0 引言
【研究意义】结球甘蓝(Brassica oleracea L. var. capitata)简称甘蓝,在中国各地广泛栽培。截止至2015年,中国审(鉴、认)定或登记的甘蓝品种共计247个[1,2,3,4,5],其中大部分为一代杂种。近年来,市场上的甘蓝品种不断增多,种植面积不断增大。但是,在种子市场上同名异物及同物异名的情况时有发生,甚至一些不合格种子混入市场,造成巨大的经济损失。因此,对品种进行快速准确鉴定,对于假种辨别和产权纠纷具有重要作用。【前人研究进展】早期的品种鉴定手段主要依赖于形态学鉴定,这种方法耗时较长且易受环境影响。随着分子生物学的发展,分子标记技术的出现为品种鉴定提供了新的手段。分子标记技术具有周期短、不受环境条件影响、可进行高通量测试分析等优点,已经在品种鉴定、种子纯度鉴定等方面得到广泛应用[6,7]。随着生物技术的进步,陆续有基于RFLP、AFLP、SSR、SNP等分子标记用于DNA指纹图谱的构建。其中,SNP分子标记是国际植物新品种权保护联盟(UPOV)BMT分子测试指南中构建DNA指纹数据库的推荐标记之一[8]。相对于传统标记,利用SNP标记构建的指纹图谱具有数量多、分布广泛、稳定性高、易于快速且高通量分型的特点[9,10]。分子标记技术在甘蓝品种指纹图谱构建中得到了广泛的应用,薄天岳等[11]通过SRAP、RAPD 2种分子标记方法,构建了甘蓝品种‘争春’、 ‘寒光2号’及其各自亲本的DNA指纹图谱,并将其用于种子纯度鉴定。宋顺华等[12]利用AFLP标记对来自全国的44份甘蓝品种进行分析,并构建了其指纹图谱。陈琛等[13]利用5对SSR标记构建了6个秋甘蓝品种及其亲本共18份材料的指纹图谱。王庆彪等[5]利用20对SSR标记构建50份中国甘蓝代表品种DNA指纹数据库并阐述了甘蓝SSR-DNA指纹鉴定的应用和技术流程。【本研究切入点】目前,甘蓝品种鉴定中常用的指纹图谱是基于AFLP、SSR等分子标记。但是,传统AFLP、SSR指纹图谱存在标记数量有限、检测位点少,位点的突变率高、对变异反应比较敏感等缺点。SNP标记作为第三代分子标记,具有数量多、分布广泛、二态性、易于快速且高通量的进行基因分型等优点。SNP标记已经在玉米[14]、油菜[15]、香菇[16]等作物指纹图谱构建中得到应用,但在甘蓝类蔬菜中尚未有基于SNP标记的指纹图谱。【拟解决的关键问题】本研究基于甘蓝高代自交系的重测序数据开发SNP标记,利用KASP技术对主要甘蓝品种进行基因分型[17],根据分型结果筛选筛选在染色体上均匀分布、多态性信息较高的SNP位点作为核心标记,并利用核心SNP标记构建甘蓝品种指纹图谱,为甘蓝品种真实性、特异性鉴定提供重要依据。1 材料与方法
1.1 供试材料
试验于2016年12月至2017年5月在中国农业科学院蔬菜花卉研究所甘蓝遗传育种实验室进行。指纹图谱构建:从国内主要育种单位搜集(鉴定、登记)市场上推广的主要代表品种59个(电子附表1)。
Table 1
附表1
附表1供试的主要甘蓝品种
Table 1The cabbage main varieties used in this study
| 编号 Code | 品种名称 Varieties name | 编号 Code | 品种名称 Varieties name | 编号 Code | 品种名称 Varieties name | ||
|---|---|---|---|---|---|---|---|
| 1 | 中甘8号 Zhonggan 8 | 21 | 绿球66 Lvqiu 66 | 41 | 中甘301 Zhonggan 301 | ||
| 2 | 京丰1号 Jingfeng 1 | 22 | 8398 8398 | 42 | 中甘101 Zhonggan 101 | ||
| 3 | 惠丰3号 Huifeng 3 | 23 | 苏甘21 Sugan 21 | 43 | 争春 Zhengchun | ||
| 4 | 惠丰4号 Huifeng 4 | 24 | 春丰 Chunfeng | 44 | 中甘96 Zhonggan 96 | ||
| 5 | 惠丰5号 Huifeng 5 | 25 | 怡春 Yichun | 45 | 中甘192 Zhonggan192 | ||
| 6 | 惠丰6号 Huifeng 6 | 26 | 中甘19 Zhonggan 19 | 46 | 中甘1305 Zhonggan 1305 | ||
| 7 | 豫甘1号 Yugan 1 | 27 | 博春 Bochun | 47 | 中甘1229 Zhonggan 1229 | ||
| 8 | 豫甘3号 Yugan3 | 28 | 中甘17 Zhonggan 17 | 48 | 中甘1327 Zhonggan 1327 | ||
| 9 | 豫甘4号 Yugan 4 | 29 | 豫生早熟牛心 Yushengzaoshuniuxin | 49 | 中甘20号 Zhonggan 20 | ||
| 10 | 西园4号 Xiyuan 4 | 30 | 争牛 Zhengniu | 50 | 中甘1280 Zhonggan 1280 | ||
| 11 | 中甘9号 Zhonggan 9 | 31 | 苏晨1号 Suchen 1 | 51 | 中甘593 Zhonggan 593 | ||
| 12 | 中甘56 Zhonggan 56 | 32 | 豫甘5号 Yugan 5 | 52 | 中甘594 Zhonggan 594 | ||
| 13 | 春甘2号 Chungan 2 | 33 | 嘉兰 Jialan | 53 | 中甘596 Zhonggan 596 | ||
| 14 | 中甘588 Zhonggan 588 | 34 | 铁头102 Tietou 102 | 54 | 晚丰 Wanfeng | ||
| 15 | 中甘828 Zhonggan 828 | 35 | 瑞甘16 Ruigan 16 | 55 | 庆丰 Qingfeng | ||
| 16 | 中甘628 Zhonggan 628 | 36 | 瑞甘17 Ruigan 17 | 56 | 中甘1268 Zhonggan 1268 | ||
| 17 | 秦甘60 Qingan 60 | 37 | 苏甘55 Sugan 55 | 57 | 中甘1266 Zhonggan 1266 | ||
| 18 | 秦甘78 Qingan 78 | 38 | 中甘18 Zhonggan 18 | 58 | 中甘21 Zhonggan 21 | ||
| 19 | 中甘11号 Zhonggan 11 | 39 | 西星甘蓝1号 Xixingganlan 1 | 59 | 秋甘1号 Qiugan 1 | ||
| 20 | 瑞甘60 Ruigan 60 | 40 | 中甘15 Zhonggan 15 |
新窗口打开
核心SNP标记验证:在上述59个主要甘蓝品种中挑选15个已推广的品种,及5个未推广的新组合。每个品种或组合取3粒种子,将种子混合构成人工虚拟混合群体。
1.2 DNA提取
将59个主要代表品种的种子放于培养皿中,采用纸上发芽,放置于25℃恒温培养箱中,催芽5—6 d。取子叶和下胚轴,每份样品中取若干个体,共计0.5 g,采用改良CTAB法[18]提取DNA,使用冷冻干燥机将样品DNA抽干后置于-80℃保存。将人工虚拟混合群体种子混合催芽,然后按单个幼苗提取DNA。
1.3 KASP标记开发及检测
对50个甘蓝高代自交系进行重测序,所测的自交系涵盖了不同茬口(春、秋、越冬)、不同熟性(早、中、晚)及不同球型(扁、圆、尖)的材料,具有很好的代表性。利用重测序数据与参考基因组(02-12)[19]进行比对,开发SNP位点。对获得的SNP位点按以下条件进行筛选:(1)位点的未测通材料数<20;(2)多态性在40%—60%;(3)在染色体中均匀分布;(4)位点前后各50 bp不存在其他变异。对于筛选获得的SNP位点,截取其前后各50 bp序列,交由LGC公司开发设计KASP引物。PCR反应体系(10.14 µL)为KASP Master mix 5 μL、KASP Primer mix 0.14 μL和模板DNA(20 ng·μL-1)5 μL。PCR反应条件为第一轮94℃ 15 min;94℃ 20 s,61—55℃ 60 s,10个Touch Down循环(每个循环降低0.6℃);第二轮94℃ 20 s,55℃ 60 s,26个循环。使用荧光微孔板检测仪检测PCR产物,用LGC公司开发的SNPviewer软件读取检测数据。所用KASP Master mix 购自英国LGC公司,货号为KBS-1016-012。
1.4 数据处理
根据分型结果计算出各个标记的等位基因频率,然后利用软件PIC_Calc 0.6计算各标记的多态性信息含量(polymorphism information content,PIC),计算公式为PIC = 1-∑fi2,其中fi为i位点基因频率。依据SNP标记具有的二态性特点,将分型数据转化为二元编码数据,将野生型(与参考基因组一致)表示为(1,0),突变体表示为(0,1),将杂合基因型表示为(1,1),缺失碱基位点记为(999,999),对每份材料进行基因型统计,利用NTSYSpc2.1软件进行相似系数计算及基于UPGMA算法的聚类分析[15]。1.5 核心SNP位点的挑选及其在品种鉴定中的应用
根据基因分型结果,筛选出多态性信息含量>0.35、无基因型数据缺失、在9条染色体上均匀分布的SNP位点作为构建甘蓝品种指纹图谱的核心位点。利用核心SNP标记位点,对人工构建的虚拟混合群体进行SNP标记分析,并与已有SNP-DNA指纹图谱进行比对,判定虚拟混合群体的60个样品的分型结果是否与指纹图谱数据库中相应品种对应。
2 结果
2.1 甘蓝SNP标记的开发
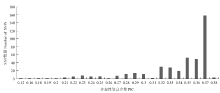
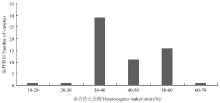
对50个甘蓝高代自交系进行重测序,平均测序深度为5×。将重测序数据与参考基因组02-12进行比对,共获得SNP位点2.54×106个。筛选位点多态性介于40%—60%、未测通材料数<20的SNP位点,共获得2.59×104个。进一步筛选在9条染色体上均匀分布、位点前后各50 bp不存在其他变异位点的SNP位点,最终获得500个SNP位点用于下一步试验,平均每条染色体上55.6个。500个SNP位点中有442个成功转化为KASP标记,转化成功率为88.4%。利用KASP平台对59个甘蓝品种进行基因型检测,442个KASP标记全部成功分型。在442个标记中,有25个标记的未分型材料数>5,为保证结果准确性,在后续分析中将这25个标记去除。在59个甘蓝品种中,全部标记的杂合位点比例大于30%的品种有57个,占所有品种的96.6%(图1)。其中,品种‘豫生早熟牛心’的杂合位点比例最高,为67.8%;‘秦甘78’杂合位点比例最低,为18.8%。所有SNP标记在全部品种中多态性信息含量(PIC)处于0.12—0.38,PIC值大于0.35的位点占所有位点的63.5%,其中PIC值为0.37的位点最多,有157个(图2)。
 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图1417个SNP标记的多态性信息含量分布情况
-->Fig. 1Distribution of polymorphism information content in 417 SNP markers
-->
 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图2基于417个SNP标记的59个主要甘蓝品种的杂合基因型比例
-->Fig. 2Heterozygous genotype ratio of 59 main varieties based on 417 SNPs
-->
2.2 核心SNP位点的挑选及DNA指纹图谱的构建
综合考虑基因分型结果,筛选PIC值大于0.35、无基因型数据缺失、在9条染色体上均匀分布的SNP位点作为构建甘蓝品种指纹图谱的核心位点,最终获得50个SNP位点用以构建主要品种的指纹图谱(表1)。50个核心SNP位点中,位点Bol2-56的PIC值最高,为0.38;位点Bol8-11的PIC值最低,为0.35;平均PIC值为0.36,表现为中度多态性。Table 1
表1
表1用于构建指纹图谱的50对核心引物
Table 1The information of 50 core primers for fingerprinting
| 名称 Name | 染色体 Chromosome | 位置 Position(bp) | 引物序列 Sequence of primer (5′-3′) | 变异类型 Alleles type | PIC |
|---|---|---|---|---|---|
| Bol1-11 | 1 | 4798641 | F: CCTGCAAGAGAGACAAGGCCAA | T/C | 0.36 |
| F: CTGCAAGAGAGACAAGGCCAG | |||||
| R: CCCATTCTCTATGACTCACTCTCCAA | |||||
| Bol1-20 | 1 | 10421763 | F: GAAGAAAAGACTACAAGAGGACTGAA | T/C | 0.36 |
| F: GAAGAAAAGACTACAAGAGGACTGAG | |||||
| R: GCAGAGATTCCATGGGTCTCTTGTT | |||||
| Bol1-31 | 1 | 17385144 | F: AAATAAACATGTGAAATTTATATATTTTGTGAC | C/T | 0.37 |
| F: AAATAAACATGTGAAATTTATATATTTTGTGAT | |||||
| R: CCAGGCAAATTAACCTCATTTAATTTTTA | |||||
| Bol1-50 | 1 | 31515139 | F: CAACATTATTATTAAAATAAAGAAACTGCAGGT | T/C | 0.37 |
| F: AACATTATTATTAAAATAAAGAAACTGCAGGC | |||||
| R: CCGAAGCCTTAGAACTCATCTGAGTA | |||||
| Bol1-57 | 1 | 37692928 | F: ACAAGTCGAAGCAGAAGACAGTATC | G/A | 0.36 |
| F: AACAAGTCGAAGCAGAAGACAGTATT | |||||
| R: CGTGACTCGAATCTGTAACATTGTTCTTA | |||||
| Bol2-20 | 2 | 11815525 | F: TGAAGTGACGAGATATTCTGG | G/A | 0.37 |
| F: CGCTTGAAGTGACGAGATATTCTGA | |||||
| R: TAAAGCGAAAGCCAACAGCGAGGTT | |||||
| Bol2-23 | 2 | 14119719 | F: CACTCATTGTTTGACGAACTTTATGTGA | T/G | 0.37 |
| F: ACTCATTGTTTGACGAACTTTATGTGC | |||||
| R: TTATGCAACAAACATAGGAGATGGCTCAA | |||||
| Bol2-42 | 2 | 29630462 | F: ATCGTGGAAACCAATCAGTTTGCG | C/T | 0.36 |
| F: ATATCGTGGAAACCAATCAGTTTGCA | |||||
| R: GGCTCCTGCACGGATCAAATACAAT | |||||
| Bol2-52 | 2 | 38362631 | F: TGCTAGAGAAGTCTCAAGAACAC | G/A | 0.36 |
| F: CCTTGCTAGAGAAGTCTCAAGAACAT | |||||
| R: GCATCATCTGGGCTCCTCTGTTTTA | |||||
| Bol2-56 | 2 | 42324031 | F: TATTATAATAAAAAGAACACAAGAAATAACTG | G/A | 0.38 |
| F: ATTATAATAAAAAGAACACAAGAAATAACTA | |||||
| R: GTCAGTTTGTGCATCCTTAATAACACAAAT | |||||
| Bol3-2 | 3 | 572949 | F: CAACTTCAGCTTCAATGGATTGTCCT | T/C | 0.37 |
| F: AACTTCAGCTTCAATGGATTGTCCC | |||||
| R: CACAAATAGCAGAACTGCAGAAAGCATT | |||||
| Bol3-12 | 3 | 10349815 | F: CCACACAGACGAACGAACTTTTGAA | T/A | 0.36 |
| F: CCACACAGACGAACGAACTTTTGAT | |||||
| R: TTTACGCAGGGGAGGGTTTGGATTA | |||||
| Bol3-21 | 3 | 19333749 | F: TCTTGCACTGATGGCAAGTCAAG | C/T | 0.37 |
| F: GTTCTTGCACTGATGGCAAGTCAAA | |||||
| R: TTGCAGGATGATAACTCTGATGGAACAAA | |||||
| 名称 Name | 染色体 Chromosome | 位置 Position(bp) | 引物序列 Sequence of primer (5′-3′) | 变异类型 Alleles type | PIC |
| Bol3-36 | 3 | 32861302 | F: CTTCAAAATTAACCAAGAAATATGAACATAC | G/C | 0.37 |
| F: CTTCAAAATTAACCAAGAAATATGAACATAG | |||||
| R: GTTATCTCCCTAATCTGTTATCTCCTCTA | |||||
| Bol3-43 | 3 | 40497522 | F: GTCGTTGATTAAGGTAGGAGAAGC | C/T | 0.37 |
| F: GTCGTTGATTAAGGTAGGAGAAGT | |||||
| R: CTGATTTCCTCCTCGGAACCAACAT | |||||
| Bol3-52 | 3 | 50577875 | F: GACACATCGTATTCTGAGGATGAAG | G/T | 0.37 |
| F: AAGACACATCGTATTCTGAGGATGAAT | |||||
| R: CCACTGATCATACAGTTCACAGTACTTT | |||||
| Bol4-1 | 4 | 2284389 | F: ATTAAGTTATTCTTAAAACTCACACATTAGTAG | G/C | 0.36 |
| F: AAGTTATTCTTAAAACTCACACATTAGTAC | |||||
| R: CCAATCTTAGAGATAATATAGCCCATGATT | |||||
| Bol4-10 | 4 | 9227821 | F: ATCGAAGTTATTGGTGGCTGTAAGG | C/T | 0.37 |
| F: GATCGAAGTTATTGGTGGCTGTAAGA | |||||
| R: TCTTTGGAACATCATCTCTACGTACCTTT | |||||
| Bol4-17 | 4 | 14041102 | F: ATCCTTGCACAAGGTCCTTGCG | C/T | 0.37 |
| F: AAATCCTTGCACAAGGTCCTTGCA | |||||
| R: AACAATTTCGAGAGTGATCCTGAGGAATT | |||||
| Bol4-30 | 4 | 23412413 | F: GGTTGAGGATTACTCTGAGGCTC | C/A | 0.37 |
| F: GGTTGAGGATTACTCTGAGGCTA | |||||
| R: CAACAGTGATCTCTTCACCTCCTGAA | |||||
| Bol4-42 | 4 | 30318761 | F: GTCATCAGTCCACGCTGGAATG | C/G | 0.37 |
| F: GTCATCAGTCCACGCTGGAATC | |||||
| R: GATGGTAGACTCCAGACGAGTTCTT | |||||
| Bol4-52 | 4 | 37907183 | F: ATGAGCAATATTAGTAATCAAAGTCATGC | C/A | 0.37 |
| F: GATGAGCAATATTAGTAATCAAAGTCATGA | |||||
| R: CTGCTCTCGATGGTATGCAATAGTTTAAA | |||||
| Bol5-1 | 5 | 179080 | F: AGGATTGAGATGGCTCGAAAATAAGAT | T/C | 0.36 |
| F: GAGATGGCTCGAAAATAAGAC | |||||
| R: CGAAATCAAGGTACCTACTACTTTGCTAA | |||||
| Bol5-23 | 5 | 13597610 | F: TATATCTTTTTGGTCAAAAGTTTATGCATTAA | A/G | 0.36 |
| F: ATATCTTTTTGGTCAAAAGTTTATGCATTAG | |||||
| R: CCATGTCCTAATCAAAAGAATCAAATCCAA | |||||
| Bol5-26 | 5 | 15649112 | F: GTTTTTGTGTGGTTCGTCTGGTCA | T/A | 0.37 |
| F: GTTTTTGTGTGGTTCGTCTGGTCT | |||||
| R: AGATACAATAGAGCCCCCACATTTGTAAT | |||||
| Bol5-32 | 5 | 19285540 | F: CAATTTTTAGTGATATACCAAAGTCTGCTTT | T/C | 0.37 |
| F: AATTTTTAGTGATATACCAAAGTCTGCTTC | |||||
| R: GACATCATTATAGCTTTCACTGATGTTGTT | |||||
| Bol5-42 | 5 | 27307916 | F: CAAATAAAGACTTGTTCAACCTCCTATC | G/T | 0.36 |
| 名称 Name | 染色体 Chromosome | 位置 Position(bp) | 引物序列 Sequence of primer (5′-3′) | 变异类型 Alleles type | PIC |
| F: ACAAATAAAGACTTGTTCAACCTCCTATA | |||||
| R: GGTCTGAGTTTGAATAAGTCCCCCTT | |||||
| Bol6-4 | 6 | 1518700 | F: GAGCTAACTACCTCGATTATTTATTTATTTAT | A/C | 0.37 |
| F: GAGCTAACTACCTCGATTATTTATTTATTTAG | |||||
| R: CTGTTGGCTTCTACCAGGTAAATTCAATA | |||||
| Bol6-13 | 6 | 8365364 | F: TATTACTATTTTGTTCATTTGTTTATTTTAA | T/A | 0.37 |
| F: CTTATTACTATTTTGTTCATTTGTTTATTTTAT | |||||
| R: CACTTTTTAACATGTACAAAATGGAAAATT | |||||
| Bol6-22 | 6 | 14958187 | F: AGAGTTAATTAATACAATAAATAGTAAATTTCA | T/C | 0.36 |
| F: AGAGTTAATTAATACAATAAATAGTAAATTTCG | |||||
| R: GGTCATCCGATTATATTGATTACGGTTAAT | |||||
| Bol6-37 | 6 | 25031015 | F: TCAACGTAAACAAACAAACATTCACATCA | T/C | 0.37 |
| F: CAACGTAAACAAACAAACATTCACATCG | |||||
| R: CTCTCTCTTTCTCTCTGTATATTCACAAAA | |||||
| Bol6-46 | 6 | 31431471 | F: CCACCTCTTCTAGGAGATGCATG | C/T | 0.37 |
| F: CCACCTCTTCTAGGAGATGCATA | |||||
| R: AGCCCCGACGTGATCATCGCAA | |||||
| Bol6-54 | 6 | 38213660 | F: GTCCAAATCCCATGGAAATATTTGAAAA | A/T | 0.37 |
| F: GTCCAAATCCCATGGAAATATTTGAAAT | |||||
| R: ATCATCATTTCACGACCGTTGTTGGTTT | |||||
| Bol7-5 | 7 | 3897328 | F: GTGTATGTATTAATGAGTCAACACATCC | C/G | 0.36 |
| F: GTGTATGTATTAATGAGTCAACACATCG | |||||
| R: GGAATTGGTCAAACCACTAATTTGATTGTA | |||||
| Bol7-21 | 7 | 18794441 | F: AGCCGCCTAGTTTATGTCGTCTT | A/G | 0.37 |
| F: GCCGCCTAGTTTATGTCGTCTC | |||||
| R: GGCTTATCGGCCCAGCGAGTTA | |||||
| Bol7-31 | 7 | 27199113 | F: AAGAATTATTACCATTTACATTATTATGTGATG | G/T | 0.36 |
| F: AAGAATTATTACCATTTACATTATTATGTGATT | |||||
| R: GTGCAACACAAATATACAGGATTAGCTGAA | |||||
| Bol7-40 | 7 | 33905766 | F: AAATTTAAAGAGGCGATTGTGGCCAA | A/G | 0.37 |
| F: AATTTAAAGAGGCGATTGTGGCCAG | |||||
| R: GAACACAATACGTAACCATAATTCTCTGAT | |||||
| BolIn-B | 7 | 38826795 | F: CCTTTAATGAGTTGAATCAAATGGTGAA | A/- | 0.37 |
| F: CCTTTAATGAGTTGAATCAAATGGTGAT | |||||
| R: GAACTTCAGAAGGATCAACGTTGTAGAAA | |||||
| Bol7-54 | 7 | 46200440 | F: ATGCCTCTTCTCTCTTTCTCCTGAA | T/C | 0.37 |
| F: GCCTCTTCTCTCTTTCTCCTGAG | |||||
| R: GAAACTAAAAGTGACACACGGAAGATGTT | |||||
| Bol8-1 | 8 | 1046990 | F: GAGGTTATCATCTTGACCTTACCATA | T/A | 0.37 |
| F: GAGGTTATCATCTTGACCTTACCATT | |||||
| 名称 Name | 染色体 Chromosome | 位置 Position(bp) | 引物序列 Sequence of primer (5′-3′) | 变异类型 Alleles type | PIC |
| R: AGCTACAAAACTCCGTCAAAAACGCTTAT | |||||
| Bol8-11 | 8 | 17295300 | F: GTCAAATGGGCTATAGATCATTAGACTTA | T/C | 0.35 |
| F: CAAATGGGCTATAGATCATTAGACTTG | |||||
| R: GAAACTGTTTCTTTGCATCTGCCAACAAA | |||||
| Bol8-26 | 8 | 27133960 | F: ACCAGTGGATGTTTCTGATGGGA | G/A | 0.36 |
| F: CCAGTGGATGTTTCTGATGGGG | |||||
| R: GCTGGAGAGATATTGGCATTCTTTAGTTT | |||||
| Bol8-35 | 8 | 34047383 | F: AATATGATCTACAACGCGCTGCC | C/T | 0.36 |
| F: GAATATGATCTACAACGCGCTGCT | |||||
| R: GTTGAAGGGACATAAGTGTTCCATACTT | |||||
| Bol8-40 | 8 | 37116007 | F: GAGAAGGAGCTCTCTGGTCTA | A/G | 0.37 |
| F: GAGAAGGAGCTCTCTGGTCTG | |||||
| R: TGATAACACGGAGAAATCAGGGGGT | |||||
| Bol8-43 | 8 | 39508393 | F: ACTATAGAAAGTGTCTATACTAATTAGAGTTA | T/G | 0.36 |
| F: CTATAGAAAGTGTCTATACTAATTAGAGTTC | |||||
| R: TGGTTTGAATGAAGATCGCTAAAGTAAATA | |||||
| Bol9-2 | 9 | 732156 | F: GGGTCTTCGACGTTTGTTTCTTGA | T/G | 0.37 |
| F: GGTCTTCGACGTTTGTTTCTTGC | |||||
| R: AACAGAGAAACAAGAGAGTTCCATTCCAA | |||||
| Bol9-12 | 9 | 8645093 | F: AAATAAAAGAAGTTTGATGAAGATGGGGT | T/A | 0.36 |
| F: AAATAAAAGAAGTTTGATGAAGATGGGGA | |||||
| R: TTTAATGGTTTTCCTTCTGTGCCTTATCAA | |||||
| Bol9-31 | 9 | 29633956 | F: ATTTACATTGTTACAAAATCAATCTCACAGTTT | T/A | 0.36 |
| F: TACATTGTTACAAAATCAATCTCACAGTTA | |||||
| R: CAAGCTAAAGCACCCACTATGAAATTGAT | |||||
| Bol9-42 | 9 | 34784844 | F: CCAAAATGACATGATTGGCTCAAAATTTT | T/C | 0.37 |
| F: CCAAAATGACATGATTGGCTCAAAATTTC | |||||
| R: GCAAAGCAGTTAAGGCAATTAACAACGAA | |||||
| Bol9-44 | 9 | 37947752 | F: CAAACTTCTTGAGATCTCTGGTCC | G/T | 0.37 |
| F: CCAAACTTCTTGAGATCTCTGGTCA | |||||
| R: TGATGAGTCACCACGTCGACAACAT |
新窗口打开
将59个主要品种的核心位点分型结果转化为二元编码数据,得到主要甘蓝品种的指纹图谱(表2)。参考甘蓝SSR-DNA指纹鉴定技术流程,品种间差异位点数≥2则可判定为不同品种。利用50个核心SNP位点构建的指纹图谱,各品种间差异位点数均≥2,因此,利用该DNA指纹图谱可对甘蓝品种进行有效区分。
Table 2
表2
表2中国59份主要甘蓝品种的SNP-DNA指纹图谱数据库
Table 2SNP-DNA Finger-printing database of 59 main cabbage varieties from China
| SNP名称 SNP allele | 01 | 02 | 03 | 04 | 05 | 06 | 07 | 08 | 09 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bol1-11 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol1-20 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | |
| Bol1-31 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 |
| 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | |
| Bol1-50 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol1-57 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol2-20 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | |
| Bol2-23 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | |
| Bol2-42 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | |
| Bol2-52 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | |
| Bol2-56 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol3-2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 |
| 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | |
| Bol3-12 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | |
| Bol3-21 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| SNP名称 SNP allele | 01 | 02 | 03 | 04 | 05 | 06 | 07 | 08 | 09 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol3-36 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 |
| 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol3-43 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 |
| 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | |
| Bol3-52 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | |
| Bol4-1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol4-10 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol4-17 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | |
| Bol4-30 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | |
| Bol4-42 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | |
| Bol4-52 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 |
| 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | |
| Bol5-1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | |
| Bol5-23 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | |
| Bol5-26 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | |
| Bol5-32 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| SNP名称 SNP allele | 01 | 02 | 03 | 04 | 05 | 06 | 07 | 08 | 09 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 |
| 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | |
| Bol5-42 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol6-4 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 |
| 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | |
| Bol6-13 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | |
| Bol6-22 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | |
| Bol6-37 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 |
| 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | |
| Bol6-46 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | |
| Bol6-54 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol7-5 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol7-21 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | |
| Bol7-31 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | |
| Bol7-40 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| BolIn-B | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | |
| SNP名称 SNP allele | 01 | 02 | 03 | 04 | 05 | 06 | 07 | 08 | 09 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 |
| Bol7-54 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | |
| Bol8-1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | |
| Bol8-11 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | |
| Bol8-26 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol8-35 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Bol8-40 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | |
| Bol8-43 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | |
| Bol9-2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | |
| Bol9-12 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | |
| Bol9-31 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 |
| 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | |
| Bol9-42 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | |
| Bol9-44 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 |
| 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
新窗口打开
2.3 主要甘蓝品种的聚类分析
根据50个核心位点的分型结果,利用NTSYS软件对59个供试品种进行聚类分析(图3)。结果表明,育成品种两两间的遗传相似系数为0.43—0.98。其中,‘秦甘78号’与‘争春’的遗传相似系数最小,为0.43,二者之间存在41个差异标记,这说明二者亲缘关系最远。而‘中甘21’与‘中甘628’的遗传相似系数最大,为0.98,二者之间的差异标记仅有2个,说明它们的遗传背景很相似。分析‘中甘21’与‘中甘628’的系谱发现,二者的父本完全相同,母本都是圆球春甘蓝材料。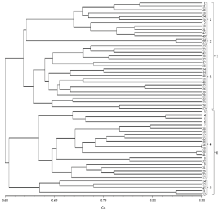 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图359个主要甘蓝品种的SNP聚类分析图
图中编号同电子附表1 The core in Fig. 3 was same as Table 1
-->Fig. 3Cluster analysis dendrogram based on SNP for 59 main cabbage varieties
-->
在遗传相似系数0.60处,可将所有供试品种分为两大类群。第一大类群共包含33个品种,在遗传相似系数0.64处可划分为3个亚类,‘中甘8号’、‘晚丰’等11个品种为第1亚类;‘春丰’、‘博春’为第2亚类;第3亚类包含‘春甘2号’、‘争春’等在内的20个品种。第二大类主要为圆球或近圆球类型甘蓝,共25个品种,在遗传相似系数0.61处可划分为2个亚类,将其命名为第4亚类与第5亚类。其中,第4亚类包含21个品种;第5亚类包含5个品种。聚类分析的结果与供试品种的来源有很强的相关性,例如:山西省农业科学院育成的惠丰系列甘蓝、河南省农业科学院育成的豫甘系列甘蓝、江苏镇江农业科学研究所育成的瑞甘系列甘蓝,上述单位培育的一系列甘蓝品种之间呈现较近的亲缘关系,聚类分析时聚在一起。结果表明,SNP聚类结果与供试品种的表型性状和地理来源相一致,能准确的反映供试品种的亲缘关系。
2.4 核心标记在品种鉴定中应用
通过人工构建的虚拟混合群体对DNA指纹图谱的核心位点进行验证,利用50个核心SNP标记对虚拟混合群体进行基因分型检测,并与已构建的DNA指纹图谱进行比对。基因分型结果显示:虚拟混合群体中来源于同一品种的3个样品其分型结果完全相同,这说明核心SNP标记具有较好的重复性。虚拟混合群体中有45个样品(15个品种)的分型结果与DNA指纹图谱中相应品种完全对应。剩余的15个样品(5个新组合),与指纹库中材料均表现出较大差异(差异位点数>2),在指纹库中无对应品种。结果表明,利用50个核心SNP标记可实现对甘蓝品种真实性和特异性的有效鉴定。3 讨论
3.1 主要甘蓝品种的代表性
目前,中国甘蓝种植面积约90万hm2[20,21],在蔬菜周年供应中占据重要地位。中国甘蓝品种选育开始于20世纪50年代、70年代以后得到快速发展。目前市场上审定推广的甘蓝品种大都为一代杂种,一代杂种的种植面积占总种植面积的90%以上[21]。本试验中所选用的59个甘蓝品种全为一代杂种。在59个甘蓝品种中,年推广面积曾达到1×104 hm2以上[23]且目前还有大面积种植的品种就有十几个,如‘京丰一号’、‘8398’、‘中甘21’、‘晚丰’、‘庆丰’、‘中甘15’、‘中甘11’、‘春丰’、‘争春’、‘西园四号’等,这些品种占到国内甘蓝种植面积的60%—70%[24]。本试验中所选的59个主要甘蓝品种涵盖了不同球型(圆球、扁球、牛心型)、不同栽培季节(春甘蓝、秋甘蓝、越冬甘蓝)及不同熟性(早、中、晚熟),这些品种具有广泛的代表性。
3.2 基于SNP位点构建DNA指纹图谱
DNA指纹图谱技术(DNA-Fingerprinting)是由英国科学家Jeffreys于1986年开发,具有快速、准确等优点,是鉴别品种、品系的有力工具,已广泛应用于很多作物的品种资源多样性和纯度鉴定研究,陆续将RFLP、AFLP、SSR、SNP等标记技术用于构建DNA指纹图谱。其中,SSR指纹技术由于其重复性好、简单易于操作、且大多数为共显性标记,常用于品种鉴定分析,已在水稻[25]、玉米[26]、柑橘[27]、甜瓜[28]等多种作物中得到应用。在甘蓝中,基于SSR标记已建立了50个代表品种的指纹图谱,并制定了SSR分子标记法进行甘蓝品种鉴定的技术规程(标准号:NY/T 2473-2013)。但SSR标记在实际使用中也暴露出一些不足,如标记数量有限、检测位点少,位点存在一定的突变率、对变异反应比较敏感等。SNP指纹技术是继RFLP和SSR之后发展起来的第3代标记技术,具有数量多、分布广泛、稳定性高、易于快速且高通量地进行基因型分型等优点[8]。与SSR相比,SNP是基于单核苷酸的突变,突变频率更低,遗传稳定性更高。SNP标记是构建DNA指纹数据库的推荐标记之一,但是目前甘蓝中尚没有基于SNP标记的指纹图谱。因此,构建基于SNP标记技术的指纹图谱对于甘蓝品种特异性和真实性鉴别、种子纯度鉴定具有重要意义。随着SNP检测技术的不断进步,SNP标记的检测成本也逐步降低。相对于SSR标记,目前高通量大样本的SNP标记检测已显现出优势。今后随着品种数量的增多、品种指纹图谱数据库的进一步丰富,高通量的SNP检测技术在新育成品种的真实性、特异性鉴定方面将具有非常广阔的应用前景。
3.3 利用核心标记构建SNP-DNA指纹图谱
指纹图谱核心标记的选择视物种基因组的复杂程度、标记类型、标记检测技术、品种数量等情况而定。匡猛等[29]利用36对SSR引物作为核心标记,构建了32个棉花主栽品种的指纹图谱。王立新[30]在小麦品种鉴定中提出使用21对核心引物、84对备用引物进行指纹研究。王庆彪等[5]使用20对核心引物构建了50个甘蓝品种的EST-SSR指纹图谱,利用其中8对多态性较好的引物便可将50个品种全部区分开。因此对于不同物种,构建指纹图谱的核心标记数量应视物种基因组复杂程度而定。SSR标记数量丰富、多态性高、呈共显性,可鉴别杂合子和纯合子,可用少量标记鉴别大量物种。与SSR标记不同,SNP标记虽然稳定性高但呈二态性,单个标记的多态性信息含量较低,鉴别相同数量品种所需要的标记数多于SSR标记。本研究中,筛选出了50个核心SNP位点用于主要甘蓝品种的指纹图谱构建,由于SNP标记的二态性,理论上每个标记可区分的杂交种数N1=3,本研究中选取得50个标记可区分的最大杂交种数N=3^50=7.18×1023,出现相同指纹图谱的概率P=1.39×10-24。因此,理论上而言,利用50个核心SNP标记构建甘蓝主要品种的指纹图谱是完全可行的。SNP-DNA指纹图谱的构建方式也与SNP检测方法有关。目前常用的SNP检测及分型的方法主要有以下几种,基于凝胶电泳检测的等位基因特异性PCR[31]、单链构象多态性[32]、酶切扩增多态性序列法[33]等;高通量自动化检测的直接测序法、DNA芯片技术[34]、竞争性等位基因特异性PCR[17]等。等位基因特异性PCR、酶切扩增多态性序列法等SNP检测方法由于其操作繁琐、准确率较低,不适用于构建指纹图谱。目前,利用DNA 芯片技术是检测 SNP 的最常用方法,已在玉米[14]、油菜[15]等作物的指纹图谱构建中得到应用。利用DNA芯片技术可直接进行指纹图谱构建,不需要进行核心标记筛选,但该方法制作DNA指纹图谱的成本较高。相比于DNA芯片技术,利用KASP技术进行SNP检测的成本较低,其成本与检测的SNP位点数呈正相关。从发展趋势来看,近年来随着甘蓝基因组重测序的陆续开展,越来越多的SNP标记得到开发,加上SNP标记相比SSR标记的一些优势,将来会更多地依赖于SNP标记进行品种的特异性、真实性鉴定。本研究中,利用KASP技术进行SNP检测并筛选出50个核心SNP位点用于构建59个主要甘蓝品种的指纹图谱,随着育种技术的发展、育成品种数量的增多,也有可能还需要增加SNP标记,尤其是与重要农艺性状紧密关联的SNP标记,以更准确、高效地检测出品种的真实性、特异性。同时,希望可以为新品种保护授权提供前期的鉴定工作,利用指纹图谱对品种进行初步鉴定,最终结合田间表型鉴定判定品种的特异性、真实性。
4 结论
利用50份甘蓝高代自交系重测序数据进行比对,共开发2.54×106个SNP位点。将442个SNP位点成功转化为KASP标记,并从中开发出一套适用于中国甘蓝品种指纹图谱构建的核心SNP组合,构建了中国59个甘蓝品种指纹图谱数据库。通过人工构建模拟群体验证,证明核心SNP组合可实现对甘蓝品种真实性和特异性的有效鉴定。The authors have declared that no competing interests exist.
