 ,, 王省芬
,, 王省芬 ,河北农业大学农学院/教育部华北作物种质资源研究与利用重点实验室,河北保定071001
,河北农业大学农学院/教育部华北作物种质资源研究与利用重点实验室,河北保定071001Cloning and Functional Characterization of Gene CRVW Involved in Cotton Resistance to Verticillium Wilt
WANG QiuYing, WANG WeiQiao, ZHANG Yan, WANG GuoNing, WU LiQiang, ZHANG GuiYin, MA ZhiYing, YANG Jun ,, WANG XingFen
,, WANG XingFen ,College of Agronomy, Hebei Agricultural University/North China Key Laboratory for Crop Germplasm Resources, Ministry of Education, Baoding 071001, Hebei
,College of Agronomy, Hebei Agricultural University/North China Key Laboratory for Crop Germplasm Resources, Ministry of Education, Baoding 071001, Hebei通讯作者:
责任编辑: 李莉
收稿日期:2019-01-8接受日期:2019-03-22网络出版日期:2019-06-01
| 基金资助: |
Received:2019-01-8Accepted:2019-03-22Online:2019-06-01
作者简介 About authors
王秋莹,Tel:0312-7528415;E-mail:wangqiuying9308@163.com。

摘要
关键词:
Abstract
Keywords:
PDF (3296KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王秋莹, 王伟巧, 张艳, 王国宁, 吴立强, 张桂寅, 马峙英, 杨君, 王省芬. 棉花CRVW的克隆与抗黄萎病功能分析[J]. 中国农业科学, 2019, 52(11): 1858-1869 doi:10.3864/j.issn.0578-1752.2019.11.002
WANG QiuYing, WANG WeiQiao, ZHANG Yan, WANG GuoNing, WU LiQiang, ZHANG GuiYin, MA ZhiYing, YANG Jun, WANG XingFen.
0 引言
【研究意义】棉花是重要的纤维作物和油料作物,也是关系国计民生的重要战略物资。在生产上,黄萎病(Verticillium wilt)对棉花产量和品质危害严重,药剂难于防治。实践表明,培育和种植抗病品种是防治该病的有效途径[1]。因此,筛选和鉴定抗病基因,不仅有利于棉花抗黄萎病机制的解析,还能为棉花分子育种提供候选基因。【前人研究进展】黄萎病是由大丽轮枝菌(Verticillium dahliae)引起的土传真菌维管束病害[2]。组织结构抗性和生理生化抗性被认为在棉花抗黄萎病过程中发挥重要作用[3-5]。随着分子生物学的发展,越来越多的抗性基因获得鉴定,使得在分子水平解析棉花黄萎病抗性机制有了长足的发展。在组织结构抗性方面,棉花主要通过提高木质素积累和改变细胞壁组成增强对黄萎病菌的抗性,其中一些重要参与基因包括:与果胶代谢相关的GhPMEI3(pectin methylesterase inhibitor)[6]、与木质素合成相关的GhLAC1(laccase)[7]和GhLAC15[8]、与细胞壁组成相关的HyPRP1(hybrid proline-rich protein)[9]等。在生理生化抗性方面,棉花能够通过GhPAO(polyamine oxidase)调控对黄萎病菌具有抑制活性的精胺(spermine)和植保素(camalexin)的积累[10],依赖GbEDS1(enhanced disease susceptibility 1)调控水杨酸(salicylic acid,SA)信号通路、引发活性氧爆发(reactive oxygen species,ROS)和酚类物质积累等[11]。由南京农业大学等单位完成的陆地棉(Gossypium hirsutum)TM-1基因组scaffold总长度为2.4 Gb,包含70 478个编码蛋白基因,通过与其他植物基因组进行比对,其中66 434个基因获得了注释[12]。相对于其他已测序植物,棉花基因组较大,基因多倍化明显,并且生长周期长,以至于通过棉花遗传转化大规模分析基因功能明显滞后于拟南芥、烟草、水稻等植物[12]。病毒诱导的基因沉默(virus-induced gene silencing,VIGS)技术避免植物转化,能够在侵染植物当代对目标基因进行功能分析[13]。因此,VIGS技术一经建立,即成为植物基因功能分析的强有力工具,得到了广泛的应用[14]。目前,该技术已经成功应用于棉花生长发育、抗病反应、代谢调控等功能基因研究[15-17]。棉花基因组研究已进入功能基因组时代,快速鉴定目标基因功能已成为未来重要的研究内容之一。【本研究切入点】河北农业大学棉花品种创新与产业化团队前期利用RNA-Seq技术构建了黄萎病菌侵染处理的海岛棉(G. barbadense)和陆地棉表达谱[11, 18],发现其中一个无功能注释的基因表达受黄萎病菌显著诱导,可能参与棉花抗黄萎病菌反应过程,故将其命名为CRVW(cotton resistance to Verticillium Wilt)。【拟解决的关键问题】本研究通过对CRVW进行克隆、生物信息学分析、亚细胞定位、组织表达特异性分析、SA诱导和黄萎病菌胁迫处理后的表达模式分析、以及抗病功能鉴定等方面的研究,为进一步揭示其抗病机制奠定基础。1 材料与方法
试验于2017—2019年在河北农业大学教育部华北作物种质资源研究与利用重点实验室进行。1.1 试验材料
陆地棉抗病品种农大601(ND601)和感病品种中棉所8号(CCRI8)由河北农业大学棉花品种创新与产业化团队提供,种植于光照周期16 h(光照)/8 h(黑暗),温度25℃,相对湿度60%—70%的植物生长室中。试验所用黄萎病菌为临西2-1,由河北农业大学棉花品种创新与产业化团队分离鉴定并继代保存于PDA(potato dextrose agar)培养基[19]。
植物亚细胞定位载体pCamE由河北农业大学棉花品种创新与产业化团队构建并保存[20]。棉花VIGS载体pTRV1和pTRV2由清华大学刘玉乐教授馈赠[13]。沉默对照载体pTRV2-CLA1(cloroplastos alterados 1 gene)和农杆菌GV3101由河北农业大学棉花品种创新与产业化团队保存。
EASYspin Plus植物RNA提取试剂盒购自北京艾德莱生物科技有限公司;EasyScript? First-Strand cDNA Synthesis SuperMix购自北京北京全式金生物技术有限公司;2×PhantaTM Master Mix购自南京诺唯赞生物科技有限公司;pGM-T载体、T4 DNA连接酶、2×Taq PCR Master Mix、大肠杆菌DH5α感受态细胞、琼脂糖凝胶DNA回收试剂盒、质粒小提试剂盒购自天根生化科技有限公司;DNA Marker、PrimeScriptTM RT Reagent Kit with gDNA Eraser和EraserSYBR?Premix Ex TaqTM试剂盒购自宝生物工程有限公司。
引物合成和测序工作由生工生物工程股份有限公司完成。
1.2 基因克隆
按照ZHANG等[18]的方法将ND601培养于MS培养基(Murashige and Skoog medium)上。取约14日龄的棉苗,按照EASYspin Plus植物RNA提取试剂盒说明书进行总RNA提取。依据EasyScript? First- Strand cDNA Synthesis SuperMix试剂盒说明书合成cDNA。根据已测序陆地棉TM-1编号为Gh_A06G1566的基因上下游序列设计扩增引物CRVW-F(5'- GATTCTTTGATTTTCAAGAGGTG-3')和CRVW-R(5'-CGATTAAAAGTAACAAATTATCGT-3')。目的基因PCR扩增体系(20 μL)为1 μL cDNA、1 μL CRVW-F、1 μL CRVW-R、10 μL 2×PhantaTM Master Mix和7 μL去离子水。扩增程序为94℃ 5 min;94℃ 30 s,56℃ 30 s,72℃ 1 min,35个循环;72℃ 10 min。1%琼脂糖凝胶电泳检测后,按照琼脂糖凝胶DNA回收试剂盒操作说明对目的片段进行回收,与pGM-T载体过夜连接(16℃)后,通过热激法转化大肠杆菌DH5α感受态细胞,菌落PCR筛选阳性克隆并送测序。1.3 生物信息学分析
基因序列处理软件为BioEdit 7.0.4(http://www. mbio.ncsu.edu/bioedit/bioedit.html);陆地棉TM-1基因组序列信息和RNA-seq数据来源于CottonFGD网站(https://cottonfgd.org/);使用在线工具ProtParam(http://web.expasy.org/protparam)预测蛋白氨基酸组成、分子量、理论等电点、不稳定指数和总平均亲水性等性质;应用PSIPRED v3.3(www.psipred.net)预测蛋白二级结构;应用ProtComp v. 9.0(http://www. softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc)预测亚细胞定位;应用PlantCARE在线软件(http://bioinformatics.psb.ugent. be/webtools/plantcare/html/)预测分析顺式作用元件。1.4 亚细胞定位载体构建与荧光观察
利用引物CRVW-SF(5'-TCTAGAATGCAACAA ATCAAAGATTCATGG-3',下划线处为XbaⅠ酶切位点)和CRVW-SR(5'-GGTACCTTTCCACCCTAAA ACCTTAAGC-3',下划线处为KpnⅠ酶切位点)扩增CRVW的ORF(去除终止密码子),并插入植物表达载体pCamE多克隆位点,由CaMV 35S启动子驱动,与编码绿色荧光蛋白(green fluorescent protein,GFP)基因融合表达。洋葱上皮细胞的制备、基因枪转化和荧光观察等操作参照YANG等[21]方法。1.5 荧光定量PCR分析
按照EASYspin Plus植物RNA提取试剂盒说明书对棉苗组织进行总RNA提取。根据PrimeScript? RT reagent Kit with gDNA Eraser试剂盒所提供方法合成cDNA。以棉花PP2A1(catalytic subunit of protein phosphatase 2A)作为内参(PP2A1-F:5'-GATCCTTG TGGAGGAGTGGA-3';PP2A1-R:5'-GCGAAACAGTT CGACGAGAT-3')[22]。CRVW表达检测引物设计为qCRVW-F(5'-TCCCGCTCCTCCTTCGATTT-3')和qCRVW-R(5'-TGCATTTCAGCCTCTGGGATT-3')。棉花SA合成和信号通路标志基因检测引物按照SUN等[23]研究报道进行合成。荧光定量PCR(real-time quantitative polymerase chain reaction,qRT-PCR)反应体系为cDNA模版1.0 μL、正反向引物各0.8 μL(10 μmol·L-1)、2×SYBR mix 10 μL和7.4 μL灭菌双蒸水。使用Bio-Rad CFX96 Real-Time PCR Detection System进行荧光定量检测,具体程序为95℃ 15 s;95℃ 10 s,58℃ 10 s,72℃ 15 s,40个循环。采用2-ΔCt法进行相对定量分析。3次生物学重复检测。1.6 SA诱导处理
参照YANG等[9]的方法,配制100 μmol·L-1的SA溶液,用喷雾器均匀喷洒于7日龄棉苗子叶,并以透明罩覆盖保湿。分别于处理后6、12、24和36 h后取子叶,-80℃保存备用。以喷洒蒸馏水组作为对照。每个时间点取3株棉苗混样,共重复3次。1.7 黄萎病菌胁迫处理
按照YANG等[9]的方法制备浓度为1×107 cfu/mL的黄萎病菌孢子悬浮液。采用“六棱塑料钵定量 接种法”对棉苗进行接菌处理[24],接种菌液体积为钵体的1/10。按5级标准统计病情,并计算病情指数[9]。1.8 棉花VIGS
设计引物CRVW-VF(5'-GAATTCTTCCCGCTCC TCCTTCGATT-3',下划线处为EcoRⅠ酶切位点)和CRVW-VR(5'-GGTACCAACCTCCACAAACCCTTG CC-3',下划线处为Kpn Ⅰ酶切位点)。通过PCR扩增400 bp的CRVW片段并插入pTRV2多克隆位点EcoRⅠ和KpnⅠ间。利用冻融法将构建好的载体转入农杆菌GV3101[25]。农杆菌介导的棉花VIGS操作按照GAO等[26]方法进行。1.9 SA含量测定
通过对约15日龄的棉苗进行接菌处理,在0、12和24 hpi(hours post inoculation)时,收集3棵长势一致的棉苗第一片和第二片真叶进行混样,作为一个生物学重复,每个处理共采集3个生物学重复。采集的组织于液氮中研磨,按照VERBERNE等[27]方法提取SA,应用Agilent HPLC 1200 Series检测SA(流动相为甲醇﹕水﹕乙酸=80﹕19﹕1,流速为1 mL·min-1,上样量为20 μL,检测波长302 nm)。根据标准曲线计算SA含量。1.10 数据分析
应用Graphpad Prism? 6.02软件进行统计分析并作图。基因在不同时间和不同样本内的相对表达量进行两因素方差分析(Two-way ANOVA)和Sidak-test(P<0.05)检测。2 结果
2.1 CRVW克隆与生物信息学分析
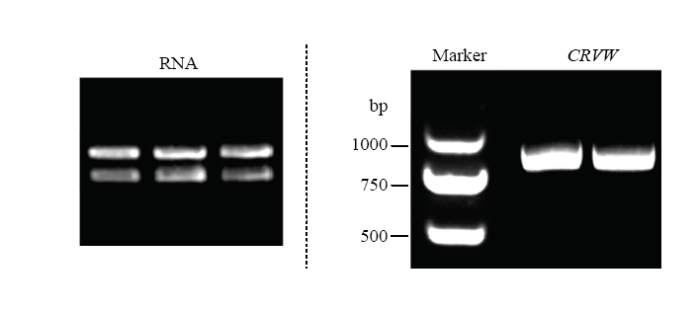
提取陆地棉ND601总RNA(图1),反转录后的cDNA作为模板,利用引物CRVW-F和CRVW-R进行PCR扩增,产物经电泳检测显示约为800 bp(图1)。PCR产物经胶回收后与T载体连接,转化大肠杆菌并提取质粒。测序后,目的基因ORF全长780 bp,与参考基因组TM-1编号为Gh_A06G1566的序列相似性100%,编码蛋白含259个氨基酸残基,尚无法预测明显的功能域,为putative uncharacterized protein。后续研究表明该基因/蛋白与棉花黄萎病抗性相关,故命名为CRVW(cotton resistance to Verticillium wilt)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1棉苗RNA提取与CRVW的PCR扩增
Fig. 1RNA extraction from cotton seedlings and PCR amplication of CRVW
经预测,CRVW分子量约为30.2 kD,理论等电点9.59,分子式C1332H2061N383O395S14;在组成蛋白的20种氨基酸中,丝氨酸(Ser)所占比例最高(12.0%),而组氨酸(His)所占的比例最低(0.8%);含32个负电荷氨基酸残基,42个正电荷氨基酸残基;不稳定指数为61.41,属于不稳定蛋白;总平均亲水性-0.975,属于疏水蛋白。二级结构预测显示CRVW含有69.50%不规则卷曲、17.76% α-螺旋、11.20%延伸链和1.54% β-折叠。
2.2 CRVW亚细胞定位
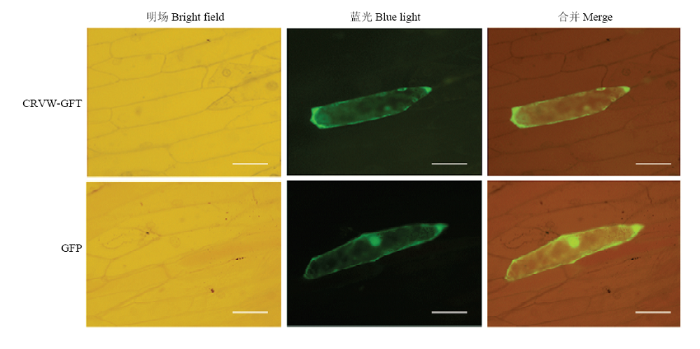
ProtComp v. 9.0在线工具预测显示,CRVW最有可能位于细胞膜(基于Neural Nets算法)和细胞质(基于Pentamers算法)。为进一步确定CRVW在细胞中的位置,将其与GFP蛋白进行融合表达。荧光显微观察显示,单独表达GFP时,荧光信号存在于在细胞核、细胞质和细胞膜中;而对于CRVW与GFP融合表达的细胞,荧光主要出现在细胞膜和细胞质(图2)。综合生物信息学预测和荧光显微观察结果,表明CRVW主要存在于植物细胞膜和细胞质。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2CRVW在洋葱表皮细胞中的亚细胞定位
Fig. 2Subcellular localization of CRVW in onion epidermal cells
2.3 CRVW的组织表达特异性
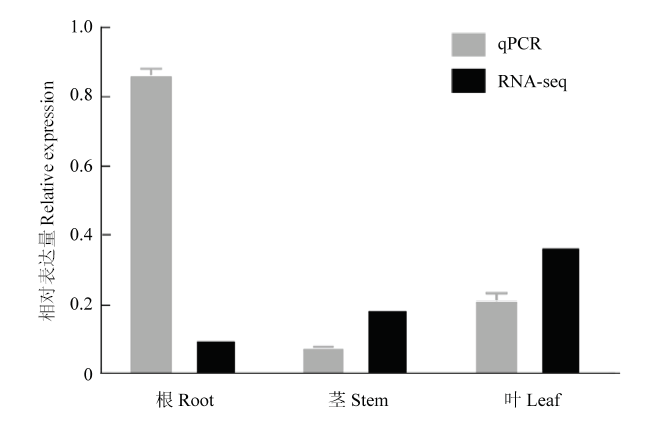
已有RNA-seq数据显示,CRVW在棉花根、茎和叶中均有表达,且叶中最高。为进一步明确CRVW的组织表达情况,利用qPCR技术对该基因在陆地棉ND601根、茎和叶中的表达进行了检测。结果显示,CRVW在以上3种组织中均有表达,但在根中的表达量最高,大约是茎中的10倍和叶中的4倍(图3)。RNA-seq和qPCR检测结果均确认CRVW在棉花根、茎和叶中都有表达,但存在组织表达差异。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3棉花不同组织中CRVW的表达分析
Fig. 3Expression analysis of CRVW in different organs of cotton plants
2.4 CRVW受SA诱导表达
从已测序棉花TM-1基因组调取CRVW ORF上游2 000 bp序列(染色体A06:100117602—100119601)作为潜在的启动子,命名为CRVW-P。预测分析显示,CRVW-P含有响应4种激素信号的顺式作用元件,包括乙烯(ethylene,ET)、SA、生长素(auxin)和脱落酸(abscisic acid,ABA)。另外,还包括一些与伤害、防御、胁迫、病菌、干旱和低温等相关的作用元件(表1)。根据TM-1 CRVW上游序列设计引物,对抗病品种ND601和感病品种CCRI8中CRVW上游序列进行同源克隆,并未发现抗感品种在所含顺式作用元件的位置和数量上存在差异(结果未显示)。Table 1
表1
表1CRVW-P中的顺式作用元件预测
Table 1
| 名称 Name | 序列 Sequence | 位置 Position | 功能 Function |
|---|---|---|---|
| ERE | ATTTTAAA | +175,+725,+1470,+1530 | 乙烯响应Ethylene responsiveness |
| TCA | TCATCTTCAT | +1935 | 水杨酸响应Salicylic acid responsiveness |
| AuxRR-core | GGTCCAT | +441 | 生长素响应Auxin responsiveness |
| ABRE | ACGTG | +67,-566 | 脱落酸响应Abscisic acid responsiveness |
| WUN-motif | AAATTTCTT | -305 | 伤害响应Wound responsiveness |
| WRE3 | CCACCT | +1761,-1987 | 伤害响应Wound responsiveness |
| STRE | AGGGG | -911 | 胁迫响应Stress responsiveness |
| TC-rich repeats | ATTCTCTAAC | +550 | 防御和胁迫响应Defense and stress responsiveness |
| box S | AGCCACC | +1759 | 伤害和病菌响应Wound and pathogen responsiveness |
| LTR | CCGAAA | -347 | 低温响应Low-temperature responsiveness |
| MBS | CAACTG | +1494 | 干旱响应Drought responsiveness |
新窗口打开|下载CSV
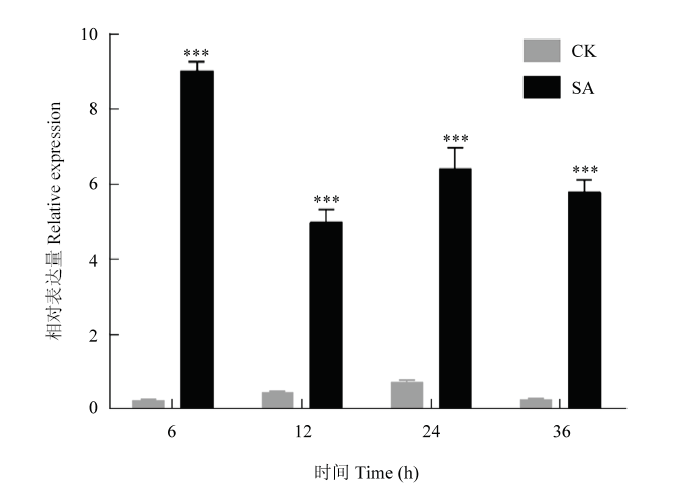
进一步对棉苗进行SA诱导处理,结果显示,与对照相比,CRVW在所有4个时间点的表达量均发生显著上调(图4),表明CRVW受SA信号通路调控。
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4水杨酸诱导棉花CRVW的表达分析
CK:对照组;SA:水杨酸处理组;***表示差异在P<0.001水平上具有显著性。下同
Fig. 4Analysis of salicylic acid-induced expression of CRVW in cotton
CK: Control group; SA: Salicylic acid induced group; *** indicate that the differences are significant at 0.001 level. The same as below
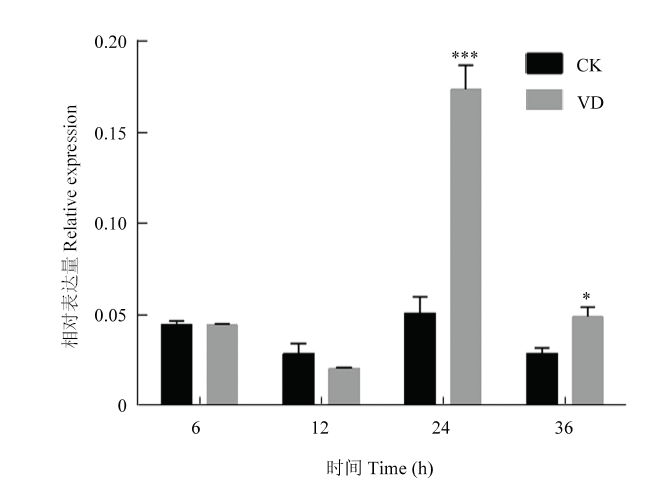
2.5 CRVW受黄萎病菌诱导显著上调表达
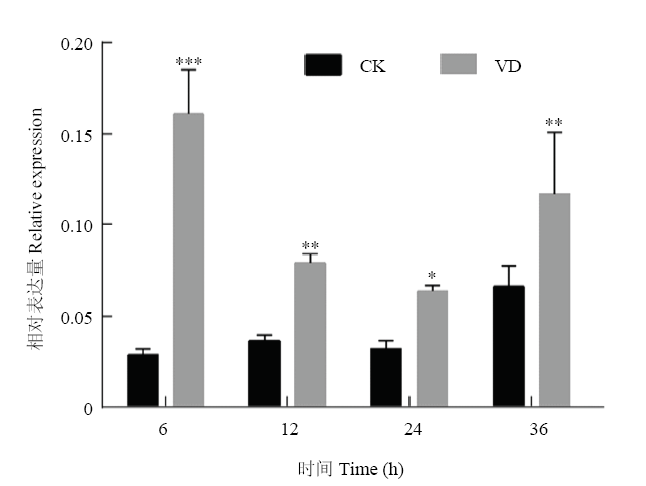
以抗病品种ND601和感病品种CCRI8根组织为分析对象,利用qPCR检测CRVW在黄萎病菌胁迫下的表达模式。黄萎病菌胁迫后,CRVW在抗病品种中显著上调表达(与同时间水处理对照相比)(图5)。而对于感病品种CCRI8,在黄萎病菌胁迫后的6和12 hpi,CRVW的表达水平未发生显著变化,直到24 hpi其表达才发生显著上调(图6)。由此可见,黄萎病菌胁迫处理后,不管是抗病品种还是感病品种,CRVW均发生显著上调表达,但其在抗病品种中发生响应的时间早于感病品种。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5黄萎病菌胁迫下CRVW在ND601根部的表达分析
VD:大丽轮枝菌处理组;CK:对照组;*表示差异在P<0.05水平上具有显著性;**表示差异在P<0.01水平上具有显著性。下同
Fig. 5Expression analysis of CRVW in ND601 roots under Verticillium dahliae stress
VD: Verticillium dahliae treatment group; CK: Control group; * indicates that the difference is significant at 0.05 level; ** indicate that the differences are significant at 0.01 level. The same as below
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6黄萎病菌胁迫下CRVW在CCRI8根部的表达分析
VD:大丽轮枝菌处理组;CK:对照组
Fig. 6Expression analysis of CRVW in CCRI8 roots under Verticillium wilt stress
VD: Verticillium dahliae treatment group; CK: Control group
2.6 沉默CRVW显著降低棉花黄萎病抗性
对尚未长出真叶的棉苗进行VIGS处理,以沉默CLA1(cloroplastos alterados 1)作为技术对照。在VIGS操作7 d后,CLA1发生沉默并表现为新生真叶白化(图7-A),表明技术体系成功建立。同时对CRVW沉默情况通过qRT-PCR确认,并进行黄萎病菌胁迫处理。在接菌处理20 d后,野生(wild type,WT)组(既无VIGS处理也未接菌)生长正常;CK组(VIGS处理中注射pTRV2空载体,同时接菌处理)表现为叶片黄化和植株萎蔫;CRVW沉默组(注射携带CRVW序列片段的pTRV2载体,同时接菌处理)表现出比CK组更明显的黄化、萎蔫和落叶等黄萎病病症(图7-B)。病指统计分析显示,CRVW沉默组病指显著高于CK组(图7-C),进一步表明CRVW沉默显著降低了棉苗对黄萎病菌的抗性。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7CRVW沉默降低了棉苗对黄萎病菌的抗性
A:CLA1沉默后棉花的白化表型;B:黄萎病菌侵染CRVW沉默后棉花的萎蔫表型;C:黄萎病菌侵染CRVW沉默后棉花的病指。WT:野生型组;CK:对照组;CRVW:CRVW沉默组
Fig. 7Silencing of CRVW reduced cotton seedlings resistance to Verticillium dahlia
A: Albino phenotype of CLA1-silenced cotton plants; B: Wilt symptom of CRVW-silenced cotton plants inoculated with V. dahliae; C: The disease indices of CRVW-silenced cotton plants with infection by V. dahliae. WT: Wild type group; CK: Control group; CRVW: CRVW-silenced group
2.7 沉默CRVW导致SA合成和信号通路相关基因表达显著降低
在黄萎病菌侵染后的12和24 hpi,对照棉苗中SA含量显著增加(与0 hpi对照相比)。而对于CRVW被沉默的棉苗,其体内SA含量仅在12 hpi显著增加(与0 hpi CRVW沉默棉苗相比)。与对照相比,在0、12和24 hpi 3个时间点,CRVW沉默棉苗中SA含量均出现显著降低(图8),约为对照的1/2。为进一步确认CRVW是否参与SA信号通路,对与SA合成和信号通路相关的5个标志基因进行表达检测。结果显示,ICS1(isochorismate synthase 1)、EDS1(enhanced disease susceptibility 1)、PAD4(phytoalexin deficient 4)、NPR1(nonexpresser of PR gene 1)和PR1(pathogenesis-related protein 1)在CRVW沉默组中的表达均显著降低,特别是在12 hpi,这些基因在沉默组中的表达比对照降低得最为明显(图9)。以上结果表明CRVW的沉默能够显著影响SA的积累,降低SA信号通路相关基因的表达。图8
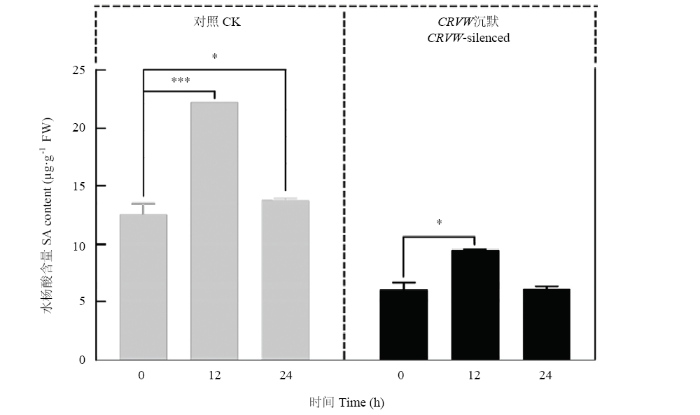 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8黄萎病菌侵染后棉苗体内水杨酸的积累
Fig. 8Accumulation of SA in V. dahliae-infected cotton seedlings
图9
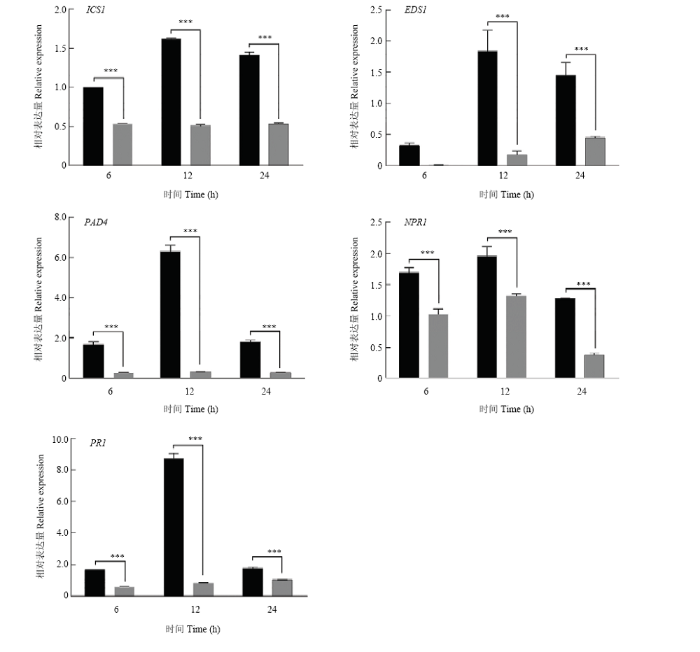 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9黄萎病菌胁迫后SA合成和信号通路相关基因在对照和CRVW沉默棉苗中的表达分析
Fig. 9Expression analysis of SA biosynthesis and signaling pathway-related genes in the CK and CRVW-silenced cotton seedlings after inoculation with the V. dahliae
3 讨论
3.1 CRVW是潜在重要的棉花抗黄萎病基因
高通量转录组测序(RNA-seq)技术为棉花抗病基因挖掘提供了新的平台。应用该技术,大量潜在的与黄萎病菌胁迫相关的棉花基因被发现[11,18,28]。棉花基因组测序的完成更是为克隆这些基因提供了极大地便利[12],使得鉴定这些基因功能成为科研工作者重要的研究内容。最初,河北农业大学棉花品种创新与产业化团队从棉花受黄萎病菌胁迫处理后的RNA-aeq数据中发现,CRVW为一个差异表达基因,但无功能注释[11,18]。本研究通过qRT-PCR对该基因的表达进行了分析,确认CRVW受黄萎病菌诱导后显著上调表达,且其在抗病品种中表达变化早于感病品种(图5和图6),表明CRVW参与棉花和黄萎病菌间的互作。通过VIGS技术对CRVW进行沉默,发现棉花对黄萎病的抗性显著降低(图7),进一步证明CRVW参与棉花对黄萎病菌的抗病过程。本研究虽然成功克隆了CRVW(图1),但其编码蛋白无明显的功能域。因此,目前还无法通过生物信息学对其进行注释。亚细胞定位和组织表达特异性分析明确了CRVW在棉花组织中的分布(图2和图3),为进一步注释其生理生化功能提供了依据。3.2 CRVW通过SA信号通路参与棉花对黄萎病菌的抗性
植物抵御病原菌侵染是一个复杂的免疫过程,激素是调控该过程的重要信号分子[29]。在已知的植物激素中,SA、茉莉酸(jasmonic acid,JA)和ET是参与免疫反应最为核心的成员[29]。在CRVW-P中存在与ET、SA、auxin和ABA等激素信号响应相关的的顺式作用元件(表1),表明CRVW可能通过以上4种激素信号通路参与棉花的生长发育和抗逆反应。棉花被黄萎病菌侵染后,体内SA会发生显著积累[11]。SA信号通路上的标志性基因EDS1发生沉默后,棉花对黄萎病的抗性也会显著降低[11],表明SA在棉花抗黄萎病过程中发挥重要的调控功能。病原菌诱导的SA合成主要经异分支酸合酶(isochorismate synthase,ICS)途径,发生在叶绿体中[30]。另外,与SA积累相关的重要标志基因有EDS1和PAD4[31]。NPR1是SA信号通路的重要调节基因。NPR1接收到SA信号后会由多聚体(oligomer)变为单体(monomer),从胞质进入细胞核,与TGA(TGACG motif-binding factor)互作,进而调节相关抗病基因的表达,特别是病程相关蛋白PR1的积累[32]。本研究检测了SA诱导后CRVW的表达情况,发现SA能够诱导CRVW显著上调表达(图4)。CRVW沉默后,棉苗体内SA含量显著降低(图8),ICS1、EDS1、PAD4、NPR1和PR1等5个与SA积累和信号调控相关的标志基因均发生了显著下调表达(图9)。据此,推测CRVW可能通过SA信号通路参与棉花对黄萎病菌的抗性。3.3 植物未知功能基因研究展望
目前,在蛋白家族数据库(Pfam)中未知功能结构域蛋白家族约占26.5%[33],意味着在植物界存在数量巨大的未知功能基因。大量组学数据分析都表明,这些未知功能基因在植物生长发育和逆境胁迫过程中发挥重要作用。因此,对这些基因功能的研究将有助于在全新层面上透彻地了解生物体复杂的生命活动机制[34]。本文对CRVW进行了表达模式分析,并证明其参与了棉花对黄萎病胁迫的响应,但其具体的调控机制还需深入研究。为进一步完善CRVW的功能注释,解析CRVW蛋白三维构象、寻找其互作蛋白、明确其参与的信号通路等都将是未来重要的研究内容。4 结论
棉花CRVW无明显结构域,且无生理生化功能注释。CRVW定位于细胞质和细胞膜,主要在棉花根部表达;CRVW可能通过SA信号通路参与棉花抗黄萎病,是一个潜在重要的抗病基因。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
Magsci

综述了我国棉花抗枯、黄萎病育种研究进展 ,内容包括 5 0年来取得的成就、经验 ,存在的问题及对策 ,是对我国棉花抗病育种工作较为全面的总结。
Magsci

综述了我国棉花抗枯、黄萎病育种研究进展 ,内容包括 5 0年来取得的成就、经验 ,存在的问题及对策 ,是对我国棉花抗病育种工作较为全面的总结。
DOI:10.1146/annurev-phyto-080508-081748URL
DOI:10.1093/jxb/err245URLPMID:21862479

The incompatible pathosystem between resistant cotton (Gossypium barbadensecv. 7124) andVerticillium dahliaestrain V991 was used to study the cotton transcriptome changes after pathogen inoculation by RNA-Seq. Of 32 774 genes detected by mapping the tags to assembly cotton contigs, 3442 defence-responsive genes were identified. Gene cluster analyses and functional assignments of differentially expressed genes indicated a significant transcriptional complexity. Quantitative real-time PCR (qPCR) was performed on selected genes with different expression levels and functional assignments to demonstrate the utility of RNA-Seq for gene expression profiles during the cotton defence response. Detailed elucidation of responses of leucine-rich repeat receptor-like kinases (LRR-RLKs), phytohormone signalling-related genes, and transcription factors described the interplay of signals that allowed the plant to fine-tune defence responses. On the basis of global gene regulation of phenylpropanoid metabolism-related genes, phenylpropanoid metabolism was deduced to be involved in the cotton defence response. A closer look at the expression of these genes, enzyme activity, and lignin levels revealed differences between resistant and susceptible cotton plants. Both types of plants showed an increased level of expression of lignin synthesis-related genes and increased phenylalanine-ammonia lyase (PAL) and peroxidase (POD) enzyme activity after inoculation withV. dahliae, but the increase was greater and faster in the resistant line. Histochemical analysis of lignin revealed that the resistant cotton not only retains its vascular structure, but also accumulates high levels of lignin. Furthermore, quantitative analysis demonstrated increased lignification and cross-linking of lignin in resistant cotton stems. Overall, a critical role for lignin was believed to contribute to the resistance of cotton to disease.
DOI:10.1111/mpp.2006.7.issue-2URL
DOI:10.1104/pp.17.01399URL
DOI:10.1104/pp.17.01628URL
DOI:10.1111/mpp.2019.20.issue-3URL
DOI:10.1186/s12870-018-1565-1 [本文引用: 3]
DOI:10.1111/tpj.12941URLPMID:26221980

Summary Verticillium dahliae is a destructive, soil-borne fungal pathogen that causes vascular wilt disease in many economically important crops worldwide. A polyamine oxidase ( PAO ) gene was identified and cloned by screening suppression subtractive hybridisation and cDNA libraries of cotton genotypes tolerant to Verticillium wilt and was induced early and strongly by inoculation with V.02dahliae and application of plant hormone. Recombinant cotton polyamine oxidase (GhPAO) was found to catalyse the conversion of spermine (Spm) to spermidine (Spd) in02vitro . Constitutive expression of GhPAO in Arabidopsis thaliana produced improved resistance to V.02dahliae and maintained putrescine, Spd and Spm at high levels. Hydrogen peroxide (H2O2), salicylic acid and camalexin (a phytoalexin) levels were distinctly increased in GhPAO -overexpressing Arabidopsis plants during V.02dahliae infection when compared with wild-type plants, and Spm and camalexin efficiently inhibited growth of V.02dahliae in02vitro . Spermine promoted the accumulation of camalexin by inducing the expression of mitogen-activated protein kinases and cytochrome P450 proteins in Arabidopsis and cotton plants. The three polyamines all showed higher accumulation in tolerant cotton cultivars than in susceptible cotton cultivars after inoculation with V.02dahliae . GhPAO silencing in cotton significantly reduced the Spd level and increased the Spm level, leading to enhanced susceptibility to infection by V.02dahliae , and the levels of H2O2 and camalexin were distinctly lower in GhPAO -silenced cotton plants after V.02dahliae infection. Together, these results suggest that GhPAO contributes to resistance of the plant against V.02dahliae through the mediation of Spm and camalexin signalling.
DOI:10.1094/MPMI-03-17-0067-RURL [本文引用: 4]
[本文引用: 1]
DOI:10.1046/j.1365-313X.2002.01297.xURL [本文引用: 1]
DOI:10.3969/j.issn.1672-416X.2009.01.026

病毒诱导基因沉默(virus induced gene silencing,VIGS)技术是一种RNA介导的抗病毒防御反应机制,目前在植物反向遗传学领域已经表现出巨大的潜力。VIGS技术不仅优于传统的植物转基因技术,方法简便,高效耐用,而且具有高通量特性。在功能基因组学领域的研究中,这些优越性已经使VIGS技术成为最具吸引力的首选技术手段。目前,VIGS体系应用最成功的植物是病毒学家常用的模式植物-本氏烟草(Nicotiana benthamiana),与此同时,也在努力改良VIGS技术,使其能够在包括单子叶植物在内的其它物种中得到广泛应用。我们讨论的重点是针对利用VIGS技术来确定基因功能,该技术已经在抗病性,逆境胁迫,细胞信号传导以及次生代谢生物合成途径研究中显示出相关基因功能的多样性,并随之构建出一系列新的相关模型。
DOI:10.3969/j.issn.1672-416X.2009.01.026

病毒诱导基因沉默(virus induced gene silencing,VIGS)技术是一种RNA介导的抗病毒防御反应机制,目前在植物反向遗传学领域已经表现出巨大的潜力。VIGS技术不仅优于传统的植物转基因技术,方法简便,高效耐用,而且具有高通量特性。在功能基因组学领域的研究中,这些优越性已经使VIGS技术成为最具吸引力的首选技术手段。目前,VIGS体系应用最成功的植物是病毒学家常用的模式植物-本氏烟草(Nicotiana benthamiana),与此同时,也在努力改良VIGS技术,使其能够在包括单子叶植物在内的其它物种中得到广泛应用。我们讨论的重点是针对利用VIGS技术来确定基因功能,该技术已经在抗病性,逆境胁迫,细胞信号传导以及次生代谢生物合成途径研究中显示出相关基因功能的多样性,并随之构建出一系列新的相关模型。
DOI:10.1111/tpj.2011.66.issue-2URL
DOI:10.1186/1471-2164-14-637URL [本文引用: 3]
DOI:1002-7807(2012)04-0348-10Magsci [本文引用: 1]

从河北省17个主要植棉县采集棉花黄萎病株,分离获得52个黄萎病菌单孢菌系,对其培养特性、致病性和ISSR(Inter-simple sequence repeat)遗传分化进行了研究。菌系培养性状研究表明,在采集的52个菌系中,菌核型菌系最多,其次是菌丝型,最少的是中间型,3种类型菌系分别占总菌系的51.9%,38.5%和9.6%。利用7个抗、感不同的鉴别寄主在光、温、湿可控的生长室鉴定了病菌的致病性,供试菌系可分为致病力强、中、弱3种类型(Ⅰ型、Ⅱ型和Ⅲ型),分别占供试菌系的51.9%,21.2%和26.9%。在供试菌系中存在比落叶型菌系致病力还要强的非落叶型菌系。基于病情指数的聚类分析结果表明,河北省棉花黄萎病菌系存在明显致病力分化,但与地理来源无关。菌核型菌系和中间型菌系多表现为强致病力或中等致病力,而菌丝型菌系的致病力变化较大。在136个ISSR标记中,80个属于多态性标记,多态性位点百分率达58.8%。基于ISSR的聚类分析结果表明,河北省棉花黄萎病菌的遗传分化较小,并且遗传分化与地理来源相关。
DOI:1002-7807(2012)04-0348-10Magsci [本文引用: 1]

从河北省17个主要植棉县采集棉花黄萎病株,分离获得52个黄萎病菌单孢菌系,对其培养特性、致病性和ISSR(Inter-simple sequence repeat)遗传分化进行了研究。菌系培养性状研究表明,在采集的52个菌系中,菌核型菌系最多,其次是菌丝型,最少的是中间型,3种类型菌系分别占总菌系的51.9%,38.5%和9.6%。利用7个抗、感不同的鉴别寄主在光、温、湿可控的生长室鉴定了病菌的致病性,供试菌系可分为致病力强、中、弱3种类型(Ⅰ型、Ⅱ型和Ⅲ型),分别占供试菌系的51.9%,21.2%和26.9%。在供试菌系中存在比落叶型菌系致病力还要强的非落叶型菌系。基于病情指数的聚类分析结果表明,河北省棉花黄萎病菌系存在明显致病力分化,但与地理来源无关。菌核型菌系和中间型菌系多表现为强致病力或中等致病力,而菌丝型菌系的致病力变化较大。在136个ISSR标记中,80个属于多态性标记,多态性位点百分率达58.8%。基于ISSR的聚类分析结果表明,河北省棉花黄萎病菌的遗传分化较小,并且遗传分化与地理来源相关。
Magsci [本文引用: 1]

为了探索通用型植物表达载体的快捷构建,本研究通过PCR克隆、酶切连接和PCR扩增获得CaMV 35S启动子序列及pUC19多克隆位点部分限制性内切酶位点和NOS终止子,构建成完整的表达组件,并将该组件插入pCAMBIA1300的多克隆位点,构建成通用型植物表达载体pCamE (GenBank登录号: JX841315)。pCamE表达组件的启动子上游、多克隆位点和终止子下游分别含有4、8和3个单拷贝限制性内切酶位点,利于外源基因的克隆和插入以及对载体启动子或终止子的修饰和更换。以绿色荧光蛋白基因(enhanced green fluorescent protein, eGFP)为报告基因,分别构建了表达载体pCam::GFP、pCam::SalGFP和pCam::DAHPS-GFP,转化洋葱(Allium cepa L.)表皮细胞和拟南芥(Arabidopsis thaliana L.),绿色荧光蛋白(GFP)和融合蛋白DAHPS-GFP在洋葱表皮细胞中均成功稳定表达;在pCam::GFP转基因拟南芥株系的根、根尖、根毛、茎表皮毛、幼叶基部、叶柄微管组织和未成熟花药等组织均能观察到强烈的绿色荧光。表明,植物表达载体pCamE是可将不同目的基因与植物染色体整合并稳定遗传的通用型植物表达载体,不受外源片段大小及插入位点的影响,具有经济实用、适用广泛、操作简便和外源基因在转基因植物中遗传稳定且正常表达等特点。该载体可用于不同基因和不同功能表达载体的快捷构建,为植物的遗传转化和基因功能研究提供了新的通用型基因表达载体工具。
Magsci [本文引用: 1]

为了探索通用型植物表达载体的快捷构建,本研究通过PCR克隆、酶切连接和PCR扩增获得CaMV 35S启动子序列及pUC19多克隆位点部分限制性内切酶位点和NOS终止子,构建成完整的表达组件,并将该组件插入pCAMBIA1300的多克隆位点,构建成通用型植物表达载体pCamE (GenBank登录号: JX841315)。pCamE表达组件的启动子上游、多克隆位点和终止子下游分别含有4、8和3个单拷贝限制性内切酶位点,利于外源基因的克隆和插入以及对载体启动子或终止子的修饰和更换。以绿色荧光蛋白基因(enhanced green fluorescent protein, eGFP)为报告基因,分别构建了表达载体pCam::GFP、pCam::SalGFP和pCam::DAHPS-GFP,转化洋葱(Allium cepa L.)表皮细胞和拟南芥(Arabidopsis thaliana L.),绿色荧光蛋白(GFP)和融合蛋白DAHPS-GFP在洋葱表皮细胞中均成功稳定表达;在pCam::GFP转基因拟南芥株系的根、根尖、根毛、茎表皮毛、幼叶基部、叶柄微管组织和未成熟花药等组织均能观察到强烈的绿色荧光。表明,植物表达载体pCamE是可将不同目的基因与植物染色体整合并稳定遗传的通用型植物表达载体,不受外源片段大小及插入位点的影响,具有经济实用、适用广泛、操作简便和外源基因在转基因植物中遗传稳定且正常表达等特点。该载体可用于不同基因和不同功能表达载体的快捷构建,为植物的遗传转化和基因功能研究提供了新的通用型基因表达载体工具。
DOI:10.1007/s00299-015-1798-5URL [本文引用: 1]
DOI:10.1186/1471-2229-10-49URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1002/pca.615URLPMID:11899606 [本文引用: 1]

Salicylic acid (SA) is a signalling compound in plants which is able to induce systemic acquired resistance. In the analysis of SA in plant tissues, the extraction recovery is often very low and variable. This is mainly caused by sublimation of SA, especially during evaporation of organic solvents. Techniques have been designed in order to overcome this problem. In the first part of the extraction procedure, sublimation of SA was prevented by addition of 0.2 M sodium hydroxide. At a later stage of the extraction procedure, sublimation of SA during solvent evaporation was controlled by the addition of a small amount of HPLC eluent. In this way, recoveries in the range of 71-91% for free SA and 65-79% for acid-hydrolysed SA were obtained. Recoveries could be further optimised by the use of an internal standard to correct for volume changes after the addition of the HPLC eluent. Copyright ? 2001 John Wiley & Sons, Ltd.
DOI:10.1186/1471-2164-14-852URL [本文引用: 1]
DOI:10.1038/nchembio.164PMID:19377457 [本文引用: 2]

Abstract Plants live in complex environments in which they intimately interact with a broad range of microbial pathogens with different lifestyles and infection strategies. The evolutionary arms race between plants and their attackers provided plants with a highly sophisticated defense system that, like the animal innate immune system, recognizes pathogen molecules and responds by activating specific defenses that are directed against the invader. Recent advances in plant immunity research have provided exciting new insights into the underlying defense signaling network. Diverse small-molecule hormones play pivotal roles in the regulation of this network. Their signaling pathways cross-communicate in an antagonistic or synergistic manner, providing the plant with a powerful capacity to finely regulate its immune response. Pathogens, on the other hand, can manipulate the plant's defense signaling network for their own benefit by affecting phytohormone homeostasis to antagonize the host immune response.
[本文引用: 1]
DOI:10.1073/pnas.1511182112URL [本文引用: 1]
[本文引用: 1]
DOI:10.1093/nar/gkr1065URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
