 ,1, 于文涛
,1, 于文涛 ,2, 蔡春平2, 林浥1, 王泽涵1, 房婉萍3, 张见明4, 叶乃兴
,2, 蔡春平2, 林浥1, 王泽涵1, 房婉萍3, 张见明4, 叶乃兴 ,1
,1Construction of Molecular ID for Tea Cultivars by Using of Single- nucleotide Polymorphism (SNP) Markers
FAN XiaoJing ,1, YU WenTao
,1, YU WenTao ,2, CAI ChunPing2, LIN Yi1, WANG ZeHan1, FANG WanPing3, ZHANG JianMing4, YE NaiXing
,2, CAI ChunPing2, LIN Yi1, WANG ZeHan1, FANG WanPing3, ZHANG JianMing4, YE NaiXing ,1
,1通讯作者:
责任编辑: 李莉
收稿日期:2020-08-28接受日期:2020-09-27网络出版日期:2021-04-16
| 基金资助: |
Corresponding authors:
Received:2020-08-28Accepted:2020-09-27Online:2021-04-16
作者简介 About authors
樊晓静,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1745KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
樊晓静, 于文涛, 蔡春平, 林浥, 王泽涵, 房婉萍, 张见明, 叶乃兴. 利用SNP标记构建茶树品种资源分子身份证[J]. 中国农业科学, 2021, 54(8): 1751-1760 doi:10.3864/j.issn.0578-1752.2021.08.014
FAN XiaoJing, YU WenTao, CAI ChunPing, LIN Yi, WANG ZeHan, FANG WanPing, ZHANG JianMing, YE NaiXing.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】茶树(Camellia sinensis (L.) O. Kuntze)为多年生异花授粉植物,具有高度异质性和杂合性[1]。中国西南地区是茶树的起源地[2],茶树从起源地向中国其他地区和国外的自然传播和人为传播过程中,积累了自然演化和人工选择的变异,从而在各茶区形成了丰富的茶树品种资源[3]。然而在茶树品种资源的大量引种及频繁的品种资源交换过程中,造成同名异物、同物异名的现象[4,5],不仅给茶树品种资源保护利用带来诸多困难,也损害了消费者的权益。因而科学准确地区分和鉴定茶树品种资源具有重要意义。【前人研究进展】传统使用形态标记、细胞标记等方法来区分、鉴定植物品种均有一定局限性。如形态学标记容易受环境和植物发育阶段等影响;细胞学标记容易受制片技术的影响,且对于染色体小数量多的物种,其染色体核型不易区分清楚[6]。随着生物信息技术的进步,以生物的遗传物质核酸的多态性为基础的分子标记得到了快速发展,DNA分析技术开始大量应用于植物学研究。RAPD、SSR等分子标记技术在研究茶树遗传多样性、指纹图谱及分子身份证构建等方面已有较多运用[7,8,9,10,11,12,13]。如安徽茶树群体中的SSR遗传多样性分析[8]、茶树SSR指纹图谱的构建[9,10]、野生茶树的RAPD分子标记鉴定[11]和名山茶树基因身份证的构建[13]。LANDER[14]在1996年提出SNP的遗传标记技术,作为继SSR为代表的第二代分子标记技术之后发展起来的第三代分子标记技术,是指个体间基因组DNA序列同一位置单个核苷酸变异所引起的多态性。SNP标记具有高密度性,且广泛存在于基因组中,如人类基因组中平均每1 000 bp就会出现一个SNP,玉米中平均每57 bp出现一个SNP[15]。SNP作为第三代分子标记,其优点在于快速、自动化、高通量[16],其多态性几乎在所有被研究物种中都被用作有效的遗传标记,包括动物[17]和植物[18]。【本研究切入点】目前,关于茶树SNP分子标记的研究,主要集中在品种资源鉴定、遗传多样性、遗传关系的分析及指纹图谱的构建[19,20],对茶树品种资源的分子身份证进行构建鲜见报道。【拟解决的关键问题】本研究通过收集国内外103份茶树种质,利用SNP标记并结合茶树品种资源基本信息构建茶树品种资源分子身份证,以期为茶树品种资源保护鉴定提供一种新思路。1 材料与方法
1.1 供试材料
供试茶树品种资源来源于武夷学院茶树种质资源圃(福建省武夷山市)和福建省农业科学院茶叶研究所茶树种质资源圃(福建省福安市),其中,中国茶树品种资源有101份,来自华南、西南、江南、江北四大茶产区的10个茶叶主产区省份;国外茶树品种2份,包括日本的玉绿和格鲁吉亚的格鲁吉亚1号。茶树品种资源详细信息见电子附表1。Table 1
表1
表1103份茶树品种资源的分子身份证
Table 1
| 品种资源 Cultivars | 分子身份证 Molecular ID | 品种资源 Cultivars | 分子身份证 Molecular ID | |
|---|---|---|---|---|
| 蕉城苦茶1号 Jiaocheng Kucha 1 | 1351232113232131123233232332 | 紫牡丹 Zimudan | 1353112323322131312131212111 | |
| 蕉城苦茶4号 Jiaocheng Kucha 4 | 1351232123232331223233232332 | 凤圆春 Fengyuanchun | 1353111111111121311211112211 | |
| 蕉城苦茶5号 Jiaocheng Kucha 5 | 1351132121112131123213212131 | 大叶乌龙 Dayewulong | 1353112112313211113231111111 | |
| 寿宁地洋1号 Shouning Diyang 1 | 1351321113222111121122111223 | 蜀永1号 Shuyong 1 | 1503323111113131121212111223 | |
| 寿宁地洋2号 Shouning Diyang 1 | 1351311131323131112113111211 | 蜀永2号 Shuyong 2 | 1503321313212133222232312222 | |
| 寿宁芎坑1号 Shouning Xiongkeng 1 | 1351121132321111233112213221 | 蜀永3号 Shuyong 2 | 1503113223211111211211122312 | |
| 寿宁芎坑2号 Shouning Xiongkeng 2 | 1351113211321131113113213212 | 蜀永808 Shuyong 808 | 1503123223212131221131112322 | |
| 凤凰苦茶 Fenghuangkucha | 1441213233232111323233211111 | 蜀永703 Shuyong 703 | 1503123213221121132231121221 | |
| 佛手 Foshou | 1353211232221111311213311211 | 名山白毫131 Mingshanbaihao 131 | 1513111231313121112112313212 | |
| 白鸡冠 Baijiguan | 1352113111121111211121213113 | 安徽7号 Anhui 7 | 1343311131232121211111313113 | |
| 慢奇兰 Manqilan | 1352122113122111323311311121 | 蒙山9号 Mengshan 9 | 1343111131311111112122113212 | |
| 金面奇兰 Jinmianqilan | 1352111131122131331111312111 | 名山早311 Mingshanzao 311 | 1513131132311121332112313232 | |
| 寿宁桃眉 Shouning Taomei | 1352131121122111332131111232 | 川茶2号 Chuancha 2 | 1513123223121111111211312322 | |
| 寿宁黄叶茶 Shouning Huangyecha | 1352113331331111113211312213 | 川茶3号 Chuancha 2 | 1513121233221131112221311221 | |
| 吴山清明茶 Wushanqingmingcha | 1352321132333131321212311221 | 紫嫣 Ziyan | 1513112211311131311211113212 | |
| 水古茶 Shuigucha | 1332323211323111312332112222 | 云抗10号 Yunkang 10 | 1533113123211131123211112332 | |
| 夜来香单丛 Yelaixiang Dancong | 1442232122121111311233113131 | 云茶1号 Yuncha 1 | 1533311332323121312331313211 | |
| 芝兰香单丛 Zhilanxiang Dancong | 1442132223232121223233211331 | 长叶白毫 Changyebaihao | 1533223123232111223233212322 | |
| 八仙香单丛 Baxianxiang Dancong | 1442233123132121123233232131 | 紫娟 Zijuan | 1533111111211231121211112132 | |
| 老仙翁单丛 Langxianweng Dancong | 1442232223212121111231232331 | 鄂茶11号 Echa 11 | 1423123131311121132113321123 | |
| 红帝单丛 Hongdi Dancong | 1442231123232221113232212331 | 千年雪 Qiannianxue | 1333311212121131312311333113 | |
| 城门单丛 Chengmen Dancong | 1442113123211111312211112111 | 平阳特早 Pingyangtezao | 1333123233121121132132112123 | |
| 探春香单丛 Tanchunxiang Dancong | 1442233123232121113231213331 | 中茶102 Zhongcha 102 | 1333121111321111332311113221 | |
| 贡香单丛 Gongxiang Dancong | 1442211211232111323212212111 | 碧云 Biyun | 1333131231332131311322311233 | |
| 棕榈香单丛 Zonglvxiang Dancong | 1442111121211211121211111111 | 白叶1号 Baiye 1 | 1333113131113131112131111211 | |
| 鸭屎香单丛 Yashaxiang Dancong | 1442131133132311113232211331 | 龙井43 Longjing 43 | 1333113313213111331132312113 | |
| 姜母香单丛 Jiangmuxiang Dancong | 1442233331232321121233232331 | 中茶108 Zhongcha 108 | 1333311331313111112222311211 | |
| 青心大冇 Qingxindamao | 1712112121132111312213112311 | 嘉茗1号 Jiaming 1 | 1333322233111111332112113223 | |
| 软枝乌龙 Ruanzhiwulong | 1712112321312131312233112112 | 龙井长叶 Longjingchangye | 1333313131123121211111313212 | |
| 四季春 Sijichun | 1712122231322111312223111223 | 黄金芽 Huangjinya | 1333123111322111131212311122 | |
| 福鼎大白茶 Fuding Dabaicha | 1353131132311121332112313231 | 安徽3号 Anhui 3 | 1343111231313121112112313211 | |
| 政和大白茶 Zhenghe Dabaicha | 1353123111121111121212111221 | 凫早2号 Fuzao 2 | 1343122332123121331231313121 | |
| 霞浦春波绿 Xiapu Chunbolv | 1353321131321211122123311121 | 舒茶早 Shuchazao | 1343121231311111322212313123 | |
| 早逢春 Zaofengchun | 1353111221212111223212211333 | 农抗早 Nongkangzao | 1343313211121121132112311211 | |
| 霞浦元宵茶 Xiapu Yuanxiaolv | 1353313233121131211311311211 | 白毫早 Baihaozao | 1433332232121111111312113232 | |
| 福云6号 Fuyun 6 | 1353111111111111111231111111 | 槠叶齐 Chuyeqi | 1433112123223111221222122112 | |
| 福云7号 Fuyun 7 | 1353213211132111111111111131 | 桃源大叶 Taoyuandaye | 1433121133223111111211311121 | |
| 福云10号 Fuyun 10 | 1353113311131111111211311131 | 涟源奇曲 Lianyuanqiqu | 1432121123211131111112112121 | |
| 福云20号 Fuyun 20 | 1353133131231111332213131131 | 湘波绿 Xiangbolv | 1433213111132211111111113112 | |
| 福云595 Fuyun 595 | 1353211211112211111111113111 | 尖波黄 Jianbohuang | 1433132111111111111212111232 | |
| 大红袍 Dahongpao | 1353322212313131322213112223 | 保靖黄金茶1号 Baojing Huangjincha 1 | 1433323212322131211211212221 | |
| 铁观音 Tieguanyin | 1353112131312111332211311112 | 乌叶单丛 Wuye Dancong | 1443132121232121113233212331 | |
| 黄棪 Huangdan | 1353212221113111313231213111 | 金萱 Jinxuan | 1713122222121111322212211122 | |
| 肉桂 Rougui | 1353211133312321333111212211 | 翠玉 Cuiyu | 1713321321231111331213312321 | |
| 本山 Benshan | 1353212331312111331111311112 | 福云8号 Fuyun 8 | 1354213311111111111231111132 | |
| 梅占 Meizhan | 1353122122112111311112312121 | 福云591 Fuyun 591 | 1354111331311211311212111111 | |
| 毛蟹 Maoxie | 1353132331311121331321311131 | 寿宁凤阳种 Shouning Fengyangzhong | 1354112332321121112112311111 | |
| 白芽奇兰 Baiyaqilan | 1353122321322331311231112221 | 金茗早 Jinmingzao | 1354112222111121113232213111 | |
| 九龙大白茶 Jiulongdabaicha | 1353213311111111111231111131 | 长乐种 Changlezhong | 1354213211132111111311111131 | |
| 九龙袍 Jiulongpao | 1353322212313131322213112123 | 玉绿 Yulv | 1005112231111131231112111211 | |
| 福建水仙 Fujian Shuixian | 1353311131311311211111311211 | 格鲁吉亚1号 Gelujiya 1 | 1005221323212111212112312121 | |
| 八仙茶 Baxiancha | 1353111123232121222232213211 |
新窗口打开|下载CSV
1.2 DNA的提取
采用新型植物基因组DNA提取试剂盒(TIANGEN,DP320,北京)提取基因组DNA,利用超微量紫外分光光度计(Implen,S60716,德国)测定DNA浓度和纯度。提取的基因组DNA于-80℃保存备用。1.3 SNP基因分型
中国乌龙茶产业协同创新中心课题组前期从国家生物信息中心(national center of biotechnology information,NCBI)的数据库(图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图124个茶树SNP位点多态性读取图
蓝色:杂合子(XY);红色:纯合子(XX);绿色:纯合子(YY)
Fig. 1Polymorphism plots of 24 SNP loci
Blue: Heterozygote (XY); Red: Homozygote (XX); Green: Homozygote (YY)
1.4 数据处理
利用Fluidigm SNP Genotyping Analysis软件(2 结果
2.1 茶树SNP标记多态性描述统计
采用的96个SNP标记位点较均匀地分布于茶树全基因组15条染色体上(电子附表2)。对103份茶树品种资源进行分析,剔除10个不具多态性的引物后,剩余86个SNP位点多态性的相关信息列于电子附表3。根据统计,这些SNP标记多态性信息指数(I)为0.071—0.693,平均值为0.517。观测杂合度(Ho)范围为0.027—0.982,平均值为0.370。期望杂合度(He)的范围在0.026—0.500,平均值为0.346。固定指数(F)为-0.964—0.462,平均值为-0.036。次等位基因频率(MAF)范围在0.013—0.500,平均值为0.269。2.2 茶树品种资源最佳SNP位点的筛选及指纹图谱的构建
信息指数常用来评价群落的遗传多样性,如果每一个体都属于不同的种,多样性指数就最大;如果每一个体都属于同一种,则其多样性指数就最小,即多样性指数越高,其区分品种能力却强。因此,基于103份茶树品种资源材料,对SNP数据进行统计分析,从86个SNP位点中优化筛选了24个多态性较高的SNP位点(电子附表4,图1),可将103份茶树品种资源完全区分开。DNA指纹图谱(图2)显示每个SNP位点处的样品是杂合子(XY)或纯合子(XX,YY)。图2
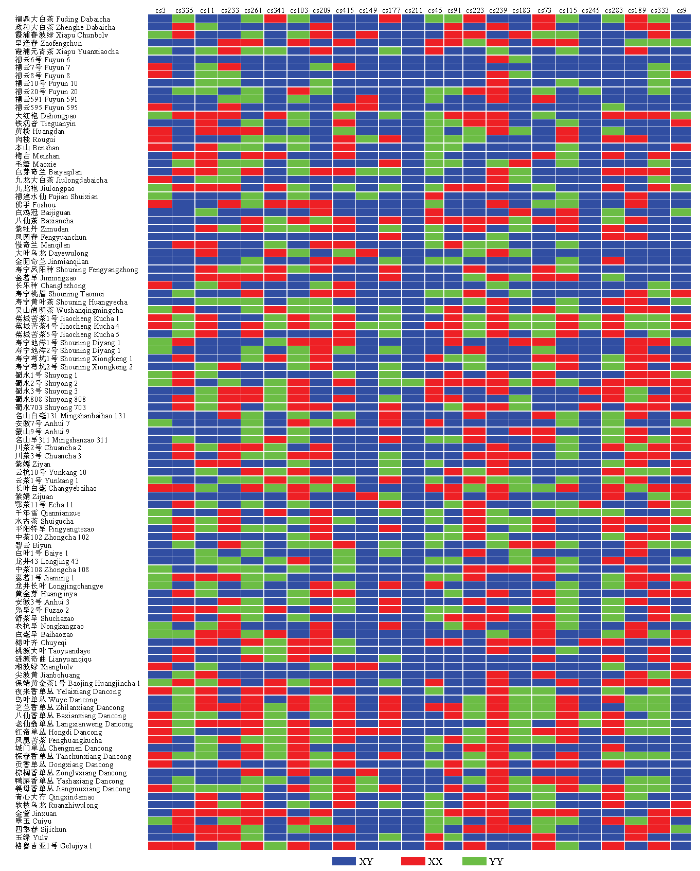 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2103份茶树品种资源SNP指纹图谱
蓝色:杂合子(XY);红色:纯合子(XX);绿色:纯合子(YY)
Fig. 2SNP fingerprints of 103 tea germplasms
Blue: Heterozygote (XY); Red: Homozygote (XX); Green: Homozygote (YY)
2.3 茶树DNA指纹图谱编码
对筛选获得的最佳SNP数据进行数字编码,作为构建分子身份证基本信息。在24个SNP标记,共有24种基因型,3种等位基因(XY、XX和YY),因此,每个位点分别用1—3代表等位基因多态性。以福鼎大白茶为例,在CS3处的基因型为TC,对应XY,编码为1,以此类推,将基因型全部转换成24数字编码为131132311121332112313231。2.4 茶树品种资源信息编码
茶树的基本信息由4位数字组成,其中,茶树品种资源类型分类参考陈亮等[27,28]方法。第1位数字代表茶组植物具体物种,茶树(Camellia sinensis (L.) O.Kuntze)为1,大厂茶(C. tachangensis F. C. Zhang)为2,厚轴茶(C. crassicolumna Chang)为3,大理茶(C. taliensis (W. W. Smith) Melchior)为4,秃房茶(C. gymnogyna Chang)为5;第2—3位数字代表行政区划代码,如福建为35,浙江为33,云南为53,国外为00;第4位数字代表茶树品种资源类型,野生茶树为1,地方品种和地方种质为2,优良品种(含育成品种)为3,新选品系、株系为4,国外品种为5。以福鼎大白茶举例,4位数字商品码为1353,其中“1”表示是分类上茶组植物种类,“35”代表该茶树种质原产地是福建,“3”表示为优良品种。2.5 茶树品种资源分子身份证的构建
基本商品信息和DNA指纹图谱共同组成由28位数字编码的茶树品种资源分子身份证(表1)。以国家级品种福鼎大白茶举例,其品种资源基本信息为:属茶组植物中的茶树(C. sinensis),原产于福建,品种资源类型为优良品种,转换成数字码为1353;其24个SNP分子标记的基因型分别为TC、TT、AT、CT、GG、GG、CC、TC、TG、TC、TT、AG、GG、TT、CC、AG、CT、TT、CC、AT、GG、GG、CC、TC,转换成24位数字码为131132311121332112313231。则福鼎大白茶的分子身份证为1353131132311121332112313231(图3-A),将其转化为条形码和二维码如图3-B所示。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3福鼎大白茶分子身份证
Fig. 3Molecular ID of Camellia sinensis Fuding Dabaicha
3 讨论
3.1 茶树品种资源SNP标记的基因分型
本研究103份茶树品种资源样品来源于中国的四大茶产区,以及日本和格鲁吉亚,来源范围广,茶树品种资源遗传多样性丰富,其中也有遗传背景相近的茶树品种亲子代,如福鼎大白茶和云南大叶种的自然杂交后代福云6号、福云7号和福云595,及以四川中叶种为母本、云南大叶种为父本的蜀永2号、蜀永3号和蜀永703。本研究中所选用的96个SNP标记位点中,多态性标记为86个,占比为89.6%。SNP标记一般只有2种碱基组成,因此,被认为具有二态性;由于具有等位基因性的特点,其等位基因在任何种群中都可被估算出来。除此之外,SNP标记在不同条件下具有较高的重复性和准确性[29,30],这是指纹图谱和分子身份证建立的重要前提。本研究通过SNP技术对这些茶树品种资源进行基因分型及统计分析,使每份品种资源具有唯一的SNP基因型,验证了SNP标记技术的准确性。与SSR相比,SNP分析不基于DNA大小片段的分离,因此,可通过高通量的形式自动化检测。目前常用的高通量、自动化程度较高的检测分析SNP的方法之一是DNA芯片法[31]。本研究所采用的高通量微流体芯片法具有高通量、试剂和样品用量少、IFC技术自动操作、试验重复性好的优点[32]。3.2 茶树最佳引物的筛选确认
筛选较少的引物,可以缩短茶树品种资源鉴定的时间,降低应用成本。最佳引物的选择是构建指纹图谱和分子身份证的重要步骤。PAN[33]利用21对引物对1 025份甘蔗种质构建了分子身份证。李春花等[34]采用15对引物构建了48份苦荞资源的分子身份证。冉昆等[35]利用10对引物构建了45份山东地方种质梨的分子身份证。高源等[36]利用TP-M13-SSR分子标记技术,通过筛选引物,对苹果分子身份证进行了构建。在本研究的96个SNP标记中,去除10个在试验样品中表现出多态性差的标记位点,再依据引物的信息指数(I),通过从高到低排序,逐步增加引物组合数量,使其能够区分更多的品种资源,以获得最佳的引物。经过分析,本研究最终筛选了24个SNP标记位点能够将103份茶树品种资源完全区分,24个SNP标记位点位于茶树全基因组的12条染色体上,染色体覆盖度为80%,覆盖度较高,有利于茶树品种资源的有效鉴别。3.3 茶树品种资源DNA分子身份证的价值与应用
作物品种资源的分子身份证与DNA指纹图谱的功能相同,都是为了区分不同生物个体。相对于指纹图谱,分子身份证是将作物品种资源的基本信息用特定的数字编码为数字串,并且辅以条形码、二维码的形式展现,更加简明直观地区分品种资源之间的差异。分子身份证因其自身所具有特性,可利用机器进行扫描,达到方便快捷地识别大量品种资源,提高品种资源鉴定和评价的效率。另外,分子身份证的唯一性,也可有效甄别市场上同名异物、同物异名现象,有利于品种识别与保护。许多植物已利用分子标记技术构建分子身份证,如水稻[37]、百合[38]、桃[39]、枸杞[40]等。本研究首次利用SNP标记技术将DNA指纹图谱与品种资源基本信息相结合,可为每份茶树品种资源构建独有的分子身份证。该身份证信息包括了茶树品种资源的分子指纹码和属性码,可快速了解其分子信息、来源等基本信息。本研究结果对于茶树品种资源区分和精准鉴定、分子数据数字化建立具有重要的意义和实际应用价值,为茶树品种资源DNA分子身份证的构建提供了思路。4 结论
筛选出24个最佳SNP位点组合,可精准区分全部103份供试茶树野生种、地方品种和地方种质、优良品种、新选品系和株系以及国外品种。将24个SNP位点组成茶树品种资源DNA指纹图谱编码,与茶树品种资源的基本属性信息编码组成28位数字的茶树品种资源DNA分子身份证,并生成相应的条形码和二维码,可快速被扫码设备识别。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.1126/science.274.5287.536URLPMID:8928008 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:19921517 [本文引用: 1]
DOI:10.1007/s12038-012-9225-3URLPMID:23107918 [本文引用: 1]

Genotyping with large numbers of molecular markers is now an indispensable tool within plant genetics and breeding. Especially through the identification of large numbers of single nucleotide polymorphism (SNP) markers using the novel high-throughput sequencing technologies, it is now possible to reliably identify many thousands of SNPs at many different loci in a given plant genome. For a number of important crop plants, SNP markers are now being used to design genotyping arrays containing thousands of markers spread over the entire genome and to analyse large numbers of samples. In this article, we discuss aspects that should be considered during the design of such large genotyping arrays and the analysis of individuals. The fact that crop plants are also often autopolyploid or allopolyploid is given due consideration. Furthermore, we outline some potential applications of large genotyping arrays including high-density genetic mapping, characterization (fingerprinting) of genetic material and breeding-related aspects such as association studies and genomic selection.
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:32709943 [本文引用: 1]
DOI:10.1016/j.molp.2020.04.010URLPMID:32353625 [本文引用: 2]

Tea plant is an important economic crop, which is used to produce the world's oldest and most widely consumed tea beverages. Here, we present a high-quality reference genome assembly of the tea plant (Camellia sinensis var. sinensis) consisting of 15 pseudo-chromosomes. LTR retrotransposons (LTR-RTs) account for 70.38% of the genome, and we present evidence that LTR-RTs play critical roles in genome size expansion and the transcriptional diversification of tea plant genes through preferential insertion in promoter regions and introns. Genes, particularly those coding for terpene biosynthesis proteins, associated with tea aroma and stress resistance were significantly amplified through recent tandem duplications and exist as gene clusters in tea plant genome. Phylogenetic analysis of the sequences of 81 tea plant accessions with diverse origins revealed three well-differentiated tea plant populations, supporting the proposition for the southwest origin of the Chinese cultivated tea plant and its later spread to western Asia through introduction. Domestication and modern breeding left significant signatures on hundreds of genes in the tea plant genome, particularly those associated with tea quality and stress resistance. The genomic sequences of the reported reference and resequenced tea plant accessions provide valuable resources for future functional genomics study and molecular breeding of improved cultivars of tea plants.
DOI:10.1016/S0022-2836(05)80360-2URLPMID:2231712 [本文引用: 1]

A new approach to rapid sequence comparison, basic local alignment search tool (BLAST), directly approximates alignments that optimize a measure of local similarity, the maximal segment pair (MSP) score. Recent mathematical results on the stochastic properties of MSP scores allow an analysis of the performance of this method as well as the statistical significance of alignments it generates. The basic algorithm is simple and robust; it can be implemented in a number of ways and applied in a variety of contexts including straightforward DNA and protein sequence database searches, motif searches, gene identification searches, and in the analysis of multiple regions of similarity in long DNA sequences. In addition to its flexibility and tractability to mathematical analysis, BLAST is an order of magnitude faster than existing sequence comparison tools of comparable sensitivity.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00122-007-0570-9URLPMID:17639299 [本文引用: 1]

We report on the comparative utilities of simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) markers for characterizing maize germplasm in terms of their informativeness, levels of missing data, repeatability and the ability to detect expected alleles in hybrids and DNA pools. Two different SNP chemistries were compared; single-base extension detected by Sequenom MassARRAY, and invasive cleavage detected by Invader chemistry with PCR. A total of 58 maize inbreds and four hybrids were genotyped with 80 SSR markers, 69 Invader SNP markers and 118 MassARRAY SNP markers, with 64 SNP loci being common to the two SNP marker chemistries. Average expected heterozygosity values were 0.62 for SSRs, 0.43 for SNPs (pre-selected for their high level of polymorphism) and 0.63 for the underlying sequence haplotypes. All individual SNP markers within the same set of sequences had an average expected heterozygosity value of 0.26. SNP marker data had more than a fourfold lower level of missing data (2.1-3.1%) compared with SSRs (13.8%). Data repeatability was higher for SNPs (98.1% for MassARRAY SNPs and 99.3% for Invader) than for SSRs (91.7%). Parental alleles were observed in hybrid genotypes in 97.0% of the cases for MassARRAY SNPs, 95.5% for Invader SNPs and 81.9% for SSRs. In pooled samples with mixtures of alleles, SSRs, MassARRAY SNPs and Invader SNPs were equally capable of detecting alleles at mid to high frequencies. However, at low frequencies, alleles were least likely to be detected using Invader SNP markers, and this technology had the highest level of missing data. Collectively, these results showed that SNP technologies can provide increased marker data quality and quantity compared with SSRs. The relative loss in polymorphism compared with SSRs can be compensated by increasing SNP numbers and by using SNP haplotypes. Determining the most appropriate SNP chemistry will be dependent upon matching the technical features of the method within the context of application, particularly in consideration of whether genotypic samples will be pooled or assayed individually.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
