 ,1,2, 顾婷
,1,2, 顾婷 ,1
,1Histone H3K27me3 in the regulation of skeletal muscle development
Yanmin Gan1, Jian Zhou1, Rong Quan1, Linjun Hong1, Zicong Li1, Enqin Zheng1, Dewu Liu1, Zhenfang Wu1,2, Gengyuan Cai ,1,2, Ting Gu
,1,2, Ting Gu ,1
,1通讯作者:
编委: 李明洲
收稿日期:2018-09-27修回日期:2019-02-26网络出版日期:2019-04-20
| 基金资助: |
Received:2018-09-27Revised:2019-02-26Online:2019-04-20
| Fund supported: |
作者简介 About authors
甘炎民,硕士研究生,专业方向:动物遗传与繁育E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (627KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
甘炎民, 周健, 全绒, 洪林君, 李紫聪, 郑恩琴, 刘德武, 吴珍芳, 蔡更元, 顾婷. 组蛋白H3K27me3对骨骼肌发育调控研究进展[J]. 遗传, 2019, 41(4): 285-292 doi:10.16288/j.yczz.18-272
Yanmin Gan, Jian Zhou, Rong Quan, Linjun Hong, Zicong Li, Enqin Zheng, Dewu Liu, Zhenfang Wu, Gengyuan Cai, Ting Gu.
在自然界中,哺乳动物的个体生长、肌肉质量与骨骼肌的生长发育相关。骨骼肌由束状肌纤维组成,是动物个体中体积最大、质量占比最高的组织,对运动系统、姿势行为、支撑作用至关重要。骨骼肌的生长发育受经典遗传和表观遗传学的共同精细调控,其中骨骼肌成肌细胞的增殖、分化及肌管融合和肌纤维体积增大是研究骨骼肌发育的核心问题。近年来组蛋白甲基化修饰成为表观遗传学领域的研究热点,能通过改变核小体结构/细胞周期蛋白基因表达以及增殖和分化相关关键蛋白的表达,与其他表观因子互作等途径参与骨骼肌发育。其中,组蛋白H3亚基第27位赖氨酸三甲基化(histone H3 lysine 27 tri-methylation, H3K27me3)是抑制性组蛋白修饰,在骨骼肌发育过程中扮演着重要角色。结合近年来与骨骼肌发育相关的H3K27三甲基化的研究进展,本文主要对组蛋白结构和甲基化修饰类型、H3K27位点的甲基化和去甲基化的调控机制及其调控骨骼肌发育的过程进行了综述,以期为科研工作者了解H3K27三甲基化对骨骼肌发育过程的调控提供帮助及进一步解析骨骼肌发育机制提供借鉴和参考。
1 组蛋白结构和甲基化修饰类型
在真核生物细胞核中,DNA链缠绕在核心组蛋白外,形成染色质的基本单位—核小体。由于组蛋白富含精氨酸和赖氨酸,带有正电荷,因此能与带有负电荷的DNA密切结合。组蛋白主要由H1、H2A、H2B、H3和H4 等5种类型蛋白质亚基组成,而其中的4种组蛋白亚基—H2A、H2B、H3和H4由一球形结构域及暴露在核小体表面的N端尾区组成,每个亚基各两拷贝组成八聚体(histone octamer),形成核小体基本结构中的核心组蛋白。两个核小体由组蛋白H1亚基与连接DNA (linker DNA)串联起来,彼此靠拢、紧密相连[1]。核心组蛋白氨基酸链两端分别为C末端和N末端:N末端富含碱性氨基酸,如精氨酸和赖氨酸;C末端富含疏水氨基酸,如异亮氨酸和缬氨酸。C端氨基酸由于其疏水性质聚集在组蛋白中心,N端碱性氨基酸暴露在八聚体外周,形成“组蛋白的尾巴”,这些暴露的碱基容易被不同的基团修饰。1964年,Murray[2]在鼠伤寒沙门菌(Salmonella typhimurium)鞭毛蛋白中发现了N-甲基化赖氨酸,这是最早发现的组蛋白甲基化修饰。组蛋白甲基化是指蛋白侧链氨基酸在各甲基化酶的催化下,以S-腺苷甲硫氨酸(S-adenosyl methionine, SAM)作为甲基供体,获得不同数目甲基的一种翻译后修饰(post-translational modification, PTM)[3]。目前为止,已经发现了多种特异性组蛋白甲基化酶,其中不同的赖氨酸甲基转移酶(histone lysine methyltransferases, HKMTs)能将赖氨酸残基分别进行单甲基化(mono-/ -me1)、双甲基化(di-/ -me2)以及三甲基化(tri-/ -me3)修饰,而精氨酸甲基转移酶(protein arginine methyltransferase, PRMT)催化精氨酸残基,对其进行单甲基化或者双甲基化修饰[1,4]。
组蛋白H3是发生修饰最多的亚基,其第4、9、27、36和79位赖氨酸残基是甲基化修饰的热点[5],对基因表达起着重要的调控作用。一般来说,组蛋白H3不同位点的赖氨酸甲基化修饰能够调节染色质结构,使染色质处于疏松或紧密状态,从而对基因转录活性进行调控[6];此外,组蛋白H3不同位点赖氨酸的甲基化与其他类型的修饰方式相互作用,如H3K4me1和H3K27ac,分别对基因启动子和增强子的活性调节发挥关键的调控作用[7,8]。
2 H3K27动态修饰过程及其作用机制
H3K27修饰是一个动态的过程,包括主动甲基化和被动去甲基化。在主动甲基化过程中,H3K27由Zeste基因增强子同源物2 (Zeste gene enhancer homolog2, EZH2)识别,并将甲基基团(-CH3)转移到组蛋白H3的27位赖氨酸残基上[9]。因此,EZH2的表达与H3K27me3水平成正比。此外,EZH2是组成多梳蛋白抑制复合物2 (polycomb repressive complex2, PRC2)的3个核心亚基之一,而PRC2是动植物机体中两大重要的多梳蛋白抑制复合物之一,其组成成分较PRC1更为保守,由EZH1/EZH2、zeste12基因抑制因子(suppressor of zeste12, Suz12)以及胚胎外胚层发育因子(embryonic ectoderm development, EED)组成。而H3K27me3去甲基化由其去甲基化酶—赖氨酸特异性脱甲基酶6B (lysine- specific demethylase 6B, Kdm6B/JMJD3)和赖氨酸特异性脱甲基酶6A (lysine-specificdemethylase 6A, Kdm6A/UTX)催化,两者皆为含有Jumonji C(JmjC)结构域的双加氧酶。H3K27me3被去甲基化后能解除转录抑制活性,启动靶基因表达[10]。研究表明,UTX作为H3K27特异性去甲基转移酶,不仅能够使H3K27me2去除二甲基化,还能使H3K27me3去除三甲基化[11]。Karl等[12]在小鼠胚胎细胞中抑制H3K27去甲基化酶UTX和JMJD3的表达后,发现Hox基因启动子区H3K27me3的表达水平升高。H3K27me3修饰是组蛋白H3亚基最稳定的表观修饰标记之一,对基因转录、DNA复制和修复具有调控作用,参与干细胞分化、肌肉分化和发育等生命活动[13]。H3K27me3修饰位点主要位于基因启动子和转录起始位点附近,是抑制性组蛋白修饰,参与基因表达、胚胎发育和细胞分化等过程[14,15]。H3K27me3通过3种方式抑制基因的表达:(1) EZH2催化组蛋白H3K27三甲基化(形成H3K27me3),被组成PRC1的色素框-结构域蛋白(chromobox- domain protein, CBX)亚基识别并进一步募集PRC1复合物,其RING1亚基将组蛋白H2A亚基第119位赖氨酸单泛素化(H2AK119ub1),使染色质结构更加致密,基因转录起始位点无法与转录酶RNA Pol Ⅱ结合(图1A)[10,16,17],这是目前发现的H3K27me3最经典的调控方式;(2) H3K27me3募集其他抑制性调控因子,如DNA甲基化转移酶(DNA methyltransferase, DNMT),使与之结合的DNA序列甲基化(图1B);(3) H3K27me3在受精卵中特异结合于印记基因的母源等位基因,以不依赖DNA-甲基化的形式对不同亲本来源的等位基因(主要为母本来源)进行印记(imprinting) (图1C)[18]。
图1
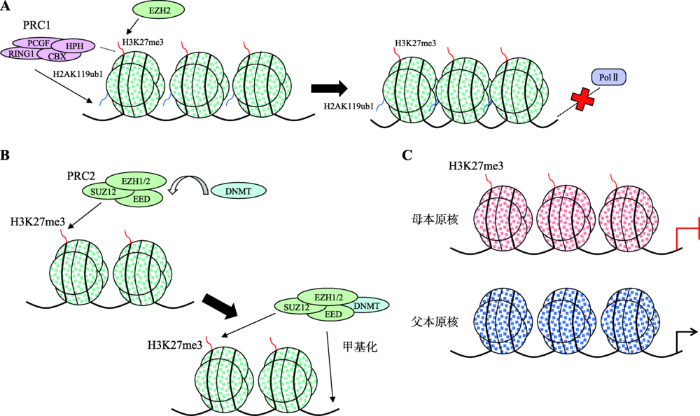 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1H3K27me3抑制基因表达的作用机制
A:H3K27me3募集PRC1复合物抑制转录;B:H3K27me3促进DNA序列甲基化;C:H3K27me3对等位基因进行印记。
Fig. 1The mechanism of H3K27me3 inhibiting gene expression
3 H3K27me3与骨骼肌发育
3.1 H3K27me3及其修饰酶对成肌细胞增殖的调控
肌肉质量通常由肌纤维的数目和横截面积决定,肌纤维数目的增加表现为肌肉增生,肌纤维横截面积的增加表现为肌肉肥大[19]。肌纤维数目的增加依赖于肌细胞的增殖,而细胞能否维持增殖状态受细胞周期蛋白和转录调控因子的共同作用。研究表明,细胞周期蛋白(如细胞周期蛋白激酶Cdk6)需要维持在较高的表达水平,以及生肌调节因子(myogenic regulatory factors, MRFs),如MyoG、MyoD、Myf5和Myf6需要维持在较低的表达水平才能保证骨骼肌细胞处于增殖状态[19]。肌细胞生成素(Myogenin, MyoG)是含螺旋-环-螺旋结构的转录因子,在成肌分化中发挥关键作用。H3K27me3能直接抑制MyoG基因的表达,促进骨骼肌细胞的增殖。Asp等[20]发现成肌细胞系C2C12分化前基因组中总体的H3K27me3修饰水平显著高于分化后,特异性抑制MyoG基因的表达,维持成肌细胞增殖状态。这与小鼠胚胎肌肉发育的过程一致。在小鼠早期胚胎(d9.5)的肌节中,H3K27me3的甲基化酶EZH2及H3K27me3表达较高,抑制MyoG的表达,同时成肌分化因子MHCⅡb的启动子和MCK的增强子等调控区域也被EZH2和转录因子YY1识别,形成H3K27me3修饰,然后进一步募集组蛋白去乙酰化酶HDAC1,促进成肌细胞增殖[21]。H3K27me3修饰同样促进家畜骨骼肌的增殖。Byrne等[22]发现在出生前后的绵羊骨骼肌基因组中H3K27me3修饰大量存在于启动子中,抑制基因的转录活性。肌骨素(myostatin, MSTN)基因能抑制骨骼肌增殖,抑制MSTN的表达能提高家畜的肌肉量。研究表明,在原代分离的绵羊成肌细胞中敲除MSTN可上调H3K27me3的甲基化酶EZH2的表达,促进肌细胞增殖[23]。
H3K27me3除了可直接靶向调节成肌分化因子及细胞周期蛋白的表达外,还可与长链非编码RNA (lncRNA)互作,调控骨骼肌细胞的增殖。lncRNA是近年来新发现的一类非编码RNA,能通过与H3K27me3的互作对骨骼肌增殖分化发挥重要的调控作用。Jin等[24]发现lncRNA SYISL能通过募集H3K27me3甲基化酶EZH2,进行H3K27me3修饰,进而抑制细胞周期蛋白激酶Cdk6的抑制因子p21基因的表达及激活p21靶基因Cdk6的表达,促进成肌细胞增殖,提高肌纤维数、肌肉密度和肌肉质量(图2)。Andresini等[25]发现lncRNA Kcnq1ot1在骨骼肌细胞C2.7中通过促进细胞周期抑制因子p57基因上MyoD结合位点处H3K27me3的累积量,阻碍转录因子MyoD的结合,从而抑制细胞周期抑制因子p57的表达,促进骨骼肌增殖。
图2
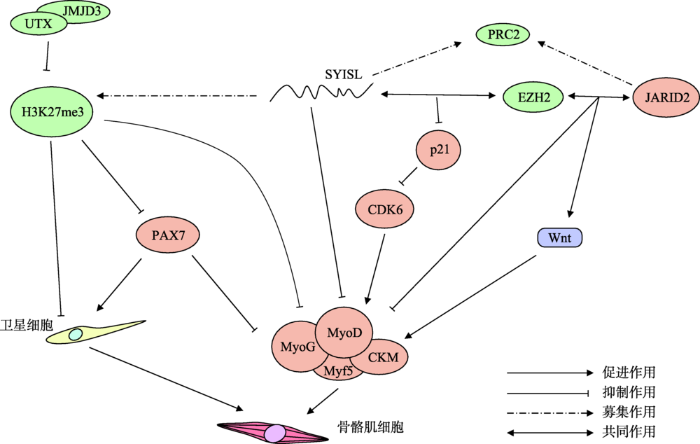 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2H3K27me3参与调控成肌细胞增殖和分化的过程
H3K27me3通过抑制Pax7和生肌调节因子的表达从而调节骨骼肌的增殖和分化;在卫星细胞的增殖和分化过程中,H3K27me3会促进卫星细胞的增殖但抑制卫星细胞的分化。H3K27me3的甲基化酶EZH2能分别与JARID2和lncRNA SYISL作用,调控生肌调节因子的表达。
Fig. 2The mechanism of H3K27me3 regulating proliferation and differentiation of myoblasts
骨骼肌卫星细胞是一类特殊的具有干细胞性质的肌细胞,在肌肉损伤修复等过程中发挥重要作用。H3K27me3对于骨骼肌卫星细胞的发育同样是必不可缺。研究表明,EZH2是维持骨骼肌卫星细胞标记基因Pax7表达的重要调控因子。在小鼠卫星细胞中将EZH2在骨骼肌中特异性敲除后,小鼠的肌肉量显著下降,卫星细胞增殖受抑制,且肌肉的损伤修复功能发生缺陷[26],说明卫星细胞中基因组H3K27me3水平的下调影响其增殖。该结果得到了其他研究的证实。Woodhouse等[27]同样制备骨骼肌卫星细胞特异敲除EZH2小鼠,发现小鼠肌肉量下降,肌纤维横截面积下降,且存活率显著下降,仅54%的条件敲除小鼠存活到50日龄,但静息状态的卫星细胞数目未发生变化,推断H3K27me3水平的降低对维持骨骼肌自身数目无影响,但抑制卫星细胞的生长增殖。
H3K27me3及其甲基化酶EZH2对其他类型的肌细胞增殖的作用与骨骼肌细胞不完全一致。在横纹肌肉瘤细胞中用siRNA敲低EZH2的表达,细胞周期抑制因子p21的表达上调,抑制横纹肌肉瘤细胞的增殖[28]。而Yu等[29]发现,在小鼠鼻滴灰尘诱导哮喘模型中加入H3K27me3去甲基化酶抑制剂GSK-J4,即维持H3K27me3水平能显著抑制气管平滑肌的增殖和迁移以及炎症反应,从而抑制哮喘的发生。LncRNA TINCR能将EZH2募集到靶基因CaMKII启动子区,提高H3K27me3水平从而抑制CaMKII的转录,抑制心肌细胞增殖和肥大[30]。此外,Akerberg等[31]构建了缺失H3K27me3去甲基化酶JMJD3和UTX的斑马鱼(Barchydanio rerio),与野生型斑马鱼的心脏组织对比,发现共表达JMJD3和UTX能显著降低H3K27me3表达量,并促进心肌细胞的增殖。
以上研究表明,H3K27me3主要促进成肌细胞和骨骼肌卫星细胞的增殖,但具有细胞类型特异性。
3.2 H3K27me3及其修饰酶对成肌细胞分化的调控
H3K27me3及其修饰酶不仅促进骨骼肌细胞的增殖,还对成肌细胞分化起重要调控作用。Adrian等[32]通过全基因组定位分析,发现在人的胚胎成纤维细胞中H3K27me3能与多种诱导细胞特异分化的基因,包括各种成肌调节因子和肌酸激酶(creatine kinase, CKM)等的调控区域结合,H3K27me3被去甲基化使骨骼肌成肌分化因子去抑制,从而诱导骨骼肌分化。骨骼肌中H3K27me3的去甲基化对启动骨骼肌分化过程至关重要。骨骼肌中H3K27me3的去甲基化的过程分为两步:在肌肉分化早期,反式激活蛋白Six4募集去甲基化酶UTX到骨骼肌分化相关基因的启动子和编码区部分区域,使转录起始位点附近区域发生H3K27me3去甲基化;转录复合物RNA Pol Ⅱ与成肌分化因子结合并向下游延伸,辅助UTX将基因编码区下游的H3K27me3标记去除[33]。肌肉的分化过程主要由生肌调节因子调控,大量研究表明这些生肌调节因子与EZH2的表达趋势相反。在小鼠肌肉分化过程中,EZH2显著下调表达,促进肌细胞分化[21]。同样,在山羊骨骼肌卫星细胞的分化过程中,EZH2的表达量在分化过程中显著下调[34]。此外,Asp等[20]发现H3K27me3的下调是控制MyoG基因表达和触动成肌细胞分化的开关。而在C2C12和骨骼肌卫星细胞中过表达EZH2基因,从而提高H3K27me3的水平能抑制这两种肌细胞的分化[21]。
H3K27me3除了可直接调控成肌细胞的分化过程外,也能与lncRNA及siRNA互作,进而调控成肌细胞的分化。最近研究发现,敲除lcnRNA SYISL能显著降低成肌分化因子MyoG、Myh4以及MCK启动子上H3K27me3的水平,促进成肌细胞分化[24]。而SYISL含有多个潜在的miRNA结合位点,如miR-1、miR-125、miR-214、miR-133和miR-124位点,它们参与调节成肌分化[35]。此外,在横纹肌肉瘤细胞中用siRNA敲低EZH2的表达,即降低H3K27me3的水平,能上调MyoG/MCK/MHC的 表达,促进横纹肌肉瘤细胞的分化[35]。同样,H3K27me3在虹鳟鱼(Oncorhynchus mykiss)发育过程中下调表达,促进pax7的同源基因pax7a2和pax7b在发育4~8天分化过程中上调表达,促进其肌肉分化[36]。在C2C12细胞中敲除H3K27me3去甲基化酶UTX,即提高H3K27me3的水平能抑制骨骼肌分化[33]。实际上在分化中的成肌细胞,UTX会被招募到MyoG和CKM基因的转录调节区域,使H3K27me3下调,进一步促进成肌细胞的分化。此外,组蛋白伴侣Spt6可促进UTX在染色质上富集并降低MyoG调节区域的H3K27me3水平[37]。
MicroRNA (miRNA)能通过影响H3K27me3的修饰水平从而调控骨骼肌分化。研究表明,miR-214在骨骼肌分化中发挥重要作用。在成肌细胞中,miR-214前体所在的内含子上募集了大量PRC2复合物,使H3K27me3高表达,miR-214转录受到抑制,抑制细胞分化;在成肌细胞分化时,EZH2的表达量下调,miRNA调控区域的H3K27me3结合量降低,使MyoD和MyoG能与miR-214转录前体调控区域结合,促进miR-214表达,而miR-214又能与EZH2的3′非翻译区结合,负调控EZH2的表达,进一步促进分化的进行[38]。类似的调控模式在横纹肌肉瘤细胞中也有报道。EZH2和H3K27me3能与miR-101的转录前体序列miR-101-2结合,抑制其表达,同时miR-101也能负调控EZH2的表达[39]。此外,有研究表明miR-26a也能通过靶向EZH2基因,促进C2C12的分化[40]。
虽然上述研究表明H3K27me3的下调是促进骨骼肌分化的,但也有研究表明H3K27me3在某些基因上的修饰可能通过信号通路的级联反应抑制肌肉细胞的分化。JARID2是jumonji蛋白家族中起抑制性作用的主要成员,为PRC2复合体中非催化亚基[41]。2018年,Adhikari和Davie[42]发现JARID能通过抑制Wnt通路的拮抗剂Sfrp1从而激活Wnt通路,Wnt通路的非经典途径被激活,即Wnt蛋白与细胞膜FZD受体(frizzled)结合,β-连环蛋白(β-catenin)降解复合体分解,释放β-catenin进入细胞核,与成肌细胞分化因子myoD和MyoG的调控区结合,促进其表达,从而促进骨骼肌分化。这说明H3K27me3也可能促进骨骼肌分化,与前人的报道结果相反。
综上所述,H3K27me3通过以下几个方面对骨骼肌发育过程进行调控:(1) H3K27me3及其甲基化酶EZH2能够直接特异性结合细胞周期蛋白及成肌分化相关基因,如Cdk6、Myogenin、MyoD等因子,从而促进成肌细胞增殖抑制肌肉的分化,而当H3K27me3受到去甲基化酶UTX、JMJD3作用,发生去甲基化,则促进其分化;(2) H3K27me3与lncRNA及miRNA等互作,调控肌肉发育;(3) H3K27me3能调控骨骼肌发育重要的信号通路,如Wnt信号通路,以启动骨骼肌分化过程(图2)。
4 结语与展望
哺乳动物骨骼肌发育是一个非常复杂的过程,受经典遗传和表观遗传共同调控。H3K27me3作为组蛋白甲基化修饰类型的重要一员,其本身能特异性结合成肌分化相关基因,起到调节骨骼肌增殖分化过程的作用。H3K27me3还参与调控骨骼肌发育信号通路,并能与lncRNA及miRNA等互作,以调控骨骼肌发育。目前已有大量研究表明H3K27me3在骨骼肌发育过程中发挥了重要的调控作用,但其对骨骼肌发育机制的具体调控过程尚不清楚。因此,本文主要概述了H3K27me3及其修饰酶对骨骼肌成肌细胞增殖分化的调控。然而,H3K27me3在骨骼肌发育过程的分子调控机制尚无进一步研究,其与骨骼肌发育的更深层次关联也有待阐明。H3K27me3对骨骼肌的调控为人们对于骨骼肌发育的认识提供一个新角度,为更深入了解和阐明骨骼肌分化和生长发育过程提供新的研究方向。随着对H3K27me3调控骨骼肌发育机制的深入研究,将为促进骨骼肌生长发育提供理论基础,不仅可以提高个体生长水平及运动能力,也可应用于畜禽生产中提高肌肉质量及肌肉产量以产生更多经济效益。因此,深入研究H3K27me3对骨骼肌发育的调控机制有着重要的价值和意义,为哺乳动物促进个体生长、提高肌肉质量及肌肉损伤后修复提供解决方案。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URL [本文引用: 2]

Epigenetics refers to non-coding sequence changes such as DNA methylation, histone modifications, chromosome remodeling and non-coding RNA regulation. Histone modifications include acetylation, phosphorylation, methylation, ubiquitination and ADP ribosylation. The combinations of different histone modifications, known as "histone code", are dynamic during development and differentiation and play important roles in the regulation of gene expressions in spatial-temporal manners. The modification on a particular residue in a histone affects not only the modifications at different residues in its own protein but also other histones. The histone modifications is a complicated network and the regulation remains elusive.
URL [本文引用: 2]

Epigenetics refers to non-coding sequence changes such as DNA methylation, histone modifications, chromosome remodeling and non-coding RNA regulation. Histone modifications include acetylation, phosphorylation, methylation, ubiquitination and ADP ribosylation. The combinations of different histone modifications, known as "histone code", are dynamic during development and differentiation and play important roles in the regulation of gene expressions in spatial-temporal manners. The modification on a particular residue in a histone affects not only the modifications at different residues in its own protein but also other histones. The histone modifications is a complicated network and the regulation remains elusive.
URLPMID:14114491 [本文引用: 1]

Biochemistry. 1964 Jan;3:10-5.
URLPMID:5846984 [本文引用: 1]

PubMed comprises more than 23 million citations for biomedical literature from MEDLINE, life science journals, and online books. Citations may include links to full-text content from PubMed Central and publisher web sites.
[本文引用: 1]
URLMagsci [本文引用: 1]

作为一种重要的表观遗传学调控机制, 组蛋白甲基化修饰在多种生命过程中发挥了重要的作用。细胞内有多种组蛋白甲基化酶和去甲基化酶共同调节组蛋白的修饰状态, 在组蛋白甲基化状态确定后, 多种效应分子特异的读取修饰信息, 从而参与基因转录调控过程。文章从组蛋白甲基化效应分子的作用机制方面综述了这一领域的研究进展。
URLMagsci [本文引用: 1]

作为一种重要的表观遗传学调控机制, 组蛋白甲基化修饰在多种生命过程中发挥了重要的作用。细胞内有多种组蛋白甲基化酶和去甲基化酶共同调节组蛋白的修饰状态, 在组蛋白甲基化状态确定后, 多种效应分子特异的读取修饰信息, 从而参与基因转录调控过程。文章从组蛋白甲基化效应分子的作用机制方面综述了这一领域的研究进展。
URL [本文引用: 1]
URLPMID:21106759 [本文引用: 1]

Developmental programs are controlled by transcription factors and chromatin regulators, which maintain specific gene expression programs through epigenetic modification of the genome. These regulatory events at enhancers contribute to the specific gene expression programs that determine cell state and the potential for differentiation into new cell types. Although enhancer elements are known to be associated with certain histone modifications and transcription factors, the relationship of these modifications to gene expression and developmental state has not been clearly defined. Here we interrogate the epigenetic landscape of enhancer elements in embryonic stem cells and several adult tissues in the mouse. We find that histone H3K27ac distinguishes active enhancers from inactive/poised enhancer elements containing H3K4me1 alone. This indicates that the amount of actively used enhancers is lower than previously anticipated. Furthermore, poised enhancer networks provide clues to unrealized developmental programs. Finally, we show that enhancers are reset during nuclear reprogramming.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:2017713478 [本文引用: 2]

Abstract The trithorax and the polycomb group proteins are chromatin modifiers, which play a key role in the epigenetic regulation of development, differentiation and maintenance of cell fates. The polycomb repressive complex 2 (PRC2) mediates transcriptional repression by catalysing the di- and tri-methylation of Lys 27 on histone H3 (H3K27me2/me3). Owing to the essential role of the PRC2 complex in repressing a large number of genes involved in somatic processes, the H3K27me3 mark is associated with the unique epigenetic state of stem cells. The rapid decrease of the H3K27me3 mark during specific stages of embryogenesis and stem-cell differentiation indicates that histone demethylases specific for H3K27me3 may exist. Here we show that the human JmjC-domain-containing proteins UTX and JMJD3 demethylate tri-methylated Lys 27 on histone H3. Furthermore, we demonstrate that ectopic expression of JMJD3 leads to a strong decrease of H3K27me3 levels and causes delocalization of polycomb proteins in vivo. Consistent with the strong decrease in H3K27me3 levels associated with HOX genes during differentiation, we show that UTX directly binds to the HOXB1 locus and is required for its activation. Finally mutation of F18E9.5, a Caenorhabditis elegans JMJD3 orthologue, or inhibition of its expression, results in abnormal gonad development. Taken together, these results suggest that H3K27me3 demethylation regulated by UTX/JMJD3 proteins is essential for proper development. Moreover, the recent demonstration that UTX associates with the H3K4me3 histone methyltransferase MLL2 (ref. 8) supports a model in which the coordinated removal of repressive marks, polycomb group displacement, and deposition of activating marks are important for the stringent regulation of transcription during cellular differentiation.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:2020850016 [本文引用: 1]

78 MS-based proteomics screen identifies novel histone lysine trimethylation readers 78 Readers are assembled into complexes and their genome-wide occupancy is determined 78 Sgf29 links the SAGA complex to H3K4me3 via a double tudor domain 78 Trimethyl reader complexes determine the biological function of their epigenetic mark
[本文引用: 1]
URLPMID:17320510 [本文引用: 1]

Polycomb group (PcG) and trithorax group (trxG) proteins are critical regulators of numerous developmental genes. To silence or activate gene expression, respectively, PcG and trxG proteins bind to specific regions of DNA and direct the posttranslational modification of histones. Recent work suggests that PcG proteins regulate the nuclear organization of their target genes and that PcG-mediated gene silencing involves noncoding RNAs and the RNAi machinery.
In: Radzioch D.
[本文引用: 1]
URLPMID:26762983 [本文引用: 1]

Trimethylated histone H3 lysine 27 (H3K27me3) is linked to gene silencing, whereas H3K4me3 is associated with gene activation. These two marks frequently co-occupy gene promoters, forming bivalent domains. Bivalency signifies repressed but activatable states of gene expression and can be resolved to active, H3K4me3-prevalent states during multiple cellular processes, including differentiation, development and epithelial mesenchymal transition. However, the molecular mechanism underlying bivalency resolution remains largely unknown. Here, we show that the H3K27 demethylase UTX (also called KDM6A) is required for the resolution and activation of numerous retinoic acid (RA)-inducible bivalent genes during the RA-driven differentiation of mouse embryonic stem cells (ESCs). Notably, UTX loss in mouse ESCs inhibited the RA-driven bivalency resolution and activation of most developmentally critical homeobox (Hox)a genes. The UTX-mediated resolution and activation of many bivalentHoxgenes during mouse ESC differentiation were recapitulated during RA-driven differentiation of human NT2/D1 embryonal carcinoma cells. In support of the importance of UTX in bivalency resolution,Utx-null mouse ESCs and UTX-depleted NT2/D1 cells displayed defects in RA-driven cellular differentiation. Our results define UTX as a bivalency-resolving histone modifier necessary for stem cell differentiation.
URLPMID:28723896 [本文引用: 1]

Mammalian sperm and oocytes have different epigenetic landscapes and are organized in different fashions. After fertilization, the initially distinct parental epigenomes become largely equalized with the exception of certain loci, including imprinting control regions. How parental chromatin becomes equalized and how imprinting control regions escape from this reprogramming is largely unknown. Here we profile parental allele-specific DNase I hypersensitive sites in mouse zygotes and morula embryos, and investigate the epigenetic mechanisms underlying these allelic sites. Integrated analyses of DNA methylome and tri-methylation at lysine 27 of histone H3 (H3K27me3) chromatin immunoprecipitation followed by sequencing identify 76 genes with paternal allele-specific DNase I hypersensitive sites that are devoid of DNA methylation but harbour maternal allele-specific H3K27me3. Interestingly, these genes are paternally expressed in preimplantation embryos, and ectopic removal of H3K27me3 induces maternal allele expression. H3K27me3-dependent imprinting is largely lost in the embryonic cell lineage, but at least five genes maintain their imprinted expression in the extra-embryonic cell lineage. The five genes include all paternally expressed autosomal imprinted genes previously demonstrated to be independent of oocyte DNA methylation. Thus, our study identifies maternal H3K27me3 as a DNA methylation-independent imprinting mechanism.
URLPMID:21602905 [本文引用: 2]

Skeletal muscle is the dominant organ system in locomotion and energy metabolism. Postnatal muscle grows and adapts largely by remodelling pre-existing fibres, whereas embryonic muscle grows by the proliferation of myogenic cells. Recently, the genetic hierarchies of the myogenic transcription factors that control vertebrate muscle development - by myoblast proliferation, migration, fusion and functional adaptation into fast-twitch and slow-twitch fibres - have become clearer. The transcriptional mechanisms controlling postnatal hypertrophic growth, remodelling and functional differentiation redeploy myogenic factors in concert with serum response factor (SRF), JUNB and forkhead box protein O3A (FOXO3A). It has also emerged that there is extensive post-transcriptional regulation by microRNAs in development and postnatal remodelling.
URL [本文引用: 2]
URL [本文引用: 3]
URLPMID:24673416 [本文引用: 1]

The ruminant developmental transition from late foetus to lamb is associated with marked changes in skeletal muscle structure and function that reflect programming for new physiological demands following birth. To determine whether epigenetic changes are involved in this transition, we investigated the genomic architecture of the chromatin modification, histone 3 lysine 27 trimethylation (H3K27me3), which typically regulates early life developmental processes; however, its role in later life processes is unclear. Chromatin immunoprecipitation coupled with next-generation sequencing was used to map H3K27me3 nucleosomes in ovine longissimus lumborum skeletal muscle at 100 days of gestation and 12 weeks post-partum. In both states, H3K27me3 modification was associated with genes, transcription start sites and CpG islands and with transcriptional silencing. The H3K27me3 peaks consisted of two major categories, promoter specific and regional, with the latter the dominant feature. Genes encoding homeobox transcription factors regulating early life development and genes involved in neural functions, particularly gated ion channels, were strongly modified by H3K27me3. Gene promoters differentially modified by H3K27me3 in the foetus and lamb were enriched for gated ion channels, which may reflect changes in neuromuscular function. However, most modified genes showed no changes, indicating that H3K27me3 does not have a large role in late muscle maturation. Notably, promyogenic transcription factors were strongly modified with H3K27me3 but showed no differences between the late gestation foetus and lamb, likely reflecting their lack of involvement in the myofibre fusion process occurring in this transition. H3K27me3 is a major architectural feature of the epigenetic landscape of ruminant skeletal muscle, and it comments on gene transcription and gene function in the context of late skeletal muscle development.
URL [本文引用: 1]
URL [本文引用: 2]
URL [本文引用: 1]
URLPMID:214985680 [本文引用: 1]

Satellite cells (SCs) sustain muscle growth and empower adult skeletal muscle with vigorous regenerative abilities. Here, we report that EZH2, the enzymatic subunit of the Polycomb-repressive complex 2 (PRC2), is expressed in both Pax7+/Myf5 stem cells and Pax7+/Myf5+ committed myogenic precursors and is required for homeostasis of the adult SC pool. Mice with conditional ablation of Ezh2 in SCs have fewer muscle postnatal Pax7+ cells and reduced muscle mass and fail to appropriately regenerate. These defects are associated with impaired SC proliferation and derepression of genes expressed in nonmuscle cell lineages. Thus, EZH2 controls self-renewal and proliferation, and maintains an appropriate transcriptional program in SCs.
[本文引用: 1]
URLPMID:4016511 [本文引用: 1]

Background Embryonal Rhabdomyosarcoma (RMS) is a pediatric soft-tissue sarcoma derived from myogenic precursors that is characterized by a good prognosis in patients with localized disease. Conversely, metastatic tumors often relapse, leading to a dismal outcome. The histone methyltransferase EZH2 epigenetically suppresses skeletal muscle differentiation by repressing the transcription of myogenic genes. Moreover, de-regulated EZH2 expression has been extensively implied in human cancers. We have previously shown that EZH2 is aberrantly over-expressed in RMS primary tumors and cell lines. Moreover, it has been recently reported that EZH2 silencing in RD cells, a recurrence-derived embryonal RMS cell line, favors myofiber-like structures formation in a pro-differentiation context. Here we evaluate whether similar effects can be obtained also in the presence of growth factor-supplemented medium (GM), that mimics a pro-proliferative microenvironment, and by pharmacological targeting of EZH2 in RD cells and in RD tumor xenografts. Methods Embryonal RMS RD cells were cultured in GM and silenced for EZH2 or treated with either the S-adenosylhomocysteine hydrolase inhibitor 3-deazaneplanocin A (DZNep) that induces EZH2 degradation, or with a new class of catalytic EZH2 inhibitors, MC1948 and MC1945, which block the catalytic activity of EZH2. RD cell proliferation and myogenic differentiation were evaluated both in vitro and in vivo. Results Here we show that EZH2 protein was abnormally expressed in 19 out of 19 (100%) embryonal RMS primary tumors and cell lines compared to their normal counterparts. Genetic down-regulation of EZH2 by silencing in GM condition reduced RD cell proliferation up-regulating p21Cip1. It also resulted in myogenic-like differentiation testified by the up-regulation of myogenic markers Myogenin, MCK and MHC. These effects were reverted by enforced over-expression of a murine Ezh2, highlighting an EZH2-specific effect. Pharmacological inhibition of EZH2 using either DZNep or MC inhibitors phenocopied the genetic knockdown of EZH2 preventing cell proliferation and restoring myogenic differentiation both in vitro and in vivo. Conclusions These results provide evidence that EZH2 function can be counteracted by pharmacological inhibition in embryonal RMS blocking proliferation even in a pro-proliferative context. They also suggest that this approach could be exploited as a differentiation therapy in adjuvant therapeutic intervention for embryonal RMS.
URL [本文引用: 1]
URLPMID:28548932 [本文引用: 1]

In the previous study, we established a mouse model of cardiac hypertrophy using transverse aortic constriction (TAC) and found that the expression of long non-coding RNAs TINCR was downregulated in myocardial tissue. The present study was designed to determine the potential role of TINCR in the pathogenesis of cardiac hypertrophy. Our results showed that enforced expression of TINCR could attenuate cardiac hypertrophy in TAC mice. Angiotensin II (Ang-II) was found to be associated with reduced TINCR expression and increased hypertrophy in cultured neonatal cardiomyocytes. RNA-binding protein immunoprecipitation assay confirmed that TINCR could directly bind with EZH2 in cardiomyocytes. The results of chromatin immunoprecipitation assay revealed that EZH2 could directly bind to CaMKII promoter region and mediate H3K27me3 modification. Knockdown of TINCR was found to reduce EZH2 occupancy and H3K27me3 binding in the promoter of CaMKII in cardiomyocytes. In addition, enforced expression of TINCR was found to decrease CaMKII expression and attenuate Ang-II-induced cardiomyocyte hypertrophy. Furthermore, our results also showed that Ang-II could increase CaMKII expression in cardiomyocytes, which consequently contributed to cellular hypertrophy. In conclusion, our findings demonstrated that TINCR could attenuate myocardial hypertrophy by epigenetically silencing of CaMKII, which may provide a novel therapeutic strategy for cardiac hypertrophy.
URL [本文引用: 1]
URL [本文引用: 1]

The Polycomb group (PcG) proteins form chromatin-modifying complexes that are essential for embryonic development and stem cell renewal and are commonly deregulated in cancer. Here, we identify their target genes using genome-wide location analysis in human embryonic fibroblasts. We find that Polycomb-Repressive Complex 1 (PRC1), PRC2, and tri-methylated histone H3K27 co-occupy >1000 silenced genes with a strong functional bias for embryonic development and cell fate decisions. We functionally identify 40 genes derepressed in human embryonic fibroblasts depleted of the PRC2 components (EZH2, EED, SUZ12) and the PRC1 component, BMI-1. Interestingly, several markers of osteogenesis, adipogenesis, and chrondrogenesis are among these genes, consistent with the mesenchymal origin of fibroblasts. Using a neuronal model of differentiation, we delineate two different mechanisms for regulating PcG target genes. For genes activated during differentiation, PcGs are displaced. However, for genes repressed during differentiation, we paradoxically find that they are already bound by the PcGs in nondifferentiated cells despite being actively transcribed. Our results are consistent with the hypothesis that PcGs are part of a preprogrammed memory system established during embryogenesis marking certain key genes for repressive signals during subsequent developmental and differentiation processes.
URL [本文引用: 2]
URL [本文引用: 1]

本实验室前期研究发现,miR-101a对山羊骨骼肌卫星细胞(skeletal muscle satellite cells,SMSCs)分化有促进作用,但其具体作用机制并不清楚。本研究利用Pic Tar、Target Scan和miRanda软件在线预测miR-101a的靶基因,并通过双荧光素酶报告基因进行实验验证;检测了山羊SMSCs分化不同时期miR-101a和靶基因的表达关系,同时分析了超表达和抑制miR-101a对靶基因表达水平的影响。结果证实,zeste增强子同源物2(enhancer of zeste homologue 2,EZH2)基因m RNA的3¢UTR具有miR-101a结合位点,是miR-101a的一个靶基因。在SMSCs分化过程中,随着miR-101a表达水平的升高,EZH2的表达在m RNA和蛋白水平均下调。抑制miR-101a后,EZH2的表达极显著升高(P0.05)。以上研究结果表明,miR-101a能通过抑制EZH2的表达来促进山羊SMSCs分化,为进一步阐明miR-101a对SMSCs的调控机制提供了理论依据。
URL [本文引用: 1]

本实验室前期研究发现,miR-101a对山羊骨骼肌卫星细胞(skeletal muscle satellite cells,SMSCs)分化有促进作用,但其具体作用机制并不清楚。本研究利用Pic Tar、Target Scan和miRanda软件在线预测miR-101a的靶基因,并通过双荧光素酶报告基因进行实验验证;检测了山羊SMSCs分化不同时期miR-101a和靶基因的表达关系,同时分析了超表达和抑制miR-101a对靶基因表达水平的影响。结果证实,zeste增强子同源物2(enhancer of zeste homologue 2,EZH2)基因m RNA的3¢UTR具有miR-101a结合位点,是miR-101a的一个靶基因。在SMSCs分化过程中,随着miR-101a表达水平的升高,EZH2的表达在m RNA和蛋白水平均下调。抑制miR-101a后,EZH2的表达极显著升高(P0.05)。以上研究结果表明,miR-101a能通过抑制EZH2的表达来促进山羊SMSCs分化,为进一步阐明miR-101a对SMSCs的调控机制提供了理论依据。
URLPMID:23522383 [本文引用: 2]

MicroRNAs (miRNAs) negatively regulate gene expression by promoting degradation of target mRNAs or inhibiting their translation. Previous studies have expanded our understanding that miRNAs play an important role in myogenesis and have a big impact on muscle mass, muscle fiber type and muscle-related diseases. The muscle-specific miRNAs, miR-206, miR-1 and miR-133, are among the most studied and best characterized miRNAs in skeletal muscle differentiation. They have a profound influence on multiple muscle differentiation processes, such as alternative splicing, DNA synthesis, and cell apoptosis. Many non-muscle-specific miRNAs are also required for the differentiation of muscle through interaction with myogenic factors. Studying the regulatory mechanisms of these miRNAs in muscle differentiation will extend our knowledge of miRNAs in muscle biology and will improve our understanding of the myogenesis regulation.
URLPMID:4344431 [本文引用: 1]

The extraordinary muscle growth potential of teleost fish, particular those of the Salmoninae clade, elicits questions about the regulation of the relatively highly conserved transcription factors of the myogenic program. The pseudotetraploid nature of the salmonid genome adds another layer of regulatory complexity that must be reconciled with epigenetic data to improve our understanding of the achievement of lifelong muscle growth in these fish. We identify three paralogous pax7 genes (pax7a1, pax7a2 and pax7b) in the rainbow trout genome. During in vitro myogenesis, pax7a1 transcripts remain stable, whereas pax7a2 and pax7b mRNAs increase in abundance, similarly to myogenin mRNAs but in contrast to the expression pattern of the mammalian ortholog. We also profile the distribution of repressive H3K27me3 and H3K9me3 and permissive H3K4me3 marks during in vitro myogenesis across these loci and find that pax7a2 expression is associated with decreased H3K27 trimethylation, whereas pax7b expression is correlated with decreased H3K9me3 and H3K27me3. These data link the unique differential expression of pax7 paralogs with epigenetic histone modifications in a vertebrate species displaying growth divergent from that of mammals and highlight an important divergence in the regulatory mechanisms of pax7 expression among vertebrates. The system described here provides a more comprehensive picture of the combinatorial control mechanisms orchestrating skeletal muscle growth in a salmonid, leading to a better understanding of myogenesis in this species and across Vertebrata more generally.
URL [本文引用: 1]

Histone chaperones affect chromatin structure and gene expression through interaction with histones and RNA polymerase II (PolII). Here, we report that the histone chaperone Spt6 counteracts H3K27me3, an epigenetic mark deposited by the Polycomb Repressive Complex 2 (PRC2) and associated with transcriptional repression. By regulating proper engagement and function of the H3K27 demethylase KDM6A (UTX), Spt6 effectively promotes H3K27 demethylation, muscle gene expression, and cell differentiation. ChIP-Seq experiments reveal an extensive genome-wide overlap of Spt6, PolII, and KDM6A at transcribed regions that are devoid of H3K27me3. Mammalian cells and zebrafish embryos with reduced Spt6 display increased H3K27me3 and diminished expression of the master regulator MyoD, resulting in myogenic differentiation defects. As a confirmation for an antagonistic relationship between Spt6 and H3K27me3, inhibition of PRC2 permits MyoD re-expression in myogenic cells with reduced Spt6. Our data indicate that, through cooperation with PolII and KDM6A, Spt6 orchestrates removal of H3K27me3, thus controlling developmental gene expression and cell differentiation.
URLPMID:2761245 [本文引用: 1]

Polycomb group (PcG) proteins exert essential functions in the most disparate biological processes. The contribution of PcG proteins to cell commitment and differentiation relates to their ability to repress transcription of developmental regulators in embryonic stem (ES) cells and in committed cell lineages, including skeletal muscle cells (SMC). PcG proteins are preferentially removed from transcribed regions, but the underlying mechanisms remain unclear. Here, PcG proteins are found to occupy and repress transcription from an intronic region containing the microRNA miR-214 in undifferentiated SMC. Differentiation coincides with PcG disengagement, recruitment of the developmental regulators MyoD and myogenin, and activation of miR-214 transcription. Once transcribed, miR-214 negatively feeds back on PcG by targeting the Ezh2 3 TR, the catalytic subunit of the PRC2 complex. miR-214-mediated Ezh2 protein reduction accelerates SMC differentiation and promotes unscheduled transcription of developmental regulators in ES cells. Thus, miR-214 and Ezh2 establish a regulatory loop controlling PcG-dependent gene expression during differentiation.
URLPMID:4527101 [本文引用: 1]

Background Rhabdomyosarcoma (RMS) is a pediatric soft tissue sarcoma arising from myogenic precursors that have lost their capability to differentiate into skeletal muscle. The polycomb-group protein EZH2 is a Lys27 histone H3 methyltransferase that regulates the balance between cell proliferation and differentiation by epigenetically silencing muscle-specific genes. EZH2 is often over-expressed in several human cancers acting as an oncogene. We previously reported that EZH2 inhibition induces cell cycle arrest followed by myogenic differentiation of RMS cells of the embryonal subtype (eRMS). MiR-101 is a microRNA involved in a negative feedback circuit with EZH2 in different normal and tumor tissues. To that, miR-101 can behave as a tumor suppressor in several cancers by repressing EZH2 expression. We, therefore, evaluated whether miR-101 is de-regulated in eRMS and investigated its interplaying with EZH2 as well as its role in the in vitro tumorigenic potential of these tumor cells. Results Herein, we report that miR-101 is down-regulated in eRMS patients and in tumor cell lines compared to their controls showing an inverse pattern of expression with EZH2. We also show that miR-101 is up-regulated in eRMS cells following both genetic and pharmacological inhibition of EZH2. In turn, miR-101 forced expression reduces EZH2 levels as well as restrains the migratory potential of eRMS cells and impairs their clonogenic and anchorage-independent growth capabilities. Finally, EZH2 recruitment to regulatory region of miR-101-2 gene decreases in EZH2-silenced eRMS cells. This phenomenon is associated to reduced H3K27me3 levels at the same regulatory locus, indicating that EZH2 directly targets miR-101 for repression in eRMS cells. Conclusions Altogether, our data show that, in human eRMS, miR-101 is involved in a negative feedback loop with EZH2, whose targeting has been previously shown to halt eRMS tumorigenicity. They also demonstrate that the re-induction of miR-101 hampers the tumor features of eRMS cells. In this scenario, epigenetic dysregulations confirm their crucial role in the pathogenesis of this soft tissue sarcoma.
URLPMID:18281287 [本文引用: 1]

Abstract MicroRNA (miRNA) are important regulators of many biological processes, but the targets for most miRNA are still poorly defined. In this study, we profiled the expression of miRNA during myogenesis, from proliferating myoblasts through to terminally differentiated myotubes. Microarray results identified six significantly differentially expressed miRNA that were more than 2-fold different in myotubes. From this list, miRNA-26a (miR-26a), an up-regulated miRNA, was further examined. Overexpression of miR-26a in murine myogenic C2C12 cells induced creatine kinase activity, an enzyme that markedly increases during myogenesis. Further, myoD and myogenin mRNA expression levels were also up-regulated. These results suggest that increased expression of miR-26a promotes myogenesis. Through a bioinformatics approach, we identified the histone methyltransferase, Enhancer of Zeste homolog 2 (Ezh2), as a potential target of miR-26a. Overexpression of miR-26a suppressed the activity of a luciferase reporter construct fused with the 3'-untranslated region of Ezh2. In addition, miR-26a overexpression decreased Ezh2 mRNA expression. These results reveal a model of regulation during myogenesis whereby the up-regulation of miR-26a acts to post-transcriptionally repress Ezh2, a known suppressor of skeletal muscle cell differentiation.
URLPMID:20123894 [本文引用: 1]

Abstract The Polycomb group proteins foster gene repression profiles required for proper development and unimpaired adulthood, and comprise the components of the Polycomb-Repressive Complex 2 (PRC2) including the histone H3 Lys 27 (H3K27) methyltransferase Ezh2. How mammalian PRC2 accesses chromatin is unclear. We found that Jarid2 associates with PRC2 and stimulates its enzymatic activity in vitro. Jarid2 contains a Jumonji C domain, but is devoid of detectable histone demethylase activity. Instead, its artificial recruitment to a promoter in vivo resulted in corecruitment of PRC2 with resultant increased levels of di- and trimethylation of H3K27 (H3K27me2/3). Jarid2 colocalizes with Ezh2 and MTF2, a homolog of Drosophila Pcl, at endogenous genes in embryonic stem (ES) cells. Jarid2 can bind DNA and its recruitment in ES cells is interdependent with that of PRC2, as Jarid2 knockdown reduced PRC2 at its target promoters, and ES cells devoid of the PRC2 component EED are deficient in Jarid2 promoter access. In addition to the well-documented defects in embryonic viability upon down-regulation of Jarid2, ES cell differentiation is impaired, as is Oct4 silencing.
URL [本文引用: 1]

JARID2 is a non-catalytic member of the polycomb repressive complex 2 (PRC2), which is known to regulate developmental target genes in embryonic stem cells. Here, we provide mechanistic insight into the modulation of Wnt signaling by JARID2 during murine skeletal muscle differentiation. We show that JARID2 is expressed in proliferating myoblasts, but downregulated upon muscle differentiation. Unexpectedly, depletion of JARID2 or the catalytic subunit of the PRC2 complex, EZH2, inhibited differentiation, suggesting that JARID2 and the PRC2 complex are required to initiate this process. Expression of the myogenic regulatory factors required to promote differentiation, MYOD and MYOG, was downregulated in the absence of JARID2, even though decreases in the methylation of histone H3 lysine 27 (H3K27me3) were observed on both promoters. We found that activation of the Wnt signaling pathway upregulated MYOD and restored differentiation. Activation of the Wnt pathway in JARID2 depleted cells caused -catenin to translocate to the nucleus, where it bound to and activated theMyod1promoter. We show that the Wnt antagonist SFRP1 is highly upregulated in the absence of JARID2 and is a direct target of JARID2 and the PRC2 complex. Ectopic expression of SFRP1 blocked MYOD and late muscle gene expression and inhibited the translocation of -catenin to the nucleus. Finally, we show that JARID2 and SFRP1 are inversely correlated in melanoma, confirming that the JARID2-mediated repression of SFRP1 extends beyond skeletal muscle and has important implications in many cellular systems, including cancer. We show that JARID2 and the PRC2 complex regulate muscle differentiation by modulating Wnt signaling through the direct repression of Wnt antagonists. The online version of this article (10.1186/s13072-018-0217-x) contains supplementary material, which is available to authorized users.
