 ,, 沈佳, 张跃建, 李国景, 牛晓伟, 寿伟松
,, 沈佳, 张跃建, 李国景, 牛晓伟, 寿伟松 ,浙江省农业科学院蔬菜研究所,杭州 310021
,浙江省农业科学院蔬菜研究所,杭州 310021Fine Mapping of an Immature Rind Color Gene GR in Melon
XU XinYang ,, SHEN Jia, ZHANG YueJian, LI GuoJing, NIU XiaoWei, SHOU WeiSong
,, SHEN Jia, ZHANG YueJian, LI GuoJing, NIU XiaoWei, SHOU WeiSong ,Institute of Vegetables, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021
,Institute of Vegetables, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021通讯作者:
责任编辑: 赵伶俐
收稿日期:2020-09-20接受日期:2020-12-16
| 基金资助: |
Received:2020-09-20Accepted:2020-12-16
作者简介 About authors
许昕阳,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2273KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
许昕阳, 沈佳, 张跃建, 李国景, 牛晓伟, 寿伟松. 甜瓜幼果果皮颜色基因GR的精细定位[J]. 中国农业科学, 2021, 54(15): 3308-3319 doi:10.3864/j.issn.0578-1752.2021.15.014
XU XinYang, SHEN Jia, ZHANG YueJian, LI GuoJing, NIU XiaoWei, SHOU WeiSong.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】甜瓜(Cucumis melo L.)是我国重要的园艺和经济作物。果皮颜色作为甜瓜最直观的外观品质性状,直接影响其商品价值和消费者的选择,开展甜瓜果皮颜色性状的遗传研究对甜瓜育种有重要意义。【前人研究进展】瓜类植物的果皮颜色调节开始于果实发育的早期,在幼果期表现为绿色,反映的是叶绿素含量[1]。随着果实发育的进行,甜瓜外果皮颜色在成熟过程中会发生较大转变[2]。成熟甜瓜果皮颜色丰富多样,主要有白色、黄色、橙色、绿色以及多种颜色混杂或者斑纹类型。早期对甜瓜果皮颜色是由单基因或者多基因控制存在较大的争议,但该争议多基于不同品种间组合的差异[3,4]。KUBICKI[5]认为幼果果皮白色对绿色为显性,并且是受单基因Wi遗传控制。PEREIRA等[6]通过构建‘Védrantais’和‘Piel de Sapo’的高密度GBS遗传图谱发现浅皮为显性性状,并且将该基因定位在7号染色体,该位置与Wi相近,推测为同一个基因。BURGER等[7]利用两个杂交群体:深皮‘Dulce’和浅皮‘TAM-Dew’,深绿‘Krymka’和浅绿‘Eshkolit Ha’Amaqim’探究甜瓜幼果果皮颜色遗传模式时发现浅皮亲本受隐性单基因控制,并且该基因与之前报道的浅皮显性基因Wi不等位。OREN等[8]利用‘Tam Dew’和‘Dulce’的重组自交系以及‘Dulce’和‘Noy Amid’的F3:4家系将一个影响甜瓜幼果果皮颜色的基因定位在4号染色体上290 kb的区段内,后通过差异性分析确定基因CmAPRR2是影响该幼果果皮颜色的基因。在成熟果中,HUGHES[9]发现‘Honeydew’品种的白色相对于‘Smiths’ Perfect cantaloupe’品种的绿色为隐性。TADMOR等[1]在黄皮‘Noy Amid’和绿皮‘TVT’杂交的F2分离群体中发现柚皮素查尔酮含量的分离比符合孟德尔3﹕1的遗传定律,证实了该色素受单基因调控。FEDER等[10]进一步利用该F2群体进行了基因定位,并结合遗传转化鉴定出影响成熟果果皮颜色的基因CmKFB。另外,利用‘Piel de Sapo’和‘PI 161375’的杂交组合,研究者们也鉴定了果皮颜色相关的QTLs[11,12]。【本研究切入点】随着分子生物学技术的发展,许多甜瓜果皮颜色相关的QTLs和基因被分离和鉴定。但是,多数基因的精细定位仍存在定位区间较大、候选基因众多等情况。【拟解决的关键问题】本研究构建薄皮甜瓜两个幼果果皮颜色差异的纯系甜瓜‘MR-1’和厚皮甜瓜‘LGR’杂交组合,对其进行表型观察、群体构建、遗传模式分析、基因图位克隆等,精细定位幼果果皮颜色基因GR,为深入揭示甜瓜幼果果皮颜色的形成机制奠定基础。1 材料与方法
1.1 试验材料
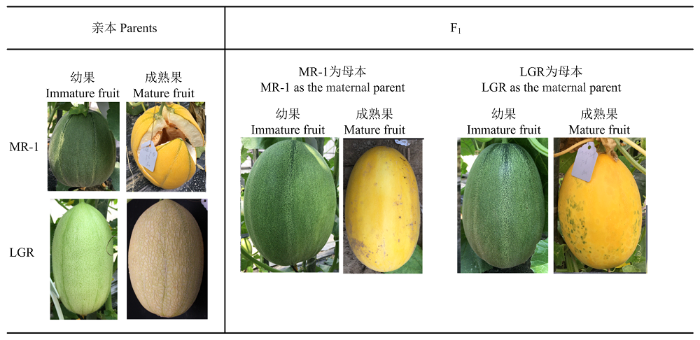
本研究所使用的材料‘MR-1’由国外引进,属于薄皮甜瓜亚种泡瓜变种(C. agrestis var. momordica),果实为近圆形;‘LGR’由浙江省农业科学院蔬菜所甜瓜研究室提供,属于厚皮甜瓜亚种冬甜瓜变种(C. melo var. inodorous),果实为长椭圆型(图1),成熟果有网纹[13,14]。利用两个材料杂交构建了正反交F1群体、F1与‘LGR’回交BC1F1群体,主要用于遗传分析;BC1F1和F2群体主要用于BSA-seq和遗传定位。杂交和回交构建于2017年春季,定位群体种植于2017年秋季、2018年春秋、2019年春秋以及2020年春季。所有材料均种植在浙江海宁甜瓜实验基地,采用常规大田生产管理。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1亲本以及正反交F1果皮颜色表型
Fig. 1Rind colors of parental and reciprocal cross F1
1.2 果皮颜色性状调查
两亲本甜瓜‘LGR’和‘MR-1’的幼果果皮颜色调查以花后10 d左右的果实为对象,利用目测对比颜色深浅,分别定为“浅绿色”和“深绿色”。遗传分析和定位群体中单株的表型调查也以花后10 d左右的果实为对象,对照两亲本的幼果果皮颜色,经3次不同时间(花后8、10和12 d)的目测定义表型。成熟果颜色调查和测色仪定量测定取自花后35 d左右的果实。利用美能达CR-400/410色彩色差计(Konica Minolta Camera Co., Ltd, Osaka, Japan)对果皮颜色定量,其中L*值代表亮度从黑(0)至白(100),a*值代表从红(+a)至绿(-a),b*值代表从黄(+b)至蓝(-b)。利用SPSS Statistics V21.0进行相关性分析。1.3 BSA-seq分析
从BC1F1群体中挑选深绿皮和浅绿皮植株各20株,利用CTAB法提取DNA。将20个深绿皮和20个浅绿皮单株DNA分别等量混合,形成两个混池。2个混池和2个亲本DNA按照样品检测、文库构建、质量检测以及上机测序等流程送由公司操作。参考基因组为甜瓜基因组Harukei3 genome reference ver1.41(1.4 分子标记开发
基于亲本重测序信息,在初定位区段的双侧和内部挑选有多态性的Indel标记,利用凝胶电泳鉴定分子标记的结合能力和特异性。后利用合适的引物对F2和BC1F1群体中的隐性单株个体进行基因型鉴定,验证初步连锁结果以及缩小定位区间。1.5 DNA提取、PCR扩增与聚丙烯酰胺凝胶电泳
取新鲜的甜瓜叶片于冷冻干燥机中放置1 d,采用CTAB法提取DNA:将叶片磨成粉末状,加入600 μL预热的CTAB提取液,摇匀,65℃水浴30 min;加入600 μL氯仿﹕异戊醇(24﹕1)抽提液,用力摇晃2 min后,12 000 r/min离心5 min;取上清液,加入360 μL(0.6倍体积上清液)-20℃预冷的异丙醇,-20℃中放置30 min;12 000 r/min离心5 min,弃上清液,加入700 μL 70%无水乙醇洗涤2次,晾干后溶于去离子水中,-20℃保存。PCR扩增体系为10 μL,其中包括:2×buffer mix 5 μL,上下游引物各0.3 μL,DNA 2 μL,加去离子水2.4 μL。PCR反应程序:98℃预变性2 min,然后扩增33个循环(包括:94℃ 30 s;55℃ 30 s;72℃ 40 min),接着72℃ 5 min,4℃ 10 min。扩增好的样品于4℃保存。
2 结果
2.1 亲本果皮颜色表型
通过对花后10 d的果皮颜色观察,发现薄皮甜瓜‘MR-1’幼果果皮表现为深绿色,花后35 d的成熟果果皮表现为亮黄色;而厚皮甜瓜‘LGR’幼果果皮表现为浅绿色,成熟果果皮表现为灰色有网纹(图1)。2.2 幼果果皮颜色遗传模式分析
为探究幼果果皮颜色的遗传规律,以‘MR-1’和‘LGR’为亲本进行正反交组配,所得F1代的幼果果皮颜色均与‘MR-1’表型相同,表明幼果果皮颜色受核基因控制,且深绿对浅绿为显性(图1)。F2群体植株的幼果果皮颜色发生分离,其中132个果实为深绿色果皮,44个果实为浅绿色果皮,符合3﹕1的孟德尔遗传分离比,BC1F1代群体表现为1﹕1的分离比(表1)。这些结果表明幼果果皮颜色是受单基因控制的质量性状。因该基因主要控制果皮绿色的深浅,因此将其命名为GR(Green Rid)。Table 1
表1
表1幼果果皮颜色表型的遗传学分析
Table 1
| 组合 Cross | 深绿皮植株 Dark green individual | 浅绿皮植株 Light green individual | 理论比 Expected ratio | Chi-square | Pa |
|---|---|---|---|---|---|
| P1 (MR-1) | 10 | 0 | |||
| P2 (LGR) | 0 | 10 | |||
| F1 (P1×P2) | 25 | 0 | |||
| F1 (P2×P1) | 25 | 0 | |||
| F2 (F1 ?) | 132 | 44 | 3﹕1 | 0.000 | 1.000 |
| BC1F1 (F1×LGR) | 98 | 89 | 1﹕1 | 0.433 | 0.510 |
新窗口打开|下载CSV
2.3 GR的初步定位
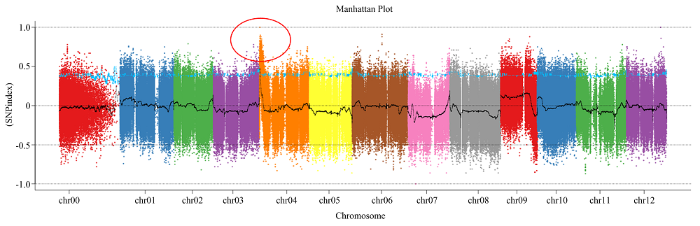
为了分离和克隆GR,首先通过从BC1F1群体中挑选浅绿色和深绿色果皮植株各20株进行DNA混池测序(BSA-seq)。参照两亲本的重测序信息,全基因组连锁分析显示4号染色体长臂上存在一个目标性状区域(图2)。该基因座的物理位置为203 744—1 974 725 bp,置信区间跨越1.8 Mb。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2GR的初定位(红圈代表目的区段)
Fig. 2Manhattan plot for mapping of GR (The red circle is the target interval)
2.4 GR的精细定位
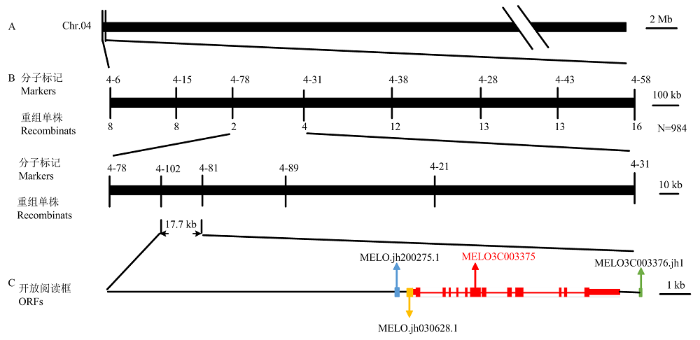
基于亲本重测序得到的覆盖了13条染色体,共1 320 497个多态性分子标记,在4号染色体上开发位于初定位区间两侧的标记4-6和4-58,对BC1F1群体中的隐性单株(浅绿皮)进行基因型鉴定。结果显示浅绿皮单株与标记4-6有十分明显的共分离趋势(图3-A),证实了BSA初定位的结果。为了精细定位该基因,开发了初定位结果内特异性好的引物(图3-B,表2),并对扩大的BC1F1和F2群体共984株(BC1F1:877株,F2:107株)进行基因型鉴定,结果共筛选出24个重组交换单株(图4-B)。通过对重组交换单株进行后代表型和基因型鉴定,最终将GR精细定位于标记4-102和4-81之间,物理距离为17.7 kb(Chr04: 602468-620197)(图4-A、B)。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3GR连锁分析
A:隐性单株在分子标记4-6的基因型。泳道1为‘MR-1’,2为‘LGR’,8、19、58为重组交换单株;B:引物多态性。上方标记为引物名,每个引物左边泳道为‘MR-1’,右边为‘LGR’。M为DNA marker
Fig. 3Linkage analysis of GR
A: Genotype of recessive single plant DNAs at marker 4-6; Lane 1 is the parent ‘MR-1’, Lane 2 is the parent ‘LGR’, Lane 8, 19, 58 are the recombination individuals; B: Polymorphism identification of markers. The mark represents the primers, the left lane of each primer is the parent ‘MR-1’, the right is the ‘LGR’. M: DNA size ladder
Table 2
表2
表2定位所用引物序列
Table 2
| 引物名 Primer | 正向引物 Forward primer (5'-3') | 反向引物 Reverse primer (5'-3') |
|---|---|---|
| 4-6 | GGTGATCTAGGGCTTCTTTT | GGGCTTACCCTTGATTTAAC |
| 4-15 | CGGATAATGGGTTCTATGTG | CAATTCAATCCCCTCACTAC |
| 4-78 | TTTTCTTTTAACTTGGCCTG | TTTTTGGAAGAGGATACAACA |
| 4-31 | TGACATCAATAATGCCTCTTT | CTTGACGTAGGACAAATGGT |
| 4-38 | GAGGATTTGTTTGGCTTTTA | CATCTCTCATGTTGATGTCG |
| 4-28 | TAATCTAAGCTCCTTCGTGG | AGTTTATGAAGGCTTTGTGG |
| 4-43 | CTACGTGGTATCACAAGCAA | AGCCAAAGTGTAATGCAACT |
| 4-58 | ACGACAAATAAATCGGTGAT | TAGGAAATCAACCAAATGCT |
| 4-102 | TGTATTATCCTCCGGACAAC | AATTACCAACCAATCCAATG |
| 4-81 | TCGGTTAAAAAGAGCCATTA | GAAGATGATTATCGGATTGC |
| 4-89 | TGAGCAAAGGTTTACAACAA | GTAATTTCAAGATCGCCTTC |
| 4-21 | AACGGTACCAAACTAGGCTT | CTTATGTTGGGAATGTGACC |
新窗口打开|下载CSV
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4GR的精细定位
A:GR初定位于4号染色体4-6和4-58标记之间;B:利用984个单株精细定位GR于4-102和4-81标记约17.7 kb范围内。标记下面的数字代表交换单株数;C:定位区间内的候选基因
Fig. 4Fine mapping of GR
A: The GR gene was located on the chromosome 4 between marker 4-6 and 4-58; B: The GR gene was mapped to 17.7 kb region between marker 4-102 and 4-81, number of recombinant individuals are shown under the markers; C: ORFs was identified as the candidate gene in the mapping interval
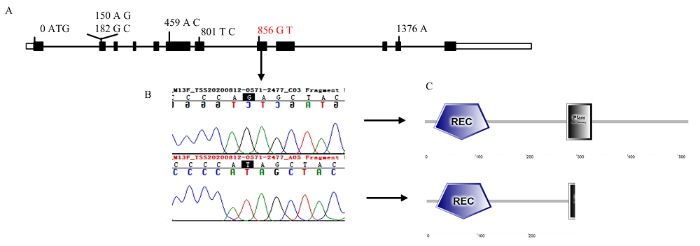
由于重组交换单株的缺乏,对17.7 kb区间内包含的基因进行序列比对分析。根据最新的甜瓜参考基因组的基因注释发现该区间包含了4个基因:2个新注释基因(MELO.jh200275.1和MELO.jh030628.1)、1个完整注释基因(MELO3C003375)以及MELO3C003376的一部分(图4-C)。其中,MELO. jh200275.1编码未知蛋白;MELO.jh030628.1预测编码一个类生长素蛋白;MELO3C003375编码一个双组分反应调节蛋白,该蛋白属于Golden2-like类转录因子,故被命名为CmAPRR2;MELO3C003376编码一个烟草花叶病毒复制蛋白。通过序列比对发现MELO.jh030628.1、MELO3C003375和MELO3C003376等3个基因的编码区序列在两亲本中存在差异(表3)。其中,MELO.jh030628.1和MELO3C003376仅存在由单碱基替换造成的错义变异,MELO3C003375除了3处同义突变和1处错义突变,还存在1处单碱基替换造成的无义变异(表3)。通过对亲本‘MR-1’和‘LGR’测序,证明‘LGR’中MELO3C003375编码区第856位碱基(G变为T)产生了终止密码子,致使MELO3C003375蛋白翻译提前终止,导致了Myb-DNA结合结构域大部分缺失(图5)。在黄瓜、番茄以及西瓜中,MELO3C003375的同源基因Csa3G904140.3、Solyc08g077230以及ClCG09G012330 被报道参与叶绿素和类胡萝卜素的合成以及质体发育等过程[8,15-16]。由此推测,MELO3C003375(CmAPRR2)为控制甜瓜幼果果皮颜色的基因GR,而单碱基替换造成的无义突变为造成两亲本幼果果皮颜色深浅的关键差异位点。
Table 3
表3
表3定位区段内基因编码区变异
Table 3
| 染色体 CHROM | 位置 POS | 参考碱基 REF | 比对碱基 ALT | 变异类型 Variant-classification | 基因号 Gene_ID | 参考氨基酸 REF (aa) | 比对氨基酸 ALT (aa) |
|---|---|---|---|---|---|---|---|
| chr04 | 612438 | T | G | 错义突变 Missense_variant | MELO.jh030628.1 | K | Q |
| chr04 | 612476 | T | C | 错义突变 Missense_variant | MELO.jh030628.1 | N | S |
| chr04 | 612484 | G | A | 同义突变 Synonymous_variant | MELO.jh030628.1 | A | A |
| chr04 | 612488 | G | T | 错义突变 Missense_variant | MELO.jh030628.1 | T | K |
| chr04 | 612491 | C | G | 错义突变 Missense_variant | MELO.jh030628.1 | R | P |
| chr04 | 612501 | C | T | 错义突变 Missense_variant | MELO.jh030628.1 | E | Q |
| chr04 | 612505 | A | G | 同义突变 Synonymous_variant | MELO.jh030628.1 | Y | S |
| chr04 | 612519 | A | T | 错义突变 Missense_variant | MELO.jh030628.1 | I | I |
| chr04 | 613526 | A | G | 同义突变 Synonymous_variant | MELO3C003375 | E | E |
| chr04 | 613558 | G | C | 错义突变 Missense_variant | MELO3C003375 | R | P |
| chr04 | 614480 | A | C | 同义突变 Synonymous_variant | MELO3C003375 | L | L |
| chr04 | 614898 | T | C | 同义突变 Synonymous_variant | MELO3C003375 | G | G |
| chr04 | 615687 | G | T | 终止突变 Stop_gained | MELO3C003375 | E | * |
| chr04 | 617619 | A | G | 错义突变 Missense_variant | MELO3C003375 | Q | R |
| chr04 | 620196 | T | G | 错义突变 Missense_variant | MELO3C003376 | L | R |
新窗口打开|下载CSV
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5MELO3C003375的基因结构、变异位点和蛋白结构域分析
A:MELO3C003375的基因结构。数字代表到转录起始位点的距离。前面的碱基代表‘MR-1’中的碱基,后面代表‘LGR’中的碱基;B:引起LGR蛋白截短的变异位点;C:‘MR-1’和‘LGR’中MELO3C003375的蛋白结构域
Fig. 5Analysis of gene structure, variant site, and protein domain of MELO3C003375
A: Gene structure of MELO3C003375. The number represents the distance to the transcription start site. The former number represents the base in the ‘MR-1’, the latter number represents the base in the ‘LGR’; B: The variant site which generating a truncated protein in ‘LGR’; C: The function domains of MELO3C003375 in ‘MR-1’ and ‘LGR’
2.5 幼果果皮颜色与成熟果皮颜色存在显著相关性
为探究甜瓜幼果果皮颜色与成熟果皮颜色之间的相关性,对BC1F1群体共153个单株的幼果进行目测,并对照亲本将其划分深绿(1)和浅绿(2)两类。因成熟果果皮颜色变化差异较大,使用测色仪定量分析成熟果颜色变异(表4)。通过统计分析和相关性检测,发现幼果果皮颜色与成熟果皮颜色的亮度指标和绿色强度指标的相关系数分别为0.849和0.857,呈极显著相关。这说明幼果果皮颜色与成熟果果皮颜色存在显著相关性。Table 4
表4
表4BC1F1群体植株果皮颜色表型
Table 4
| 单株 Individual | 目测分型 Visual phenotype | 测色仪分型 Colorimeter phenotype | 单株 Individual | 目测分型 Visual phenotype | 测色仪分型 Colorimeter phenotype | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| L* | a* | b* | -a*/b* | L* | a* | b* | -a*/b* | ||||
| 1938-10 | 1 | 34.13 | -9.27 | 12.92 | 0.72 | 1938-23 | 2 | 64.23 | -21.15 | 37.73 | 0.56 |
| 1938-102 | 1 | 35.93 | -11.70 | 15.61 | 0.75 | 1938-108 | 2 | 64.65 | -21.53 | 39.58 | 0.54 |
| 1938-7 | 1 | 37.88 | -11.34 | 15.67 | 0.72 | 1938-103 | 2 | 64.92 | -18.34 | 35.51 | 0.52 |
| 1938-118 | 1 | 38.22 | -11.56 | 15.74 | 0.73 | 1938-85 | 2 | 65.01 | -21.73 | 38.89 | 0.56 |
| 1938-145 | 1 | 39.05 | -14.92 | 19.32 | 0.77 | 1938-171 | 2 | 65.56 | -16.07 | 30.12 | 0.53 |
| 1938-121 | 1 | 39.61 | -11.60 | 16.06 | 0.72 | 1938-178 | 2 | 66.27 | -19.51 | 36.34 | 0.54 |
| 1938-54 | 1 | 39.99 | -12.87 | 18.36 | 0.70 | 1938-94 | 2 | 66.33 | -20.65 | 37.11 | 0.56 |
| 1938-6 | 1 | 40.02 | -13.92 | 19.04 | 0.73 | 1938-197 | 2 | 66.35 | -20.05 | 37.66 | 0.53 |
| 1938-157 | 1 | 40.29 | -14.03 | 18.26 | 0.77 | 1938-179 | 2 | 66.75 | -18.38 | 35.02 | 0.52 |
| 1938-142 | 1 | 40.31 | -14.97 | 20.42 | 0.73 | 1938-61 | 2 | 67.49 | -18.07 | 33.77 | 0.54 |
| 1938-136 | 1 | 40.76 | -12.21 | 16.60 | 0.74 | 1938-82 | 2 | 67.50 | -20.62 | 37.68 | 0.55 |
| 1938-151 | 1 | 40.93 | -13.66 | 19.46 | 0.70 | 1938-15 | 2 | 67.67 | -21.07 | 41.99 | 0.50 |
| 1938-169 | 1 | 41.14 | -16.18 | 22.67 | 0.71 | 1938-30 | 2 | 67.78 | -20.14 | 38.48 | 0.52 |
| 1938-25 | 1 | 41.19 | -12.37 | 16.92 | 0.73 | 1938-44 | 2 | 67.99 | -18.63 | 34.39 | 0.54 |
| 1938-14 | 1 | 41.87 | -12.74 | 17.96 | 0.71 | 1938-24 | 2 | 68.09 | -18.93 | 35.43 | 0.53 |
| 1938-80 | 1 | 42.00 | -13.50 | 18.71 | 0.72 | 1938-42 | 2 | 68.11 | -14.18 | 26.98 | 0.53 |
| 1938-84 | 1 | 42.02 | -13.71 | 18.85 | 0.73 | 1938-74 | 2 | 68.24 | -16.08 | 30.06 | 0.53 |
| 1938-55 | 1 | 42.28 | -16.38 | 23.00 | 0.71 | 1938-41 | 2 | 68.28 | -19.11 | 36.31 | 0.53 |
| 1938-97 | 1 | 42.50 | -12.92 | 18.75 | 0.69 | 1938-192 | 2 | 69.22 | -18.21 | 35.16 | 0.52 |
| 1938-155 | 1 | 42.50 | -13.65 | 19.51 | 0.70 | 1938-78 | 2 | 69.43 | -16.80 | 34.09 | 0.49 |
| 1938-123 | 1 | 42.60 | -15.64 | 22.26 | 0.70 | 1938-127 | 2 | 69.47 | -18.10 | 35.94 | 0.50 |
| 1938-106 | 2 | 42.73 | -15.78 | 32.13 | 0.49 | 1938-47 | 2 | 69.84 | -12.30 | 25.56 | 0.48 |
| 1938-105 | 1 | 43.01 | -14.05 | 20.31 | 0.69 | 1938-34 | 2 | 70.05 | -13.82 | 27.42 | 0.50 |
| 1938-90 | 1 | 43.11 | -15.87 | 22.53 | 0.70 | 1938-36 | 2 | 70.16 | -16.87 | 31.90 | 0.53 |
| 1938-196 | 1 | 43.13 | -15.91 | 23.06 | 0.69 | 1938-56 | 2 | 70.91 | -18.35 | 36.30 | 0.51 |
| 1938-73 | 1 | 43.20 | -14.87 | 20.70 | 0.72 | 1938-32 | 2 | 71.22 | -20.39 | 39.66 | 0.51 |
| 1938-172 | 1 | 43.63 | -15.79 | 22.55 | 0.70 | 1938-37 | 2 | 71.61 | -14.19 | 29.57 | 0.48 |
| 1938-138 | 1 | 43.66 | -19.06 | 27.68 | 0.69 | 1938-174 | 2 | 72.01 | -17.03 | 33.74 | 0.50 |
| 1938-158 | 1 | 43.80 | -15.26 | 21.35 | 0.71 | 1938-100 | 2 | 72.10 | -16.69 | 33.75 | 0.49 |
| 1938-96 | 1 | 44.14 | -13.62 | 19.23 | 0.71 | 1938-148 | 2 | 72.40 | -12.86 | 26.55 | 0.48 |
| 1938-183 | 1 | 44.20 | -15.36 | 21.77 | 0.71 | 1938-43 | 2 | 72.51 | -16.45 | 33.17 | 0.50 |
| 1938-110 | 1 | 44.29 | -14.86 | 20.62 | 0.72 | 1938-137 | 2 | 72.85 | -17.16 | 33.69 | 0.51 |
| 1938-66 | 1 | 44.30 | -14.96 | 21.47 | 0.70 | 1938-170 | 2 | 73.15 | -10.46 | 22.55 | 0.46 |
| 1938-8 | 1 | 44.50 | -15.71 | 22.95 | 0.68 | 1938-164 | 2 | 73.16 | -15.30 | 30.24 | 0.51 |
| 1938-135 | 1 | 44.59 | -16.82 | 23.88 | 0.70 | 1938-93 | 2 | 73.48 | -10.44 | 23.49 | 0.44 |
| 1938-119 | 1 | 44.61 | -14.32 | 20.77 | 0.69 | 1938-128 | 2 | 73.53 | -10.25 | 23.35 | 0.44 |
| 1938-72 | 1 | 44.61 | -15.18 | 21.38 | 0.71 | 1938-131 | 2 | 73.74 | -12.46 | 26.94 | 0.46 |
| 1938-181 | 1 | 44.64 | -17.28 | 24.91 | 0.69 | 1938-198 | 2 | 74.25 | -15.37 | 30.34 | 0.51 |
| 单株 Individual | 目测分型 Visual phenotype | 测色仪分型 Colorimeter phenotype | 单株 Individual | 目测分型 Visual phenotype | 测色仪分型 Colorimeter phenotype | ||||||
| L* | a* | b* | -a*/b* | L* | a* | b* | -a*/b* | ||||
| 1938-86 | 1 | 44.69 | -16.33 | 23.36 | 0.70 | 1938-133 | 2 | 74.31 | -7.57 | 23.46 | 0.32 |
| 1938-186 | 1 | 44.86 | -17.29 | 24.84 | 0.70 | 1938-160 | 2 | 74.51 | -13.13 | 27.28 | 0.48 |
| 1938-18 | 1 | 45.09 | -17.55 | 25.70 | 0.68 | 1938-176 | 2 | 74.58 | -11.32 | 23.60 | 0.48 |
| 1938-129 | 1 | 45.55 | -14.07 | 19.89 | 0.71 | 1938-182 | 2 | 74.86 | -14.14 | 30.47 | 0.46 |
| 1938-116 | 1 | 45.69 | -19.19 | 29.01 | 0.66 | 1938-51 | 2 | 75.18 | -9.40 | 22.19 | 0.42 |
| 1938-91 | 1 | 45.81 | -14.98 | 21.89 | 0.68 | 1938-58 | 2 | 75.23 | -14.90 | 32.67 | 0.46 |
| 1938-154 | 1 | 45.87 | -17.03 | 24.69 | 0.69 | 1938-109 | 2 | 75.44 | -10.67 | 22.89 | 0.47 |
| 1938-146 | 1 | 45.96 | -15.47 | 22.17 | 0.70 | 1938-166 | 2 | 75.78 | -8.28 | 24.49 | 0.34 |
| 1938-147 | 1 | 46.04 | -17.91 | 26.64 | 0.67 | 1938-45 | 2 | 75.92 | -9.62 | 21.11 | 0.46 |
| 1938-67 | 1 | 46.07 | -16.66 | 24.57 | 0.68 | 1938-165 | 2 | 76.03 | -10.21 | 22.82 | 0.45 |
| 1938-199 | 1 | 46.18 | -16.44 | 24.74 | 0.66 | 1938-112 | 2 | 76.14 | -9.10 | 21.58 | 0.42 |
| 1938-12 | 1 | 46.36 | -13.68 | 20.22 | 0.68 | 1938-168 | 2 | 76.22 | -9.99 | 23.75 | 0.42 |
| 1938-75 | 1 | 46.45 | -14.91 | 21.76 | 0.69 | 1938-50 | 2 | 76.59 | -8.39 | 23.53 | 0.36 |
| 1938-76 | 1 | 46.76 | -19.53 | 27.56 | 0.71 | 1938-22 | 2 | 77.06 | -7.11 | 22.34 | 0.32 |
| 1938-153 | 1 | 46.80 | -13.74 | 19.54 | 0.70 | 1938-5 | 2 | 77.33 | -9.11 | 22.33 | 0.41 |
| 1938-98 | 1 | 46.92 | -17.90 | 26.62 | 0.67 | 1938-177 | 2 | 77.33 | -13.14 | 27.97 | 0.47 |
| 1938-40 | 1 | 47.11 | -20.92 | 30.50 | 0.69 | 1938-115 | 2 | 78.12 | -9.06 | 23.98 | 0.38 |
| 1938-120 | 1 | 47.14 | -17.81 | 27.06 | 0.66 | 1938-159 | 2 | 78.33 | -7.92 | 21.01 | 0.38 |
| 1938-57 | 1 | 47.61 | -13.86 | 20.00 | 0.69 | 1938-114 | 2 | 78.38 | -9.95 | 22.76 | 0.44 |
| 1938-11 | 1 | 47.64 | -17.65 | 26.57 | 0.66 | 1938-35 | 2 | 78.40 | -8.03 | 23.03 | 0.35 |
| 1938-69 | 1 | 47.71 | -14.40 | 20.78 | 0.69 | 1938-60 | 2 | 78.41 | -8.91 | 23.92 | 0.37 |
| 1938-107 | 1 | 48.32 | -14.43 | 21.45 | 0.67 | 1938-77 | 2 | 78.64 | -9.13 | 22.56 | 0.40 |
| 1938-113 | 1 | 48.62 | -16.57 | 24.84 | 0.67 | 1938-173 | 2 | 78.74 | -8.24 | 22.42 | 0.37 |
| 1938-59 | 1 | 48.94 | -17.18 | 25.16 | 0.68 | 1938-200 | 2 | 78.87 | -14.35 | 30.49 | 0.47 |
| 1938-162 | 1 | 48.95 | -16.55 | 24.53 | 0.67 | 1938-38 | 2 | 79.10 | -9.19 | 22.90 | 0.40 |
| 1938-193 | 1 | 49.07 | -14.29 | 20.99 | 0.68 | 1938-31 | 2 | 79.14 | -7.07 | 21.17 | 0.33 |
| 1938-48 | 1 | 49.71 | -16.18 | 23.58 | 0.69 | 1938-144 | 2 | 79.14 | -9.61 | 23.88 | 0.40 |
| 1938-33 | 1 | 51.37 | -12.38 | 19.50 | 0.63 | 1938-83 | 2 | 79.18 | -5.58 | 20.69 | 0.27 |
| 1938-49 | 1 | 51.39 | -16.94 | 25.05 | 0.68 | 1938-188 | 2 | 79.65 | -7.15 | 19.66 | 0.36 |
| 1938-52 | 1 | 51.56 | -16.89 | 24.72 | 0.68 | 1938-143 | 2 | 79.73 | -8.08 | 21.52 | 0.38 |
| 1938-180 | 1 | 51.58 | -19.16 | 30.58 | 0.63 | 1938-39 | 2 | 80.58 | -6.52 | 21.68 | 0.30 |
| 1938-167 | 1 | 52.73 | -18.01 | 28.87 | 0.62 | 1938-111 | 2 | 80.87 | -5.44 | 21.39 | 0.25 |
| 1938-125 | 1 | 53.74 | -19.25 | 33.86 | 0.57 | 1938-62 | 2 | 81.17 | -5.93 | 18.66 | 0.32 |
| 1938-195 | 1 | 56.00 | -21.92 | 35.41 | 0.62 | 1938-124 | 2 | 81.51 | -6.67 | 18.92 | 0.35 |
| 1938-79 | 1 | 59.12 | -19.87 | 39.01 | 0.51 | 1938-149 | 2 | 81.98 | -7.08 | 25.33 | 0.28 |
| 1938-140 | 2 | 61.54 | -21.20 | 38.29 | 0.55 | 1938-194 | 2 | 82.38 | -4.25 | 20.43 | 0.21 |
| 1938-16 | 2 | 61.87 | -22.45 | 39.16 | 0.57 | 1938-65 | 2 | 82.81 | -6.49 | 26.53 | 0.24 |
| 1938-26 | 2 | 61.95 | -23.03 | 40.14 | 0.57 | 1938-2 | 2 | 83.14 | -4.57 | 21.21 | 0.22 |
| 1938-141 | 2 | 64.21 | -19.76 | 37.03 | 0.53 | 1938-202 | 2 | 77.91 | -6.22 | 33.89 | 0.18 |
新窗口打开|下载CSV
3 讨论
随着甜瓜基因组研究的深入,功能基因组学取得了快速发展[17]。作为重要的农艺性状,果皮颜色的遗传模式一直是研究者关注的热点。4号染色体长臂被广泛报道含有一个调控甜瓜果皮颜色的基因[8,18-20]。OREN等[8]利用3个亲本‘Tam Dew’‘Dulce’和‘Noy Amid’将幼果果皮颜色基因定组位在4号染色体290 kb区段内;ZHAO等[18]利用全基因组关联分析(GWAS)将果皮颜色基因定位于4号染色体500 kb的范围内。因该定位范围内含有一个编码双组分反应调节蛋白CmAPRR2的基因MELO3C003375,且该基因的同源基因APRR2曾在黄瓜、番茄和胡椒中被报道可调控果皮的叶绿素代谢和色素积累,因此MELO3C003375(CmAPRR2)被前人认定为候选基因[8,18]。有别于前人使用厚皮甜瓜进行亚种内组配,本研究利用厚皮与薄皮甜瓜开展亚种间杂交;同时,本研究的亲本在果形、网纹以及成熟果果皮颜色等方面与前人选材存在较大差异,这不仅给该性状的遗传定位选材增添了新类型,也暗示了该基因功能在亚种间的保守性[8,13-14]。另外,本研究采用BSA-seq确定初定位区间、重测序开发新标记和图位克隆缩小区段这一思路清晰且完整的研究方案,通过多代表型和基因型的验证,将GR精细定位在17.7 kb的区间内。但重组交换单株的缺乏使该区间较难进行进一步的缩小。通过对区间内注释基因的测序分析发现MELO3C003375(CmAPRR2)在‘LGR’中存在1处无义变异,致使该转录因子调控下游基因的重要结构域(Myb-DNA结合结构域)缺失,因此,MELO3C003375(CmAPRR2)被认为是最佳候选基因。相比以往报道,本研究极大地缩短了幼果果皮颜色性状基因的定位区间,提高了MELO3C003375(CmAPRR2)作为候选基因的可信度。此外,本研究还发现‘MR-1’亲本中MELO3C003375第二个外显子处的单碱基变异与已报道的材料都不相同,这也为甜瓜中MELO3C003375的变异类型增添了新认知。
甜瓜果皮颜色丰富多样,FALLIK等[21]对不同成熟度的‘Galia’甜瓜进行了果皮颜色变化调查并将其分成6个阶段:(1)深绿色;(2)绿色;(3)淡黄色夹杂绿色;(4)淡黄色;(5)黄色;(6)深黄色至橙色。REID等[22]通过评估‘Crenshaw’‘Persian’和‘PMR45’3个甜瓜品种发现果实的成熟会伴随着叶绿素含量的下降和类胡萝卜素的上升。而FLÜGEL和GROSS[23]在‘Galia’甜瓜中观察到类胡萝卜素含量在发育期间并没有变化,外果皮变黄主要是因为在成熟过程中叶绿素的降解增加。因此,果皮颜色变化被认为主要由叶绿素和类胡萝卜素(主要是β-胡萝卜素)引起[24]。在本研究中,MELO3C003375(CmAPRR2)作为调控甜瓜幼果果皮颜色的基因,其是否参与了单一色素或者组合色素的合成或者分解过程?在黄瓜中,APRR2被发现影响质体发育和叶绿素的合成[15]。PAN等[16]发现番茄和辣椒果实中过表达APRR类转录因子会增加幼果的叶绿素和成熟果的类胡萝卜素。OREN等[8]发现CmAPRR2的表达与β-类胡萝卜素的含量存在紧密的相关性。另外,与APRR2相关的GLK2转录因子也曾被报道影响质体的发育以及调节植物的叶绿素和类胡萝卜素的含量[25,26,27]。这些报道都为下一步比较分析亲本色素含量提供了依据和方向。
作为双组分反应调节蛋白(ARR)的一类,APRR2属于B型响应调节蛋白,该类型的蛋白被广泛报道参与细胞分裂素的信号转导[28]。在arr1 arr10 arr12的三突变体中,ARGYROS等[29]发现叶绿素的含量显著减少并且大部分的细胞分裂素调节基因显著下调。在拟南芥中,CHOI等[30]发现ARR2可通过Myb-DNA结合结构域与TGA3互作,进而通过结合植物防御相关基因Pr1启动子的ACGTCATAGA基序来调控Pr1的表达。这些展现了ARRs类转录因子对下游基因的调控作用以及Myb-DNA结合结构域对ARRs类转录因子功能的重要影响。在本研究中,‘LGR’与‘MR-1’的MELO3C003375(CmAPRR2)共有5处SNP差异,其中仅有无义突变在蛋白功能结构域中发生,造成‘LGR’中MELO3C003375(CmAPRR2)的Myb-DNA结合结构域大部分缺失。类似地,OREN等[8]在甜瓜‘Tam Dew’中也发现了相同的无义突变,并且通过等位性测验证实了该变异是造成‘Tam Dew’幼果浅色的原因。基于Myb-DNA结合结构域的功能特点,猜测该无义突变会导致MELO3C003375(CmAPRR2)蛋白无法识别和结合下游基因,进而无法调控色素代谢,以致产生浅绿色果皮。对于APRR2或者ARRs类转录因子调控色素相关基因的方式(直接或者有其他基因介导),暂时未有相关深入的研究。鉴于拟南芥中转座酶类的转录因子FHY3和FAR1,以及ABRE类转录因子ABF2、ABF3和ABF4常通过直接调节叶绿素代谢途径中的结构基因来影响叶绿素水平[31,32],推测MELO3C003375(CmAPRR2)通过直接调控色素代谢途径中的结构基因来影响幼果果皮颜色,而缺失了Myb-DNA结合结构域的MELO3C003375(CmAPRR2)则极有可能因为无法直接识别与结合结构基因的启动子,进而无法调节结构基因表达,造成‘LGR’无法积累色素,产生浅绿色幼果。
作为对消费者有吸引力的性状,果皮颜色一直是甜瓜育种的重要目标。相较于成熟果,幼嫩果实颜色相对单一,主要由叶绿素影响。但成熟果与幼嫩果实之间是否存在关联性,研究却很少涉及。本研究中,两亲本幼嫩果果皮颜色差异明显,定位群体(BC1F1)单株表型也分离显著,随着发育进程,亲本以及单株果皮颜色发生变化,但有趣的是,浅绿皮植株和深绿皮成熟果果皮颜色的绿色强度却形成了显著的差异分化,这表明即使果实在发育过程中会发生转变,但成熟果果皮颜色仍然与幼嫩果果皮颜色存在紧密的相关性,该结论为未来进行甜瓜外观品质育种提供了思路。另外,本研究发现在BC1F1群体幼果为深绿皮的植株中,部分单株的成熟果会表现出黄色,而部分单株仍然表现为深绿色。这表明虽然成熟果颜色与幼果颜色有显著相关性,但两者之间的内在联系以及它们与果实发育进程之间的关系还需进一步的探索和解析。这为后续进行成熟果的相关研究提供了思路,也暗示选择合适时期的材料对果实皮色的研究十分重要。
4 结论
甜瓜幼果果皮颜色(深绿和浅绿)是受单个核基因GR控制的质量性状,且深绿对浅绿为显性。通过遗传定位将该GR定位于4号染色体分子标记4-102和4-81之间,物理距离约为17.7 kb。此区间内MELO3C003375(CmAPRR2)的第8个外显子在‘LGR’中发生单碱基替换(G856T),造成蛋白翻译提前终止,推测MELO3C003375(CmAPRR2)即为调控甜瓜幼果果皮颜色的基因GR。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1021/jf1021797URL [本文引用: 2]
DOI:10.21273/HORTSCI.40.6.1928URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1186/s12870-018-1537-5URL [本文引用: 1]
[本文引用: 1]
DOI:10.1093/jxb/erz182URL [本文引用: 8]
[本文引用: 1]
[本文引用: 1]
DOI:10.21273/JASHS.132.1.80URL [本文引用: 1]
DOI:10.21273/JASHS.133.1.139URL [本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
DOI:10.1007/s00122-016-2700-8URL [本文引用: 2]
DOI:10.1104/pp.112.212654URL [本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/s41588-019-0522-8URL [本文引用: 3]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S0925-5214(00)00185-XURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/jhered/esm108URL [本文引用: 1]
[本文引用: 1]
DOI:10.1111/tpj.2008.56.issue-3URL [本文引用: 1]
DOI:10.1105/tpc.108.065250URL [本文引用: 1]
DOI:10.1105/tpc.105.035451URL [本文引用: 1]
DOI:10.1105/tpc.108.059584URL [本文引用: 1]
DOI:10.1016/j.devcel.2010.07.011URL [本文引用: 1]
DOI:10.1105/tpc.112.097022URL [本文引用: 1]
DOI:10.1016/j.molp.2016.06.006URL [本文引用: 1]
