2.
Molecular mechanisms of meiotic recombination suppression in plants
Fan Li1, Rongpei Yu1, Dan Sun1, Jihua Wang1, Shenchong Li1, Jiwei Ruan1, Qinli Shan1, Pingli Lu2, Guoxian Wang11. 2.
编委: 张宪省
收稿日期:2018-06-22修回日期:2018-08-31网络出版日期:2019-01-20
| 基金资助: |
Received:2018-06-22Revised:2018-08-31Online:2019-01-20
| Fund supported: |
作者简介 About authors
李帆,博士,助理研究员,研究方向:植物遗传与分子育种E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (675KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李帆, 余蓉培, 孙丹, 王继华, 李绅崇, 阮继伟, 单芹丽, 陆平利, 汪国鲜. 抑制植物减数分裂重组的分子机理[J]. 遗传, 2019, 41(1): 52-65 doi:10.16288/j.yczz.18-165
Fan Li, Rongpei Yu, Dan Sun, Jihua Wang, Shenchong Li, Jiwei Ruan, Qinli Shan, Pingli Lu, Guoxian Wang.
减数分裂(meiosis)是生物细胞中染色体数目减半的一种特殊的细胞分裂方式,在该过程中DNA只复制一次,但细胞连续分裂两次,从而形成染色体数目减半的配子[1,2,3]。在减数第一次分裂过程中,为了确保同源染色体的精确分离和染色体数目的减半,同源染色体间需要形成至少一个物理连接点,称为交叉结(chiasmata)[4,5]。交叉通过修复作用产生同源染色体间遗传物质的相互交换,即同源染色体间的重组(recombination),进而形成具有遗传多样性的配子[6,7]。减数分裂同源重组不仅保证了物种染色体的精确分离,同时又促进了父母亲本之间遗传物质的相互交换,从而在配子中形成遗传变异[8,9]。因此,减数分裂同源重组对生物进化和物种形成至关重要,也是植物新品种培育和开发的基础生物学过程。特别是在全球气候变化的背景下,人类面临各种挑战,减数分裂同源重组为充分利用植物的遗传多样性进行新品种的培育和创新提供了基础。
从植物进化的角度,重组率是生物在重组成本和重组优势之间维持的一种特定平衡,是物种长期以来对环境变化不断进化和演变的一种适应和自然选择[10]。在大多数真核生物中,由于调控重组基因的高度保守性,减数分裂重组率被维持在一个相对较低的水平,并且远低于其自身的自然潜力,但对其调控网络和抑制形成的分子机理还知之甚少[11,12]。减数分裂同源重组是真核生物有性生殖过程中的基本生物学过程,其相关研究一直是遗传学领域的核心科学问题,受到世界****的广泛关注[13,14,15,16]。近年来,植物减数分裂同源重组的分子调控研究取得了重大进展,特别是多个减数分裂重组抑制基因陆续在拟南芥(Arabidopsis thaliana)中被发现和鉴定,进一步增加了对这一复杂生物学过程的认识。本文以拟南芥为对象,综述了植物减数分裂重组抑制基因研究的重要进展。
1 DNA双链断裂和交叉形成
1.1 DNA双链断裂的产生
减数分裂同源重组起始于DNA双链断裂(double strand break, DSB),其由高度保守的拓扑异构酶SPO11 (sporulation 11)蛋白催化形成(图1)[6,17]。SPO11蛋白结构类似于古细菌中的TopoⅥ (Topoisomerase Ⅵ) A亚基,而TopoⅥ是由两个A亚基和两个B亚基组成的异源四聚体酶(A2B2 heterotetramer)[5]。最近,古细菌TopoⅥ复合体B亚基的同源蛋白MTOPVIB (meiotic topoisomerase VIB-like)在拟南芥和水稻(Oryza sativa)中被鉴定,研究显示其在减数分裂中对诱导DSB形成和重组启动至关重要[18,19]。图1
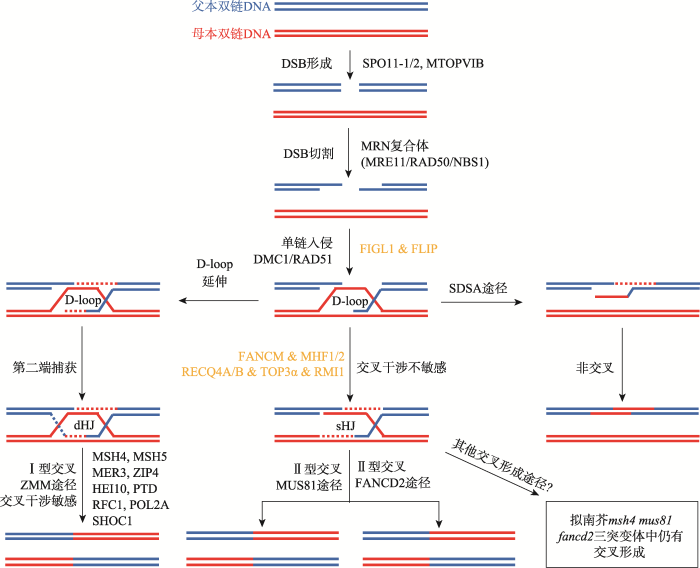 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1拟南芥减数分裂重组途径模型
在该模型图中,蓝色和红色线条分别描绘了两条父母亲本双链DNA。减数分裂重组起始于双链DNA的双链断裂(DSB),然后单链核酸内切酶切割断裂双链DNA的5′末端以形成3′端单链DNA (ssDNA),在重组酶的促进作用下,3′单链DNA尾部侵入同源双链DNA形成重组中间体D-loop。重组中间体或者形成dHJ(double Holliday Junction)结构,并通过ZMM途径(MSH4、MSH5、MER3、ZIP4、SHOC1、HEI10、RFC1、PTD和POL2A)形成Ⅰ型交叉重组,或者形成sHJ(single Holliday junction)结构由MUS81和FANCD2交叉干涉不敏感途径形成Ⅱ型交叉重组。但绝大多数D-loop重组中间体是通过合成型依赖性退火反应(SDSA)途径由重组抑制基因(FANCM、MHF1/2、TOP3α、RECQ4A/B、RMI1、FIGL1和FLIP)调控形成非交叉(NCOs)。
Fig. 1A meiotic recombination pathway model in Arabidopsis
拟南芥基因组存在3个SPO11同源基因(SPO11-1、SPO11-2和SPO11-3),但只有SPO11-1和SPO11-2作为异源二聚体参与调控减数分裂重组的启动,而SPO11-3只涉及体细胞的有丝分裂,不具有调控减数分裂的功能[20,21]。在植物中,其他一些基因也参与了诱导减数分裂DSB的形成,如PRD1 (putative recombination initiation defect 1)、PRD2、AtPRD3/OsPAIR1 (homologous pairing aberration in rice meiosis 1)、DFO (DSB formation)和CRC1 (central region component 1)[22,23,24,25,26]。不同于大部分植物中含有多个SPO11同源基因,在动物和酵母中只含有1个SPO11基因[27,28]。酵母中的DSB形成除了需要SPO11蛋白以外,还有其他9个蛋白参与调控,即RED50 (radiation sensitive 50)、MER2 (meiotic recombination 2)、MEI4 (meiosis defective4)、MRE11 (meiotic recombination 11)、REC102 (recombination-deficient 102)、REC104 (recombination-deficient 104)、REC114 (recombination-deficient 114)、SKI8 (superkiller 8)和XRS2 (X-ray sensitive 2)[2,29]。然而,这些参与酵母DSB形成的蛋白在物种间存在蛋白序列或者功能上的变异。如拟南芥中DSB的形成并不需要MRE11、RAD50和XRS2蛋白的参与,但这些蛋白直接作用于断裂双链5′末端的切除;SKI8尽管在几种真菌中非常保守,但在拟南芥中并不保守,且不参与减数分裂重组过程[30,31]。
1.2 重组中间体的形成与分解
DNA双链断裂产生之后,MRN复合体(MRE11、RAD50和NBS1)对断裂双链任一侧的5′末端进行切割,并产生3′端单链DNA(ssDNA)尾巴的突出端[32,33]。随后在DMC1 (DNA meiotic recombinase 1)和RAD51 (radiation sensitive 51)重组酶的促进作用下,这些ssDNAs启动同源序列搜索并入侵同源染色体或者非同源染色体的姐妹染色单体形成稳定的单链侵入中间体[34,35]。DMC1是首次在酵母中发现的减数分裂特异基因,只在减数分裂过程中发挥作用,而RAD51参与了有丝分裂和减数分裂的重组。Kurzbauer等[36]通过细胞学研究发现DMC1和RAD51重组酶倾向于定位于减数分裂DSB的相反两端,表明其在DSB修复过程中承担着不同的生物学功能,这也与DSB两端不同的修复结果兼容。在单链入侵形成中间体后,3′末端入侵链作为引物纵向延伸到同源双链DNA (dsDNA)中形成D-loop (displacement loop)结构[37]。D-loop的形成是减数分裂同源重组的重要中间体,其形成表明3′端入侵单链成功定位到了同源DNA参考序列[38],该早期中间体在之后可以通过不同的修复途径形成同源染色体交叉或者非交叉(non-crossovers, NCOs)[39,40]。D-loop在酶催化作用下进一步被修复形成Holliday junction (HJ)中间体结构,HJ是由两个同源双链DNA分子互换配对并相互连接形成的一种“十字交叉”中间体(four-way junction)[41]。HJ中间体的形成被认为是同源染色体交叉产生的关键结构,其两种类型的中间体(sHJ和dHJ)通过不同的修复途径产生两种类型的交叉[42,43]。在单链入侵形成D-loop结构后,如果D-loop的入侵链没有继续纵向延伸,则形成一个“十字交叉”中间体sHJ (single Holliday junction),进一步被Mus81 (methyl methane sulfonate and ultraviolet sensitive 81)蛋白分解产生Ⅱ型交叉(class Ⅱ CO)或者非交叉[39]。如果D-loop入侵链继续深入延伸到同源断裂双链中,并捕获断裂双链的第二端进行退火、合成与连接,则形成独特的异源双链DNA结构dHJ (double Holliday junction),并通过ZMM (ZIP-MSH-MER)途径分解形成Ⅰ型交叉(class Ⅰ CO)[40]。
1.3 交叉的形成被严格限制
在减数分裂开始初期,DNA双链产生大量双链断裂,但不管基因组的大小或者染色体数目的多少,只有极少数的断裂双链被修复形成交叉,其余的大量DSBs通过不同的途径和机制修复形成了非交叉。在模式植物拟南芥中,细胞学分析认为每个减数分裂的细胞大约形成200个双链断裂,但只有约10个断裂双链被修复形成交叉,其余的断裂双链则被修复产生非交叉,但到目前为止抑制交叉形成的机理尚不清楚[44,45,46,47,48]。目前,多项研究表明交叉的形成主要受多个机制的共同影响,如交叉保障(obligate CO)、交叉干涉(CO interference)、交叉稳态(CO homeostasis)和抗交叉因子(anti-CO factor)[49,50,51,52,53,54,55,56,57,58,59]。交叉保障是指每个配对同源染色体之间需要至少形成一个交叉以保障同源染色体后期的准确分离[49]。但是,在大部分生物中,一个交叉的形成会抑制其两侧相邻位置中另一个交叉的产生,最终导致交叉在染色体上非随机分布,这种现象被称为交叉干涉[50,51]。而交叉稳态则作为系统性缓冲机制,在早期交叉前体DSB数量急剧变化的情况下保持交叉数量的稳定[52,53]。近年来,减数分裂重组抑制基因在拟南芥中陆续被发现,揭示了重组中间体如何通过合成型依赖性退火反应(synthesis-dependent strand annealing, SDSA)途径分解为非交叉的机制[54,55,56,57,58,59]。
1.4 交叉形成的遗传途径
在大多数真核生物的减数分裂重组中至少存在两种不同的交叉形成途径,根据对交叉干扰是否敏感将其分为Ⅰ型交叉和Ⅱ型交叉[60,61,62]。其中,Ⅰ型交叉为干涉敏感型交叉,约占交叉总数的80%~ 85%,主要受保守的ZMM途径的调控,包括MSH4 (mutS homolog 4)[63]、MSH5 (mutS homolog 5)[64]、MER3 (meiotic recombination 3)[65,66]、ZIP4 (zinc transporter 4 precursor)[67]、SHOC1 (shortage of crossovers 1)[68]、HEI10 (human enhancer of cell invasion No.10)[69]、RFC1 (replication factor C1)[70]、PTD (parting dancers)[1,71]和POL2A (DNA polymerase 2A)[72]等蛋白。而与Ⅰ型交叉对应的是干涉不敏感的Ⅱ型交叉,该交叉的形成依赖于两条平行的途径:MUS81途径和FANCD2途径[73,74,75]。通常情况下,两种交叉形成途径广泛存在于在大多数真核生物中,例如酿酒酵母(Saccharomyces cerevisiae),哺乳动物和植物[9,76]。但也有例外,如在裂殖酵母(Schizosaccharomyces pombe)中,其减数分裂期间只形成sHJ中间体,故只存在Ⅱ型交叉形成途径[39]。而在秀丽隐杆线虫(Caenorhabditis elegans)中,所有的交叉均表现为干扰敏感,表明其交叉的产生均通过Ⅰ型交叉形成途径[77]。此外,值得注意的是,在拟南芥msh4 mus81 fancd2三突变体中,虽然同时缺乏形成Ⅰ型和Ⅱ类型交叉的所有关键基因,但仍有交叉形成,这说明阻断Ⅰ型和Ⅱ型交叉形成途径后触发了其他未知的交叉形成途径产生交叉[74,75]。这种现象也被证实存在于果蝇和酵母中,这些证据表明其他交叉形成途径的存在,且与已知的交叉形成途径同时共存或是互斥独存[78,79]。
2 减数分裂重组抑制基因
在大多数真核生物的减数分裂过程中,双链断裂与交叉形成的比率(DSBs/COs)存在极大差异,如拟南芥中DSBs/COs比率约为200∶10,这表明生物进化过程中存在着遗传机制限制大多数断裂双链修复形成交叉[80]。近年来,许多调控减数分裂过程的基因已经被克隆,但抑制减数分裂同源重组的分子机理仍不太清楚[20,25,81,82]。多种模式植物(如拟南芥和水稻)基因组测序的完成和全基因组测序技术的成熟,加速了植物减数分裂重组抑制基因的鉴定与功能研究。2012年,为了揭示减数分裂重组抑制基因,法国科学家Crismani等[54]利用正向遗传学通过EMS诱变拟南芥zmm突变体种子和大规模突变体筛选,并获得多个重组恢复系,最终鉴定出抑制Ⅱ型交叉形成的9个基因(FANCM、MHF1/2、TOP3α、RECQ4A/B、FIGL1、RMI1和FLIP,图2)[54,55,56,57]。该研究巧妙利用了zmm突变体短角果的表型(因缺乏Ⅰ型交叉形成的ZMM基因而育性降低)进行果夹表型恢复系的筛选,其原理是重组抑制基因的突变会降低或者丧失重组抑制作用,这能促进交叉的形成和重组率的提高,进而恢复zmm突变体的育性,使植株的果夹变长甚至恢复原有长度。这样的重组恢复植株能非常容易的通过果夹长度表型筛选出来,最后通过全基因组测序鉴定突变基因。例如,拟南芥FANCM基因突变后,恢复了zip4突变体的育性,使zip4突变体的短果夹增长,进而筛选获得fancm突变体。
图2
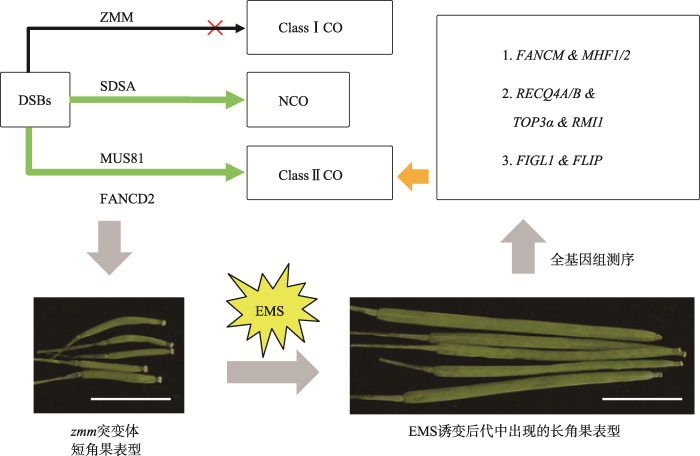 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2利用拟南芥zmm突变体短角果表型筛选重组抑制基因
拟南芥减数分裂重组抑制突变体筛选试验利用减数分裂ZMM交叉形成途径被关闭后,使得交叉只能通过Ⅱ型交叉形成途径产生,且导致短角果表型的形成。而EMS对zmm种子的随机诱变导致重组抑制基因的突变和抑制功能的丧失,进而增加了减数分裂重组率,弥补甚至恢复了zmm背景下的减数分裂交叉形成,在后代中产生长角果表型,进而筛选获得重组抑制突变体,最后通过全基因组测序鉴定出减数分裂重组抑制基因。图中拟南芥角果的标尺为1 cm。
Fig. 2Using short silique phenotype of zmm mutants to screen for meiotic recombination suppressors in Arabidopsis
2.1 FANCM联合MHF1与MHF2抑制减数分裂重组
FANCM (fanconi anemia complementation group M)解旋酶是在拟南芥中发现的首个减数分裂重组抑制蛋白,研究认为其通过SDSA途径分解D-loops中间体产生非交叉,从而抑制Ⅱ型交叉的形成[54,83,84]。拟南芥fancm突变体的交叉数量在雌雄两性的减数分裂过程中都得到极大提高,重组率也比野生型平均增加了3倍,但其植株的生长和生育情况与野生型无异,表明重组率的增加并不会影响植物的生长发育,证明植物在自然选择和进化过程中形成了遗传抑制机制限制过多的交叉形成[54]。由于Ⅰ型交叉特异性指示蛋白MLH1不能标记fancm突变体中增加的交叉,且花粉荧光标记四分体分析显示fancm突变体中不存在交叉干涉,证明FANCM不是通过Ⅰ型交叉形成途径抑制重组。而fancm mus81双突变体表现出严重的生长缺陷且缺乏二价体,表明fancm突变体中增加的交叉形成依赖MUS81蛋白。因此,FANCM是通过Ⅱ型交叉形成途径抑制减数分裂重 组。进一步研究发现,双突变体fancm-1 spo11-1和fancm-1 dmc1中的交叉未能恢复,表明fancm突变体中增加的交叉需要SPO11和DMC1蛋白的参与,即FANCM的抑制作用发生在DNA双链断裂和单链入侵之后[54]。在酵母中,FANCM的同源基因MPH1和FML1分别在芽殖酵母(Saccharomyces cerevisiae)和裂殖酵母(Schizosaccharomyces pombe)中同样被证实通过分解D-loops中间体促进SDSA途径的非交叉形成[84,85]。通过对所有FA(fanconi anemia)途径相关蛋白的研究,发现只有FANCM的DNA结合辅助因子MHF1和MHF2具有抑制减数分裂重组的功能,其通过形成异源四聚体来增强FANCM解旋酶的活性,促进FANCM与DNA结合,从而抑制Ⅱ型交叉的形成[55,86~89]。在拟南芥多个突变体中,双突变体msh5 mhf1、msh5 mhf2、hei10 mhf2和三突变体hei10 mhf1 mhf2中二价体的形成没有差异,证明MHF1和MHF2通过相同的途径抑制减数分裂重组[55]。另一方面,mhf2突变体仅能提高1.6倍重组率,但fancm和fancm mhf2突变体却能达3倍的增加,这表明MHF2与FANCM抑制重组的途径是一致的,但MHF2的抑制作用弱于FANCM[55]。此外,在mhf1 mus81和mhf2 mus81双突变体中表现出严重的减数分裂缺陷,但单突变体mhf1、mhf2和mus81中未观察到明显的减数分裂缺陷,表明MHF1和MHF2对减数分裂重组的抑制作用依赖于MUS81,这与FANCM的抑制途径相同[55]。因此,MHF1和MHF2作为辅助因子参与FANCM的Ⅱ型交叉形成途径抑制减数分裂重组。
2.2 BTR重组抑制途径
在减数分裂过程中,为了避免染色体间的纠缠和断裂,DNA双链断裂及其修复过程中产生的重组中间体需要通过不同的途径分解成为交叉或者非交叉[90]。高度保守的BTR复合体(bloom syndrome- Topoisomerase 3α-RecQ-mediated genome instability 1)在拟南芥(BLM-TOP3α-RMI1)和酵母(SGS1-TOP3α- RMI1)中通过限制减数分裂重组中间体形成交叉,进而保障了染色体的完整[91,92,93,94]。例如,BTR复合体中的RECQ4A/B、TOP3α和RMI1蛋白通过同一途径抑制Ⅱ型交叉的形成,但与FANCM抑制途径不同[58]。RECQ4A和RECQ4B属于哺乳动物BLM (酵母中为SGS1)中的两个冗余同源蛋白,且RECQ4B只存在于十字花科植物中[95,96],而TOP3α和RMI1为BTR复合体中的单拷贝基因[96],其相互作用在减数分裂重组中发挥多种功能。首先,RECQ4A/B、TOP3α和RMI1蛋白均能通过D-loop重组中间体分解途径阻止Ⅱ型交叉的形成,促进非交叉的产生,但花粉荧光标记四分体对不同突变体重组率的检查显示不同基因及组合对重组提高的强度不同[58]。例如,单突变体recq4a和recq4b中的重组率没有显著增加,而双突变体recq4a recq4b中的重组率平均提高了5倍,突变体recq4a recq4b fancm的重组率甚至提高了9倍。这样的重组叠加效应也发生在top3α和top3α fancm、rmi1和rmi1 fancm突变体中,其重组率分别平均提高了3倍和5倍、4倍和5倍,这表明BTR重组抑制途径与FANCM途径相互独立但并非功能冗余,这可能与BTR复合体在减数分裂重组过程中发挥多种功能有关[56,58]。由于RECQ4A/B、TOP3α和RMI1两两组合的突变体中(recq4ab top3α、recq4ab rmi1和top3α rmi1)均表现出严重的减数分裂缺陷,导致不能直接测量这些基因型组合的重组率,但推测其可能通过同一途径协同抑制交叉形成,因为:(1) RECQ4A/B、TOP3α和RMI1同属于BTR蛋白复合体,且均从拟南芥和其他物种的体细胞中共同纯化形成;(2) recq4ab和rmi1突变体具有相似的重组增加情况;(3) 其与fancm形成的双突变体均表现重组叠加效应。
其次,TOP3α和RMI1在拟南芥、酿酒酵母和秀丽隐杆线虫(Caenorhabditis elegans)的减数分裂重组过程中具有双重作用:除了限制多余交叉形成以外,还具有分解重组中间体或者预防形成不可分解的重组中间体的作用,但RECQ4A/B不参与该过程。同时研究证实TOP3α和RMI1的C-末端结构域中分别包含的锌指结构域(zinc finger domain)和OB样折叠结构域(oligo-binding fold domain)是抑制Ⅱ型交叉形成的关键。最近多项研究表明,BTR复合体可促进一部分I型交叉的形成[56,58,93,97,98]。在拟南芥中,TOP3α和RMI1的突变基因虽然能增加Ⅱ型交叉,并恢复zmm突变体中的大部分交叉,但top3α和rmi1突变体在第一次减数分裂中期仍然出现一些单价体,这表明在大量Ⅱ型交叉形成的背景下,交叉保障并未完全严格执行,这导致了在单突变体中出现这一微小的重组缺陷[58]。这也与Jagut等[97,98]对秀丽隐杆线虫中RMI-1和HIM-6 (BLM)能促进Ⅰ型交叉形成的研究结果一致。
2.3 FIGL1-FLIP复合体通过调节单链入侵抑制Ⅱ型交叉形成
FIGL1 (fidgetin-like-1)和FLIP (fidgetin-like-1 interacting protein)通过形成具有广泛保守性的复合体,与负责催化同源重组中DNA链交换的DMC1和RAD51重组酶相互作用,共同调节重组过程中单链入侵的关键步骤,从而抑制Ⅱ型交叉的形成[59]。在真核生物中,FIGL1和FLIP的同源基因在所有脊椎动物和陆地植物中均具有保守性,但在拟南芥和人类中并非完全保守[59]。例如,FIGL1直接与RAD51和DMC1重组酶相互作用,且在植物和哺乳动物中保守,但在FLIP中只有人类的同源蛋白与DMC1进行了互动,且在拟南芥和人类中均未检测到FLIP和RAD51之间的相互作用。从更广的范围来看,虽然FIGL1和FLIP均在其他遥远进化枝的物种中被检测到,表明该复合体在真核生物进化过程的早期就已经形成,但是其他进化枝的一些物种中已经失去了FIGL1和FLIP。同样,具有FLIP的物种中包含FIGL1,但在多个包含FIGL1的物种中并没有检测到FLIP的存在,这可能与FIGL1是FIGL1-FLIP复合体的核心活性因子,而FLIP是FIGL1活性功能中可有可无的因素有关。最新研究也表明,真核生物中FLIP在序列水平上显示出低保守性,如在人类和拟南芥中只有12%的序列相同。但是,这些同源蛋白都包含了一个未知功能的DUF4487结构域,如在水稻中鉴定的FLIP的同源蛋白MEICA1 (meiotic chromosome association 1)也包含该结构域,其在DMC1催化单链入侵之后,与BTR复合体中 的TOP3α相互作用,抑制水稻减数分裂Ⅰ型交叉形成,这也与拟南芥中FLIP抑制Ⅱ型交叉形成的途径不同[59,99]。FIGL1与FLIP作为复合体共同抑制减数分裂同源重组,但FIGL1抑制程度要高于FLIP,如figl1突变体中的重组率提高了1.7倍,而flip突变体中仅提高了1.3倍。进一步研究发现,figl1 flip双突变体与figl1单突变体相比并没有显著提高重组率,表明FIGL1与FLIP通过同一途径抑制减数分裂交叉形成,其中FIGL1是FIGL1-FLIP复合体的核心,而FLIP是不可缺少的因素[57,59]。酵母双杂交检测显示,FIGL1-FLIP复合体通过FRBD结构域与RAD51和DMC1相互作用,其可能通过限制单链入侵调节RAD51和DMC1重组酶的活性:即FIGL1和FLIP蛋白的缺失导致RAD51和DMC1重组酶活性的增强或酶功能作用时间的延长,产生异常重组中间体,如多单链侵入结合分子[59]。因此,FLIP-FIGL1复合体通过阻止异常重组中间体的形成来调控单链入侵的质量。与BTR复合体相关突变体相似,在figl1和flip突变体中观察到少量单价体的出现,表明交叉保障的正常实施在缺失FIGL1与FLIP蛋白的情况下受到了轻微影响,其可能是由于RAD51和DMC1重组酶活性受影响而产生了异常重组中间体,而这些本应形成交叉的异常重组中间体未能成功转化形成交叉,导致交叉保障没有严格执行[59]。
与野生型相比,figl1和fancm单突变体的重组率分别增加了1.7倍和3倍,而figl1 fancm双突变体的重组率则显著提高了6倍,这表明figl1和fancm对增加减数分裂重组具有叠加效应[57]。进一步研究表明,fancm突变体仅能在纯合背景下提高重组率,而杂合背景下的重组率增加受到抑制,如Col/Ler F1代杂合背景下fancm突变体重组率仅提高了22%,而在Col纯合背景下能增加300%的重组率[57,100]。相反,figl1突变体不存在这种情况,在纯合和杂合背景下均能同样的提高重组率[57]。FANCM突变基因在杂合背景下低效的重组提高能力可能与碱基对错配导致的父母亲本染色体间的序列差异有关[100]。值得注意的是,虽然figl1和fancm突变基因的叠加能极大的提高重组率,但这些突变体中交叉增加的区域均集中在染色体两侧的端粒,而着丝粒附近的异染色质仍是减数分裂重组的“冷区”[57,100,101]。
2.4 抑制Ⅰ型交叉形成的基因
由于以上重组抑制突变体的筛选是建立在zmm等突变体背景之下,虽然能快速高效的筛选来获得大量重组恢复系,但这些恢复系中增加的交叉均来自Ⅱ型交叉形成途径,因此该研究不能揭示Ⅰ型交叉抑制基因。然而,Ⅰ型交叉形成途径调控着80%~85%的交叉形成,是最主要的减数分裂重组调控途径。在秀丽隐杆线虫中,其减数分裂过程中产生的交叉均来自于Ⅰ型交叉形成途径,研究发现联会复合体(synaptonemal complex, SC)的一些元件蛋白对交叉形成具有双重作用(促进和抑制)。如在秀丽隐杆线 虫中,SYPs (synaptonemal complex central region proteins, SYPs)对减数分裂交叉形成是至关重要的。但最近的研究发现利用RNA干扰部分降低SYPs (SYP-1、SYP-2和SYP-3)蛋白的表达水平(削弱60%~70%)能减弱交叉干扰,增加交叉数量,并减少交叉干扰有效作用距离,这表明SYPs限制了秀丽隐杆线虫中Ⅰ型交叉的形成[102]。相同的现象也在水稻联会复合体中央元件蛋白ZEP1 (synaptonemal complex central element protein)的功能研究中被证实。在水稻ZEP1部分功能丧失的突变体中,交叉干扰强度减弱,交叉数量也提高了1.8倍,进一步的细胞学和zip4 zep1双突变体的遗传分析证明,这些额外增加的交叉主要来自于I型交叉形成途径,这表明水稻ZEP1是Ⅰ型交叉抑制基因[103]。最近,利用拟南芥Col/Ler染色体替换系(chromosome substitution lines, CSLs)和花粉荧光标记系(fluorescent tagged lines, FTLs)研究发现,HEI10蛋白的多态性(R264G, Col/Ler)导致Col/Ler杂合体中的重组率显著低于Col/Col纯合体,表明HEI10基因的自然变异调控植物减数分裂重组[104]。研究还发现hei10/null杂合体的重组率显著低于hei10/hei10纯合体,说明HEI10等位基因对重组的调控具有剂量敏感性。进一步研究表明,增加HEI10基因的拷贝数能提高两倍的重组率,但交叉干扰程度降低,更为重要的是HEI10双拷贝突变体与recq4a recq4b突变体结合互作,通过Ⅰ型和Ⅱ型交叉形成途径显著提高重组率4倍[104,105]。
3 展望
植物减数分裂重组抑制基因是植物在自然选择过程中适应环境变化进化形成的,是在特定生境条件下为维持重组成本和重组优势间平衡的一种保护机制。这种保护机制体现在植物在感知环境变化过程中对减数分裂重组调控的变化,为后代适应新的环境提前做好准备。例如,植物减数分裂重组对环境温度的变化就极为敏感,在植物和动物中的研究表明减数分裂重组率随着温度的升高而增加[106,107,108,109]。而对植物群体而言,过高的重组率并不利于种群的稳定遗传,这可能与高水平的重组率在减数分裂过程中会产生同源染色体分离异常,进而导致生育缺陷[110]。同时,在稳定的生境条件下,高重组率将破坏植物中存在的有利等位基因组合,对植物的稳定遗传产生不利影响。因此,绝大多数真核生物选择限制减数分裂过程中过多的交叉形成。然而,对于植物育种学家来说,由于植物减数分裂重组过程被严格的限制,这极大的制约了植物育种的效率和质量。而减数分裂重组抑制基因的应用能打破减数分裂重组的自然限制,极大的提高杂交后代的重组率,丰富遗传多样性和创造新的等位基因组合,这样大大提高了获得理想植物表型的概率,从而提高植物育种的效率和质量。例如在花卉新品种培育中,获得花色奇特、花型优美的品种是花卉育种工作的重要目标,但由于传统杂交育种中存在重组率低和遗传连锁的现象,难以获得理想的表型和新颖奇特的品种。而重组率的提高能打破基因的连锁,产生更为丰富的基因组合类型,从而选(培)育出“新奇特”的花卉新品种。因此,植物减数分裂同源重组抑制机制的深入研究对植物育种具有十分重要的意义,也能从分子水平上揭示物种适应环境变化不断进化和演变的机制。
Blary等[111]将FANCM抑制基因在芜菁(Brassica rapa)和甘蓝型油菜(Brassica napus)中进行功能缺失突变,发现重组率在芜菁和甘蓝型油菜fancm突变体中分别提高了3倍和1.3倍。该研究为减数分裂重组抑制基因在植物育种中的应用提供了基础和方法。首先,敲除目标植物中重组抑制基因,获得重组抑制突变体。然而,由于很难预测错义突变对蛋白质功能的影响,即使这些突变位点位于重组抑制基因高度保守的结构域中,这可能导致突变体中重组抑制基因功能并未完全丧失。因此,改良获得重组抑制基因无义突变的方法尤为重要。近年来,快速发展的CRISPR/Cas9技术能在基因的多个同源拷贝中产生稳定和可遗传的突变,为植物靶向诱变提供了新的方法,也为重组抑制基因的转化研究开辟了新的途径。其次,在育种中如何利用超重组植物进行新品种选育也是一个巨大的挑战。目前育种家主要通过杂交育种来选育优秀和理想性状表型的新品种,但目前所有的重组抑制基因均表现为隐性性状,这制约了重组抑制基因在育种中的应用。因此,育种策略和显性育种系统的开发也是重组抑制基因育种应用中值得重点研究的方向。
近年来,对植物减数分裂重组抑制基因的研究取得了突破性进展,但是对抑制同源重组的调控网络和关键环节仍然不太清楚,如是否存在与3条交叉形成途径之外的其他类型的途径?是否存在显性重组抑制基因调控交叉形成?新技术和新方法的问世和应用能加快减数分裂重组抑制基因的筛选与鉴定研究。例如,利用流式细胞仪和花粉荧光标记系高效快速的测定染色体特定区间的重组率,进而筛选获得重组率提高或降低的突变体,为植物减数分裂重组调控机制的研究提供科研材料[112,113,114]。因此,应该加大对重组抑制基因及其调控途径的研究,为充分利用植物的遗传潜力进行创新育种奠定理论基础。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:24656236 [本文引用: 2]

In eukaryotes, crossovers together with sister chromatid cohesion maintain physical association between homologous chromosomes, ensuring accurate chromosome segregation during meiosis I and resulting in exchange of genetic information between homologues. The Arabidopsis PTD (Parting Dancers) gene affects the level of meiotic crossover formation, but its functional relationships with other core meiotic genes, such as AtSPO11-1, AtRAD51, and AtMSH4, are unclear; whether PTD has other functions in meiosis is also unknown. To further analyze PTD function and to test for epistatic relationships, we compared the meiotic chromosome behaviors of Atspo11-1 ptd and Atrad51 ptd double mutants with the relevant single mutants. The results suggest that PTD functions downstream of AtSPO11-1 and AtRAD51 in the meiotic recombination pathway. Furthermore, we found that meiotic defects in rck ptd and Atmsh4 ptd double mutants showed similar meiotic phenotypes to those of the relevant single mutants, providing genetic evidences for roles of PTD and RCK in the type I crossovers pathway. Moreover, we employed a pollen tetrad-based fluorescence method and found that the meiotic crossover frequencies in two genetic intervals were significantly reduced from 6.63% and 22.26% in wild-type to 1.14% and 6.36%, respectively, in the ptd-2 mutant. These results revealed new aspects of PTD function in meiotic crossover formation.
URL [本文引用: 2]

Meiosis is the crucial process in eukaryotic sexual reproduction in which one diploid germ cell divides to produce four haploid gametes. Several key events, including homologous chromosomes pairing, synapsis, recombination, and segregation occur sequentially during this process. However, although all these events are widely conserved in species, and they are controlled by both genetic and epigenetic factors, the detailed molecular mechanisms remain obscure up to now. As the major classical genetic materials, plants also have inherent advantages in meiotic studies. Furthermore, the genomes of several model plants have been sequenced which could greatly accelerate meiotic research using molecular tools. In this review, we give an overview of the discovery of meiotic genes mainly in Arabidopsis and rice, with a particular focus on their functions in homologous recombination.
URLPMID:26849908 [本文引用: 1]

During the first division of meiosis, segregation of homologous chromosomes reduces the chromosome number by half. In most species, sister chromatid cohesion and reciprocal recombination (crossing-over) between homologous chromosomes are essential to provide tension to signal proper chromosome segregation during the first meiotic division. Crossovers are not distributed uniformly throughout the genome and are repressed at and near the centromeres. Rare crossovers that occur too near or in the centromere interfere with proper segregation and can give rise to aneuploid progeny, which can be severely defective or inviable. We review here how crossing-over occurs and how it is prevented in and around the centromeres. Molecular mechanisms of centromeric repression are only now being elucidated. However, rapid advances in understanding crossing-over, chromosome structure, and centromere functions promise to explain how potentially deleterious crossovers are avoided in certain chromosomal regions while allowing beneficial crossovers in others.
URLPMID:23733849 [本文引用: 1]

Meiotic crossovers facilitate the segregation of homologous chromosomes and increase genetic diversity. The formation of meiotic crossovers was previously posited to occur via two pathways, with the relative use of each pathway varying between organisms; however, this paradigm could not explain all crossovers, and many of the key proteins involved were unidentified. Recent studies that identify some of these proteins reinforce and expand the model of two meiotic crossover pathways. The results provide novel insights into the evolutionary origins of the pathways, suggesting that one is similar to a mitotic DNA repair pathway and the other evolved to incorporate special features unique to meiosis.
URLPMID:25494464 [本文引用: 2]

Abstract Meiosis is the cell division that reshuffles genetic information between generations. Recently, much progress has been made in understanding this process; in particular, the identification and functional analysis of more than 80 plant genes involved in meiosis have dramatically deepened our knowledge of this peculiar cell division. In this review, we provide an overview of advancements in the understanding of all aspects of plant meiosis, including recombination, chromosome synapsis, cell cycle control, chromosome distribution, and the challenge of polyploidy. Expected final online publication date for the Annual Review of Plant Biology Volume 66 is April 29, 2015. Please see http://www.annualreviews.org/catalog/pubdates.aspx for revised estimates.
URL [本文引用: 2]
URLPMID:4060054 [本文引用: 1]

Faithful chromosome segregation in meiosis is essential for ploidy stability over sexual life cycles. In plants, defective chromosome segregation caused by gene mutations or other factors leads to the formation of unbalanced or unreduced gametes creating aneuploid or polyploid progeny, respectively. Accurate segregation requires the coordinated execution of conserved processes occurring throughout the two meiotic cell divisions. Synapsis and recombination ensure the establishment of chiasmata that hold homologous chromosomes together allowing their correct segregation in the first meiotic division, which is also tightly regulated by cell-cycle dependent release of cohesin and monopolar attachment of sister kinetochores to microtubules. In meiosis II, bi-orientation of sister kinetochores and proper spindle orientation correctly segregate chromosomes in four haploid cells. Checkpoint mechanisms acting at kinetochores control the accuracy of kinetochore-microtubule attachment, thus ensuring the completion of segregation. Here we review the current knowledge on the processes taking place during chromosome segregation in plant meiosis, focusing on the characterization of the molecular factors involved.
URLPMID:28211906 [本文引用: 1]

Meiotic homologous recombination plays a central role in creating genetic variability, making it an essential biological process relevant to evolution and crop breeding. In this study, we used pollen-specific fluorescent tagged lines (FTLs) to measure male meiotic recombination frequency during the development ofArabidopsis thaliana. Interestingly, a subset of pollen grains consistently shows loss of fluorescence expression in tested lines. Using nine independent FTL intervals, the spatio-temporal dynamics of male recombination frequency was assessed during plant development, considering both shoot type and plant age as independent parameters. In most genomic intervals assayed, male meiotic recombination frequency is highly consistent during plant development, showing no significant change between different shoot types and during plant aging. However, in some genomic regions, such as I1a and I5a, a small but significant effect of either developmental position or plant age were observed, indicating that the meiotic CO frequency in those intervals varies during plant development. Furthermore, from an overall view of all nine genomic intervals assayed, both primary and tertiary shoots show a similar dynamics of increasing recombination frequency during development, while secondary and lateral shoots remain highly stable. Our results provide new insights in the dynamics of male meiotic recombination frequency during plant development.
URLPMID:4665078 [本文引用: 2]

The study of homologous recombination has its historical roots in meiosis. In this context, recombination occurs as a programmed event that culminates in the formation of crossovers, which are essential for accurate chromosome segregation and create new combinations of parental alleles. Thus, meiotic recombination underlies both the independent assortment of parental chromosomes and genetic linkage. This review highlights the features of meiotic recombination that distinguish it from recombinational repair in somatic cells, and how the molecular processes of meiotic recombination are embedded and interdependent with the chromosome structures that characterize meiotic prophase. A more in-depth review presents our understanding of how crossover and noncrossover pathways of meiotic recombination are differentiated and regulated. The final section of this review summarizes the studies that have defined defective recombination as a leading cause of pregnancy loss and congenital disease in humans.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:21282472 [本文引用: 1]

Meiotic crossovers are essential for ensuring correct chromosome segregation as well as for creating new combinations of alleles for natural selection to take place. During meiosis, excess meiotic double-strand breaks (DSBs) are generated; a subset of these breaks are repaired to form crossovers, whereas the remainder are repaired as non-crossovers. What determines where meiotic DSBs are created and whether a crossover or non-crossover will be formed at any particular DSB remains largely unclear. Nevertheless, several recent papers have revealed important insights into the factors that control the decision between crossover and non-crossover formation in meiosis, including DNA elements that determine the positioning of meiotic DSBs, and the generation and processing of recombination intermediates. In this review, we focus on the factors that influence DSB positioning, the proteins required for the formation of recombination intermediates and how the processing of these structures generates either a crossover or non-crossover in various organisms. A discussion of crossover interference, assurance and homeostasis, which influence crossing over on a chromosome-wide and genome-wide scale - in addition to current models for the generation of interference - is also included. This Commentary aims to highlight recent advances in our understanding of the factors that promote or prevent meiotic crossing over.
URLPMID:16025405 [本文引用: 1]

Abstract: A brief introduction is presented with some thought on the origin of meiosis. Subsequently, a sequential overview of the diverse processes that take place during meiosis is provided, with an eye to similarities and differences between the different eukaryotic systems. In the final part, we try to summarize the available core meiotic mutants and make a comprehensive comparison for orthologous genes between fungal, plant, and animal systems.
URLPMID:24136506 [本文引用: 1]

During meiosis, a programmed induction of DNA double-strand breaks (DSBs) leads to the exchange of genetic material between homologous chromosomes. These exchanges increase genome diversity and are essential for proper chromosome segregation at the first meiotic division. Recent findings have highlighted an unexpected molecular control of the distribution of meiotic DSBs in mammals by a rapidly evolving gene, PR domain-containing 9 (PRDM9), and genome-wide analyses have facilitated the characterization of meiotic DSB sites at unprecedented resolution. In addition, the identification of new players in DSB repair processes has allowed the delineation of recombination pathways that have two major outcomes, crossovers and non-crossovers, which have distinct mechanistic roles and consequences for genome evolution.
URLPMID:25651968 [本文引用: 1]

Meiotic recombination is a deeply conserved process within eukaryotes that has a profound effect on patterns of natural genetic variation. During meiosis homologous chromosomes pair and undergo DNA double strand breaks generated by the Spo11 endonuclease. These breaks can be repaired as crossovers that result in reciprocal exchange between chromosomes. The frequency of recombination along chromosomes is highly variable, for example, crossovers are rarely observed in heterochromatin and the centromeric regions. Recent work in plants has shown that crossover hotspots occur in gene promoters and are associated with specific chromatin modifications, including H2 A.Z. Meiotic chromosomes are also organized in loop-base arrays connected to an underlying chromosome axis, which likely interacts with chromatin to organize patterns of recombination.Therefore, epigenetic information exerts a major influence on patterns of meiotic recombination along chromosomes, genetic variation within populations and evolution of plant genomes.
URLPMID:28258986 [本文引用: 1]

Meiotic recombination ensures the fertility of gametes and creates novel genetic combinations. Although meiotic crossover (CO) frequency is under homeostatic control, CO frequency is also plastic in nature and can respond to environmental conditions. Most investigations have focused on temperature and recombination, but other external and internal stimuli also have important roles in modulating CO frequency. Even less is understood about the molecular mechanisms that underly these phenomenon, but recent work has begun to advance our knowledge in this field. In this review, we identify and explore potential mechanisms including changes in: the synaptonemal complex, chromatin state, DNA methylation, and RNA splicing.
URLPMID:26917764 [本文引用: 1]

Eukaryotes generate germ cells through meiotic recombination. This process initiates through breaks in genomic DNA catalyzed by the SPO11 protein. Vrielynck et al. and Robert et al. discover that SPO11, like topoisomerase VI enzymes, interacts with a partner protein (see the Perspective by Bouuaert and Keeney). This partner is required for proper meiotic recombination and is found in a wide range of eukaryotes, suggesting that it is a universal feature of the essential recombination step.Science , this issue p. [939][1], [943][2]; see also p. [916][3] [1]: /lookup/doi/10.1126/science.aad5196 [2]: /lookup/doi/10.1126/science.aad5309 [3]: /lookup/doi/10.1126/science.aaf2509
URLPMID:26917763 [本文引用: 1]

The SPO11 protein catalyzes the formation of meiotic DNA double strand breaks (DSBs) and is homologous to the A subunit of an archaeal topoisomerase (topo VI). Topo VI are heterotetrameric enzymes comprising two A and two B subunits; however, no topo VIB involved in meiotic recombination had been identified. We characterized a structural homolog of the archaeal topo VIB subunit [meiotic topoisomerase VIB–like (MTOPVIB)], which is essential for meiotic DSB formation. It forms a complex with the two Arabidopsis thaliana SPO11 orthologs required for meiotic DSB formation (SPO11-1 and SPO11-2) and is absolutely required for the formation of the SPO11-1/SPO11-2 heterodimer. These findings suggest that the catalytic core complex responsible for meiotic DSB formation in eukaryotes adopts a topo VI–like structure.
URLPMID:27477684 [本文引用: 1]
URLPMID:17018031 [本文引用: 2]

Summary Top of page Summary Introduction Results Discussion Experimental procedures Acknowledgements References The Spo11 protein is a eukaryotic homologue of the archaeal DNA topoisomerase VIA subunit (topo VIA). In archaea it is involved, together with its B subunit (topo VIB), in DNA replication. However, most eukaryotes, including yeasts, insects and vertebrates, instead have a single gene for Spo11/topo VIA and no homologues for topo VIB. In these organisms, Spo11 mediates DNA double-strand breaks that initiate meiotic recombination. Many plant species, in contrast to other eukaryotes, have three homologues for Spo11/topo VIA and one for topo VIB. The homologues in Arabidopsis, AtSPO11-1, AtSPO11-2 and AtSPO11-3, all share 20 30% sequence similarity with other Spo11/topo VIA proteins, but their functional relationship during meiosis or other processes is not well understood. Previous genetic evidence suggests that AtSPO11-1 is a true orthologue of Spo11 in other eukaryotes and is required for meiotic recombination, whereas AtSPO11-3 is involved in DNA endo-reduplication as a part of the topo VI complex. In this study, we show that plants homozygous for atspo11-2 exhibit a severe sterility phenotype. Both male and female meiosis are severely disrupted in the atspo11-2 mutant, and this is associated with severe defects in synapsis during the first meiotic division and reduced meiotic recombination. Further genetic analysis revealed that AtSPO11-1 and AtSPO11-2 genetically interact, i.e. plants heterozygous for both atspo11-1 and atspo11-2 are also sterile, suggesting that AtSPO11-1 and AtSPO11-2 have largely overlapping functions. Thus, the three Arabidopsis Spo11 homologues appear to function in two discrete processes, i.e. AtSPO11-1 and AtSPO11-2 in meiotic recombination and AtSPO11-3 in DNA replication.
.
[本文引用: 1]
URLPMID:23943860 [本文引用: 1]

In meiosis, homologous recombination entails programmed DNA double-strand break (DSB) formation and synaptonemal complex (SC) assembly coupled with the DSB repair. Although SCs display extensive structural conservation among species, their components identified are poorly conserved at the sequence level. Here, we identified a novel SC component, designated CENTRAL REGION COMPONENT1 (CRC1), in rice (Oryza sativa). CRC1 colocalizes with ZEP1, the rice SC transverse filament protein, to the central region of SCs in a mutually dependent fashion. Consistent with this colocalization, CRC1 interacts with ZEP1 in yeast two-hybrid assays. CRC1 is orthologous to Saccharomyces cerevisiae pachytene checkpoint2 (Pch2) and Mus musculus THYROID RECEPTOR-INTERACTING PROTEIN13 (TRIP13) and may be a conserved SC component. Additionally, we provide evidence that CRC1 is essential for meiotic DSB formation. CRC1 interacts with HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 (PAIR1) in vitro, suggesting that these proteins act as a complex to promote DSB formation. PAIR2, the rice ortholog of budding yeast homolog pairing1, is required for homologous chromosome pairing. We found that CRC1 is also essential for the recruitment of PAIR2 onto meiotic chromosomes. The roles of CRC1 identified here have not been reported for Pch2 or TRIP13.
[本文引用: 1]
[本文引用: 1]
URLPMID:22694475 [本文引用: 2]

DNA double-strand break (DSB) formation is the initial event for meiotic recombination catalyzed by the conserved Spo11 protein. In Arabidopsis, several proteins have been reported to be involved in DSB formation. Here, we report an Arabidopsis DSB forming (DFO) gene in Arabidopsis that is involved in DSB formation. The dfo mutant exhibits reduced fertility, producing polyads with an abnormal number of microspores, unlike the tetrads in the wild type. The dfo meiocytes were defective in homologous chromosome synapsis and segregation. Genetic analysis revealed that the homologous recombination of Atdfo-1 is severely affected in meiotic prophase I. DFO encodes a protein without any known conserved domain. There was no homologue identified outside the plant kingdom, indicating that AtDFO is a plant-specific protein. AtMRE11 has been reported to be responsible for processing SPO11-generated DSBs. The Atmre11 mutant displays chromosome fragmentation during meiosis. However, the Atdfo Atmre11 double mutant had no such chromosome fragmentation, indicating that AtDFO is required for DSB formation.
URLPMID:20551173 [本文引用: 1]

Abstract Meiotic recombination is initiated by the programmed induction of DNA double-strand breaks (DSBs) catalyzed by the evolutionarily conserved Spo11 protein. Studies in yeast have shown that DSB formation requires several other proteins, the role and conservation of which remain unknown. Here we show that two of these Saccharomyces cerevisiae proteins, Mei4 and Rec114, are evolutionarily conserved in most eukaryotes. Mei4(-/-) mice are deficient in meiotic DSB formation, thus showing the functional conservation of Mei4 in mice. Cytological analyses reveal that, in mice, MEI4 is localized in discrete foci on the axes of meiotic chromosomes that do not overlap with DMC1 and RPA foci. We thus propose that MEI4 acts as a structural component of the DSB machinery that ensures meiotic DSB formation on chromosome axes. We show that mouse MEI4 and REC114 proteins interact directly, and we identify conserved motifs as required for this interaction. Finally, the unexpected, concomitant absence of Mei4 and Rec114, as well as of Mnd1, Hop2, and Dmc1, in some eukaryotic species (particularly Neurospora crassa, Drosophila melanogaster, and Caenorhabditis elegans) suggests the existence of Mei4-Rec114-dependent and Mei4-Rec114-independent mechanisms for DSB formation, and a functional relationship between the chromosome axis and DSB formation.
URLPMID:3172816 [本文引用: 1]

Meiotic recombination is carried out through a specialized pathway for the formation and repair of DNA double-strand breaks made by the Spo11 protein, a relative of archaeal topoisomerase VI. This review summarizes recent studies that provide insight to the mechanism of DNA cleavage by Spo11, functional interactions of Spo11 with other proteins required for break formation, mechanisms that control the timing of recombination initiation, and evolutionary conservation and divergence of these processes.
URLPMID:4292169 [本文引用: 1]

Meiotic recombination involves the formation and repair of programmed DNA double-strand breaks (DSBs) catalyzed by the conserved Spo11 protein. This review summarizes recent studies pertaining to the formation of meiotic DSBs, including the mechanism of DNA cleavage by Spo11, proteins required for break formation, and mechanisms that control the location, timing, and number of DSBs. Where appropriate, findings in different organisms are discussed to highlight evolutionary conservation or divergence.
URLPMID:21220780 [本文引用: 1]

ABSTRACT Meiosis is an essential process for sexually reproducing organisms, leading to the formation of specialized generative cells. This review intends to highlight current knowledge of early events during meiosis derived from various model organisms, including plants. It will particularly focus on cis- and trans-requirements of meiotic DNA double strand break (DSB) formation, a hallmark event during meiosis and a prerequisite for recombination of genetic traits. Proteins involved in DSB formation in different organisms, emphasizing the known factors from plants, will be introduced and their functions outlined. Recent technical advances in DSB detection and meiotic recombination analysis will be reviewed, as these new tools now allow analysis of early meiotic recombination in plants with incredible accuracy. To anticipate future directions in plant meiosis research, unpublished results will be included wherever possible.
URLPMID:3804616 [本文引用: 1]

The evolutionary conserved Mre11/Rad50/Nbs1 complex functions as one of the guardians of genome integrity in eukaryotes; it is required for the double-strand break repair, meiosis, DNA checkpoint, and telomere maintenance. To better understand the role of the MRE11 gene in Arabidopsis, we performed comparative analysis of several mre11 alleles with respect to genome stability and meiosis. The mre11-4 and mre11-2 alleles presumably produce truncated MRE11 proteins composed of the first 499 and 529 amino acids, respectively. Although the putative MRE11 truncated proteins differ only by 30 amino acids, the mutants exhibited strikingly different phenotypes in regards to growth morphology, genome stability and meiosis. While the mre11-2 mutants are fully fertile and undergo normal meiosis, the mre11-4 plants are sterile due to aberrant repair of meiotic DNA breaks. Structural homology analysis suggests that the T-DNA insertion in the mre11-4 allele probably disrupted the putative RAD50 interaction and/or homodimerization domain, which is assumed to be preserved in mre11-2 allele. Intriguingly, introgression of the atm-2 mutant plant into the mre11-2 background renders the double mutant infertile, a phenotype not observed in either parent line. This data indicate that MRE11 partially compensates for ATM deficiency in meiosis of Arabidopsis.
URLPMID:16716192 [本文引用: 1]

Meiotic recombination involves the formation and repair of DNA double-strand breaks (DSB). One of the genes required for DSB formation in the yeast Saccharomyces cerevisiae , Ski8/Rec103 , is intriguing because it also plays a role in cytoplasmic RNA metabolism, a function difficult to relate to DSB formation. The meiotic role of Ski8 is conserved in several fungi, but has not been investigated outside this kingdom. We identified the Ski8 homolog in Arabidopsis thaliana and isolated two mutants. We showed that the Arabidopsis Ski8 homolog was required for normal plant development and growth, suggesting a conserved somatic function, but that it was not required for meiotic recombination or progression. The data presented here provide strong evidence that the meiotic role of Ski8 is not conserved in Arabidopsis and sequence analysis suggests that this may also be the case in a range of other species.
URLPMID:15790808 [本文引用: 1]

The ataxia-telangiectasia mutated (ATM) kinase signals the presence of DNA double-strand breaks in mammalian cells by phosphorylating proteins that initiate cell-cycle arrest, apoptosis, and DNA repair. We show that the Mre11-Rad50-Nbs1 (MRN) complex acts as a double-strand break sensor for ATM and recruits ATM to broken DNA molecules. Inactive ATM dimers were activated in vitro with DNA in the presence of MRN, leading to phosphorylation of the downstream cellular targets p53 and Chk2. ATM autophosphorylation was not required for monomerization of ATM by MRN. The unwinding of DNA ends by MRN was essential for ATM stimulation, which is consistent with the central role of single-stranded DNA as an evolutionarily conserved signal for DNA damage.
URLPMID:4252570 [本文引用: 1]

Author information: (1)The Howard Hughes Medical Institute, The Department of Molecular Biosciences, The Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX 78712, USA. Electronic address: tpaull@utexas.edu. (2)The Howard Hughes Medical Institute, The Department of Molecular Biosciences, The Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, TX 78712, USA.
URLPMID:3784562 [本文引用: 1]

Recombination establishes the chiasmata that physically link pairs of homologous chromosomes in meiosis, ensuring their balanced segregation at the first meiotic division and generating genetic variation. The visible manifestation of genetic crossing-overs, chiasmata are the result of an intricate and tightly regulated process involving induction of DNA double-strand breaks and their repair through invasion of a homologous template DNA duplex, catalysed by RAD51 and DMC1 in most eukaryotes. We describe here a RAD51-GFP fusion protein that retains the ability to assemble at DNA breaks but has lost its DNA break repair capacity. This protein fully complements the meiotic chromosomal fragmentation and sterility of Arabidopsis rad51, but not rad51 dmc1 mutants. Even though DMC1 is the only active meiotic strand transfer protein in the absence of RAD51 catalytic activity, no effect on genetic map distance was observed in complemented rad51 plants. The presence of inactive RAD51 nucleofilaments is thus able to fully support meiotic DSB repair and normal levels of crossing-over by DMC1. Our data demonstrate that RAD51 plays a supporting role for DMC1 in meiotic recombination in the flowering plant, Arabidopsis. Recombination ensures coordinated disjunction of pairs of homologous chromosomes and generates genetic exchanges in meiosis and, with some exceptions, involves the co-operation of the RAD51 and DMC1 strand-exchange proteins. We describe here a RAD51-GFP fusion protein that has lost its DNA break repair capacity but retains the ability to assemble at DNA breaks in the plant, Arabidopsis - fully complementing the meiotic chromosomal fragmentation and sterility of rad51 mutants, and this depends upon DMC1. No effect on genetic map distance was observed in complemented rad51 plants even though DMC1 is the only active strand transfer protein. The inactive RAD51 nucleofilaments are thus able to fully support meiotic DSB repair and normal levels of crossing-over by DMC1 in Arabidopsis. The RAD51-GFP protein confers a dominant-negative inhibition of RAD51-dependent mitotic recombination, while remaining fully fertile - a novel and valuable tool for research in this domain. These phenotypes are equivalent to those of the recently reported yeast rad51-II3A mutant, (Cloud et al. 2012), carrying the implication of their probable generality in other eukaryotes and extending them to a species with a very different relation between numbers of meiotic DNA double-strand breaks and crossing-overs ( 2 DSB/CO in yeast; 25 30 DSB/CO in Arabidopsis; 15 DSB/CO in mice).
URLPMID:26720282 [本文引用: 1]

PLOS Genetics is an open-access
URL [本文引用: 1]

Meiosis ensures the reduction of the genome before the formation of generative cells and promotes the exchange of genetic information between homologous chromosomes by recombination. Essential for these events are programmed DNA double strand breaks (DSBs) providing single-stranded DNA overhangs after their processing. These overhangs, together with the RADiation sensitive51 (RAD51) and DMC1 Disrupted Meiotic cDNA1 (DMC1) recombinases, mediate the search for homologous sequences. Current models propose that the two ends flanking a meiotic DSB have different fates during DNA repair, but the molecular details remained elusive. Here we present evidence, obtained in the model plant Arabidopsis thaliana, that the two recombinases, RAD51 and DMC1, localize to opposite sides of a meiotic DSB. We further demonstrate that the ATR kinase is involved in regulating DMC1 deposition at meiotic DSB sites, and that its elimination allows DMC1-mediated meiotic DSB repair even in the absence of RAD51. DMCI's ability to promote interhomolog DSB repair is not a property of the protein itself but the consequence of an ASYNAPTIC1 (Hopi)-mediated impediment for intersister repair. Taken together, these results demonstrate that DMC1 functions independently and spatially separated from RAD51 during meiosis and that ATR is an integral part of the regular meiotic program.
URLPMID:4059524 [本文引用: 1]

Homologous recombination (HR) is critical for genome integrity. The displacement loop (D loop) constitutes a key HR intermediate. Rad51 forms D loops in concert with the Rad54 motor protein. Employing Rad51 nucleoprotein filaments that mimic the length and structure of in vivo substrates, Wright and Heyer analyze Rad54 function in D loop reaction and suggest a heteroduplex DNA (hDNA) pump model consolidating seemingly disparate activities: Rad54 drives D loop formation while removing Rad51 from hDNA.
URLPMID:20690856 [本文引用: 1]

Abstract Homologous recombination (HR) is required for accurate chromosome segregation during the first meiotic division and constitutes a key repair and tolerance pathway for complex DNA damage, including DNA double-strand breaks, interstrand crosslinks, and DNA gaps. In addition, recombination and replication are inextricably linked, as recombination recovers stalled and broken replication forks, enabling the evolution of larger genomes/replicons. Defects in recombination lead to genomic instability and elevated cancer predisposition, demonstrating a clear cellular need for recombination. However, recombination can also lead to genome rearrangements. Unrestrained recombination causes undesired endpoints (translocation, deletion, inversion) and the accumulation of toxic recombination intermediates. Evidently, HR must be carefully regulated to match specific cellular needs. Here, we review the factors and mechanistic stages of recombination that are subject to regulation and suggest that recombination achieves flexibility and robustness by proceeding through metastable, reversible intermediates.
URLPMID:17174892 [本文引用: 3]

Crossing-over between homologous chromosomes facilitates their accurate segregation at the first division of meiosis. Current models for crossing-over invoke an intermediate in which homologs are connected by two crossed-strand structures called Holliday junctions. Such double Holliday junctions are a prominent intermediate in Saccharomyces cerevisiae meiosis, where they form preferentially between homologs rather than between sister chromatids. In sharp contrast, we find that single Holliday junctions are the predominant intermediate in Schizosaccharomyces pombe meiosis. Furthermore, these single Holliday junctions arise preferentially between sister chromatids rather than between homologs. We show that Mus81 is required for Holliday junction resolution, providing further in vivo evidence that the structure-specific endonuclease Mus81-Eme1 is a Holliday junction resolvase. To reconcile these observations, we present a unifying recombination model applicable for both meiosis and mitosis in which single Holliday junctions arise from single- or double-strand breaks, lesions postulated by previous models to initiate recombination.
URLPMID:28108697 [本文引用: 2]

Meiosis is a specialized cell division, essential in most reproducing organisms to halve the number of chromosomes, thereby enabling the restoration of ploidy levels during fertilization. A key step of meiosis is homologous recombination, which promotes homologous pairing and generates crossovers (COs) to connect homologous chromosomes until their separation at anaphase I. These CO sites, seen cytologically as chiasmata, represent a reciprocal exchange of genetic information between two homologous nonsister chromatids. This gene reshuffling during meiosis has a significant influence on evolution and also plays an essential role in plant breeding, because a successful breeding program depends on the ability to bring the desired combinations of alleles on chromosomes. However, the number and distribution of COs during meiosis is highly constrained. There is at least one CO per chromosome pair to ensure accurate segregation of homologs, but in most organisms, the CO number rarely exceeds three regardless of chromosome size. Moreover, their positions are not random on chromosomes but exhibit regional preference. Thus, genes in recombination-poor regions tend to be inherited together, hindering the generation of novel allelic combinations that could be exploited by breeding programs. Recently, much progress has been made in understanding meiotic recombination. In particular, many genes involved in the process in Arabidopsis (Arabidopsis thaliana) have been identified and analyzed. With the coming challenges of food security and climate change, and our enhanced knowledge of how COs are formed, the interest and needs in manipulating CO formation are greater than ever before. In this review, we focus on advances in understanding meiotic recombination and then summarize the attempts to manipulate CO formation. Last, we pay special attention to the meiotic recombination in polyploidy, which is a common genomic feature for many crop plants.
[本文引用: 1]
URLPMID:11741546 [本文引用: 1]

Mus81, a protein with homology to the XPF subunit of the ERCC1-XPF endonuclease, is important for replicational stress tolerance in both budding and fission yeast. Human Mus81 has associated endonuclease activity against structure-specific oligonucleotide substrates, including synthetic Holliday junctions. Mus81-associated endonuclease resolves Holliday junctions into linear duplexes by cutting across the junction exclusively on strands of like polarity. In addition, Mus81 protein abundance increases in cells following exposure to agents that block DNA replication. Taken together, these findings suggest a role for Mus81 in resolving Holliday junctions that arise when DNA replication is blocked by damage or by nucleotide depletion. Mus81 is not related by sequence to previously characterized Holliday junction resolving enzymes, and it has distinct enzymatic properties that suggest it uses a novel enzymatic strategy to cleave Holliday junctions.
URLPMID:19020614 [本文引用: 1]

Abstract Four-way DNA intermediates, also known as Holliday junctions, are formed during homologous recombination and DNA repair, and their resolution is necessary for proper chromosome segregation. Here we identify nucleases from Saccharomyces cerevisiae and human cells that promote Holliday junction resolution, in a manner analogous to that shown by the Escherichia coli Holliday junction resolvase RuvC. The human Holliday junction resolvase, GEN1, and its yeast orthologue, Yen1, were independently identified using two distinct experimental approaches: GEN1 was identified by mass spectrometry following extensive fractionation of HeLa cell-free extracts, whereas Yen1 was detected by screening a yeast gene fusion library for nucleases capable of Holliday junction resolution. The eukaryotic Holliday junction resolvases represent a new subclass of the Rad2/XPG family of nucleases. Recombinant GEN1 and Yen1 resolve Holliday junctions by the introduction of symmetrically related cuts across the junction point, to produce nicked duplex products in which the nicks can be readily ligated.
[本文引用: 1]
URLPMID:3271061 [本文引用: 1]

In this study we have analysed AtASY3, a coiled-coil domain protein that is required for normal meiosis inArabidopsis. Analysis of anAtasy3-1mutant reveals that loss of the protein compromises chromosome axis formation and results in reduced numbers of meiotic crossovers (COs). Although the frequency of DNA double-strand breaks (DSBs) appears moderately reduced inAtasy3-1, the main recombination defect is a reduction in the formation of COs. Immunolocalization studies in wild-type meiocytes indicate that the HORMA protein AtASY1, which is related to Hop1 in budding yeast, forms hyper-abundant domains along the chromosomes that are spatially associated with DSBs and early recombination pathway proteins. Loss of AtASY3 disrupts the axial organization of AtASY1. Furthermore we show that the AtASY3 and AtASY1 homologs BoASY3 and BoASY1, from the closely related speciesBrassica oleracea, are co-immunoprecipitated from meiocyte extracts and that AtASY3 interacts with AtASY1 via residues in its predicted coiled-coil domain. Together our results suggest that AtASY3 is a functional homolog of Red1. Since studies in budding yeast indicate that Red1 and Hop1 play a key role in establishing a bias to favor inter-homolog recombination (IHR), we propose that AtASY3 and AtASY1 may have a similar role inArabidopsis. Loss of AtASY3 also disrupts synaptonemal complex (SC) formation. InAtasy3-1the transverse filament protein AtZYP1 forms small patches rather than a continuous SC. The few AtMLH1 foci that remain inAtasy3-1are found in association with the AtZYP1 patches. This is sufficient to prevent the ectopic recombination observed in the absence of AtZYP1, thus emphasizing that in addition to its structural role the protein is important for CO formation. Homologous recombination (HR) during prophase I of meiosis leads to the formation of physical connections, known as chiasmata, between homologous chromosomes (homologs). Chiasmata are essential for accurate homolog segregation at the first meiotic division. HR is initiated by the formation of DNA double-strand breaks (DSBs). As DNA replication prior to meiosis results in the duplication of each homolog to form two identical sister chromatids, a DSB in one sister chromatid could potentially be repaired using the other as the repair template rather than one of the two non-sister chromatids of the homolog. If this route were predominant, the formation of chiasmata would be disfavored and chromosome segregation would be compromised. However, during meiosis there is a strong bias towards inter-homolog recombination (IHR). In this study we have identified AtASY3, a component of the proteinaceous axes that organize the chromosomes during meiosis inArabidopsis. We find that AtASY3 interacts with AtASY1, a previously identified axis protein that is essential for crossover formation. We show that loss of AtASY3 disrupts the axis-organization of AtASY1. This results in a substantial reduction in chiasmata, and there is extensive chromosome mis-segregation. We propose that loss of AtASY3 affects the efficiency of the inter-homolog bias.
URLPMID:3290786 [本文引用: 1]

Meiotic recombination, including crossovers (COs) and gene conversions (GCs), impacts natural variation and is an important evolutionary force. COs increase genetic diversity by redistributing existing variation, whereas GCs can alter allelic frequency. Here, we sequenced Arabidopsis Landsberg erecta (Ler) and two sets of all four meiotic products from a Columbia (Col)/Ler hybrid to investigate genome-wide variation and meiotic recombination at nucleotide resolution. Comparing Ler and Col sequences uncovered 349,171 Single Nucleotide Polymorphisms (SNPs), 58,085 small and 2315 large insertions/deletions (indels), with highly correlated genome-wide distributions of SNPs, and small indels. A total of 443 genes have at least 10 nonsynonymous substitutions in protein-coding regions, with enrichment for disease-resistance genes. Another 316 genes are affected by large indels, including 130 genes with complete deletion of coding regions in Ler. Using the Arabidopsis qrt1 mutant, two sets of four meiotic products were generated and analyzed by sequencing for meiotic recombination, representing the first tetrad analysis with whole-genome sequencing in a nonfungal species. We detected 18 COs, six of which had an associated GC event, and four GCs without COs (NCOs), and revealed that Arabidopsis GCs are likely fewer and with shorter tracts than those in yeast. Meiotic recombination and chromosome assortment events dramatically redistributed genome variation in meiotic products, contributing to population diversity. In particular, meiosis provides a rapid mechanism to generate copy-number variation (CNV) of sequences that have different chromosomal positions in Col and Ler.
URLPMID:3464199 [本文引用: 1]

Gene conversion, the non-reciprocal exchange of genetic information, is one of the potential products of meiotic recombination. It can shape genome structure by acting on repetitive DNA elements, influence allele frequencies at the population level, and is known to be implicated in human disease. But gene conversion is hard to detect directly except in organisms, like fungi, that group their gametes following meiosis. We have developed a novel visual assay that enables us to detect gene conversion events directly in the gametes of the flowering plant Arabidopsis thaliana. Using this assay we measured gene conversion events across the genome of more than one million meioses and determined that the genome-wide average frequency is 3.5x10(-4) conversions per locus per meiosis. We also detected significant locus-to-locus variation in conversion frequency but no intra-locus variation. Significantly, we found one locus on the short arm of chromosome 4 that experienced 3-fold to 6-fold more gene conversions than the other loci tested. Finally, we demonstrated that we could modulate conversion frequency by varying experimental conditions.
URLPMID:24958856 [本文引用: 1]

DNA polymorphisms are important markers in genetic analyses and are increasingly detected by using genome resequencing. However, the presence of repetitive sequences and structural variants can lead to false positives in the identification of polymorphic alleles. Here, we describe an analysis strategy that minimizes false positives in allelic detection and present analyses of recently published resequencing data from Arabidopsis meiotic products and individual humans. Our analysis enables the accurate detection of sequencing errors, small insertions and deletions (indels), and structural variants, including large reciprocal indels and copy number variants, from comparisons between the resequenced and reference genomes. We offer an alternative interpretation of the sequencing data of meiotic products, including the number and type of recombination events, to illustrate the potential for mistakes in single-nucleotide polymorphism calling. Using these examples, we propose that the detection of DNA polymorphisms using resequencing data needs to account for nonallelic homologous sequences.
URLPMID:16873056 [本文引用: 2]

During meiosis, crossing-over—the exchange of genetic material between maternal and paternal chromosomes—is stringently controlled to restrict the number of crossovers per chromosome pair. In this issue of Cell, Martini et al. (2006) report that the reduction of crossover-initiating events does not result in fewer crossovers. These results have important implications for our understanding of crossover control.
[本文引用: 2]
[本文引用: 2]
URLPMID:26668366 [本文引用: 2]

Abstract During meiotic recombination, double-strand breaks (DSBs) are formed in chromosomal DNA and then repaired as either crossovers (COs) or non-crossovers (NCOs). In most taxa, the number of DSBs vastly exceeds the number of COs. COs are required for generating genetic diversity in the progeny, as well as proper chromosome segregation. Their formation is tightly controlled so that there is at least one CO per pair of homologous chromosomes whereas the maximum number of COs per chromosome pair is fairly limited. One of the main mechanisms controlling the number of recombination events per meiosis is CO homeostasis, which maintains a stable CO number even when the DSB number is dramatically altered. The existence of CO homeostasis has been reported in several species, including mouse, yeast, and Caenorhabditis elegans. However, it is not known whether homeostasis exists in the same form in all species. In addition, the studies of homeostasis have been conducted using mutants and/or transgenic lines exhibiting fairly severe meiotic phenotypes, and it is unclear how important homeostasis is under normal physiological conditions. We found that, in maize, CO control is robust only to ensure one CO per chromosome pair. However, once this limit is reached, the CO number is linearly related to the DSB number. We propose that CO control is a multifaceted process whose different aspects have a varying degree of importance in different species.
URLPMID:25590558 [本文引用: 2]

During meiosis, crossover recombination is tightly regulated. A spatial patterning phenomenon known as interference ensures that crossovers are well-spaced along the chromosomes. Additionally, every pair of homologs acquires at least one crossover. A third feature, crossover homeostasis, buffers the system such that the number of crossovers remains steady despite decreases or increases in the number of earlier recombinational interactions. Here we summarize recent work from our laboratory supporting the idea that all 3 of these aspects are intrinsic consequences of a single basic process and suggesting that the underlying logic of this process corresponds to that embodied in a particular (beam-film) model.
URL [本文引用: 7]
URLPMID:4132730 [本文引用: 7]

Meiotic crossovers (COs) have two important roles, shuffling genetic information and ensuring proper chromosome segregation. Despite their importance and a large excess of precursors (i.e., DNA double-strand breaks, DSBs), the number of COs is tightly regulated, typically one to three per chromosome pair. The mechanisms ensuring that most DSBs are repaired as non-COs and the evolutionary... [Show full abstract]
URLPMID:25825745 [本文引用: 5]

Abstract Meiotic crossovers (COs) have two important roles, shuffling genetic information and ensuring proper chromosome segregation. Despite their importance and a large excess of precursors (i.e., DNA double-strand breaks, DSBs), the number of COs is tightly regulated, typically one to three per chromosome pair. The mechanisms ensuring that most DSBs are repaired as non-COs and the evolutionary forces imposing this constraint are poorly understood. Here we identified Topoisomerase302± (TOP302±) and the RECQ4 helicases--the Arabidopsis slow growth suppressor 1 (Sgs1)/Bloom syndrome protein (BLM) homologs--as major barriers to meiotic CO formation. First, the characterization of a specific TOP302± mutant allele revealed that, in addition to its role in DNA repair, this topoisomerase antagonizes CO formation. Further, we found that RECQ4A and RECQ4B constitute the strongest meiotic anti-CO activity identified to date, their concomitant depletion leading to a sixfold increase in CO frequency. In both top302± and recq4ab mutants, DSB number is unaffected, and extra COs arise from a normally minor pathway. Finally, both TOP302± and RECQ4A/B act independently of the previously identified anti-CO Fanconi anemia of complementation group M (FANCM) helicase. This finding shows that several parallel pathways actively limit CO formation and suggests that the RECQA/B and FANCM helicases prevent COs by processing different substrates. Despite a ninefold increase in CO frequency, chromosome segregation was unaffected. This finding supports the idea that CO number is restricted not because of mechanical constraints but likely because of the long-term costs of recombination. Furthermore, this work demonstrates how manipulating a few genes holds great promise for increasing recombination frequency in plant-breeding programs.
URLPMID:4498898 [本文引用: 8]

Meiotic crossovers (COs) generate genetic diversity and are critical for the correct completion of meiosis in most species. Their occurrence is tightly constrained but the mechanisms underlying this limitation remain poorly understood. Here we identified the conserved AAA-ATPase FIDGETIN-LIKE-1 (FIGL1) as a negative regulator of meiotic CO formation. We show that Arabidopsis FIGL1 limits CO formation genome-wide, that FIGL1 controls dynamics of the two conserved recombinases DMC1 and RAD51 and that FIGL1 hinders the interaction between homologous chromosomes, suggesting that FIGL1 counteracts DMC1/RAD51-mediated inter-homologue strand invasion to limit CO formation. Further, depleting both FIGL1 and the previously identified anti-CO helicase FANCM synergistically increases crossover frequency. Additionally, we showed that the effect of mutating FANCM on recombination is much lower in F1 hybrids contrasting from the phenotype of inbred lines, while figl1 mutation equally increases crossovers in both contexts. This shows that the modes of action of FIGL1 and FANCM are differently affected by genomic contexts. We propose that FIGL1 and FANCM represent two successive barriers to CO formation, one limiting strand invasion, the other disassembling D-loops to promote SDSA, which when both lifted, leads to a large increase of crossovers, without impairing meiotic progression. Sexually reproducing species produce offspring that are genetically unique from one another, despite having the same parents. This uniqueness is created by meiosis, which is a specialized cell division. After meiosis each parent transmits half of their DNA, but each time this occurs, the 'half portion' of DNA transmitted to offspring is different from the previous. The differences are due to resorting the parental chromosomes, but also recombining them. Here we describe a gene IDGETIN-LIKE 1 hich limits the amount of recombination that occurs during meiosis. Previously we identified a gene with a similar function, FANCM. FIGL1 and FANCM operate through distinct mechanisms. This discovery will be useful to understand more, from an evolutionary perspective, why recombination is naturally limited. Also this has potentially significant applications for plant breeding which is largely about sampling many 'recombinants' to find individuals that have heritable advantages compared to their parents.
URLPMID:27965412 [本文引用: 7]

At meiosis, hundreds of programmed DNA doublestrand breaks (DSBs) form and are repaired by homologous recombination. From this large number of DSBs, only a subset yields crossovers (COs), with a minimum of one CO per chromosome pair. All DSBs must be repaired and every recombination intermediate must be resolved to avoid subsequent entanglement and chromosome breakage. The conserved BLM-TOP3 alpha-RMI1 (BTR) complex acts on early and late meiotic recombination intermediates to both limit CO outcome and promote chromosome integrity. In Arabidopsis, the BLM homologues RECQ4A and RECQ4B act redundantly to prevent meiotic extra COs, but recombination intermediates are fully resolved in their absence. In contrast, TOP3 alpha is needed for both processes. Here we show through the characterization of specific mutants that RMI1 is a major anti-CO factor, in addition to being essential to prevent chromosome breakage and entanglement. Further, our findings suggest a specific role of the C-terminal domains of RMI1 and TOP3 alpha, that respectively contain an Oligo Binding domain (OB2) and ZINC finger motifs, in preventing extraCO. We propose that these domains of TOP3 alpha and RMI1 define a sub-domain of the BTR complex which is dispensable for the resolution of recombination intermediates but crucial to limit extra-COs.
URLPMID:29608566 [本文引用: 8]

Les crossing-overs (CO) sont issus d’échange réciproque de matériel génétique entre les chromosomes homologues. Les COs produisent de la diversité génétique et sont essentiels chez la plupart des eucaryotes, pour la distribution équilibrée des chromosomes lors de la méiose. Malgré leur importance, et un large excès de précurseurs moléculaires, le nombre de CO est très limité dans la grande... [Show full abstract]
URLPMID:4867269 [本文引用: 1]

Meiosis presents many important mysteries that await elucidation. Here we discuss two such aspects. First, we consider how the current meiotic program might have evolved. We emphasize the central feature of this program: how homologous chromosomes find one another (“pair”) so as to create the connections required for their regular segregation at Meiosis I. Points of emphasis include the facts that: (i) the classical “bouquet stage” is not required for initial homolog contacts in the current evolved meiotic program; and (ii) diverse observations point to commonality between molecules that mediate meiotic inter-homolog interactions and molecules that are integral to centromeres and/or to microtubule organizing centers (a.k.a. spindle pole bodies or centrosomes). Second, we provide an overview of the classical phenomenon of crossover (CO) interference in an effort to bridge the gap between description on the one hand versus logic and mechanism on the other.
URLPMID:4830878 [本文引用: 1]

Whole genome duplication is a prominent feature of many highly evolved organisms, especially plants. When duplications occur within species, they yield genomes comprising multiple identical or very similar copies of each chromosome ( utopolyploids ). Such genomes face special challenges during meiosis, the specialized cellular program that underlies gamete formation for sexual reproduction. Comparisons between newly formed (neo)-autotetraploids and fully evolved autotetraploids suggest that these challenges are solved by specific restrictions on the positions of crossover recombination events and, thus, the positions of chiasmata, which govern the segregation of homologs at the first meiotic division. We propose that a critical feature in the evolution of these more effective chiasma patterns is an increase in the effective distance of meiotic crossover interference, which plays a central role in crossover positioning. We discuss the findings in several organisms, including the recent identification of relevant genes inArabidopsis arenosa, that support this hypothesis. The online version of this article (doi:10.1007/s00412-015-0571-4) contains supplementary material, which is available to authorized users.
URLPMID:27989672 [本文引用: 1]

Abstract In most sexually reproducing organisms, crossover formation between homologous chromosomes is necessary for proper chromosome disjunction during meiosis I. During meiotic recombination, a subset of programmed DNA double-strand breaks (DSBs) are repaired as crossovers, with the remainder becoming noncrossovers [1]. Whether a repair intermediate is designated to become a crossover is a highly regulated decision that integrates several crossover patterning processes, both along chromosome arms (interference and the centromere effect) and between chromosomes (crossover assurance) [2]. Because the mechanisms that generate crossover patterning have remained elusive for over a century, it has been difficult to assess the relationship between crossover patterning and meiotic chromosome behavior. We show here that meiotic crossover patterning is lost in Drosophila melanogaster mutants that lack the Bloom syndrome helicase. In the absence of interference and the centromere effect, crossovers are distributed more uniformly along chromosomes. Crossovers even occur on the small chromosome 4, which normally never has meiotic crossovers [3]. Regulated distribution of crossovers between chromosome pairs is also lost, resulting in an elevated frequency of homologs that do not receive a crossover, which in turn leads to elevated nondisjunction.
[本文引用: 1]
URLPMID:18318687 [本文引用: 1]

MSH5 , a meiosis-specific member of the MutS-homologue family of genes, is required for normal levels of recombination in budding yeast, mouse and Caenorhabditis elegans . In this paper we report the identification and characterization of the Arabidopsis homologue of MSH5 ( AtMSH5 ). Transcripts of AtMSH5 are specific to reproductive tissues, and immunofluorescence studies indicate that expression of the protein is abundant during prophase I of meiosis. In a T-DNA tagged insertional mutant ( Atmsh5-1 ), recombination is reduced to about 13% of wild-type levels. The residual chiasmata are randomly distributed between cells and chromosomes. These data provide further evidence for at least two pathways of meiotic recombination in Arabidopsis and indicate that AtMSH5 protein is required for the formation of class I interference-sensitive crossovers. Localization of AtMSH5 to meiotic chromosomes occurs at leptotene and is dependent on DNA double-strand break formation and strand exchange. Localization of AtMSH5 to the chromatin at mid-prophase I is dependent on expression of AtMSH4 . At late zygotene/early pachytene a proportion of AtMSH5 foci co-localize with AtMLH1 which marks crossover-designated sites. Chromosome synapsis appears to proceed normally, without significant delay, in Atmsh5-1 but the pachytene stage is extended by several hours, indicative of the operation of a surveillance system that monitors the progression of prophase I.
URL [本文引用: 1]
URLPMID:15854901 [本文引用: 1]

Background: Crossovers are essential for the completion of meiosis. Recently, two pathways of crossover formation have been identified on the basis of distinct genetic controls. In one pathway, crossover inhibits the occurrence of another such event in a distance-dependent manner. This phenomenon is known as interference. The second kind of crossover is insensitive to interference. The two pathways function independently in budding yeast. Only interference-insensitive crossovers occur in Schizosaccharomyces pombe. In contrast, only interference-sensitive crossovers occur in Caenorabditis elegans. The situation in mammals and plants remains unclear. Mer3 is one of the genes shown to be required for the formation of interference-sensitive crossovers in Saccharomyces cerevisiae. Results: To unravel the crossover status in the plant Arabidopsis thaliana, we investigated the role of the A. thaliana MER3 gene through the characterization of a series of allelic mutants. All mer3 mutants showed low levels of fertility and a significant decrease (about 75%) but not a total disappearance of meiotic crossovers, with the number of recombination events initiated in the mutants being similar to that in the wild-type. Genetic analyses showed that the residual crossovers in mer3 mutants did not display interference in one set of adjacent intervals. Conclusions: Mutation in MER3 in Arabidopsis appeared to be specific to recombination events resulting in interference-sensitive crossovers. Thus, MER3 function is conserved from yeast to plants and may exist in other metazoans. Arabidopsis therefore has at least two pathways for crossover formation, one giving rise to interference-sensitive crossover and the other to independently distributed crossovers.
URLPMID:1877879 [本文引用: 1]

Zip4/Spo22 is required for CO formation but not for synapsis completion in Arabidopsis thaliana.In budding yeast meiosis, the formation of interference-sensitive crossovers requires the ZMM proteins. These ZMM proteins are essential in forming a mature synaptonemal complex, and a subset of these (, , and ) has been proposed to compose the core of synapsis initiation complexes (). Zip4/Spo22 functions with to promote polymerization of along chromosomes, making it a crucial component. In higher eukaryotes, synapsis and recombination have often been correlated, but it is totally unknown how these two processes are linked. In this study, we present the characterization of a higher eukaryote component homologue: Arabidopsis AtZIP4. We show that mutations in AtZIP4 belong to the same epistasis group as Atmsh4 and eliminate approximately 85% of crossovers (COs). Furthermore, genetic analyses on two adjacent intervals of Chromosome I established that the remaining COs in Atzip4 do not show interference. Lastly, immunolocalization studies showed that polymerization of the central element of the synaptonemal complex is not affected in Atzip4 background, even if it may proceed from fewer sites compared to wild type. These results reveal that function in CO formation is conserved from budding yeast to Arabidopsis. On the other hand, and contrary to the situation in yeast, mutation in AtZIP4 does not prevent synapsis, showing that both aspects of the function (i.e., CO maturation and synapsis) can be uncoupled.
URLPMID:18812090 [本文引用: 1]

Crossovers (COs) are essential for the completion of meiosis in most species and lead to new allelic combinations in gametes [1]. Two pathways of meiotic crossover formation have been distinguished. Class I COs, which are the major class of CO in budding yeast, mammals, Caenorhabditis elegans, and Arabidopsis, depend on a group of proteins called ZMM and rely on specific DNA structure intermediates that are processed to form COs [2 6]. We identified a novel gene, SHOC1, involved in meiosis in Arabidopsis. Shoc1 mutants showed a striking reduction in the number of COs produced, a similar phenotype to the previously described Arabidopsis zmm mutants. The early steps of recombination, revealed by DMC1 foci, and completion of synapsis are not affected in shoc1 mutants. Double mutant analysis showed that SHOC1 acts in the same pathway as AtMSH5, a conserved member of the ZMM group [7]. SHOC1 is thus a novel gene required for class I CO formation in Arabidopsis. Sequence similarity studies detected putative SHOC1 homologs in a large range of eukaryotes including human. SHOC1 appears to be related to the XPF endonuclease protein family, which suggests that it is directly involved in the maturation of DNA intermediates that lead to COs.
URLPMID:22844245 [本文引用: 1]

In numerous species, the formation of meiotic crossovers is largely under the control of a group of proteins known as ZMM. Here, we identified a new ZMM protein, HEI10, a RING finger-containing protein that is well conserved among species. We show that HEI10 is structurally and functionally related to the yeast Zip3 ZMM and that it is absolutely required for class I crossover (CO) formation in Arabidopsis thaliana. Furthermore, we show that it is present as numerous foci on the chromosome axes and the synaptonemal complex central element until pachytene. Then, from pachytene to diakinesis, HEI10 is retained at a limited number of sites that correspond to class I COs, where it co-localises with MLH1. Assuming that HEI10 early staining represents an early selection of recombination intermediates to be channelled into the ZMM pathway, HEI10 would therefore draw a continuity between early chosen recombination intermediates and final class I COs.
URLPMID:3493451 [本文引用: 1]

Abstract During meiotic recombination, induced double-strand breaks (DSBs) are processed into crossovers (COs) and non-COs (NCO); the former are required for proper chromosome segregation and fertility. DNA synthesis is essential in current models of meiotic recombination pathways and includes only leading strand DNA synthesis, but few genes crucial for DNA synthesis have been tested genetically for their functions in meiosis. Furthermore, lagging strand synthesis has been assumed to be unnecessary. Here we show that the Arabidopsis thaliana DNA replication factor C1 (RFC1) important for lagging strand synthesis is necessary for fertility, meiotic bivalent formation, and homolog segregation. Loss of meiotic RFC1 function caused abnormal meiotic chromosome association and other cytological defects; genetic analyses with other meiotic mutations indicate that RFC1 acts in the MSH4-dependent interference-sensitive pathway for CO formation. In a rfc1 mutant, residual pollen viability is MUS81-dependent and COs exhibit essentially no interference, indicating that these COs form via the MUS81-dependent interference-insensitive pathway. We hypothesize that lagging strand DNA synthesis is important for the formation of double Holliday junctions, but not alternative recombination intermediates. That RFC1 is found in divergent eukaryotes suggests a previously unrecognized and highly conserved role for DNA synthesis in discriminating between recombination pathways.
URLPMID:21771883 [本文引用: 1]

Two distinct pathways for meiotic crossover formation coexist in most eukaryotes. The Arabidopsis SHOC1 protein is required forclass I crossovers and shows sequence similarity with the XPF endonuclease family. Active XPF endonucleases form a heterodimerwith ERCC1 proteins. Here, we show that PTD, an ERCC1-like protein, is required for class-I-interfering crossovers along withSHOC1, MSH4, MSH5, MER3 and MLH3. SHOC1 interacts with PTD in a two-hybrid assay, through its XPF-like nuclease-(HhH)2domain. We propose that a XPF-ERCC1-like heterodimer, represented by SHOC1 and PTD in Arabidopsis, involving Zip2 inSaccharomyces cerevisiae and C9orf84 in human, is required for formation of class I crossovers.
URLPMID:26392549 [本文引用: 1]

Meiosis halves diploid genomes to haploid and is essential for sexual reproduction in eukaryotes. Meiotic recombination ensures physical association of homologs and their subsequent accurate segregation and results in the redistribution of genetic variations among progeny. Most organisms have two classes of cross-overs (COs): interference-sensitive (type I) and -insensitive (type...
URLPMID:17696612 [本文引用: 1]

Abstract MUS81 is conserved among plants, animals, and fungi and is known to be involved in mitotic DNA damage repair and meiotic recombination. Here we present a functional characterization of the Arabidopsis thaliana homolog AtMUS81, which has a role in both mitotic and meiotic cells. The AtMUS81 transcript is produced in all tissues, but is elevated greater than 9-fold in the anthers and its levels are increased in response to gamma radiation and methyl methanesulfonate treatment. An Atmus81 transfer-DNA insertion mutant shows increased sensitivity to a wide range of DNA-damaging agents, confirming its role in mitotically proliferating cells. To examine its role in meiosis, we employed a pollen tetrad-based visual assay. Data from genetic intervals on Chromosomes 1 and 3 show that Atmus81 mutants have a moderate decrease in meiotic recombination. Importantly, measurements of recombination in a pair of adjacent intervals on Chromosome 5 demonstrate that the remaining crossovers in Atmus81 are interference sensitive, and that interference levels in the Atmus81 mutant are significantly greater than those in wild type. These data are consistent with the hypothesis that AtMUS81 is involved in a secondary subset of meiotic crossovers that are interference insensitive.
URLPMID:18182028 [本文引用: 2]

Meiotic crossovers/chiasmata, that are required to ensure chromosome disjunction, arise via the class I interference-dependent pathway or via the class II interference-free pathway. The proportions of these two classes vary considerably between different organisms. In Arabidopsis, about 85% of chiasmata are eliminated in Atmsh4 mutants, denoting that these are class I events. In budding and fission yeasts Msh4-independent crossovers arise largely or entirely via a Mus81-dependent pathway. To investigate the origins of the 15% residual (AtMSH4-independent) chiasmata in Arabidopsis we conducted a cytological and molecular analysis of AtMUS81 meiotic expression and function. Although AtMUS81 functions in somatic DNA repair and recombination, it is more highly expressed in reproductive tissues. The protein is abundantly present in early prophase I meiocytes, where it co-localizes, in a double-strand break-dependent manner, with the recombination protein AtRAD51. Despite this, an Atmus81 mutant shows normal growth and has no obvious defects in reproductive development that would indicate meiotic impairment. A cytological analysis confirmed that meiosis was apparently normal in this mutant and its mean chiasma frequency was similar to that of wild-type plants. However, an Atmsh4 / Atmus81 double mutant revealed a significantly reduced mean chiasma frequency (0.85 per cell), compared with an Atmsh4 single mutant (1.25 per cell), from which we conclude that AtMUS81 accounts for some, but not all, of the 15% AtMSH4-independent residual crossovers. It is possible that other genes are responsible for these residual chiasmata. Alternatively the AtMUS81 pathway coexists with an alternative parallel pathway that can perform the same functions.
[本文引用: 2]
URLPMID:3925842 [本文引用: 1]

Meiotic recombination plays a critical role in achieving accurate chromosome segregation and increasing genetic diversity. Many studies, mostly in yeast, have provided important insights into the coordination and interplay between the proteins involved in the homologous recombination pathway, especially the recombinase RAD51 and the meiosis-specific DMC1. Here we summarize the current progresses on the function of both recombinases and the CX3 complex encoded byAtRAD51paralogs, in the plant model speciesArabidopsis thaliana. Similarities and differences respect to the function of these proteins in other organisms are also indicated.
URLPMID:13678597 [本文引用: 1]

A central event in sexual reproduction is the reduction in chromosome number that occurs at the meiosis I division. Most eukaryotes rely on crossing over between homologs, and the resulting chiasmata, to direct meiosis I chromosome segregation, yet make very few crossovers per chromosome pair. This indicates that meiotic recombination must be tightly regulated to ensure that each chromosome pair enjoys the crossover necessary to ensure correct segregation. Here, we investigate control of meiotic crossing over in Caenorhabditis elegans, which averages only one crossover per chromosome pair per meiosis, by constructing genetic maps of end-to-end fusions of whole chromosomes. Fusion of chromosomes removes the requirement for a crossover in each component chromosome segment and thereby reveals a propensity to restrict the number of crossovers such that pairs of fusion chromosomes composed of two or even three whole chromosomes enjoy but a single crossover in the majority of meioses. This regulation can operate over physical distances encompassing half the genome. The meiotic behavior of heterozygous fusion chromosomes further suggests that continuous meiotic chromosome axes, or structures that depend on properly assembled axes, may be important for crossover regulation.
URLPMID:12504024 [本文引用: 1]

MEI-9 is the Drosophila homolog of the human structure-specific DNA endonuclease XPF. Like XPF, MEI-9 functions in nucleotide excision repair and interstrand crosslink repair. MEI-9 is also required to generate meiotic crossovers, in a function thought to be associated with resolution of Holliday junction intermediates. We report here the identification of MUS312, a protein that physically interacts with MEI-9. We show that mutations in mus312 elicit a meiotic phenotype identical to that of mei-9 mutants. A missense mutation in mei-9 that disrupts the MEI-9 US312 interaction abolishes the meiotic function of mei-9 but does not affect the DNA repair functions of mei-9. We propose that MUS312 facilitates resolution of meiotic Holliday junction intermediates by MEI-9.
[本文引用: 1]
URL [本文引用: 1]
URLPMID:19781752 [本文引用: 1]

Meiotic crossover (CO) recombination facilitates evolution and accurate chromosome segregation. CO distribution is tightly regulated: homolog pairs receive at least one CO, CO spacing is nonrandom, and COs occur preferentially in short genomic intervals called hotspots. We show that CO number and distribution are controlled on a chromosome-wide basis at the level of DNA double-strand break (DSB) formation by a condensin complex composed of subunits from two known condensins: the C. elegans dosage compensation complex and mitotic condensin II. Disruption of any subunit of the CO-controlling condensin dominantly changes DSB distribution, and thereby COs, and extends meiotic chromosome axes. These phenotypes are cosuppressed by disruption of a chromosome axis element. Our data implicate higher-order chromosome structure in the regulation of CO recombination, provide a model for the rapid evolution of CO hotspots, and show that reshuffling of interchangeable molecular parts can create independent machines with similar architectures but distinct biological functions.
URLPMID:23017241 [本文引用: 1]

Sexual eukaryotes reproduce via the meiotic cell division, where ploidy is halved and homologous chromosomes undergo reciprocal genetic exchange, termed crossover (CO). CO frequency has a profound effect on patterns of genetic variation and species evolution. Relative CO rates vary extensively both within and between plant genomes. Plant genome size varies by over 1000-fold, largely due to differential expansion of repetitive sequences, and increased genome size is associated with reduced CO frequency. Gene versus repeat sequences associate with distinct chromatin modifications, and evidence from plant genomes indicates that this epigenetic information influences CO patterns. This is consistent with data from diverse eukaryotes that demonstrate the importance of chromatin structure for control of meiotic recombination. In this review I will discuss CO frequency patterns in plant genomes and recent advances in understanding recombination distributions.
URLPMID:18206976 [本文引用: 1]

Fanconi anemia (FA) is a genetically heterogeneous cancer-prone disorder associated with chromosomal instability and cellular hypersensitivity to DNA crosslinking agents. The FA pathway is suspected to play a crucial role in the cellular response to DNA replication stress. At a molecular level, however, the function of most of the FA proteins is unknown. FANCM displays DNA-dependent ATPase activity and promotes the dissociation of DNA triplexes, but the physiological significance of this activity remains elusive. Here we show that purified FANCM binds to Holliday junctions and replication forks with high specificity and promotes migration of their junction point in an ATPase-dependent manner. Furthermore, we provide evidence that FANCM can dissociate large recombination intermediates, via branch migration of Holliday junctions through 2.6 kb of DNA. Our data suggest a direct role for FANCM in DNA processing, consistent with the current view that FA proteins coordinate DNA repair at stalled replication forks.
URLPMID:3399777 [本文引用: 2]

A homolog of a human Fanconi anemia complementation group protein is involved in controlling crossing over during meiosis.
URLPMID:19136626 [本文引用: 1]

Abstract Eukaryotes possess mechanisms to limit crossing over during homologous recombination, thus avoiding possible chromosomal rearrangements. We show here that budding yeast Mph1, an ortholog of human FancM helicase, utilizes its helicase activity to suppress spontaneous unequal sister chromatid exchanges and DNA double-strand break-induced chromosome crossovers. Since the efficiency and kinetics of break repair are unaffected, Mph1 appears to channel repair intermediates into a noncrossover pathway. Importantly, Mph1 works independently of two other helicases-Srs2 and Sgs1-that also attenuate crossing over. By chromatin immunoprecipitation, we find targeting of Mph1 to double-strand breaks in cells. Purified Mph1 binds D-loop structures and is particularly adept at unwinding these structures. Importantly, Mph1, but not a helicase-defective variant, dissociates Rad51-made D-loops. Overall, the results from our analyses suggest a new role of Mph1 in promoting the noncrossover repair of DNA double-strand breaks.
URLPMID:2847587 [本文引用: 1]

FANCM remodels branched DNA structures and plays essential roles in the cellular response to DNA replication stress. Here, we show that FANCM forms a conserved DNA-remodeling complex with a histone-fold heterodimer, MHF. We find that MHF stimulates DNA binding and replication fork remodeling by FANCM. In the cell, FANCM and MHF are rapidly recruited to forks stalled by DNA interstrand crosslinks, and both are required for cellular resistance to such lesions. In vertebrates, FANCM-MHF associates with the Fanconi anemia (FA) core complex, promotes FANCD2 monoubiquitination in response to DNA damage, and suppresses sister-chromatid exchanges. Yeast orthologs of these proteins function together to resist MMS-induced DNA damage and promote gene conversion at blocked replication forks. Thus, FANCM-MHF is an essential DNA-remodeling complex that protects replication forks from yeast to human.Graphical AbstractView high quality image (221K)
URLPMID:20347429

78 MHF1 and MHF2 are histone-fold-containing components of the FA core complex 78 MHF1 is essential for the stability and chromatin association of FANCM 78 MHF1 is required for the normal function of the FA pathway 78 MHF1-MHF2 form a complex that binds DNA and stimulates the branch migration activity of FANCM
URLPMID:22325783

78 S.02cerevisae MHF1 and MHF2 assume a typical histone fold 78 S.02cerevisae MHF1-MHF2 complex has a heterotetrameric architecture 78 Loop L3 and helices α2 and α3 of MHF1 contribute most to heterotetramer interfaces 78 The heterotetramer assembly is essential for the function of the complex
URLPMID:3646547 [本文引用: 1]

Fanconi anemia (FA) is a rare genetic disease characterized by chromosomal instability and cancer susceptibility. The Fanconi anemia complementation group protein M (FANCM) forms an evolutionarily conserved DNA-processing complex with MHF1/MHF2 (histone-fold-containing proteins), which is essential for DNA repair in response to genotoxic stress. Here we present the crystal structures of the MHF1-MHF2 complex alone and bound to a fragment of FANCM (FANCM661-800, designated FANCM-F). The structures show that MHF1 and MHF2 form a compact tetramer to which FANCM-F binds through a “dual-V” shaped structure. FANCM-F and (MHF1-MHF2)2cooperate to constitute a new DNA-binding site that is coupled to the canonical L1L2 region. Perturbation of the MHF-FANCM-F structural plasticity changes the localization of FANCM in vivo. The MHF-FANCM interaction and its subcellular localization are altered by a disease-associated mutant of FANCM. These findings reveal the molecular basis of MHF-FANCM recognition and provide mechanistic insights into the pathway leading to FA.
URLPMID:22500800 [本文引用: 1]

At the final step of homologous recombination, Holliday junction-containing joint molecules (JMs) are resolved to form crossover or noncrossover products. The enzymes responsible for JM resolution in02vivo remain uncertain, but three distinct endonucleases capable of resolving JMs in02vitro have been identified: Mus81-Mms4(EME1), Slx1-Slx4(BTBD12), and Yen1(GEN1). Using physical monitoring of recombination during budding yeast meiosis, we show that all three endonucleases are capable of promoting JM resolution in02vivo. However, in mms4 slx4 yen1 triple mutants, JM resolution and crossing over occur efficiently. Paradoxically, crossing over in this background is strongly dependent on the Blooms helicase ortholog Sgs1, a component of a well-characterized anticrossover activity. Sgs1-dependent crossing over, but not JM resolution per se, also requires XPG family nuclease Exo1 and the MutLγ complex Mlh1-Mlh3. Thus, Sgs1, Exo1, and MutLγ together define a previously undescribed meiotic JM resolution pathway that produces the majority of crossovers in budding yeast and, by inference, in mammals.
URLPMID:22500736 [本文引用: 1]

78 Sgs1 is required for normal noncrossover and crossover formation during meiosis 78 Noncrossovers and crossovers form via different pathways in wild-type 78 Noncrossovers and crossovers form via a common pathway in sgs1 mutants 78 Polo kinase Cdc5 triggers joint molecule resolution in both wild-type and sgs1
URLPMID:4338413 [本文引用: 1]

Homologous recombination is critical for genome separation during meiosis. Kaur et al. show that topoisomerase III, Rmi1, and the helicase Sgs1 act together to prevent formation of aberrant recombination intermediates and to ensure that recombination intermediates are completely resolved during meiosis.
URLPMID:25699709 [本文引用: 2]

Topoisomerases are essential for chromosome metabolism. Tang et al. show that the Top3-Rmi1 complex must decatenate DNA strands to process intermediates during homologous recombination. Top3-Rmi1 partners with BLM/Sgs1 to prevent abnormal recombination. Top3-Rmi1 is also essential for disentangling chromosomes, enabling their complete separation.
URLPMID:25699708 [本文引用: 1]

Mutations in the topoisomerase Top3 lead to an extreme hyper-recombination phenotype, but a mechanistic explanation remained elusive. Fasching et02al. provide in02vitro biochemical evidence that Top3 has anti-recombination activity that it uses to dissolve D loops that form during recombination.
URLPMID:16371241 [本文引用: 1]

RecQ helicases are conserved throughout all kingdoms of life regarding their overall structure and function. They are 3 5 DNA helicases resolving different recombinogenic DNA structures. The RecQ helicases are key factors in a number of DNA repair and recombination pathways involved in the maintenance of genome integrity. In eukaryotes the number of RecQ genes and the structure of RecQ proteins vary strongly between organisms. Therefore, they have been named RecQ-like genes. Knockouts of several RecQ-like genes cause severe diseases in animals or harmful cellular phenotypes in yeast. Until now the largest number of RecQ-like genes per organism has been found in plants. Arabidopsis and rice possess seven different RecQ-like genes each. In the almost completely sequenced genome of the moss Physcomitrella patens at least five RecQ-like genes are present. One of the major present and future research aims is to define putative plant-specific functions and to assign their roles in DNA repair and recombination pathways in relation to RecQ genes from other eukaryotes. Regarding their intron positions, the structures of six RecQ-like genes of dicots and monocots are virtually identical indicating a conservation over a time scale of 150 million years. In contrast to other eukaryotes one gene (RecQsim) exists exclusively in plants. It possesses an interrupted helicase domain but nevertheless seems to have maintained the RecQ function. Owing to a recent gene duplication besides the AtRecQl4A gene an additional RecQ-like gene (AtRecQl4B) exists in the Brassicaceae only. Genetic studies indicate that a AtRecQl4A knockout results in sensitivity to mutagens as well as an hyper-recombination phenotype. Since AtRecQl4B was still present, both genes must have non-redundant roles. Analysis of plant RecQ-like genes will not only increase the knowledge on DNA repair and recombination, but also on the evolution and radiation of protein families.
URLPMID:18000056 [本文引用: 2]

RecQ helicases are involved in the processing of DNA structures arising during replication, recombination, and repair throughout all kingdoms of life. Mutations of different RecQ homologues are responsible for severe human diseases, such as Blooms (BLM) or Werner (WRN) syndrome. The loss of RecQ function is often accompanied by hyperrecombination caused by a lack of crossover suppression. In the Arabidopsis genome seven different RecQ genes are present. Two of them (AtRECQ4A and 4B) arose because of a recent duplication and are still nearly 70% identical on a protein level. Knockout of these genes leads to antagonistic phenotypes: the RECQ4A mutant shows sensitivity to DNA-damaging agents, enhanced homologous recombination (HR) and lethality in a mus81 background. Moreover, mutation of RECQ4A partially suppresses the lethal phenotype of an AtTOP3 mutant, a phenomenon that had previously been demonstrated for RecQ homologues of unicellular eukaryotes only. Together, these facts strongly suggest that in plants RECQ4A is functionally equivalent to SGS1 of Saccharomyces cerevisiae and the mammalian BLM protein. In stark contrast, mutants of the closely related RECQ4B are not mutagen-sensitive, not viable in a mus81 background, and unable to suppress the induced lethality caused by loss of TOP3 . Moreover, they are strongly impaired in HR. Thus, AtRECQ4B is specifically required to promote but not to suppress crossovers, a role in which it differs from all eukaryotic RecQ homologues known.
[本文引用: 2]
URLPMID:27011106 [本文引用: 2]

During the first meiotic division, crossovers (COs) between homologous chromosomes ensure their correct segregation. COs are produced by homologous recombination (HR)-mediated repair of programmed DNA double strand breaks (DSBs). As more DSBs are induced than COs, mechanisms are required to establish a regulated number of COs and to repair remaining intermediates as non-crossovers (NCOs). We show that theCaenorhabditis elegansRMI1 homolog-1 (RMH-1) functions during meiosis to promote both CO and NCO HR at appropriate chromosomal sites. RMH-1 accumulates at CO sites, dependent on known pro-CO factors, and acts to promote CO designation and enforce the CO outcome of HR-intermediate resolution. RMH-1 also localizes at NCO sites and functions in parallel with SMC-5 to antagonize excess HR-based connections between chromosomes. Moreover, RMH-1 also has a major role in channeling DSBs into an NCO HR outcome near the centers of chromosomes, thereby ensuring that COs form predominantly at off-center positions. A nematode homolog of the conserved DNA repair factor RMI1 plays multiple genetically separable roles that together ensure the faithful inheritance of intact genomes during sexual reproduction. During meiosis, faithful separation of chromosomes into gametes is essential for fertility and healthy progeny. During the first meiotic division, crossovers (CO) between parental homologs ensure their correct segregation. Programmed DNA double strand breaks (DSBs) and resection steps generate single-stranded overhangs that invade a sister chromatid of the homolog to initiate homologous recombination. This culminates in the generation of a DNA double Holliday junction (dHJ). This can be acted upon by resolvases to produce CO and non-crossover (NCO) products, depending on where the resolvases cut the DNA. Alternatively, NCOs can also be produced by decatenation via the RecQ helicase opoisomeraseIII mi1 (RTR) complex. The mammalian RTR contains a topoisomerase, Bloom helicase, and RMI1/2 scaffolding components. It disassembles dHJs in vitro and contributes the major NCO activity in mitosis. Here, we provide evidence that theCaenorhabditis elegansRMH-1 functions in distinct complexes during meiosis to produce both COs and NCOs in an in vivo animal model system. Strikingly, RMH-1 spatially regulates the distribution of COs on chromosomes, demonstrating that the RTR complex can act locally within specific chromosome domains.
URLPMID:28696221 [本文引用: 1]

Abstract Homologous recombination plays a central role in guaranteeing chromosome segregation during meiosis. The precise regulation of the resolution of recombination intermediates is critical for the success of meiosis. Many proteins, including the RECQ DNA helicases (Sgs1/BLM) and Topoisomerase 302± (TOP302±), have essential functions in managing recombination intermediates. However, many other factors involved in this process remain to be defined. Here, we report the isolation of meiotic chromosome association 1 (MEICA1), a novel protein participating in meiotic recombination in rice ( Oryza sativa ). Loss of MEICA1 leads to nonhomologous chromosome association, the formation of massive chromosome bridges, and fragmentation. MEICA1 interacts with MSH7, suggesting its role in preventing nonallelic recombination. In addition, MEICA1 has an anticrossover activity revealed by suppressing the defects of crossover formation in msh5 meica1 compared with that in msh5 , showing the similar function with its interacted protein TOP302±. Thus, our data establish two pivotal roles for MEICA1 in meiosis: preventing aberrant meiotic recombination and regulating crossover formation. 0008 2017 American Society of Plant Biologists. All rights reserved.
URLPMID:25815584 [本文引用: 3]

10.7554/eLife.03708.001During meiosis homologous chromosomes undergo crossover recombination. Sequence differences between homologs can locally inhibit crossovers. Despite this, nucleotide diversity and population-scaled recombination are positively correlated in eukaryote genomes. To investigate interactions between heterozygosity and recombination we crossed Arabidopsis lines carrying fluorescent crossover reporters to 32 diverse accessions and observed hybrids with significantly higher and lower crossovers than homozygotes. Using recombinant populations derived from these crosses we observed that heterozygous regions increase crossovers when juxtaposed with homozygous regions, which reciprocally decrease. Total crossovers measured by chiasmata were unchanged when heterozygosity was varied, consistent with homeostatic control. We tested the effects of heterozygosity in mutants where the balance of interfering and non-interfering crossover repair is altered. Crossover remodeling at homozygosity-heterozygosity junctions requires interference, and non-interfering repair is inefficient in heterozygous regions. As a consequence, heterozygous regions show stronger crossover interference. Our findings reveal how varying homolog polymorphism patterns can shape meiotic recombination.DOI: http://dx.doi.org/10.7554/eLife.03708.001
URLPMID:26494791 [本文引用: 1]

During meiosis, homologous chromosomes undergo crossover recombination, which is typically concentrated in narrow hot spots that are controlled by genetic and epigenetic information. Arabidopsis chromosomes are highly DNA methylated in the repetitive centromeres, which are also crossover-suppressed. Here we demonstrate that RNA-directed DNA methylation is sufficient to locally silence Arabidopsis euchromatic crossover hot spots and is associated with increased nucleosome density and H3K9me2. However, loss of CG DNA methylation maintenance in met1 triggers epigenetic crossover remodeling at the chromosome scale, with pericentromeric decreases and euchromatic increases in recombination. We used recombination mutants that alter interfering and noninterfering crossover repair pathways (fancm and zip4) to demonstrate that remodeling primarily involves redistribution of interfering crossovers. Using whole-genome bisulfite sequencing, we show that crossover remodeling is driven by loss of CG methylation within the centromeric regions. Using cytogenetics, we profiled meiotic DNA double-strand break (DSB) foci in met1 and found them unchanged relative to wild type. We propose that met1 chromosome structure is altered, causing centromere-proximal DSBs to be inhibited from maturation into interfering crossovers. These data demonstrate that DNA methylation is sufficient to silence crossover hot spots and plays a key role in establishing domains of meiotic recombination along chromosomes.
URLPMID:3920622 [本文引用: 1]

Crossover recombination events between homologous chromosomes are required to form chiasmata, temporary connections between homologues that ensure their proper segregation at meiosis I-1. Despite this requirement for crossovers and an excess of the double-strand DNA breaks that are the initiating events for meiotic recombination, most organisms make very few crossovers per chromosome pair(2). Moreover, crossovers tend to inhibit the formation of other crossovers nearby on the same chromosome pair, a poorly understood phenomenon known as crossover interference(3,4). Here we show that the synaptonemal complex, a meiosis-specific structure that assembles between aligned homologous chromosomes, both constrains and is altered by crossover recombination events. Using a cytological marker of crossover sites in Caenorhabditis elegans(5), we show that partial depletion of the synaptonemal complex central region proteins attenuates crossover interference, increasing crossovers and reducing the effective distance over which interference operates, indicating that synaptonemal complex proteins limit crossovers. Moreover, we show that crossovers are associated with a local 0.4-0.5-micrometre increase in chromosome axis length. We propose that meiotic crossover regulation operates as a self-limiting system in which meiotic chromosome structures establish an environment that promotes crossover formation, which in turn alters chromosome structure to inhibit other crossovers at additional sites.
URLPMID:25936677 [本文引用: 1]
URLPMID:28223312 [本文引用: 2]

During meiosis, homologous chromosomes undergo crossover recombination, which creates genetic diversity and balances homolog segregation. Despite these critical functions, crossover frequency varies extensively within and between species. Although natural crossover recombination modifier loci have been detected in plants, causal genes have remained elusive. Using natural Arabidopsis thaliana accessions, we identified two major recombination quantitative trait loci (rQTLs) that explain 56.9% of crossover variation in ColxLer F2 populations. We mapped rQTL1 to semidominant polymorphisms in HEI10, which encodes a conserved ubiquitin E3 ligase that regulates crossovers. Null hei10 mutants are haploinsufficient, and, using genome-wide mapping and immunocytology, we show that transformation of additional HEI10 copies is sufficient to more than double euchromatic crossovers. However, heterochromatic centromeres remained recombination-suppressed. The strongest HEI10-mediated crossover increases occur in subtelomeric euchromatin, which is reminiscent of sex differences in Arabidopsis recombination. Our work reveals that HEI10 naturally limits Arabidopsis crossovers and has the potential to influence the response to selection.
URLPMID:29463699 [本文引用: 1]

During meiosis homologous chromosomes undergo reciprocal crossovers, which generates genetic diversity and underpins classical crop improvement. Meiotic recombination initiates from DNA double strand breaks, which are processed into single-stranded DNA that can invade a homologous chromosome. The resulting joint molecules can ultimately be resolved as crossovers. In Arabidopsis competing pathways balance the repair of ~100-200 meiotic DSBs into ~10 crossovers per meiosis, with the excess DSBs repaired as non-crossovers. In order to bias DSB repair towards crossovers, we simultaneously increased dosage of the pro-crossover E3 ligase gene HEI10 and introduced mutations in the anti-crossover helicase genes RECQ4A and RECQ4B. As HEI10 and recq4a recq4b increase interfering and non-interfering crossover pathways respectively, they combine additively to yield a massive meiotic recombination increase. Interestingly, we also show that increased HEI10 dosage increases crossover coincidence, which indicates an effect of HEI10 on interference. We also show that patterns of interhomolog polymorphism and heterochromatin drive recombination increases towards the sub-telomeres in both HEI10 and recq4a recq4b backgrounds, while the centromeres remain crossover-suppressed. These results provide a genetic framework for engineering meiotic recombination landscapes in plant genomes.
URLPMID:17360452 [本文引用: 1]

Recombination, in the form of cross-overs (COs) and gene conversion (GC), is a highly conserved feature of meiosis from fungi to mammals. Recombination helps ensure chromosome segregation and promotes allelic diversity. Lesions in the recombination machinery are often catastrophic for meiosis, resulting in sterility. We have developed a visual assay capable of detecting Cos and GCs and measuring CO interference in Arabidopsis thaliana. This flexible assay utilizes transgene constructs encoding pollen-expressed fluorescent proteins of three different colors in the qrt1 mutant background. By observing the segregation of the fluorescent alleles in 92,489 pollen tetrads, we demonstrate (i) a correlation between developmental position and CO frequency, (ii) a temperature dependence for CO frequency, (iii) the ability to detect meiotic GC events, and (iv) the ability to rapidly assess CO interference.
URLPMID:23104831 [本文引用: 1]

Meiosis involves reciprocal exchange of genetic information between homologous chromosomes to generate new allelic combinations. In cereals, the distribution of genetic crossovers, cytologically visible as chiasmata, is skewed toward the distal regions of the chromosomes. However, many genes are known to lie within interstitial/proximal regions of low recombination, creating a limitation for breeders. We investigated the factors underlying the pattern of chiasma formation in barley (Hordeum vulgare) and show that chiasma distribution reflects polarization in the spatiotemporal initiation of recombination, chromosome pairing, and synapsis. Consequently, meiotic progression in distal chromosomal regions occurs in coordination with the chromatin cycles that are a conserved feature of the meiotic program. Recombination initiation in interstitial and proximal regions occurs later than distal events, is not coordinated with the cycles, and rarely progresses to form chiasmata. Early recombination initiation is spatially associated with early replicating, euchromatic DNA, which is predominately found in distal regions. We demonstrate that a modest temperature shift is sufficient to alter meiotic progression in relation to the chromosome cycles. The polarization of the meiotic processes is reduced and is accompanied by a shift in chiasma distribution with an increase in interstitial and proximal chiasmata, suggesting a potential route to modify recombination in cereals.
URL [本文引用: 1]

Summary Barley ( Hordeum vulgare ) is a crop of global significance. However, a third of the genes of barley are largely inaccessible to conventional breeding programmes as crossovers are localised to the ends of the chromosomes. This work examines whether crossovers can be shifted to more proximal regions simply by elevating growth temperature. We utilised a genome-wide marker set for linkage analysis combined with cytological mapping of crossover events to examine the recombination landscape of plants grown at different temperatures. We found that barley shows heterochiasmy, that is, differences between female and male recombination frequencies. In addition, we found that elevated temperature significantly changes patterns of recombination in male meiosis only, with a repositioning of Class I crossovers determined by cytological mapping of HvMLH3 foci. We show that the length of synaptonemal complexes in male meiocytes increases in response to temperature. The results demonstrate that the distribution of crossover events are malleable and can be shifted to proximal regions by altering the growth temperature. The shift in recombination is the result of altering the distribution of Class I crossovers, but the higher recombination at elevated temperatures is potentially not the result of an increase in Class I events.
URLPMID:25664766 [本文引用: 1]

Summary Numerous studies have argued that environmental variations may contribute to evolution through the generation of novel heritable variations via meiotic recombination, which plays a crucial role in crop domestication and improvement. Rice is one of the most important staple crops, but no direct estimate of recombination events has yet been made at a fine scale. Here, we address this limitation by sequencing 41 rice individuals with high sequencing coverage and c . 90002000 accurate markers. An average of 33.9 crossover ( c . 4.5302cM02Mb611) and 2.47 non-crossover events were detected per F2 plant, which is similar to the values in Arabidopsis . Although not all samples in the stress treatment group showed an increased number of crossover events, environmental stress increased the recombination rate in c . 28.5% of samples. Interestingly, the crossovers showed a highly uneven distribution among and along chromosomes, with c . 13.9% of the entire genome devoid of crossovers, including 11 of the 12 centromere regions, and c . 0.72% of the genome containing large numbers of crossovers (>025002cM02Mb611). The gene ontology (GO) categories showed that genes clustered within the recombination hot spot regions primarily tended to be involved in responses to environmental stimuli, suggesting that recombination plays an important role for adaptive evolution in rapidly changing environments.
URLPMID:29183972 [本文引用: 1]

Abstract Meiotic crossovers shuffle parental genetic information, providing novel combinations of alleles on which natural or artificial selection can act. However, crossover events are relatively rare, typically one to three exchange points per chromosome pair. Recent work has identified three pathways limiting meiotic crossovers in Arabidopsis thaliana that rely on the activity of FANCM [Crismani W, et al. (2012) Science 336:1588-1590], RECQ4 [S gu la-Arnaud M, et al. (2015) Proc Natl Acad Sci USA 112:4713-4718], and FIGL1 [Girard C, et al. (2015) PLoS Genet 11:e1005369]. Here we analyzed recombination in plants in which one, two, or all three of these pathways were disrupted in both pure line and hybrid contexts. The greatest effect was observed when combining recq4 and figl1 mutations, which increased the hybrid genetic map length from 389 to 3,037 cM. This corresponds to an unprecedented 7.8-fold increase in crossover frequency. Disrupting the three pathways did not further increase recombination, suggesting that some upper limit had been reached. The increase in crossovers is not uniform along chromosomes and rises from centromere to telomere. Finally, although in wild type recombination is much higher in male meiosis than in female meiosis (490 cM vs. 290 cM), female recombination is higher than male recombination in recq4 figl1 (3,200 cM vs. 2,720 cM), suggesting that the factors that make wild-type female meiosis less recombinogenic than male wild-type meiosis do not apply in the mutant context. The massive increase in recombination observed in recq4 figl1 hybrids opens the possibility of manipulating recombination to enhance plant breeding efficiency.
URLPMID:29628933 [本文引用: 1]

Meiotic crossovers (COs) are essential for proper chromosome segregation and the reshuffling of alleles during meiosis. In WT plants, the number of COs is usually small, which limits the genetic variation that can be captured by plant breeding programs. Part of this limitation is imposed by proteins like FANCM, the inactivation of which results in a 3-fold increase in COs inArabidopsis thaliana. Whether the same holds true in crops needed to be established. In this study, we identified EMS induced mutations in FANCM in two species of economic relevance within the genusBrassica. We showed that CO frequencies were increased infancmmutants in both diploid and tetraploidBrassicas, Brassica rapaandBrassica napusrespectively. InB. rapa, we observed a 3-fold increase in the number of COs, equal to the increase observed previously inArabidopsis. InB. napuswe observed a lesser but consistent increase (1.3-fold) in both euploid (AACC) and allohaploid (AC) plants. Complementation tests inA. thalianasuggest that the smaller increase in crossover frequency observed inB. napusreflects residual activity of the mutant C copy of FANCM. Altogether our results indicate that the anti-CO activity of FANCM is conserved across theBrassica, opening new avenues to make a wider range of genetic diversity accessible to crop improvement.
URLPMID:18193020 [本文引用: 1]

Abstract In most organisms, one crossover (CO) event inhibits the chances of another nearby event. The term used to describe this phenomenon is 'CO interference'. Here, we describe a protocol for quickly generating large data sets that are amenable to CO interference analysis in the flowering plant, Arabidopsis thaliana. We employ a visual assay that utilizes transgenic marker constructs encoding pollen-expressed fluorescent proteins of three colors in the quartet mutant background. In this genetic background, male meiotic products--the pollen grains--remain physically attached thereby facilitating tetrad analysis. We have developed a library of mapped marker insertions that, when crossed together, create adjacent intervals that can be rapidly and simultaneously screened for COs. This assay system is capable of detecting and differentiating single COs as well as two-, three- and four-strand double COs. We also describe how to analyze the data that are produced by this method. To generate and score a double interval in a wild-type and mutant background using this protocol will take 22-27 weeks.
URLPMID:24113785 [本文引用: 1]

During meiosis, reciprocal exchange between homologous chromosomes occurs as a result of crossovers (COs). CO frequency varies within genomes and is subject to genetic, epigenetic and environmental control. As robust measurement of COs is limited by their low numbers, typically 1-2 per chromosome, we adapted flow cytometry for use with Arabidopsis transgenic fluorescent protein-tagged lines (FTLs) that express eCFP, dsRed or eYFP fluorescent proteins in pollen. Segregation of genetically linked transgenes encoding fluorescent proteins of distinct colors can be used to detect COs. The fluorescence of up to 80,000 pollen grains per individual plant can be measured in 10-15 min using this protocol. A key element of CO control is interference, which inhibits closely spaced COs. We describe a three-color assay for the measurement of CO frequency in adjacent intervals and calculation of CO interference. We show that this protocol can be used to detect changes in CO frequency and interference in the fancm zip4 double mutant. By enabling high-throughput measurement of CO frequency and interference, these methods will facilitate genetic dissection of meiotic recombination control.
[本文引用: 1]
