 ,1,2,3,4
,1,2,3,4Synergistic regulation of the erythroid differentiation of K562 cells by KLF1 and KLF9
Lan Ren1,2, Rudan Xiao1,2, Qian Zhang1,2, Xiaomin Lou1,3, Zhaojun Zhang1,3,4, Xiangdong Fang ,1,2,3,4
,1,2,3,4通讯作者:
编委: 杨昭庆
收稿日期:2018-06-29修回日期:2018-09-30网络出版日期:2018-11-20
| 基金资助: |
Received:2018-06-29Revised:2018-09-30Online:2018-11-20
| Fund supported: |
作者简介 About authors
任岚,硕士研究生,专业方向:疾病组学与转化医学研究E-mail:eico.renlan@gmail.com。
肖茹丹,硕士研究生,专业方向:疾病组学与转化医学研究E-mail:xiaorudan@big.ac.cn,任岚和肖茹丹并列第一作者。

摘要
关键词:
Abstract
Keywords:
PDF (763KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
任岚, 肖茹丹, 张倩, 娄晓敏, 张昭军, 方向东. KLF1和KLF9对K562细胞红系分化的协同调控作用[J]. 遗传, 2018, 40(11): 998-1006 doi:10.16288/j.yczz.18-174
Lan Ren, Rudan Xiao, Qian Zhang, Xiaomin Lou, Zhaojun Zhang, Xiangdong Fang.
Krüppel样因子(Krüppel-like factors, KLFs)是锌指蛋白超家族的一个亚家族[1],参与细胞内的多种生理、病理过程[2, 3],与红系分化过程密切相关[4]。KLF1通过结合β-珠蛋白启动子和位点控制区(locus control region, LCR),促进成年期β-珠蛋白的表达和胎儿期γ-珠蛋白到β-珠蛋白基因的转换[5, 6]。KLF2、KLF11、KLF13等主要是促进胚胎期ε-珠蛋白与胎儿期γ-珠蛋白基因的表达,同时这些珠蛋白的表达还受KLF3、KLF8等因子的抑制[7,8,9,10]。KLF17 参与调控早期红系分化过程[11]。在小鼠中敲除KLF1、KLF2、KLF3、KLF6和 KLF13等基因都可观察到造血缺陷或贫血等异常表型[7, 12-14]。此外,还发现多个KLF因子对红系分化具有协同作用,但具体作用机制目前仍不清楚。如KLF1和KLF2通过控制以Myc为中心的作用网络调控胚胎早期珠蛋白基因表达和红细胞生成过程[15]。在已分化的红系细胞中,KLF3的表达依赖于KLF1,在KLF1-/-小鼠胎肝红细胞中KLF3表达水平明显降低[16, 17]。在KLF1-/-小鼠胎肝中,KLF9的表达水平显著降低[17],但是KLF9调控红系分化的方式及功能目前尚未见报道。
本课题组前期进行了人脐带血造血干细胞(hematopoietic stem cells, HSC)向红系分化过程中特定阶段细胞(HSC、P2、P3、P4、P5、RBC)的转录组研究[18],通过分析KLF家族成员在上述时期转录组数据中的表达丰度,发现KLF1、KLF9、KLF15这3个因子在由HSC分化的红系细胞中的表达水平显著高于未分化的HSC,提示这3个因子可能发挥了协同调控细胞红系分化的作用。由于KLF15在这些细胞中的表达水平极低,并且该基因不仅仅在红系细胞中发挥作用,其在人胚胎干细胞中的表达水平也高于已分化的红系细胞[19],因此并不支持其作为促进红系分化的调控因子。KLF9由于在其非编码基因组区域存在红系特异增强子元件,并且已被证明是潜在的红系分化的调控因子[19]。因此,本研究主要关注KLF1和KLF9对红系分化的协同调控作用。
本研究利用K562细胞模型,首先确认了KLF1和KLF9基因表达的正相关性,发现二者共表达显著促进红系分化和β-珠蛋白的表达;通过对KLF1和KLF9分别和共同过表达、敲低表达的K562细胞的转录组数据进行分析,探讨了二者对红系分化协同调控的可能机制。
1 材料和方法
1.1 材料
大肠杆菌DH5α菌株、质粒载体pCDH-CD713B、pRNAT-U6.1/Neo由本实验室保存;K562细胞来源于ATCC细胞库;Trizol试剂盒、反转录试剂盒、实时定量PCR试剂盒、限制性内切酶等试剂购自美国Thermo Scientific公司。1.2 过表达细胞株的构建
根据NCBI上提供的KLF1、KLF9共识编码序列(consensus coding sequence, CCDS),设计二者全长表达的引物(KLF1引物两端带有XhoⅠ和NotⅠ酶切位点、KLF9引物两端带有XbaⅠ和EcoRⅠ酶切位点)。提取K562细胞的cDNA作为模板,PCR扩增分别得到全长的KLF1、KLF9片段,并与pCDH- CD713B载体分别经过XhoⅠ和NotⅠ或XbaⅠ和EcoRⅠ双酶切后,连接得到过表达质粒。随后,利用电转染方法将过表达质粒分别转染K562细胞24~48 h后,利用G418 (800 μg/mL)分别筛选出KLF1和KLF9稳定过表达的细胞,将对照组、KLF1过表达细胞株、KLF9过表达细胞株和KLF1、KLF9共同过表达细胞株分别命名为Ctrl-OE、KLF1-OE、KLF9-OE、KLF(1+9)-OE。1.3 敲低表达细胞株的构建
设计并合成KLF1的siRNA (吉玛基因,苏州),利用Lipofectamine LTX试剂盒转染K562细胞。根据RNAi Designer网站设计KLF9的shRNA,并将shRNA连接到pRNAT-U6.1/Neo载体后,采用电转的方法将重组质粒转染K562细胞。转染后的K562细胞,通过G418 (800 μg/mL)的筛选获得稳定敲低表达的细胞株,将对照组、KLF1敲低表达细胞株、KLF9敲低表达细胞株和KLF1、KLF9共同敲低表达细胞株分别命名为Ctrl-KD、KLF1-KD、KLF9-KD、KLF(1+9)-KD。1.4 实时荧光定量PCR (qRT-PCR)反应
利用Trizol法提取K562细胞总RNA,,采用TaqMan反转录试剂盒对1 mg总RNA进行反转录。引物序列见表1。利用CFX96TM Real-Time系统检测上述基因mRNA表达丰度。扩增条件:95℃ 5 min;95℃ 30 s,60℃ 15 s,72℃ 10 s,78℃ 40 s,40个循环;78℃ 10 min。溶解曲线的程序为95℃ 15 s;60℃ 15 s;95℃ 15 s。上述基因均被检测3次,其表达水平以GAPDH的丰度作为内参,用CFXManager软件分析基因表达结果,并采用T-test方法分析差异显著性。Table 1
表1
表1 本研究所使用的引物序列信息
Table 1
| 基因 | 引物序列(5′→3′) |
|---|---|
| KLF1 | F:TTGCGGCAAGAGCTACACC |
| R:GTCAGAGCGCGAAAAAGCAC | |
| KLF9 | F:ACAGTGGCTGTGGGAAAGTC |
| R: AACTGCTTTTCCCCAGTGTG | |
| HBB | F:TTGAGTCCTTTGGGGATCTG |
| R:AGCTCACTCAGTGTGGCAAA | |
| HBG | F:CCCAGAGGTTCTTTGACAGG |
| R:TTCTCAGGATCCACATGCAG | |
| HBE | F:TTTGGAAACCTGTCGTCTCC |
| R:GGCTTGAGGTTGTCCTCTCC | |
| GAPDH | F:TGTTGCCATCAATGACCCCTT |
| R:CTCCACGACGTACTCAGCG |
新窗口打开|下载CSV
1.5 K562细胞红系分化诱导方法
取1×106个K562细胞置于6 cm培养皿中,添加5 mL RPMI 1640完全培养基和25 μL 10 mmol/L 的Hemin试剂后,摇动培养皿使之混匀;置于37℃、5% CO2的培养箱中培养,加Hemin诱导72 h后进行后续实验。1.6 流式细胞术(FACS)检测红细胞分化方法
将Hemin试剂诱导的K562细胞收集到新的15 mL离心管中,300 g离心5 min,弃上清;用1×PBS (含2% FBS,2 mmol/L EDTA)重悬细胞,300 g离心5 min,弃上清后再用PBS洗一次,用500 μL 1× PBS重悬,将细胞转移到流式管中;加入抗体CD235a- PE和CD71-APC染色,每管各0.5 μL,混匀后放4℃黑暗静置10 min;300 g离心5 min,弃上清后用1×PBS洗一次,再重悬于500 μL 1×PBS中;用流式细胞仪分析CD235a和CD71双阳细胞的比例。1.7 转录组建库与测序方法
Ctrl-OE、KLF1-OE、KLF9-OE、KLF(1+9)-OE、Ctrl-KD、KLF1-KD、KLF9-KD和KLF(1+9)-KD细胞分别经Hemin诱导72 h后,分别收集细胞提取细胞的总RNA,由北京基因组研究所所级中心测序平台进行转录组建库,通过HiSeq2000测序平台进行RNA-seq转录组测序。1.8 转录组数据分析方法
1.8.1 RNA-Seq组学数据在参考基因组上的比对RNA-Seq组学测序得到的序列首先用FastQC评估测序质量。使用HISAT2对原始数据进行基因组比对。参考基因组用GRCh37。
1.8.2 转录本组装和定量分析
用samtools工具处理HISAT2输出的结果sam文件,转换成可供基因组定量分析的bam文件。使用StringTie组装转录本、注释并估测转录本的丰度等。序列数目根据基因长度和总的比对上的序列数进行标准化,以FPKM作为统一的度量标准[20]。FPKM置信区间估算是通过Bayesian推理方法来实现的[21]。
1.8.3 差异表达基因的鉴定
用Edge R软件包进行基因差异表达分析。筛选标准为fold-change≥2.0,且P≤0.05。
1.8.4 基因的功能注释和网络分析
用DAVID平台分析差异表达基因的Gene Ontology和差异显著性通路。分子相互作用网络通过STRING公共知识数据库分析获得。
2 结果与分析
2.1 KLF1和KLF9基因表达的相关性
如果KLF1与KLF9协同调控红系分化,那么在红细胞中这两个基因的表达可能会存在相关性。本研究在KLF1、KLF9分别过表达和敲低表达的K562细胞中,利用qRT-PCR技术检测这两个基因的表达情况。结果发现,过表达一个KLF基因后,另一个基因的表达水平会显著升高;反之亦然(图1)。因此,KLF1与KLF9基因表达呈正相关。图1
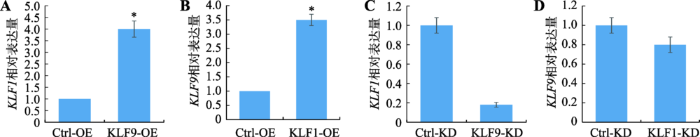 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1K562细胞中KLF1和KLF9基因表达的相关性分析
A:K562细胞中KLF9过表达后KLF1的表达量;B:K562细胞中KLF1过表达后KLF9的表达量;C:K562细胞中KLF9敲低表达后KLF1的表达量;D:K562细胞中KLF1过表达后KLF9的表达量。Ctrl-OE:对照;KLF1-OE:KLF1过表达细胞株;KLF9-OE:KLF9过表达细胞株;KLF1-KD:KLF1敲低表达细胞株;KLF9-KD:KLF9敲低表达细胞株。*表示 P<0.05,差异显著。
Fig. 1Correlation analysis of KLF1 and KLF9 gene expression in K562 cells
2.2 KLF1和KLF9协同促进K562细胞红系分化
为研究KLF1和KLF9是否协同调控红系分化,本研究分别利用Hemin对KLF1、KLF9及共同过表达、敲低表达的K562稳定细胞株进行体外红系诱导分化,利用FACS技术分析红细胞表面红系特异标志物CD235a和CD71的表达变化。通过对3次实验结果的统计分析,发现KLF1、KLF9分别高表达均可以显著增加CD71+、CD 235a+双阳性K562细胞的比例,反之亦然(图2)。KLF1和KLF9共同高表达时CD235a+和CD71+双阳性细胞的比例更高(图2A),表明KLF1和KLF9对红系分化有协同促进作用。KLF1和KLF9共同敲低表达虽也能部分抑制红系分化,但抑制作用并不如单一基因敲低表达的作用明显(图2B),可能是由于二者共同敲低表达使得细胞启动了其他基因参与红系分化。图2
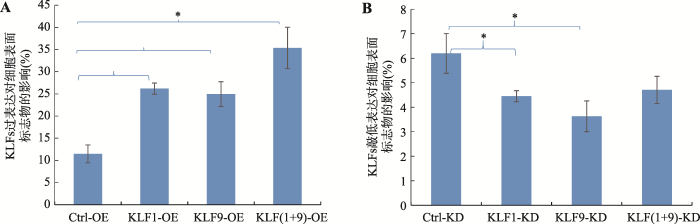 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2 KLF1和KLF9对Hemin诱导K562细胞红系分化的影响
A:FACS分析KLF1、KLF9分别和共同过表达对K562细胞红系分化的影响;B:FACS分析KLF1、KLF9分别和共同敲低表达对K562细胞红系分化的影响。KLF(1+9)-OE:KLF1、KLF9共同过表达细胞株;KLF(1+9)-KD:KLF1、KLF9共同敲低表达细胞株。*表示P<0.05,差异显著;**表示P<0.01,差异极显著。
Fig. 2The effects of KLF1 and KLF9 on the erythroid differentiation of K562 cells induced by Hemin
2.3 KLF1和KLF9协同促进HBB的表达
珠蛋白基因的表达变化也是评估红系分化的重要标志之一,不同类型的珠蛋白基因(HBE、HBG和HBB)在红系分化的不同阶段呈现特征性表达。在K562细胞的红系诱导分化模型中,本研究分别检测了KLF1、KLF9及其共同过表达或敲低表达对β珠蛋白基因家族中HBE、HBG、HBB基因表达的影响。结果表明,KLF1和KLF9两个基因单独和共同过表达均可促进HBB表达(P<0.05),且共同过表达促进效果更显著(P<0.01),进一步表明二者具有协同促进红系分化的作用(图3A)。除单独过表达 KLF1可显著抑制HBG的表达外[22],其他过表达方式对HBE和HBG表达的影响不显著,且敲低表达实验并不能显著下调HBB、HBE和HBG的表达(图3B)。图3
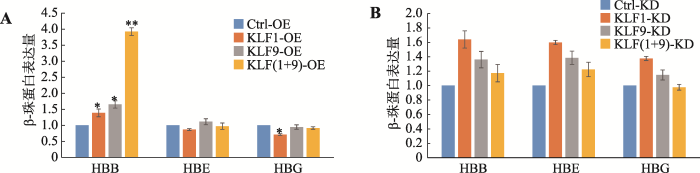 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3KLF1和KLF9对不同类型珠蛋白基因表达的影响
A:不同类型珠蛋白基因在KLF1、KLF9分别和共同过表达细胞中的表达丰度;B:不同类型珠蛋白基因在KLF1、KLF9分别和共同敲低表达细胞中的表达丰度。*表示P<0.05,差异显著;**表示 P<0.01,差异极显著。
Fig. 3The effects of KLF1 and KLF9 on the gene expression of different β-globins
2.4 KLF1和KLF9协同调控红系分化的分子机制分析
为深入探究KLF1和KLF9协同调控红系分化的分子机制,本研究对KLF1、KLF9及共同过表达或敲低表达细胞株进行转录组测序分析,按照fold-change≥2.0且P≤0.05的筛选标准,各组差异基因的上下调数目,结果如表2所示。敲低表达组的差异基因数目多于过表达组,KLF1和KLF9共同过表达或敲低表达组的红系分化相关的差异基因数目多于其单独敲低表达或过表达组。Table 2
表2
表2 不同处理组差异基因上调和下调数目
Table 2
| KLF1-OE vs. Ctrl-OE | KLF9-OE vs. Ctrl-OE | KLF(1+9)-OE vs. Ctrl-OE | KLF1-KD vs. Ctrl-KD | KLF9-KD vs. Ctrl-KD | KLF(1+9)-KD vs. Ctrl-KD | |
|---|---|---|---|---|---|---|
| 差异基因数目 (上调/下调) | 280/553 | 166/151 | 262/305 | 538/408 | 390/532 | 497/427 |
| 红系分化相关 差异基因数目 | 45 | 29 | 45 | 79 | 66 | 88 |
新窗口打开|下载CSV
利用DAVID数据库对差异基因进行KEGG分析,结果表明差异基因所富集的通路,有些在KLF1、KLF9单独和共同过表达、敲低表达中均受到了影响(图4)。PI3K-Akt通路和FoxO通路是KLF1过表达和二者共同过表达的共有通路,且均与红系分化相关[23]。受KLF9过表达影响的TNF通路与PI3K-Akt通路有直接关系,暗示KLF1和KLF9可能通过这几个通路协同调控K562细胞的红系分化进程。此外,在所有敲低表达组中共有的NF-KappaB通路也与TNF通路有着密切关系,这些通路的相互关系如图5所示。
图4
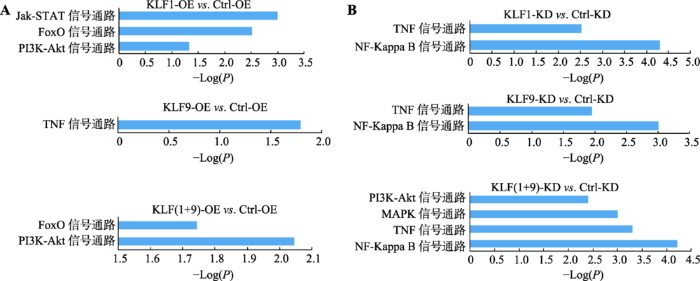 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4利用DAVID分析红系分化过程中受KLF1和KLF9过表达和敲低表达影响的主要信号通路
A:KLF1和KLF9过表达影响的主要信号通路;B:KLF1和KLF9敲低表达影响的主要信号通路。
Fig. 4The disturbed pathways were enhanced during K562 cells erythroid differentiation with KLF1 and KLF9 overexpression and knockdown
图5
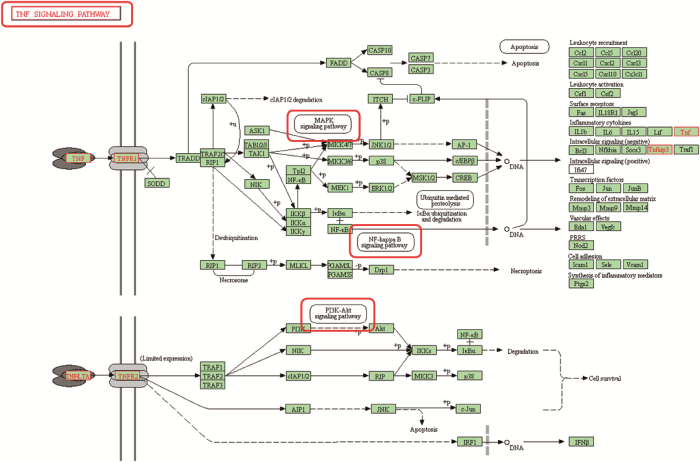 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5KEGG分析显示TNF通路、NF-KappaB通路、MAPK通路和PI3K-Akt通路有相互关联
Fig. 5KEGG analysis shows a relationship among the TNF, NF-KappaB, MAPK, and PI3K-Akt signaling pathways
此外,将差异表达基因提交至STRING分子网络分析平台,构建PPI分子调控网络(图6)。对比网络核心基因,发现共有的潜在靶基因是FOS、TF、IL8,其中TF与KLF1有关,而IL8、FOS被证实与KLF9有关[23],TNF和相关凋亡诱导配体TRAIL作用可抑制正常红细胞生成[24]。此外,共同存在的一些血红蛋白HBA1、HBD、HBE1、HBG2和HBZ的差异也提示KLF1和KLF9在红系分化发育过程中发挥着重要作用。
图6
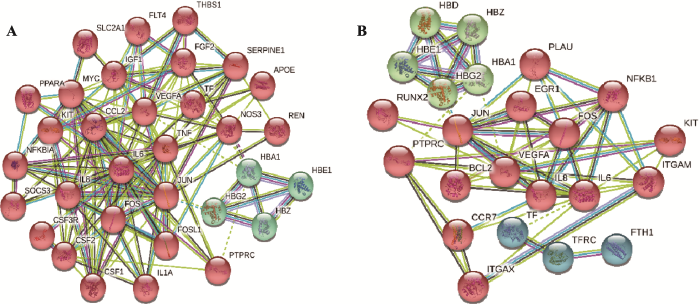 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6KLF1和KLF9过表达和敲低表达导致的蛋白互作关系网络
A:KLF1和KLF9过表达导致的蛋白互作关系网络;B:KLF1和KLF9敲低表达导致的蛋白互作关系网络。
Fig. 6Protein-protein interaction networks induced by KLF1 and KLF9 overexpression and knockdown
3 讨 论
本研究利用K562细胞红系分化模型证明了两个KLF家族成员KLF1和KLF9对红系分化的协同促进作用,发现了潜在受KLF1和KLF9协同调控的信号通路与候选靶基因。这些信息在很大程度上丰富了KLF家族因子调控红系分化的分子网络,有助于拓展对KLF家族成员与红系造血相关疾病的病理机制之间可能联系的认识。K562细胞是胎儿期红系细胞株,尽管在体外红系诱导分化过程中不易评估胎儿期向成人期转换过程而具有局限性,但因其易于常规培养、便于基因遗传操作与表型评估等特征,仍作为评估未知因子调控红系分化的较佳模型[25, 26]。目前虽尚未有KLF9可调控成人期红系细胞分化的报道,但在成人急性淋巴细胞白血病患者中KLF9显著上调[27],且有研究表明在成人红系祖细胞中降低KLF1水平可再次激活胎儿β-珠蛋白基因表达[28],所以推测KLF1和KLF9可能在成人期红系细胞分化中也发挥一定的作用,但还有待进一步探究。本研究发现KLF1和KLF9在K562细胞模型共同过表达可显著促进红系分化,说明这两个基因在调控K562细胞红系分化进程中存在一定程度上互相促进或者相辅相成的关系。
共同过表达KLF1和KLF9对HBE和HBG的表达并没有影响,表明KLF1和KLF9并不通过调控这两个珠蛋白基因促进红系分化。但是,这两个KLF因子却可以特异性地协同促进HBB基因的表达水平,明显高于KLF1和KLF9单独的调控作用。HBB的异常表达可导致β地中海贫血[29],通过转录因子调控并促进HBB编码基因表达一直是探索β-地贫临床治疗策略之一,本研究为提高HBB基因的表达水平提供了新的思路。因此,KLF1和KLF9协同促进HBB的表达依然具有重要理论意义。
通过对KLF1、KLF9表达水平改变所引起的差异表达基因的信号通路和蛋白质互作网络分析,鉴定了FOS、TF、JUN、FOXO3、TNF、IL8和RUNX2等几个可能发挥关键作用的重点基因。在后续研究中,我们会进一步利用染色质免疫共沉淀及荧光双报告基因等技术验证这些候选靶基因,结合原代造血干细胞、成人期HUDEP-2红系细胞模型和斑马鱼等模型,通过调节靶基因表达水平或改变通路等策略探索KLF1和KLF9调控HBB珠蛋白及红系分化的分子机制。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URL [本文引用: 1]
URL [本文引用: 1]
URLMagsci [本文引用: 1]

<p>Sp1/Krüppel样因子(Sp1-like and Krüppel-like factors, Sp1/KLFs)是一组与真核细胞转录调控密切相关的锌指蛋白。Sp1/KLFs的羧基末端高度保守, 含有3个串联的Cys<sub>2</sub>His<sub>2</sub>锌指结构, 用于结合DNA; 其氨基末端在不同的家族成员间存在较大差异, 主要是通过结合辅助因子发挥转录调控作用。Sp1/KLFs的表达具有组织、细胞分布以及发育时期的特异性, 它们通过调控多种富含GC或CACCC的启动子的基因的表达, 参与细胞增殖、分化、凋亡和肿瘤发生、发展等多种生理、病理过程。文章综述了Sp1/KLFs的结构特征、作用机制及生物学功能。</p>
URLMagsci [本文引用: 1]

<p>Sp1/Krüppel样因子(Sp1-like and Krüppel-like factors, Sp1/KLFs)是一组与真核细胞转录调控密切相关的锌指蛋白。Sp1/KLFs的羧基末端高度保守, 含有3个串联的Cys<sub>2</sub>His<sub>2</sub>锌指结构, 用于结合DNA; 其氨基末端在不同的家族成员间存在较大差异, 主要是通过结合辅助因子发挥转录调控作用。Sp1/KLFs的表达具有组织、细胞分布以及发育时期的特异性, 它们通过调控多种富含GC或CACCC的启动子的基因的表达, 参与细胞增殖、分化、凋亡和肿瘤发生、发展等多种生理、病理过程。文章综述了Sp1/KLFs的结构特征、作用机制及生物学功能。</p>
Magsci [本文引用: 1]

Krüppel样因子(Krüppel-like factors, KLFs)是一组与真核基因转录调控密切相关的锌指蛋白.KLFs高度保守的羧基末端含3个串联的Cys2His2型锌指结构,用于结合GC盒和CACCC盒等DNA序列. 红细胞中特异表达的珠蛋白基因和许多红系调控因子中都含有CACCC盒.已有研究发现,多个KLFs通过结合CACCC盒参与调控珠蛋白基因表达和红系分化,例如,KLF1通过结合β-珠蛋白启动子和位点控制区(locus control region, LCR),促进β-珠蛋白的表达、γ-向β-珠蛋白基因的转换和红系分化;KLF2、KLF11和KLF13分别促进ε-和γ-珠蛋白基因的表达;KLF4促进α-和γ-珠蛋白基因的表达;KLF3和KLF8则抑制ε-和γ-珠蛋白基因的表达. 本文综述了KLFs调控珠蛋白基因表达和红系分化的研究进展.
Magsci [本文引用: 1]

Krüppel样因子(Krüppel-like factors, KLFs)是一组与真核基因转录调控密切相关的锌指蛋白.KLFs高度保守的羧基末端含3个串联的Cys2His2型锌指结构,用于结合GC盒和CACCC盒等DNA序列. 红细胞中特异表达的珠蛋白基因和许多红系调控因子中都含有CACCC盒.已有研究发现,多个KLFs通过结合CACCC盒参与调控珠蛋白基因表达和红系分化,例如,KLF1通过结合β-珠蛋白启动子和位点控制区(locus control region, LCR),促进β-珠蛋白的表达、γ-向β-珠蛋白基因的转换和红系分化;KLF2、KLF11和KLF13分别促进ε-和γ-珠蛋白基因的表达;KLF4促进α-和γ-珠蛋白基因的表达;KLF3和KLF8则抑制ε-和γ-珠蛋白基因的表达. 本文综述了KLFs调控珠蛋白基因表达和红系分化的研究进展.
URL [本文引用: 1]
URLPMID:20508144 [本文引用: 1]

Abstract KLF1 regulates a diverse suite of genes to direct erythroid cell differentiation from bipotent progenitors. To determine the local cis-regulatory contexts and transcription factor networks in which KLF1 operates, we performed KLF1 ChIP-seq in the mouse. We found at least 945 sites in the genome of E14.5 fetal liver erythroid cells which are occupied by endogenous KLF1. Many of these recovered sites reside in erythroid gene promoters such as Hbb-b1, but the majority are distant to any known gene. Our data suggests KLF1 directly regulates most aspects of terminal erythroid differentiation including production of alpha- and beta-globin protein chains, heme biosynthesis, coordination of proliferation and anti-apoptotic pathways, and construction of the red cell membrane and cytoskeleton by functioning primarily as a transcriptional activator. Additionally, we suggest new mechanisms for KLF1 cooperation with other transcription factors, in particular the erythroid transcription factor GATA1, to maintain homeostasis in the erythroid compartment.
URLPMID:15947087 [本文引用: 2]

Abstract The Kr ppel-like factors (KLFs) are a family of C2/H2 zinc finger DNA-binding proteins that are important in controlling developmental programs. Erythroid Kr ppel-like factor (EKLF or KLF1) positively regulates the beta-globin gene in definitive erythroid cells. KLF2 (LKLF) is closely related to EKLF and is expressed in erythroid cells. KLF2-/- mice die between embryonic day 12.5 (E12.5) and E14.5, because of severe intraembryonic hemorrhaging. They also display growth retardation and anemia. We investigated the expression of the beta-like globin genes in KLF2 knockout mice. Our results show that KLF2-/- mice have a significant reduction of murine embryonic Ey- and beta h1-globin but not zeta-globin gene expression in the E10.5 yolk sac, compared with wild-type mice. The expression of the adult beta(maj)- and beta(min)-globin genes is unaffected in the fetal livers of E12.5 embryos. In mice carrying the entire human globin locus, KLF2 also regulates the expression of the human embryonic epsilon-globin gene but not the adult beta-globin gene, suggesting that this developmental-stage-specific role is evolutionarily conserved. KLF2 also plays a role in the maturation and/or stability of erythroid cells in the yolk sac. KLF2-/- embryos have a significantly increased number of primitive erythroid cells undergoing apoptotic cell death.
URL [本文引用: 1]
URLPMID:1907364 [本文引用: 1]

Sp/KLF family of factors regulates gene expression by binding to the CACCC/GC/GT boxes in the DNA through their highly conserved three zinc finger domains. To investigate the role of this family of factors in erythroid differentiation and globin gene expression, we first measured the expression levels of selected Sp/KLF factors in primary cells of fetal and adult stages of erythroid development. This quantitative analysis revealed that their expression levels vary significantly in cells of either stages of the erythroid development. Significant difference in their expression levels was observed between fetal and adult erythroid cells for some Sp/KLF factors. Functional studies using RNA interference revealed that the silencing of Sp1 and KLF8 resulted in elevated level of 纬 globin expression in K562 cells. In addition, K562 cells become visibly red after Sp1 knockdown. Benzidine staining revealed significant hemoglobinization of these cells, indicating erythroid differentiation. Moreover, the expression of PU.1, ETS1 and Notch1 is significantly down-regulated in the cells that underwent erythroid differentiation following Sp1 knockdown. Overexpression of PU.1 or ETS1 efficiently blocked the erythroid differentiation caused by Sp1 knockdown in K562 cells. The expression of c-Kit, however, was significantly up-regulated. These data indicate that Sp1 may play an important role in erythroid differentiation.
URLPMID:22711990 [本文引用: 1]

The CACCC-box binding protein erythroid Kruppel-like factor (EKLF/KLF1) is a master regulator that directs the expression of many important erythroid genes. We have previously shown that EKLF drives transcription of the gene for a second KLF, basic Kruppel-like factor, or KLF3. We have now tested the in vivo role of KLF3 in erythroid cells by examining Klf3 knockout mice. KLF3-deficient adults exhibit a mild compensated anemia, including enlarged spleens, increased red pulp, and a higher percentage of erythroid progenitors, together with elevated reticulocytes and abnormal erythrocytes in the peripheral blood. Impaired erythroid maturation is also observed in the fetal liver. We have found that KLF3 levels rise as erythroid cells mature to become TER119(+). Consistent with this, microarray analysis of both TER119(-) and TER119(+) erythroid populations revealed that KLF3 is most critical at the later stages of erythroid maturation and is indeed primarily a transcriptional repressor. Notably, many of the genes repressed by KLF3 are also known to be activated by EKLF. However, the majority of these are not currently recognized as erythroid-cell-specific genes. These results reveal the molecular and physiological function of KLF3, defining it as a feedback repressor that counters the activity of EKLF at selected target genes to achieve normal erythropoiesis.
URLPMID:16460907 [本文引用: 1]

The Sp/KLF transcription factors perform a variety of biological functions, but are related in that they bind GC-box and CACCC-box sequences in DNA via a highly conserved DNA-binding domain. A database homology search, using the zinc finger DNA-binding domain characteristic of the family, has identified human KLF17 as a new family member that is most closely related to KLFs 1 8 and 12. Zfp393), although it has diverged significantly. The DNA-binding domain is the most conserved region, suggesting that both the murine and the human forms recognize the same binding sites in DNA and may retain similar functions. We show that human KLF17 can bind G/C-rich sites via its zinc fingers and is able to activate transcription from CACCC-box elements. This is the first report of the DNA-binding characteristics and transactivation activity of human KLF17, which, together with the homology it displays to other KLF proteins, put it in the Sp/KLF family.
URLPMID:22711990 [本文引用: 1]

The CACCC-box binding protein erythroid Kruppel-like factor (EKLF/KLF1) is a master regulator that directs the expression of many important erythroid genes. We have previously shown that EKLF drives transcription of the gene for a second KLF, basic Kruppel-like factor, or KLF3. We have now tested the in vivo role of KLF3 in erythroid cells by examining Klf3 knockout mice. KLF3-deficient adults exhibit a mild compensated anemia, including enlarged spleens, increased red pulp, and a higher percentage of erythroid progenitors, together with elevated reticulocytes and abnormal erythrocytes in the peripheral blood. Impaired erythroid maturation is also observed in the fetal liver. We have found that KLF3 levels rise as erythroid cells mature to become TER119(+). Consistent with this, microarray analysis of both TER119(-) and TER119(+) erythroid populations revealed that KLF3 is most critical at the later stages of erythroid maturation and is indeed primarily a transcriptional repressor. Notably, many of the genes repressed by KLF3 are also known to be activated by EKLF. However, the majority of these are not currently recognized as erythroid-cell-specific genes. These results reveal the molecular and physiological function of KLF3, defining it as a feedback repressor that counters the activity of EKLF at selected target genes to achieve normal erythropoiesis.
URL
URLPMID:16234353 [本文引用: 1]

Abstract Kr ppel-like factor 6 (KLF6) is a member of a growing family of transcription factors that share a common 3 C2H2 zinc finger DNA binding domain and have broad activity in regulating proliferation and development. We have previously established that Klf6 is expressed in neuronal tissue, hindgut, heart, lung, kidney, and limb buds during midgestation. To explore the potential role of Klf6 in mouse development, we analyzed Klf6-/- mice and found that the homozygous mutation is embryonic lethal by embryonic day (E) 12.5 and associated with markedly reduced hematopoiesis and poorly organized yolk sac vascularization. Additionally, mRNA levels of Scl and Gata1 were reduced by approximately 80% in Klf6-/- yolk sacs. To further analyze this phenotype, we generated Klf6-/- embryonic stem (ES) cells by homologous recombination, and compared their capacity to differentiate into the hematopoietic lineage with that of either Klf6+/- or Klf6+/+ ES cells. Consistent with the phenotype in the early embryo, Klf6-/- ES cells displayed significant hematopoietic defects following differentiation into EBs. Prolongation of epiblast-like cells and delays in mesoderm induction were also observed in the Klf6-/- EBs, associated with delayed expression of Brachyury, Klf1, and Gata1. Forced expression of KLF6 using a tet-inducible system enhanced the hematopoietic potential of wild-type EBs. Collectively, these findings implicate Klf6 in ES-cell differentiation and hematopoiesis.
URLPMID:3434496 [本文引用: 1]

The Kr ppel-like factor 1 (KLF1) and KLF2 positively regulate embryonic -globin expression and have additional overlapping roles in embryonic (primitive) erythropoiesis. KLF1(-/-) KLF2(-/-) double knockout mice are anemic at embryonic day 10.5 (E10.5) and die by E11.5, in contrast to single knockouts. To investigate the combined roles of KLF1 and KLF2 in primitive erythropoiesis, expression profiling of E9.5 erythroid cells was performed. A limited number of genes had a significantly decreasing trend of expression in wild-type, KLF1(-/-), and KLF1(-/-) KLF2(-/-) mice. Among these, the gene for Myc (c-Myc) emerged as a central node in the most significant gene network. The expression of the Myc gene is synergistically regulated by KLF1 and KLF2, and both factors bind the Myc promoters. To characterize the role of Myc in primitive erythropoiesis, ablation was performed specifically in mouse embryonic proerythroblast cells. After E9.5, these embryos exhibit an arrest in the normal expansion of circulating red cells and develop anemia, analogous to KLF1(-/-) KLF2(-/-) embryos. In the absence of Myc, circulating erythroid cells do not show the normal increase in - and -like globin gene expression but, interestingly, have accelerated erythroid cell maturation between E9.5 and E11.5. This study reveals a novel regulatory network by which KLF1 and KLF2 regulate Myc to control the primitive erythropoietic program.
URL [本文引用: 1]

CACCC boxes are among the critical sequences present in regulatory elements of genes expressed in erythroid cells, as well as in selected other cell types. While an erythroid cell-specific CACCC-box-binding protein, EKLF, has been shown to be required in vivo for proper expression of the adult beta-globin gene, it is dispensable for the regulation of several other globin and nonglobin erythroid cell-expressed genes. In the work described here, we searched for additional CACCC-box transcription factors that might be active in murine erythroid cells. We identified a major gel shift activity (termed BKLF), present in yolk sac and fetal liver erythroid cells, that could be distinguished from EKLF by specific antisera. Through relaxed-stringency hybridization, we obtained the cDNA encoding BKLF, a highly basic, novel zinc finger protein that is related to EKLF and other Kr ppel-like members in its DNA-binding domain but unrelated elsewhere. BKLF, which is widely but not ubiquitously expressed in cell lines, is highly expressed in the midbrain region of embryonic mice and appears to correspond to the gel shift activity TEF-2, a transcriptional activator implicated in regulation of the simian virus 40 enhancer and other CACCC-box-containing regulatory elements. Because BKLF binds with high affinity and preferentially over Sp1 to many CACCC sequences of erythroid cell expressed genes, it is likely to participate in the control of many genes whose expression appears independent of the action of EKLF.
URLPMID:12020 [本文引用: 2]

Erythroid Kruppel-like factor (EKLF, KLF1) plays an important role in definitive erythropoiesis and beta-globin gene regulation but failure to rectify lethal fetal anemia upon correction of globin chain imbalance suggested additional critical EKLF target genes. We employed expression profiling of EKLF-null fetal liver and EKLF-null erythroid cell lines containing an inducible EKLF-estrogen receptor (EKLF-ER) fusion construct to search for such targets. An overlapping list of EKLF-regulated genes from the 2 systems included alpha-hemoglobin stabilizing protein (AHSP), cytoskeletal proteins, hemesynthesis enzymes, transcription factors, and blood group antigens. One EKLF target gene, dematin, which encodes an erythrocyte cytoskeletal protein (band 4.9), contains several phylogenetically conserved consensus CACC motifs predicted to bind EKLF. Chromatin immunoprecipitation demonstrated in vivo EKLF occupancy at these sites and promoter reporter assays showed that EKLF activates gene transcription through these DNA elements. Furthermore, investigation of EKLF target genes in the yolk sac led to the discovery of unexpected additional defects in the embryonic red cell membrane and cytoskeleton. In short, EKLF regulates global erythroid gene expression that is critical for the development of primitive and definitive red cells.
URLPMID:27272188 [本文引用: 1]

在 erythroid 开发期间编码基因, miRNAs,和 lncRNAs 上的研究在最近的年里被执行了。然而,集中于三种 RNA 类型的集成的分析还得被做。在现在的学习,我们比较了编码基因, miRNA,和 lncRNA 表示侧面的动力学。为了在在 transcriptome 控制这些变化的红血球生成和潜在的机制探索动态变化,铺平,我们利用了定序技术从绳索血获得 transcriptome 数据的高产量造血的干细胞和下列四个 erythroid 区别阶段,以及从成熟的红血房间。结果显示 lncRNAs 为 erythroid 区别正在答应房间标记候选人。聚类分析分类差别表示基因进在 stemness 维护期间对应于动态变化的四种子类型,中间区别,并且成熟。综合分析揭示了 RNA 潜在地参予了控制血细胞成熟的那 noncoding,并且特别与到氧种类和 DNA 损坏的 heme 新陈代谢和回答联系了。这些规章的相互作用在一个全面网络,在 RNA 之间的从而推断的关联和他们的联系功能被显示。这些数据为正常红血球生成的学习提供了一个大量的资源,它将允许 erythroid 开发的进一步的调查和理解并且获得 erythroid 混乱。
URLPMID:23985037 [本文引用: 2]

Abstract BACKGROUND: Mapping of DNase I hypersensitive sites (DHSs) is a powerful tool to experimentally identify cis-regulatory elements (CREs). Among CREs, enhancers are abundant and predominantly act in driving cell-specific gene expression. Kr ppel-like factors (KLFs) are a family of eukaryotic transcription factors. Several KLFs have been demonstrated to play important roles in hematopoiesis. However, transcriptional regulation of KLFs via CREs, particularly enhancers, in erythroid cells has been poorly understood. RESULTS: In this study, 23 erythroid-specific or putative erythroid-specific DHSs were identified by DNase-seq in the genomic regions of 17 human KLFs, and their enhancer activities were evaluated using dual-luciferase reporter (DLR) assay. Of the 23 erythroid-specific DHSs, the enhancer activities of 15 DHSs were comparable to that of the classical enhancer HS2 in driving minimal promoter (minP). Fifteen DHSs, some overlapping those that increased minP activities, acted as enhancers when driving the corresponding KLF promoters (KLF-Ps) in erythroid cells; of these, 10 DHSs were finally characterized as erythroid-specific KLF enhancers. These 10 erythroid-specific KLF enhancers were further confirmed using chromatin immunoprecipitation coupled to sequencing (ChIP-seq) data-based bioinformatic and biochemical analyses. CONCLUSION: Our present findings provide a feasible strategy to extensively identify gene- and cell-specific enhancers from DHSs obtained by high-throughput sequencing, which will help reveal the transcriptional regulation and biological functions of genes in some specific cells.
URL [本文引用: 1]
URLPMID:19244387 [本文引用: 1]

SUMMARY: The development of RNA sequencing (RNA-Seq) makes it possible for us to measure transcription at an unprecedented precision and throughput. However, challenges remain in understanding the source and distribution of the reads, modeling the transcript abundance and developing efficient computational methods. In this article, we develop a method to deal with the isoform expression estimation problem. The count of reads falling into a locus on the genome annotated with multiple isoforms is modeled as a Poisson variable. The expression of each individual isoform is estimated by solving a convex optimization problem and statistical inferences about the parameters are obtained from the posterior distribution by importance sampling. Our results show that isoform expression inference in RNA-Seq is possible by employing appropriate statistical methods.
URL [本文引用: 1]
URLPMID:3973089 [本文引用: 2]

Genetic studies have identified common variants within the intergenic region (HBS1L-MYB) between GTP-binding elongation factor HBS1L and myeloblastosis oncogene MYB on chromosome 6q that are associated with elevated fetal hemoglobin (HbF) levels and alterations of other clinically important human erythroid traits. It is unclear how these noncoding sequence variants affect multiple erythrocyte characteristics. Here, we determined that several HBS1L-MYB intergenic variants affect regulatory elements that are occupied by key erythroid transcription factors within this region. These elements interact with MYB, a critical regulator of erythroid development and HbF levels. We found that several HBS1L-MYB intergenic variants reduce transcription factor binding, affecting long-range interactions with MYB and MYB expression levels. These data provide a functional explanation for the genetic association of HBS1L-MYB intergenic polymorphisms with human erythroid traits and HbF levels. Our results further designate MYB as a target for therapeutic induction of HbF to ameliorate sickle cell and -thalassemia disease severity.
[本文引用: 1]
URLPMID:8462662 [本文引用: 1]

Abstract The human erythroleukemia cell line K562 can be induced to differentiate along the erythroid and megakaryocytic lineages. Here we demonstrate that hexamethylene bisacetamide (HMBA) induced K562 cells to differentiate along a third pathway. This was accompanied by downregulation of two transcription factors normally expressed in erythroid, mast and megakaryocyte lineages. Northern analysis demonstrated coordinate downregulation of alpha globin and gamma globin in addition to the two lineage-restricted transcription factors, SCL and GATA-1. Proliferation of the K562 cells was also suppressed. Clonal assay showed that the suppression was irreversible and appeared analogous to the commitment of murine erythroleukemia (MEL) cells to terminal differentiation. In contrast to MEL cells, however, K562 cells acquired a macrophage-like morphology and exhibited a complete failure to generate benzidine-positive cells. Electron microscopy revealed a marked increase in granules resembling those specific for eosinophils. Surface marker analysis showed that HMBA-induced cells expressed reduced levels of glycophorin A, CD5, CD7 and CD11b. No upregulation of megakaryocyte or lymphoid markers occurred. Thus the response of K562 cells to HMBA may provide a useful experimental system for studying the molecular mechanisms responsible for downmodulation of lineage-restricted transcription factors during hemopoietic lineage commitment.
URLPMID:9299849 [本文引用: 1]

Abstract BACKGROUND AND OBJECTIVE: Human leukemic K562 cells are able to undergo erythroid differentiation in vitro when cultured with a variety of inducers, leading to increased expression of embryo-fetal globin genes such as the zita, epsilon and gamma-globin genes. Therefore the K562 cell line has been proposed as a very useful in vitro model system for determining the therapeutical potential of new differentiating compounds as well as for studying the molecular mechanism(s) that regulate changes in the expression of embryonic and fetal human globin genes. In this study we explored whether nucleoside triphosphates and related compounds are able to induce differentiation of K562 cells. METHODS: K562 cell differentiation was studied using the benzidine test; hemoglobins were characterized by cellulose acetate gel electrophoresis and mRNA accumulation was investigated by Northern blot analysis. RESULTS: The main conclusion of this paper is that guanine, guanosine and guanine ribonucleotides are effective inducers of K562 cell differentiation. Expression of both Hb Portland and Hb Gower 1 is increased in GTP-induced K562 cells. This increase is associated with greater gamma-globin mRNA accumulation. By contrast, ATP, CTP and UTP are not able to induce erythroid differentiation. INTERPRETATION AND CONCLUSIONS: These findings suggest that guanine, guanosine and guanine ribonucleotides are inducers of erythroid differentiation of K562 cells. This is of some relevance since differentiating compounds have been proposed as antitumor agents. In addition, inducers of erythroid differentiation that stimulate gamma-globin synthesis might be considered in the experimental therapy of hematological diseases associated with a failure in the expression of adult beta-globin genes.
URLPMID:22848414 [本文引用: 1]

Background Deletions of IKAROS (IKZF1) frequently occur in B-cell precursor acute lymphoblastic leukemia (B-ALL) but the mechanisms by which they influence pathogenesis are unclear. To address this issue, a cohort of 144 adult B-ALL patients (106 BCR-ABL1-positive and 38 B-ALL negative for known molecular rearrangements) was screened for IKZF1 deletions by single nucleotide polymorphism (SNP) arrays; a sub-cohort of these patients (44%) was then analyzed for gene expression profiling. Principal Findings Total or partial deletions of IKZF1 were more frequent in BCR-ABL1-positive than in BCR-ABL1-negative B-ALL cases (75% vs 58%, respectively, p = 0.04). Comparison of the gene expression signatures of patients carrying IKZF1 deletion vs those without showed a unique signature featured by down-regulation of B-cell lineage and DNA repair genes and up-regulation of genes involved in cell cycle, JAK-STAT signalling and stem cell self-renewal. Through chromatin immunoprecipitation and luciferase reporter assays we corroborated these findings both in vivo and in vitro, showing that Ikaros deleted isoforms lacked the ability to directly regulate a large group of the genes in the signature, such as IGLL1, BLK, EBF1, MSH2, BUB3, ETV6, YES1, CDKN1A (p21), CDKN2C (p18) and MCL1. Conclusions Here we identified and validated for the first time molecular pathways specifically controlled by IKZF1, shedding light into IKZF1 role in B-ALL pathogenesis.
URL [本文引用: 1]
URL [本文引用: 1]

正红细胞是具有重要生理功能的 血液细胞。成年人的红细胞主要来源于分布在胸骨、椎骨、肋骨等扁骨内的红骨髓。红骨髓中的髓系多能干细胞分化成红系定向祖细胞,红系定向祖细胞在促红细胞 生成素(EPO)的作用下,能增殖分化为原红细胞。原红细胞经过有丝分裂、增殖成为网织红细胞,进而发育成为成熟红细胞。红细胞成熟时没有细胞核和细胞 器,胞质内充满血红蛋白,具有携带氧气和二氧化碳的功能。
URL [本文引用: 1]

正红细胞是具有重要生理功能的 血液细胞。成年人的红细胞主要来源于分布在胸骨、椎骨、肋骨等扁骨内的红骨髓。红骨髓中的髓系多能干细胞分化成红系定向祖细胞,红系定向祖细胞在促红细胞 生成素(EPO)的作用下,能增殖分化为原红细胞。原红细胞经过有丝分裂、增殖成为网织红细胞,进而发育成为成熟红细胞。红细胞成熟时没有细胞核和细胞 器,胞质内充满血红蛋白,具有携带氧气和二氧化碳的功能。
