 ,1,2
,1,2Advances in m 6A modification and its regulation of viral replication
Peng Xue1, Tao Jiang1, Xingjia Shen ,1,2
,1,2通讯作者:
编委: 岑山
收稿日期:2019-02-20修回日期:2019-04-8网络出版日期:2019-05-20
| 基金资助: |
Received:2019-02-20Revised:2019-04-8Online:2019-05-20
| Fund supported: |
作者简介 About authors
薛鹏,硕士研究生,专业方向:基因表达调控E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (468KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
薛鹏, 蒋涛, 沈兴家. m 6A修饰及其对病毒复制过程调控研究进展 [J]. 遗传, 2019, 41(5): 404-412 doi:10.16288/j.yczz.19-042
Peng Xue, Tao Jiang, Xingjia Shen.
RNA不仅能将遗传信息翻译成蛋白质,而且能调控多种生物学进程,因而其在生物系统中具有重要的作用。作为重要的遗传介质,mRNA的化学修饰是一种重要的RNA转录后调控机制,在众多已知的RNA修饰方式中,m6A甲基化修饰是真核生物mRNA中最普遍的一种修饰方式,于1974年首次被发现,并引起了人们的极大兴趣[1,2]。m6A修饰在从酵母、植物、果蝇到哺乳动物等真核生物到RNA病毒都具有较高的保守性[3]。在体外对甲基转移酶突变和底物偏好性的研究发现,m6A修饰主要集中在终止密码子附近及3′端非翻译区,其共有基序为[G/A/U][G>A]m6AC[U>A>C],而在拟南芥的起始密码子附近也发现了m6A甲基化修饰[4,5]。mRNA上m6A普遍存在而含量极少并且是非随机分布的,这意味着m6A修饰可能在转录后调控中具有重要的作用,而已有的研究表明,m6A修饰可以影响mRNA代谢过程,包括mRNA的加工、出核运输、翻译和稳定性,以及在RNA转录后水平调控基因的表达,进而影响各种生理过程,如生长、发育、生殖、细胞多能性、减数分裂、昼夜节律和疾病发生等[6,7]。
病毒被称为细胞的“寄生虫”,致病病毒对人类的生命健康构成了巨大威胁。病毒的生命周期是依赖宿主细胞的相关机制和途径进行的,从表观遗传学角度研究病毒活动过程对促进抗病毒机制的研究具有重要意义。人们最先在腺病毒(Ad)和甲型流感病毒(IAV)的mRNA上检测到m6A[8,9]。随后,在单纯疱疹病毒1型(HSV-1)、劳斯氏肉瘤病毒(RSV)、猴空泡病毒40 (SV40)、B77肉瘤病毒、禽流感病毒和猫白血病病毒等病毒RNA中也检测到了m6A[10]。研究发现m6A甲基化表观遗传修饰在病毒的侵染复制过程中具有重要作用。本文对m6A修饰的相关概念、m6A在病毒复制中的作用及对免疫反应的影响等方面进行了阐述,以期更深入地了解m6A修饰在病毒感染中的作用,为抗病毒药物的研发提供参考。
1 m6A修饰的分子机制
m6A是腺苷酸(A)在甲基转移酶复合物的催化下,第6位的N发生甲基化的修饰方式。RNA的m6A修饰是一个可逆的动态过程,此过程由甲基转移酶复合物、去甲基化酶和读取蛋白共同完成(图1)。图1
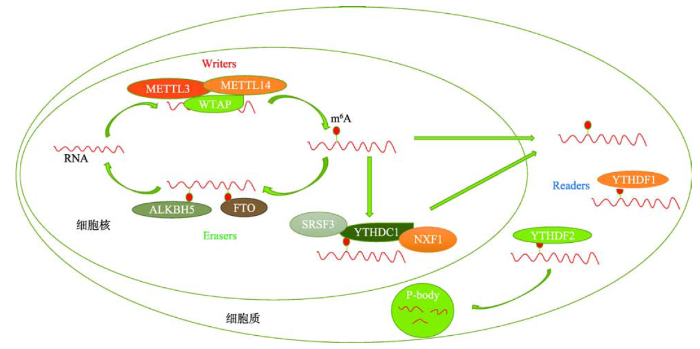 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1m6A修饰的分子机制
METTL3、METTL14和WTAP组成甲基转移酶复合物(Writers)。ALKBH5和FTO是常见的去甲基化酶(Erasers)。YTHDF1、YTHDF2和YTHDC1是常见的读取蛋白(Readers)。SRSF3是剪接因子。NXF1是核RNA输出因子。P-body是RNA降解的场所。核内RNA在甲基转移酶复合物的作用下,形成m6A;而核内RNA上的m6A也可在去甲基化酶的作用下发生去甲基化。随后,在核内RNA的进一步加工过程中,核内的读取蛋白会与m6A位点结合;而当成熟的RNA出核后,细胞质内的读取蛋白会与RNA上的m6A位点结合。不同的读取蛋白结合到m6A位点后执行不同的功能,如YTHDC1能够识别RNA上的m6A位点,同时与SRSF3及NXF1相互作用,从而促进RNA的核输出;YTHDF1能够促进mRNA的翻译;YTHDF2识别RNA上的m6A位点后,能够引导RNA至P-body,从而促进RNA的降解。
Fig. 1The molecular mechanism of m6A modification
1.1 m6A甲基转移酶复合物
甲基转移酶复合物(Writers)由多种甲基转移酶(如METTL3和METTL14)和相关蛋白质亚基(如WTAP)组成。甲基转移酶样3 (METTL3)有催化活性,但METTL3需要和甲基转移酶样14 (METTL14)按 1:1的比例结合,形成异源二聚体,并在肾母细胞瘤1相关蛋白(WTAP)的引导下到达RNA上需要被修饰的位点,由METTL3将S-腺苷甲硫氨酸(SAM)上的甲基在转移到腺苷酸(A)上第6位的N上[11]。近年来又陆续发现并鉴定了KIAA1429[12]、RBM15、HAKAI[13]和METTL16[14]等m6A甲基转移酶复合物的新组分。1.2 m6A去甲基化酶
m6A去甲基化酶(Erasers)负责去除RNA上的甲基化基团。迄今为止,在哺乳动物中只发现了两种m6A去甲基酶:肥胖相关蛋白(FTO)和ALKB同源蛋白5(ALKBH5),它们都属于Fe2+/α-酮戊二酸依赖的非血红素双加氧酶AlkB蛋白家族[15,16]。FTO首先氧化m6A生成氧化中间体N6-羟甲基腺苷(hm6A),再进一步氧化生成N6-甲酰腺苷(f6A),最终在其他酶的作用下,还原成腺苷。相较于FTO,ALKBH5直接将m6A还原成腺苷,不产生中间产物[17]。近期有研究表明,FTO更多的是在另一种甲基化修饰——N6,2°-O二甲基腺苷(m6Am)的去甲基化过程中行使功能[18]。1.3 m6A读取蛋白
m6A的功能是通过RNA结合蛋白介导的,此类蛋白被称为m6A“阅读器”(Readers),Readers可以选择性地结合到含有m6A的RNA上。m6A读取蛋白通过YT521-B同源域(YTH)直接与m6A结合,从而识别发生m6A修饰的RNA[19]。人类中已发现5种含YTH域蛋白,包括两种亚型——YTHDFs和YTHDCs,其中,YTHDFs包括YTHDF1、YTHDF2和YTHDF3[20],YTHDCs包括YTHDC1和YTHDC2[21,22]。YTHDF2是第一个被鉴定的具有生物学功能的YTH家族蛋白[23,24],它选择性结合m6A修饰的mRNA,然后通过招募CCR4-NOT去腺苷酸酶复合物的CNOT亚基,促进mRNA的去腺苷酸化和降解[25]。YTHDF2主要存在于细胞质中,但在热休克压力下,YTHDF2会转移到细胞核中以保护5′非翻译区的m6A免受FTO去甲基化作用的影响,并促进mRNA的非帽依赖性翻译[26]。YTHDC1主要存在于细胞核中,能够通过与多种剪接因子的互作,调控mRNA的剪接[27,28]。YTHDC2在提高其靶mRNA的翻译效率和降低mRNA丰度方面具有双重作用[29]。2 m6A修饰与病毒复制
目前有关m6A修饰对病毒复制的调控及其机制的研究相对较少,m6A修饰作为一种表观遗传修饰,对不同病毒的复制具有完全相反的调控作用(表1)。Table 1
表1
表1 m6A对病毒复制的调控及其机制
Table 1
| 病毒 | m6A对病毒复制的调控 | 作用机制 | 参考文献 |
|---|---|---|---|
| HIV-1 | 存在分歧 | 促进病毒mRNA出核运输 | [30,33] |
| 影响减少HIV-1病毒gRNA丰度 | |||
| 降低转录本稳定性 | |||
| IVA | 正调控 | 未知 | [35] |
| ZIKV | 负调控 | 促进病毒裂解转录本的降解来抑制ZIKV的裂解复制 | [36] |
| HCV | 负调控 | YTHDF蛋白可能通过与病毒RNA竞争核衣壳蛋白来抑制感染性病毒粒子的产生 | [37] |
| 病毒 | m6A对病毒复制的调控 | 作用机制 | 参考文献 |
| HBV | 未知 | HBV转录本pgRNA 5°端茎环上的m6A修饰促进pgRNA的逆转录 | [38] |
| HBV转录本pgRNA 3°端茎环上的m6A修饰降低HBV RNA的稳定性 | |||
| KSHV | 存在分歧 | 调节ORF50 pre-RNA的剪接 | [39,40] |
| 影响病毒转录本的稳定性 | |||
| SV40 | 正调控 | 促进晚期转录本的翻译 | [42] |
| AMV | 负调控 | 引发病毒mRNA的沉默和衰减来抑制病毒的复制 | [43] |
新窗口打开|下载CSV
2.1 m6A修饰对RNA病毒复制的调控
2.1.1 m6A修饰对人类免疫缺陷病毒Ⅰ型病毒(HIV-1)复制的调控研究发现,HIV-1的感染会引起宿主和病毒mRNA的m6A丰度升高。Lichinchi等[30]通过shRNA介导METTL3、METTL14和ALKBH5的沉默,然后对HIV-1包膜糖蛋白GP120的RNA水平进行定量分析,并对感染后72 h的病毒衣壳蛋白p24进行免疫印迹,发现METTL3和METTL14的沉默,使GP120和p24表达水平降低,ALKBH5沉默使GP120和p24表达水平显著增加,表明m6A的修饰丰度与GP120和p24表达水平呈正相关。此外,HIV-1 Rev响应元件(RRE) RNA茎环Ⅱ区域的保守腺苷(A7883)发生m6A修饰,增加了HIV-1 Rev蛋白与RRE的结合,促进了RNA的出核运输,从而增强了HIV-1的复制。Kennedy等[31]发现YTHDF蛋白尤其是YTHDF2蛋白的过表达显著增强了293T细胞感染HIV-1病毒24和48 h后Nef、Tat和Rev的mRNA以及病毒基因组RNA (gRNA)的表达。Tirumuru等[32]观察到HeLa细胞中m6A识别蛋白YTHDF1-3的过表达使HIV-1感染被抑制,而在YTHDF1-3沉默后,增加了HIV-1的感染,尤其是YTHDF2,其过表达对HIV-1感染的抑制效果与叠氮胸苷(AZT)对HIV-1的抑制效果相近,推测YTHDF1-3可能通过抑制HIV-1的逆转录活性来抑制HIV-1的感染。Lu等[33]对m6A识别蛋白YTHDF1-3抑制HIV-1的逆转录进而抑制HIV-1的感染的机制进行了深入研究,发现YTHDF1-3降低了病毒gRNA的水平并抑制了早期和晚期的逆转录产物的产生,推测YTHDF1-3可能是通过影响病毒gRNA和逆转录产物的稳定性实现对HIV-1感染的调控。
2.1.2 m6A修饰对甲型流感病毒(IAV)复制的调控
IAV含有一个分段的、负性的单链RNA基因组,在其mRNA上发现了大约24个m6A修饰位点,其中8个位点集中在血凝素(HA) mRNA片段上[34]。Courtney等[35]发现,人肺上皮细胞系A549中关键的m6A甲基转移酶METTL3的突变能够抑制IAV复制,相反地,过表达m6A读取蛋白YTHDF2能够促进IAV复制和感染性病毒颗粒的产生。通过检测HA的mRNA和vRNA上的m6A位点并将其mRNA和vRNA上的甲基化位点突变,作者发现HA基因表达水平和IAV致病性降低。尽管实验结果表明m6A修饰能够增强IAV的复制,但具体的作用机制还有待进一步研究。
2.1.3 m6A修饰对寨卡病毒(ZIKV)复制的调控
ZIKV的RNA中含有丰富的m6A修饰位点,其复制受宿主甲基转移酶METTL3和METTL14以及去甲基化酶ALKBH5和FTO的调控,当METTL3和METTL14的表达量降低时,ZIKV的复制增加,而将ALKBH5和FTO沉默,会减少ZIKV的复制[36]。研究人员在分析读取蛋白YTHDF1-3对ZIKV复制的影响时发现,相较于YTHDF1和YTHDF3,YTHDF2的沉默引起ZIKV复制增加的程度最大;同样,YTHDF2的过表达降低ZIKV RNA的表达水平的效果最显著,进一步研究发现YTHDF2可能通过促进病毒裂解转录本的降解来抑制ZIKV的裂解复制,这表明m6A修饰在ZIKV病毒的复制过程中起负调控作用(图2)。
图2
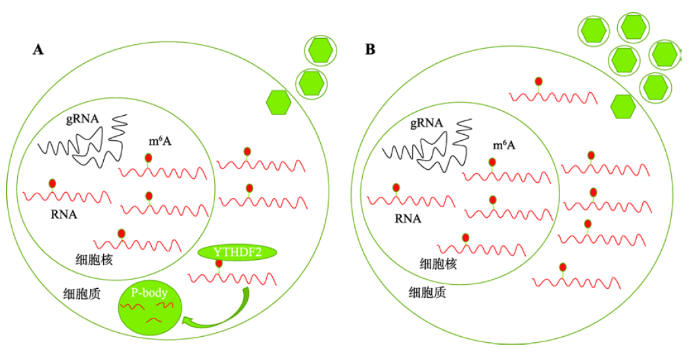 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2m6A修饰在ZIKV裂解增殖中的作用
A:正常情况下,发生m6A修饰的裂解转录本会被YTHDF2识别并降解,产生的病毒粒子较少;B:在YTHDF2沉默的情况下,发生m6A修饰的裂解转录本的降解减少,产生的病毒粒子增多。gRNA是ZIKV的基因组RNA。
Fig.2The role of m6A in lytic replication of ZIKV
2.1.4 m6A修饰对丙型肝炎病毒(HCV)复制的调控
Gokhale等[37]在研究m6A修饰对HCV复制的影响时发现,METTL3和METTL14的表达量的变化对HCV RNA的复制和翻译没有影响,但能调控感染性HCV病毒粒子的产生及释放,并且这种调控作用是与m6A修饰丰度是负相关的。此外,在HCV感染过程中,YTHDF蛋白重新定位到脂滴——病毒装配位点,调节病毒粒子的产生,而YTHDF的沉默增加了感染性病毒颗粒的产生。HCV病毒E1基因上m6A修饰位点的突变,导致病毒RNA与核衣壳蛋白(或称核心蛋白)结合增加,而与YTHDF蛋白的结合减少,进而导致感染性病毒颗粒的增加,因此,Gokhale等[37]推测YTHDF蛋白可能通过与病毒RNA竞争核衣壳蛋白来抑制感染性病毒粒子的产生。
2.2 m6A修饰对DNA病毒复制的调控
2.2.1 m6A修饰对乙型肝炎病毒(HBV)复制的调控与HCV不同,HBV是一种DNA病毒,通过一种称为前基因组RNA (pgRNA)的中间RNA完成其生命周期。Imam等[38]研究发现,pgRNA 5′端ε茎环和HBV转录本3′端都存在m6A修饰。METTL3和METTL14的沉默导致HBc和HBs两种病毒蛋白的表达增加,而ALKBH5和FTO的沉默降低了HBV病毒蛋白的表达。同样地,YTHDF2或YTHDF3的沉默显著延长了HBV转录本的半衰期,并增加了HBV蛋白HBs和HBc的表达,表明YTHDF蛋白也负向调控HBV蛋白的表达。在HBV转录本中,pgRNA 5′端茎环上的m6A修饰对pgRNA的逆转录起着正向调控的作用,而HBV 转录本3′端茎环上的m6A修饰对HBV RNA的稳定性进行负向调控,这表明m6A修饰位点的差异性可引起后续调控功能的差异。
2.2.2 m6A修饰对卡波氏肉瘤相关疱疹病毒(KSHV)复制的调控
KSHV在宿主细胞中建立潜伏感染仅需表达几个潜伏基因。当潜伏感染的细胞被重新激活时,进行裂解复制,表达大多数病毒基因并产生病毒粒子[39]。m6A修饰的阻断抑制了编码关键KSHV裂解开关蛋白复制转录激活因子(RTA)的pre-mRNA的剪接,阻断了病毒的裂解复制。RTA诱导m6A并增强其自身的pre-mRNA剪接。Ye等[39]的实验结果不仅证明了m6A在调控RTA pre-mRNA剪接中的重要作用,而
且表明KSHV已经进化出一种通过操纵宿主m6A修饰,来使其在促进裂解复制方面发挥优势的机制。Tan等[40]过表达YTHDF3,发现病毒转录本的水平下降但相关病毒蛋白的水平没有显著变化。KiSLK细胞中YTHDF2沉默导致病毒粒子的产生增加到了原来的4倍,同时病毒mRNA ORF50、ORF57、ORFK8和ORF65的表达量上升了2~6倍进而导致蛋白质水平的升高,而YTHDF2过表达能够扭转这些影响,这表明YTHDF2可能通过促进病毒裂解转录本的降解来抑制KSHV的裂解复制,而这可能是细胞抑制病毒复制的一种防御机制。Hesser等[41]报道,在感染致癌的人类DNA病毒卡波西肉瘤相关疱疹病毒(KSHV)的细胞中,m6A水平显著升高,但发现m6A修饰机制的沉默对不同细胞中病毒基因的表达有不同的影响。在iSLK.219和iSLK.BAC16细胞中,METTL3和YTHDF2的沉默使病毒裂解转激活因子ORF50发生转录后积累,进而显著减少了病毒粒子的产生,这表明m6A起着促进病毒产生的作用。相比之下,在KSHV感染的B细胞中,METTL3或YTHDF2缺失时,ORF50蛋白表达增加,进而增加了病毒粒子的产生。虽然结果存在差异,但却都表明m6A修饰通过调控ORF50来调控KSHV的裂解复制。
2.2.3 m6A修饰对猴空泡病毒40 (SV40)复制的调控
SV40是一种DNA病毒,属于多瘤病毒家族。Tsai等[42]研究发现,过表达m6A读取蛋白质YTHDF2能够诱发更快速的病毒复制,而YTHDF2基因的突变失活或METTL3的沉默则会抑制病毒的复制。利用同义突变,使SV40晚期mRNA上的大多数m6A修饰位点发生突变,可以观察到突变型SV40比野生型SV40复制速度减慢,相较于野生型病毒,突变型病毒的转录本水平并没有显著变化,而VP1蛋白水平显著下降,因而Tsai等[42]推测m6A修饰主要通过促进晚期转录本的翻译来增强病毒基因表达,进而促进SV40病毒的复制。
2.3 m6A修饰对植物病毒复制的调控
植物病毒对于植物的正常生长与发育构成重大威胁。m6A表观修饰不仅发生在影响动物的病毒中,在植物病毒中也有报道。Martinez-Perez等[43]在拟南芥多功能紫花苜蓿花叶病毒(AMV)与衣壳蛋白(CP)相互作用的酵母双杂交筛选中,鉴定出了去甲基化酶AlkB家族成员atALKBH9B。在雀麦花叶病毒科的两个成员,AMV和CMV(黄瓜花叶病毒)的基因组中也发现了m6A的存在,并发现atALKBH9B的突变导致AMV基因组中m6A水平上升了35%,而超甲基化降低了AMV的感染效率。进一步的研究发现atALKBH9B与小干扰RNA(siRNA)的组成部分SGS3和处理小体(P-body)中的一种降解酶DCP1完全重叠,这表明m6A修饰可能通过引发病毒mRNA的沉默和衰减来负向调节AMV的复制。3 m6A修饰对免疫反应的影响
免疫系统是高等生物抵御病毒侵染的重要屏障。目前,对于m6A在免疫系统中的作用及其在宿主病原体相互作用中的作用知之甚少。先天免疫提供对病毒感染的第一反应,入侵的病原体核酸被细胞质甲酸诱导基因Ⅰ (RIG-Ⅰ)样受体和膜结合的Toll样受体(TLRs)识别并结合,从而激活先天免疫反应。先天免疫必须区分宿主和病原体的核酸,以便在不激活自身免疫反应的情况下产生保护性免疫反应。当修饰过的核苷酸存在于RNA分子中时,会降低先天免疫反应的幅度,如含有m6A修饰的病毒RNA与RIG-Ⅰ结合较差,因而无法触发强烈的先天免疫反应[44]。Karikó等[45]的研究表明,经过m6A修饰的RNA刺激DC细胞只能引起少量的细胞因子的释放,同样地,当甲基化的RNA刺激表达TLRs的细胞时,也只引起少量免疫激活标志物的释放,而无法触发强烈的先天免疫反应。Lichinchi等[30]用HIV-1 (LAI病毒株)感染CD4+ T细胞,发现细胞中总RNA中m6A的水平上升了约30%,这表明HIV-1感染T细胞可促进病毒和宿主RNA的甲基化,从而抑制免疫监视或扰乱宿主遗传网络,从而成功复制[46]。此外,ZIKV在潜伏期时,通过提高自身m6A水平来逃避宿主细胞的免疫监视,在裂解复制期间,ZIKV可动态修改宿主免疫相关转录本,通过篡夺m6A修饰机制来抑制宿主的抗病毒反应[10]。在IAV感染过程中过表达YTHDF2可提高感染性病毒粒子的释放,而YTHDF2介导的mRNA降解可能降低宿主抗病毒基因转录本,从而增强病毒复制[35]。总之,病毒通过借助宿主的m6A修饰机制,来逃避免疫系统的监察或抑制免疫系统的功能等,使病毒自身能够存活。4 结语与展望
随着对m6A修饰机制研究的逐渐深入,发现其对RNA水平的调控作用变得更加复杂和多样。m6A修饰的调控功能通过读取蛋白来执行,一方面,读取蛋白能够促进经m6A修饰的mRNA的翻译,另一方面,也会通过降低靶mRNA的稳定性而促进其降解,有关这两种调控功能的选择的机制还有待研究。关于m6A修饰在病毒复制过程中的调控机制的研究表明,m6A修饰对病毒复制的调控是通过影响病毒mRNA或基因组RNA的稳定性来实现的,众多研究都证实了读取蛋白YTHDF2对病毒复制的影响,实验结果也表明YTHDF2的过表达能抑制多种病毒的复制。另外,也有实验表明YTHDF2能够促进病毒的复制。这些研究结果提示可以将靶向调控m6A修饰的药物作为抗病毒药物的开发方向,如3-脱氮腺苷(3-deazaadenosine, DAA)已经被证明可以通过阻断的S-腺苷高半胱氨酸(SAC)的水解,使S-腺苷甲硫氨酸(SAM)降解,从而阻断mRNA底物中m6A的加入[10]。Kennedy等[31]用DAA处理HIV-1感染的CEM-SS细胞,发现HIV-1的复制受到抑制,证明DAA对HIV-1复制的有效抑制作用。同时,有报道称DAA可以抑制RSV、IAV等病毒的复制[47,48]。在研究m6A修饰对病毒复制的调控时,也应对m6A修饰的动态变化对宿主细胞的影响给予高度关注,这或许可以为新型抗病毒药物的开发提供参考。此外,病毒RNA的高甲基化使部分病毒成功躲过免疫系统的监察,那么生物体内是否存在某些机制能够监测到这些异常的甲基化,进而反馈给免疫系统进行处理,这有待进一步的研究。(责任编委: 岑山)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:4372599 [本文引用: 1]

The poly(A) tract found in eukaryotic mRNA was used to study methylation in mRNA obtained from Novikoff hepatoma cells. Methyl labeling of RNA was achieved with L-[methyl-3H]methionine under conditions that suppress radioactive incorporation into the purine ring. RNA that contains a poly(A) segment was obtained from polysomal RNA by chromatography on oligo(dT)-cellulose. Sucrose density gradient centrifugation of this RNA revealed a pattern expected for mRNA. The composition of the methyl-labeld nucleosides in the RNA was analyzed after complete enzymatic degradation to nucleosides. By use of DEAE-cellulose (borate) chromatography, which separates 2′-O-methylnucleosides from normal and base-methylated nucleosides, about 50% of the radioactivity was recovered in the 2′-O-methylnucleoside fraction and 50% in the base-methylnucleoside fraction. High-speed liquid chromatography (Aminex A-5) of the 2′-O-methylnucleoside fraction produced four peaks coincident with the four 2′-O-methylnucleoside standards. Analysis of the base-methylnucleoside fraction revealed a unique pattern. While ribosomal RNA and tRNA possessed complex base-methylnucleoside patterns, the distribution in mRNA was quite simple, consisting predominantly of N6-methyladenosine. These results demonstrate a unique distribution of methylated nucleosides in mRNA. By analogy to ribosomal RNA synthesis, the presence of methylnucleosides in mRNA may reflect a cellular mechanism for the selective processing of certain mRNA sequences.
URL [本文引用: 1]

Messenger RNA of mouse L cells is methylated in both base and ribose moieties. On the average there are about 2.2 methyl groups per 1000 nucleotides in mRNA, a proportion which is about one-sixth that of mammalian ribosomal RNA. The variety of methylated bases in mRNA is more limited than in ribosomal RNA. A very low level of methylation is detected in heterogeneous nuclear RNA, suggesting that methylation, like polyadenylation, may constitute a post-transcriptional modification of messenger RNA precursor in eucaryotic cells.
URLPMID:24662220 [本文引用: 1]

Cellular RNAs carry diverse chemical modifications that used to be regarded as static and having minor roles in 'fine-tuning' structural and functional properties of RNAs. In this Review, we focus on reversible methylation through the most prevalent mammalian mRNA internal modification, N6-methyladenosine (m6A). Recent studies have discovered protein 'writers', 'erasers' and 'readers' of this RNA chemical mark, as well as its dynamic deposition on mRNA and other types of nuclear RNA. These findings strongly indicate dynamic regulatory roles that are analogous to the well-known reversible epigenetic modifications of DNA and histone proteins. This reversible RNA methylation adds a new dimension to the developing picture of post-transcriptional regulation of gene expression.
URLMagsci [本文引用: 1]

<p>RNA酶促共价修饰研究, 尤其是m<sup>6</sup>A(6-甲基腺嘌呤), 是RNA生物学研究的一个新兴领域。m<sup>6</sup>A是真核生物mRNA内部序列中最常见的一种转录后修饰形式, 由包含3个独立组分的复合物mRNA: m<sup>6</sup>A甲基转移酶催化生成。最新研究发现肥胖相关蛋白FTO可以脱掉m<sup>6</sup>A上的甲基, 表明该甲基化过程是可逆的。抑制或敲除m<sup>6</sup>A甲基转移酶会引起重要的表型变化, 但是由于过去的检测方法受限, m<sup>6</sup>A确切的作用机制目前为止还不甚清楚。二代测序技术结合免疫沉淀方法为大规模检测m<sup>6</sup>A修饰并研究其作用机制提供了可能。文章主要综述了m<sup>6</sup>A的发现史、生成机制、组织和基因组分布、检测方法、生物学功能等及其最新研究进展, 并通过比较3种IP-seq技术和数据分析的异同及优缺点, 对m<sup>6</sup>A这种RNA表观修饰研究中尚未解决的问题进行了讨论。</p>
URLMagsci [本文引用: 1]

<p>RNA酶促共价修饰研究, 尤其是m<sup>6</sup>A(6-甲基腺嘌呤), 是RNA生物学研究的一个新兴领域。m<sup>6</sup>A是真核生物mRNA内部序列中最常见的一种转录后修饰形式, 由包含3个独立组分的复合物mRNA: m<sup>6</sup>A甲基转移酶催化生成。最新研究发现肥胖相关蛋白FTO可以脱掉m<sup>6</sup>A上的甲基, 表明该甲基化过程是可逆的。抑制或敲除m<sup>6</sup>A甲基转移酶会引起重要的表型变化, 但是由于过去的检测方法受限, m<sup>6</sup>A确切的作用机制目前为止还不甚清楚。二代测序技术结合免疫沉淀方法为大规模检测m<sup>6</sup>A修饰并研究其作用机制提供了可能。文章主要综述了m<sup>6</sup>A的发现史、生成机制、组织和基因组分布、检测方法、生物学功能等及其最新研究进展, 并通过比较3种IP-seq技术和数据分析的异同及优缺点, 对m<sup>6</sup>A这种RNA表观修饰研究中尚未解决的问题进行了讨论。</p>
URLPMID:27808276 [本文引用: 1]

The recent discovery of reversible mRNA methylation has opened a new realm of post-transcriptional gene regulation in eukaryotes. The identification and functional characterization of proteins that specifically recognize RNA N(6)-methyladenosine (m(6)A) unveiled it as a modification that cells utilize to accelerate mRNA metabolism and translation. N(6)-adenosine methylation directs mRNAs to distinct fates by grouping them for differential processing, translation and decay in processes such as cell differentiation, embryonic development and stress responses. Other mRNA modifications, including N(1)-methyladenosine (m(1)A), 5-methylcytosine (m(5)C) and pseudouridine, together with m(6)A form the epitranscriptome and collectively code a new layer of information that controls protein synthesis.
URLPMID:28499622 [本文引用: 1]

Abstract Modifications in mRNA constitute ancient mechanisms to regulate gene expression post-transcriptionally. N 6 -methyladenosine (m 6 A) is the most prominent mRNA modification, and is installed by a large methyltransferase complex (the m 6 A 'writer'), not only specifically bound by RNA-binding proteins (the m 6 A 'readers'), but also removed by demethylases (the m 6 A 'erasers'). m 6 A mRNA modifications have been linked to regulation at multiple steps in mRNA processing. In analogy to the regulation of gene expression by miRNAs, we propose that the main function of m 6 A is post-transcriptional fine-tuning of gene expression. In contrast to miRNA regulation, which mostly reduces gene expression, we argue that m 6 A provides a fast mean to post-transcriptionally maximize gene expression. Additionally, m 6 A appears to have a second function during developmental transitions by targeting m 6 A-marked transcripts for degradation. Copyright 漏 2017 Elsevier Ltd. All rights reserved.
URLPMID:27179969 [本文引用: 1]

N (6)-methyladenosine (m(6)A) is the most abundant and reversible internal modification ubiquitously occurring in eukaryotic mRNA, albeit the significant biological roles of m(6)A methylation have remained largely unclear. The well-known DNA and histone methylations play crucial roles in epigenetic modification of biologic processes in eukaryotes. Analogously, the dynamic and reversible m(6)A RNA modification, which is installed by methyltransferase (METTL3, METTL14, and WTAP), reversed by demethylases (FTO, ALKBH5) and mediated by m(6)A-binding proteins (YTHDF1-3, YTHDC1), may also have a profound impact on gene expression regulation. Recent discoveries of the distributions, functions, and mechanisms of m(6)A modification suggest that this methylation functionally modulates the eukaryotic transcriptome to influence mRNA transcription, splicing, nuclear export, localization, translation, and stability. This reversible mRNA methylation shed light on a new dimension of post-transcriptional regulation of gene expression at the RNA level. m(6)A methylation also plays significant and broad roles in various physiological processes, such as development, fertility, carcinogenesis, stemness, early mortality, meiosis and circadian cycle, and links to obesity, cancer, and other human diseases. This review mainly describes the current knowledge of m(6)A and perspectives on future investigations.
[本文引用: 1]
URLPMID:1255846 [本文引用: 1]

RNA labeled with [methyl-3H] methionine and [14C]uridine was isolated from the cytoplasm of adenovirus-infected cells and purified by poly(U)-Sepharose chromatography and hybridization to filters containing immobilized adeovirus DNA. Analysis by dimethyl sulfoxide-sucrose gradient sedimentation suggested that the major mRNA species were methylated. 7-Methylguanosine was identified at the 5'-terminus of the advenovirus-specific RNA and could be removed by periodate oxidation and beta-elimination. Structures of the type m7G(5')ppp(5')Nm containing the unusual nucleoside N6, O2'-dimethyladenosine, and smaller amounts of 2'-O-methyladenosine were isolated by DEAE-cellulose chromatography after P1 nuclease digestion of the RNA. Evidence for some 5'-terminal sequences, m7G(5')ppp(5')m6AmpNm, with additional 2'-O-methylribonucleosides was also obtained. A base-methylated nucleoside, N6-methyladenosine, is located within the RNA chain and is released as a mononucleotide by alkali hydrolysis.
[本文引用: 3]
[本文引用: 1]
[本文引用: 1]
URLPMID:4142486 [本文引用: 1]

N6-methyladenosine (m6A) is a highly abundant modification of mRNA. Schwartz et聽al. identify and validate a network of proteins required for mRNA methylation in mammalian cells. They define two distinct classes of methylation sites. The majority of sites depend on the identified proteins, are located at internal positions in transcripts, and inversely correlate with mRNA stability. Sites independent of these proteins form at the first transcribed base as part of the cap structure, forming a previously unappreciated layer of transcriptome complexity.
URLPMID:3829175 [本文引用: 1]

Background: WTAP is a ubiquitously expressed nuclear protein that is required for mammalian early embryo development and cell cycle progression. Results: WTAP forms a complex with several splicing regulators. Conclusion: WTAP regulates both the cell cycle and alternative splicing by the formation of a protein complex. Significance: Characterization of this protein complex will help to elucidate the critically important function of WTAP in alternative splicing and cell proliferation.Wilms' tumor 1-associating protein (WTAP) is a putative splicing regulator that is thought to be required for cell cycle progression through the stabilization of cyclin A2 mRNA and mammalian early embryo development. To further understand how WTAP acts in the context of the cellular machinery, we identified its interacting proteins in human umbilical vein endothelial cells and HeLa cells using shotgun proteomics. Here we show that WTAP forms a novel protein complex including Hakai, Virilizer homolog, KIAA0853, RBM15, the arginine/serine-rich domain-containing proteins BCLAF1 and THRAP3, and certain general splicing regulators, most of which have reported roles in post-transcriptional regulation. The depletion of these respective components of the complex resulted in reduced cell proliferation along with G(2)/M accumulation. Double knockdown of the serine/arginine-rich (SR)-like proteins BCLAF1 and THRAP3 by siRNA resulted in a decrease in the nuclear speckle localization of WTAP, whereas the nuclear speckles were intact. Furthermore, we found that the WTAP complex regulates alternative splicing of the WTAP pre-mRNA by promoting the production of a truncated isoform, leading to a change in WTAP protein expression. Collectively, these findings show that the WTAP complex is a novel component of the RNA processing machinery, implying an important role in both posttranscriptional control and cell cycle regulation.
URLPMID:28525753 [本文引用: 1]

Maintenance of proper levels of the methyl donor S-adenosylmethionine (SAM) is critical for a wide variety of biological processes. We demonstrate that the N 6 -adenosine methyltransferase METTL16 regulates expression of human MAT2A, which encodes the SAM synthetase expressed in most cells. Upon SAM depletion by methionine starvation, cells induce MAT2A expression by enhanced splicing of a retained intron. Induction requires METTL16 and its methylation substrate, a vertebrate conserved hairpin (hp1) in the MAT2A 3′ UTR. Increasing METTL16 occupancy on the MAT2A 3′ UTR is sufficient to induce efficient splicing. We propose that, under SAM-limiting conditions, METTL16 occupancy on hp1 increases due to inefficient enzymatic turnover, which promotes MAT2A splicing. We further show that METTL16 is the long-unknown methyltransferase for the U6 spliceosomal small nuclear RNA (snRNA). These observations suggest that the conserved U6 snRNA methyltransferase evolved an additional function in vertebrates to regulate SAM homeostasis.
URLPMID:22002720 [本文引用: 1]

Abstract We report here that fat mass and obesity-associated protein (FTO) has efficient oxidative demethylation activity targeting the abundant N6-methyladenosine (m(6)A) residues in RNA in vitro. FTO knockdown with siRNA led to increased amounts of m(6)A in mRNA, whereas overexpression of FTO resulted in decreased amounts of m(6)A in human cells. We further show the partial colocalization of FTO with nuclear speckles, which supports the notion that m(6)A in nuclear RNA is a major physiological substrate of FTO.
URLPMID:23177736 [本文引用: 1]

78 ALKBH5 is a mammalian m6A RNA demethylase 78 RNA demethylation affects mRNA export and RNA metabolism 78 RNA demethylation is important for mouse fertility 78 Reversible mammalian messenger RNA methylation affects gene expression
URLPMID:24489119 [本文引用: 1]

ALKBH5 is a 2-oxoglutarate (2OG) and ferrous iron-dependent nucleic acid oxygenase (NAOX) that catalyzes the demethylation of N(6)-methyladenine in RNA. ALKBH5 is upregulated under hypoxia and plays a role in spermatogenesis. We describe a crystal structure of human ALKBH5 (residues 66-292) to 2.0 03 resolution. ALKBH5646469606760 has a double-stranded β-helix core fold as observed in other 2OG and iron-dependent oxygenase family members. The active site metal is octahedrally coordinated by an HXD…H motif (comprising residues His204, Asp206 and His266) and three water molecules. ALKBH5 shares a nucleotide recognition lid and conserved active site residues with other NAOXs. A large loop (βIV-V) in ALKBH5 occupies a similar region as the L1 loop of the fat mass and obesity-associated protein that is proposed to confer single-stranded RNA selectivity. Unexpectedly, a small molecule inhibitor, IOX3, was observed covalently attached to the side chain of Cys200 located outside of the active site. Modelling substrate into the active site based on other NAOX-nucleic acid complexes reveals conserved residues important for recognition and demethylation mechanisms. The structural insights will aid in the development of inhibitors selective for NAOXs, for use as functional probes and for therapeutic benefit.
URLPMID:5513158 [本文引用: 1]

Abstract Internal bases in mRNA can be subjected to modifications that influence the fate of mRNA in cells. One of the most prevalent modified bases is found at the 5' end of mRNA, at the first encoded nucleotide adjacent to the 7-methylguanosine cap. Here we show that this nucleotide, N 6 ,2'-O-dimethyladenosine (m 6 A m ), is a reversible modification that influences cellular mRNA fate. Using a transcriptome-wide map of m 6 A m we find that m 6 A m -initiated transcripts are markedly more stable than mRNAs that begin with other nucleotides. We show that the enhanced stability of m 6 A m -initiated transcripts is due to resistance to the mRNA-decapping enzyme DCP2. Moreover, we find that m 6 A m is selectively demethylated by fat mass and obesity-associated protein (FTO). FTO preferentially demethylates m 6 A m rather than N 6 -methyladenosine (m 6 A), and reduces the stability of m 6 A m mRNAs. Together, these findings show that the methylation status of m 6 A m in the 5' cap is a dynamic and reversible epitranscriptomic modification that determines mRNA stability.
URLPMID:29103884 [本文引用: 1]

Abstract N 6 -Methyladenosine (m 6 A) is the most prevalent post-transcriptional modification of eukaryotic mRNA and long noncoding RNA. m 6 A mediates its effects primarily by recruiting proteins, including the multiprotein eukaryotic initiation factor 3 complex and a set of proteins that contain the YTH domain. Here we describe the mechanisms by which YTH domain-containing proteins bind m 6 A and influence the fate of m 6 A-containing RNA in mammalian cells. We discuss the diverse, and occasionally contradictory, functions ascribed to these proteins and the emerging concepts that are influencing our understanding of these proteins and their effects on the epitranscriptome.
URLPMID:24284625 [本文引用: 1]

Abstract N(6)-methyladenosine (m(6)A) is the most prevalent internal (non-cap) modification present in the messenger RNA of all higher eukaryotes. Although essential to cell viability and development, the exact role of m(6)A modification remains to be determined. The recent discovery of two m(6)A demethylases in mammalian cells highlighted the importance of m(6)A in basic biological functions and disease. Here we show that m(6)A is selectively recognized by the human YTH domain family 2 (YTHDF2) 'reader' protein to regulate mRNA degradation. We identified over 3,000 cellular RNA targets of YTHDF2, most of which are mRNAs, but which also include non-coding RNAs, with a conserved core motif of G(m(6)A)C. We further establish the role of YTHDF2 in RNA metabolism, showing that binding of YTHDF2 results in the localization of bound mRNA from the translatable pool to mRNA decay sites, such as processing bodies. The carboxy-terminal domain of YTHDF2 selectively binds to m(6)A-containing mRNA, whereas the amino-terminal domain is responsible for the localization of the YTHDF2-mRNA complex to cellular RNA decay sites. Our results indicate that the dynamic m(6)A modification is recognized by selectively binding proteins to affect the translation status and lifetime of mRNA.
URL [本文引用: 1]
URLPMID:5509218 [本文引用: 1]

The long non-coding RNA X-inactive specific transcript (XIST) mediates the transcriptional silencing of genes on the X chromosome. Here we show that, in human cells, XIST is highly methylated with at least 78 N-methyladenosine (mA) residues鈥攁 reversible base modification of unknown function in long non-coding RNAs. We show that mA formation in XIST, as well as in cellular mRNAs, is mediated by RNA-binding motif protein 15 (RBM15) and its paralogue RBM15B, which bind the mA-methylation complex and recruit it to specific sites in RNA. This results in the methylation of adenosine nucleotides in adjacent mA consensus motifs. Furthermore, we show that knockdown of RBM15 and RBM15B, or knockdown of methyltransferase like 3 (METTL3), an mA methyltransferase, impairs XIST-mediated gene silencing. A systematic comparison of mA-binding proteins shows that YTH domain containing 1 (YTHDC1) preferentially recognizes mA residues on XIST and is required for XIST function. Additionally, artificial tethering of YTHDC1 to XIST rescues XIST-mediated silencing upon loss of mA. These data reveal a pathway of mA formation and recognition required for XIST-mediated transcriptional repression.
[本文引用: 1]
URLPMID:20202062020202020202020 [本文引用: 1]

N(6)-methyladenosine (m(6)A) has been identified as the most abundant internal modification of messenger RNA in eukaryotes. m(6)A modification is involved in cell fate determination in yeast and embryo development in plants. Its mammalian function remains unknown but thousands of mammalian mRNAs and long non-coding RNAs (lncRNAs) show m(6)A modification and m(6)A demethylases are required for mammalian energy homeostasis and fertility. We identify two proteins, the putative m(6)A MTase, methyltransferase-like 3 (Mettl3; ref. ), and a related but uncharacterized protein Mettl14, that function synergistically to control m(6)A formation in mammalian cells. Knockdown of Mettl3 and Mettl14 in mouse embryonic stem cells (mESCs) led to similar phenotypes, characterized by lack of m(6)A RNA methylation and lost self-renewal capability. A large number of transcripts, including many encoding developmental regulators, exhibit m(6)A methylation inversely correlated with mRNA stability and gene expression. The human antigen R (HuR) and microRNA pathways were linked to these effects. This gene regulatory mechanism operating in mESCs through m(6)A methylation is required to keep mESCs at their ground state and may be relevant to thousands of mRNAs and lncRNAs in various cell types.
URLPMID:27558897 [本文引用: 1]

Methylation at the N6 position of adenosine (m(6)A) is the most abundant RNA modification within protein-coding and long noncoding RNAs in eukaryotes and is a reversible process with important biological functions. YT521-B homology domain family (YTHDF) proteins are the readers of m(6)A, the binding of which results in the alteration of the translation efficiency and stability of m(6)A-containing RNAs. However, the mechanism by which YTHDF proteins cause the degradation of m(6)A-containing RNAs is poorly understood. Here we report that m(6)A-containing RNAs exhibit accelerated deadenylation that is mediated by the CCR4-NOT deadenylase complex. We further show that YTHDF2 recruits the CCR4-NOT complex through a direct interaction between the YTHDF2 N-terminal region and the SH domain of the CNOT1 subunit, and that this recruitment is essential for the deadenylation of m(6)A-containing RNAs by CAF1 and CCR4. Therefore, we have uncovered the mechanism of YTHDF2-mediated degradation of m(6)A-containing RNAs in mammalian cells.
URLPMID:4851248 [本文引用: 1]

The most abundant mRNA post-transcriptional modification isN6-methyladenosine (m6A) that has broad roles in RNA biology1-5. In mammalian cells, the asymmetric distribution of m6A along mRNAs leaves relatively less methylation in the 5′ untranslated region (5′UTR) compared to other regions6,7. However, whether and how 5′UTR methylation is regulated is poorly understood. Despite the crucial role of the 5′UTR in translation initiation, very little is known whether m6A modification influences mRNA translation. Here we show that in response to heat shock stress, m6A is preferentially deposited to the 5′UTR of newly transcribed mRNAs. We found that the dynamic 5′UTR methylation is a result of stress-induced nuclear localization of YTHDF2, a well characterized m6A “reader”. Upon heat shock stress, the nuclear YTHDF2 preserves 5′UTR methylation of stress-induced transcripts by limiting the m6A “eraser” FTO from demethylation. Remarkably, the increased 5′UTR methylation in the form of m6A promotes cap-independent translation initiation, providing a mechanism for selective mRNA translation under heat shock stress. Using Hsp70 mRNA as an example, we demonstrate that a single site m6A modification in the 5′UTR enables translation initiation independent of the 5′ end m7G cap. The elucidation of the dynamic feature of 5′UTR methylation and its critical role in cap-independent translation not only expands the breadth of physiological roles of m6A, but also uncovers a previously unappreciated translational control mechanism in heat shock response.
URLPMID:26876937 [本文引用: 1]

Xiao et02al. show that m6A reader YTHDC1 promotes exon inclusion of targeted mRNAs through facilitating SRSF3 while blocking SRSF10 mRNA binding, demonstrating how m6A reader YTHDC1 directly regulates mRNA splicing by bridging interactions oftrans- andcis-regulatory elements.
URL [本文引用: 1]

表观遗传学修饰包括DNA、RNA和蛋白质的化学修饰,基于非序列改变所致基因表达和功能水平变化。近年来,在DNA和蛋白质修饰基础上,可逆RNA甲基化修饰研究引领了第3次表观遗传学修饰研究的浪潮。RNA存在100余种化学修饰,甲基化是最主要的修饰形式。鉴定RNA甲基化修饰酶及研发其转录组水平高通量检测技术,是揭示RNA化学修饰调控基因表达和功能规律的基础。本文主要总结了近年来本课题组与合作团队及国内外同行在RNA甲基化表观转录组学研究中取得的主要前沿进展,包括发现了RNA去甲基酶、甲基转移酶和结合蛋白,揭示RNA甲基化修饰调控RNA加工代谢,及其调控正常生理和异常病理等重要生命进程。这些系列研究成果证明RNA甲基化修饰类似于DNA甲基化,具有可逆性,拓展了RNA甲基化表观转录组学研究新领域,完善了中心法则表观遗传学规律。
URL [本文引用: 1]

表观遗传学修饰包括DNA、RNA和蛋白质的化学修饰,基于非序列改变所致基因表达和功能水平变化。近年来,在DNA和蛋白质修饰基础上,可逆RNA甲基化修饰研究引领了第3次表观遗传学修饰研究的浪潮。RNA存在100余种化学修饰,甲基化是最主要的修饰形式。鉴定RNA甲基化修饰酶及研发其转录组水平高通量检测技术,是揭示RNA化学修饰调控基因表达和功能规律的基础。本文主要总结了近年来本课题组与合作团队及国内外同行在RNA甲基化表观转录组学研究中取得的主要前沿进展,包括发现了RNA去甲基酶、甲基转移酶和结合蛋白,揭示RNA甲基化修饰调控RNA加工代谢,及其调控正常生理和异常病理等重要生命进程。这些系列研究成果证明RNA甲基化修饰类似于DNA甲基化,具有可逆性,拓展了RNA甲基化表观转录组学研究新领域,完善了中心法则表观遗传学规律。
[本文引用: 1]
URLPMID:27572442 [本文引用: 2]

Abstract N(6)-methyladenosine (m(6)A) is the most prevalent internal modification of eukaryotic mRNA. Very little is known of the function of m(6)A in the immune system or its role in host-pathogen interactions. Here, we investigate the topology, dynamics and bidirectional influences of the viral-host RNA methylomes during HIV-1 infection of human CD4 T cells. We show that viral infection triggers a massive increase in m(6)A in both host and viral mRNAs. In HIV-1 mRNA, we identified 14 methylation peaks in coding and noncoding regions, splicing junctions and splicing regulatory sequences. We also identified a set of 56 human gene transcripts that were uniquely methylated in HIV-1-infected T cells and were enriched for functions in viral gene expression. The functional relevance of m(6)A for viral replication was demonstrated by silencing of the m(6)A writer or the eraser enzymes, which decreased or increased HIV-1 replication, respectively. Furthermore, methylation of two conserved adenosines in the stem loop II region of HIV-1 Rev response element (RRE) RNA enhanced binding of HIV-1 Rev protein to the RRE in vivo and influenced nuclear export of RNA. Our results identify a new mechanism for the control of HIV-1 replication and its interaction with the host immune system.
URLPMID:29241043 [本文引用: 2]

Kennedy et02al. show that the HIV-1 RNA genome contains specific m6A editing sites, located in the viral 3′ UTR, that enhance RNA expression and function. These m6A residues function by recruiting cellular YTHDF proteins, and inhibiting YTHDF binding to viral RNAs therefore inhibits viral replication.
URLPMID:27371828 [本文引用: 1]

10.7554/eLife.15528.001The internal N6-methyladenosine (m6A) methylation of eukaryotic nuclear RNA controls post-transcriptional gene expression, which is regulated by methyltransferases (writers), demethylases (erasers), and m6A-binding proteins (readers) in cells. The YTH domain family proteins (YTHDF1???3) bind to m6A-modified cellular RNAs and affect RNA metabolism and processing. Here, we show that YTHDF1???3 proteins recognize m6A-modified HIV-1 RNA and inhibit HIV-1 infection in cell lines and primary CD4+ T-cells. We further mapped the YTHDF1???3 binding sites in HIV-1 RNA from infected cells. We found that the overexpression of YTHDF proteins in cells inhibited HIV-1 infection mainly by decreasing HIV-1 reverse transcription, while knockdown of YTHDF1???3 in cells had the opposite effects. Moreover, silencing the m6A writers decreased HIV-1 Gag protein expression in virus-producing cells, while silencing the m6A erasers increased Gag expression. Our findings suggest an important role of m6A modification of HIV-1 RNA in viral infection and HIV-1 protein synthesis.DOI: http://dx.doi.org/10.7554/eLife.15528.001
URL [本文引用: 1]
URLPMID:3600638 [本文引用: 1]

Abstract Influenza virus mRNA is posttranscriptionally methylated at internal adenosine residues to form N6-methyladenosine (m6A). It has been previously shown that there is an average of three m6A residues per influenza virus mRNA (R. M. Krug, M. A. Morgan, and A. J. Shatkin, J. Virol. 20:45-53, 1976). To determine the distribution of m6A in the different influenza virus mRNAs, we purified six of the mRNAs by hybrid selection, digested them with nuclease, and determined their methylation patterns by high-pressure liquid chromatography. The amount of m6A in the different mRNAs varied from one in matrix to eight in hemagglutinin.
[本文引用: 2]
URLPMID:27773536 [本文引用: 1]

Lichinchi et聽al. show that Zika virus (ZIKV) RNA is modified at N6methylation of adenosine (m6A). m6A modifications in ZIKV RNA control viral replication and are regulated by host methyltransferases and demethylases. ZIKV infection alters the location of m6A in host mRNAs, methylation motifs, and target genes modified by methyltransferases.
URL [本文引用: 2]
URL [本文引用: 1]
URLPMID:3103228 [本文引用: 2]

The life cycle of Kaposi's sarcoma-associated herpesvirus (KSHV) consists of latent and lytic replication phases. During latent infection, only a limited number of KSHV genes are expressed. However, this phase of replication is essential for persistent infection, evasion of host immune response, and induction of KSHV-related malignancies. KSHV reactivation from latency produces a wide range of viral products and infectious virions. The resulting de novo infection and viral lytic products modulate diverse cellular pathways and stromal microenvironment, which promote the development of Kaposi's sarcoma (KS). The mechanisms controlling KSHV latency and reactivation are complex, involving both viral and host factors, and are modulated by diverse environmental factors. Here, we review the cellular and molecular basis of KSHV latency and reactivation with a focus on the most recent advancements in the field.
[本文引用: 1]
URL [本文引用: 1]
URLPMID:29447282 [本文引用: 2]

Abstract Polyomaviruses are a family of small DNA tumor viruses that includes several pathogenic human members, including Merkel cell polyomavirus, BK virus and JC virus. As is characteristic of DNA tumor viruses, gene expression in polyomaviruses is temporally regulated into an early phase, consisting of the viral regulatory proteins, and a late phase, consisting of the viral structural proteins. Previously, the late transcripts expressed by the prototypic polyomavirus simian virus 40 (SV40) were reported to contain several adenosines bearing methyl groups at the N6 position (m6A), although the precise location of these m6A residues, and their phenotypic effects, have not been investigated. Here, we first demonstrate that overexpression of the key m6A reader protein YTHDF2 induces more rapid viral replication, and larger viral plaques, in SV40 infected BSC40 cells, while mutational inactivation of the endogenous YTHDF2 gene, or the m6A methyltransferase METTL3, has the opposite effect, thus suggesting a positive role for m6A in the regulation of SV40 gene expression. To directly test this hypothesis, we mapped sites of m6A addition on SV40 transcripts and identified two m6A sites on the viral early transcripts and eleven m6A sites on the late mRNAs. Using synonymous mutations, we inactivated the majority of the m6A sites on the SV40 late mRNAs and observed that the resultant viral mutant replicated more slowly than wild type SV40. Alternative splicing of SV40 late mRNAs was unaffected by the reduction in m6A residues and our data instead suggest that m6A enhances the translation of viral late transcripts. Together, these data argue that the addition of m6A residues to the late transcripts encoded by SV40 plays an important role in enhancing viral gene expression and, hence, replication.
URL [本文引用: 1]
URLPMID:27651356 [本文引用: 1]

Invading pathogen nucleic acids are recognized and bound by cytoplasmic (retinoic acid-inducible gene I [RIG-I]-like) and membrane-bound (Toll-like) pattern recognition receptors to activate innate immune signaling. Modified nucleotides, when present in RNA molecules, diminish the magnitude of these signaling responses. However, mechanisms explaining the blunted signaling have not been elucidated. In this study, we used several independent biological assays, including inhibition of virus replication, RIG-I:RNA binding assays, and limited trypsin digestion of RIG-I:RNA complexes, to begin to understand how RNAs containing modified nucleotides avoid or suppress innate immune signaling. The experiments were based on a model innate immune activating RNA molecule, the polyU/UC RNA domain of hepatitis C virus, which was transcribedin vitrowith canonical nucleotides or with one of eight modified nucleotides. The approach revealed signature assay responses associated with individual modified nucleotides or classes of modified nucleotides. For example, while bothN-6-methyladenosine (m6A) and pseudouridine nucleotides correlate with diminished signaling, RNA containing m6A modifications bound RIG-I poorly, while RNA containing pseudouridine bound RIG-I with high affinity but failed to trigger the canonical RIG-I conformational changes associated with robust signaling. These data advance understanding of RNA-mediated innate immune signaling, with additional relevance for applying nucleotide modifications to RNA therapeutics. The innate immune system provides the first response to virus infections and must distinguish between host and pathogen nucleic acids to mount a protective immune response without activating autoimmune responses. While the presence of nucleotide modifications in RNA is known to correlate with diminished innate immune signaling, the underlying mechanisms have not been explored. The data reported here are important for defining mechanistic details to explain signaling suppression by RNAs containing modified nucleotides. The results suggest that RNAs containing modified nucleotides interrupt signaling at early steps of the RIG-I-like innate immune activation pathway and also that nucleotide modifications with similar chemical structures can be organized into classes that suppress or evade innate immune signaling steps. These data contribute to defining the molecular basis for innate immune signaling suppression by RNAs containing modified nucleotides. The results have important implications for designing therapeutic RNAs that evade innate immune detection.
URLPMID:16111635 [本文引用: 1]

DNA and RNA stimulate the mammalian innate immune system through activation of Toll-like receptors (TLRs). DNA containing methylated CpG motifs, however, is not stimulatory. Selected nucleosides in naturally occurring RNA are also methylated or otherwise modified, but the immunomodulatory effects of these alterations remain untested. We show that RNA signals through human TLR3, TLR7, and TLR8, but incorporation of modified nucleosides m5C, m6A, m5U, s2U, or pseudouridine ablates activity. Dendritic cells (DCs) exposed to such modified RNA express significantly less cytokines and activation markers than those treated with unmodified RNA. DCs and TLR-expressing cells are potently activated by bacterial and mitochondrial RNA, but not by mammalian total RNA, which is abundant in modified nucleosides. We conclude that nucleoside modifications suppress the potential of RNA to activate DCs. The innate immune system may therefore detect RNA lacking nucleoside modification as a means of selectively responding to bacteria or necrotic tissue.
[本文引用: 1]
URLPMID:213879 [本文引用: 1]

http://linkinghub.elsevier.com/retrieve/pii/0042682278901915
URLPMID:9578320 [本文引用: 1]

S-Adenosylhomocysteine (AdoHcy), formed after the donation of the methyl group of S-adenosylmethionine to a methyl acceptor, is hydrolyzed to adenosine and homocysteine by AdoHcy hydrolase physiologically. The administration of the inhibitors of AdoHcy hydrolase to cells or animals normally results in an accumulation of cellular AdoHcy higher than those found in controls, which is often accompanied by a simultaneous rise in S-adenosylmethionine because of the feedback inhibition by AdoHcy on most methylation reactions. AdoHcy hydrolase has become a tantalizing pharmacological target for inhibition since its blockade can affect cellular methylation of phospholipids, proteins, small molecules, DNA, and RNA. Indeed, all of these different methylation reactions have been found to be inhibitable by the nucleoside inhibitors/substrates of AdoHcy hydrolase. Among the interesting effects are the activation of genes, induction of cellular differentiation, increased expression of transcription factors, and sometimes the repression of genes. Furthermore, some of the nucleosides show remarkable antiviral activities in vitro and in vivo. However, the mode of action of the inhibitors appears complex. Although the inhibition of methylation might account for some of the biological effects, the ability of some of the nucleoside inhibitors to undergo metabolic phosphorylation to nucleotides may account for part of their biological activities. The defining mode of action responsible for their biological effects still awaits biochemical elaboration, especially regarding their antiviral effects, induction of genes, or cellular differentiation.
