统中生成,进入实际饲养管理中;饲养管理系统基于移动端开发,通过二维码形成的地址访问相对应的笼位、小鼠信息,常规的繁育操作如配繁、生仔报告、剪尾、分笼、处死均通过“动物管家”APP进行,确保了数据库内存储的活体信息是实时更新的;表型分析项目从繁育管理系统中实际存在的小鼠开始建立,通过建立标准化的分析流程,标准化的数据录入格式,自动化的数据处理方式,保障了数据的真实性,排除人工带入的误差;品系经精子冻存后,冻存样本信息导入精子库管理系统,同时返回繁育管理系统,这样共同构成品系的真实存储状况;MDMPR前端展示数据从各个系统中进行调用,共同构成展示部分(图5)。图5
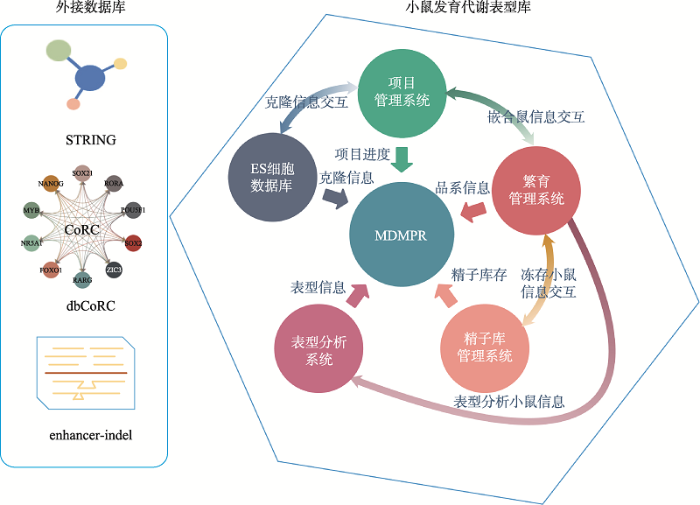 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5MDMPR系统架构
Fig. 5MDMPR system structure
4.3 MDMPR前端展示界面设计
数据合理的展示方式实际是用户最为关注的点,我们以使用者的角度,对MDMPR中来源于不同系统的数据有机组合,构成前端展示的界面,使用户能够在一个界面获取到所需数据的概览,如在检索结果的主页即能查看到细胞、小鼠、表型、相关数据库的情况,第一时间掌握资源及数据的储存情况;同时大量使用直观的图示,使用户能够在最短的时间内获取到最为重要的信息,极大的提升了使用的体验度;此外创造性的加入了关注功能,基于全部项目进度的信息化,能够在关注基因状态改变的同时自动化的向用户推送信息,掌握第一手资讯(图6)。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6MDMPR前端页面设计
Fig. 6Interface design of MDMPR
5 MDMPR应用实例
MDMPR除了对于单个基因的表型数据进行分析外,基于其数据量、产生数据方式带来的平行可比性,可以对数据进行大规模的表型与基因关联分析,筛选出关键基因。我们曾经成功的利用IMPC联盟ICS、TCP、RBRC、WTSI四个中心的小鼠饮食和活动的数据作为模型训练的数据初步开发了深度卷积神经网络方法并对少数指标进行了表型筛选,筛选了10种饮食或者活动异常的基因型小鼠,并针对Slc7a11基因敲除小鼠进行了生物节律的深入表型验证和分析,验证了其在外界光照牵引小鼠昼夜节律中的作用,进一步的功能分析暗示其可能通过调控生物钟核心SCN (Suprachiasmatic Nucleus)神经元之间的同步影响外界光照对昼夜节律的牵引,成果已发表在PLoS Genetics[15]。IMPC也已有多方面的尝试,在大数据比较的基础上找到了一些新的发育相关基因[9,16~18]。以此为例,在MDMPR数据广度和深度的不断充实下,可以通过类似的大规模表型与基因关联分析,发掘出一些新的基因功能。6 结语与展望
MDMPR最主要的核心在于其资源与数据的开放共享,这在国内属于创新性的尝试,可以将国家的研究经费转化出最大的价值,被更多的相关研究领域的人所利用。标准化的表型分析流程,信息化的流程及数据管理,保障了产生数据的真实性及及时性,能够为研究者在项目起始阶段提供初期实验数据,对立项及研究方向提供指引,是研究者开展研究的一个利器。虽然MDMPR尚处在起步阶段,资源及表型的数据尚在积累中,从数量上来说还不足,但随着项目的不断进展,更多的资源和数据将会加入,不断充实表型库,让更多用户能够获得有用信息,促进研究的进展。在系统架构上,我们也在不断的进行优化,提升用户在使用上的体验,通过整合其他的数据库,力争将MDMPR建设成一站式的资源数据平台。尽管项目投入有限,我们也将充分利用现有条件,通过技术手段的提升,压缩成本,尽可能在完成项目目标的基础上,增加更多的资源与数据。同时,我们也积极在国内外寻求合作的机会,招募有意愿的研究单位共同加入到此项目中,扩充研究的宽度,使资源的使用效率最大化。目前已有数家合作单位参与到项目中,承担如痛觉、生殖、应激行为等方面的分析,进一步拓展了表型库的覆盖范围,并且已经获得了一些有意义的数据,数据库也在同步准备中,将在不久后开放展示。相信在不久的将来,MDMPR将建设成为国内最大、最全、开放程度最高、实用性最强的资源库,极大地推进代谢、发育甚至其他领域的研究进展。致谢
感谢科技部重点研发计划“发育编程及其代谢调节”重点专项“建立小鼠发育代谢表型库”项目参与单位苏州大学、南京大学、复旦大学、中国科学院遗传与发育生物学研究所、东华大学、军事医学科学院的项目组成员对于此项目所付出的努力,感谢南京百迈斯信息科技有限公司在MDMPR数据库建设中的全力支持。
附录:
附加材料详见文章电子版Table 1
附表1
附表1 CAM-SU GRC微生物质控标准
Table 1
| 病原体 | 检测项目 | 英文名称 | 质控标准 |
|---|---|---|---|
| 病毒 | 鼠痘病毒 | ECT (Mousepox (Ectromelia) Virus) | 国标必检项 |
| 小鼠肝炎病毒 | MHV (Mouse hepatitis virus) | 国标必检项 | |
| 仙台病毒 | SV (Sendai virus) | 国标必检项 | |
| 小鼠肺炎病毒 | PVM (Pneumonia virus of Mice) | 国标必检项 | |
| 呼肠孤病毒Ⅲ型 | REO-3 (Reovirus type 3) | 国标必检项 | |
| 小鼠细小病毒(MVM株) | MVM (Minute virus of Mice) | 国标必检项 | |
| 汉坦病毒 | HV (Hantavirus) | 国标选做项 | |
| 淋巴细胞脉络丛脑膜炎病毒 | LCMV (Lymphocytic choriomeningitis virus) | 国标选做项 | |
| 小鼠脑脊髓炎病毒 | TMEV (Theiler’s mouse encephalomyelitis virus) | 国标选做项 | |
| 小鼠腺病毒 | MAV (Mouse adenovirus type 1(FL) & 2(K87)) | 国标选做项 | |
| 多瘤病毒 | Poly (Polyoma virus) | 国标选做项 | |
| 小鼠轮状病毒 | EDIM (Mouse rotavirus) | 国际标准选做项 | |
| 小鼠巨细胞瘤病毒 | MCMV (Mouse Cytomegalovirus) | 国际标准选做项 | |
| 乳酸脱氢酶增高病毒 | LDV (Lactate dehydrogenase-elevating virus) | 国际标准选做项 | |
| 小鼠细小病毒 | MPV (Mouse parvovirus) | 国际标准选做项 | |
| 小鼠细小病毒NS-1 | PARV-NS1 (Parvovirus NS-1) | 国际标准选做项 | |
| K病毒 | K virus | 国际标准选做项 | |
| 小鼠诺如病毒 | MNV (Murine norovirus) | 国际标准选做项 | |
| 病原菌 | 支原体 | MYCO (Mycoplasma) | 国标选做项 |
| 沙门菌 | SALM (Salmonella spp.) | 国标必检项 | |
| 鼠棒状杆菌 | CKUT (Corynebacterium kutscheri) | 国标必检项 | |
| 泰泽病原体 | CPIL (Clostridium piliforme (Tyzzer’s organism)) | 国标必检项 | |
| 噬肺巴斯德杆菌 | PPNE (Pasteurella pneumotropica) | 国标必检项 | |
| 金黄色葡萄球菌 | SAUR (Staphylococcus aureus) | 国标必检项 | |
| 绿脓杆菌 | PAER (Pseudomonsa aeruginosa) | 国标必检项 | |
| 肺炎克雷伯氏杆菌 | KPNE (Klebsiella pneumonia) | 国标必检项 | |
| 肺炎链球菌 | SPNE (Streptococcus pneumonia) | 国标选做项 | |
| 乙型溶血性链球菌 | BHS (Streptococcus (Beta-hemolytic)) | 国标选做项 | |
| 多杀巴斯德杆菌 | PMUL (Pasteurella multocida) | 国标选做项 | |
| 枸橼酸杆菌 | CROD (Citrobacter rodentium) | 国际标准选做项 | |
| 支气管鲍特杆菌 | BBRO (Bordetella bronchiseptica) | 国标选做项 | |
| 产酸克雷伯氏杆菌 | KOXY (Klebsiella oxytoca) | 国际标准选做项 | |
| 肺炎支原体 | MPUL (Mycoplasma pulmonis) | 国标必检项 | |
| 奇异变形杆菌 | PMIR (Proteus mirabilis) | 国际标准选做项 | |
| 乙型溶血性链球菌B群 | GBBHS (Beta strep.Sp.Group B) | 国标选做项 | |
| 乙型溶血性链球菌G群 | GGBHS (Beta strep.Sp.Group G) | 国标选做项 | |
| 寄生虫 | 体外寄生虫 | ECTO (Ectoparasites) | 国标必检项 |
| 弓形虫 | TOXO (Toxoplasma gondii) | 国标必检项 | |
| 全部蠕虫 | HELM (All Helminths) | 国标必检项 | |
| 鞭毛虫 | FLAG (Flagellates) | 国标必检项 | |
| 纤毛虫 | CILI (Ciliates) | 国标选做项 | |
| 兔脑原虫 | ECUN (Encephalitozoon cuniculi) | 国标选做项 |
新窗口打开|下载CSV
Supplementary Table 2
附表2
附表2 MARC微生物质控标准
Supplementary Table 2
| 病原体 | 检测项目 | 英文名称 | 检测方法 |
|---|---|---|---|
| 病毒 | 鼠痘病毒 | ECT (Mousepox (Ectromelia) Virus) | 国标必检项 |
| 小鼠肝炎病毒 | MHV (Mouse hepatitis virus) | 国标必检项 | |
| 仙台病毒 | SV (Sendai virus) | 国标必检项 | |
| 小鼠肺炎病毒 | PVM (Pneumonia virus of Mice) | 国标必检项 | |
| 呼肠孤病毒Ⅲ型 | REO-3 (Reovirus type 3) | 国标必检项 | |
| 汉坦病毒 | HV (Hantavirus) | 国标选做项 | |
| 淋巴细胞脉络丛脑膜炎病毒 | LCMV (Lymphocytic choriomeningitis virus) | 国标选做项 | |
| 小鼠脑脊髓炎病毒 | TMEV (Theiler’s mouse encephalomyelitis virus) | 国标选做项 | |
| 小鼠腺病毒 | MAV (Mouse adenovirus type 1(FL) & 2(K87)) | 国标选做项 | |
| 多瘤病毒 | Poly (Polyoma virus) | 国标选做项 | |
| 小鼠轮状病毒 | EDIM (Mouse rotavirus) | 国际标准选做项 | |
| 小鼠巨细胞瘤病毒 | MCMV (Mouse Cytomegalovirus) | 国际标准选做项 | |
| 小鼠细小病毒 | MPV (Mouse parvovirus) | 国际标准选做项 | |
| K病毒 | K virus | 国际标准选做项 | |
| 病原菌 | 肺支原体 | MPUL (Mycoplasma pulmonis) | 国标必检项 |
| 沙门菌 | SALM (Salmonella spp.) | 国标必检项 | |
| 鼠棒状杆菌 | CKUT (Corynebacterium kutscheri) | 国标必检项 | |
| 泰泽病原体 | CPIL (Clostridium piliforme (Tyzzer’s organism)) | 国标必检项 | |
| 噬肺巴斯德杆菌 | PPNE (Pasteurella pneumotropica) | 国标必检项 | |
| 金黄色葡萄球菌 | SAUR (Staphylococcus aureus) | 国标必检项 | |
| 绿脓杆菌 | PAER (Pseudomonsa aeruginosa) | 国标必检项 | |
| 肺炎克雷伯氏杆菌 | KPNE (Klebsiella pneumonia) | 国标必检项 | |
| 肺炎链球菌 | SPNE (Streptococcus pneumonia) | 国标选做项 | |
| 乙型溶血性链球菌 | BHS (Streptococcus (Beta-hemolytic)) | 国标选做项 | |
| 皮肤病原真菌 | Pathogenic dermal fungi | 国标选做项 | |
| 假结核耶尔森菌 | Yersinia pseudotuberculosis | 国标选做项 | |
| 小肠结肠炎耶尔森菌 | Yesinia enterocolitica | 国标选做项 | |
| 念珠状链杆菌 | Streptobacillus moniliformis | 国标选做项 | |
| 大肠埃希菌O115 a,C,K(B) | Escherichia coli O115 a,C,K(B) | 国标选做项 | |
| 纤毛相关呼吸道杆菌 | CARB (Cilia-Associated Respiratory Bacillus) | 国际标准选做项 | |
| 寄生虫 | 体外寄生虫 | ECTO (Ectoparasites) | 国标必检项 |
| 弓形虫 | TOXO (Toxoplasma gondii) | 国标必检项 | |
| 蠕虫 | HELM (Helminth) | 国标必检项 | |
| 鞭毛虫 | FLAG (Flagellates) | 国标必检项 | |
| 卡式肺孢子虫 | Pneumocystis carinii | 国标选做项 | |
| 小鼠脑炎原虫 | ECUN (Encephalitozoon cuniculi) | 国标选做项 |
新窗口打开|下载CSV
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1530/JOE-16-0258URLPMID:27535945 [本文引用: 1]

The genetic determinants of osteoporosis remain poorly understood, and there is a large unmet need for new treatments in our ageing society. Thus, new approaches for gene discovery in skeletal disease are required to complement the current genome-wide association studies in human populations. The International Knockout Mouse Consortium (IKMC) and the International Mouse Phenotyping Consortium (IMPC) provide such an opportunity. The IKMC generates knockout mice representing each of the known protein-coding genes in C57BL/6 mice and, as part of the IMPC initiative, the Origins of Bone and Cartilage Disease project identifies mutants with significant outlier skeletal phenotypes. This initiative will add value to data from large human cohorts and provide a new understanding of bone and cartilage pathophysiology, ultimately leading to the identification of novel drug targets for the treatment of skeletal disease.
DOI:10.1089/omi.2013.0158URL [本文引用: 1]

Omics research infrastructure such as databases and bio-repositories requires effective governance to support pre-competitive research. Governance includes the use of legal agreements, such as Material Transfer Agreements (MTAs). We analyze the use of such agreements in the mouse research commons, including by two large-scale resource development projects: the International Knockout Mouse Consortium (IKMC) and International Mouse Phenotyping Consortium (IMPC). We combine an analysis of legal agreements and semi-structured interviews with 87 members of the mouse model research community to examine legal agreements in four contexts: (1) between researchers; (2) deposit into repositories; (3) distribution by repositories; and (4) exchanges between repositories, especially those that are consortium members of the IKMC and IMPC. We conclude that legal agreements for the deposit and distribution of research reagents should be kept as simple and standard as possible, especially when minimal enforcement capacity and resources exist. Simple and standardized legal agreements reduce transactional bottlenecks and facilitate the creation of a vibrant and sustainable research commons, supported by repositories and databases.
DOI:10.1242/dmm.011833URL [本文引用: 1]

Identifying genes that are important for embryo development is a crucial first step towards understanding their many functions in driving the ordered growth, differentiation and organogenesis of embryos. It can also shed light on the origins of developmental disease and congenital abnormalities. Current international efforts to examine gene function in the mouse provide a unique opportunity to pinpoint genes that are involved in embryogenesis, owing to the emergence of embryonic lethal knockout mutants. Through internationally coordinated efforts, the International Knockout Mouse Consortium (IKMC) has generated a public resource of mouse knockout strains and, in April 2012, the International Mouse Phenotyping Consortium (IMPC), supported by the EU InfraCoMP programme, convened a workshop to discuss developing a phenotyping pipeline for the investigation of embryonic lethal knockout lines. This workshop brought together over 100 scientists, from 13 countries, who are working in the academic and commercial research sectors, including experts and opinion leaders in the fields of embryology, animal imaging, data capture, quality control and annotation, high-throughput mouse production, phenotyping, and reporter gene analysis. This article summarises the outcome of the workshop, including (1) the vital scientific importance of phenotyping embryonic lethal mouse strains for basic and translational research; (2) a common framework to harmonise international efforts within this context; (3) the types of phenotyping that are likely to be most appropriate for systematic use, with a focus on 3D embryo imaging; (4) the importance of centralising data in a standardised form to facilitate data mining; and (5) the development of online tools to allow open access to and dissemination of the phenotyping data.
DOI:10.5483/bmbrep.2012.45.12.242URLPMID:23261053 [本文引用: 1]

Phenotypic analysis of gene-specific knockout (KO) mice has revolutionized our understanding of in vivo gene functions. As the use of mouse embryonic stem (ES) cells is inevitable for conventional gene targeting, the generation of knockout mice remains a very time-consuming and expensive process. To accelerate the large-scale production and phenotype analyses of KO mice, international efforts have organized global consortia such as the International Knockout Mouse Consortium (IKMC) and International Mouse Phenotype Consortium (IMPC), and they are persistently expanding the KO mouse catalogue that is publicly available for the researches studying specific genes of interests in vivo. However, new technologies, adopting zinc-finger nucleases (ZFNs) or Transcription Activator-Like Effector (TALE) Nucleases (TALENs) to edit the mouse genome, are now emerging as valuable and effective shortcuts alternative for the conventional gene targeting using ES cells. Here, we introduce the recent achievement of IKMC, and evaluate the significance of ZFN/TALEN technology in mouse genetics.
DOI:10.1007/s00335-012-9428-9URL [本文引用: 1]

The International Mouse Phenotyping Consortium (IMPC) (http://www.mousephenotype.org) will reveal the pleiotropic functions of every gene in the mouse genome and uncover the wider role of genetic loci within diverse biological systems. Comprehensive informatics solutions are vital to ensuring that this vast array of data is captured in a standardised manner and made accessible to the scientific community for interrogation and analysis. Here we review the existing EuroPhenome and WTSI phenotype informatics systems and the IKMC portal, and present plans for extending these systems and lessons learned to the development of a robust IMPC informatics infrastructure.
DOI:10.1038/ng0904-925URLPMID:15340424 [本文引用: 1]

The European Mouse Mutagenesis Consortium is the European initiative contributing to the international effort on functional annotation of the mouse genome. Its objectives are to establish and integrate mutagenesis platforms, gene expression resources, phenotyping units, storage and distribution centers and bioinformatics resources. The combined efforts will accelerate our understanding of gene function and of human health and disease.
DOI:10.1038/ng0904-921URLPMID:15340423 [本文引用: 1]

Mouse knockout technology provides a powerful means of elucidating gene function in vivo, and a publicly available genome-wide collection of mouse knockouts would be significantly enabling for biomedical discovery. To date, published knockouts exist for only about 10% of mouse genes. Furthermore, many of these are limited in utility because they have not been made or phenotyped in standardized ways, and many are not freely available to researchers. It is time to harness new technologies and efficiencies of production to mount a high-throughput international effort to produce and phenotype knockouts for all mouse genes, and place these resources into the public domain.
DOI:10.1038/s41467-017-01995-2URLPMID:29348434 [本文引用: 1]

Metabolic diseases are a worldwide problem but the underlying genetic factors and their relevance to metabolic disease remain incompletely understood. Genome-wide research is needed to characterize so-far unannotated mammalian metabolic genes. Here, we generate and analyze metabolic phenotypic data of 2016 knockout mouse strains under the aegis of the International Mouse Phenotyping Consortium (IMPC) and find 974 gene knockouts with strong metabolic phenotypes. 429 of those had no previous link to metabolism and 51 genes remain functionally completely unannotated. We compared human orthologues of these uncharacterized genes in five GWAS consortia and indeed 23 candidate genes are associated with metabolic disease. We further identify common regulatory elements in promoters of candidate genes. As each regulatory element is composed of several transcription factor binding sites, our data reveal an extensive metabolic phenotype-associated network of co-regulated genes. Our systematic mouse phenotype analysis thus paves the way for full functional annotation of the genome.
DOI:10.1038/nature19356URLPMID:27626380 [本文引用: 2]

Approximately one-third of all mammalian genes are essential for life. Phenotypes resulting from knockouts of these genes in mice have provided tremendous insight into gene function and congenital disorders. As part of the International Mouse Phenotyping Consortium effort to generate and phenotypically characterize 5,000 knockout mouse lines, here we identify 410 lethal genes during the production of the first 1,751 unique gene knockouts. Using a standardized phenotyping platform that incorporates high-resolution 3D imaging, we identify phenotypes at multiple time points for previously uncharacterized genes and additional phenotypes for genes with previously reported mutant phenotypes. Unexpectedly, our analysis reveals that incomplete penetrance and variable expressivity are common even on a defined genetic background. In addition, we show that human disease genes are enriched for essential genes, thus providing a dataset that facilitates the prioritization and validation of mutations identified in clinical sequencing efforts.
DOI:10.1038/ng1105-1155URLPMID:16254554 [本文引用: 1]
DOI:10.1038/nature10163URLPMID:21677750 [本文引用: 1]

Gene targeting in embryonic stem cells has become the principal technology for manipulation of the mouse genome, offering unrivalled accuracy in allele design and access to conditional mutagenesis. To bring these advantages to the wider research community, large-scale mouse knockout programmes are producing a permanent resource of targeted mutations in all protein-coding genes. Here we report the establishment of a high-throughput gene-targeting pipeline for the generation of reporter-tagged, conditional alleles. Computational allele design, 96-well modular vector construction and high-efficiency gene-targeting strategies have been combined to mutate genes on an unprecedented scale. So far, more than 12,000 vectors and 9,000 conditional targeted alleles have been produced in highly germline-competent C57BL/6N embryonic stem cells. High-throughput genome engineering highlighted by this study is broadly applicable to rat and human stem cells and provides a foundation for future genome-wide efforts aimed at deciphering the function of all genes encoded by the mammalian genome.
DOI:10.1038/465526aURLPMID:20520665 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1371/journal.pgen.1008577URLPMID:31929527 [本文引用: 1]

Circadian systems provide a fitness advantage to organisms by allowing them to adapt to daily changes of environmental cues, such as light/dark cycles. The molecular mechanism underlying the circadian clock has been well characterized. However, how internal circadian clocks are entrained with regular daily light/dark cycles remains unclear. By collecting and analyzing indirect calorimetry (IC) data from more than 2000 wild-type mice available from the International Mouse Phenotyping Consortium (IMPC), we show that the onset time and peak phase of activity and food intake rhythms are reliable parameters for screening defects of circadian misalignment. We developed a machine learning algorithm to quantify these two parameters in our misalignment screen (SyncScreener) with existing datasets and used it to screen 750 mutant mouse lines from five IMPC phenotyping centres. Mutants of five genes (Slc7a11, Rhbdl1, Spop, Ctc1 and Oxtr) were found to be associated with altered patterns of activity or food intake. By further studying the Slc7a11tm1a/tm1a mice, we confirmed its advanced activity phase phenotype in response to a simulated jetlag and skeleton photoperiod stimuli. Disruption of Slc7a11 affected the intercellular communication in the suprachiasmatic nucleus, suggesting a defect in synchronization of clock neurons. Our study has established a systematic phenotype analysis approach that can be used to uncover the mechanism of circadian entrainment in mice.
DOI:10.1038/s41467-017-00595-4URLPMID:29026089 [本文引用: 1]

The developmental and physiological complexity of the auditory system is likely reflected in the underlying set of genes involved in auditory function. In humans, over 150 non-syndromic loci have been identified, and there are more than 400 human genetic syndromes with a hearing loss component. Over 100 non-syndromic hearing loss genes have been identified in mouse and human, but we remain ignorant of the full extent of the genetic landscape involved in auditory dysfunction. As part of the International Mouse Phenotyping Consortium, we undertook a hearing loss screen in a cohort of 3006 mouse knockout strains. In total, we identify 67 candidate hearing loss genes. We detect known hearing loss genes, but the vast majority, 52, of the candidate genes were novel. Our analysis reveals a large and unexplored genetic landscape involved with auditory function.The full extent of the genetic basis for hearing impairment is unknown. Here, as part of the International Mouse Phenotyping Consortium, the authors perform a hearing loss screen in 3006 mouse knockout strains and identify 52 new candidate genes for genetic hearing loss.
DOI:10.1038/s42003-018-0226-0URLPMID:30588515

Despite advances in next generation sequencing technologies, determining the genetic basis of ocular disease remains a major challenge due to the limited access and prohibitive cost of human forward genetics. Thus, less than 4,000 genes currently have available phenotype information for any organ system. Here we report the ophthalmic findings from the International Mouse Phenotyping Consortium, a large-scale functional genetic screen with the goal of generating and phenotyping a null mutant for every mouse gene. Of 4364 genes evaluated, 347 were identified to influence ocular phenotypes, 75% of which are entirely novel in ocular pathology. This discovery greatly increases the current number of genes known to contribute to ophthalmic disease, and it is likely that many of the genes will subsequently prove to be important in human ocular development and disease.
DOI:10.1038/s41598-019-47286-2URLPMID:31371754 [本文引用: 1]

Oculocutaneous syndromes are often due to mutations in single genes. In some cases, mouse models for these diseases exist in spontaneously occurring mutations, or in mice resulting from forward mutatagenesis screens. Here we present novel genes that may be causative for oculocutaneous disease in humans, discovered as part of a genome-wide screen of knockout-mice in a targeted single-gene deletion project. The International Mouse Phenotyping Consortium (IMPC) database (data release 10.0) was interrogated for all mouse strains with integument abnormalities, which were then cross-referenced individually to identify knockouts with concomitant ocular abnormalities attributed to the same targeted gene deletion. The search yielded 307 knockout strains from unique genes with integument abnormalities, 226 of which have not been previously associated with oculocutaneous conditions. Of the 307 knockout strains with integument abnormalities, 52 were determined to have ocular changes attributed to the targeted deletion, 35 of which represent novel oculocutaneous genes. Some examples of various integument abnormalities are shown, as well as two examples of knockout strains with oculocutaneous phenotypes. Each of the novel genes provided here are potentially relevant to the pathophysiology of human integumentary, or oculocutaneous conditions, such as albinism, phakomatoses, or other multi-system syndromes. The novel genes reported here may implicate molecular pathways relevant to these human diseases and may contribute to the discovery of novel therapeutic targets.
