 ,儿科重大疾病研究教育部重点实验室,北京 100045
,儿科重大疾病研究教育部重点实验室,北京 100045Mining and characterization of preterm birth related genes
Xuanshi Liu, Wei Li ,Genetics and Birth Defects Control Center, Beijing Children's Hospital, Capital Medical University, National Center for Children's Health, Beijing 100045, China
,Genetics and Birth Defects Control Center, Beijing Children's Hospital, Capital Medical University, National Center for Children's Health, Beijing 100045, China通讯作者:
编委: 方向东
收稿日期:2019-03-21修回日期:2019-05-8网络出版日期:2019-05-20
Received:2019-03-21Revised:2019-05-8Online:2019-05-20
作者简介 About authors
刘玄石,博士研究生,助理研究员,专业方向:生物信息学E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (830KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
刘玄石, 李巍. 早产相关基因的挖掘与特征分析[J]. 遗传, 2019, 41(5): 413-421 doi:10.16288/j.yczz.19-078
Xuanshi Liu, Wei Li.
早产是指胎儿在完成37周妊娠前出生。2010年,世界卫生组织等国际组织对全世界184个国家的调查发现,新生儿的早产率大致是5%~ 18%[1],中国的早产率大约是7%,每年约有120万早产婴儿,全球排名第二,仅低于印度[2]。除死亡风险外,早产还可能伴有脑瘫、肺部疾病、听觉和视觉缺陷等风险[1,2],甚至有研究发现早产与成年后发生的一些慢性疾病相关,如心血管疾病和糖尿病等[3]。目前,早产的发生机制尚不明确。根据双生子及家系研究的估算,遗传因素对早产风险的影响大约占15%~35%[4,5,6]。早期对早产遗传机制的研究,通常根据早产病理学特点,选择可能相关的基因展开研究。例如,与新生儿出生体重和月经期有关的PON2[7],参与炎症反应的TNF、IL10[8]和TLR2[9],与血管生成有关的VEGF[10,11]等。近年来,采用高通量测序技术对早产遗传因素的研究,发现了大量相关的位点和基因,包括采用全基因组关联分析找到的与自发早产相关的3个位点(rs17053026、rs17527054和rs3777722)[12],以及位于EBF1、EEFSEC和AGTR2基因上的与早产相关的位点[13];利用全外显子测序发现与早产最显著相关的位点落在CR1基因外显子上[14];全基因组、转录组和甲基化数据的结果提示RAB31和RBPJ基因与早产相关[15]等。虽然针对早产遗传因素的研究已经积累了大量数据,然而由于早产的遗传机制相当复杂,现有研究结果也缺乏较好的归纳和整合,如Database for Preterm Birth (dbPTB)最后一次更新是2014年,这使得后续采用生物信息学手段对早产遗传信息的挖掘和早产遗传模型的构建变得困难[16]。因此,本研究利用生物信息学方法,通过挖掘文献数据库以及疾病基因数据库中报道的早产相关基因信息,整合并分析早产相关基因的特征,为早产的遗传研究提供重要资源。
1 材料与方法
1.1 数据库和软件
(1)文献数据库:美国国家医学图书馆(PubMed,1.2 文献数据库的信息挖掘
2019年3月8日,通过计算机检索PubMed数据库,采用关键词检索式“preterm birth”AND“gene”,检索年限为建库至2019年3月。整理出所有文献的PMID,输入文本挖掘工具SciMiner。SciMiner软件通过关键字“preterm birth”,以及软件内置的正则表达规则和基因字典,挖掘文献中与早产相关基因。为避免过度匹配,对SciMiner挖掘结果设置阈值和人工审核的两层过滤方式。首先根据设置的阙值,删除了仅在2篇及以下文献中出现的基因。其次通过人工核查摘要,删除摘要中没有直接提及早产的基因。最后筛选出用于后续分析的基因列表。1.3 疾病数据库的信息挖掘
通过Shell脚本程序,搜索疾病数据库OMIM,ClinVar和CTD,查找与“preterm birth”或其同义词匹配的记录,提取记录下的基因信息,并合并进文献数据库筛选出的基因列表。1.4 基因富集分析
采用R软件包ClusterProfiler对筛选出的基因,进行了基因功能(Gene Ontology, GO)和KEGG通路(京都基因与基因组大百科全书数据库,Kyoto Encyclopedia of Genes and Genomes)以及Reactome通路[19]的富集分析,对结果进行多重检验后,获得显著的功能和通路,以FDR<0.05 (false discovery rate)作为显著性的阈值。1.5 基因特征的收集
采用Ensembl的BioMart,收集了20320个基因的长度,转录本数量,GC含量特征(人基因组版本GRCh37.p13/hg19)。根据筛选出的基因列表,采用Shell脚本程序,从BioMart数据中提取了所需基因的特征信息。2 结果与分析
2.1 早产相关基因数据库挖掘结果
通过计算机检索PubMed数据库获得来源于800种杂志的2264篇相关文献的摘要,并通过PMID和SciMiner软件挖掘出了文献中与早产可能相关的2149个基因。其中,文献数量居前5%的杂志多数是临床专业期刊(附表1)。经过阈值和人工审核的两层过滤,筛选出在1274篇文献里出现的355个基因(附表2),表1列出了在文献数量中排名前5%的基因。Supplementary Table 1
附表1
附表1 PubMed检索结果中居前5%的杂志
Supplementary Table 1
| 排序 | 期刊 | 用于过滤的文献数量 | 排序 | 期刊 | 用于过滤的文献数量 |
|---|---|---|---|---|---|
| 1 | PLoS One | 109 | 11 | Mol Hum Reprod | 25 |
| 2 | Pediatr Res | 75 | 12 | Sci Rep | 22 |
| 3 | Am J Obstet Gynecol | 71 | 13 | Endocrinology | 22 |
| 4 | J Matern Fetal Neonatal Med | 36 | 14 | Hum Mol Genet | 20 |
| 5 | Reprod Sci | 35 | 15 | Am J Physiol Lung Cell Mol Physiol | 20 |
| 6 | Placenta | 30 | 16 | J Perinatol | 19 |
| 7 | Am J Reprod Immunol | 29 | 17 | Neonatology | 18 |
| 8 | Am J Med Genet A | 29 | 18 | Am J Pathol | 18 |
| 9 | Proc Natl Acad Sci U S A | 28 | 19 | J Reprod Immunol | 17 |
| 10 | Biol Reprod | 26 | 20 | Pediatrics | 16 |
新窗口打开|下载CSV
Supplementary Table 2
附表2
附表2 早产相关基因
Supplementary Table 2
| 基因名称 | 基因ID | 基因全称 | 有基因记录的文献数 |
|---|---|---|---|
| TNF | 11892 | tumor necrosis factor (TNF superfamily, member 2) | 156 |
| IL6 | 6018 | interleukin 6 (interferon, beta 2) | 155 |
| IL1B | 5992 | interleukin 1, beta | 140 |
| IL8 | 6025 | interleukin 8 | 85 |
| NFKB1 | 7794 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | 68 |
| COL1A1 | 2197 | collagen, type I, alpha 1 | 68 |
| PTGS2 | 9605 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 63 |
| TLR4 | 11850 | toll-like receptor 4 | 57 |
| VEGFA | 12680 | vascular endothelial growth factor A | 57 |
| IL10 | 5962 | interleukin 10 | 53 |
| MT-RNR2 | 7471 | mitochondrially encoded 16S RNA | 51 |
| INS | 6081 | insulin | 46 |
| PGR | 8910 | progesterone receptor | 42 |
| IGF1 | 5464 | insulin-like growth factor 1 (somatomedin C) | 39 |
| TGFB1 | 11766 | transforming growth factor, beta 1 | 39 |
| SFTPD | 10803 | surfactant, pulmonary-associated protein D | 38 |
| MMP9 | 7176 | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | 36 |
| NR3C1 | 7978 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | 35 |
| SFTPA2B | 23441 | surfactant, pulmonary-associated protein A2B | 34 |
| IL1A | 5991 | interleukin 1, alpha | 33 |
| SFTPA1 | 10798 | surfactant, pulmonary-associated protein A1 | 32 |
| 基因名称 | 基因ID | 基因全称 | 有基因记录的文献数 |
| CCL2 | 10618 | chemokine (C-C motif) ligand 2 | 29 |
| F2 | 3535 | coagulation factor II (thrombin) | 26 |
| IL4 | 6014 | interleukin 4 | 25 |
| TLR2 | 11848 | toll-like receptor 2 | 25 |
| SFTPB | 10801 | surfactant, pulmonary-associated protein B | 24 |
| IFNG | 5438 | interferon, gamma | 24 |
| IL1RN | 6000 | interleukin 1 receptor antagonist | 24 |
| MBL2 | 6922 | mannose-binding lectin (protein C) 2, soluble (opsonic defect) | 23 |
| SFTPC | 10802 | surfactant, pulmonary-associated protein C | 23 |
| OXTR | 8529 | oxytocin receptor | 23 |
| MTHFR | 7436 | 5,10-methylenetetrahydrofolate reductase (NADPH) | 23 |
| MAPK1 | 6871 | mitogen-activated protein kinase 1 | 23 |
| NOS2A | 7873 | nitric oxide synthase 2A (inducible, hepatocytes) | 22 |
| ACE | 2707 | angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | 21 |
| REN | 9958 | renin | 21 |
| CRH | 2355 | corticotropin releasing hormone | 20 |
| ALB | 399 | albumin | 20 |
| CYP1A1 | 2595 | cytochrome P450, family 1, subfamily A, polypeptide 1 | 18 |
| MMP1 | 7155 | matrix metallopeptidase 1 (interstitial collagenase) | 18 |
| GSTT1 | 4641 | glutathione S-transferase theta 1 | 17 |
| GJA1 | 4274 | gap junction protein, alpha 1, 43kDa | 17 |
| CD14 | 1628 | CD14 molecule | 17 |
| CASP3 | 1504 | caspase 3, apoptosis-related cysteine peptidase | 17 |
| APOE | 613 | apolipoprotein E | 16 |
| NOS3 | 7876 | nitric oxide synthase 3 (endothelial cell) | 16 |
| F5 | 3542 | coagulation factor V (proaccelerin, labile factor) | 16 |
| JUN | 6204 | jun oncogene | 15 |
| IGF2 | 5466 | insulin-like growth factor 2 (somatomedin A) | 15 |
| LEP | 6553 | leptin | 15 |
| BCL2 | 990 | B-cell CLL/lymphoma 2 | 15 |
| GAPDH | 4141 | glyceraldehyde-3-phosphate dehydrogenase | 15 |
| FOS | 3796 | v-fos FBJ murine osteosarcoma viral oncogene homolog | 15 |
| MMP2 | 7166 | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | 15 |
| SERPINH1 | 1546 | serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | 14 |
| FLT1 | 3763 | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 14 |
| NFKBIA | 7797 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | 14 |
| GSTM1 | 4632 | glutathione S-transferase M1 | 13 |
| SERPINE1 | 8583 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 13 |
| 基因名称 | 基因ID | 基因全称 | 有基因记录的文献数 |
| IL6R | 6019 | interleukin 6 receptor | 13 |
| TP53 | 11998 | tumor protein p53 | 13 |
| TWIST1 | 12428 | twist homolog 1 (acrocephalosyndactyly 3; Saethre-Chotzen syndrome) (Drosophila) | 13 |
| SOD1 | 11179 | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 13 |
| IL2 | 6001 | interleukin 2 | 13 |
| CD4 | 1678 | CD4 molecule | 13 |
| AGT | 333 | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | 12 |
| PPARG | 9236 | peroxisome proliferator-activated receptor gamma | 12 |
| CAT | 1516 | catalase | 12 |
| CYP2B6 | 2615 | cytochrome P450, family 2, subfamily B, polypeptide 6 | 12 |
| PGF | 8893 | placental growth factor | 11 |
| S100A9 | 10499 | S100 calcium binding protein A9 | 11 |
| GSTA1 | 4626 | glutathione S-transferase A1 | 11 |
| KDR | 6307 | kinase insert domain receptor (a type III receptor tyrosine kinase) | 11 |
| PRKCA | 9393 | protein kinase C, alpha | 11 |
| STAT1 | 11362 | signal transducer and activator of transcription 1, 91kDa | 10 |
| MBP | 6925 | myelin basic protein | 10 |
| IL13 | 5973 | interleukin 13 | 10 |
| EDN1 | 3176 | endothelin 1 | 10 |
| LTA | 6709 | lymphotoxin alpha (TNF superfamily, member 1) | 10 |
| TFAP2A | 11742 | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | 10 |
| TLR5 | 11851 | toll-like receptor 5 | 10 |
| TBXAS1 | 11609 | thromboxane A synthase 1 (platelet, cytochrome P450, family 5, subfamily A) | 10 |
| IL1R1 | 5993 | interleukin 1 receptor, type I | 10 |
| CYP3A5 | 2638 | cytochrome P450, family 3, subfamily A, polypeptide 5 | 9 |
| IGFBP1 | 5469 | insulin-like growth factor binding protein 1 | 9 |
| EGR1 | 3238 | early growth response 1 | 9 |
| FAS | 11920 | Fas (TNF receptor superfamily, member 6) | 9 |
| ADRB2 | 286 | adrenergic, beta-2-, receptor, surface | 9 |
| MMP8 | 7175 | matrix metallopeptidase 8 (neutrophil collagenase) | 9 |
| PTGER4 | 9596 | prostaglandin E receptor 4 (subtype EP4) | 8 |
| CSF3 | 2438 | colony stimulating factor 3 (granulocyte) | 8 |
| TNFRSF1A | 11916 | tumor necrosis factor receptor superfamily, member 1A | 8 |
| NES | 7756 | nestin | 8 |
| FGFR3 | 3690 | fibroblast growth factor receptor 3 (achondroplasia, thanatophoric dwarfism) | 8 |
| MAPK3 | 6877 | mitogen-activated protein kinase 3 | 8 |
| COX8A | 2294 | cytochrome c oxidase subunit 8A (ubiquitous) | 8 |
| ESR2 | 3468 | estrogen receptor 2 (ER beta) | 8 |
| PPARA | 9232 | peroxisome proliferator-activated receptor alpha | 8 |
| FSHR | 3969 | follicle stimulating hormone receptor | 8 |
| 基因名称 | 基因ID | 基因全称 | 有基因记录的文献数 |
| MAPK14 | 6876 | mitogen-activated protein kinase 14 | 8 |
| RPS27A | 10417 | ribosomal protein S27a | 8 |
| H19 | 4713 | H19, imprinted maternally expressed transcript | 8 |
| SP1 | 11205 | Sp1 transcription factor | 7 |
| SOD2 | 11180 | superoxide dismutase 2, mitochondrial | 7 |
| MT-CO2 | 7421 | mitochondrially encoded cytochrome c oxidase II | 7 |
| IL17A | 5981 | interleukin 17A | 7 |
| IGFBP3 | 5472 | insulin-like growth factor binding protein 3 | 7 |
| PTGER3 | 9595 | prostaglandin E receptor 3 (subtype EP3) | 7 |
| IRF6 | 6121 | interferon regulatory factor 6 | 7 |
| MYD88 | 7562 | myeloid differentiation primary response gene (88) | 7 |
| PLAT | 9051 | plasminogen activator, tissue | 7 |
| ICAM1 | 5344 | intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | 7 |
| MAPK8 | 6881 | mitogen-activated protein kinase 8 | 7 |
| MMP3 | 7173 | matrix metallopeptidase 3 (stromelysin 1, progelatinase) | 7 |
| HSD11B2 | 5209 | hydroxysteroid (11-beta) dehydrogenase 2 | 7 |
| CD8A | 1706 | CD8a molecule | 7 |
| SLC6A3 | 11049 | solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 | 7 |
| SLC12A1 | 10910 | solute carrier family 12 (sodium/potassium/chloride transporters), member 1 | 6 |
| NFE2L2 | 7782 | nuclear factor (erythroid-derived 2)-like 2 | 6 |
| HPGD | 5154 | hydroxyprostaglandin dehydrogenase 15-(NAD) | 6 |
| PTGER2 | 9594 | prostaglandin E receptor 2 (subtype EP2), 53kDa | 6 |
| ABCA3 | 33 | ATP-binding cassette, sub-family A (ABC1), member 3 | 6 |
| SMN1 | 11117 | survival of motor neuron 1, telomeric | 6 |
| CXCL10 | 10637 | chemokine (C-X-C motif) ligand 10 | 6 |
| DEFB1 | 2766 | defensin, beta 1 | 6 |
| LPAL2 | 21210 | lipoprotein, Lp(a)-like 2 | 6 |
| NOS1 | 7872 | nitric oxide synthase 1 (neuronal) | 6 |
| FGFR1 | 3688 | fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | 6 |
| CASP1 | 1499 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | 6 |
| AR | 644 | androgen receptor (dihydrotestosterone receptor; testicular feminization; spinal and bulbar muscular atrophy; Kennedy disease) | 6 |
| ATM | 795 | ataxia telangiectasia mutated | 6 |
| ZMPSTE24 | 12877 | zinc metallopeptidase (STE24 homolog, S. cerevisiae) | 6 |
| CXCL1 | 4602 | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | 6 |
| NDP | 7678 | Norrie disease (pseudoglioma) | 6 |
| TLR3 | 11849 | toll-like receptor 3 | 6 |
| FLG | 3748 | filaggrin | 5 |
| SLC6A4 | 11050 | solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | 5 |
| RUNX2 | 10472 | runt-related transcription factor 2 | 5 |
| 基因名称 | 基因ID | 基因全称 | 有基因记录的文献数 |
| ABCB1 | 40 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 | 5 |
| NR3C2 | 7979 | nuclear receptor subfamily 3, group C, member 2 | 5 |
| EGFR | 3236 | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 5 |
| LOR | 6663 | loricrin | 5 |
| HDAC9 | 14065 | histone deacetylase 9 | 5 |
| TNFRSF1B | 11917 | tumor necrosis factor receptor superfamily, member 1B | 5 |
| EPO | 3415 | erythropoietin | 5 |
| NOD2 | 5331 | nucleotide-binding oligomerization domain containing 2 | 5 |
| LEPR | 6554 | leptin receptor | 5 |
| CTNNB1 | 2514 | catenin (cadherin-associated protein), beta 1, 88kDa | 5 |
| THBS1 | 11785 | thrombospondin 1 | 5 |
| TNFAIP3 | 11896 | tumor necrosis factor, alpha-induced protein 3 | 5 |
| S100A6 | 10496 | S100 calcium binding protein A6 | 5 |
| TGFB2 | 11768 | transforming growth factor, beta 2 | 5 |
| IL5 | 6016 | interleukin 5 (colony-stimulating factor, eosinophil) | 5 |
| SLC2A4 | 11009 | solute carrier family 2 (facilitated glucose transporter), member 4 | 5 |
| ACPP | 125 | acid phosphatase, prostate | 5 |
| TCEAL1 | 11616 | transcription elongation factor A (SII)-like 1 | 5 |
| COL1A2 | 2198 | collagen, type I, alpha 2 | 5 |
| CTGF | 2500 | connective tissue growth factor | 5 |
| F2R | 3537 | coagulation factor II (thrombin) receptor | 5 |
| CD163 | 1631 | CD163 molecule | 5 |
| JAG1 | 6188 | jagged 1 (Alagille syndrome) | 5 |
| IL12A | 5969 | interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) | 5 |
| TIRAP | 17192 | toll-interleukin 1 receptor (TIR) domain containing adaptor protein | 5 |
| FOXP3 | 6106 | forkhead box P3 | 5 |
| MEST | 7028 | mesoderm specific transcript homolog (mouse) | 5 |
| CFH | 4883 | complement factor H | 5 |
| IRAK1 | 6112 | interleukin-1 receptor-associated kinase 1 | 5 |
| PRKAR2A | 9391 | protein kinase, cAMP-dependent, regulatory, type II, alpha | 5 |
| TIMP2 | 11821 | TIMP metallopeptidase inhibitor 2 | 5 |
| CDKN1C | 1786 | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 4 |
| GORASP1 | 16769 | golgi reassembly stacking protein 1, 65kDa | 4 |
| HLA-G | 4964 | major histocompatibility complex, class I, G | 4 |
| PON1 | 9204 | paraoxonase 1 | 4 |
| RAF1 | 9829 | v-raf-1 murine leukemia viral oncogene homolog 1 | 4 |
| PTPN11 | 9644 | protein tyrosine phosphatase, non-receptor type 11 (Noonan syndrome 1) | 4 |
| LCN2 | 6526 | lipocalin 2 | 4 |
| CALCA | 1437 | calcitonin-related polypeptide alpha | 4 |
| 基因名称 | 基因ID | 基因全称 | 有基因记录的文献数 |
| KCNH2 | 6251 | potassium voltage-gated channel, subfamily H (eag-related), member 2 | 4 |
| TIMP1 | 11820 | TIMP metallopeptidase inhibitor 1 | 4 |
| GPX1 | 4553 | glutathione peroxidase 1 | 4 |
| SERPINB2 | 8584 | serpin peptidase inhibitor, clade B (ovalbumin), member 2 | 4 |
| NLRP3 | 16400 | NLR family, pyrin domain containing 3 | 4 |
| MIF | 7097 | macrophage migration inhibitory factor (glycosylation-inhibiting factor) | 4 |
| IL1R2 | 5994 | interleukin 1 receptor, type II | 4 |
| ERAL1 | 3424 | Era G-protein-like 1 (E. coli) | 4 |
| IFNA1 | 5417 | interferon, alpha 1 | 4 |
| PLAGL1 | 9046 | pleiomorphic adenoma gene-like 1 | 4 |
| CYP27B1 | 2606 | cytochrome P450, family 27, subfamily B, polypeptide 1 | 4 |
| ZEB1 | 11642 | zinc finger E-box binding homeobox 1 | 4 |
| CXCL12 | 10672 | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 4 |
| LBP | 6517 | lipopolysaccharide binding protein | 4 |
| WNT4 | 12783 | wingless-type MMTV integration site family, member 4 | 4 |
| IL4R | 6015 | interleukin 4 receptor | 4 |
| INSR | 6091 | insulin receptor | 4 |
| MAPK10 | 6872 | mitogen-activated protein kinase 10 | 4 |
| DES | 2770 | desmin | 4 |
| PHEX | 8918 | phosphate regulating endopeptidase homolog, X-linked (hypophosphatemia, vitamin D resistant rickets) | 4 |
| PTPRC | 9666 | protein tyrosine phosphatase, receptor type, C | 4 |
| SLC26A4 | 8818 | solute carrier family 26, member 4 | 4 |
| TEK | 11724 | TEK tyrosine kinase, endothelial (venous malformations, multiple cutaneous and mucosal) | 4 |
| TLR6 | 16711 | toll-like receptor 6 | 4 |
| TSHB | 12372 | thyroid stimulating hormone, beta | 4 |
| CCL3 | 10627 | chemokine (C-C motif) ligand 3 | 4 |
| CYP17A1 | 2593 | cytochrome P450, family 17, subfamily A, polypeptide 1 | 4 |
| CYP19A1 | 2594 | cytochrome P450, family 19, subfamily A, polypeptide 1 | 4 |
| FSHB | 3964 | follicle stimulating hormone, beta polypeptide | 4 |
| IL10RA | 5964 | interleukin 10 receptor, alpha | 4 |
| VIM | 12692 | vimentin | 4 |
| ADAMTS2 | 218 | ADAM metallopeptidase with thrombospondin type 1 motif, 2 | 4 |
| ADAMTS4 | 220 | ADAM metallopeptidase with thrombospondin type 1 motif, 4 | 4 |
| ATP2A3 | 813 | ATPase, Ca++ transporting, ubiquitous | 4 |
| CHRNA9 | 14079 | cholinergic receptor, nicotinic, alpha 9 | 4 |
| COL2A1 | 2200 | collagen, type II, alpha 1 | 4 |
| COL5A1 | 2209 | collagen, type V, alpha 1 | 4 |
| HBG2 | 4832 | hemoglobin, gamma G | 4 |
| NOX5 | 14874 | NADPH oxidase, EF-hand calcium binding domain 5 | 4 |
| 基因名称 | 基因ID | 基因全称 | 有基因记录的文献数 |
| RELA | 9955 | v-rel reticuloendotheliosis viral oncogene homolog A, nuclear factor of kappa light polypeptide gene enhancer in B-cells 3, p65 (avian) | 4 |
| TF | 11740 | transferrin | 4 |
| TLR10 | 15634 | toll-like receptor 10 | 4 |
| PLCB1 | 15917 | phospholipase C, beta 1 (phosphoinositide-specific) | 4 |
| MASP2 | 6902 | mannan-binding lectin serine peptidase 2 | 3 |
| CYP3A4 | 2637 | cytochrome P450, family 3, subfamily A, polypeptide 4 | 3 |
| GHRL | 18129 | ghrelin/obestatin preprohormone | 3 |
| GJB2 | 4284 | gap junction protein, beta 2, 26kDa | 3 |
| BGN | 1044 | biglycan | 3 |
| GHR | 4263 | growth hormone receptor | 3 |
| NEU1 | 7758 | sialidase 1 (lysosomal sialidase) | 3 |
| PSEN1 | 9508 | presenilin 1 (Alzheimer disease 3) | 3 |
| SMAD7 | 6773 | SMAD family member 7 | 3 |
| CAMP | 1472 | cathelicidin antimicrobial peptide | 3 |
| DEFB4 | 2767 | defensin, beta 4 | 3 |
| IGF1R | 5465 | insulin-like growth factor 1 receptor | 3 |
| CAP1 | 20040 | CAP, adenylate cyclase-associated protein 1 (yeast) | 3 |
| GDF9 | 4224 | growth differentiation factor 9 | 3 |
| PHOX2B | 9143 | paired-like homeobox 2b | 3 |
| CCL8 | 10635 | chemokine (C-C motif) ligand 8 | 3 |
| KCNB1 | 6231 | potassium voltage-gated channel, Shab-related subfamily, member 1 | 3 |
| SLC27A4 | 10998 | solute carrier family 27 (fatty acid transporter), member 4 | 3 |
| HMGB1 | 4983 | high-mobility group box 1 | 3 |
| FASLG | 11936 | Fas ligand (TNF superfamily, member 6) | 3 |
| FZD4 | 4042 | frizzled homolog 4 (Drosophila) | 3 |
| TXN | 12435 | thioredoxin | 3 |
| MAP2 | 6839 | microtubule-associated protein 2 | 3 |
| NGF | 7808 | nerve growth factor (beta polypeptide) | 3 |
| PROK1 | 18454 | prokineticin 1 | 3 |
| COMT | 2228 | catechol-O-methyltransferase | 3 |
| FOXO1 | 3819 | forkhead box O1 | 3 |
| FOXO3 | 3821 | forkhead box O3 | 3 |
| HGF | 4893 | hepatocyte growth factor (hepapoietin A; scatter factor) | 3 |
| KISS1 | 6341 | KiSS-1 metastasis-suppressor | 3 |
| TYMS | 12441 | thymidylate synthetase | 3 |
| RELB | 9956 | v-rel reticuloendotheliosis viral oncogene homolog B, nuclear factor of kappa light polypeptide gene enhancer in B-cells 3 (avian) | 3 |
| UGT1A1 | 12530 | UDP glucuronosyltransferase 1 family, polypeptide A1 | 3 |
| CDKN2A | 1787 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 3 |
| CYP21A2 | 2600 | cytochrome P450, family 21, subfamily A, polypeptide 2 | 3 |
| 基因名称 | 基因ID | 基因全称 | 有基因记录的文献数 |
| GATA6 | 4174 | GATA binding protein 6 | 3 |
| ITGB4 | 6158 | integrin, beta 4 | 3 |
| S100A8 | 10498 | S100 calcium binding protein A8 | 3 |
| SOD3 | 11181 | superoxide dismutase 3, extracellular | 3 |
| CD68 | 1693 | CD68 molecule | 3 |
| MIRN210 | 31587 | microRNA 210 | 3 |
| PPT1 | 9325 | palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) | 3 |
| ZEB2 | 14881 | zinc finger E-box binding homeobox 2 | 3 |
| ABCA1 | 29 | ATP-binding cassette, sub-family A (ABC1), member 1 | 3 |
| CREBBP | 2348 | CREB binding protein (Rubinstein-Taybi syndrome) | 3 |
| P2RX7 | 8537 | purinergic receptor P2X, ligand-gated ion channel, 7 | 3 |
| UCP2 | 12518 | uncoupling protein 2 (mitochondrial, proton carrier) | 3 |
| CACNA1G | 1394 | calcium channel, voltage-dependent, T type, alpha 1G subunit | 3 |
| MARK2 | 3332 | MAP/microtubule affinity-regulating kinase 2 | 3 |
| SLC27A1 | 10995 | solute carrier family 27 (fatty acid transporter), member 1 | 3 |
| TFRC | 11763 | transferrin receptor (p90, CD71) | 3 |
| TICAM1 | 18348 | toll-like receptor adaptor molecule 1 | 3 |
| FADS2 | 3575 | fatty acid desaturase 2 | 3 |
| FGF7 | 3685 | fibroblast growth factor 7 (keratinocyte growth factor) | 3 |
| OPRM1 | 8156 | opioid receptor, mu 1 | 3 |
| SPAG8 | 14105 | sperm associated antigen 8 | 3 |
| CRHR1 | 2357 | corticotropin releasing hormone receptor 1 | 3 |
| DICER1 | 17098 | dicer 1, ribonuclease type III | 3 |
| FTO | 24678 | fat mass and obesity associated | 3 |
| MMP12 | 7158 | matrix metallopeptidase 12 (macrophage elastase) | 3 |
| CAV1 | 1527 | caveolin 1, caveolae protein, 22kDa | 3 |
| ECE1 | 3146 | endothelin converting enzyme 1 | 3 |
| FBN1 | 3603 | fibrillin 1 | 3 |
| FST | 3971 | follistatin | 3 |
| IL9 | 6029 | interleukin 9 | 3 |
| MT-RNR1 | 7470 | mitochondrially encoded 12S RNA | 3 |
| SCNN1A | 10599 | sodium channel, nonvoltage-gated 1 alpha | 3 |
| SDHC | 10682 | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 3 |
| WT1 | 12796 | Wilms tumor 1 | 3 |
| ATG16L1 | 21498 | ATG16 autophagy related 16-like 1 (S. cerevisiae) | 3 |
| CASP8 | 1509 | caspase 8, apoptosis-related cysteine peptidase | 3 |
| FURIN | 8568 | furin (paired basic amino acid cleaving enzyme) | 3 |
| FUT1 | 4012 | fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) | 3 |
| HBA2 | 4824 | hemoglobin, alpha 2 | 3 |
| IFNB1 | 5434 | interferon, beta 1, fibroblast | 3 |
| 基因名称 | 基因ID | 基因全称 | 有基因记录的文献数 |
| KRT5 | 6442 | keratin 5 (epidermolysis bullosa simplex, Dowling-Meara/Kobner/Weber-Cockayne types) | 3 |
| OPRD1 | 8153 | opioid receptor, delta 1 | 3 |
| OXT | 8528 | oxytocin, prepro- (neurophysin I) | 3 |
| PDGFRA | 8803 | platelet-derived growth factor receptor, alpha polypeptide | 3 |
| SERPINC1 | 775 | serpin peptidase inhibitor, clade C (antithrombin), member 1 | 3 |
| SLC2A1 | 11005 | solute carrier family 2 (facilitated glucose transporter), member 1 | 3 |
| CSH1 | 2440 | chorionic somatomammotropin hormone 1 (placental lactogen) | 3 |
| ETS1 | 3488 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 3 |
| HDAC1 | 4852 | histone deacetylase 1 | 3 |
| ITGA6 | 6142 | integrin, alpha 6 | 3 |
| MET | 7029 | met proto-oncogene (hepatocyte growth factor receptor) | 3 |
| TRAF1 | 12031 | TNF receptor-associated factor 1 | 3 |
| CD1D | 1637 | CD1d molecule | 3 |
| CYP2E1 | 2631 | cytochrome P450, family 2, subfamily E, polypeptide 1 | 3 |
| DLL4 | 2910 | delta-like 4 (Drosophila) | 3 |
| FAM129B | 25282 | family with sequence similarity 129, member B | 3 |
| IRS1 | 6125 | insulin receptor substrate 1 | 3 |
| LTF | 6720 | lactotransferrin | 3 |
| MTRR | 7473 | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase | 3 |
| PLOD1 | 9081 | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 3 |
| VIP | 12693 | vasoactive intestinal peptide | 3 |
| ABCA12 | 14637 | ATP-binding cassette, sub-family A (ABC1), member 12 | 3 |
| ABCC2 | 53 | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | 3 |
| ABO | 79 | ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) | 3 |
| ADCY10 | 21285 | adenylate cyclase 10 (soluble) | 3 |
| APLP2 | 598 | amyloid beta (A4) precursor-like protein 2 | 3 |
| CCND1 | 1582 | cyclin D1 | 3 |
| CD69 | 1694 | CD69 molecule | 3 |
| COL5A2 | 2210 | collagen, type V, alpha 2 | 3 |
| DLL1 | 2908 | delta-like 1 (Drosophila) | 3 |
| EPAS1 | 3374 | endothelial PAS domain protein 1 | 3 |
| HAS2 | 4819 | hyaluronan synthase 2 | 3 |
| IL12RB1 | 5971 | interleukin 12 receptor, beta 1 | 3 |
| MDK | 6972 | midkine (neurite growth-promoting factor 2) | 3 |
| PLG | 9071 | plasminogen | 3 |
| PRKAA2 | 9377 | protein kinase, AMP-activated, alpha 2 catalytic subunit | 3 |
| S100A10 | 10487 | S100 calcium binding protein A10 | 3 |
| SIRT1 | 14929 | sirtuin (silent mating type information regulation 2 homolog) 1 (S. cerevisiae) | 3 |
| TPO | 12015 | thyroid peroxidase | 3 |
| 基因名称 | 基因ID | 基因全称 | 有基因记录的文献数 |
| VCAM1 | 12663 | vascular cell adhesion molecule 1 | 3 |
| AIF1 | 352 | allograft inflammatory factor 1 | 3 |
| BAX | 959 | BCL2-associated X protein | 3 |
| CCND2 | 1583 | cyclin D2 | 3 |
| CD27 | 11922 | CD27 molecule | 3 |
| CDH17 | 1756 | cadherin 17, LI cadherin (liver-intestine) | 3 |
| COL4A3 | 2204 | collagen, type IV, alpha 3 (Goodpasture antigen) | 3 |
| CREB1 | 2345 | cAMP responsive element binding protein 1 | 3 |
| CXCL9 | 7098 | chemokine (C-X-C motif) ligand 9 | 3 |
| CYCS | 19986 | cytochrome c, somatic | 3 |
| HSD11B1 | 5208 | hydroxysteroid (11-beta) dehydrogenase 1 | 3 |
| MMP13 | 7159 | matrix metallopeptidase 13 (collagenase 3) | 3 |
| MSMB | 7372 | microseminoprotein, beta- | 3 |
| NCAM1 | 7656 | neural cell adhesion molecule 1 | 3 |
| NCOA1 | 7668 | nuclear receptor coactivator 1 | 3 |
| NEFL | 7739 | neurofilament, light polypeptide 68kDa | 3 |
| NTRK2 | 8032 | neurotrophic tyrosine kinase, receptor, type 2 | 3 |
| PARP1 | 270 | poly (ADP-ribose) polymerase family, member 1 | 3 |
| PYCARD | 16608 | PYD and CARD domain containing | 3 |
| RARA | 9864 | retinoic acid receptor, alpha | 3 |
| RXRA | 10477 | retinoid X receptor, alpha | 3 |
新窗口打开|下载CSV
Table 1
表1
表1 筛选出的基因列表中排前5%的早产相关基因
Table 1
| 基因名称 | 基因ID | 基因名全称 | 有基因记录的文献数量 |
|---|---|---|---|
| TNF | 11892 | Tumor necrosis factor (TNF superfamily, member 2) | 156 |
| IL6 | 6018 | Interleukin 6 (Interferon, beta 2) | 155 |
| IL1B | 5992 | Interleukin 1 beta | 140 |
| IL8 | 6025 | Interleukin 8 | 85 |
| NFKB1 | 7794 | Nuclear factor of kappa light polypeptide gene Enhancer in B-cells 1 (p105) | 68 |
| COL1A1 | 2197 | Collagen type I alpha 1 chain | 68 |
| PTGS2 | 9605 | Prostaglandin-endoperoxide synthase 2 (Prostaglandin G/H synthase and cyclooxygenase) | 63 |
| TLR4 | 11850 | Toll-like receptor 4 | 57 |
| VEGFA | 12680 | Vascular endothelial growth factor A | 57 |
| IL10 | 5962 | Interleukin 10 | 53 |
| MT-RNR2 | 7471 | Mitochondrially encoded 16S RNA | 51 |
| INS | 6081 | Insulin | 46 |
| PGR | 8910 | Progesterone receptor | 42 |
| IGF1 | 5464 | Insulin-like growth factor 1 (Somatomedin C) | 39 |
| TGFB1 | 11766 | Transforming growth factor beta 1 | 39 |
| SFTPD | 10803 | Surfactant, pulmonary-associated protein D | 38 |
| MMP9 | 7176 | Matrix metallopeptidase 9 (Gelatinase B, 92kDa gelatinase, 92 kDa type IV collagenase) | 36 |
| NR3C1 | 7978 | Nuclear receptor subfamily 3, group C, member 1 (Glucocorticoid receptor) | 35 |
| SFTPA2B | 23441 | Surfactant, pulmonary-associated protein A2B | 34 |
| IL1A | 5991 | Interleukin 1 alpha | 33 |
新窗口打开|下载CSV
通过对疾病数据库OMIM、ClinVar和CTD的挖掘,找到1个早产相关基因(SERPINH1)。由于该基因已存在于上述355个基因中,因此最终用于分析的基因数目不变。
GO富集分析发现174种显著的生物学功能(FDR<0.05)。根据显著性由高到低排列,前10种生物学功能包括:受体配体活性(receptor ligand activity)、细胞因子受体结合(cytokine receptor binding)、细胞因子活性(cytokine activity)、生长因子活性(growth factor activity)、生长因子结合(growth factor binding)、蛋白酶结合(protease binding)、血红素结合(heme binding)、生长因子受体结合(growth factor receptor binding)、四吡咯结合(tetrapyrrole binding)和脂多糖结合(lipopolysaccharide binding) (图1,附表3)。其中具有受体配体活性功能的基因数量最多,共有61个。
图1
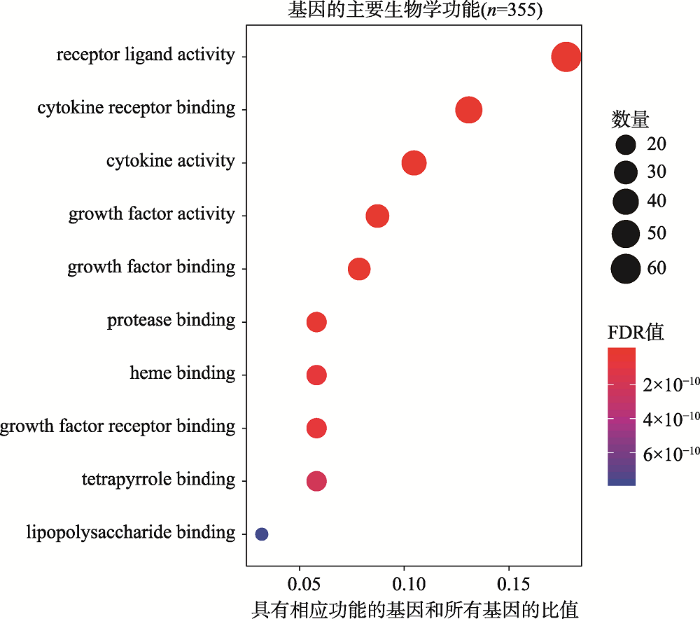 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1基因分子功能的GO富集
颜色代表FDR值的大小,由蓝色到红色FDR值逐渐变小,圆点的面积代表基因的数量。
Fig. 1GO enrichment analysis of molecular function in genes
Supplementary Table 3
附表3
附表3 早产相关基因分子功能的GO富集
Supplementary Table 3
| 分子功能 | 基因数量 | P值 | FDR |
|---|---|---|---|
| receptor ligand activity | 61 | 2.34*10-32 | 1.03*10-29 |
| cytokine receptor binding | 45 | 3.51*10-28 | 7.77*10-26 |
| cytokine activity | 36 | 4.29*10-23 | 6.32*10-21 |
| growth factor activity | 30 | 1.23*10-20 | 1.36*10-18 |
| growth factor binding | 27 | 1.03*10-19 | 9.14*10-18 |
| protease binding | 20 | 4.11*10-13 | 3.03*10-11 |
| heme binding | 20 | 1.02*10-12 | 6.44*10-11 |
| growth factor receptor binding | 20 | 1.18*10-12 | 6.52*10-11 |
| tetrapyrrole binding | 20 | 4.17*10-12 | 2.05*10-10 |
| lipopolysaccharide binding | 11 | 1.75*10-11 | 7.75*10-10 |
新窗口打开|下载CSV
KEGG富集分析发现的显著信号通路达到158个(FDR<0.05)。前10条通路根据显著性由高到低排列分别是:糖尿病并发症中的AGE-RAGE信号通路(AGE-RAGE signaling pathway in diabetic complications),Chagas病(美洲锥虫病),IL-17信号通路(IL-17 signaling pathway),TNF信号通路(TNF signaling pathway),PI3K-Akt信号通路(PI3K-Akt signaling pathway),Toll样受体信号通路(Toll-like receptor signaling pathway),结核(tuberculosis),炎症性肠病(inflammatory bowel disease (IBD)),乙型肝炎(hepatitis B)和流体剪切力和动脉粥样硬化(fluid shear stress and atherosclerosis) (图2,附表4)。
图2
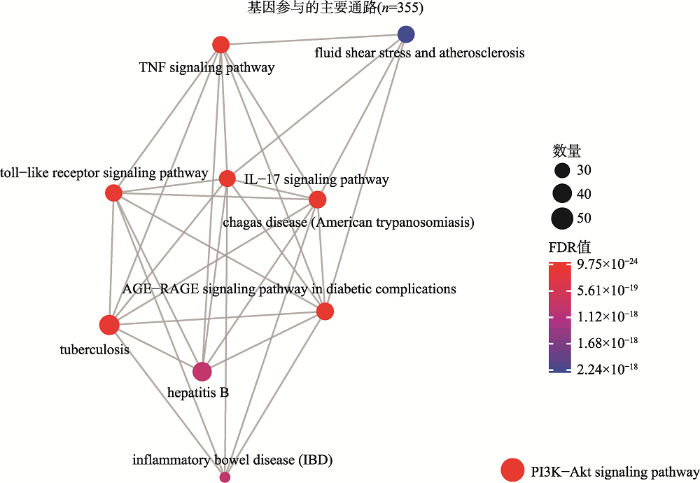 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2基因KEGG通路的富集结果
颜色代表FDR值的大小,由蓝色到红色FDR值逐渐变小,圆点的面积代表基因的数量。
Fig. 2KEGG enrichment analysis of genes
Supplementary Table 4
附表4
附表4 早产相关基因KEGG通路富集
Supplementary Table 4
| 通路 | 基因数量 | P值 | FDR |
|---|---|---|---|
| AGE-RAGE signaling pathway in diabetic complications | 35 | 1.02*10-25 | 9.75*10-24 |
| Chagas disease (American trypanosomiasis) | 35 | 3.26*10-25 | 1.56*10-23 |
| IL-17 signaling pathway | 33 | 1.63*10-24 | 5.19*10-23 |
| TNF signaling pathway | 34 | 5.68*10-23 | 1.36*10-21 |
| PI3K-Akt signaling pathway | 57 | 2.78*10-22 | 5.33*10-21 |
| Toll-like receptor signaling pathway | 32 | 1.33*10-21 | 2.13*10-20 |
| Tuberculosis | 40 | 4.46*10-21 | 6.10*10-20 |
| Inflammatory bowel disease (IBD) | 25 | 8.31*10-20 | 9.07*10-19 |
| Hepatitis B | 37 | 8.53*10-20 | 9.07*10-19 |
| Fluid shear stress and atherosclerosis | 34 | 2.34*10-19 | 2.24*10-18 |
新窗口打开|下载CSV
Reactome通路富集分析中前10个显著通路分别是:白细胞介素信号(Signaling by Interleukins),白细胞介素4和白细胞介素-13信号传导(Interleukin-4 and Interleukin-13 signaling),白细胞介素10信号传导(Interleukin-10 signaling),Toll样受体级联(Toll-like Receptor Cascades),Toll样受体4 (TLR4)级联(Toll Like Receptor 4 (TLR4) Cascade),Toll样受体TLR1:TLR2级联(Toll Like Receptor TLR1: TLR2 Cascade),Toll样受体2 (TLR2)级联(Toll Like Receptor 2 (TLR2) Cascade),免疫系统疾病(Diseases of Immune System),与TLR信号级联相关疾病(Diseases associated with the TLR signaling cascade),质膜上启动的MyD88:MAL (TIRAP)级联(MyD88:MAL (TIRAP) cascade initiated on plasma membrane) (图3,附表5)。
图3
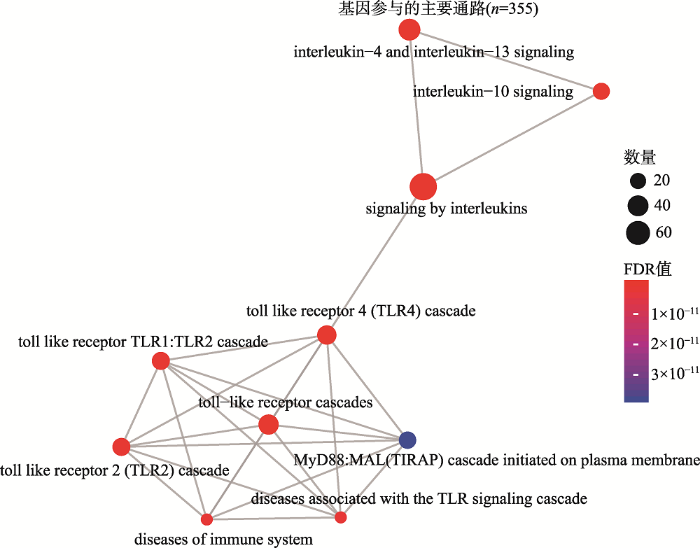 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3基因Reactome通路的富集
颜色代表FDR值的大小,由蓝色到红色FDR值逐渐变小,圆点的面积代表基因的数量。
Fig. 3Reactome enrichment analysis of genes
Supplementary Table 5
附表5
附表5 早产相关基因Reactome通路富集
Supplementary Table 5
| 通路 | 基因数量 | P值 | FDR |
|---|---|---|---|
| Signaling by Interleukins | 78 | 2.15*10-37 | 1.47*10-34 |
| Interleukin-4 and Interleukin-13 signaling | 40 | 2.20*10-33 | 7.54*10-31 |
| Interleukin-10 signaling | 20 | 1.26*10-18 | 2.87*10-16 |
| Toll-like Receptor Cascades | 32 | 3.57*10-18 | 6.12*10-16 |
| Toll Like Receptor 4 (TLR4) Cascade | 28 | 1.53*10-16 | 2.09*10-14 |
| Toll Like Receptor TLR1:TLR2 Cascade | 23 | 1.18*10-14 | 1.15*10-12 |
| Toll Like Receptor 2 (TLR2) Cascade | 23 | 1.18*10-14 | 1.15*10-12 |
| Diseases of Immune System | 13 | 2.88*10-14 | 2.19*10-12 |
| Diseases associated with the TLR signaling cascade | 13 | 2.88*10-14 | 2.19*10-12 |
| MyD88:MAL(TIRAP) cascade initiated on plasma membrane | 34 | 6.15*10-13 | 3.83*10-11 |
新窗口打开|下载CSV
2.2 基因特征的收集与分析结果
对比早产基因的每个基因转录本数量和全基因组每个基因的转录本数量,早产基因的转录本数量平均值(8.2)要高于全基因组基因的转录本数量平均值(7.5) (图4A)。在显著性水平α=0.1的情况下,差异显著(t检验:P=0.06)。针对GC含量的比较,早产基因和全基因组基因之间没有明显差异(t检验:P=0.70,α=0.1) (图4B)。图4
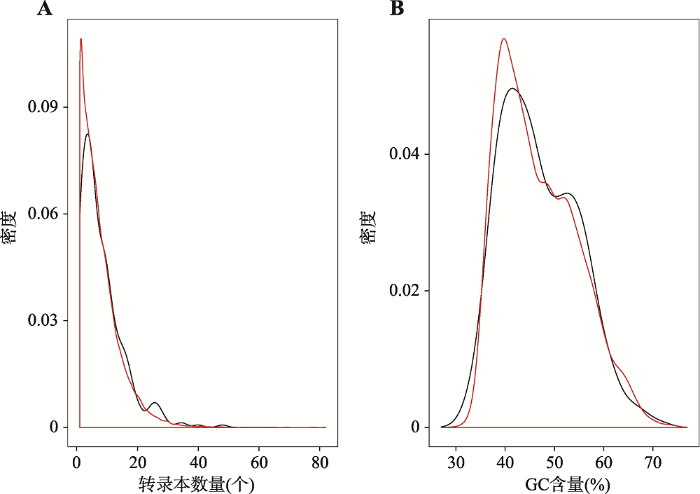 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4对比早产基因和全基因组基因的转录本数量以及GC含量
A:转录本数量分布(个);B:GC含量分布(%)。红色的曲线代表全基因组,黑色的曲线代表早产基因。
Fig. 4Comparisons between preterm birth related genes and genes in whole genome in terms of transcript numbers and GC contents
在早产基因长度和全基因组编码蛋白的基因长度的比较中发现,早产基因的平均长度为63 100 bp,而全基因组基因的长度平均为61 191 bp (图5)。在显著性水平α=0.1的情况下,差异不显著(t检验:P=0.73)。
图5
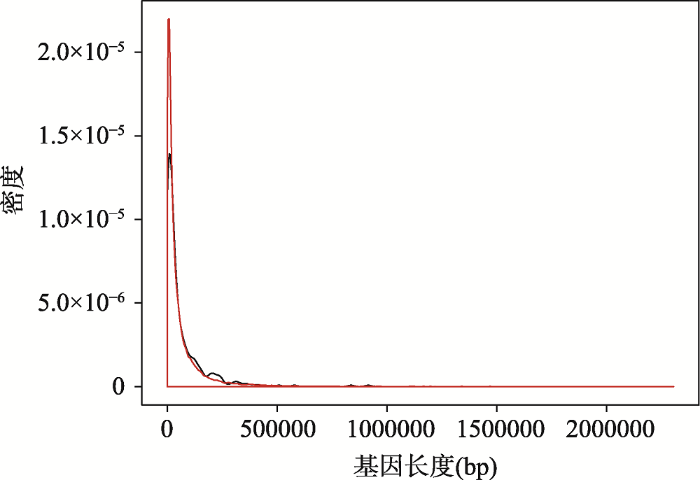 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5对比早产基因和全基因组编码蛋白基因的长度
红色的曲线代表全基因组,黑色的曲线代表早产基因。
Fig. 5Comparisons between preterm birth related genes and protein coding genes in whole genome in terms of gene lengths
3 讨论
早产是新生儿健康研究领域的一个极其重要的研究方向。虽然关于早产发生发展的分子作用机制尚不明确,但是已有大量研究表明早产的发生与遗传有关,并已产生了大量的数据。本研究通过文本挖掘工具挖掘PubMed中所检索的2264篇早产相关文献中的基因,结合阈值和人工审核的两层过滤以及疾病数据库记录,最终锁定355个早产相关基因。这是目前为止从文献中挖掘的最新的早产相关基因数据集。富集分析表明早产相关基因大多集中在免疫相关通路,基因特征分析发现早产相关基因和全基因组基因对比,GC含量和基因长度没有差异,而转录本数量有差异。以往的研究发现,免疫和炎症反应对维持妊娠和决定分娩时间起重要作用[8,20,21]。其中,由于父源和母源抗原的同时存在,母胎免疫耐受的维持在妊娠期间起重要作用,而这种稳态的破坏,可能会导致早产的发生[20]。先天免疫细胞通过释放炎性因子来影响妊娠过程和分娩时间,例如巨噬细胞释放的炎性因子可能促进催产素的产生,从而使子宫发生收缩,为分娩做准备[22]。同时,先天免疫和获得性免疫之间的失衡,也可能导致早产发生[23]。本研究采用挖掘得到的早产相关基因进行KEGG和Reactome富集分析,结果发现早产基因大多集中在免疫和炎症反应相关通路,这一点与以往的研究发现相吻合。先天免疫系统反映了对感染的应答作用,包括但不限于巨噬细胞、toll-like受体、噬中性粒细胞和细胞因子等;获得性免疫系统主要是T淋巴细胞和B淋巴细胞[24]。GO富集分析的结果也体现了早产相关基因具备与免疫过程密切相关的分子功能,包括受体配体活性、细胞因子受体活性等。本研究找到的前20个早产相关基因中,大多与免疫直接或间接相关。其中研究TNF基因的文献数目最多,研究包括胎儿肠膜发育和早产介导炎症[25]、环境内分泌物与孕期炎症生物标志物[26]。
据文献报道,人类基因组可能在疾病中具备一定特征[27,28],如慢性阻塞性肺疾病相关的基因转录本复杂度与对照组显著不同[29],内源性疾病的基因编码区具有高GC含量[30],在神经发育和神经退行性疾病中发现基因的长度扮演重要角色[31],其中在自闭症可能的候选基因中有许多长基因[32]。为进一步探索早产相关基因的基因组特征,本研究对比了早产相关基因与全基因组基因在转录本数量、GC含量和基因长度上的差异。其中,转录本数量存在差异。有研究发现,具有较多转录本数量的基因多为管家基因或必需基因,在生物学上起重要作用[33],然而针对转录本数量较多的早产相关基因,目前尚无文献报道。这些基因在早产所起的作用,仍需要进一步研究。GC含量在本研究中反映的是鸟嘌呤和胞嘧啶在每个基因中所占的比例。本研究并未发现早产相关基因与全基因组基因GC含量上存在显著差异。同时,早产基因在基因长度上与全基因组的所有基因相比,也无明显差异。
然而,本研究也有一定的局限性。首先,在数据库的甄选上,挖掘文献中早产相关基因时,也可以考虑包括中文数据库,例如CNKI,可以挖掘更多与中国人早产相关的研究和相关基因。其次,对基因的特征分析可以引入更多的变量,如种族信息等。对不同种族的研究,或许可以找到疾病相关且种族特异的遗传背景[34]。
综上所述,本研究结合文本挖掘和两层过滤方法以及疾病数据库记录,最终锁定355个早产相关基因,是截止到投稿时,最新的早产相关基因的整合记录。富集分析表明早产相关基因大多集中在免疫相关信号通路,基因特征分析提示了早产相关基因的转录本数量对比全基因组基因有一定差异。本研究对早产基因的挖掘和整合,可以为早产的遗传研究提供重要资源和提示相关研究方向。
(责任编委: 方向东)
附录
附表1~5见文章电子版参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
.
URL [本文引用: 2]
.
URL [本文引用: 2]
.
URL [本文引用: 1]
.
URLPMID:4678031 [本文引用: 1]

Preterm birth (PTB), defined as birth prior to a gestational age (GA) of 37 completed weeks, affects more than 10% of births worldwide. PTB is the leading cause of neonatal mortality and is associated with a broad spectrum of lifelong morbidity in survivors. The etiology of spontaneous PTB (SPTB) is complex and has an important genetic component. Previous studies have compared monozygotic and dizygotic twin mothers and their families to estimate the heritability of SPTB, but these approaches cannot separate the relative contributions of the maternal and the fetal genomes to GA or SPTB. Using the Utah Population Database, we assessed the heritability of GA in more than 2 million post-1945 Utah births, the largest familial GA dataset ever assembled. We estimated a narrow-sense heritability of 13.3% for GA and a broad-sense heritability of 24.5%. A maternal effect (which includes the effect of the maternal genome) accounts for 15.2% of the variance of GA, and the remaining 60.3% is contributed by individual environmental effects. Given the relatively low heritability of GA and SPTB in the general population, multiplex SPTB pedigrees are likely to provide more power for gene detection than will samples of unrelated individuals. Furthermore, nongenetic factors provide important targets for therapeutic intervention.
URLPMID:18295169 [本文引用: 1]

The objective of the study was to assess relative maternal and paternal genetic influences on birth timing. Utilizing The Netherlands Twin Registry, we examined the correlation in birth timing of infants born to monozygotic (MZ) twins and their first-degree relatives (dizygotic twins and siblings of twins). Genetic models estimated the relative influence of genetic and common environmental factors through model fitting of additive genetic (A), common environmental (C), individual-specific environmental factors, and combinations thereof. We evaluated birth timing correlation among the infants of 1390 twins and their 644 siblings. The correlation in MZ female twins ( r = 0.330) was greater than MZ male twins ( r = 610.096). Positive correlation were also found in sister-sister pairs ( r = 0.223) but not in brother-brother ( r = 610.045) or brother-sister pairs ( r = 610.038). The most parsimonious AE model indicated a significant maternal contribution of genetic and individual-specific environmental factors to birth timing, but no paternal heritability was demonstrated. Heritability of birth timing in women was 34%; and the remaining variance (66%) was caused by individual-specific environmental factors. Our data implicate a significant contribution of maternal but not paternal genetic influences on birth timing.
.
URLPMID:23568591 [本文引用: 1]

Although there is increasing evidence that genetic factors influence gestational age, it is unclear to what extent this is due to fetal and/or maternal genes. In this study, we apply a novel analytical model to estimate genetic and environmental contributions to pregnancy history records obtained from 165,952 Swedish families consisting of offspring of twins, full siblings, and half-siblings (1987-2008). Results indicated that fetal genetic factors explained 13.1% (95% confidence interval (CI): 6.8, 19.4) of the variation in gestational age at delivery, while maternal genetic factors accounted for 20.6% (95% CI: 18.1, 23.2). The largest contribution to differences in the timing of birth were environmental factors, of which 10.1% (95% CI: 7.0, 13.2) was due to factors shared by births of the same mother, and 56.2% (95% CI: 53.0, 59.4) was pregnancy specific. Similar models fit to the same data dichotomized at clinically meaningful thresholds (e. g., preterm birth) resulted in less stable parameter estimates, but the collective results supported a model of homogeneous genetic and environmental effects across the range of gestational age. Since environmental factors explained most differences in the timing of birth, genetic studies may benefit from understanding the specific effect of fetal and maternal genes in the context of these yet-unidentified factors.
URLMagsci [本文引用: 1]

探讨新生儿对氧磷酶2基因多态性(PON2148,PON2311)对早产的影响。采用横断面调查方法,使用统一的调查表,由安庆市各县医院对入院分娩孕妇及其单胎、活产、早产和对照新生儿进行调查,共得到有效样本194个母亲-新生儿对。单因素分析结果显示:PON2 Ala148Ala纯合子基因型与Gly148Gly纯合子基因型 / Ala148Gly杂合子基因型比较致早产的危险性升高且有显著意义;同样,PON2 Ser311Ser纯合子基因型致早产的危险性升高且有显著意义。进一步分析PON2148位点多态性和PON2311位点多态性是否存在交互作用,结果显示:这两个位点多态性之间无明显交互作用。对氧磷酶2基因PON2148位点多态性和PON2311位点多态性与新生儿早产相关,但PON2148位点多态性和PON2311位点多态性之间对早产的影响无明显交互作用。<br><br>Association of PON2 Gene Polymorphisms in Neonates with Preterm<br>LIANG Hong-ye1,WU Bai-yang1,CHEN Da-fang1,YANG Fan2,HU Hai-yan2,CHEN Li1,XU Xi-ping1.<br>1.Department of Biology & Genetics,Peking University Health Science Center,Beijing 100083,China;<br>2.Anqing Branch of Institute for Biomedicine,Anhui Medical University,Anqing 246000,China<br>Abstract:The objective is to investigate whether gene polymorphisms in the PON2 gene (PON2148 and PON2311) of neonates are associated with preterm. Using standard questionnaires,194 singleton live born mother-neonate pairs (include preterm cases and term controls) were investigated by the trained field workers with cross-sectional survey at the hospitals in Anqing,Anhui Province,China. Epidemiological and clinical data and blood samples were obtained from 194 mother-neonate pairs. Among neonates,PON2 Ala148Ala homozygote is significantly associated with preterm,compared with Gly148Gly homozygote / Ala148Gly heterozygote before and after adjustment confounders and the same was true for PON2 Ser311Ser homozygote. However,when PON2148 polymorphism and PON2311 polymorphism were considered jointly,no significant gene interaction between PON2148 polymorphism and PON2311 polymorphism in relation to preterm was observed. We draw a conclusion from this research that both PON2148 polymorphism and PON2311 polymorphism in neonates are significantly associated with preterm respectively. But the gene interactions between PON2148 polymorphism and PON2311 polymorphism in neonates are not significantly associated with preterm.<br>Key words:paraoxonase 2 gene (PON2 gene);gene polymorphism;preterm;genotype<br>
URLMagsci [本文引用: 1]

探讨新生儿对氧磷酶2基因多态性(PON2148,PON2311)对早产的影响。采用横断面调查方法,使用统一的调查表,由安庆市各县医院对入院分娩孕妇及其单胎、活产、早产和对照新生儿进行调查,共得到有效样本194个母亲-新生儿对。单因素分析结果显示:PON2 Ala148Ala纯合子基因型与Gly148Gly纯合子基因型 / Ala148Gly杂合子基因型比较致早产的危险性升高且有显著意义;同样,PON2 Ser311Ser纯合子基因型致早产的危险性升高且有显著意义。进一步分析PON2148位点多态性和PON2311位点多态性是否存在交互作用,结果显示:这两个位点多态性之间无明显交互作用。对氧磷酶2基因PON2148位点多态性和PON2311位点多态性与新生儿早产相关,但PON2148位点多态性和PON2311位点多态性之间对早产的影响无明显交互作用。<br><br>Association of PON2 Gene Polymorphisms in Neonates with Preterm<br>LIANG Hong-ye1,WU Bai-yang1,CHEN Da-fang1,YANG Fan2,HU Hai-yan2,CHEN Li1,XU Xi-ping1.<br>1.Department of Biology & Genetics,Peking University Health Science Center,Beijing 100083,China;<br>2.Anqing Branch of Institute for Biomedicine,Anhui Medical University,Anqing 246000,China<br>Abstract:The objective is to investigate whether gene polymorphisms in the PON2 gene (PON2148 and PON2311) of neonates are associated with preterm. Using standard questionnaires,194 singleton live born mother-neonate pairs (include preterm cases and term controls) were investigated by the trained field workers with cross-sectional survey at the hospitals in Anqing,Anhui Province,China. Epidemiological and clinical data and blood samples were obtained from 194 mother-neonate pairs. Among neonates,PON2 Ala148Ala homozygote is significantly associated with preterm,compared with Gly148Gly homozygote / Ala148Gly heterozygote before and after adjustment confounders and the same was true for PON2 Ser311Ser homozygote. However,when PON2148 polymorphism and PON2311 polymorphism were considered jointly,no significant gene interaction between PON2148 polymorphism and PON2311 polymorphism in relation to preterm was observed. We draw a conclusion from this research that both PON2148 polymorphism and PON2311 polymorphism in neonates are significantly associated with preterm respectively. But the gene interactions between PON2148 polymorphism and PON2311 polymorphism in neonates are not significantly associated with preterm.<br>Key words:paraoxonase 2 gene (PON2 gene);gene polymorphism;preterm;genotype<br>
.
URL [本文引用: 2]
.
URLPMID:17667860 [本文引用: 1]

Abstract Evidence is increasing for a role of polymorphisms in maternal or fetal innate immune response genes in preterm birth. Toll-like receptors (TLRs) are important receptors in the innate immunity. The genotype distribution of two TLR2 single nucleotide polymorphisms (SNPs) and one TLR4 SNP were determined among 524 neonates and associated with gestational age (GA). Genomic DNA was isolated from prospectively collected blood samples and polymorphisms in TLR2 (T-16934A, RS4696480 and Arg753Gln, RS5743708) and TLR4 (Thr399Ile, RS4986791) were determined using sequence specific primers by PCR. Allele frequencies of two TLR2 SNPs and one TLR4 SNP were analyzed according to prematurity. Analysis among 305 infants, after exclusion of infants born after multiple pregnancy or because of preeclampsia, revealed significantly shorter GAs for infants carrying two polymorphic TLR2 alleles (-16934TA/AA and 753ArgGln/GlnGln) compared with infants carrying one polymorphic and one wild-type allele or two wild-type alleles (median GA 30.6 wk versus 34.1-36.8 wk, respectively, p < 0.02). Carriage of two variant TLR2 alleles potentially leads to aberrant innate immune responses, which may have contributed to very preterm birth.
.
URLPMID:15059159 [本文引用: 1]

Background.68 There is convincing evidence for a central role of vascular endothelial growth factor (VEGF) in fetal and placental angiogenesis. Our present study was undertaken to examine the possible relationship between two common functional VEGF gene polymorphisms (6102634G/C and 936C/T), linked with altered VEGF gene responsiveness, and spontaneous preterm delivery. Methods.68 Genomic DNA was extracted from whole blood from 54 women with preterm labor and 79 menopausal women with at least two term spontaneous labors. DNA samples were analyzed by polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP). Results.68 Individuals with 936T/T or 936C/T genotype demonstrated a statistically significant association with preterm delivery compared with those sharing 936C/C genotype [ P 02=020.0009, risk factor 2.05, 95% confidence interval (CI) 1.37–3.06]. There were no significant associations between spontaneous preterm delivery and 6166634 genotypes. Conclusion.02 An association was demonstrated between the VEGF 936C/T polymorphism and deliveries before 3702weeks of gestation.
.
URLPMID:17676631 [本文引用: 1]

The occurrence of preterm delivery has been increasing in the U.S. Previous studies have identified risk factors for preterm delivery that may have genetic influences. We conducted a case-control study comparing the frequencies of 49 genetic polymorphisms among 62 preterm infants and 553 term infants. The polymorphisms that we examined were involved in xenobiotic-metabolism, blood pressure, coagulation, the inflammatory response, cell-cell interaction, or folate-homocysteine metabolism. Univariate ana- lyses on the individual polymorphisms revealed a statistically significant effect for the variant genotypes compared to the wildtype genotypes in SERPINE1 11053G > T (OR = 0.4, 95% CI=0.2-0.8). This finding suggests the coagulation/ thrombophilic pathway may influence the development of preterm delivery. (c) 2007 Wiley-Liss, Inc.
.
URLPMID:25599974 [本文引用: 1]

ABSTRACT Preterm birth is the leading cause of infant morbidity and mortality. Despite extensive research, the genetic contributions to spontaneous preterm birth (SPTB) are not well understood. Term controls were matched with cases by race/ethnicity, maternal age, and parity prior to recruitment. Genotyping was performed using Affymetrix SNP Array 6.0 assays. Statistical analyses utilized PLINK to compare allele occurrence rates between case and control groups, and incorporated quality control and multiple-testing adjustments. We analyzed DNA samples from mother–infant pairs from early SPTB cases (200/7–336/7 weeks, 959 women and 979 neonates) and term delivery controls (390/7–416/7 weeks, 960 women and 985 neonates). For validation purposes, we included an independent validation cohort consisting of early SPTB cases (293 mothers and 243 infants) and term controls (200 mothers and 149 infants). Clustering analysis revealed no population stratification. Multiple maternal SNPs were identified with association P -values between 10 × 10–5 and 10 × 10–6. The most significant maternal SNP was rs17053026 on chromosome 3 with an odds ratio (OR) 0.44 with a P -value of 1.0 × 10–6. Two neonatal SNPs reached the genome-wide significance threshold, including rs17527054 on chromosome 6p22 with a P -value of 2.7 × 10–12 and rs3777722 on chromosome 6q27 with a P -value of 1.4 × 10–10. However, we could not replicate these findings after adjusting for multiple comparisons in a validation cohort. This is the first report of a genome-wide case-control study to identify single nucleotide polymorphisms (SNPs) that correlate with SPTB.
.
URL [本文引用: 1]
.
URL [本文引用: 1]
.
URL [本文引用: 1]
URLPMID:3275764 [本文引用: 1]

<p id="p-2">Genome-wide association studies (GWAS) query the entire genome in a hypothesis-free, unbiased manner. Since they have the potential for identifying novel genetic variants, they have become a very popular approach to the investigation of complex diseases. Nonetheless, since the success of the GWAS approach varies widely, the identification of genetic variants for complex diseases remains a difficult problem. We developed a novel bioinformatics approach to identify the nominal genetic variants associated with complex diseases. To test the feasibility of our approach, we developed a web-based aggregation tool to organize the genes, genetic variations and pathways involved in preterm birth. We used semantic data mining to extract all published articles related to preterm birth. All articles were reviewed by a team of curators. Genes identified from public databases and archives of expression arrays were aggregated with genes curated from the literature. Pathway analysis was used to impute genes from pathways identified in the curations. The curated articles and collected genetic information form a unique resource for investigators interested in preterm birth. The Database for Preterm Birth exemplifies an approach that is generalizable to other disorders for which there is evidence of significant genetic contributions.
.
URLPMID:22455463 [本文引用: 1]

Abstract Increasing quantitative data generated from transcriptomics and proteomics require integrative strategies for analysis. Here, we present an R package, clusterProfiler that automates the process of biological-term classification and the enrichment analysis of gene clusters. The analysis module and visualization module were combined into a reusable workflow. Currently, clusterProfiler supports three species, including humans, mice, and yeast. Methods provided in this package can be easily extended to other species and ontologies. The clusterProfiler package is released under Artistic-2.0 License within Bioconductor project. The source code and vignette are freely available at http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html.
.
URLPMID:19188191 [本文引用: 1]

Summary:SciMiner is a web-based literature mining and functional analysis tool that identifies genes and proteins using a context specific analysis of MEDLINE abstracts and full texts. SciMiner accepts a free text query (PubMed Entrez search) or a list of PubMed identifiers as input. SciMiner uses both regular expression patterns and dictionaries of gene symbols and names compiled from multiple sources. Ambiguous acronyms are resolved by a scoring scheme based on the co-occurrence of acronyms and corresponding description terms, which incorporates optional user-defined filters. Functional enrichment analyses are used to identify highly relevant targets (genes and proteins), GO (Gene Ontology) terms, MeSH (Medical Subject Headings) terms, pathways and protein rotein interaction networks by comparing identified targets from one search result with those from other searches or to the full HGNC [HUGO (Human Genome Organization) Gene Nomenclature Committee] gene set. The performance of gene/protein name identification was evaluated using the BioCreAtIvE (Critical Assessment of Information Extraction systems in Biology) version 2 (Year 2006) Gene Normalization Task as a gold standard. SciMiner achieved 87.1% recall, 71.3% precision and 75.8%F-measure. SciMiner's literature mining performance coupled with functional enrichment analyses provides an efficient platform for retrieval and summary of rich biological information from corpora of users' interests. Availability:http://jdrf.neurology.med.umich.edu/SciMiner/. A server version of the SciMiner is also available for download and enables users to utilize their institution's journal subscriptions. Contact:juhur@umich.edu Supplementary information:Supplementary dataare available atBioinformaticsonline.
.
URLPMID:24243840 [本文引用: 1]

Reactome (http://www.reactome.org) is a manually curated open-source open-data resource of human pathways and reactions. The current version 46 describes 7088 human proteins (34% of the predicted human proteome), participating in 6744 reactions based on data extracted from 15 107 research publications with PubMed links. The Reactome Web site and analysis tool set have been completely redesigned to increase speed, flexibility and user friendliness. The data model has been extended to support annotation of disease processes due to infectious agents and to mutation.
URLPMID:25124429 [本文引用: 2]

Preterm birth is associated with 5 to 18% of pregnancies and is a leading cause of infant morbidity and mortality. Spontaneous preterm labor, a syndrome caused by multiple pathologic processes, leads to 70% of preterm births. The prevention and the treatment of preterm labor have been long-standing challenges. We summarize the current understanding of the mechanisms of disease implicated in this condition and review advances relevant to intra-amniotic infection, decidual senescence, and breakdown of maternal-fetal tolerance. The success of progestogen treatment to prevent preterm birth in a subset of patients at risk is a cause for optimism. Solving the mystery of preterm labor, which compromises the health of future generations, is a formidable scientific challenge worthy of investment.
.
URLPMID:15284723 [本文引用: 1]

Author information: (1)Perinatology Research Branch, National Institute of Child Health & Human Development/National Institutes of Health/Department of Health and Human Services, Bethesda, MD, USA.
.
URLPMID:10994633 [本文引用: 1]

PROBLEM: Little is known regarding the regulation of the timing of parturition. Recent evidence suggests an interaction between the immune system and uterine contractility in late gestation. METHOD: Pregnant rats were treated with LPS in vivo in attempts to establish a model of premature parturition induced by the pro-inflammatory response. Uterine explants were incubated in vitro to determine the effects of IL-6 on uterine synthesis of oxytocin (OT) and its receptor (OTR). RESULTS: LPS injection was quite toxic to pregnant rats and gave extremely variable results. In animals that delivered, there was a marked increase in the uterine concentrations of OTR and OTR mRNA. There was no consistent effect regarding the timing of parturition. IL-6 caused a significant increase in the concentration of OTR mRNA in uterine explants from pregnant rats but not in tissues from non-pregnant animals. CONCLUSION: Rat uterine concentrations of OTR are regulated by IL-6. Pro-inflammatory cytokines may stimulate uterine contractility in late gestation rat uterine tissues through a mechanism involving stimulation of OTR.
.
URLPMID:24954221 [本文引用: 1]

Abstract Labor resembles an inflammatory response that includes secretion of cytokines/chemokines by resident and infiltrating immune cells into reproductive tissues and the maternal/fetal interface. Untimely activation of these inflammatory pathways leads to preterm labor, which can result in preterm birth. Preterm birth is a major determinant of neonatal mortality and morbidity; therefore, the elucidation of the process of labor at a cellular and molecular level is essential for understanding the pathophysiology of preterm labor. Here, we summarize the role of innate and adaptive immune cells in the physiological or pathological activation of labor. We review published literature regarding the role of innate and adaptive immune cells in the cervix, myometrium, fetal membranes, decidua and the fetus in late pregnancy and labor at term and preterm. Accumulating evidence suggests that innate immune cells (neutrophils, macrophages and mast cells) mediate the process of labor by releasing pro-inflammatory factors such as cytokines, chemokines and matrix metalloproteinases. Adaptive immune cells (T-cell subsets and B cells) participate in the maintenance of fetomaternal tolerance during pregnancy, and an alteration in their function or abundance may lead to labor at term or preterm. Also, immune cells that bridge the innate and adaptive immune systems (natural killer T (NKT) cells and dendritic cells (DCs)) seem to participate in the pathophysiology of preterm labor. In conclusion, a balance between innate and adaptive immune cells is required in order to sustain pregnancy; an alteration of this balance will lead to labor at term or preterm.
.
URLPMID:3659282 [本文引用: 1]

Abstract Preterm birth occurs in 11% of live births globally and accounts for 35% of all newborn deaths. Preterm newborns have immature immune systems, with reduced innate and adaptive immunity; their immune systems may be further compromised by various factors associated with preterm birth. The immune systems of preterm infants have a smaller pool of monocytes and neutrophils, impaired ability of these cells to kill pathogens, and lower production of cytokines which limits T cell activation and reduces the ability to fight bacteria and detect viruses in cells, compared to term infants. Intrauterine inflammation is a major contributor to preterm birth, and causes premature immune activation and cytokine production. This can induce immune tolerance leading to reduced newborn immune function. Intrauterine inflammation is associated with an increased risk of early-onset sepsis and likely has long-term adverse immune consequences. Requisite medical interventions further impact on immune development and function. Antenatal corticosteroid treatment to prevent newborn respiratory disease is routine but may be immunosuppressive, and has been associated with febrile responses, reductions in lymphocyte proliferation and cytokine production, and increased risk of infection. Invasive medical procedures result in an increased risk of late-onset sepsis. Respiratory support can cause chronic inflammatory lung disease associated with increased risk of long-term morbidity. Colonization of the infant by microorganisms at birth is a significant contributor to the establishment of the microbiome. Caesarean section affects infant colonization, potentially contributing to lifelong immune function and well-being. Several factors associated with preterm birth alter immune function. A better understanding of perinatal modification of the preterm immune system will allow for the refinement of care to minimize lifelong adverse immune consequences.
[本文引用: 1]
.
URLPMID:24845688 [本文引用: 1]

Phthalate exposure during pregnancy has been linked to adverse birth outcomes such as preterm birth, and inflammation and oxidative stress may mediate these relationships. In a prospective cohort study of pregnant women recruited early in gestation in Northern Puerto Rico, we investigated the associations between urinary phthalate metabolites and biomarkers of inflammation, including C-reactive protein, IL-1尾, IL-6, IL-10, and TNF-伪, and oxidative stress, including 8-hydroxydeoxyguanosine (OHdG) and 8-isoprostane. Inflammation biomarkers were measured in plasma twice during pregnancy (N = 215 measurements, N = 120 subjects), and oxidative stress biomarkers in urine were measured three times (N = 148 measurements, N = 54 subjects) per woman. In adjusted linear mixed models, metabolites of di-2-ethylhexyl phthalate (DEHP) were associated with increased IL-6 and IL-10 but relationships were generally not statistically significant. All phthalates were associated with increases in oxidative stress markers. Relationships with OHdG were significant for DEHP metabolites as well as mono-n-butyl phthalate (MBP) and monoiso-butyl phthalate (MiBP). For 8-isoprostane, associations with nearly all phthalates were statistically significant and the largest effect estimates were observed for MBP and MiBP (49-50% increase in 8-isoprostane with an interquartile range increase in metabolite concentration). These relationships suggest a possible mechanism for phthalate action that may be relevant to a number of adverse health outcomes.
.
URLPMID:24425794 [本文引用: 1]

Increasing evidence indicates that genes containing disease causal variation have distinct functional and genomic properties. The importance of understanding these properties is highlighted by efforts to filter lists of variants from next-generation sequencing studies, where the number of potentially deleterious variants, which are in fact unrelated to disease, may be large. Available evidence indicates that the majority of disease genes are 鈥榥on-essential鈥 and their products occupy functionally peripheral positions in protein networks. They tend to be intermediate between genes that have core biological functions, particularly low mutation rates and low haplotype diversity, and genes for which high haplotype diversity and high mutation rates are advantageous (such as those involved in sensory perception and some immune system functions). Evidence presented here supports these conclusions through analysis of integrated data sets incorporating the latest mutational profiles, linkage disequilibrium structure and other genomic properties of individual genes. The analysis highlights the contrasting functions of genes predicted as least and most likely to contain disease variation and provides a basis for filtering gene variant lists to exclude the least plausible disease candidates.
.
URLPMID:28968721 [本文引用: 1]

Abstract Despite the identification of many genetic variants contributing to human disease (the 'disease genome'), establishing reliable molecular diagnoses remain challenging in many cases. The ability to sequence the genomes of patients has been transformative, but difficulty in interpretation of voluminous genetic variation often confounds recognition of underlying causal variants. There are numerous predictors of pathogenicity for individual DNA variants, but their utility is reduced because many plausibly pathogenic variants are probably neutral. The rapidly increasing quantity and quality of information on the properties of genes suggests that gene-specific information might be useful for prediction of causal variation when used alongside variant-specific predictors of pathogenicity. The key to understanding the role of genes in disease relates in part to gene essentiality, which has recently been approximated, for example, by quantifying the degree of intolerance of individual genes to loss-of-function variation. Increasing understanding of the interplay between genetic recombination, selection and mutation and their relationship to gene essentiality suggests that gene-specific information may be useful for the interpretation of sequenced genomes. Considered alongside additional distinctive properties of the disease genome, such as the timing of the evolutionary emergence of genes and the roles of their products in protein networks, the case for using gene-specific measures to guide filtering of sequenced genomes seems strong. The Author 2017. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
.
URLPMID:4610675 [本文引用: 1]

Genome-wide association studies aim to correlate genotype with phenotype. Many common diseases including Type II , Alzheimer's, Parkinson's and (COPD) are complex genetic traits with hundreds of different loci that are associated with varied disease risk. Identifying common features in the genes associated with each disease remains a challenge. Furthermore, the role of post-transcriptional regulation, and in particular alternative splicing, is still poorly understood in most multigenic diseases. We therefore compiled comprehensive lists of genes associated with Type II , Alzheimer's, Parkinson's and COPD in an attempt to identify common features of their corresponding mRNA transcripts within each gene set. The gene is a well-recognized genetic risk factor of COPD and it produces 11 transcript variants, which is exceptional for a gene. This led us to hypothesize that other genes associated with COPD, and complex disorders in general, are highly transcriptionally diverse. We found that COPD-associated genes have a statistically significant enrichment in transcript complexity stemming from a disproportionately high level of alternative splicing, however, Type II , Alzheimer's and genes were not significantly enriched. We also identified a subset of transcriptionally complex COPD-associated genes (~40%) that are differentially expressed between mild, moderate and severe COPD. Although the genes associated with other are not extensively documented, we found preliminary data that idiopathic genes, but not modulators, are also more transcriptionally complex. Interestingly, complex COPD transcripts are more often the product of alternative acceptor site usage. To verify the biological importance of these alternative transcripts, we used RNA-sequencing analyses to determine that COPD-associated genes are frequently expressed in lung and liver tissues and are regulated in a tissue-specific manner. Additionally, many complex COPD-associated genes are spliced differently between COPD and non-COPD patients. Our analysis therefore suggests that post-transcriptional regulation, particularly alternative splicing, is an important feature specific to COPD disease etiology that warrants further investigation.
.
URLPMID:28232902 [本文引用: 1]

We analyze a correlation between the GC content in genes of 12 eukaryotic species and the level of intrinsic disorder in their corresponding proteins. Comprehensive computational analysis has revealed that the disordered regions in eukaryotes are encoded by the GC-enriched gene regions and that this enrichment is correlated with the amount of disorder and is present across proteins and species characterized by varying amounts of disorder. The GC enrichment is a result of higher rate of amino acid coded by GC-rich codons in the disordered regions. Individual amino acids have the same GC-content profile between different species. Eukaryotic proteins with the disordered regions encoded by the GC-enriched gene segments carry out important biological functions including interactions with RNAs, DNAs, nucleotides, binding of calcium and metal ions, are involved in transcription, transport, cell division and certain signaling pathways, and are localized primarily in nucleus, cytosol and cytoplasm. We also investigate a possible relationship between GC content, intrinsic disorder and protein evolution. Analysis of a devised 090008age090009 of amino acids, their disorder-promoting capacity and the GC-enrichment of their codons suggests that the early amino acids are mostly disorder-promoting and their codons are GC-rich while most of late amino acids are mostly order-promoting.
.
URLPMID:25905808 [本文引用: 1]

A recent study by Gabel et al. (2015) found that Mecp2, the gene mutated in Rett syndrome, represses long (> 100 kb) genes associated with neuronal physiology and connectivity by binding to methylated CA sites in DNA. This study adds to a growing body of literature implicating gene length and transcriptional mechanisms in neurodevelopmental and neurodegenerative disorders.
.
URLPMID:23995680 [本文引用: 1]

Abstract Topoisomerases are expressed throughout the developing and adult brain and are mutated in some individuals with autism spectrum disorder (ASD). However, how topoisomerases are mechanistically connected to ASD is unknown. Here we find that topotecan, a topoisomerase 1 (TOP1) inhibitor, dose-dependently reduces the expression of extremely long genes in mouse and human neurons, including nearly all genes that are longer than 200 ilobases. Expression of long genes is also reduced after knockdown of Top1 or Top2b in neurons, highlighting that both enzymes are required for full expression of long genes. By mapping RNA polymerase II density genome-wide in neurons, we found that this length-dependent effect on gene expression was due to impaired transcription elongation. Interestingly, many high-confidence ASD candidate genes are exceptionally long and were reduced in expression after TOP1 inhibition. Our findings suggest that chemicals and genetic mutations that impair topoisomerases could commonly contribute to ASD and other neurodevelopmental disorders.
.
URLPMID:26279404 [本文引用: 1]

Alternative splicing is a process observed in gene expression that results in a multi-exon gene to produce multiple mRNA variants which might have different functions and activities. Although physiologically important, many aspects of genes with different number of transcript variants (or splice variants) still remain to be characterized. In this study, we provide bioinformatic evidence that genes with a greater number of transcript variants are more likely to play functionally important roles in cells, compared with those having fewer transcript variants. Among 21鈥983 human genes, 3728 genes were found to have a single transcript, and the remaining genes had 2 to 77 transcript variants. The genes with more transcript variants exhibited greater frequencies of acting as housekeeping and essential genes rather than tissue-selective and non-essential genes. They were found to be more conserved among 64 vertebrate species as orthologs, subjected to regulations by transcription factors and microRNAs, and showed hub node-like properties in the human protein rotein interaction network. These findings were also confirmed by metabolic simulations of 60 cancer metabolic models. All these results indicate that genes with a greater number of transcript variants play biologically more fundamental roles.
.
URLPMID:5760643 [本文引用: 1]

Abstract Preterm birth (PTB), or the delivery prior to 37 weeks of gestation, is a significant cause of infant morbidity and mortality. Although twin studies estimate that maternal genetic contributions account for approximately 30% of the incidence of PTB, and other studies reported fetal gene polymorphism association, to date no consistent associations have been identified. In this study, we performed the largest reported genome-wide association study analysis on 1,349 cases of PTB and 12,595 ancestry-matched controls from the focusing on genomic fetal signals. We tested over 2 million single nucleotide polymorphisms (SNPs) for associations with PTB across five subpopulations: African (AFR), the Americas (AMR), European, South Asian, and East Asian. We identified only two intergenic loci associated with PTB at a genome-wide level of significance: rs17591250 (P090009=0900094.55E-09) on chromosome 1 in the AFR population and rs1979081 (P090009=0900093.72E-08) on chromosome 8 in the AMR group. We have queried several existing replication cohorts and found no support of these associations. We conclude that the fetal genetic contribution to PTB is unlikely due to single common genetic variant, but could be explained by interactions of multiple common variants, or of rare variants affected by environmental influences, all not detectable using a GWAS alone.
