 ,1, 周秀文1, 梁雪2, 郭营1, 赵岩1, 李斯深1, 孔凡美
,1, 周秀文1, 梁雪2, 郭营1, 赵岩1, 李斯深1, 孔凡美 ,1,*
,1,*Genome-Wide Association Analysis for Yield and Nitrogen Efficiency Related Traits of Wheat at Seedling Stage
ZHANG PengXia ,1, ZHOU XiuWen1, LIANG Xue2, GUO Ying1, ZHAO Yan1, LI SiShen1, KONG FanMei
,1, ZHOU XiuWen1, LIANG Xue2, GUO Ying1, ZHAO Yan1, LI SiShen1, KONG FanMei ,1,*
,1,*通讯作者:
责任编辑: 李莉
收稿日期:2021-02-7接受日期:2021-04-14
| 基金资助: |
Received:2021-02-7Accepted:2021-04-14
作者简介 About authors
联系方式:张鹏霞,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (4907KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张鹏霞, 周秀文, 梁雪, 郭营, 赵岩, 李斯深, 孔凡美. 小麦苗期生物量及氮效率相关性状的全基因组关联分析. 中国农业科学, 2021, 54(21): 4487-4499 doi:10.3864/j.issn.0578-1752.2021.21.001
ZHANG PengXia, ZHOU XiuWen, LIANG Xue, GUO Ying, ZHAO Yan, LI SiShen, KONG FanMei.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】小麦是中国最重要的粮食作物之一,人类消耗热量的五分之一来自于小麦。氮元素是植物生长发育过程中最重要的矿质营养元素,目前,全球肥料使用量的62%来自于氮肥(FAO,2011),自2000年以来,氮肥消耗量以每年10%的速度急剧增长[1]。随着氮肥施用量的增多,小麦表现出耐肥的特性,使得氮肥利用率持续降低[2,3,4],中国氮肥利用率仅有21.2%—35.9%,氮肥的过量使用不仅导致了一系列环境污染问题[5,6,7],而且还增加了生产成本[8],因此,提高氮肥利用率以选育氮高效的小麦新品种是当前农业生产上面临的紧迫且重要的课题之一。【前人研究进展】国内外大量研究表明,不同基因型的小麦对矿质元素的吸收和利用存在显著差异[9,10,11],并且认为可以通过遗传改良来实现小麦氮素的高效利用。因此,对小麦氮效率的相关性状进行遗传学研究,充分挖掘其氮高效利用潜力,是提高小麦氮素利用效率、选育氮高效的小麦新品种和促进农业可持续发展的有效途径[12,13,14]。随着分子生物学技术及基因组测序技术的快速发展,全基因组关联分析成为一种获取候选基因的有效方法,由于该方法能够快速构建作图群体并绘制高分辨率的遗传图谱,通过多年多点处理快速锁定并分离出显著关联位点的候选基因,显著提高候选基因的准确度,逐渐在各类作物氮效率研究中得到应用[15,16,17]。迄今为止,基于SNP标记的全基因组关联分析(genome-wide association study,GWAS)在小麦加工品质[18,19]、生理[20,21]和抗病性[19, 22-23]等相关性状基因的研究中均有广泛报道。【本研究切入点】目前,小麦氮素相关性状的遗传学研究相对较少,尽管不同作物中氮素利用效率相关性状的QTL定位已有较多研究,然而据此克隆到的氮效率相关基因却鲜见报道。其主要原因在于,氮效率相关性状受环境影响大,表型鉴定相对繁琐;同时,受遗传图谱密度、供试群体大小及其特性的影响,鉴定的QTL覆盖基因组区间比较大,环境稳定性差,精确度有限,因此很难据此预测和分离相关候选基因。随着小麦基因组测序结果的公开,利用GWAS分析进行候选基因挖掘的可靠度不断增加,获得稳定可靠的显著关联SNP位点成为候选基因筛选和预测的关键,而群体表型性状精确鉴定和高精度SNP图谱是获得可靠SNP关联位点的前提。苗期是小麦生长的关键时期,苗期对氮素环境的适应能力在一定程度上能够反映其对养分环境的适应潜力。此外,营养液培养的试验条件可以实现精确控制,获得的表型性状精确度大幅度提高,从而为获得可靠的SNP关联位点提供关键数据基础。【拟解决的关键问题】本研究以134份小麦品种(系)组成的自然群体为供试群体,采用营养液培养的方法,连续2年分别设置低氮、正常氮和高氮处理,测定小麦苗期生物量及氮效率相关性状,并进行全基因组关联分析,期望定位到小麦苗期生物量及氮效率相关性状的重要QTL,挖掘、预测氮素高效利用的相关基因,为进一步开展小麦氮效率基因的克隆及其遗传改良提供依据。1 材料与方法
1.1 试验材料
以134份小麦品种(系)组成的自然群体为试验材料,包括国内山东省品种66个、河北省品种24个、河南省品种16个、陕西省品种14个、山西省品种8个、江苏省品种2个和四川省品种2个,以及国外澳大利亚品种1个和加拿大品种1个(电子附表1)。1.2 试验设计
1.2.1 试验处理 在山东农业大学本部玻璃温室进行营养液培养,参考Hoagland营养液配方配制试验所用营养液,并根据小麦的营养特性进行适当修改(正常营养液配方见表1),设置低氮(0.4 mmol·L-1,T1)、正常氮(4 mmol·L-1,T2)和高氮(8 mmol·L-1,T3)3个处理,各处理重复4次,随机区组设计。Table 1
表1
表1小麦苗期试验正常营养液组分
Table 1
| 大量元素 Macronutrients | 浓度 Concentration (mmol·L-1) | 微量元素 Trace elements | 浓度 Concentration (μmol·L-1) | |
|---|---|---|---|---|
| KH2PO4 | 0.2 | H3BO3 | 1.0 | |
| MgSO4·7H2O | 0.5 | (NH4)6Mo7O24·4H2O | 0.1 | |
| KCl | 1.8 | CuSO4·5H2O | 0.5 | |
| CaCl2 | 1.5 | ZnSO4·7H2O | 1.0 | |
| (NH4)2SO4·H2O | 1.0 | MnSO4·H2O | 1.0 | |
| Ca(NO3)2·4H2O | 1.0 | Fe·EDTA | 100.0 |
新窗口打开|下载CSV
1.2.2 试验方案 选择大小基本一致的134份供试小麦品种(系)种子,用10%的H2O2消毒灭菌5 min后用蒸馏水冲洗,摆放在发芽网上进行发芽。幼苗长至1叶1心时(约7 d),每个材料选择长势一致的3株幼苗,去掉幼苗胚乳后移至育苗盘中,每个品种(系)的3株幼苗放置在同一穴中。134个材料统一放置在同一个育苗盘中并置于盛有Hoagland营养液的黑色塑料水产盒中(容积25 L),添加营养液20 L。前3 d采用1/2浓度的营养液进行缓苗,之后用完全培养液(加处理)进行培养,每隔3 d更换1次营养液,用稀NaOH和0.1 mmol·L-1 HCl调节pH至6.0—6.2,24 h不间断通气,培养28 d收获。
第一次试验时间(E1)为2013年11月25日—2014年1月17日,温度为5.8—28.6℃,湿度为8.6%—85.9%,光照强度为0—57.8 klx;第二次试验时间(E2)为2014年3月2日—2014年4月22日,温度为7.5—33.9℃,湿度为10.1%—91.3%,光照强度为0—69.0 klx。
1.2.3 测定指标、方法及计算方法 用烘箱105℃将样品烘干,然后用千分之一电子天平称量地上部生物量(shoot dry weight,SDW)与根系生物量(root dry weight,RDW),计算植株总生物量(total dry weight of plant,TDW)=SDW+RDW。采用H2SO4-H2O2消煮法和凯氏定氮仪测定地上部氮含量(shoot nitrogen concentration,SNC)与根系氮含量(root nitrogen concentration,RNC),计算植株总氮含量(total nitrogen concentration of plant,TNC)=(SNC×SDW+RNC× RDW)/TDW、地上部氮累积量(shoot nitrogen accumulation,SNA)=SDW×SNC、根系氮累积量(root nitrogen accumulation,RNA)=RDW×RNC、植株氮累积量(total nitrogen accumulation of plant,TNA)=TDW×TNC、氮累积量根冠比(root/shoot ratio of nitrogen accumulation,RSNA)=RNA/SNA、地上部氮利用效率(shoot nitrogen use efficiency,SNUE)=SDW/SNC、根系氮利用效率(root nitrogen use efficiency,RNUE)=RDW/RNC和植株氮利用效率(total nitrogen use efficiency of plant,TNUE)=TDW/ TNC[24]。
1.3 数据统计方法
1.3.1 遗传多样性分析和群体结构分析 利用SPSS 19.0软件和R语言进行数据统计分析和结果可视化,计算性状广义遗传力:hB2=σg2/(σg2+σe2)[25],其中,σe2表示环境方差,σg2表示遗传方差。用Structure 2.3.1软件对供试群体进行群体遗传结构分析,并计算其相应的Q值。依据似然值最大的原则选取合适的K值作为群体数目。利用Powermarker V3.25软件对SNP的多态性信息含量、基因多样性和遗传距离进行分析,结合软件NJ phylogenetic tree绘制聚类图,利用TASSEL V5.0进行供试群体主成分分析,最终结合以上3种方法确定供试群体的群体结构和亚群数目。1.3.2 遗传图谱和全基因组关联分析 利用小麦90K(含81 587个SNP)基因芯片对134份小麦品种(系)组成的自然群体进行扫描,在得到的SNP标记位点中,删除缺失率大于15%,最小等位基因频率(minimum allele frequency,MAF)小于5%的SNP标记位点,最终剩余9 329个SNP位点,大约85.24%(7952/9329)SNP有遗传位置信息,分布在21条染色体上[26]。试验分别对2次试验环境(E1和E2)下的3个氮处理(T1E1、T2E1、T3E1、T1E2、T2E2和T3E2)以及2个环境下相同处理的平均值(T1AV、T2AV和T3AV)的14个性状(共126个性状-环境)进行关联分析。利用TASSEL V5.0软件中Analysis模块下的Link.Diseq和Kinship计算亲缘关系K矩阵,采用TASSEL V5.0软件中的MLM+Q+K混合线性模型进行全基因组关联分析,P≤0.001时认为存在显著关联。曼哈顿图和Q-Q图可视化在R环境中通过CMplot程序包完成。
2 结果
2.1 小麦苗期氮效率相关性状表型数据分析
通过对小麦苗期氮效率相关性状表型进行分析(表2),与正常氮处理相比,低氮和高氮处理下苗期生物量及氮效率相关性状均差异显著。低氮处理条件下,根系、地上部及植株氮含量和氮积累量均显著下降,而根生物量及根、植株氮效率均显著增加,高氮处理下,几乎所有指标均显著增加。各供试性状在2次试验间均表现出显著差异,如,2014年(E2)的植株生物量、植株氮累积量及植株氮利用效率均显著高于2013年(E1),这可能与试验中的其他因素(如温度、湿度以及光照的差异等)有关。Table 2
表2
表2小麦苗期生物量和氮效率相关性状的统计参数和遗传力
Table 2
| 性状 Trait | 处理 Treatment | 均值 Average | 变异系数 CV (%) | 遗传力 hB2 (%) | 性状 Trait | 处理 Treatment | 均值 Average | 变异系数 CV (%) | 遗传力 hB2 (%) | |
|---|---|---|---|---|---|---|---|---|---|---|
| 地上部干重 SDW (mg) | T1E1 | 107.95cd | 21.40 | 94.95 | 地上部氮累积量 SNA (mg/plant) | T1E1 | 5.00d | 12.77 | 93.53 | |
| T2E1 | 104.08d | 23.99 | T2E1 | 5.98c | 6.93 | |||||
| T3E1 | 124.71b | 23.51 | T3E1 | 7.64b | 10.36 | |||||
| T1E2 | 112.33c | 19.16 | T1E2 | 4.32e | 8.64 | |||||
| T2E2 | 129.49b | 22.28 | T2E2 | 7.79b | 10.23 | |||||
| T3E2 | 148.77a | 22.62 | T3E2 | 9.68a | 4.76 | |||||
| 根系干重 RDW (mg) | T1E1 | 27.80c | 35.87 | 92.76 | 根系氮累积量 RNA(mg/plant) | T1E1 | 0.99c | 22.54 | 93.49 | |
| T2E1 | 19.50e | 21.49 | T2E1 | 0.77d | 24.18 | |||||
| T3E1 | 22.40d | 22.99 | T3E1 | 0.98c | 24.21 | |||||
| T1E2 | 50.10a | 27.03 | T1E2 | 1.14b | 19.78 | |||||
| T2E2 | 27.60c | 22.30 | T2E2 | 1.00c | 23.44 | |||||
| T3E2 | 32.20b | 22.89 | T3E2 | 1.27a | 23.85 | |||||
| 植株总干重 TDW (mg) | T1E1 | 136.06d | 21.00 | 95.37 | 植株总氮累积量 TNA (mg/plant) | T1E1 | 5.97d | 26.93 | 94.31 | |
| T2E1 | 123.08e | 21.99 | T2E1 | 6.75c | 24.20 | |||||
| T3E1 | 147.10c | 22.81 | T3E1 | 8.62b | 25.48 | |||||
| T1E2 | 162.52b | 20.53 | T1E2 | 5.46d | 21.53 | |||||
| T2E2 | 157.18b | 21.77 | T2E2 | 8.79b | 25.94 | |||||
| T3E2 | 180.66a | 22.03 | T3E2 | 10.95a | 23.66 | |||||
| 生物量根冠比 RSDW | T1E1 | 0.27b | 32.68 | 89.72 | 氮累积量根冠比 RSNA | T1E1 | 0.20b | 37.13 | 84.25 | |
| T2E1 | 0.19d | 16.80 | T2E1 | 0.13c | 23.66 | |||||
| T3E1 | 0.18d | 17.48 | T3E1 | 0.13c | 26.83 | |||||
| T1E2 | 0.44a | 16.12 | T1E2 | 0.27a | 28.16 | |||||
| T2E2 | 0.22c | 13.16 | T2E2 | 0.13c | 26.20 | |||||
| T3E2 | 0.22c | 14.00 | T3E2 | 0.13c | 27.63 | |||||
| 地上部氮含量 SNC (g·kg-1) | T1E1 | 46.31d | 6.42 | 86.39 | 地上部氮利用效率 SNUE (g·g-1kg-1) | T1E1 | 2.35bc | 26.93 | 94.79 | |
| T2E1 | 57.55c | 3.86 | T2E1 | 1.82e | 24.20 | |||||
| T3E1 | 61.21b | 3.85 | T3E1 | 2.04d | 25.48 | |||||
| T1E2 | 38.66e | 10.41 | T1E2 | 3.00a | 21.53 | |||||
| T2E2 | 59.79b | 4.75 | T2E2 | 2.18cd | 25.94 | |||||
| T3E2 | 64.96a | 4.47 | T3E2 | 2.29b | 23.66 | |||||
| 根系氮含量 RNC (g·kg-1) | T1E1 | 35.32c | 8.21 | 85.71 | 根系氮利用效率 RNUE (g·g-1kg-1) | T1E1 | 0.80b | 27.23 | 89.44 | |
| T2E1 | 40.48b | 8.09 | T2E1 | 0.47c | 22.32 | |||||
| T3E1 | 43.74a | 12.5 | T3E1 | 0.51c | 18.75 | |||||
| T1E2 | 22.64d | 7.25 | T1E2 | 2.26a | 19.18 | |||||
| T2E2 | 35.51c | 10.1 | T2E2 | 0.78b | 16.92 | |||||
| T3E2 | 39.44b | 9.37 | T3E2 | 0.81b | 17.52 | |||||
| 植株总氮含量 TNC (g·kg-1) | T1E1 | 44.07d | 5.26 | 23.68 | 植株总氮利用效率 TNUE (g·g-1kg-1) | T1E1 | 3.09b | 22.66 | 41.34 | |
| T2E1 | 54.87c | 6.12 | T2E1 | 2.25e | 24.62 | |||||
| T3E1 | 58.56b | 4.25 | T3E1 | 2.51d | 23.62 | |||||
| T1E2 | 33.75e | 5.10 | T1E2 | 4.92a | 22.15 | |||||
| T2E2 | 55.50c | 11.20 | T2E2 | 2.84c | 21.40 | |||||
| T3E2 | 60.44a | 5.26 | T3E2 | 2.99b | 22.11 |
新窗口打开|下载CSV
此外,供试群体生物量及氮效率相关性状也表现出显著的基因型差异(表2)。供试群体变异系数范围为3.41%(地上部氮含量)—37.13%(氮累积量根冠比),生物量及氮效率相关性状均表现出连续变异,为典型的数量性状,遗传率为41.34%(植株总氮利用效率)—95.37%(植株总干重)。对2个环境下的均值进行相关性分析(图1),结果表明,除了根系干重与地上部累积量,植株总干重与植株总氮利用效率,植株总氮累积量与根系氮利用效率不相关,苗期生物量与氮效率相关性状间均呈显著正相关或负相关。
图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1苗期生物量及氮效率相关性状之间的相关系数
SDW:地上部干重;TDW:植株总干重;RSDW:生物量根冠比;SNC:地上部氮含量;RNC:根系氮含量;TNC:植株总氮含量;SNA:地上部氮累积量;RNA:根系氮累积量;TNA:植株总氮累积量;RSNC:氮累积量根冠比;SNUE:地上部氮利用效率;RNUE:根系氮利用效率;TNUE:植株总氮利用效率。下同
Fig. 1Correlation coefficients (r) between traits related to biomass and nitrogen efficiency at seedling
SDW: Shoot dry weight; TDW: Total dry weight of plant; RSDW: Root/shoot ratio of biomass; SNC: Shoot nitrogen concentration; RNC: Root nitrogen concentration; TNC: Total nitrogen concentration of plant; SNA: Shoot nitrogen accumulation; RNA: Root nitrogen accumulation; TNA: Total nitrogen accumulation of plant; RSNC: Root/shoot ratio of nitrogen accumulation; SNUE: Shoot nitrogen use efficiency; RNUE: Root nitrogen use efficiency; TNUE: Total nitrogen use efficiency of plant. The same as below
2.2 群体结构分析
用Structure 22.3.4软件进行群体结构分析(图2-a和图2-c),结果表明,K=2时,ΔK达到峰值。利用Power Marker V3.25分析SNP的多态性信息含量和基因多样性,计算自然群体两两间的遗传距离,构建NJ聚类图(图2-b)。利用TASSEL V5.0软件进行供试群体主成分分析,结果显示,前2个主成分可解释14.85%的群体信息。综合以上3种方法来研究供试群体结构,最终将供试群体确定为2个亚群,分别为13个小麦品种/系(亚群1)和121个小麦品种/系(亚群2)[26]。图2
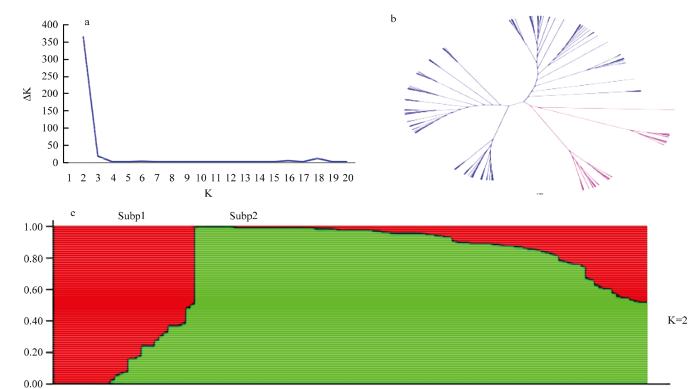 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2供试134个小麦品种的群体结构
a:供试材料的K值与ΔK的关系;b:基于遗传距离的NJ聚类图;c:群体结构;2个亚群用不同颜色表示
Fig. 2Population structure analysis of 134 cultivars based on unlinked SNP markers
a: Graphical relationship between K and ΔK for wheat accessions; b: Neighbor-joining tree of 134 wheat accessions; c: Population structure of wheat accessions; The two subgroups identified from the tree are in different colors
2.3 小麦苗期氮效率相关性状的稳定位点
共调查了14个生物量及氮效率相关性状,分别对2次试验(E1和E2)的3个氮处理(T1E1、T2E1、T3E1、T1E2、T2E2和T3E2)以及2次试验相同处理的平均值(T1AV、T2AV和T3AV)共9个关联分析环境的126个环境-性状进行关联分析,共定位到838个显著(P≤0.001)关联的SNP标记,分布在小麦21条染色体上(电子附表2和电子附图1)。在定位到的838个显著关联的SNP标记位点中,仅在一个关联分析环境中被检测到SNP标记位点有435个(51.91%),至少在2个关联分析环境中被检测到SNP标记位点有403个,其中有8个SNP标记位点(RAC875_c29540_391、BobWhite_c9881_1312、GENE-0427_442、wsnp_RFL_Contig3060_2967139、Kukri_c65481_121、tplb0025f09_1052、wsnp_Ku_c1765_ 3452586、BS00022498_51)至少在3个关联分析环境中被检测到,分别与根系氮利用效率、根系干重、地上部累积量、植株总干重、植株总氮累积量、植株总氮利用效率、地上部干重和生物量根冠比显著关联。其中,Kukri_c65481_121和tplb0025f09_1052分别与植株总氮利用效率(TNUE)和根系总氮利用效率(RNUE)显著关联并在4个环境下(包括均值环境)被检测到,tplb0025f09_1052对根系氮利用效率的贡献率达到36.09%(表3)。Table 3
表3
表3与苗期生物量及氮效率相关性状显著关联(P≤0.001)的环境稳定SNP标记
Table 3
| 性状 Trait | 环境 Environment | 标记 Marker | 染色体 Chr. | 位置 Position (bp) | P | 贡献率 R2(%) | |
|---|---|---|---|---|---|---|---|
| 最大值 Max | 最小值 Min | ||||||
| RNUE | T1E1, T3AV, T3E2 | RAC875_c29540_391 | 1A | 8290703 | 9.95E-04 | 1.39E-05 | 11.05—18.34 |
| 根系干重RDW | T2E1, T3AV, T3E2 | BobWhite_c9881_1312 | 1B | 10256166 | 3.58E-04 | 3.49E-04 | 10.09—10.13 |
| 根系氮利用效率RNUE | T2AV, T2E1, T3E1 | BobWhite_c9881_1312 | 1B | 10256166 | 7.45E-04 | 3.75E-05 | 9.04—13.69 |
| 地上部氮含量SNC | T1E1, T3AV, T3E2 | GENE-0427_442 | 1B | 5688119 | 5.79E-04 | 8.62E-05 | 9.35—12.35 |
| 植株总干重TDW | T1E1, T3AV, T3E2 | GENE-0427_442 | 1B | 5688119 | 6.54E-04 | 1.88E-04 | 9.17—11.11 |
| 植株总氮含量TNC | T1E1, T3AV, T3E2 | GENE-0427_442 | 1B | 5688119 | 6.25E-04 | 1.08E-04 | 9.23—11.98 |
| 地上部氮含量SNC | T2AV, T2E1, T1E2 | wsnp_RFL_Contig3060_2967139 | 2A | 9928033 | 8.05E-04 | 1.87E-04 | 11.41—13.91 |
| 植株总氮含量TNC | T2AV, T2E1, T1E2 | wsnp_RFL_Contig3060_2967139 | 2A | 9928033 | 2.95E-04 | 2.05E-04 | 12.57—13.74 |
| 植株总氮利用效率TNUE | T2AV, T2E1, T1AV, T1E2 | Kukri_c65481_121 | 3B | 13483435 | 7.19E-04 | 2.08E-04 | 9.09—11.40 |
| 根系氮利用效率RNUE | T1AV, T1E1, T3AV, T3E2 | tplb0025f09_1052 | 4B | 1591267 | 7.25E-04 | 1.32E-09 | 11.49—36.09 |
| 地上部干重SDW | T2E2, T1AV, T1E1 | wsnp_Ku_c1765_3452586 | 6A | 14086572 | 6.09E-04 | 1.39E-04 | 9.07—11.58 |
| 生物量根冠比RSDW | T2AV, T2E2, T3E2 | BS00022498_51 | 7B | 6251008 | 5.32E-04 | 3.25E-04 | 12.01—12.85 |
新窗口打开|下载CSV
2.4 小麦苗期氮效率相关性状共同关联位点和单倍型分析
试验筛选出211个同时与至少3个性状显著关联的SNP标记,其中同时与至少6个性状显著关联的SNP标记有5个(表4),1A染色体的BS00077492_51与SDW、TDW、SNC、TNC、SNUE和TNUE均显著关联,1B染色体的BS00065737_51、IACX16577和GENE-0427_442与SDW、TDW、SNC、TNC、SNUE和TNUE均显著关联,2A染色体的wsnp_RFL_ Contig3060_2967139与SDW、TDW、SNC、TNC、SNUE和TNUE均显著关联,这些位点相对更为可靠,可用于分子标记辅助育种和QTL精细定位。对共定位的5个位点进行单倍型分析(图3),结果表明,在SNP位点BS00065737_51、IACX16577和GENE-0427_442存在3个单倍型(1B-Hap1、1B-Hap2和1B-Hap3),单倍型为1B-Hap3材料的地上部干重和地上部氮含量均显著高于1B-Hap2的,在SNP位点wsnp_RFL_Contig3060_2967139存在3个单倍型(2A-Hap1、2A-Hap2和2A-Hap3),单倍型为2A-Hap3材料的地上部干重均显著高于2A-Hap2的地上部干重,单倍型为2A-Hap2材料的植株总干重、地上部氮含量、植株总氮含量、地上部氮利用效率和植株总氮利用效率均显著高于2A-Hap1和2A-Hap3。Table 4
表4
表4小麦苗期同时与6个性状显著关联的SNP位点
Table 4
| 标记和染色体 Marker and Chr. | 性状 Trait | 分析环境 Analysis environment | 贡献率 R2 (%) |
|---|---|---|---|
| BS00077492_51 1A | 地上部干重SDW | T1E1 | 12.63 |
| 植株总干重TDW | T1E1 | 11.31 | |
| 地上部氮含量SNC | T1E1 | 11.73 | |
| 植株总氮含量TNC | T1E1 | 11.76 | |
| 地上部氮利用效率SNUE | T1E1 | 12.42 | |
| 植株总氮利用效率TNUE | T1E1 | 11.80 | |
| BS00065737_51 1B | 地上部干重SDW | T1E1, T3E2 | 8.93—11.37 |
| 植株总干重TDW | T1E1 | 8.60 | |
| 地上部氮含量SNC | T1E1 | 9.80 | |
| 植株总氮含量TNC | T1E1 | 8.87 | |
| 地上部氮利用效率SNUE | T1E1, T3E2 | 10.11—10.75 | |
| 植株总氮利用效率TNUE | T3E2 | 9.01 | |
| GENE-0427_442 1B | 地上部干重SDW | T1E1, T3E2 | 10.86—11.65 |
| 植株总干重TDW | T1E1, T3AV, T3E2 | 9.17—11.11 | |
| 地上部氮含量SNC | T1E1, T3AV, T3E2 | 9.35—12.35 | |
| 植株总氮含量TNC | T1E1, T3AV, T3E2 | 9.23—11.98 | |
| 地上部氮利用效率SNUE | T3E2 | 10.44 | |
| 植株总氮利用效率TNUE | T3AV, T3E2 | 9.65—11.33 | |
| IACX16577 1B | 地上部干重SDW | T1E1, T3E2 | 9.49—11.83 |
| 植株总干重TDW | T1E1, T3E2 | 8.82—8.97 | |
| 地上部氮含量SNC | T1E1, T3E2 | 8.92—9.96 | |
| 植株总氮含量TNC | T1E1, T3E2 | 8.72—9.30 | |
| 地上部氮利用效率SNUE | T1E1, T3E2 | 10.82—11.02 | |
| 植株总氮利用效率TNUE | T1E1, T3E2 | 9.19—9.81 | |
| wsnp_RFL_Contig3060_2967139 2A | 地上部干重SDW | T2AV, T2E1 | 13.39—13.77 |
| 植株总干重TDW | T2AV, T2E1 | 12.85—13.57 | |
| 地上部氮含量SNC | T2AV, T2E1, T1E2 | 13.25—13.91 | |
| 植株总氮含量TNC | T2AV, T2E1, T1E2 | 13.33—13.74 | |
| 地上部氮利用效率SNUE | T2AV, T2E1 | 12.99—13.69 | |
| 植株总氮利用效率TNUE | T2AV, T2E1 | 12.04—12.73 |
新窗口打开|下载CSV
2.5 候选基因预测
根据小麦基因组注释及LD衰减水平,对与6个性状共同定位的5个SNP位点和2个多环境(4个环境)关联的SNP位点进行了候选基因的筛选。据目标SNP附近214 kb的基因组区域中共鉴定出候选基因84个,根据已知克隆氮效率基因的编码蛋白类型、候选基因功能注释信息及利用植物比较基因组学资源库蛋白序列同源比对分析,筛选到3个候选基因与氮效率相关(表5和图3),其中,TraesCS1B02G058300编码的Agamous MADS-box transcription factor已经有研究报道参与小麦氮效率调控,该基因主要在根部表达;TraesCS2A02G122000编码Membrane-associated 30 kD protein;TraesCS4B02G007100编码R3h domain containing protein。图3
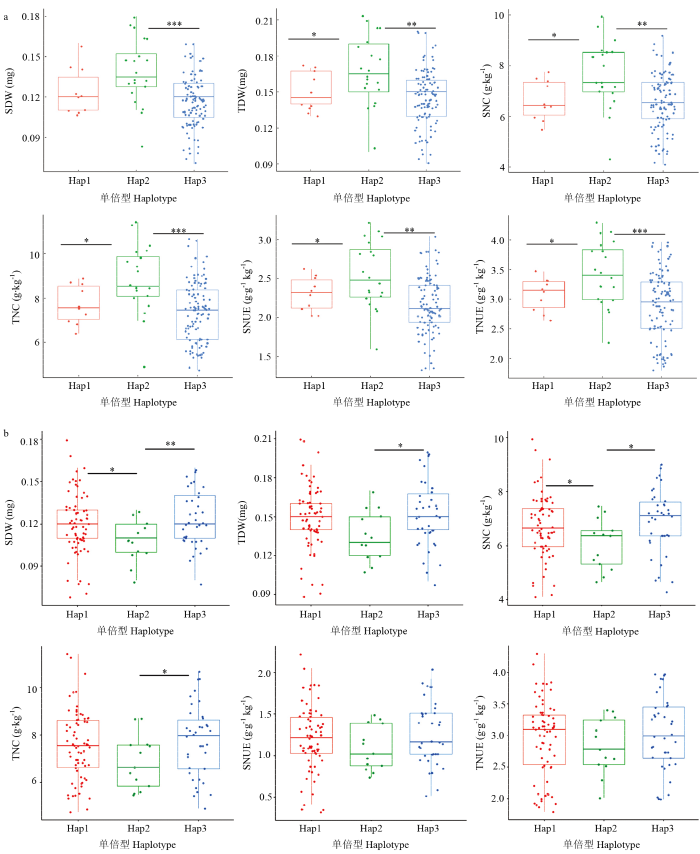 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3SNP位点的单倍型分析
a:SNP位点BS00065737_51、IACX16577和GENE-0427_442的单倍型分析;b:SNP位点snp_RFL_Contig3060_2967139的单倍型分析。*、**、***分别表示在0.05、0.01、0.001水平差异显著
Fig. 3Haplotype analysis of SNP markers
a: Haplotype analysis of BS00065737_51, IACX16577 and GENE-0427_442; b: Haplotype analysis of snp_RFL_Contig3060_2967139. *, **, ***represent significant at the 0.05, 0.01, 0.001 probability levels, respectively
Table 5
表5
表5候选基因预测
Table 5
| 染色体 Chr. | 基因 Gene | 位置 Position (Mb) | 标记 Marker | 基因注释或编码蛋白 Gene annotation or coding protein |
|---|---|---|---|---|
| 1B | TraesCS1B02G058300 | 183.39 | BS00065737_51 | 无性的MADS盒子转录因子 Agamous MADS-box transcription factor |
| 2A | TraesCS2A02G122000 | 332.13 | wsnp_RFL_Contig3060_2967139 | 与膜相关的30 kD蛋白 Membrane-associated 30 kD protein |
| 4B | TraesCS4B02G007100 | 46.43 | tplb0025f09_1052 | 含R3h结构域的蛋白质 R3h domain containing protein |
新窗口打开|下载CSV
根据3个目标候选基因的不同时期表达模式和氮响应情况的分析(电子附图4),表明TraesCS1B02G058300和TraesCS4B02G007100在根系中具有较高的表达量,而TraesCS2A02G122000在叶中具有较高的表达量。TraesCS1B02G058300在氮缺乏处理下表达,而TraesCS4B02G007100和TraesCS2A02G122000在氮缺乏处理下表达量相对较低。
3 讨论
氮素是植物叶绿素、蛋白质、核酸、酶类、生物碱、植物激素以及维生素类等物质的重要组成成分,是决定作物产量的首要因素,选育氮高效品种是维持我国小麦生产安全和可持续发展的重要途径。长期以来,由于育种群体大,选择频率高,氮素测定消耗大,导致氮素利用效率评价困难,育种家主要依据地上部表型性状来判断品种氮素利用效率[27,28]。近年来,小麦基因芯片的大量开发和应用,促进了数量性状遗传研究的进程,为育种过程中氮素效率的评价提供了新的思路[19,29]。GWAS分析是解析数量性状遗传特性的重要工具,一方面,与双亲群体相比,自然群体积累了广泛的重组事件,可以提供更多有效重组信息,可以更加精细定位QTL,且可短时间内发现更多关联位点。此外,GWAS分析具有更高的分辨率,有利于了解目标基因的位置、遗传效应和基因网络等信息以及遗传位点的精细定位和图位克隆。本研究利用小麦90K SNP芯片,在不同的氮素水平下,通过GWAS分析对小麦苗期的生物量及氮效率相关性状进行相关研究,以期发掘与氮素利用效率相关的SNP标记和遗传位点,预测相关性状的候选基因,为小麦氮效率相关性状的基因克隆及其在育种中的应用提供参考。目前,在小麦不同氮处理试验中已经有大量的QTL和QTL簇被检测到[30,31,32,33,34],其中大部分QTL只能在单一的环境中被检测到。本研究对14个生物量及氮效率相关性状进行全基因组关联分析,共检测到838个显著关联的SNP位点,分布在21条染色体上。其中,435个SNP位点仅在一个关联分析环境中被检测到,占总数的51.91%。这些SNP位点的相关基因极有可能与不同氮素环境下的特异调控有关,但是由于数量众多、贡献率小和稳定性差,难以进行这些SNP位点相关基因的下一步研究。但与此同时,有8个SNP位点(RAC875_c29540_391、BobWhite_c9881_1312、GENE-0427_442、wsnp_RFL_Contig3060_2967139、Kukri_c65481_121、tplb0025f09_1052、wsnp_Ku_ c1765_3452586、BS00022498_51)至少在3个关联分析环境中被检测到,这些SNP位点存在氮素利用效率相关基因的可能性较大,但迄今尚未有进一步的研究,无法探讨其遗传机制。尽管前人针对小麦氮素利用效率遗传机制开展过多项研究,也在小麦2A、2B、4A、5A、7A和7B染色体上发现一批控制小麦氮素利用效率相关遗传位点[15, 35-37],但是由于试验群体、标记类型和标记来源的差异较大,无法进行可靠的对比,无法确定是否与本研究发现位点一致。
另外,本研究筛选出5个多性状共定位的SNP位点(BS00077492_51、BS00065737_51、GENE-0427_ 442、IACX16577和wsnp_RFL_Contig3060_2967139),分布于1A、1B(2)和2A(2)染色体上,共同定位的性状主要有地上部干重(SDW)、植株总干重(TDW)、地上部氮累积量(SNC)、植株总氮累积量(TNC)、地上部氮利用效率(SNUE)和植株总氮利用效率(TNUE)。相关性分析结果表明,这6个性状均在P<0.01水平呈极显著相关,表明同一SNP位点可同时与多个性状显著关联,同时也表明这些性状(生物量和氮效率)之间存在着密切关系,可能受相同的SNP/基因调控。大量QTL分析结果也表明,在染色体上存在着热点QTL区域[32, 38]。张国华[39]研究结果表明多个性状之间存在相互关联现象,这种现象可能与QTL相互连锁或一因多效有关。在5个共定位SNP位点中,4个SNP位点在低氮或高氮条件下表达,而在正常施氮条件下不表达,表明这些SNP位点与小麦对不同氮素环境的适应性密切相关。这些与小麦氮效率相关的高频表达SNP位点和多性状共定位SNP位点可能对小麦氮效率利用有重要的调控作用,在进行小麦氮效率遗传分子机制的研究中值得进一步探讨。通过对5个位点的单倍型分析,发现SNP位点wsnp_RFL_Contig3060_2967139的单倍型2A-Hap2为地上部氮利用效率和植株总氮利用效率的优异单倍型,在所有试验材料中,济麦20、鲁麦14、洛旱6号、新麦26、衡0628、冀5265、金麦1号、石麦18号、石新618、陕农534、西农85、西农213、淮麦16和澳大利亚红麦等品种具有较高的氮素利用效率,含有优异等位基因,可作为氮高效品种推广或作为氮高效育种的优异种质材料应用。
根据小麦基因组注释及连锁不平衡(linkage disequilibrium,LD)衰减水平,在5个与6个性状共同定位的SNP位点和2个多环境(4个环境)关联的SNP位点214 kb的基因组区域中共鉴定出候选基因84个,初步筛选得到3个候选基因与氮效率相关。其中,在1B染色体上检测到一个编码Agamous MADS-box transcription factor的候选基因(TraesCS1B02G058300)。MADS-box基因家族在植物生长发育和信号传导过程中起着不可替代的作用[40],它的功能主要是参与调控植物生殖生长阶段,如控制开花时间、决定分生组织特性、花器官形态建成和果实成熟等,同时调节着植物根、叶、花和果实的发育,尤其在生殖发育中控制花分生组织和花器官中的基因表达,此外,也有研究表明MADS-box基因的表达也参与胚形态建成[41]、抗逆境[42]、以及决定开花时间[43]等过程的调控。最新的研究表明[44,45],MADS-box基因家族参与调控小麦氮效率,在根部硝酸盐供应中起重要作用,这为进一步发掘该基因的功能提供了新线索。以上研究结果也为本试验鉴定得到的氮效率相关候选基因提供了可靠依据。此外。在4B染色体上检测到一个编码R3h domain containing protein的候选基因(TraesCS4B02G007100),在含有优异单倍型的SNP位点wsnp_RFL_Contig3060_ 2967139检测到一个编码Membrane-associated 30 kDa protein的候选基因(TraesCS2A02G122000),尚未有关小麦氮效率的研究,值得深入探讨。
随着小麦基因组序列的释放,基因功能验证与发掘成为小麦后基因组时代的重要课题。生物体中普遍存在的一因多效现象也让基因功能的确定成为一大难题,仍需要扎实的基因功能验证工作。目前基于可靠的关联标记的生物信息分析是发掘复杂农艺性状相关候选基因的有效方法,然而,由于小麦氮效率涉及到多个复杂的生理、生化过程,要确定特定基因的功能还需要后续进一步的功能验证工作。
4 结论
不同氮素处理显著影响小麦苗期生物量、氮效率相关性状及其相关QTL的表达,大多数SNP位点仅在1个氮素检测环境中被检测到,但也存在环境稳定性较强的位点。生物量及氮效率相关性状之间存在显著相关关系,并在一定程度上受到相同的QTL/基因控制,相关候选基因的功能有待进一步验证。(责任编辑 李莉)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1007/s10681-017-1869-5URL [本文引用: 1]
DOI:10.1007/s10113-016-1101-5URL [本文引用: 1]
DOI:10.1016/j.agrformet.2018.07.019URL [本文引用: 1]
[本文引用: 1]
DOI:10.1038/ngeo608URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.biortech.2004.05.007URL [本文引用: 1]
DOI:10.1016/j.fcr.2016.10.001URL [本文引用: 1]
DOI:10.1007/s11104-015-2694-zURL [本文引用: 1]
DOI:10.1016/j.plantsci.2011.03.020URL [本文引用: 1]
DOI:10.1016/j.fcr.2017.12.004URL [本文引用: 1]
DOI:10.1007/s40009-017-0553-6URL [本文引用: 1]
DOI:10.1111/nph.2005.168.issue-2URL [本文引用: 1]
DOI:10.1007/s00122-004-1902-7URL [本文引用: 2]
DOI:10.1534/genetics.105.048603URL [本文引用: 1]
DOI:10.1038/90135URL [本文引用: 1]
DOI:10.1371/journal.pone.0164293URL [本文引用: 1]
DOI:10.1186/s12870-017-1167-3URL [本文引用: 3]
DOI:10.1186/s12870-018-1234-4URL [本文引用: 1]
DOI:10.1186/1471-2229-14-128URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1080/01904168109362919URL [本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 2]
[D].
[本文引用: 2]
[本文引用: 1]
DOI:10.1038/nplants.2015.117URL [本文引用: 1]
DOI:10.1038/srep41247URL [本文引用: 1]
DOI:10.1007/s00122-013-2249-8URL [本文引用: 1]
DOI:10.1007/s00122-009-1076-4URL [本文引用: 1]
DOI:10.1007/s00122-011-1749-7URL [本文引用: 2]
DOI:10.1007/s00122-006-0429-5URL [本文引用: 1]
DOI:10.1007/s00122-013-2201-yURL [本文引用: 1]
DOI:10.1007/s11104-006-0030-3URL [本文引用: 1]
DOI:10.1186/1471-2156-15-57URL
DOI:10.1007/s00122-009-1076-4URL [本文引用: 1]
DOI:10.1007/s11104-013-1844-4URL [本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1016/S0378-1119(03)00747-9URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1104/pp.117.1.91URL [本文引用: 1]
DOI:10.1111/pbi.2018.16.issue-6URL [本文引用: 1]
DOI:10.1104/pp.19.01482URL [本文引用: 1]
