 ,1,21
,1,21 2
3
Genome Wide Identification and Expression Analysis of the U-box Gene Family in Citrus
LI QiuYue1, ZHANG YaFei1, PENG Jie1, WANG Xu1, ZHANG ZhiQiang1, DAI XiangSheng3, JIANG Dong ,1,21
,1,21 2
3
通讯作者:
责任编辑: 赵伶俐
收稿日期:2018-12-29接受日期:2019-02-18网络出版日期:2019-06-01
| 基金资助: |
Received:2018-12-29Accepted:2019-02-18Online:2019-06-01
作者简介 About authors
李秋月,E-mail:437010037@qq.com。

摘要
关键词:
Abstract
Keywords:
PDF (7808KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李秋月, 张亚飞, 彭洁, 王旭, 张志强, 戴祥生, 江东. 柑橘U-box基因家族的鉴定及表达分析[J]. 中国农业科学, 2019, 52(11): 1942-1960 doi:10.3864/j.issn.0578-1752.2019.11.009
LI QiuYue, ZHANG YaFei, PENG Jie, WANG Xu, ZHANG ZhiQiang, DAI XiangSheng, JIANG Dong.
0 引言
【研究意义】柑橘是世界范围内重要的热带、亚热带经济类果树之一,但由于生物胁迫、非生物胁迫等原因严重影响了其产量和品质,造成较大的经济损失。U-box基因参与生物胁迫与非生物胁迫的应答,并对不同的激素处理具有不同的应答模式。本研究利用生物信息学的方法对柑橘全基因组的U-box基因家族进行全面分析,深入了解柑橘生长发育和逆境响应的分子基础,研究结果对于认识柑橘U-box的功能和进一步培育或改良柑橘抗性品种具有重要意义。【前人研究进展】泛素26s蛋白酶体途径(ubiquity-26s proteasomepathway,UPP)是目前己知所有真核生物体内具有高度选择性的蛋白降解途径[1],早期的泛素化主要由E1、E2和E3这3种酶来完成。E1激活酶负责激活泛素,E2耦联酶可直接将泛素转移到底物蛋白质,或者同泛素一起转移给E3连接酶[2]。决定底物蛋白特异性识别的E3连接酶,根据作用机理和亚基的组成可以分为4类:单亚基泛素连接酶,包括HECT(homologous to E6-AP COOH-Terminus)、U-box、RING(really interesting new gene)、多亚基泛素连接酶,如cullin-RING(CRLs)[3]。U-box域在植物、动物和酵母等真核生物中高度保守,由70多个氨基酸残基构成,最早从酵母UFD2(ubiquitin fusion degradation protein-2)中发现[4]。U-box蛋白参与蛋白泛素化降解,同时也对细胞进行自身功能调控,并对生物胁迫、非生物胁迫、生长发育以及对激素做出响应[5,6,7,8]。GONZALEZ-LAMOTHE等[9]表明CMPG1在烟草与番茄中参与多个抗病基因介导的抗性反应;同时,U-box泛素连接酶在低温、干旱、盐胁迫及光胁迫等非生物胁迫的过程中发挥重要作用,拟南芥中的CHIP、AtPUB18、AtPUB19、AtPUB30,辣椒的PUB1及大豆中的GmPUB8响应植物对低温、高盐和干旱的相应过程[10,11,12];除此之外,一些研究还发现U-box泛素连接酶调控植物的生长发育,BRAUMANN等[13]发现缺失U-box E3泛素连接酶能够导致brh2和ari-l突变体发生矮化;U-box蛋白可参与植物激素调控通路,拟南芥U-box泛素连接酶AtPUB9与脱落酸(ABA)信号通路有关[14],在ABA处理下,U-box蛋白在细胞中重新分配,与野生型相比,AtPUB9突变体种子在萌发时对ABA耐受性增强。综上所述,U-box蛋白具有多种生物学功能,在调节植物的生长发育及生物胁迫与非生物胁迫方面具有重要作用,因此备受研究者的关注。【本研究切入点】U-box基因家族广泛参与植物生物胁迫与非生物胁迫响应,但目前对U-box家族的研究主要集中在拟南芥、棉花、番茄、水稻和苜蓿等模式植物中,在柑橘中对U-box基因家族进行鉴定和分析尚未见报道。柑橘克里曼丁(Citrus clementina)的全基因组测序已经完成[15],为进一步研究柑橘相关基因的功能及其相互之间的关系奠定了基础。【拟解决的关键问题】本研究利用生物信息学的方法对柑橘U-box基因家族成员进行鉴定,并对该家族的基本信息、保守结构域、Scafflod定位等进行预测分析,利用qRT-PCR技术进行组织表达模式及对非生物胁迫和激素的响应进行表达分析,为阐明柑橘U-box基因家族成员的生物学功能奠定基础。1 材料与方法
试验于2018年5—10月在中国农业科学院柑桔研究所资源育种室进行。1.1 柑橘U-box基因家族成员鉴定
柑橘克里曼丁橘(Citrus clementina)的基因组和蛋白组数据下载于公共数据库Phytozome(https:// phytozome.jgi.doe.gov);在拟南芥数据库(http://www. arabidopsis.org/browse/genefamily/pub.jsp)下载拟南芥U-box基因家族基因序列和蛋白序列。利用3种方法鉴定柑橘中的U-box基因。(1)首先以拟南芥中已经鉴定出的64个U-box基因家族成员的蛋白序列在克里曼丁蛋白质组数据库进行在线BLAST搜索;(2)同时在Pfam数据库(http://pfam.xfam.org/)中下载所有物种U-box结构域序列(PF04564),利用Hmmer2.3.2(http://hmmer.janelia.org/)构建隐马氏模型,在Phytozome的克里曼丁蛋白数据库中搜索含有U-box结构域的序列;(3)合并(1)和(2)的结果,去除无完整读码框的序列。将得到的结果使用在线工具SMART(http://smart.embl-heidelberg.de/)进一步分析结构域,去除不包含U-box结构域的序列,最终得到柑橘U-box家族所有基因。将得到的U-box成员按照Scafflod定位进行命名,如CcPUB1:Cc为Citrus clementina缩写,PUB是基因家族的缩写,1是根据在Scafflod上的位置给这个成员的序号。对柑橘U-box蛋白的分子量和等电点预测使用ExPASy Proteomics Server(http://www.expasy.org/proteomics)。同时用在线软件MBC(http://cello.life.nctu.edu.tw)对柑橘U-box蛋白进行亚细胞定位预测。1.2 柑橘U-box家族系统进化及蛋白结构域分析
利用MEGA5.2中的MUSCLE程序将鉴定到的柑橘U-box家族蛋白序列进行多重序列比对,并利用邻接法(Neighbor-Joining,NJ)构建系统进化树。U-box基因家族蛋白的结构域使用在线工具SMART。1.3 柑橘U-box家族基因结构及Scafflod定位分析
在phytozome数据库中下载已鉴定的柑橘U-box基因的DNA序列,用GSDS(http://gsds.cbi.pku.edu. cn/)制作基因结构图。同时获得基因的位置信息,基因的Scafflod定位图用MapChart软件展示。1.4 柑橘U-box基因家族的启动子分析
从克里曼丁全基因组数据库中提取每个柑橘U-box基因起始密码子上游2 000 bp基因组序列,顺式作用元件预测使用PlantCARE(http://bioinformatics. psb.ugent.be/webtools/plantcare/html/)。1.5 试验材料与处理
选用锦橙(Citrus sinensis Osbeck.)为试材。2018年10月在中国农业科学院柑橘研究所国家果树种质(重庆)柑橘圃采集锦橙果实,将种子去皮,放在湿润的培养皿中;然后将其放入28℃的培养箱中进行催芽。选取萌发整齐一致的种子,播种于填装有混合基质的盆中,置人工气候箱培养(温度28℃,光照16 h/黑暗8 h),当第一片真叶充分展开后,选取长势一致的幼苗进行非生物胁迫和激素处理。激素包括GA3(0.5 mmol·L-1)、ABA(100 μmol·L-1)、IAA(20 μmol·L-1);非生物胁迫包括高盐NaCl(300 mmol·L-1)、10%(ω)PEG6000。具体步骤如下:在室温下,将幼苗洗净,放入盛有激素或10 μmol·L-1 DMSO(激素处理的平行对照)或H2O(非生物胁迫的对照)的灭菌瓶中摇匀,使根系充分接触液体。分别在处理后的0、3、6、12和24 h收集幼苗,3次生物学重复,放入液氮速冻,-80℃保存备用。为说明CcPUB4在NaCl条件下是受Na+还是Cl-影响,选用对盐敏感的大果枳(Poncirus trifoliate Raf.),用Na2CO3(6.25 g·L-1)和CaCl2(10.1 g·L-1)进行处理。处理方式同上。
采集同一时期罗浮金柑(Fortunella margarita Swing)的茎、嫩叶、花、幼果,液氮速冻,-80℃保存,用于分析U-box家族成员表达的组织特异性。
1.6 柑橘U-box家族基因的冷胁迫表达模式分析
在NCBI数据库中下载枳在冷胁迫、干旱胁迫和盐胁迫下及4个冷处理下的文库(GSE67439_pooled- Unigene.fa.gz)并建立本地数据库,用柑橘U-box家族基因的CDS序列在数据库中blast,找到相应基因的RNA-Seq编号,然后下载4个冷处理下表达数据,提取出U-box家族基因的表达量数据,利用在线软件Morpheus(https://software.broadinstitute.org/morpheus/绘制表达量热图。1.7 总RNA提取与cDNA合成
使用RNAprep pure植物总RNA提取试剂盒(DP432,天根)提取植物总RNA。使用PrimeScriptTM RT Reagent Kit With gDNA Eraser(Perfect Real Time)(RR047,TaKaKa)试剂盒,将RNA反转录成cDNA供荧光定量使用。1.8 实时荧光定量 PCR
使用Prime3设计引物,引物信息见表1。内参为柑橘β-Actin,在CFX96 TouchTM荧光定量PCR仪上对柑橘U-box家族的部分成员的表达量进行分析。扩增体系含2 μL cDNA,上、下游引物各0.5 μL,SYBR 6.25 μL反应Mix,3.25 μL ddH2O,总体系12.5 μL。反应程序为:95℃ 30 s,95℃ 5 s,60℃ 34 s,95℃ 15 s,60℃ 60 s,95℃ 15 s,共40个循环。每个处理3次重复。用SPSS软件进行差异显著性分析,P<0.05表示差异显著。Table 1
表1
表1本试验所用引物
Table 1
| 引物名称 Primer name | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| CcPUB4 | CCGGTGACTTTATCCACTGGG | GAATGGGTTCAAAACTCGTCAGG |
| CcPUB9 | GCCAGCTGAGGTCCCGGATT | TGCCTTGTATGCCCAACCATGT |
| CcPUB10 | CAAAACCCGTAGTCCGAAAA | AATTCACCATCCGAGTCTGC |
| CcPUB41 | ACGCCGTCCAGTATCCTTC | AATTCCAGCATTCCCATCCA |
| CcPUB48 | CTGAGCGAAGGTACCAGAG | AGTGTTCATCCACCATTCC |
| β-Actin | CCCCATCGTTACCGTCCAG | CGCCTTGCCAGTTGAATATCC |
新窗口打开|下载CSV
2 结果
2.1 柑橘U-box基因家族成员信息
通过在线BLAST比对以及Hmmer搜索,经过SMART服务器分析去除不含U-box结构域的序列,从柑橘基因组中鉴定出56个U-box基因,并且56个柑橘U-box蛋白均含有62—68个氨基酸的U-box结构域,其中40个U-box蛋白的U-box保守结构域都含有63个氨基酸,表2所示。通过ExPASy工具分析,柑橘中最长的U-box蛋白(Ciclev10024300m)包含1 441个氨基酸残基,分子量也为56个蛋白质中最大,为160.62 kD,最短的U-box蛋白(Ciclev10032341m)包含281个氨基酸残基,分子量是56个蛋白中最小的。等电点范围为5.19(Ciclev10030837m)—9.14(Ciclev10011217m)。利用Cello软件对柑橘U-box家族成员进行亚细胞定位分析,结果显示该基因家族成员位于细胞不同位置,其中CcPUB15和CcPUB43定位于质膜,CcPUB9、CcPUB11、CcPUB17、CcPUB33、CcPUB34存在于细胞质,其他成员位于细胞核或叶绿体。Table 2
表2
表2柑橘基因组中的U-box基因
Table 2
| 基因名称 Gene name | 基因组登录号 Gene accession No. | 蛋白质大小 Protein length (aa) | 分子量 Molecular mass (KD) | 等电点 Isoelectric point (PI) | box位置 U-box domain location | 细胞定位 Subcellular localizations |
|---|---|---|---|---|---|---|
| CcPUB1 | Ciclev10007255m | 1380 | 152.75 | 5.46 | 449-513 | 细胞核 Nuclear |
| CcPUB2 | Ciclev10007527m | 769 | 85.86 | 6.02 | 700-762 | 叶绿体 Chloroplast |
| CcPUB3 | Ciclev10018144m | 1088 | 121.83 | 6.16 | 613-677 | 细胞核 Nuclear |
| CcPUB4 | Ciclev10015253m | 444 | 49.84 | 8.1 | 36-100 | 叶绿体 Chloroplast |
| CcPUB5 | Ciclev10014584m | 636 | 69.48 | 6.56 | 257-320 | 叶绿体 Chloroplast |
| CcPUB6 | Ciclev10014430m | 715 | 78.42 | 7.01 | 291-354 | 叶绿体 Chloroplast |
| CcPUB7 | Ciclev10019643m | 537 | 58.77 | 6.17 | 28-91 | 叶绿体 Chloroplast |
| CcPUB8 | Ciclev10019147m | 683 | 75.51 | 8.48 | 279-342 | 叶绿体 Chloroplast |
| CcPUB9 | Ciclev10032341m | 281 | 31.97 | 6.03 | 207-270 | 细胞质 Cytoplasmic |
| CcPUB10 | Ciclev10024300m | 1441 | 160.62 | 5.78 | 506-570 | 细胞核 Nuclear |
| CcPUB11 | Ciclev10024335m | 433 | 47.91 | 5.77 | 79-142 | 细胞质 Cytoplasmic |
| CcPUB12 | Ciclev10018951m | 775 | 85.47 | 6.91 | 239-302 | 细胞核 Nuclear |
| CcPUB13 | Ciclev10018671m | 1008 | 112.36 | 5.75 | 265-328 | 细胞核 Nuclear |
| CcPUB14 | Ciclev10018795m | 888 | 99.09 | 5.89 | 821-884 | 细胞核 Nuclear |
| CcPUB15 | Ciclev10033932m | 687 | 75.23 | 6.63 | 284-347 | 质膜 Plasma |
| CcPUB16 | Ciclev10030962m | 627 | 68.42 | 5.49 | 249-312 | 叶绿体 Chloroplast |
| CcPUB17 | Ciclev10031496m | 456 | 50.85 | 8.39 | 75-138 | 细胞质 Cytoplasmic |
| CcPUB18 | Ciclev10030608m | 1019 | 113.74 | 6.24 | 233-300 | 细胞核 Nuclear |
| CcPUB19 | Ciclev10030759m | 775 | 86 | 5.57 | 282-345 | 细胞核 Nuclear |
| CcPUB20 | Ciclev10030837m | 713 | 79.13 | 5.19 | 220-283 | 细胞核 Nuclear |
| CcPUB21 | Ciclev10030762m | 775 | 86 | 5.57 | 282-345 | 细胞核 Nuclear |
| CcPUB22 | Ciclev10001406m | 395 | 44.04 | 6.57 | 6-69 | 细胞核 Nuclear |
| CcPUB23 | Ciclev10001380m | 398 | 44.51 | 8.54 | 13-76 | 叶绿体 Chloroplast |
| CcPUB24 | Ciclev10000413m | 728 | 80.97 | 6.79 | 248-311 | 细胞核 Nuclear |
| CcPUB25 | Ciclev10000389m | 744 | 82.67 | 6.62 | 264-327 | 细胞核 Nuclear |
| CcPUB26 | Ciclev10003715m | 643 | 71.66 | 5.37 | 274-337 | 叶绿体 Chloroplast |
| CcPUB27 | Ciclev10000306m | 813 | 89.10 | 5.38 | 32-100 | 细胞核 Nuclear |
| CcPUB28 | Ciclev10011217m | 683 | 74.80 | 9.14 | 276-339 | 叶绿体 Chloroplast |
| CcPUB29 | Ciclev10011844m | 416 | 46.12 | 8.38 | 9-74 | 叶绿体 Chloroplast |
| CcPUB30 | Ciclev10011847m | 414 | 46.93 | 8.16 | 12-78 | 叶绿体 Chloroplast |
| CcPUB31 | Ciclev10011415m | 546 | 59.20 | 8.42 | 45-108 | 叶绿体 Chloroplast |
| CcPUB32 | Ciclev10010958m | 1049 | 118.38 | 5.48 | 954-1017 | 叶绿体 Chloroplast |
| CcPUB33 | Ciclev10011388m | 564 | 64.68 | 5.22 | 469-532 | 细胞质 Cytoplasmic |
| CcPUB34 | Ciclev10011387m | 564 | 64.68 | 5.22 | 469-532 | 细胞质 Cytoplasmic |
| CcPUB35 | Ciclev10011113m | 779 | 88.36 | 5.43 | 684-747 | 叶绿体 Chloroplast |
| CcPUB36 | Ciclev10025062m | 679 | 75.18 | 5.65 | 210-273 | 叶绿体Chloroplast |
| 基因名称 Gene name | 基因组登录号 Gene accession No. | 蛋白质大小 Protein length (aa) | 分子量 Molecular mass (KD) | 等电点 Isoelectric point (PI) | box位置 U-box domain location | 细胞定位 Subcellular localizations |
| CcPUB37 | Ciclev10024987m | 737 | 81.58 | 5.84 | 268-331 | 叶绿体Chloroplast |
| CcPUB38 | Ciclev10024899m | 828 | 90.51 | 5.92 | 239-302 | 叶绿体Chloroplast |
| CcPUB39 | Ciclev10025589m | 450 | 49.00 | 7.01 | 69-132 | 细胞质Cytoplasmic |
| CcPUB40 | Ciclev10025214m | 597 | 65.21 | 6.98 | 39-101 | 叶绿体Chloroplast |
| CcPUB41 | Ciclev10024952m | 760 | 85.82 | 6.21 | 688-751 | 细胞核Nuclear |
| CcPUB42 | Ciclev10024909m | 808 | 91.47 | 6.18 | 736-799 | 叶绿体Chloroplast |
| CcPUB43 | Ciclev10025637m | 435 | 48.23 | 8 | 13-76 | 质膜Plasma |
| CcPUB44 | Ciclev10028003m | 643 | 71.65 | 6.54 | 272-335 | 叶绿体Chloroplast |
| CcPUB45 | Ciclev10028522m | 418 | 45.74 | 6.21 | 17-80 | 细胞核Nuclear |
| CcPUB46 | Ciclev10028557m | 407 | 45.49 | 8.78 | 9-76 | 叶绿体Chloroplast |
| CcPUB47 | Ciclev10028564m | 405 | 45.68 | 8.92 | 9-74 | 叶绿体Chloroplast |
| CcPUB48 | Ciclev10027966m | 661 | 71.37 | 5.43 | 261-324 | 叶绿体Chloroplast |
| CcPUB49 | Ciclev10028111m | 564 | 60.47 | 5.46 | 164-227 | 叶绿体Chloroplast |
| CcPUB50 | Ciclev10027788m | 887 | 99.83 | 8.08 | 820-883 | 细胞核Nuclear |
| CcPUB51 | Ciclev10029840m | 1048 | 115.56 | 6.56 | 265-333 | 叶绿体Chloroplast |
| CcPUB52 | Ciclev10004235m | 1012 | 112.79 | 5.91 | 261-324 | 叶绿体Chloroplast |
| CcPUB53 | Ciclev10004724m | 526 | 56.90 | 6.07 | 1-66 | 叶绿体Chloroplast |
| CcPUB54 | Ciclev10004393m | 757 | 84.621 | 5.93 | 692-754 | 细胞核Nuclear |
| CcPUB55 | Ciclev10004338m | 810 | 90.50 | 6.31 | 745-807 | 细胞核Nuclear |
| CcPUB56 | Ciclev10006472m | 780 | 88.56 | 6.55 | 713-776 | 叶绿体Chloroplast |
新窗口打开|下载CSV
2.2 柑橘U-box家族系统进化及蛋白结构域分析
为研究柑橘U-box基因家族系统进化关系,对56个柑橘U-box蛋白构建了系统进化树,经过SMART蛋白结构域分析,结果显示除U-box结构域外,柑橘U-box蛋白还含有其他结构域。根据进化树以及蛋白结构域(图1),将柑橘中的56个U-box蛋白分为7种类型,即U-box only、U-box+ARM-1、U-box+ARM-2、Kinase+U-box、U-box+WD40-1、U-box+WD4-2和TPR+U-box类,每一类型分别有17、16、5、8、3、2和5个U-box蛋白。U-box only结构的成员在柑橘中最多,共17个。ARM亚家族在植物中研究较多,并在水稻和拟南芥中都为最大亚类[16],而在柑橘中为第二大亚类。Kinase亚家族有8个成员,其中3个包含有丝/苏/酪蛋白激酶结构域(STYKc domain),另外5个为丝/苏氨酸蛋白激酶结构域(S_TKc domain)。同时本研究发现其中76%的U-box家族成员的U-box结构域位于N末端,而U-box结构域位于C末端的成员比较少,U-box结构域位于N末端的聚在一起,位于C端的聚为另一类。位于同一亚家族U-box蛋白含有的结构域种类和数量具有较高的一致性。图1
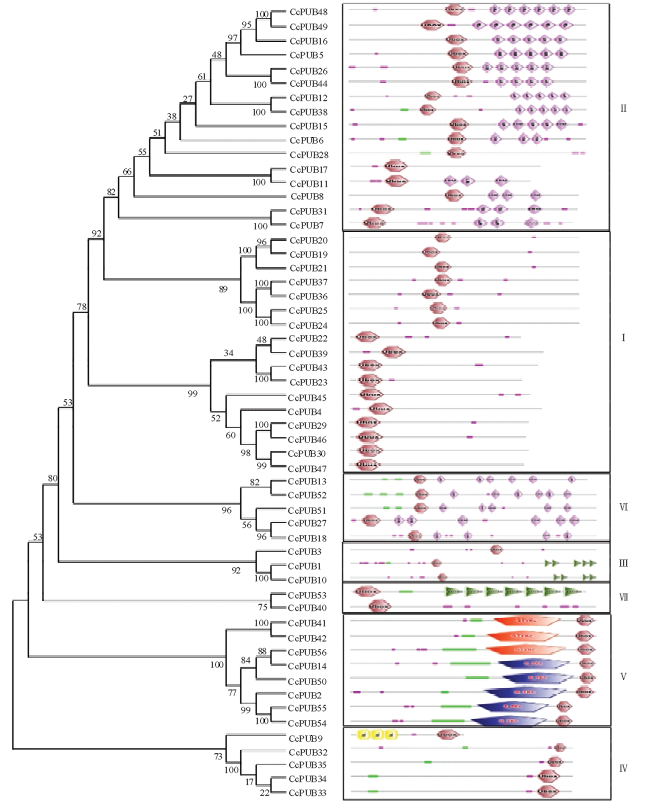 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1柑橘U-box家族进化树与蛋白结构
Ⅰ:U-box only;Ⅱ:U-box+ARM;Ⅲ:U-box+WD40_1;Ⅳ:TPR+U-box;Ⅴ:Kinase+U-box;Ⅵ:U-box+ARM_2;Ⅶ:U-box+WD40_2
Fig. 1The phylogenetic tree and structure of U-box proteins in citrus
为深入研究单子叶植物水稻U-box基因家族与双子叶植物柑橘和拟南芥U-box基因家族的进化关系,对柑橘(56个)、水稻(77个)和拟南芥(64个)共197个U-box蛋白进行了系统进化树构建(图2)。根据亲缘关系远近可将197个U-box蛋白分为7个亚家族,分别含有52、59、46、13、2、3和20个U-box基因。双子叶和单子叶植物的U-box家族成员在7个亚家族内均有分布。第Ⅰ亚家族和第Ⅱ亚家族含有的结构域较单一,而其他亚家族含有多种不同的结构域。CcPUB23和OsPUB39单独聚在一个小分支上,在拟南芥中未发现与其高度同源的序列。AtPUB49和CcPUB40进化距离最近,推测其在功能上具有一定的相似性。
图2
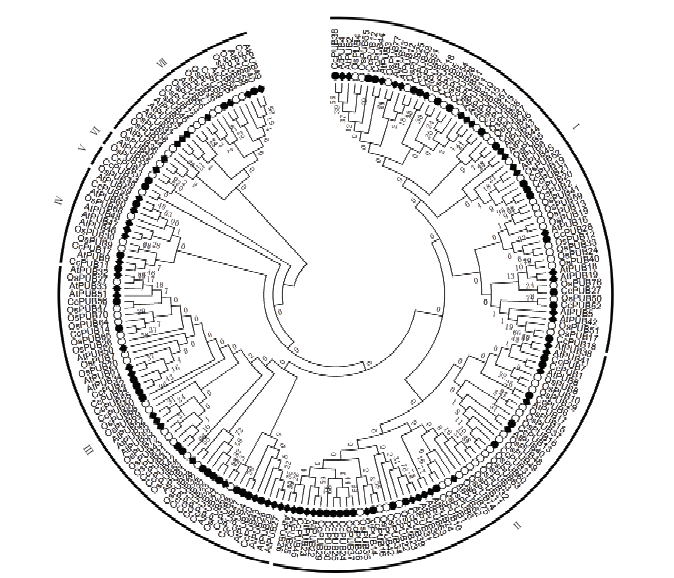 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2柑橘、拟南芥和水稻U-box家族蛋白的系统进化树
Fig. 2The phylogenetic tree of U-box protein between Citrus, Arabidopsis and rice
2.3 柑橘U-box家族基因结构及染色体定位分析
利用GSDS软件对U-box家族各成员的基因结构进行了分析。结果显示(图3),柑橘U-box家族的基因结构存在较大的差异,外显子数目为1—16个,内含子数目为0—15个。对基因结构进一步分析发现,U-box only亚家族的基因结构较简单,一半成员都不含内含子;含有U-box和ARM结构域的成员中多数基因含有3个内含子;含有U-box和Kinase结构域的成员的基因结构极为相似,内含子数目在6—9个;具有U-box和WD40结构域的成员所含内含子数目最多,内含子数在12—15个;含有TPR和U-box结构域的成员含有7个内含子。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图356个柑橘U-box基因的结构分析
Fig. 3Gene structure of 56 U-box gene in citrus
根据U-box家族在Scafflod的位置信息,利用MapChart软件获得了56个U-box基因在柑橘Scafflod上的分布图(图4)。由图可知,柑橘U-box家族成员在Scafflod上呈不均分布。在3号、6号和8号Scafflod上分布最多,含有8个U-box基因,而1号Scafflod上分布的U-box基因最少,只含有2个U-box成员。进一步分析发现,U-box在Scafflod上呈现区域性,从4号、5号、6号、7号和8号Scafflod中可以看到,在Scafflod上的某个区域家族成员比较集中。
图4
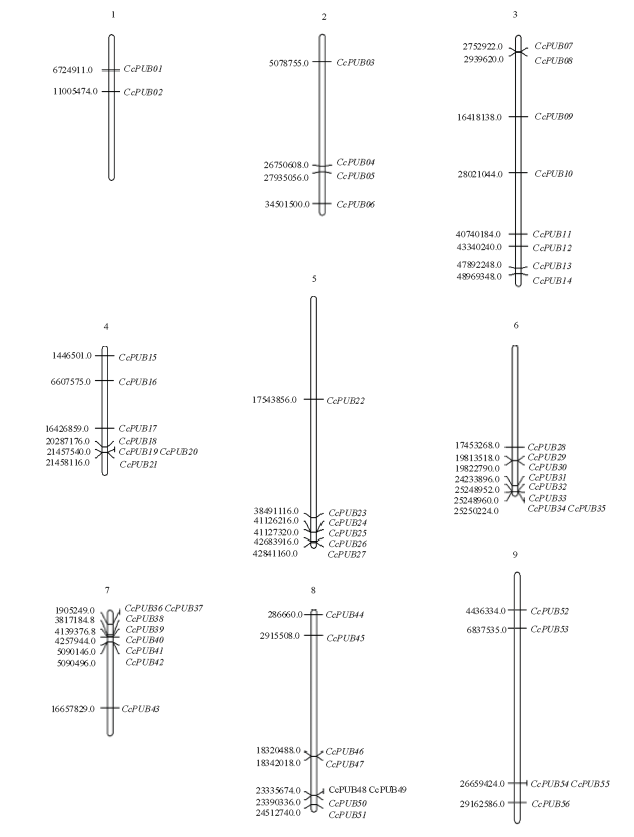 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4柑橘U-box基因在Scafflod上的定位(Mb)
Fig. 4Scafflod locations of U-box gene in Citrus (Mb)
2.4 柑橘U-box基因家族的启动子分析
分析基因启动子的作用元件可以预测基因的潜在功能,为了解柑橘U-box家族应答生物胁迫和非生物胁迫反应的潜在机制,本研究分析了柑橘U-box家族的启动子的顺式作用元件。结果表明,该家族的启动子区域富含响应植物激素和逆境胁迫的顺式作用元件(表3)。几乎所有柑橘U-box基因启动子区至少含有一个植物激素响应元件,包括赤霉素响应元件GARE-motif,乙烯响应元件ERE,MeJA响应元件TGACG-motif和脱落酸(ABA)响应元件ABRE等,51个U-box基因(占总基因数91.1%)至少含有一种生物或非生物胁迫响应元件,非生物响应元件包括光响应元件G-box、低温胁迫响应元件LTR、冷和脱水响应元件DRE等。Table 3
表3
表3PlantCARE预测CcPUB基因家族启动子区顺式作用原件
Table 3
| 顺式元件 Cis-element | 典型序列 Typical sequence | 特性 Characteristic | 基因 Gene |
|---|---|---|---|
| G-box | CACGT | 光响应 Cis-acting regulatory element involved in light responsiveness | CcPUB1、CcPUB3—CcPUB11、CcPUB13—CcPUB17、CcPUB21— CcPUB23、CcPUB25、CcPUB26、CcPUB28—CcPUB37、CcPUB39、CcPUB42— CcPUB45、CcPUB47—CcPUB56 |
| ATCT-motif | AATCT | 光响应 Part of a conserved DNA module involved in light responsiveness | CcPUB7、CcPUB18、CcPUB22、CcPUB23、CcPUB28、CcPUB30、CcPUB31、CcPUB39、CcPUB53、CcPUB55、CcPUB56 |
| ACE | AAAACGTTTA | 光响应 Cis-acting element involved in light responsiveness | CcPUB3、CcPUB10、CcPUB21、CcPUB26、CcPUB32、CcPUB39 |
| LTR | CCGAAA | 低温反应 Cis-acting element involved in low- temperature responsiveness | CcPUB2、CcPUB5、CcPUB8、CcPUB10、CcPUB16、CcPUB17、CcPUB23、CcPUB25、CcPUB26、CcPUB28、CcPUB29、CcPUB32、CcPUB38、CcPUB39、CcPUB42—CcPUB44、CcPUB46、CcPUB56 |
| TGACG-motif | TGACG | MeJA响应 Cis-acting regulatory element involved in the MeJA-responsiveness | CcPUB1—CcPUB6、CcPUB8、CcPUB9、CcPUB11、CcPUB13— CcPUB18、CcPUB23、CcPUB27、CcPUB29、CcPUB31—CcPUB38、CcPUB40—CcPUB44、CcPUB46、CcPUB47、CcPUB50、CcPUB52、CcPUB55、CcPUB56 |
| ABRE | GACACGTGGC TACGTG | 脱落酸响应 Cis-acting element involved in the abscisic acid responsiveness | CcPUB1—CcPUB11、CcPUB13—CcPUB17、CcPUB22、CcPUB23、CcPUB26、CcPUB28—CcPUB30、CcPUB32—CcPUB37、CcPUB39、CcPUB42—CcPUB45、CcPUB47—CcPUB53、CcPUB55、CcPUB56 |
| GARE-motif | TCTGTTG, AAACAGA | 赤霉素响应 Gibberellin-responsive element | CcPUB2、CcPUB5、CcPUB6、CcPUB8、CcPUB9、CcPUB13、CcPUB15、CcPUB16、CcPUB27、CcPUB31、CcPUB45、CcPUB47、 |
| MBS | TAACTG | MYB结合位点参与干旱诱导 MYB binding site involved in drought- induction | CcPUB1、CcPUB4—CcPUB9、CcPUB11、CcPUB14、CcPUB16、CcPUB17、CcPUB21、CcPUB23、CcPUB25—CcPUB27、CcPUB31— CcPUB37、CcPUB39、CcPUB44、CcPUB50、CcPUB51、CcPUB55、CcPUB56 |
| DRE | TGGCCGAC | 冷和脱水 Regulatory element involved in cold- and dehydration-responsiveness | CcPUB4、CcPUB5、CcPUB7、CcPUB8、CcPUB13、CcPUB21、CcPUB23、CcPUB26、CcPUB29、CcPUB37、CcPUB40—CcPUB42、CcPUB44、CcPUB46、CcPUB51、CcPUB56 |
| ERE | ATTTTAAA | 乙烯响应 Ethylene responsive element | CcPUB2—CcPUB9、CcPUB11、CcPUB12、CcPUB14、CcPUB17、CcPUB18、CcPUB21—CcPUB23、CcPUB25、CcPUB26、CcPUB28、CcPUB29、CcPUB31、CcPUB32、CcPUB37—CcPUB42、CcPUB44、CcPUB45、CcPUB47、CcPUB50—CcPUB53、CcPUB56 |
新窗口打开|下载CSV
2.5 U-box家族基因的表达分析
2.5.1 组织特异性 为了解柑橘U-box基因家族5种结构域(U-box only、U-box+ARM、U-box+WD40、Kinase+U-box、TPR+U-box)的组织表达模式,分别从5种结构域中选取1个代表基因进行组织特异性分析,利用qRT-PCR分析其在金柑嫩叶、幼果、茎和花中的相对表达量。由图(5)可看出,这5个基因在各个组织中均有表达。其中CcPUB4、CcPUB9、CcPUB41和CcPUB48在幼果中表达量都较低,而CcPUB10主要在幼果中表达,CcPUB4主要在花中表达,CcPUB9、CcPUB41和CcPUB48主要在茎中表达。图5
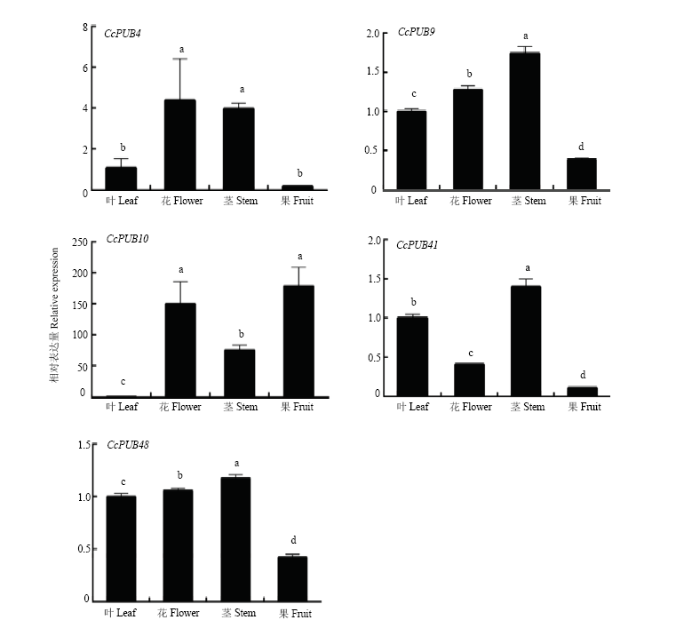 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5CcPUB4、CcPUB9、CcPUB10、CcPUB41和CcPUB48在不同组织中的相对表达量
不同小写字母表示差异显著(P<0.05)。下同
Fig. 5Relative expression of CcPUB4, CcPUB9, CcPUB10, CcPUB41 and CcPUB48 in different tissues
Different lowercase letters indicate significant difference (P<0.05). The same as below
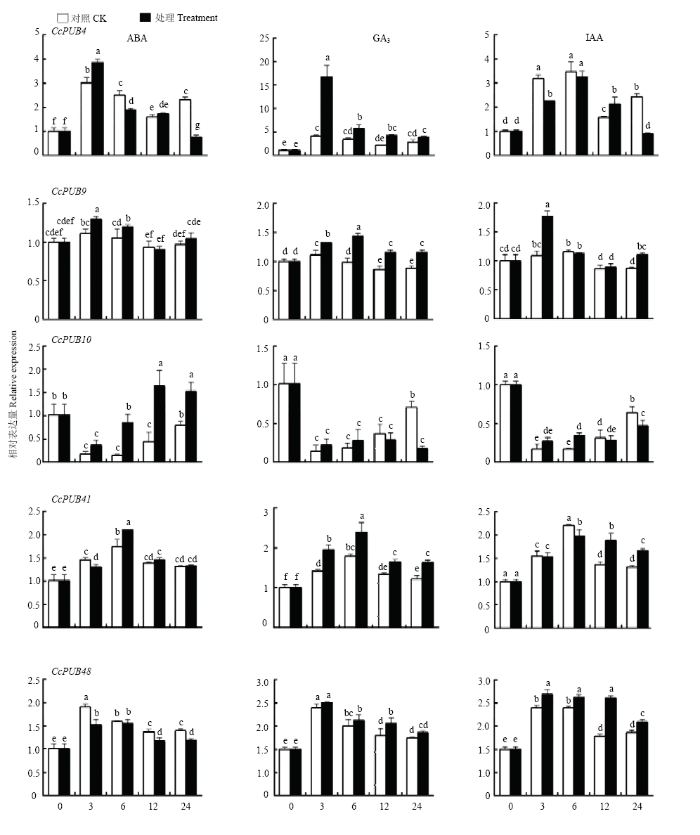
2.5.2 激素对CcPUBs表达的影响 利用qRT-PCR分析柑橘U-box基因家族中5个代表基因在ABA、GA3和IAA处理下的相对表达量(图6)。除了CcPUB9、CcPUB41和CcPUB48不响应ABA、GA3和IAA外,CcPUB4和CcPUB10对各处理表现出不同的表达模式。在ABA和IAA处理下,CcPUB4的表达量在0—12 h无明显变化,在处理24 h后CcPUB4的表达明显受到抑制,而在GA3处理3 h后CcPUB4表现为上调,CcPUB4的表达量是对照的4倍。CcPUB10在ABA的处理下表达呈现上升的趋势,而在GA3处理24 h后表达 下调,在IAA的处理下无明显变化。上述结果表明,激素能够诱导CcPUB4和CcPUB10基因的表达。
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6不同外源激素处理条件下CcPUB4、CcPUB9、CcPUB10、CcPUB41和CcPUB48的相对表达量
Fig. 6Relative expressions of CcPUB4, CcPUB9, CcPUB10, CcPUB41 and CcPUB48 under different treatments of exogenous plant hormones
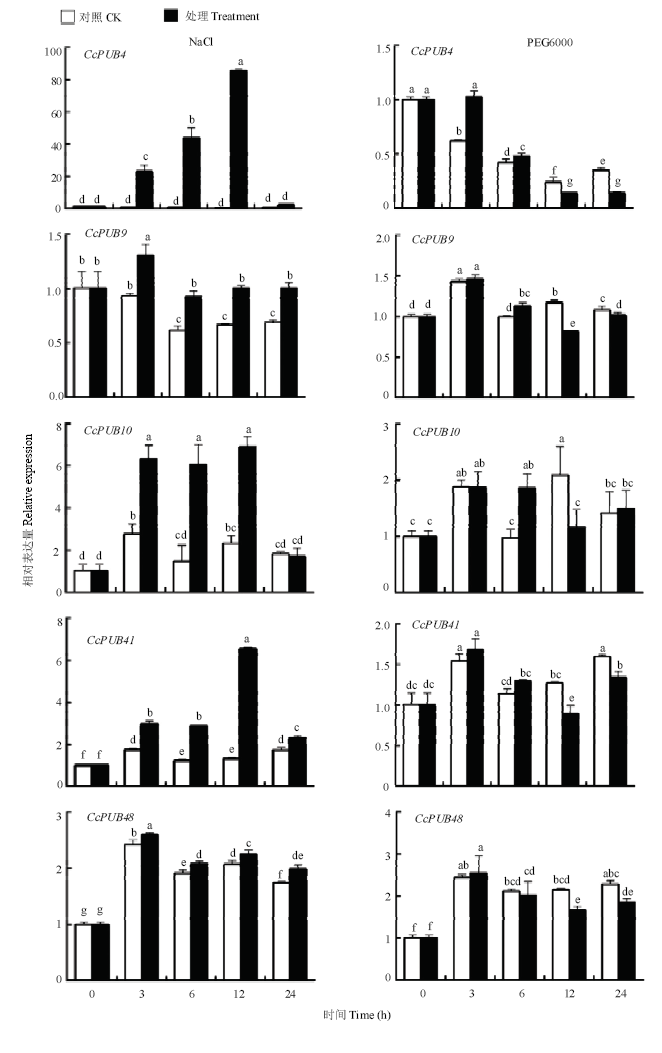
2.5.3 非生物胁迫对CcPUBs表达的影响 在盐胁迫条件下,CcPUB4、CcPUB9、CcPUB10和CcPUB41表达均上调,且CcPUB4、CcPUB10、CcPUB41呈现相似的表达趋势,而CcPUB48的表达不受NaCl的诱导(图7)。在PEG6000的处理条件下,CcPUB4的表达量呈现先上调后下降,而CcPUB9、CcPUB41和CcPUB48在PEG6000的处理下无明显变化,CcPUB10在PEG6000条件下无规律变化。
图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7非生物胁迫下CcPUB4、CcPUB9 、CcPUB10、CcPUB41和CcPUB48的相对表达量
Fig. 7Relative expressions of CcPUB4, CcPUB9, CcPUB10, CcPUB41 and CcPUB48 under different abiotic stresses
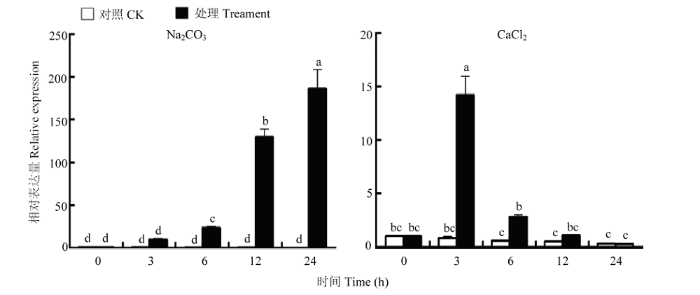
2.5.4 Na2CO3和CaCl2条件下,CcPUB4的相对表达量 由于植物在盐胁迫的条件下会改变细胞中已有的Na+和Cl-平衡,通过上述的非生物胁迫的结果发现,CcPUB4在NaCl条件下显著表达,但是并不能确定该基因是受NaCl中哪种离子的影响,因此,采用相同浓度的Na+和Cl-对枳壳进行处理。结果显示CcPUB4在Na2CO3条件下显著上调,并且与在NaCl条件下的表达趋势相似,在CaCl2条件下CcPUB4的表达趋势却与在NaCl条件下的表达趋势不同(图8)。
图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8在Na2CO3和CaCl2胁迫下CcPUB4的相对表达量
Fig. 8Relative expression of CcPUB4 under Na2CO3 and CaCl2 stress
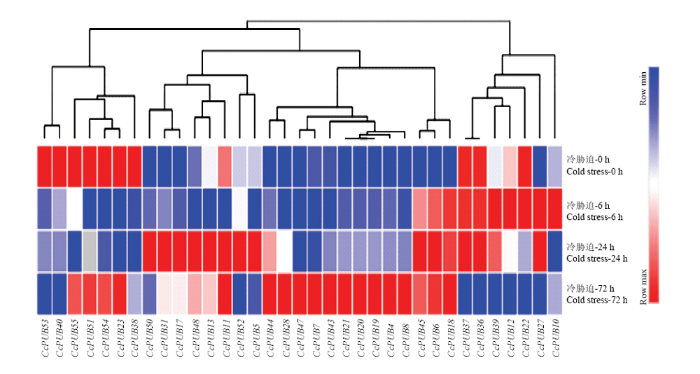
2.5.5 CcPUBs在冷胁迫下的表达模式分析 柑橘U-box蛋白可参与调节植物对非生物胁迫的应答,因此,利用枳的RNA-Seq数据分析该基因家族在冷胁迫下的表达模式。在干旱胁迫、盐胁迫和冷胁迫的RNA-Seq文库中,找到47个U-box基因序列,其中35个基因在0、6、24和72 h冷胁迫下有表达数据,其他12个在冷胁迫下没有表达数据(CcPUB1、CcPUB2、CcPUB9、CcPUB14、CcPUB16、CcPUB26、CcPUB32、CcPUB33、CcPUB34、CcPUB35、CcPUB42、CcPUB56)。利用Morpheus绘制35个U-box基因的表达热图,分析显示,这35个基因在冷胁迫下具有表达差异,并且在冷胁迫下具有4种不同的应答模式(图9),CcPUB38、CcPUB23、CcPUB54、CcPUB51、CcPUB55聚在一分支,在冷胁迫下它们的表达量是先下降再上升;CcPUB10、CcPUB27、CcPUB22、CcPUB12、CcPUB39、CcPUB36和CcPUB37聚在一分支,在冷胁迫下它们的表达量降低;CcPUB5、CcPUB52、CcPUB11、CcPUB13、CcPUB48、CcPUB17、CcPUB31和CcPUB50聚在一分支,在冷胁迫下它们的表达量先上升再下降;CcPUB18、CcPUB6、CcPUB8、CcPUB4、CcPUB19、CcPUB20、CcPUB21、CcPUB43、CcPUB7、CcPUB47、CcPUB28和CcPUB44聚在一分支,该分支的基因都响应冷胁迫,所有基因的表达量都上升。进一步的分析发现,一些进化关系比较接近的U-box基因,在冷胁迫下具有相同的表达模式并聚在一起,如CcPUB40和CcPUB53,CcPUB36和CcPUB37,CcPUB19、CcPUB20和CcPUB21。
图9
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9柑橘U-box家族在冷胁迫下的表达谱分析
Fig. 9Expression profile analysis of Citrus U-box family under cold Stress
3 讨论
U-box基因家族是一类具有U-box结构域的基因家族,其所编码蛋白大部分是泛素系统中决定底物识别特性的泛素连接酶E3。从酵母到人类,几乎所有真核生物都含有U-box蛋白,尤其是植物体内大量存在该类蛋白,如苜蓿中有41个U-box基因[16],拟南芥基因组中存在64个U-box基因[17],水稻(Oryza sativa)中已鉴定出77个带有U-box结构的基因[18],番茄中鉴定出56个U-box基因[19],雷蒙德氏棉中有93个GrPUBs基因家族成员[20],人有21个U-box基因[21]。本研究从克里曼丁全基因组数据库中获得了56个U-box基因成员,与番茄中U-box成员数相同,而少于拟南芥和水稻中的成员数,表明不同物种的U-box成员数存在差异。虽然番茄的U-box数量与柑橘中的数量一样多,但其基因组大小约为900 Mb[22],远大于柑橘301.4 Mb[15],由此可见U-box基因家族成员的多少与基因组大小没有直接关系,这与郑兴卫等[16]在苜蓿U-box基因家族研究中的结论一致。柑橘U-box基因家族的蛋白质大小在31.97—160.62 kD之间,PI在5.19—9.14之间,表明柑橘U-box基因家族成员间的蛋白质大小、PI等特征差异较大。除了含有U-box结构域外,U-box蛋白中还存在ARM、WD40和TPR二级结构域,这些结构域主要用来介导U-box蛋白与底物蛋白的特异性识别。为了对柑橘中的U-box家族进行功能归类,利用聚类分析法构建柑橘、水稻和拟南芥的U-box家族系统进化树,由于U-box基因的保守性,具有相似或者相同功能的基因位于同一组,这为研究该基因家族相关基因的功能提供了可靠的依据。在拟南芥中一些成员的功能已经得到验证:TPR+U-box亚家族中的AtCHIP参与非生物胁迫调节,在低温和黑暗条件下,AtCHIP改变应激反应信号转导中PP2A的活性,从而对非生物胁迫做出响应[23],柑橘CcPUB9与拟南芥AtCHIP聚在一起,具有较近的亲缘关系,猜测TPR+U-box亚家族其他成员在柑橘中起着同样的作用。U-box+ARM是研究最多的亚家族,属于U-box+ARM亚家族的AtPUB18、AtPUB19、AtPUB46和AtPUB48在干旱胁迫中起着重要作用[24,25],AtPUB17能正调控细胞凋亡和植物防御的过程[26],同时AtPUB9在磷酸盐饥饿条件下调控侧根的发育[27],由此推测U-box+ARM亚家族其他基因具有相同或相似的功能。U-box only亚家族的基因结构简单,在植物生长发育和非生物胁迫中发挥重要作用,齐晨辉等[28]发现MdPUB24过量表达的苹果愈伤组织和异位表达的拟南芥幼苗在盐胁迫条件下,生长势与野生型相比明显变弱,表明MdPUB24负调控盐胁迫。根据进化关系中基因的同源性,可以推测位于同一组的拟南芥在柑橘中的直系同源基因也可能参与相似的调控途径。
本研究对单子叶和双子叶植物中的U-box基因家族成员的进化关系进行分析,单子叶植物和双子叶植物在7个亚家族中均有分布,说明U-box家族在进化上较保守,U-box家族的起源出现在单双子叶植物分化之前。本研究发现第Ⅰ亚家族和第Ⅱ亚家族的U-box成员数较多,推测该亚家族中的U-box基因可能在植物的生命进程中发挥了重要的作用。在第Ⅴ亚家族中没有拟南芥的成员,可能是该亚家族的基因发生了丢失现象。CcPUB32、CcPUB33、CcPUB34和CcPUB35位于同一聚类组中并且位于相同的染色体上,CcPUB19、CcPUB20和CcPUB21也出现这种紧密连锁,推测在进化的过程中这些基因可能通过染色体内的复制发生了特异性扩张,这些基因的功能有待进一步研究。
U-box基因参与了叶、花、茎和果的生长发育[29,30],本试验中的CcPUB10主要在果和花中表达,在叶中几乎不表达;CcPUB4主要在花和茎中表达,而CcPUB9、CcPUB41和CcPUB48在茎中显著表达,表明柑橘U-box基因调控叶、花、茎和果生长发育。近年来,大量研究表明,E3泛素连接酶是一种极其活跃的激素感知成分,在激素途径的抑制解除以及激素生物合成的调控中具有重要作用[31]。本试验通过外施IAA、GA3和ABA激素处理,实时荧光定量结果显示,在GA3的处理下,CcPUB4上调,在ABA和IAA处理24 h表达受到抑制;CcPUB10在ABA的处理下呈现逐渐上调,在GA3和IAA的处理下无明显规律。U-box only亚家族的CcPUB4、TPR+U-box亚家族的CcPUB9、U-box+WD40-1亚家族的CcPUB10以及Kinase+U-box亚家族的CcPUB41均受NaCl的影响呈现上调,且CcPUB4在NaCl处理12 h后其表达量是对照的270倍,说明该基因可能参与了植物的盐胁迫响应,HWANG等[32]抑制拟南芥AtPUB30导致发芽期间对盐胁迫的耐受性增加,同时,CHO等[33]发现在拟南芥中超表达辣椒PUB1的植株耐盐性增加。在盐胁迫的条件下可破坏细胞中已有的Na+和Cl-平衡,因此本试验用Na2CO3和CaCl2对枳壳进行处理,进一步探明是外源的Na+还是Cl-会引起CcPUB4表达量上升,结果显示在Na2CO3处理下CcPUB4表达明显上调且和NaCl呈现相似的表达模式,而CaCl2处理只在处理后3 h有较高的表达水平,之后CcPUB4的表达水平降低到正常值,证实了盐胁迫下,细胞中的Na+平衡被打破。CcPUB4如何在盐胁迫下发挥作用,是下一步研究的重点内容。CcPUB4在PEG6000下的条件下表达量先上升再下降,其结果与拟南芥的同源基因AtPUB20在干旱胁迫下的结果一致[34],SEO等[35]研究证实U-box家族中的AtPUB18/AtPUB19和AtPUB22/ AtPUB23在干旱胁迫下分别是ABA依赖和ABA非依赖途径的负调控因子。CcPUBs不同成员间在冷胁迫下具有不同的应答模式。通过对这35个基因的启动子分析发现,其中17个基因包含冷胁迫响应元件DRE或低温响应元件LTR,表明CcPUBs对冷胁迫具有响应。其中,CcPUB4、CcPUB19、CcPUB20、CcPUB21、CcPUB43在冷胁迫下是上调表达的并且都属于U-box only亚族,由此预测U-box only亚族在冷胁迫过程中存在着正调控作用,与CcPUB8同源的AtPUB19在冷胁迫下上调[11],YEE等[36]对拟南芥的U-box家族在冷胁迫下的表达进行分析,发现大多数U-box基因对冷胁迫都具有响应,该结果与本试验结果一致。在冷胁迫下只有35个U-box基因有表达数据,其他12个没有在冷胁迫下的表达数据,可能是由于该测序的混合池中包括盐胁迫和干旱胁迫下的表达数据[37],这12个基因只在盐胁迫和干旱胁迫下特定响应。综上可知,U-box基因家族可能参与了植物激素以及逆境响应调控。对CcPUBs基因家族的启动子分析发现,其启动子均含有逆境、激素、温度等响应顺式作用元件如G-box光响应元件、DRE冷和脱水响应元件、MBS、MYB结合位点参与干旱诱导响应元件等,该结果为柑橘U-box基因对逆境和激素的应答机制提供了理论依据。
4 结论
本研究从柑橘全基因组上鉴定出56个U-box基因成员,其蛋白均含有U-box保守结构域。除U-box only亚家族和U-box+ARM-1亚家族部分成员不含内含子外,其余成员均含有内含子,内含子数在0—15个。亚细胞定位显示柑橘U-box家族基因成员在细胞质、叶绿体、细胞核、细胞质膜中均存在。56个U-box基因分布在1—9号Scaffold且分布不均,U-box基因家族参与植物对冷胁迫的应答,并表现出4种不同的应答模式。实时荧光定量PCR结果表明,柑橘U-box基因家族表达具有组织特异性,且对激素和非生物胁迫有不同程度的响应。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1146/annurev.arplant.55.031903.141801URLPMID:15377232 [本文引用: 1]

Abstract Much of plant physiology, growth, and development is controlled by the selective removal of short-lived regulatory proteins. One important proteolytic pathway involves the small protein ubiquitin (Ub) and the 26S proteasome, a 2-MDa protease complex. In this pathway, Ub is attached to proteins destined for degradation; the resulting Ub-protein conjugates are then recognized and catabolized by the 26S proteasome. This review describes our current understanding of the pathway in plants at the biochemical, genomic, and genetic levels, using Arabidopsis thaliana as the model. Collectively, these analyses show that the Ub/26S proteasome pathway is one of the most elaborate regulatory mechanisms in plants. The genome of Arabidopsis encodes more than 1400 (or >5% of the proteome) pathway components that can be connected to almost all aspects of its biology. Most pathway components participate in the Ub-ligation reactions that choose with exquisite specificity which proteins should be ubiquitinated. What remains to be determined is the identity of the targets, which may number in the thousands in plants.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S0959-440X(02)00389-5URL [本文引用: 1]
DOI:10.1074/jbc.M102755200URL [本文引用: 1]
DOI:10.1126/stke.2002.116.pe4URLPMID:11805346 [本文引用: 1]

Abstract Ubiquitin ligases determine protein stability in a highly regulated manner by coordinating the addition of polyubiquitin chains to proteins that are then targeted to the proteasome for degradation. Ubiquitin ligases have generally been separated into two groups--those containing HECT domains and those with RING finger domains. Recently, a third group of ubiquitin ligases has emerged: those containing a U-box domain. Patterson discusses what is known about the few U-box-containing proteins that have been characterized, although the general properties of U-box proteins that distinguish them from other ubiquitin ligases are still a matter of speculation.
DOI:10.1105/tpc.109.072660URLPMID:20086187 [本文引用: 1]

The ubiquitin (Ub)/26S proteasome system (UPS) directs the turnover of numerous regulatory proteins, thereby exerting control over many aspects of plant growth, development, and survival. The UPS is directed in part by a group of Ub-like/Ub-associated (UBL/UBA) proteins that help shuttle ubiquitylated proteins to the 26S proteasome for breakdown. Here, we describe the collection of UBL/UBA proteins in Arabidopsis thaliana, including four isoforms that comprise the RADIATION SENSITIVE23 (RAD23) family. The nuclear-enriched RAD23 proteins bind Ub conjugates, especially those linked internally through Lys-48, via their UBA domains, and associate with the 26S proteasome Ub receptor RPN10 via their N-terminal UBL domains. Whereas homozygous mutants individually affecting the four RAD23 genes are without phenotypic consequences (rad23a, rad23c, and rad23d) or induce mild phyllotaxy and sterility defects (rad23b), higher-order mutant combinations generate severely dwarfed plants, with the quadruple mutant displaying reproductive lethality. Both the synergistic effects of a rad23b-1 rpn10-1 combination and the response of rad23b plants to mitomycin C suggest that RAD23b regulates cell division. Taken together, RAD23 proteins appear to play an essential role in the cell cycle, morphology, and fertility of plants through their delivery of UPS substrates to the 26S proteasome.
[本文引用: 1]
DOI:10.1105/tpc.106.040998URL [本文引用: 1]
DOI:10.1104/pp.106.087965URL [本文引用: 1]
DOI:10.1093/mp/ssr030URLPMID:3221247 [本文引用: 2]

Ubiquitination 是重要蛋白质 translational 以后修正,它在更高的植物,和 U 盒子 E3 ligases 玩涉及各种各样的细胞的过程在在优核质的多样的功能的重要角色。这里,我们描述 Arabidopsis thaliana PUB19 (AtPUB19 ) 的功能,我们在它示威了一在里面编码 U 盒子类型 E3 ubiquitin ligase 的 vitro 试金。AtPUB19 由干旱,盐,寒冷,和 abscisic 酸(骆驼毛的织物) 是起来调整的。AtPUB19 的下面规定导致了超敏性到骆驼毛的织物,提高了导致骆驼毛的织物的有气孔的关门,并且提高了干旱忍耐,当 AtPUB19 overexpression 导致了反向的显型时。分子的分析证明很多骆驼毛的织物和压力标记基因的表示层次在两 AtPUB19 overexpressing 和 atpub19-1 异种植物被改变。在,我们 AtPUB19 否定地调整的数据表演在 A 的骆驼毛的织物和干旱回答。thaliana。
DOI:10.1007/s00299-014-1706-4URL [本文引用: 1]
DOI:10.1007/s10725-018-0423-3 [本文引用: 1]
DOI:10.1104/pp.108.123380URL [本文引用: 1]
DOI:10.1038/nature25447PMID:29414943 [本文引用: 2]

The genus Citrus, comprising some of the most widely cultivated fruit crops worldwide, includes an uncertain number of species. Here we describe ten natural citrus species, using genomic, phylogenetic and biogeographic analyses of 60 accessions representing diverse citrus germ plasms, and propose that citrus diversified during the late Miocene epoch through a rapid southeast Asian radiation that correlates with a marked weakening of the monsoons. A second radiation enabled by migration across the Wallace line gave rise to the Australian limes in the early Pliocene epoch. Further identification and analyses of hybrids and admixed genomes provides insights into the genealogy of major commercial cultivars of citrus. Among mandarins and sweet orange, we find an extensive network of relatedness that illuminates the domestication of these groups. Widespread pummelo admixture among these mandarins and its correlation with fruit size and acidity suggests a plausible role of pummelo introgression in the selection of palatable mandarins. This work provides a new evolutionary framework for the genus Citrus. (R sum d'auteur)
DOI:10.11686/cyxb2015102Magsci [本文引用: 3]

植物基因组中广泛存在U-box基因,其编码蛋白大部分作为泛素系统中决定底物特异性识别的E<sub>3</sub>泛素连接酶,广泛地调控植物生长生殖发育以及响应逆境胁迫等过程。本文利用蒺藜苜蓿基因组数据库,通过生物信息学手段,鉴定蒺藜苜蓿U-box家族基因;采用MEGA6软件进行系统进化树分析;通过GSDS在线工具和Mapinspect软件进行基因结构及染色体定位分析;利用已有的蒺藜苜蓿芯片数据进行组织表达和胁迫响应表达分析。结果表明,蒺藜苜蓿基因组中含有41个U-box基因,不均匀地分布于蒺藜苜蓿的8条染色体上。根据结构域组成和系统进化分析将这些U-box蛋白分成6类。基因表达模式分析发现,该家族成员的表达具有一定的组织特异性,并能响应盐、干旱和氮素胁迫。这些研究结果为蒺藜苜蓿U-box基因家族的功能分析奠定了基础。
DOI:10.11686/cyxb2015102Magsci [本文引用: 3]

植物基因组中广泛存在U-box基因,其编码蛋白大部分作为泛素系统中决定底物特异性识别的E<sub>3</sub>泛素连接酶,广泛地调控植物生长生殖发育以及响应逆境胁迫等过程。本文利用蒺藜苜蓿基因组数据库,通过生物信息学手段,鉴定蒺藜苜蓿U-box家族基因;采用MEGA6软件进行系统进化树分析;通过GSDS在线工具和Mapinspect软件进行基因结构及染色体定位分析;利用已有的蒺藜苜蓿芯片数据进行组织表达和胁迫响应表达分析。结果表明,蒺藜苜蓿基因组中含有41个U-box基因,不均匀地分布于蒺藜苜蓿的8条染色体上。根据结构域组成和系统进化分析将这些U-box蛋白分成6类。基因表达模式分析发现,该家族成员的表达具有一定的组织特异性,并能响应盐、干旱和氮素胁迫。这些研究结果为蒺藜苜蓿U-box基因家族的功能分析奠定了基础。
DOI:10.1042/BJ20071568URLPMID:18393940 [本文引用: 1]

The variance of the U-box domain in sixtyfour Arabidopsis thaliana ubiquitin-protein ligases (E3s) was used to examine the interactions between E3s and ubiquitin-conjugating enzymes (E2s). E2s and E3s are components of the ubiquitin protein degradation pathway. Seven U-box proteins were analyzed for their ability to ubiquitinate proteins in vitro in cooperation with different E2s. All U-box domains exhibited ubiquitination activity and interacted productively with UBC4/5-type E2s. Three and four of the U-box domains mediated ubiquitin addition in the presence of UBC13 and UBC7 E2s, respectively, but no productive interaction was observed with the UBC15 E2 tested. The activity of AtPUB54 was dependent on Trp266 in the E2-binding cleft, and the E2 selectivity was changed by substitution of this position. The function of the distant U-box protein, AtPUB49, representing a large family of eukaryotic proteins containing a U-box linked to a cyclophilin-like peptidyl-prolyl cis-trans isomerase domain, was characterized biochemically. AtPUB49 functioned both as a prolyl isomerase and a chaperone by catalyzing cis/trans isomerization of peptidyl-prolyl bonds and dissolving protein aggregates. In conclusion, both typical and atypical Arabidopsis U-box proteins were active E3s. The overlap in the E3/E2 selectivity suggests that in vivo specificity is not determined only by the E3/E2 interactions but also by other parameters, e.g. co-existence or interactions with additional domains. The biochemical functions of AtPUB49 suggest that the protein can be involved in folding or degradation of protein substrates. Similar functions can also be retained within a protein complex with separate chaperone and U-box proteins.
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[本文引用: 1]

泛素蛋白质的降解途径是植物应答发育阶段信息、适应环境胁迫的重要信号途径调节机制。E3泛素连接酶基因是泛素蛋白质降解途径中决定蛋白质降解特异性的关键因子。本研究利用番茄的基因组和基因芯片数据,在番茄基因组中鉴定出56个U-box类基因。番茄U-box基因家族成员间大小、等电点、结构域等特征差异很大,并无显著的聚集特征。与此不同,番茄U-box基因的结构虽然同样差异巨大,但按照系统进化关系,大致分为三类:无内含子、有3-5个内含子、10个或更多内含子。番茄U-box基因的染色体分布位置也非常不均衡。在一号染色体上,5个U-box基因首尾相接连续分布在一起,这5个基因序列也非常相似,系统进化树分析表明这些基因亲缘关系非常接近。利用番茄功能基因组学数据库,观察发现番茄U-box基因成员不仅参与响应干旱、病菌侵染等外界胁迫。丛枝菌根真菌侵染也会导致番茄U-box基因Solyc01g00-7000.2.1在根部表达量显著上调。本研究团队计划对U-box基因Solyc01g007000.2.1在丛枝菌根真菌与番茄互作过程中的作用,进行进一步的分析。
[本文引用: 1]

泛素蛋白质的降解途径是植物应答发育阶段信息、适应环境胁迫的重要信号途径调节机制。E3泛素连接酶基因是泛素蛋白质降解途径中决定蛋白质降解特异性的关键因子。本研究利用番茄的基因组和基因芯片数据,在番茄基因组中鉴定出56个U-box类基因。番茄U-box基因家族成员间大小、等电点、结构域等特征差异很大,并无显著的聚集特征。与此不同,番茄U-box基因的结构虽然同样差异巨大,但按照系统进化关系,大致分为三类:无内含子、有3-5个内含子、10个或更多内含子。番茄U-box基因的染色体分布位置也非常不均衡。在一号染色体上,5个U-box基因首尾相接连续分布在一起,这5个基因序列也非常相似,系统进化树分析表明这些基因亲缘关系非常接近。利用番茄功能基因组学数据库,观察发现番茄U-box基因成员不仅参与响应干旱、病菌侵染等外界胁迫。丛枝菌根真菌侵染也会导致番茄U-box基因Solyc01g00-7000.2.1在根部表达量显著上调。本研究团队计划对U-box基因Solyc01g007000.2.1在丛枝菌根真菌与番茄互作过程中的作用,进行进一步的分析。
[本文引用: 1]
[本文引用: 1]
DOI:10.1186/1471-2148-10-331URLPMID:20979629 [本文引用: 1]

Background The patterns of emergence and diversification of the families of ubiquitin ligases provide insights about the evolution of the eukaryotic ubiquitination system. U-box ubiquitin ligases (UULs) are proteins characterized by containing a peculiar protein domain known as U box. In this study, the origin of the animal UUL genes is described. Results Phylogenetic and structural data indicate that six of the seven main UUL-encoding genes found in humans (UBE4A, UBE4B, UIP5, PRP19, CHIP and CYC4) were already present in the ancestor of all current metazoans and the seventh (WDSUB1) is found in placozoans, cnidarians and bilaterians. The fact that only 4 - 5 genes orthologous to the human ones are present in the choanoflagellate Monosiga brevicollis suggests that several animal-specific cooptions of the U box to generate new genes occurred. Significantly, Monosiga contains five additional UUL genes that are not present in animals. One of them is also present in distantly-related protozoans. Along animal evolution, losses of UUL-encoding genes are rare, except in nematodes, which lack three of them. These general patterns are highly congruent with those found for other two families (RBR, HECT) of ubiquitin ligases. Conclusions Finding that the patterns of emergence, diversification and loss of three unrelated families of ubiquitin ligases (RBR, HECT and U-box) are parallel indicates that there are underlying, linage-specific evolutionary forces shaping the complexity of the animal ubiquitin system.
DOI:10.1038/nature11119 [本文引用: 1]
DOI:10.1111/tpj.2006.46.issue-4URL [本文引用: 1]
DOI:10.1186/s12870-016-0963-5URL [本文引用: 1]
DOI:10.1111/plb.2011.13.issue-5URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1104/pp.112.199430URL [本文引用: 1]
[本文引用: 1]
DOI:10.1038/nchembio.165URLPMID:19377456 [本文引用: 1]

Abstract The plant hormones are a structurally unrelated collection of small molecules derived from various essential metabolic pathways. These compounds are important regulators of plant growth and mediate responses to both biotic and abiotic stresses. During the last ten years there have been many exciting advances in our understanding of plant hormone biology, including new discoveries in the areas of hormone biosynthesis, transport, perception and response. Receptors for many of the major hormones have now been identified, providing new opportunities to study the chemical specificity of hormone signaling. These studies also reveal a surprisingly important role for the ubiquitin-proteasome pathway in hormone signaling. In addition, recent work confirms that hormone signaling interacts at multiple levels during plant growth and development. In the future, a major challenge will be to understand how the information conveyed by these simple compounds is integrated during plant growth.
DOI:10.1007/s00299-014-1706-4URL [本文引用: 1]
DOI:10.1104/pp.106.087965URL [本文引用: 1]
DOI:10.1371/journal.pone.0049207URL [本文引用: 1]
DOI:10.1104/pp.112.202143URL [本文引用: 1]
DOI:10.1093/jxb/ern369URLPMID:19196749 [本文引用: 1]

Abstract Ubiquitin-mediated proteolysis is an integral part of diverse cellular functions, and of the three enzymes involved in linking ubiquitin to protein targets, the E3 ubiquitin ligases are of particular interest as they confer substrate specificity during this process. The E3 ubiquitin ligases can be categorized based on mechanism of action and on the presence of specific domains such as RING, HECT, F-box, and U-box. In plants, the U-box family has undergone a large gene expansion that may be attributable to biological processes unique to the plant life cycle. For example, there are 64 predicted plant U-box (PUB) proteins in Arabidopsis, and the biological roles of many of these have yet to be determined. Research on PUB genes from several different plants has started to elucidate a range of functions for this family, from self-incompatibility and hormone responses to defence and abiotic stress responses. Expression profiling has also been used as a starting point to elucidate PUB function, and has uncovered a strong connection of PUB genes to various stress responses. Finally, some PUB proteins have been linked to receptor kinases as upstream activators, and downstream target substrates are also starting to emerge. The mechanisms of action range from the observation of mono-ubiquitination during non-proteolytic signalling to directed regulation of proteasomal components during stress responses, and cell death appears to be a theme underlying many PUB functions.
DOI:10.1186/s12864-015-1629-7URL [本文引用: 1]
