 ,2,3, 陈嘉瑜
,2,3, 陈嘉瑜 ,1,2, 高绍荣
,1,2, 高绍荣 ,1,2
,1,2Historical review of reprogramming and pluripotent stem cell research in China
Lan Kang ,2,3, Jiayu Chen
,2,3, Jiayu Chen ,1,2, Shaorong Gao
,1,2, Shaorong Gao ,1,2
,1,2通讯作者:
第一联系人:
编委: 王晓群
收稿日期:2018-07-20修回日期:2018-08-30网络出版日期:2018-10-20
| 基金资助: |
Received:2018-07-20Revised:2018-08-30Online:2018-10-20
| Fund supported: |
作者简介 About authors
康岚,教授,研究方向:干细胞与体细胞重编程E-mail:
陈嘉瑜,副教授,研究方向:干细胞与体细胞重编程E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (428KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
康岚, 陈嘉瑜, 高绍荣. 中国细胞重编程和多能干细胞研究进展[J]. 遗传, 2018, 40(10): 825-840 doi:10.16288/j.yczz.18-209
Lan Kang, Jiayu Chen, Shaorong Gao.
在国家“七五”、“八五”、“十一五”、“十二五”、“十三五”、“863计划”、“973计划”、“重大科学研究计划”、“重点研发计划”及自然科学基金等国家科技项目和各省部级项目的大力支持下,中国在生殖发育与干细胞领域的研究从早期跟随性研究,逐渐成长为引领世界的分子机制研究和临床转化性探索。2018年,在中国诞生了世界第一例非人灵长类体细胞核移植克隆猴,成为该领域发展中的又一重大里程碑。中国科学家结合我国的实际情况,通过几代人的不懈努力取得了一系列重大进展,已经在干细胞研究领域跻身于世界第一梯队。本文从体细胞核移植、诱导多能干细胞、单倍体多能干细胞和胚胎早期发育研究4个方面对中国的重编程和多能干细胞研究进展进行了历史性回顾。
1 体细胞核移植的前世今生
核移植(nuclear transfer, NT),又称为体细胞核移植(somatic cell nuclear transfer, SCNT),更被人们所熟知的名字是克隆(cloning),是指将供体细胞核移入去除遗传物质的卵母细胞中,使细胞重获全能性的一种方法(图1A)。值得注意的是,核移植技术是目前已知的唯一一种可以高效地、快速地使分化的细胞获得全能性的方式。该技术在过去的60多年里,对核质关系、细胞分化、细胞多能性、表观遗传学、发育生物学和生殖生物学等方面的研究以及转化医学和遗传资源保存等方面做出了巨大的贡献[1,2,3,4]。图1
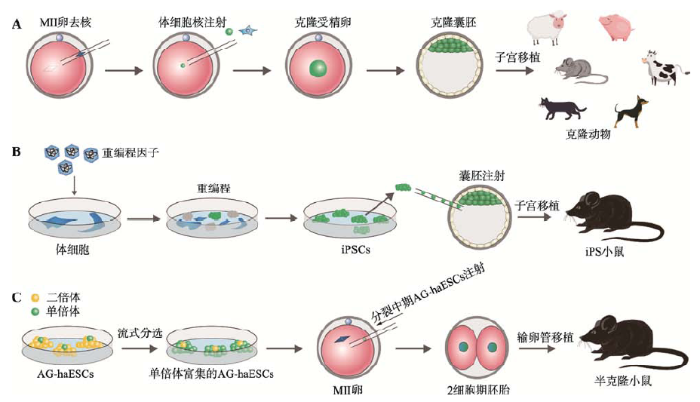 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1体细胞核移植、iPSC和单倍体ESC的原理模式图
A:体细胞核移植获得克隆动物。将第二次减数分裂中期(MII)的卵细胞去核,然后注入体细胞核,从而得到克隆胚胎,继而可以发育到克隆动物。B:由iPSC获得iPS小鼠。通过将重编程因子导入到体细胞中并使其表达,可将体细胞重编程为iPSC,将iPSC注射到囊胚或者4倍体囊胚,可得到嵌合体或者全iPS小鼠。C:由单倍体ESC获得半克隆小鼠。孤雄单倍体胚胎干细胞(androgenetic haploid embryonic stem cells, AG-haESCs)在体外通过流式分选纯化和维持,将分裂中期的AG-haESCs注射到MII卵细胞中,可使其正常发育,继而得到半克隆小鼠。
Fig. 1Schematic representation of somatic cell nuclear transfer, iPSCs and haploid embryonic stem cells
1.1 两栖类和鱼类核移植
1952年,美国费城Lankenau医学研究所的科学家Briggs和King证明,将发育至囊胚期豹蛙(Rana pipiens)的卵细胞核移植入去核的豹蛙卵母细胞后,所产生的胚胎可以正常发育并孵出蝌蚪[5]。这是人类第一次培育出克隆动物。然而,当他们利用囊胚阶段后期产生的更为分化的细胞作为细胞核供体进行核移植实验时,却发现这些核移植后的胚胎发育异常,最终不能获得蝌蚪[6]。该实验也表明,供体细胞核的差异会影响体细胞重编程的能力(这一现象在几十年后的哺乳动物的克隆实验中也被进一步证实)。20世纪60年代,英国牛津大学Gurdon等[7, 8]将内胚层细胞来源的蝌蚪肠道细胞的细胞核移植入去核的爪蛙(Xenopus laevis)卵母细胞中,发现核移植后约1%的胚胎可以正常发育并形成蝌蚪和爪蛙,由此获得了全世界首例发育至性成熟的成体克隆动物。Gurdon教授也因此项突破性的工作,与另一位日本生物学家Shinya Yamanaka教授(建立了iPS技术)分享了2012年诺贝尔生理学或医学奖。该项研究也证明早期胚胎发育所需要的基因,即使在分化的细胞中也可以被再激活,并且暗示了卵母细胞中具有非常重要的重编程因子,且不同品系的卵母细胞的重编程能力可能不同。在此后几年的研究中,他们将更为分化的成体细胞(adult somatic cell)的细胞核进行核移植时,仅能够获得蝌蚪,却不能得到发育至性成熟时期的蛙[9, 10]。同一时期,在历经战火和正在重生的中国,也有一位胚胎发育学家进行了相似的研究,但却鲜被国际所知。中国早期的发育生物学家、实验胚胎学主要奠基人童第周先生,师从比利时比京大学(今法语布鲁塞尔自由大学)Albert Brachet教授和Albert Dalcq教授后获得动物学博士学位,于1934年毅然回国,在同济大学进行教学与研究工作。抗日战争时期,同济大学迁往四川李庄办学,在极其艰苦的工作环境和简陋的设备条件下,童第周先生与其夫人叶毓芬教授合作,以其超高的实验技巧进行了两栖类和鱼胚的发育和可塑性研究,通过切割、移植、离心等方法,他们以融合的胚胎、半胚胎以及离心后的胚胎为来源,获得多个异常发育的鲫鱼。新中国成立后,童第周先生于1956年在青岛组织举行了遗传学研讨会。该会议促使他进一步利用两栖类和鱼类的胚胎系统,研究细胞核与细胞质对胚胎命运的影响,并以自然辩证法的角度阐述核质关系。1962年,童第周先生领导的团队将鲤鱼及鲫鱼的囊胚细胞核分离,并相互进行核移植,培育出第一尾属间核质杂种鱼,开创了异种核移植的先河,并发现了脊椎动物远缘物种间细胞核和细胞质之间的可配合性。该实验还发现,异种核移植后的卵发育到成体后有些性状介于两种鱼之间,由此也证明细胞质存在可影响胚胎命运的物质,并希望进一步利用该特性产生更强壮的鱼类,以便适合生产繁殖。但由于历史原因,该工作直到1973年才得以发表[11]。童第周先生在极其简陋的条件下,用一台显微镜、一套基本手术器械和双手,以一己之力,穷毕生心血,创立了中国的克隆事业,被誉为“中国克隆之父”。此后,1981年中国科学院水生生物研究所陈宏溪研究员领导的课题组将成年三倍体鲫鱼的肾脏细胞核移植到二倍体鲫鱼的去核卵子里,获得了三倍体的克隆鱼。
1.2 哺乳动物核移植
哺乳动物的核移植研究相对于两栖类动物更为困难,因为哺乳动物卵母细胞的直径小,卵母细胞取材困难,且数量相对较少。1975年,英国牛津大学Bromhall教授[12]将桑椹胚期细胞的细胞核移植入去核的兔卵母细胞中,证实了这些核移植后的胚胎可以发育至桑椹胚时期。1983年,美国费城Wistar研究所McGrath教授和Solter教授通过将受精后合子的细胞核移植入去核的小鼠卵母细胞中,最终获得了发育至成年的小鼠[13]。1986年,英国剑桥大学AFRC研究所Willadsen教授[14]将发育至8-细胞或16-细胞时期胚胎中的细胞与去核的羊卵母细胞融合后,成功得到了健康的克隆动物。随后的几年中,以胚胎发育期的细胞为供核细胞的核移植实验,在兔(Oryctolagus cuniculus)[15]、猪(Sus scrofa)[16]、小鼠(Mus musculus)[17]、牛(Bos taurus)[18]以及猴(Rhesuls macaque)[19]等动物上均取得了成功。然而,以终末分化的体细胞为供核细胞的核移植实验,一直难以获得存活的克隆动物。
直到1996年,核移植领域取得了重大的突破。英国爱丁堡大学Roslin研究所生物学家Campbell教授和Wilmut爵士(教授)等以胚胎来源的、发育至第9天的分化细胞为供核细胞,成功获得了健康的克隆羊[20]。该核移植实验的成功,主要是由于在核移植操作前,供核细胞经过药物处理后停滞在了特定的时期。使用类似的技术,Wilmut爵士等利用来自成年羊的乳腺上皮细胞的细胞核,获得了健康存活的、可发育至成年的绵羊,即著名的“多莉”(Dolly)羊[21]。由此,以终末分化的体细胞为供核细胞的核移植研究迈入了一个新的篇章。随后,体细胞核移植先后在牛(1998年)[22]、小鼠(1998年)[23]、山羊(Capra hircus) (1999年)[24]、猪(2000年)[25, 26]、猫(Felis domesticus) (2002年)[27]和兔(2002年)[28]等多种动物中获得了成功(表1)。中国科学院动物研究所周琪研究员在法国国家农业科学研究院(INRA)完成了世界上第一例克隆大鼠(Rattus norvegicus)的制作[29]。2005年,韩国国立首尔大学黄禹锡教授领导的研究团队完成了狗(Canis lupus familiaris)的克隆[30]。这些重要的研究成果均证明,许多哺乳动物的体细胞基因组,甚至那些终末分化的细胞在进行核移植后,是能够被重新激活的,它们可以表达正常发育所需的全部基因并最终产生存活的克隆动物。同时,这些研究也表明在分化过程中细胞基因组上的发育限制是可逆的遗传学和表观遗传修饰变化,而卵母细胞中一定含有某些重要的物质能够有效地将终末分化的体细胞进行重编程。
Table 1
表1
表1 代表性哺乳动物克隆汇总
Table 1
| 年份 | 物种 | 细胞来源 |
|---|---|---|
| 1997 | 绵羊(Ovis aries) | 成体乳腺上皮细胞[21] |
| 1998 | 奶牛(Bos taurus) | 胚胎成纤维细胞[31]、 成体卵丘颗粒细胞[22] |
| 1998 | 小鼠(Mus musculus) | 成体卵丘颗粒细胞[23] |
| 1998 | 山羊(Capra hircus) | 胚胎成纤维细胞[24] |
| 1999 | 猪(Sus scrofa) | 胚胎成纤维细胞[26]、 壁层颗粒细胞[25] |
| 2002 | 兔(Oryctolagus cuniculus) | 成体卵丘颗粒细胞[28] |
| 2002 | 猫(Felis domesticus) | 成体卵丘颗粒细胞[27] |
| 2003 | 马(Equus caballus) | 成体成纤维细胞[32] |
| 2003 | 大鼠(Rattus norvegicus) | 胚胎成纤维细胞[29] |
| 2005 | 狗(Canis lupus familiaris) | 成体成纤维细胞[30] |
| 2006 | 雪貂(Mustela putorius furo) | 成体卵丘颗粒细胞[33] |
| 2007 | 水牛(Bubalus bubalis) | 胚胎成纤维细胞和 壁层颗粒细胞[34] |
| 2010 | 骆驼(Camelus dromedarius) | 成体卵丘颗粒细胞[35] |
| 2018 | 食蟹猴(Macaca fasciculari)s | 胚胎成纤维细胞[36] |
新窗口打开|下载CSV
自1990年后,中国科学家也扎根于祖国大地进行了多种哺乳动物的克隆研究工作,并利用最新的技术获得了胚胎细胞或体细胞克隆动物。在克隆羊领域,1991年西北农林科技大学张涌教授团队成功获得了胚胎细胞核移植的山羊,并在2000年成功获得了存活的、有生育能力的体细胞克隆山羊“阳阳”。在克隆牛领域,1995年中国第一头胚胎细胞克隆牛在广西农业大学完成。2002年,中国科学院动物研究所陈大元研究员在国家自然科学基金重点项目支持下,利用牛耳上皮成纤维细胞为供体细胞,获得了14头存活的克隆牛,由此真正实现了中国体细胞克隆牛的零突破。在克隆猪方面,2005年中国农业大学李宁教授研究团队成功获得了体细胞克隆香猪;2007年,东北农业大学刘忠华教授研究团队成功完成了东北农民猪的体细胞克隆猪的制作[37]。
1999年之后,陈大元研究员进一步继承并发展了其恩师童第周先生的异种克隆工作,展开了包括大熊猫在内的多种哺乳动物之间的异种克隆研究,并对异种克隆胚胎的体外发育潜能、异种着床情况及供体和受体线粒体含量的动态变化特征等展开了系统研究[38,39,40,41,42,43,44,45,46,47]。陈大元研究员所领导的团队将大熊猫的体细胞核移植到去核的兔卵母细胞中,发现这些异种克隆胚胎有一定比例可在体外发育至囊胚阶段。他们还进一步将这些异种克隆胚胎移植入假孕兔或猫的输卵管内,观察并记录了异种胚胎的着床和发育情况[40, 41]。
1.3 核移植中的表观遗传研究
21世纪,随着克隆技术和相关仪器的进一步优化以及测序技术尤其是微量组学技术的发展,克隆胚胎的研究进入了分子水平和人类疾病治疗探索阶段。继童第周先生之后的中国第三代克隆胚胎研究的科学家,也登上了世界舞台。多年以来,体细胞核移植的效率一直较低,其中供体细胞DNA甲基化和组蛋白修饰的不完全重编程被认为是造成低效率的重要原因。英国剑桥大学Babraham研究所Reik教授领导的团队发现核移植胚胎中存在异常的甲基化水平,原第二军医大学刘厚奇教授团队和东北农业大学刘忠华教授团队也分别证实降低核移植胚胎中的DNA甲基化有助于提高核移植胚胎的发育潜能[48,49,50,51]。此外,许多报道也证实了核移植胚胎中存在异常的组蛋白修饰,而组蛋白乙酰化酶抑制剂TSA和Scriptaid可显著提高核移植胚胎的发育率[52]。2007年,北京生命科学研究所高绍荣教授课题组揭示了核移植胚胎发育第一个细胞分裂周期中,组蛋白H3K9、H3K14、H4K16、H4K8和H4K12上乙酰化修饰的动态变化规律,并进一步发现H3K9me3和H3K9me2在核移植后逐渐去甲基化[53]。2011年,中国科学院生物化学与细胞生物学研究所李劲松研究员课题组将克隆囊胚的内细胞团与正常胚胎融合后的四倍体胚胎进行聚合,由此替换了克隆胚胎的滋养外胚层细胞,不仅将克隆小鼠的生率提高了6倍,也同时证实了克隆囊胚的滋养外胚层存在缺陷[54]。此后,高绍荣教授课题组还证实克隆技术相比于iPS技术,能更好地恢复体细胞中受损的端粒长度及线粒体功能[55],并进一步利用超高难度的单胚胎多次显微操作技术,深入探讨了小鼠受精卵中雌雄原核的不对称重编程能力,证明了重编程因子主要集中在雄原核内[56]。2014年,哈佛大学霍华德休斯医学研究所张毅教授提出体细胞基因组H3K9me3是体细胞核移植的一个主要障碍,过表达H3K9去甲基化酶或降低体细胞中H3K9me3水平能极大改善核移植的效率[57]。此后,同济大学高绍荣教授课题组和张勇教授课题组联合通过建立早期克隆胚胎命运追溯系统,结合单细胞/微量细胞测序技术,绘制了不同发育命运胚胎细胞的基因表达图谱,也进一步证实H3K9me3是体细胞核移植胚胎表观重编程的一个主要障碍,同时也证明体细胞基因组上H3K4me3的存在也是体细胞基因组不能被完美重编程的一个障碍[58]。2018年,张毅教授和高绍荣教授课题组再次发表相关工作,分别证明H3K27me3和DNA再甲基化是克隆胚胎发育的表观遗传障碍[59, 60]。1.4 人克隆胚胎干细胞研究
多种哺乳动物的卵母细胞均具有重编程体细胞的能力,那么建立人(Homo sapiens)的克隆胚胎干细胞系,似乎为胚胎干细胞及其分化细胞的应用提供了非常好的供体细胞来源。然而,出于道德及伦理学上的考虑,许多国家法律明令禁止人类克隆实验。人类克隆胚胎干细胞研究相对于传统的人类胚胎干细胞研究,面临着更大的道德挑战,因为建立人类克隆胚胎干细胞系需要他人胚胎的供给,这可能造成人类胚胎的非法买卖,使得人类胚胎变为建立克隆细胞系的工具。此外,有限的人类卵母细胞数量也是人类体细胞核移植研究中不得不面临的难题。人类克隆胚胎干细胞的研究至关重要的一点既是建立人类克隆胚胎干细胞系。过去的十几年里,全世界的科学家一直在致力于人类细胞相关的克隆研究,却均未能成功获得人类克隆胚胎干细胞系。虽然在2005年韩国黄禹锡教授领导的团队声称建立了病人特异的胚胎干细胞,但这一研究却被最终定性为欺诈丑闻[61]。人类克隆胚胎面临的一个主要问题是克隆胚胎的早期发育阻滞,进而阻止了稳定的胚胎干细胞系的建立,这可能是由于作为供体的成体细胞核不能激活胚胎发育的关键基因所导致,表现为大部分的克隆胚胎在到达8-细胞期之后就失去了进一步发育的潜能[62]。在一些极少的案例中,即使克隆胚胎能够发育至囊胚阶段,稳定的胚胎干细胞系依然不能被成功建立[63]。虽然早期胚胎发育阻滞的根本原因仍不清楚,绝大多数涉及到人类卵母细胞的核移植实验,采用的却是为非灵长类动物设计的核移植实验方法。因此,实验技术方法的不完善也可能是造成相关人类核移植胚胎干细胞建立失败的原因之一。2007年,美国俄勒冈健康与科学大学Mitalipov教授所领导的科研团队以恒河猴的皮肤成纤维细胞作为供核细胞,成功建立了灵长类动物的克隆胚胎干细胞系[64]。因此,有理由推断包括人类在内的灵长类动物的卵母细胞应该与其他哺乳类动物一样,也具有重编程体细胞的能力。在随后几年的科学工作中,Mitalipov教授课题组以恒河猴为实验动物模型,探索灵长类动物核移植技术的操作方法以提高核移植后胚胎的囊胚形成率和建系效率。他们通过调整细胞融合方法、移植后胚胎激活方法以及在培养液中添加一些特殊的因子,最终在2013年建立了世界上首例人类克隆胚胎干细胞系,并发现人类体细胞重编效率与受体人类卵母细胞的质量息息相关[65]。由此,人类体细胞核移植研究迈入了一个崭新的阶段。随后研究人员利用成年人和老年人的体细胞核作为供体,成功构建了人的核移植胚胎干细胞系,并发现在核移植操作后激活之前,略微延长孵育期可以提高克隆胚胎的发育效率[66]。此后,由Ⅰ型糖尿病患者得到的核移植胚胎干细胞也被成功建立,并被证明可以分化形成能产生胰岛素的β细胞[67]。以体细胞核移植技术为基础获得的克隆胚胎干细胞中,几乎所有的线粒体均来自于受体卵母细胞,在面对线粒体功能缺陷相关的疾病时,人核移植技术为患有此类疾病的患者提供了一个可能的治疗手段。然而,传统核移植方法携带的少量供体线粒体可能存在竞争优势,最终在发育过程中取代受体线粒体并发挥功能[68]。2014年,复旦大学医学神经生物学国家重点实验室沙红英博士等利用极体作为供体DNA,在两种不同线粒体遗传背景的小鼠之间进行了核移植研究,证明了极体移植可以极大程度防止母源性遗传疾病的发生,为阻断线粒体遗传病研究提供了思路[69]。2017年,Mitalipov教授课题组实现了人胚胎第一极体的核移植[70]。
1.5 克隆猴的诞生
在1997年“Dolly”羊出生后的20年间,体细胞核移植无论在技术还是理论、细胞还是分子水平上都取得了多项重量级成果,已有超过20种哺乳动物通过体细胞核移植技术,获得了可存活的克隆动物。然而,通过该技术实现非人灵长类动物的克隆却一直未能实现。早在克隆羊工作被报道当年,美国俄勒冈灵长类动物研究中心孟励博士就以恒河猴4-细胞胚胎的卵裂球为供体细胞核,获得了第一只“克隆猴”[71](非体细胞克隆)。2000年,该中心利用胚胎分裂法获得了“克隆猴”Tetra[72](非体细胞克隆)。然而,这两项工作并未获得学术界的较大关注,因为其实际并非体细胞核移植!2018年1月,中国科学院上海神经科学研究所孙强研究员领导的团队,利用TSA和Kdm4d的组合处理,首次真正意义上利用体细胞核移植技术完成了体细胞克隆猴(食蟹猴)的制作,两只存活的食蟹猴分别被命名为“中中”和“华华”(均由胚胎成纤维细胞核为供体进行核移植后获得)[36]。但是,利用更为终末分化的卵丘颗粒细胞作为供体时,克隆猴出生后却未能继续存活,提示非人灵长类的体细胞核移植技术仍然有待进一步改善,更提醒人们不仅在伦理上,现在的技术瓶颈也不允许进行克隆人的尝试。
经过几十年的努力,中国科学家在体细胞核移植、灵长类动物模型研究和早期胚胎发育机制方面取得了一系列重大突破,已经处于世界领先水平。然而,动物克隆依然是一项成功率低、成本高昂的实验,其短期内转化应用的前景仍不明朗。克隆技术虽然被认为可以用于挽救濒临灭绝的动物(如前述陈大元研究员的克隆熊猫研究),甚至可能将那些已经灭绝在历史长河中的动物再度“复活”,但是目前这些尝试仍处于早期的科学研究阶段。然而,就宠物市场而言,克隆狗似乎是目前已知最好的克隆技术的商业化应用。目前,韩国多家单位已经利用该技术,将优质警犬进行克隆用于安保,而竞赛型犬的保种与克隆也具有极大市场。此外,近年来基因编辑技术的发展及其与克隆技术的逐渐结合,也将推进克隆技术的推广和应用。
2 从胚胎干细胞到诱导多能干细胞
干细胞是一类具有自我更新和多向分化能力的细胞,这一“干性”使它们具有巨大的应用潜力。干细胞在生物体内和发育过程中广泛存在,根据其所在的组织和功能可分为造血干细胞、肌肉干细胞和神经干细胞等;根据发育阶段可分为胚胎干细胞和成体干细胞;根据分化潜能可分为全能干细胞、多能干细胞和单能干细胞。这些细胞在组织器官发育、功能维持和损伤修复上起到重要作用。2.1 早期干细胞研究
造血干细胞最早被应用于临床。早在20世纪50年代,美国华盛顿大学医学院E. Donnall Thomas成功地应用双胞胎间的骨髓移植治疗白血病。1969年,经过组织配型和抗抑制剂药物的作用,首次成功地进行了异体骨髓移植。1964年,陆道培院士成功主持完成了亚洲首例骨髓移植,用于治疗再生障碍性贫血,并在1981年成功完成中国首例异基因骨髓移植。吴祖泽院士作为中国造血干细胞研究的奠基人,在国际上首次获得人源性干细胞生长因子,完成了世界首例胎肝造血干细胞移植治疗急性重度骨髓型放射病人,被誉为“中国造血干细胞之父”。
1981年,英国剑桥大学Evans和Kaufman[73]首次通过体外培养从囊胚期小鼠胚胎得到具有多能性的胚胎干细胞。1998年,美国威斯康辛大学Thomson实验室成功建立了人胚胎干细胞[74],使以细胞治疗为基础的再生医学得到更加广泛的关注。从2000年开始,在“973计划”和“863计划”项目的支持下,中国干细胞研究得以蓬勃发展,陆续建立起各个干细胞研究中心,培养了大批人才,并开始在国际上发声。1994年成立的中国科学院分子发育生物学重点实验室(现分子发育生物学国家重点实验室)是中国在发育生物学领域建立最早的重点实验室,通过细胞核移植和转基因技术得到中国第一批克隆羊和转基因羊。1999年,盛慧珍博士从美国NIH回国,在上海市科委和上海第二医科大学的支持下,建立了上海市发育生物学重点实验室,2003年发表了关于异种细胞融合的文章,通过将人皮肤细胞与兔卵细胞融合,获得胚胎干细胞,首次将人类体细胞重编程,证明“治疗性克隆”的可行性[75]。2000年,李凌松博士从美国斯坦福大学回国并创建北京大学干细胞研究中心,该中心长期从事干细胞功能基因及相关疾病应用的研究。此后,李凌松教授作为首席科学家主持了中国胚胎干细胞研究领域最早的“973计划”项目(之一),通过该项目支持了一批目前中国胚胎干细胞研究的中坚力量(周琪院士与裴端卿研究员是该973项目骨干)。此后,李凌松教授还参与创建了中国第一家干细胞公司——“北科生物”。
2.2 诱导多能干细胞
2006年是干细胞研究领域最振奋人心的一年。基于对胚胎干细胞基因表达模式以及其特异性基因的研究,日本京都大学Shinya Yamanaka实验室发现了本世纪干细胞领域最激动人心的研究之一——诱导多能干细胞(induced pluripotent stem cell, iPSC) (图1B)[76]。他们用4个转录因子——Oct4、Sox2、Klf4和c-Myc成功地完成体细胞向iPSC的重编程。这一发现打破了多能干细胞临床应用中细胞来源和伦理问题的局限,可以有效实现病人特异性iPSC的建立及个性治疗,开辟了重编程和再生医学的全新领域。Yamanaka也因为此项贡献与John Gurdon分享了2012年的诺贝尔生理学与医学奖。2007年,Yamanaka和Thomson实验室分别成功建立了人的iPSC[77, 78]。同年,麻省理工学院怀特黑德生物医学研究所Jaenisch实验室将iPS技术应用于镰刀型贫血的小鼠模型的治疗上,提出利用iPSC治疗单基因遗传疾病的策略[79]。2009年,同济大学生命科学与技术学院高绍荣教授(时任北京生命科学研究所研究员)课题组将这一策略应用于地中海贫血症患者来源的细胞,获得病人特异的功能恢复的造血细胞[80]。此后,大量工作报道了利用iPS技术获得病人特异性多能性细胞,可应用于单基因遗传病治疗模型和患者特异性病理筛查。作为多能性细胞,iPSC是否真的具有多能性,是iPS技术投入应用前必须回答的问题,尤其是2006年Yamanaka发表的工作中iPSC甚至没能产生嵌合体小鼠。2009年,周琪院士和高绍荣教授两个课题组同时发表独立工作,通过四倍体胚胎互补得到了完全由iPSC来源的iPS小鼠,证明iPSC能够具备完全多能性,至此打破了对iPSC质量的质疑,使iPS技术的发展更进一步,该成果被美国TIMES评为2009年世界十大医学突破之一[81, 82]。2010年,通过对具备和不具备完全多能性的小鼠iPSC的基因表达对比,美国哈佛医学院Hochedlinger和周琪院士课题组都找到了位于染色体12qF1区域的Dlk1-Dio3基因簇,该基因簇的转录产物,尤其是Gtl2和Rian,在嵌合体实验中嵌合率很低,并且在不具备完全多能性的iPS细胞系中被异常沉默,但在具有完全多能性的iPSC中则表达正常,因此该区域的表达情况有潜力作为完全多能性iPSC的候选标记[83, 84]。2017年,邓宏魁教授课题组成功建立同时具有胚胎和胚外组织分化能力的EPSC(extended pluripotent stem cells),更加拓宽了人们对多能细胞的认识和应用期望[85]。
经典的iPSC建立需要利用逆转录病毒将重编程因子导入体细胞使其实现过表达,而无论转基因还是逆转录病毒的使用都是阻碍iPS技术走上临床的重要因素。在数年的时间里,国内外科学家进行了大量尝试,影响基因组稳定性的逆转录病毒可以用腺病毒、瞬时表达质粒甚至mRNA或重组蛋白来代替,而部分重编程因子也不断被发现可以被化学小分子所取代。2013年,北京大学干细胞研究中心邓宏魁教授课题组成功实现了完全利用小分子化合物诱导小鼠体细胞重编程为CiPSC,实现了iPS技术的革命性突破[86]。结合过去研究中业已成熟的培养技术,获得临床使用标准的病人特异性多能干细胞的目标已经触手可及。
重编程过程实现了两种完全不同的细胞类型的转变以及多能性的重新建立,深入探究该过程的机制对于理解“细胞命运如何决定”这一细胞生物学关键科学问题具有重要意义。在十几年的发展中,本领域科学家从基因表达模式、细胞分群分期、动力学模型、信号通路调控、表观遗传变化特点和调控等方面对iPSC建立过程中的分子机制进行了大量深入探究。在此期间,中国科学家也做出了卓越贡献。2010年,裴端卿研究员课题组首次发现维生素C可以大幅提高iPSC的诱导效率[87],该方法在国际上受到广泛认可和应用;同年,该课题组又与加拿大Wrana实验室同时发表独立文章,首次发现重编程早期的MET现象[88]。之后,裴端卿研究员课题组和中国科学院上海生物化学与细胞生物学研究所徐国良研究员课题组合作发表两篇论文,深入阐述了在维生素C大幅提高iPSC诱导效率和重编程MET过程中表观修饰蛋白TET、TDG发挥的重要功能[89, 90]。2013年,裴端卿研究员课题组又发现表观修饰H3K9甲基化是完全重编程的屏障之一[91]。同年,高绍荣教授课题组首次揭示了TET1及5hmC参与了细胞多能性的获取和整体表观体系的重建[92]。邓宏魁教授课题组发现促进分化的关键基因可以替代多能性关键基因,实现体细胞重编程,为研究细胞命运决定提供了全新的视角[93]。2017年和2018年,裴端卿研究员课题组利用ATAC (Assay for Transposase-Accessible Chromatin)测序分别阐述了经典重编程和小分子药物诱导重编程过程中的染色质开放状态的动态变化特点[94, 95]。2018年,利用单细胞RNA测序,邓宏魁教授课题组发现小分子药物诱导重编程过程中存在早期胚胎特点,为理解该过程提供新的线索[96]。
虽然iPSC的机制并未完全解析,多能干细胞的分化方法也没有完全成熟,但基于未来iPSC在临床上获得大量应用的预期前景,中国与日本、英国都开始建立临床级的HLA配型的人iPS细胞库,以期在技术和政策都成熟以后,获得迅速广泛的临床应用。
3 为应用而生的单倍体多能干细胞
所有哺乳动物的体细胞都具有来自双亲的两套基因组,只有一套染色体的单倍性只存在于配子(gamete)中。自然界中很少有单倍体动物,只在少数非脊椎动物中存在单倍体动物,如螨虫(Sarcoptes scabiei)、黄蜂(Wasp)。在脊椎动物中,单倍体动物仅存在于实验研究中。例如,单倍体的斑马鱼胚胎能发育到器官形成但不能发育到成熟阶段[97]。在哺乳动物中,通过激活成熟卵母细胞或者将精子注入到去核卵母细胞中能够分别得到孤雌或者孤雄的单倍体胚胎[98, 99],然而小鼠单倍体胚胎发育到囊胚的效率较低,胚胎植入之后出现明显的发育缺陷,只能发育到卵圆柱期(egg cylinder stage)[100]。2009年,Yi等[101]首次获得青鳉鱼(Oryzias latipes)的单倍体胚胎干细胞。2011年,Elling和Leeb等研究小组同时报道了小鼠孤雌单倍体胚胎干细胞的建立和体外维持并用于遗传筛选[102,103,104]。随后,李劲松研究员和周琪院士课题组同时报道了两种建立孤雄单倍体胚胎干细胞的方法,并突破性地将该孤雄单倍体胚胎干细胞注入到成熟的卵母细胞,成功制作了可发育至成年并具有生殖系传递能力的半克隆小鼠(图1C)[105, 106]。此外,李劲松研究员课题组还发现同时敲除H19和Gtl2差异甲基化区域,可显著提高制作半克隆小鼠的效率[107]。随后,李劲松研究员和周琪院士课题组又分别成功建立了食蟹猴[108]和大鼠[109]的孤雄单倍体胚胎干细胞系。2016年,人孤雌单倍体胚胎干细胞也终于被成功建立[110, 111]。单倍体胚胎干细胞具有干细胞的多能性、多分化潜能和单倍性,通过与基因组编辑系统的联合,具有巨大的基因功能研究和表型筛选的意义,在生物医学研究和隐性基因功能研究方面具有二倍体研究工具不能相比的优势。4 单细胞测序技术支持下的胚胎早期发育研究
早期胚胎发育经历了从全能性受精卵到多能性囊胚的过程,是胚胎干细胞的来源,也是理解细胞命运决定的重要生物过程,其分子基础是基因转录及表观遗传修饰的精确调控。DNA甲基化、组蛋白修饰、核小体定位及染色质高级结构等表观遗传状态,在这些命运转变过程中均扮演着重要角色。尽管国内外科学家已经取得了一系列进展,但目前国际上对于该类命运转变过程中细胞间表观遗传信息的差异、相互作用及其与细胞命运关联的研究仍然比较初步。传统方式使用大量细胞产生的表观遗传组学数据掩盖了细胞间非对称性信息,仅能得到众多细胞的平均信号。因此,近几年单细胞测序技术的长足发展极大地促进了胚胎早期发育的研究。近几年,基于第三代测序技术的单细胞技术,特别是以多重退火和成环循环扩增(multiple annealing and looping-based amplification cycles, MALBAC)技术为代表的单细胞全基因组测序技术,以及以SMART-seq技术为代表的单细胞mRNA测序技术等得到了快速的发展,并被广泛地应用于稀缺材料或细胞个体差异巨大的研究领域中,极大地扩展了人们对于发育、生殖以及癌症等相关领域的了解。无论是技术还是科研成果,中国多家研究单位已在该领域达到了国际顶尖水平。2013年,北京大学汤富酬教授课题组和同济大学范国平教授课题组,率先利用微量RNA-seq技术,分别报道了人和鼠胚胎发育过程中转录组的动态变化[112, 113]。2014年,北京大学谢晓亮教授课题组、汤富酬教授课题组和北京大学第三医院乔杰院士课题组联合利用单细胞DNA检测技术,在不破坏卵和胚胎本身结构的情况下,建立了母源性遗传病胚胎诊断的新方法,世界上第一例“MALBAC婴儿”在北医三院诞生,标志着中国胚胎植入前遗传诊断技术处于世界领先水平[114]。此外,汤富酬教授课题组首次建立了单细胞RRBS (reduced representation bisulfite sequencing)方法[115],并利用微量甲基化测定技术,研究了人类早期胚胎发育和原始生殖细胞发育中的甲基化组改变[116, 117]。中国科学院北京基因组研究所刘江研究员使用斑马鱼[118]和小鼠[119]为模型,揭示了子代甲基化图谱继承和重编程的规律,研究成果首次证明除了基因组DNA外,表观遗传信息也可以遗传到子代。2016年,高绍荣教授课题组和清华大学颉伟教授课题组同时发表论文,利用微量细胞ChIP-seq技术,从全基因组水平上揭示了小鼠从配子到植入前胚胎发育过程中组蛋白修饰H3K4me3和H3K27me3遗传和重编程的模式和分子机制[120, 121]。2018年,高绍荣教授课题组的研究进一步揭示了异染色质的标志组蛋白修饰H3K9me3在配子细胞以及受精后和早期胚胎发育过程中的重编程与其在逆转座子沉默中的作用及调控机制[122]。2017年,刘江研究员课题组和颉伟教授课题组同时发表文章,揭示染色体3D高级结构在配子中的特殊状态以及如何在胚胎发育过程中变化而影响胚胎的发育[123, 124]。同年,汤富酬教授课题组又开发出单细胞多组学测序技术,首次在单细胞分辨率,解析了人类着床前胚胎发育过程中DNA甲基化组和染色质状态组的重编程过程,以及染色质状态与DNA甲基化之间的相互关系等关键生物学特征[125]。
由于单细胞测序技术的简化和成本大幅降低,对组织中数以万计的细胞进行单细胞测序成为可能。2018年,浙江大学郭国骥教授课题组发表了里程碑式的研究成果,利用自主开发的一套完全国产化的Microwell-seq高通量单细胞测序平台,对来自小鼠近50种器官和组织40余万个细胞进行了系统性的单细胞转录组分析,并构建了首个哺乳动物细胞图谱[126]。同年,汤富酬教授和中国科学院生物物理研究所王晓群研究员课题组绘制了人胚胎前额皮层和消化系统发育过程的单细胞图谱[127, 128]。相信在不久的将来,解析胚胎发育中各个谱系细胞分化过程中的表达图谱和分子机制将被解析,并促使干细胞体外分化体系不断完善,助力干细胞治疗真正走上临床。
5 结语与展望
近几十年是干细胞领域飞速发展的时期,优秀成果不胜枚举。随着中国经济实力的发展壮大,科研实力也在稳步增强,加之经费和政策的支持,干细胞研究领域达到了国际并跑甚至领跑的水平,在体细胞核移植、诱导多能干细胞、单倍体多能干细胞和胚胎早期发育方面都做出了巨大的贡献。近年来,单细胞多水平测序技术的发展极大地推动了人们对重编程和胚胎发育的理解,也将对重编程和干细胞的机制做出更为细致的解读。同时,对组织器官发育过程全细胞的单细胞测序,对细胞谱系的产生、维持、分化以及功能形成都提供了巨大信息,为干细胞体外分化技术的成熟起到重要的促进作用。相信在不久的将来,干细胞将在许多临床应用方向上大放异彩,而中国也将是其高速发展的舞台。(责任编委: 王晓群)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:23166393 [本文引用: 1]

Somatic cell nuclear transfer (SCNT) cloning is the sole reproductive engineering technology that endows the somatic cell genome with totipotency. Since the first report on the birth of a cloned sheep from adult somatic cells in 1997, many technical improvements in SCNT have been made by using different epigenetic approaches, including enhancement of the levels of histone acetylation in the chromatin of the reconstructed embryos. Although it will take a considerable time before we fully understand the nature of genomic programming and totipotency, we may expect that somatic cell cloning technology will soon become broadly applicable to practical purposes, including medicine, pharmaceutical manufacturing and agriculture. Here we review recent progress in somatic cell cloning, with a special emphasis on epigenetic studies using the laboratory mouse as a model.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:20535199 [本文引用: 1]

The stable states of differentiated cells are now known to be controlled by dynamic mechanisms that can easily be perturbed. An adult cell can therefore be reprogrammed, altering its pattern of gene expression, and hence its fate, to that typical of another cell type. This has been shown by three distinct experimental approaches to nuclear reprogramming: nuclear transfer, cell fusion and transcription-factor transduction. Using these approaches, nuclei from 'terminally differentiated' somatic cells can be induced to express genes that are typical of embryonic stem cells, which can differentiate to form all of the cell types in the body. This remarkable discovery of cellular plasticity has important medical applications.
URLPMID:16589125 [本文引用: 1]

Proc Natl Acad Sci U S A. 1952 May;38(5):455-63.
URL [本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URLPMID:5530658 [本文引用: 1]

IT has yet to be proved that somatic cells of an adult animal possess genes other than those necessary for their own growth and differentiation. A particularly stringent test of the genetic content of the nucleus of a somatic cell is provided by transplanting it to an enucleated egg. The development of the egg tests the capacity of the genes in the transplanted nucleus to elicit normal early development and differentiation. Although this method has been applied to certain cells of embryos, larvae 1,2 and newly metamorphosed froglets 3 , it has not been possible, until now, to transfer nuclei successfully from the cells of normal adult animals.
URLPMID:1102625 [本文引用: 1]

Nuclei from keratinized skin cells of adult Xenopus foot-webs have been transplanted to enucleated eggs of the same species. 2. The cells used to provide donor nuclei were obtained as a monolayer outgrowth from cultured foot-web explants. When explants were cultured without plasma for 3 days, over 99-9% of the outgrowth cells contained keratin as revealed by the binding of monospecific fluorescent antibody prepared against purified Xenopus keratin. Nuclei were transplanted from cells which had been cultured for 31/2 days. 3. None of the first transfer embryos developed as far as tadpoles. Eleven clones of embryos were prepared from the nuclei of partial first-transfer blastulae by use of serial nuclear transplantation. Eight of these clones contained swimming tadpoles with functional muscle and nerve cells, and six clones contained tadpoles with beating hearts, well differentiated eyes, and other organs. 4. To prove that the nuclei of nuclear-transplant tadpoles were derived from the transplanted skin cell nuclei and not from a failure of ultraviolet light to inactivate the recipient egg nucleus, 1-nu skin cell nuclei were transplanted to eggs laid by 2-nu frogs. Several advanced tadpoles from six clones were analysed for nucleolar and chromosome number and found to be 1-nu diploids. 5. The six clones of advanced tadpoles which were proved to carry the donor nuclear marker represent six first-transfer nuclei in a total sample of 129 skin cell nuclei originally transplanted. The probability that all six nuclei were derived from the 0-1% of the donor cell population not proved to contain keratin is less than one in 10(10). 6. We conclude that cell specialization does not involve any loss, irreversible inactivation or permanent change in chromosomal genes required for development.
[本文引用: 1]
[本文引用: 1]
URLPMID:1207752 [本文引用: 1]

SINCE the experiments of Briggs and King 1 , in which nuclei from blastula cells were transplanted by microinjection into enucleated frogs' eggs, the technique of nuclear transplantation has been used extensively to study interactions between nucleus and cytoplasm during differentiation in amphibians 2–5 . But severe difficulties are encountered in attempts to apply this technique to the mammalian egg. The rabbit egg, with a diameter of 100 08m, is 1,000 times smaller in volume than a frog's egg and the mouse egg has a volume one-third that of a rabbit's egg. An alternative and technically easier method of transplanting a nucleus is by inducing fusion of an egg and a cell with Sendai virus. The technique of virus-induced fusion, originally developed for hybridising somatic cells 6–7 , has been adapted to fuse somatic cells with unfertilised or fertilised mouse eggs 8–13 . Although Sendai virus was shown not to harm mouse embryos in these experiments and cells were successfully fused, the transplanted nuclei did not survive. Unfertilised mouse eggs which were fused with cells from embryos at the 2–8-cell stage failed to cleave 8 .
URLPMID:6141214 [本文引用: 1]

Abstract The ability of foreign nuclei to support development in nuclear transplantation manipulations has proven an effective means to assess the consequences of nuclear differentiation. In addition, nuclear transplantation might serve to define the persistence and role of maternally inherited cytoplasmic constitutents during embryogenesis. We have extended the use of a technique that enables the efficient transfer of one-cell-stage pronuclei into the cytoplasm of enucleated mouse embryos, and have successfully transferred two-, four-, eight-cell-stage and inner cell mass (ICM) cell nuclei. We have also used this technique as a means to determining that the stage-specific embryonic antigen, SSEA-3, is a cytoplasmic contribution of the unfertilized ovum. The potential value of this technique in determining the developmental capacity of nuclei from various embryonic stages, and in determining nuclear/cytoplasmic origins of early embryonic gene products, is discussed.
URLPMID:3951549 [本文引用: 1]

Nuclear transplantation and cell fusion techniques have proved valuable for embryological studies in several non-mammalian animal species 1 . More recently these procedures have been used successfully in small laboratory mammals, notably the mouse, to investigate the ability of nuclei and cytoplasm from various sources to produce viable embryos when combined 2鈥6 . The use of a similar approach to study the developmental biology of large domestic animals presents a number of technical and practical difficulties and so far there has been no report of attempts to perform nuclear transplantation in sheep embryos. Here I describe such a procedure and its use to investigate the development of embryos in which whole blastomeres from 8- and 16-cell embryos were combined with enucleated or nucleated halves of unfertilized eggs. The procedure involves bisection of single-cell eggs in a medium containing cytochalasin; fusion of egg halves with single blastomeres, induced using Sendai virus or an electrofusion apparatus; and embedding in agar, followed by culture of the reconstituted embryos in the ligated oviducts of ewes in dioestrus. I show that fully viable embryos may be obtained by this procedure.
URLPMID:3196797 [本文引用: 1]

The first six genetically verified nuclear transplant have been produced in this study. Individual eight-cell stage embryo blastomeres were transferred and fused with enucleated mature oocytes of which six full-term offspring were produced out of 164 manipulated eggs. The following efficiency rates were determined for the nuclear transplantation procedure: chromosomal removal from oocytes, 92%; fusion rate, 84%; activation rate, 46%; embryo transfer rate, 27%. Additional reasons for the low efficiency rate of nuclear transplant embryos may include limited due to in recipient oocytes and asynchronous transfers of manipulated embryos to recipient females. The successful to term may have been due to the ability of the mature oocyte to reprogram the eight-cell stage nuclei. The number of cells in blastocysts derived from isolated eight-cell blastomeres (18 +/-.08) was lower than that of nonmanipulated pronuclear (106 +/- 5.1) and nuclear transplant embryos derived from eight-cell stage nuclei (91 +/- 10.2) (p less than 0.001). This evidence along with the significant amount of nuclear swelling in nuclear transplant embryos and a delay in the time of indicate that nuclear reprogramming had taken place in these embryos. Successful nuclear reprogramming indicates that serial transfers could result in the expanded multiplication of mammalian embryos.
URLPMID:2590712 [本文引用: 1]

Nuclear transfer was evaluated in early porcine embryos. Pronuclear stage embryos were centrifuged, treated with cytoskeletal inhibitors, and subsequently enucleated. Pronuclei containing karyoplasts were placed in the perivitelline space of the enucleated zygote and fused to the enucleated zygote with electrofusion. The resulting pronuclear exchange embryos were either monitored for cleavage in vitro (9/13 cleaved and contained 2 nuclei after 24 h, 69%) or for in vivo development. In vivo development after 3 days resulted in 14/15 (93%) of the embryos transferred cleaving to the greater than or equal to 4-cell stage and after 7 days 6/16 (38%) reaching the expanded blastocyst stage. A total of 56 pronuclear exchange embryos were allowed to go to term, and 7 piglets were born. A similar manipulation procedure was used to transfer 2-, 4- or 8-cell nuclei to enucleated, activated meiotic metaphase II oocytes. Enucleation was effective in 74% (36/49) of the contemporary oocytes. Activation was successful in 81% (37/46) of nonmanipulated but pulsed oocytes versus 13% (4/31) of control oocytes (p less than 0.01). After 6 days in vivo, 9% (1/11) of the 2-cell nuclei, 8% (7/83) of the 4-cell nuclei, and 19% (11/57) of the 8-cell nuclei transferred to enucleated, activated meiotic metaphase II oocytes resulted in development to the compact morula or blastocyst stage (p less than 0.01). A total of 88 nuclear transfer embryos were transferred to recipient gilts for continued development. A single piglet was born after the transfer of a 4-cell nucleus to an enucleated, activated metaphase II oocyte and subsequent in vivo development.(ABSTRACT TRUNCATED AT 250 WORDS)
URL [本文引用: 1]
URLPMID:8016127 [本文引用: 1]

We report here the isolation and in vitro culture of bovine inner cell mass (ICM) cells and the use of ICM cells in nuclear transfer to produce totipotent blastocysts that resulted in calves born. Of 15 cell lines represented in this study, 13 were derived from immunosurgically isolated ICM of 3 in vitro produced day 9-10 bovine blastocysts, while 2 lines were derived from single blastocysts. Approximately 70% of attempted cell lines became established cell lines when started from 3 ICMs. The ability to establish cell lines was dependent on the number of ICMs starting the line. Sire differences were noted in the ability of ICMs to establish cell lines and to form blastocysts. The cell lines were cultured as a low cell density suspension in the medium CR1aa plus selenium, insulin, and transferrin (SIT) and 5% fetal calf serum (FCS) for 6-101 days before use in nuclear transfer, at which time some had multiplied to more than 2000 cells. If allowed to aggregate, cells of established cell lines formed embryoid bodies. A total of 659 nuclear transfer clones were made by fusing the ES cells into enucleated oocytes with polyethylene glycol; 460 of these fused, based on cleavage (70%). After culture of the clones for 7 days in vitro in CR1aa/SIT/5% FCS, 109 (24%) of those fused became blastocysts. Thirty-four blastocysts were transferred into uteri of 27 cows, and 13 cows (49%) became pregnant. Four of the 13 cows gave birth to 4 normal calves. DNA typing showed the calves to be derived from the respective sires of the cell lines. The calves were derived from cultures of less than 28 days.
URLPMID:9241063 [本文引用: 1]

Genetically identical nonhuman primates can provide a powerful animal model for gene therapy and research activities where the physiological parameters directly or indirectly under study are heritable. Here we demonstrate that nuclear transfer is a viable technology for the production of identical rhesus macaques. Oocytes recovered from gonadotropin-treated females were enucleated by aspiration of the first polar body and underlying ooplasm, then activated by cycloheximide exposure. Individual diploid blastomeres, recovered from in vitro-fertilization-produced embryos (either fresh or frozen-thawed) and used as nuclear donors, were injected under the zona pellucida of enucleated (chromosome-free) oocytes and fused by electric pulses. The reconstituted embryos were cocultured on buffalo rat liver cells before cryostorage and transfer to synchronized host mothers. Of the 9 females receiving a total of 29 reconstituted embryos, 3 became pregnant, with two live births resulting, one male and one female. The parentage of both infants was established unequivocally by genotype analysis at 7 highly variable short tandem repeat loci.
URLPMID:8598906 [本文引用: 1]

Abstract Nuclear transfer has been used in mammals as both a valuable tool in embryological studies and as a method for the multiplication of 'elite' embryos. Offspring have only been reported when early embryos, or embryo-derived cells during primary culture, were used as nuclear donors. Here we provide the first report, to our knowledge, of live mammalian offspring following nuclear transfer from an established cell line. Lambs were born after cells derived from sheep embryos, which had been cultured for 6 to 13 passages, were induced to quiesce by serum starvation before transfer of their nuclei into enucleated oocytes. Induction of quiescence in the donor cells may modify the donor chromatin structure to help nuclear reprogramming and allow development. This approach will provide the same powerful opportunities for analysis and modification of gene function in livestock species that are available in the mouse through the use of embryonic stem cells.
URL [本文引用: 2]
URLPMID:9851933 [本文引用: 2]

Eight calves were derived from differentiated cells of a single adult cow, five from cumulus cells and three from oviductal cells out of 10 embryos transferred to surrogate cows (80 percent success). All calves were visibly normal, but four died at or soon after birth from environmental causes, and postmortem analysis revealed no abnormality. These results show that bovine cumulus and oviductal epithelial cells of the adult have the genetic content to direct the development of newborn calves.
URLPMID:9690471 [本文引用: 2]

Until recently, fertilization was the only way to produce viable mammalian offspring, a process implicitly involving male and female gametes. However, techniques involving fusion of embryonic or fetal somatic cells with enucleated oocytes have become steadily more successful in generating cloned young. Dolly the sheep was produced by electrofusion of sheep mammary-derived cells with enucleated sheep oocytes. Here we investigate the factors governing embryonic development by introducing nuclei from somatic cells (Sertoli, neuronal and cumulus cells) taken from adult mice into enucleated mouse oocytes. We found that some enucleated oocytes receiving Sertoli or neuronal nuclei developed in vitro and implanted following transfer, but none developed beyond 8.5 days post coitum; however, a high percentage of enucleated oocytes receiving cumulus nuclei developed in vitro. Once transferred, many of these embryos implanted and, although most were subsequently resorbed, a significant proportion (2 to 2.8%) developed to term. These experiments show that for mammals, nuclei from terminally differentiated, adult somatic cells of known phenotype introduced into enucleated oocytes are capable of supporting full development.
URLPMID:10331804 [本文引用: 2]

In this study, we demonstrate the production of transgenic by nuclear transfer of fetal somatic cells. Donor karyoplasts were obtained from a primary fetal somatic cell line derived from a 40-day transgenic female fetus produced by artificial of a nontransgenic adult female with semen from a transgenic male. Live offspring were produced with two nuclear transfer procedures. In one protocol, oocytes at the arrested II stage were enucleated, electrofused with donor somatic cells, and simultaneously activated. In the second protocol, activated in vivo oocytes were enucleated at the II stage, electrofused with donor somatic cells, and simultaneously activated a second time to induce genome reactivation. Three healthy identical female offspring were born. Genotypic analyses confirmed that all cloned offspring were derived from the donor cell line. Analysis of the milk of one of the transgenic cloned showed high-level production of , similar to the parental transgenic line.
URLPMID:10993078 [本文引用: 2]

Since the first report of live mammals produced by nuclear transfer from a cultured differentiated cell population in 1995 (ref. 1), successful development has been obtained in sheep, cattle, mice and goats using a variety of somatic cell types as nuclear donors. The methodology used for embryo reconstruction in each of these species is essentially similar: diploid donor nuclei have been transplanted into enucleated MII oocytes that are activated on, or after transfer. In sheep and goat pre-activated oocytes have also proved successful as cytoplast recipients. The reconstructed embryos are then cultured and selected embryos transferred to surrogate recipients for development to term. In pigs, nuclear transfer has been significantly less successful; a single piglet was reported after transfer of a blastomere nucleus from a four-cell embryo to an enucleated oocyte; however, no live offspring were obtained in studies using somatic cells such as diploid or mitotic fetal fibroblasts as nuclear donors. The development of embryos reconstructed by nuclear transfer is dependent upon a range of factors. Here we investigate some of these factors and report the successful production of cloned piglets from a cultured adult somatic cell population using a new nuclear transfer procedure.
URLPMID:10947985 [本文引用: 2]

Pig cloning will have a marked impact on the optimization of meat production and xenotransplantation. To clone pigs from differentiated cells, we microinjected the nuclei of porcine (Sus scrofa) fetal fibroblasts into enucleated oocytes, and development was induced by electroactivation. The transfer of 110 cloned embryos to four surrogate mothers produced an apparently normal female piglet. The clonal provenance of the piglet was indicated by her coat color and confirmed by DNA microsatellite analysis.
URLPMID:11859353 [本文引用: 2]

Sheep, mice, cattle, goats and pigs have all been cloned by transfer of a donor cell nucleus into an enucleated ovum, and now we add the successful cloning of a cat (Felis domesticus) to this list. However, this cloning technology may not be readily extendable to other mammalian species if our understanding of their reproductive processes is limited or if there are species-specific obstacles.
URLPMID:11923842 [本文引用: 2]

Abstract We have developed a method to produce live somatic clones in the rabbit, one of the mammalian species considered up to now as difficult to clone. To do so, we have modified current cloning protocols proven successful in other species by taking into account both the rapid kinetics of the cell cycle of rabbit embryos and the narrow window of time for their implantation after transfer into foster recipients. Although our method still has a low level of efficiency, it has produced several clones now proven to be fertile. Our work indicates that cloning can probably be carried out successfully in any mammalian species by taking into account physiological features of their oocytes and embryos. Our results will contribute to extending the use of rabbit models for biomedical research.
URL [本文引用: 2]
URLPMID:16079832 [本文引用: 2]

Abstract Several mammals--including sheep, mice, cows, goats, pigs, rabbits, cats, a mule, a horse and a litter of three rats--have been cloned by transfer of a nucleus from a somatic cell into an egg cell (oocyte) that has had its nucleus removed. This technology has not so far been successful in dogs because of the difficulty of maturing canine oocytes in vitro. Here we describe the cloning of two Afghan hounds by nuclear transfer from adult skin cells into oocytes that had matured in vivo. Together with detailed sequence information generated by the canine-genome project, the ability to clone dogs by somatic-cell nuclear transfer should help to determine genetic and environmental contributions to the diverse biological and behavioural traits associated with the many different canine breeds.
URL [本文引用: 1]
URLPMID:12904778 [本文引用: 1]

Abstract Several animal species, including sheep, mice, cattle, goats, rabbits, cats, pigs and, more recently, mules have been reproduced by somatic cell cloning, with the offspring being a genetic copy of the animal donor of the nuclear material used for transfer into an enucleated oocyte. Here we use this technology to clone an adult horse and show that it is possible to establish a viable, full-term pregnancy in which the surrogate mother is also the nuclear donor. The cloned offspring is therefore genetically identical to the mare who carried it, challenging the idea that maternal immunological recognition of fetal antigens influences the well-being of the fetus and the outcome of the pregnancy.
URLPMID:1892907 [本文引用: 1]

Abstract Somatic cell nuclear transfer (SCNT) offers great potential for developing better animal models of human disease. The domestic ferret (Mustela putorius furo) is an ideal animal model for influenza infections and potentially other human respiratory diseases such as cystic fibrosis, where mouse models have failed to reproduce the human disease phenotype. Here, we report the successful production of live cloned, reproductively competent, ferrets using species-specific SCNT methodologies. Critical to developing a successful SCNT protocol for the ferret was the finding that hormonal treatment, normally used for superovulation, adversely affected the developmental potential of recipient oocytes. The onset of Oct4 expression was delayed and incomplete in parthenogenetically activated oocytes collected from hormone-treated females relative to oocytes collected from females naturally mated with vasectomized males. Stimulation induced by mating and in vitro oocyte maturation produced the optimal oocyte recipient for SCNT. Although nuclear injection and cell fusion produced mid-term fetuses at equivalent rates (approximately 3-4%), only cell fusion gave rise to healthy surviving clones. Single cell fusion rates and the efficiency of SCNT were also enhanced by placing two somatic cells into the perivitelline space. These species-specific modifications facilitated the birth of live, healthy, and fertile cloned ferrets. The development of microsatellite genotyping for domestic ferrets confirmed that ferret clones were genetically derived from their respective somatic cells and unrelated to their surrogate mother. With this technology, it is now feasible to begin generating genetically defined ferrets for studying transmissible and inherited human lung diseases. Cloning of the domestic ferret may also aid in recovery and conservation of the endangered black-footed ferret and European mink.
URLPMID:17475931 [本文引用: 1]

Cloning of buffalos (Bubalus bubalis) through nuclear transfer is a potential alternative approach in genetic improvement of buffalos. However, to our knowledge, cloned offspring of buffalos derived from embryonic, fetal, or somatic cells have not yet been reported. Thus, factors affecting the nuclear transfer of buffalo somatic cells were examined, and the possibility of cloning buffalos was explored in the present study. Treatment of buffalo fibroblasts and granulosa cells with aphidicolin plus serum starvation resulted in more cells being arrested at the G0/G1 phase, the proportion of cells with DNA fragmentation being less, and the number of embryos derived from these cells that developed to blastocysts being greater. In addition, a difference was found in the development of embryos reconstructed with fetal fibroblasts from different individuals (P < 0.001). Forty-two blastocysts derived from granulosa cells and fetal fibroblasts were transferred into 21 recipient swamp buffalos, and 4 recipients were confirmed to be pregnant by rectal palpation on Day 60 of gestation. One recipient received two embryos from fetal fibroblasts aborted on Day 300 of gestation and delivered two female premature calves. Three recipients maintained pregnancy to term and delivered three female cloned calves after Days 338-349 of gestation. These results indicate that buffalo embryos derived from either fetal fibroblasts or granulosa cells can develop to the term of gestation and result in newborn calves.
URLPMID:19812298 [本文引用: 1]

Objective: In this study, we demonstrate the use of somatic cell nuclear transfer to produce the first cloned camelid, a dromedary camel (Camelus dromedarius) belonging to the family Camelidae.Materials and Methods: Donor karyoplasts were obtained from adult skin fibroblasts, cumulus cells, or fetal fibroblasts, and in vivo-matured oocytes, obtained from preovulatory follicles of superstimulated female camels by transvaginal ultrasound guided ovum pick-up, were used as cytoplasts. Reconstructed embryos were cultured in vitro for 7 days up to the hatching/hatched blastocyst stage before they were transferred to synchronized recipients on Day 6 after ovulation.Results: Pregnancies were achieved from the embryos reconstructed from all cell types, and a healthy calf, named Injaz, was born from the pregnancy by an embryo reconstructed with cumulus cells. Genotype analyses, using 25 dromedary camel microsatellite markers, confirmed that the cloned calf was derived from the donor cell line and the ovarian tissue.Conclusion: The present study reports, for the first time, establishment of pregnancies and birth of the first cloned camelid, a dromedary camel (C. dromedarius), by use of somatic cell nuclear transfer. This has opened doors for the amelioration and preservation of genetically valuable animals like high milk producers, racing champions, and males of high genetic merit in camelids. We also demonstrated, for the first time, that adult and fetal fibroblasts can be cultured, expanded, and frozen without losing their ability to support the development of nuclear transfer embryos, a technology that may potentially be used to modify fibroblast genome by homologous recombination so as to generate genetically altered cloned animals.
URLPMID:29395327 [本文引用: 2]

Generation of genetically uniform non-human primates may help to establish animal models for primate biology and biomedical research. In this study, we have successfully cloned cynomolgus monkeys ( Macaca fascicularis ) by somatic cell nuclear transfer (SCNT). We found that injection of H3K9me3 demethylase Kdm4d mRNA and treatment with histone deacetylase inhibitor trichostatin A at one-cell stage following SCNT greatly improved blastocyst development and pregnancy rate of transplanted SCNT embryos in surrogate monkeys. For SCNT using fetal monkey fibroblasts, 6 pregnancies were confirmed in 21 surrogates and yielded 2healthy babies. For SCNT using adult monkey cumulus cells, 22 pregnancies were confirmed in 42surrogates and yielded 2 babies that were short-lived. In both cases, genetic analyses confirmed that the nuclear DNA and mitochondria DNA of the monkey offspring originated from the nucleus donor cell and the oocyte donor monkey, respectively. Thus, cloning macaque monkeys by SCNT is feasible using fetal fibroblasts.
[本文引用: 1]
URLPMID:17034044 [本文引用: 1]

Abstract Interspecies nuclear transfer is an invalulable tool for studying nucleus–cytoplasm interactions; and at the same time, it provides a possible alternative to clone endangered animals whose oocytes are difficult to obtain. In the present study, we investigated the possibility of cloning Tibetan antelope embryos using abattoir-derived caprine oocytes as recipients. Effects of culture conditions, enucleation timing, and donor cell passages on the in vitro development of Tibetan antelope-goat cloned embryos were studied. Maternal to zygotic transition timing of interspecies Tibetan antelope embryos was also investigated using two types of cloned embryos, Tibetan antelope-rabbit and Tibetan antelope-goat embryos. Our results indicate that: (1) goat oocyte is able to reprogram somatic cells of different genus and supports development to blastocyst in vitro. (2) Coculture system supported the development of Tibetan antelope-goat embryos to blastocyst rate stage (4.0%), while CR1aa alone did not. (3) When MII phase enucleated caprine cytoplast and TII phase enucleated caprine cytoplast were used as recipients, the fusion rate and blastocyst rate of hybrid embryos were not statistically different (73.9% vs. 67.4%; 4.0% vs. 1.1%). (4) When donor cells at 3–8 passages were used, 2.9% hybrid embryos developed to blastocysts, while none developed to blastocysts when cells at 10–17 passages were used. (5) There may be a morula-to-blastocyst block for Tibetan antelope-goat, while there may be an 8- to 16-cell block for Tibetan antelope-rabbit embryos. Mol. Reprod. Dev. 74: 412–419, 2007. 08 2006 Wiley-Liss, Inc.
URL [本文引用: 1]
URL [本文引用: 2]
[本文引用: 2]
URLPMID:16368565 [本文引用: 1]

Previous reports have indicated that failure in cloning is attributed to the removal of nuclear mitotic apparatus (NuMA) during and subsequent abnormal organization of mitotic apparatus. This study investigated the transformation and assembly of tubulin and during the first of cloned embryos reconstructed by using enucleated oocytes as recipients. After the oocyte fused with a fibroblast, extensive organization was observed around the introduced in most reconstructed embryos, suggesting the introduction of a somatic . A high proportion of fibroblast nuclei transferred into non-activated oocytes underwent premature (PCC), transient and , followed by the of two -like structures. In contrast, fibroblast nuclei in pre-activated ooplasm rarely underwent PCC, but formed a swollen -like structure. Normal were observed in about one third of the cloned embryos reconstructed by both methods. After transferring fibroblasts into NuMA-removed enucleated oocytes, NuMA was localized in pseudo-pronuclei and gradually moved to at the first . NuMA microinjection resulted in disorganization and misalignment, but did not significantly affect early cleavage. Our findings indicate that: 1. NuMA in donor fibroblast may contribute to form a normal in enucleated oocyte; 2. when non-activated cytoplasts and pre-activated cytoplasts are used as recipients, the donor nuclei undergo different morphological changes, but yield similar early ; 3. although abnormal and alignment may cause low efficiency of animal cloning, these abnormalities do not significantly affect early cleavage.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:15349841 [本文引用: 1]

Abstract Interspecies nuclear transfer (INT) has been used as an invaluable tool for studying nucleus-cytoplasm interactions; and it may also be a method for rescuing endangered species whose oocytes are difficult to obtain. In the present study, we investigated interaction of the chicken genome with the rabbit oocyte cytoplasm. When chicken blastodermal cells were transferred into the perivitelline space of rabbit oocytes, 79.3% of the couplets were fused and 9.7% of the fused embryos developed to the blastocyst stage. Both M199 and SOF medium were used for culturing chicken-rabbit cloned embryos; embryo development was arrested at the 8-cell stage obtained in SOF medium, while the rates of morulae and blastocysts were 12.1 and 9.7%, respectively, in M199 medium. Polymerase chain reaction (PCR) amplification of nuclear DNA and karyotype analyses confirmed that genetic material of morulae and blastocysts was derived from the chicken donor cells. Analysis mitochondrial constitution of the chicken-rabbit cloned embryos found that mitochondria, from both donor cells and enucleated oocytes, co-existed. Our results suggest that: (1) chicken genome can coordinate with rabbit oocyte cytoplasm in early embryo development; (2) there may be an 8- to 16-cell stage block for the chicken-rabbit cloned embryos when cultured in vitro; (3) mitochondrial DNA from the chicken donor cells was not eliminated until the blastocyst stage in the chicken-rabbit cloned embryos; (4) factors existing in ooplasm for somatic nucleus reprogramming may be highly conservative.
URL [本文引用: 1]
URLPMID:12840813 [本文引用: 1]

Abstract Interspecies cloning may be used as an effective method to conserve highly endangered species and to support the development of non-human primate animal models for studying therapeutic cloning and nuclear–cytoplasm interaction. The use of the monkey model for biomedical research can avoid legal, ethical, and experimental limitations encountered in a clinical situation. We describe in this study the in vitro development of macaca–rabbit embryos produced by fusing macaca fibroblasts with enucleated rabbit oocytes and examine the fate of mitochondrial DNA in these embryos. We show that macaca–rabbit cloned embryos can develop to the blastocyst stage when cultured in vitro in HECM 10 +10% FBS and that mitochondrial DNA derived from donor somatic cells was detectable in cloned embryos throughout preimplantation development. These results suggest that (1) macaca fibroblast nuclei can dedifferentiate in enucleated metaphase II rabbit oocytes; (2) HECM 10 +10% FBS can break through the development block and support the development of macaca–rabbit cloned embryos to blastocysts; and (3) donor-cell-derived mitochondrial DNA is not eliminated until blastocyst stage. Mol. Reprod. Dev. 65: 396–401, 2003. 08 2003 Wiley-Liss, Inc.
URL [本文引用: 1]
URLPMID:22932499 [本文引用: 1]

The cloning of Dolly the sheep was a remarkable demonstration of the oocyte's ability to reprogram a specialized nucleus. However, embryos derived from such somatic cell nuclear transfer (SCNT) very rarely result in live births-a fate that may be linked to observed epigenetic defects. A new genome-wide study shows that epigenetic reprogramming in SCNT embryos does not fully recapitulate the natural DNA demethylation events occurring at fertilization, resulting in aberrant methylation at some promoters and repetitive elements that may contribute to developmental failure.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:2757202 [本文引用: 1]

Abstract Epigenetic reprogramming plays a central role in the development of cloned embryos generated by somatic cell nuclear transfer, and it is believed that aberrant reprogramming leads to the abnormal development of most cloned embryos. Recent studies show that trimethylation of H3K27 (H3K27me3) contributes to the maintenance of embryonic stem cell pluripotency because the differentiation genes are always occupied by nucleosomes trimethylated at H3K27, which represses gene expression. Here, we provide evidence that differential H3K27me3 modification exists between normal fertilization-produced blastocysts and somatic cell nuclear transfer cloned blastocysts; H3K27me3 was specifically found in cells of the inner cell mass (ICM) of normal blastocysts, whereas there was no modification of H3K27me3 in the ICM of cloned blastocysts. Subsequently, we demonstrated that the differentiation-related genes, which are marked by H3K27me3 in embryonic stem cells, were expressed at significantly higher levels in cloned embryos than in normal embryos. The polycomb repressive complex 2 (PRC2) component genes (Eed, Ezh2, and Suz12), which are responsible for the generation of H3K27me3, were expressed at lower levels in the cloned embryos. Our results suggest that reduced expression of PRC2 component genes in cloned embryos results in defective modification of H3K27me3 to the differentiation-related genes in pluripotent ICM cells. This results in premature expression of developmental genes and death of somatic cloned embryos shortly after implantation. Taken together, these studies suggest that H3K27me3 might be an important epigenetic marker with which to evaluate the developmental potential of cloned embryos.
URL [本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URLPMID:24630997 [本文引用: 1]

Abstract It has been demonstrated that reprogramming factors are sequestered in the pronuclei of zygotes after fertilization, because zygotes enucleated at the M phase instead of interphase of the first mitosis can support the development of cloned embryos. However, the contribution of the parental pronucleus derived from either the sperm or the oocyte in reprogramming remains elusive. Here, we demonstrate that the parental pronuclei have asymmetric reprogramming capacities and that the reprogramming factors reside predominantly in the male pronucleus. As a result, only female pronucleus-depleted (FPD) mouse zygotes can reprogram somatic cells to a pluripotent state and support the full-term development of cloned embryos; male pronucleus-depleted (MPD) zygotes fail to support somatic cell reprogramming. We further demonstrate that fusion of an additional male pronucleus into a zygote greatly enhances reprogramming efficiency. Our data provide a clue to further identify critical reprogramming factors in the male pronucleus. Copyright 2014 The Authors. Published by Elsevier Inc. All rights reserved.
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URLPMID:22120211 [本文引用: 1]

The exchange of the oocyte's genome with the genome of a somatic cell, followed by the derivation of pluripotent stem cells, could enable the generation of specific cells affected in degenerative human diseases. Such cells, carrying the patient's genome, might be useful for cell replacement. Here we report that the development of human oocytes after genome exchange arrests at late cleavage stages in association with transcriptional abnormalities. In contrast, if the oocyte genome is not removed and the somatic cell genome is merely added, the resultant triploid cells develop to the blastocyst stage. Stem cell lines derived from these blastocysts differentiate into cell types of all three germ layers, and a pluripotent gene expression program is established on the genome derived from the somatic cell. This result demonstrates the feasibility of reprogramming human cells using oocytes and identifies removal of the oocyte genome as the primary cause of developmental failure after genome exchange.
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:24746675 [本文引用: 1]

Chung et02al. show that recently developed technology that enables human SCNT reprogramming from embryonic cells is also effective with donor cells from adults, enhancing the therapeutic potential of this approach.
URL [本文引用: 1]
URLPMID:27281217 [本文引用: 1]

Abstract Mitochondrial DNA (mtDNA) mutations are maternally inherited and are associated with a broad range of debilitating and fatal diseases. Reproductive technologies designed to uncouple the inheritance of mtDNA from nuclear DNA may enable affected women to have a genetically related child with a greatly reduced risk of mtDNA disease. Here we report the first preclinical studies on pronuclear transplantation (PNT). Surprisingly, techniques used in proof-of-concept studies involving abnormally fertilized human zygotes were not well tolerated by normally fertilized zygotes. We have therefore developed an alternative approach based on transplanting pronuclei shortly after completion of meiosis rather than shortly before the first mitotic division. This promotes efficient development to the blastocyst stage with no detectable effect on aneuploidy or gene expression. After optimization, mtDNA carryover was reduced to <2% in the majority (79%) of PNT blastocysts. The importance of reducing carryover to the lowest possible levels is highlighted by a progressive increase in heteroplasmy in a stem cell line derived from a PNT blastocyst with 4% mtDNA carryover. We conclude that PNT has the potential to reduce the risk of mtDNA disease, but it may not guarantee prevention.
URLPMID:24949971 [本文引用: 1]

Abstract Inherited mtDNA diseases transmit maternally and cause severe phenotypes. Currently, there is no effective therapy or genetic screens for these diseases; however, nuclear genome transfer between patients' and healthy eggs to replace mutant mtDNAs holds promises. Considering that a polar body contains few mitochondria and shares the same genomic material as an oocyte, we perform polar body transfer to prevent the transmission of mtDNA variants. We compare the effects of different types of germline genome transfer, including spindle-chromosome transfer, pronuclear transfer, and first and second polar body transfer, in mice. Reconstructed embryos support normal fertilization and produce live offspring. Importantly, genetic analysis confirms that the F1 generation from polar body transfer possesses minimal donor mtDNA carryover compared to the F1 generation from other procedures. Moreover, the mtDNA genotype remains stable in F2 progeny after polar body transfer. Our preclinical model demonstrates polar body transfer has great potential to prevent inherited mtDNA diseases. Copyright 2014 Elsevier Inc. All rights reserved.
URLPMID:27840020 [本文引用: 1]

Abstract Oocyte defects lie at the heart of some forms of infertility and could potentially be addressed therapeutically by alternative routes for oocyte formation. Here, we describe the generation of functional human oocytes following nuclear transfer of first polar body (PB1) genomes from metaphase II (MII) oocytes into enucleated donor MII cytoplasm (PBNT). The reconstructed oocytes supported the formation of de novo meiotic spindles and, after fertilization with sperm, meiosis completion and formation of normal diploid zygotes. While PBNT zygotes developed to blastocysts less frequently (42%) than controls (75%), genome-wide genetic, epigenetic, and transcriptional analyses of PBNT and control ESCs indicated comparable numbers of structural variations and markedly similar DNA methylation and transcriptome profiles. We conclude that rescue of PB1 genetic material via introduction into donor cytoplasm may offer a source of oocytes for infertility treatment or mitochondrial replacement therapy for mtDNA disease. Copyright 2017 Elsevier Inc. All rights reserved.
URLPMID:9241063 [本文引用: 1]

Genetically identical nonhuman primates can provide a powerful animal model for gene therapy and research activities where the physiological parameters directly or indirectly under study are heritable. Here we demonstrate that nuclear transfer is a viable technology for the production of identical rhesus macaques. Oocytes recovered from gonadotropin-treated females were enucleated by aspiration of the first polar body and underlying ooplasm, then activated by cycloheximide exposure. Individual diploid blastomeres, recovered from in vitro-fertilization-produced embryos (either fresh or frozen-thawed) and used as nuclear donors, were injected under the zona pellucida of enucleated (chromosome-free) oocytes and fused by electric pulses. The reconstituted embryos were cocultured on buffalo rat liver cells before cryostorage and transfer to synchronized host mothers. Of the 9 females receiving a total of 29 reconstituted embryos, 3 became pregnant, with two live births resulting, one male and one female. The parentage of both infants was established unequivocally by genotype analysis at 7 highly variable short tandem repeat loci.
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:18063756 [本文引用: 1]

Abstract It has recently been demonstrated that mouse and human fibroblasts can be reprogrammed into an embryonic stem cell-like state by introducing combinations of four transcription factors. However, the therapeutic potential of such induced pluripotent stem (iPS) cells remained undefined. By using a humanized sickle cell anemia mouse model, we show that mice can be rescued after transplantation with hematopoietic progenitors obtained in vitro from autologous iPS cells. This was achieved after correction of the human sickle hemoglobin allele by gene-specific targeting. Our results provide proof of principle for using transcription factor-induced reprogramming combined with gene and cell therapy for disease treatment in mice. The problems associated with using retroviruses and oncogenes for reprogramming need to be resolved before iPS cells can be considered for human therapy.
URL [本文引用: 1]
URLPMID:19631602 [本文引用: 1]

To our knowledge, for the first time, we demonstrate that induced pluripotent stem cells (iPSCs) can autonomously generate full-term mice via tetraploid blastocysts complementation. Differentiated somatic cells can be reprogrammed into iPSCs by forced expression of four transcription factors ct4, Sox2, Klf4, and c-Myc. However, it has been unclear whether reprogrammed iPSCs are fully pluripotent, resembling normal embryonic stem cells (ESCs), as no iPSC lines have shown the ability to autonomously generate full-term mice after injection into tetraploid blastocysts. Here we provide evidence demonstrating that an iPSC line induced by the four transcription factors can be used to generate full-term mice from complemented tetraploid blastocysts and thus appears to be fully pluripotent. This work serves as a proof of principle that iPSCs can in fact generate full-term embryos by tetraploid complementation.
URLPMID:19672241 [本文引用: 1]

We used viral vectors to introduce four genes into mouse fibroblast cells to reprogram these somatic cells into pluripotent cell in order to create iPS cells.After carrying out a standard set of tests to check whether the reprogramming had worked,our research group confirmed that we have generated 37 iPS cell lines.Then we tested the pluripotency of these iPS cell lines by tetraploid complementation and 6 of 37 iPS cell lines generated 27 live mice.12 mice that were mated produced healthy offspring and for now,they have got hundreds of second generation,and more than 100 third-generation mice which were healthy and viable.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:28388409 [本文引用: 1]

Of all known cultured stem cell types, pluripotent stem cells (PSCs) sit atop the landscape of developmental potency and are characterized by their ability to generate all cell types of an adult organism. However, PSCs show limited contribution to the extraembryonic placental tissues invivo. Here, we show that a chemical cocktail enables the derivation of stem cells with unique functional and molecular features from mice and humans, designated as extended pluripotent stem (EPS) cells, which are capable of chimerizing both embryonic and extraembryonic tissues. Notably, a single mouse EPS cell shows widespread chimeric contribution to both embryonic and extraembryonic lineages invivo and permits generating single-EPS-cell-derived mice by tetraploid complementation. Furthermore, human EPS cells exhibit interspecies chimeric competency in mouse conceptuses. Our findings constitute a first step toward capturing pluripotent stem cells with extraembryonic developmental potentials in culture and open new avenues for basic and translational research.
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:24162740 [本文引用: 1]

Vitamin C, a micronutrient known for its anti-scurvy activity in humans, promotes the generation of induced pluripotent stem cells (iPSCs) through the activity of histone demethylating dioxygenases. TET hydroxylases are also dioxygenases implicated in active DNA demethylation. Here we report that TET1 either positively or negatively regulates somatic cell reprogramming depending on the absence or presence of vitamin C. TET1 deficiency enhances reprogramming, and its overexpression impairs reprogramming in the context of vitamin C by modulating the obligatory mesenchymal-to-epithelial transition (MET). In the absence of vitamin C, TET1 promotes somatic cell reprogramming independent of MET. Consistently, TET1 regulates 5-hydroxymethylcytosine (5hmC) formation at loci critical for MET in a vitamin C-dependent fashion. Our findings suggest that vitamin C has a vital role in determining the biological outcome of TET1 function at the cellular level. Given its benefit to human health, vitamin C should be investigated further for its role in epigenetic regulation.
URL [本文引用: 1]
URLPMID:23202127 [本文引用: 1]

The induction of pluripotent stem cells (iPSCs) by defined factors is poorly understood stepwise. Here, we show that histone H3 lysine 9 (H3K9) methylation is the primary epigenetic determinant for the intermediate pre-iPSC state, and its removal leads to fully reprogrammed iPSCs. We generated a panel of stable pre-iPSCs that exhibit pluripotent properties but do not activate the core pluripotency network, although they remain sensitive to vitamin C for conversion into iPSCs. Bone morphogenetic proteins (BMPs) were subsequently identified in serum as critical signaling molecules in arresting reprogramming at the pre-iPSC state. Mechanistically, we identified H3K9 methyltransferases as downstream targets of BMPs and showed that they function with their corresponding demethylases as the on/off switch for the pre-iPSC fate by regulating H3K9 methylation status at the core pluripotency loci. Our results not only establish pre-iPSCs as an epigenetically stable signpost along the reprogramming road map, but they also provide mechanistic insights into the epigenetic reprogramming of cell fate.
URL [本文引用: 1]
URLPMID:23706735 [本文引用: 1]

Abstract The reprogramming factors that induce pluripotency have been identified primarily from embryonic stem cell (ESC)-enriched, pluripotency-associated factors. Here, we report that, during mouse somatic cell reprogramming, pluripotency can be induced with lineage specifiers that are pluripotency rivals to suppress ESC identity, most of which are not enriched in ESCs. We found that OCT4 and SOX2, the core regulators of pluripotency, can be replaced by lineage specifiers that are involved in mesendodermal (ME) specification and in ectodermal (ECT) specification, respectively. OCT4 and its substitutes attenuated the elevated expression of a group of ECT genes, whereas SOX2 and its substitutes curtailed a group of ME genes during reprogramming. Surprisingly, the two counteracting lineage specifiers can synergistically induce pluripotency in the absence of both OCT4 and SOX2. Our study suggests a "seesaw model" in which a balance that is established using pluripotency factors and/or counteracting lineage specifiers can facilitate reprogramming. Copyright 2013 Elsevier Inc. All rights reserved.
.
URLPMID:29220666 [本文引用: 1]

Abstract Cell-fate decisions remain poorly understood at the chromatin level. Here, we map chromatin remodeling dynamics during induction of pluripotent stem cells. ATAC-seq profiling of MEFs expressing Oct4-Sox2-Klf4 (OSK) reveals dynamic changes in chromatin states shifting from open to closed (OC) and closed to open (CO), with an initial burst of OC and an ending surge of CO. The OC loci are largely composed of genes associated with a somatic fate, while the CO loci are associated with pluripotency. Factors/conditions known to impede reprogramming prevent OSK-driven OC and skew OC-CO dynamics. While the CO loci are enriched for OSK motifs, the OC loci are not, suggesting alternative mechanisms for chromatin closing. Sap30, a Sin3A corepressor complex component, is required for the OC shift and facilitates reduced H3K27ac deposition at OC loci. These results reveal a chromatin accessibility logic during reprogramming that may apply to other cell-fate decisions.
URLPMID:29625068 [本文引用: 1]

Abstract Despite its exciting potential, chemical induction of pluripotency (CIP) efficiency remains low and the mechanisms are poorly understood. We report the development of an efficient two-step serum- and replating-free CIP protocol and the associated chromatin accessibility dynamics (CAD) by assay for transposase-accessible chromatin (ATAC)-seq. CIP reorganizes the somatic genome to an intermediate state that is resolved under 2iL condition by re-closing previously opened loci prior to pluripotency acquisition with gradual opening of loci enriched with motifs for the OCT/SOX/KLF families. Bromodeoxyuridine, a critical ingredient of CIP, is responsible for both closing and opening critical loci, at least in part by preventing the opening of loci enriched with motifs for the AP1 family and facilitating the opening of loci enriched with SOX/KLF/GATA motifs. These changes differ markedly from CAD observed during Yamanaka-factor-driven reprogramming. Our study provides insights into small-molecule-based reprogramming mechanisms and reorganization of nuclear architecture associated with cell-fate decisions.
[本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:1176880 [本文引用: 1]

A pronucleus can be microsurgically removed from the fertilized mouse egg. Out of 145 haploid eggs obtained by this method and transplanted into the oviduct of pseudopregnant recipients, 36 multicellular embryos were recovered on the 4th or 5th day. On the 4th day all embryos were morulae composed of 8-50 cells, with the majority containing 8-16 cells. After an additional 24 h in vivo or in vitro the cell number increased considerably, sometimes up to as many as 80. Out of 36 multicellular embryos only one developed into a blastocyst while the others remained at the morula stage. Karyological investigations confirmed that the embryos were haploid and revealed that all were gynogenetic. Possible reasons for the absence of the androgenones and for the scarcity of blastocysts are discussed.
URLPMID:670867 [本文引用: 1]

Abstract The chromosome constitution of early postimplantation presumptive haploid parthenogenetic mouse embryos was examined. All the embryos isolated were at the egg-cylinder stage and seven contained dividing cells. In two of the apparently healthy embryos only haploid mitoses were seen, whereas in five others an approximately equal proportion of haploid and diploid mitoses was observed. Out of 52 cells in which unequivocal counts could be made, only one contained more than the euploid number of chromosomes (mouse, n = 20). Possible reasons for the poorer development of haploid compared to diploid parthenogenetic embryos are discussed.
URL [本文引用: 1]
URLPMID:22136931 [本文引用: 1]

Abstract All somatic mammalian cells carry two copies of chromosomes (diploidy), whereas organisms with a single copy of their genome, such as yeast, provide a basis for recessive genetics. Here we report the generation of haploid mouse ESC lines from parthenogenetic embryos. These cells carry 20 chromosomes, express stem cell markers, and develop into all germ layers in vitro and in vivo. We also developed a reversible mutagenesis protocol that allows saturated genetic recessive screens and results in homozygous alleles. This system allowed us to generate a knockout cell line for the microRNA processing enzyme Drosha. In a forward genetic screen, we identified Gpr107 as a molecule essential for killing by ricin, a toxin being used as a bioweapon. Our results open the possibility of combining the power of a haploid genome with pluripotency of embryonic stem cells to uncover fundamental biological processes in defined cell types at a genomic scale. Copyright 2011 Elsevier Inc. All rights reserved.
URLPMID:3209452 [本文引用: 1]

Most animals are diploid, but haploid-only and male-haploid (such as honeybee and ant) species have been described. The diploid genomes of complex organisms limit genetic approaches in biomedical model species such as mice. To overcome this problem, experimental induction of haploidy has been used in fish. Haploid development in zebrafish has been applied for genetic screening. Recently, haploid pluripotent cell lines from medaka fish (Oryzias latipes) have also been established. In contrast, haploidy seems less compatible with development in mammals. Although haploid cells have been observed in egg cylinder stage parthenogenetic mouse embryos, most cells in surviving embryos become diploid. Here we describe haploid mouse embryonic stem cells and show their application in forward genetic screening.
URLPMID:22136917 [本文引用: 1]

Elling et al., 2011, this issue of Leeb and Wutz, 2011, Nature), report the isolation of haploid pluripotent mouse ESCs, thus enabling efficient functional screening for genes involved in diverse cellular and developmental processes.
URLPMID:23023130 [本文引用: 1]

Abstract Haploids and double haploids are important resources for studying recessive traits and have large impacts on crop breeding, but natural haploids are rare in animals. Mammalian haploids are restricted to germline cells and are occasionally found in tumours with massive chromosome loss. Recent success in establishing haploid embryonic stem (ES) cells in medaka fish and mice raised the possibility of using engineered mammalian haploid cells in genetic studies. However, the availability and functional characterization of mammalian haploid ES cells are still limited. Here we show that mouse androgenetic haploid ES (ahES) cell lines can be established by transferring sperm into an enucleated oocyte. The ahES cells maintain haploidy and stable growth over 30 passages, express pluripotent markers, possess the ability to differentiate into all three germ layers in vitro and in vivo, and contribute to germlines of chimaeras when injected into blastocysts. Although epigenetically distinct from sperm cells, the ahES cells can produce viable and fertile progenies after intracytoplasmic injection into mature oocytes. The oocyte-injection procedure can also produce viable transgenic mice from genetically engineered ahES cells. Our findings show the developmental pluripotency of androgenentic haploids and provide a new tool to quickly produce genetic models for recessive traits. They may also shed new light on assisted reproduction.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:23856644 [本文引用: 1]

Abstract Recent success in the derivation of haploid embryonic stem cells (haESCs) from mouse via parthenogenesis and androgenesis has enabled genetic screening in mammalian cells and generation of gene-modified animals. However, whether haESCs can be derived from primates remains unknown. Here, we report the derivation of haESCs from parthenogenetic blastocysts of Macaca fascicularis monkeys. These cells, termed as PG-haESCs, are pluripotent and can differentiate to cells of three embryonic germ layers in vitro or in vivo. Interestingly, the haploidy of one monkey PG-haESC line (MPH1) is more stable compared with that of the other one (MPH2), as shown by the existence of haploid cells for more than 140 days without fluorescence-activated cell sorting (FACS) enrichment of haploid cells. Importantly, transgenic monkey PG-haESC lines can be generated by lentivirus- and piggyBac transposon-mediated gene transfer. Moreover, genetic screening is feasible in monkey PG-haESCs. Our results demonstrate that PG-haESCs can be generated from monkeys, providing an ideal tool for genetic analyses in primates.
URLPMID:24360884 [本文引用: 1]

Li et02al. show here that androgenetic rat haploid embryonic stem cells can be used for genetic screening, random or homologous mutagenesis, and CRISPR-Cas-mediated gene editing.
URLPMID:26982723 [本文引用: 1]

Abstract Diploidy is a fundamental genetic feature in mammals, in which haploid cells normally arise only as post-meiotic germ cells that serve to ensure a diploid genome upon fertilization. Gamete manipulation has yielded haploid embryonic stem (ES) cells from several mammalian species, but haploid human ES cells have yet to be reported. Here we generated and analysed a collection of human parthenogenetic ES cell lines originating from haploid oocytes, leading to the successful isolation and maintenance of human ES cell lines with a normal haploid karyotype. Haploid human ES cells exhibited typical pluripotent stem cell characteristics, such as self-renewal capacity and a pluripotency-specific molecular signature. Moreover, we demonstrated the utility of these cells as a platform for loss-of-function genetic screening. Although haploid human ES cells resembled their diploid counterparts, they also displayed distinct properties including differential regulation of X chromosome inactivation and of genes involved in oxidative phosphorylation, alongside reduction in absolute gene expression levels and cell size. Surprisingly, we found that a haploid human genome is compatible not only with the undifferentiated pluripotent state, but also with differentiated somatic fates representing all three embryonic germ layers both in vitro and in vivo, despite a persistent dosage imbalance between the autosomes and X chromosome. We expect that haploid human ES cells will provide novel means for studying human functional genomics and development.
URL [本文引用: 1]
URLPMID:23934149 [本文引用: 1]

Abstract Measuring gene expression in individual cells is crucial for understanding the gene regulatory network controlling human embryonic development. Here we apply single-cell RNA sequencing (RNA-Seq) analysis to 124 individual cells from human preimplantation embryos and human embryonic stem cells (hESCs) at different passages. The number of maternally expressed genes detected in our data set is 22,687, including 8,701 long noncoding RNAs (lncRNAs), which represents a significant increase from 9,735 maternal genes detected previously by cDNA microarray. We discovered 2,733 novel lncRNAs, many of which are expressed in specific developmental stages. To address the long-standing question whether gene expression signatures of human epiblast (EPI) and in vitro hESCs are the same, we found that EPI cells and primary hESC outgrowth have dramatically different transcriptomes, with 1,498 genes showing differential expression between them. This work provides a comprehensive framework of the transcriptome landscapes of human early embryos and hESCs.
URLPMID:23892778 [本文引用: 1]

Abstract Mammalian pre-implantation development is a complex process involving dramatic changes in the transcriptional architecture. We report here a comprehensive analysis of transcriptome dynamics from oocyte to morula in both human and mouse embryos, using single-cell RNA sequencing. Based on single-nucleotide variants in human blastomere messenger RNAs and paternal-specific single-nucleotide polymorphisms, we identify novel stage-specific monoallelic expression patterns for a significant portion of polymorphic gene transcripts (2565to 53%). By weighted gene co-expression network analysis, we find that each developmental stage can be delineated concisely by a small number of functional modules of co-expressed genes. This result indicates a sequential order of transcriptional changes in pathways of cell cycle, gene regulation, translation and metabolism, acting in a step-wise fashion from cleavage to morula. Cross-species comparisons with mouse pre-implantation embryos reveal that the majority of human stage-specific modules (765out of659) are notably preserved, but developmental specificity and timing differ between human and mouse. Furthermore, we identify conserved key members (or hub genes) of the human and mouse networks. These genes represent novel candidates that are likely to be key in driving mammalian pre-implantation development. Together, the results provide a valuable resource to dissect gene regulatory mechanisms underlying progressive development of early mammalian embryos.
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:26046443 [本文引用: 1]

Abstract Germ cells are vital for transmitting genetic information from one generation to the next and for maintaining the continuation of species. Here, we analyze the transcriptome of human primordial germ cells (PGCs) from the migrating stage to the gonadal stage at single-cell and single-base resolutions. Human PGCs show unique transcription patterns involving the simultaneous expression of both pluripotency genes and germline-specific genes, with a subset of them displaying developmental-stage-specific features. Furthermore, we analyze the DNA methylome of human PGCs and find global demethylation of their genomes. Approximately 10 to 11 weeks after gestation, the PGCs are nearly devoid of any DNA methylation, with only 7.8% and 6.0% of the median methylation levels in male and female PGCs, respectively. Our work paves the way toward deciphering the complex epigenetic reprogramming of the germline with the aim of restoring totipotency in fertilized oocytes. Copyright 2015 Elsevier Inc. All rights reserved.
URLPMID:25079557 [本文引用: 1]

Abstract DNA methylation is a crucial element in the epigenetic regulation of mammalian embryonic development. However, its dynamic patterns have not been analysed at the genome scale in human pre-implantation embryos due to technical difficulties and the scarcity of required materials. Here we systematically profile the methylome of human early embryos from the zygotic stage through to post-implantation by reduced representation bisulphite sequencing and whole-genome bisulphite sequencing. We show that the major wave of genome-wide demethylation is complete at the 2-cell stage, contrary to previous observations in mice. Moreover, the demethylation of the paternal genome is much faster than that of the maternal genome, and by the end of the zygotic stage the genome-wide methylation level in male pronuclei is already lower than that in female pronuclei. The inverse correlation between promoter methylation and gene expression gradually strengthens during early embryonic development, reaching its peak at the post-implantation stage. Furthermore, we show that active genes, with the trimethylation of histone H3 at lysine 4 (H3K4me3) mark at the promoter regions in pluripotent human embryonic stem cells, are essentially devoid of DNA methylation in both mature gametes and throughout pre-implantation development. Finally, we also show that long interspersed nuclear elements or short interspersed nuclear elements that are evolutionarily young are demethylated to a milder extent compared to older elements in the same family and have higher abundance of transcripts, indicating that early embryos tend to retain higher residual methylation at the evolutionarily younger and more active transposable elements. Our work provides insights into the critical features of the methylome of human early embryos, as well as its functional relation to the regulation of gene expression and the repression of transposable elements.
URLPMID:4081501 [本文引用: 1]

After fertilization, the zebrafish sperm DNA methylation pattern remains stable, whereas the oocyte pattern is progressively modified via demethylation and de novo methylation to match the sperm pattern by the midblastula stage.
URLPMID:24813617 [本文引用: 1]

Abstract The reprogramming of parental methylomes is essential for embryonic development. In mammals, paternal 5-methylcytosines (5mCs) have been proposed to be actively converted to oxidized bases. These paternal oxidized bases and maternal 5mCs are believed to be passively diluted by cell divisions. By generating single-base resolution, allele-specific DNA methylomes from mouse gametes, early embryos, and primordial germ cell (PGC), as well as single-base-resolution maps of oxidized cytosine bases for early embryos, we report the existence of 5hmC and 5fC in both maternal and paternal genomes and find that 5mC or its oxidized derivatives, at the majority of demethylated CpGs, are converted to unmodified cytosines independent of passive dilution from gametes to four-cell embryos. Therefore, we conclude that paternal methylome and at least a significant proportion of maternal methylome go through active demethylation during embryonic development. Additionally, all the known imprinting control regions (ICRs) were classified into germ-line or somatic ICRs. Copyright 2014 Elsevier Inc. All rights reserved.
URLPMID:27626379 [本文引用: 1]

Abstract Histone modifications have critical roles in regulating the expression of developmental genes during embryo development in mammals. However, genome-wide analyses of histone modifications in pre-implantation embryos have been impeded by the scarcity of the required materials. Here, by using a small-scale chromatin immunoprecipitation followed by sequencing (ChIP-seq) method, we map the genome-wide profiles of histone H3 lysine 4 trimethylation (H3K4me3) and histone H3 lysine 27 trimethylation (H3K27me3), which are associated with gene activation and repression, respectively, in mouse pre-implantation embryos. We find that the re-establishment of H3K4me3, especially on promoter regions, occurs much more rapidly than that of H3K27me3 following fertilization, which is consistent with the major wave of zygotic genome activation at the two-cell stage. Furthermore, H3K4me3 and H3K27me3 possess distinct features of sequence preference and dynamics in pre-implantation embryos. Although H3K4me3 modifications occur consistently at transcription start sites, the breadth of the H3K4me3 domain is a highly dynamic feature. Notably, the broad H3K4me3 domain (wider than 5 kb) is associated with higher transcription activity and cell identity not only in pre-implantation development but also in the process of deriving embryonic stem cells from the inner cell mass and trophoblast stem cells from the trophectoderm. Compared to embryonic stem cells, we found that the bivalency (that is, co-occurrence of H3K4me3 and H3K27me3) in early embryos is relatively infrequent and unstable. Taken together, our results provide a genome-wide map of H3K4me3 and H3K27me3 modifications in pre-implantation embryos, facilitating further exploration of the mechanism for epigenetic regulation in early embryos.
URLPMID:27626382 [本文引用: 1]

Abstract Histone modifications are fundamental epigenetic regulators that control many crucial cellular processes. However, whether these marks can be passed on from mammalian gametes to the next generation is a long-standing question that remains unanswered. Here, by developing a highly sensitive approach, STAR ChIP-seq, we provide a panoramic view of the landscape of H3K4me3, a histone hallmark for transcription initiation, from developing gametes to post-implantation embryos. We find that upon fertilization, extensive reprogramming occurs on the paternal genome, as H3K4me3 peaks are depleted in zygotes but are readily observed after major zygotic genome activation at the late two-cell stage. On the maternal genome, we unexpectedly find a non-canonical form of H3K4me3 (ncH3K4me3) in full-grown and mature oocytes, which exists as broad peaks at promoters and a large number of distal loci. Such broad H3K4me3 peaks are in contrast to the typical sharp H3K4me3 peaks restricted to CpG-rich regions of promoters. Notably, ncH3K4me3 in oocytes overlaps almost exclusively with partially methylated DNA domains. It is then inherited in pre-implantation embryos, before being erased in the late two-cell embryos, when canonical H3K4me3 starts to be established. The removal of ncH3K4me3 requires zygotic transcription but is independent of DNA replication-mediated passive dilution. Finally, downregulation of H3K4me3 in full-grown oocytes by overexpression of the H3K4me3 demethylase KDM5B is associated with defects in genome silencing. Taken together, these data unveil inheritance and highly dynamic reprogramming of the epigenome in early mammalian development.
URLPMID:29686265 [本文引用: 1]

Maternal imprinting at the Xist gene is essential to achieve paternal allele-specific imprinted X-chromosome inactivation (XCI) in female mammals. However, the mechanism underlying Xist imprinting is unclear. Here we show that the Xist locus is coated with a broad H3K27me3 domain that is established during oocyte growth and persists through preimplantation development in mice. Loss of maternal... [Show full abstract]
URL [本文引用: 1]
URLPMID:28703188 [本文引用: 1]

Abstract In mammals, chromatin organization undergoes drastic reprogramming after fertilization. However, the three-dimensional structure of chromatin and its reprogramming in preimplantation development remain poorly understood. Here, by developing a low-input Hi-C (genome-wide chromosome conformation capture) approach, we examined the reprogramming of chromatin organization during early development in mice. We found that oocytes in metaphase II show homogeneous chromatin folding that lacks detectable topologically associating domains (TADs) and chromatin compartments. Strikingly, chromatin shows greatly diminished higher-order structure after fertilization. Unexpectedly, the subsequent establishment of chromatin organization is a prolonged process that extends through preimplantation development, as characterized by slow consolidation of TADs and segregation of chromatin compartments. The two sets of parental chromosomes are spatially separated from each other and display distinct compartmentalization in zygotes. Such allele separation and allelic compartmentalization can be found as late as the 8-cell stage. Finally, we show that chromatin compaction in preimplantation embryos can partially proceed in the absence of zygotic transcription and is a multi-level hierarchical process. Taken together, our data suggest that chromatin may exist in a markedly relaxed state after fertilization, followed by progressive maturation of higher-order chromatin architecture during early development.
URLPMID:28621329 [本文引用: 1]

Single-cell epigenome sequencing techniques have recently been developed. However, the combination of different layers of epigenome sequencing in an individual cell has not yet been achieved. Here, we developed a single-cell multi-omics sequencing technology (single-cell COOL-seq) that can analyze the chromatin state/nucleosome positioning, DNA methylation, copy number variation and ploidy simultaneously from the same individual mammalian cell. We used this method to analyze the reprogramming of the chromatin state and DNA methylation in mouse preimplantation embryos. We found that within < 12 h of fertilization, each individual cell undergoes global genome demethylation together with the rapid and global reprogramming of both maternal and paternal genomes to a highly opened chromatin state. This was followed by decreased openness after the late zygote stage. Furthermore, from the late zygote to the 4-cell stage, the residual DNA methylation is preferentially preserved on intergenic regions of the paternal alleles and intragenic regions of maternal alleles in each individual blastomere. However, chromatin accessibility is similar between paternal and maternal alleles in each individual cell from the late zygote to the blastocyst stage. The binding motifs of several pluripotency regulators are enriched at distal nucleosome depleted regions from as early as the 2-cell stage. This indicates that thecis-regulatory elements of such target genes have been primed to an open state from the 2-cell stage onward, long before pluripotency is eventually established in the ICM of the blastocyst. Genes may be classified into homogeneously open, homogeneously closed and divergent states based on the chromatin accessibility of their promoter regions among individual cells. This can be traced to step-wise transitions during preimplantation development. Our study offers the first single-cell and parental allele-specific analysis of the genome-scale chromatin state and DNA methylation dynamics at single-base resolution in early mouse embryos and provides new insights into the heterogeneous yet highly ordered features of epigenomic reprogramming during this process.
URLPMID:29474909 [本文引用: 1]

Single-cell RNA sequencing (scRNA-seq) technologies are poised to reshape the current cell-type classification system. However, a transcriptome-based single-cell atlas has not been achieved for complex mammalian systems. Here, we developed Microwell-seq, a high-throughput and low-cost scRNA-seq platform using simple, inexpensive devices. Using Microwell-seq, we analyzed more than 400,000 single cells covering all of the major mouse organs and constructed a basic scheme for a mouse cell atlas (MCA). We reveal a single-cell hierarchy for many tissues that have not been well characterized previously. We built a web-based “single-cell MCA analysis” pipeline that accurately defines cell types based on single-cell digital expression. Our study demonstrates the wide applicability of the Microwell-seq technology and MCA resource.
URLPMID:29539641 [本文引用: 1]

Abstract The mammalian prefrontal cortex comprises a set of highly specialized brain areas containing billions of cells and serves as the centre of the highest-order cognitive functions, such as memory, cognitive ability, decision-making and social behaviour. Although neural circuits are formed in the late stages of human embryonic development and even after birth, diverse classes of functional cells are generated and migrate to the appropriate locations earlier in development. Dysfunction of the prefrontal cortex contributes to cognitive deficits and the majority of neurodevelopmental disorders; there is therefore a need for detailed knowledge of the development of the prefrontal cortex. However, it is still difficult to identify cell types in the developing human prefrontal cortex and to distinguish their developmental features. Here we analyse more than 2,300 single cells in the developing human prefrontal cortex from gestational weeks 8 to 26 using RNA sequencing. We identify 35 subtypes of cells in six main classes and trace the developmental trajectories of these cells. Detailed analysis of neural progenitor cells highlights new marker genes and unique developmental features of intermediate progenitor cells. We also map the timeline of neurogenesis of excitatory neurons in the prefrontal cortex and detect the presence of interneuron progenitors in early developing prefrontal cortex. Moreover, we reveal the intrinsic development-dependent signals that regulate neuron generation and circuit formation using single-cell transcriptomic data analysis. Our screening and characterization approach provides a blueprint for understanding the development of the human prefrontal cortex in the early and mid-gestational stages in order to systematically dissect the cellular basis and molecular regulation of prefrontal cortex function in humans.
[本文引用: 1]
