 ,中国农业科学院北京畜牧兽医研究所,北京 100193
,中国农业科学院北京畜牧兽医研究所,北京 100193Applications of genome selection in sheep breeding
Zhida Zhao, Li Zhang ,Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing 100193, China
,Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing 100193, China通讯作者:
编委: 蒋思文
收稿日期:2018-11-14修回日期:2019-02-5网络出版日期:2019-04-20
| 基金资助: |
Received:2018-11-14Revised:2019-02-5Online:2019-04-20
| Fund supported: |
作者简介 About authors
赵志达,硕士研究生,专业方向:羊分子育种及生产E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (359KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
赵志达, 张莉. 基因组选择在绵羊育种中的应用[J]. 遗传, 2019, 41(4): 293-303 doi:10.16288/j.yczz.18-251
Zhida Zhao, Li Zhang.
畜禽育种通常通过估算育种值(estimated breeding values, EBVs)来预测后代性状,以期得到符合人类需求的产品。EBV的准确度决定了育种工作效率。随着高通量测序技术和生物信息学的发展,利用各种基因组学手段挖掘重要经济性状功能基因已成为畜禽育种的研究热点。目前,已鉴定出一批和畜禽经济性状具有显著相关的数量性状基因座(quantitative trait locus, QTL)。标记辅助选择(marker assisted selection, MAS)就是利用这些已知的QTL进行选育,在一定程度上提高了育种工作效率,但目前所挖掘到的QTL尚不足总量的20%,且MAS难以解释复杂的数量性状,存在一定的局限性。2001年,Meuwissen等[1]首次提出基因组选择(genome selection, GS),该方法使用覆盖全基因组的全部遗传标记进行育种值估计,相比传统方法具有更高的准确性,不仅可以实现早期选种、缩短世代间隔,还有降低近交、加速遗传进展的优点。
目前,GS已成为动植物育种领域的研究热点。我国已在奶牛、生猪和肉鸡中开展,在其他畜禽上应用较少。中国作为养羊大国,对羊及其产品的需求逐年增长,如何选育和培育产量更高、品质更好的优良品种,是当前羊产业发展亟需解决的问题。目前,英国、法国、澳大利亚和新西兰等国家已将GS技术应用于羊的育种中并取得了良好的育种进展,而我国在这方面仍是空白。本文对绵羊(Ovis aries)基因组育种技术和GS的发展及应用进行了综述,以期为今后在绵羊上开展GS研究提供借鉴与参考。
1 分子育种技术的发展现状
传统育种主要利用表型信息和系谱信息,通过最佳线性无偏估计(best linear unbiased predication, BLUP)计算育种值,并以此作为畜禽选育的基础从而获得理想的遗传进展。BLUP法在畜禽一些重要经济性状的选择上取得了较好的效果,极大地推动了我国畜禽育种工作。但是通过系谱得出的只是期望的遗传关系,实际中可能因孟德尔抽样离差而失准;同时,对于一些低遗传力、受性别限制、不能早期测定、测定成本高,以及测量难度大的性状,传统育种方法存在较大的局限性。随着分子遗传学和数量遗传学的发展,畜禽育种逐渐聚焦到分子水平和基因组水平,即基因组育种技术。限制性片段长度多态性技术(restriction fragment length polymorphism, RFLP)于1974年面世,该技术推动了人类对DNA多态性的研究。MAS通过RFLP标记基因组并利用标记的基因型估计性状的表型,以此进行选种。该方法挖掘了与畜禽重要经济性状具有显著相关的QTL或主效基因并进行选种选育,当起始基因位点之间连锁不平衡(linkage disequilibrium, LD)很强时效果更佳,在清除隐性有害基因时尤为明显,并且遗传标记遗传稳定,不易受环境影响,无性别、年龄限制,通过早期选择能够节省成更多的成本[2]。
目前,遗传标记的挖掘手段现已有十几种成熟的技术,如扩增片段长度多态性标记辅助选择(amplified fragment length polymorphism, AFLP)、RFLP、随机扩增多态性标记辅助选择(random amplified polymorphic DNA, RAPD)、微卫星标记辅助选择(simple sequence repeat, SSR)和单核苷酸多态性标记辅助选择(single nucleotide polymorphism, SNP)等。由于SNP是基于单核苷酸水平的多态性,比其他标记遗传稳定性更高,而且因其密度高、分布广、检出率高,更适合快速高通量检测。因此,SNP已成为动植物育种和生命科学研究领域重要工具之一[3]。MAS通过挖掘与重要经济性状显著相关的分子标记,并利用这些标记实现性状的定向选择。该方法需要找到与性状LD的分子标记,但LD状态可能因重组而打破,在不同群体中LD程度亦不相同,所以该方法有时并不可靠。
随着高通量测序技术的发展、商用SNP芯片的问世及分型技术的诞生,测序成本大大降低。全基因组关联分析(genome-wide association study, GWAS)、拷贝数变异(copy number variation, CNV)等方法相继出现,并且用于畜禽经济性状主效基因的挖掘与鉴定。1996年,Risch等[4]提出GWAS方法,该方法以LD为基础,在全基因组水平上识别高密度的SNPs标记并与复杂性状的表型变异进行关联分析,从而分析这些SNPs对表型的遗传效应[5]。GWAS最早应用于人类疾病研究。2005年,Science杂志首次报道了与年龄具有相关性的黄斑变性GWAS研究[6];之后,又有****对冠心病[7]、肥胖[8,9]等疾病进行GWAS研究。目前GWAS已广泛应用于挖掘畜禽重要经济性状的遗传标记研究中。张莉等[10]利用50K芯片对329只绵羊(苏尼特羊、德国肉用美利奴羊和杜泊羊)开展体重性状的GWAS分析,检测到与断奶后日增重相关并达到基因组显著水平的SNPs位点10个,达到染色体显著水平的关联位点22个,获得了MEF2B、RFXANK和CAMKMT等影响绵羊体重性状的重要候选基因。Demars等[11]对Grvette和Olkuska两个绵羊品种进行GWAS分析,得到与绵羊产羔数相关的重要候选基因BMP15。Johnston[12]等对Soay绵羊进行GWAS分析,得到控制有无角性状的基因RXFP2。Doreen等[13]对23只特克赛尔羊的疾病性状进行GWAS分析,发现了影响小眼畸形病的主要候选基因PITX3,该基因位于22号染色体上。此外,还有许多针对绵羊多角、毛和毛色、生产发育、繁殖、乳用性状、疾病等不同经济性状的GWAS研究。
CNV是指DNA片段在kb~Mb范围内的突变,包括由插入、缺失、重复和复合多位点导致的变异。2004年,Sebat等[14]提出CNVs多态性在分析与判定人群间的遗传变异上有重要作用。2006年,Redon等[15]发表了人类基因组第一代CNV图谱。由于CNV与某些人类疾病的致病机理存在密切相关性,因此一直是研究的热点。CNV对培育性能优秀、抗病力强的家禽也具有重要意义,刘佳森等[16]利用50K羊芯片对71只苏尼特羊进行基因分型,共检测到134个拷贝数变异区域(copy number variation regions, CNVR),对CNVR覆盖的基因进行注释和功能分析,结果表明这些基因与嗅觉感官知觉、化学刺激感官知觉和识别等环境应答有关,对5个CNVR进行qRT-PCR验证,其中3个CNVR得到验证。侯成林等[17]采用比较基因组杂交芯片技术对3种绵羊(蒙古羊、哈萨克羊和藏羊)进行CNV研究,共检测到28个CNVR,通过对CNVR覆盖的基因进行注释和代谢通路分析,发现在蒙古羊和藏羊的血红蛋白基因存在CNV扩张,该现象可能与两种绵羊高原低氧的生活环境有关。经过验证,83.3%的qRT-PCR结果与芯片检测结果一致。
尽管GWAS和CNV等方法是挖掘畜禽经济性状主效基因的重要方法,但由于GWAS实验结果中存在大量假阳性问题,并且目前研究所得到的具有显著相关的QTL数量有限,同时畜禽大多数重要的经济性状都是受多基因与环境共同调控的复杂的数量性状,因此仅仅依靠以上方法进行选择还存在一定的不足。
2 基因组选择技术及其应用
随着畜禽基因组测序工作的相继完成,测序成本越来越低,加之计算机运算能力不断提升,这为全新育种技术的发展创造了技术条件。GS直接利用全基因组所有的标记效应去估计育种值,为现代育种工作提供了新的思路。2.1基因组选择的原理
畜禽性状受一个或多个基因控制,尽管已证实存在一些主效基因,但很多性状的功能基因尚不明确,仍在探索之中。DNA上存在大量的已知位置的SNP标记,它们和某些畜禽重要性状的基因存在一个或多个标记LD。由于每个SNP与主效基因间存在一个或多个LD,多个SNP就可能在不同程度上解释同一个基因的效应,即通过已知SNPs的多态性和表型来估计其效应。假定影响数量性状的每一个QTL和基因芯片中的SNPs存在多个LD[18],通过对全基因组范围内所有与QTL有关的SNP位点进行连锁检测,由参考群的表型估计出每个SNPs的效应,并根据其估计效应对候选群育种值进行估计,由此得到的育种值称为基因组育种值(genomic estimated breeding value, GEBV)[1]。该方法可以追溯到所有影响目标性状的QTL,进一步克服了传统MAS中标记揭示遗传方差较少的问题,提高了育种值的准确性[19],对育种工作起到至关重要的作用。利用SNP分型和SNP效应预测育种值公式:$GEBV={{m}_{1}}{{\hat{q}}_{1}}+{{m}_{2}}{{\hat{q}}_{2}}+\ldots +{{m}_{p}}{{\hat{q}}_{p}}=\underset{i=1}{\overset{p}{\mathop \sum }}\,{{m}_{i}}{{\hat{q}}_{i}}$
其中,${{m}_{i}}$表示第i位点的标记基因型,${{\hat{q}}_{i}}$表示第$i$位点的标记估计效应值,$p$表示标记数量。
Meuwissen等[1]通过模拟实验得出,仅通过标记预测育种值的准确性就可以达到0.85。2006年,牛全基因组序列信息公布,Schaeffer等[20]对GS在奶牛育种中的应用进行经济学分析,得出GS技术降低了92%的奶牛育种成本。此外,科研人员对GS在猪[21,22]、羊[22,23]和鸡[24,25,26]等畜禽中的应用策略也进行了探讨。
2.2 基因组育种值的常见算法
2.2.1 间接法间接法是通过构建参考群,利用参考群个体的表型和全基因组的分型信息,估计全基因组中每一个SNP在不同性状的效应值。之后通过对候选群进行SNPs分型,利用参考群SNPs的标记效应,累加获得候选群的GEBVs(图1)。
图1
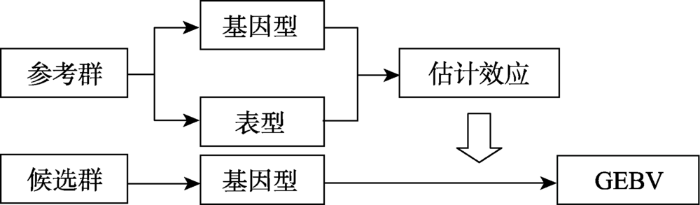 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1GS间接法模式计算图
Fig. 1Calculation diagram of GS indirect method
间接法常用的估计方法包括岭回归最佳线性无偏预测法(ridge regression best linear unbiased prediction, RRBLUP)和贝叶斯法[1,27,28](BayesA、BayesB、BayesC、BayesS和fBayesB)。
RRBLUP法假设所有标记都有效应,且方差相同;BayesA假设所有标记都有效应,且效应方差服从逆卡方分布(inversed chisquare distribution);BayesB假定标记效应方差以π(π为引入的一个新参数)的概率值为0,以(1-π)的概率服从逆卡方分布[1]。BayesS和BayesA的标记效应方差的分布假设相同,但在效应估计是运用压缩算法[27]。因BayesB计算时间长,Meuwissen等[29]通过条件期望迭代(iteration condition expectation)的算法改进得到fBayesB(fast BayesB)。Verbyla等[30]通过随机搜索变量选择(stochastic search variable selection, SSVS)对π在估计标记效应时进行求解。另外,Meuwissen等[31]将标记效应假定为两个方差不等的正态分布的混合分布,将方差参数和π作为模型变量求解,对BayesB改进后的算法称为BayesC,相较于前者,BayesC计算效率更高。
此外,还有主成分分析(principle component analysis)[32]、机器学习(machine learning)[33]、最小二乘回归(least square regression)[1]、半参数法(semiparametric)[34]、非参数法(nonparametric)[35]和贝叶斯LASSO法(least absolute shrinkage and selection operator)[36]等。
2.2.2 直接法
直接法是通过SNPs构建个体间关系矩阵,将关系矩阵放入混合模型方程组(mid model equations, MME)直接获得个体的基因组估计育种值(图2)。
图2
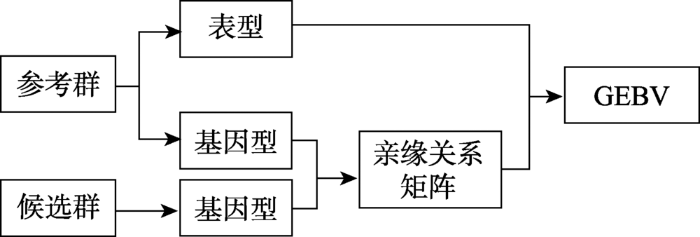 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2GS直接法模式计算图
Fig. 2Calculation diagram of GS direct method
直接法常用的估计方法包括:VanRaden等[37]和Habier等[38]提出的利用标记构建个体间关系矩阵G的基因组最佳线性无偏估计法(genome best linear unbiased prediction, GBLUP);Misztal等[39,40]、Legarra等[41]和Christensen等[42]先后提出的将系谱矩阵A和基因组关系矩阵G构建为H矩阵的一步法(single step GBLUP, SSBLUP);Zhang等[43]提出的给予不同标记不同权重构建关系矩阵的TABLUP法。
2.3 GEBV准确性的影响因素
GEBV的准确性是GEBV和真实育种值(true breeding values, TBV)的相关系数[1,44],计算公式为:$r=\frac{\text{GEBV}}{\text{TBV}}$
GEBV的准确性直接影响育种工作效率。几种影响GEBV准确性的常见因素如下。
2.3.1 计算模型
对于间接法来说,BayesB方法在多数情况下优于其他方法[1,44],Calus[45]认为这可能与模拟数据中有限的QTL数量有关,与BayesB方法的理论假设比较吻合。Meuwissen等[29]通过对BayesB进行改进得到了计算效率更好的BayesC,fBayesB的准确性比BayesB略低,但运算时间极大缩短。
对于直接法来说,Goddard等[46]和Hayes等[47]通过理论分析,认为GBLUP和RRBLUP使用相同的信息且假设相同,并得出以上两个方法等价的结论。Zhang等[43]认为,考虑到影响育种值评估时性状的所有基因的位置在全基因组中分布不均匀、基因效应不等的情况,应对标记先进行加权再构建个体关系矩阵,提出了构建TA矩阵(trait-specific relationship matrix, TA矩阵)的TABLUP法。该方法充分考虑了目标性状的遗传结构,因此优于RRBLUP和GBLUP法[48]。
另外,Garrick等[49]认为,对于由少数QTL控制的性状,或来自多品种、远缘相关基因型的数据,Bayes Lasso和MIXTURE算法有效地利用了LD信息,对GEBV的准确性优于GBLUP算法。Ostersen等[50]认为用deregressed EBV代替EBV作为响应变量(response variable),可提高GEBV的准确性,而算法应按照实际情况合理选择。
2.3.2 LD和标记数目
考虑到GS的假设前提为影响数量性状的每一个QTL和基因芯片中的SNPs存在多个LD,即SNPs与QTL必须要存在足够LD,即标记数目越多,准确度越高。Meuwissen等[1]通过类比计算得到相邻标记之间r2≥0.2才能进行GS;Calus等[45]则认为r2越大,准确度越大,即增加芯片的密度和容量,可提升准确性。但芯片密度越大,会增加成本和计算时长,因此在GS实际工作中,需要充分根据所选性状遗传力的高低和实验成本,选择合适的芯片。Erbe等[51]研究发现,考虑SNP间的LD程度可提升3.6%的准确性,该结果与增加芯片密度结果类似(芯片所含SNP个数由39 745增加到624 214)。此外,利用基因填充技术将低密度填充为中高密度芯片[52,53]和定制个性化芯片,在成本允许的情况下也可以提升GEBV的准确性。
2.3.3 参考群的规模和结构
在对标记效应进行估计时,需要对观察到的表型和基因型进行比较,实际情况中可能会出现各种误差。在概率论中,实验次数越多,频率越趋于概率。因此在减少测定误差的同时,要尽可能记录更多个体的表型信息,即参考群数量越大,标记效应估计越准确。Meuwissen等[1]通过模拟实验得出结论,在对遗传力为0.3的性状进行育种值估计时,样本数大于2000时准确性较高。Goddard等[54]研究表明,性状遗传力越低,所需样本越多,遗传力为0.1的性状需要10 000以上的群体数量才能保证准确度达到0.4。Jeremy等[55]认为,在分析混合性别模型时,应考虑性染色体上的LD,按性别估计GEBV。因此,为了提高GEBV的准确性,需要考虑群体结构,设计足够大的参考群体或选择遗传力较高的目标性状。
2.3.4 非加性效应
大部分重要性状都由多个基因共同调控,基因间的交互作用会影响表型,这种现象称为非加性效应(non-additive allelic effect),主要包括显性效应、上位效应和互作效应,不能稳定遗传。对于非加性效应的计算,Xu等[27,56]通过引入参数,利用近似方法简化计算并在大麦(Hordeum vulgare)回交实验中得到应用。Gianola等[34]提出半参数方法并使用模拟数据对一些互作效应进行了研究,认为该方法可能适用于高密度标记数据估计GEBV。
2.3.5 亲缘距离
LD在生殖细胞减数分裂过程中,配子中的DNA可能发生重组或者片段丢失等突变,使SNPs失去与原QTL的LD状态。Meuwissen等[1]通过研究发现,随世代增加,GEBV的准确性会逐渐降低,在经历5个世代后尤为明显,因此每经3个世代就需要重新估计标记效应。Habier等[38]发现随参考群与验证群间世代增加,GEBV的准确性却有所下降。另外,在不同群体间,QTL和SNPs间的LD不尽相同,因此在对同一品种间多场联合测定时,必须要考虑不同场区的LD和环境效应。由此可见,在绵羊实际育种工作中,需要不断完善和优化参考群体。每年可以从基因分型过的候选群中选出一部分进行表型测定,在拥有表型和基因分型时,该群体就可以并入参考群。
2.4 GS技术在羊育种中的应用
目前,国内外在奶牛上实施GS技术已取得明显的遗传进展,但在羊上的应用很少。羊是重要的经济家畜,也是当前我国实施乡村振兴战略和开展精准扶贫的适宜畜种。根据中国畜牧业信息网和FAO (联合国粮食及农业组织,http://www.fao.org)的最新统计,2009~2016年我国绵羊的数量从1.28亿头上升到1.62亿头,羊肉消费量也由433.7万吨增加到468.0万吨,预计今后还会逐年稳步增加。随着羊肉、羊毛、羊奶等羊产品的消费增长,开展羊高效育种和群体改良迫在眉睫,GS技术的实施也是必然的趋势。羊常规育种分为纯种繁育和杂交育种,其中以杂交育种居多。关于混合品种的GS,Dodds等[57]通过GBLUP计算方法对Romney、Coopworth和Perendale 3种肉毛兼用羊进行GEBV估计与验证,认为GS技术可以在混合品种的群体中进行GEBV估计,该研究考虑了品种结构并得出与预测结果相似的结论,并认为考虑品种结构的效应可以增加GEBV估计的准确度;另外,考虑动物的基因分型会减小偏差,提高准确性。Daetwyler等[58]使用50K芯片运用BayesA和GBLUP两种算法,以纯种美利奴羊和与终端杂交的美利奴羊作为参考群,研究了GS对肉质和羊毛预测的准确性,其中GBLUP和BayesA结论差异明显,说明当前SNP密度不足以跨物种使用标记效应,可能由不同品种中相同QTL的位置不同导致;该研究认为随着参考群样本含量以及SNP芯片密度增加,将会进一步提高GEBV的准确性。
繁殖性状直接影响养羊的经济效益,是育种中需要重点关注的经济性状,但由多基因控制且受环境影响,遗传力极低。Newton等[59]利用基因组信息评估澳大利亚绵羊繁殖性状的遗传进展,研究发现不同年龄的基因组信息会影响遗传增益,公羊1岁时的遗传增益显著高于2岁。Pickering等[60]对4237只Romney羊进行GS预测,发现繁殖性状的准确性在0.16~0.52之间。
胴体性状和羊毛性状也是绵羊的重要经济性状。Auvray等[61]对Romney、Coopworth、Perendale绵羊进行GS分析,并得出GS技术可以在以上3种羊中进行断奶重、胴体重和粪便虫卵数的育种值预测,在Romney中可以对产羔数进行预测。Slack-Smith等[62]研究发现,GS在澳大利亚绵羊胴体、生长性状、冻后眼肌面积和肌间脂肪含量的准确性分别为0.71、0.82、0.78和0.61。Brito等[63]在新西兰绵羊活体性状准确性范围为(0.18±0.07)~(0.33±0.10),胴体性状为(0.28±0.09)~(0.55±0.05),肉质性状为(0.21±0.07)~(0.36±0.08)。Moghaddar等[64]对Poll Dorset、White Suffolk和Border Leicester 3种羊的体重进行GEBV估计,Poll Dorset 的估计准确性为0.11~0.27,后两个品种的准确性在0.25~0.63之间。Shumbusho等[65]认为,在优化育种方案或使用全基因组信息均比传统方法得到更高的绵羊产肉性状遗传度进展。Daetwyler等[66]指出,仅针对纯种美利奴羊来说,1岁时的油性羊毛重量(greasy fleece weigh, GFW)和1岁时羊毛纤维直径(fibre diameter, FD)的准确度均超过0.70,短纤维强度(staple strength, SS)的准确性低于0.70,这可能与SS记录的数量少或者SS比GFW和FD的遗传力更低相关。Moghaddar等[64]发现GS对1岁和成年的Merino羊毛性状的估计准确性为0.33~0.75。Bolormaa等[67]对3种羊毛质量性状进行GS分析,发现BayesR和GBLUP算法的平均GEBV精确度相似,约为0.22,BayesR对羊毛产量和FD准确度均大于0.40,而对羊皮质量和污浊程度的准确度较差。
羊的泌乳性状与羔羊的成活率密切相关,且羊奶也具有重要的经济价值,因此对羊泌乳性状和乳房性状的研究也极为重要。Duchemin等[68]对法系Lacaune绵羊群体(约2500只)的泌乳性状进行了研究,认为GS技术可以使公羔的GEBV精确度提升18%~25%,BayesC计算时间过长,sPLS法对于GEBV的估计具有稳健性,其结果非常接近GBLUP结果。McLaren等[69]通过分析29个群体2957头纯种Texel母羊,认为其乳房形态和乳房炎有较高的相关性,并影响羊肉产量,该研究认为可以利用GS技术通过分析影响乳房形态的基因提高羊肉产量,并认为这是肉羊育种的一种新思路。Mucha等[70]对1960只英国奶山羊的产奶量和泌乳量进行GS试验,得出SSBLUP和GBLUP的预测准确性分别为0.61和0.32,SSBLUP具有更高的准确性。Carillier等[71]对法国Saanen奶山羊和Alpine羊进行GS选择育种,GEBV交叉验证的准确性为36%~53%。Larroque等[72]研究发现,GBLUP在奶山羊和奶绵羊上的准确性比荷斯坦奶牛差,但SSBLUP的准确性比GBLUP高,通过交叉验证发现,法国Lacaune奶绵羊GEBV准确性增加了0.47,Lacaune奶山羊增加了0.43。Baloche等[73]通过对比BLUP和SSBLUP对Lacaune奶山羊的选择效果,发现GS比传统育种方法的准确性更高,达到52%;此外,SSBLUP比BLUP准确性高58%。Molina等[74]使用55K芯片估计西班牙Florida奶山羊的GEBV,结果显示SSBLUP平均增加5.86%的准确性。
在验证不同设计模型中GS的效益研究中,Van der Werf等[23]在肉羊和细毛羊群体中进行GS模拟实验,结果表明,相比传统方法,GS方法在肉羊和细毛羊中经济选择指数分别提高了30%和40%。Raoul等[75]采用低密度SNP芯片(very low-density SNP panel)对Lacaune绵羊进行GS,结果表明:对雄性参考群使用中等密度基因型(medium-density genotypes)时,基因组设计(genomic design)的遗传效益提升26%,对同时包含雄性和雌性的参考群使用中等密度的基因型,其遗传效益提升54%;无论基因组情况如何,基因组设计的近亲繁殖率都会比传统设计低。对雌性和雄性候选绵羊采用极低密度的基因型与归集过程(imputation process)相结合导致小型的绵羊育种计划的遗传效益大幅增加。Santos等[76]通过比较不同的绵羊育种策略的准确性及经济收益,认为显性+选择性基因组选择(pheno + selective GS)可以在最短时间内实现盈亏平衡且效益最大,并有效降低了遗传间隔。
3 结语与展望
GS育种比传统育种技术具有更高的准确性,可实现早期选种、缩短世代间隔,还具有降低近交、加速遗传进展等优点,成为国内外畜禽育种工作的研究热点[77]。在我国,GS已在奶牛、生猪和肉鸡上大量应用,一些企业和实验室开始对绵羊育种进行GS探索,但至今尚未有系统的研究和结果。国外GS的案例已经证明该技术能显著提高绵羊的生长性状、羊毛性状和泌乳性状等选择的准确性。也有研究表明在考虑基因分型、增加样本量和群体构成时可获得更高的准确性,而采用极低密度芯片和归集过程相结合、显性和选择性GS相结合也可以增加遗传效益。GS技术对畜禽育种的贡献很大,但在理论研究和实际操作中也存在一些问题,主要表现在以下4个方面:(1)GS需要种羊场有完整的系谱、生产性能测定资料,同时需要持续的测定,测定费用高,回报慢;(2)GS对低遗传力性状选择效果较差,需要大规模的参考群;(3)GS依据参考群的SNPs效应估计候选群的GEBV,但不适用与新出现的精英个体和远缘个体[55];(4)计算时间较长,仍需进一步研究算法。
目前,生长速度慢、繁殖效率低是我国肉羊产业发展中的瓶颈问题,因此,在制定育种目标时应结合产业的需求进行合理的选育。生产大数据是开展GS研究的基础,羊场必须要持续开展生产性能测定并建立核心群生产信息数据平台。为降低GS成本,可以先采用传统方法进行选择,然后再进行SSBLUP选择。以某羊场为例,假设对3000只候选羊进行GS选育,育种目标是提高生长速度和增加产羔数,可以先用BLUP对候选群体进行育种值估计并排名,然后对排名前30%(选择强度视实际情况)的个体再进行GEBV估计。对这批羊进行后续生长性能测定,得到表型+系谱+基因型信息,该群体又可并入参考群中(节省参考群测定费用)。此外通过和其他相同品种的羊场进行联合育种[78],羊场之间可以互相导血,使开展联合育种的羊存在一定的亲缘关系,既可使联合场区的参考群的信息得到共享(另需考虑场区效应),同时还扩大了参考群的规模,提高了选择的准确性和效率,并极大程度地降低了测定成本。同时,还应结合对候选群体的表型选择,以减少精英个体的遗漏。此外,我国羊品种资源丰富,自主研发覆盖与绵羊重要经济性状相关联位点的中低密度育种芯片,亦能提高GBEV的准确性。
BLUP和MAS在羊育种工作中虽取得一定的成效,但面临由多基因控制的复杂数量性状时,只能在很小的层面去解释,而GS通过关联基因组内所有标记进行位点效应估计,很好地解决了传统方法难以解决的问题,是今后畜禽育种的大趋势,也是育种发展史上的一个重要里程碑。中国农业科学院北京畜牧兽医研究所对肉鸡和肉牛进行了GS研究并获得较好的成效,同时自主研发出鸡芯片“IASCHICK”。本研究团队将对已有的绵羊育种结果进行梳理和总结,研发更加适合我国绵羊育种的基因芯片,并开展湖羊的GS育种研究,探索GS技术在羊育种中的方法和效率。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 11]
URLMagsci [本文引用: 1]

随着分子数量遗传学及其相关学科的发展,有关动物遗传标记辅助选择方面的研究也在不断深入,且已经在动物遗传改良中有了一些成功应用的示例。就如何综合利用表型、系谱和遗传标记信息进行育种值估计的统计学方法研究方面,目前已基本形成了较为完善的统计学方法。同时,在标记辅助选择相对效率及其影响因素,以及标记辅助选择实施方案的研究上也取得了不少喜人的成果。本文综述了动物遗传标记辅助选择研究的一些进展,并对标记辅助选择在动物遗传改良中应用的有关问题进行了讨论。<br>Abstract:With the development of molecular and quantitative genetics and its related subjects,it made a great progress on the research about animal genetic marker-assisted selection (MAS).There were also some successful examples on the application of MAS to animal genetic improvement.The statistical method which using phenotypic,pedigree and genetic marker information to predict individual breeding values has already been developed.Many achievements were obtained from the researches,which carried on MAS relative efficiency and its affecting factors and selection schemes.The present paper reviewed some progresses of MAS research and discussed some problems about MAS application to animal breeding.
URLMagsci [本文引用: 1]

随着分子数量遗传学及其相关学科的发展,有关动物遗传标记辅助选择方面的研究也在不断深入,且已经在动物遗传改良中有了一些成功应用的示例。就如何综合利用表型、系谱和遗传标记信息进行育种值估计的统计学方法研究方面,目前已基本形成了较为完善的统计学方法。同时,在标记辅助选择相对效率及其影响因素,以及标记辅助选择实施方案的研究上也取得了不少喜人的成果。本文综述了动物遗传标记辅助选择研究的一些进展,并对标记辅助选择在动物遗传改良中应用的有关问题进行了讨论。<br>Abstract:With the development of molecular and quantitative genetics and its related subjects,it made a great progress on the research about animal genetic marker-assisted selection (MAS).There were also some successful examples on the application of MAS to animal genetic improvement.The statistical method which using phenotypic,pedigree and genetic marker information to predict individual breeding values has already been developed.Many achievements were obtained from the researches,which carried on MAS relative efficiency and its affecting factors and selection schemes.The present paper reviewed some progresses of MAS research and discussed some problems about MAS application to animal breeding.
.
URL [本文引用: 1]

SNP (single nucleotide polymorphism) marker, as a molecular marker of the third generation, plays an important role in molecular genetics, pharmacogenetics, forensic medicine, diagnosis and treatment of diseases recently. In this paper, the definition and characteristics of SNP molecular marker were both introduced. And the application profiles of Bin map, high throughput SNP-chip classification and population evolution research on the basis of SNP molecular marker was presented respectively. Finally, the problems of SNP molecular marker at the present stage were proposed, and the prospects of SNP molecular marker was predicted as well.
URL [本文引用: 1]

SNP (single nucleotide polymorphism) marker, as a molecular marker of the third generation, plays an important role in molecular genetics, pharmacogenetics, forensic medicine, diagnosis and treatment of diseases recently. In this paper, the definition and characteristics of SNP molecular marker were both introduced. And the application profiles of Bin map, high throughput SNP-chip classification and population evolution research on the basis of SNP molecular marker was presented respectively. Finally, the problems of SNP molecular marker at the present stage were proposed, and the prospects of SNP molecular marker was predicted as well.
URLPMID:9583430 [本文引用: 1]

The identification of the genetic basis of complex human diseases such as schizophrenia and diabetes has proven difficult. In their Perspective, Risch and Merikangas propose that we can best accomplish this goal by combining the power of the human genome project with association studies, a method for determining the basis of a genetic disease.
URLPMID:15716906 [本文引用: 1]

Abstract Genetic factors strongly affect susceptibility to common diseases and also influence disease-related quantitative traits. Identifying the relevant genes has been difficult, in part because each causal gene only makes a small contribution to overall heritability. Genetic association studies offer a potentially powerful approach for mapping causal genes with modest effects, but are limited because only a small number of genes can be studied at a time. Genome-wide association studies will soon become possible, and could open new frontiers in our understanding and treatment of disease. However, the execution and analysis of such studies will require great care.
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:17434869 [本文引用: 1]

Obesity is a serious international health problem that increases the risk of several common diseases. The genetic factors predisposing to obesity are poorly understood. A genome-wide search for type 2 diabetes-susceptibility genes identified a common variant in the FTO (fat mass and obesity associated) gene that predisposes to diabetes through an effect on body mass index (BMI). An additive association of the variant with BMI was replicated in 13 cohorts with 38,759 participants. The 16% of adults who are homozygous for the risk allele weighed about 3 kilograms more and had 1.67-fold increased odds of obesity when compared with those not inheriting a risk allele. This association was observed from age 7 years upward and reflects a specific increase in fat mass.
[本文引用: 1]

Motivated by mining major candidate genes across Ovine genome, the present study is to perform genome-wide association studies(GWAS) to detect genes associated with body weight traits. Using Illumina OvineSNP50 BeadChip, we performed a GWA study in 329 purebred sheep phenotyped for 6 body weight traits(birth weight, weaning weight, 6-month weight, pre-weaning gain, post-weaning gain, daily weight gain). Statistics and data analysis were based on TASSEL program,mixed linear model and the latest Ovis_aries_v3.1 genome sequence (released October 2012). The results indicated that 10 SNPs consistently reached genome-wise significant level for post-weaning gain and 22 SNPs reached chromosome-wise significant level for other body weight traits. The SNPs were within (MEF2B,RFXANK,et al) or close to some ovine genes, which were thought to be the most important candidate genes associated with body weight traits. The results will contribute to identify candidate genes for ovine body weight traits, and facilitate the potential utilization of genes involved production traits in sheep in future.
[本文引用: 1]

Motivated by mining major candidate genes across Ovine genome, the present study is to perform genome-wide association studies(GWAS) to detect genes associated with body weight traits. Using Illumina OvineSNP50 BeadChip, we performed a GWA study in 329 purebred sheep phenotyped for 6 body weight traits(birth weight, weaning weight, 6-month weight, pre-weaning gain, post-weaning gain, daily weight gain). Statistics and data analysis were based on TASSEL program,mixed linear model and the latest Ovis_aries_v3.1 genome sequence (released October 2012). The results indicated that 10 SNPs consistently reached genome-wise significant level for post-weaning gain and 22 SNPs reached chromosome-wise significant level for other body weight traits. The SNPs were within (MEF2B,RFXANK,et al) or close to some ovine genes, which were thought to be the most important candidate genes associated with body weight traits. The results will contribute to identify candidate genes for ovine body weight traits, and facilitate the potential utilization of genes involved production traits in sheep in future.
URL [本文引用: 1]
URLPMID:21651634 [本文引用: 1]

Understanding the genetic architecture of phenotypic variation in natural populations is a fundamental goal of evolutionary genetics. Wild Soay sheep (Ovis aries) have an inherited polymorphism for horn morphology in both sexes, controlled by a single autosomal locus, Horns. The majority of males have large normal horns, but a small number have vestigial, deformed horns, known as scurs; females have either normal horns, scurs or no horns (polled). Given that scurred males and polled females have reduced fitness within each sex, it is counterintuitive that the polymorphism persists within the population. Therefore, identifying the genetic basis of horn type will provide a vital foundation for understanding why the different morphs are maintained in the face of natural selection. We conducted a genome-wide association study using 鈭36 000 single nucleotide polymorphisms (SNPs) and determined the main candidate for Horns as RXFP2, an autosomal gene with a known involvement in determining primary sex characters in humans and mice. Evidence from additional SNPs in and around RXFP2 supports a new model of horn-type inheritance in Soay sheep, and for the first time, sheep with the same horn phenotype but different underlying genotypes can be identified. In addition, RXFP2 was shown to be an additive quantitative trait locus (QTL) for horn size in normal-horned males, accounting for up to 76% of additive genetic variation in this trait. This finding contrasts markedly from genome-wide association studies of quantitative traits in humans and some model species, where it is often observed that mapped loci only explain a modest proportion of the overall genetic variation.
URLPMID:2805710 [本文引用: 1]

Microphthalmia in sheep is an autosomal recessive inherited congenital anomaly found within the Texel breed. It is characterized by extremely small or absent eyes and affected lambs are absolutely blind. For the first time, we use a genome-wide ovine SNP array for positional cloning of a Mendelian trait in sheep. Genotyping 23 cases and 23 controls using Illumina's OvineSNP50 BeadChip allowed us to localize the causative mutation for microphthalmia to a 2.4 Mb interval on sheep chromosome 22 by association and homozygosity mapping. ThePITX3gene is located within this interval and encodes a homeodomain-containing transcription factor involved in vertebrate lens formation. An abnormal development of the lens vesicle was shown to be the primary event in ovine microphthalmia. Therefore, we consideredPITX3a positional and functional candidate gene. An ovine BAC clone was sequenced, and after full-length cDNA cloning thePITX3gene was annotated. Here we show that the ovine microphthalmia phenotype is perfectly associated with a missense mutation (c.338G>C, p.R113P) in the evolutionary conserved homeodomain ofPITX3. Selection against this candidate causative mutation can now be used to eliminate microphthalmia from Texel sheep in production systems. Furthermore, the identification of a naturally occurringPITX3mutation offers the opportunity to use the Texel as a genetically characterized large animal model for human microphthalmia.
URLPMID:15273396 [本文引用: 1]

The extent to which large duplications and deletions contribute to human genetic variation and diversity is unknown. Here, we show that large-scale copy number polymorphisms (CNPs) (about 100 kilobases and greater) contribute substantially to genomic variation between normal humans. Representational oligonucleotide microarray analysis of 20 individuals revealed a total of 221 copy number differences representing 76 unique CNPs. On average, individuals differed by 11 CNPs, and the average length of a CNP interval was 465 kilobases. We observed copy number variation of 70 different genes within CNP intervals, including genes involved in neurological function, regulation of cell growth, regulation of metabolism, and several genes known to be associated with disease.
[本文引用: 1]
URL [本文引用: 1]

In this study, 71 Sunite sheep were genotyped using Illumina OvineSNP50 BeadChip array, a total of 134 CNV regions (CNVR) across the genome were identified, which covered 25.95 Mb of the sheep genome.CNVR size on autosomes range from 29.48 kb to 1.30 Mb. Functional indicated that the most genes overrepresented in environmental responses, which involved in sensory perception of smell, sensory perception of chemical stimulus, sensory perception, cognition. To confirm these findings, quantitative PCR (qPCR) was performed for 5 CNVR and 3 of them were successfully validated. Our findings would be of help to understand the sheep genome and provide preliminary foundation for investigating the association between important economic traits and CNV.
URL [本文引用: 1]

In this study, 71 Sunite sheep were genotyped using Illumina OvineSNP50 BeadChip array, a total of 134 CNV regions (CNVR) across the genome were identified, which covered 25.95 Mb of the sheep genome.CNVR size on autosomes range from 29.48 kb to 1.30 Mb. Functional indicated that the most genes overrepresented in environmental responses, which involved in sensory perception of smell, sensory perception of chemical stimulus, sensory perception, cognition. To confirm these findings, quantitative PCR (qPCR) was performed for 5 CNVR and 3 of them were successfully validated. Our findings would be of help to understand the sheep genome and provide preliminary foundation for investigating the association between important economic traits and CNV.
URLMagsci [本文引用: 1]

为寻找绵羊基因组中可能的遗传性状相关标记,本试验采用比较基因组杂交(comparative genomic hybridization,CGH)芯片技术,构建了蒙古羊、哈萨克羊、藏羊的拷贝数变异(copy number variation,CNV)多样性图谱。试验结果显示,共检测出28个CNV区域(CNV region,CNVRs),包括11个扩增型、15个缺失型和2个扩增—缺失型。通过功能注释和代谢通路分析发现,在蒙古羊和藏羊基因组中血红蛋白基因存在拷贝数扩张,可能与两种绵羊长期生活在高原低氧环境中产生的适应性有关。对CNVRs和CNV相关基因进行实时荧光定量PCR检验,83.3%的实时荧光定量PCR结果与芯片检测结果一致。通过对中国北方3种绵羊的基因组CNV的研究,为不同绵羊品种间遗传变异的研究奠定了基础。
URLMagsci [本文引用: 1]

为寻找绵羊基因组中可能的遗传性状相关标记,本试验采用比较基因组杂交(comparative genomic hybridization,CGH)芯片技术,构建了蒙古羊、哈萨克羊、藏羊的拷贝数变异(copy number variation,CNV)多样性图谱。试验结果显示,共检测出28个CNV区域(CNV region,CNVRs),包括11个扩增型、15个缺失型和2个扩增—缺失型。通过功能注释和代谢通路分析发现,在蒙古羊和藏羊基因组中血红蛋白基因存在拷贝数扩张,可能与两种绵羊长期生活在高原低氧环境中产生的适应性有关。对CNVRs和CNV相关基因进行实时荧光定量PCR检验,83.3%的实时荧光定量PCR结果与芯片检测结果一致。通过对中国北方3种绵羊的基因组CNV的研究,为不同绵羊品种间遗传变异的研究奠定了基础。
[本文引用: 1]
URLPMID:19109259 [本文引用: 1]

Genetic progress will increase when breeders examine genotypes in addition to pedigrees and phenotypes. Genotypes for 38,416 markers and August 2003 genetic evaluations for 3,576 Holstein bulls born before 1999 were used to predict January 2008 daughter deviations for 1,759 bulls born from 1999 through 2002. Genotypes were generated using the Illumina BovineSNP50 BeadChip and DNA from semen contributed by US and Canadian artificial-insemination organizations to the Cooperative Dairy DNA Repository. Genomic predictions for 5 yield traits, 5 fitness traits, 16 conformation traits, and net merit were computed using a linear model with an assumed normal distribution for marker effects and also using a nonlinear model with a heavier tailed prior distribution to account for major genes. The official parent average from 2003 and a 2003 parent average computed from only the subset of genotyped ancestors were combined with genomic predictions using a selection index. Combined predictions were more accurate than official parent averages for all 27 traits. The coefficients of determination (R2) were 0.05 to 0.38 greater with nonlinear genomic predictions included compared with those from parent average alone. Linear genomic predictions had R2 values similar to those from nonlinear predictions but averaged just 0.01 lower. The greatest benefits of genomic prediction were for fat percentage because of a known gene with a large effect. The R2 values were converted to realized reliabilities by dividing by mean reliability of 2008 daughter deviations and then adding the difference between published and observed reliabilities of 2003 parent averages. When averaged across all traits, combined genomic predictions had realized reliabilities that were 23% greater than reliabilities of parent averages (50 vs. 27%), and gains in information were equivalent to 11 additional daughter records. Reliability increased more by doubling the number of bulls genotyped than the number of markers genotyped. Genomic prediction improves reliability by tracing the inheritance of genes even with small effects.
URLPMID:16882088 [本文引用: 1]

Summary Animals can be genotyped for thousands of single nucleotide polymorphisms (SNPs) at one time, where the SNPs are located at roughly 1-cM intervals throughout the genome. For each contiguous pair of SNPs there are four possible haplotypes that could be inherited from the sire. The effects of each interval on a trait can be estimated for all intervals simultaneously in a model where interval effects are random factors. Given the estimated effects of each haplotype for every interval in the genome, and given an animal's genotype, a ‘genomic’ estimated breeding value is obtained by summing the estimated effects for that genotype. The accuracy of that estimator of breeding values is around 80%. Because the genomic estimated breeding values can be calculated at birth, and because it has a high accuracy, a strategy that utilizes these advantages was compared with a traditional progeny testing strategy under a typical Canadian-like dairy cattle situation. Costs of proving bulls were reduced by 92% and genetic change was increased by a factor of 2. Genome-wide selection may become a popular tool for genetic improvement in livestock.
URL [本文引用: 1]

This article describes the current status of sequencing of the pig genome and how the findings will benefit pig production in terms of genetic improvement through selection for better performance traits.
[本文引用: 2]
[本文引用: 2]
URLPMID:21255418 [本文引用: 1]

Background Genomic selection involves breeding value estimation of selection candidates based on high-density SNP genotypes. To quantify the potential benefit of genomic selection, accuracies of estimated breeding values (EBV) obtained with different methods using pedigree or high-density SNP genotypes were evaluated and compared in a commercial layer chicken breeding line. Methods The following traits were analyzed: egg production, egg weight, egg color, shell strength, age at sexual maturity, body weight, albumen height, and yolk weight. Predictions appropriate for early or late selection were compared. A total of 2,708 birds were genotyped for 23,356 segregating SNP, including 1,563 females with records. Phenotypes on relatives without genotypes were incorporated in the analysis (in total 13,049 production records). The data were analyzed with a Reduced Animal Model using a relationship matrix based on pedigree data or on marker genotypes and with a Bayesian method using model averaging. Using a validation set that consisted of individuals from the generation following training, these methods were compared by correlating EBV with phenotypes corrected for fixed effects, selecting the top 30 individuals based on EBV and evaluating their mean phenotype, and by regressing phenotypes on EBV. Results Using high-density SNP genotypes increased accuracies of EBV up to two-fold for selection at an early age and by up to 88% for selection at a later age. Accuracy increases at an early age can be mostly attributed to improved estimates of parental EBV for shell quality and egg production, while for other egg quality traits it is mostly due to improved estimates of Mendelian sampling effects. A relatively small number of markers was sufficient to explain most of the genetic variation for egg weight and body weight.
[本文引用: 1]
URLPMID:21406356 [本文引用: 1]

Salmonella propagation by apparently healthy chickens could be decreased by the selection and use of chicken lines that are more resistant to carrier state. Using a reduced set of markers, this study investigates, for the first time to the authors' knowledge, the feasibility of a genomic selection approach for resistance to carrier state in hen lines. In this study, commercial laying hen lines were divergently selected for resistance to Salmonella carrier state at 2 different ages: young chicks and adults at the peak of lay. A total of 600 birds were typed with 831 informative SNP markers and artificially infected with Salmonella Enteritidis. Phenotypes were collected 28 d (389 young animals) or 38 d (208 adults) after infection. Two types of variance component analyses, including SNP data or not, were performed and compared. The set of SNP used was efficient in capturing a large part of the genetic variation. Average accuracies from mixed model equations did not change between analyses, showing that using SNP data does not increase information in this data set. These results confirm that genomic selection for Salmonella carrier state resistance in laying hens is promising. Nevertheless, a denser SNP coverage of the genome on a greater number of animals is still needed to assess its feasibility and efficiency.
URLPMID:12618414 [本文引用: 3]

Molecular markers have been used to map quantitative trait loci. However, they are rarely used to evaluate effects of chromosome segments of the entire genome. The original interval-mapping approach and various modified versions of it may have limited use in evaluating the genetic effects of the entire genome because they require evaluation of multiple models and model selection. Here we present a Bayesian regression method to simultaneously estimate genetic effects associated with markers of the entire genome. With the Bayesian method, we were able to handle situations in which the number of effects is even larger than the number of observations. The key to the success is that we allow each marker effect to have its own variance parameter, which in turn has its own prior distribution so that the variance can be estimated from the data. Under this hierarchical model, we were able to handle a large number of markers and most of the markers may have negligible effects. As a result, it is possible to evaluate the distribution of the marker effects. Using data from the North American Barley Genome Mapping Project in double-haploid barley, we found that the distribution of gene effects follows closely an L-shaped Gamma distribution, which is in contrast to the bell-shaped Gamma distribution when the gene effects were estimated from interval mapping. In addition, we show that the Bayesian method serves as an alternative or even better QTL mapping method because it produces clearer signals for QTL. Similar results were found from simulated data sets of F2 and backcross (BC) families.
URLMagsci [本文引用: 1]

<p>基因组育种值估计是基因组选择的重要环节, 基因组育种值的准确性是基因组选择成功应用的关键, 而其准确性在很大程度上取决于估计方法。目前研究和应用最多的基因组育种值估计方法是贝叶斯(Bayes)和最佳线性无偏预测(BLUP)两大类方法。文章系统介绍了目前已提出的各种Bayes方法, 并总结了该类方法的估计效果和各方面的改进。模拟数据和实际数据研究结果都表明, Bayes类方法估计基因组育种值的准确性优于BLUP类方法, 特别对于存在较大效应QTL的性状其优势更明显。由于Bayes方法的理论和计算过程相对复杂, 目前其在实际育种中的运用不如BLUP类方法普遍, 但随着快速算法的开发和计算机硬件的改进, 计算问题有望得到解决; 另外, 随着对基因组和性状遗传结构研究的深入开展, 能为Bayes方法提供更为准确的先验信息, 从而使Bayes方法估计基因组育种值准确性的优势更加突出, 应用将会更加广泛。</p>
URLMagsci [本文引用: 1]

<p>基因组育种值估计是基因组选择的重要环节, 基因组育种值的准确性是基因组选择成功应用的关键, 而其准确性在很大程度上取决于估计方法。目前研究和应用最多的基因组育种值估计方法是贝叶斯(Bayes)和最佳线性无偏预测(BLUP)两大类方法。文章系统介绍了目前已提出的各种Bayes方法, 并总结了该类方法的估计效果和各方面的改进。模拟数据和实际数据研究结果都表明, Bayes类方法估计基因组育种值的准确性优于BLUP类方法, 特别对于存在较大效应QTL的性状其优势更明显。由于Bayes方法的理论和计算过程相对复杂, 目前其在实际育种中的运用不如BLUP类方法普遍, 但随着快速算法的开发和计算机硬件的改进, 计算问题有望得到解决; 另外, 随着对基因组和性状遗传结构研究的深入开展, 能为Bayes方法提供更为准确的先验信息, 从而使Bayes方法估计基因组育种值准确性的优势更加突出, 应用将会更加广泛。</p>
PMID:2637029 [本文引用: 2]

Genomic selection uses genome-wide dense SNP marker genotyping for the prediction of genetic values, and consists of two steps: (1) estimation of SNP effects, and (2) prediction of genetic value based on SNP genotypes and estimates of their effects. For the former step, BayesB type of estimators have been proposed, which assume a priori that many markers have no effects, and some have an effect coming from a gamma or exponential distribution, i.e. a fat-tailed distribution. Whilst such estimators have been developed using Monte Carlo Markov chain (MCMC), here we derive a much faster non-MCMC based estimator by analytically performing the required integrations. The accuracy of the genome-wide breeding value estimates was 0.011 (s.e. 0.005) lower than that of the MCMC based BayesB predictor, which may be because the integrations were performed one-by-one instead of for all SNPs simultaneously. The bias of the new method was opposite to that of the MCMC based BayesB, in that the new method underestimates the breeding values of the best selection candidates, whereas MCMC-BayesB overestimated their breeding values. The new method was computationally several orders of magnitude faster than MCMC based BayesB, which will mainly be advantageous in computer simulations of entire breeding schemes, in cross-validation testing, and practical schemes with frequent re-estimation of breeding values.
URLPMID:19922694 [本文引用: 1]

Genomic selection describes a selection strategy based on genomic breeding values predicted from dense single nucleotide polymorphism (SNP) data. Multiple methods have been proposed but the critical issue is how to decide whether an SNP should be included in the predictive set to estimate breeding values. One major disadvantage of the traditional Bayes B approach is its high computational demands caused by the changing dimensionality of the models. The use of stochastic search variable selection (SSVS) retains the same assumptions about the distribution of SNP effects as Bayes B, while maintaining constant dimensionality. When Bayesian SSVS was used to predict genomic breeding values for real dairy data over a range of traits it produced accuracies higher or equivalent to other genomic selection methods with significantly decreased computational and time demands than Bayes B.
URLPMID:2708128 [本文引用: 1]

Background Recent developments in SNP discovery and high throughput genotyping technology have made the use of high-density SNP markers to predict breeding values feasible. This involves estimation of the SNP effects in a training data set, and use of these estimates to evaluate the breeding values of other 'evaluation' individuals. Simulation studies have shown that these predictions of breeding values can be accurate, when training and evaluation individuals are (closely) related. However, many general applications of genomic selection require the prediction of breeding values of 'unrelated' individuals, i.e. individuals from the same population, but not particularly closely related to the training individuals. Methods Accuracy of selection was investigated by computer simulation of small populations. Using scaling arguments, the results were extended to different populations, training data sets and genome sizes, and different trait heritabilities. Results Prediction of breeding values of unrelated individuals required a substantially higher marker density and number of training records than when prediction individuals were offspring of training individuals. However, when the number of records was 2*Ne*L and the number of markers was 10*Ne*L, the breeding values of unrelated individuals could be predicted with accuracies of 0.88 ??? 0.93, where Ne is the effective population size and L the genome size in Morgan. Reducing this requirement to 1*Ne*L individuals, reduced prediction accuracies to 0.73???0.83. Conclusion For livestock populations, 1NeL requires about ~30,000 training records, but this may be reduced if training and evaluation animals are related. A prediction equation is presented, that predicts accuracy when training and evaluation individuals are related. For humans, 1NeL requires ~350,000 individuals, which means that human disease risk prediction is possible only for diseases that are determined by a limited number of genes. Otherwise, genotyping and phenotypic recording need to become very common in the future.
URLPMID:2671482 [本文引用: 1]

Partial least square regression (PLSR) and principal component regression (PCR) are methods designed for situations where the number of predictors is larger than the number of records. The aim was to compare the accuracy of genome-wide breeding values (EBV) produced using PLSR and PCR with a Bayesian method, 'BayesB'. Marker densities of 1, 2, 4 and 8 Ne markers/Morgan were evaluated when the effective population size (Ne) was 100. The correlation between true breeding value and estimated breeding value increased with density from 0.611 to 0.681 and 0.604 to 0.658 using PLSR and PCR respectively, with an overall advantage to PLSR of 0.016 (s.e = 0.008). Both methods gave a lower accuracy compared to the 'BayesB', for which accuracy increased from 0.690 to 0.860. PLSR and PCR appeared less responsive to increased marker density with the advantage of 'BayesB' increasing by 17% from a marker density of 1 to 8Ne/M. PCR and PLSR showed greater bias than 'BayesB' in predicting breeding values at all densities. Although, the PLSR and PCR were computationally faster and simpler, these advantages do not outweigh the reduction in accuracy, and there is a benefit in obtaining relevant prior information from the distribution of gene effects.
URLPMID:18076475 [本文引用: 1]

Summary Genome-wide association studies using single nucleotide polymorphisms (SNPs) can identify genetic variants related to complex traits. Typically thousands of SNPs are genotyped, whereas the number of phenotypes for which there is genomic information may be smaller. When predicting phenotypes, options for statistical model building range from incorporating all possible markers into the specification to including only sets of relevant SNPs (features). In the latter case, an efficient method of selecting influential features is required. A two-step feature selection method for binary traits was developed, which consisted of filtering (using information gain), and wrapping (using na07ve Bayesian classification). The filter reduces the large number of SNPs to a much smaller size, to facilitate the wrapper step. As the procedure is tailored for discrete outcomes, an approach based on discretization of phenotypic values was developed, to enable feature selection in a classification framework. The method was applied to chick mortality rates (0–1402days of age) on progeny from 201 sires in a commercial broiler line, with the goal of identifying SNPs (over 5000) related to progeny mortality. To mimic a case–control study, sires were clustered into two groups, low and high, according to two arbitrarily chosen mortality rate cut points. By varying these thresholds, 11 different ‘case–control’ samples were formed, and the SNP selection procedure was applied to each sample. To compare the 11 sets of chosen SNPs, predicted residual sum of squares (PRESS) from a linear model was used. The two-step method improved na07ve Bayesian classification accuracy over the case without feature selection (from around 50 to above 90% without and with feature selection in each case–control sample). The best case–control group (63 sires above or below the thresholds) had the smallest PRESS statistic among groups with model p-values below 0.003. The 17 SNPs selected using this group accounted for 31% of the variation in raw mortality rates between sire families.
URL [本文引用: 2]
[本文引用: 1]
URLPMID:19293140 [本文引用: 1]

Abstract The availability of genomewide dense markers brings opportunities and challenges to breeding programs. An important question concerns the ways in which dense markers and pedigrees, together with phenotypic records, should be used to arrive at predictions of genetic values for complex traits. If a large number of markers are included in a regression model, marker-specific shrinkage of regression coefficients may be needed. For this reason, the Bayesian least absolute shrinkage and selection operator (LASSO) (BL) appears to be an interesting approach for fitting marker effects in a regression model. This article adapts the BL to arrive at a regression model where markers, pedigrees, and covariates other than markers are considered jointly. Connections between BL and other marker-based regression models are discussed, and the sensitivity of BL with respect to the choice of prior distributions assigned to key parameters is evaluated using simulation. The proposed model was fitted to two data sets from wheat and mouse populations, and evaluated using cross-validation methods. Results indicate that inclusion of markers in the regression further improved the predictive ability of models. An R program that implements the proposed model is freely available.
URLPMID:18946147 [本文引用: 1]

Efficient methods for processing genomic data were developed to increase reliability of estimated breeding values and to estimate thousands of marker effects simultaneously. Algorithms were derived and computer programs tested with simulated data for 2,967 bulls and 50,000 markers distributed randomly across 30 chromosomes. Estimation of genomic inbreeding coefficients required accurate estimates of allele frequencies in the base population. Linear model predictions of breeding values were computed by 3 equivalent methods: 1) iteration for individual allele effects followed by summation across loci to obtain estimated breeding values, 2) selection index including a genomic relationship matrix, and 3) mixed model equations including the inverse of genomic relationships. A blend of first- and second-order Jacobi iteration using 2 separate relaxation factors converged well for allele frequencies and effects. Reliability of predicted net merit for young bulls was 63% compared with 32% using the traditional relationship matrix. Nonlinear predictions were also computed using iteration on data and nonlinear regression on marker deviations; an additional (about 3%) gain in reliability for young bulls increased average reliability to 66%. Computing times increased linearly with number of genotypes. Estimation of allele frequencies required 2 processor days, and genomic predictions required <1 d per trait, and traits were processed in parallel. Information from genotyping was equivalent to about 20 daughters with phenotypic records. Actual gains may differ because the simulation did not account for linkage disequilibrium in the base population or selection in subsequent generations.
URLPMID:20170500 [本文引用: 2]

Background The impact of additive-genetic relationships captured by single nucleotide polymorphisms (SNPs) on the accuracy of genomic breeding values (GEBVs) has been demonstrated, but recent studies...
URL [本文引用: 1]
URLPMID:20105546 [本文引用: 1]

The first national single-step, full-information (phenotype, pedigree, and marker genotype) genetic evaluation was developed for final score of US Holsteins. Data included final scores recorded from 1955 to 2009 for 6,232,548 Holsteins cows. BovineSNP50 (Illumina, San Diego, CA) genotypes from the Cooperative Dairy DNA Repository (Beltsville, MD) were available for 6,508 bulls. Three analyses used a repeatability animal model as currently used for the national US evaluation. The first 2 analyses used final scores recorded up to 2004. The first analysis used only a pedigree-based relationship matrix. The second analysis used a relationship matrix based on both pedigree and genomic information (single-step approach). The third analysis used the complete data set and only the pedigree-based relationship matrix. The fourth analysis used predictions from the first analysis (final scores up to 2004 and only a pedigree-based relationship matrix) and prediction using a genomic based matrix to obtain genetic evaluation (multiple-step approach). Different allele frequencies were tested in construction of the genomic relationship matrix. Coefficients of determination between predictions of young bulls from parent average, single-step, and multiple-step approaches and their 2009 daughter deviations were 0.24, 0.37 to 0.41, and 0.40, respectively. The highest coefficient of determination for a single-step approach was observed when using a genomic relationship matrix with assumed allele frequencies of 0.5. Coefficients for regression of 2009 daughter deviations on parent-average, single-step, and multiple-step predictions were 0.76, 0.68 to 0.79, and 0.86, respectively, which indicated some inflation of predictions. The single-step regression coefficient could be increased up to 0.92 by scaling differences between the genomic and pedigree-based relationship matrices with little loss in accuracy of prediction. One complete evaluation took about 2h of computing time and 2.7 gigabytes of memory. Computing times for single-step analyses were slightly longer (2%) than for pedigree-based analysis. A national single-step genetic evaluation with the pedigree relationship matrix augmented with genomic information provided genomic predictions with accuracy and bias comparable to multiple-step procedures and could account for any population or data structure. Advantages of single-step evaluations should increase in the future when animals are pre-selected on genotypes.
URLPMID:22818478 [本文引用: 1]

The single-step genomic BLUP (SSGBLUP) is a method that can integrate pedigree and genotypes at molecular markers in an optimal way. However, its present form (regular SSGBLUP) has a high computational cost (cubic in the number of genotyped animals) and may need extensive rewriting of genetic evaluation software. In this work, we propose several strategies to implement the single step in a simpler manner. The first one expands the single-step mixed-model equations to obtain equivalent equations from which the regular (including pedigree and records only) mixed-model equations are a subset. These new equations (unsymmetric extended SSGBLUP) have low computational cost, but require a nonsymmetric solver such as the biconjugate gradient stabilized method or successive underrelaxation, which is a variant of successive overrelaxation, with a relaxation factor lower than 1. In addition, we show a new derivation of the single-step method, which includes, as an extra effect, deviations from strictly polygenic breeding values. As a result, the same set of equations as above is obtained. We show that, whereas the new derivation shows apparent problems of nonpositive definiteness for certain covariance matrices, a proper equivalent model including imaginary effects always exists, leading always to the regular SSGBLUP mixed model equations. The system of equations can be solved (iterative SSGBLUP) by iterating between a pedigree and records evaluation and a genomic evaluation (each one solved by any iterative or direct method), whereas global iteration can use a block version of successive underrelaxation, which ensures convergence. The genomic evaluation can explicitly include marker or haplotype effects and possibly involve nonlinear (e.g., Bayesian by Markov chain Monte Carlo) methods. In a simulated example with 28,800 individuals and 1,800 genotyped individuals, all methods converged quickly to the same solutions. Using existing efficient methods with limited memory requirements to compute the products Gt and A22t for any t (where G and A22 are genomic and pedigree relationships for genotyped animals, and t is a vector), all strategies can be converted to iteration on data procedures for which the total number of operations is linear in the number of animals + number of genotyped animals number of markers.
URLPMID:2834608 [本文引用: 1]

Background The use of genomic selection in breeding programs may increase the rate of genetic improvement, reduce the generation time, and provide higher accuracy of estimated breeding values (EBVs). A number of different methods have been developed for genomic prediction of breeding values, but many of them assume that all animals have been genotyped. In practice, not all animals are genotyped, and the methods have to be adapted to this situation. Results In this paper we provide an extension of a linear mixed model method for genomic prediction to the situation with non-genotyped animals. The model specifies that a breeding value is the sum of a genomic and a polygenic genetic random effect, where genomic genetic random effects are correlated with a genomic relationship matrix constructed from markers and the polygenic genetic random effects are correlated with the usual relationship matrix. The extension of the model to non-genotyped animals is made by using the pedigree to derive an extension of the genomic relationship matrix to non-genotyped animals. As a result, in the extended model the estimated breeding values are obtained by blending the information used to compute traditional EBVs and the information used to compute purely genomic EBVs. Parameters in the model are estimated using average information REML and estimated breeding values are best linear unbiased predictions (BLUPs). The method is illustrated using a simulated data set. Conclusions The extension of the method to non-genotyped animals presented in this paper makes it possible to integrate all the genomic, pedigree and phenotype information into a one-step procedure for genomic prediction. Such a one-step procedure results in more accurate estimated breeding values and has the potential to become the standard tool for genomic prediction of breeding values in future practical evaluations in pig and cattle breeding.
[本文引用: 2]
URL [本文引用: 2]

Genomic selection (GS) is a marker-assisted selection method, in which high density markers covering the whole genome are used simultaneously for individual genetic evaluation via genomic estimated breeding values (GEBVs). GS can increase the accuracy of selection, shorten the generation interval by selecting individuals at the early stage of life, and accelerate genetic progress. With the availability of high density whole genome SNP (single nucleotide polymorphism) chips for livestock, GS is reshaping the conventional animal breeding systems. In many countries, GS is becoming the major genetic evaluation method for bull selection in dairy cattle and GS may soon completely replace the traditional genetic evaluation system. In recent years, GS has become an important research topic in animal, plant and aquiculture breeding and many exciting results have been reported. In this paper, the methods for obtaining GEBVs, factors affecting the accuracy of GEBVs, and the current status of implementation of GS in livestock are reviewed. Some unresolved issues related to GS in livestock are also discussed.
URLPMID:22443868 [本文引用: 2]

Animal breeding faces one of the most significant changes of the past decades the implementation of genomic selection. Genomic selection uses dense marker maps to predict the breeding value of animals with reported accuracies that are up to 0.31 higher than those of pedigree indexes, without the need to phenotype the animals themselves, or close relatives thereof. The basic principle is that because of the high marker density, each quantitative trait loci (QTL) is in linkage disequilibrium (LD) with at least one nearby marker. The process involves putting a reference population together of animals with known phenotypes and genotypes to estimate the marker effects. Marker effects have been estimated with several different methods that generally aim at reducing the dimensions of the marker data. Nearly all reported models only included additive effects. Once the marker effects are estimated, breeding values of young selection candidates can be predicted with reported accuracies up to 0.85. Although results from simulation studies suggest that different models may yield more accurate genomic estimated breeding values (GEBVs) for different traits, depending on the underlying QTL distribution of the trait, there is so far only little evidence from studies based on real data to support this. The accuracy of genomic predictions strongly depends on characteristics of the reference populations, such as number of animals, number of markers, and the heritability of the recorded phenotype. Another important factor is the relationship between animals in the reference population and the evaluated animals. The breakup of LD between markers and QTL across generations advocates frequent re-estimation of marker effects to maintain the accuracy of GEBVs at an acceptable level. Therefore, at low frequencies of re-estimating marker effects, it becomes more important that the model that estimates the marker effects capitalizes on LD information that is persistent across generations.
URL [本文引用: 1]
URLPMID:19220931 [本文引用: 1]

Abstract Dense marker genotypes allow the construction of the realized relationship matrix between individuals, with elements the realized proportion of the genome that is identical by descent (IBD) between pairs of individuals. In this paper, we demonstrate that by replacing the average relationship matrix derived from pedigree with the realized relationship matrix in best linear unbiased prediction (BLUP) of breeding values, the accuracy of the breeding values can be substantially increased, especially for individuals with no phenotype of their own. We further demonstrate that this method of predicting breeding values is exactly equivalent to the genomic selection methodology where the effects of quantitative trait loci (QTLs) contributing to variation in the trait are assumed to be normally distributed. The accuracy of breeding values predicted using the realized relationship matrix in the BLUP equations can be deterministically predicted for known family relationships, for example half sibs. The deterministic method uses the effective number of independently segregating loci controlling the phenotype that depends on the type of family relationship and the length of the genome. The accuracy of predicted breeding values depends on this number of effective loci, the family relationship and the number of phenotypic records. The deterministic prediction demonstrates that the accuracy of breeding values can approach unity if enough relatives are genotyped and phenotyped. For example, when 1000 full sibs per family were genotyped and phenotyped, and the heritability of the trait was 0.5, the reliability of predicted genomic breeding values (GEBVs) for individuals in the same full sib family without phenotypes was 0.82. These results were verified by simulation. A deterministic prediction was also derived for random mating populations, where the effective population size is the key parameter determining the effective number of independently segregating loci. If the effective population size is large, a very large number of individuals must be genotyped and phenotyped in order to accurately predict breeding values for unphenotyped individuals from the same population. If the heritability of the trait is 0.3, and N(e)=100, approximately 12474 individuals with genotypes and phenotypes are required in order to predict GEBVs of un-phenotyped individuals in the same population with an accuracy of 0.7 [corrected].
URL [本文引用: 1]
URLPMID:20043827 [本文引用: 1]

Background Genomic prediction of breeding values involves a so-called training analysis that predicts the influence of small genomic regions by regression of observed information on marker genotypes for a given population of individuals. Available observations may take the form of individual phenotypes, repeated observations, records on close family members such as progeny, estimated breeding values (EBV) or their deregressed counterparts from genetic evaluations. The literature indicates that researchers are inconsistent in their approach to using EBV or deregressed data, and as to using the appropriate methods for weighting some data sources to account for heterogeneous variance. Methods A logical approach to using information for genomic prediction is introduced, which demonstrates the appropriate weights for analyzing observations with heterogeneous variance and explains the need for and the manner in which EBV should have parent average effects removed, be deregressed and weighted. Results An appropriate deregression for genomic regression analyses is EBV/r2 where EBV excludes parent information and r2 is the reliability of that EBV. The appropriate weights for deregressed breeding values are neither the reliability nor the prediction error variance, two alternatives that have been used in published studies, but the ratio (1 - h2)/[(c + (1 - r2)/r2)h2] where c > 0 is the fraction of genetic variance not explained by markers. Conclusions Phenotypic information on some individuals and deregressed data on others can be combined in genomic analyses using appropriate weighting.
[本文引用: 1]
URL [本文引用: 1]
URLPMID:20412938 [本文引用: 1]

The availability of dense single nucleotide polymorphism (SNP) genotypes for dairy cattle has created exciting research opportunities and revolutionized practical breeding programs. Broader application of this technology will lead to situations in which genotypes from different low-, medium-, or high-density platforms must be combined. In this case, missing SNP genotypes can be imputed using family- or population-based algorithms. Our objective was to evaluate the accuracy of imputation in Jersey cattle, using reference panels comprising 2,542 animals with 43,385 SNP genotypes and study samples of 604 animals for which genotypes were available for 1, 2, 5, 10, 20, 40, or 80% of loci. Two population-based algorithms, fastPHASE 1.2 (P. Scheet and M. Stevens; University of Washington TechTransfer Digital Ventures Program, Seattle, WA) and IMPUTE 2.0 (B. Howie and J. Marchini; Department of Statistics, University of Oxford, UK), were used to impute genotypes on Bos taurus autosomes 1, 15, and 28. The mean proportion of genotypes imputed correctly ranged from 0.659 to 0.801 when 1 to 2% of genotypes were available in the study samples, from 0.733 to 0.964 when 5 to 20% of genotypes were available, and from 0.896 to 0.995 when 40 to 80% of genotypes were available. In the absence of pedigrees or genotypes of close relatives, the accuracy of imputation may be modest (generally 40,000 SNP) from a reference population. Accurate imputation of high-density genotypes from inexpensive low- or medium-density platforms could greatly enhance the efficiency of whole-genome selection programs in dairy cattle.
URLPMID:20965364 [本文引用: 1]

The availability of high-density bovine genotyping arrays made implementation of genomic selection possible in dairy cattle. Development of low-density single nucleotide polymorphism (SNP) panels will allow the extension of genomic selection to a larger portion of the population. Prediction of ungenotyped markers, called imputation, is a strategy that allows using the same low-density chips for all traits (and for different breeds). In the present study, we evaluated the accuracy of imputation with low-density genotyping arrays in the Dutch Holstein population. Five different sizes of genotyping arrays were tested, from 384 to 6,000 SNP. According to marker density, the overall allelic imputation error rate obtained with the program DAGPHASE, which relies on linkage disequilibrium and linkage, ranged from 11.7 to 2.0%, and that obtained with the program CHROMIBD, which relies on linkage and the set of all genotyped ancestors, ranged from 10.7 to 3.3%. However, imputation efficiency was influenced by the relationship between low-density and high-density genotyped animals. Animals with both parents genotyped had particularly low imputation error rates:
URLPMID:19448663 [本文引用: 1]

Genome-wide panels of SNPs have recently been used in domestic animal species to map and identify genes for many traits and to select genetically desirable livestock. This has led to the discovery of the causal genes and mutations for several single-gene traits but not for complex traits. However, the genetic merit of animals can still be estimated by genomic selection, which uses genome-wide SNP panels as markers and statistical methods that capture the effects of large numbers of SNPs simultaneously. This approach is expected to double the rate of genetic improvement per year in many livestock systems.
URL [本文引用: 2]

78 Equivalent models for Genomic Selection (GS) using GBLUP are demonstrated. 78 SNP effects are estimated by regression on genomic breeding values. 78 Genomic relationship matrices are biased due to the use of only common SNP. 78 Sex-linked loci are incorporated into GS and GWAS analyses. 78 Accuracy of GS depends on relatedness of individuals to training population members.
URL [本文引用: 1]

The doubled-haploid (DH) barley population (Harrington x TR306) developed by the North American Barley Genome Mapping Project (NABGMP) for QTL mapping consisted of 145 lines and 127 markers covering a total genome length of 1270 cM. These DH lines were evaluated in approximately 25 environments for seven quantitative traits: heading, height, kernel weight, lodging, maturity, test weight, and yield. We applied an empirical Bayes method that simultaneously estimates 127 main effects for all markers and 127(127-1)/2=8001 interaction effects for all marker pairs in a single model. We found that the largest main-effect QTL (single marker) and the largest epistatic effect (single pair of markers) explained approximately 18 and 2.6% of the phenotypic variance, respectively. On average, the sum of all significant main effects and the sum of all significant epistatic effects contributed 35 and 6% of the total phenotypic variance, respectively. Epistasis seems to be negligible for all the seven traits. We also found that whether two loci interact does not depend on whether or not the loci have individual main effects. This invalidates the common practice of epistatic analysis in which epistatic effects are estimated only for pairs of loci of which both have main effects.
In:
[本文引用: 1]
URLPMID:22585798 [本文引用: 1]

In genome-wide association studies, failure to remove variation due to population structure results in spurious associations. In contrast, for predictions of future phenotypes or estimated breeding values from dense SNP data, exploiting population structure arising from relatedness can actually increase the accuracy of prediction in some cases, for example, when the selection candidates are offspring of the reference population where the prediction equation was derived. In populations with large effective population size or with multiple breeds and strains, it has not been demonstrated whether and when accounting for or removing variation due to population structure will affect the accuracy of genomic prediction. Our aim in this study was to determine whether accounting for population structure would increase the accuracy of genomic predictions, both within and across breeds. First, we have attempted to decompose the accuracy of genomic prediction into contributions from population structure or linkage disequilibrium (LD) between markers and QTL using a diverse multi-breed () data set, genotyped for 48,640 SNP. We demonstrate that SNP from a single can achieve up to 86% of the accuracy for genomic predictions using all SNP. This result suggests that most of the prediction accuracy is due to population structure, because a single is expected to capture relationships but is unlikely to contain all QTL. We then explored principal component analysis () as an approach to disentangle the respective contributions of population structure and LD between SNP and QTL to the accuracy of genomic predictions. Results showed that fitting an increasing number of principle components (PC; as covariates) decreased within breed accuracy until a lower plateau was reached. We speculate that this plateau is a measure of the accuracy due to LD. In conclusion, a large proportion of the accuracy for genomic predictions in our data was due to variation associated with population structure. Surprisingly, accounting for this structure generally decreased the accuracy of across breed genomic predictions.
URL [本文引用: 1]

Genomic selection could be useful in sheep-breeding programs, especially if rams and ewes are first mated at an earlier age than is the current industry practice. However, young-ewe (1 year old) fertility rates are known to be lower and more variable than those of mature ewes. The aim of the present study was to evaluate how young-ewe fertility rate affects risk and expected genetic gain in Australian sheep-breeding programs that use genomic information and select ewes and rams at different ages. The study used stochastic simulation to model different flock age structures and young-ewe fertility levels with and without genomic information for Merino and maternal sheep-breeding programs. The results from 10 years of selection were used to compare breeding programs on the basis of the mean and variation in genetic gain. Ram and ewe age, availability of genomic information on males and young-ewe fertility level all significantly (P < 0.05) affected expected genetic gain. Higher young-ewe fertility rates significantly increased expected genetic gain. Low fertility rate of young ewes (10%) resulted in net genetic gain similar to not selecting ewes until they were 19 months old and did not increase breeding-program risk, as the likelihood of genetic gain being lower than the range of possible solutions from a breeding program with late selection of both sexes was zero. Genomic information was of significantly (P < 0.05) more value for 1-year-old rams than for 2-year-old rams. Unless genomic information was available, early mating of rams offered no greater gain in Merino breeding programs and increased breeding-program risk. It is concluded that genomic information decreases the risk associated with selecting replacements at 7 months of age. Genetic progress is unlikely to be adversely affected if fertility levels above 10% can be achieved. Whether the joining of young ewes is a viable management decision for a breeder will depend on the fertility level that can be achieved in their young ewes and on other costs associated with the early mating of ewes.
In:
URL [本文引用: 1]

ABSTRACT To identify the impact of using molecular breeding values (mBVs) on the New Zealand sheep dual-purpose (DP) index, genomic selection (GS) accuracies were estimated using a training and validation data set consisting of 4,237 genotyped and pedigree recorded Romney animals. Molecular BVs and their accuracies for a range of DP production traits including live weight, fleece weight, faecal egg count, dagginess, reproduction and survival were estimated. The Romney mBV accuracies ranged from 0.16 to 0.52. For the majority of production traits the accuracies of the mBVs contributed information equivalent to having 1 to 8 measured progeny. For the traits: number of lambs born, lamb survival and lamb survival maternal the mBVs contributed between 11 to 145 measured progeny, albeit lamb survival maternal had a large error estimate. Combined with reducing the generation interval of rams used, from 2 years to 1 year, the potential increase in genetic gain in using mBVs in a New Zealand DP index was estimated to be 84%.
URLPMID:25149326 [本文引用: 1]

The aim of genomic prediction is to predict breeding value from genomic data. We describe the development of genomic prediction equations and accuracies for molecular breeding values (MBV) for industry use, focusing on the methodology used to deal with predictions for the New Zealand sheep population structure. This is made up of a mixture of pure and crossbred animals, but principally Romney based. In particular, we used pedigree-based EBV for 8 traits (weaning weight as a direct effect, weaning weight as a maternal effect, live weight at 8 mo, live weight at 12 mo, greasy fleece weight at 12 mo, lamb fleece weight, adult fleece weight, and number of lambs born) and Illumina OvineSNP50 BeadChip genotypes from 13,420 animals to investigate BLUP with different genomic relationship matrices (GRM) based on SNP markers and to investigate varying sets of older animals (training sets) to predict the MBV of younger animals (validation sets). The GRM tested included modifications to account for allele frequency differences between breeds, rescaling so that the mean GRM is equal to the mean of the traditional pedigree numerator relationship matrix A: , and combining of the GRM with A: using a convex combination with a weight estimated by maximizing a conditional restricted likelihood. We found that these modifications were beneficial and recommend using a breed-adjusted GRM combined with A: . Training data sets with Romney, Coopworth, and Perendale animals all together usually predicted better than using just a pure breed training data set for all traits. But predictions for the breed Perendale were more accurate with a Perendale training set for 3 of the 8 traits. We concluded that using a mixed-breed training set for all combinations of traits and breeds was best but advise that increasing the number of Perendale animals genotyped should be a priority to increase the MBV accuracies obtained for that breed.
[本文引用: 1]
URLPMID:5267438 [本文引用: 1]

New Zealand has some unique Terminal Sire composite sheep breeds, which were developed in the last three decades to meet commercial needs. These composite breeds were developed based on crossing various Terminal Sire and Maternal breeds and, therefore, present high genetic diversity compared to other sheep breeds. Their breeding programs are focused on improving carcass and meat quality traits. There is an interest from the industry to implement genomic selection in this population to increase the rates of genetic gain. Therefore, the main objectives of this study were to determine the accuracy of predicted genomic breeding values for various growth, carcass and meat quality traits using a HD SNP chip and to evaluate alternative genomic relationship matrices, validation designs and genomic prediction scenarios. A large multi-breed population (n=6514,845) was genotyped with the HD SNP chip (60002K) and phenotypes were collected for a variety of traits. The average observed accuracies (± SD) for traits measured in the live animal, carcass, and, meat quality traits ranged from 0.1865±650.07 to 0.3365±650.10, 0.2865±650.09 to 0.5565±650.05 and 0.2165±650.07 to 0.3665±650.08, respectively, depending on the scenario/method used in the genomic predictions. When accounting for population stratification by adjusting for 2, 4 or 6 principal components (PCs) the observed accuracies of molecular breeding values (mBVs) decreased or kept constant for all traits. The mBVs observed accuracies when fitting bothGandAmatrices were similar to fitting onlyGmatrix. The lowest accuracies were observed for k-means cross-validation and forward validation performed within each k-means cluster. The accuracies observed in this study support the feasibility of genomic selection for growth, carcass and meat quality traits in New Zealand Terminal Sire breeds using the Ovine HD SNP chip. There was a clear advantage on using a mixed training population instead of performing analyzes per genomic clusters. In order to perform genomic predictions per breed group, genotyping more animals is recommended to increase the size of the training population within each group and the genetic relationship between training and validation populations. The different scenarios evaluated in this study will help geneticists and breeders to make wiser decisions in their breeding programs. The online version of this article (doi:10.1186/s12863-017-0476-8) contains supplementary material, which is available to authorized users.
URL [本文引用: 2]
URLPMID:23736059 [本文引用: 1]

In conventional small ruminant breeding programs, only pedigree and phenotype records are used to make selection decisions but prospects of including genomic information are now under consideration. The objective of this study was to assess the potential benefits of genomic selection on the genetic gain in French sheep and goat breeding designs of today. Traditional and genomic scenarios were modeled with deterministic methods for 3 breeding programs. The models included decisional variables related to male selection candidates, progeny testing capacity, and economic weights that were optimized to maximize annual genetic gain (AGG) of i) a meat sheep breeding program that improved a meat trait of heritability (h(2)) = 0.30 and a maternal trait of h(2) = 0.09 and ii) dairy sheep and goat breeding programs that improved a milk trait of h(2) = 0.30. Values of +/- 0.20 of genetic correlation between meat and maternal traits were considered to study their effects on AGG. The Bulmer effect was accounted for and the results presented here are the averages of AGG after 10 generations of selection. Results showed that current traditional breeding programs provide an AGG of 0.095 genetic standard deviation (sigma(a)) for meat and 0.061 sigma(a) for maternal trait in meat breed and 0.147 sigma(a) and 0.120 sigma(a) in sheep and goat dairy breeds, respectively. By optimizing decisional variables, the AGG with traditional selection methods increased to 0.139 sigma(a) for meat and 0.096 sigma(a) for maternal traits in meat breeding programs and to 0.174 sigma(a) and 0.183 sigma(a) in dairy sheep and goat breeding programs, respectively. With a medium-sized reference population (nref) of 2,000 individuals, the best genomic scenarios gave an AGG that was 17.9% greater than with traditional selection methods with optimized values of decisional variables for combined meat and maternal traits in meat sheep, 51.7% in dairy sheep, and 26.2% in dairy goats. The superiority of genomic schemes increased with the size of the reference population and genomic selection gave the best results when nref > 1,000 individuals for dairy breeds and nref > 2,000 individuals for meat breed. Genetic correlation between meat and maternal traits had a large impact on the genetic gain of both traits. Changes in AGG due to correlation were greatest for low heritable maternal traits. As a general rule, AGG was increased both by optimizing selection designs and including genomic information.
URL [本文引用: 1]

Estimated breeding values for the selection of more profitable sheep for the sheep meat and wool industries are currently based on pedigree and phenotypic records. With the advent of a medium-density DNA marker array, which genotypes ~50 000 ovine single nucleotide polymorphisms, a third source of information has become available. The aim of this paper was to determine whether this genomic information can be used to predict estimated breeding values for wool and meat traits. The effects of all single nucleotide polymorphism markers in a multi-breed sheep reference population of 7180 individuals with phenotypic records were estimated to derive prediction equations for genomic estimated breeding values (GEBV) for greasy fleece weight, fibre diameter, staple strength, breech wrinkle score, weight at ultrasound scanning, scanned eye muscle depth and scanned fat depth. Five hundred and forty industry sires with very accurate Australian sheep breeding values were used as a validation population and the accuracies of GEBV were assessed according to correlations between GEBV and Australian sheep breeding values . The accuracies of GEBV ranged from 0.15 to 0.79 for wool traits in Merino sheep and from 0.07 to 0.57 for meat traits in all breeds studied. Merino industry sires tended to have more accurate GEBV than terminal and maternal breeds because the reference population consisted mainly of Merino haplotypes. The lower accuracy for terminal and maternal breeds suggests that the density of genetic markers used was not high enough for accurate across-breed prediction of marker effects. Our results indicate that an increase in the size of the reference population will increase the accuracy of GEBV.
URLPMID:5558709 [本文引用: 1]

The application of genomic selection to sheep breeding could lead to substantial increases in profitability of wool production due to the availability of accurate breeding values from single nucleotide polymorphism (SNP) data. Several key traits determine the value of wool and influence a sheep susceptibility to fleece rot and fly strike. Our aim was to predict genomic estimated breeding values (GEBV) and to compare three methods of combining information across traits to map polymorphisms that affect these traits. GEBV for 5726 Merino and Merino crossbred sheep were calculated using BayesR and genomic best linear unbiased prediction (GBLUP) with real and imputed 510,174 SNPs for 22 traits (at yearling and adult ages) including wool production and quality, and breech conformation traits that are associated with susceptibility to fly strike. Accuracies of these GEBV were assessed using fivefold cross-validation. We also devised and compared three approximate multi-trait analyses to map pleiotropic quantitative trait loci (QTL): a multi-trait genome-wide association study and two multi-trait methods that use the output from BayesR analyses. One BayesR method used local GEBV for each trait, while the other used the posterior probabilities that a SNP had an effect on each trait. BayesR and GBLUP resulted in similar average GEBV accuracies across traits (~0.22). BayesR accuracies were highest for wool yield and fibre diameter (>0.40) and lowest for skin quality and dag score (<0.10). Generally, accuracy was higher for traits with larger reference populations and higher heritability. In total, the three multi-trait analyses identified 206 putative QTL, of which 20 were common to the three analyses. The two BayesR multi-trait approaches mapped QTL in a more defined manner than the multi-trait GWAS. We identified genes with known effects on hair growth (i.e.FGF5,STAT3,KRT86, andALX4) near SNPs with pleiotropic effects on wool traits. The mean accuracy of genomic prediction across wool traits was around 0.22. The three multi-trait analyses identified 206 putative QTL across the ovine genome. Detailed phenotypic information helped to identify likely candidate genes. The online version of this article (doi:10.1186/s12711-017-0337-y) contains supplementary material, which is available to authorized users.
URLPMID:22541502 [本文引用: 1]

Genomic selection aims to increase accuracy and to decrease generation intervals, thus increasing genetic gains in animal breeding. Using real data of the French Lacaune dairy sheep breed, the purpose of this study was to compare the observed accuracies of genomic estimated breeding values using different models (infinitesimal only, markers only, and joint estimation of infinitesimal and marker effects) and methods [BLUP, Bayes C , partial least squares (PLS), and sparse PLS]. The training data set included results of progeny tests of 1,886 rams born from 1998 to 2006, whereas the validation set had results of 681 rams born in 2007 and 2008. The 3 lactation traits studied (milk yield, fat content, and somatic cell scores) had heritabilities varying from 0.14 to 0.41. The inclusion of molecular information, as compared with traditional schemes, increased accuracies of estimated breeding values of young males at birth from 18 up to 25%, according to the trait. Accuracies of genomic methods varied from 0.4 to 0.6, according to the traits, with minor differences among genomic approaches. In Bayes C , the joint estimation of marker and infinitesimal effects had a slightly favorable effect on the accuracies of genomic estimated breeding values, and were especially beneficial for somatic cell counts, the less heritable trait. Inclusion of infinitesimal effects also improved slopes of predictive regression equations. Methods that select markers implicitly (Bayes C and sparse PLS) were advantageous for some models and traits, and are of interest for further quantitative trait loci studies.
URL [本文引用: 1]

Mastitis can prove expensive in sheep reared for meat production due to costs associated with treatment methods, poor lamb growth and premature culling of ewes. The most commonly used method to detect mastitis, in dairy systems, is somatic cell counts. However, in many meat-producing sheep flocks ewes are not routinely handled, thus regular milk sampling is not always possible. It is therefore worthwhile to investigate alternative phenotypes, such as those associated with udder conformation and methods of evaluating somatic cell counts in the milk, such as the California Mastitis Test. The main objectives of this study were therefore, a) to estimate genetic parameters of traits relating to mastitis and udder conformation in a meat sheep breed; b) estimate the level of association between somatic cell counts and the California Mastitis Test and c) assess the relationships between mastitis and both udder conformation and lamb live weights. Data were collected from Texel ewes based on 29 flocks, throughout the UK, during 2015 and 2016. The ewes were scored twice each year, at mid- and late-lactation. Eight different conformation traits, relating to udder and teat characteristics, and milk samples were recorded. The data set comprised of data available for 2 957 ewes. The pedigree file used contained sire and dam information for 31 775 individuals. The animal models used fitted relevant fixed and random effects. Heritability estimates for traits relating to mastitis (somatic cell score and the California Mastitis Test), ranged from 0.08 to 0.11 and 0.07 to 0.11 respectively. High genetic correlations were observed between somatic cell score and the California Mastitis Test (0.76 to 0.98), indicating the California Mastitis Test to be worthwhile for assessing infection levels, particularly at mid-lactation. The strongest correlations observed between the mastitis traits and the udder conformation traits were associated with udder depth (0.61 to 0.75) also at mid-lactation. Moderately negative correlations were also observed between the mastitis traits and teat angle, with those estimated at mid-lactation ranging from -0.41 to -0.55. Negative phenotypic correlations were estimated between mastitis and the weight of lamb reared by the ewe (-0.15 to -0.23), suggesting that lamb weights fell as infection levels rose. Genetic correlations were not significantly different from zero. Reducing mastitis will lead to improvements in flock productivity and the health and welfare of the animals. It will also improve the efficiency of production and the resilience to disease challenge. The economic benefits, therefore, of these results combined could be substantial not only in this breed but also in the overall meat sheep industry.
URLPMID:26342984 [本文引用: 1]

The objective of this study was to estimate genomic breeding values for milk yield in crossbred dairy goats. The research was based on data provided by 2 commercial goat farms in the UK comprising 590,409 milk yield records on 14,453 dairy goats kidding between 1987 and 2013. The population was created by crossing 3 breeds: Alpine, Saanen, and Toggenburg. In each generation the best performing animals were selected for breeding, and as a result, a synthetic breed was created. The pedigree file contained 30,139 individuals, of which 2,799 were founders. The data set contained test-day records of milk yield, lactation number, farm, age at kidding, and year and season of kidding. Data on milk composition was unavailable. In total 1,960 animals were genotyped with the Illumina 50K caprine chip. Two methods for estimation of genomic breeding value were compared LUP at the single nucleotide polymorphism level (BLUP-SNP) and single-step BLUP. The highest accuracy of 0.61 was obtained with single-step BLUP, and the lowest (0.36) with BLUP-SNP. Linkage disequilibrium (r2, the squared correlation of the alleles at 2 loci) at 50 kb (distance between 2 SNP) was 0.18. This is the first attempt to implement genomic selection in UK dairy goats. Results indicate that the single-step method provides the highest accuracy for populations with a small number of genotyped individuals, where the number of genotyped males is low and females are predominant in the reference population.
URL [本文引用: 1]
In:
URL [本文引用: 1]

ABSTRACT Abstract Text: Genomic selection opens new perspectives for breeding programs of dairy ruminants. In France, several projects have enabled the creation of reference populations in dairy sheep and goats. Early studies have shown that the reliability of genomic evaluations using a GBLUP method in these species is lower than in Holstein dairy cattle breed. The single-step approach gives best predictions for candidates at birth (genomic evaluation accuracy obtained by cross validation for milk yield of 0.47 in Lacaune dairy sheep and 0.43 in goats breeds). The multi-breed approach is effective in goats by blending Alpine and Saanen breeds, but not in sheep. Finally, switching to genomic selection is planned in Lacaune dairy sheep and is under consideration for other sheep breeds. In goats, inclusion of major genes in genomic evaluations should be explored before switching to genomic breeding programs. Keywords: Dairy goats Dairy sheep Genomic selection
In:
URL [本文引用: 1]

Abstract: Genomic selection experiment in Lacaune dairy sheep: progeny test results of rams initially selected either on parent average or on genomic prediction (10th World Congress on Genetics Applied to Livestock Production)
URL [本文引用: 1]
URLPMID:29065868 [本文引用: 1]

Building an efficient reference population for genomic selection is an issue when the recorded population is small and phenotypes are poorly informed, which is often the case in sheep breeding programs. Using stochastic simulation, we evaluated a genomic design based on a reference population with medium-density genotypes [around 4502K single nucleotide polymorphisms (SNPs)] of dams that were imputed from very low-density genotypes (≤021000 SNPs). A population under selection for a maternal trait was simulated using real genotypes. Genetic gains realized from classical selection and genomic selection designs were compared. Genomic selection scenarios that differed in reference population structure (whether or not dams were included in the reference) and genotype quality (medium-density or imputed to medium-density from very low-density) were evaluated. The genomic design increased genetic gain by 26% when the reference population was based on sire medium-density genotypes and by 54% when the reference population included both sire and dam medium-density genotypes. When medium-density genotypes of male candidates and dams were replaced by imputed genotypes from very low-density SNP genotypes (1000 SNPs), the increase in gain was 22% for the sire reference population and 42% for the sire and dam reference population. The rate of increase in inbreeding was lower (from 61 20 to 61 34%) for the genomic design than for the classical design regardless of the genomic scenario. We show that very low-density genotypes of male candidates and dams combined with an imputation process result in a substantial increase in genetic gain for small sheep breeding programs.
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLMagsci [本文引用: 1]

品种选育在农业生产中占有十分重要的地位, 育种值估计是品种选育的核心。随着遗传标记的发展, 尤其是高通量的基因分型技术, 使得从基因组水平估计育种值成为可能, 即基因组选择。文章将基因组选择的方法分为两类:一是基于估计等位基因效应来预测基因组估计育种值(GEBV), 如最小二乘法, 随机回归-最佳线性无偏预测(RR-BLUP)、Bayes、主成分分析等方法; 二是基于遗传关系矩阵来预测GEBV, 通过采用高通量标记构建个体间的遗传关系矩阵, 然后用线性混合模型来预测育种值, 即GBLUP法, 并以这两种分类简要介绍了基因组选择各种方法的大致原理。影响基因组选择准确性的因素主要有标记类型和密度、单倍型长度、参考群体大小和标记-数量性状基因座(QTL)连锁不平衡(LD)大小等; 在基因组选择的各种方法中, 一般说来Bayes方法和GBLUP方法具有较高的准确性, 最小二乘法最差; GBLUP计算速度快, 能够将标记和系谱结合起来, 因而比其他方法更具优势。尽管基因组选择取得了很大进展, 但在理论方面还面临着一些挑战, 如联合育种、长期选择的遗传进展及如何解析与性状有关和无关的标记等。基因组选择在一些动植物育种上已经开始应用, 在人类遗传倾向预测和进化动力学研究中也有潜在的应用前景。基因组选择在个体间亲缘关系的量化上有了突破, 比传统方法更加精确, 因此, 基因组选择将会是动植物育种史上革命性的事件。
URLMagsci [本文引用: 1]

品种选育在农业生产中占有十分重要的地位, 育种值估计是品种选育的核心。随着遗传标记的发展, 尤其是高通量的基因分型技术, 使得从基因组水平估计育种值成为可能, 即基因组选择。文章将基因组选择的方法分为两类:一是基于估计等位基因效应来预测基因组估计育种值(GEBV), 如最小二乘法, 随机回归-最佳线性无偏预测(RR-BLUP)、Bayes、主成分分析等方法; 二是基于遗传关系矩阵来预测GEBV, 通过采用高通量标记构建个体间的遗传关系矩阵, 然后用线性混合模型来预测育种值, 即GBLUP法, 并以这两种分类简要介绍了基因组选择各种方法的大致原理。影响基因组选择准确性的因素主要有标记类型和密度、单倍型长度、参考群体大小和标记-数量性状基因座(QTL)连锁不平衡(LD)大小等; 在基因组选择的各种方法中, 一般说来Bayes方法和GBLUP方法具有较高的准确性, 最小二乘法最差; GBLUP计算速度快, 能够将标记和系谱结合起来, 因而比其他方法更具优势。尽管基因组选择取得了很大进展, 但在理论方面还面临着一些挑战, 如联合育种、长期选择的遗传进展及如何解析与性状有关和无关的标记等。基因组选择在一些动植物育种上已经开始应用, 在人类遗传倾向预测和进化动力学研究中也有潜在的应用前景。基因组选择在个体间亲缘关系的量化上有了突破, 比传统方法更加精确, 因此, 基因组选择将会是动植物育种史上革命性的事件。
